Abstract
5′- and 3′-untranslated regions (UTRs) are important regulators of gene expression and play key roles in disease progression and susceptibility. The 5′-UTR of the fragile X mental retardation 1 (FMR1) gene contains a CGG repeat element that is expanded (>200 CGG repeats; full mutation) and methylated in fragile X syndrome (FXS), the most common form of inherited intellectual disability (ID) and known cause of autism. Significant phenotypic involvement has also emerged in some individuals with the premutation (55–200 CGG repeats), including fragile X-associated premature ovarian insufficiency (FXPOI) in females, and the neurodegenerative disorder, fragile X-associated tremor/ataxia syndrome (FXTAS), in older adult carriers. Here, we show that FMR1 mRNA in human and mouse brain is expressed as a combination of multiple isoforms that use alternative transcriptional start sites and different polyadenylation sites. Furthermore, we have identified a novel human transcription start site used in brain but not in lymphoblastoid cells, and have detected FMR1 isoforms generated through the use of both canonical and non-canonical polyadenylation signals. Importantly, in both human and mouse, a specific regulation of the UTRs is observed in brain of FMR1 premutation alleles, suggesting that the transcript variants may play a role in premutation-related pathologies.
INTRODUCTION
Fragile X syndrome (FXS) is the most common form of inherited cognitive impairment, and the most common single gene mutation associated with autism (1–3). FXS is caused by a trinucleotide repeat expansion (CGG)n in the 5′-untranslated region (5′-UTR) of the fragile X mental retardation 1 (FMR1) gene located at Xq27.3. Full mutation CGG-repeat expansions (>200 repeats) are generally accompanied by methylation-coupled transcriptional silencing of the FMR1 gene, and consequent absence of the encoded protein (FMRP) (1,4,5). Substantially diminished or absent FMRP results in aberrant brain development and function due to the importance of FMRP in synaptic stabilization and plasticity (2,3,6–8).
Normal alleles have approximately 12–44 CGG repeats; while premutation alleles have between 55 and 200 CGG repeats, are typically unmethylated, and usually do not result in gene inactivation. Over the past several years, there has been an expanding awareness of the phenotypes associated with the premutation (1,9,10). Two abnormal phenotypes specific to premutation alleles have been described: fragile X-associated premature ovarian insufficiency (FXPOI), defined by cessation of menses prior to age 40 (11,12); and fragile X-associated tremor/ataxia syndrome (FXTAS), a neurodegenerative disorder which affects older adult carriers of the FMR1 premutation alleles and is characterized by progressive cerebellar gait ataxia, intention tremor, cognitive decline and some psychiatric involvement (9,10,13). In addition, the most consistent deficits seen in premutation carriers include shyness, anxiety, social deficits, Attention Deficit Hyperactivity Disorder (ADHD) and executive function deficits (14,15).
Although the molecular mechanisms underlying the premutation pathologies are not well understood, a consistent molecular feature is the elevation of FMR1 mRNA levels (16–18), as a consequence of an increased rate of FMR1 transcription (19). However, carriers of premutation alleles show decreased levels of FMRP (17,20–22), due to a reduced translational efficiency of FMR1 mRNA containing the expanded CGG repeat (20). These observations, coupled with the absence of cases of either FXPOI or FXTAS in the full mutation range (where protein levels are low–absent), led to the hypothesis that the premutation-specific disorders are due to an RNA toxic gain-of-function mechanism (23–25). Additionally, FMR1 mRNA has been detected in the ubiquitin-positive intranuclear inclusions found in neurons and astrocytes throughout the brain of FXTAS patients (26,27).
A variety of mRNA transcripts arise from a combination of alternative splicing, alternative transcriptional start sites selection and differential usage of polyadenylation sites. In fact, these events generate the vast majority of diversity of gene expression and have been described for over 180 000 mouse transcripts (28). Besides alternative splicing, which can produce extraordinary protein diversity, regulation at the level of the 5′- and 3′-UTRs modulates mRNA processing, nuclear export, stability, subcellular localization and translational efficiency (29). Such processes are crucial for differential expression of a gene during development, tissue differentiation and under certain pathological conditions (29,30).
In the case of the FMR1 gene, which in human spans ∼38 Kb of genomic DNA and contains 17 exons (31), extensive alternative splicing has been demonstrated, with splicing variants changing during neuronal differentiation both in human and mouse (32–36). In addition, multiple transcriptional initiation sites, whose distribution appears to be modulated by the number of CGG repeats, have been detected in cultured cells from both normal controls and premutation carriers (37). However, little information is yet available on the transcription sites and polyadenylation signal distribution and selection in both human and mouse tissues.
Thus, it is important to investigate how the variety of different FMR1 transcripts and splice variants may be related to protein function and to the clinical phenotype. Here, we report a detailed study on the structure and differential usage of the 5′- and 3′-UTRs of the FMR1 gene in both human and mouse brain, showing a different expression in expanded premutation alleles compared to normal alleles. In addition, we provide evidence of the potential role played by the 3′-UTR region in translational regulation of the FMR1 gene, and that could explain, at least in part, the reduced FMRP levels observed in premutation alleles.
MATERIALS AND METHODS
Animal care and human tissues
Animal care was conducted in conformance with the institutional guidelines that are in compliance with Italian (DL N116, GU, suppl 40, 18-2-1992) and international laws and policies (European Community Council Directive 86/609, OJa L 358, 1, December 12, 1987; National Institutes of Health Guide for the Care and Use of Laboratory Animals, US National Research Council, 1996, Belgian law of August 14th, 1986, concerning the protection and well-being of animals, and the following K.B. of November 14th, 1993 and K.B of September 13th, 2004, concerning the protection of all laboratory animals, as well as to the European Community Council Directive 86/609, Oja L 358, 1, December 12, 1987). All mice (5-months-old CGG knock-in (KI) and Wild Type (WT) littermates) had a C57BL/6 genetic background (38).
All studies of ‘post mortem’ human tissue were performed following informed consent in accordance with University of California, Davis, IRB approved protocols.
RNA isolation
Total RNA was isolated from human brain regions (cortex, hippocampus and cerebellum) derived from a subject carrying an FMR1 premutation allele (#334-03KC, 85 CGG) and from an age matched normal control (#946,27 CGG). Total RNA was also isolated from total brain or cerebellum and hippocampus derived from wild-type (WT) (8 CGG) and CGG KI mice (98 CGG) (38). Isolation of total RNA was performed using Trizol reagent (Invitrogen) as recommended by the manufacturer. Poly(A)+ RNAs from the samples were prepared using the Oligotex mRNA mini kit (Qiagen). The integrity of the total RNA was checked by monitoring the presence of intact ribosomal RNAs, 28 S and 18 S using a Bioanalyzer (Agilent Technologies).
RLM-RACE analysis
5′ and 3′ RLM-RACE (RNA ligase-mediated rapid amplification of cDNA ends) analysis was performed on 1 µg of total RNA from human or mouse tissues using the Generacer Kit™ (Invitrogen), as suggested by the manufacturer. Briefly, mRNA was treated with calf intestinal phosphatase (CIP) to remove the 5′ phosphates from any truncated (i.e. non-capped) mRNA. Dephosphorylated RNA was treated with tobacco acid pyrophosphatase (TAP) to remove the 5′ cap from full-length mRNA, leaving a 5′ phosphate. The GeneRacer™ RNA oligomer was then ligated to the 5′-end of the mRNA using T7 RNA ligase. RNA obtained from this step of the 5′-RACE and the total RNA for the 3′ reaction were reverse transcribed using Avian Myeloblastosis Virus Reverse Transcriptase with the GeneRacer™oligo-dT primer. The regions corresponding to the legitimate 5′-ends of the capped RNA species were PCR amplified from cDNA templates using the GeneRacer™ 5′ primer (5′-CGACTGGAGCACGAGGACACTGA-3′) and a FMR1 gene-specific primer (human, 5′-CCTCCACCGGAAGTGAAACCGAA-3′; mouse, 5′-TCGCCGTCCGTTTGCTTCAC-3′).
To obtain 3′-ends, we amplified the first-strand cDNA using a FMR1 gene-specific primer (human, 5′-CTAAATGTTAAAGATGTAGCAAACCCTG-3′; mouse 5′-GGAAACGACGATCATTCCCGAACAGA-3′) and the GeneRacer™ 3′ Primer (5′-GCTGTCAACGATACGCTACGTAACG-3′).
