Abstract
The nuclear factor of activated T-cells (NFAT) c1 has been shown to be essential for Ca2+-dependent upregulation of myosin heavy chain (MyHC) I/β expression during skeletal muscle fiber type transformation. Here, we report activation of extracellular signal-regulated kinase (ERK) 1/2 in Ca2+-ionophore-treated C2C12 myotubes and electrostimulated soleus muscle. Activated ERK1/2 enhanced NFATc1-dependent upregulation of a −2.4 kb MyHCI/β promoter construct without affecting subcellular localization of endogenous NFATc1. Instead, ERK1/2-augmented phosphorylation of transcriptional coactivator p300, promoted its recruitment to NFATc1 and increased NFATc1–DNA binding to a NFAT site of the MyHCI/β promoter. In line, inhibition of ERK1/2 signaling abolished the effects of p300. Comparison between wild-type p300 and an acetyltransferase-deficient mutant (p300DY) indicated increased NFATc1–DNA binding as a consequence of p300-mediated acetylation of NFATc1. Activation of the MyHCI/β promoter by p300 depends on two conserved acetylation sites in NFATc1, which affect DNA binding and transcriptional stimulation. NFATc1 acetylation occurred in Ca2+-ionophore treated C2C12 myotubes or electrostimulated soleus. Finally, endogenous MyHCI/β gene expression in C2C12 myotubes was strongly inhibited by p300DY and a mutant deficient in ERK phosphorylation sites. In conclusion, ERK1/2-mediated phosphorylation of p300 is crucial for enhancing NFATc1 transactivation function by acetylation, which is essential for Ca2+-induced MyHCI/β expression.
INTRODUCTION
The nuclear factor of activated T-cells (NFAT) comprises a family of five transcription factors, which are located in a phosphorylated, inactive state in the cytoplasm of many cell types including skeletal myotubes. Four members of this family, NFATc1(2/c), NFATc2(1/p), NFATc3(4/x) and NFATc4(3), are downstream targets of the Ca2+-/calmodulin-dependent phosphatase calcineurin (1,2). During periods of elevated intracellular calcium concentration ([Ca2+]i), NFAT is dephosphorylated by calcineurin, translocates into the nucleus, binds to consensus DNA sites and stimulates gene transcription. Upon cessation of the Ca2+-signal, termination of NFAT signaling occurs through rephosphorylation of NFAT by protein kinases, resulting in its retrograde translocation to the cytoplasm (3,4). Several distinct sequences, including the serine-rich region (SRR) and the serine–proline (SP)-rich boxes, are not only important for NFAT regulation by calcineurin, but are also major targets for phosphorylation by various protein kinases (1).
Based on their contraction speed, force development, fatigability and metabolic functions, skeletal muscle fibers have been classified into distinct fiber types (5). The four different fiber types are characterized as fast-twitch glycolytic (type IIB and IID/X), fast-twitch oxidative/glycolytic (type IIA) and slow-twitch oxidative (type I), which express myosin heavy chain (MyHC) isoforms IIB, IID/X, IIA and I/β, respectively. In response to altered physiological demands transformation of fiber types occurs (6,7). The transformation process includes functional, biochemical and morphological changes of skeletal muscle cells which are a consequence of different fiber type-specific gene expression patterns. The resulting profound changes in muscle fiber type are termed fast-to-slow or slow-to-fast transformation. Elevations of [Ca2+]i are thought to underlie fast-to-slow shifts in muscle gene expression (8–10).
Calcineurin and the NFAT family member c1 are essential for the upregulation of MyHCI/β promoter activity and mRNA expression during fast-to-slow transformation of primary skeletal myotubes as well as C2C12 myotubes (10–13). C2C12 myotubes have been previously shown to be a suitable system for investigating fiber type transformation (13–15). The differentiated C2C12 cells express a fast fiber type-like character in terms of high expression of endogenous fast MyHCIId/x protein and mRNA as well as exogenous MyHCIId/x promoter activity, and low expression of slow MyHCI/β protein and mRNA as well as MyHCI/β promoter activity. The pattern of MyHC expression and promoter activities can be switched to a slow fiber type-like character (8,13,16) by electrostimulation with a slow fiber type pattern, or by addition of Ca2+-ionophore A23187. As shown previously, the resting [Ca2+]i of primary skeletal myotubes treated with 0.1 µM of Ca2+-ionophore A23187 (10) tended to be in the range of the resting [Ca2+]i of the extensor digitorum longus muscle, expressing mainly fast fibers, when subjected to low-frequency stimulation (17) to induce fast-to-slow fiber transformation. Consistent with these findings in cultured myotubes, calcineurin has been shown to promote the slow muscle phenotype in animal models (18–20), and NFATc1 has been identified as a key transcription factor for the activity-dependent MyHCI/β expression in slow-twitch soleus muscle (21). NFATc1 but not c2 or c3 undergoes nuclear translocation in response to increased intracellular Ca2+-levels in primary skeletal myotubes (22). We have previously demonstrated by protein–DNA binding analysis a Ca2+-ionophore-inducible and calcineurin-dependent binding of NFATc1 to a NFAT consensus binding site within the proximal MyHCI/β promoter that results in the subsequent recruitment of transcriptional coactivator p300 in rabbit primary skeletal myotubes (13).
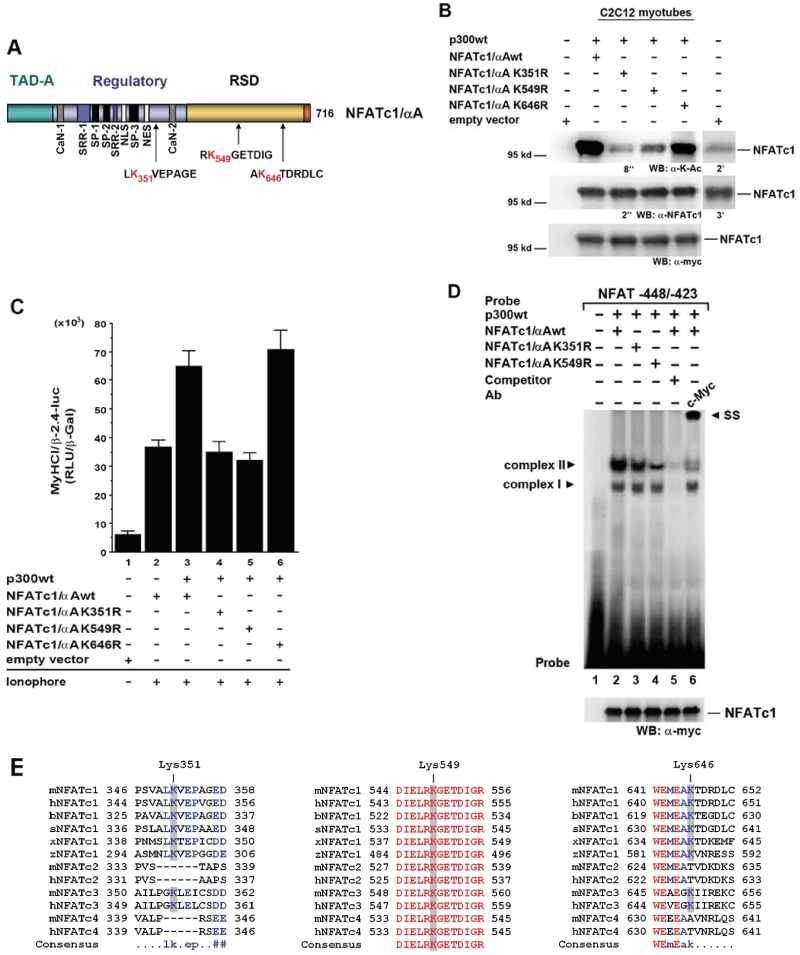
NFATc1 is synthesized in six isoforms that differ in their N- and/or C-termini due to two different promoter and poly(A) site usage as well as alternative splicing events (23,24). The N-terminal α peptide consists of 42 amino acids while the N-terminal β peptide comprises 29 amino acids. The NFATc1/A isoform contains a relatively short C-terminus, whereas the isoforms NFATc1/B and C spans longer extra C-terminal peptides. Consistent with other members of the NFAT family, NFATc1/αA shares a conserved central region that harbors an N-terminal strong transactivation domain A (TAD-A) and a regulatory domain (Figure 7A). In addition, the DNA binding domain shares >70% sequence homology among NFAT proteins and <20% sequence homology to the DNA binding domain of Rel/NF-κB factors, but it adopts a very similar conformation and is thus designated as Rel similarity (or homology) domain (RSD or RHD) (1,22).
Figure 7.
Identification of lysine residues of NFATc1/αA acetylated by p300. (A) Structure of NFATc1/αA. NFATc1/αA is one of the six NFATc1 isoforms which differ in their N- and/or C-termini due to two different promoter and poly(A) site usage as well as alternative splicing events. The structure shows the NFAT-homology region that comprises the amino-terminal transactivation domain (TAD), the calcineurin binding site (CaN), the nuclear localization sequence (NLS), the nuclear exit sequence (NES), the serine-rich regions SRR1 and 2, as well as the SP1, SP2 and SP3 motifs (Ser-Pro rich) that are targeted by maintenance and export kinases. The Rel similarity (or homology) domain (RSD or RHD) comprises the DNA binding motif. Sequences containing possible acetylation sites, lysines 351, 549 and 646 (K351, K549 and K646), investigated in this study are shown below. (B) C2C12 cells were transfected with Myc-tagged NFATc1/αAwt, or mutant NFATc1/αA K351R, or K549R, or K646R, or empty vector, respectively, and cotransfected with p300wt or empty vector. Cells were grown for 24 h in GM and then for 4 days in DM with Ca2+-ionophore A23187 (0.1 µM). Cell lysates were probed with anti-K-Ac and anti-NFATc1 antibodies. Exposure times for enhanced chemiluminescence detection were indicated. Longer exposure times (2 and 3 min) demonstrate endogenous NFATc1 and NFATc1-K-Ac levels in cells transfected with empty vectors. The expression levels of the transfected Myc-tagged NFATc1wt and mutants were detected by immunoblotting with anti-c-Myc-antibody. (C) C2C12 cells were transiently transfected with a −2.4-kb MyHCI/β promoter construct and cotransfected with expression vectors coding for Myc-tagged wild-type NFATc1αA (NFATc1αAwt), or mutant NFATc1αA K351R, or K549R, or K646R, or empty vector. Cells were additionally cotransfected with expression vectors for wild-type p300 (p300wt) or empty vector. Cells were grown for 24 h in GM and then for 2 days in DM with or without Ca2+-ionophore A23187 (0.1 µM). The promoter activity is expressed as relative light units per unit β-galactosidase (RLU/β-Gal). The data represent the mean ± SD of triplicate data points. (D) EMSA demonstrates that acetylation-defective NFATc1/αA mutants have reduced DNA-binding activity. Nuclear extracts from C2C12 myotubes transfected with Myc-His-tagged NFATc1/αAwt, or K351R, or K549R mutants (all expressed at similar levels, as revealed by western blot analysis with anti-c-Myc-antibody) and grown for 3 days in DM in the presence of Ca2+-ionophore were mixed with an excess of radiolabeled or cold competitor NFAT probe. After incubation, samples were fractionated at 4°C on a 3.0 % nondenaturing polyacrylamide gel for 2 h at 33 mA. Complexes are indicated by an arrowhead. Ab, antibody; SS, supershift. (E) NFATc1 isoforms and other members of the NFAT family from several vertebrates were aligned using MuliAlign 5.4.1 to show sequences including the two functional important lysines (equivalent to K351 and K549 in murine NFATc1), which can be acetylated by p300, and lysine 646 (K646), which is not acetylated. Blue, conserved amino acids in at least all NFATc1 of several vertebrates; red, conserved amino acids in all members of the NFAT family. b, Bos taurus (bovine); h, human; m, mouse; s, Sus scrota (boar); x, Xenopus; z, zebra fish.
The transcriptional coactivator p300 and its homolog, the cAMP responsive element binding protein (CREB)-binding protein (CBP) are both direct targets of cellular signaling pathways, resulting in their post-translational modification (25). Several studies reported that their function can be regulated by direct phosphorylation, and phosphorylation sites have been identified (26–28). Phosphorylation and activation of CBP and p300 by extracellular signal-regulated kinase (ERK) 1/2 has been demonstrated (29,30), with enhanced histone acetyltransferase (HAT) activity resulting in the activation of gene expression (31). Despite a high degree of homology, CBP and p300 are not completely redundant but display also unique roles (32). HATs are not only responsible for histone acetylation but can also acetylate a variety of non-histone proteins including several transcription factors (33). With regard to NFAT, p300/CBP were shown to interact with different family members in vitro, in T-cells and cardiomyocytes (34–37).
