Abstract
The 5′ leader of the human immunodeficiency virus type 1 (HIV-1) genomic RNA harbors an internal ribosome entry site (IRES) that is functional during the G2/M phase of the cell cycle. Here we show that translation initiation mediated by the HIV-1 IRES requires the participation of trans-acting cellular factors other than the canonical translational machinery. We used ‘standard’ chemical and enzymatic probes and an ‘RNA SHAPE’ analysis to model the structure of the HIV-1 5′ leader and we show, by means of a footprinting assay, that G2/M extracts provide protections to regions previously identified as crucial for HIV-1 IRES activity. We also assessed the impact of mutations on IRES function. Strikingly, mutations did not significantly affect IRES activity suggesting that the requirement for pre-formed stable secondary or tertiary structure within the HIV-1 IRES may not be as strict as has been described for other viral IRESes. Finally, we used a proteomic approach to identify cellular proteins within the G2/M extracts that interact with the HIV-1 5′ leader. Together, data show that HIV-1 IRES-mediated translation initiation is modulated by cellular proteins.
INTRODUCTION
Initiation of protein synthesis in the eukaryotic cell leads to the assembly of the 80S ribosome at the start codon of the mRNA. At least two mechanisms for recruiting and positioning ribosomes on the mRNA have been described (1,2). The primary mechanism involves the recognition of the 5′ cap structure (m7GpppN) by eukaryotic translation initiation factors (eIFs), followed by binding of the 40S ribosomal subunit and scanning downstream to the initiation codon (1,2). Alternatively, in some mRNAs a structural element, the internal ribosome entry site (IRES), allows assembly of the translational machinery at a position close to or directly at the initiation codon (3,4). IRES elements were first described in the uncapped poliovirus and encephalomyocarditis virus mRNAs (5,6). Functionally, the IRESes were identified by inserting the 5′-untranslated region (UTR) of the viral mRNA into the intercistronic spacer of a bicistronic construct encoding two proteins (5,6). In this context, expression of the second cistron documented the ability of the inserted sequence to promote internal ribosome binding and translation independent of the first cistron. Since the initial characterization of IRES elements in Picornaviridae, viruses from other families including the Retroviridae have been shown to initiate translation via an IRES (7–9).
The study of the mechanism of translation initiation of the full length RNA of the human immunodeficiency virus type 1 (HIV-1) revealed that this capped and polyadenylated mRNA can initiate protein synthesis through the canonical cap-dependent or by the alternative IRES-dependent mechanism (8–15). The HIV-1 full-length RNA harbors two IRESes, the first in the mRNAs 5′-UTR (here referred to as the HIV-1 IRES) (10,13), and the second within the Gag open reading frame (the HIV-1 gag IRES) (11,12,14). Translation initiation of the viral structural proteins, Gag and GagPol can thus be driven by three independent mechanisms, the canonical cap-dependant process (8,15), or by two internal ribosome entry events dependant on the HIV-1 IRES, or the HIV-1 gag IRES (8,10–12). In addition, the translation of a shorter 40K-Gag isoform of currently unknown function is directed by the HIV-1 gag IRES (8,9,11,12).
The observed redundancy and the conservation of the different mechanisms for the initiation of protein synthesis among primate lentiviruses suggest that translation initiation of HIV-1 mRNA is a key step during the viral life cycle (7–9,12). Alternative initiation may allow the viral mRNA to bypass the constraints of global cellular translation repression that normally target cap-dependent translation initiation, a proposal given credence by evidence that HIV-1 IRES supports translation initiation during osmotic stress (13,16). Additionally, HIV-1 gene expression is influenced by the cell cycle as evidenced by the observation that HIV-1-infected cells arrested in G2/M by the viral protein Vpr or by chemicals, exhibit enhanced levels of viral mRNA transcription and translation (17,18). Notably, the HIV-1 IRES supports translation of viral mRNA in HeLa cells that have been arrested in the G2/M phase of the cell cycle (10), when global cellular cap-dependent translation initiation is suppressed (19). IRES-mediated translation initiation may also ensure synthesis of viral structural proteins during the late stages of the replication cycle, when the eIF4G and the poly(A) binding protein (PABP), both required for cap-dependent translation initiation, are targeted by the viral protease (20–24).
To date the molecular mechanisms that determine the function of the IRESes harbored within the HIV-1 full-length mRNA are not clearly understood. However, recent reports suggest that translation initiation driven by the HIV-1 IRES can be modulated by cellular proteins (16,25,26). The heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1), eIF5A, the human rev-interacting protein (hRIP) and DEAD (Asp–Glu–Ala–Asp) box polypeptide 3 (DDX3) have been identified as a cellular factor that enhance HIV-1 IRES activity (16,26), while the human embryonic lethal abnormal vision (ELAV)-like protein, HuR, has been describe as a negative modulator of HIV-1 IRES activity (25). These reports are in keeping with existing evidence that IRES-dependent translation for a number of viral and cellular mRNAs requires the presence of an additional and sometimes complex set of trans-acting factors for full translational activity (4,27,28). IRES trans-acting factors (ITAFs) are thought to bind to the mRNA, inducing conformational changes needed to structurally form the IRES, thereby facilitating ribosome recruitment (4,27,28).
In this study, we show that translation mediated by the HIV-1 IRES requires G2/M-specific cellular factors. We show that cell extracts alter the accessibility of chemical reagents to single-stranded regions present within the HIV-1 5′ leader region. Close analysis of these data reveal that cell factors protect a region of the HIV-1 5′ leader known to participate in IRES activity (10). A mutational analysis revealed that the HIV-1 IRES function is resistant to the introduction of mutations that were predicted to disrupt local RNA structures (29). This observation suggests that the requirement for a single, stable pre-formed secondary or tertiary structure may not be as rigid as has been described for other viral IRESes. Finally, using a proteomic approach we identify proteins present in cell extracts that interact with the HIV-1 5′ leader. Together our data suggest that the translational activity from the HIV-1 IRES is most probably modulated in trans by a group of proteins that specifically interact with the HIV-1 5′ leader during the different stages of the cell cycle.
MATERIALS AND METHODS
Plasmid
The dlΔEMCV and dl HIV-1 IRES plasmids were as previously described (10,25). The long distance interactions (LDI)/branched multiple hairpin (BMH) stabilizing mutations previously described by Abbink et al. (29) were introduced in the 5′ leader of the proviral clone pNL4.3 by overlapping extension PCR (30), using primers described in Table 1. In each case, the amplicon was digested with EcoRI and NcoI (both restriction sites added by PCR) and inserted into the intercistronic region of dl HIV-1 IRES plasmid as described (10), previously digested with the same enzymes (Fermentas, Vilnius, Lithuania). Upon sequencing additional mutations that were not originally included in the primers were identified in four constructs (namely Mut L5, Mut L6, Mut L7 and Mut L8); these mutants were included in the study. Mutant L9 was constructed by digesting Mut L8 with PauI and XbaI (Fermentas) and cloning the PauI–XbaI fragment into the Mut L7 digested with the same enzymes. As before, the generated mutant HIV-1 5′ leader was inserted into the intercistronic region of dl HIV-1 IRES plasmid as described (10). The authenticity of all plasmids used in this study was confirmed by sequencing (Macrogen Corp, Rockville, MD, USA).
Table 1.
Primers used to generate the HIV-1 Leader mutants
| Mutant | Sense primer | Antisense primer |
|---|---|---|
| Mut L1 | 5′-GGTGCGCACΔCCAAAAATTTTGACTAGCGGAGGCT-3′ | 5′-AAAATTTTTGGΔGTGCGCACCAGTCGCCGCCCC-3′ |
| Mut L2 | 5′-GAGTACGCCATCCTTTTTGACTAGCGGAGGCTAGAAGG-3′ | 5′-GTCAAAAAGGATGGCGTACTCACCAGTCGCC3′ |
| Mut L3 | 5′-ACGCCAGGGGTTTTGACTAGCGGAGGCTAGAAGG-3′ | 5′-AGTCAAAACCCCTGGCGTACTCACCAGTCGCC-3′ |
| Mut L4 | 5′-TTTTTCTAGCGGAGGCTAGAAGGAG-3′ | 5′-AGCCTCCGCTAGAAAAAATTTTTGGCGTACTCACC-3′ |
| Mut L5–L10 | 5′-TTTAGCTAGCGGAGGCTAGAAGGAG-3′ | 5′-AGCCTCCGCTAGCTAAAATTTTTGGCGTACTCACC-3′ |
| Mut L6 | 5′-GACGCTGGTGAGTACGCCAAAAATTTTG-3′ | 5′-TTTTTGGCGTACTCACCAGΔCGTCGCCCCTCGCCTCTTG-3′ |
| Mut L7 | 5′-CGACGCACCCCTCGGCTTGCTGAAGCGCGC-3′ | 5′-CAGCAAGCCGAGGGGTGCGTCGAGAGATCTCCTC-3′ |
| Mut L8–L11 | 5′-CCCGGCGACTGGTGAGTACGCC-3′ | 5′-ACTCACCAGTCGCCGGGACTCGCCTCTTGCCGTG-3′ |
Δ, deleted nucleotide with respect to pNL4.3 (AF 324493); bold, nucleotide changes with respect to pNL4.3.
Cell culture
HeLa cells were cultured in Dulbecco’s modified Eagle’s medium (Gibco-BRL) with 100 U/ml of penicillin–streptomycin (HyClone) and 10% fetal bovine serum (HyClone) at 37°C in a 5% CO2 atmosphere. Nocodazol (400 ng/ml; Sigma-Aldrich) and l-mimosine (20 mM; Sigma-Aldrich) were used to enrich cells in the G2/M or G1 phase of the cell cycle, respectively. Cell cycle arrest was confirmed by flow cytometry as previously described (10). Cytoplasmic cell extracts were prepared following a previously described protocol (10,25). Upon preparation, extracts were tittered to determine the concentration that should be used in the assay. The adequate experimental concentration varied from one cell extract to another.
