Abstract
Phage-mediated transfer of microbial genetic elements plays a crucial role in bacterial life style and evolution. In this study, we identify the RinA family of phage-encoded proteins as activators required for transcription of the late operon in a large group of temperate staphylococcal phages. RinA binds to a tightly regulated promoter region, situated upstream of the terS gene, that controls expression of the morphogenetic and lysis modules of the phage, activating their transcription. As expected, rinA deletion eliminated formation of functional phage particles and significantly decreased the transfer of phage and pathogenicity island encoded virulence factors. A genetic analysis of the late promoter region showed that a fragment of 272 bp contains both the promoter and the region necessary for activation by RinA. In addition, we demonstrated that RinA is the only phage-encoded protein required for the activation of this promoter region. This region was shown to be divergent among different phages. Consequently, phages with divergent promoter regions carried allelic variants of the RinA protein, which specifically recognize its own promoter sequence. Finally, most Gram-postive bacteria carry bacteriophages encoding RinA homologue proteins. Characterization of several of these proteins demonstrated that control by RinA of the phage-mediated packaging and transfer of virulence factor is a conserved mechanism regulating horizontal gene transfer.
INTRODUCTION
Horizontal gene transfer (HGT) between microorganisms has a great impact on the evolution of bacterial pathogens. Phages, plasmids and pathogenicity islands have all been recognized as mobile carriers of virulence-associated gene clusters. In HGT between bacteria, phages play important roles by carrying accessory virulence factors and by acting as gene transfer vehicles. Both functions are well documented in Staphylococcus aureus phages. Many clinically relevant toxins from staphylococci, including Panton–Valentine leukocidin (PVL), staphylokinase, enterotoxin A and exfoliative toxin A, are phage-encoded [recently reviewed in (1)]. In addition, a number of staphylococcal superantigens are encoded on a family of pathogenicity islands (SaPIs) that exploit certain helper bacteriophages for high frequency horizontal transfer. In the absence of a helper phage, SaPIs reside stably in the chromosomes of their host bacteria under the control of the master repressor Stl (2). Following helper phage infection, SaPIs are specifically de-repressed by phage-encoded antirepressors (3) (M.D. Harwich et al., in preparation), leading to their excision from the chromosome and their autonomous replication (4). The SaPI-encoded small terminase subunit then directs encapsidation of the replicated SaPI DNA (5) in phage-like particles composed of phage virion proteins (6,7) but with smaller capsids that accommodate the SaPI genome (generally 15–18 kb) while excluding the larger genome of the helper phage (8,9).
Relatively little is known about the biology of staphylococcal phages, despite their relevance in the pathogenic process of S. aureus and the availability of more than a hundred S. aureus phage and prophage genome sequences in public databases. Most known staphylococcal phages are temperate, tailed bacterial viruses belonging to the family Siphoviridae. The genomes of these phages are organized in discrete functional modules, including those for lysogeny, DNA replication, phage assembly, DNA packaging and host cell lysis. Like other bacteriophages, these staphylococcal phages are predicted to exert temporal control of gene expression during infection of a sensitive host, or during induction from a lysogenic strain. This temporal transcription of phage genes is expected to be divided into at least three stages; early (expression of the lysogeny and control modules), middle (replication) and late (packaging, assembly and lysis modules).
In this work, we have characterized genes controlling expression of the phage late genes. Previous results from our group, using lysogenic, SaPI-positive strains, demonstrated that in the absence of phage induction, no phage or SaPI particles were produced and there was no horizontal transfer of the SaPI-encoded virulence factors. Since SaPIs are encapsidated in phage-encoded particles, we expected that control of expression of the morphogenetic and lysis modules of the staphylococcal phages is an essential step in phage-mediated transfer of virulence factors encoded on these mobile genetic elements (MGE) as well as for the transfer of toxins carried on the phage genomes themselves. In this study we have examined transcriptional control of the highly conserved morphogenetic gene cluster present in phages ϕ11 and 80α, which serve as helpers for some of the well-studied SaPIs, as well as in the clinically relevant phage ϕSLT, which encodes the PVL toxin. Phage ϕ11 mobilizes SaPIbov1 while 80α induces several SaPIs, including SaPI1, SaPI2, SaPIn1, SaPIbov1 and SaPIbov2 (10). We demonstrate that the phage rinA gene, previously reported to be a regulator of int transcription (11), is actually an activator of phage late gene transcription and not of int. Phages lacking rinA did not generate either phage or SaPI functional particles, and were defective in horizontal transfer of MGE-encoded virulence factors. Furthermore, since RinA homologs are also present in phages from other clinically relevant Gram-positive bacteria, including Streptococcus suis, Clostridium botulinum, Listeria monocytogenes and Enterococcus faecalis, we extended our results by demonstrating that these RinA homologues are functionally identical to those characterized from staphylococci, which indicates that control of phage morphogenetic genes by RinA is a widespread mechanism used by phages infecting Gram-positive bacteria.
MATERIAL AND METHODS
Bacterial strains and growth conditions
Bacterial strains used in these studies are listed in Supplementary Table S1. Bacteria were grown at 37°C overnight on Trypticase Soy (TSA) agar medium, supplemented with antibiotics as appropriate. Broth cultures were grown at 37°C in Trypticase Soy Broth (TSB) broth with shaking (240 rpm).
For prophage induction, bacteria were grown in TSB to OD540 = 0.4 and induced by adding mitomycin C (2 ug/ml). Cultures were grown at 32°C with slow shaking (80 rpm). Lysis usually occurred within 3 h. Samples were removed at various time points after phage induction, and standard SDS minilysates were prepared and separated on 0.7% agarose gels, as described previously (12,13). Standard procedures were used for preparation and analysis of phage lysates, lysogens and transduction in S. aureus, as described previously (12).
DNA methods
General DNA manipulations were performed by standard procedures (14,15). Oligonucleotides used in this study are listed in Supplementary Table S3. Labeling of the probes and DNA hybridization were performed according to the protocol supplied with the PCR-DIG DNA-labeling and chemiluminescent detection kit (Roche). To produce the strains carrying mutant prophages, allelic exchange was performed using derivatives of plasmid pMAD (16) carrying the desired mutations, as described previously (8). Plasmid constructs (Supplementary Table S2) were prepared by cloning PCR products obtained from oligonucleotide primers as listed in Supplementary Table S3. All constructs were sequenced by the Institute core sequencing laboratory.
Enzyme assays
β-Lactamase assays, using nitrocefin as substrate, were performed as described (17), using a Thermomax (Molecular Devices) microtiter plate reader. Cells were obtained in exponential phase. β-Lactamase units are defined as (Vmax)/OD650.
Real-time quantitative PCR
Total S. aureus RNA was prepared using the Fast RNA-Blue kit (Bio101) according to the manufacturer's instructions. Two micrograms of each RNA were subjected, in duplicate, to DNase I (Invitrogen) treatment for 30 min at 37°C. The enzyme was inactivated at 65°C in the presence of EDTA. To verify the absence of genomic DNA in every sample, the RNA duplicates were reverse transcribed in the presence and absence of M-MLV Reverse Transcriptase (Invitrogen). All preparations were purified using QIAquick PCR purification kit (Qiagen). Twenty-five nanograms of each reaction product was used for a real-time quantitative PCR using the iCycler machine (Bio-Rad) and the LC-DNA Master SYBR Green I mix (Biorad). The different genes were amplified using oligonucleotides listed in Supplementary Table S2. The gyrB transcripts that are constitutively expressed were amplified as an endogenous control. The level of expression of the different genes were normalized with respect to gyrB expression. Only samples with no amplification of gyrB in the minus reverse transcriptase aliquot were included in the study. To monitor the specificity, the final PCR products were analyzed by melting curves and electrophoresis. In each experiment, all the reactions were performed in triplicate. The relative transcriptional levels within distinct experiments were determined by using the 2−ΔΔCT method (18). The results show the average ± SEM of at least four independent experiments.