Cloning and sequencing
PCR products obtained by 5′- and 3′-RLM-RACE reactions were cloned into pCR 4.1 vector (TA Cloning Kit, Invitrogen) or into pGEM-T-Easy vector (Promega, Madison, WI, USA), and ligated products were transfected into One-Shot chemically competent cells (Invitrogen). Plasmid DNAs were isolated and purified using plasmid mini kits (Promega), and were screened for insertions following EcoRI digestion. Positive clones for each transfection reaction were sequenced by an automated sequencer (ABI PRISM 310 Genetic Analyzer, Applied Biosystems) using vector-specific primers. The number of clones sequenced for each brain region is indicated for the mouse and human 5′-UTRs in Figure 3A and Figure 6A, respectively. The statistical significance was assessed by using the Pearson's chi-square test (χ2 test) and the significance level was defined at P-values <0.05 (*P < 0.05, **P < 0.01, ***P < 0.001).
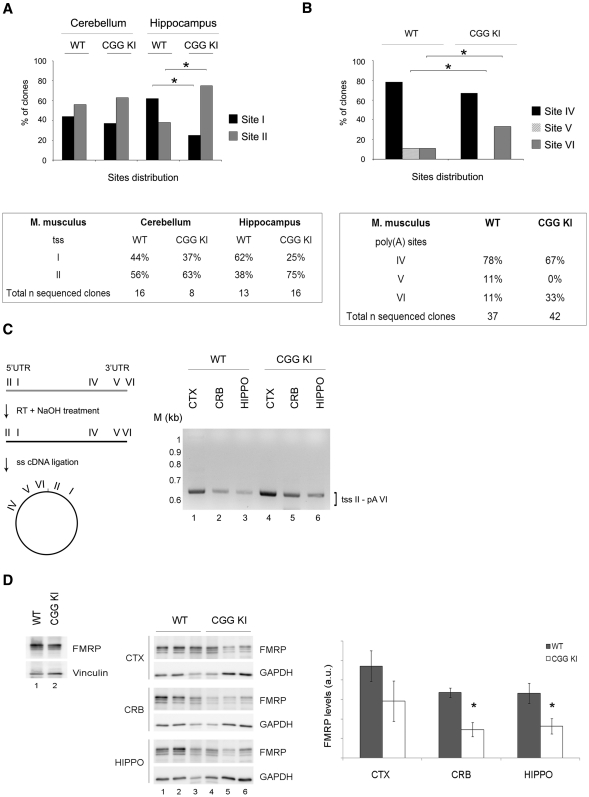
Figure 3.
Brain specific UTRs in a mouse model for the premutation syndromes. (A) Top panel: Histograms showing the distribution of the two tss in the cerebellum and hippocampus from WT and CGG KI mice. χ2 test, *P < 0.05. Bottom panel: percentage of independent sequenced clones for WT and CGG KI mice. (B) Top panel: Histograms showing the distribution of the three poly(A) site usage in total brain from WT and CGG KI mice. χ2 test, *P < 0.05. Bottom panel: percentage of independent sequenced clones for WT and CGG KI mice. (C) Left panel: scheme of the technique used to detect simultaneously the 5′- and 3′-UTRs from single transcript variants. Briefly, the RNA is retrotranscribed, the cDNA is self-ligated and the 3′ and 5′ regions of the circular cDNA amplified by PCR using a forward primer in the 3′-UTR and a reverse primer in the 5′-UTR. Right panel: nested PCR on the circular cDNAs in cortex, cerebellum and hippocampus from WT and CGG KI mice. M, DNA ladder (1 kb). (D) Left panel: FMRP expression in total mouse brain from WT (lane 1) and CGG KI (lane 2). Central panel: FMRP levels in cortex, hippocampus and cerebellum from WT (lanes 1–3) and CGG KI mice (lanes 4–6). Vinculin and GAPDH are used as loading control. Right panel: quantification of FMRP normalized for GAPDH (n = 3, Student's t-test, *P < 0.05).
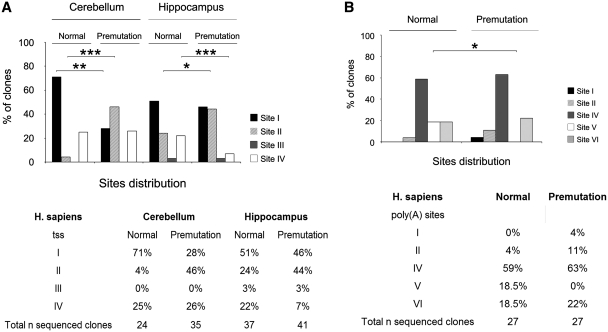
Figure 6.
UTRs detection in the human premutation alleles. (A) Top panel: Histograms showing the distribution of the four tss in normal and premutation alleles in human cerebellum and hippocampus. χ2 test, *P < 0.05, **P < 0.01, ***P < 0.001. Bottom panel: Table showing the number of independent sequenced clones containing the different tss expressed as percentage in the two brain regions analyzed for both normal and premutation alleles. (B) Top panel: Histograms showing the distribution of the five poly(A) sites identified in normal and premutation alleles in human cerebral cortex. χ2 test, *P < 0.05. Bottom panel: The total number of sequenced clones for the normal and premutation allele is expressed as percentage in the five poly(A) sites.
CGG repeat allele sizing
DNA was isolated from both mouse and human brain tissue (∼100 mg) using standard conditions (Qiagen) and analyzed by PCR with oligonucleotide primers specific for a portion of the Fmr1 genomic DNA sequence flanking the CGG repeat region. PCR products were separated on a 6% polyacrylamide sequencing gel and transferred to a nylon membrane (Roche) and the hybridization was performed with DIG labeled oligonucleotide probe (CGG)8 using conditions as described in (39).
UTRs detection through cDNA circularization and PCR
RNAs isolated from mouse cortex, cerebellum and hippocampus were subjected to retrotranscription using Oligo(dT)12–18 (Invitrogen) and Superscript™ III RT (Invitrogen). After removal of the template RNA with NaOH hydrolysis, the 5′-end of the cDNA was phosphorylated using a polynucleotide kinase (New England Biolabs) and self-ligated using the CircligaseTM II ssDNA ligase (Epicentre Biotech.), according to manufacturer's instructions. The cDNA was then used as template to amplify the 3′ and 5′ ends using a forward primer (CB378, 5′-CCAAGGTAGAATGACCTTGTA-3′) and a reverse primer (CB375, 5′-GCGCAGCCCCTCCCCTTTG-3′). The PCR products were analyzed on 2% agarose gels and the images acquired with a gel scanning (ImageQuant300, GE Healthcare) using the ImageQuant software (GE Healthcare). The identity of the PCR products was assessed by a nested PCR using the forward primer CB379 (5′-GCTCTTGGGCAATATTCTCTG-3′) and sequencing of the product.
Western blotting
Western blotting analysis was performed according to standard procedures. Briefly, total mouse brain or brain areas (cortex, hippocampus and cerebellum) from WT (8 CGG) and CGG KI (98–182 CGG) were homogenized in Laemmli buffer as described in (40). Proteins from total brain (10 μg) or from brain areas (5 μg) were loaded onto a SDS-PAGE gel and transferred on a polyvinylidene fluoride (PVDF) membrane (Millipore). Membranes were probed with an anti-FMRP polyclonal antibody (41) (1:1000), anti-GAPDH (mouse monoclonal, Chemicon, 1:5000) or anti-Vinculin (mouse monoclonal, Sigma, 1:2000). The signals were developed using a chemiluminescence technique (ECL Basic or Plus, Amersham Pharmacia) using Fuji film LAS-3000 (Fuji). Quantification was performed normalizing FMRP levels to GAPDH using the AIDA software (Raytest Isotopenmeßgeräte GmbH). The statistical significance was assessed by using Student's t-test and the significance level was defined at P-values <0.05.
Polyadenylation test (PAT)
The PAT assay was performed as described in di Penta et al. (42). Briefly, 1 μg of total RNA extracted as previously described was tagged by annealing oligo RACE4 (5′-GCTTCAGATCAAGGTGACC(T)12-aminolink) to the end of the mRNA and extending it with Klenow polymerase (Roche) at 37°C for 60 min. The tagged RNAs were reverse transcribed by Superscript™ III RT (Invitrogen) at 48°C for 60 min using an oligo similar to RACE4 (CB125, 5′-GCTTCAGATCAAGGTGACCTTTTT-3′). The cDNA was then amplified using a forward primer able to detect the three mRNA species, using the poly(A) sites IV, V and VI (BMN284, 5′-GGTAGAATGACCTTGTAATGT-3′), a forward primer able to detect only the transcripts containing the poly(A) site VI (CB246, 5′-TCTGAAGATTGTTTATCTTATC-3′), or a forward primer for β-actin mRNA (AD48, 5′-AGGTGACAGCATTGCTTCTG-3′), and the reverse CB125. As a negative control, RNAs were deadenylated by incubating with Oligo(dT)12–18 (Invitrogen) and RNase H (Fermentas) at 37°C for 90 min, purified by phenol-chloroform and subjected to the same tagging reaction. Graphs shown in Figure 4B–D were elaborated following a gel scanning (ImageQuant300, GE Healthcare) using the ImageQuant software (GE Healthcare).