In vitro transcription
Capped RNAs were synthesized using the mMESSAGE mMACHINE High Yield Capped RNA Transcription Kit (Applied Biosystems/Ambion, Austin, TX, USA), while capped and polyadenylated RNA transcripts were synthesized using the complete mMessage mMachine T7 Ultra Kit (Applied Biosystems/Ambion) according to the manufacturer’s protocol. Uncapped RNA was synthesized by in vitro transcription conducted in a final volume of 200 µl using T7 RNA polymerase, 5 mM DTT, 5 mM rNTP’s, 1X transcription buffer (40 mM Tris–HCl pH 8.0, 25 mM MgCl2, 1 mM spermidine) and 0.04 U RNase Inhibitor (Applied Biosystems/Ambion) and incubated 2 h at 37°C. Upon synthesis, RNAs were treated with DNAse RQ1 (Promega, Madison, WI, USA) for 20 min at 37°C. RNA was precipitated with 2.5 M LiCl, centrifuged at 16 000g, 30 min at 4°C, washed with 70% ethanol and resuspended in 50 µl of nuclease-free water. RNA concentrations were determined spectrophotometrically (NanoDrop Technology, Wilmington, DE, USA) and RNA integrity was monitored by electrophoresis on denaturing agarose gels.
In vitro translation
In vitro transcribed dl HIV-1 IRES RNAs (8 ng/µl) were translated in 25% (v/v) nuclease-treated rabbit reticulocyte lysate (RRL; Promega), supplemented or not with cell extracts prepared as previously described (25). Final concentrations of extract used in each experiment are indicated in figure legends. Cell extracts were pre-incubated with RNA for 5 min prior to addition of the RRL mix as previously described (25). Luciferase activities were measured using the DLR™ Assay System (Promega) according to manufacturer’s instructions on a Sirius Luminometer (Berthold Detection Systems GmbH, Pforzheim, Germany) as previously described (25,31).
For 35S-methionine labeling, bicistronic RNA (1 pmol) was translated in 50% of RRL Flexi® (Promega) in the presence of 20 µM amino acids (lacking methionine), 0.5 mM MgOAc2, 160 mM KOAc, 0.8 U/µl de RNasin® Plus RNase Inhibitor (Promega) and 0.6 mCi/ml [35S]-methionine. Translation reactions, conducted in final volume of 10 µl, were incubated for 90 min at 30°C. The reaction was stopped with 90 µl of protein loading buffer as previously described (12). Ten microliter of the final mix was loaded and resolved by SDS–PAGE (12%), and bands visualized using a BAS-5000 phosphorimager (Fujifilm).
Oocyte microinjection
Oocytes were isolated from Xenopus laevis ovarian fragments and microinjected with glass micropipettes calibrated to deliver a final volume of 50 nl, as previously described (25). To assess the effect of G2/M HeLa extracts on IRES activity, oocytes were first microinjected with 6.25 ng of in vitro transcribed, capped and polyadenylated bicistronic RNA generated from either dlΔEMCV or dl HIV-1 IRES plasmids. Fifteen minutes later oocytes were microinjected with 200 ng of G2/M HeLa extracts. Oocytes were incubated for 24 h at 15°C in a standard Barth’s solution supplemented with 10 UI/l penicillin–streptomicine and 2 mM pyruvate. Oocyte lysates were prepared in 1× passive lysis buffer (Promega Corporation), centrifuged at 16000g for 5 min and 1–5 µl of supernatant was used in the detection of luciferase as described above.
RNA probing
The secondary structure of the HIV-1 5′-UTR was probed using DiMethyl Sulfate (DMS, Across Organics), N-cyclohexyl-N-[N-methylmorpholino)-ethyl]-carbodiimid-4-toluolsulfonate (CMCT, Merck) and RNAse V1 (Applied Biosystems/Ambion) as described previously (12,32). RNA Selective 2′ Hydroxyl Acylation analysis by Primer Extension (SHAPE) analysis was conducted using 1-methyl-7-nitroisatoic anhydride (1M7) as a modifying agent as previously described (33,34). In brief, 10 pmol of in vitro transcribed RNA, which included the 5′ leader of HIV-1 (pNL4.3) and the first 58 nt of fluc (recovered from the dl HIV-1 IRES plasmid using a primer
T7HIVF 5′-CCATATGTAATACGACTCACTATAGGTCTCTCTGGTTAGA-3′ and Fluc30bp 5′-CATCTTCCAGCGGATAGAATG-3′) were resuspended in 30 µl of 80 mM HEPES pH 7.5 (or 50 mM borate potassium pH 8 for CMCT), denatured for 2 min at 80°C, and then 2 μl of 3 M KCl and 2 μl of 40 mM MgCl2 were added. Upon a 10 min incubation at 30°C DMS (0.2 mM final), CMCT (25 mM final), RNAse V1 (0.01 or 0.025 U) or 1M7 was added and the mixture was incubated for 5 min (10 min for CMCT). Mock controls, where the chemical was replaced by water or DMSO (for the 1M7 probing) were also included. The modification reaction was stopped in ice by addition of 10 μg of yeast tRNA. As previously described (32,34), the time and concentration of modification agent were established to generate at the most one modification per molecule. The reaction was then immediately precipitated in dry ice with ethanol and 5 M ammonium acetate. RNA was then resuspended in 0.5 M ammonium acetate, ethanol precipitated in presence of 20 µg of glycogen, washed with 70% ethanol and resuspended in 6 μl of nuclease-free water. For DMS, CMCT and V1 probing, modifications were revealed by reverse transcription (AMV RT; Promega) using a 32P-labeled primers (Fluc30bp; HIV1-336 R 5′-TTTGAAAAACACGAATTCGGTCTCTCTG-3′; 100pbHIV-1 5′-ACTTTGAGCACTCAAGGCAAG-3′; 200pbHIV-1 5′-TTCGCTTTCAAGTCCCTGTTC-3′) according to the manufacturer’s instructions (Promega). Reverse transcription products were resolved by 8% denaturing PAGE; the resulting gel was scanned on a Typhoon trio variable mode imager (Amersham Biosciences). The relative proportion of each product was determined, drawing profiles with Multi Gauge V3 software (Fujifilm). For 1M7 probing, modifications were revealed using RNAse H− M-MLV RT (Promega) and the Fluc30pb primer labeled with WellRed D2, D3, D4 (Sigma), or IR-800 (MWG Eurofins) fluorophores, cDNA fragments were resolved by capillary electrophoresis (Beckman Coulter CEQ 8000). Data were then interpreted and analyzed using the software ‘shapefinder’ (35) (http://bioinfo.unc.edu).
1M7 footprinting experiments were carried as described above except that non-synchronized, G1 or G2/M synchronized, Hela extracts (6 µg of total proteins) were added after RNA renaturation, the mixture was then incubated for 10 min at 30°C before 1M7 addition. Profiles were compared to the profile obtained with a mocked control containing the equivalent amount of buffer instead of extracts. Increases or decreases of 2-fold of the reactivity, with the higher reactivity being at least 0.3, was considered significant and reported.
Synthesis of 1-methyl-7-nitroisatoic anhydride was as described by Mortimer and Weeks (34).
DNA transfection
Cells were seeded at 1 × 105 cell/well in 12-well plates and DNA transfection (200 ng/well) was performed at 60% confluence using the JetPEI transfection system (PolyPlus transfection, France) according to the manufacturer’s protocol. After 24 h, the culture medium was removed and cells lysed with 1× passive lysis buffer (Promega) as described in the DLR™ Assay System manual (Promega) (10,25,31). Protein concentration was determined by a Bradford assay using the Bio-Rad Protein Assay (Bio-Rad Laboratories, Inc., Hercules, CA, USA). FLuc and RLuc activities were measured as described above.
RNA constructs and cellular extracts preparation for pull-down experiments
HeLa cells were grown to confluency in standard media or media supplemented with 400 ng/ml nocodazole. Cells were detached with trypsin/EDTA, and pelleted. Following a cold phosphate buffered saline (PBS) wash, the cell pellet was resuspended in cold buffer A (10 mM Hepes, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, supplemented with protease inhibitors) and incubated on ice for 5 min. Cells were lysed with a pre-chilled Dounce homogenizer for 20 strokes using a tight fitting pestle. Dounced cells were centrifuged at 228g for 5 min at 4°C to pellet nuclei and other fragments. The supernatant was retained as the cytoplasmic fraction and the pellet was retained as the nuclear fraction. The cytoplasmic fraction was supplemented with 5× RIPA buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1% NP-40, 0.5% deoxycholate, supplemented with protease inhibitors) to a 1× final concentration and centrifuged at 2800g for 10 min at 4°C to pellet solids. The nuclear pellet was resuspended in buffer S1 (0.25 M Sucrose, 10 mM MgCl2), layered over a sucrose cushion (0.8 M Sucrose, 0.5 mM MgCl2) and centrifuged at 2800 × g for 10 min at 4°C. The pellet was resuspended in 10 pellet volumes 1× RIPA buffer and sonicated on ice. The lysate was centrifuged at 2800 × g for 10 min at 4°C to pellet solids. Protein concentration was determined using a Bradford assay (BioRad).
Templates for in vitro transcription of strep-aptamer tagged HIV-1 5′ leader (nucleotides 1–384) and the reverse complement HIV-1 5′ leader (RC) were generated by PCR amplification from plasmid HIV 1–384 described by Brasey et al. (10). A T7 promoter was added to the 5′-end, and the sequence for a streptomycin binding RNA aptamer (36) was added on the 3′ end using the primer pair 5′TAATACGACTCACTATAGggtctctctggttagaccag3′ (forward) and 5′GGATCCGACCGTGGTGCCCTTGCGGGGCAGAAGTCCAAATGCGATCCcccatttatctaattctccc3′ (reverse) for the HIV-1 5′ leader and primers 5′TAATACGACTCACTATAGGcccatttatctaattctccc3′ (forward) and 5′GGATCCGACCGTGGTGCCCTTGCGGGGCAGAAGTCCAAATGCGATCCggtctctctggttagaccag3′ (reverse) for the HIV-1 RC 5′ leader. RNA was in vitro transcribed from PCR templates using T7 RNA polymerase as previously described (37). RNA was purified on a denaturing polyacrylamide gel, passively eluted in diethylpyrocarbonate treated water, and concentrated using spin concentrator columns (Amicon). RNA integrity and purity was verified by denaturing polyacrylamide gel electrophoresis, and concentration was determined by ultraviolet (UV) spectroscopy.
