Abstract
RNA polymerase III recognizes and pauses at its terminator, an oligo(dT) tract in non-template DNA, terminates 3′ oligo(rU) synthesis within this sequence, and releases the RNA. The pol III subunit Rpc11p (C11) mediates RNA 3′–5′ cleavage in the catalytic center of pol III during pausing. The amino and carboxyl regions of C11 are homologous to domains of the pol II subunit Rpb9p, and the pol II elongation and RNA cleavage factor, TFIIS, respectively. We isolated C11 mutants from Schizosaccharomyces pombe that cause pol III to readthrough terminators in vivo. Mutant RNA confirmed the presence of terminator readthrough transcripts. A predominant mutation site, F32, resides in the C11 Rpb9-like domain. Another mutagenic approach confirmed the F32 mutation and also isolated I34 and Y30 mutants. Modeling Y30, F32 and I34 of C11 in available cryoEM pol III structures predicts a hydrophobic patch that may interface with C53/37. Another termination mutant, Rpc2-T455I, appears to reside internally, near the RNA–DNA hybrid. We show that the Rpb9 and TFIIS homologous mutants of C11 reflect distinct activities, that differentially affect terminator recognition and RNA 3′ cleavage. We propose that these C11 domains integrate action at the upper jaw and center of pol III during termination.
INTRODUCTION
Eukaryote genomes are transcribed by three nuclear RNA polymerases (pols) that share some subunits, exhibit homology in others, and also have unique subunits (1). While much structure–function information is available on transcription initiation (1,2), less is known for termination, which is important to the gene targeted for expression as well as downstream DNA (3). Pols I, II and III terminate transcription by different mechanisms (reviewed in 3). Termination signals for pols I and II are somewhat distant from the site at which RNA chain elongation is terminated. In contrast, the termination signal for pol III and the site at which RNA elongation is terminated are coincident. A termination-related feature of pol III is facilitated reinitiation, which enables production of a large number of RNA molecules (e.g. 105) from a single gene (4–7).
The termination signal for pol III is an oligo(dT) tract which is copied into the 3′-terminal oligo(rU) tract on nascent pol III transcripts (8,9, reviewed in 10). Pol III pauses within the oligo(dT) terminator and this can be uncoupled from release of the RNA (11). Pol III harbors a ribonuclease activity that attacks the 3′-end of RNA in or near the catalytic center of pol III at pause sites (12), similar to the RNA 3′ cleavage activity that is facilitated by elongation factor TFIIS which helps pol II escape from states of pausing and backtracking (13,14). Several point mutations in the pol III core subunit, Rpc2p (a.k.a. RET1) that cause pol III to read through oligo(dT) terminators (15,16) also affect its 3′–5′ RNA cleavage activity (17). Three regions of RET1 that affect 3′ RNA cleavage, elongation and pausing, contribute to termination (17–19).
With 17 integral subunits, pol III is the most complex of the nuclear RNA pols. In addition to its 10-subunit core, pol III contains three peripheral subcomplexes, the C17/25 stalk, the C82/34/31 initiation complex and the C53/37 termination complex, which also participates in promoter opening (2,20). The initiation subcomplex can be dissociated from core pol III by solvent (21–23). The C53/37 subcomplex appears to be unstably associated since its subunits are substoichiometric relative to core subunits in purified pol III, ∼35% of which some preparations lacks only C53/37 (21,24,25, see also 26). Loss of C53/37 from pol III appears to more readily occur when the core subunit, C11, is deficient, since a mutant C11 with decreased affinity for pol III also lacks C53/37, designated pol III Δ (13). Deletion of the C-terminal region of C37 causes loss of C53 and C11. Although these observations led to the suggestion that C11 and C53/37 interact in a trimeric complex (27), this could not be substantiated by direct analysis (26), suggesting that their interdependent associations may reflect some degree of allosteric mechanism of pol III.
Understanding pol III termination was much advanced by studying pol III Δ, which is defective for terminator recognition (13). The absence of C11 impairs termination due to loss of C53/37 since adding back recombinant C53/37 to pol III Δ restores terminator recognition in the absence of C11 (20,27). C11 is required in addition to C53/C37 for facilitated reinitiation in vitro, a termination-related process (27). The demonstration that C53/C37 lies near the pol III active site and participates in promoter opening is consistent with this (20). Since the termination activity of C53/37 can operate in vitro without C11 (27), the extent to which C11 may influence this function, especially in vivo, is unclear.
C11 is a 12.5 kDa polypeptide comprised of a region homologous to the N-terminal Zn binding domain of the pol II subunit Rpb9p, followed by a linker and a region homologous to the TFIIS Zn ribbon motif that promotes pol II-associated RNA 3′ cleavage (13). Indeed, C11 mediates RNA 3′ cleavage at pause sites (13). Data from Schizosaccharomyces pombe indicate that RNA 3′ cleavage is also active during the pausing phase of termination since C11 mutations that decrease 3′ cleavage activity results in nascent pol III transcripts that bear lengthened 3′ oligo(rU) tracts (28). The cumulative data suggest that C11 may link terminator recognition to RNA 3′ cleavage although our understanding is based mostly on in vitro studies in which the whole C11 subunit is missing from pol III.
Similar structures for pols II and III suggest that the N-terminal Rpb9p-homologous region of C11 would interact with the Rpc2p pol III subunit very similar to the Rpb9p–Rpb2p interaction that comprises part of the upper jaw (29, 30). This expectation is supported by a small deletion in S. pombe Rpc2p that causes loss of C11 (31) and consistent with very good fit of the Rpb9p N-terminal domain in the cryoEM pol III density (2,32).
A prior screen for C11 mutants in S. pombe used a suppressor tRNA gene that requires accurate termination and efficient post-transcriptional processing (28). That screen produced C11 mutants deficient in RNA 3′ cleavage activity that exhibited alterations in tRNA 3′ end metabolism in vivo (28). For the present study, we developed a dimeric tRNA reporter gene from which the downstream suppressor tRNA will be produced only if pol III fails to recognize an upstream terminator. This led to a new class of C11 mutants, that read through otherwise efficient dT(7) and dT(6) terminators. The most predominant mutation isolated, C11-F32S, resides in the Rpb9-like domain, on a surface that comprises part of the upper jaw, that is predicted to interface with C53/37. Another mutagenesis method, that produces greater mutagenic diversity, led to isolation of additional C11 mutants, with mutations of the invariant residues, Y30 and I34, as well as F32. These mutations reflect a hydrophobic patch on C11 whose integrity appear to be important for efficient termination. That the Rpb9-homologous and TFIIS-homologous domains of C11 mediate separable activities in termination and termination-associated RNA 3′ cleavage is a major finding of this work, and suggests that these are linked and may be coordinately active during termination. Since the Rpb9-homologous domain of C11 is anchored at the periphery of pol III as part of the upper jaw, and the TFIIS-homologous domain presumably inserts into the catalytic center, we suggest that C11 may coordinate action at the periphery and center of the enzyme, and may serve as a model of termination-related information integration for other RNA polymerases.
MATERIALS AND METHODS
The S. pombe strains used are listed in Table 1. The suppressor tRNA in yJI1 contains the dimeric construct pDRT6T at the leu1+ locus, comprised of a wild-type tRNASerUGA followed by a linker and dT(6) terminator, CTAGATTTTTT, followed by the suppressor tRNASerUCA-G37:10,C40,C47:3,C47:6, a GAAGATC trailer and 21T terminator, cloned in the PstI and SacI sites of pJK148 (33). This suppressor was developed for high-specific activity relative to other tRNASerUCA alleles (28,34) (data not shown), to cause suppression if only a fraction of pol III reads through the dT(6) terminator.
Table 1.
S. pombe strains used
| Strain name | Genotype | Reference |
|---|---|---|
| yAS99 | h- ade6-704 ura4-D18 leu1-32 | (34,36) |
| yYH1 | h- ade6-704 ura4- leu1-32::[tRNAmSer7T-leu1+] | (28) |
| yJI1 | h- ade6-704 ura4- leu1-32::[DRT6T-leu1+] | This report |
| yAS68 | h- ade6-704 ura4- leu1-32::[tRNAmSer3T-leu1+] | This report |
| yAS77 | h- ade6-704 ura4- leu1-32::[tRNAmSer6T-leu1+] | This report |
Both for the initial genetic screening and for subsequent TMS assays and northern blotting, the C11 and Rpc2 alleles tested were expressed from plasmid in S. pombe in the presence of the wild-type endogenous alleles. The quantitative suppression assay used for Figure 1 was as described (33). Briefly, the dimeric suppressor tRNA constructs linearized with NdeI were used to transform yAS99. Transformants were selected on EMM media lacking leucine with 10 mg/l adenine. At least 300 colonies of each transformation were scored for red (no suppression) or pink-white (suppression). All other suppression assays were done in strains already carrying a suppressor tRNA reporter gene. In these cases, the cells were transformed with pRep4X expression plasmids containing C11 or Rpc2 as indicated and either spread or streaked on plates containing EMM lacking leucine and uracil, and containing 10 mg/l adenine.
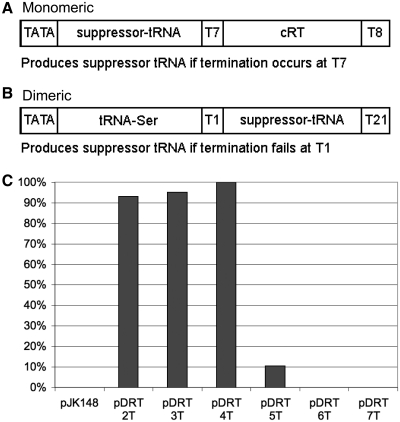
Figure 1.
