Abstract
CCTG tetranucleotide repeat expansion is associated with a hereditary neurological disease called myotonic dystrophy type 2 (DM2). The underlying reasons that lead to genetic instability and thus repeat expansion during DNA replication remains elusive. Here, we have shown CCTG repeats have a high propensity to form metastable hairpin and dumbbell structures using high-resolution nuclear magnetic resonance (NMR) spectroscopy. When the repeat length is equal to three, a hairpin with a two-residue CT loop is formed. In addition to the hairpin, a dumbbell structure with two CT-loops is formed when the repeat length is equal to four. Nuclear Overhauser effect (NOE) and chemical shift data reveal both the hairpin and dumbbell structures contain a flexible stem comprising a C-bulge and a T·T mismatch. With the aid of single-site mutation samples, NMR results show these peculiar structures undergo dynamic conformational exchange. In addition to the intrinsic flexibility in the stem region of these structures, the exchange process also serves as an origin of genetic instability that leads to repeat expansion during DNA replication. The structural features provide important drug target information for developing therapeutics to inhibit the expansion process and thus the onset of DM2.
INTRODUCTION
Approximately 30 hereditary disorders in human have been found to be caused by the expansion of unstable DNA repeating sequences (1). Among them, myotonic dystrophy (DM) is the most common muscular dystrophy in adults, and is characterized by hyperexcitability of skeletal muscle (myotonia) and muscle degeneration (myopathy), a conduction defect in cardiac muscle cells and cataracts (2,3). There are two major types of DM, namely, myotonic dystrophy type 1 (DM1) and myotonic dystrophy type 2 (DM2). DM1 is induced by the unstable trinucleotide CTG repeat expansion in the DMPK gene (4,5), whereas DM2 is related to tetranucleotide CCTG repeat expansion in the first intron of the ZNF9 gene (3). Both DM1 and DM2 are caused by the expanded repeats transcribed into RNA but not translated into protein (6), with the pathogenic effect of the RNA transcribed from the expanded allele containing the long tracts of (CUG)n and (CCUG)n repeats, respectively (7,8).
Although the clinical myotonia in DM2 is usually milder and less pathognomonic than in DM1 (9,10), CCTG expansion can be much larger than that of CTG. In fact, CCTG expansion is by far the largest expansion observed (11), with alleles ranging in size from ∼75 to 11 000 repeats (9,12), whereas CTG expansion only involves 50–2000 repeats. Earlier biochemical studies have suggested CCTG repeats lack the capacity to adopt a defined base-paired hairpin structure, contrary to the complementary CAGG repeats (13,14). Nevertheless, recent gel mobility and genetic assays have shown that both CCTG and CAGG repeats can form slipped-strand structures (11,15). Yet, no detailed features of the hairpin or slipped-strand structures have been reported. In addition, the underlying reasons for how these repeats can contribute to genetic instability and cause such highly variable and multifaceted diseases are still unresolved (16,17).
Despite all DNA repeating sequences known to undergo expansions and lead to human neurological diseases can form one or several alternative conformations such as hairpin, slipped-strand triplex, quadruplex or unwound DNA structures (18), structural studies using, for example, X-ray crystallographic or NMR solution methods are not always successful. For X-ray crystallographic studies, the growth of diffraction-quality crystals has been limited by the intrinsic flexibility of these repeating sequences. For NMR solution structure studies, the difficulties come from the severe signal overlap due to the repetitive nature of repeating sequences and the serious peak broadening due to the presence of conformational exchange and the large molecular size of longer repeats. The dynamic nature of these repeats may also affect the structural studies using biochemical methods such as enzymatic and chemical probing. To circumvent these problems, our group previously differentiated the NMR signals from different conformers of CTG repeats through alternating the experimental conditions in order to disturb the conformational populations (19). We successfully determined the solution structural features of CTG repeating sequences and revealed that the repeat length governs the CTG hairpin structures. Here, through the use of single-site mutation samples, we have successfully identified the presence of dynamically exchanging hairpin and dumbbell structures in CCTG repeating sequences by 1H and 31P NMR spectroscopy. In addition to conformational exchange, these structures also contain a flexible stem comprising a C-bulge and a T·T mismatch. These exchange processes and flexible structural elements provide an account for the origin of genetic instability in CCTG repeats that leads to repeat expansion during DNA replication.
MATERIALS AND METHODS
DNA samples were synthesized using an Applied Biosystems model 394 DNA synthesizer and purified using denaturing polyacrylamide gel electrophoresis and diethylaminoethyl Sephacel anion exchange column chromatography. The samples were then desalted using Amicon Ultra-4 centrifugal filter devices. NMR samples were prepared by dissolving 0.5 µmol of purified DNA into 500 µl of buffer solution containing 150 mM sodium chloride, 10 mM sodium phosphate (pH 7.0) and 0.1 mM 2,2-dimethyl-2-silapentane-5-sulfonic acid (DSS).