Pull-down experiments
Pull-down experiments were performed using aptamer tagged RNA and a streptomycin-conjugated sepharose column (36). Aptamer tagged RNAs were folded over a series of incubations (65°C for 5 min, 37°C for 5 min, room temperature for 5 min) and added to 1 ml of column buffer (50 mM Tris HCl, pH 7.5, 5 mM MgCl2, 250 mM NaCl). Folded RNA was loaded onto a column packed with 1 ml bed volume of streptomycin conjugated sepharose that had been equilibrated three times with column buffer and blocked with 20 µg tRNA. The RNA was incubated on the column for 10 min, and washed twice with 1 ml column buffer. 150 µg of protein extract (supplemented with RNASE inhibitor) was loaded onto the column and incubated for 10 min. The column was washed 10× with column buffer, and RNA–protein complexes were eluted three times with 1 ml column buffer supplemented with streptomycin to 10 µM. Elutions were concentrated using spin concentrator columns (Amicon) according to the manufacturer’s protocol.
Mass spectrometry analysis
Eluted protein samples were reduced with 5 mM DTT (30 min at 70°C) and alkylated with 15 mM iodoacetamide (30 min at room temperature in the dark). Samples were digested with 1.2 µg trypsin overnight at room temperature and peptides desalted using C18 tips (Omix) according to the manufacturer’s protocol. Acetonitrile was evaporated and the samples were brought up to a final volume of 15 µl with 1% formic acid. Tryptic peptides were analyzed by the UC-Denver proteomics core on an Agilent 1200 nanoLC system directly infused for MS/MS analysis on a LTQ-FT Ultra hybrid mass spectrometer (ThermoFisher). Peptides were separated using a 90 min gradient of increasing acetonitrile (8–35%) with 0.1% formic acid as a pairing agent. Electrospray ionization was performed at 200 V on the column eluent. Parent scans (MS) were acquired in the ICR cell at 50 000 resolution. Collision induced dissociation was performed in the ion trap and product ions recorded (MS/MS). Ion peak lists were created using PAVA (UCSF) and searched against the human Swiss Prot database using the Mascot server (Version 2.2, Matrix Science). Mass tolerances of ±10 ppm were used for MS peaks, and ±0.8 Da for MS/MS fragment ions. The modifications of cysteine carbamidomethylation, methionine oxidation, N-terminal acetylation of protein, N-terminal pyroglutamic acid formation and phosphorylation of serine, threonine, and tyrosine residues were allowed for. Protein identifications were considered significant if two or more peptides matched with an expect value of below 0.01.
RESULTS
Cap-independent translation initiation from the HIV-1 IRES in RRL requires cytoplasmic cellular factors
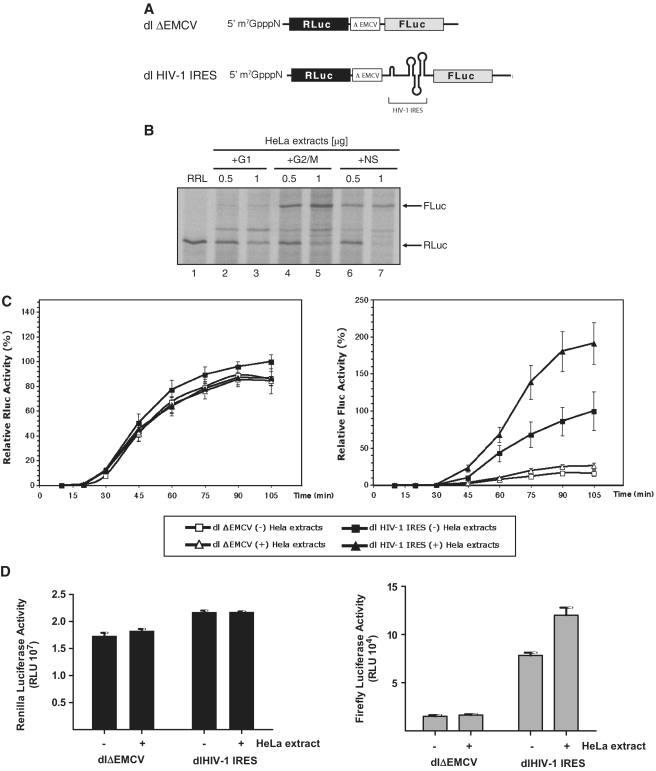
The HIV-1 IRES was identified by cloning the 5′ leader region of the laboratory adapted HIV-1 infectious recombinant proviral clone pNL4.3 or the CXCR4 (X4)-tropic HIV-1 primary isolate HIV-LAI in the intercistronic region of a dual luciferase (dl) reporter construct (10,13). In the context of this bicistronic RNA the HIV-1 IRES was shown to be cell-cycle regulated and preferentially active during G2/M (10), and to be functional during osmotic stress (13,16). In addition, the HIV-1 IRES is functional in HeLa cell translational extracts (10), in transfected HeLa or Jurkat T cells (10,13,16,25), and in X. laevis oocytes (25); however, it is poorly active in RRL (8,10).
Poor activity of the HIV-1 IRES in RRL suggests that, similar to poliovirus (PV) and human rhinovirus (hRV) IRES elements (38–40), additional factors entirely absent or present only at low concentration in the RRL may be required for HIV-1 IRES activity. To assess this possibility, RRL programmed with the dl HIV-1 IRES RNA (depicted in Figure 1A) was supplemented with 0.5 or 1 µg of cytoplasmic HeLa extracts generated from non-synchronized, mimosine-G1 or nocodazole-G2/M arrested cells. Cell arrest in G1 or G2/M was confirmed by flow cytometry (10). Translation reactions were conducted in the presence of 35S-methionine and resolved and visualized as indicated in ‘Materials and Methods’ section. One representative experiment is shown in Figure 1B. It should be noted that optimal translation of the upstream reporter occurred at a narrow range of cell extract concentrations, since translation of the first cistron was consistently abolished with high concentrations of cell extract (Figure 1B compare lanes 2, 4, 6 with lanes 3, 5, 7). In contrast, the addition of 0.5 µg of cell extracts to the RRL had no significant impact on RLuc synthesis, thus no effect on the translation of the upstream message of the bicistronic reporter (Figrue 1B lanes 2, 4 and 6). RNA did not significantly vary among the different assays suggesting that extracts did not affect RNA integrity (data not shown). Based on this observation, in this particular experiment, data generated with 1 µg of HeLa cell extracts were not taken into account in further analysis. In agreement with previous reports (8,10), HIV-1 IRES activity was negligible in non-supplemented or G1-supplemented RRL (Figure 1B, lanes 1 and 2). When RRL was supplemented with 0.5 µg of cytoplasmic extracts generated from non-synchronized (NS; Figure 1B, lane 6) or G2/M (Figure 1B, lane 4) cells, HIV-1 IRES activity increased markedly as evidenced by the appearance of FLuc protein. IRES activity was always greatest when RRL was supplemented with G2/M extract (Figure 1B). These observations echoed previous studies conducted in HeLa cells or HeLa cell based translational extracts (10).
Figure 1.
HeLa cell cytoplasmic factors are required for HIV-1 IRES activity. (A) Schematic representation of the dl HIV-1 IRES and dlΔEMCV RNAs used in this study. (B) RRL alone or supplemented with cytoplasmic extracts (0.5 or 1 µg of total protein) generated from NS HeLa cells or cells arrested in G1 or in G2/M were programmed with the dl HIV-1 IRES RNA. [35S]-methionine-labeled proteins were resolved by SDS–PAGE and visualized as indicated in ‘Materials and Methods’ section. (C) Kinetics of HIV-1 IRES translation in the presence of G2/M HeLa extracts. The capped dlΔEMCV (open shapes) or dl HIV-1 IRES (filled shapes) RNAs (8 ng/µl) were used to program RRL. In vitro translation reactions were supplemented with 160 ng/µl of G2/M cytoplasmic extracts (Δ). Renilla luciferase (RLuc) and Firefly luciferase (FLuc) activities were measured at the indicated times. The RLuc and FLuc activities of the dl HIV-1 IRES measured at 105 min of in vitro translation in non-suplemented RRL (filled square) were arbitrary set to 100%. Relative RLuc activity (left panel) and relative FLuc activity (right panel) are shown. Values are the means ± SEM (error bars) of five independent experiments. (D) Capped and polyadenylated RNA corresponding to the dlΔEMCV or dl HIV-1 IRES vectors (6.25 ng) were microinjected into X. laevis oocytes with (+) or without (−) cytoplasmic extracts generated from G2/M arrested HeLa cells (200 ng) as described in ‘Materials and Methods’ section. Oocytes were harvested 24 h after the microinjection and processed and RLuc and FLuc activities were determined RLU. The RLuc (left panel) and FLuc (right panel) activities for each RNA are shown. Each value is the mean ± SEM from at least three oocytes obtained from different animals.
To further extend these observations, we next studied the kinetics of protein synthesis using luciferase activity as the experimental readout. The RLuc/FLuc bicistronic RNA dlΔEMCV, that harbors a defective encephalomyocarditis virus (ΔEMCV) IRES known to inhibit ribosome reinitiation and read-through, inserted upstream of the FLuc reporter, was used as a negative control (10,31,41). In Figure 1C, RLuc and FLuc activities are independently displayed. The maximal RLuc and FLuc activity obtained in non-supplemented translation reactions in RRL was arbitrarily set to 100%. In agreement with data presented in Figure 1B, the activity of the HIV-1 IRES increases when RRL is supplemented with G2/M extracts (Figure 1C). As before (Figure 1B), the addition of G2/M extracts to the RRL had little or no impact on translation initiation from the first cistron (see RLuc, Figure 1C). Together, the observed enhancement by G2/M cell extracts which only impacts translation from the second cistron implies that one or more cellular factor(s) present in G2/M HeLa extracts may act in trans to specifically overcome the translational inefficiency of the HIV-1 IRES in RRL.
Next, we evaluated the effect of G2/M extracts on HIV-1 IRES activity in X. laevis oocytes, a system known to support its function (25). To this end the control dlΔEMCV or the dl HIV-1 IRES RNA were microinjected into X. Laevis oocytes either alone or with G2/M extract as previously described by others (42). Once more, no effect was evident on RLuc translation as indicated by a constant luciferase activity (Figure 1D). In contrast FLuc activity increased >50% in the presence of cell extracts (Figure 1D). Just as in RRL, factors present in G2/M cell extracts seem to be capable of specifically stimulating HIV-1 IRES activity.