Schematic representations of the tRNA-mediated suppression (TMS) systems, that use (A) monomeric and (B) dimeric tRNA reporter genes, the latter designated pDRT. The complementary read through (cRT) region in the monomeric construct is highly complementary to the sequence of the tRNA, so that if synthesized, it can fold back and interfere with processing of the tRNA (33). Note that for B, the upstream tRNA provides the pol III upstream promoter element (37). (C) Relative TMS activity of dimeric (pDRT) constructs bearing oligo(dT) tracts of varying length, T2-T7, in the T1 region, assayed in strain yAS99 as described (33) (actual activity of pDRT-4T was ∼75%). pJK148 is a negative control that contains no suppressor tRNA gene.
The library of randomly mutagenized rpc11+ in pRep4X was described (28). rpc2+ was amplified from S. pombe DNA and cloned into pRep4X. Site-directed mutagenesis was by QuikChange XL (Stratagene). All constructs were verified by sequencing.
QuikChange (Stratagene) was used for position-specific codon randomization, with pRep4X-Rpc11 as template, using the following primers:
Val15: cacttaattgtcgcgNNNgacgaggaaggaaggaatgcgttcg;
Val15R: cgaacgcattccttccttcctcgtcNNNcgcgacaattaagtg;
Asp16 : cttaattgtcgcggttNNNgaggaaggaaggaatgcgttcg;
Asp16R: cgaacgcattccttccttcctcNNNaaccgcgacaattaag;
Glu18: cgcggttgacgagNNNggaaggaatgcgttcgactgtag;
Glu18R: ctacagtcgaacgcattccttccNNNctcgtcaaccgcg;
Gly19: gcggttgacgaggaaNNNaggaatgcgttcgactgtagaacatgt;
Gly19R: acatgttctacagtcgaacgcattcctNNNttcctcgtcaaccgc;
Arg20: cggttgacgaggaaggaNNNaatgcgttcgactgtagaa;
Arg20R: ttctacagtcgaacgcattNNNtccttcctcgtcaaccg;
Cys28: ggaaggaatgcgttcgactgtagaacaNNNccttatcattttcca;
Cys28R: tggaaaatgataaggNNNtgttctacagtcgaacgcattccttcc;
Pro29: ggaaggaatgcgttcgactgtagaacatgtNNNtatcattttcc;
Pro29R: ggaaaatgataNNNacatgttctacagtcgaacgcattccttcc;
Tyr30: cgttcgactgtagaacatgtcctNNNcattttccaatttctacttttc;
Tyr30R: gaaaagtagaaattggaaaatgNNNaggacatgttctacagtcgaacg;
His31: gcgttcgactgtagaacatgtccttatNNNtttccaatttctac;
His31R: gtagaaattggaaaNNNataaggacatgttctacagtcgaacgc;
Phe32: cgttcgactgtagaacatgtccttatcatNNNccaatttctac;
Phe32R: gtagaaattggNNNatgataaggacatgttctacagtcgaacg;
Pro33: catgtccttatcattttNNNatttctacttttctctacagtcgtcacg;
Pro33R: cgtgacgactgtagagaaaagtagaaatNNNaaaatgataaggacatg;
Ile34: catgtccttatcattttccaNNNtctacttttctctacagtcgtcacg;
Ile34R: cgtgacgactgtagagaaaagtagaNNNtggaaaatgataaggacatg;
Reactions were thermal cycled: 95°C for 1.5 min, followed by 18 cycles of 95°C for 30 s, 50°C for 1 min, and 68°C for 9 min. Products were digested with DpnI and transformed into DH5α cells. Between 5 and 10 colonies were selected from each position-specific transformation and the isolated plasmids sequenced to determine approximate mutation rate at each codon: V15 80%, D16 100%, E18 40%, G19 80%, R20 70%, C28 100%, P29 100%, Y30 80%, H31 60%, F32 40%, P33 40%, I34 100%. All colonies from each transformation were collected and stored as glycerol stocks.
The cRT region of the suppressor tRNA allele in yAS68, yAS77 and yYH1 follows the dT(7) terminator; three single-stranded 32P-end labeled probes complementary to the cRT readthrough transcript were combined for northern blotting: CCTGCTGGTGACGGAGACC, CTAGTCTTGATTGGTTC and GTTAGACTTCAATCCTG.
Structure modeling was by MacPyMol (DeLano Scientific LLC) and the UCSF Chimera package (Resource for Biocomputing, Visualization and Informatics, University of California, San Francisco) (35) using publicly available protein data base (PDB) and Electron microscopy data base (EMDB) files as indicated in the figure legends.
RESULTS
Our laboratory has used tRNA-mediated suppression (TMS) in S. pombe to study transcriptional and post-transcriptional aspects of tRNA biogenesis (28,33,34,36–41). In this method, a pol III-dependent suppressor tRNA suppresses a premature stop codon in ade6-704 and alleviates accumulation of red pigment. We showed that S. pombe pol III terminates with high efficiency at a tract of five or more dT residues but will mostly read through four or fewer dTs (33). We have characterized suppressor tRNA alleles that vary in their dependence on the 3′ riboexonuclease, Rrp6, the oligo(rU) 3′-OH end-binding protein, La and C11 (28,34). In all of these tRNA alleles, a dT(7) terminator resided at the 3′-end of a monomeric suppressor tRNA gene (T7 in Figure 1A), followed by a complementary readthrough (cRT) region that stabilizes readthrough transcripts by fold-back basepairing to the tRNA sequence and interferes with processing to a functional tRNA, in vivo (33). Positive TMS (white colonies) occurs when termination at dT(7) is accurate and efficient and the nascent transcript undergoes efficient post-transcriptional processing.
A dimeric tRNA gene that yields TMS if pol III reads through an oligo(dT) tract
For the present work, we first developed a dimeric tRNA reporter gene similar to natural dimeric tRNA genes in S. pombe (42,43). In the natural S. pombe gene, a tRNASer sequence is followed by a linker GTATTTG and the second tRNA sequence (42). Our dimeric tRNA gene contains an upstream wild-type tRNASer followed by a linker that contains a dT(n) tract and the suppressor tRNA. The suppressor tRNA is downstream of the dT(n) terminator of the upstream tRNA (designated T1 in Figure 1B). Only if pol III fails to stop at the dT(n) terminator at T1 and elongates an additional 120 bp, will the suppressor tRNA be produced, in this case from a dimeric tRNA precursor that can be processed to mature tRNA (42,44). Production of suppressor tRNA will require initiation and elongation by a pol III that is especially defective for termination.
Various tRNA sequences were examined in vivo to obtain a reliable dimeric reporter with high-specific activity (data not shown). As a test, we modified the T1 region by making constructs that differ only in the number of dT residues (2T, 3T, 7T etc.), and examined their TMS activities using a described quantitative assay (33). Constructs with 2T, 3T and 4T at T1 were active for TMS (Figure 1C), indicating pol III readthrough in agreement with prior data (33). The 5T construct was less active for TMS, consistent with ∼10% of pol III reading through the dT(5) terminator (33). The 6T and 7T constructs produced much less TMS activity indicating that they were highly effective at stopping pol III from transcribing into the downstream suppressor tRNA.
Isolation of novel C11 mutants
We created a S. pombe strain that contains the dT(6) terminator located at T1 in the dimeric tRNA gene and used it to screen rpc11-mutants for TMS. We screened a library of randomly mutagenized rpc11+ for white colonies that appeared among a mass of red background colonies. For this as for our prior screen, the mutagenized C11 library was expressed ectopically in the presence of chromosomal encoded native C11 (28). When the same C11 library was used previously in the monomeric suppressor tRNA strain, yYH1, we obtained 104 suppressed colonies at a rate of 0.06% (28). In contrast, the screen in the dimeric host strain was reproducibly less yielding, producing only six suppressed colonies at a rate of ∼0.004%.
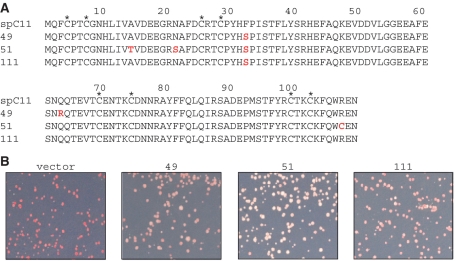
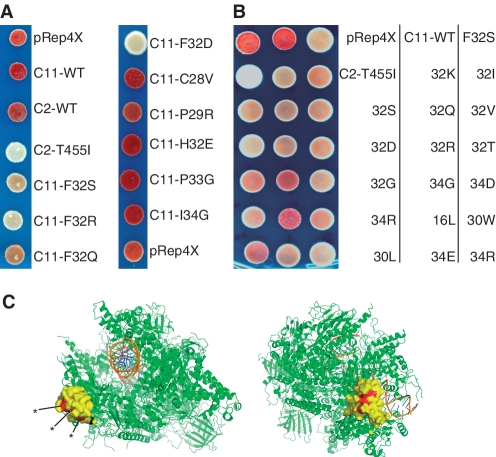
The C11 plasmids recovered from the mutants were purified, sequenced and reintroduced into the dimeric host strain for verification. Four of the six plasmids led to increased TMS of which three had multiple mutations (Figure 2A, one was identical to another and is not shown). All of the C11 mutants isolated contained the F32S mutation in the Rpb9p N-terminal Zn binding homology domain, and for clone 111 this was the only mutation (Figure 2A). The three C11 mutants exhibited different degrees of TMS with clone 51 exhibiting the greatest suppression activity in this assay (Figure 2B).