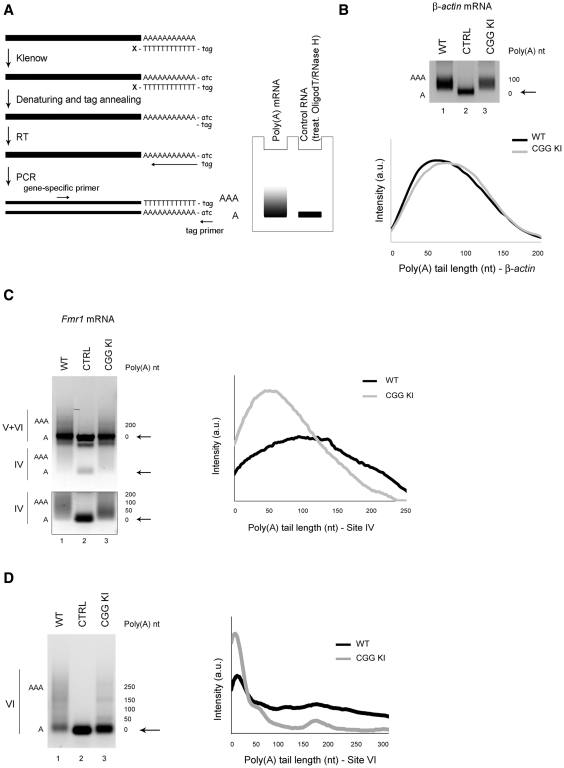
Figure 4.
Polyadenylation state of Fmr1 mRNA variants in the CGG KI mouse brain. (A) Left panel: schematic view of the polyadenylation assay (PAT). According to di Penta et al. (42), the poly(A) tails are tagged by incubating the RNA with a (T)12-tag oligonucleotide, blocked at the 3′-end, in the presence of dNTPs and Klenow enzyme to fill in the complementary tag sequence. The RNA is then denatured and annealed to a DNA primer, identical to the tag, to start a reverse transcription (RT). The cDNA is then amplified using a gene-specific forward primer and the reverse tag oligo. Right panel: cartoon of a polyadenylation profile obtained with a PAT assay. The PCR of a polyadenylated mRNA gives rise to a smear while the same mRNA deadenylated with oligodT and RNase H prior to poly(A) tagging is used as a negative control and gives a sharp band. (B) Upper panel: β-actin mRNA in WT (lane 1) and CGG KI (lane 3). Deadenylated RNA is shown as negative control (lane 2) and the deadenylated form is indicated by black arrows. Lower panel: dispersion graph representing the distribution of the β-actin polyadenylated transcripts in WT (black line) and CGG KI (grey line). The signal intensity along the lane has been plotted against the poly(A) tail length, estimated from the molecular markers loaded on the same gel. (C) PAT for all three poly(A) Fmr1 mRNA variants. Because of close proximity, the transcripts containing sites V and VI cannot be discriminated and therefore they are not taken into exam (upper panel). Black arrows points to the deadenylated form. The polyadenylation of transcripts using site IV from WT (lane 1) and CGG KI (lane 3) has been independently acquired and highlighted in the box below. Deadenylated RNA treated as mentioned above, is shown as negative control (lane 2). Right panel: dispersion graph for Fmr1 variants using site IV in WT (black line) and CGG KI (grey line). (D) Left panel: PAT for Fmr1 variants using site VI in WT (lane 1) and CGG KI (lane 3) brain. Deadenylated RNA as above is used as negative control (lane 2). Right panel: Dispersion graph representing the distribution of the polyadenylated transcripts using site VI in WT (black line) and CGG KI (grey line).
Polysome/mRNPs gradient
Polysome/mRNPs gradients were performed according to (40,43,44), with minor modifications. Total mouse brains from WT (8 CGG) and CGG KI (181–184 CGG) were homogenized in lysis buffer (10 mM NaCl, 10 mM MgCl2, 10 mM Tris–HCl pH = 7.5, 1% Triton X100, 1% DOC, 1 mM DTT, 100 μg/mL Cycloheximide, 40 U/ml RNasin). For the polysome/mRNP analysis of the mRNAs strong detergent like DOC 1% was used as in (44). These stringent conditions are only used for RNA analysis. The lysates were centrifuged 8 min at 12 000 g at 4°C and one-fourth of the supernatant was centrifuged through a 5–60% (w/v) sucrose gradient for 135 min at 37 000g in a Beckman SW41 rotor. Each gradient was collected in 11 fractions. RNA was extracted from the fractions and analyzed by RT-PCR using gene specific primers for αCaMKII mRNA (BMN 190, 5′-ACCCTGGCCTGGTCCTTCAATG-3′; BMN 191, 5′-AGCCATCCTCACCACTATGCTGG-3′), β-actin mRNA (CD27, 5′-AGCAAGAGAGGTATCCTGACC-3′; CD28 5′-GCCAATAGTGATGACCTGGCC-3′), Fmr1 coding sequence mRNA (BMN 12, 5′-GGGTTGGACCTAACTCCTC-3′; BMN 13 5′- GTCTCTGTGGTCAGATTCTG-3′) and Fmr1 isoform VI mRNA (BMN 186, 5′- CGTTAAAATATGCACTAAGTCTC-3′; BMN 187 5′- GCTTTTTCATAAGTGCAACTTTTT-3′). The amount of template and the number of amplification cycles were preliminary optimized for each PCR reaction to avoid saturation as follows: αCaMKII mRNA (Tm = 70°C, 35 cycles), β-actin mRNA (Tm = 64°C, 35 cycles), Fmr1 coding sequence mRNA (Tm = 60°C, 38 cycles), and Fmr1 isoform VI mRNA (Tm = 60°C, 40 cycles). The PCR products were analyzed on a 1.2% agarose gel and the images acquired with a gel scanning (ImageQuant300, GE Healthcare) using the ImageQuant software (GE Healthcare). Percentage of messenger on polysome (PMP) was calculated as described in the ‘Results’ section.
RESULTS
FMR1 mRNA UTR structures in human and mouse brain
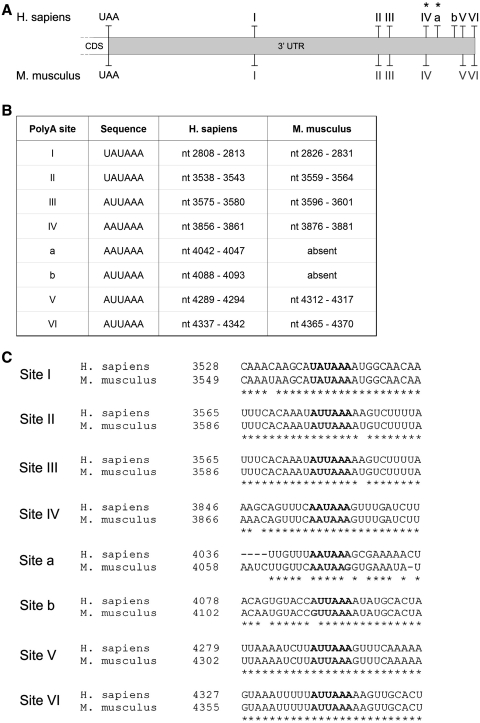
The structure of the 5′-UTR of FMR1 mRNA has been studied thus far only in lymphoblastoid cell lines and primary cultured astrocytes, where the presence of three transcription start sites (tss) has been reported (37). In order to study the brain-specific features of the 5′-UTR of the mRNA, we investigated the tss in both human and mouse brain. For this purpose, we isolated the 5′-end of FMR1 mRNA from brain using a 5′ RNA ligase-mediated RACE (RLM-RACE). The resulting PCR products (four major bands for human and two for mouse, data not shown) were cloned and sequenced. As shown in Figure 1, we obtained four tss in human, three of them corresponding to tss I, II and III previously identified in lymphoblastoid cells and astrocytes (37), confirming and extending earlier findings to brain tissue. In addition, we identified a novel tss (site IV) located between the previously identified sites I and II, not identified in lymphoblastoid cells, suggesting the presence of a novel start site in brain (Figure 1A–B, marked with an asterisk). Similarly to sites I, II and III, the new site corresponds to an Inr-like sequence (consensus YYAN(T/A)YY) (45).