It has been demonstrated that increased [Ca2+]i, including membrane depolarization-induced rise in [Ca2+]i in neuronal and skeletal muscle cells, activates mitogen-activated protein kinase (MAPK)/ERK kinase 1/2 (MEK1/2), the upstream kinase of ERK1/2, and ERK 1/2 (38–40). Until now, only a limited number of data available in the literature investigates ERK1/2 signaling on myosin heavy-chain gene expression and shows opposing effects. For example, during muscle regeneration following muscle damage the onset of the Ras-MEK1/2-ERK1/2 pathway promotes slow MyHCI/β gene expression (41). Other studies revealed that activated ERK2 downregulates a rat MyHCI/β promoter in the murine C2C12 cell line while the same promoter construct was not affected by ERK2 in slow type soleus or fast type gastrocnemius muscle in rats (42). Thus, further studies are needed to define the role of MEK1/2-ERK1/2 signaling for fiber type-specific MyHC gene expression. Concerning MyHCI/β expression, a possible relevance of ERK1/2 on NFATc1 nuclear translocation has not been investigated yet. So far, analysis on subcellular localization, DNA binding and transactivation function of different NFAT family members in T-cells and cardiomyocytes revealed different modes of action of the MEK1/2-ERK1/2 pathway on NFAT transcriptional function depending on the kind of family member and cell type (43–46).
The present article now investigates how activation of the MEK1–ERK1/2-signaling pathway modulates the NFATc1-dependent stimulation of MyHCI/β promoter activity and gene expression induced by increased intracellular Ca2+. We demonstrate that MEK1–ERK1/2 signaling induces the recruitment of p300 to the MyHCI/β promoter and that acetylation of NFATc1 by p300 is crucial for enhanced transcriptional activity of NFATc1.
MATERIALS AND METHODS
Plasmid construction
The −2.4 kb MyHCI/β and the −2.8 kb MyHCIId/x promoter luciferase reporter constructs have been described previously (13). To generate a−2.4 kb MyHCI/β promoter construct mutated in the −439/−432-bp NFAT consensus binding site (13), nucleotides from −438 to −436 (GGA) were changed to TTC by using the QuikChange II Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s instructions. Constitutively nuclear mutant NFATc1, NFATc1ΔSRR, lacking amino acids 180–183 (Ser180, Ser181, Arg182, Ser183) in the SRR has also been described (13). The NFATc1 isoform used here is the short isoform αA (23,24). An c-Myc-His-tagged NFATc1 expression vector was generated by subcloning NFATc1 from pcDNA3–NFATc1, a gift from M. A. Brown (47), into pcDNA3.1/Myc-His(-) (Invitrogen, Karlsruhe, Germany). Single lysine-to-arginine (K-to-R) NFATc1 mutants (K351R, K549R, K646R) were generated by PCR-based site-directed mutagenesis using the Phusion Site-Directed Mutagenesis Kit (New England Biolabs GmbH, Frankfurt, Germany) according to the manufacturer’s instructions. Constitutively active mutant MEK1 (pMCL–HA–MEK1–R4F, ΔN3/S218E/S222D; MEK1ca) and dominant negative mutant MEK1 (pMCL–HA–MEK1-8 E, K97M; MEK1dn) (48) were gifts from Dr N. G. Ahn. The human wild-type p300 (pCMVβ-p300-Myc; p300wt) and the acetyltransferase-deficient p300 mutant (pCMVβ-p300DY-Myc; p300DY) (49) were gifts from Dr T.-P. Yao. To generate p300 with mutations in three possible ERK-specific phosphorylation motifs (p300SA3, with serine residues 2279, 2315 and 2366 in pCMVβ-p300SA3-Myc replaced by alanine), again the Phusion Site-Directed Mutagenesis Kit was used. The CBP expression vector pRc/RSV-mCBP-HA (50) was kindly provided by Dr R. H. Goodman.
Cell culture and transfections and luciferase reporter gene assays
Mouse C2C12 myoblasts or HEK 293 cells were cultured in growth medium (GM), consisting of complete Dulbecco’s-modified Eagle’s medium with high glucose (4.5 g/l) (DMEM) (Invitrogen) supplemented with 10% foetal bovine serum (FCS) (Sigma, Taufkirchen, Germany), 2 mM l-glutamine, 100 U/ml penicillin and 100 mg/ml streptomycin. C2C12 cells were transfected as described previously (13), using 0.5 µg of signal molecule expression vector. Transfected cells were treated 24 h post-transfection with of 0.1 µM Ca2+-ionophore A23187 and/or 10 µM of 1,4-bis[2-aminophenylthio]butadiene (U0126, Sigma) for 2 days in differentiation medium (DM, DMEM plus 5% horse serum). Transient transfections of HEK 293 cells were performed at 50–80% confluence. Non-modified branched polyethylenimine (Sigma-Aldrich) (4.5 µl of a 1 mg/ml solution) was added to a mixture of 1.5 µg DNA and 90 µl serum- and antibiotics-free DMEM, with subsequent addition of 900 µl serum- and antibiotics-free DMEM.
Luciferase reporter gene assays were described previously (13). Cells were cotransfected with pCMV-Gal as an internal reference. The β-galactosidase activity was determined by a standard colorimetric assay (51). Alternatively, the Luminescent β-galactosidase Reporter System 3 (Clontech, Takara Bio Europe, St Germain-en-laye, France) was used according to the manufacturer’s instructions with minor modifications.
RNA interference assays
C2C12 myoblasts transfected with MyHCI/β promoter DNA and co-transfected with an expression vector coding for constitutively nuclear NFATc1ΔSRR or empty expression vector pME18S, were additionally transfected after 24 h in GM with 0.825 µg of a pool of doublestranded 20–25 nt siRNA that specifically targets mouse p300 (siRNAp300) or a non-specific doublestranded control siRNA (Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), using siRNA Transfection Reagent (8 µl/µg of respective siRNA; Santa Cruz Biotechnology, Inc.) according to the manufacturer’s instructions. Cells were grown for a further 2 days in DM.
Intracellular fractionation and western blot analysis
For intracellular fractionation, C2C12 cells transfected with NFATc1ΔSRR or the empty vector and grown for 2 days in DM in the presence or absence of 0.1 µM Ca2+-ionophore A23187 and/or 10 µM U0126 were suspended and incubated in cell fractionation buffer containing 5 mM Tris–HCl (pH 7.4), 5 mM KCl, 1.5 mM MgCl2, 0.1 mM EGTA, 1 mM DTT, 0.2 mM PMSF, 2 mM benzamidine and 5% (w/v) Trasylol for 5 min. For cell fractionation, 0.1% Nonidet P-40 (NP40) was added into suspension. Cell extracts were centrifuged at 850g. Pellet fraction was lysed in and aliquots of supernatant fraction were added to gel loading buffer [160 mM Tris–HCL, pH 6.8, 4% (w/v) sodium dodecyl sulphate (SDS), 20% (v/v) glycerol, 0.5% β-mercaptoethanol (v/v), 0.008% (w/v) bromophenol blue].
For western blot analysis, HEK 293 were lysed 2 days and C2C12 cells 3 days post-transfection in 150 µl of gel loading buffer, and then incubated for 10 min at 95°C. Proteins were separated by SDS–PAGE as described previously (52), and blotted on a polyvinylidene diflouride (PVDF) membrane (Sigma). Primary antibodies used were anti-ERK2 (sc-81459), anti-histone H3 (sc-10809), anti-NFATc1 (sc-13033), anti-p53 (sc-98), anti-p300 (sc-584), anti-c-Myc (sc-40), anti-α-tubulin (sc-8035) (Santa Cruz Biotechnology, Inc.), anti-ERK1/2 (p44/42; #9102), anti-phospho-ERK1/2 (Thr202/Tyr204; #9106), anti-phospho-serine-MAPK/CDK (#2325) (Cell Signaling, New England Biolabs), anti-acetylated-lysine (ICP0380) (Immunechem, Burnaby, BC, Canada) and anti-phospho-serine/threonine-proline (Ser/Thr-Pro, minimal ERK consensus phosphoacceptor motif (53); #05-368) (Upstate Biotechnologies, Millipore, Schwalbach/Ts., Germany) antibodies. An anti-phospho-serine-MAPK/cyclin-dependent kinase (CDK) antibody chosen with regard to previously reported findings of C-terminal serine phosphorylation of p300 by ERK2 in keratinocytes (31) can detect MAPK but also CDK substrates. The acetylation levels of c-Myc-tagged NFATc1 were determined in the presence of general histone deacetylase (HDAC) inhibitors trichostatin A (TSA, 300 nM; Sigma) and nicotinamide (NIA, 5 mM; Sigma) added 24 h before lysis (HEK 293 cells) or present in the lysis buffer (C2C12 myotubes). Bound antibodies were detected with either anti-goat, anti-mouse or anti rabbit IgG conjugated to horseradish peroxidase (HRP) (Promega). Signals were visualized by enhanced chemiluminescence detection (Santa Cruz Biotechnology, Inc.) with the LAS-3000 imaging system (Fujifilm Europe GmbH, Düsseldorf, Germany).
Immunoprecipitation assays
C2C12 myotubes from a 60-mm Petri dish were washed twice with 200 µl PBS, scraped, collected by centrifugation and lysed in immunoprecipitation (IP) buffer (50 mM HEPES (pH 7.5), 1% Triton X-100, 10% (v/v) glycerol, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA). Soleus muscles were isolated from mice as described previously (54) and immediately frozen in liquid nitrogen. Frozen muscles were dounce-homogenized on ice in lysis buffer with addition of complete protease inhibitor cocktail (Roche Diagnostics GmbH, Mannheim, Germany) using glass-on-glass homogenizer. For IP, pooled lysates from eight equally treated muscles were used. Protein was quantitated using the Bradford assay (BioRad Laboratories GmbH, München, Germany). Nuclear extracts (2.5 mg of protein) were incubated overnight with 30 µl of anti-p300 or anti-NFATc1 antibodies under gentle shaking at 4°C, and subsequently incubated with 25 µl of protein G Sepharose (Amersham Biosciences Europe, Freiburg, Germany) for 1.5 h at 4°C. Immunoprecipitated beads were pelleted and washed five times with cold washing buffer (1x PBS containing 0.5% Nonidet P-40). Protein was removed from the beads by boiling in 2× SDS loading buffer for 5 min and western blotting was carried out using anti-phospho-serine-MAPK/CDK or anti-NFATc1 antibodies.
In vitro kinase assay
Wild type p300 or mutant p300SA3 protein was incubated in 20 µl of kinase buffer [20 mM HEPES (pH 7.4), 10 mM MgCl2, 25 mM NaCl, 5 mM glycerol phosphate, 0.2 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol, 0.1 mM Na3VO4, 10 µM ATP] at 30°C for 20 min with recombinant activated ERK2 (Sigma). About 0.2–2 µg of substrate and 1 µCi of [γ-33P] ATP (3000 Ci/mmol) were used. The reaction was stopped by adding an equal volume of 5× SDS loading buffer and by heating at 95°C for 5 min. After separation through 10% SDS–PAGE, the gel was dried and radioactivity was visualized by phosphorimaging using a Fuji Bas-1500 (Fujifilm Europe). For western blot analysis, a separate reaction using non-radioactive ATP was run and the blot was probed with anti-phospho-Ser/Thr-Pro, anti-p300 and anti-ERK2 antibodies.
Immunofluorescence studies
For detection of endogenous NFATc1, C2C12 myoblasts were seeded on glass cover slips and cultured for 4 days in DM. After 2 days cells were treated with 0.1 µM Ca2+-ionophore A23187 and/or 10 µM U0126 for 2 days. For detection of MyHCI/β gene expression, C2C12 myoblasts transfected with 0.75 µg of expression plasmid were grown for 24 h in GM and then for 3 or 4 days in DM in the presence or absence of 0.1 µM Ca2+-ionophore A23187. Fixation and staining of cells were performed as described previously (12), using primary goat polyclonal anti-NFATc1 (Santa Cruz Biotechnology, Inc.), or mouse anti-MyHCI/β antibodies (Sigma), and secondary fluorescein isothiocyanate (FITC)-labeled antibodies (Santa Cruz Biotechnology, Inc.). Nuclei were stained with DAPI (Sigma). Stained cells were photographed on an inverted fluorescence microscope (Leica Microsystems, Wetzlar, Germany; magnification ×400).
Preparation of nuclear extracts and electrophoretic mobility shift assays
C2C12 cells transfected with expression plasmids were grown for 24 h in GM and then for 3 days in DM with or without Ca2+-ionophore A23187 (0.1 µM). Nuclear extract preparation and EMSAs were performed as previously described (55). Oligonucleotides (−448/−423, 5′-CTC CAG GCC AGG AAA GCA GGG AAA TT-3′) used as a probe contain a NFAT consensus binding site of the rabbit slow MyHCI/β promoter (GenBank accession number AF192306) at −439/−432 (13). In competition experiments, unlabeled annealed oligonucleotide (200-fold excess) of self DNA was added. Antibody supershift assays were performed by pre-incubating nuclear extracts (NEs) or in vitro translated NFATc1 with preimmune, or anti-NFATc1, or anti-p300, or anti-c-Myc antisera (Santa Cruz Biotechnology, Inc.).