Purification of RinA protein
The rinA gene was amplified by PCR using primers orf28phi11-5cX and orf28phi11-7mB (Supplementary Table S3), digested with XbaI and BamHI, and cloned into plasmid pGEX-4 T-1 (GE Healthcare). Expression and purification of the GST-tagged fusion RinA protein was performed following the instructions of the manufacturer. The purity of the purified GST-tagged RinA fusion protein was confirmed by sodium dodecyl sulphate (SDS) gels stained with Commassie Brilliant Blue R-250. The purified protein was found to be more than 98% pure in an SDS-12% polyacrylamide gel. The concentration of the purified proteins was determined by the Bradford protein assay (Bio-Rad, Hercules, CA), using bovine serum albumin as the standard.
Mobility shift assays
Electrophoretic mobility shift assays (EMSAs) were performed as described before (3,5) using purified RinA protein and a DIG-labeled DNA fragment obtained by PCR using the oligonucleotides listed in Supplementary Table S3.
Southern blot analysis of phage late promoters
RNA samples isolated during a time course of 80α infection were pooled to yield 100 µg of total RNA which was concentrated to 12 µl using an RNeasy MinElute column (Qiagen). This RNA was then used as a substrate for vaccinia virus capping enzyme, using the ScriptCap m7G Capping System (Epicenter Biotechnologies). The RNA was heated at 65°C for 10 min, placed immediately on ice, and then incubated at 37°C for 1 h in a reaction containing 1× reaction buffer (provided by the manufacturer), 1 mM GTP (including 1 mCi of α-32P GTP), 0.1 mM SAM, 50 U of RNase inhibitor and 25 U of capping enzyme. The labeled RNA was purified using a RNeasy Minelute column, and an aliquot was checked on a gel to ensure that most of the labeled RNA was small enough (>200 bp) to hybridize only to fragments containing the initial transcribed region. The RNA probe was incubated at 70°C for 10 min, and added to 10 ml of UltraHyb (Ambion) for hybridization to Southern blots. A positively charged nylon membrane (ICN) containing restriction fragments of 80α DNA that had been resolved by agarose gel electrophoresis was prepared by capillary transfer using standard methods (15) and pre-hybridized in UltraHyb for 1 h at 42°C prior to addition of the probe. After hybridization overnight at 42°C, membranes were washed twice at 42°C for 10 min in 2× SSC, 0.1% SDS and twice at 42°C for 10 min in 0.1× SSC, 0.1% SDS prior to exposure to a phosphor screen (Molecular Dynamics).
Rapid amplification of cDNA 5′-ends
Amplification of the terS cDNA 5′-end of ϕ11 was performed using the 5′/3′ RACE kit (Roche), according to the manufacturer's protocol. First-strand cDNA synthesis was performed using the oligo ORF29-phi11-sp1c (Supplementary Table S3). The cDNA mixtures were amplified by PCR using the oligo(dT) anchor primer and the gene-specific primers ORF29-phi11-sp2c and ORF29-phi11-sp3c (Supplementary Table S3). The PCR products were purified from 1.2% agarose gels and subjected to DNA sequence analysis. The 5′-end of 80α terS was amplified using the FirstChoice RLM-RACE kit (Ambion) according to the manufacturer’s instructions, after treatment of RNA with Terminator enzyme (Epicentre Biotechnologies) to degrade fragments containing 5′ monophosphate termini. First strand cDNA synthesis was performed using random decamers, after ligation of the 5′ rapid amplification of cDNA 5′/3′ ends (RACE) adapter. Following cDNA synthesis, nested PCR was performed using Pfu Turbo (Stratagene) with terS specific primers MH22 and MH23 (Supplementary Table S3) and the 5′ outer and inner adapter specific primers provided with the kit. Amplicons from the MH22/inner adapter primer reaction were ligated into NruI-digested pBR322 and individual clones were sequenced.
RESULTS
Deletion of rinA eliminates production of both phage and SaPI particles
The virion proteins of ϕ11 and 80α, which are involved in the production of both phage and SaPI particles (6,7,19), are encoded by a cluster of genes that are all oriented in the same direction on the phage genome (Figure 1). We expected phage-encoded factor(s) to be required for expression of these morphogenetic genes. Two attractive candidates for late gene regulation were RinA and RinB, homologous of which are found in the vast majority of S. aureus Siphoviridae and which are encoded by genes immediately upstream of the morphogenetic gene cluster. These two genes had been reported previously to be involved in activation of expression of the cloned phage ϕ11 int gene (11), consistent with a regulatory function. To analyze their possible role as regulators of the phage morphogenetic genes, we separately inactivated either rinA or rinB in a ϕ11 prophage by constructing in-frame deletions. We also introduced SaPIbov1 containing a tetracycline marker inserted in the tst locus (SaPIbov1 tst::tetM) into the strains carrying the prophage mutants, in order to examine effects on production of SaPIbov1 transducing particles as well as infectious phage. The different phage mutants (with or without SaPIs) were analyzed for definable stages of the phage and SaPI life cycles following induction with mitomycin C.
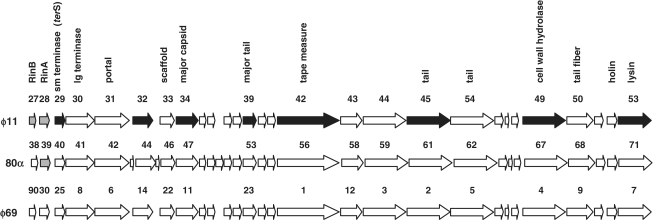
Figure 1.
Location of the genes controlled by RinA. Partial genetic maps of ϕ11 (GenBank accession number AF424781), 80α (accession number DQ517338) and ϕ69 (accession number AY954951), showing the location of rinA and the genes under rinA control. The three phages share 96% nucleotide identity across this entire region. Arrows indicate predicted Open Reading Frames (ORFs), numbered as annotated. Gray arrows indicate the regulatory genes deleted in this study, while black arrows indicate genes for which rinA dependence was directly demonstrated by qRT-PCR.
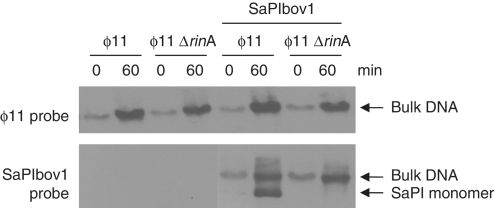
Deletion of rinA eliminated or significantly reduced the production of functional phage or SaPI particles, as assayed by plaque formation or transduction, respectively (Table 1). Furthermore, there was no lysis of the rinA mutant after induction, suggesting that these mutants were defective in expression of the lysis module. The rinA mutant was not impaired in either ϕ11 or SaPIbov1 DNA replication; Southern blot analysis of minilysates prepared 60 min after induction showed that the phage and the SaPI signals in the bulk DNA were amplified to essentially the same degree in the mutant as in the wild-type strains (Figure 2). However, the rinA mutant failed to produce a linear SaPI extrachromosomal band. This is consistent with an encapsidation defect, as the SaPI band is believed to arise from the release of packaged SaPI DNA by disruption of intracellular SaPI capsids (5,6). Similar results were obtained following deletion of the rinA gene from 80α (Table 1), which was expected because both rinA (Supplementary Figure S1) and the entire morphogenetic gene cluster of 80α and ϕ11 (Figure 1) are essentially identical. In order to determine the generality of this mechanism, we also constructed a rinA mutant of phage ϕSLT, which encodes a RinA homolog that is only 23% identical to that of ϕ11 (Supplementary Figure S1), although similar to RinA proteins encoded by other staphylococcal phages (data not shown). We used a detoxified derivative of phage ϕSLT (20) containing a tetracycline-resistant marker (tetM) inserted in the PVL locus (21). This mutant was also deficient in generating functional phage particles (Table 1), and in lysing the bacteria after prophage induction.
Table 1.
