Abstract
Human immunodeficiency virus-1 (HIV-1) infection leads to changes in cellular gene expression, which in turn tend to modulate viral gene expression and replication. Cellular heat shock proteins (HSPs) are induced upon heat shock, UV irradiation and microbial or viral infections. We have reported earlier Nef-dependent induction of HSP40 leading to increased HIV-1 gene expression; however, the mechanism of induction remained to be elucidated. As expression of HSPs is regulated by heat shock factors (HSFs), we have now studied the role of HSF1 not only in Nef-dependent HSP40 induction but also in HIV-1 gene expression. Our results show that HSF1 is also induced during HIV-1 infection and it positively regulates HIV-1 gene expression by two distinct pathways. First, along with Nef it activates HSP40 promoter which in turn leads to increased HIV-1 gene expression. Second, HSF1 directly interacts with newly identified HSF1 binding sequence on HIV-1 LTR promoter and induces viral gene expression and replication. Thus, the present work not only identifies a molecular basis for HSF1-mediated enhancement of viral replication but also provides another example of how HIV-1 uses host cell machinery for its successful replication in the host.
INTRODUCTION
Cellular heat shock proteins (HSPs) are molecular chaperones primarily involved in protein folding, transport and assembly. In addition, some of these proteins are specifically induced during stress conditions like heat shock, UV irradiation and microbial/viral infection (1,2). Recent studies have revealed that HSPs are also involved in apoptosis and immune response (3,4). Viruses modulate expression of many cellular proteins for their successful replication and induction of HSPs has been reported as one of the earliest change following viral infection (5). Human immunodeficiency virus-1 (HIV-1) was reported to induce HSP27 and HSP70 expression during early infection (6).
HIV-1 Nef, a 27–30 kDa myristoylated phosphoprotein, contributes to viral pathogenesis by modulating cellular gene expression and signaling pathways (7). Nef has been also implicated in the activation of T cells, making the cells permissible to the virus (8). Although initially reported as a negative factor for HIV-1 replication in T-cell lines (9,10), Nef has been later demonstrated to be an enhancer of virus replication (11–14). However, the molecular mechanism of this positive effect remains to be clearly understood. We have earlier shown that Nef interacts with HSP40, and this interaction was necessary for Nef mediated increase in viral gene expression and replication. Furthermore, it was also shown that HSP40 expression increased in HIV-1 NL4-3 transfected cells in a Nef-dependent manner (15). However, the mechanism of HSP40 upregulation during HIV-1 infection remains to be elucidated.
The inducible expression of HSPs is primarily regulated by heat shock factors (HSFs). HSF1 is the major transcription factor that regulates the transcription of HSP genes in response to stress. It binds to conserved regulatory sequences in the HSP promoters known as heat shock elements (HSE), which is represented by two or three inverted repeats of the sequence nGAAn (16). Normally, HSF1 is predominantly present in a cytoplasmic monomeric inactive form; however, upon stress it gets homo-trimerized and translocated to nucleus and acquires high affinity HSE binding and transcription enhancing activity. Recent studies indicate that phosphorylation also plays a major role in regulation of HSF1 activity; specifically Ser230 and Ser326 are inducibly phosphorylated during stress resulting in increased transcriptional activity (17,18). The response of HIV-1 to various stress proteins, including HSPs, could also lead to modulation of HIV-1 long terminal repeat promoter (LTR)-driven gene expression (19). Several studies have suggested that heat shock could activate the LTR-driven transcription in cells (20,21); however, the mechanism of activation has not been clearly understood (22).
During our efforts to understand the mechanism of Nef-dependent upregulation of HSP40 in HIV-1 infection, we have identified the importance of HSF1 in HIV-1 gene expression and replication in the present study. Our results clearly show that HSF1 positively regulates HIV-1 gene expression and replication by two distinct pathways. First, it induces HSP40 expression in association with viral protein Nef, both of which has been earlier shown to be required for increased viral gene expression (15). Second, activated HSF1 directly interacts with LTR to induce viral gene expression and replication.
MATERIALS AND METHODS
Cell lines, plasmids and reagents
HIV-1 NL4-3 Nef expression vector pcDNA-Nef was obtained from Dr M. Federico (23). NL4-3 Nef and its point mutants tagged with HA (HA-Nef) plasmids were obtained from Dr W.C. Greene (24). HSF1 encoding plasmid pCMV-HSF1-Flag was a gift of Dr J. Goldman. This HSF1 was further subcloned in pET-28a(+) vector (Novagen, USA). HIV-1 NL4-3 molecular clone (pNL4-3) was obtained from the NIH AIDS repository (25). The nef-deleted NL4-3 molecular clone (pNL4–3ΔNef) was obtained from Dr J.C. Guatelli (26). The HIV-1 LTR reporter vector, pLTR luc was cloned in our laboratory as reported earlier (27). HIV-1 LTR and its deletion mutant constructs of CD series were obtained from Dr T. Okamoto (28). HSP40 promoter construct was obtained from Dr K. Ohtsuka (29), which was used to subclone HSP40 promoter in pGL3 luciferase vector. HEK-293T and Jurkat cells were obtained from the NCCS Cell Repository, India. CEM-GFP, a CD4+ human T-cell line, was obtained from the NIH AIDS repository, USA (30). A polyclonal anti-Nef serum was obtained from Dr S. Jameel (31). Monoclonal and polyclonal Nef antibodies were obtained from Chemicon, USA and NIH AIDS Repository, respectively. Antibodies against HSP40, HA-tag and phospho-HSF1 were obtained from Santa Cruz Biotechnology, USA. Monoclonal and polyclonal HSF1 antibodies were obtained from Chemicon and Cell Signaling, USA, respectively. HSF1 Ab-4 cocktail (Clones 4B4 + 10H4 + 10H8 Rat monoclonal antibody) was obtained from Thermo-Fisher Scientific, USA. Mouse monoclonal Flag-tag and Tubulin antibodies were obtained from Sigma, USA. Control and Sp1 siRNA pools were from Santa Cruz Biotechnology, USA and HSF1 siRNA pool was obtained from Dharmacon, USA.
Transient transfection and luciferase assay
HEK-293T cells were transfected with reporter constructs along with other expression vectors using calcium phosphate precipitation method and cells were harvested 36 h post-transfection for luciferase assay. For siRNA experiments, cells were first transfected with siRNA using Lipofectamine 2000, followed by second transfection with other vectors after 24 h. The cells were then lysed and analyzed for luciferase activity using Luclite substrate (PerkinElmer Life Sciences, USA). Luciferase assays were analyzed using TopCount microplate reader (PerkinElmer Life Sciences, USA). The results were normalized with EGFP expression using Fluoroskan Ascent microplate reader (Thermo Labsystems, USA).
HIV-1 infection and virus quantitation
CEM-GFP or Jurkat cells (5 × 106) were infected with HIV-1 NL4-3 virus at 0.1 multiplicity of infection (MOI) in the presence of Polybrene (1 µg/ml) as described earlier (15). Human peripheral blood was collected from normal seronegative donors and PBMCs were isolated using Ficoll-Hypaque (Amersham Bioscience, USA). The cells were activated with PHA and later infected at 0.5 MOI of NL4-3 virus as described in detail earlier (15). The culture supernatants from NL4-3 infected and transfected cells were used to determine virus production by p24gag antigen capture ELISA (PerkinElmer Life Sciences, USA).
Immunoprecipitation, His pull-down and immunoblotting
HEK-293T cells overexpressing HA-Nef and HSF1 were lysed in lysis buffer (50 mM Tris–HCl pH 7.4, 5 mM EDTA, 0.12 M NaCl, 0.5% NP40, 0.5 mM NaF, 1 mM DTT, 0.5 mM PMSF) on ice for 45 min. Clarified lysates were incubated with anti-Flag antibody and the antigen–antibody complex was pulled down by an equal mixture of protein A and G agarose beads followed by resolution on 12% SDS–PAGE. Proteins were transferred on to PVDF membrane and probed with HA antibody. The blots were developed by the ECL Plus system (Amersham Biosciences, USA). Similar co-immunoprecipitation experiment was performed with HIV-1-infected CEM-GFP cell lysates, which were immunoprecipitated with Nef antibody followed by immunoblotting with HSF1 antibody.