Semiquantitative reverse transcription PCR
Total RNA was isolated using NucleoSpin RNAII kit (Macherey & Nagel, Düren, Germany). mRNA was reverse transcribed and cDNA amplified with Phusion reverse transcription (RT)–PCR Kit (New England Biolabs) according to the manufacturer’s instruction. PCR was performed in a reaction of 50 µl with maximum cycle number of 35. PCR products were analyzed by 2% agarose gel electrophoresis and visualized with ethidium bromide staining. Primer pairs used were: mouse MyHCI/β forward (F): 5′-GCT GAG GCC CAG AAA CAA G-3′, and reverse (R): 5′-TTC CAC GAT GGC GAT GTT C-3′; mouse MyHCIId/x F: 5′-ACG CTG GAT GCT GAG ATT AG-3′, and R: 5′-GAG TGG TTC AGC TGG ATT TC-3′; human p300 F: 5′-ATCAGCAGCGACTCCTTCAG-3′, and R: 5′-CAGATCCTCTTCTGAGATGAGTT-3′; mouse 18 s rRNA F: 5′-GGA CCA GAG CGA AAG CAT TT-3′ and R: 5′-TGC CAG AGT CTC GTT CGT TAT-3′. PCR with primers specific for 18 s rRNA was performed for normalization of transcript levels. The mRNA expression of transfected Myc-tagged p300 expression vectors was evaluated using a Myc-specific reverse and the p300 forward primer.
Muscle preparation and stimulation
Soleus muscles were isolated from mice as described previously (54). Pairs of solei were placed in bathing solutions (120 mM NaCl, 3.3 mm KCl, 1.2 mM MgSO4, 1.2 mM KH2PO4, 1.3 mM CaCl2, 25 mM NaHCO3, 100 mg/100 ml glucose) equilibrated with 95% O2 and 5% CO2 immediately upon removal. The muscles were mounted between an external force transducer (Harvard, Bioscience 529503) and a rigid hook inside bathing chambers to give isometric conditions. After initial equilibration of soleus muscle pairs in solution (120 min), one of the muscles was kept unstimulated in solution for the entire duration of the experiment. The other muscle was stimulated directly via platinum electrodes placed on either side of the muscle for 30 min with 15 Hz (5 s every minute, pulse duration 1 ms).
Multiple sequence alignment
Amino acid sequences of human NFATc1 (NP_765978), NFATc2 (NP_036472), NFATc3 (NP_775186), and NFATc4 (NP_001185894); mouse NFATc1 (NP_001157581), NFATc2 (NP_001129545), NFATc3 (NP_035031) and NFATc4 (NP_001161818); Bos taurus NFATc1 (NP_001160087); Sus scrofa NFATc1 (NP_999326); Xenopus NFATc1 (NP_001085919); and zebra fish NFATc1 (NP_001038624) were aligned with MultAlin 5.4.1.
RESULTS
MEK1–ERK1/2 signaling increased Ca2+-ionophore-induced slow MyHCI/β promoter activation
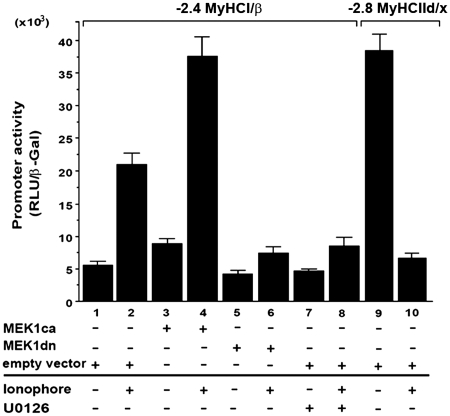
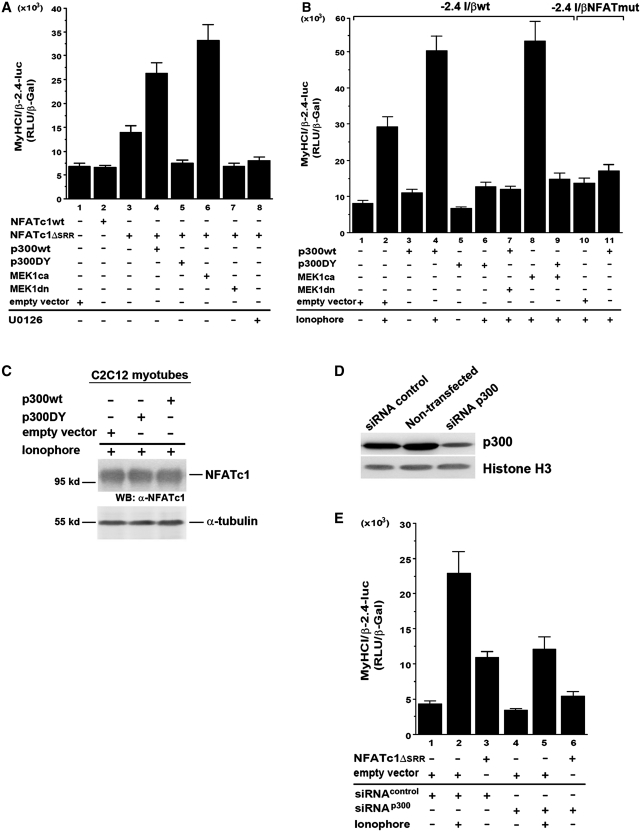
Previous studies indicate that the Ras-MEK1/2–ERK1/2 signaling pathway promotes MyHCI/β gene expression during regeneration processes in slow-twitch soleus muscles fibers (41). To investigate a possible role of MEK1–ERK1/2 signaling for MyHCI/β promoter regulation, we used a genetic and a pharmacological approach. U0126 (10 µM), a specific inhibitor of direct upstream activators of ERK1/2, MEK1/2, or overexpression of a dominant-negative MEK1 (MEK1-8 E, MEK1dn) decreased, and overexpression of a constitutively active MEK1 (MEK1–R4F, MEK1ca) increased basal activity of a −2.4-kb MyHCI/β promoter luciferase reporter construct (Figure 1, lanes 1, 3, 5 and 7), indicating an activating impact of MEK1–ERK1/2 on the basal promoter activity.
Figure 1.
MEK1–ERK1/2 signaling increases Ca2+-ionophore-induced slow MyHCI/β promoter activation in C2C12 myotubes. C2C12 cells were transiently transfected with a −2.4-kb MyHCI/β or with a −2.8-kb MyHCIId/x promoter luciferase reporter construct alone or cotransfected with expression vectors for constitutively active MEK1 (MKK1ca), or dominant negative MEK1 (MEK1dn), or empty vector. Cells were grown for 24 h in GM and then for 2 days in DM with or without Ca2+-ionophore A23187 (0.1 µM) and/or U0126 (10 µM). The promoter activity is expressed as relative light units per unit β-galactosidase (RLU/β-Gal). The data represent the mean ± SD of triplicate data points.
Consistent with a fast fiber type-like character of C2C12 myotubes in terms of MyHC isoform expression (13–15), the slow MyHCI/β promoter has a lower activity in untreated myotubes compared with a −2.8-kb fast MyHCIId/x promoter construct (Figure 1, lanes 1 and 9). Upregulation of the low basal MyHCI/β promoter activity was induced by treatment with 0.1 µM Ca2+-ionophore A23187, while MyHCIId/x promoter activity was diminished (lanes 1, 2, 9 and 10), demonstrating a fast-to-slow transformation on the level of MyHC promoter. MEK1ca increased the MyHCI/β promoter activity in Ca2+-ionophore-treated C2C12 myotubes, whereas U0126 or MMK1dn reduced the Ca2+-ionophore-induced increase (lanes 2, 4, 6 and 8). These data suggest a role for the MEK1–ERK1/2 pathway in the upregulation of the MyHCI/β promoter activity.
MEK1–ERK1/2 signaling increased Ca2+-ionophore-induced NFATc1 transcriptional activity depending on a specific NFAT binding site
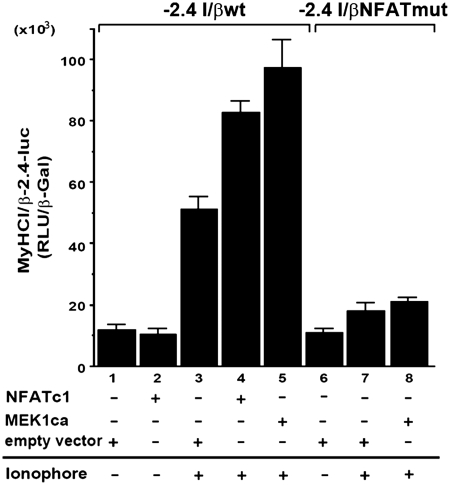
The importance of the transcription factor NFATc1 for MyHCI/β promoter activation (13,21) led us to investigate a possible interaction between MEK1-ERK1/2 signaling and NFATc1. The short isoform NFATc1/αA was used here in transient transfection assays (23,24). Although the activating effect of 0.1 µM Ca2+-ionophore on wildtype (wt) MyHCI/β (−2.4MyHCI/βwt) promoter activity was further increased by overexpression of NFATc1 (Figure 2, lanes 1, 3 and 4), the relatively moderate effect might reflect the need for additional but endogenously limited factor(s) for robust MyHCI/β promoter activation. A putative NFAT binding site at −439/−432 bp in the MyHCI/β upstream regulatory region had been previously identified (13). When the −439/−432-bp NFAT binding site was mutated (−2.4MyHCI/βNFATmut), no effect on the basal MyHCI/β promoter activity was found in line with NFATc1 not activating the wild-type promoter under control conditions (Figure 2, lanes 1, 2 and 6). However, the Ca2+-ionophore-induced increase in promoter activity was reduced in −2.4MyHCI/βNFATmut (lanes 1, 3, 6 and 7), indicating functional significance of the −439/−432-bp NFAT binding site for MyHCI/β promoter activation. The activating effect of MEK1ca coexpression on the wild-type promoter activity under Ca2+-ionophore was abolished with −2.4MyHCI/βNFATmut (Figure 2, lanes 3, 5, 7 and 8), indicating that the NFAT binding site is crucial for the activating effect of MEK1–ERK1/2 signaling.
Figure 2.
MEK1–ERK1/2 signaling increases Ca2+-ionophore-induced NFATc1 transcriptional activity depending on a specific NFAT binding site. C2C12 cells were transiently transfected with a −2.4-kb wild-type MyHCI/β (−2.4 I/βwt) promoter construct or a −2.4-kb MyHCI/β promoter construct mutated in the −439/−432-bp NFAT binding site (−2.4 I/βmut) alone, or were cotransfected with expression vectors for NFATc1, or MEK1ca, or empty vector. Cells were grown for 24 h in GM and then for 2 days in DM with or without Ca2+-ionophore A23187 (0.1 µM). The promoter activity is expressed as relative light units per unit β-galactosidase (RLU/β-Gal). The data represent the mean ± SD of triplicate data points.
The MEK1/ERK1/2 pathway activates the MyHCI/β promoter via nuclear NFATc1
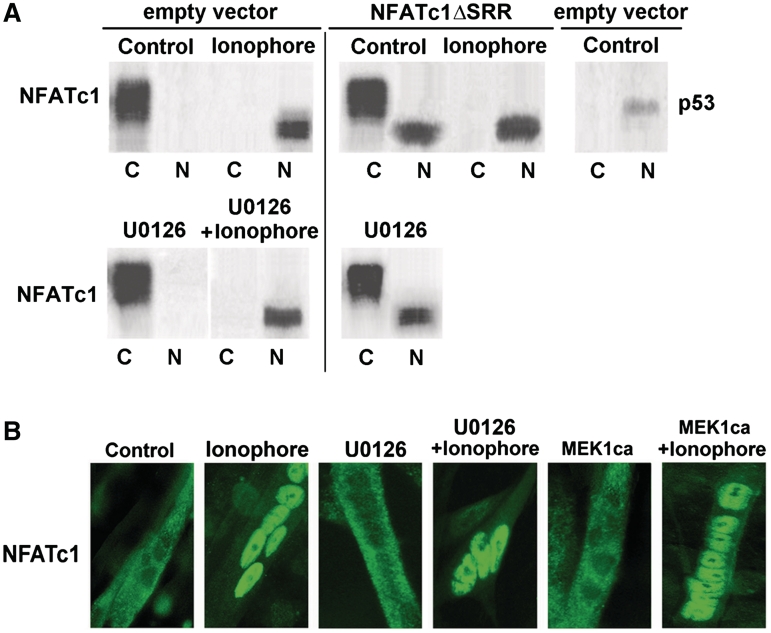
To investigate interaction between NFATc1 and MEK1–ERK1/2 signaling without interfering with other factors activated by Ca2+-ionophore, we took advantage of mutant NFATc1, NFATc1ΔSRR (13). A deletion in the regulatory SRR resulted in nuclear localization of transfected NFATc1ΔSRR in untreated fast fiber type-like C2C12 myotubes, whereas endogenous NFATc1 was almost exclusively cytoplasmic, as demonstrated by western blot and immunofluorescence analysis [Figure 3A (upper panel) and B]. The absence of a distinct NFATc1 band in western blots corresponds to the previously reported existence of multiple phosphorylation states of cytoplasmic and nuclear NFATc1 (56–58). In accordance with the subcellular localization, NFATc1ΔSRR, but not wild-type NFATc1 (NFATc1wt) was able to activate the MyHCI/β promoter in control cells (Figure 4A, lanes 1–3). Cotransfection of MEK1ca increased NFATc1ΔSRR-induced MyHCI/β promoter activity (lanes 1, 3 and 6). Conversely, addition of MEK1dn or inhibitor U0126 (10 µM) reduced NFATc1ΔSRR-induced MyHCI/β promoter upregulation (lanes 1, 3, 7 and 8). Neither ERK1/2 inhibition via U0126 (10 µM) nor activation via MEK1ca altered the subcellular localization of endogenous NFATc1 (cytoplasmic) or transfected NFATc1ΔSRR (nuclear) in untreated C2C12 myotubes [Figure 3A (compare lower with upper panel) and B]. Similarly, the nuclear translocation of endogenous NFATc1 induced by 0.1 µM Ca2+-ionophore was not affected. The data show that MEK1–ERK1/2 signaling is not affecting NFATc1 subcellular localization, but exerts its activating function on MyHCI/β promoter activity via nuclear NFATc1.