Effect of phage mutations on phage titre and SaPI transfera
| Donor strain | ϕ | Plasmid | SaPI | Phage titreb | Transduction titrec |
|---|---|---|---|---|---|
| RN451 | ϕ11 wt | – | – | 1.3 × 109 | – |
| JP1794 | ϕ11 wt | – | SaPIbov1 | 1.4 × 107 | 1.6 × 108 |
| JP4028 | ϕ11 ΔrinA | – | – | <10 | – |
| JP4221 | ϕ11 ΔrinA | pCN51-rinAϕ11 | – | 2.4 × 108 | – |
| JP4128 | ϕ11 ΔrinA | – | SaPIbov1 | <10 | 8.0 × 103 |
| JP5961 | ϕ11 ΔrinA | pCN51-rinAϕ11 | SaPIbov1 | 2.2 × 106 | 2.4 × 107 |
| RN10359 | 80α wt | – | – | 2.9 × 1010 | – |
| JP3603 | 80α wt | – | SaPIbov1 | 6.9 × 108 | 7.8 × 107 |
| JP3602 | 80α wt | – | SaPI1 | 4.5 × 108 | 8.2 × 108 |
| JP4717 | 80α ΔrinA | – | – | <10 | – |
| JP5418 | 80α ΔrinA | pCN51-rinAϕ11 | – | 6.1 × 108 | – |
| JP5293 | 80α ΔrinA | – | SaPIbov1 | <10 | 770 |
| JP5294 | 80α ΔrinA | – | SaPI1 | <10 | 22 |
| JP5419 | 80α ΔrinA | pCN51-rinAϕ11 | SaPIbov1 | 9.0 × 107 | 4.0 × 107 |
| JP5420 | 80α ΔrinA | pCN51-rinAϕ11 | SaPI1 | 2.8 × 107 | 6.9 × 107 |
| JP5011 | ϕSLT wt | – | – | 1.7 × 105 | 8.7 × 104 |
| JP6895 | ϕSLT ΔrinA | – | – | <10 | <10 |
| JP6391 | ϕSLT ΔrinA | pCN51-rinAϕSLT | – | 9.0 × 104 | 7.7 × 103 |
| JP7188 | ϕ11-ϕ69 chimera | – | – | <10 | – |
| JP7242 | ϕ11-ϕ69 chimera | – | SaPIbov1 | <10 | 1.2 × 103 |
| JP7218 | ϕ11-ϕ69 chimera | pCN51-rinAϕ69 | – | ND | – |
| JP7243 | ϕ11-ϕ69 chimera | pCN51-rinAϕ69 | SaPIbov1 | ND | 4.2 × 106 |
aThe means of results from three independent experiments are presented. Variation was within ±5% in all cases.
bPfu/ml of induced culture, using RN4220 (or pCN51-complemented RN4200) as recipient.
cTransductants/ml of induced culture, using RN4220 as recipient.
Figure 2.
Characterization of the ϕ11 rinA mutant. Southern blot of ϕ11 wt and rinA mutant lysates from strains with or without SaPIbov1 tst::tetM. Samples were taken before or 60 min after MC induction, minilysates were prepared, the DNA was separated on agarose and blotted with a phage- or SaPIbov1-specific probe, as indicated. The upper band is ‘bulk’ DNA, including chromosomal, phage and replicating SaPI DNA; the lower band is SaPI linear monomers released from phage heads.
To confirm that the observed phenotypes were due to the rinA mutations, plasmids pJP740 (pCN51-rinAϕ11), and pJP838 (pCN51-rinAϕSLT), carrying the rinA genes from ϕ11 and ϕSLT, respectively, under the control of an inducible promoter, were introduced into strains carrying the corresponding rinA mutant prophage. Complementation with plasmid-encoded RinA restored packaging, lysis and production of viable phage (Table 1), confirming its crucial role in phage gene expression. Similarly, complementation of SaPI-positive ϕ11 and 80α rinA lysogens with plasmid pJP740 restored SaPI packaging and transfer (Table 1).
Mutation of rinB had no effect on either phage production or transfer of SaPI (data not shown). Although an earlier report implicated RinB in the expression of the ϕ11 int gene (11), our results suggest that expression of integrase at levels needed for prophage induction must be RinB-independent. Regulation of int thus appears to be more complex than initially suggested by the report of Ye et al. (11), and is the subject of further investigation. In addition, RinB does not appear to play a role in regulation of late gene expression.
RinA activates transcription of the packaging, assembly and lysis genes
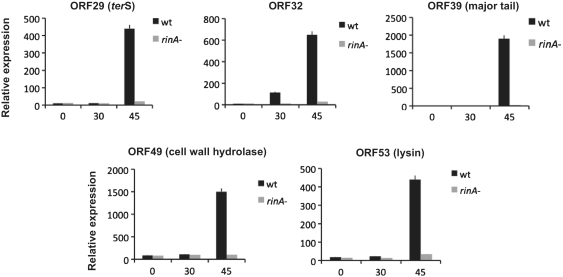
The phenotype of the rinA mutants suggested that RinA might control phage and SaPI packaging through expression of the phage morphogenetic genes. The mRNA levels of several ϕ11 late genes were measured by quantitative real-time PCR at various times (0, 30 and 45 min) after mitomycin C induction, comparing expression levels in the wild-type phage and the rinA mutant. As shown in Figure 3, the rinA mutation resulted in a significant (P < 0.05) decrease in the expression of genes involved in DNA packaging, capsid assembly and cell lysis. Similar results were obtained for the genes encoding the major capsid protein and several components of the phage tail (data not shown). These results indicate that RinA is required for transcription of the phage-encoded packaging and lysis modules, and also confirm the expected temporal expression of these genes.
Figure 3.
RinA controls expression of the morphogenetic and lysis clusters of ϕ11. Real-time quantification of expression of different genes in ϕ11 wild type and the rinA mutant at different times (0, 30 or 45 min) after MC induction of the phage lytic cycle. Expression was normalized to gyrB.
Identification of a RinA-inducible promoter upstream of terS
Based on genome organization, we predicted the existence of at least one promoter directly controlled by RinA and located just upstream of the terminase small subunit gene (terS; ORF29 ϕ11), the first gene in the morphogenetic gene cluster. To determine the transcription start site and locate this putative promoter, 5′ RACE analysis was performed with total RNA isolated after induction of the ϕ11 prophage in strain RN451. A rinA-dependent 5′-end was identified in RNA isolated 60 min after MC induction of the wild-type (wt) strain RN451, which mapped to a C residue located 23 bp upstream of the terS ATG initiation codon (Supplementary Figure S2). In the case of 80α late mRNA, the 5′-end was mapped to the adjacent G residue 24 bp upstream of the terS gene.
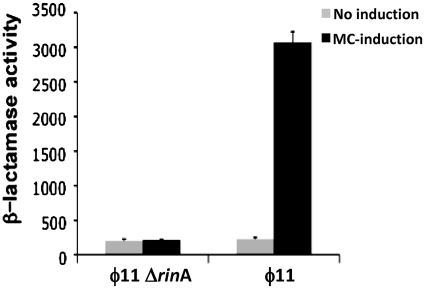
To determine whether the observed 5′-end corresponded to a promoter that was indeed regulated by RinA, a 425 bp fragment spanning the rinA-terS junction was fused to the β-lactamase reporter gene present in plasmid pCN41, generating plasmid pJP742. This plasmid was introduced into strains RN451 (ϕ11 lysogen) and JP4028 (RN451 ΔrinA), and, after prophage induction, the transcription of the β-lactamase reporter was analyzed. As shown in Figure 4, β-lactamase expression increased significantly after induction of the wt strain RN451 but not in its derivative rinA mutant, clearly confirming the existence of a promoter regulated by RinA in this region of the ϕ11 genome.
Figure 4.
Effect of rinA on transcription of terS. RN451 or its derivative rinA mutant strain JP4028 containing plasmid pJP742 (terS–blaZ transcriptional fusion) were assayed 90 min after prophage induction for β-lactamase activity under standard conditions, as described in ‘Material and Methods’ section. Samples were normalized for total cell mass (β-lactamase units/OD650).