Figure 2.
S. pombe C11-mutants isolated from a library of PCR-mutagenized C11 (28), using the pDRT-6T dimeric tRNA reporter gene. (A) Sequence alignment of C11 and the three unique C11-mutants numbered 49, 51 and 111. Asterisks indicate invariant cysteines in the Zn-binding motifs. (B) tRNA-mediated suppression phenotypes of the C11-mutants 49, 51 and 111 shown in A and vector control as indicated; red colony color reflects no suppression.
Characteristics of these mutants differed from those isolated using the monomeric reporter (28). In addition to lower rate, none of the mutations, A14T, N21S and Q63R in the new mutants corresponded to any of the 215 mutations in the 104 C11-mutants previously obtained (R.M., unpublished data) (28). The new C11 mutations were enriched in the Rpb9-homologous region of C11, in contrast to the prior mutations, which were enriched in the TFIIS-homologous domain (28). To further examine this, we independently screened the dimeric tRNA strain with a mixture of all 104 of the C11-mutant plasmids isolated from the monomeric tRNA screen (28). This yielded no suppressed colonies, indicating that the dimeric tRNA reporter is quite insensitive to C11 mutations enriched in the TFIIS-homology domain that activate the monomeric reporter.
Structure modeling of the N-terminal homologous regions of C11 and Rpb9
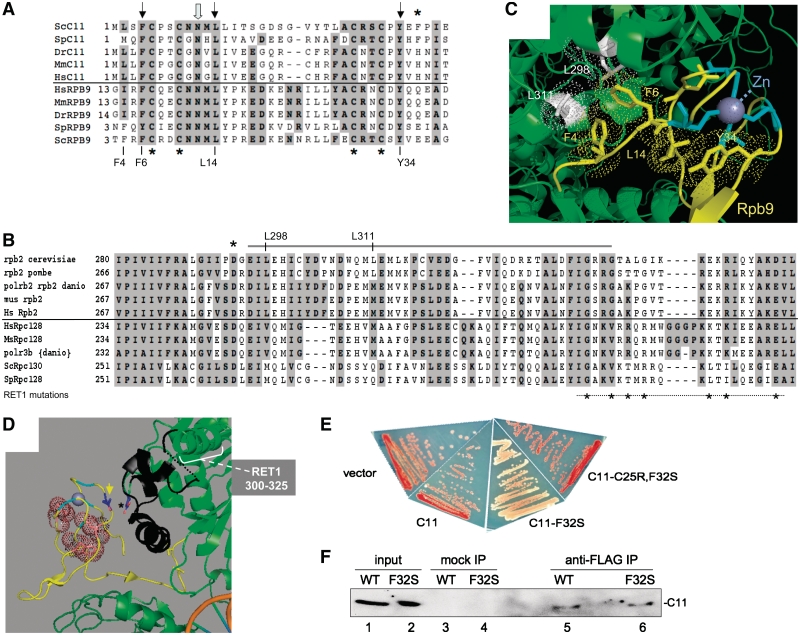
Since the first description of C11 as a sequence homolog of Rpb9p (13), cryoEM densities of yeast pol III have become available and have been fitted with high resolution crystal structures of core pol II that include Rpb9p (2,21,29,30,32). To gain insight into the C11-F32S mutation, we did sequence alignment of the homologous regions of C11 and Rpb9p from phylogenetically distant organisms (Figure 3A). The N-terminal regions of C11 and Rpb9p contain four invariant cysteines (13), which in Rpb9p coordinate Zn (45), and as expected, C11 isolated from pol III contains Zn (21). Evidence that support similar Zn binding motifs in the N-terminal domains of C11 and Rpb9p, with similar docking to their core polymerase is presented in Figure 3A–D. In addition to Cysteines, the Phe (F), Asn (N), Leu (L) and Tyr (Y) are highly conserved in Rpb9p and C11 (Figure 3A). We also aligned Rpc2p and Rpb2p sequences (Figure 3B). As can be seen from the pol II crystal structure, the F, L and Y contribute to a hydrophobic network juxtaposed to the Zn binding Cys cluster in the N-terminal domain of Rpb9p (Figure 3C, Cys backbone in cyan, hydrophobic side chains as yellow colored dots). The hydrophobic network extends to three other conserved residues, F4 in Rpb9, and L298 and L311 in Rpb2 (Figure 3C) all of which are conserved as hydrophobic in C11 and Rpc2, respectively. This suggests similar packing of the N-terminal domains of Rpb9p and C11.
Figure 3.
(A) Sequence alignment of the N-terminal regions of C11 and Rpb9p, above and below horizontal line, respectively, from several organisms. Invariant Cysteines are indicated by asterisks below the sequences. Highly conserved residues are indicated by downward arrows above the sequences, and for S. cerevisiae Rpb9 these are numbered below the sequence. The C11 F32 position is indicated by an asterisk above. (B) Sequence alignment of homologous regions of Rpb2p and Rpc2p from several organisms, above and below horizontal line, respectively. The asterisk indicates position D-294 (S. cerevisiae numbering). The gray line above the sequences spans the region that when deleted in S. pombe Rpc2p leads to loss of C11 (31). The asterisks below the alignment indicate positions of mutations in S. cerevisiae RET1 (Rpc2) that cause decreased termination (18). C) Structure view of a segment of the yeast pol II core (PDB ID 3M40) that includes the N-terminal domain of Rpb9p shown as yellow ribbon. The highly conserved F, L andY highlighted in A above as well as F4 are represented as side chains with space-filling dots. Side chains of L298 and L311 of Rpb2 are colored white with space-filling dots. The backbone positions and side chains of the clustered cysteines in Rpb9 are shown in cyan, and the coordinated Zn as a purple sphere. (D) Structure model (PDB ID 3M40) showing the Rpb9p N-12 side chain, which is highly conserved in C11, indicated by a yellow arrow. The side chain of the highly conserved D-294 in Rpb2p is indicated by an asterisk; the N12-D94 distance is <4 Å. The region of Rpb2 that corresponds to the deletion in S. pombe Rpc2p that leads to loss of C11 (31) is colored black. (E) TMS phenotypes of the site-directed mutants, C11-F32S and C11-C25R, F32SC11, as well as wild-type C11 and vector control (see text). (F) Coimmunoprecipitation of HA-tagged Rpc11p-WT and -F32S with Flag-Rpc2p. Strain yYH3272 carrying Flag-Rpc2p (37) was transformed with HA-C11-WT or HA-C11-F32S, and extracts made. Inputs (lanes 1 and 2), the products of mock IP using nonimmune IgG (lanes 3 and 4), and products of IP using anti-Flag IgG were blotted and probed with anti-HA Ab. The lanes between 4 and 5, and 5 and 6, were not loaded.
The conserved N12 of the N-terminal domain of Rpb9p makes contact with D294 in Rpb2p (3.2 Å, Figure 3D, yellow arrow/black asterisk). Sequence alignment reveals that D294 is conserved in Rpc2p as well as Rpb2p (asterisk, Figure 3B). The Rpb2p and Rpc2p alignment also shows the sequence tract of spRpc2p which when deleted causes loss of C11 (31) (gray line above the sequences, Figure 3B). This ‘deleted’ sequence corresponds to the region colored black in Figure 3D which is in proximity to the N-terminal domain of Rpb9p and overlaps with the RET1 300–325 regions (Figure 3B, white bracket in Figure 3D), comprising part of the lobe-jaw adjacent to the DNA binding cleft. These observations support other evidence that indicate that the homologous N-terminal regions of the core subunits Rpb9p and C11 are analogously positioned on pols II and III, respectively.
The point mutation F32S in spC11 compromises termination by pol III
One C11 mutant had a single substitution, F32S, which was also present in the other mutants (Figure 2A). To verify the importance of this substitution, we independently created it by site-directed mutagenesis. This mutant, C11-F32S, exhibited increased TMS, whereas wild-type C11 and control plasmid were inactive (Figure 3E). We suspected that in order to exhibit the readthrough TMS phenotype, the mutated C11 would have to associate with pol III, displacing wild-type C11 as was apparent for the previous C11 mutants (28), and that this would be dependent on the integrity of its Rpb9-homologous Zn binding domain. Consistent with this, none of the invariant cysteines in the N-terminal domain were found mutated in any of the previously characterized C11-mutants (28). We tested if mutation of invariant Cys-25 which is predicted to disrupt the N-terminal Zn binding domain, as in the double mutant, C11-C25R, F32S, would inactivate C11-F32S for TMS. Indeed, C11-C25R, F32S was inactive for TMS (Figure 3E), consistent with the idea that C11-F32S associates with pol III to exert its phenotype. We do not know how much C11-F32S is incorporated into pol III in the mutant cells because we cannot distinguish C11-F32S from C11-WT subunits in isolated pol III. However, by using HA-tagged versions of C11, the experiment in Figure 3F indicates that HA-tagged C11-F32S can be coimmunoprecipitated with Flag-tagged Rpc2p, in agreement with data in Figure 3E.