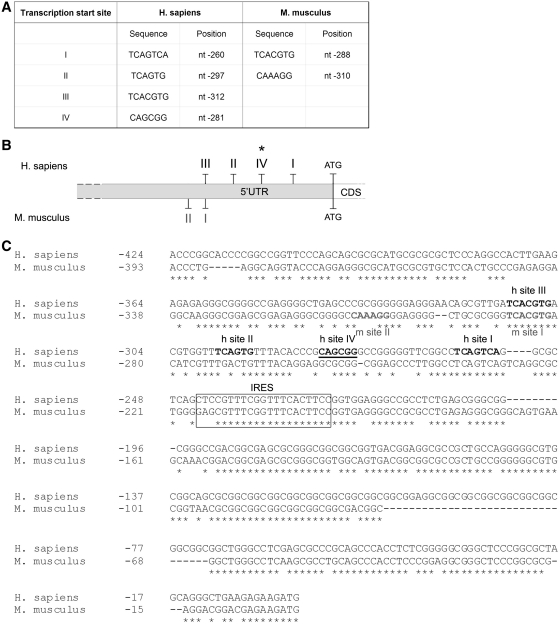
Figure 1.
Structure of the FMR1 5′-UTR in human and mouse brain. (A) Nucleotide position of the tss in both human and mouse. The tss are numbered to the left with respect to the translation codon (+1 ATG, 13967 nt) on a sequence reference (for human access number: L29074: for mouse access number AY630338). (B) Scheme of the tss in human (I, II, III and IV) and mouse (I and II). The asterisk marks the new tss. (C) Sequence comparison between human and mouse 5′-UTRs. The four human (h site I, II, III and IV) and the two mouse (m sites I and II) tss are bolded, in black and grey, respectively. The newly identified human transcription start site IV is underlined. The box represents the previously described Internal Ribosome Entry Site (IRES) (61,62). The alignment was performed using the ClustalW software.
The 5′-RLM-RACE analysis performed on mouse brain, revealed a different functional organization of the 5′-UTR. Two tss were detected: site I corresponds to the human tss III, while mouse start site II is located further upstream. The nucleotide positions of the two mouse tss are shown in Figure 1A. A diagram of the human and mouse 5′-UTRs is shown in Figure 1B, while the alignment of the promoter region with the position of the different start sites and the relative sequences identified is shown in Figure 1C.
In addition to tss selection, alternative polyadenylation strongly contributes to transcript variability; in fact, almost 50% of human genes have more than one polyadenylation site (46,47). Since the structures of human and mouse FMR1 3′-UTRs have not been characterized, we decided to investigate, via sequence alignment, the presence of both canonical and non-canonical polyadenylation sequences (48). As shown in Figure 2A, we found eight putative polyadenylation sites in human (I–VI and a, b); among which only two sites contain the canonical polyadenylation signal represented by the conserved hexamer AAUAAA (sites IV and a, Figure 2A–B; asterisk). The other sites are non-canonical and include the single-nucleotide variant of the consensus sequence for the first base (UAUAAA, sites I and II) and the second nucleotide (AUUAAA, sites III, b, V, VI) (Figure 2B). The alignment between the human and the murine sequences revealed that six of the eight human sites were also present in mouse (I–VI), while human sites a and b were not conserved (Figure 2C). 3′-RACE performed on both human and mouse brain revealed that only three of the putative poly(A) sites were used in normal brain, specifically the IV, V, VI (Figures 3B and 6B).
Figure 2.
Structure of the FMR1 3′-UTR in human and mouse brain. (A) Schematic representation of the putative poly(A) sites, taking into account both canonical and not-canonical sites (48) in human and mouse. Asterisks mark canonical poly(A) sites. (B) Nucleotide position and consensus sequence of the poly(A) sites in both human and mouse. (C) Alignment between the regions containing the putative poly(A) signals in human (accession number NM_002024) and mouse (accession number NM_008031). Consensus sequences are bolded. The alignment was performed using the ClustalW software.
Differential usage of tss and polyadenylation sites in a mouse model for the premutation
To investigate whether the variants of the mouse Fmr1 mRNA change according to the CGG repeat length, we took advantage of the mouse model developed to study triplet instability generated by replacing the murine (CGG)8 element with a human (CGG)98 repeat (CGG KI) (38). Since alternative transcriptional start site usage in the FMR1 gene depends on CGG repeat size in human lymphoblastoid cell lines and cultured astrocytes (37), we determined the role of the CGG repeat length on start site selection in the CGG KI mouse. For this purpose, we performed a 5′-RLM-RACE on cerebellum and hippocampus; the resulting PCR products were cloned and sequenced. A total of 24 clones from cerebellum (WT, 16 clones; CGG KI, 8 clones) and 29 clones from hippocampus (WT, 13 clones; CGG KI, 16 clones) were analyzed for the presence of the two alternative transcriptional start sites above described (Figure 1 and 3A). A differential usage of tss I and II was observed in the hippocampus of the CGG KI mice compared to WT, indicating that in mice, as in humans, the usage of the different tss appears to be CGG repeat number dependent. Specifically, the expression of the short Fmr1 transcript using tss I was higher in WT compared to CGG KI mouse (44% and 62% in WT cerebellum and hippocampus versus 37% and 25% in CGG KI cerebellum and hippocampus, χ2 test, P < 0.05 in the hippocampus, not significant in cerebellum, P > 0.05) (Figure 3A). Conversely, Fmr1 mRNAs containing site II were mostly used in the hippocampus from CGG KI mice (63% and 75% in the cerebellum and hippocampus CGG KI versus 56% and 38% of the corresponding WT brain regions, χ2 test, P < 0.05 in the hippocampus, not significant in cerebellum, P > 0.05) (Figure 3A).
The structure of the 3′-UTR of Fmr1 mRNA and its expression in premutation alleles are completely unknown. As mentioned, the in silico analysis revealed six putative polyadenylation signals in the mouse (Figure 2), while using the 3′-RACE on total brain we could detect only three of them (Figure 3B). To investigate a possible difference with normal alleles, we performed a 3′-RACE using total RNA isolated from WT and CGG KI mice littermates, and cloned and sequenced the PCR products as previously reported (three major bands, data not shown). As shown in Figure 3B, two poly(A) sites were observed in the CGG KI (sites IV and VI). Site IV, the canonical poly(A) signal, was the most utilized in both WT and CGG KI mice (78% in WT and 67% in the CGG KI brain). Site V, which was virtually absent in the brain from the CGG KI mouse, was detected in WT although at low levels (11% in WT and no clones in CGG KI, χ2 test, P < 0.05). Differently, site VI was increased in CGG KI brain (33% in the CGG KI versus 11% in the WT, χ2 test, P < 0.05), possibly suggesting a compensation mechanism for the absence of the isoform generated with poly(A) site V.
Additionally, we aimed to study how different 5′- and 3′-UTRs are assorted in multiple Fmr1 transcripts and whether this variety changes in premutation alleles. We applied an approach based on retro-transcription of the RNA followed by self-ligation of the cDNA. RNAs from cortex, cerebellum and hippocampus from WT and CGG KI were retrotranscribed, the cDNA circularized by ligation and then subjected to PCR using pairs of primers able to amplify only the circularized cDNA (Figure 3C, left panel). The resulting PCR products include both 5′- and 3′-ends of a single mRNA molecule joined in the point of ligation (Figure 3C, left panel).
We used several primers downstream the tss I and tss II as well as upstream poly(A) sites IV and V. Possibly due to the Fmr1 mRNA structure, only the combination of primers upstream tss II (CB375) and poly(A) site IV were able (CB378) to successfully amplify a PCR product corresponding to the Fmr1 mRNA isoform with the longest 5′- (tss II) and 3′-UTR poly(A) VI (Figure 3C, right panel). Sequencing assessed the identity of the PCR products. Our data show that the long 5′-UTR was combined with the long 3′-UTR, although we cannot exclude the existence of other combinations. Of note, the longest UTRs are the ones that change in CGG KI animals.
The mouse model for the premutation shows reduced FMRP levels and altered polyadenylation
To understand whether the Fmr1 mRNA variants could contribute to differential FMRP expression, we measured the protein levels in different mouse brain regions (comparing CGG KI and WT), as well as the polyadenylation state of different Fmr1 transcripts, which normally reflects the stability and the translational efficiency of the mRNA itself.
As previously shown with two different methods (21,22), FMRP levels were decreased in total brain from CGG KI compared to WT mice (Figure 3D, left panel). Moreover, FMRP expression levels were measured in three brain regions (i.e. cortex, hippocampus and cerebellum) of both CGG KI and WT. A significant decrease in FMRP expression was detected in both hippocampus and cerebellum (Figure 3D, central and right panels, n = 3, Student's t-test, P < 0.05), while no significant difference was observed in the cortex. Our findings are in agreement with the observations by Usdin and collaborators on another mouse model for the Fragile X premutation (22) in which a differential deficit of FMRP in different brain areas was observed. These observations raise the question whether in premutation carriers clinical symptoms result from an FMRP deficit in specific brain regions.