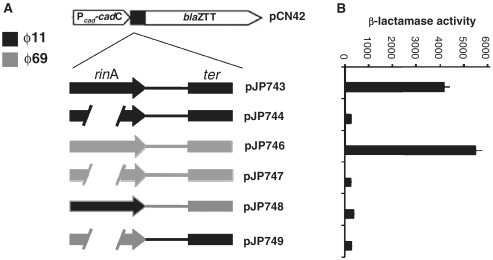
RinA is sufficient to activate ter expression
We next examined whether expression of RinA was enough to induce expression of the terS gene in the absence of other phage-encoded proteins. To test this we made use of plasmid pCN42, which contains a Pcad inducible promoter and a β-lactamase reporter (Figure 5). We introduced into this plasmid a PCR fragment containing the rinA coding sequence (expression of which depends on the Pcad promoter present in the plasmid), the terS promoter region, and the 5′ coding region of the terS gene, which was fused to the β-lactamase reporter. The resulting plasmid, pJP743, is illustrated in Figure 5. As a control, we amplified this region using as a template the DNA from the ϕ11 rinA mutant, generating plasmid pJP744, which does not express RinA. These plasmids were introduced into strain RN4220, and the expression of the β-lactamase reporter was measured. As shown in Figure 5, only the plasmid expressing RinA also expressed the β-lactamase reporter, confirming the previous results and indicating that RinA is the only phage-encoded protein required for the expression of terS. Furthermore, the lack of β-lactamase expression from the rinA mutant plasmid indicates the presence of a transcriptional terminator between the rinA and terS genes. One potential sequence in the region with the characteristics of a factor-independent terminator is indicated in Supplementary Figure S2.
Figure 5.
Specificity of RinA proteins. (A) Schematic representation of the different blaZ transcriptional fusions. (B) Derivatives of strain RN4220 containing each of the indicated plasmids were assayed at mid-exponential phase for β-lactamase activity under standard conditions. Samples were normalized for total cell mass (β-lactamase units/OD650).
Specificity of RinA activation
The ϕ69 RinA homolog has limited similarity to that of ϕ11 (Supplementary Figure S1), although the packaging module of both phages is basically identical (Figure 1). Using the same β-lactamase expression system described above, we showed that RinAϕ69 was also necessary and sufficient to activate ϕ69 terS expression (plasmids pJP746, pJP747; Figure 5). We next tested for possible cross-reactivity between the ϕ11 and ϕ69 RinA proteins by determining whether either could activate terS expression from the other phage. Plasmid pJP748 carries the coding region of rinAϕ11 under the control of the inducible promoter present in pCN42, and the promoter region and terS 5′ coding region from ϕ69 fused to blaZ. In contrast, plasmid pJP749 carries the coding region of rinAϕ69, and the promoter region and partial terS gene from ϕ11. As shown in Figure 5, the two different RinA proteins uniquely activated expression of their cognate terS genes. Since the sequences 5′ to terS are highly similar until ∼120 bp upstream of the presumptive transcription start and then diverge (Supplementary Figure S2B), we speculate that the different RinA proteins recognize these divergent regions to regulate phage late gene expression.
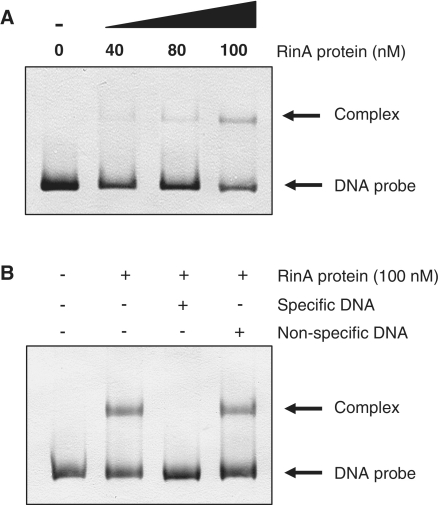
RinA binds to the terS promoter
The results described above suggested that RinA should bind specifically to the terS promoter region. To test this, GST-tagged ϕ11 RinA was expressed in Escherichia coli, purified, and incubated with an end-labeled 272-bp PCR fragment spanning the entire presumptive terS promoter region (see Supplementary Figure S2 for the sequence of this region). Complexes were examined by electrophoretic mobility shift assays. As shown in Figure 6A, the recombinant RinA protein retarded mobility of the terS promoter fragment in a dose-dependent manner. Detectable binding to the labeled fragment was eliminated in the presence of an excess (500-fold) of unlabeled terS fragment, but not when a 500-fold excess of non-specific unlabeled DNA fragment was used as a competitor (Figure 6B). This confirms the specific binding of RinA protein to the terS promoter region, consistent with its role as an activator of terS transcription.
Figure 6.
Binding of RinA to the terS promoter. (A) Electrophoretic mobility of a DIG-labeled promoter fragment was measured in the presence or absence of increasing amounts of purified ϕ11 RinA protein, as indicated on the top line. (B) In the competition assays, 500-fold excesses of specific- or non-specific unlabeled DNA fragments were added, as indicated. In all cases, the concentration of the Dig-labeled probe was 21 nM.
To examine whether the divergent part of the region was involved in the observed DNA-binding activity of RinA, smaller DNA fragments lacking 87 or 50 bp from the 5′-end of this 272 bp fragment were constructed (Supplementary Figure S2). When EMSA was performed using these DNA fragments, no protein–DNA binding was detected (data not shown), consistent with a requirement for virtually all of this upstream divergent region in RinA binding.
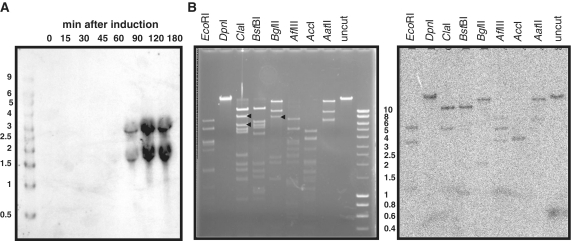
Characterization of the rinA-dependent transcription unit
The terS gene lies just downstream of rinA, and at the beginning of the phage morphogenetic gene cluster. All of the morphogenetic genes are encoded on the same strand, with little intergenic space between them, suggesting that they might comprise a single large transcription unit. In an attempt to characterize the phage transcription units, independent Northern blot analyses were performed on RNA isolated during both 80α and ϕ11 infections. In both cases, although specific hybridization was obtained at appropriate time points, only small RNAs in the 2–4 kb size range could be detected, using probes from several different genes across this region. A representative example is shown in Figure 7A. This suggests that either there are multiple transcription start sites throughout the late gene cluster or that the RNA is rapidly processed. Multiple transcription start sites should yield multiple RNAs containing 5′ triphosphate ends. To estimate the number of promoters present in the phage genome, Southern blot analysis of 80α DNA was performed with pooled phage RNA collected at various times after infection and labeled specifically at the 5′ triphosphate termini arising from initiation of transcription. This analysis does not allow unambiguous identification of unique promoter-containing fragments, but did indicate that the total number of promoters in the phage genome is relatively small (Figure 7B). Furthermore, several unique large restriction fragments that included only late region DNA downstream of terS were not detected in this analysis, consistent with the hypothesis that there is a single promoter for the late genes.
Figure 7.
Detection of transcription start sites in the 80α genome. (A) Northern blot of RNA isolated at the indicated time points after MC induction of an 80α lysogen and probed with a biotinylated terS oligonucleotide. Sizes in kilobase of the markers (Riboladder Long, Bioline) are shown on the left. (B) Agarose gel (left) and Southern blot (right) of 80α virion DNA, digested with the indicated restriction enzymes and probed with total RNA that was isolated from infected cells at various time points, pooled, and 5′-end-labeled at triphosphate termini using vaccinia virus capping enzyme. Sizes in kilobase of the markers (HyperLadder I, Bioline) are indicated to the right of the gel. Black arrowheads on the gel denote examples of unique large fragments from the late region that are not detected in the blot, but to which hybridization would be expected if there were additional transcription start sites downstream of terS.