Figure 3.
The MEK1-ERK1/2 pathway is not affecting NFATc1 subcellular localization. (A) Western blot analysis of the localization of endogenous NFATc1 (empty vector) and transfected constitutively nuclear NFATc1ΔSRR in C2C12 myotubes. C2C12 cells transfected with NFATc1ΔSRR or the empty vector were grown for 24 h in GM and for 2 days in DM in the presence or absence (Control) of Ca2+-ionophore A23187 (0.1 µM) and/or U0126 (10 µM). After cell fractionation, aliquots of the pellet (nuclear, N) and supernatant (cytoplasmic, C) fraction were analyzed by western blotting using anti-NFATc1 and anti-p53 antibodies. Detection of p53 serves as a control for nuclear localization. (B) Immunofluorescence analysis of the localization of endogenous NFATc1 in C2C12 myotubes. Cells transfected with MEK1ca expression vector were grown for 24 h in GM and then for 3 days in DM, untransfected C2C12 myotubes for 4 days in DM (Control) or for 2 days in DM and then for additional 2 days in DM in the presence of Ca2+-ionophore (0.1 µM) and/or U0126 (10 µM). Cells were stained with an anti-NFATc1-antibody. NFATc1 was visualized by a FITC-labeled secondary antibody. Fluorescence was detected by using an inverted fluorescence photomicroscope at a magnification of ×400.
Figure 4.
MEK1–ERK1/2 synergy with NFATc1 depends on transcriptional coactivator p300. C2C12 cells were transiently transfected with wildtype −2.4 kb MyHCI/β (−2.4 I/βwt) promoter construct and cotransfected (A) with or (B) without expression vectors coding for wild-type NFATc1 (NFATc1wt), or constitutively nuclear NFATc1ΔSRR, or empty vector. Cells were additionally cotransfected with expression vectors for wild-type p300 (p300wt), or acetyltransferase-deficient p300 (p300DY), or MEK1ca or MEK1dn, or empty vector. In (B) mutant (NFAT binding site) −2.4 kb MyHCI/β (−2.4 I/βNFATmut) promoter construct was also transfected. Cells were grown for 24 h in GM and then for 2 days in DM with or without U0126 (10 µM), or Ca2+-ionophore A23187 (0.1 µM). (C) Western blot analysis of NFATc1 expression in Ca2+-ionophore-treated C2C12 cells transfected with p300wt, p300DY or the empty vector, using an anti-NFATc1 antibody. The blot was reprobed with anti-α-tubulin antibody as loading control. (D) Western blot analysis of p300 expression in non-transfected C2C12 myotubes, or cells transfected with a pool of double-stranded 20–25-nt siRNA that specifically target mouse p300 (siRNA p300), or with non-specific doublestranded control siRNA (siRNA control), using an anti-p300 antibody. The blot was reprobed with anti-histone H3 antibody as loading control. (E) C2C12 cells were transiently transfected with a −2.4-kb MyHCI/β promoter construct and cotransfected with NFATc1ΔSRR or empty vector. After grown for 24 h in GM, cells were cotransfected with p300 siRNA (siRNA p300) or a non-specific control siRNA (siRNA control), and then grown for 2 d in DM. The promoter activity in (A), (B) and (E) is expressed as relative light units per unit β-galactosidase (RLU/β-Gal). The data represent the mean ± SD of triplicate data points.
MEK1–ERK1/2 synergy with NFATc1 depends on transcriptional coactivator p300
Using chromatinimmunoprecipitation, we previously demonstrated NFATc1 binding to the −439/−432-bp site in the MyHCI/β promoter and recruitment of the transcriptional coactivator p300 (13). Transfection of p300 wildtype (p300wt) caused a moderate increase of basal MyHCI/β promoter activity in control and a further increase in Ca2+-ionophore-treated C2C12 myotubes alone (Figure 4B, lanes 1–4). In addition, coexpression of p300wt with NFATc1ΔSRR in untreated cells further increased the promoter activity as compared with NFATc1ΔSRR alone (Figure 4A, lanes 1, 3 and 4), indicating that NFATc1-mediated MyHCI/β promoter activation depends on an interaction with p300. Consistently, mutating the NFAT binding site nearly abolished the effect of p300 on the MyHCI/β promoter in Ca2+-ionophore-treated C2C12 myotubes (Figure 4B, lanes 2, 4, 10 and 11). The data demonstrate MyHCI/β promoter upregulation by p300, which is associated with an intact −439/−432-bp NFAT binding site.
Cotransfection with p300DY, a p300 mutant deficient in its acetyltransferase function, led to a reduction of the Ca2+-ionophore effect on MyHCI/β promoter activity, abolishing also the effect of p300wt (Figure 4B, lanes 1, 2, 4 and 6). Moreover, p300DY almost completely abolished the activating effect of cotransfection of NFATc1ΔSRR and p300 (Figure 4A, lanes 1 and 3–5). Thus, the activating effect of p300 on the MyHCI/β promoter correlates with the acetyltransferase function. Western blot analysis demonstrated that the effect of exogenous p300wt or p300DY was not associated with a change in endogenous NFATc1 protein expression (Figure 4C).
Next, immunoblotting revealed a significant decrease in p300 protein expression in myotubes transfected with specific p300 siRNA but not in cells transfected with non-specific double-stranded control siRNA, or non-transfected cells (Figure 4D). Knock-down of p300 led to a small decrease in basal MyHCI/β promoter activity and a reduction in the Ca2+-ionophore-induced increase in promoter activity (Figure 4E, lanes 1, 2, 4 and 5), and the activating effect of NFATc1ΔSRR was also diminished (lanes 1, 3, 4 and 6). These data further support a role of p300 for NFATc1-mediated MyHCI/β promoter activation by Ca2+-ionophore that depends on the intact −439/−432-bp NFAT binding site. The small effect of p300 on MyHCI/β promoter basal activity may be due to the interaction of p300 with transcription factors like MyoD known to mediate the basal activity (13).
MEK1–ERK1/2 signaling indirectly regulates NFATc1–p300 complex formation at the NFAT site via p300
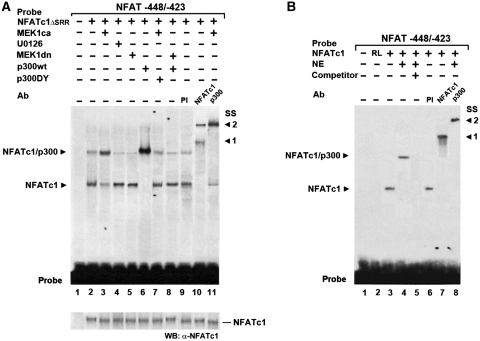
We further examined whether DNA binding of NFATc1 is altered by MEK1–ERK1/2 signaling. NFATc1ΔSRR was comparably expressed in nuclear extracts (NEs) from C2C12 myotubes not treated with Ca2+-ionophore (with endogenous NFATc1 nearly completely cytoplasmic, cf. Figure 3) used in electrophoretic mobility shift assays (EMSAs) (Figure 5A, WB). No complex formation was found with a probe containing the −439/−432-bp NFAT binding site and NE from untransfected control cells (lane 1). Overexpression of NFATc1ΔSRR led to formation of two complexes, both supershifted (SS 1 and 2) by an anti-NFATc1 antibody but not preimmune serum (PI) (lanes 2, 9 and 10), indicating binding of NFATc1ΔSRR to the probe. The faster mobility complex (lower complex) was similar to that formed by the binding of in vitro translated NFATc1 to the probe (Figure 5B, lanes 1–3, 6 and 7, with supershift SS 1). The slower mobility complex (upper complex) in Figure 5A was similar to that observed when NE of untreated C2C12 myotubes was added to the in vitro translated NFATc1 (Figure 5B, lane 4). An antibody against p300 supershifted (SS 2) the upper complex (lanes 4 and 8), indicating that the complex included endogenous p300. Consistently, coexpression of NFATc1ΔSRR and p300wt led to increased formation of the upper complex and disappearance of the lower complex (Figure 5A, compare lanes 2 and 6). Thus, these data demonstrate binding of NFATc1ΔSRR alone to the probe in the lower complex, presumably due to limited availability of endogenous p300, and additional recruitment of p300 in the upper complex.
Figure 5.
MEK1–ERK1/2 signaling indirectly regulates NFATc1-p300 complex formation at the NFAT site via p300. EMSAs demonstrate that NFATc1–p300 DNA binding complex stability depends on MEK1–ERK1/2 signaling. Radiolabeled oligonucleotide probes containing the −439/−432-bp NFAT binding site from the MyHCI/β promoter were incubated (A) with nuclear extracts (NEs) from C2C12 myotubes grown for 3 days in DM or (B) with in vitro translated NFATc1 or unprogrammed rabbit reticulocyte lysate (RL), with or without NEs from C2C12 myotubes grown for 3 days in DM. In (A) cells were transfected 24 h prior NE preparation with or without constitutively nuclear NFATc1ΔSRR expression vector alone, or were cotransfected with expression vectors for MEK1ca, or MEK1dn or p300wt, or p300DY, or empty vector. In addition, cells were grown with or without U0126 (10 µM). In supershift experiments, preimmune serum (PI), or anti-NFATc1, or anti-p300 antibodies (Ab) were added. A 200-fold excess of unlabeled wild-type competitor DNA (competitor +) was used in (B) for determination of specific protein–DNA binding reaction (lane 5). After the incubation, samples were fractionated on 5% polyacrylamide gels. Complexes are indicated by an arrowhead. SS: supershift, with (A) arrowhead 1 indicating the supershifted NFATc1ΔSRR–probe complex, and arrowhead 2 indicating the NFATc1ΔSRR/p300–probe complex and (B) arrowhead 1 indicating the supershifted NFATc1–probe complex, and arrowhead 2 indicating the NFATc1/p300–probe complex. Probe: bottom of the gel indicates excess probe. WB: western blot analysis of transfected NFATc1ΔSRR in NEs by probing aliquots with an anti-NFATc1 antibody.
MEK1ca increased while U0126 (10 µM) or MEK1dn decreased upper complex formation, and lower complex was reduced by MEK1ca while increased by U0126 or MEK1dn (Figure 5A, lanes 2–5). An anti-p300 antibody supershifted (SS 2) the upper complex completely while leaving the lower complex unaffected, demonstrating additional recruitment of p300 induced by activated MEK1–ERK1/2 (lanes 2, 3 and 11). The data suggest that complex formation of NFATc1ΔSRR with p300 but not binding of NFATc1ΔSRR to the NFAT site per se was augmented by activated MEK1–ERK1/2 signaling. In line, the p300wt-induced increase in upper complex formation was nearly abolished by coexpression of MEK1dn, with additional reappearance of the lower complex (lanes 6 and 8). In addition, the limited amount of lower complex formed in the presence of MEK1dn compared with the much more intensive upper band in the absence of MEK1dn (compare lanes 6 and 8) indicates that NFATc1 alone has a limited affinity to the NFAT site-containing probe, while p300 obviously greatly increases the affinity of NFATc1 to the site. Furthermore, a decrease of the upper complex and an additional increase of the lower complex was observed when p300DY was coexpressed with MEK1ca, compared with MEK1ca expression alone (lanes 3 and 7), indicating that the acetyltransferase function of p300 may also be important for NFATc1ΔSRR/p300 complex stability. Taken together, the data suggest an effect of MEK1–ERK1/2 MAPK signaling on complex stability of NFATc1ΔSRR/p300 at the NFAT site mediated by p300 (acetyltransferase activity) that can account for MEK1–ERK1/2 and p300 action on NFATc1 transcriptional function and hence for MyHCI/β promoter upregulation as found in transient transfection assays (cf. Figures 1, 2 and 4).