The differing characteristics of the mutants isolated using the monomeric and dimeric tRNA genes suggested that dysfunction of the Rpb9-homologous and the TFIIS-homologous domains might exhibit differential activity in the two reporter systems.
The Rpb9- and TFIIS-homologous domains of C11 differentially affect two pol III termination phenotypes
We examined C11-F32S and C11-C102S, the latter a previously characterized 3′ cleavage mutant (28), in side-by-side comparisons in the monomeric and dimeric reporter strains, yYH1 and yJI1. For a control, we made a T455I mutation in S. pombe rpc2+, the gene that encodes the second largest subunit of pol III. Saccharomyces cerevisiae Rpc2-T455I is a pol III termination mutant that causes readthrough of an oligo(dT) terminator (19). The data below confirm this for S. pombe Rpc2-T455I.
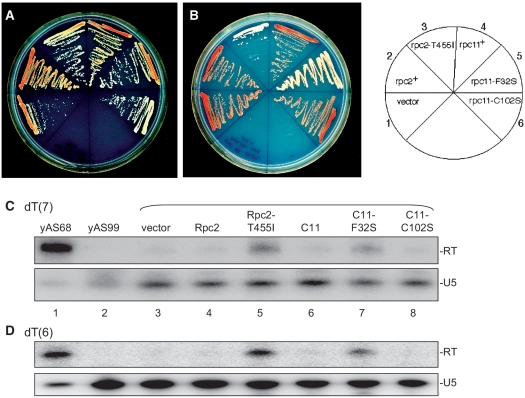
The comparisons in yYH1 and yJI1 are shown in Figure 4A and B, respectively. Baseline suppression for each strain is shown in Sector 1, empty vector. While wild-type Rpc2 had no effect in either strain, Rpc2-T455I was clearly more active (white) in yJI1 than in yYH1, consistent with dT(6) readthrough as expected. Likewise, C11-F32S was more active in yJI1 than in yYH1, while wild-type C11 had no effect (Sector 4). In sharp contrast, C11-C102S was far more active in yYH1 than in yJI1. In summary, C11-F32S exhibited the dT(6) readthrough phenotype in yJI1 while C11-C102S did not but did exhibit the RNA 3′ cleavage deficiency phenotype in yYH1, while C11-F32S did not.
Figure 4.
Side-by-side analyses in yYH1 and yJI1 of multiple S. pombe pol III mutants. Strains yYH1 (A) and yJI1 (B) bear monomeric and dimeric suppressor tRNA genes, respectively. The expression plasmids carrying the alleles to be tested (C11, Rpc2, etc.) were assayed in both strains and displayed according to the schematic to the right. (C) Northern blot of RNAs isolated from yYH1, which carries a reporter tRNA gene with a dT(7) terminator, after transformation with the plasmid-borne expression alleles indicated above lanes 3–8; lanes 1 and 2 contain RNA from positive and negative control strains. Upper panel shows blot probed for the readthrough transcript from the suppressor tRNA dT(7) gene. Lower panel shows same blot probed for U5 snRNA as a loading control. Lane 1 was intentionally under loaded so that the intensity of the yAS68 RT band would be closer to the linear range of detection, and to avoid contamination of other lanes. (D) Northern blot of RNAs isolated from yAS77, which carries a reporter tRNA gene with a dT(6) terminator; lanes are as in C above. Quantitation of C and D is reported in Table 2.
Although Rpc2-T455I was active in yJI1, it was less active in yYH1 (compare Sectors 3, 1 and 2, Figure 4A). This is not unexpected since TMS in yYH1 results from 1 to 2 nt lengthening of pre-tRNA 3′ oligo(rU) (28), and Rpc2-T455I was shown to produce 1–2 nt longer 3′ oligo(rU) on its terminated RNA relative to wild-type Rpc2 [see Figure 7 in (18)].
C11-F32S causes pol III to read through dT(6) and dT(7) terminators in vivo
In order to examine effects of C11-mutants on pre-tRNA transcripts, we performed northern blots, using a probe specific for the readthrough transcripts. For this, we examined yYH1 whose suppressor tRNA gene ends with a dT(7) terminator followed by a complementary readthrough (cRT) region (Figure 1A) that stabilizes readthrough transcripts in vivo (33). A positive control is yAS68, whose suppressor tRNA gene is identical to yYH1 except that it has dT(3) in place of the dT(7) terminator and accumulates high levels of readthrough transcripts [3T lane, Figure 3A in (33)] that serve as a marker (Figure 4C, lane 1). Lane 1 was under loaded so that the RT band was in the same range of detection as the other RT bands and to avoid contamination of other lanes, for quantitation (below). The negative control is yAS99, which does not carry a suppressor tRNA and should not hybridize with the probe (lane 2). While vector, wild-type Rpc2, and wild-type C11 showed background levels of readthrough transcripts (-RT, lanes 3, 4 and 6), Rpc2-T455I and C11-F32S revealed higher levels (lanes 5 and 7). C11-C102S showed only background levels of RT transcripts, confirming that this mutant does not exhibit significant readthrough (28). Rpc2-T455I and C11-F32S produced significantly more readthrough of the dT(7) terminator as compared with vector and their wild-type controls (Figure 4C).
While Figure 4C shows readthrough of a dT(7) terminator, the yJI1 strain assay carries a dT(6) terminator. Therefore to evaluate readthrough of a dT(6) terminator the northern blot experiment was performed again using recipient strain yAS77 whose tRNA reporter is identical to that in yYH1 and yAS68 except that it contains a dT(6) terminator (Figure 4D). Again, Rpc2-T455I and C11-F32S produced more readthrough of the dT(6) terminator as compared with vector and their wild-type controls (Figure 4D, Table 2). These data confirmed that C11-F32S causes pol III to read through dT(6) and dT(7) terminators in vivo, accounting for the TMS phenotype observed in yJI1.
Table 2.
Quantitation of terminator readthrough (relative to yAS68 cRT/U5)
| Sample | dT(7); %RTa | dT(6); %RT | Relative increase in RT dT(7) to dT(6)b |
|---|---|---|---|
| yAS68-dT(3)c | 100 | 100 | – |
| yAS99 | – | – | – |
| +vector | 0 | <1 | – |
| +Rpc2 | <1 | <1 | – |
| +Rpc2 T455I | 7.4 | 21 | 2.8 |
| +Rpc11 | <1 | <1 | – |
| +Rpc11 F32S | 5.9 | 15 | 2.6 |
| +Rpc11 C102S | <1 | <1 | – |
aReadthrough in yYH1; cRT signal/U5 signal, background corrected against +vector sample.
byAS77 {dT(6)} relative to yYH1 {dT(7)}.
ccRT/U5 signal of yAS68 is presumed to represent 100% readthrough.
Quantitation of the data in Figure 4C and D with calibration of transcript levels in yAS68 set as 100% readthrough (Table 2), led to two deductions. Reducing oligo(dT) length from dT(7) to dT(6) led to 2.8- and 2.6-fold increases in readthrough transcripts by Rpc2-T455I and C11-F32S, respectively. Second, it appears that only ∼15% of the pol III in the C11-F32S cells fails to terminate at the dT(6) terminator and reads through into the downstream suppressor tRNA in yJI1. This is ∼21% for Rpc2-T455I (Table 2).
Structure modeling suggests interface of C11-F32 and C53/37
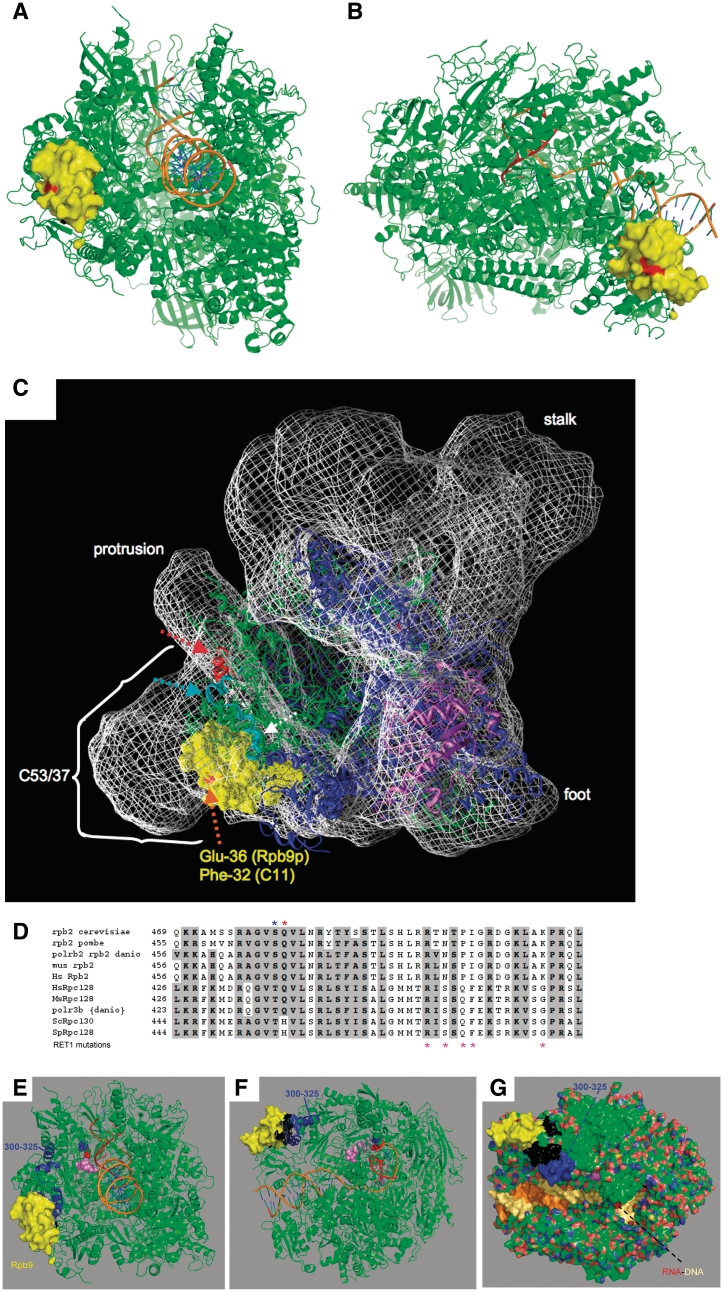
The crystal structure of the Rpb9p N-terminal domain of the core pol II structure can be fit into the 9.9 Å cryoEM pol III density (32). Two recent cryoEM structures into which the C53/37 dimerization domains have been localized provide high-resolution models that suggest juxtaposition of the N-terminal domain of C11 with a part of the C53/37 density (2,32). From the alignment of Rpb9 and C11, we modeled the F32-homologous residue of Rpb9p (Glu-36) onto the pol II crystal structure. This revealed that the F32-homologous position is located on an outward facing surface of Rpb9 (Figure 5A and B). We then fitted the core pol II structure with the C11 F32-homologous residue of Rpb9p (Glu-36, colored red) into the 9.9 Å cryoEM envelope (Figure 5C). From this, the F32 of C11 would appear to face into the C53/37 density (Figure 5C). Extrapolating from a similar view, C11 F32 appears to be near the C-terminal extension of C37 [see Figure 2B in(2)].
