Abstract
The EKC/KEOPS complex is universally conserved in Archaea and Eukarya and has been implicated in several cellular processes, including transcription, telomere homeostasis and genomic instability. However, the molecular function of the complex has remained elusive so far. We analyzed the transcriptome of EKC/KEOPS mutants and observed a specific profile that is highly enriched in targets of the Gcn4p transcriptional activator. GCN4 expression was found to be activated at the translational level in mutants via the defective recognition of the inhibitory upstream ORFs (uORFs) present in its leader. We show that EKC/KEOPS mutants are defective for the N6-threonylcarbamoyl adenosine modification at position 37 (t6A37) of tRNAs decoding ANN codons, which affects initiation at the inhibitory uORFs and provokes Gcn4 de-repression. Structural modeling reveals similarities between Kae1 and bacterial enzymes involved in carbamoylation reactions analogous to t6A37 formation, supporting a direct role for the EKC in tRNA modification. These findings are further supported by strong genetic interactions of EKC mutants with a translation initiation factor and with threonine biosynthesis genes. Overall, our data provide a novel twist to understanding the primary function of the EKC/KEOPS and its impact on several essential cellular functions like transcription and telomere homeostasis.
INTRODUCTION
Subunits of the Endopeptidase-like Kinase Chromatin (EKC)-associated complex [also called Kinase putative Endopeptidase and Other Proteins of Small size (KEOPS)] were first identified by genetic analyses (1,2). We isolated Pcc1p, a protein of small size with no predicted functional domains in a search for suppressors of a splicing defect and later demonstrated that its function is not related to splicing (2). Additional genetic, biochemical and cell biology experiments led to the definition of a five subunits complex (Pcc1p, Pcc2p/Gon7p, Cgi121p, Bud32p and Kae1p) the integrity of which is required for normal induction of pheromone- and galactose-responsive genes. Components of the EKC/KEOPS have been found to associate with transcribed chromatin and the biochemical defect in transcription activation of GAL genes was mapped to the recruitment of transcriptional co-activators (2). A parallel genome-wide screen led to the identification of the cgi121 null mutant as a suppressor of the telomere-capping defect of cdc13-1 cells. Deletion of CGI121 or other EKC/KEOPS subunits suppresses telomeres resection and single strand DNA production due to defective protection by the capping complex (1). All the tested EKC/KEOPS mutants have short telomeres in an otherwise wild-type context, an observation seemingly at odds with the telomere protective effect in the context of telomeres capping deficiencies. The molecular mechanism linking the involvement of the complex in transcription and telomere maintenance is unclear but it has been suggested to be related to effects on chromatin structure (1,2). Finally, DNA instability has also been associated with mutation of EKC/KEOPS components and the bacterial counterparts of Kae1p [our unpublished data; (3)]
Of the five subunits, the only ones containing predicted functional domains are Kae1p and Bud32p. Kae1p was originally proposed to be a candidate endopeptidase, based on sequence similarities with bacterial zinc-dependent metalloproteases of the M22 family (4). However, such a function could never be demonstrated and it does not appear to be compatible with the structure of Kae1p homologues from Archae and Eukaryotes (5–7). Structural studies (5,7) have revealed the existence of an actin-like ATPase domain and a metal (iron) binding site. It has been shown that archaeal PaKae1 has DNA binding capabilities and apurinic site endonuclease activities (7).
Bud32p is an atypical kinase that lacks some of the canonical domains required for substrate recognition (5,6). In vitro phosphorylation assays have demonstrated that this protein is a functional kinase that is also responsible for its autophosphorylation (8). It has been suggested that the association of Kae1p with Bud32p inhibits the phosphorylation activity of the latter (6).
One of the most striking features of EKC/KEOPS is its conservation and Kae1p belongs to the category of universal protein families (9). With the exception of Pcc2p/Gon7p that is restricted to Fungi, the whole complex is conserved in Archaea and Eukarya (2). With roughly 60% identity from Saccharomyces cerevisiae to man, Kae1p is the most conserved subunit and is also the most ancient member of the complex, with clear homologues (YgjD) present in Bacteria (3). Widespread conservation is however at odd with a primary function that would be limited to features restricted to eukaryotes, such as telomere maintenance or the establishment of specific chromatin structures. Therefore, in spite of the large number of phenotypes described for the mutants of the EKC/KEOPS complex, the molecular function of this set of proteins remains elusive. To address this important question we undertook genetic and transcriptome analyses of EKC/KEOPS mutants. We describe unexpected links to translation and tRNA metabolism as well as a specific transcriptome profile characterized by the inappropriate expression of amino acid biosynthesis genes. These phenotypes are causally linked by the translational misregulation of GCN4, encoding the transcriptional activator responsible for regulation of most amino acid biosynthesis genes. Phylogenetic and biochemical analyses strongly suggest a direct role of the EKC/KEOPS complex in N6-threonylcarbamoyl adenosine modification at position 37 (t6A37) of the anticodon stem-loop of all tRNAs decoding ANN codons. We show that the molecular mechanism underlying the translational defects relates to defective t6A37 formation, which affects the codon–anticodon interaction at the first AUG codon. The universality of this modification fits the evolutionary conservation of Kae1p, providing a novel twist to understanding the impact of this important set of proteins on several essential cellular functions.
MATERIAL AND METHODS
Manipulation of yeast strains and genetic screens
All the strains used in this study are listed in Supplementary Table S1. Genetic crossing and analysis were performed with standard methods. The synthetic lethal screen with the pcc1-4 mutant was based on a colorimetric assay to detect the ability of double mutant cells to grow in the absence of a wild-type copy of PCC1. Briefly, an ade8Δ, ade2-1 W303 strain containing an integrated pcc1-4 allele was transformed with a centromeric plasmid containing a wild-type copy of PCC1, ADE8 and the URA3 markers (pCM188-PCC1-ADE8). This strain is red because of the accumulation of a pigment in ade2-1/ADE8 cells. Random loss of the plasmid leads to the appearance of white sectors (the phenotype of ade2-1/ade8Δ cells). Cells were UV-mutagenized to 10% viability and mutants that failed to lose the plasmid (i.e. that were unable to live without a wt copy of PCC1) were retained for further analysis. The tif5-K55E mutation was isolated with standard genetic methods. Genomewide genetic interaction mapping (GIM) was performed as described (10).
Microarray experiments
Expression-profiling of mutant strains was performed using two-channel, long oligomer arrays as previously described (11). In brief, each strain was cultured twice and amplified, labeled cRNA was hybridized twice in dye-swap replicate against a single common reference wt cRNA, yielding a total of four estimates of changes in gene expression for each strain versus wt. Normalization was performed using print-tip Loess followed by dye-bias correction as described (11). Statistical analysis was performed by LIMMA (12) versus a collection of wt profiles hybridized versus the same common reference. A gene was regarded as significantly changed if the P-value was <0.01 and the fold-change >1.5. Microarray data have been submitted to the ArrayExpress database (http://www.ebi.ac.uk/microarray) under accession number E-TABM-1070 (username Reviewer_E-TABM-1070, password 1283942451014)
Polysome profile analysis
Polysomes extracts were prepared and analyzed by sedimentation in 10–50% sucrose gradients as previously described (13). Gradients were fractionated with an ISCO gradient fractionator and the absorbance profiles at 254 nm were monitored within an ISCO UA-5 absorbance monitor. Cellular extract for ribosomal subunit quantification were prepared and analyzed on low-Mg++ sucrose gradients as described (14).