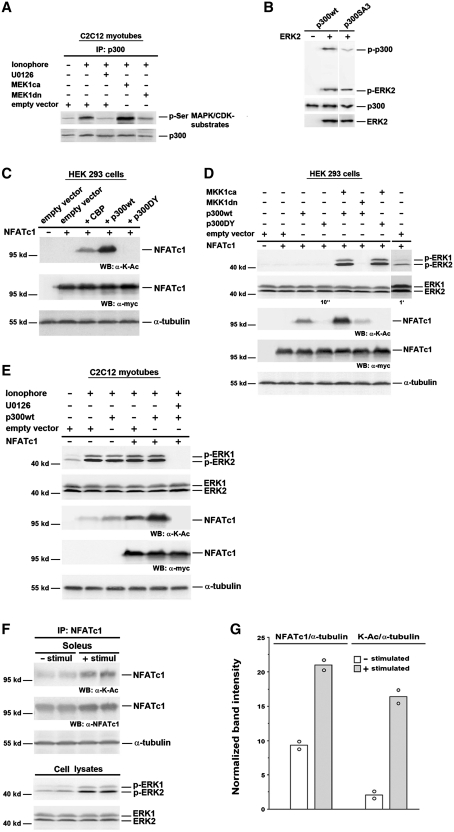
MEK1–ERK1/2 signaling-dependent p300 phosphorylation in vivo and in vitro
We then investigated whether p300 can be a substrate of ERK1/2 in C2C12 myotubes. Protein immunoprecipitated from cell lysates with anti-p300 antibodies was immunoblotted with antibodies specific for phospho-serine (p-Ser) followed by proline (Pro), a sequence motif found in MAPK or cyclin-dependent kinase (CDK) substrates. The level of p300 serine phosphorylation (p300-p-Ser) was increased in C2C12 myotubes treated with 0.1 µM Ca2+-ionophore compared with untreated cells (Figure 6A). MEK1ca further increased p300-p-Ser levels in Ca2+-ionophore-treated C2C12 myotubes, whereas MEK1dn or 10 µM U0126 nearly abolished the Ca2+-ionophore effect. These results indicate that the Ca2+-ionophore-induced activation of MEK1–ERK1/2 signaling increases the proline-directed phosphorylation of p300. To confirm that p300 can be direct substrate of ERK, an in vitro kinase assay was performed. Wild-type p300 protein was phosphorylated by recombinant activated ERK2 in vitro as detected by anti-p-Ser/Threonine (Thr)-Pro antibodies (Figure 6B), demonstrating that ERK2 can phosphorylate p300. In addition, autophosphorylation of the kinase (p-ERK2) is often observed in this kind of assay. Previous data indicate that the ERK-specific phosphorylation motif features serine–proline or threonine–proline phosphorylation sites in the vicinity of an ERK binding site (Phe-Xaa-Phe-Pro) (59). Phosphorylation by ERK2 was reduced in p300SA3, a mutant with serine residues 2279, 2315 and 2366 being replaced by alanine (Figure 6B). These sites play major roles in epidermal growth factor (EGF)-induced ERK2-mediated phosphorylation of p300 in keratinocytes (28). Taken together, these data indicate that p300 is a direct target of MEK1-ERK1/2 signaling via serine-phosphorylation by ERK1/2 in Ca2+-ionophore-treated C2C12 myotubes.
Figure 6.
NFATc1 is acetylated by p300 in a MEK1–ERK1/2-dependent manner. (A) Analysis of p300 phosphorylation by IP and western blot. C2C12 cells were transfected with MEK1ca, or MEK1dn, or empty vector. After 24 h in GM cells were further grown in DM. Two days after transfection cells were pretreated with or without U0126 (10 µM) for 30 min, followed by treatment with Ca2+-ionophore A23187 (0.1 µM) for 3 h. Protein from cell lysates was immunoprecipitated (IP) with anti-p300 antibody. Phosphorylation of p300 was analyzed using an anti-phospho-serine (p-Ser) MAPK/CDK antibody, and p300 protein expression was detected by reprobing with an anti-p300 antibody. (B) In vitro kinase assay with p300wt or p300SA3 incubated with recombinant activated ERK2. The phosphorylation level of p300 was analyzed by western blot using anti-phospho-Ser/Thr-Pro antibodies. The input of proteins was analyzed with anti-p300 and anti-ERK2 antibodies. Analysis of NFATc1 lysine acetylation and ERK1/2 activation in (C) and (D) HEK 293 cells; (E) C2C12 myotubes; and (F) mouse soleus muscle by western blot (WB). In (C–E) HEK 293 or C2C12 cells were transiently transfected with or without NFATc1-c-Myc expression vector alone, or with p300wt, or p300DY, or CBP, or MEK1ca, or MEK1dn, or empty vector. HEK 293 cells were then grown for 2 days in GM, C2C12 cells for 24 h in GM and for 2 days in DM. HDAC inhibitors (300 nM TSA and 5 mM NIA) were added 24 h before lysis in (C) and (D) and to the lysis buffer in (E). C2C12 myotubes were grown in the presence or absence of Ca2+-ionophore (0.1 µM) and/or U0126 (10 µM) for 12 h before lysis. (F) Isolated mouse soleus muscles were electrostimulated (30 min, 15 Hz; + stimul) or not stimulated (− stimul). Eight solei were pooled per each group and proteins were immunoprecipitated (IP) from nuclear extracts with anti-NFATc1 antibodies. Expression of Myc-tagged NFATc1 was monitored by an anti-c-Myc antibody, and NFATc1 lysine acetylation was detected by using an anti acetyl-lysine antibody (K-Ac). Expression of endogenous NFATc1 was analyzed by reprobing with an anti-NFATc1 antibody. Expression of phosphorylated ERK1 and 2 (p-ERK1 and 2) was analyzed using anti-phospho-ERK1/2 antibodies, and expression of total ERK1 and 2 with anti-ERK1/2 antibodies, directly analyzed from cell lysates in (F). Exposure times for enhanced chemiluminescence detection with the LAS-3000 imaging system in (D) were 10 s or 1 min. Detection of α-tubulin served as a loading control. Molecular weights are indicated. (G) Band intensities from western blot analysis as shown in (F) were normalized to α-tubulin and presented as mean of 2 and single values (open circle).
MEK1–ERK1/2 signaling-dependent p300 phosphorylation induces NFATc1 acetylation in HEK 293 cells
The functional importance of p300 acetyltransferase function for NFATc1-dependent MyHCI/β promoter activation prompted us to investigate the possible acetylation of NFATc1 by p300. Lysine (K)-acetylation of Myc-tagged NFATc1 (NFATc1-K-Ac) was detected, when p300wt but not when p300DY was coexpressed in HEK 293 cells (Figure 6C). In contrast to p300wt, expression of CBP led to a much smaller increase in the level of NFATc1-K-Ac. Lysine acetylation of NFATc1 was further increased by coexpression of MEK1ca and p300wt (Figure 6D). In addition, MEK1ca robustly increased the small level of basal ERK1/2 phosphorylation (see 1 min exposure time in Figure 6D), indicating activation without changes in the level of total ERK1/2 expression. In contrast, coexpression of MEK1dn abolished ERK1/2 phosphorylation and severely reduced NFATc1 lysine acetylation. The activating effect of MEK1ca on NFATc1-K-Ac but not on ERK1/2 phosphorylation was abolished by coexpression of p300DY. Taken together, the data demonstrate that p300 can acetylate NFATc1, and acetylation is MEK1–ERK1/2-dependent.
MEK1–ERK1/2 signaling-dependent p300 phosphorylation induces NFATc1 acetylation in C2C12 myotubes and soleus muscle
In C2C12 myotubes treated with 0.1 µM Ca2+-ionophore, a small amount of acetylation of endogenous NFATc1 could be detected as compared with untreated myotubes (Figure 6E). Increased acetylation of endogenous NFATc1 was found in cells treated with Ca2+-ionophore and overexpressing p300wt, indicating the ability of p300 to acetylate endogenous NFATc1. The level of NFATc1-K-Ac was also increased by overexpressing Myc-tagged NFATc1 alone and further enhanced by coexpression of p300wt. Western blot analysis revealed an increase in ERK1/2 phosphorylation in Ca2+-ionophore-treated myotubes, while total ERK1/2 expression was not altered, indicating stimulation of MEK1–ERK1/2 signaling by Ca2+-ionophore (Figure 6E). U0126 (10 µM) inhibited ERK1/2 activation as demonstrated by abolished phosphorylation of ERK1/2, and blocked lysine acetylation of NFATc1 in Ca2+-ionophore-treated cells coexpressing NFATc1 and p300wt. Together, the results indicate that the Ca2+-ionophore-induced phosphorylation of p300, which is dependent on activated ERK1/2 signaling, mediates acetylation of NFATc1 in vivo.
To test whether NFATc1 lysine acetylation can occur in whole muscle, mouse soleus was stimulated. Mouse soleus has in contrast to other species a significant (nearly 50%) proportion of fast fibers (60). When isolated soleus was stimulated for 30 min with 15 Hz, an increase of nuclear NFATc1 was found (Figure 6F), in accordance with previous findings in isolated fibers from fast flexor digitorum brevis in culture using a similar pattern (61). Stimulation also increased the level of total lysine-acetylated NFATc1 as well as the level relative to nuclear NFATc1 (Figure 6G). Furthermore, activation of ERK1 and 2 was enhanced by stimulation, as demonstrated by increased phosphorylation, while total ERK1/2 expression was not altered (Figure 6F). The data show that activity-dependent ERK1/2 activation and NFATc1 lysine acetylation can both occur in whole muscle.
Identification of acetylation sites in NFATc1 and effects on transactivation function and DNA binding
To prove that acetylation of NFATc1 is indeed mediated by p300, we searched for relevant acetylation sites in NFATc1/αA. The spectrum of modifications potentially affecting a given lysine residue is defined by the presence/absence of several adjacent residues, including a glutamic acid, a proline, and/or a serine–proline motif. For example, a G/SKXXP motif has been shown to be a consensus site for the acetylation of the Brm proteins by CBP/p300 (62) while Sp3 acetylation occurs at a motif that contains three glutamic acid residues, KEEEP (63) We identified two potential acetylation sites, lysines 351 and 549 (K351, K549) as part of a KXEP and KXE motif, respectively, and compared those with lysine 646 (K646), not comprising such a motif (Figure 7A). K351 is located in the regulatory and K549 as well as K646 in the RSD domain. We generated mutants by replacing lysine residues with arginine and transfected C2C12 cells with Myc-tagged wild-type NFATc1/αA and mutants to test if acetylation can occur in vivo (Figure 7B). Acetylation of wild-type NFATc1/αA in the presence of p300 and Ca2+-ionophore was clearly detected. Compared to wild-type, acetylation of the K351R and K549R mutants was significantly impaired while no significant decrease in acetylation occurred for the K646R mutant. By utilizing these NFATc1/αA mutants, we tested whether or not p300 activates the MyHCI/β promoter through NFATc1 acetylation. MyHCI/β promoter activation was reduced by the acetylation-defective NFATc1/αA mutants K351R and K549R, but not by K646R when compared to wild-type in Ca2+-ionophore treated cells coexpressing p300wt (Figure 7C). Thus, we conclude that K351 and K549 are major acetylation sites and that p300 is acting on the MyHCI/β promoter via acetylation of NFATc1/αA rather than merely being recruited to acetylate nucleosomal tails of histones.
We also tested whether acetylation affects NFATc1 DNA binding. Using the NFAT binding site probe (Figure 5), complex formation in nuclear extracts from Ca2+-ionophore-treated C2C12 cells expressing Myc-His-tagged wild type or mutant NFATc1/αA together with p300wt was analyzed by EMSA. Two NFATc1-specific complexes were detected: one containing mainly the endogenous NFATc1 (complex I) and one containing mainly the exogenously transfected Myc-NFATc1/αAwt (complex II) (Figure 7C, lane 2). The specificity of complex formation was verified both by competition with a 200-fold excess of cold probe (lane 5) and by supershifting only complex II using an anti-c-Myc antibody (lane 6). Importantly, lesser complex II formation together with unchanged complex I formation was found with NFATc1/αAK351R and K549R, demonstrating that both mutants had a lower DNA binding activity than wild-type. These findings can account for an acetylation-mediated increase in NFATc1 affinity to the MHCI/β promoter and NFATc1-mediated transcriptional activation.
Interestingly, multiple sequence alignment revealed that one acetylated lysine in murine NFATc1 (K351) and adjacent amino acids are fully conserved in NFATc1 of boar, bovine, fish, frog, human and mouse origin, and in NFATc3 of mouse and human origin (Figure 7E, in blue). The second major acetylated lysine (K549) and adjacent amino acids are fully conserved in NFATc1-4 of boar, bovine, fish, frog, human and mouse (Figure 7E, in red).