NMR experiments were performed using Bruker AV-500 and AV-700 spectrometers operating at 500.30 MHz and 700.21 MHz, respectively. Imino proton spectra were acquired with samples in 10% D2O using the water suppression by gradient-tailored excitation (WATERGATE) pulse sequence (20,21). 2D WATERGATE-nuclear Overhauser effect spectroscopy (NOESY) experiments were performed with a mixing time of 300 ms. For studying non-labile proton signals, 2D NOESY with a mixing time of 300 ms, total correlation spectroscopy (TOCSY) with a mixing time of 75 ms and double quantum filtered-correlation spectroscopy (DQF-COSY) were performed with samples in 100% D2O. For studying conformational exchange, 2D rotating-frame Overhauser effect spectroscopy (ROESY) and 31P–31P exchange spectroscopy (EXSY) (22) with a mixing time of ∼100−200 ms were performed. A 4 k × 180 dataset with at least 96 scans was collected for each EXSY spectrum. The 31P spectral width was set to 9 ppm with the carrier frequency positioned at −3.8 ppm. Proton decoupling was executed by WALTZ-16 (23) composite pulse decoupling sequence during the EXSY acquisition period. The data matrix was finally zero-filled to give a 4 k × 1 k dataset with exponential multiplication window function applied to both dimensions. Backbone 31P signals were assigned using 2D TOCSY and 1H–31P heteronuclear single quantum coherence spectroscopy (HSQC) experiments. 31P chemical shifts were indirectly referenced to DSS using the derived nucleus-specific ratio of 0.404 808 636 (24).
RESULTS
CCTG hairpin with a two-residue CT-loop
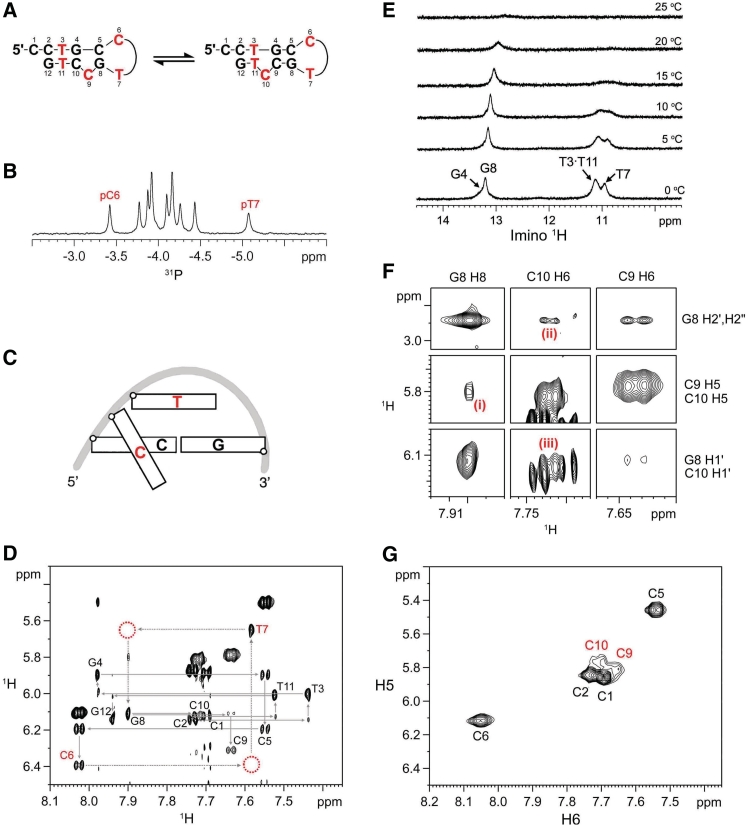
The formation of a hairpin structure containing a two-residue CT-loop (Figure 1A) could be observed when the CCTG repeat length became three as revealed by the 31P and 1H NMR data. The presence of CT-loop is supported by the unusually downfield pC6 (−3.42 ppm) and upfield pT7 (−5.07 ppm) 31P chemical shifts (Figure 1B), when compared to those of B-DNA (−4.6 to −3.0 ppm) (25) and random coil DNA (−4.15 to −3.87 ppm) (26). The CT-loop adopts the type II conformation (Figure 1C) in which the unusual shifts of C6 and T7 arise from the different backbone orientation with its preceding C5 in the stem and the local sharp turn in the backbone of the two nucleotides in the loop, respectively (27).
Figure 1.
(CCTG)3 adopts a hairpin structure. (A) The hairpin structure contains a two-residue CT-loop and a flexible stem comprising a shifting C-bulge and a T·T mismatch. (B) The downfield pC6 and upfield pT7 31P shifts at 25°C support the presence of CT-loop. (C) A schematic representation of type II loop. The second cytosine (in red) is positioned in the minor groove and perpendicular to the loop closing base pair, whereas the thymine stacks over the loop closing base pair. (D) NOESY H1′–H6/H8 fingerprint region of (CCTG)3 at 25°C. The unusually downfield C6 H5, H6 and H1′, and upfield T7 H1′ indicate the formation of CT-loop. The open circles show the missing sequential internucleotide NOEs. (E) Two weak guanine imino signals appeared at lower temperatures in G·C Watson−Crick region at ∼13.2 ppm. Owing to the end-fraying of the terminal base pair, G12 imino signal was not observed. The imino signal at ∼10.95 ppm was assigned to T7, of which it has a similar imino chemical shift of the CT-loop. (F) The C9-bulge conformer is supported by (i) G8 H8–C10 H5, (ii) G8 H2′/H2″–C10 H6, and (iii) G8 H1′–C10 H6 NOEs at 25°C. The C10-bulge conformer is less populated and supported by the sequential NOEs between G8 and C9. (G) TOCSY spectrum at 15°C shows broadening of C9 and C10 signals due to the shifting C-bulge.