β-Galactosidase assays with Gcn4-LacZ reporters
Wild-type or mutant strains containing the reporter plasmids p180, p227 or p4164 (kind gift of A. G. Hinnebusch) were grown at 30°C in CSM-Ura to OD600 0.3–0.6. Cells pellets were resuspended in 500 µl of buffer Z (100 mM HNaPO4, pH 7.2; 10 mM KCl; 1 mM MgSO4; 50 mM β-mercaptoethanol) and permeabilized by the addition of 200 µl of ether. After ether evaporation, 100 µl of ONPG (4 mg/ml in buffer Z) was added and the reaction incubated at 30°C until the development of a yellow color. The reaction was stopped by the addition of 250 µl of 1M Na2CO3 and the OD420 of the supernatants was measured. β-Galactosidase activity was expressed in arbitrary units (similar to Miller units) as follow: activity = 1000 × OD420 / [OD6OO of cultured sample × volume of cultured sample (ml) × reaction time (s)].
t6A analysis
Bulk tRNA was prepared, hydrolyzed, and analyzed by liquid chromatography-tandem mass spectrometry (LC–MS–MS) as described in (15). Levels of t6A were measured by integrating the peak area from the extraction ion chromatograms. The ratios of Ψ-modified base/m1G was used to normalize for tRNA concentration across samples. Levels for mutant strains were expressed relatively to wild-type levels. The MS–MS fragmentation data were also used to confirm the presence of t6A.
RESULTS
Genetic interactions of EKC/KEOPS mutants
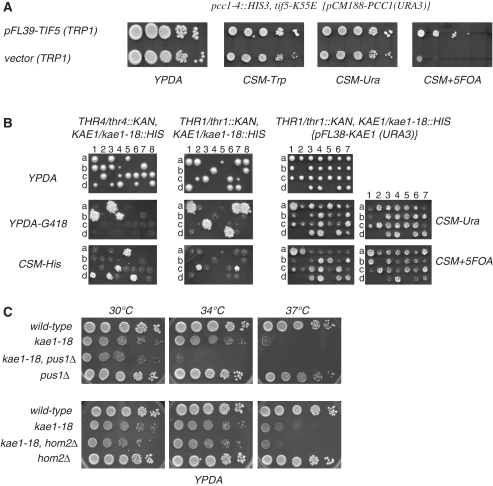
To elucidate the molecular mechanism of action of the complex we analyzed the genetic interaction profile of kae1 and pcc1 EKC/KEOPS mutants. A genetic screen for additional mutations preventing growth of thermosensitive pcc1-4 cells at permissive temperature resulted, as expected, in the isolation of alleles of two other subunits of the complex, Kae1p and Pcc2/Gon7. We also isolated a mutant allele of the translation initiation factor Tif5p/eIF5, an essential GTPase activating protein (GAP) (Figure 1A). This factor is required for stimulating GTP hydrolysis by eIF2 in the 43S pre-initiation complex (PIC) upon AUG codon–anticon base-pairing, thereby allowing the transition from initiation to translation elongation. By virtue of its physical interactions with other translation factors, eIF5 is also required for the formation of the PIC and the selection of the starting AUG codon [for recent reviews see (16,17)]. The sequence of the mutated allele revealed that a single A to G change is present in the coding sequence, leading to mutation of K55 to E. Accordingly, we named this allele tif5-K55E.
Figure 1.
Genetic interactions of EKC mutants. (A) Synthetic lethality of pcc1-4 and tif5-K55E cells. Double pcc1-4/tif5-K55E mutant is inviable when a URA3 marked plasmid expression the wt PCC1 gene is cured by selection on 5-FOA plates (rightmost panel). (B) Dissection of kae1-18/KAE1, thr4Δ/THR4 and kae1-18/KAE1, thr1Δ/THR4 diploids reveals the inviability of kae1-18/thr1Δ and kae1-18/thr4Δ spores. Both lethal phenotypes can be saved by expression of wild-type KAE1 gene (shown for the kae1-18/thr1Δ mutant in the rightmost set of panels). (C) Synthetic slow growth phenotype of double kae1-18/pus1Δ and kae1-18/hom2Δ strains. Growth was for 2 days at the indicated temperature.
To extend these results we performed a genomewide genetic interaction mapping (GIM) (10). For better sensitivity and specificity we used a hypomorphic PCC1 allele that contains a substitution of its terminator with a CYC1 terminator (unpublished data). To validate the results obtained and to filter out false positives, the deletion strains identified in the screen were individually crossed to the EKC/KEOPS mutant kae1-18 that is more severely affected than PCC1-CYC1ter cells. Strong negative genetic interaction leading to inviability at 30°C was observed for thr1 and thr4 deletion mutants (Figure 1B). Thr1p and Thr4p are the enzymes required for the conversion of homoserine to threonine in the two last steps of the biosynthesis of this amino acid. Genetic interactions (although not leading to lethality) were also confirmed with PUS1, required for pseudouridylation of tRNAs and U2 snRNA, and, to a lesser extent, HOM2, involved in an earlier step of threonine biosynthesis (Figure 1C) as well as TRM1 encoding the enzyme responsible for N2,N2-dimethylguanosine modification in both cytoplasmic and mitochondrial tRNAs (data not shown).
Together, these results suggested that the EKC/KEOPS might be involved in translation and/or tRNA metabolism. The stronger genetic interaction with genes involved in threonine biosynthesis also suggests a specific relationship between this pathway and the function of the EKC/KEOPS (see below).
A peculiar transcriptome profile in EKC complex mutants
The unexpected results of the genetic screens prompted us to perform transcriptome analysis of EKC/KEOPS mutants. We expected that a transcriptome profile similar to that of mutants altering known cellular functions would reveal important aspects of the role of the complex. Since most EKC null mutants are extremely sick (2), we integrated less affected thermosensitive mutants of Kae1p (kae1-18), Pcc1p (pcc1-4) and Pcc2/Gon7p (pcc2-ts1) in the BY4742 strain and analyzed the transcriptome of these cells using DNA microarrays at the semi-permissive temperature of 30°C (‘Methods’ section). Isogenic bud32 and cgi121 null mutants from the yeast deletion strain collection were also profiled. In several of the independent isolates analyzed we noticed the presence of whole chromosomal duplications, suggesting the occurrence of genomic instability in these mutants. Only euploid isolates were considered for further analyses. Since different isolates of bud32 null mutants, obtained from different sources, all contained chromosomal duplications, we did not further pursue analysis of this mutant.
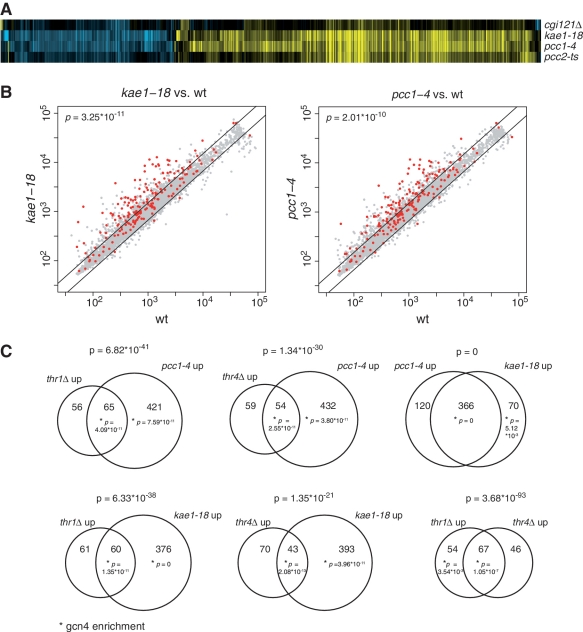
The EKC/KEOPS mutants show significant changes in gene expression, with a total of 579 genes upregulated and 282 genes downregulated collectively in all single mutants compared with wt (P < 0.01, fold-change >1.5, Figure 2 and Supplementary Figure S1). As is often observed for mutants derived from subunits of the same protein complex (18), the profiles of the EKC/KEOPS mutants are related to each other (Figure 2A). The kae1-18 and pcc1-4 mutants show the strongest expression signature, consisting of essentially the same set of mRNA expression changes, with the pcc2 and cgi121 mutants affecting a subset of the same genes, in most cases to a lower degree (Figure 2 and Supplementary Figure S1). Strikingly, the expression-profile signature of the EKC/KEOPS mutants is highly enriched for upregulated genes involved in amino acid biosynthesis (Gene Ontology biological process, cellular amino acid biosynthetic process, enrichment P = 4×10−34), as well as related metabolic pathways (Supplementary Table S2). Genes encoding enzymes involved in most cellular amino acid biosynthesis pathways are upregulated in the pcc1-4 mutant (Supplementary Figures S1 and S2). These results were independently confirmed by quantitative RT-PCR analysis using a subset of the upregulated genes. Moreover, we showed that upregulation was suppressed by expression of the corresponding wt gene in mutant cells, demonstrating that the expression profile is indeed a consequence of the EKC/KEOPS mutations (data not shown). Surprisingly, such a massive upregulation of amino acid biosynthesis genes was not observed in the collection of over 1500 mutants for which the transcriptome was analyzed during growth in identical conditions (F.C.P.H., unpublished results), indicating that the EKC/KEOPS mutant profiles are highly specific, at least among the mutants analyzed.