Figure 5.
Modeling termination-relevant mutations in the pol III cyroEM structure. (A and B) Views of pol II in which the surface of Rpb9 N-terminal domain is yellow with the Glu-36 position that corresponds to C11-F32 colored red (PDB ID 3M40). (C) The cryoEM structure (EMD-1802) (32) was fit with the core pol II crystal structure (PDB 1WCM) (52) lacking the stalk and the C-terminal domain of Rpb9p. Fit was by manual manipulation followed by automated fit using UCSF Chimera (35) (final lowest average value = 0.9628 for 23833 atoms, 4042 outside contour). The view is similar to Figure 2A and B in (2) and the front view in Figure 2B in (32). Pol II is depicted as ribbon except for the Rpb9 N-terminal domain which is a yellow surface view, with Glu-36 (corresponding to C11-F32) colored red and indicated by an orange arrow. Rpb2p is colored light green except for two regions proximal to the C53/37 density, which are cyan and red, indicated by dashed arrows. Cyan corresponds to a region in S. pombe Rpc2 that when deleted causes loss of C11 (31), and the red helix corresponds to the region containing RET1 decreased termination mutations indicated in Fig. 3B. Rpb1 is colored blue. Envelope densities corresponding to the protrusion, stalk and foot, and C53/37 are indicated (32). (D) Alignment of sequences from several organisms of a central homologous region of Rpb2 and Rpc2. Blue asterisk above the sequences indicates S480 in Rpb2 and T455 in Rpc2, the latter when mutated as part of this study led to decreased termination. The red asterisk indicates Rpb2 position Q481 (see text). Pink asterisks below Rpc2 indicate mutations in RET1 that decrease termination (18). (E–G) Pol II (PDB ID 3M40) with an RNA–DNA hybrid; RNA backbone is red. Rpb2 S480 and Q481 are shown as blue and red spheres. RET1 mutations indicated by asterisks in B are shown as pink spheres. The N-terminal domain of Rpb9 is shown as yellow surface. In F and G, the sequence tract corresponding to the region in spRpc2 that when deleted causes loss of C11 is colored black and the region corresponding to the 300–325 regions of RET1 (see Figure 3B) is colored blue (31).
We also modeled other pol III analogous parts of the fitted EM structure that are relevant to C11 association and/or termination (Figure 5C). The Rpb2-homologous region of S. pombe Rpc2 that when deleted caused loss of the C11 subunit is shown in cyan (Figure 5C) (this region is shown in black in Figure 3D). The alignment in Figure 3B shows sequence overlap between this region of Rpc2 and a tract containing point mutations in S. cerevisiae Rpc2 (RET1) referred to as the 300–325 regions, that cause decreased termination (18) (asterisks below Figure 3B). The Rpb2 sequences corresponding to the 300–325 regions of RET1 is a red colored helix in Figure 5C. This helix comprises cryoEM pol III density that bridges the protrusion and the C53/37 density. This modeling suggests that F32 of C11 interfaces with part of the C53/37 heterodimer. The model also illustrates that the N-terminal domain of C11 appears to contact the Rpc2p segment that when deleted causes loss of C11 (31), and that this segment is contiguous with Rpc2p (RET1) 300–325, a region previously implicated in termination.
The Rpc2p-T455I termination mutation is probably near the RNA–DNA hybrid
Pols III from human, S. cerevisiae and S. pombe differ in the minimal length of the oligo(dT) tract required for efficient termination (33), matching the distribution of oligo(dT) lengths observed for tRNA gene terminators assessed by genome-wide analysis (46). The 455I mutation of S. pombe Rpc2p caused pol III to readthrough the dT(6) and dT(7) terminators, similar to reports on S. cerevisiae (19). At the time, this mutation was analysed in S. cerevisiae (18), a relevant crystal structure had not been determined. We therefore modeled this mutation onto a pol II complex containing a DNA–RNA hybrid (Figure 5D–G). Rpc2p T455 corresponds to Rpb2p S480 (Figure 5D). In pol II, this residue immediately precedes Q481 which is within 4 Å of the RNA in the RNA–DNA hybrid (47). In pol II, Q481 makes contact with the ribose between ribonucleotides six and seven of the hybrid (47). While this Q is conserved in vertebrate pols III, it is replaced by H in the yeasts’ Rpc2p (Figure 5D). S480 and Q481 are shown as blue and red spheres, respectively, in Figure 5E and F. Thus, it would appear that the mechanism by which Rpc2p-455I decreases pol III termination may be through its effects on the RNA–DNA hybrid (48, 49). Other RET1 mutation positions in this region that decreased termination (18) are shown as pink asterisks below in Figure 5D and pink spheres in Figure 5E–G.
Additional mutants suggest Y30, F32 and I34 as a hydrophobic patch on yeast C11
As noted above, the dimeric tRNA screen yielded many fewer mutants than the screen using the monomeric suppressor tRNA gene. In an attempt to find additional mutants, we performed 12 site-directed mutageneses, each randomizing a specific codon in S. pombe C11. The targeted positions in S. pombe C11 were V15, D16, E18, G19, R20, C28, P29, Y30, H31, F32, P33 and I34. We verified a distribution of changes at each position (data not shown) before the constructs were tested in our dimeric tRNA strain by two approaches. First, plasmids with sequence-verified specific mutations were tested singly. Of the position 32 replacements, R and D produced the strongest suppression phenotypes, while S, G and Q were less suppressed but more than the control, confirming sensitivity of F32 to phenotypic mutation (Figure 6A and Table 3). This also identified mutations of positions 30 and 34, both of which are invariant in C11 (Figure 3A), as causing the readthrough phenotype. In contrast, mutations at numerous other positions did not produce significant suppression phenotype (Figure 6A and Table 3).
Figure 6.
Codon randomization leads to additional C11 termination readthrough mutants. (A) TMS spot assay comparing suppression activities of various individual position specific mutations of C11. See Table 3 for complete list. (B) TMS spot assay showing activities of mutants isolated by screening using the mini-library containing a mixture of all codon randomizations of 12 codons (see text). The identity key for the three columns is to the right. See Table 4 for complete list. (C) Pol II homology model with the three most frequent C11 mutants in red, each indicated by an asterisk, as reflected on the Rpb9p surface (yellow) as in Figure 5A.
Table 3.
TMS results of site-directed mutagenesis
| Plasmid | SSa | Plasmid | SS | Plasmid | SS |
|---|---|---|---|---|---|
| Rep4X | 0 | D16L | 0 | H31R | 0 |
| C11 WTb | 0 | E18H | 0 | H31T | 0 |
| C2 WTb | 0 | E18T | 0 | H31E | 0 |
| C2 T455Ib | 5 | G19C | 0 | F32D | 4 |
| C11 F32Sb | 3 | G19E | 0 | F32R | 4 |
| V15L | 0 | R20L | 0 | F32Q | 2 |
| V15K | 0 | R20Y | 0 | F32G | 1 |
| V15W | 0 | C28V | 0 | P33G | 0 |
| V15S | 0 | C28D | 0 | I34S | 2 |
| V15E | 0 | C28R | 0 | I34L | 0 |
| V15E | 0 | P29R | 1 | I34T | 0 |
| V15I | 0 | P29E | 0 | I34G | 0 |
| D16V | 0 | P29D | 0 | H31R | 0 |
| D16R | 0 | Y30W | 3 | H31T | 0 |
| D16P | 0 | Y30N | 0 | H31E | 0 |
aSuppression score.
bC11-WT, RpC2-WT, C2-T455I & C11-F32S were included as controls.