Cytoplasmic cellular factors alter the accessibility of chemical reagents to single-stranded regions present within the HIV-1 5′ leader
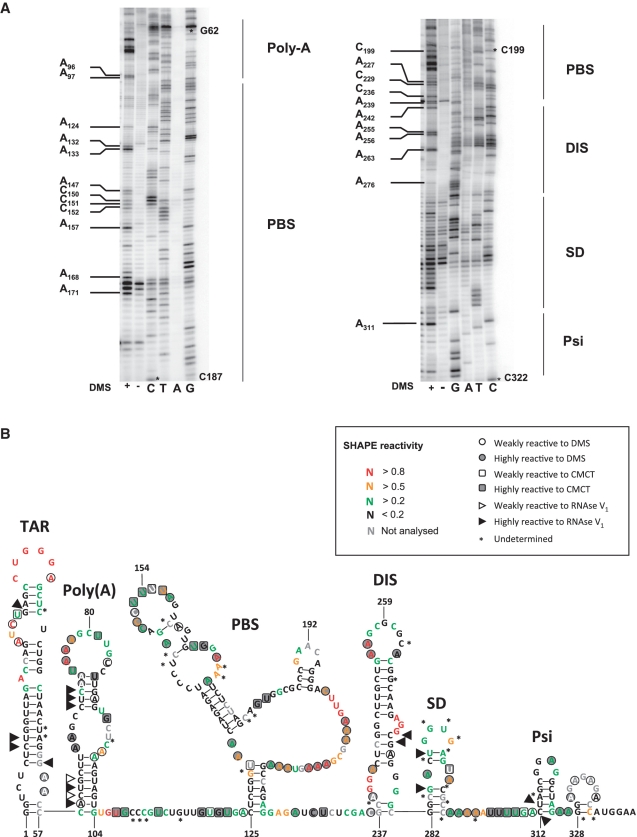
We next sought to establish whether cytoplasmic cell extracts altered the chemical modification profile of the HIV-1 5′ leader. Before addressing this specific question, the secondary structure of the HIV-1 5′ leader recovered from the dl HIV IRES construct (nucleotides 1–336 from clone pNL4.3 followed by 58 nt of fluc gene) was probed using DiMethyl sulfate (DMS), N-Cyclohexyl-N-[N-Methylmorpholino)-ethyl]-Carbodiimide-4-Toluolsulfonate (CMCT) and RNAse V1 as described (12,32). CMCT and DMS were used to detect accessible RNA functional groups consistent with single-stranded regions, while RNAse V1 revealed stacked or paired nucleotides. Modifications were mapped by reverse transcription using a 32P-labeled primer. For each run the reverse transcription pattern of the modified RNA was compared to the profile obtained with a non-modified RNA. Modifications were classified into two categories according to the relative intensity of the induced stop by comparison with the control. Modifications were classified as ‘weak’ when inducing a 2- to 3-fold increase in intensity of the RT stop, and as ‘highly reactive’ for higher intensities. A typical example of our results using DMS and covering the full HIV-1 5′ leader is shown (Figure 2A). The strong hits for DMS and CMCT were considered in the initial secondary-structure modeling using the Mfold algorithm as previously described (12,32). The structure obtained was then fitted onto a model structure of the HIV-1 leader (43), taking into account the V1 data and the weak DMS and CMCT hits. To further validate our probing data and to gain information on the intrinsic nucleotide flexibilities that characterize the secondary structure we also performed a detailed RNA Selective 2′Hydroxyl Acylation analysis by Primer Extension (SHAPE) analysis using the same RNA and 1-methyl-7-nitroisatoic anhydride (1M7) as a modifying agent (33,34). 1M7 reacts with flexible ribose groups of nucleotides that are not in a strong Watson–Crick pair or any other rigid tertiary interaction (33). In these experiments, modifications were mapped by reverse transcription using a fluorescent primer and the raw data were processed using ‘Shapefinder’ (35). The secondary structure of the HIV-1 5′ leader was modeled using ‘RNA structure 5.03′ and the pseudo-energy constraints derived from the probing analysis (44). The integrated data, DMS/CMCT/RNAse V1 probing and RNA SHAPE analysis, were fitted onto a model structure of the HIV-1 leader (Figure 2B). Both methods, DMS/CMCT/RNAse V1 probing and RNA SHAPE analysis (Figure 3A), showed high consistency and yield essentially the same structure model. Most of the discrepancies observed when comparing both models consist in nucleotides reactive with DMS or CMCT but unreactive to 1M7 (for example A66–A67 and G92). As DMS and CMCT probe the availability of ‘Watson–Crick’ position and 1M7 the flexibility of the ribose, such results could indicate bases involved in non-canonical base pairs. This is particularly interesting in the case of A66–A67 and G92 which are located within an asymmetrical bulge (Figure 2B). The observed probing signatures and the established model are similar to those obtained by others, in vitro or ex vivo (43,45–47). As notable differences, the regions involved in the LDI with the gag open reading frame (ORF) (U5 G108–C114 and part of the polyA loop G79–C85) were accessible to single-strand probes. This observation is expected because the construct used in the study lacks the gag coding sequence. Interestingly, 3 nt within the palindrome in the DIS loop are reactive to 1M7, suggesting that the kissing-loop interaction that initiates non-covalent dimerization of the genomic RNA (48), was not formed under the experimental conditions used for probing.
Figure 2.
Secondary structure model of the HIV-1 leader. The HIV-1 5′ leader recovered from the dl HIV IRES construct (nucleotide 1–336 from clone pNL4.3 followed by 58 nts of fLuc gene) was probed using DiMethyl Sulfate (DMS), N-Cyclohexyl-N- [N-Methylmorpholino)-ethyl]-Carbodiimide-4-Toluolsulfonate (CMCT) and RNAse V1 as previously described (12,32) or using 1-methyl-7-nitroisatoic anhydride (1M7) as a modifying agent. (A) Typical examples of probing DMS probing. The HIV-1 5′ leader was probed using (+) DMS. Reverse transcription (RT) products were separated on a 8% gel as indicated in the ‘Materials and Methods’ section. Sequencing lanes were also included. Note that DMS induces a premature RT stop 1 nt before the hit. Therefore the DMS induced stops migrate faster than the corresponding sequence product (12,32). The RT pattern of the modified RNA was compared to the profile obtained with an unmodified RNA. Some hits are indicated in the figure. The asterisks on the gel denote the nucleotide position. (B) Results were fitted in a model of the HIV-1 5′ leader (43), the respective reactivity of the different probes is indicated as motioned in the box. The main HIV-1 structural elements present in the 5′ leader the TAR and poly(A) loops, PBS, DIS, SD and Psi are indicated (43).
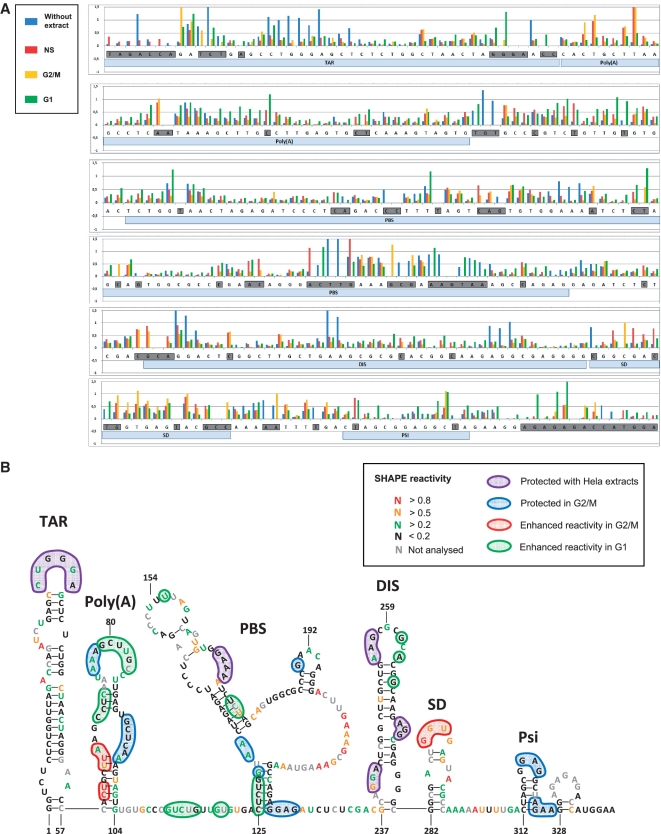
Figure 3.
HeLa cell extracts alter the accessibility of 1M7 to single-stranded regions present within the HIV-1 5′ leader. The HIV-1 5′ leader RNA was probed using 1M7 in the presence or absence of HeLa cell cytoplasmic extracts generated from NS, or cells arrested in the G1, or in the G2/M phase of the cell cycle. (A) Histogram representing the ‘SHAPE’ reactivity for each nucleotides of the 5′-UTR in absence of extracts [(-) blue bars] in presence of NS (red bars), G1 arrested [(G1) green bars] or G2/M arrested [(G2M) yellow bars] HeLa extracts. Nucleotides which reactivity is undetermined in at least one of the tested conditions are boxed in grey. Local RNA structures are indicated as hallmark. (B) Data presented in A, are incorporated in the model depicted in Figure 2, the nucleotides protected by NS, G1 and G2/M extracts are boxed in purple, those specifically protected by G2/M extracts are boxed in blue, while those which reactivity is enhanced in presence of G2/M cytoplasmic extracts are boxed in red, nucleotides which reactivity is enhanced in presence of G1 extracts are boxed in green. The reactivity of each nucleotide in presence of G2/M extract is encoded by a specific colour, the reactivity are thus also valid for the area protected by all extracts (boxed in purple), but not for the nucleotides which reactivity is enhanced in presence of G1 extracts.