Figure 2.
Transcriptome analysis of EKC mutants. (A) Heatmap of all 861genes with significant changes in gene expression (fold change >1.5, P < 0.01) in either cgi121Δ, pcc1-4, kae1-18 and pcc2-ts. Each profile is the average of four measurements of mutant versus wt, with yellow indicating upregulation, blue downregulation and black no change. Genes not depicted do not change in any mutant. See Supplementary Figure S1 for a version annotated with gene names (B) Scatterplots of normalized intensities of gene expression of all genes in pcc1-4 and kae1-18 versus wild-type. Gcn4p targets, derived from genome-wide Gcn4p chromatin immunoprecipitation (20) are colored red. P-values indicate significance of the Gcn4p target enrichment by a hypergeometric test. (C) Venn diagrams of the upregulated genes of pcc1-4, kae1-18, thr1Δ and thr4Δ. The significance of the overlap and of Gcn4p target enrichment (marked by asterisk) is shown above and in the Venn diagram, respectively.
To investigate how the genetic interactions revealed by the genome-wide screen relate to the expression-signature of EKC/KEOPS mutants, the thr1, thr4, trm1 and pus1 null mutants were also expression-profiled. Interestingly, the gene expression changes observed in thr1 and thr4 mutants overlap highly significantly with pcc1 and kae1 mutants (P ranges from 10−21 to 7×10−41), and includes genes involved in amino acid biosynthesis (Figure 2 and Supplementary Figure S1). The expression-profile of trm1 and pus1 cells do not show a significant overlap with EKC/KEOPS or thr1/4 mutants (data not shown), suggesting the existence of different mechanisms underlying the observed genetic interactions.
Translation of the transcription activator Gcn4 is de-repressed in EKC mutants
Amino acid biosynthesis genes are generally activated in conditions of amino acids starvation. We observed upregulation of these genes in EKC mutants despite growth in the presence of normal levels of amino acids, suggesting that integrity of the EKC is required for their regulation. The transcriptional activator Gcn4p is involved in the activation of amino acid biosynthesis genes [for a review see (19)] and its binding site targets (20) are highly enriched in the EKC/KEOPS mutant signature genes (P = 3.25 × 10−11 and 2 × 10−10, respectively, for kae1 and pcc1 mutants, Figure 2B) as well as in the signature genes shared by EKC/KEOPS mutants and thr1/4 mutants (P ranging from 4 × 10−11 to 2 × 10−13) (Figure 2C). Overall, the most prominent feature of the transcriptome profile shared by EKC/KEOPS and thr1/4 mutants is the highly significant presence of genes regulated by the transcriptional activator Gcn4p (Figure 2C). Consistent with the notion that upregulated genes are Gcn4p targets, genes upregulated in EKC/KEOPS mutants are generally downregulated in a gcn4Δ mutant (data not shown). We first surmised that mutation of EKC/KEOPS might affect expression of Gcn4p at the level of mRNA abundance by impacting transcription and/or turnover of the GCN4 RNA. However, the levels of this RNA were unaffected in mutants relative to the wild-type, ruling out this simple hypothesis (data not shown).
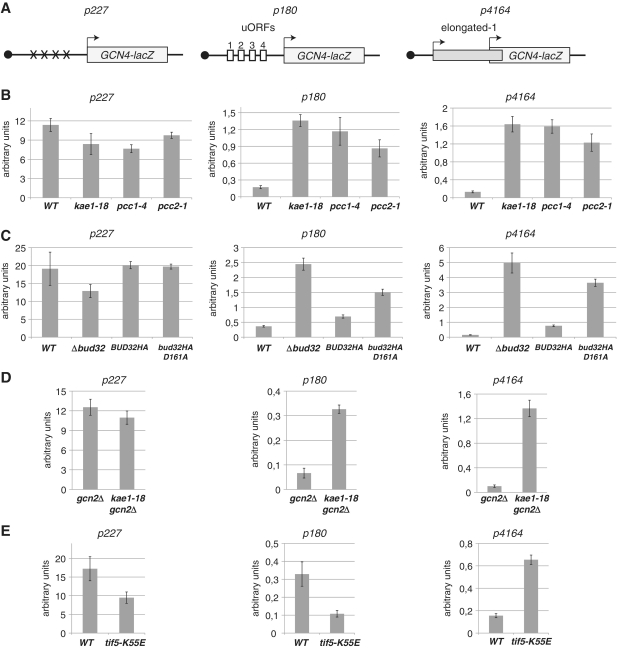
GCN4 is regulated at the translation level by the presence of four short ORFs (uORFs1–4, Figure 3A) upstream of the AUG of the Gcn4p coding sequence (19). In normal conditions, translation starts at the AUG of uORF1 but a large fraction of the 40S ribosomal particle can resume scanning after terminating translation of uORF1. In the presence of normal levels of amino acids and charged tRNAs, translation resumes preferentially at the downstream uORFs2-4 preventing normal expression of Gcn4p. Under amino acid shortage, the low level of charged tRNAs leads to phosphorylation of eIF2α by the Gcn2 kinase, which decreases the efficiency of ternary complex formation. This leads to ribosome scanning through uORF2-4 allowing translation initiation at the downstream Gcn4p AUG. Mutations that affect PIC formation, ribosome scanning or AUG recognition often lead to a General Control De-repressed (Gcd−) phenotype whereby Gcn4p expression is produced constitutively (19). We surmised that increased expression of amino acid genes might result from increased Gcn4p translation. To assess translational de-repression of GCN4, we employed LacZ fusion reporter genes described by the Hinnebush laboratory (21,22). In these well-characterized constructs (Figure 3A), the 5′ region of the GCN4 gene, containing the four uORFs is fused to the β-galactosidase coding frame so that only use of the GCN4 AUG leads to expression of the enzyme (Figure 3A, construct p180). To control for the global efficiency of translation (and mRNA production) in the different strains we used a similar construct that lacked all four uORFs (Figure 3A, p227). Expression of LacZ from the construct containing the unmodified GCN4 leader was strongly increased in the mutant strains relative to the wild-type (Figure 3B). This was not due to a global increase in the amount of RNA or the efficiency of translation in kae1-18 and pcc1-4 cells since β-galactosidase production from the construct lacking the uORFs (p227) was even modestly decreased relative to the wild-type (Figure 3B).
Figure 3.