To measure the poly(A) length of the different Fmr1 mRNA isoforms, we used the poly(A) test assay (PAT, Figure 4A). Briefly, the poly(A) tail of total mRNAs was tagged as previously described (42) (Figure 4A; see ‘Materials and Methods’ section). The mRNAs were reverse transcribed and amplified by PCR with a forward gene-specific primer. Usually, a polyadenylated mRNA will produce PCR products appearing as a smear, while the same mRNA which goes through an enzymatic shortening of the poly(A) tail (OligodT/RNAse H treated) produces a single PCR product detected as a sharp band (Figure 4A, right panel). β-Actin mRNA, known to have a short poly(A) tail (49), was used as negative control and indeed displayed a polyadenylation tail within 100 nucleotides (nt) in both WT and CGG KI (Figure 4B, upper panel, compare lane 1 to lane 2). To quantify the results, the signal intensity along the lane was plotted as function of the poly(A) tail length (Figure 4C, lower panel) and no major changes in the polyadenylation of β-actin mRNA were detected between WT and CGG KI. Using a gene-specific primer upstream of site IV, we detected all three variants (Figure 4B, left panel), although the mRNAs using sites V and VI could not be distinguished. However, transcripts using site IV had a poly(A) tail at least up to 250 nt in WT (Figure 4C, left panel, compare lane 1 and 2, which represents the control deadenylated mRNA). Poly(A) tails longer than 250 nt could not be detected due to the poly(A) interference of isoforms V and VI. The mRNAs using site IV showed a different distribution between the two genotypes, with an increased population of short poly(A) mRNAs (mainly <100 nt) in the CGG KI compared to WT (Figure 4C, compare lane 3 to 1 in the left panel and grey to black line in the right panel).
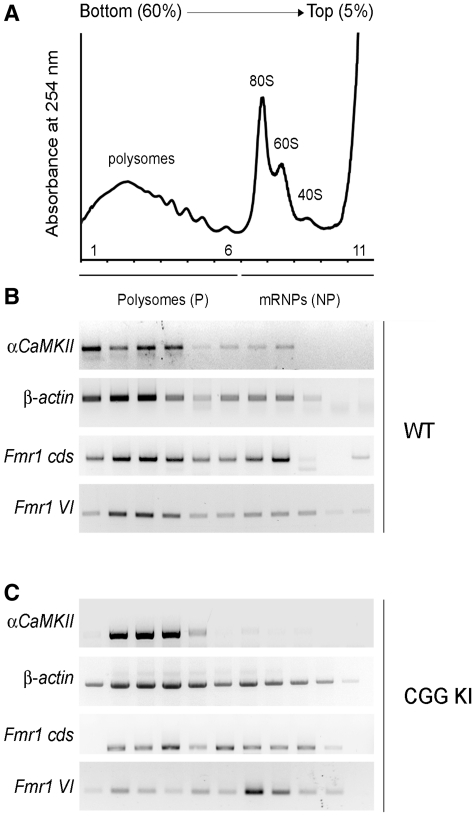
The poly(A) length of transcripts using site V could not be analyzed because of its close vicinity to site VI. Using this technique, Fmr1 mRNA generated with the use of poly(A) site V was detected, although the same mRNA was not revealed using the cloning strategy, possibly due to its very low abundance. However, we were able to discriminate the polyadenylation state of transcripts using site VI, by using a forward primer within sites V and VI. As shown in Figure 4D, left panel, these mRNAs exhibited a poly(A) tail up to 300 nt (compare lane 1 to 2, which represents the deadenylated form). As for transcripts using site IV, the polyadenylation state of this isoform was altered in CGG KI mouse, with an increased population of short poly(A) mRNAs (mainly <50 nt) (Figure 4D, left panel, compare lane 3 to 1, right panel, compare grey to black line). Our findings indicate that the CGG repeat element affects the polyadenylation of at least two 3′-UTR variants (site IV and VI). Specifically, a reduction in the length of the poly(A) tail species was observed in CGG KI mice. Since it has been widely reported that poorly polyadenylated mRNAs are not efficiently translated (50,51), the observed difference in poly(A) tail length could account, at least for a portion, of the reduced FMRP levels found in premutation alleles (Figure 3D). In order to verify this hypothesis, we tested the translational efficiency of the Fmr1 mRNA isoforms by analyzing their partitioning between actively translating polysomes and silent messenger ribonucleoparticles (mRNPs) (representative profile, Figure 5A). Brain extracts from WT and CGG KI mice were fractionated along a sucrose gradient, RNA was extracted from each fraction and subjected to RT-PCR analysis as previously reported (44, 52). As shown in Figure 5B, in WT brain mRNAs with a high translational efficiency, such as αCaMKII and β-actin mRNAs, are predominantly distributed on heavy polysomes rather than light mRNPs. Using a gene specific oligo for the Fmr1 coding sequence (Fmr1 cds), we detected all Fmr1 isoforms actively translated and associated to polysomes, similarly to controls (compare to β-actin mRNA). To quantify the efficiency of mRNA translation, we used the percentage of messenger on polysomes (PMP), calculated as the intensity of the signal on polysome fractions over the total. This analysis showed that Fmr1 mRNA was highly translated, with a PMP of 65%. In addition, we investigated the polyadenylation variant VI (Fmr1 VI), which in agreement with its long poly (A) tail (Figure 4D), was indeed mainly detected on the polysomes in WT brains (PMP = 70%) (Figure 5B). The same studies performed on the CGG KI mice (Figure 5C) showed that while the translation of control mRNAs was comparable between WT and KI, the translational efficiency of Fmr1 mRNAs (cds) was reduced in the KI mice (PMP = 47% in CGG KI versus a PMP = 65% in WT). Our data confirm and extend to brain previous reports of an inverse correlation between translational efficiency and CGG length in premutation (97–195 CGG) (20), unmethylated full mutation (400 CGG) (53) and unmethylated full mutation (266–285 CGG) (54).
Figure 5.
Translational efficiency of Fmr1 mRNA variants in WT and CGG KI mouse brain. (A) Representative polysome/mRNPs profile, showing the fractionation of polysomes (P) and light mRNPs (non-polysomes, NP). The absorbance at 254 nm has been plotted for 11 fractions obtained from a 5–60% sucrose gradient. Ribosomal subunits 40 S and 60 S, the monomer 80 S and the polyribosomes are indicated. (B) RT-PCR on the polysome/mRNPs fractions from WT brains to detect αCaMKII, β-Actin, Fmr1 coding sequence (cds) and Fmr1 isoform VI mRNAs. (C) Same as in (B) from the CGG KI mice.
Isoform VI, consistent with its shorter poly(A) tail in the CGG KI mice (Figure 4D), was shifted from polysomes to lighter fractions (Figure 5C), indicating a reduction in its translation efficiency (PMP = 25% in CGG KI versus 70% in WT). Remarkably, in the CGG KI, the polysome/mRNP distribution of isoform VI changes to a larger extent compared to all Fmr1 transcripts. All together, these data indicate that in WT brain the Fmr1 isoform VI contributes mainly to the FMRP synthesis, but its translation is reduced in the premutation allele, possibly due to the combinatorial effect of the CGG expansion and altered polyadenylation. In the CGG KI allele, specific variants seem to display a different pattern of regulation compared to WT alleles.
Differential usage of tss and polyadenylation sites in human premutation carriers
To extend our findings to human brain, we investigated the structure of both 5′ and 3′ ends of FMR1 mRNA from ‘post mortem’ brain tissues. 5′-RLM-RACE was performed on total RNA isolated from cerebellum and hippocampus of normal (27 CGG repeats) and premutation (85 CGG repeats) male subjects. As shown in Figure 6A, a total of 24 clones from normal cerebellum, 35 from premutation cerebellum, 37 from normal hippocampus, and 41 from premutation hippocampus were sequenced and analyzed for the presence of alternative tss. In human brain, as well as in lymphoblastoid cell lines and primary astrocytes (37), the usage of tss appears to be dependent on the CGG repeat length, such that in normal FMR1 alleles site I represents the major transcription start site, at least in cerebellum (71% in normal cerebellum and 51% in normal hippocampus). This site is selected preferentially in normal cerebellum compared to premutation tissue (71% versus 28%, χ2 test, P < 0.01). In contrast, transcripts containing site II are more abundant in premutation tissues (46% and 44% in cerebellum and hippocampus from premutation patient versus 4% and 24% from normal individual, χ2 test, P < 0.001 and P < 0.05, respectively) (Figure 6A). FMR1 transcripts containing site III were not identified in cerebellum and detected at very low level in hippocampus (both premutation and normal). Interestingly, in lymphoblasts and astrocytes this site was found to be highly used in expanded alleles with CGG repeat number in the upper premutation range (∼180 CGG repeats) (37), while the brain premutation sample analyzed here had only 85 CGG repeats. The distribution of site IV did not change between normal and premutation cerebellum (25% and 26%, respectively), but decreased in the premutation hippocampus compared to normal (7% versus 22%, χ2 test, P < 0.001). It is noteworthy that although the three initiation sites were also present in premutation alleles, their distribution was significantly different from the distribution observed for the normal allele. Moreover, there was a significant difference in the distribution of site I and II between normal and premutation alleles (χ2 test, P < 0.01 and P < 0.001, respectively) in cerebellum, demonstrating that the downstream CGG repeat element influences the start site selection of the FMR1 gene, extending the previous findings to brain tissue.