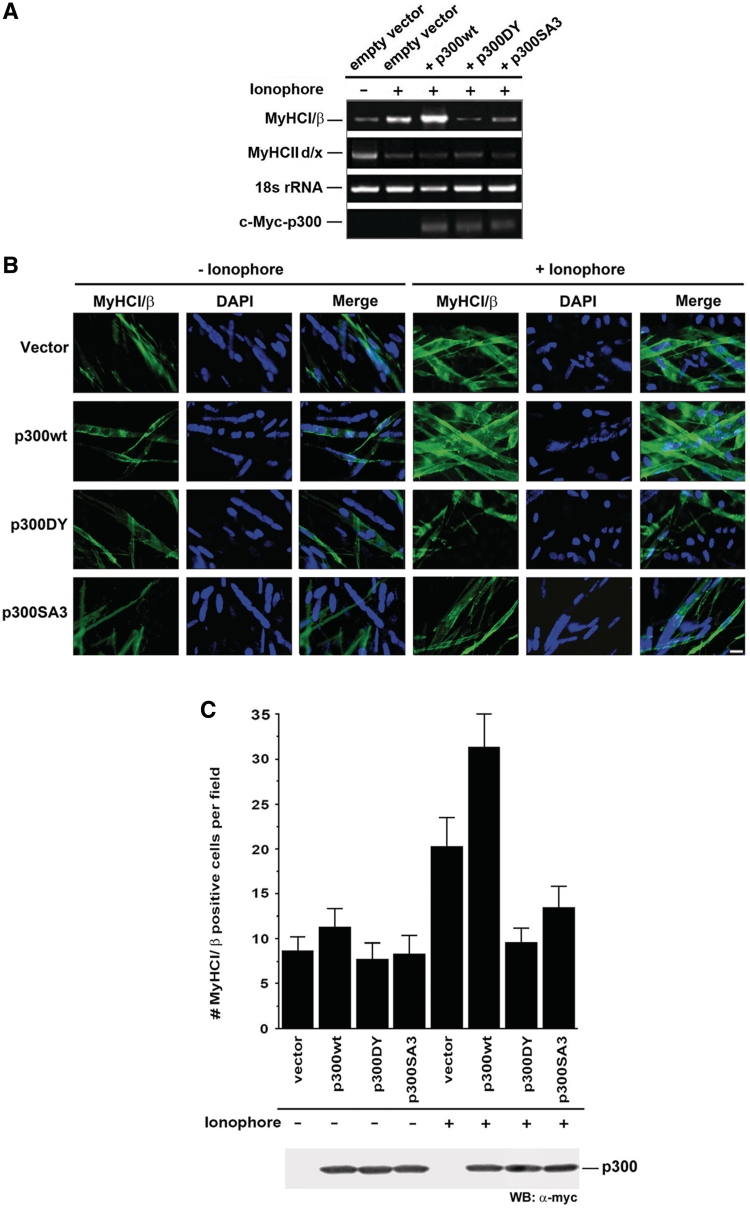
Ca2+-ionophore-induced MyHCI/β gene expression depends on phosphorylation of p300 and its acetyltransferase function
We further assessed the effect of p300 on endogenous MyHCI/β mRNA and protein levels in C2C12 myotubes by semiquantitative RT–PCR and immunofluorescence analysis. Low levels of MyHCI/β and high levels MyHCIId/x mRNA were detected in untreated cells (Figure 8A). Ca2+-ionophore (0.1 µM) decreased MyHCIId/x mRNA and increased MyHCI/β mRNA and protein (Figure 8B and C) levels, demonstrating the fast-to-slow transformation at the mRNA level. A further increase in expression of MyHCI/β mRNA and protein was apparent in cells transfected with p300wt while p300DY completely and p300SA3 largely abolished the Ca2+-ionophore-induced increase. Thus, the activating effect of p300 on MyHCI/β endogenous gene expression correlates with its acetyltransferase function, in line with data on promoter activity (Figure 4B), and depends at least in part on the phosphorylation of serines 2279, 2315 and 2366. Taken together, the results implicate that ERK-dependent serine phosphorylation of p300 is a regulatory mechanism pivotal for a strong MyHCI/β gene expression induced by increased intracellular Ca2+-levels. Furthermore, decreased MyHCIId/x mRNA levels in Ca2+-ionophore-treated C2C12 cells were unaffected by p300wt, p300DY or p300SA3, suggesting that downregulation of MyHCIId/x expression was not mediated via p300.
Figure 8.
Ca2+-ionophore-induced MyHCI/β gene expression depends on phosphorylation of p300 and its acetyltransferase function. (A) Semiquantitative RT–PCR analysis of endogenous MyHCI/β and IId/x expression. Total RNA was isolated from C2C12 cells transfected with p300wt-, or p300DY-, or p300SA3-Myc, or empty Myc-tag expression vector. Cells were grown for 24 h in GM and then for 2 days in DM with or without Ca2+-ionophore. PCR products were separated on a 2% agarose gel and visualized with ethidium bromide staining. 18 s rRNA levels were used for normalization. The mRNA expression of transfected Myc-tagged p300, p300DY and p300SA3 was evaluated using a Myc-specific reverse and a p300 forward primer. (B) Immunofluorescence analysis of endogenous MyHCI/β expression in C2C12 myotubes. C2C12 cells were transiently transfected with p300wt-, or p300DY-, or p300SA3-Myc, or empty Myc-tag expression vector. Cells were grown for 24 h in GM and then for 4 days in DM with or without Ca2+-ionophore A23187 (0.1 µM). Cells were stained with an anti-MyHCI/β-antibody. MyHCI/β was visualized by a FITC-labeled secondary antibody. Fluorescence was detected by using an inverted fluorescence photomicroscope. MyHCI/β positive cells appear green. Nuclei were stained with DAPI. Scale bar, 100 µm. (C) Histogram illustrating the mean number of MyHCI/β positive cells per vision field (n = 6) as examined by immunofluorescence analysis shown in (B). Western blot analysis using anti-c-Myc antibody illustrating p300 expression levels in cells transfected with Myc-tagged p300 expression vectors.
DISCUSSION
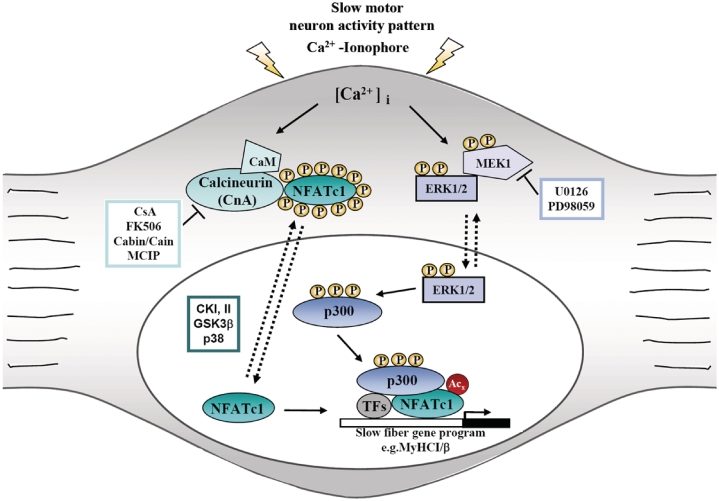
In the present study, the role of MEK1–ERK1/2 signaling pathway and transcriptional coactivator p300 in regulating NFATc1-dependent slow MyHCI/β promoter activity and endogenous gene expression was investigated. We previously found that increased [Ca2+]i in primary skeletal muscle cells induces the recruitment of p300 to NFATc1 bound to the MyHCI/β promoter (13). The main findings presented now about a mechanism of signaling leading to slow fiber-specific gene expression are summarized in a model (Figure 9). Briefly (i) phosphorylation of p300 via MEK1-ERK1/2 signaling is a major effect caused by elevated [Ca2+]i; (ii) phosphorylation of p300 stimulates its binding to NFATc1; (iii) binding of p300 promotes lysine acetylation of NFATc1; and (iv) enhances NFATc1 interaction with and transactivation of the MyHCI/β promoter.
Figure 9.
Proposed model of the interaction of calcineurin-NFATc1, MEK1–ERK1/2 and p300signaling leading to slow skeletal muscle fiber-specific gene expression. Stimuli that increase [Ca2+]i result in translocation of dephosphorylated NFAT to the nucleus where it participates in mediating Ca2+-inducible gene expression of slow fiber genes, such as MyHCI/β. This induction of MyHCI/β can be further stimulated by Ca2+-mediated activation of the MEK1–ERK1/2 pathway resulting in the phosphorylation of transcriptional coactivator p300, leading to its recruitment to NFATc1 bound to the MyHCI/β promoter. Once recruited, phosphorylated p300 acetylates NFATc1 and enhances NFATc1–DNA binding. As a consequence, p300 enhances gene expression of MyHCI/β mediated by increased intracellular Ca2+-concentration. CaM, calmodulin; CKI/II, casein kinase I/II; CsA, cyclosporine A; GSK3β, glycogensynthase kinase-3β; MCIP (myocyte-enriched calcineurin interacting protein).
HATs like p300 or CBP not only acetylate histones but also target a variety of non-histone proteins including several transcription factors (33). For example, HAT activity of p300 was shown to be crucial for transactivation of the keratin 16 promoter. The C-terminus of the transcription factor c-Jun, which plays an important role for keratin 16 expression, can be acetylated by p300 (64). During myogenesis, acetylation of myocyte enhancer factor 2 (MEF-2) by p300 enhances the transcriptional activity by increasing DNA binding activity (65). Here, the acetyltransferase function of p300 is shown to be important for NFATc1 acetylation and subsequent MyHCI/β promoter activation by NFATc1 in myotubes, at least in part by sustaining complex formation at the NFAT binding site of the promoter. Transcription factor binding to DNA is thought to be affected by acetylation presumably through conformational changes (65). In addition to these findings in skeletal myotubes, p300 also affects the expression of MyHCβ in cardiomyocytes. A previous study showed that oncogene E1A-mediated transcriptional repression of MyHCβ promoter activity in primary cardiomyocytes was relieved by overexpression of p300 (66).
Besides possessing intrinsic HAT activity, p300/CBP can also mediate the transactivation function of transcription factors to enhance gene transcription via creating a bridge between transcription factors and the basal transcription machinery, or acting as a scaffold for the assembly of multiprotein complexes, including transcription factors and cofactors (67). The latter two functions may also contribute to p300 induced increases in NFATc1 transcriptional activity. In accordance with data presented in skeletal myotubes, the mechanism underlying increased NFAT transcriptional activation function is also achieved through interaction with transcriptional coactivators in other cell types. Both the N-terminal and the C-terminal TAD of NFATc4 are necessary for interaction with CBP in COS cells, and potentiation of NFATc4 transcriptional activity was meditated by CBP (68). In cardiomyocytes, p300 markedly potentiates the binding of NFATc1 to the B cell leukemia-2 (bcl-2) NFAT element by interacting with NFATc1 (69). The N-terminal TAD, TAD-A, of NFATc1 and c2 binds p300/CBP and enhances TAD-A activity in T-cells (34,35). Complex formation of p300 with NFATc1 in myotubes was previously shown by us (13). Here, we provide further elucidation of the mechanism revealing that p300-mediated lysine acetylation of NFATc1 is at least one factor that markedly increases NFATc1-dependent transactivation function. Our data do not exclude acetylation of other proteins such as histones by p300 that may synergize in regulating NFAT function during skeletal muscle fiber transformation.
The functional cooperation between cofactors and transcription factors can require phosphorylation of the transcription factor (29,70). Several NFAT family members have phosphorylation sites for protein kinases, including MAPKs, within the NH2-terminal domain (1,44). The NFATc1–TAD-A was at least shown to be an in vitro target of ERK2, and ERK2 increased TAD-A activity in T-cells (35). Moreover, MEK1 overexpression has been shown to increase NFATc3 DNA binding in cardiomyocytes, with NFATc3 directly phosphorylated by activated ERK2 (44). In addition to this direct effect, indirect enhancement of NFAT-dependent gene expression occurs through induction of AP-1 activity via MEK1–ERK1/2 signaling. Our data do not rule out phosphorylation of NFATc1 by MEK1–ERK1/2 signaling in the C2C12 myotubes, but are in favor of the phosphorylation of p300 with subsequent acetylation of NFATc1. Clearly, the role of MEK1–ERK1/2 is not to function as a regulator of NFATc1 nuclear shuttling.
Indeed, coactivators like p300 or CBP themselves are controlled by an array of various covalent modifications, leading to changes in HAT activity or protein–protein interactions (71). Direct phosphorylation of p300/CBP can control their recruitment to the transcriptional complex (27,72), and a number of phosphorylatable, evolutionary conserved residues are present in p300 and CBP. As several different phosphorylation sites are available to MAPKs (67,73), phosphorylation and subsequent activation of CBP by MAPKs (29) including ERK1 have been shown, resulting in stimulation of HAT enzymatic activity (74). EGF-dependent C-terminal serine phosphorylation by ERK2 regulates recruitment of p300 to the keratin 16 promoter in an epidermal keratinocyte cell line, thereby stimulating acetyltransferase activity and interaction with transcription factor Sp1 (31). We have now demonstrated that phosphorylation of serine residues in ERK-specific phosphorylation motifs of p300 induced by Ca2+-ionophore affected the control of MyHCI/β gene expression.
The two major p300 acetylation sites in NFATc1/αA identified here are within the regulatory domain (K351) and the Rel DNA binding domain (RSD; K549). We show that acetylation at these sites can affect NFATc1 DNA binding (Figure 7). The NFAT DNA binding domain is very similar in its conformation to the Rel DNA binding domain of Rel/NF-κB factors, and acetylation of the RelA subunit by p300/CBP or PCAF (p300/CBP-associated factor) at distinct sites either enhances or diminishes DNA binding and regulates nuclear function of NF-κB (75). Even though one acetylation site (K351) on NFATc1 is not within the DNA binding domain it also affects DNA binding. Likewise, p53 acetylation by p300 on multiple sites at the C-terminal regulatory domain affects the function of its DNA binding domain in the middle of the protein, presumably through conformational changes (76,77). The exact underlying mechanism for this effect remains unclear so far. It is reasonable that acetylation at K351 can alter the conformation of NFATc1 such that NFATc1 efficiently recruits the transcriptional coactivator with concomitant increase in complex stability. To conclude, changes in DNA binding are involved in acetylation mediated increases in NFATc1 transactivation function. Our data do neither exclude a role of other lysines for the transactivation function of NFATc1/αA nor other possible effects of acetylation like modulation of protein–protein interactions.