Translational regulation of Gnc4 expression is altered in EKC and tif5 mutants. (A) Schematic of the GCN4-lacZ fusions reporters. The 5′ region of GCN4 RNA contains four upstream reading frames (uORFs 1-4, empty boxes) upstream of the main GCN4-lacZ ORF (light gray box, the arrow indicates its start codon). Construct P180 contains the unmodified region. P227 contains point mutations (X) in the four upstream AUG codons. p4164 contains an elongated version of uORF1 (thin gray box, the arrow indicates its start codon) that is not in frame with the GCN4 ORF. (B) EKC/KEOPS mutants kae1, pcc1 and pcc2 de-repress GCN4 translation and increase AUG leaky scanning. β-Galactosidase activity (arbitrary units) was measured from wild-type or mutant cells transformed with the GCN4-LacZ reporters indicated above each panel. Average of four independent experiments each one performed in triplicate. Errors bars represent standard deviations. (C) As in (B) GCN4-LacZ analysis of Δbud32 and the catalytic mutant bud32D161A. Since the latter is also HA tagged, analysis was performed in parallel with a tagged version of wild-type BUD32. (D) Gcn4 activation by EKC mutants is independent of the Gcn2-dependent amino acids shortage pathway. Double gcn2Δ/kae1-18 mutants transformed with the Gcn4-LacZ reporters were analyzed as in (B). Note that in the absence of Gcn2p, the overall levels of β-galactosidase obtained from the p180 reporter are decreased suggesting that the Gcn2 pathway is partially activated in these growth conditions. The relative levels of activation of the reporter are however similar in the presence and absence of Gcn2 (E). Leaky scanning in tif5-K55E mutant cells. β-Galactosidase activities were measured as in (B) from wild-type or tif1-5 cells containing the GCN4-LacZ reporters grown at the permissive (30°C) temperature. Average of four independent experiments each one performed in triplicate. Errors bars represent standard deviations.
Defective recognition of initiator AUG codons in EKC and tif5-K55E mutants
Activation of GCN4 translation in pcc1 and kae1 mutant cells could be due to defective recognition of the inhibitory uORFs 2–4 after post-termination re-initiation (see above), to activation of the Gcn2 kinase pathway, for instance as a consequence of some molecular event that creates or mimics amino acid shortage or both. To assess whether defective recognition of AUG codons occurs in EKC/KEOPS mutants, we employed a construct containing an out of frame AUG upstream of the LacZ ORF (Figure 3A, p4164). In this construct, β-galactosidase can only be produced by leaky scanning through the upstream AUG allowing the occurrence of translation initiation at the downstream start codon (note that leaky scanning at the second AUG would still allow a positive read out of the reporter) (22). Interestingly, a strong increase in β-galactosidase production was observed with this construct in pcc1 and kae1 mutants relative to the wild-type, indicating the occurrence of leaky AUG recognition during scanning. To extend these results we analyzed the implication of another EKC/KEOPS component, the Bud32 kinase, in translational repression of Gcn4p. Bud32p is closely related to the RIO family of protein kinases and the Bud32-D161A mutant has been shown to be catalytically inactive in vitro (8). De-repression of Gcn4p translation and leaky AUG scanning were observed both in bud32 cells and the catalytically inactive mutant (Figure 3C, constructs p180 and p4164), although the effect was less prominent in the latter.
These results indicate that de-repression of Gcn4 translation occurs as a result of defective recognition of the upstream inhibitory ORFs. However, we cannot formally rule out that the amino acids shortage response is also somehow activated in mutants. As outlined earlier, the latter depends critically on the phosphorylation of eIF2α by the Gcn2 kinase and gcn2Δ cells have been shown to be defective for Gcn4 activation under amino acids shortage (19). A direct activation of Gcn4 translation by defective AUG recognition in EKC/KEOPS mutants would imply shortcutting the Gcn2p pathway. Consistent with this notion, mutation of Kae1p and Pcc1p activated β-galactosidase expression from the construct containing the wild-type Gcn4 leader even in a gcn2Δ context (Figure 3D, construct p180 and Supplementary Figure S3). As expected, expression of LacZ from the construct containing the out of frame upstream AUG was not affected by the absence of Gcn2p (Figure 3D, construct p4164). Note that in a gcn2Δ background the overall levels of β-galactosidase expressed from the p180 construct are generally lower, possibly because the Gcn2 pathway is activated to low levels in the growth conditions employed. However the relative effect (fold activation) of EKC mutations is similar in gcn2Δ and GCN2 background.
Overall, these results support the notion that integrity of the EKC/KEOPS (and to some extent of the kinase activity of Bud32p) is required to maintain Gcn4 translational repression. They strongly suggest that de-repression of GCN4 in EKC/KEOPS mutants results from skipping non-productive translation of uORFs in the GCN4 leader via the defective recognition of the respective upstream AUGs.
Finally, we wanted to assess whether a translational phenotype underlay the synthetic lethality of tif5-K55E/pcc1-4 cells. Consistent with this possibility, several alleles of TIF5 have been isolated that affect translation initiation at different steps (16,23). Therefore, we transformed the isolated tif5-K55E strain with the Gcn4-LacZ constructs. In contrast to EKC mutants, a global effect on translation was observed in tif5-K55E cells (Figure 3E, p227), possibly reflecting a global decrease of translation initiation efficiency.
Importantly, β-galactosidase colorimetric assays revealed the occurrence of leaky scanning in tif5-K55E cells (Figure 3E, construct p4164), which is even more prominent considering the global effect observed with the construct lacking the out of frame AUGs. Interestingly, we did not observe activation of Gcn4p translation in the construct containing all four uORFs (Figure 3C, construct p180) in tif5-K55E. This result might either imply that tif5-K55E cells are also defective for initiation at uORF1 [which is required for Gcn4 regulation, (19)], or that TC recruitment to the post-termination scanning ribosome is not affected in this mutant (see ‘Discussion’ section).
Sucrose gradient analysis of polysomes in tif5-K55E cells (Supplementary Figure S4A, left panels) revealed a profile very similar to the ones previously observed for eIF5-depleted or mutant cells that are defective in translation initiation (24,25). The marked decrease in polyribosome content relative to the free ribosomal particles (that most likely constitute the bulk of the 80S peak) is characteristic of a defect in translation initiation. A similar profile was also observed in kae1Δ cells (Supplementary Figure S4). The translational phenotype of tif5 and kae1 is not due to a defect in the biogenesis of the 40S and 60S ribosomal particles as a parallel analysis of total dissociated ribosomes (Supplementary Figure S4A, right panels) revealed very similar levels of the two particles in the mutant and the wild-type strains (Supplementary Figure S4). Finally, no apparent differences were observed in the levels of ribosomal 25S and 18S mature ribosomal RNAs (Supplementary Figure S4B).
Overall, these results strongly suggest that the stringent requirement for eIF5 function in EKC/KEOPS mutant cells relates to the efficient selection of the initiator AUG. Overall, they strongly support the involvement of EKC/KEOPS in AUG start site selection and translation initiation.
Phylogenetic and biochemical analyses implicate EKC/KEOPS in t6A37 synthesis
Deregulation of Gcn4p translation was recently observed in sua5 mutants (26). Sua5p was originally isolated because its inactivation allowed expression of a cyc1 allele containing an out-of-frame upstream AUG codon (27). The molecular mechanism of this suppression was recently elucidated by showing that Sua5p is required for the N6-threonylcarbamoyl modification present at adenine 37 (t6A37) in the anticodon stem-loop of all tRNAs decoding ANN codons (15). Defective modification of the initiator tRNAMet in sua5 cells leads to a Gcd- phenotype, i.e. constitutive expression of the GCN4 gene (26). The Gcd− phenotype and the genetic interaction of EKC mutants with threonine biosynthesis (threonine is part of the t6A37 modification) and tRNA modification genes are suggestive of similarities with Sua5p.