Finally, we investigated the structure of the human FMR1 3′-UTR. We compared the poly(A) signal usage in normal and premutation carriers by sequencing a total of 54 clones (27 for each condition) (Figure 6B). Site IV, the canonical poly(A) signal, was predominantly used compared to the other sites in both alleles (59% in the normal and 63% in the premutation brain). Site VI exhibited lower usage than site IV and was comparable for the two alleles (18.5% in the normal and 22% in the premutation brain). Interestingly, site V usage was only detected in normal brain (18.5% of usage) with no evidence of usage in the premutation tissue (χ2 test, P < 0.05). Finally, sites I and II already rarely used in the premutation brain (4% and 11%, respectively), were almost not detected in the normal condition (no clones and 4%, respectively).
DISCUSSION
The UTRs of mRNAs play a critical role in many steps of post-transcriptional control of gene expression. In fact, alternative transcripts using different 5′- or 3′-UTRs may account for different mRNA stability, translational efficiency and localization (29). Mutations or perturbations of these regions may alter the RNA interactions with specific proteins and may shift the physiological balance from health to disease, resulting in conditions such as cancer and bipolar affective disorder (30,55). Mutations in the 5′-UTRs, which alter mRNA translation have been reported to be involved in a number of human diseases including breast cancer, Alzheimer's Disease, Hemoglobin H disease and congenital heart disease (30,56). These observations support a crucial role of the UTRs in health and disease (30).
In the current study, we have investigated the structure of the UTRs of the FMR1 gene responsible for two severe neurological pathologies, FXS and FXTAS. Through in silico and RACE analyses, we have characterized both the 5′- and 3′-UTRs of FMR1 mRNA in mouse and human brain with respect to transcription initiation and to polyadenylation site selection. We found that in human brain the FMR1 mRNA is expressed as different isoforms that contain alternative transcriptional start and polyadenylation sites (Figures 1, 2 and 6). Specifically, three transcription start sites, previously described in human cultured cells (37), were also identified in brain together with a new transcriptional start site (site IV, Figures 1 and 6 A), the latter suggesting the presence of a brain FMR1 isoform that could account for specific neuronal functions. These findings were also extended to the mouse brain, where two sites were identified, the first one corresponding to a human site and the other located upstream of the human site III (Figures 1 and 3 A). Of note, both human and mouse FMR1 promoters lack of a functional TATA box or TATA-like sequences element; all the tss are located within Inr-like motifs.
Besides the variety generated by the 5′-UTR, FMR1 mRNA has three different variants due to the selection of different polyadenylation signals in the 3′-UTR (Figure 2). The canonical poly(A) (site IV, sequence AAUAAA) is predominant, while two single-nucleotide variants of this consensus sequence (sites V and VI) are less used (Figures 3B and 6B).
To determine whether diverse UTRs play a role in premutation alleles, we have studied the FMR1 variants in the brain of CGG KI mice and in ‘post mortem’ brain tissues from a human premutation carrier. Noteworthy, the transcript variants change when the CGG triplets are expanded over the normal threshold, namely in the CGG KI mouse (Figure 3) and in the premutation carrier (Figure 6). In both species, the premutation allele favors the selection of long 5′-UTR, indicating that also in neurons the expanded CGG element affects upstream transcription events. In the mouse, the longest 5′-UTR variant is more expressed in the hippocampus from CGG KI compared to WT; in human tissues, the percentage of the longest variant (tss III) does not change in premutation carriers, but this site is rarely used. On the contrary, in both cerebellum and hippocampus, the other long variant (site II) is significantly more represented in premutation carriers, while shorter variants (sites I and IV in cerebellum and hippocampus, respectively) are less expressed. Additional variations occur in the 3′-UTR of premutation alleles, where the isoform generated by the poly(A) site V decreases in both species (Figures 3B and 6B) and only in human two short variants appear (Figure 6B). The combination of alternative 5′- and 3′-UTRs can potentially generate a variety of FMR1 transcripts. Using a method based on self-ligation of single mRNA species, we could identify at least one variant harboring the longest 5′- and 3′-UTRs (Figure 3C). Evidence for a direct effect of the CGG expansion on FMR1 translation efficiency was initially reported in both fibroblast (54) and lymphoblastoid cell lines (20,53). In particular, Feng et al. (54) showed that FMR1 mRNAs harboring an expansion of 266–285 CGG expansion (full mutation range) are associated with stalled 40 S ribosomal subunits, thus leading to translational inhibition. Additional effects can arise from other events affecting mRNA translation, such as polyadenylation (50,51). Indeed, we found that the premutation has an effect on the polyadenylation state for at least two Fmr1 mRNA variants: the transcripts generated using poly(A) site IV and VI have reduced polyadenylation in the CGG KI genotype (Figure 4C, D). Since site IV is the most used in both normal and premutation alleles and site VI is preferentially selected in CGG KI brain, the decreased polyadenylation of both isoforms could contribute to the observed reduction of FMRP expression levels observed in premutation alleles (Figure 3D). Consistently, Fmr1 mRNAs, in particular isoform VI, show an altered polysome/mRNP distribution in CGG KI brain, suggesting reduced translation efficiency (Figure 5C). Thus, the overall effect of 5′ and 3′ variations, including the expanded CGG element, results in a significant reduction of FMRP levels in the brain of CGG KI mice, especially in the cerebellum and the hippocampus (Figure 3D).
The combination of alternative 5′- and 3′-UTRs increases the complexity of FMR1 expression and the possible roles of RNA isoforms in neuronal physiology, as well as in FMR1 related disorders. Alternative FMR1 transcripts generated with different UTRs may account for differential FMRP expression in various cell types. FMRP is expressed in both neuron and glia (41,43,57) and specific FMR1 mRNA isoforms could also be differentially expressed in these two cell types. Intriguingly, the tss usage seems to display a tissue specificity, with differences between cerebellum and hippocampus in both species. This opens the possibility that specific FMR1 transcripts may contribute to a tissue-specific profiling of different neuronal cell types. Furthermore, differential UTR usage may regulate the FMR1 mRNA translational efficiency. In organisms ranging from viruses to humans, protein-mediated interactions between transcript termini result in the formation of an RNA loop. Such RNA ‘circularization’ is thought to increase translational efficiency (58). It is tempting to hypothesize that unfavorable combination of different 5′-UTRs and 3′-UTRs could contribute to the lower efficiency of translation observed in premutation alleles (20). Moreover, mRNAs with short poly(A) tail are translated less efficiently (50,51). Indeed, the decreased poly(A) tail length of Fmr1 mRNA using poly(A) site IV and VI points towards the mechanism of an impaired translation, possibly due to a non-efficient circularization of the mRNA; consistently, the transcripts using poly(A) site VI have a reduced translational efficiency in the CGG KI brains. Finally, UTR utilization is likely to be involved in the correct sub-cellular localization of FMR1 mRNA. It has been shown that Fmr1 is transported along the dendrites (41,59), so alternative 3′-UTRs may be involved in its localization to specific sub-cellular compartments. Interestingly, it has been shown that Bdnf mRNA containing the longest 3′-UTR is localized at the dendrites and it has a distinct role in spine morphology and synaptic plasticity (60). Further studies are required to determine if a differential sub-cellular localization and function of the alternative transcripts occur in the FMR1 gene and if it may have a role in the pathology of FMR1 related disorders including FXS, FXTAS and FXPOI.
FUNDING
National Fragile X Foundation (to F.T.); UC Davis Health System Research Award (to F.T.); National Institute of Child Health and Human Development HD02274 (to F.T.). the National Institutes of Health (HD040661) (to P.J.H.); COFIN-2003 and 2008 (to C.B.), VIB and Telethon (to C.B.), Associazione Italiana X Fragile (to S.D.R.), Methusalem grant to Bart De Strooper (supporting S.D.R. and G.L.F.) VIB. Funding for open access charge: National Fragile X Foundation (to F.T.) and VIB (to C.B.). The charges should be equally divided between F.T. and C.B.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Tilmann Achsel for his help with the PAT assay and for critical reading of the manuscript. We are extremely grateful to Elien Theuns for her support with the polysome/mRNPs gradients. We are thankful to Katrien Gobien and Jonathan Royaert for technical help and Eliane Cherretté for assistance.