Owing to the type II loop conformation, the sequential inter-nucleotide C6 H1′–T7 H6 and T7 H1′–G8 H8 NOEs were not observed in the NOESY H1′–H6/H8 fingerprint region (Figure 1D). In addition, the H5 (6.11 ppm) and H6 (8.03 ppm) chemical shifts of C6 were unusually downfield than those of B-DNA (5.25–5.95 and 7.15–7.60 ppm, respectively) (25) and random coil DNA (5.80–6.05 and 7.40–7.80 ppm, respectively) (28) because the C6 base is positioned in the minor groove and perpendicular to the loop closing base pair. The T7 H1′ chemical shift was unusually upfield (5.65 ppm) due to its sugar stacks over the base plane of the closing base pair. All these unusual shifts agree well with the characteristic peaks of CT-loop (27). The formation of a CT-loop in (CCTG)3 suggests C5 and G8 form a C·G closing base pair. From the 1H NMR imino region, two weak signals were observed in the Watson–Crick region (∼13–14 ppm) at lower temperatures (Figure 1E). The presence of an NOE between the sharper imino at 13.20 ppm and C5 amino (Supplementary Figure S1A) indicates the formation of a G8·C5 Watson–Crick base pair, confirming (CCTG)3 forms a hairpin structure with a CT-loop.
A flexible stem in CCTG hairpin
Interestingly, this hairpin structure does not resemble any of the proposed and predicted structures (13,29). It forms a two-residue CT-loop and contains neither tandem C·T nor G·T mismatches in the stem region. Instead, it has a flexible stem comprising a C-bulge and a T·T mismatch (Figure 1A) as revealed by the NOE and imino data. The complete sequential walk from C1 to C5 (Figure 1D) shows that the nucleotides along the 5′-end were well-stacked before reaching the CT-loop. On the contrary, the 3′-end was more dynamic due to its realignment with the 5′-end to form a T·T mismatch and a C-bulge shifting between C9 and C10 positions (Figure 1A). The presence of C9-bulge conformer is supported by the NOEs between G8 and C10 including (i) G8 H8–C10 H5, (ii) G8 H2′/H2″–C10 H6 and (iii) G8 H1′–C10 H6 NOEs (Figure 1F). The C10-bulge conformer is supported by the sequential NOEs between G8 and C9 despite the C9 H1′–T11 H6 NOE is weak which suggests it is less populated. The shifting C-bulge is further supported by the severe peak broadening of C9 and C10 in their TOCSY (Figure 1G) and NOESY spectra (Supplementary Figure S1B) at 15°C while the signals of G8, T11 and G12 remain sharp. The dynamics of this shifting C-bulge also accounts for the unusually weak imino signals (Figure 1E), at which the small broad signal at ∼13.26 ppm was assigned to the averaged G4 imino of the two conformers.
The presence of T·T mismatch in the hairpin stem was evidenced by the broad imino signal at ∼11.12 ppm (Figure 1E), which was assigned to the averaged signals of T3 and T11 of a wobble T3·T11 mismatch with two possible pairing modes (Supplementary Figure S1C) (30–32). This averaged signal was broad owing to the shifting C-bulge which continuously affect the electronic environment of the T·T mismatch. On the contrary, the formation of tandem C·T mismatches in the stem region is less likely since their thymine imino signals would not be observed (33,34).
To verify the formation of a C-bulge and a T·T mismatch in the hairpin stem region, the hairpin structures of (CCTG)3 were stabilized by the addition of a G at the 3′-end (Supplementary Figure S2A). With such stabilization, the imino signals of G4, G8 and G12 were observed and successfully assigned (Supplementary Figure S2B), indicating the formation of G4·C10, G8·C5 and G12·C2 Watson–Crick base pairs and thus the presence of C-bulge and T·T mismatch in the hairpin stem region. As revealed from G4·C10 imino-amino NOE (Supplementary Figure S2B) and the less broadened C10 H5−H6 TOCSY signal (Supplementary Figure S2C), the stabilized hairpin stem reduced the dynamics of the C-bulge, causing the C9-bulge conformer became predominant and the T3 and T11 imino signals were resolved at ∼10.9 and 11.3 ppm, respectively (Supplementary Figure S2D). Based on the sequential NOE connectivities (Supplementary Figure S2E), the distinctive NOEs from the T11–G12–G13 fragment allow unambiguous assignment of T11 H6 and other base proton signals. The presence of neighboring C-bulge in the stem region causes T11 H6 to be more downfield than T3 H6, which has also been observed in the sequential NOE assignment of (CCTG)3 (Figure 1D).