In the second approach, all C11 plasmids randomized at the 12 codons were combined into a mini-library and used to screen for the readthrough TMS phenotype. The rate of suppressed colonies obtained from this mini-library screen was ∼19%. Thirty-five of these C11 mutants were characterized and sequenced. The suppression phenotypes of a range of these are shown in Figure 6B. The most frequent readthrough mutants obtained from the mini-library contained mutations at position 32 (77%), followed by position 34 (14%) and position 30 (7%), as detailed in Table 4.
Table 4.
C11 position-specific randomized codon mini-library
| Position (#)a | Mut identity | Position (#) | Mut identity | |
|---|---|---|---|---|
| V15 (0) | –b | P29 (0) | – | |
| D16 (1) | L | Y30 (2) | W, L | |
| E18 (0) | – | H31 (0) | – | |
| G19 (0) | – | F32 (27) | D(4)a, G, I(4), K(2), Q(6), R(5), S, T, V(3) | |
| R20 (0) | – | P33 (0) | – | |
| C28 (0) | – | I34 (5) | – |
aThe number of mutants obtained in parentheses.
b–Not applicable, none recovered.
The side chains of Y30, F32 and I34 emerge as a contiguous hydrophobic surface in the Rpb9p-C11 homology model (Figure 6C).
DISCUSSION
A clear conclusion that can be drawn from this work is that mutation of F32 in spC11 causes significant readthrough by S. pombe pol III of otherwise efficient terminators, dT(6) and dT(7) in vivo. C11-F32S was isolated from a PCR-mediated mutagenesis-based genetic screen from which only two other unique mutants were isolated and these also had the F32 mutation. Another mutagenesis approach, position-specific codon randomization of 12 codons in the Rpb9-homologous domain of C11, confirmed position 32 as the most frequently mutated residue in the readthrough mutants, and also identified mutations of the invariant residues Y30 and I34 as conferring the pol III readthrough phenotype. Indeed, modeling suggests that Y30, F32 and I34 form a contiguous hydrophobic patch on C11. We propose that the integrity of this region of C11 is important for terminator recognition.
Terminator readthrough was confirmed in F32S mutants by detecting transcripts that extended beyond dT(6) and dT(7) terminators in RNA isolated from C11-F32S mutants but not the negative control strains tested (Figure 4C and D). For this we used northern blotting with quantitation that included a positive control construct with dT(3) in place of an efficient terminator to represent 100% readthrough. After obtaining the F32S mutant, we created a Rpc2 mutant, Rpc2-T455I by site-directed mutagenesis, to serve as a control for terminator readthrough based on prior characterization of S. cerevisiae Rpc2-T455I (18,19). Including the Rpc2-T455I mutant in subsequent analyses provided assurance that the C11-F32S mutation also caused terminator readthrough. The C11-F32S and Rpc2-T455I mutants behaved similarly as analyzed in vivo by TMS and by northern blotting whereas the wild-type versions of these subunits behaved similar to the empty vector control in both assays when analyzed in parallel (Figure 4).
It was expected that the C11-F32S and Rpc2-T455I mutations would cause more readthrough of a dT(6) than a dT(7) terminator. This was confirmed by quantitation of readthrough transcripts which estimated that C11-F32S caused ∼2.6-fold and Rpc2-T455I caused ∼2.8-fold more readthrough of a dT(6) than a dT(7) terminator (Table 2). We note that while the absolute quantities may not be precise, as described below we are confident that they reflect relative differences, both for dT(6) and dT(7) readthrough, and for comparison of C11-F32S and Rpc2-T455I.
Quantitation of readthrough transcripts also allowed us to estimate the fraction of pols III that read through the dT(7) and dT(6) terminators to be 5.9% and 15%, respectively, for C11-F32S, and 7.4% and 21% for Rpc2-T455I (Table 2). That Rpc2-T455I produced more readthrough transcripts than C11-F32S is in agreement with its reproducibly stronger suppression phenotype (Figure 4B, data not shown). The quantitative estimates are also consistent with the fact that the suppressor tRNA allele in yJI1 exhibits high specific activity for TMS relative to all other suppressor tRNA alleles examined (data not shown), as it has no debilitating mutations as do our other alleles (34) (see ‘Materials and Methods’ section). The estimate that ∼15% of pol III reads through the dT(6) terminator in yJI1 C11-F32S cells is in agreement with our unpublished quantitative comparison of TMS-specific activities of various suppressor tRNA alleles.
Although no significant difference was detected in the amount of C11 that copurified with epitope-tagged pol III isolated from the C11-F32S and C11-WT cells (Figure 3F), we cannot determine what fraction of the pol III contains the C11-F32S subunit. While it is possible that C11-F32S may cause dissociation of C53/37 from pol III we have been unable to document this. Around 15–20% loss of C53/37 from pol III would be beyond our ability to confidently distinguish. Nor can we determine how much the Rpc2p-T455I subunit replaces endogenous Rpc2p in the cellular pol III pool. Although we cannot rule out that the small fraction of pol III in the C11-F32S cells that reads through dT(6) and dT(7) terminators is deficient in C53/37, the data also leave open the possibility that C11-F32S and other mutations affect termination by another mechanism.
It seems reasonable to assume that <100% of the pol III in C11-F32S cells actually contains the C11-F32S subunit with the remainder containing wild-type endogenous C11. Accordingly, 15% terminator readthrough likely represents an underestimate of the potential strength of the F32S substitution. Unfortunately, the S. pombe in vitro transcription system required to examine this has not been developed.
Position-specific codon randomization mutagenesis led to additional C11 mutants
PCR-mediated mutagenesis, as performed to create our original C11 library of 150 000 independent clones (28), is limited in the diversity of mutations that lead to amino acid changes. We therefore used position-specific codon randomization of 12 positions in the Rpb9-homologous domain of C11, including position 32. This revealed that the readthrough phenotype was caused by the F32S mutation as well as mutation to other amino acids, confirming and extending the results obtained with PCR-mediated mutagenesis. Moreover, the codon randomization approach led to isolation of additional mutants, including several that changed invariant Y30 and I34 to other amino acids, whereas mutations at multiple other positions did not produce the readthrough phenotype.
The cumulative results indicate that only a small subset of C11 mutations lead to termination failure in our system. Other mutations in C11 that would cripple pol III for initiation and/or elongation would not be detectable by our dimeric tRNA gene assay. Accordingly, compromising termination specifically may explain why so few mutations could be obtained.
Two termination-related activities of C11
The in vivo data reported here demonstrate that the F32S and C102S mutations in C11, located in distinct domains of C11 separated by a linker, differentially affect two termination-related phenotypes reflected by different suppressor tRNA reporter constructs. Suppression in yJI1 reports failure of pol III to terminate at an oligo(dT) terminator. Suppression in yYH1 reports decreased 3′–5′ cleavage at the RNA 3′ terminus during the pausing phase of termination. In yJI1, not only must pol III fail to stop at the dT(6) test terminator but it must then continue ∼120 bp beyond this to synthesize the suppressor tRNA. F32S causes this while C102S does not. In contrast, this type of termination defect, skipping an oligo(dT) terminator and continuing, is detrimental to the suppression phenotype in yYH1 (33). In yYH1, increased suppression occurs as a result of deficiency of 3′ cleavage of the 3′ oligo(rU) terminus of the RNA during pausing. Thus, the two different activities of C11 monitored here are: (i) terminator recognition and pausing, mediated by the C11 N-terminal domain possibly involving C53/37, and (ii) terminal oligo(rU) 3′ cleavage that occurs during terminator pausing, mediated by the C-terminal TFIIS-homologous domain.
The quantitative extent to which the two activities distinguished here are manifested should not be expected to be equal. In addition to the different positions of the test terminator relative to the suppressor tRNA sequence in the reporters, the suppressor tRNAs have different specific activities due to sequence differences, and are not directly comparable because monomeric and dimeric tRNA precursors use different processing pathways (42). Also, the C102S and F32S C11 subunits may be incorporated into pol III with different efficiencies. Thus, the apparent specificity of C11-F32S in the readthrough assay does not exclude the possibility that this mutation might affect RNA 3′–5′ cleavage activity and 3′ oligo(rU) length with resulting phenotype in yYH1 if it were incorporated into pol III more efficiently. While only ∼15% of pol III in C11-F32S cells leads to dT(6) readthrough, we believe that more C11-C102S is incorporated into pol III. The RNA 3′–5′ cleavage activity of C11-C102S pol III measured in vitro was ∼20% of wild-type pol III suggesting that at least 80% of pol III contained the C11-C102S subunit (28). Further analysis must await the ability to analyse pol III that homogeneously contains C11-F32S. Nonetheless, this does not detract from the ability and specificity of C11-F32S to promote terminator readthrough in yJI1 as determined by TMS and northern blotting, especially since C11-C102S does not do so by either assay.
RNA polymerase-specific use of RNA 3′–5′ cleavage activities?
The fact that the TFIIS-homologous domain of C11 could not be accounted for in cryoEM structures suggests that it is mobile. Indeed, residence of the RNA cleavage finger of C11 in the catalytic site is not expected to be present during polymerase elongation but to be used intermittently, as is TFIIS. Yet, pol III carries the C11 cleavage domain as part of the core enzyme, presumably so it can be summoned to the catalytic site quickly and efficiently. C11 cleavage activity may contribute to termination by increasing pause time at a terminator, perhaps summoned via signals from C53/37 or other parts of the polymerase that undergo allosteric movement during termination.