Once we had a model secondary structure for the HIV-1 5′ leader we probed the RNA using 1M7 in the presence of cytoplasmic cell extract (see ‘Materials and Methods’ section) and compared the data with those generated in absence of extracts (Figures 2B and 3A). A position was considered as protected (or exposed) when the reactivity value was at least 2-fold lower (or higher) than in the mock control. We tested RNA with or without cell extracts generated from non-synchronized cells (NS) (red bars in Figure 3A), cells arrested in G1 (green bars in Figure 3A), or cells arrested in G2/M (yellow bars in Figure 3A). The modification of the pattern upon addition of NS cell extracts or extracts generated from cells blocked in G1 or G2/M is summarized on Figure 3B. Interestingly, distinct 1M7 accessibility RNA patterns are observed when NS, G1 or G2/M extract is used (Figure 3A and B). However, significant protections are seen upon addition of cell extracts independently of the stage where the cells were blocked. Specifically, we observe a strong protection of the TAR apical loop (C29–A34), in the PBS (G167–A170) and in three occurrences in the DIS element (G240–A242, A255–A256 and A271–G273). As depicted in Figure 3, some positions are protected in the presence of G2/M extracts while few others are highly reactive. These reactivity modifications map to four specific regions: the poly(A) loop (increased reactivity of C60–U61 and U64–A66; protection of A76–A78 and C95–A97), the PBS structure (protection of G130–A133, G190 and G223–G226), the SD loop (increased reactivity of G289 and the Psi stem–loop immediately upstream from the initiation codon (protection of G318–G320 andA324–A327). It is interesting to highlight that the PBS structure, the SD loop, and the Psi stem–loop are regions previously identified as crucial for HIV-1 IRES activity (10). As reported in Figure 3, G1-specific alterations of the modification pattern exclusively consist of positions that become highly reactive. Most interestingly, sequences modeled as double stranded in all conditions (C70–C72 in the poly(A) stem–loop, C125–G129 and U174–U176 in the PBS structure) clearly react as unpaired nucleotides upon G1 extract addition.
In summary, we find that the addition of HeLa cell extracts alters the accessibility of 1M7 to discrete regions of the HIV-1 5′ leader. Most interestingly, we observed a G2/M-specific pattern most probably confirming that one or more proteins delivered with those cell extracts interacts with the HIV-1 5′ leader. It should be stated, however, that at this point we cannot tell if our observation results from the footprint of proteins, or from a structural rearrangement, or from both.
Cap-independent translation initiation driven from HIV-1 IRES is resistant to point mutations
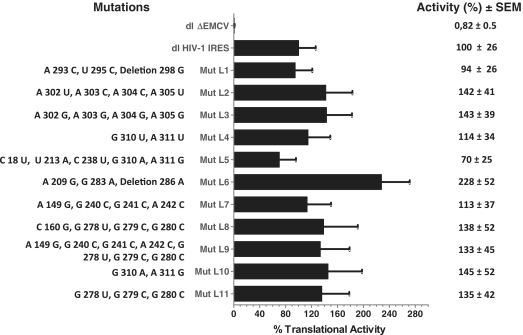
The HIV-1 5′ leader is capable of forming a complex secondary structure with multiple junctions, internal loops and stem–loop elements that is involved in many steps of the viral life cycle including translation (10,43). The most probable secondary structure model of the HIV-1 5′ leader, known as the Branched Multiple Hairpin (BMH, see Figure 2), comprises the PBS, dimer-initiation site (DIS), splice donor (SD) and hairpin loops, all structural elements involved in genomic RNA dimerization, reverse transcription, splicing and packaging (46,47,49). An alternative secondary structure model of the 5′ leader known as the LDI would engender alternative base pairing that disrupts the DIS hairpin loop (49–51). When comparing the LDI and the BMH models the region embedding the Gag start codon is contained in different secondary structure elements, but the initiation codon itself is mostly occluded in both structures (49–51). Abbink et al. (2005) reported a series of mutations designed to alter the LDI-BMH equilibrium (29). However, one important caveat of the study was that the specific secondary structure of the described mutants was not probed and thus the predicted changes to the structure of the HIV-1 leader remain largely speculative (29). Nonetheless, we decided to evaluate the effect of the mutations described by Abbink et al. (2005) on translation initiation driven by the HIV-1 IRES. The rational for this experiment was based on the observation that the introduction of point mutations within the sequence of a viral IRES can have a profound impact on its function (4,52–55). Therefore, the mutations reported by Abbink et al. (2005) were introduced into the 5′ leader of the HIV-1 clone pNL4.3 and examined in the context of bicistronic mRNAs (similar to those depicted in Figure 1A). Based on what has been previously reported (29), mutants leaders (Mut L) Mut L7, Mut L8 and Mut L11 are expected to favor the BMH conformation, Mut L1, Mut L2, Mut L4, Mut L5, Mut L6 and Mut L10 favor the LDI conformer while with the wild-type and mutants Mut L3 and Mut L9 the two conformers are expected to be in equilibrium.
Vectors were transfected into HeLa cells and the RLuc and FLuc activities were monitored as described in previous studies (10,16,25). Vector dl HIV-1 IRES, harboring the leader (nucleotide 1–336) of the HIV-1 infectious recombinant proviral clone pNL4.3 (10), was used as a positive control and sets the 100% of activity, while construct dlΔEMCV, used as a negative control (10,31,41), showed only 0.8% of the activity of the HIV-1 IRES. As in previous studies (10,16,25,31), the FLuc/RLuc ratio was used as an index of IRES activity and the activity of the mutants are expressed as relative translation (%) with respect to the wild-type construct. As shown in Figure 4, the most affected mutant was Mut L6 which exhibits an increase (1.3-fold) in IRES activity with respect to the control HIV-1 IRES (pNL4.3). However, none of the relative translation efficiencies were significantly different from the control IRES, as determined by a one-way ANOVA (P > 0.05). Therefore, echoing previous reports (13,29), no correlation between the IRES activity and the putative BMH/LDI switch of conformation is evidenced. To this respect, it is important to note that the existence of the LDI conformation is not supported in vivo (46,47). Somewhat surprisingly, data do show that the activity of the HIV-1 IRES is resistant to mutations spread all along the 5′ leader sequence, a feature that directly contrast what has been described for most viral IRES elements (4,52–55).
Figure 4.
Translational activity of the mutant HIV-1 IRES elements. HIV-1 mutant leaders generated as described in ‘Materials and Methods’ section and arbitrarily named Mut L1–L11 were cloned into the dual luciferase bicistronic vector (dl-vector). The dl HIV-1 mutant vectors (Mut L1–L11) were transfected into HeLa cells and IRES-mediated translational activity was evaluated in comparison with the dl HIV-1 IRES vector and the dlΔEMCV control (10,41). The nucleotide changes with respect to the pNL4.3 (AF 324493) leader included in the dl HIV-1 IRES vector, are shown (left). RLuc and FLuc activities were measured and the [(FLuc/RLuc)] ratio was used as an index of IRES activity. The [(FLuc/RLuc)] ratio of the dl HIV-1 IRES vector (10), was arbitrarily set to 100%. Values are the means ±SEM (error bars) of three independent experiments.
Specific cellular factors from G2/M extracts bind the HIV-1 5′ leader
Results presented in Figures 3 and 4 suggest that IRES-mediated translation initiation is most probably modulated by a distinct group of proteins that specifically interact with the HIV-1 5′ leader during the G2/M phase of the cell cycle. If indeed the case, different sets of proteins would be expected to bind the HIV-1 5′ leader in different stages of the cell cycle. To evaluate this possibility we used a proteomic approach to identify the proteins present in HeLa cell extracts generated from NS or G2/M arrested cells that interact with the HIV-1 5′ leader. In these assays, an uncapped streptomycin binding RNA aptamer (strep-aptamer)-tagged HIV-1 5′ leader was used as bait (36), while the reverse complement HIV-1 5′ leader was used as a control RNA to discard non-specific RNA-binding proteins from the subsequent analysis. Pull-down experiments were performed using extracts from either NS or G2/M arrested cells. Mass Spectrometry analysis, conducted as indicated in the ‘Materials and Methods’ section, allowed the identification of 54 proteins that pulled-down with the HIV-1 5′ leader when cytoplasmic extracts from non-synchronized cells were used as the protein source (data not shown). Interestingly, only 18 proteins were identified as pulling-down with the viral 5′ leader RNA when G2/M extracts were used (Table 2). From these, only two proteins where common between NS and G2/M extracts (Table 2) indicating that a distinct set of proteins interact with the leader during G2/M. This result correlates with biochemical assays demonstrating that the accessibility of 1M7 to discrete regions of the HIV-1 5′ leader changes when extracts from NS or G2/M cells are used.
Table 2.
Cellular factors from G2/M extracts identified to that were pulldown bind the HIV-1 5′ leader
| Accession No. | Protein Name | Score | Mass (Da) | Matches | Coverage (%) | emPAI | References |
|---|---|---|---|---|---|---|---|
| P53999 | Activated RNA polymerase II transcriptional coactivator p15a | 157 | 14 386 | 5 | 26 | 1.05 | (62) |
| Q07021 | Complement component 1 Q subcomponent-binding proteina | 174 | 31 742 | 4 | 5 | 0.25 | (63) |
| P11387 | DNA topoisomerase 1a | 391 | 91 125 | 25 | 17.3 | 0.43 | (64–69) |
| P14866 | Heterogeneous nuclear ribonucleoprotein L (hnRNP L) | 127 | 60 719 | 6 | 12.9 | 0.06 | |
| P07196 | Neurofilament triplet L protein | 122 | 61 536 | 4 | 4.1 | 0.12 | |
| P19338 | Nucleolin (Protein C23)a | 105 | 76 625 | 4 | 3.4 | 0.1 | (70,71) |
| P06454 | Prothymosin αa | 113 | 12 196 | 2 | 12.6 | 0.32 | (72) |
| P37108 | Signal recognition particle 14 kDa protein (SRP14)b | 146 | 14 675 | 4 | 28.7 | 0.61 | |
| P55072 | Transitional endoplasmic reticulum ATPase (TER ATPase) | 444 | 89 950 | 11 | 8.4 | 0.33 | |
| P09234 | U1 small nuclear ribonucleoprotein C (U1 snRNP protein C) | 191 | 17 552 | 4 | 18.9 | 0.49 | (73) |
| P62861 | 40 S ribosomal protein S30 | 124 | 6644 | 13 | 20.3 | 6.08 | |
| P16989 | DNA-binding protein A (Cold shock domain-containing protein A) | 225 | 40 066 | 10 | 14.8 | 0.31 | |
| P62873 | Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit β 1 | 85 | 38 151 | 3 | 12.1 | 0.1 | |
| Q9BUJ2 | Heterogeneous nuclear ribonucleoprotein U | 79 | 91 164 | 33 | 25.5 | 0.89 | |
| P17096 | High mobility group protein HMG-I/HMG-Y (HMG-I(Y))a | 232 | 11 669 | 6 | 15 | 1.41 | (74,75) |
| Q04837 | Single-stranded DNA binding protein, mitochondrial precursor (Mt-SSB)a | 71 | 17 249 | 2 | 20.3 | 0.23 | (76) |
| P07951 | Tropomyosin β-chain (β-tropomyosin)a | 61 | 32 945 | 5 | 10.6 | 0.24 | (77) |
| Q13748 | Tubulin α-2 chain (α-tubulin 2)a,b | 143 | 50 612 | 6 | 16.9 | 0.24 | (78–82) |
Score = protein identification score, reflects amount of peptide matches and percent coverage obtained for the protein; Mass = protein mass; Matches = number of peptides corresponding to the protein; percent coverage = sequence coverage of identified peptides over the protein; emPAI = exponentially modified protein abundance index.
aProteins known to play a role in the HIV-1 replication.
bProteins also found in non-synchronized cytoplasmic cell extracts.