Escherichia coli BL21 (DE3) cells expressing His-HSF1 were induced with isopropyl β-d-thiogalactoside followed by purification of His-HSF1 protein using Ni-NTA beads (Qiagen, Germany). Transfected 293T cells, overexpressing wild-type and different mutants of Nef were lysed in cold lysis buffer (25 mM HEPES, pH 7.3, 0.1 M NaCl, 5 mM EDTA, 0.5% Triton X-100 and 1 mM DTT) with protease inhibitor cocktail (Roche Applied Bioscience, Germany). The clarified lysates were incubated with His-HSF1 protein in binding buffer pH 8.0 (50 mM NaH2PO4, 300 mM NaCl, 10 mM Imidazole and 0.1% Tween-20) overnight at 4°C followed by pull down with Ni-NTA beads. The complexes were then resolved on 12% SDS–PAGE. Proteins were transferred onto PVDF membrane and were probed with polyclonal HA-Nef antibody. Furthermore, HIV-1-infected CEM-GFP and Jurkat cell lysates were run on SDS–PAGE, followed by immunoblotting for HSP40 and HSF1, respectively.
Reverse transcription PCR
RNA was prepared from 2 × 106 HIV-1 NL4-3-infected CEM-GFP cells using TRIzol Reagent (Invitrogen, USA). The cDNA was prepared using MMLV Reverse Transcriptase (Invitrogen, USA) followed by PCR amplification for HSP40 and human β-Actin with Taq polymerase (Invitrogen, USA) using standard conditions and gene-specific primers listed in Supplementary Table S1.
Construction of HSP40 promoter point mutant reporter vectors by site-directed mutagenesis
HSP40 wild-type promoter construct was used as a template for creating mutation at different transcription factor binding sites by PCR amplification using different mutagenic primers (see Supplementary Table S2) and Quik change site directed mutagenesis kit (Stratagene, USA) following manufacturer's instructions.
Preparation of nuclear and cytoplasmic fraction
Uninfected and HIV-1-infected Jurkat cells were used to prepare nuclear and cytoplasmic extracts using ProteoJet Cytoplasmic and Nuclear Extraction Kit (Fermentas, Germany). These extracts were used to study phosphorylation of HSF1 using pHSF1 (Ser230) antibody.
Chemical cross-linking by ethylene glycol bis(succinimidylsuccinate)
Uninfected and HIV-1-infected CEM-GFP cells (5 × 106) were resuspended in 100–200 µl cold lysis buffer followed by lysis on ice for 30 min. The cell lysate was spun at 12 000 rpm for 10 min at 4°C. About 100 µg of protein was used for cross-linking using ethylene glycol bis(succinimidylsuccinate) (EGS) as described in details elsewhere (32).
Electrophoretic mobility shift assay
Various synthetic oligonucleotide probes spanning the NF-κB-Sp1 region of HIV-1 LTR were labeled using [γ32P] dATP by T4 polynucleotide kinase (NEB, USA) for 30 min at 37°C and were purified by Probequant G-50 columns (Amersham Biosciences, USA). Binding reactions were set up with both recombinant His-HSF1 protein and nuclear extract from HIV-1-infected Jurkat cells. Binding reactions were incubated at 37°C for 15 min and loaded on 6% polyacrylamide gel. The nucleotide sequence of probes P1, P2, P3 and P4 are listed in Supplementary Table S3.
Chromatin immunoprecipitation
CEM-GFP cells (2 × 107) were infected with NL4-3 virus at 0.1 MOI. On Day 5 post-infection, cells were harvested and fixed with 1% formaldehyde. Following fixation, chromatin immunoprecipitation (ChIP) was performed by primary immunoprecipitation with Nef polyclonal antibody using ChIP assay kit (Upstate Biotechnology, USA) according to manufacturer's instructions. In sequential ChIP experiment, the eluate after Nef immunoprecipitation was re-incubated with either rat monoclonal HSF1 antibody or rabbit polyclonal Sp1 antibody overnight at 4°C. The occupancy on HSP40 promoter was checked using different primers as schematically shown in Figure 2C. The primers used for PCR amplification of HSP40 promoter region is given in Supplementary Table S4. Similar ChIP analysis was also performed for recruitment of HSF1 on the HIV-1 LTR promoter using HSF1 antibody. The sequence of primers used in the LTR ChIP is listed in Supplementary Table S5.
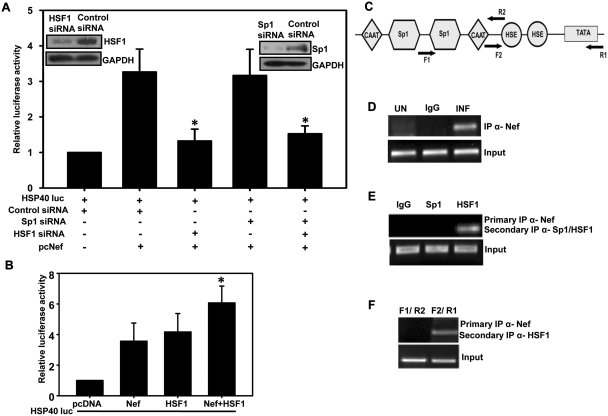
Figure 2.
Nef and HSF1 proteins are co-recruited on HSE of HSP40 promoter in HIV-1-infected cells. (A) HSF1 is required for Nef-mediated activation of HSP40 promoter. HSF1 and Sp1 depleted 293T cells were co-transfected with HSP40-luc and Nef expression vectors and analyzed for luciferase activity. Gene silencing efficiency of HSF1 and Sp1 siRNAs are shown as inset. (B) Both HSF1 and Nef activate HSP40 promoter-driven gene expression in 293T cells as analyzed by luciferase assay. (C) Schematic representation of HSP40 promoter showing the position of primers used in ChIP analysis. (D) Nef is recruited on HSP40 promoter during HIV-1 infection. ChIP analysis was performed with HIV-1-infected CEM-GFP cells using Nef antibody followed by PCR amplification using F2 and R1 primers. (E) Nef and HSF1 are co-recruited on HSP40 promoter as analyzed by sequential ChIP. Primary immunoprecipitation was performed with Nef and secondary immunoprecipitaion with HSF1 or Sp1 antibody followed by PCR analysis using F2 and R1 primers. (F) Nef and HSF1 is recruited specifically at HSE elements on HSP40 promoter. Sequential ChIP analysis was performed as in E above using either F1 and R2 or F2 and R1 primer sets. The error bars represent the mean ± SEM of three independent experiments. Statistical analysis was performed using Student's t-test, with the levels of significance defined as *P < 0.05.
Immunofluorescence microscopy
HEK-293T cells grown on cover slips were transfected with both pcDNA-Nef and pCMV-HSF1 or pNL4-3 by Lipofectamine 2000 and harvested 24 h post-transfection for immunofluorescence studies. Paraformaldehyde-fixed and permeabilized cells were blocked with 10% FCS and stained with HSF1 and Nef antibody, respectively. The secondary antibodies used for HSF1 and Nef were Cy3-conjugated goat anti-rabbit IgG and Cy2-conjugated to goat anti-mouse IgG, respectively. After washing, cells were counterstained with DAPI present in the mounting media and the slides were analyzed with a confocal microscope (Zeiss LSM 510, Germany). Similarly, uninfected and infected Jurkat cells were fixed and were spun on a glass slide to obtain a monolayer of cells. Then the cells were stained with polyclonal HSF1 antibody or pHSF1 (Ser230) antibody. The secondary antibody used was Cy3-conjugated goat anti-rabbit IgG. Cells were counterstained with DAPI and analyzed with a confocal microscope.