It is noteworthy that one acetylatable lysine and adjacent amino acids (K351) in murine NFATc1 are fully conserved across several different species and the second acetylatable lysine (K549) and adjacent amino acids are further conserved between NFATc1-4 family members (Figure 7E). This suggests that NFAT acetylation by p300 at these sites might at least be part of a general mechanism conserved in vertebrates. Although we have studied the acetylation of NFATc1/αA in the context of NFATc1-dependent MyHCI/β gene regulation as a key event of slow fiber type-specific gene expression upon increased intracellular Ca2+ concentrations, it is conceivable that NFATc1 isoforms and/or the other NFAT family members in various tissues are similarly regulated by acetylation and that such an event may also take place in other biological processes, including NFAT roles in the immune, nervous, vascular and endocrine systems, in heart and in cancer progression (78).
In summary, interaction of the MEK1–ERK1/2 pathway with the transcriptional coactivator p300 was found to play a key role in NFATc1-dependent MyHCI/β promoter upregulation. The data presented here provide a molecular basis for driving slow fiber-specific MyHCI/β gene expression and point to a role of MEK1–ERK1/2 signaling in the regulation of muscle fiber type. Consistent with our findings, (Ras-)MEK1/2-ERK signaling has been shown to play a pivotal role in slow MyHCI/β gene expression in regenerating rat soleus muscle (40) and in rat primary skeletal muscle cells (79). Interestingly, further studies in C2C12 and primary myotubes revealed that Ca2+-ionophore-induced activity of a fast MyHCIIa promoter construct is also mediated by calcineurin/NFAT and MEK1-dependent signaling (80), while another MAPK, p38α/β, is essential for fast MyHCIId/x gene expression (81,82), and reduced p38α/β signaling is involved in Ca2+-ionophore-mediated MyHCIId/x downregulation (81), whereas MEK1–ERK1/2 is not (data not shown). It has been proposed earlier (83) that different subset of genes are regulated by different signal transduction pathways to achieve muscle fiber type-specific changes in gene expression. In view of the data emerging so far, different signaling pathways can also converge to regulate gene expression during fast-to-slow fiber type transformation.
FUNDING
Deutsche Forschungsgemeinschaft. Funding for open access charge: Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to Dr N. G. Ahn, Dr M. A. Brown and Dr T.-P. Yao for their generous gift of plasmids (pMCL-HA-MEK1-R4F and pMCL-HA-MEK1-8 E; pcDNA3-NFATc1; pCMVβ-p300-Myc and pCMVβ-p300DY-Myc). We thank Dr M. Gaestel for critical reading of the manuscript and helpful discussions. We wish to thank Dr C. Geers-Knörr for help with muscle preparation and stimulation, and W. Zingel for technical assistance.
REFERENCES
- 1.Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003;17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- 2.Im SH, Rao A. Activation and deactivation of gene expression by Ca2+/calcineurin-NFAT-mediated signaling. Mol. Cells. 2004;18:1–9. [PubMed] [Google Scholar]
- 3.Masuda ES, Imamura R, Amasaki Y, Arai K, Arai N. Signalling into the T-cell nucleus: NFAT regulation. Cell Signal. 1998;10:599–611. doi: 10.1016/s0898-6568(98)00019-9. [DOI] [PubMed] [Google Scholar]
- 4.Zhu J, McKeon F. Nucleocytoplasmic shuttling and the control of NF-AT signaling. Cell Mol. Life Sci. 2000;57:411–420. doi: 10.1007/PL00000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Booth FW, Baldwin KM. Exercise: Regulation and Integration of Multiple Systems. In: Rowell LB, Shepherd JT, editors. Handbook of Physiology. Oxford: Oxford University Press; 1996. pp. 1075–1123. [Google Scholar]
- 6.Pette D, Staron RS. Transitions of muscle fiber phenotypic profiles. Histochem. Cell Biol. 2001;115:359–372. doi: 10.1007/s004180100268. [DOI] [PubMed] [Google Scholar]
- 7.Goldspink G. Gene expression in skeletal muscle. Biochem. Soc. Trans. 2002;30:285–290. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- 8.Kubis HP, Haller EA, Wetzel P, Gros G. Adult fast myosin pattern and Ca2+-induced slow myosin pattern in primary skeletal muscle culture. Proc. Natl Acad. Sci. USA. 1997;94:4205–4210. doi: 10.1073/pnas.94.8.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams RS, Rosenberg P. Calcium-dependent gene regulation in myocyte hypertrophy and remodeling. Cold Spring Harb. Symp. Quant. Biol. 2002;67:339–344. doi: 10.1101/sqb.2002.67.339. [DOI] [PubMed] [Google Scholar]
- 10.Kubis HP, Hanke N, Scheibe RJ, Meissner JD, Gros G. Ca2+ transients activate calcineurin/NFATc1 and initiate fast-to-slow transformation in a primary skeletal muscle culture. Am. J. Physiol. Cell Physiol. 2003;285:C56–C63. doi: 10.1152/ajpcell.00377.2002. [DOI] [PubMed] [Google Scholar]
- 11.Meissner JD, Gros G, Scheibe RJ, Scholz M, Kubis HP. Calcineurin regulates slow myosin, but not fast myosin or metabolic enzymes, during fast-to-slow transformation in rabbit skeletal muscle cell culture. J. Physiol. 2001;533:215–226. doi: 10.1111/j.1469-7793.2001.0215b.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kubis HP, Scheibe RJ, Meissner JD, Hornung G, Gros G. Fast-to-slow transformation and nuclear import/export kinetics of the transcription factor NFATc1 during electrostimulation of rabbit muscle cells in culture. J. Physiol. 2002;541:835–847. doi: 10.1113/jphysiol.2002.017574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meissner JD, Umeda PK, Chang KC, Gros G, Scheibe RJ. Activation of the beta myosin heavy chain promoter by MEF-2D, MyoD, p300, and the calcineurin/NFATc1 pathway. J. Cell Physiol. 2007;211:138–148. doi: 10.1002/jcp.20916. [DOI] [PubMed] [Google Scholar]
- 14.Zebedin E, Sandtner W, Galler S, Szendroedi J, Just H, Todt H, Hilber K. Fiber type conversion alters inactivation of voltage-dependent sodium currents in murine C2C12 skeletal muscle cells. Am. J . Physiol. Cell Physiol. 2004;287:C270–C280. doi: 10.1152/ajpcell.00015.2004. [DOI] [PubMed] [Google Scholar]
- 15.Nedachi T, Fujita H, Kanzaki M. Contractile C2C12 myotube model for studying exercise-inducible responses in skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 2008;295:E1191–E1204. doi: 10.1152/ajpendo.90280.2008. [DOI] [PubMed] [Google Scholar]
- 16.Meissner JD, Kubis HP, Scheibe RJ, Gros G. Reversible Ca2+-induced fast-to-slow transition in primary skeletal muscle culture cells at the mRNA level. J. Physiol. 1999;523:19–28. doi: 10.1111/j.1469-7793.2000.t01-1-00019.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carroll S, Nicotera P, Pette D. Calcium transients in single fibers of low-frequency stimulated fast-twitch muscle of rat. Am. J. Physiol. 1999;277:C1122–C1129. doi: 10.1152/ajpcell.1999.277.6.C1122. [DOI] [PubMed] [Google Scholar]
- 18.Chin ER, Olson EN, Richardson JA, Yang Q, Humphries C, Shelton JM, Wu H, Zhu W, Bassel-Duby R, Williams RS. A calcineurin-dependent transcriptional pathway controls skeletal muscle fiber type. Genes Dev. 1998;12:2499–2509. doi: 10.1101/gad.12.16.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naya FJ, Mercer B, Shelton J, Richardson JA, Williams RS, Olson EN. Stimulation of slow skeletal muscle fiber gene expression by calcineurin in vivo. J. Biol. Chem. 2000;275:4545–4548. doi: 10.1074/jbc.275.7.4545. [DOI] [PubMed] [Google Scholar]
- 20.Serrano AL, Murgia M, Pallafacchina G, Calabria E, Coniglio P, Lomo T, Schiaffino S. Calcineurin controls nerve activity-dependent specification of slow skeletal muscle fibers but not muscle growth. Proc. Natl Acad Sci. USA. 2001;98:13108–13113. doi: 10.1073/pnas.231148598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCullagh KJ, Calabria E, Pallafacchina G, Ciciliot S, Serrano AL, Argentini C, Kalhovde JM, Lomo T, Schiaffino S. NFAT is a nerve activity sensor in skeletal muscle and controls activity-dependent myosin switching. Proc. Natl Acad. Sci. USA. 2004;101:10590–10595. doi: 10.1073/pnas.0308035101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abbott KL, Friday BB, Thaloor D, Murphy TJ, Pavlath GK. Activation and cellular localization of the cyclosporine A-sensitive transcription factor NF-AT in skeletal muscle cells. Mol. Biol. Cell. 1998;9:2905–2916. doi: 10.1091/mbc.9.10.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Serfling E, Chuvpilo S, Liu J, Hofer T, Palmetshofer A. NFATc1 autoregulation: a crucial step for cell-fate determination. Trends Immunol. 2006;27:461–469. doi: 10.1016/j.it.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 24.Vihma H, Pruunsild P, Timmusk T. Alternative splicing and expression of human and mouse NFAT genes. Genomics. 2008;92:279–291. doi: 10.1016/j.ygeno.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Legube G, Trouche D. Regulating histone acetyltransferases and deacetylases. EMBO Rep. 2003;4:944–947. doi: 10.1038/sj.embor.embor941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan LW, Gambee JE. Phosphorylation of p300 at serine 89 by protein kinase C. J. Biol. Chem. 2000;275:40946–40951. doi: 10.1074/jbc.M007832200. [DOI] [PubMed] [Google Scholar]
- 27.Zanger K, Radovick S, Wondisford FE. CREB binding protein recruitment to the transcription complex requires growth factor-dependent phosphorylation of its GF box. Mol. Cell. 2001;7:551–558. doi: 10.1016/s1097-2765(01)00202-7. [DOI] [PubMed] [Google Scholar]
- 28.Impey S, Fong AL, Wang Y, Cardinaux JR, Fass DM, Obrietan K, Wayman GA, Storm DR, Soderling TR, Goodman RH. Phosphorylation of CBP mediates transcriptional activation by neural activity and CaM kinase IV. Neuron. 2002;34:235–244. doi: 10.1016/s0896-6273(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 29.Janknecht R, Nordheim A. MAP kinase-dependent transcriptional coactivation by Elk-1 and its cofactor CBP. Biochem. Biophys. Res. Commun. 1996;228:831–837. doi: 10.1006/bbrc.1996.1740. [DOI] [PubMed] [Google Scholar]
- 30.Gusterson R, Brar B, Faulkes D, Giordano A, Chrivia J, Latchman D. The transcriptional co-activators CBP and p300 are activated via phenylephrine through the p42/p44 MAPK cascade. J. Biol. Chem. 2002;277:2517–2524. doi: 10.1074/jbc.M104626200. [DOI] [PubMed] [Google Scholar]
- 31.Chen YJ, Wang YN, Chang WC. ERK2-mediated C-terminal serine phosphorylation of p300 is vital to the regulation of epidermal growth factor-induced keratin 16 gene expression. J. Biol. Chem. 2007;282:27215–27228. doi: 10.1074/jbc.M700264200. [DOI] [PubMed] [Google Scholar]
- 32.Kalkhoven E. CBP and p300: HATs for different occasions. Biochem. Pharmacol. 2004;68:1145–1155. doi: 10.1016/j.bcp.2004.03.045. [DOI] [PubMed] [Google Scholar]
- 33.Glozak MA, Sengupta N, Zhang X, Seto E. Acetylation and deacetylation of non-histone proteins. Gene. 2005;363:15–23. doi: 10.1016/j.gene.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Rodriguez C, Rao A. Nuclear factor of activated T cells (NFAT)-dependent transactivation regulated by the coactivators p300/CREB-binding protein (CBP) J. Exp. Med. 1998;187:2031–2036. doi: 10.1084/jem.187.12.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Avots A, Buttmann M, Chuvpilo S, Escher C, Smola U, Bannister AJ, Rapp UR, Kouzarides T, Serfling E. CBP/p300 integrates Raf/Rac-signaling pathways in the transcriptional induction of NF-ATc during T cell activation. Immunity. 1999;10:515–524. doi: 10.1016/s1074-7613(00)80051-5. [DOI] [PubMed] [Google Scholar]