To investigate further the transcription of the morphogenetic genes, we isolated RNA late in infection with 80α, converted it to cDNA and used that cDNA in endpoint RT-PCR primer pairs flanking intergenic regions throughout the late gene cluster, from terminase through the lysis genes. A product was obtained with all primer pairs from terL to ORF69 (data not shown). The presence of RNA spanning each of the late gene intergenic regions also suggests that these genes are all cotranscribed. The alternative explanation for the endpoint RT-PCR results, i.e. the presence of multiple promoters within the late gene coding sequences reading through into adjacent genes, is not consistent with the absence of transcription start sites in the late region revealed by the Southern blot analysis.
We did not obtain amplification of the intergenic regions between ORF69 and holin or between holin and lysin, although the genomic DNA in this region was readily amplified with the same primer pairs (data not shown). The intergenic regions between ORF69 and holin and between holin and lysin are 58 and 12 nt, respectively, making it unlikely that new transcription initiates in these regions. We were unable to demonstrate the existence of a promoter in this region of the genome by 5′ RACE (data not shown). It is possible that RNA processing explains the lack of PCR amplification across these two intergenic regions, or that RNA secondary structure interfered with cDNA synthesis.
We felt it necessary to further investigate the possibility of an additional promoter in this region, however, because studies on ϕSa3ms, a related temperate siphovirus that carries a gene for staphylococcal enterotoxin SEA just upstream of the lysis genes, suggested that this phage had a promoter near the end of the tail gene cluster (22). We replaced the terS upstream region in strain RN451, which is lysogenic for ϕ11, with the corresponding region from ϕ69, generating strain JP7188. We also introduced SaPIbov1 tst::tetM into this strain, generating strain JP7242. We predicted that if an additional RinA-dependent promoter controls the lysis module, these strains will lyse after MC-induction, although no functional particles will be generated, since the RinA from ϕ11 will not bind to the new RinAϕ69-binding region controlling terS expression. In contrast, if all genes are cotranscribed from the promoter situated 5′ of the terS gene, these strains not only will fail to produce functional SaPI or phage particles, but also will not lyse. As shown in Table 1, the latter was the case; no lysis was observed when the prophage carried the heterologous rinA binding region upstream of terS. In addition, qRT-PCR demonstrated that the expression of genes from the packaging and lysis modules were significantly reduced compared with wild-type strains (data not shown). Finally, strains JP7188 and JP7242 were complemented with plasmid pJP741, which overexpresses the ϕ69 RinA. The complemented strains lysed and produced functional SaPI particles after MC-induction (Table 1). Taken together, the simplest interpretation of all of these results is that there is a single late gene transcription unit, initiating from the RinA-dependent promoter just upstream of terS.
RinA does not control int expression
As discussed earlier, rinA was initially identified as a positive regulatory gene required for expression of the cloned ϕ11int gene (11). To investigate this further, we analyzed int expression in ϕ11 wt and rinA mutant strains, using qRT-PCR. Expression of int was not affected in the rinA mutant (Supplementary Figure S3), suggesting that the RinA protein does not have a direct role controlling the expression of int, at least under conditions of prophage induction.
To confirm this, we analyzed the activity of the Int protein in the rinA mutant. Previous work had identified Int as essential for phage integration and excision (23). We predicted that prophage excision would be reduced in the rinA mutant if RinA is required for int expression. We measured by qPCR the excision activities of prophages ϕ11, ϕ11 ΔrinA and ϕ11 Δint (as a control), using primers hybridizing to the regions flanking the ϕ11 attB site. As expected, excision of the ϕ11 rinA mutant was similar to wt, while no excision was observed in the ϕ11 Δint mutant (data not shown). This confirms the previously reported requirement for Int in prophage excision as well as the lack of involvement of RinA in regulation of int expression during this process.
RinA does not control SaPI-encoded operon I expression
We have previously reported that SaPI packaging depends on expression of the SaPI-encoded homologue of the phage terminase small subunit (terS). In addition, SaPIs remodel assembly of the phage capsid proteins to generate capsids one-third the size of the helper phage capsids, which accommodate their smaller genomes while excluding complete helper phage genomes. This requires two SaPI genes, cp1 and cp2, which are adjacent in a six-gene LexA-regulated operon, operon I, that also encodes the SaPI-specific terS (5). In order to determine if RinA controls expression of SaPIbov1 operon I, we analyzed SaPIbov1 terS expression in ϕ11 wt and rinA mutant strains, using qRT-PCR. Expression of SaPIbov1 terS was not impaired in the rinA mutant, even under conditions where LexA-dependent SaPI operon I expression was blocked by mutation (data not shown), suggesting that the RinA protein does not have a direct role controlling the expression of SaPI-encoded operon I.
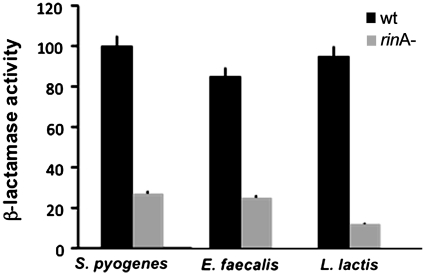
RinA homologues control packaging in phages from other Gram-positive bacteria
RinA from ϕ11 is the archetype for a widespread family of conserved phage proteins. Since RinA homologs are present in phages from a number of other Gram-positive bacteria (Table 2), we analyzed the possibility that the mechanism described here, implicating RinA in control of phage packaging and lysis, is a common one. To test the role of additional RinA homologs in expression of phage packaging genes, we introduced into plasmid pCN42, carrying the blaZ transcription fusions, fragments including the rinA gene, the putative promoter region located upstream of the terS gene, and the 5′ region of the terS gene from prophages found in E. faecalis, Streptococcus pyogenes or Lactococcus lactis (Table 2), generating plasmids pJP535 (E. faecalis), pJP543 (S. pyogenes) and pJP564 (L. lactis). As a control, we also generated plasmids carrying in-frame deletions in each of the rinA homolog genes (plasmids pJP537, pJP544 and pJP565, respectively). We introduced these plasmids into S. aureus strain RN4220 and assayed expression of the β-lactamase reporter gene. Not only were the phage late promoter regions present in these plasmids recognized by the S. aureus RNA polymerase enzyme, but also expression of each was dependent upon the rinA homolog carried on the cloned fragment (Figure 8). This confirms that RinA activation of late transcription is a widespread strategy used to control packaging and lysis by phages infecting Gram-positive bacteria.
Table 2.
RinA homologs in phages from Gram-positive bacteria
| Species | Accession number |
|---|---|
| Lactobacillus casei | YP_001987048 |
| Lactobacillus reuteri | ZP_03073841 |
| Bacillus thuringiensis serovar pakistani str. | ZP_04123836 |
| Geobacillus sp. G11MC16 | ZP_03149736 |
| Lactobacillus plantarum | ACT61868 |
| Enterococcus faecalis | AAO81076 |
| Lactococcus lactis subsp. lactis | YP_003353517 |
| Clostridium thermocellum | ABN53671 |
| Streptococcus suis | ZP_03626078 |
| Clostridium botulinum | EES48701 |
| Listeria monocytogenes | ADB68097 |
| Streptococcus gallolyticus | YP_003429885 |
| Clostridium phage phiSM101 | YP_699949 |
| Streptococcus equi subsp. equi | YP_002747279 |
| Streptococcus pneumoniae | ACA36251 |
| Lactococcus phage phiLC3 | AAS66802 |
| Streptococcus pyogenes | ABF36520 |
| Lactobacillus jensenii | ZP_05861716 |
| Bacillus cereus | ZP_04240708 |
| Bacillus clausii | YP_176339 |
| Streptococcus phage NZ131.2 | YP_002285775 |
| Listeria welshimeri | YP_849416 |
| Streptococcus agalactiae | ZP_00782383 |
Figure 8.