Quantitation of HSF1 expression by RT–PCR
HSF1 expression was analyzed by quantitative real-time RT–PCR in a 10 µl reaction mixture containing SYBR Green IQ supermix (Bio-Rad, USA) and 10 pmol concentration of each of the human GAPDH and HSF1 primer pairs (see Supplementary Table S6) using the Realplex4 Mastercycler (Eppendorf, Germany). The amplification was performed using one cycle of 95°C for 2 min and 40 cycles of 94°C for 1 min, 60°C for 30 s and 68°C for 1 min followed by melt curve analysis. The changes in the threshold cycle (CT) values were calculated by the equation ΔCT = CT,target−CT,input. The fold difference was calculated as follows:
Statistical analysis
All experiments were repeated at least three times. The error bars represent the mean ± SEM of three independent experiments. Statistical analysis of the experimental data was performed using Student's t-test, with the levels of significance defined as *P < 0.05.
RESULTS
HIV-1 Nef induces HSP40 promoter activity
We have reported earlier that HSP40 is induced in Nef-dependent manner in HIV-1 NL4-3 virus transfected HEK-293T cells (15). In order to confirm this finding in HIV-1-infected T cells, we first infected CEM-GFP cells with wild-type and nef-deleted NL4-3 virus and analyzed HSP40 expression on different days post-infection by both RT–PCR and immunoblotting. As shown in Figure 1A and B, HSP40 was induced in HIV-1 NL4-3-infected cells in a time-dependent manner. However, this increase was not observed in Nef deleted NL4-3-infected cells (Figure 1C and D) confirming that HSP40 upregulation during HIV-1 infection is dependent on the Nef protein.
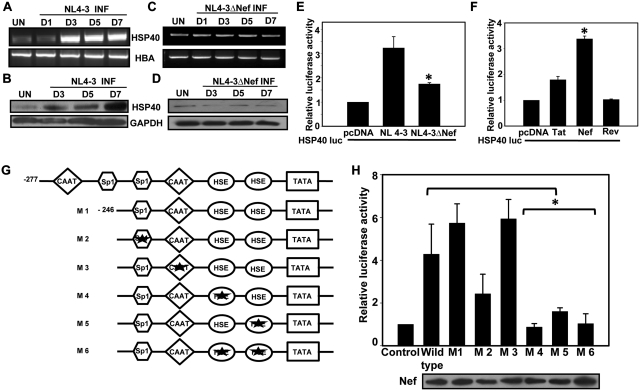
Figure 1.
Heat shock elements are required for Nef-dependent upregulation of HSP40 promoter-driven gene expression. (A and B) Expression profile of HSP40 in HIV-1 NL4-3-infected CEM-GFP cells as analyzed by RT–PCR and immunoblotting. (C and D) Expression profile of HSP40 in HIV-1 NL4-3ΔNef-infected CEM-GFP cells as analyzed by RT–PCR and immunoblotting. (E) Nef is required for HSP40 promoter activation by NL4-3 virus in transfected 293T cells. (F) HSP40 promoter is specifically activated by HIV-1 Nef but not Tat and Rev in 293T cells. (G) Schematic representation of HSP40 promoter and its mutants. Star marks indicate the mutated site. (H) Nef induces HSP40 promoter through HSE elements. 293T cells co-transfected with various HSP40 promoter-luc construct along with Nef expression vector were analyzed for luciferase activity (top) and Nef expression (bottom). The error bars represent the mean ± SEM of three independent experiments. Statistical analysis was performed using Student's t-test, with the levels of significance defined as *P < 0.05.
In order to understand the mechanism of HSP40 induction, we first looked at the role of HIV-1 and Nef on the HSP40 promoter activation, if any. HEK-293T cells were co-transfected with HSP40 promoter-luciferase construct along with wild-type or nef deleted NL4-3 molecular clone followed by analysis of luciferase activity. The result clearly shows that HSP40 promoter activity was significantly induced by wild-type NL4-3 but not with the nef deleted clone (Figure 1E) suggesting thereby that Nef was responsible for induction of HSP40 promoter activity. In order to confirm this observation, we then co-transfected some individual HIV-1 protein expressing vectors along with HSP40-luciferase construct in 293T cells. The results clearly show that Nef specifically induces HSP40 promoter activity whereas Tat and Rev did not show any significant effect (Figure 1F). We then wanted to identify the region of HSP40 promoter (29) involved in Nef-mediated induction of its activity, for which we created different deletion and point mutations in the promoter as represented in Figure 1G. These mutant constructs were then used in co-transfection assay along with Nef in 293T cells followed by luciferase activity analysis. As there was no significant change in the activity of −246 promoter mutant with Nef as compared to the wild-type HSP40 (−277) promoter-luc construct, the point mutants were made in the −246 HSP40 promoter sequence. The activity analysis of mutants show that HSE elements in the HSP40 promoter were necessary for Nef-mediated upregulation, whereas CAAT box did not seem to play any role (Figure 1H). Mutation at Sp1 sites also seems to moderately inhibit Nef-induced HSP40 promoter activity (Figure 1H). Thus above results indicate the possible involvement of HSF1 and Sp1 transcription factors in Nef-mediated upregulation of HSP40 promoter activity.
HSF1 is required for Nef-mediated upregulation of HSP40 in HIV-1-infected cells
In order to establish the requirement of HSF1 and Sp1 for Nef-mediated induction of HSP40 promoter, endogenous HSF1 and Sp1 expression was then silenced by gene specific siRNA transfection in 293T cells (Figure 2A, inset). These cells were then co-transfected with Nef and HSP40 promoter-luciferase construct. Our results show that silencing of Sp1 did not significantly reduce Nef-mediated induction of HSP40 promoter, whereas HSF1 knockdown resulted in significant reduction in promoter activity as compared to control siRNA transfected cells (Figure 2A). When both Sp1 and HSF1 were silenced, the HSP40 promoter activity was the same as that observed with HSF1 alone silenced cells. These results suggest that HSF1 plays an important role in Nef induced HSP40 expression. As HSF1 was known to be involved in regulation of HSP promoters by interacting with HSE elements and Nef was also observed to mediate its effects through HSE elements above, we then co-transfected HSF1 or Nef along with HSP40 promoter-luciferase construct to analyze the promoter activity. The results indicate that although Nef or HSF1 alone can induce the promoter activity but when expressed together, they show significantly more effect (Figure 2B).
As the inducible effect of Nef on HSP40 promoter seems to be associated with HSF1, we then looked at the recruitment of Nef on HSP40 promoter at HSE elements as shown in Figure 2C by ChIP assay. Cross-linked chromatin from HIV-1-infected cells was pulled down with Nef antibody followed by PCR amplification using primers F2 and R1 (Figure 2C) spanning HSE region of HSP40 promoter. Strong PCR signal was observed only in infected cells (Figure 2D, lane 3) indicating recruitment of Nef on HSP40 promoter sequence encompassing HSE elements. Since HSE is the consensus site for HSF1, we then checked the co-occupancy of Nef/HSF1 or Nef/Sp1 on the HSE elements (Figure 2E) by sequential ChIP assay. Co-recruitment of Nef and Sp1 was not detected (Figure 2E, lane 2), whereas recruitment of both Nef and HSF1 on HSE elements was clearly observed in HIV-1-infected cells (Figure 2E, lane 3). Also, no detectable amplification was observed in PCR using F1 and R2 primers encompassing a region upstream to HSE elements (Figure 2F, lane 1) whereas strong PCR signal was observed with primers F2 and R1 encompassing the region containing just HSE elements (Figure 2F, lane 2), indicating thereby specific recruitment of HSF1 and Nef at this site. These results further potentiate our finding that HSF1 is necessary for Nef induced HSP40 gene expression.