CCTG dumbbell with two CT-loops
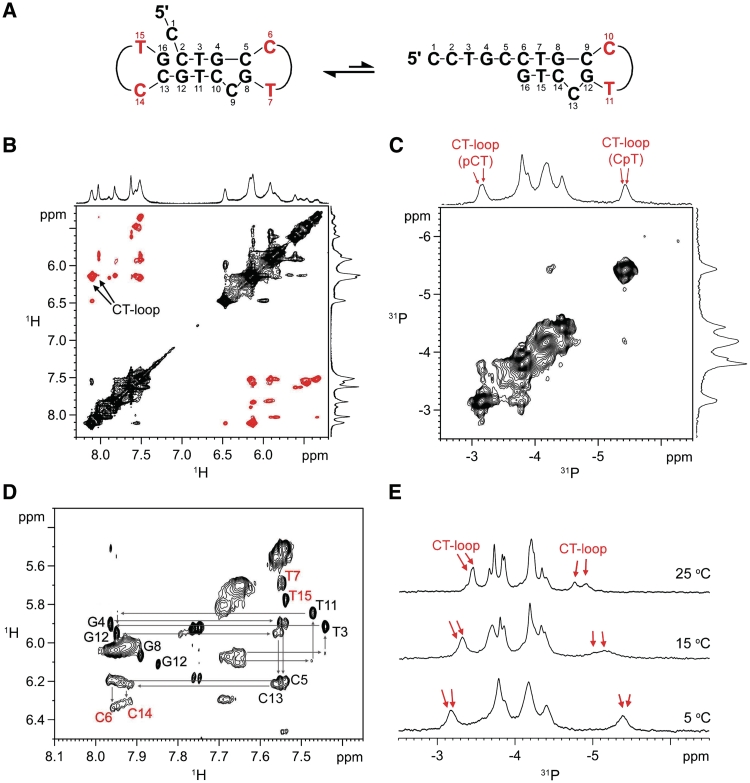
In addition to the formation of hairpin, CCTG repeats can also adopt a dumbbell structure, as observed in (CCTG)4 (Figure 2A). The identification of the dumbbell structure was less straightforward due to the presence of conformational exchange. This was evidenced by the exchange cross peaks in 2D 1H-1H ROESY (Figure 2B) and 31P-31P EXSY (Figure 2C) spectra at 0°C. The NMR features of CT-loop (27) including (i) the unusually downfield signals of cytosine H5 (∼6.13 ppm) and H6 (∼8.11 ppm) (Figure 2B), and (ii) the characteristic 31P signals of pCT (−3.14 ppm) and CpT (−5.42 ppm) (Figure 2C) were also observed. Meanwhile, upon increasing the temperature to 25°C, only one set of peaks was observed and the sequential NOE assignment (Figure 2D) shows two CT-loops were formed by C6 and T7 of the second repeat, and C14 and T15 of the fourth repeat, respectively, resulting in a dumbbell structure. The characteristic 31P CT-loop signals (Figure 2E) also support the presence of two CT-loops. The characteristic 1H and 31P chemical shifts of CT-loops were quite distinctly retained at 25°C, suggesting a prominent population of the dumbbell structure in the exchange equilibrium.
Figure 2.
CCTG repeats can also adopt a dumbbell structure. (A) The dumbbell structure contains two CT-loops. It undergoes conformational exchange with the hairpin conformer in (CCTG)4. (B) At 0°C, ROESY exchange cross peaks (in black) were observed. (C) 2D 31P-31P EXSY spectrum also shows exchange cross peaks at 0°C. (D) 2D NOESY fingerprint region of (CCTG)4 shows the two CT-loops were formed by C6 and T7, and C14 and T15, respectively. Only one single set of peaks was observed at 25°C. (E) Two upfield and two downfield 31P signals were observed.
In order to unravel the solution structural features of the exchanging conformers and verify the dumbbell structure being predominant, four single-site mutational (CCTG)4 samples were prepared, in which the second cytosine of each repeat was substituted by a thymine. Since both CT- and TT-loops adopt the same type II loop (27,35,36), such substitutions would not affect much of the overall structure. More importantly, they help identify which CCTG repeat would preferentially form a CT-loop as TT-loops can be easily distinguished by their (i) characteristic downfield methyl 1H signals of the first thymine residue (27,35), and (ii) 31P chemical shifts of the second thymine residue which is more upfield than those of CT-loops (35).
First repeat not involved in forming loop
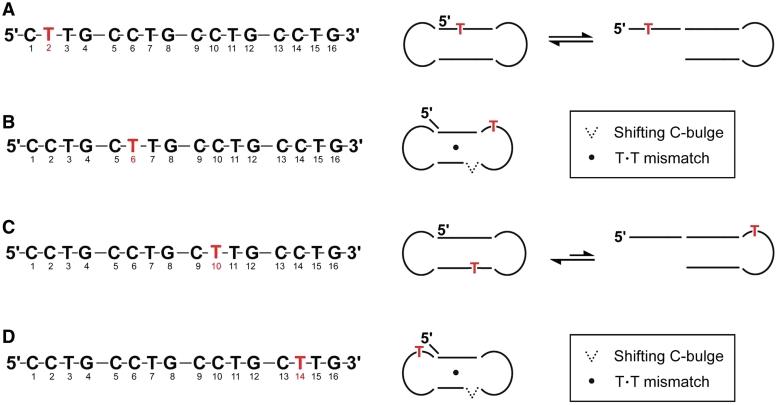
In (CCTG)4-C2T, the second cytosine of the first repeat was substituted with a thymine. The absence of an unusually downfield thymine methyl 1H signal at ∼2.05 ppm (Supplementary Figure S3A) and an enormously upfield 31P signal at approximately −6.0 ppm (Supplementary Figure S3B) indicates no TT-loop was formed, suggesting the first repeat of (CCTG)4 is less prone to form CT-loop. The spectral features in the ROESY (Supplementary Figure S3C) and EXSY (Supplementary Figure S3D) spectra were similar to those of (CCTG)4, suggesting a dumbbell conformer is in exchange with another conformer. Since the first repeat was less prone to form CT-loop, it is likely that this conformer is a hairpin with a CT-loop formed by the third repeat (Figure 3A).
Figure 3.
Dumbbell conformer also occurs in single-site mutation samples. (A) (CCTG)4-C2T. (B) (CCTG)4-C6T. (C) (CCTG)4-C10T. (D) (CCTG)4-C14T. A single dumbbell structure comprising a CT- and TT-loop was observed in (CCTG)4-C6T and (CCTG)4-C14T. The stem region contains a one-residue 5′-overhang, a T·T mismatch and a shifting C-bulge.
Second repeat involved in forming loop
When the second cytosine of the second repeat of (CCTG)4 was substituted with a thymine, a single set of peaks was observed (Supplementary Figure S4). As revealed in the EXSY spectrum (Supplementary Figure S4A), no conformational exchange was present. The characteristic downfield T6 H6 (Supplementary Figure S4B) and T6 methyl (Supplementary Figure S4C) 1H peaks, and the enormously upfield pT7 31P signal (Supplementary Figure S4D) indicate the formation of a TT-loop by T6 and T7. In addition, the downfield C14 H5 and H6 chemical shifts (Supplementary Figure S4B) and characteristic 31P signals (Supplementary Figure S4D) reveal the formation of a CT-loop by C14 and T15 in the fourth repeat, resulting in a dumbbell structure with a TT- and a CT-loop (Figure 3B). These loops were further supported by the formation of G8·C5 and G16·C13 Watson−Crick base pairs as revealed by their imino-amino and imino-imino NOEs (Supplementary Figure S4E and F).