RinA homologues control expression of the morphogenetic clusters of bacteriophages from other Gram-positive bacteria. Expression of β-lactamase was assayed in RN4220-derivative strains containing plasmids carrying terS-blaZ fusions and expressing or lacking the cognate rinA gene from phages found in S. pyogenes (pJP543 and pJP544), E. faecalis (pJP535 and pJP537) and L. lactis (pJP564 and pJP565). Samples were normalized for total cell mass.
DISCUSSION
Staphylococcus aureus can adapt to different niches and hosts, producing a wide spectrum of acute and chronic diseases. Most of the variation between different S. aureus strains is due to the presence of MGE such as plasmids, prophages, pathogenicity islands, transposons and insertion sequences (24–27). These MGEs are clearly relevant in the pathogenic process of this bacterium, since many virulence factors are encoded on such mobile elements. In particular, bacteriophages play an important role in the pathogenicity of S. aureus either by carrying accessory virulence factors such as PVL (encoded by the luk-PV operon), staphylokinase (encoded by sak), enterotoxin A (encoded by sea) and exfoliative toxin A (encoded by eta) or by interrupting chromosomal virulence genes such as those for β-hemolysin (hlb) and lipase (geh) upon insertion [reviewed in (1)]. Phages are also the primary vehicle of lateral gene transfer between S. aureus strains, providing the species with the potential for broad genetic variation. It has been reported recently that phages increase the genome plasticity of S. aureus during infection, facilitating the adaptation of the pathogen to various host conditions (28). Additionally, certain staphylococcal phages carry out a highly specialized example of phage-mediated HGT, the mobilization of SaPIs. SaPI replication and high frequency transduction is closely linked to the phage lytic cycle, which induces SaPI replication and provides the proteins required for SaPI packaging and transfer [for review see (10)]. Despite the clear importance of staphylococcal phages to S. aureus pathogenesis, and the large number of sequenced staphylococcal phage genomes now available in public genome databases, the number of studies characterizing the biology of these relevant elements is limited.
To ensure tight control of genes required in the later stages of infection, bacteriophages have evolved a variety of mechanisms involving synthesis of phage-encoded control factors during the early stages of infection. In this work, we have identified the activator of phage late transcription and localized the promoter responsible for the expression of the morphogenetic genes in several representatives from a large family of related S. aureus phages. This promoter is active only during the late phase of the lytic cycle and requires the phage-encoded RinA protein for activity. The late promoter is located upstream of the morphogenetic cluster and controls the synthesis of a single large operon encoding the proteins responsible for assembly of phage particles, DNA packaging and host cell lysis. Expression of this gene cluster is essential not only for propagation of the phage, but also for transfer of phage-borne toxin genes, generalized transduction and the horizontal transmission of other MGEs that exploit the phage for their own transfer, such as SaPIs. As would be expected for an activator of late transcription, preliminary analysis of ϕ11-encoded genes after SOS induction, using tiling arrays, indicate that rinA is expressed earlier than terS, in a transcription unit containing genes involved in phage replication (data not shown).
Activation of late transcription by positive regulatory proteins is a strategy employed by a variety of phages with dsDNA genomes. Enterobacteriophage P2 and related phages encode a small zinc-binding protein, Ogr, that binds to conserved inverted repeats centered at about −55 relative to the transcription initiation sites of P2 late promoters (29–32). Protein–protein interaction between Ogr and the α subunit of the host RNA polymerase is required to stimulate P2 late gene transcription (33). Activation of phage T4 late promoters requires two phage-encoded proteins that function as core RNA polymerase subunits. T4 gp55 functions as a sigma factor that allows recognition of the 8 bp late promoter consensus motif, while gp33 is a coactivator for high level late transcription (34,35). T4 late transcription also requires a unique transcriptional activator, gp45, which is a sliding clamp that interacts with gp55, gp33 and T4 DNA polymerase to couple transcription to phage DNA replication (36). Phage P1 late transcription requires the phage-encoded Lpa protein, which binds to a conserved sequence centered at −22, as well as host-encoded coactivator, SspA (37). The phage Mu late activator protein, C, binds to a region of Mu late promoters from −30 to −55 and stimulates both binding of RNA polymerase and subsequent promoter release (38–40). Much less is known about the mechanism of transcription activation in the dsDNA phages of Gram-positive bacteria. The classic example is Bacillus subtilis phage SP01, which employs successive phage-encoded sigma factors to direct the host RNA polymerase to phage middle and late promoters (41) and also requires a T4 gp45-like protein for late transcription (42). The regulatory protein p4 from B. subtilis phage ϕ29 binds between −58 and −104 upstream of the late promoter transcription start site and activates through contact with the α subunit of RNA polymerase (43,44). An activator of late gene transcription (Alt) has also been identified in the L. lactis phage TP901-1 (45). Alt binds to a promoter region located upstream of the terS gene, recognizing a series of repeats −76 to −32 relative to the transcription start site (46). A common feature of all of these mechanisms is the recognition of sequences within or just upstream of the RNA polymerase binding site by a protein involved in activation of late transcription. RinA, in contrast, appears to require, for binding, sequences located more than 155 bp upstream of the terS transcription start site. Since essentially the same +1 was independently mapped by RACE for both ϕ11 and 80α, and the 80α RNA was treated to eliminate molecules with 5′ monophosphate termini, we have high confidence that this site corresponds to the actual start of transcription and not an end generated by processing. The large distance between the transcription start and sequences needed for RinA binding therefore suggests a mechanism for transcription activation that is more complex than a direct interaction between RinA and an adjacent RNA polymerase.
RinA was initially reported as an activator of the ϕ11 int gene (11). In earlier work, Ye et al. (23) identified the putative promoter region for the ϕ11 int gene and observed that the DNA sequence of the region between the translation start sites of the int and xis genes, which are transcribed divergently, was conserved between phages L54a and phage ϕ11, suggesting that this conserved region could contain the potential regulatory sites for expression of the int and xis genes. Using a reporter plasmid containing the gene xylE under control of this intervening sequence, including the ϕ11 int promoter, different cloned regions of the ϕ11 genome were screened for activation of expression. It was found that regulatory proteins RinA and RinB were both required to activate expression of the ϕ11int gene, while RinA alone was capable of activating L54a int gene transcription (11). Surprisingly, our results with the effects of rinA and rinB mutants do not confirm this initial characterization, suggesting that control of int expression is more complicated than predicted. Furthermore, RinA from phage L54a is even more divergent from ϕ11 than the RinA of ϕ69, sharing only 25% amino acid identity. Since we have demonstrated that the RinA proteins from ϕ11 and ϕ69 recognize and bind to different specific DNA regulatory sequences, it is difficult to explain how the ϕ11 and L54a proteins could bind to a conserved DNA sequence between int and xis to activate ϕ11 int expression.
RinA homologues can be found in bacteriophages infecting a variety of Gram-positive bacteria. We tested several of these and found that all were transcriptional activators that controlled expression of terS, the first gene of the morphogenetic cluster, in their respective phages. Thus, we propose that this family of proteins represents a conserved strategy for regulating the packaging and transfer of many bacteriophages in Gram-positive bacteria. Interestingly, an NCBI Conserved Domain Database search indicates that the RinA family of phage proteins also shows weak similarity to another large family of transcriptional regulators, ArpU. ArpU was described as a regulator of cellular muramidase-2 of Enterococcus hirae (47). Although initially reported as a chromosomally encoded gene, ArpU appears to have been cloned from a prophage. ArpU homologues are also present in a variety of different Gram-positive bacteriophages, and, like RinA, are localized upstream of the first gene of the morphogenetic cluster of the phage. This suggests that the ArpU-related proteins have a similar role in activating bacteriophage late gene expression, although this has not been tested directly. The C-terminal half of RinA also shows limited similarity to a portion of several sigma factors corresponding to conserved region 4. The significance of this is unclear. This region of sigma is involved in recognition of the −35 element of promoters (48). While it seems quite unlikely that RinA is functioning as a sigma factor, this may indicate that the C-terminal part of the protein contains the DNA recognition determinants. On the other hand, sigma region 4 is also a target for a number of transcription factors (49) so this could be a protein–protein interaction domain. Further investigation of the structure and function of RinA is clearly needed to determine its mechanism of action in activation of transcription.