Nef interacts with human HSF1 both in vitro and in vivo
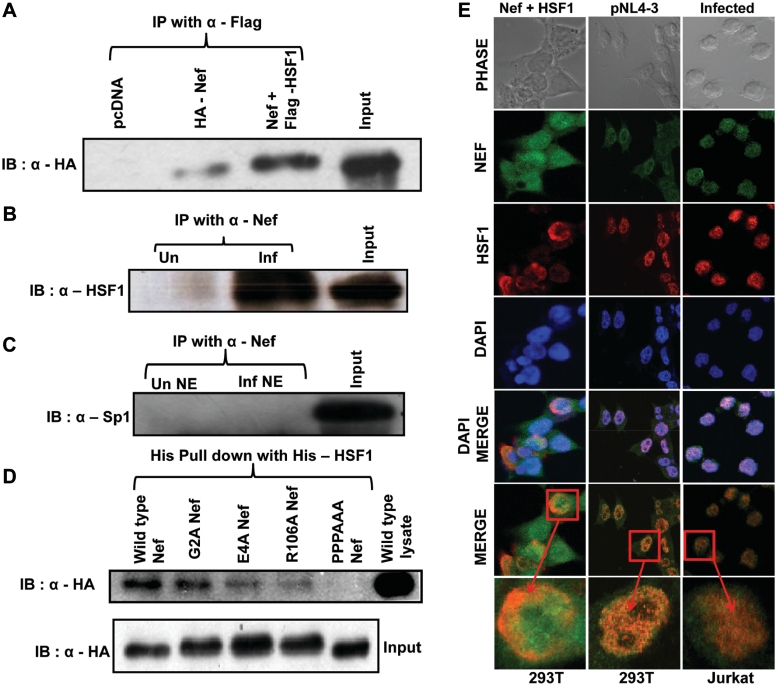
In order to test whether the co-recruitment of Nef and HSF1 on HSP40 promoter in vivo was a result of their physical interaction, we performed co-immunoprecipitation with lysates of 293T cells expressing Flag-HSF1 and HA-Nef. The Nef protein co-immunoprecipitated with human HSF1 (Figure 3A, lane 3) in cells expressing both the proteins. Similar co-immunoprecipitation experiment with HIV-1-infected CEM-GFP cell lysates also showed interaction of HSF1 with Nef in infected cells (Figure 3B). We have also looked at the possible interaction of Nef with Sp1 in HIV-1-infected CEM-GFP nuclear extract, but we did no find any interaction between Nef and Sp1 (Figure 3C). Thus both HSF1 and Nef seem to interact in HIV-1-infected cells and might exist as a complex on HSP40 promoter for its induction. To further identify the HSF1 interacting domain of Nef, purified His-HSF1 was incubated with 293T cell lysates expressing Nef wild-type or mutant proteins followed by pull down of complex by immobilization on Ni-NTA beads (Figure 3D). As expected Nef protein was specifically pulled down by His-HSF1. Although there was some reduction in binding with all the nef mutants but mutation in proline-rich motif of Nef between amino acids 69–78 completely abolished its interaction with HSF1 (lane 5, Figure 3D). This result further confirms that Nef and HSF1 specifically interact with each other and proline rich motif of Nef seems to play an important role in this interaction.
Figure 3.
HIV-1 Nef physically interacts and co-localize with HSF1 both in vitro and in vivo. (A) HSF1 and Nef interact in 293T cells overexpressing HA-Nef and Flag-HSF1 as analyzed by co-immunoprecipitation. The input indicates lysate prepared from 293T cells co-transfected with HA-Nef and Flag-HSF1 expression vectors. (B) Nef co-immunoprecipitates with HSF1 in HIV-1-infected CEM-GFP cells. The input indicates lysate prepared from CEM-GFP cells infected with HIV-1. (C) Nef and Sp1 do not interact in HIV-1-infected CEM-GFP cells. The input indicates lysate prepared from CEM-GFP cells infected with HIV-1. (D) Proline-rich motif of Nef is important for interaction with HSF1. Lysates of HEK-293T cells expressing different HA-tagged Nef and its mutants were used for His-pull down with purified His-tagged HSF1. The input indicates the lysates prepared from wild-type and mutant Nef transfected 293T cells. (E) Nef and HSF1 co-localize in Nef and HSF1 expressing cells. Immunofluorescence studies were performed with 293T cells transfected with Nef and HSF1 vectors (left column), pNL4-3 transfected 293T cells (middle column) and HIV-1-infected Jurkat cells using Nef and HSF1 antibody (right column). The bottom panel in each column is a magnified image of one cell from the merged image panel. Arrows indicate co-localization of Nef and HSF1.
HSF1 has been reported to be localized in the nucleus after heat shock or stress (16,33), whereas Nef was reported to be a predominantly cytoplasmic protein but was also shown to be present in the nucleus of HIV-1-infected cells (34,35). As these proteins were found to physically interact, we then performed immunofluorescence staining for both HSF1 and Nef in transfected 293T and HIV-1-infected Jurkat cells. As shown in Figure 3E, confocal microscopic analysis clearly indicate that both proteins co-localize in the nucleus of Nef and HSF1 cotransfected, pNL4-3 transfected and HIV-1-infected cells (Figure 3E). All these results further confirm that Nef and HSF1 not only interact with each other but they also co-localize in nucleus and get recruited on HSP40 promoter during HIV-1 infection as a complex.
HSF1 is induced in HIV-1-infected cells
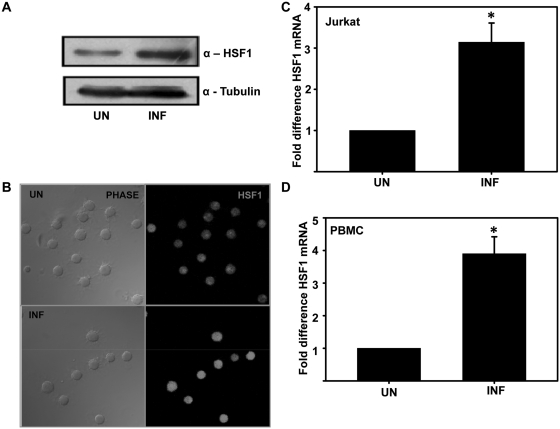
As HSF1 expression status has not been studied during HIV-1 infection, we then assessed HSF1 expression in HIV-1-infected Jurkat cells by both immunoblotting and immunofluorescence. The results show that HSF1 was upregulated in HIV-1-infected Jurkat cells (Figure 4A and B). We further confirmed this finding by quantitative real-time PCR using RNA prepared from uninfected and HIV-1-infected (Day 5) Jurkat cells and human PBMCs. As shown in Figure 4C and D, HSF1 expression was induced in HIV-1-infected Jurkat and PBMCs. All these data clearly indicate upregulation of HSF1 during HIV-1 infection.
Figure 4.
HSF1 is upregulated during HIV-1 infection. (A) HSF1 expression in HIV-1 NL4-3-infected Jurkat cells on Day 5 post-infection as analyzed by immunoblotting. (B) Immunofluorescence analysis of HSF1 expression in HIV-1-infected Jurkat cells. (C) qRT–PCR analysis of HSF1 mRNA expression in HIV-1-infected Jurkat cells on Day 5 post-infection. (D) qRT–PCR analysis of HSF1 mRNA expression in HIV-1-infected PBMCs on Day 5 post-infection. GAPDH was used as internal control for normalization. The error bars represent the mean ± SEM of three independent experiments. Statistical analysis was performed using Student's t-test, with the levels of significance defined as *P < 0.05.
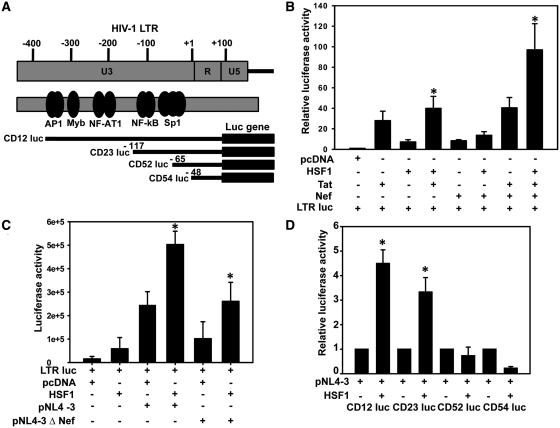
HSF1 enhances HIV-1 LTR-driven gene expression
As HSF1 expression is induced during HIV-1 infection and it has been implicated in LTR-mediated gene expression, we then analyzed the effect of HSF1 on LTR-driven gene expression (Figure 5A). HSF1 overexpression along with Tat or Nef significantly induced LTR-driven gene expression, as compared to Tat or Nef alone or together (Figure 5B). This result indicates that HSF1 activates LTR-driven gene expression.