Taken together, the results of the pull-down experiments suggest that the assembly of protein factors on the HIV-1 leader varies when non-synchronized or G2/M arrested extracts are used. Combined with the results from Figures 1 and 4, these data support the notion that G2/M-specific proteins bind the HIV-1 leader to support IRES-mediated translation initiation.
Overall, the biological significance of the pull-down analysis is still to be determined, work is being performed to determine the role of these and other RNA binding proteins in the control of HIV-1 IRES-mediated translation initiation.
DISCUSSION
In this study, we report that in common with most cellular IRESes and some viral IRESes translation initiation from the HIV-1 IRES in RRL requires the addition of exogenous proteins (Figure 1). Echoing previous reports suggesting that activity of the HIV-1 IRES is enhanced in the G2/M phase of the cell cycle (10), G2/M extracts were more effective at stimulation of IRES-mediated translation initiation than G1 or extracts generated from no-synchronous cells (NS in Figure 1). This could be due to the presence of specific G2/M factors, or simply reflect the reduced competition for the translation machinery due to the impairment of cap-dependent translation during the G2/M phase of the cell cycle. The latter possibility was disregarded as the addition of G2/M extracts did not significantly impact translation initiation from the first cistron in two independent translation systems, the RRL and the X. laevis oocytes (Figure 1). In an attempt to resolve this issue and to further characterize the molecular events involved in this process, we show that the addition of cell extracts induces an alteration of the 1M7 modification profile of the HIV-1 leader (Figure 3). The sequences directly upstream the initiation codon appear to be protected with the extracts generated from G2/M arrested cells, possibly reflecting bound G2/M-specific proteins or perhaps even the presence of the initiation complex on the start codon. In good agreement with the functional data, we observe a specific G2/M footprint on the sequences spanning from the poly(A) stem–loop to the SD stem–loop. It should be noted that Brasey et al. (10) previously reported that the poly(A) loop (nucleotide 58–104) is not part of the minimal IRES (nucleotide 104–336), but the PBS structure, the SD loop and the Psi stem–loop are regions previously identified as crucial for HIV-1 IRES activity (10). Our data do not formally allow us to conclude if the protection of specific position within the HIV-1 leader is due to protein or ribosome binding or if we observe some local RNA structural rearrangement. However, beside the case of the poly(A) loop, protections in loop or bulges suggest the presence of proteins on those nucleotides rather than a structural modification. In contrast, the modification pattern obtained with G1 cell extracts suggests a significant alteration of the global structure of the HIV-1 5′ leader. This observation raises the possibility that IRES activity is inhibited in G1 due to a disruption of the active structure. In conclusion, our observations suggest that the G2/M cellular extracts contain proteins that stimulate HIV-1 translation, and that one or several G2/M-specific ITAFs bind within regions known to be critical for the IRES function.
To gain information on the set of proteins from G2/M cell extracts that interact with the HIV-1 5′ leader a proteomic approach was used. Surprisingly, a discrete set of proteins from G2/M arrested extracts pull-down with the HIV-1 leader (Table 2). Interestingly, many of the proteins identified have been previously implicated in HIV-1 replication (Table 2), although their direct role in HIV-1 IRES-mediated translation initiation is still unknown. Together, our data (Figures 1, 3 and 4; Table 2) warrants the notion that a protein complex that forms on the viral RNA somehow imprints the information required to determine IRES activity.
An additional and unexpected finding reported in this study is that the HIV-1 IRES activity is resistant to a number of mutations designed to disrupt RNA structure (29). Mutations introduced in the TAR stem–loop or within the region spanning between the major 5′ splice site and the initiation codon (ML1-ML6) had no significant effect on translation [Figure 4 and references (13,29)]. This confirms the results of a precedent study that shows the TAR element, and the nucleotides beyond the splice site (U291) could be deleted without significantly affecting the IRES-mediated translational activity (10). More surprisingly, mutations within the PBS or the DIS stem–loop which are known to be essential for HIV-1 IRES do not significantly alter the translation efficiency in our bicistronic assay (Figure 4). Even though unexpected, these results are consistent with a recently published report that conducted a similar set of experiments in the context of a bicistronic mRNA (13). Together, these observations are in direct contrast to what is observed with other viral IRESes such as those present within picornavirus, HCV and CrPV RNAs, where simple point mutations that alter their secondary/tertiary RNA structure can totally abolish IRES activity (4,52–55). However, it remains possible that point mutations to as-of-yet undiscovered critical elements in the HIV-1 IRES may have a stronger effect than those explored here.
Interestingly, point mutations or deletions within many cellular IRESes have little impact on translation initiation, suggesting that the structure–function relationship in cellular IRESes is not as rigid as that observed for viral IRESes (4,28,56). In this respect, it is tempting to speculate that the HIV-1 IRES is an atypical viral IRES as it seems to shares certain properties normally ascribed to cellular IRESes (4,28,57,58), but that are absent from their viral counterparts. Moreover, a striking difference between the HIV-1 full-length mRNA and most viral mRNAs that harbor an IRES, is that the former possesses both a 5′ cap and the poly(A) tail. Incorporation of a cap structure onto a picornavirus mRNA inhibits IRES-mediated translation, suggesting that in the context of viral mRNAs the two mechanisms are mutually exclusive (59). Yet, as cellular mRNAs that harbor IRES elements, the HIV-1 mRNA is also capped. However, at this point, we cannot tell if the two mechanisms are used simultaneously or at two different stages of the virus life cycle (8). Additionally, and in sharp contrast to most (+) RNA viruses that harbor IRES elements that are synthesized by a viral RNA-dependent RNA polymerase in the cell cytoplasm where it is translated (27), transcription of the HIV-1 mRNA takes place in the host cell nucleus. As nascent cellular mRNAs, the HIV-1 mRNA would be expected to first encounter RNA binding proteins in the nucleus structuring a distinct ribonucleoprotein (RNP) complex with nuclear RNA binding factors (60,61). These RNA binding proteins of nuclear origin might be part of an ‘IRES RNP’-specific signal that is further modified by interaction with cytoplasmic proteins prior to its associating with the translation apparatus; as would be the case of cellular mRNA that harbor IRES elements (4,27,28).
In summary, in this study, we describe that G2/M extracts harbor factors capable of enhancing HIV-1 IRES activity. Furthermore, we describe that a discrete set of proteins present within G2/M extracts bind the HIV-1 5′ leader. Even though the role on IRES function of the identified proteins is presently unknown, our data are consistent with the notion that cellular proteins are directly involved in the regulation of HIV-1 IRES activity. In fact, current evidence indicate that cellular proteins cannot only stimulate HIV-1 IRES-mediated translation initiation [Figure 1, Table 2 and reference (16)], but they can also inhibit it (25). Thus, we propose a mechanism by which G2/M-specific proteins bind to local structures within the 5′ leader, and subsequently recruit, or stimulate the internal recruitment of the initiation complex. Together, findings presented herein give new insights into the RNA structure/function relationship and provide a valuable framework for further dissection of the molecular mechanism involved in HIV-1 IRES activity.
FUNDING
Grant FONDECYT No. 1090318 and the Proyecto Núcleo Milenio P-07-088-F to M.L.L., grant PICS 5283 (CNRS-CONICYT) (to B.S. and M.L.L.); the Escuela de Medicina de la Pontificia Universidad Católica de Chile (PMD 10/09) and CONICYT travel grant (to M.V.); grant PFB12/2007 to CARE; the Agence Nationale de Recherche contre le SIDA (ANRS) for B.S. Laboratory work; an ‘ATIP’ from CNRS and by SIDACTION. The National Institutes of Health (GM-72560 and GM-81346 to IRES work in JSK’s Lab). CONICYT Doctoral Fellowship (to M.V. and F.V.); le Ministére de l’Éducation et de la Recherche (PhD fellowship to J.D.); VRI-Pontificia Universidad Católica de Chile fellowship (to A.L.); Doctoral Fellow of MECE(2)SUP-UNAB (to P.R.); T-DMP is a Pre-doctoral Fellow of the American Heart Association. Funding for open access charge: Comisión Nacional de Investigación Científica y Tecnológica, Gobierno de Chile, CONICYT, through grant FONDECYT No 1090318 and PICS N°5238 from CNRS-DRI.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr M. Rau (Oxford, UK) for critical reading and editing of the manuscript. We are grateful to Dr Nahum Sonenberg (Department of Biochemistry, McGill University, Canada) for providing some of the plasmids used in this study. We thank Dr M. Ocqueteau and Mr M. Galleguillos for their help with flow cytometry (Departamento de Hemato-Oncología, Facultad de Medicina, Pontificia Universidad Católica de Chile) and Mr F.E. Rodríguez and Dr J.P. Huidobro-Toro for their assistance with the experiments using X. laevis oocytes. We thank the University of Colorado Cancer Center Mass Spectrometry Core for assistance. B.S. and J.D. would like to thank Dr K. Weeks for making available all the methods and software necessary for the ‘SHAPE’ analysis, and Dr L. Micouin (CNRS UMR 8638) for helping in the synthesis of 1M7. M.V. and A.L. conducted this work as part of their PhD Thesis, programa de Doctorado en Ciencias Médicas, Facultad de Medicina, and programa de Doctorado en Ciencias Biológicas, Pontificia Universidad Católica de Chile, respectively. P.R. is a student of the program Doctorado en Biotecnología, at the Universidad Andres Bello. J.S.K. is a Howard Hughes Medical Institute Early Career Scientist.