Horizontal transfer of DNA can occur in bacteria by transformation, conjugation and transduction. In S. aureus, there is little evidence that transformation occurs. In addition, conjugative plasmids are not widespread and conjugative transposons are even less common. In contrast, many of the known S. aureus bacteriophages are generalized transducing phages that can package S. aureus chromosomal DNA as well as MGEs, and transfer them to other S. aureus strains (12). Transduction is likely the predominant mechanism of HGT in S. aureus. Since transfer of the different MGEs requires exploitation of the phage mechanisms involved in virion assembly and DNA packaging, characterization of the phage proteins controlling these processes are important in understanding how MGEs are transferred. In this work we have identified and characterized a key regulator of genes involved in phage assembly and packaging. This activator, RinA, is not only present in most of the S. aureus phages, but also in phages infecting a wide variety of other Gram-positive bacteria. Deletion of this activator resulted not only in elimination of the phage titre, but also significantly reduced transfer of SaPI-encoded virulence factors, and is predicted to show a similar reduction in transmission of plasmid-encoded virulence factors. Thus, we have identified an important mechanism involved in phage-mediated HGT, which plays a key role in bacterial evolution and pathogenesis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Consolider-Ingenio CSD2009-00006, BIO2005-08399-C02-02, BIO2008-05284-C02-02 and BIO2008-00642-E/C from the Ministerio de Ciencia e Innovación (MICINN), and grants from the Cardenal Herrera-CEU University (PRCEU-UCH39/10 and Copernicus-Banco Santander program), from the Conselleria de Agricultura, Pesca i Alimentació (CAPiA) and from the Generalitat Valenciana (ACOMP07/258) to J.R.P., grants [BFU2008-01078] from the MICINN and [2009SGR1106] from the Generalitat de Catalunya to J.B., NIH grants [R21 AI067654 and R56 AI081837] and a grant-in-aid from the A.D. Williams Trust and the Baruch Foundation Trust to G.E.C., a VCU Graduate School Thesis and Dissertation Award to M.D.H., and NIH grant [R01AI022159-23A2] to R.P.N. Funding for open access charge: Consolider-Ingenio CSD2009-00006, BIO2005-08399-C02-02, BIO2008-05284-C02-02 and BIO2008-00642-E/C from the Ministerio de Ciencia e Innovación (MICINN), Spain.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Łoś M, Kuzio J, McConnell MR, Kropinski AM, Węgrzyn G, Christie GE. Lysogenic conversion in bacteria of importance to the food industry. In: Sabour MP, Griffiths M, editors. Bacteriophage in the Detection and Control of Foodborne Pathogens. Washington, DC: ASM Press; 2010. pp. 157–198. [Google Scholar]
- 2.Ubeda C, Maiques E, Barry P, Matthews A, Tormo MA, Lasa I, Novick RP, Penades JR. SaPI mutations affecting replication and transfer and enabling autonomous replication in the absence of helper phage. Mol. Microbiol. 2008;67:493–503. doi: 10.1111/j.1365-2958.2007.06027.x. [DOI] [PubMed] [Google Scholar]
- 3.Tormo-Más MA, Mir I, Shrestha A, Tallent SM, Campoy S, Lasa I, Barbé J, Novick RP, Christie GE, Penadés JR. Moonlighting phage proteins de-repress staphylococcal pathogenicity islands. Nature. 2010;465:779–782. doi: 10.1038/nature09065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ubeda C, Barry P, Penades JR, Novick RP. A pathogenicity island replicon in Staphylococcus aureus replicates as an unstable plasmid. Proc. Natl Acad. Sci. USA. 2007;104:14182–14188. doi: 10.1073/pnas.0705994104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ubeda C, Maiques E, Tormo MA, Campoy S, Lasa I, Barbe J, Novick RP, Penades JR. SaPI operon I is required for SaPI packaging and is controlled by LexA. Mol. Microbiol. 2007;65:41–50. doi: 10.1111/j.1365-2958.2007.05758.x. [DOI] [PubMed] [Google Scholar]
- 6.Tormo MA, Ferrer MD, Maiques E, Ubeda C, Selva L, Lasa I, Calvete JJ, Novick RP, Penades JR. Staphylococcus aureus pathogenicity island DNA is packaged in particles composed of phage proteins. J. Bacteriol. 2008;190:2434–2440. doi: 10.1128/JB.01349-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tallent SM, Langston TB, Moran RG, Christie GE. Transducing particles of Staphylococcus aureus pathogenicity island SaPI1 are comprised of helper phage-encoded proteins. J. Bacteriol. 2007;189:7520–7524. doi: 10.1128/JB.00738-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ubeda C, Maiques E, Knecht E, Lasa I, Novick RP, Penades JR. Antibiotic-induced SOS response promotes horizontal dissemination of pathogenicity island-encoded virulence factors in staphylococci. Mol. Microbiol. 2005;56:836–844. doi: 10.1111/j.1365-2958.2005.04584.x. [DOI] [PubMed] [Google Scholar]
- 9.Ruzin A, Lindsay J, Novick RP. Molecular genetics of SaPI1 - a mobile pathogenicity island in Staphylococcus aureus. Mol. Microbiol. 2001;41:365–377. doi: 10.1046/j.1365-2958.2001.02488.x. [DOI] [PubMed] [Google Scholar]
- 10.Novick RP, Christie GE, Penades JR. The phage-related chromosomal islands of Gram-positive bacteria. Nat. Rev. Microbiol. 2010;8:541–551. doi: 10.1038/nrmicro2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye ZH, Lee CY. Cloning, sequencing, and genetic characterization of regulatory genes, rinA and rinB, required for the activation of staphylococcal phage phi 11 int expression. J. Bacteriol. 1993;175:1095–1102. doi: 10.1128/jb.175.4.1095-1102.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Novick RP. Genetic systems in staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- 13.Lindsay JA, Ruzin A, Ross HF, Kurepina N, Novick RP. The gene for toxic shock toxin is carried by a family of mobile pathogenicity islands in Staphylococcus aureus. Mol. Microbiol. 1998;29:527–543. doi: 10.1046/j.1365-2958.1998.00947.x. [DOI] [PubMed] [Google Scholar]
- 14.Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York, NY: John Wiley & Sons; 1990. [Google Scholar]
- 15.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a Laboratory Manual. NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 16.Arnaud M, Chastanet A, Debarbouille M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 2004;70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji G, Beavis R, Novick RP. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Poliakov A, Chang JR, Spilman MS, Damle PK, Christie GE, Mobley JA, Dokland T. Capsid size determination by Staphylococcus aureus pathogenicity island SaPI1 involves specific incorporation of SaPI1 proteins into procapsids. J. Mol. Biol. 2008;380:465–475. doi: 10.1016/j.jmb.2008.04.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narita S, Kaneko J, Chiba J, Piemont Y, Jarraud S, Etienne J, Kamio Y. Phage conversion of Panton-Valentine leukocidin in Staphylococcus aureus: molecular analysis of a PVL-converting phage, phiSLT. Gene. 2001;268:195–206. doi: 10.1016/s0378-1119(01)00390-0. [DOI] [PubMed] [Google Scholar]
- 21.Labandeira-Rey M, Couzon F, Boisset S, Brown EL, Bes M, Benito Y, Barbu EM, Vazquez V, Hook M, Etienne J, et al. Staphylococcus aureus Panton-Valentine leukocidin causes necrotizing pneumonia. Science. 2007;315:1130–1133. doi: 10.1126/science.1137165. [DOI] [PubMed] [Google Scholar]