REFERENCES
- 1.Jacquemont S, Hagerman RJ, Hagerman PJ, Leehey MA. Fragile-X syndrome and fragile X-associated tremor/ataxia syndrome: two faces of FMR1. Lancet Neurol. 2007;6:45–55. doi: 10.1016/S1474-4422(06)70676-7. [DOI] [PubMed] [Google Scholar]
- 2.Lightbody AA, Reiss AL. Gene, brain, and behavior relationships in fragile X syndrome: evidence from neuroimaging studies. Dev. Disabil. Res. Rev. 2009;15:343–352. doi: 10.1002/ddrr.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassell GJ, Warren ST. Fragile X syndrome: loss of local mRNA regulation alters synaptic development and function. Neuron. 2008;60:201–214. doi: 10.1016/j.neuron.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pieretti M, Zhang FP, Fu YH, Warren ST, Oostra BA, Caskey CT, Nelson DL. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991;66:817–822. doi: 10.1016/0092-8674(91)90125-i. [DOI] [PubMed] [Google Scholar]
- 5.Verkerk AJ, Pieretti M, Sutcliffe JS, Fu YH, Kuhl DP, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang FP, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 6.De Rubeis S, Bagni C. Fragile X mental retardation protein control of neuronal mRNA metabolism: Insights into mRNA stability. Mol. Cell Neurosci. 2010;43:43–50. doi: 10.1016/j.mcn.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 7.Zukin RS, Richter JD, Bagni C. Signals, synapses, and synthesis: how new proteins control plasticity. Front Neural. Circuits. 2009;3:14. doi: 10.3389/neuro.04.014.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pfeiffer BE, Huber KM. The state of synapses in fragile X syndrome. Neuroscientist. 2009;15:549–567. doi: 10.1177/1073858409333075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hagerman PJ, Hagerman RJ. The fragile-X premutation: a maturing perspective. Am. J. Hum. Genet. 2004;74:805–816. doi: 10.1086/386296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bourgeois JA, Coffey SM, Rivera SM, Hessl D, Gane LW, Tassone F, Greco C, Finucane B, Nelson L, Berry-Kravis E, et al. A review of fragile X premutation disorders: expanding the psychiatric perspective. J. Clin. Psychiatry. 2009;70:852–862. doi: 10.4088/JCP.08m04476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allingham-Hawkins DJ, Babul-Hirji R, Chitayat D, Holden JJ, Yang KT, Lee C, Hudson R, Gorwill H, Nolin SL, Glicksman A, et al. Fragile X premutation is a significant risk factor for premature ovarian failure: the International Collaborative POF in Fragile X study–preliminary data. Am. J. Med. Genet. 1999;83:322–325. [PMC free article] [PubMed] [Google Scholar]
- 12.Toniolo D, Rizzolio F. X chromosome and ovarian failure. Semin. Reprod. Med. 2007;25:264–271. doi: 10.1055/s-2007-980220. [DOI] [PubMed] [Google Scholar]
- 13.Berry-Kravis E, Abrams L, Coffey SM, Hall DA, Greco C, Gane LW, Grigsby J, Bourgeois JA, Finucane B, Jacquemont S, et al. Fragile X-associated tremor/ataxia syndrome: clinical features, genetics, and testing guidelines. Mov. Disord. 2007;22:2018–2030. doi: 10.1002/mds.21493. [DOI] [PubMed] [Google Scholar]
- 14.Cornish KM, Kogan C, Turk J, Manly T, James N, Mills A, Dalton A. The emerging fragile X premutation phenotype: evidence from the domain of social cognition. Brain Cogn. 2005;57:53–60. doi: 10.1016/j.bandc.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 15.Farzin F, Perry H, Hessl D, Loesch D, Cohen J, Bacalman S, Gane L, Tassone F, Hagerman P, Hagerman R. Autism spectrum disorders and attention-deficit/hyperactivity disorder in boys with the fragile X premutation. J. Dev. Behav. Pediatr. 2006;27:S137–S144. doi: 10.1097/00004703-200604002-00012. [DOI] [PubMed] [Google Scholar]
- 16.Tassone F, Hagerman RJ, Taylor AK, Gane LW, Godfrey TE, Hagerman PJ. Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am. J. Hum. Genet. 2000;66:6–15. doi: 10.1086/302720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenneson A, Zhang F, Hagedorn CH, Warren ST. Reduced FMRP and increased FMR1 transcription is proportionally associated with CGG repeat number in intermediate-length and premutation carriers. Hum. Mol. Genet. 2001;10:1449–1454. doi: 10.1093/hmg/10.14.1449. [DOI] [PubMed] [Google Scholar]
- 18.Allen EG, He W, Yadav-Shah M, Sherman SL. A study of the distributional characteristics of FMR1 transcript levels in 238 individuals. Hum. Genet. 2004;114:439–447. doi: 10.1007/s00439-004-1086-x. [DOI] [PubMed] [Google Scholar]
- 19.Tassone F, Beilina A, Carosi C, Albertosi S, Bagni C, Li L, Glover K, Bentley D, Hagerman PJ. Elevated FMR1 mRNA in premutation carriers is due to increased transcription. RNA. 2007;13:555–562. doi: 10.1261/rna.280807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Primerano B, Tassone F, Hagerman RJ, Hagerman P, Amaldi F, Bagni C. Reduced FMR1 mRNA translation efficiency in fragile X patients with premutations. RNA. 2002;8:1482–1488. [PMC free article] [PubMed] [Google Scholar]
- 21.Brouwer JR, Huizer K, Severijnen LA, Hukema RK, Berman RF, Oostra BA, Willemsen R. CGG-repeat length and neuropathological and molecular correlates in a mouse model for fragile X-associated tremor/ataxia syndrome. J. Neurochem. 2008;107:1671–1682. doi: 10.1111/j.1471-4159.2008.05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Entezam A, Biacsi R, Orrison B, Saha T, Hoffman GE, Grabczyk E, Nussbaum RL, Usdin K. Regional FMRP deficits and large repeat expansions into the full mutation range in a new Fragile X premutation mouse model. Gene. 2007;395:125–134. doi: 10.1016/j.gene.2007.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brouwer JR, Willemsen R, Oostra BA. The FMR1 gene and fragile X-associated tremor/ataxia syndrome. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2009;150B:782–798. doi: 10.1002/ajmg.b.30910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garcia-Arocena D, Hagerman PJ. Advances in understanding the molecular basis of FXTAS. Hum. Mol. Genet. 2010;19:R83–R89. doi: 10.1093/hmg/ddq166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tan H, Li H, Jin P. RNA-mediated pathogenesis in fragile X-associated disorders. Neurosci. Lett. 2009;466:103–108. doi: 10.1016/j.neulet.2009.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tassone F, Iwahashi C, Hagerman PJ. FMR1 RNA within the intranuclear inclusions of fragile X-associated tremor/ataxia syndrome (FXTAS) RNA Biol. 2004;1:103–105. doi: 10.4161/rna.1.2.1035. [DOI] [PubMed] [Google Scholar]
- 27.Greco CM, Hagerman RJ, Tassone F, Chudley AE, Del Bigio MR, Jacquemont S, Leehey M, Hagerman PJ. Neuronal intranuclear inclusions in a new cerebellar tremor/ataxia syndrome among fragile X carriers. Brain. 2002;125:1760–1771. doi: 10.1093/brain/awf184. [DOI] [PubMed] [Google Scholar]
- 28.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 29.Mignone F, Gissi C, Liuni S, Pesole G. Untranslated regions of mRNAs. Genome Biol. 2002;3:REVIEWS0004. doi: 10.1186/gb-2002-3-3-reviews0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chatterjee S, Pal JK. Role of 5′- and 3′-untranslated regions of mRNAs in human diseases. Biol. Cell. 2009;101:251–262. doi: 10.1042/BC20080104. [DOI] [PubMed] [Google Scholar]
- 31.Eichler EE, Richards S, Gibbs RA, Nelson DL. Fine structure of the human FMR1 gene. Hum. Mol. Genet. 1993;2:1147–1153. doi: 10.1093/hmg/2.8.1147. [DOI] [PubMed] [Google Scholar]
- 32.Sittler A, Devys D, Weber C, Mandel JL. Alternative splicing of exon 14 determines nuclear or cytoplasmic localisation of fmr1 protein isoforms. Hum. Mol. Genet. 1996;5:95–102. doi: 10.1093/hmg/5.1.95. [DOI] [PubMed] [Google Scholar]