In this dumbbell structure, an unusual NOE was interestingly observed between G16 and C2 (Supplementary Figure 4B), which suggest G16 and C2 stacked well upon each other via intra-molecular 3′−5′ terminal stacking interaction (37,38), while C1 extruded from the stem and formed a one-residue 5′-overhang (Figure 3B). Similar to the stem region of (CCTG)3 hairpin, the peak broadening in C9 and C10 TOCSY H5−H6 cross peaks (Supplementary Figure S4G) and the mismatch T·T imino signals (Supplementary Figure S4H) support the presence of a shifting C-bulge and T·T mismatch in the stem region, respectively. The T imino signals at ∼11 ppm were contributed by the T3·T11 mispair which overlapped with those of T7 and T15 in the two-residue loops.
A small population of hairpin with the third repeat involved in loop
Upon substituting the second cytosine in the third repeat of (CCTG)4, conformational exchange between a major dumbbell structure and a minor hairpin structure with the third repeat forming a TT-loop was present (Figure 3C). A small but distinctively upfield 31P signal appeared at approximately −6.0 ppm (Supplementary Figure S5A), indicating the formation of a minor conformer with a TT-loop. The characteristic downfield T10 methyl 1H signal was also observed at ∼2.08 ppm (Supplementary Figure S5B), which was partially overlapped with other signals. While the minor conformer contains the TT-loop, the major conformer was a dumbbell containing two CT-loops as revealed by two intense downfield cytosine H5−H6 TOCSY signals (Supplementary Figure S5C). Conformational exchange was present between the two conformers, as shown in 2D EXSY (Supplementary Figure S5D) and 2D ROESY (Supplementary Figure S5E and F) spectra.
Fourth repeat also involved in forming loop
A stable dumbbell structure was also observed at 25°C upon substituting the second cytosine in the fourth repeat of (CCTG)4 with a thymine (Figure 3D). The dumbbell structure contains a CT-loop and a TT-loop, as evidenced by its characteristic 1H (Supplementary Figure S6A) and 31P (Supplementary Figure S6B) signals. Similar to (CCTG)4-C6T, the EXSY spectrum shows no conformational exchange in (CCTG)4-C14T (Supplementary Figure S6C). A one-residue 5′-overhang and intra-molecular 3′−5′ terminal stacking interaction was also present between G16 and C2 as supported by the G16−C2 NOE (Supplementary Figure S6D). In addition, a shifting C-bulge and a T·T mismatch were also observed in the stem region of the dumbbell structure as evidenced by the peak broadening of C9 and C10 H5−H6 TOCSY cross peaks (Supplementary Figure S6E) and the mismatch imino signals (Supplementary Figure S6F), respectively.
The results from the single-site mutational studies show that the first repeat of (CCTG)4 has low propensity to form CT-loop, inferring the two exchanging conformers in (CCTG)4 include a peculiar dumbbell structure containing two CT-loops in the second and fourth repeats, and a hairpin structure containing a CT-loop in the third repeat. Both of the conformers contain a flexible stem region comprising a shifting C-bulge and a T·T mismatch. The dynamics from the stem regions probably contributes to the conformational exchange process between these conformers.
Solution structures of longer CCTG repeats
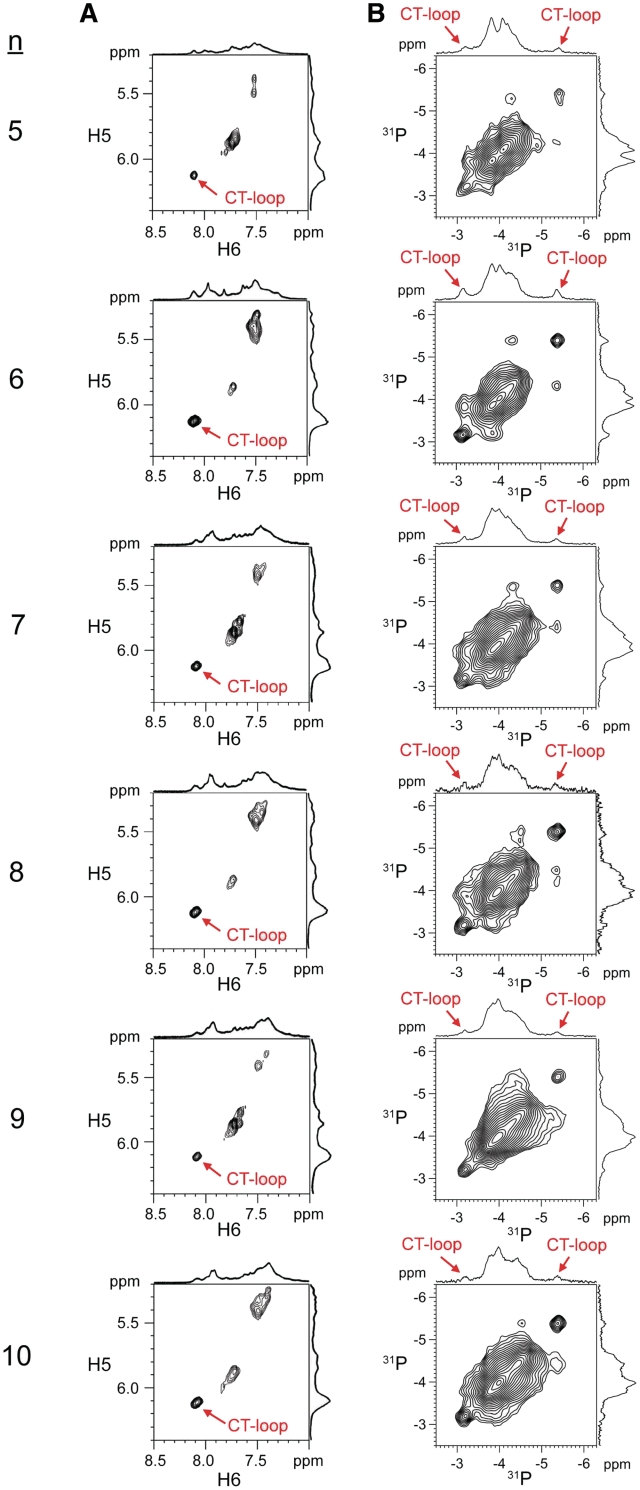
In order to investigate if the dumbbell and hairpin structures were also present in longer CCTG repeats, 1H and 31P NMR studies were extended to CCTG samples containing five to ten repeats. Although peak broadening and overlap were severe, the NMR features of CT-loop were still observed from the characteristic 1H (Figure 4A) and 31P signals (Figure 4B), suggesting the formation of dumbbell and hairpin conformers is possible. For longer repeats, it is likely that several forms of the hairpin and dumbbell conformers are present. The increase in conformational space also allows a more feasible exchange among different conformers. As evidenced by the 31P−31P EXSY spectra, the distinctive 31P signals show exchange cross peaks with the main band 31P signals (Figure 4B), indicating conformational exchange is also present in these longer repeats.