While this work was in progress, a connection between Sua5p and Kae1p was revealed. Sua5 contains an N-terminal YrdC domain (residues 1–250) that is also present in a number of bacterial enzymes, such as NodU, HypF, NovN and CmcH. Remarkably these enzymes are all involved in the carbamoylation of different substrates (28)
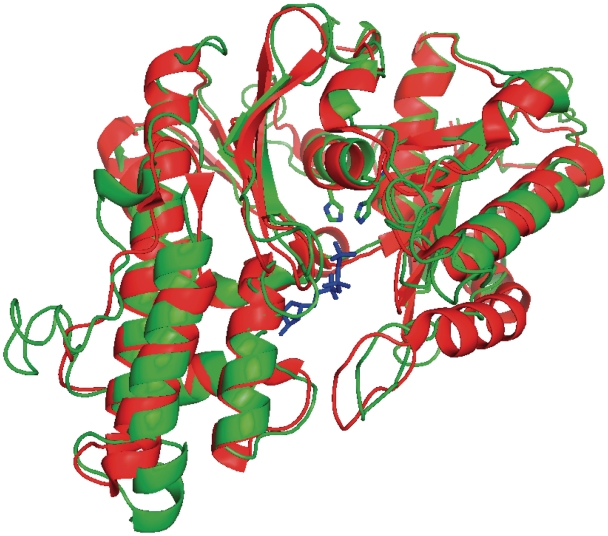
Three-dimensional (3D) atomic models from multiple threading alignments and iterative structural assembly simulations indicates that NodU, NovN, HypF and CmcH all have a similar domain architecture that also contains an additional homologous module in addition to the YrdC module. In spite of low sequence similarity (16% identity on average), this module can be convincingly predicted to have the same fold as Pyrococcus abyssii Kae1 (PaKae1). Using the I-tasser meta server, we constructed a 3D model for this module and analyzed conserved amino acid positions between the carbamoyl transferases and Kae1. The structure of PaKae1 was obtained in the presence of the non-hydrolysable AMPPNP nucleotide, which is bound in a tunnel between two domains (7). The gamma phosphate group of AMPPNP interacts with the side chains of His107, Tyr127, Ser129 and Asp285. The phosphate oxygens of the nucleotide are further liganded to a Fe ion that is bound to Kae1 through the side chains of His107, Tyr127, His111 and Asp285 in a configuration that is very similar to that of the Fe ions found in the active site of acid phosphatases. All these residues are absolutely conserved in eukaryotic and archaeal Kae1 sequences. The 3D model of the Kae1 modules of NodU (Figure 4) shows that both histidines are conserved in the structure and that the groove could easily accommodate a nucleotide as is the case for Kae1. These data indicate that the carbamoylating enzymes contain distinct modules that have active site configurations respectively similar to Kae1p and Sua5p, suggesting that they catalyze similar chemical reactions.
Figure 4.
Superposition of the crystal structure of Pa-Kae1 (red) and a model for the Kae1 module of NodU (green) as generated by the structure prediction metaserver I-tasser. The AMPPNP nucleotide observed in the crystal structure of Pa-Kae1 is shown in sticks (blue). The two conserved histidines of the putative active site of NodU are also represented.
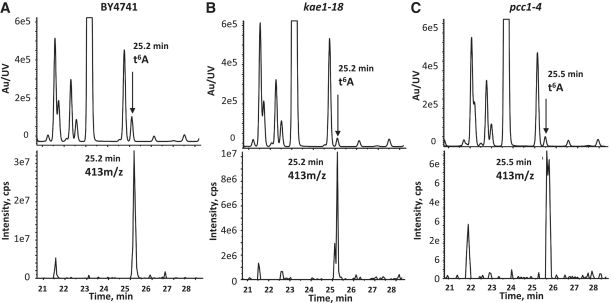
Inspired by these considerations, we performed LC–MS analysis of tRNAs extracted from wt, kae1-18 and pcc1-4 strains. To minimize indirect effects the experiment was performed at the permissive temperature (30°C) for the thermosensitive alleles. The characteristic t6A37 25.5 min peak was strongly reduced in kae1-18 and pcc1-4 cells to, respectively, 25 and 30% of wild-type levels, which is highly significant considering that the analysis was performed with an allele that is only partially defective at this temperature. The absence of t6A37 in EKC mutants was independently reported by two other groups while this work was under revision (28,29).
These experiments together with the data reported by El Yacoubi et al. and Srinivasan et al. (28,29) indicate that EKC mutants are defective for t6A37 modification of ANN decoding tRNAs. Since this modification affects the efficiency of codon–anticodon recognition and tRNAMet is also t6A37 modified, these findings strongly suggest that translational de-repression of Gcn4p in EKC mutants is mechanistically linked to defective recognition of uORF AUG start codons by unmodified tRNAMet. Overall, these data set the basis for understanding the molecular events associated with alteration of EKC/KEOPS integrity.
DISCUSSION
In spite of several phenotypes associated to mutants of the EKC/KEOPS, the primary function of this complex that is conserved in Eukarya and Archaea has remained elusive so far. The conservation of one subunit of the complex, Kae1p, in Bacteria adds a universal twist to this important question. Several theoretical possibilities exist to relate universal conservation and function. First, it is possible, that an ancient function, which was conserved in Bacteria, has been diverted in Eukarya and Archaea to serve evolutionary novel and distinct purposes and that this requires the other EKC/KEOPS components. In this view, the ancestral function of Kae1-like proteins might be lost in Eukarya and Archaea. Another possibility is that the ancestral function is universally present, but requires the additional complexity of the whole EKC/KEOPS in Eukarya and Archaea. A corollary of this possibility is that the phenotypes observed in EKC/KEOPS mutants are indirect effects of the perturbation of this ancestral function. Finally, both the ancestral and the archaeal/eukaryotic specific functions might co-exist, and depend on the same or different biochemical forms of the complex. The results presented here allow ruling out the first possibility by providing evidence for a biochemical function of the complex that is universally conserved and impacts the efficiency and specificity of translation.
We have shown that EKC/KEOPS mutants are defective in the translational regulation of Gcn4p, which mechanistically relates to defective t6A37 modification of ANN-decoding tRNAs. These findings are strengthened by the genetic interactions of pcc1 and kae1 mutants with genes involved in the synthesis of threonine, an amino acid that is part of the t6A37 modification (30,31), and by the upregulation of Gcn4p targets in thr1 and thr4 mutants. One additional strong connection to translation is provided by the synthetic lethality of EKC mutants with a mutant in the translation factor eIF5, a protein that is at the heart of the translation initiation process and the selection of the initiator AUG, which is also impacted by the t6A37 modification. Finally, structural modeling underscores the similarities between Kae1p and bacterial enzymes catalyzing similar chemical reactions as t6A37 synthesis (Figure 4). While this work was in progress El Yacoubi et al. and Srinivasan et al. revealed a phylogenetic connection between Sua5p and Kae1p and showed that kae1Δ cells are defective for t6A37 modification (28,29). A defect in t6A37 synthesis was also reported in pcc1Δ, bud32Δ and the catalytic mutant allele of Bud32, bud32-D161A (29), which is fully consistent with our genetic assays on Gcn4 activation and our LC–MS data.
Deregulation of Gcn4p expression in EKC/KEOPS mutants is linked to defective t6A synthesis
In the quest for a molecular function of the EKC/KEOPS that could relate the different phenotypes associated to its alterations, we first performed genetic screens and transcriptome analyses. The specific transcriptome profile of EKC/KEOPS mutants was particularly striking as a large majority of genes involved in the synthesis of amino acids were found to be upregulated (Supplementary Figures S1 and S2). We show that this is due to increased expression of the Gcn4p transcription factor that controls a large majority of the upregulated genes. Regulation of the GCN4 locus occurs at the translational level (19). Translation starts in the GCN4 leader at an upstream ORF (uORF1) but most ribosomes resume scanning after termination. Regulation occurs by altering the efficiency of recognition of three additional inhibitory upstream ORFs (uORFs2-4): whenever these are skipped, translation can occur at the downstream in frame Gcn4 AUG (Figure 3). Under amino acids shortage, phosphorylation of eIF2α by the Gcn2p kinase decreases the levels of the ternary complex (containing the charged tRNAiMet and eIF2-GTP), which impairs recognition of the inhibitory uORFs and allows expression of Gcn4p. Mutations altering formation of the PIC, the scanning process or the recognition of the AUG by tRNAMet, allows reading through ORFs2-4 AUGs and expression of Gcn4p, i.e. a Gcd− phenotype [for a review see (19)].