Figure 5.
HSF1 induces HIV-1 LTR-driven gene expression in Nef independent manner. (A) Schematic representation of HIV-1 LTR and the LTR-luc mutants used in the present study. (B) HSF1 enhances HIV-1 LTR-driven luciferase expression in 293T cells co-transfected with Tat and Nef. (C) HSF1 induces LTR-driven luciferase expression independent of Nef. 293T cells were transfected with pNL4-3 or Nef deleted pNL4-3 along with LTR-luc and HSF1 vectors and were analyzed for luciferase activity. (D) HSF1 induces HIV-1 LTR activity through its enhancer region. 293T cells were transfected with different LTR-luc mutants along with HSF1 and pNL4-3 and luciferase activity was analyzed 36 h post-transfection. The error bars represent the mean ± SEM of three independent experiments. Statistical analysis was performed using Student's t-test, with the levels of significance defined as *P < 0.05.
As HSP40 gene expression was induced in Nef-dependent manner during HIV-1 infection, we then investigated role of Nef in HSF1-mediated induction of LTR-driven gene expression. As shown in Figure 5C, HSF1 induced LTR-driven gene expression in both wild-type and nef-deleted NL4-3 transfected cells, indicating that activity of HSF1 on LTR was Nef independent. We then wanted to identify the region of LTR which was involved in HSF1-mediated induction of LTR activity. We used different LTR deletion mutants (schematically presented in Figure 5A) for co-transfection with HSF1 and pNL4-3 in 293T cells followed by reporter assay. The results indicate that, region encompassing NF-κB and Sp1 elements (−117 to −65) is involved in HSF1-mediated induction of LTR activity (Figure 5D). Deletion of this region, as present in CD52 luc (−65 luc) did not show any induction with HSF1, suggesting the role of NF-κB-Sp1 enhancer region of LTR in HSF1-mediated activation.
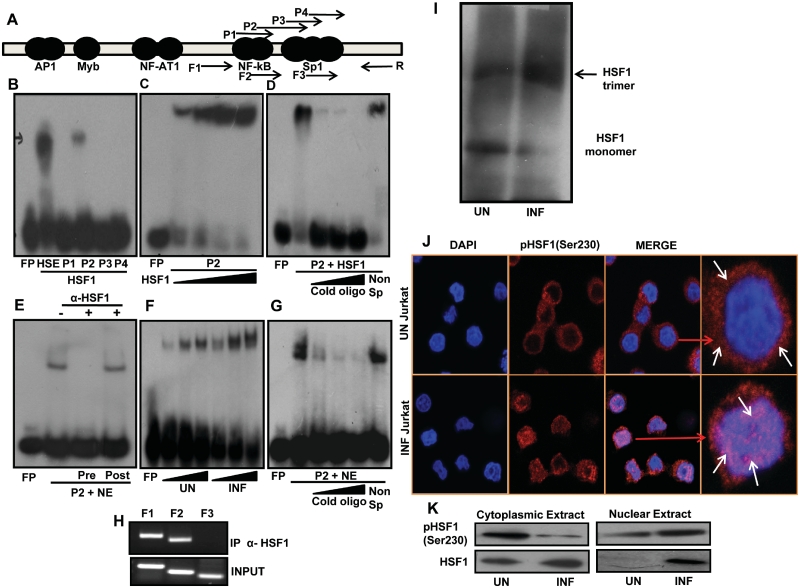
HSF1 induces HIV-1 gene expression by interacting with newly identified HSF1 binding sequence in the LTR
We have shown above that HSF1-mediated induction of LTR is dependent upon the enhancer region. In order to identify the HSF1 binding sequence in HIV-1 LTR, we then analyzed the LTR nucleotide sequence using TFSEARCH program version 1.3 (36), which revealed putative HSF1 binding site in LTR enhancer region (−69 to −91, data not shown). Based on this prediction, we made four overlapping oligonucleotide probes P1, P2, P3 and P4 encompassing the enhancer region as shown in Figure 6A. Recombinant His-HSF1 protein was used in electrophoretic mobility shift assay (EMSA) with these four probes along with consensus HSE probe as a positive control. Interestingly, probes P1, P3 and P4 failed to form any complex whereas P2 formed nucleoprotein complex with His-HSF1 (Figure 6B). P2 comprises of nucleotide sequence from −69 to −91 of LTR promoter. To further confirm the specificity of HSF1 binding, we analyzed P2 binding with increasing amount of His-HSF1 protein, which resulted in a dose dependent increase in complex formation (Figure 6C). Binding specificity of P2 for HSF1 was also confirmed by competition experiments with specific and non-specific oligo (Figure 6D). In order to see the binding with cellular HSF1, we then used nuclear extract from HIV-1-infected Jurkat cells. P2 formed nucleoprotein complex with HIV-1-infected nuclear extract (Figure 6E). Loss of P2 binding was observed in pre-incubation with HSF1 antibody; however, post-incubation with anti-HSF1 did not show any effect on binding (Figure 6E). This shows that HSF1 antibody specifically blocks the binding between P2 and HSF1. Furthermore, increased P2 binding was observed with infected nuclear extract (Figure 6F), which could be explained by increased HSF1 expression during HIV-1 infection. P2 binding specificity was further checked by competition experiment that showed loss of binding with excess cold oligo but not with non-specific oligo (Figure 6G). We also analyzed the in vivo HSF1 recruitment on HIV-1 promoter by ChIP assay. Cross-linked chromatin from HIV-1-infected Jurkat cells was pulled down with HSF1 antibody and recruitment on LTR was checked by PCR amplification using forward primers (F1, F2, F3) and reverse primer (R) spanning the enhancer region as shown in Figure 6A. Strong signal in PCR with primers F1/R and F2/R was obtained, which includes a region downstream to NF-AT1 from −144 to +12 and −93 to +12, respectively. However in the same experiment no PCR signal was obtained with F3/R primer set encompassing a region downstream to third Sp1 site between −75 to +12 (Figure 6H). Thus our in vitro and cellular studies have shown for the first time binding and recruitment of HSF1 on a newly identified HSF1 binding site in HIV-1 LTR (−69 to −91, GGGACTTTCCAGGGAGGTGTGGC).
Figure 6.
HSF1 interacts with novel HSF1 binding site on LTR and gets functionally activated during infection. (A) Schematic representation of HIV-1 LTR showing the positions of oligonucleotide probes (P1 to P4) and primers (F1–F3 and R) used in EMSA and ChIP analysis, respectively. (B) His-HSF1 specifically binds to probe P2 in EMSA analysis. HSE consensus sequence was used as positive control (lane 2). (C) His-HSF1 binds to probe P2 in dose-dependent manner. (D) His-HSF1 binding to P2 is inhibited by cold P2 oligo but not by non-specific P1 oligo. (E) Probe P2 binds to Jurkat nuclear extract which is inhibited by HSF1 antibody pre-incubation. P2 binding remains unaffected by post-incubation with HSF1 antibody. (F) P2 probe binds with nuclear extract in dose-dependent manner. (G) P2 binding to nuclear extract is competitively inhibited by cold P2 oligo but not by non-specific P1 oligo. (H) HSF1 is recruited on the putative HSF1 binding sequence in HIV-1 LTR during HIV-1 infection of Jurkat cells. ChiP analysis was performed with Nef antibody followed by PCR amplification using F1 to F3 and R primers. (I) HSF1 trimerization increases during HIV-1 infection. HIV-1-infected CEM-GFP cell lysates were used for chemical cross-linking by EGS followed by gel electrophoresis as described in the text. (J) HSF1 is phosphorylated and translocated in to the nucleus of HIV-1-infected cells. HIV-1-infected Jurkat cells were used for immunostaining with polyclonal phospho-HSF1 (Ser230) antibody. The extreme right panel in each row is a magnified image of one cell from the merged image panel. Arrows indicate localization of phospho-HSF1 in the cell. (K) HSF1 is phosphorylated in HIV-1-infected Jurkat cells as analyzed by immunoblotting of nuclear and cytoplasmic fractions.