Figure 4.
CT-loops and conformational exchange are also present in longer CCTG repeats. (A) TOCSY cytosine H5–H6 cross peaks of (CCTG)5–10 at 0°C. The unusually downfield H5 and H6 cross peaks indicate the presence of CT-loop. (B) Conformational exchange was evidenced by the 31P−31P EXSY cross peaks at 5°C.
DISCUSSION
Significance of the dumbbell conformer
As revealed in the structural features of (CCTG)4, both the dumbbell and hairpin conformers contain the CT-loop. The formation of dumbbell conformer can be considered by folding the last repeat of a CCTG hairpin structure with a 3′-overhanging stem (Figure 5A). This 3′-terminal folding process may serve as a pathway to further stabilize the hairpin structure through the intra-molecular 3′−5′ terminal stacking interaction. To identity if folding of the last repeat would also occur in longer repeats, the second cytosine in the last repeat of (CCTG)5 and (CCTG)6 was substituted with a thymine to form the single-site mutation samples, (CCTG)5-C18T and (CCTG)6-C22T, respectively. The co-existence of both TT- and CT-loops was observed in both cases as evidenced by their characteristic 1H and 31P signals (Supplementary Figures S7 and S8). 31P−31P EXSY spectra also show these distinctive 31P signals are in exchange with the main band 31P signals (Supplementary Figures S7D and S8D), suggesting conformational exchange is also present in longer repeats.
Figure 5.
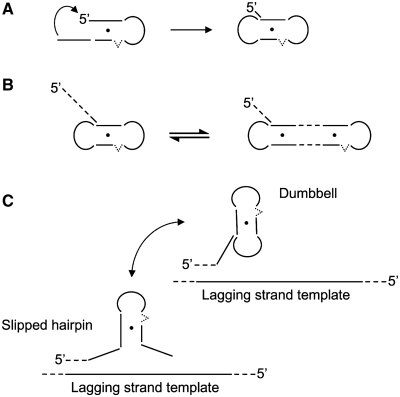
Significance of CCTG dumbbell structure. (A) Folding of the last repeat of a CCTG hairpin structure with a 3′-overhanging stem results in a dumbbell structure. (B) Possible dumbbell structures in longer repeats. The dumbbell structure can be formed by the last four repeats with a long 5′-overhang. Alternatively, realignment of the repeat strand can occur, leading to the formation of a dumbbell structure with shorter 5′-overhang but a longer stem. (C) Formation of the dumbbell structure in CCTG repeats stabilizes the slipped hairpin structure and thereby increasing the formation propensity of unusual structure during DNA replication.
Implication from the 5′-overhang of (CCTG)4
Owing to the increase in complexity of the exchange process, detailed identification of the exchanging conformers is not feasible. Nevertheless, the presence of TT-loop in (CCTG)5-C18T and (CCTG)6-C22T suggests the formation of dumbbell conformer is possible. The one-residue 5′-overhang observed in the dumbbell structure of (CCTG)4 infers that formation of 5′-overhang in longer repeats may be essential in maintaining dumbbell structures. In longer repeats, it is possible that the dumbbell structure can be formed by the last four repeats, with a long 5′-overhang (Figure 5B). If realignment of the repeat strand occurs, this can lead to the formation of a dumbbell structure with a shorter 5′-overhang but a longer stem region containing more than one T·T mismatch. Thereby, it is expected that the genetic instability will increase with repeat length. Based on these results, we propose that when fragments of CCTG repeats are synthesized opposite to the lagging strand template during DNA replication, expansion of CCTG repeats will be resulted if slippage of these fragments leads to the formation of CCTG hairpins. As these hairpins contain shifting C-bulge and T·T mismatches in the stem region, the internal dynamics will contribute to instability and realignment of these repeats. The likelihood of CCTG repeat expansion increases as these metastable hairpins will further be stabilized by folding of the last CCTG repeat to form dumbbell structure, thereby increasing the formation propensity of unusual structures (Figure 5C).