The Gcd- phenotype we observed for EKC/KEOPS mutants could in principle be due to the activation of the Gcn2p pathway by some events causing or mimicking amino acid shortage. For instance, it has been shown that t6A37 is an important determinant of the recognition of tRNAIle by the cognate isoleucyl-tRNA synthetase (32) and the presence of uncharged tRNAs is known to activate the Gcn2 pathway. We show, however, that Gcn4p translation is activated in EKC mutants even in the absence of Gcn2p (Figure 3D), which does not support this hypothesis. The Gcd- phenotype of EKC mutants is unlikely due to a defect in ribosomal biogenesis as previously described for an rpl16BΔ mutant (33) as the levels of 40S and the 60S particles and their ratio was very similar to the wild-type even in the severely affected kae1Δ mutant (Supplementary Figure S4 and data not shown)
The most likely mechanisms underlying Gcn4p activation in EKC mutants is the occurrence of scanning through the inhibitory uORFs2-4 in the GCN4 leader after translation of uORF1 (which is required for regulation). We have shown that leaky scanning indeed occurs in EKC mutants by using a reporter gene that allows expression of β-galactosidase only upon skipping of an upstream AUG (Figure 3B, construct p4164).
The most likely mechanism underlying leaky scanning in EKC mutants is the faulty recognition of the starting AUG codon, as a consequence of the strong reduction in t6A37 modified initiator tRNAMet that we observed in kae1-18 cells (Figure 5). Indeed, the presence of t6A37 has been shown to confer a particular structure to the anticodon loop that stabilizes the codon–anticodon interaction and increases the binding to the ribosome (34–37). Similar findings have been reported for the Sua5p protein that has also been shown to induce constitutive activation of the same Gcn4-LacZ reporters we used in this study and to be defective for the t6A37 modification (15,26). Gcn2-independent activation of Gcn4p translation was previously shown for mutants or conditions leading to altered tRNA metabolism (38,39).
Figure 5.
kae1-18 and pcc1-4 cells are defective for t6A37 formation. LC–MS–MS analysis of tRNAs extracted from wild-type (A) and mutant kae1-18 (B) and pcc1-4 (C) strains as indicated. The UV traces are shown in the top panels and the extraction ion chromatograms in the bottom panels for each strain. t6A37 levels in kae1-18 and pcc1-4 mutants were, respectively, 25% and 30% of wild-type after normalization to the ratios of Ψ/m1G to account for differences in tRNA concentrations. Ψ/m1G ratios were not significantly different (0.21, 0.24 and 0.23 for BY4742, kae1-18 and pcc1-4, respectively). Extraction and analysis were performed twice independently.
The genetic interactions of EKC mutants provide strong support for the proposed function of the complex in t6A synthesis
The finding that integrity of the threonine synthesis pathway is essential in a context of defective EKC/KEOPS substantiates the implication of EKC/KEOPS in t6A37 synthesis. Threonine is part of the t6A37 and has been shown to be required, together with carbonate and ATP, for in vitro synthesis of this modification (40,41). Deletion of THR1 or THR4, involved in the two last steps of threonine synthesis, leads to lethality in a kae1-18 mutant. This finding is particularly striking considering that all genetic analyses have been performed in rich, threonine-containing medium, which allows normal growth of thr1 or thr4 threonine auxotrophs. This suggests that impairment of endogenous synthesis in thr mutants leads to a decrease in threonine availability to levels that do not significantly affect normal cell metabolism and protein synthesis still strongly enhance the effect of partial alterations in t6A synthesis due to EKC/KEOPS mutations. The stronger effect of thr1 and thr4 deletion on kae1-18 mutants compared to upstream mutations in the threonine pathway (e.g. hom2) might suggest the alternative, provocative, hypothesis that Thr1p and Thr4p directly take part in t6A synthesis, which might for instance imply the existence of a homoserine intermediate that would be converted to threonine by the sequential action of the two enzymes. This homoserine pathway might be a preferred but not an obligatory pathway, which would be consistent with early in vivo and in vitro studies reporting the incorporation of threonine in t6A (30,31,40,41).
An additional piece of evidence that the EKC/KEOPS impacts translation via the t6A modification, is the strong genetic interaction with the translation initiation GTPase-activating protein (GAP) Tif5p/eIF5. This protein is involved in several steps of translation initiation (16,17). After the stable interaction of initiator tRNAMet with the starting AUG eIF5 enhances eIF2 GTPase activity, which prompts structural rearrangements in the scanning ribosome and allows the transition from initiation to elongation. By virtue of the multiple interactions of its carboxy-terminal domain (CTD) with other translation initiation factors, eIF5 also functions as an assembly platform that favors the interaction of the TC with the scanning ribosome and the formation of the translation PIC (16). Consistent with this function, several recessive tif5 alleles have been isolated in the CTD inducing a Gcd−phenotype that can be mechanistically associated with defective recruitment of the TC to the re-initiating ribosome on the GCN4 RNA (23). We show that the recessive tif5 allele that we isolated based on its synthetic lethality with pcc1-4 (tif5-K55E) has a polysome profile similar to the one of previously characterized tif5 alleles and consistent with a defect in translation initiation (24,25). Consistent with this notion, tif5-K55E have a strong leaky scanning phenotype, which parallels the one observed with pcc1 and kae1 cells. In contrast to EKC mutants, however, this particular allele does not induce a Gcd- phenotype (i.e. it does not constitutively activate translation of the wild-type Gcn4-LacZ reporter, Figure 3E, p180). This might be due to the fact that efficient Gcn4 activation requires translation of uORF1 and that tif5-K55E cells might severely affect recognition of the uORF1 start codon. Alternatively, this mutation of eIF5 might not affect TC recruitment to the post-termination 40S particle, which would allow translation of the inhibitory uORFs2-4. This would be consistent with the finding that the mutation isolated is not in the CTD but in the N-terminal GAP domain. Dominant mutations in this region have been isolated that have a relaxed AUG recognition and enhanced GAP activity (SUI phenotype) (42). Alleles with reduced GAP activity have also been described that have a reduced rate of translation initiation (43), but, generally, mutations in the N-terminal domain do not lead to a leaky scanning phenotype (23). Therefore, the K55E allele behaves differently from described mutations in the GAP domain, possibly implicating GTP hydrolysis in scanning and AUG selection. We suggest that, while EKC mutants affect the efficiency of AUG recognition via the unmodified initiator tRNAMet, the tif5-K55E allele slows down GTP hydrolysis after AUG codon–anticon interaction, delaying (or preventing) the structural rearrangements required for the interruption of scanning and the transition to elongation. The future biochemical characterization of this allele might reveal novel interesting features of the mechanism of translation initiation and eIF5 function.