- 22.Sumby P, Waldor MK. Transcription of the toxin genes present within the Staphylococcal phage phiSa3ms is intimately linked with the phage's life cycle. J. Bacteriol. 2003;185:6841–6851. doi: 10.1128/JB.185.23.6841-6851.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye ZH, Buranen SL, Lee CY. Sequence analysis and comparison of int and xis genes from staphylococcal bacteriophages L54a and phi 11. J. Bacteriol. 1990;172:2568–2575. doi: 10.1128/jb.172.5.2568-2575.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herron-Olson L, Fitzgerald JR, Musser JM, Kapur V. Molecular correlates of host specialization in Staphylococcus aureus. PLoS One. 2007;2:e1120. doi: 10.1371/journal.pone.0001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lowder BV, Guinane CM, Ben Zakour NL, Weinert LA, Conway-Morris A, Cartwright RA, Simpson AJ, Rambaut A, Nubel U, Fitzgerald JR. Recent human-to-poultry host jump, adaptation, and pandemic spread of Staphylococcus aureus. Proc. Natl Acad. Sci. USA. 2009;106:19545–19550. doi: 10.1073/pnas.0909285106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guinane CM, Ben Zakour NL, Tormo-Mas MA, Weinert LA, Lowder BV, Cartwright RA, Smyth DS, Smyth CJ, Lindsay JA, Gould KA, et al. Evolutionary genomics of Staphylococcus aureus reveals insights into the origin and molecular basis of ruminant host adaptation. Genome Biol. Evol. 2010;2:454–466. doi: 10.1093/gbe/evq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Viana D, Blanco J, Tormo-Mas MA, Selva L, Guinane CM, Baselga R, Corpa JM, Lasa I, Novick RP, Fitzgerald R, et al. Adaptation of Staphylococcus aureus to ruminant and equine hosts involves SaPI-carried variants of von Willebrand factor-binding protein. Mol. Microbiol. 2010;77:1583–1594. doi: 10.1111/j.1365-2958.2010.07312.x. [DOI] [PubMed] [Google Scholar]
- 28.Goerke C, Wirtz C, Fluckiger U, Wolz C. Extensive phage dynamics in Staphylococcus aureus contributes to adaptation to the human host during infection. Mol. Microbiol. 2006;61:1673–1685. doi: 10.1111/j.1365-2958.2006.05354.x. [DOI] [PubMed] [Google Scholar]
- 29.Christie GE, Calendar R. Bacteriophage P2 late promoters. II. Comparison of the four late promoter sequences. J. Mol. Biol. 1985;181:373–382. doi: 10.1016/0022-2836(85)90226-8. [DOI] [PubMed] [Google Scholar]
- 30.Grambow NJ, Birkeland NK, Anders DL, Christie GE. Deletion analysis of a bacteriophage P2 late promoter. Gene. 1990;95:9–15. doi: 10.1016/0378-1119(90)90407-i. [DOI] [PubMed] [Google Scholar]
- 31.Birkeland NK, Lindqvist BH, Christie GE. Control of bacteriophage P2 gene expression: analysis of transcription of the ogr gene. J. Bacteriol. 1991;173:6927–6934. doi: 10.1128/jb.173.21.6927-6934.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Bokkelen GB, Dale EC, Halling C, Calendar R. Mutational analysis of a bacteriophage P4 late promoter. J. Bacteriol. 1991;173:37–45. doi: 10.1128/jb.173.1.37-45.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wood LF, Tszine NY, Christie GE. Activation of P2 late transcription by P2 Ogr protein requires a discrete contact site on the C terminus of the alpha subunit of Escherichia coli RNA polymerase. J. Mol. Biol. 1997;274:1–7. doi: 10.1006/jmbi.1997.1390. [DOI] [PubMed] [Google Scholar]
- 34.Kassavetis GA, Geiduschek EP. Defining a bacteriophage T4 late promoter: bacteriophage T4 gene 55 protein suffices for directing late promoter recognition. Proc. Natl Acad. Sci. USA. 1984;81:5101–5105. doi: 10.1073/pnas.81.16.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Herendeen DR, Williams KP, Kassavetis GA, Geiduschek EP. An RNA polymerase-binding protein that is required for communication between an enhancer and a promoter. Science. 1990;248:573–578. doi: 10.1126/science.2185541. [DOI] [PubMed] [Google Scholar]
- 36.Nechaev S, Geiduschek EP. Dissection of the bacteriophage T4 late promoter complex. J. Mol. Biol. 2008;379:402–413. doi: 10.1016/j.jmb.2008.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hansen AM, Lehnherr H, Wang X, Mobley V, Jin DJ. Escherichia coli SspA is a transcription activator for bacteriophage P1 late genes. Mol. Microbiol. 2003;48:1621–1631. doi: 10.1046/j.1365-2958.2003.03533.x. [DOI] [PubMed] [Google Scholar]
- 38.Margolin W, Howe MM. Localization and DNA sequence analysis of the C gene of bacteriophage Mu, the positive regulator of Mu late transcription. Nucleic Acids Res. 1986;14:4881–4897. doi: 10.1093/nar/14.12.4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun W, Hattman S, Kool E. Interaction of the bacteriophage Mu transcriptional activator protein, C, with its target site in the mom promoter. J. Mol. Biol. 1997;273:765–774. doi: 10.1006/jmbi.1997.1349. [DOI] [PubMed] [Google Scholar]
- 40.Chakraborty A, Nagaraja V. Dual role for transactivator protein C in activation of mom promoter of bacteriophage Mu. J. Biol. Chem. 2006;281:8511–8517. doi: 10.1074/jbc.M512906200. [DOI] [PubMed] [Google Scholar]
- 41.Talkington C, Pero J. Promoter recognition by phage SP01-modified RNA polymerase. Proc. Natl Acad. Sci. USA. 1978;75:1185–1189. doi: 10.1073/pnas.75.3.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Greene JR, Chelm BK, Geiduschek EP. SP01 gene 27 is required for viral late transcription. J. Virol. 1982;41:715–720. doi: 10.1128/jvi.41.2.715-720.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barthelemy I, Lazaro JM, Mendez E, Mellado RP, Salas M. Purification in an active form of the phage phi 29 protein p4 that controls the viral late transcription. Nucleic Acids Res. 1987;15:7781–7793. doi: 10.1093/nar/15.19.7781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mencia M, Monsalve M, Rojo F, Salas M. Transcription activation by phage phi29 protein p4 is mediated by interaction with the alpha subunit of Bacillus subtilis RNA polymerase. Proc. Natl Acad. Sci. USA. 1996;93:6616–6620. doi: 10.1073/pnas.93.13.6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brondsted L, Pedersen M, Hammer K. An activator of transcription regulates phage TP901-1 late gene expression. Appl. Environ. Microbiol. 2001;67:5626–5633. doi: 10.1128/AEM.67.12.5626-5633.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pedersen M, Kilstrup M, Hammer K. Identification of DNA-binding sites for the activator involved in late transcription of the temperate lactococcal phage TP901-1. Virology. 2006;345:446–456. doi: 10.1016/j.virol.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 47.Lleo MM, Fontana R, Solioz M. Identification of a gene (arpU) controlling muramidase-2 export in Enterococcus hirae. J. Bacteriol. 1995;177:5912–5917. doi: 10.1128/jb.177.20.5912-5917.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gross C, Lonetto M, Losick R. Bacterial Sigma Factors. In: McKnight SYK, editor. Transcriptional Regulation. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1992. pp. 129–176. [Google Scholar]
- 49.Lonetto MA, Rhodius V, Lamberg K, Kiley P, Busby S, Gross C. Identification of a contact site for different transcription activators in region 4 of the Escherichia coli RNA polymerase sigma70 subunit. J. Mol. Biol. 1998;284:1353–1365. doi: 10.1006/jmbi.1998.2268. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.