HSF1 is functionally activated during HIV-1 viral infection
Earlier reports have shown induction of HSF1 transactivation activity by its phosphorylation, trimerization and nuclear translocation (17,18). We then looked at the trimerization of HSF1 by chemical cross-linking using EGS followed by immunoblotting with monoclonal HSF1 antibody (Figure 6I). HSF1 was seen as an inactive monomer in uninfected cells but upon infection it was predominantly present as a trimer (Figure 6I). Furthermore, immunostaining with phospho-HSF1 (S230) antibody showed cytoplasmic localization of pHSF1 in uninfected cells but following infection, it was predominantly localized in the nucleus (Figure 6J). Notably, co-localization of pHSF1 with DAPI was distinctly visible in infected Jurkat cells (Figure 6J). This was further confirmed by immunoblotting of nuclear and cytoplasmic fraction of HIV-1-infected Jurkat cells using pHSF1(S230) antibody. Phospho-HSF1 was predominantly observed in the cytoplasm of uninfected cells whereas most of it was present in the nucleus of infected cells (Figure 6K). Put together all these data clearly suggest that HSF1 is functionally activated during HIV-1 infection and in turn regulates HIV-1 gene expression and replication.
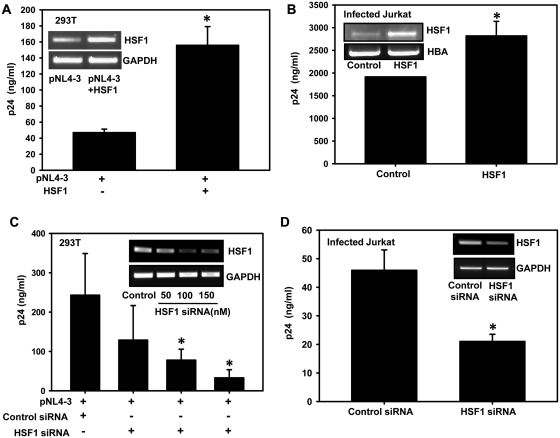
HSF1 regulates HIV-1 gene expression and replication
Our observation of HSF1 induced HSP40 promoter and LTR-driven gene expression led us to further investigate whether expression of HSF1 directly modulates HIV-1 replication or virus production. We thus performed a single cycle replication study in 293T cells by co-transfecting pNL4-3 along with HSF1 and virus production was analyzed in culture supernatants by p24 antigen capture ELISA. HSF1 overexpression resulted in enhanced virus production as compared to control cells (Figure 7A). Similar result was also obtained with Jurkat cells, transfected with HSF1 first, followed by infection with NL4-3 virus (Figure 7B). This finding was further validated by looking at the effect of HSF1 silencing on virus production. HSF1 silencing was followed by transfection with pNL4-3 in 293T cells and infection in Jurkat cells and virus production was analyzed by p24 ELISA assay. HSF1 silencing in both 293T and Jurkat cells lead to significant reduction in virus production (Figure 7C and D), clearly indicating the importance of HSF1 in the viral life cycle.
Figure 7.
HSF1 overexpression increases HIV-1 replication whereas it's silencing leads to inhibition of HIV-1 replication. (A) HSF1 overexpression increases virus production. Culture supernatants of 293T cells transfected with pNL4-3 and HSF1 vectors were analyzed for virus production using p24 antigen capture ELISA. HSF1 overexpression is shown in the inset (B) HSF1 overexpression leads to increased virus production in HIV-1 NL4-3-infected Jurkat cells. (C) HSF1 downregulation reduces HIV-1 virus production. Culture supernatants from 293T cells co-transfected with pNL4-3 and increasing concentrations of HSF1 siRNA were analyzed for virus production using p24 ELISA. Efficiency of gene silencing was checked by RT–PCR (inset). (D) HSF1 silencing leads to inhibition of virus production in HIV-1-infected Jurkat cells. Jurkat cells were first transfected with HSF1 siRNA followed by infection with NL4-3 virus. Cells were analyzed for HSF1 silencing by RT–PCR as shown in (inset) and culture supernatant was used for p24 ELISA. The error bars represent the mean ± SEM of two independent experiments. Statistical analysis was performed using Student's t-test, with the levels of significance defined as *P < 0.05.
Taken together, all these results indicate that HSF1 positively regulates HIV-1 gene expression and replication by two distinct pathways. Firstly, it induces HSP40 promoter activity along with Nef and secondly it directly interacts with HIV-1 LTR to induce viral gene expression and replication.
DISCUSSION
Expression of HSP family members is modulated in various disease conditions like cancer (37) and sepsis (38). Acute infection of cells with viruses also induces expression of stress proteins (39,40). In case of acute HIV-1 infection modulation of HSP27 and HSP70, expression in CD4+ T cells have been reported earlier (6); however, the molecular mechanism of this modulation remains obscure. We have shown earlier that HSP40 is upregulated in presence of Nef (15). Here, we first confirm this Nef-dependent phenomenon in HIV-1-infected T cells followed by the identification of the mechanism. Normally the gene expression of HSPs is regulated by heat shock elements (HSE) present on the promoter (41) and HSF protein, which specifically bind to these HSE sequences and enhance HSP gene expression (42). In case of HIV-1 infection, there is one report showing Vpr-dependent modulation of HSP27 expression through HSF1 (43). We report here for the first time that Nef induces HSP40 expression by forming a complex with HSF1. Nef, initially reported to be a transcriptional repressor of HIV-1 LTR (9,44) has been later shown to act as a positive regulator of LTR by a variety of mechanisms (12,45). In our effort to understand mechanistic details of Nef-dependent HSP40 upregulation, we find that positive effect of Nef on HSP40 expression was mediated by its interaction with HSF1. As, HSF1 is known to regulate expression of HSPs by being recruited on the promoter of HSPs, we also investigated the recruitment of Nef on HSP40 promoter. Nef seems to be recruited at HSE elements on HSP40 promoter along with HSF1. This finding was further supported by overexpression and gene silencing studies, where overexpression of HSF1 enhanced HSP40 expression in Nef-dependent manner whereas its silencing resulted in inhibition of HSP40 promoter activity in presence of Nef. Our results thus imply that the co-occupancy of Nef and HSF1 on HSP40 promoter activates the promoter to increase HSP40 expression in HIV-1-infected cells, which in turn enhances viral gene expression and production as elucidated earlier (15).
In addition to stress response, HSF1 is also reported to be involved in developmental processes by regulating expression of some other genes like inflammatory cytokines (46). It can act both as transcriptional activator as well as repressor depending on the presence of HSE elements and cohort of additional factors which it recruits (46). In view of the role that HSF1 plays for regulating the transcription of non-HSP genes, in second part of our study, we have analyzed direct role of HSF1 in LTR-driven gene expression. This study was also supported by several previous observations where activation of LTR-driven gene expression was reported under hyperthermic condition (20–22). Suppression of HIV-1 LTR by a mutant HSF has been also reported (47); however, the modulation of LTR activity by HSF1 remains to be clearly elucidated. Our transient transfection studies clearly show that HSF1 enhances Tat induced LTR-driven gene expression. Furthermore, our studies with mutant LTR-luc constructs indicate the role of enhancer region of the LTR in positive regulation by HSF1. Computational analysis of HIV-1 LTR enhancer region resulted in identification of a putative HSF1 binding site in this region. We then confirmed binding of HSF1 with the putative binding site (−69 to −91) in the enhancer region on HIV-1 LTR both in vitro and in vivo. Furthermore, enhanced expression of HSF1 observed in HIV-1-infected cells may play a role in enhancing the positive effect of HSF1 on LTR-driven gene expression.