Relationship to other EKC phenotypes
Several phenotypes have been described for mutants of the EKC/KEOPS complex, ranging from defective transcription (2) to telomere homeostasis (1) and genomic instability (our unpublished data; 3). It is obviously possible that these phenotypes result from indirect effects due to defective t6A modification in several tRNAs that affect the translation of transcription or telomere maintenance factors. One obvious candidate would be the Est3p component of telomerase that requires a frameshift for its expression (44). However, defective codon recognition due to lack of t6A modification rather favors Est3p production by enhancing frameshifting [as was shown for sua5Δ cells, (26)] and it has been shown that a frameshift independent EST3-fsc allele has normal growth and telomere lengths (44). Another possibility is that the EKC/KEOPS has additional functions that might or might not be related to carbamoylation. This might correlate to the increased complexity of the archaeal/eukaryotic enzymes relative to the prokaryotic counterparts and to the physical separation of the Sua5 and Kae1 modules. It is important to point out, however, that both sua5 and EKC mutants have telomeric defects (1,45), suggesting that, if a eukaryotic specific effect in telomere homeostasis exists, this might involve a similar chemistry as t6A37 formation. One interesting possibility is the existence of an additional target for a carbamoylation reaction analogous to t6A37 formation, which might be a protein factor or an RNA with similar or different characteristics from the tRNA anticodon stem loop. In the context of the whole EKC the carbamoylation reaction might acquire additional substrate specificity and even be regulated based on cellular physiology or cell cycle stage. In the absence of reported physical interactions between Sua5p and EKC components, the molecular mechanism ensuring the coordination of their activities remains, however, elusive.
The long sought elucidation of at least one of the EKC/KEOPS primary functions is an important step for understanding the impact of these conserved factors on several cellular processes. Future challenges include the identification of additional targets or the discovery of the mechanistic pathways linking tRNA modification, transcription and telomere homeostasis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Centre National pour la Recherche Scientifique (CNRS), the Agence Nationale pour la Recherche (ANR) (grant ANR-09-BLAN-0349-03 to D.L. and H.vT. and ANR-08-JCJC-0019-01 to C.S.) Netherlands Organization of Scientific Research (NWO) (grants 021.002.035, 817.02.015, 050.71.057, 911.06.009, 016.108.607 to T.L.L.) Netherlands Bioinformatics Centre (NBIC), the U.S. Department of Energy (grant no. DEFG0207ER64498 to V.deC.-L.) and by the National Institutes of Health (grant no. R01 GM70641-01 to V.deC.-L.). Fondation pour la Recherche Médicale (FRM) to X.L. Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank A. Hinnebusch for the kind gift of Gcn4-LacZ constructs, J. Boulay M. G. Koerkamp, D. van Leenen, C. Ko, P. Kemmeren, L. Bakker and S. van Hooff for expert technical assistance, F. Lacroute for help with yeast genetics and P. Forterre, C. Mann and H. Grosjean for fruitful discussions.
REFERENCES
- 1.Downey M, Houlsworth R, Maringele L, Rollie A, Brehme M, Galicia S, Guillard S, Partington M, Zubko MK, Krogan NJ, et al. A genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell. 2006;124:1155–1168. doi: 10.1016/j.cell.2005.12.044. [DOI] [PubMed] [Google Scholar]
- 2.Kisseleva-Romanova E, Lopreiato R, Baudin-Baillieu A, Rousselle JC, Ilan L, Hofmann K, Namane A, Mann C, Libri D. Yeast homolog of a cancer-testis antigen defines a new transcription complex. EMBO J. 2006;25:3576–3585. doi: 10.1038/sj.emboj.7601235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oberto J, Breuil N, Hecker A, Farina F, Brochier-Armanet C, Culetto E, Forterre P. Qri7/OSGEPL, the mitochondrial version of the universal Kae1/YgjD protein, is essential for mitochondrial genome maintenance. Nucleic Acids Res. 2009;37:5343–5352. doi: 10.1093/nar/gkp557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mellors A, Lo RY. O-sialoglycoprotease from Pasteurella haemolytica. Methods Enzymol. 1995;248:728–740. doi: 10.1016/0076-6879(95)48049-8. [DOI] [PubMed] [Google Scholar]
- 5.Mao DY, Neculai D, Downey M, Orlicky S, Haffani YZ, Ceccarelli DF, Ho JS, Szilard RK, Zhang W, Ho CS, et al. Atomic structure of the KEOPS complex: an ancient protein kinase-containing molecular machine. Mol. Cell. 2008;32:259–275. doi: 10.1016/j.molcel.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hecker A, Lopreiato R, Graille M, Collinet B, Forterre P, Libri D, van Tilbeurgh H. Structure of the archaeal Kae1/Bud32 fusion protein MJ1130: a model for the eukaryotic EKC/KEOPS subcomplex. EMBO J. 2008;27:2340–2351. doi: 10.1038/emboj.2008.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hecker A, Leulliot N, Gadelle D, Graille M, Justome A, Dorlet P, Brochier C, Quevillon-Cheruel S, Le Cam E, van Tilbeurgh H, et al. An archaeal orthologue of the universal protein Kae1 is an iron metalloprotein which exhibits atypical DNA-binding properties and apurinic-endonuclease activity in vitro. Nucleic Acids Res. 2007;35:6042–6051. doi: 10.1093/nar/gkm554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Facchin S, Lopreiato R, Stocchetto S, Arrigoni G, Cesaro L, Marin O, Carignani G, Pinna LA. Structure-function analysis of yeast piD261/Bud32, an atypical protein kinase essential for normal cell life. Biochem. J. 2002;364:457–463. doi: 10.1042/BJ20011376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galperin MY, Koonin EV. 'Conserved hypothetical' proteins: prioritization of targets for experimental study. Nucleic Acids Res. 2004;32:5452–5463. doi: 10.1093/nar/gkh885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Decourty L, Saveanu C, Zemam K, Hantraye F, Frachon E, Rousselle JC, Fromont-Racine M, Jacquier A. Linking functionally related genes by sensitive and quantitative characterization of genetic interaction profiles. Proc. Natl Acad. Sci. USA. 2008;105:5821–5826. doi: 10.1073/pnas.0710533105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Margaritis T, Lijnzaad P, van Leenen D, Bouwmeester D, Kemmeren P, van Hooff SR, Holstege FC. Adaptable gene-specific dye bias correction for two-channel DNA microarrays. Mol. Syst. Biol. 2009;5:266. doi: 10.1038/msb.2009.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 13.Daugeron MC, Prouteau M, Lacroute F, Seraphin B. The highly conserved eukaryotic DRG factors are required for efficient translation in a manner redundant with the putative RNA helicase Slh1. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq898. doi:10.1093/nar/gkq898 [Epub ahead of print, 13 November 2010] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kressler D, de la Cruz J, Rojo M, Linder P. Fal1p is an essential DEAD-box protein involved in 40S-ribosomal-subunit biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1997;17:7283–7294. doi: 10.1128/mcb.17.12.7283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El Yacoubi B, Lyons B, Cruz Y, Reddy R, Nordin B, Agnelli F, Williamson JR, Schimmel P, Swairjo MA, de Crecy-Lagard V. The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res. 2009;37:2894–2909. doi: 10.1093/nar/gkp152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lorsch JR, Dever TE. Molecular view of 43 S complex formation and start site selection in eukaryotic translation initiation. J. Biol. Chem. 2010;285:21203–21207. doi: 10.1074/jbc.R110.119743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benschop JJ, Brabers N, van Leenen D, Bakker LV, van Deutekom HW, van Berkum NL, Apweiler E, Lijnzaad P, Holstege FC, Kemmeren P. A consensus of core protein complex compositions for Saccharomyces cerevisiae. Mol. Cell. 2010;38:916–928. doi: 10.1016/j.molcel.