Genetic instability in CCTG repeats
The structural outcomes from (CCTG)3 and (CCTG)4 reveal that CCTG repeats form exchanging dumbbell and hairpin structures. Nevertheless, the shifting C-bugle and T·T mismatch in the stem regions of these structures provide the internal dynamics that contributes to the inherent flexibility in these structure. For longer repeats, we have shown that they also adopt structures containing CT-loops and undergo conformational exchange. Thereby, it is likely that the genetic instability of CCTG repeats originates from the conformational exchange process and internal dynamics that comes from the shifting C-bugle and T·T mismatch in the stem region.
As conformational space increases with repeat length, it is possible that multiple forms of hairpin and dumbbell structures can be adopted by long CCTG repeats. Earlier enzymatic and chemical probing studies on CCTG repeats showed equal intensities of cleavage along the repeats and thus suggested CCTG repeats do not adopt any stable secondary structures (13). These cleavage patterns were probably due to exchange among different conformers, which originated from the flexible stem within each conformer. The results of our present work reveal CCTG repeats can adopt the dumbbell and hairpin conformers. More importantly, the CT-loop and the shifting C-bugle and T·T mismatch in the stem region of these conformers may serve as important therapeutic targets to hamper the repeat expansion process and thus the onset of DM2.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Research Grants Council of the Hong Kong Special Administrative Region (Project No. CUHK401206); UGC Special Equipment Grant CUHK/09. Funding for open access charge: The Chinese University of Hong Kong.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Professors H.K Lee, H.N.C. Wong and J.C. Yu for their continuing support on the research activities of our group.
REFERENCES
- 1.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 2.Mankodi A, Logigian E, Callahan L, McClain C, White R, Henderson D, Krym M, Thornton CA. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 3.Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, Day JW, Ranum LP. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- 4.Mahadevan M, Tsilfidis C, Sabourin L, Shutler G, Amemiya C, Jansen G, Neville C, Narang M, Barcelo J, O'Hoy K, et al. Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science. 1992;255:1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- 5.Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, Hunter K, Stanton VP, Thirion JP, Hudson T, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 6.Cho DH, Tapscott SJ. Myotonic dystrophy: emerging mechanisms for DM1 and DM2. Biochim. Biophys. Acta. 2007;1772:195–204. doi: 10.1016/j.bbadis.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 7.Lee JE, Cooper TA. Pathogenic mechanisms of myotonic dystrophy. Biochem. Soc. Trans. 2009;37:1281–1286. doi: 10.1042/BST0371281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ranum LP, Day JW. Myotonic dystrophy: RNA pathogenesis comes into focus. Am. J. Hum. Genet. 2004;74:793–804. doi: 10.1086/383590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salisbury E, Schoser B, Schneider-Gold C, Wang GL, Huichalaf C, Jin B, Sirito M, Sarkar P, Krahe R, Timchenko NA, et al. Expression of RNA CCUG repeats dysregulates translation and degradation of proteins in myotonic dystrophy 2 patients. Am. J. Pathol. 2009;175:748–762. doi: 10.2353/ajpath.2009.090047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vihola A, Bassez G, Meola G, Zhang S, Haapasalo H, Paetau A, Mancinelli E, Rouche A, Hogrel JY, Laforet P, et al. Histopathological differences of myotonic dystrophy type 1 (DM1) and PROMM/DM2. Neurology. 2003;60:1854–1857. doi: 10.1212/01.wnl.0000065898.61358.09. [DOI] [PubMed] [Google Scholar]
- 11.Edwards SF, Hashem VI, Klysik EA, Sinden RR. Genetic instabilities of (CCTG).(CAGG) and (ATTCT).(AGAAT) disease-associated repeats reveal multiple pathways for repeat deletion. Mol. Carcinog. 2009;48:336–349. doi: 10.1002/mc.20534. [DOI] [PubMed] [Google Scholar]
- 12.Ranum LP, Day JW. Pathogenic RNA repeats: an expanding role in genetic disease. Trends Genet. 2004;20:506–512. doi: 10.1016/j.tig.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Dere R, Napierala M, Ranum LP, Wells RD. Hairpin structure-forming propensity of the (CCTG•CAGG) tetranucleotide repeats contributes to the genetic instability associated with myotonic dystrophy type 2. J. Biol. Chem. 2004;279:41715–41726. doi: 10.1074/jbc.M406415200. [DOI] [PubMed] [Google Scholar]