REFERENCES
- 1.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lopez-Lastra M, Rivas A, Barria MI. Protein synthesis in eukaryotes: the growing biological relevance of cap-independent translation initiation. Biol. Res. 2005;38:121–146. doi: 10.4067/s0716-97602005000200003. [DOI] [PubMed] [Google Scholar]
- 4.Fitzgerald KD, Semler BL. Bridging IRES elements in mRNAs to the eukaryotic translation apparatus. Biochim. Biophys. Acta. 2009;1789:518–528. doi: 10.1016/j.bbagrm.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 7.Balvay L, Lopez Lastra M, Sargueil B, Darlix JL, Ohlmann T. Translational control of retroviruses. Nat. Rev. Microbiol. 2007;5:128–140. doi: 10.1038/nrmicro1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ricci EP, Soto Rifo R, Herbreteau CH, Decimo D, Ohlmann T. Lentiviral RNAs can use different mechanisms for translation initiation. Biochem. Soc. Trans. 2008;36:690–693. doi: 10.1042/BST0360690. [DOI] [PubMed] [Google Scholar]
- 9.Chamond N, Locker N, Sargueil B. The different pathways of HIV genomic RNA translation. Biochem. Soc. Trans. 2011;38:1548–1552. doi: 10.1042/BST0381548. [DOI] [PubMed] [Google Scholar]
- 10.Brasey A, Lopez-Lastra M, Ohlmann T, Beerens N, Berkhout B, Darlix JL, Sonenberg N. The leader of human immunodeficiency virus type 1 genomic RNA harbors an internal ribosome entry segment that is active during the G2/M phase of the cell cycle. J. Virol. 2003;77:3939–3949. doi: 10.1128/JVI.77.7.3939-3949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buck CB, Shen X, Egan MA, Pierson TC, Walker CM, Siliciano RF. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J. Virol. 2001;75:181–191. doi: 10.1128/JVI.75.1.181-191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weill L, James L, Ulryck N, Chamond N, Herbreteau CH, Ohlmann T, Sargueil B. A new type of IRES within gag coding region recruits three initiation complexes on HIV-2 genomic RNA. Nucleic Acids Res. 2010;38:1367–1381. doi: 10.1093/nar/gkp1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gendron K, Ferbeyre G, Heveker N, Brakier-Gingras L. The activity of the HIV-1 IRES is stimulated by oxidative stress and controlled by a negative regulatory element. Nucleic Acids Res. 2011;39:902–912. doi: 10.1093/nar/gkq885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Locker N, Chamond N, Sargueil B. A conserved structure within the HIV gag open reading frame that controls translation initiation directly recruits the 40 S subunit and eIF3. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkq1118. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berkhout B, Arts K, Abbink TE. Ribosomal scanning on the 5′-untranslated region of the human immunodeficiency virus RNA genome. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr113. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monette A, Ajamian L, Lopez-Lastra M, Mouland AJ. Human immunodeficiency virus type 1 (HIV-1) induces the cytoplasmic retention of heterogeneous nuclear ribonucleoprotein A1 by disrupting nuclear import: implications for HIV-1 gene expression. J. Biol. Chem. 2009;284:31350–31362. doi: 10.1074/jbc.M109.048736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goh WC, Rogel ME, Kinsey CM, Michael SF, Fultz PN, Nowak MA, Hahn BH, Emerman M. HIV-1 Vpr increases viral expression by manipulation of the cell cycle: a mechanism for selection of Vpr in vivo. Nat. Med. 1998;4:65–71. doi: 10.1038/nm0198-065. [DOI] [PubMed] [Google Scholar]
- 18.Thierry S, Marechal V, Rosenzwajg M, Sabbah M, Redeuilh G, Nicolas JC, Gozlan J. Cell cycle arrest in G2 induces human immunodeficiency virus type 1 transcriptional activation through histone acetylation and recruitment of CBP, NF-kappaB, and c-Jun to the long terminal repeat promoter. J. Virol. 2004;78:12198–12206. doi: 10.1128/JVI.78.22.12198-12206.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pyronnet S, Dostie J, Sonenberg N. Suppression of cap-dependent translation in mitosis. Genes Dev. 2001;15:2083–2093. doi: 10.1101/gad.889201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alvarez E, Castello A, Menendez-Arias L, Carrasco L. HIV protease cleaves poly(A)-binding protein. Biochem. J. 2006;396:219–226. doi: 10.1042/BJ20060108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ventoso I, Blanco R, Perales C, Carrasco L. HIV-1 protease cleaves eukaryotic initiation factor 4 G and inhibits cap-dependent translation. Proc. Natl Acad. Sci. USA. 2001;98:12966–12971. doi: 10.1073/pnas.231343498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohlmann T, Prevot D, Decimo D, Roux F, Garin J, Morley SJ, Darlix JL. In vitro cleavage of eIF4GI but not eIF4GII by HIV-1 protease and its effects on translation in the rabbit reticulocyte lysate system. J. Mol. Biol. 2002;318:9–20. doi: 10.1016/S0022-2836(02)00070-0. [DOI] [PubMed] [Google Scholar]
- 23.Perales C, Carrasco L, Ventoso I. Cleavage of eIF4G by HIV-1 protease: effects on translation. FEBS Lett. 2003;533:89–94. doi: 10.1016/s0014-5793(02)03764-x. [DOI] [PubMed] [Google Scholar]
- 24.Castello A, Franco D, Moral-Lopez P, Berlanga JJ, Alvarez E, Wimmer E, Carrasco L. HIV-1 protease inhibits Cap- and poly(A)-dependent translation upon eIF4GI and PABP cleavage. PLoS One. 2009;4:e7997. doi: 10.1371/journal.pone.0007997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rivas-Aravena A, Ramdohr P, Vallejos M, Valiente-Echeverria F, Dormoy-Raclet V, Rodriguez F, Pino K, Holzmann C, Huidobro-Toro JP, Gallouzi IE, et al. The Elav-like protein HuR exerts translational control of viral internal ribosome entry sites. Virology. 2009;392:178–185. doi: 10.1016/j.virol.2009.06.050. [DOI] [PubMed] [Google Scholar]
- 26.Liu J, Henao-Mejia J, Liu H, Zhao Y, He JJ. Translational Regulation of HIV-1 Replication by HIV-1 Rev Cellular Cofactors Sam68, eIF5A, hRIP, and DDX3. J. Neuroimmune Pharmacol. 2011 doi: 10.1007/s11481-011-9265-8. In Press. [DOI] [PubMed] [Google Scholar]
- 27.Semler BL, Waterman ML. IRES-mediated pathways to polysomes: nuclear versus cytoplasmic routes. Trends Microbiol. 2008;16:1–5. doi: 10.1016/j.tim.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 28.King HA, Cobbold LC, Willis AE. The role of IRES trans-acting factors in regulating translation initiation. Biochem. Soc. Trans. 2010;38:1581–1586. doi: 10.1042/BST0381581. [DOI] [PubMed] [Google Scholar]
- 29.Abbink TE, Ooms M, Haasnoot PC, Berkhout B. The HIV-1 leader RNA conformational switch regulates RNA dimerization but does not regulate mRNA translation. Biochemistry. 2005;44:9058–9066. doi: 10.1021/bi0502588. [DOI] [PubMed] [Google Scholar]
- 30.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 31.Vallejos M, Ramdohr P, Valiente-Echeverria F, Tapia K, Rodriguez FE, Lowy F, Huidobro-Toro JP, Dangerfield JA, Lopez-Lastra M. The 5′-untranslated region of the mouse mammary tumor virus mRNA exhibits cap-independent translation initiation. Nucleic Acids Res. 2010;38:618–632. doi: 10.1093/nar/gkp890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.James L, Sargueil B. RNA secondary structure of the feline immunodeficiency virus 5′UTR and Gag coding region. Nucleic Acids Res. 2008;36:4653–4666. doi: 10.1093/nar/gkn447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Low JT, Weeks KM. SHAPE-directed RNA secondary structure prediction. Methods. 2010;52:150–158. doi: 10.1016/j.ymeth.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J. Am. Chem. Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 35.Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. ShapeFinder: a software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA. 2008;14:1979–1990. doi: 10.1261/rna.1166808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Windbichler N, Schroeder R. Isolation of specific RNA-binding proteins using the streptomycin-binding RNA aptamer. Nat. Protoc. 2006;1:637–640. doi: 10.1038/nprot.2006.95. [DOI] [PubMed] [Google Scholar]
- 37.Pfingsten JS, Castile AE, Kieft JS. Mechanistic role of structurally dynamic regions in Dicistroviridae IGR IRESs. J. Mol. Biol. 2010;395:205–217. doi: 10.1016/j.jmb.2009.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borman A, Howell MT, Patton JG, Jackson RJ. The involvement of a spliceosome component in internal initiation of human rhinovirus RNA translation. J. Gen. Virol. 1993;74(Pt 9):1775–1788. doi: 10.1099/0022-1317-74-9-1775. [DOI] [PubMed] [Google Scholar]
- 39.Brown BA, Ehrenfeld E. Translation of poliovirus RNA in vitro: changes in cleavage pattern and initiation sites by ribosomal salt wash. Virology. 1979;97:396–405. doi: 10.1016/0042-6822(79)90350-7. [DOI] [PubMed] [Google Scholar]
- 40.Dorner AJ, Semler BL, Jackson RJ, Hanecak R, Duprey E, Wimmer E. In vitro translation of poliovirus RNA: utilization of internal initiation sites in reticulocyte lysate. J. Virol. 1984;50:507–514. doi: 10.1128/jvi.50.2.507-514.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilson JE, Powell MJ, Hoover SE, Sarnow P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol. Cell. Biol. 2000;20:4990–4999. doi: 10.1128/mcb.20.14.4990-4999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gamarnik AV, Andino R. Exploring RNA virus replication in Xenopus oocytes. Methods Mol. Biol. 2006;322:367–378. doi: 10.1007/978-1-59745-000-3_26. [DOI] [PubMed] [Google Scholar]
- 43.Berkhout B. Structure and function of the human immunodeficiency virus leader RNA. Prog. Nucleic Acid Res. Mol. Biol. 1996;54:1–34. doi: 10.1016/s0079-6603(08)60359-1. [DOI] [PubMed] [Google Scholar]