- 36.Falvo JV, Lin CH, Tsytsykova AV, Hwang PK, Thanos D, Goldfeld AE, Maniatis T. A dimer-specific function of the transcription factor NFATp. Proc. Natl Acad. Sci. USA. 2008;105:19637–19642. doi: 10.1073/pnas.0810648105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Granja AG, Perkins ND, Revilla Y. A238L inhibits NF-ATc2, NF-kappa B, and c-Jun activation through a novel mechanism involving protein kinase C-theta-mediated up-regulation of the amino-terminal transactivation domain of p300. J. Immunol. 2008;180:2429–2442. doi: 10.4049/jimmunol.180.4.2429. [DOI] [PubMed] [Google Scholar]
- 38.Powell JA, Carrasco MA, Adams DS, Drouet B, Rios J, Muller M, Estrada M, Jaimovich E. IP(3) receptor function and localization in myotubes: an unexplored Ca(2+) signaling pathway in skeletal muscle. J. Cell Sci. 2001;114:3673–3683. doi: 10.1242/jcs.114.20.3673. [DOI] [PubMed] [Google Scholar]
- 39.Espinosa A, Leiva A, Pena M, Muller M, Debandi A, Hidalgo C, Carrasco MA, Jaimovich E. Myotube depolarization generates reactive oxygen species through NAD(P)H oxidase; ROS-elicited Ca2+ stimulates ERK, CREB, early genes. J. Cell Physiol. 2006;209:379–388. doi: 10.1002/jcp.20745. [DOI] [PubMed] [Google Scholar]
- 40.Pearson G, Robinson F, Beers GT, Xu BE, Karandikar M, Berman K, Cobb MH. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev. 2001;22:153–183. doi: 10.1210/edrv.22.2.0428. [DOI] [PubMed] [Google Scholar]
- 41.Murgia M, Serrano AL, Calabria E, Pallafacchina G, Lomo T, Schiaffino S. Ras is involved in nerve-activity-dependent regulation of muscle genes. Nat. Cell Biol. 2000;2:142–147. doi: 10.1038/35004013. [DOI] [PubMed] [Google Scholar]
- 42.Shi H, Scheffler JM, Pleitner JM, Zeng C, Park S, Hannon KM, Grant AL, Gerrard DE. Modulation of skeletal muscle fiber type by mitogen-activated protein kinase signaling. FASEB J. 2008;22:2990–3000. doi: 10.1096/fj.07-097600. [DOI] [PubMed] [Google Scholar]
- 43.Ichida M, Finkel T. Ras regulates NFAT3 activity in cardiac myocytes. J. Biol. Chem. 2001;276:3524–3530. doi: 10.1074/jbc.M004275200. [DOI] [PubMed] [Google Scholar]
- 44.Sanna B, Bueno OF, Dai YS, Wilkins BJ, Molkentin JD. Direct and indirect interactions between calcineurin-NFAT and MEK1-extracellular signal-regulated kinase 1/2 signaling pathways regulate cardiac gene expression and cellular growth. Mol. Cell. Biol. 2005;25:865–878. doi: 10.1128/MCB.25.3.865-878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Porter CM, Havens MA, Clipstone NA. Identification of amino acid residues and protein kinases involved in the regulation of NFATc subcellular localization. J. Biol. Chem. 2000;275:3543–3551. doi: 10.1074/jbc.275.5.3543. [DOI] [PubMed] [Google Scholar]
- 46.Yang TT, Xiong Q, Graef IA, Crabtree GR, Chow CW. Recruitment of the extracellular signal-regulated kinase/ribosomal S6 kinase signaling pathway to the NFATc4 transcription activation complex. Mol. Cell. Biol. 2005;25:907–920. doi: 10.1128/MCB.25.3.907-920.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hock MB, Brown MA. Nuclear factor of activated T cells 2 transactivation in mast cells: a novel isoform-specific transactivation domain confers unique FcepsilonRI responsiveness. J. Biol. Chem. 2003;278:26695–26703. doi: 10.1074/jbc.M301007200. [DOI] [PubMed] [Google Scholar]
- 48.Mansour SJ, Matten WT, Hermann AS, Candia JM, Rong S, Fukasawa K, Vande Woude GF, Ahn NG. Transformation of mammalian cells by constitutively active MAP kinase kinase. Science. 1994;265:966–970. doi: 10.1126/science.8052857. [DOI] [PubMed] [Google Scholar]
- 49.Ito A, Lai CH, Zhao X, Saito S, Hamilton MH, Appella E, Yao TP. p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 2001;20:1331–1340. doi: 10.1093/emboj/20.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kwok RP, Lundblad JR, Chrivia JC, Richards JP, Bachinger HP, Brennan RG, Roberts SG, Green MR, Goodman RH. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. pp. 16.66–16.67. [Google Scholar]
- 52.Scheibe RJ, Moeller-Runge I, Mueller WH. Retinoic acid induces the expression of alkaline phosphatase in P19 teratocarcinoma cells. J. Biol. Chem. 1991;266:21300–21305. [PubMed] [Google Scholar]
- 53.Yang SH, Sharrocks AD, Whitmarsh AJ. Transcriptional regulation by the MAP kinase signaling cascades. Gene. 2003;320:3–21. doi: 10.1016/s0378-1119(03)00816-3. [DOI] [PubMed] [Google Scholar]
- 54.Geers C, Gros G. Effects of carbonic anhydrase inhibitors on contraction, intracellular pH and energy-rich phosphates of rat skeletal muscle. J. Physiol. 1990;423:279–297. doi: 10.1113/jphysiol.1990.sp018022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Darby TG, Meissner JD, Ruhlmann A, Mueller WH, Scheibe RJ. Functional interference between retinoic acid or steroid hormone receptors and the oncoprotein Fli-1. Oncogene. 1997;15:3067–3082. doi: 10.1038/sj.onc.1201503. [DOI] [PubMed] [Google Scholar]
- 56.Dunn SE, Chin ER, Michel RN. Matching of calcineurin activity to upstream effectors is critical for skeletal muscle fiber growth. J. Cell Biol. 2000;151:663–672. doi: 10.1083/jcb.151.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swoap SJ, Hunter RB, Stevenson EJ, Felton HM, Kansagra NV, Lang JM, Esser KA, Kandarian SC. The calcineurin-NFAT pathway and muscle fiber-type gene expression. Am. J. Physiol. Cell Physiol. 2000;279:C915–C924. doi: 10.1152/ajpcell.2000.279.4.C915. [DOI] [PubMed] [Google Scholar]
- 58.Calabria E, Ciciliot S, Moretti I, Garcia M, Picard A, Dyar KA, Pallafacchina G, Tothova J, Schiaffino S, Murgia M. NFAT isoforms control activity-dependent muscle fiber type specification. Proc. Natl Acad. Sci. USA. 2009;106:13335–13340. doi: 10.1073/pnas.0812911106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kolch W. Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem. J. 2000;351(Pt 2):289–305. [PMC free article] [PubMed] [Google Scholar]
- 60.Asmussen G, Schmalbruch I, Soukup T, Pette D. Contractile properties, fiber types, and myosin isoforms in fast and slow muscles of hyperactive Japanese waltzing mice. Exp. Neurol. 2003;184:758–766. doi: 10.1016/S0014-4886(03)00294-2. [DOI] [PubMed] [Google Scholar]
- 61.Liu Y, Cseresnyes Z, Randall WR, Schneider MF. Activity-dependent nuclear translocation and intranuclear distribution of NFATc in adult skeletal muscle fibers. J. Cell Biol. 2001;155:27–39. doi: 10.1083/jcb.200103020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu. Rev. Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 63.Braun H, Koop R, Ertmer A, Nacht S, Suske G. Transcription factor Sp3 is regulated by acetylation. Nucleic Acids Res. 2001;29:4994–5000. doi: 10.1093/nar/29.24.4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang YN, Chen YJ, Chang WC. Activation of extracellular signal-regulated kinase signaling by epidermal growth factor mediates c-Jun activation and p300 recruitment in keratin 16 gene expression. Mol. Pharmacol. 2006;69:85–98. doi: 10.1124/mol.105.016220. [DOI] [PubMed] [Google Scholar]
- 65.Ma K, Chan JK, Zhu G, Wu Z. Myocyte enhancer factor 2 acetylation by p300 enhances its DNA binding activity, transcriptional activity, and myogenic differentiation. Mol. Cell. Biol. 2005;25:3575–3582. doi: 10.1128/MCB.25.9.3575-3582.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hasegawa K, Meyers MB, Kitsis RN. Transcriptional coactivator p300 stimulates cell type-specific gene expression in cardiac myocytes. J. Biol. Chem. 1997;272:20049–20054. doi: 10.1074/jbc.272.32.20049. [DOI] [PubMed] [Google Scholar]
- 67.Chan HM, La Thangue NB. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 2001;114:2363–2373. doi: 10.1242/jcs.114.13.2363. [DOI] [PubMed] [Google Scholar]
- 68.Yang T, Davis RJ, Chow CW. Requirement of two NFATc4 transactivation domains for CBP potentiation. J. Biol. Chem. 2001;276:39569–39576. doi: 10.1074/jbc.M102961200. [DOI] [PubMed] [Google Scholar]
- 69.Kawamura T, Ono K, Morimoto T, Akao M, Iwai-Kanai E, Wada H, Sowa N, Kita T, Hasegawa K. Endothelin-1-dependent nuclear factor of activated T lymphocyte signaling associates with transcriptional coactivator p300 in the activation of the B cell leukemia-2 promoter in cardiac myocytes. Circ. Res. 2004;94:1492–1499. doi: 10.1161/01.RES.0000129701.14494.52. [DOI] [PubMed] [Google Scholar]
- 70.Chawla S, Bading H. CREB/CBP and SRE-interacting transcriptional regulators are fast on-off switches: duration of calcium transients specifies the magnitude of transcriptional responses. J. Neurochem. 2001;79:849–858. doi: 10.1046/j.1471-4159.2001.00645.x. [DOI] [PubMed] [Google Scholar]
- 71.Gamble MJ, Freedman LP. A coactivator code for transcription. Trends Biochem. Sci. 2002;27:165–167. doi: 10.1016/s0968-0004(02)02076-5. [DOI] [PubMed] [Google Scholar]
- 72.Huang WC, Chen CC. Akt phosphorylation of p300 at Ser-1834 is essential for its histone acetyltransferase and transcriptional activity. Mol. Cell Biol. 2005;25:6592–6602. doi: 10.1128/MCB.25.15.6592-6602.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- 74.Ait-Si-Ali S, Carlisi D, Ramirez S, Upegui-Gonzalez LC, Duquet A, Robin P, Rudkin B, Harel-Bellan A, Trouche D. Phosphorylation by p44 MAP Kinase/ERK1 stimulates CBP histone acetyl transferase activity in vitro. Biochem. Biophys. Res. Commun. 1999;262:157–162. doi: 10.1006/bbrc.1999.1132. [DOI] [PubMed] [Google Scholar]
- 75.Perkins ND. Post-translational modifications regulating the activity and function of the nuclear factor kappa B pathway. Oncogene. 2006;25:6717–6730. doi: 10.1038/sj.onc.1209937. [DOI] [PubMed] [Google Scholar]
- 76.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 77.Brooks CL, Gu W. Ubiquitination, phosphorylation and acetylation: the molecular basis for p53 regulation. Curr. Opin. Cell Biol. 2003;15:164–171. doi: 10.1016/s0955-0674(03)00003-6. [DOI] [PubMed] [Google Scholar]
- 78.Wu H, Peisley A, Graef IA, Crabtree GR. NFAT signaling and the invention of vertebrates. Trends Cell Biol. 2007;17:251–260. doi: 10.1016/j.tcb.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 79.Higginson J, Wackerhage H, Woods N, Schjerling P, Ratkevicius A, Grunnet N, Quistorff B. Blockades of mitogen-activated protein kinase and calcineurin both change fibre-type markers in skeletal muscle culture. Pflugers Arch. 2002;445:437–443. doi: 10.1007/s00424-002-0939-1. [DOI] [PubMed] [Google Scholar]
- 80.Allen DL, Leinwand LA. Intracellular calcium and myosin isoform transitions. Calcineurin and calcium-calmodulin kinase pathways regulate preferential activation of the IIa myosin heavy chain promoter. J. Biol. Chem. 2002;277:45323–45330. doi: 10.1074/jbc.M208302200. [DOI] [PubMed] [Google Scholar]
- 81.Meissner JD, Chang KC, Kubis HP, Nebreda AR, Gros G, Scheibe RJ. The p38alpha/beta MAP kinases mediate recruitment of CBP to preserve fast myosin heavy chain IId/x gene activity in myotubes. J. Biol. Chem. 2007;282:7265–7275. doi: 10.1074/jbc.M609076200. [DOI] [PubMed] [Google Scholar]
- 82.Perdiguero E, Ruiz-Bonilla V, Gresh L, Hui L, Ballestar E, Sousa-Victor P, Baeza-Raja B, Jardi M, Bosch-Comas A, Esteller M, et al. Genetic analysis of p38 MAP kinases in myogenesis: fundamental role of p38alpha in abrogating myoblast proliferation. EMBO J. 2007;26:1245–1256. doi: 10.1038/sj.emboj.7601587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Spangenburg EE, Booth FW. Molecular regulation of individual skeletal muscle fibre types. Acta. Physiol. Scand. 2003;178:413–424. doi: 10.1046/j.1365-201X.2003.01158.x. [DOI] [PubMed] [Google Scholar]