The activation of HSF1 is associated with transition of the monomeric inactive to trimeric active form and concomitant post-translational modification like phosphorylation and translocation into the nucleus (42). So to further explore the mechanism of HSF1-driven LTR gene expression we have tried to correlate its LTR DNA binding activity with the transcriptionally active state. We observed that following HIV-1 infection, HSF1 not only shows increased trimerization but also shows increased phosphorylarion and nuclear re-localization in infected cells. This functional activation seems to facilitate the binding of HSF1 on both HSP40 promoter and LTR to enhance viral gene expression. However, it is still unclear how this binding activates the promoters. This binding might induce some molecular and physical changes in promoter at the chromatin level that allow binding of some activator molecules to mediate activation as observed in case of IL-6 gene, where the binding of HSF1 on its promoter led to opening of chromatin for binding of activator or repressor molecules (48). Future studies on the molecular details of this phenomenon at chromatin level may unravel novel mechanistic details about HIV-1 gene regulation.
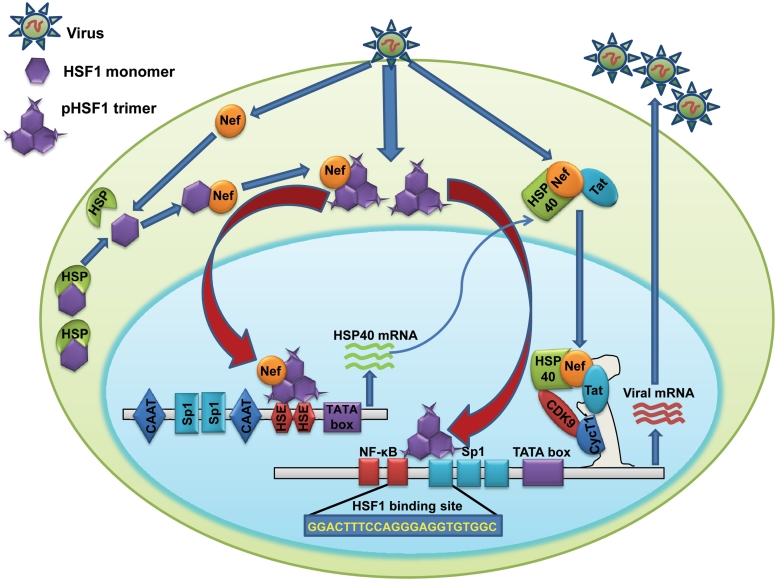
Activation of HSF1-mediated stress response in reaction to microbial infection is functionally important for elevated expression of HSPs, which generally act as an alert signal for host to elicit anti-microbial immune response (49). We have attempted to understand the functional relevance of increased HSF1 expression and activation during HIV infection and our results clearly show that HSF 1 positively regulates HIV-1 gene expression and replication as HSF1 overexpression enhanced viral gene expression whereas its knockdown reduced virus production. As regulation of HIV-1 gene expression involves interplay between viral and cellular host factors, our present study adds another complex layer of regulation in HIV-1 gene expression by HSF1. In summary, we conclude that HSF1 regulates viral replication by two distinct pathways. It interacts with Nef and both are recruited on the HSP40 promoter to enhance HSP40 expression in infected cells. HSP40 enhances viral replication by binding to Cdk9 and modulating the activity of P-TEFb (15). Second, it also regulates the viral transcription by directly interacting with newly identified HSF1 binding site on HIV-1 LTR independent of Nef (Figure 8). Increased expression, hyper-phosphorylation and increased nuclear translocation of HSF1 in HIV-1-infected cells contribute to its enhanced activity. So our present work not only identifies HSF1 as a pro-viral cellular factor but also provides a molecular mechanism for HSF1-mediated enhancement of viral gene expression and replication. Finally, the study also provides another example of how HIV-1 uses host cell proteins for its successful replication in the host and identifies a possible anti-viral target for future studies.
Figure 8.
Mechanistic model showing HSF1-mediated regulation of HIV-1 transcription and replication by two distinct pathways. First pathway involves Nef-dependent activation of HSP40 promoter activity leading to increased viral gene expression as detailed in the text whereas the second pathway involves Nef independent direct activation of HIV-1 LTR promoter activity to increase viral gene expression and replication.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Department of Biotechnology, Government of India; the National Centre for Cell Science, Pune. P.R. is a senior research fellow of the Department of Biotechnology, Government of India. Funding for open access charge: The Department of Biotechnology, Government of India; the National Centre for Cell Science, Pune.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are thankful to Dr G.C. Mishra, Director, National Centre for Cell Science (NCCS) for his support and encouragement. pNL4-3 plasmid, CEM-GFP cell line and Nef antiserum reagents were obtained from AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. We thank Drs W.C. Greene, J. Goldman, J.C. Guatelli, T. Okamoto, K. Ohtsuka and M. Federico for providing different plasmids. We also thank Dr Manish Kumar for his technical support.
REFERENCES
- 1.Brenner BG, Wainberg Z. Heat shock proteins: novel therapeutic tools for HIV-infection? Expert. Opin. Biol. Ther. 2001;1:67–77. doi: 10.1517/14712598.1.1.67. [DOI] [PubMed] [Google Scholar]
- 2.Creagh EM, Sheehan D, Cotter TG. Heat shock proteins–modulators of apoptosis in tumour cells. Leukemia. 2000;14:1161–1173. doi: 10.1038/sj.leu.2401841. [DOI] [PubMed] [Google Scholar]
- 3.Takayama S, Reed JC, Homma S. Heat-shock proteins as regulators of apoptosis. Oncogene. 2003;22:9041–9047. doi: 10.1038/sj.onc.1207114. [DOI] [PubMed] [Google Scholar]
- 4.van Eden W, van der ZR, Prakken B. Heat-shock proteins induce T-cell regulation of chronic inflammation. Nat. Rev. Immunol. 2005;5:318–330. doi: 10.1038/nri1593. [DOI] [PubMed] [Google Scholar]
- 5.Asea A, Kraeft SK, Kurt-Jones EA, Stevenson MA, Chen LB, Finberg RW, Koo GC, Calderwood SK. HSP70 stimulates cytokine production through a CD14-dependant pathway, demonstrating its dual role as a chaperone and cytokine. Nat. Med. 2000;6:435–442. doi: 10.1038/74697. [DOI] [PubMed] [Google Scholar]
- 6.Wainberg Z, Oliveira M, Lerner S, Tao Y, Brenner BG. Modulation of stress protein (hsp27 and hsp70) expression in CD4+ lymphocytic cells following acute infection with human immunodeficiency virus type-1. Virology. 1997;233:364–373. doi: 10.1006/viro.1997.8618. [DOI] [PubMed] [Google Scholar]
- 7.Simmons A, Aluvihare V, McMichael A. Nef triggers a transcriptional program in T cells imitating single-signal T cell activation and inducing HIV virulence mediators. Immunity. 2001;14:763–777. doi: 10.1016/s1074-7613(01)00158-3. [DOI] [PubMed] [Google Scholar]
- 8.Fackler OT, Baur AS. Live and let die: Nef functions beyond HIV replication. Immunity. 2002;16:493–497. doi: 10.1016/s1074-7613(02)00307-2. [DOI] [PubMed] [Google Scholar]
- 9.Niederman TM, Thielan BJ, Ratner L. Human immunodeficiency virus type 1 negative factor is a transcriptional silencer. Proc. Natl Acad. Sci. USA. 1989;86:1128–1132. doi: 10.1073/pnas.86.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arora VK, Fredericksen BL, Garcia JV. Nef: agent of cell subversion. Microbes. Infect. 2002;4:189–199. doi: 10.1016/s1286-4579(01)01527-1. [DOI] [PubMed] [Google Scholar]
- 11.Glushakova S, Grivel JC, Suryanarayana K, Meylan P, Lifson JD, Desrosiers R, Margolis L. Nef enhances human immunodeficiency virus replication and responsiveness to interleukin-2 in human lymphoid tissue ex vivo. J. Virol. 1999;73:3968–3974. doi: 10.1128/jvi.73.5.3968-3974.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joseph AM, Ladha JS, Mojamdar M, Mitra D. Human immunodeficiency virus-1 Nef protein interacts with Tat and enhances HIV-1 gene expression. FEBS Lett. 2003;548:37–42. doi: 10.1016/s0014-5793(03)00725-7. [DOI] [PubMed] [Google Scholar]