- 14.Dere R, Wells RD. DM2 CCTG•CAGG repeats are crossover hotspots that are more prone to expansions than the DM1 CTG•CAG repeats in Escherichia coli. J. Mol. Biol. 2006;360:21–36. doi: 10.1016/j.jmb.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 15.Edwards SF, Sirito M, Krahe R, Sinden RR. A Z-DNA sequence reduces slipped-strand structure formation in the myotonic dystrophy type 2 (CCTG) • (CAGG) repeat. Proc. Natl Acad. Sci. USA. 2009;106:3270–3275. doi: 10.1073/pnas.0807699106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakamori M, Thornton C. Epigenetic changes and non-coding expanded repeats. Neurobiol. Dis. 2010;39:21–27. doi: 10.1016/j.nbd.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gatchel JR, Zoghbi HY. Diseases of unstable repeat expansion: mechanisms and common principles. Nat. Rev. Genet. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 18.Voineagu I, Freudenreich CH, Mirkin SM. Checkpoint responses to unusual structures formed by DNA repeats. Mol. Carcinog. 2009;48:309–318. doi: 10.1002/mc.20512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chi LM, Lam SL. Structural roles of CTG repeats in slippage expansion during DNA replication. Nucleic Acids Res. 2005;33:1604–1617. doi: 10.1093/nar/gki307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 21.Sklenar V, Piotto M, Leppik R, Saudek V. Gradient-tailored water suppression for 1H-15N HSQC experiments optimized to retain full sensitivity. J. Magn. Reson. Ser. A. 1993;102:241–245. [Google Scholar]
- 22.Jeener J, Meier BH, Bachmann P, Ernst RR. Investigation of exchange processes by two-dimensional NMR spectroscopy. J. Chem. Phys. 1979;71:4546–4563. [Google Scholar]
- 23.Shaka AJ, Keeler J, Frenkiel T, Freeman R. An improved sequence for broadband decoupling: WALTZ-16. J. Magn. Reson. 1983;52:335–338. [Google Scholar]
- 24.Markley JL, Bax A, Arata Y, Hilbers CW, Kaptein R, Sykes BD, Wright PE, Wüthrich K. Recommendations for the presentation of NMR structures of proteins and nucleic acids. IUPAC-IUBMB-IUPAB Inter-Union Task Group on the standardization of data bases of protein and nucleic acid structures determined by NMR spectroscopy. J. Biomol. NMR. 1998;12:1–23. doi: 10.1023/a:1008290618449. [DOI] [PubMed] [Google Scholar]
- 25.Lam SL, Chi LM. Use of chemical shifts for structural studies of nucleic acids. Prog. Nucleic Magn. Reson. Spectros. 2010;56:289–310. doi: 10.1016/j.pnmrs.2010.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Ho CN, Lam SL. Random coil phosphorus chemical shift of deoxyribonucleic acids. J. Magn. Reson. 2004;171:193–200. doi: 10.1016/j.jmr.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 27.Ippel HH, van den Elst H, van der Marel GA, van Boom JH, Altona C. Structural similarities and differences between H1- and H2-family DNA minihairpin loops: NMR studies of octameric minihairpins. Biopolymers. 1998;46:375–393. [Google Scholar]
- 28.Lam SL, Ip LN, Cui X, Ho CN. Random coil proton chemical shifts of deoxyribonucleic acids. J. Biomol. NMR. 2002;24:329–337. doi: 10.1023/a:1021671531438. [DOI] [PubMed] [Google Scholar]
- 29.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gervais V, Cognet JA, Le Bret M, Sowers LC, Fazakerley GV. Solution structure of two mismatches A•A and T•T in the K-ras gene context by nuclear magnetic resonance and molecular dynamics. Eur. J. Biochem. 1995;228:279–290. [PubMed] [Google Scholar]
- 31.Kouchakdjian M, Li BF, Swann PF, Patel DJ. Pyrimidine•pyrimidine base-pair mismatches in DNA. A nuclear magnetic resonance study of T•T pairing at neutral pH and C•C pairing at acidic pH in dodecanucleotide duplexes. J. Mol. Biol. 1988;202:139–155. doi: 10.1016/0022-2836(88)90526-8. [DOI] [PubMed] [Google Scholar]
- 32.Arnold FH, Wolk S, Cruz P, Tinoco I., Jr Structure, dynamics, and thermodynamics of mismatched DNA oligonucleotide duplexes d(CCCAGGG)2 and d(CCCTGGG)2. Biochemistry. 1987;26:4068–4075. doi: 10.1021/bi00387a049. [DOI] [PubMed] [Google Scholar]
- 33.Jaishree TN, Wang AH. NMR studies of pH-dependent conformational polymorphism of alternating (C-T)n sequences. Nucleic Acids Res. 1993;21:3839–3844. doi: 10.1093/nar/21.16.3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jaishree TN, Wang AH. Conformations of the alternating (C-T)n sequence under neutral and low pH. FEBS Lett. 1994;337:139–144. doi: 10.1016/0014-5793(94)80261-0. [DOI] [PubMed] [Google Scholar]
- 35.Ippel JH, Lanzotti V, Galeone A, Mayol L, van den Boogaart JE, Pikkemaat JA, Altona C. An NMR study of the conformation and thermodynamics of the circular dumbbell d<pCGC-TT-CGC-TT> slow exchange between two- and four-membered hairpin loops. J. Biomol. Struct. Dyn. 1992;9:821–836. doi: 10.1080/07391102.1992.10507961. [DOI] [PubMed] [Google Scholar]
- 36.van Dongen MJ, Wijmenga SS, van der Marel GA, van Boom JH, Hilbers CW. The transition from a neutral-pH double helix to a low-pH triple helix induces a conformational switch in the CCCG tetraloop closing a Watson-Crick stem. J. Mol. Biol. 1996;263:715–729. doi: 10.1006/jmbi.1996.0611. [DOI] [PubMed] [Google Scholar]
- 37.Singh S, Patel PK, Hosur RV. Structural polymorphism and dynamism in the DNA segment GATCTTCCCCCCGGAA: NMR investigations of hairpin, dumbbell, nicked duplex, parallel strands, and i-motif. Biochemistry. 1997;36:13214–13222. doi: 10.1021/bi970819y. [DOI] [PubMed] [Google Scholar]
- 38.Kozerski L, Mazurek AP, Kawecki R, Bocian W, Krajewski P, Bednarek E, Sitkowski J, Williamson MP, Moir AJ, Hansen PE. A nicked duplex decamer DNA with a PEG6 tether. Nucleic Acids Res. 2001;29:1132–1143. doi: 10.1093/nar/29.5.1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.