It is intriguing that the RNA 3′–5′ cleavage activity of pol III is mediated by an integral core subunit whereas the orthologous activities for pol II and bacterial RNA polymerase rely on the TFIIS and GreB/A factors, which transiently associate with their respective enzyme as needed at pause sites. While TFIIS and GreB/A act at pause sites they have not been proposed to function during termination. Moreover, the C11 subunit and its RNA 3′–5′ cleavage activity is essential for viability (13,28) whereas its TFIIS and GreA/B counterparts are not.
Remarkably, TFIIS was found at pol III-transcribed genes in vivo and has been shown to affect transcription start site by pol III in vitro (50). In this case, recombinant TFIIS added to in vitro transcription reactions suppressed inaccurate start site selection by pol III, at a +4/+8 site, and promoted initiation at the correct start site (50). TFIIS containing a point mutation (E291A) that inhibits intrinsic RNA 3′ cleavage by pol II failed to promote the correct start site initiation by pol III. Yet, wild-type TFIIS did not promote RNA 3′ cleavage when added to pol III Δ stalled due to lack of NTPs, nor did it inhibit pol III intrinsic RNA 3′ cleavage mediated by C11 (50). These intriguing results (50) suggest that the TFIIS effects may be limited to a pol III conformation during start site selection, but not while pol III is stalled during elongation, which might be more specific to the TFIIS-homologous domain of C11.
Subtle mutations in C11 appear to mimic termination failure due to loss of C53/37
It had been unknown if the termination failure observed for pol III Δ, which lacks C53/37 and C11, results from a structural disfigurement in pol III caused by loss of the entire core subunit, C11. The in vivo terminator readthrough data presented here are significant because they arise from subtle mutations, single point substitutions, and suggest that dysfunction of C11 and loss of C53/37 produce similar termination failure, although that has not been directly tested.
Different classes of termination mutants localize differently in pol III
While the pols III from distant species share similar oligo(dT)-dependent mechanisms of termination, they also exhibit a subtle but distinguishable difference. Vertebrate pol III executes very efficient termination at dT(4), whereas dT(4) does not cause termination by S. pombe and S. cerevisiae pols III which require successively longer dT tracts (33) as reflected in the oligo(dT) lengths of pol III terminators in the genomes of these species (46). It is therefore noteworthy that the Rpc2-T455I mutation in S. pombe causes termination failure at dT(6) and dT(7), similar to that reported for S. cerevisiae, arguing that these yeast pols III are similar in this aspect of termination.
Y30, F32S and I34 are in the N-terminal Rpb9-homologous domain of C11 which appears to reside on a peripheral surface of the polymerase, close to an Rpc2p (RET1) helix whose mutation causes decreased termination in S. cerevisiae (18). This helix, comprising residues 300–325 would appear to be located on the upper jaw on incoming DNA (Figure 5C–E). It was hypothesized that this region, comprising amino acids 300–325 of RET1 is involved in RNA release (18). Based on the position on the upper jaw, it may be involved in DNA release during termination.
Quite distinct from the rather peripheral location of the N-terminal domain of C11 and the more proximal Rpc2p 300–325 regions in the pol II homology model, is T455 in Rpc2p. The opportunity to map these mutations onto a model pol II complex containing a RNA–DNA hybrid may provide additional insight. Based on the structural homology model, Rpc2p-455 would appear to be in close proximity to the RNA strand of the RNA–DNA hybrid and this suggests that it may affect hybrid stability or dynamics. This location is indeed consistent with the earlier prediction that RET1 455–521 would involve the RNA–DNA hybrid (18). Hybrid destabilization appears to contribute to intrinsic termination by bacterial RNA polymerase (48,49,51). Our modeling suggests that the Rpc2p 472–485 regions of RET1 lies along the DNA just downstream of the hybrid, possibly close to the unwound non-template DNA (Figure 5C and D).
A third region of RET1 mutations, 1061–1082, caused increased termination and were hypothesized to contact DNA (18) and this is also consistent with our modeling. One of these RET1 mutations, at M1065, corresponds to Rpb2-M1133 which lies within 4 Å of the template DNA in the RNA–DNA hybrid (data not shown). Although Pols II and III execute termination differently in response to different signals, and it is therefore unlikely that a detailed mechanism for Pol III termination can be precisely inferred from the available Pol II structures, the modeling nonetheless suggests that two of the three previously identified RET1 regions, 455–485 and 1061–1082, involved in termination would appear to be relatively close to the catalytic center of pol III, whereas the 300–325 regions is more peripheral, located on the upper jaw and close to C11 and C53/37 (20). By this analogy it is tempting to speculate that the 300–325 regions represents an allosteric, i.e. structural class of Rpc2p termination mutant while the other two RET1 regions are more directly involved in cessation of RNA synthesis. By association, RET1 300–325 may be part of an allosteric signaling network that involves C53/37 and C11.
A model that emerges is that the TFIIS-homologous Zn-ribbon domain of C11 can sample the catalytic center while the Rpb9p-homologous Zn binding domain is attached at the periphery and somewhat close to the upper jaw that holds DNA, potentially situated to transmit signals to and from the catalytic center and periphery of the complex. Termination by pols I and II appear also to involve signals from the periphery, e.g. according to the torpedo model, to the catalytic center, where presumably the enzyme must ultimately give up its hold on the RNA–DNA hybrid in order to terminate (3).
FUNDING
Funding for open access charge: Intramural Research Program of the NICHD, NIH.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank E. P. Geiduschek and G. A. Kassavetis for helpful comments, as well as R. Intine, M. Bayfield and others for assistance and/or comments.
REFERENCES
- 1.Geiger SR, Lorenzen K, Schreieck A, Hanecker P, Kostrewa D, Heck AJ, Cramer P. RNA polymerase I contains a TFIIF-related DNA-binding subcomplex. Mol. Cell. 2010;39:583–594. doi: 10.1016/j.molcel.2010.07.028. [DOI] [PubMed] [Google Scholar]
- 2.Vannini A, Ringel R, Kusser AG, Berninghausen O, Kassavetis GA, Cramer P. Molecular basis of RNA polymerase III transcription repression by Maf1. Cell. 2010;143:59–70. doi: 10.1016/j.cell.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Richard P, Manley JL. Transcription termination by nuclear RNA polymerases. Genes Dev. 2009;23:1247–1269. doi: 10.1101/gad.1792809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottlieb E, Steitz JA. The RNA binding protein La influences both the accuracy and the efficiency of RNA polymerase III transcription in vitro. EMBO J. 1989;8:841–850. doi: 10.1002/j.1460-2075.1989.tb03445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dieci G, Sentenac A. Facilitated recycling by RNA polymerase III. Cell. 1996;84:245–252. doi: 10.1016/s0092-8674(00)80979-4. [DOI] [PubMed] [Google Scholar]
- 6.Fan H, Sakulich AL, Goodier JL, Zhang X, Qin J, Maraia RJ. Phosphorylation of the human La antigen on serine 366 can regulate recycling of RNA polymerase III transcription complexes. Cell. 1997;88:707–715. doi: 10.1016/s0092-8674(00)81913-3. [DOI] [PubMed] [Google Scholar]
- 7.Maraia RJ. Transcription termination factor La is also an initiation factor for RNA polymerase III. Proc. Natl Acad. Sci. USA. 1996;93:3383–3387. doi: 10.1073/pnas.93.8.3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bogenhagen DF, Brown DD. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981;24:261–270. doi: 10.1016/0092-8674(81)90522-5. [DOI] [PubMed] [Google Scholar]
- 9.Cozzarelli NR, Gerrard S, Schlissel M, Brown DD, Bogenhagen DF. Purified RNA polymerase III accurately and efficiently terminates transcription of 5S RNA genes. Cell. 1983;34:829–35. doi: 10.1016/0092-8674(83)90540-8. [DOI] [PubMed] [Google Scholar]
- 10.Huang Y, Maraia RJ. Comparison of the RNA polymerase III transcription machinery in S. pombe, S. cerevisiae and humans (Review) Nucleic Acids Res. 2001;29:2675–2690. doi: 10.1093/nar/29.13.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Campbell FE, Setzer DR. Transcription termination by RNA polymerase III: uncoupling of polymerase release from termination signal recognition. Mol. Cell. Biol. 1992;12:2260–2272. doi: 10.1128/mcb.12.5.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitehall SK, Bardeleben C, Kassavetis GA. Hydrolytic cleavage of nascent RNA in RNA polymerase III ternary transcription complexes. J. Biol. Chem. 1994;269:2299–2306. [PubMed] [Google Scholar]