- 44.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc. Natl Acad. Sci. USA. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Damgaard CK, Andersen ES, Knudsen B, Gorodkin J, Kjems J. RNA interactions in the 5′ region of the HIV-1 genome. J. Mol. Biol. 2004;336:369–379. doi: 10.1016/j.jmb.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 46.Paillart JC, Dettenhofer M, Yu XF, Ehresmann C, Ehresmann B, Marquet R. First snapshots of the HIV-1 RNA structure in infected cells and in virions. J. Biol. Chem. 2004;279:48397–48403. doi: 10.1074/jbc.M408294200. [DOI] [PubMed] [Google Scholar]
- 47.Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Paillart JC, Shehu-Xhilaga M, Marquet R, Mak J. Dimerization of retroviral RNA genomes: an inseparable pair. Nat. Rev. Microbiol. 2004;2:461–472. doi: 10.1038/nrmicro903. [DOI] [PubMed] [Google Scholar]
- 49.Huthoff H, Berkhout B. Two alternating structures of the HIV-1 leader RNA. RNA. 2001;7:143–157. doi: 10.1017/s1355838201001881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abbink TE, Berkhout B. A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J. Biol. Chem. 2003;278:11601–11611. doi: 10.1074/jbc.M210291200. [DOI] [PubMed] [Google Scholar]
- 51.Huthoff H, Berkhout B. Multiple secondary structure rearrangements during HIV-1 RNA dimerization. Biochemistry. 2002;41:10439–10445. doi: 10.1021/bi025993n. [DOI] [PubMed] [Google Scholar]
- 52.Fernandez-Miragall O, Lopez de Quinto S, Martinez-Salas E. Relevance of RNA structure for the activity of picornavirus IRES elements. Virus Res. 2009;139:172–182. doi: 10.1016/j.virusres.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 53.Fraser CS, Doudna JA. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat. Rev. Microbiol. 2007;5:29–38. doi: 10.1038/nrmicro1558. [DOI] [PubMed] [Google Scholar]
- 54.Kieft JS. Viral IRES RNA structures and ribosome interactions. Trends Biochem. Sci. 2008;33:274–283. doi: 10.1016/j.tibs.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martinez-Salas E. The impact of RNA structure on picornavirus IRES activity. Trends Microbiol. 2008;16:230–237. doi: 10.1016/j.tim.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Le Quesne JP, Stoneley M, Fraser GA, Willis AE. Derivation of a structural model for the c-myc IRES. J. Mol. Biol. 2001;310:111–126. doi: 10.1006/jmbi.2001.4745. [DOI] [PubMed] [Google Scholar]
- 57.Chappell SA, Edelman GM, Mauro VP. A 9-nt segment of a cellular mRNA can function as an internal ribosome entry site (IRES) and when present in linked multiple copies greatly enhances IRES activity. Proc. Natl Acad. Sci. USA. 2000;97:1536–1541. doi: 10.1073/pnas.97.4.1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chappell SA, Mauro VP. The internal ribosome entry site (IRES) contained within the RNA-binding motif protein 3 (Rbm3) mRNA is composed of functionally distinct elements. J. Biol. Chem. 2003;278:33793–33800. doi: 10.1074/jbc.M303495200. [DOI] [PubMed] [Google Scholar]
- 59.Hambidge SJ, Sarnow P. Terminal 7-methyl-guanosine cap structure on the normally uncapped 5′ noncoding region of poliovirus mRNA inhibits its translation in mammalian cells. J. Virol. 1991;65:6312–6315. doi: 10.1128/jvi.65.11.6312-6315.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell. Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 61.Cochrane AW, McNally MT, Mouland AJ. The retrovirus RNA trafficking granule: from birth to maturity. Retrovirology. 2006;3:18. doi: 10.1186/1742-4690-3-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Holloway AF, Occhiodoro F, Mittler G, Meisterernst M, Shannon MF. Functional interaction between the HIV transactivator Tat and the transcriptional coactivator PC4 in T cells. J. Biol. Chem. 2000;275:21668–21677. doi: 10.1074/jbc.M909058199. [DOI] [PubMed] [Google Scholar]
- 63.Zheng YH, Yu HF, Peterlin BM. Human p32 protein relieves a post-transcriptional block to HIV replication in murine cells. Nat. Cell. Biol. 2003;5:611–618. doi: 10.1038/ncb1000. [DOI] [PubMed] [Google Scholar]
- 64.Berro R, Kehn K, de la Fuente C, Pumfery A, Adair R, Wade J, Colberg-Poley AM, Hiscott J, Kashanchi F. Acetylated Tat regulates human immunodeficiency virus type 1 splicing through its interaction with the splicing regulator p32. J. Virol. 2006;80:3189–3204. doi: 10.1128/JVI.80.7.3189-3204.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fridell RA, Harding LS, Bogerd HP, Cullen BR. Identification of a novel human zinc finger protein that specifically interacts with the activation domain of lentiviral Tat proteins. Virology. 1995;209:347–357. doi: 10.1006/viro.1995.1266. [DOI] [PubMed] [Google Scholar]
- 66.Shoya Y, Tokunaga K, Sawa H, Maeda M, Ueno T, Yoshikawa T, Hasegawa H, Sata T, Kurata T, Hall WW, et al. Human topoisomerase I promotes HIV-1 proviral DNA synthesis: implications for the species specificity and cellular tropism of HIV-1 infection. Proc. Natl Acad. Sci. USA. 2003;100:8442–8447. doi: 10.1073/pnas.1430827100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takahashi H, Matsuda M, Kojima A, Sata T, Andoh T, Kurata T, Nagashima K, Hall WW. Human immunodeficiency virus type 1 reverse transcriptase: enhancement of activity by interaction with cellular topoisomerase I. Proc. Natl Acad. Sci. USA. 1995;92:5694–5698. doi: 10.1073/pnas.92.12.5694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Takahashi H, Sawa H, Hasegawa H, Shoya Y, Sata T, Hall WW, Nagashima K, Kurata T. Topoisomerase I and ATP activate cDNA synthesis of human immunodeficiency virus type 1. Biochem. Biophys. Res. Commun. 2002;294:509–517. doi: 10.1016/S0006-291X(02)00503-X. [DOI] [PubMed] [Google Scholar]
- 69.Tange TO, Jensen TH, Kjems J. In vitro interaction between human immunodeficiency virus type 1 Rev protein and splicing factor ASF/SF2-associated protein, p32. J. Biol. Chem. 1996;271:10066–10072. doi: 10.1074/jbc.271.17.10066. [DOI] [PubMed] [Google Scholar]
- 70.Nisole S, Krust B, Hovanessian AG. Anchorage of HIV on permissive cells leads to coaggregation of viral particles with surface nucleolin at membrane raft microdomains. Exp. Cell. Res. 2002;276:155–173. doi: 10.1006/excr.2002.5522. [DOI] [PubMed] [Google Scholar]
- 71.Ueno T, Tokunaga K, Sawa H, Maeda M, Chiba J, Kojima A, Hasegawa H, Shoya Y, Sata T, Kurata T, et al. Nucleolin and the packaging signal, psi, promote the budding of human immunodeficiency virus type-1 (HIV-1) Microbiol. Immunol. 2004;48:111–118. doi: 10.1111/j.1348-0421.2004.tb03496.x. [DOI] [PubMed] [Google Scholar]
- 72.Kubota S, Adachi Y, Copeland TD, Oroszlan S. Binding of human prothymosin alpha to the leucine-motif/activation domains of HTLV-I Rex and HIV-1 Rev. Eur. J. Biochem. 1995;233:48–54. doi: 10.1111/j.1432-1033.1995.048_1.x. [DOI] [PubMed] [Google Scholar]
- 73.Ashe MP, Pearson LH, Proudfoot NJ. The HIV-1 5′ LTR poly(A) site is inactivated by U1 snRNP interaction with the downstream major splice donor site. EMBO J. 1997;16:5752–5763. doi: 10.1093/emboj/16.18.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li L, Yoder K, Hansen MS, Olvera J, Miller MD, Bushman FD. Retroviral cDNA integration: stimulation by HMG I family proteins. J. Virol. 2000;74:10965–10974. doi: 10.1128/jvi.74.23.10965-10974.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Miller MD, Farnet CM, Bushman FD. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 1997;71:5382–5390. doi: 10.1128/jvi.71.7.5382-5390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pocernich CB, Poon HF, Boyd-Kimball D, Lynn BC, Nath A, Klein JB, Butterfield DA. Proteomic analysis of oxidatively modified proteins induced by the mitochondrial toxin 3-nitropropionic acid in human astrocytes expressing the HIV protein tat. Brain Res. Mol. Brain Res. 2005;133:299–306. doi: 10.1016/j.molbrainres.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 77.Shoeman RL, Sachse C, Honer B, Mothes E, Kaufmann M, Traub P. Cleavage of human and mouse cytoskeletal and sarcomeric proteins by human immunodeficiency virus type 1 protease. Actin, desmin, myosin, and tropomyosin. Am. J. Pathol. 1993;142:221–230. [PMC free article] [PubMed] [Google Scholar]
- 78.Campbell GR, Pasquier E, Watkins J, Bourgarel-Rey V, Peyrot V, Esquieu D, Barbier P, de Mareuil J, Braguer D, Kaleebu P, et al. The glutamine-rich region of the HIV-1 Tat protein is involved in T-cell apoptosis. J. Biol. Chem. 2004;279:48197–48204. doi: 10.1074/jbc.M406195200. [DOI] [PubMed] [Google Scholar]
- 79.Canani RB, De Marco G, Passariello A, Buccigrossi V, Ruotolo S, Bracale I, Porcaro F, Bifulco G, Guarino A. Inhibitory effect of HIV-1 Tat protein on the sodium-D-glucose symporter of human intestinal epithelial cells. AIDS. 2006;20:5–10. doi: 10.1097/01.aids.0000198088.85572.68. [DOI] [PubMed] [Google Scholar]
- 80.de Mareuil J, Carre M, Barbier P, Campbell GR, Lancelot S, Opi S, Esquieu D, Watkins JD, Prevot C, Braguer D, et al. HIV-1 Tat protein enhances microtubule polymerization. Retrovirology. 2005;2:5. doi: 10.1186/1742-4690-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Valenzuela-Fernandez A, Alvarez S, Gordon-Alonso M, Barrero M, Ursa A, Cabrero JR, Fernandez G, Naranjo-Suarez S, Yanez-Mo M, Serrador JM, et al. Histone deacetylase 6 regulates human immunodeficiency virus type 1 infection. Mol. Biol. Cell. 2005;16:5445–5454. doi: 10.1091/mbc.E05-04-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Watts NR, Sackett DL, Ward RD, Miller MW, Wingfield PT, Stahl SS, Steven AC. HIV-1 rev depolymerizes microtubules to form stable bilayered rings. J. Cell. Biol. 2000;150:349–360. doi: 10.1083/jcb.150.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]