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 19.Hinnebusch AG. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 20.MacIsaac KD, Wang T, Gordon DB, Gifford DK, Stormo GD, Fraenkel E. An improved map of conserved regulatory sites for Saccharomyces cerevisiae. BMC Bioinformatics. 2006;7:113. doi: 10.1186/1471-2105-7-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mueller PP, Harashima S, Hinnebusch AG. A segment of GCN4 mRNA containing the upstream AUG codons confers translational control upon a heterologous yeast transcript. Proc. Natl Acad. Sci. USA. 1987;84:2863–2867. doi: 10.1073/pnas.84.9.2863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong J, Nanda JS, Rahman H, Pruitt MR, Shin BS, Wong CM, Lorsch JR, Hinnebusch AG. Genetic identification of yeast 18S rRNA residues required for efficient recruitment of initiator tRNA(Met) and AUG selection. Genes Dev. 2008;22:2242–2255. doi: 10.1101/gad.1696608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh CR, Curtis C, Yamamoto Y, Hall NS, Kruse DS, He H, Hannig EM, Asano K. Eukaryotic translation initiation factor 5 is critical for integrity of the scanning preinitiation complex and accurate control of GCN4 translation. Mol. Cell. Biol. 2005;25:5480–5491. doi: 10.1128/MCB.25.13.5480-5491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maiti T, Das S, Maitra U. Isolation and functional characterization of a temperature-sensitive mutant of the yeast Saccharomyces cerevisiae in translation initiation factor eIF5: an eIF5-dependent cell-free translation system. Gene. 2000;244:109–118. doi: 10.1016/s0378-1119(99)00570-3. [DOI] [PubMed] [Google Scholar]
- 25.Maiti T, Maitra U. Characterization of translation initiation factor 5 (eIF5) from Saccharomyces cerevisiae. Functional homology with mammalian eIF5 and the effect of depletion of eIF5 on protein synthesis in vivo and in vitro. J. Biol. Chem. 1997;272:18333–18340. doi: 10.1074/jbc.272.29.18333. [DOI] [PubMed] [Google Scholar]
- 26.Lin CA, Ellis SR, True HL. The Sua5 protein is essential for normal translational regulation in yeast. Mol. Cell. Biol. 2010;30:354–363. doi: 10.1128/MCB.00754-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Na JG, Pinto I, Hampsey M. Isolation and characterization of SUA5, a novel gene required for normal growth in Saccharomyces cerevisiae. Genetics. 1992;131:791–801. doi: 10.1093/genetics/131.4.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El Yacoubi B, Hatin I, Deutsch C, Kahveci T, Rousset JP, Iwata-Reuyl D, Murzin AG, de Crecy-Lagard V. A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification. EMBO J. 2011;30:882–893. doi: 10.1038/emboj.2010.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Srinivasan M, Mehta P, Yu Y, Prugar E, Koonin EV, Karzai AW, Sternglanz R. The highly conserved KEOPS/EKC complex is essential for a universal tRNA modification, t6A. EMBO J. 2011;30:873–881. doi: 10.1038/emboj.2010.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chheda GB, Hong CI, Piskorz CF, Harmon GA. Biosynthesis of N-(purin-6-ylcarbamoyl)-L-threonine riboside. Incorporation of L-threonine in vivo into modified nucleoside of transfer ribonucleic acid. Biochem J. 1972;127:515–519. doi: 10.1042/bj1270515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Powers DM, Peterkofsky A. Biosynthesis and specific labeling of N-(purin-6-ylcarbamoyl)threonine of Escherichia coli transfer RNA. Biochem. Biophys. Res. Commun. 1972;46:831–838. doi: 10.1016/s0006-291x(72)80216-x. [DOI] [PubMed] [Google Scholar]
- 32.Nureki O, Niimi T, Muramatsu T, Kanno H, Kohno T, Florentz C, Giege R, Yokoyama S. Molecular recognition of the identity-determinant set of isoleucine transfer RNA from Escherichia coli. J. Mol. Biol. 1994;236:710–724. doi: 10.1006/jmbi.1994.1184. [DOI] [PubMed] [Google Scholar]
- 33.Foiani M, Cigan AM, Paddon CJ, Harashima S, Hinnebusch AG. GCD2, a translational repressor of the GCN4 gene, has a general function in the initiation of protein synthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1991;11:3203–3216. doi: 10.1128/mcb.11.6.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stuart JW, Gdaniec Z, Guenther R, Marszalek M, Sochacka E, Malkiewicz A, Agris PF. Functional anticodon architecture of human tRNALys3 includes disruption of intraloop hydrogen bonding by the naturally occurring amino acid modification, t6A. Biochemistry. 2000;39:13396–13404. doi: 10.1021/bi0013039. [DOI] [PubMed] [Google Scholar]
- 35.Stuart JW, Koshlap KM, Guenther R, Agris PF. Naturally-occurring modification restricts the anticodon domain conformational space of tRNA(Phe) J. Mol. Biol. 2003;334:901–918. doi: 10.1016/j.jmb.2003.09.058. [DOI] [PubMed] [Google Scholar]
- 36.Weissenbach J, Grosjean H. Effect of threonylcarbamoyl modification (t6A) in yeast tRNA Arg III on codon-anticodon and anticodon-anticodon interactions. A thermodynamic and kinetic evaluation. Eur. J. Biochem. 1981;116:207–213. doi: 10.1111/j.1432-1033.1981.tb05320.x. [DOI] [PubMed] [Google Scholar]
- 37.Yarian C, Marszalek M, Sochacka E, Malkiewicz A, Guenther R, Miskiewicz A, Agris PF. Modified nucleoside dependent Watson-Crick and wobble codon binding by tRNALysUUU species. Biochemistry. 2000;39:13390–13395. doi: 10.1021/bi001302g. [DOI] [PubMed] [Google Scholar]
- 38.Qiu H, Hu C, Anderson J, Bjork GR, Sarkar S, Hopper AK, Hinnebusch AG. Defects in tRNA processing and nuclear export induce GCN4 translation independently of phosphorylation of the alpha subunit of eukaryotic translation initiation factor 2. Mol. Cell. Biol. 2000;20:2505–2516. doi: 10.1128/mcb.20.7.2505-2516.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vazquez de Aldana CR, Wek RC, Segundo PS, Truesdell AG, Hinnebusch AG. Multicopy tRNA genes functionally suppress mutations in yeast eIF-2 alpha kinase GCN2: evidence for separate pathways coupling GCN4 expression to unchanged tRNA. Mol. Cell. Biol. 1994;14:7920–7932. doi: 10.1128/mcb.14.12.7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elkins BN, Keller EB. The enzymatic synthesis of N-(purin-6-ylcarbamoyl)threonine, an anticodon-adjacent base in transfer ribonucleic acid. Biochemistry. 1974;13:4622–4628. doi: 10.1021/bi00719a024. [DOI] [PubMed] [Google Scholar]
- 41.Korner A, Soll D. N-(purin-6-ylcarbamoyl)threonine: biosynthesis in vitro in transfer RNA by an enzyme purified from Escherichia coli. FEBS Lett. 1974;39:301–306. doi: 10.1016/0014-5793(74)80135-3. [DOI] [PubMed] [Google Scholar]
- 42.Huang HK, Yoon H, Hannig EM, Donahue TF. GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Dev. 1997;11:2396–2413. doi: 10.1101/gad.11.18.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Asano K, Shalev A, Phan L, Nielsen K, Clayton J, Valasek L, Donahue TF, Hinnebusch AG. Multiple roles for the C-terminal domain of eIF5 in translation initiation complex assembly and GTPase activation. EMBO J. 2001;20:2326–2337. doi: 10.1093/emboj/20.9.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morris DK, Lundblad V. Programmed translational frameshifting in a gene required for yeast telomere replication. Curr. Biol. 1997;7:969–976. doi: 10.1016/s0960-9822(06)00416-7. [DOI] [PubMed] [Google Scholar]
- 45.Meng FL, Hu Y, Shen N, Tong XJ, Wang J, Ding J, Zhou JQ. Sua5p a single-stranded telomeric DNA-binding protein facilitates telomere replication. EMBO J. 2009;28:1466–1478. doi: 10.1038/emboj.2009.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.