- 13.Chedin S, Riva M, Schultz P, Sentenac A, Carles C. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev. 1998;12:3857–3871. doi: 10.1101/gad.12.24.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung AC, Cramer P. Structural basis of RNA polymerase II backtracking, arrest and reactivation. Nature. 2011;471:249–253. doi: 10.1038/nature09785. [DOI] [PubMed] [Google Scholar]
- 15.James P, Whelen S, Hall BD. The RET1 gene of yeast encodes the second-largest subunit of RNA polymerase III. J. Biol. Chem. 1991;266:5616–5624. [PubMed] [Google Scholar]
- 16.James P, Hall BD. ret1-1, a yeast mutant affecting transcription termination by RNA polymerase III. Genetics. 1990;125:293–303. doi: 10.1093/genetics/125.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bobkova EV, Habib N, Alexander G, Hall BD. Mutational analysis of the hydrolytic activity of yeast RNA polymerase III. J. Biol. Chem. 1999;274:21342–21348. doi: 10.1074/jbc.274.30.21342. [DOI] [PubMed] [Google Scholar]
- 18.Shaaban SA, Bobkova EV, Chudzik D, Hall BD. In vitro analysis of elongation and termination by mutant RNA polymerases with altered termination behavior. Mol. Cell. Biol. 1996;16:6468–76. doi: 10.1128/mcb.16.11.6468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shaaban SA, Krupp BM, Hall BD. Termination-altering mutations in the second-largest subunit of yeast RNA polymerase III. Mol. Cell. Biol. 1995;15:1467–1478. doi: 10.1128/mcb.15.3.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kassavetis GA, Prakash P, Shim E. The C53/C37 subcomplex of RNA polymerase III lies near the active site and participates in promoter opening. J. Biol. Chem. 2010;285:2695–2706. doi: 10.1074/jbc.M109.074013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lorenzen K, Vannini A, Cramer P, Heck AJ. Structural biology of RNA polymerase III: mass spectrometry elucidates subcomplex architecture. Structure. 2007;15:1237–1245. doi: 10.1016/j.str.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 22.Wang Z, Roeder RG. Three human RNA polymerase III-specific subunits form a subcomplex with a selective function in specific transcription initiation. Genes Dev. 1997;11:1315–1326. doi: 10.1101/gad.11.10.1315. [DOI] [PubMed] [Google Scholar]
- 23.Werner M, Chaussivert N, Willis IM, Sentenac A. Interaction between a complex of RNA polymerase III subunits and the 70- kDa component of transcription factor IIIB. J. Biol. Chem. 1993;268:20721–20724. [PubMed] [Google Scholar]
- 24.Sadhale PP, Woychik NA. C25, an essential RNA polymerase III subunit related to the RNA polymerase II subunit RPB7. Mol. Cell. Biol. 1994;14:6164–6170. doi: 10.1128/mcb.14.9.6164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sentenac A. Eukaryotic RNA polymerases. CRC critical reviews in biochemistry. 1985;18:31–90. doi: 10.3109/10409238509082539. [DOI] [PubMed] [Google Scholar]
- 26.Lane LA, Fernandez-Tornero C, Zhou M, Morgner N, Ptchelkine D, Steuerwald U, Politis A, Lindner D, Gvozdenovic J, Gavin AC, et al. Mass spectrometry reveals stable modules in holo and apo RNA polymerases I and III. Structure. 2011;19:90–100. doi: 10.1016/j.str.2010.11.009. [DOI] [PubMed] [Google Scholar]
- 27.Landrieux E, Alic N, Ducrot C, Acker J, Riva M, Carles C. A subcomplex of RNA polymerase III subunits involved in transcription termination and reinitiation. EMBO J. 2006;25:118–128. doi: 10.1038/sj.emboj.7600915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Y, Intine RV, Mozlin A, Hasson S, Maraia RJ. Mutations in the RNA Polymerase III Subunit Rpc11p That Decrease RNA 3′ Cleavage Activity Increase 3′-Terminal Oligo(U) Length and La-Dependent tRNA Processing. Mol. Cell. Biol. 2005;25:621–636. doi: 10.1128/MCB.25.2.621-636.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fernandez-Tornero C, Bottcher B, Riva M, Carles C, Steuerwald U, Ruigrok RW, Sentenac A, Muller CW, Schoehn G. Insights into transcription initiation and termination from the electron microscopy structure of yeast RNA polymerase III. Mol. Cell. 2007;25:813–823. doi: 10.1016/j.molcel.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 30.Cramer P. Recent structural studies of RNA polymerases II and III. Biochem. Soc. Trans. 2006;34:1058–1061. doi: 10.1042/BST0341058. [DOI] [PubMed] [Google Scholar]
- 31.Yee NS, Gong W, Huang Y, Lorent K, Dolan AC, Maraia RJ, Pack M. Mutation of RNA polymerase III subunit rpc2/polr3b leads to deficiency of the RNA cleavage subunit, Rpc11/Polr3k, and disrupts zebrafish digestive system development. PLoS Biol. 2007;5:2484–2492. doi: 10.1371/journal.pbio.0050312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fernandez-Tornero C, Bottcher B, Rashid UJ, Steuerwald U, Florchinger B, Devos DP, Lindner D, Muller CW. Conformational flexibility of RNA polymerase III during transcriptional elongation. EMBO J. 2010;29:3762–3772. doi: 10.1038/emboj.2010.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamada M, Sakulich AL, Koduru SB, Maraia R. Transcription termination by RNA polymerase III in fission yeast: A genetic and biochemical model system. J. Biol. Chem. 2000;275:29076–29081. doi: 10.1074/jbc.M003980200. [DOI] [PubMed] [Google Scholar]
- 34.Huang Y, Bayfield MA, Intine RV, Maraia RJ. Separate RNA-binding surfaces on the multifunctional La protein mediate distinguishable activities in tRNA maturation. Nat. Struct. Mol. Biol. 2006;13:611–618. doi: 10.1038/nsmb1110. [DOI] [PubMed] [Google Scholar]
- 35.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 36.Intine RVA, Sakulich AL, Koduru SB, Huang Y, Pierstorrf E, Goodier JL, Phan L, Maraia RJ. Control of transfer RNA maturation by phosphorylation of the human La antigen on serine 366. Mol. Cell. 2000;6:339–348. doi: 10.1016/s1097-2765(00)00034-4. [DOI] [PubMed] [Google Scholar]
- 37.Hamada M, Huang Y, Lowe TM, Maraia RJ. Widespread Use of TATA Elements in the Core Promoters for RNA Polymerases III, II, and I in Fission Yeast. Mol. Cell. Biol. 2001;21:6870–6881. doi: 10.1128/MCB.21.20.6870-6881.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Intine RV, Dundr M, Misteli T, Maraia RJ. Aberrant nuclear trafficking of La protein leads to disordered processing of associated precursor tRNAs. Mol. Cell. 2002;9:1113–1123. doi: 10.1016/s1097-2765(02)00533-6. [DOI] [PubMed] [Google Scholar]
- 39.Huang Y, McGillicuddy E, Weindel M, Dong S, Maraia R. The fission yeast TFIIB-related factor limits RNA polymerase III to a TATA-dependent pathway of TBP recruitment. Nucleic Acids Res. 2003;31:2108–2116. doi: 10.1093/nar/gkg301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bayfield MA, Kaiser TE, Intine RV, Maraia RJ. Conservation of a masked nuclear export activity of La proteins and its effects on tRNA maturation. Mol. Cell. Biol. 2007;27:3303–3312. doi: 10.1128/MCB.00026-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park JM, Intine RV, Maraia RJ. Mouse and Human La Proteins Differ in Kinase Substrate Activity and Activation Mechanism for tRNA Processing. Gene Expr. 2007;14:71–81. doi: 10.3727/105221607783417619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mao J, Schmidt O, Soll D. Dimeric transfer RNA precursors in S. pombe. Cell. 1980;21:509–516. doi: 10.1016/0092-8674(80)90488-2. [DOI] [PubMed] [Google Scholar]
- 43.Willis I, Schmidt P, Soll D. A selection for mutants of the RNA polymerase III transcription apparatus: PCF1 stimulates transcription of tRNA and 5S RNA genes. EMBO J. 1989;8:4281–4288. doi: 10.1002/j.1460-2075.1989.tb08614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmidt O, Mao J, Ogden R, Beckmann J, Sakano H, Abelson J, Soll D. Dimeric tRNA precursors in yeast. Nature. 1980;287:750–752. doi: 10.1038/287750a0. [DOI] [PubMed] [Google Scholar]
- 45.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 46.Braglia P, Percudani R, Dieci G. Sequence context effects on oligo(dT) termination signal recognition by Saccharomyces cerevisiae RNA polymerase III. J. Biol. Chem. 2005;280:19551–19562. doi: 10.1074/jbc.M412238200. [DOI] [PubMed] [Google Scholar]
- 47.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: an RNA polymerase II elongation complex at 3.3 A resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 48.Komissarova N, Becker J, Solter S, Kireeva M, Kashlev M. Shortening of RNA:DNA hybrid in the elongation complex of RNA polymerase is a prerequisite for transcription termination. Mol. Cell. 2002;10:1151–1162. doi: 10.1016/s1097-2765(02)00738-4. [DOI] [PubMed] [Google Scholar]
- 49.Santangelo TJ, Roberts JW. Forward translocation is the natural pathway of RNA release at an intrinsic terminator. Mol. Cell. 2004;14:117–126. doi: 10.1016/s1097-2765(04)00154-6. [DOI] [PubMed] [Google Scholar]
- 50.Ghavi-Helm Y, Michaut M, Acker J, Aude JC, Thuriaux P, Werner M, Soutourina J. Genome-wide location analysis reveals a role of TFIIS in RNA polymerase III transcription. Genes Dev. 2008;22:1934–1947. doi: 10.1101/gad.471908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Larson MH, Greenleaf WJ, Landick R, Block SM. Applied force reveals mechanistic and energetic details of transcription termination. Cell. 2008;132:971–982. doi: 10.1016/j.cell.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Armache KJ, Mitterweger S, Meinhart A, Cramer P. Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J. Biol. Chem. 2005;280:7131–7134. doi: 10.1074/jbc.M413038200. [DOI] [PubMed] [Google Scholar]