- 33.Ashley CT, Sutcliffe JS, Kunst CB, Leiner HA, Eichler EE, Nelson DL, Warren ST. Human and murine FMR-1: alternative splicing and translational initiation downstream of the CGG-repeat. Nat. Genet. 1993;4:244–251. doi: 10.1038/ng0793-244. [DOI] [PubMed] [Google Scholar]
- 34.Verheij C, Bakker CE, de Graaff E, Keulemans J, Willemsen R, Verkerk AJ, Galjaard H, Reuser AJ, Hoogeveen AT, Oostra BA. Characterization and localization of the FMR-1 gene product associated with fragile X syndrome. Nature. 1993;363:722–724. doi: 10.1038/363722a0. [DOI] [PubMed] [Google Scholar]
- 35.Huang T, Li LY, Shen Y, Qin XB, Pang ZL, Wu GY. Alternative splicing of the FMR1 gene in human fetal brain neurons. Am. J. Med. Genet. 1996;64:252–255. doi: 10.1002/(SICI)1096-8628(19960809)64:2<252::AID-AJMG3>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 36.Xie W, Dolzhanskaya N, LaFauci G, Dobkin C, Denman RB. Tissue and developmental regulation of fragile X mental retardation 1 exon 12 and 15 isoforms. Neurobiol. Dis. 2009;35:52–62. doi: 10.1016/j.nbd.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 37.Beilina A, Tassone F, Schwartz PH, Sahota P, Hagerman PJ. Redistribution of transcription start sites within the FMR1 promoter region with expansion of the downstream CGG-repeat element. Hum. Mol. Genet. 2004;13:543–549. doi: 10.1093/hmg/ddh053. [DOI] [PubMed] [Google Scholar]
- 38.Bontekoe CJ, Bakker CE, Nieuwenhuizen IM, van der Linde H, Lans H, de Lange D, Hirst MC, Oostra BA. Instability of a (CGG)98 repeat in the Fmr1 promoter. Hum. Mol. Genet. 2001;10:1693–1699. doi: 10.1093/hmg/10.16.1693. [DOI] [PubMed] [Google Scholar]
- 39.Tassone F, Pan R, Amiri K, Taylor AK, Hagerman PJ. A rapid polymerase chain reaction-based screening method for identification of all expanded alleles of the fragile X (FMR1) gene in newborn and high-risk populations. J. Mol. Diagn. 2008;10:43–49. doi: 10.2353/jmoldx.2008.070073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Napoli I, Mercaldo V, Boyl PP, Eleuteri B, Zalfa F, De Rubeis S, Di Marino D, Mohr E, Massimi M, Falconi M, et al. The fragile X syndrome protein represses activity-dependent translation through CYFIP1, a new 4E-BP. Cell. 2008;134:1042–1054. doi: 10.1016/j.cell.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 41.Ferrari F, Mercaldo V, Piccoli G, Sala C, Cannata S, Achsel T, Bagni C. The fragile X mental retardation protein-RNP granules show an mGluR-dependent localization in the post-synaptic spines. Mol. Cell. Neurosci. 2007;34:343–354. doi: 10.1016/j.mcn.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 42.di Penta A, Mercaldo V, Florenzano F, Munck S, Ciotti MT, Zalfa F, Mercanti D, Molinari M, Bagni C, Achsel T. Dendritic LSm1/CBP80-mRNPs mark the early steps of transport commitment and translational control. J. Cell Biol. 2009;184:423–435. doi: 10.1083/jcb.200807033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, Bagni C. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 44.Bagni C, Mannucci L, Dotti CG, Amaldi F. Chemical stimulation of synaptosomes modulates alpha -Ca2+/calmodulin-dependent protein kinase II mRNA association to polysomes. J. Neurosci. 2000;20:RC76. doi: 10.1523/JNEUROSCI.20-10-j0004.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Javahery R, Khachi A, Lo K, Zenzie-Gregory B, Smale ST. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol. Cell. Biol. 1994;14:116–127. doi: 10.1128/mcb.14.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan J, Marr TG. Computational analysis of 3′-ends of ESTs shows four classes of alternative polyadenylation in human, mouse, and rat. Genome Res. 2005;15:369–375. doi: 10.1101/gr.3109605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tian B, Mukhopadhyay R, Mathews MB. Polymorphic CUG repeats in human mRNAs and their effects on gene expression. RNA Biol. 2005;2:149–156. doi: 10.4161/rna.2.4.2446. [DOI] [PubMed] [Google Scholar]
- 48.Sheets MD, Ogg SC, Wickens MP. Point mutations in AAUAAA and the poly (A) addition site: effects on the accuracy and efficiency of cleavage and polyadenylation in vitro. Nucleic Acids Res. 1990;18:5799–5805. doi: 10.1093/nar/18.19.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meijer HA, Bushell M, Hill K, Gant TW, Willis AE, Jones P, de Moor CH. A novel method for poly(A) fractionation reveals a large population of mRNAs with a short poly(A) tail in mammalian cells. Nucleic Acids Res. 2007;35:e132. doi: 10.1093/nar/gkm830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gallie DR. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- 51.Sonenberg N, Dever TE. Eukaryotic translation initiation factors and regulators. Curr. Opin. Struct. Biol. 2003;13:56–63. doi: 10.1016/s0959-440x(03)00009-5. [DOI] [PubMed] [Google Scholar]
- 52.Gu W, Pan F, Singer RH. Blocking beta-catenin binding to the ZBP1 promoter represses ZBP1 expression, leading to increased proliferation and migration of metastatic breast-cancer cells. J. Cell Sci. 2009;122:1895–1905. doi: 10.1242/jcs.045278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pietrobono R, Tabolacci E, Zalfa F, Zito I, Terracciano A, Moscato U, Bagni C, Oostra B, Chiurazzi P, Neri G. Molecular dissection of the events leading to inactivation of the FMR1 gene. Hum. Mol. Genet. 2005;14:267–277. doi: 10.1093/hmg/ddi024. [DOI] [PubMed] [Google Scholar]
- 54.Feng Y, Zhang F, Lokey LK, Chastain JL, Lakkis L, Eberhart D, Warren ST. Translational suppression by trinucleotide repeat expansion at FMR1. Science. 1995;268:731–734. doi: 10.1126/science.7732383. [DOI] [PubMed] [Google Scholar]
- 55.Lopez de Silanes I, Quesada MP, Esteller M. Aberrant regulation of messenger RNA 3′-untranslated region in human cancer. Cell Oncol. 2007;29:1–17. doi: 10.1155/2007/586139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pickering BM, Willis AE. The implications of structured 5′ untranslated regions on translation and disease. Semin. Cell Dev. Biol. 2005;16:39–47. doi: 10.1016/j.semcdb.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 57.Wang H, Ku L, Osterhout DJ, Li W, Ahmadian A, Liang Z, Feng Y. Developmentally-programmed FMRP expression in oligodendrocytes: a potential role of FMRP in regulating translation in oligodendroglia progenitors. Hum. Mol. Genet. 2004;13:79–89. doi: 10.1093/hmg/ddh009. [DOI] [PubMed] [Google Scholar]
- 58.Mazumder B, Seshadri V, Fox PL. Translational control by the 3′-UTR: the ends specify the means. Trends Biochem. Sci. 2003;28:91–98. doi: 10.1016/S0968-0004(03)00002-1. [DOI] [PubMed] [Google Scholar]
- 59.Antar LN, Dictenberg JB, Plociniak M, Afroz R, Bassell GJ. Localization of FMRP-associated mRNA granules and requirement of microtubules for activity-dependent trafficking in hippocampal neurons. Genes Brain Behav. 2005;4:350–359. doi: 10.1111/j.1601-183X.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- 60.An JJ, Gharami K, Liao GY, Woo NH, Lau AG, Vanevski F, Torre ER, Jones KR, Feng Y, Lu B, et al. Distinct role of long 3′ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell. 2008;134:175–187. doi: 10.1016/j.cell.2008.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chiang PW, Carpenter LE, Hagerman PJ. The 5′-untranslated region of the FMR1 message facilitates translation by internal ribosome entry. J. Biol. Chem. 2001;276:37916–37921. doi: 10.1074/jbc.M105584200. [DOI] [PubMed] [Google Scholar]
- 62.Dobson T, Kube E, Timmerman S, Krushel LA. Identifying intrinsic and extrinsic determinants that regulate internal initiation of translation mediated by the FMR1 5′ leader. BMC Mol. Biol. 2008;9:89. doi: 10.1186/1471-2199-9-89. [DOI] [PMC free article] [PubMed] [Google Scholar]