- 13.Lundquist CA, Zhou J, Aiken C. Nef stimulates human immunodeficiency virus type 1 replication in primary T cells by enhancing virion-associated gp120 levels: coreceptor-dependent requirement for Nef in viral replication. J. Virol. 2004;78:6287–6296. doi: 10.1128/JVI.78.12.6287-6296.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wolf D, Witte V, Clark P, Blume K, Lichtenheld MG, Baur AS. HIV Nef enhances Tat-mediated viral transcription through a hnRNP-K-nucleated signaling complex. Cell Host. Microbe. 2008;4:398–408. doi: 10.1016/j.chom.2008.08.013. [DOI] [PubMed] [Google Scholar]
- 15.Kumar M, Mitra D. Heat shock protein 40 is necessary for human immunodeficiency virus-1 Nef-mediated enhancement of viral gene expression and replication. J. Biol. Chem. 2005;280:40041–40050. doi: 10.1074/jbc.M508904200. [DOI] [PubMed] [Google Scholar]
- 16.Wu C. Heat shock transcription factors: structure and regulation. Annu. Rev. Cell Dev. Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 17.Holmberg CI, Hietakangas V, Mikhailov A, Rantanen JO, Kallio M, Meinander A, Hellman J, Morrice N, MacKintosh C, Morimoto RI, et al. Phosphorylation of serine 230 promotes inducible transcriptional activity of heat shock factor 1. EMBO J. 2001;20:3800–3810. doi: 10.1093/emboj/20.14.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guettouche T, Boellmann F, Lane WS, Voellmy R. Analysis of phosphorylation of human heat shock factor 1 in cells experiencing a stress. BMC Biochem. 2005;6:4. doi: 10.1186/1471-2091-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stanley SK, Bressler PB, Poli G, Fauci AS. Heat shock induction of HIV production from chronically infected promonocytic and T cell lines. J. Immunol. 1990;145:1120–1126. [PubMed] [Google Scholar]
- 20.Re MC, Furlini G, La Placa M. Rapid detection of HIV-1 in clinical samples by co-culture with heat-shocked cells. J. Virol. Methods. 1989;26:313–317. doi: 10.1016/0166-0934(89)90113-4. [DOI] [PubMed] [Google Scholar]
- 21.Hashimoto K, Baba M, Gohnai K, Sato M, Shigeta S. Heat shock induces HIV-1 replication in chronically infected promyelocyte cell line OM10.1. Arch. Virol. 1996;141:439–447. doi: 10.1007/BF01718308. [DOI] [PubMed] [Google Scholar]
- 22.Kretz-Remy C, Munsch B, Arrigo AP. NFkappa B-dependent transcriptional activation during heat shock recovery. Thermolability of the NF-kappaB.Ikappa B complex. J. Biol. Chem. 2001;276:43723–43733. doi: 10.1074/jbc.M010821200. [DOI] [PubMed] [Google Scholar]
- 23.Olivetta E, Federico M. HIV-1 Nef protects human-monocyte-derived macrophages from HIV-1-induced apoptosis. Exp. Cell Res. 2006;312:890–900. doi: 10.1016/j.yexcr.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Geleziunas R, Xu W, Takeda K, Ichijo H, Greene WC. HIV-1 Nef inhibits ASK1-dependent death signalling providing a potential mechanism for protecting the infected host cell. Nature. 2001;410:834–838. doi: 10.1038/35071111. [DOI] [PubMed] [Google Scholar]
- 25.Adachi A, Gendelman HE, Koenig S, Folks T, Willey R, Rabson A, Martin MA. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chowers MY, Spina CA, Kwoh TJ, Fitch NJ, Richman DD, Guatelli JC. Optimal infectivity in vitro of human immunodeficiency virus type 1 requires an intact nef gene. J. Virol. 1994;68:2906–2914. doi: 10.1128/jvi.68.5.2906-2914.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dandekar DH, Kumar M, Ladha JS, Ganesh KN, Mitra D. A quantitative method for normalization of transfection efficiency using enhanced green fluorescent protein. Anal. Biochem. 2005;342:341–344. doi: 10.1016/j.ab.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 28.Takada N, Sanda T, Okamoto H, Yang JP, Asamitsu K, Sarol L, Kimura G, Uranishi H, Tetsuka T, Okamoto T. RelA-associated inhibitor blocks transcription of human immunodeficiency virus type 1 by inhibiting NF-kappaB and Sp1 actions. J. Virol. 2002;76:8019–8030. doi: 10.1128/JVI.76.16.8019-8030.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hata M, Ohtsuka K. Characterization of HSE sequences in human Hsp40 gene: structural and promoter analysis. Biochim. Biophys. Acta. 1998;1397:43–55. doi: 10.1016/s0167-4781(97)00208-x. [DOI] [PubMed] [Google Scholar]
- 30.Gervaix A, West D, Leoni LM, Richman DD, Wong-Staal F, Corbeil J. A new reporter cell line to monitor HIV infection and drug susceptibility in vitro. Proc. Natl Acad. Sci. USA. 1997;94:4653–4658. doi: 10.1073/pnas.94.9.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chaudhry A, Das SR, Hussain A, Mayor S, George A, Bal V, Jameel S, Rath S. The Nef protein of HIV-1 induces loss of cell surface costimulatory molecules CD80 and CD86 in APCs. J. Immunol. 2005;175:4566–4574. doi: 10.4049/jimmunol.175.7.4566. [DOI] [PubMed] [Google Scholar]
- 32.Westerheide SD, Anckar J, Stevens SM, Jr, Sistonen L, Morimoto RI. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science. 2009;323:1063–1066. doi: 10.1126/science.1165946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pirkkala L, Nykanen P, Sistonen L. Roles of the heat shock transcription factors in regulation of the heat shock response and beyond. FASEB J. 2001;15:1118–1131. doi: 10.1096/fj00-0294rev. [DOI] [PubMed] [Google Scholar]
- 34.Ovod V, Lagerstedt A, Ranki A, Gombert FO, Spohn R, Tahtinen M, Jung G, Krohn KJ. Immunological variation and immunohistochemical localization of HIV-1 Nef demonstrated with monoclonal antibodies. AIDS. 1992;6:25–34. doi: 10.1097/00002030-199201000-00003. [DOI] [PubMed] [Google Scholar]
- 35.Murti KG, Brown PS, Ratner L, Garcia JV. Highly localized tracks of human immunodeficiency virus type 1 Nef in the nucleus of cells of a human CD4+ T-cell line. Proc. Natl Acad. Sci. USA. 1993;90:11895–11899. doi: 10.1073/pnas.90.24.11895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel AE, Kel OV, Ignatieva EV, Ananko EA, Podkolodnaya OA, Kolpakov FA, et al. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaattela M. Escaping cell death: survival proteins in cancer. Exp. Cell Res. 1999;248:30–43. doi: 10.1006/excr.1999.4455. [DOI] [PubMed] [Google Scholar]
- 38.Hashiguchi N, Ogura H, Tanaka H, Koh T, Nakamori Y, Noborio M, Shiozaki T, Nishino M, Kuwagata Y, Shimazu T, et al. Enhanced expression of heat shock proteins in activated polymorphonuclear leukocytes in patients with sepsis. J. Trauma. 2001;51:1104–1109. doi: 10.1097/00005373-200112000-00015. [DOI] [PubMed] [Google Scholar]
- 39.Jindal S, Malkovsky M. Stress responses to viral infection. Trends Microbiol. 1994;2:89–91. doi: 10.1016/0966-842x(94)90540-1. [DOI] [PubMed] [Google Scholar]
- 40.Santoro MG. Heat shock proteins and virus replication: hsp70s as mediators of the antiviral effects of prostaglandins. Experientia. 1994;50:1039–1047. doi: 10.1007/BF01923459. [DOI] [PubMed] [Google Scholar]
- 41.Amin J, Ananthan J, Voellmy R. Key features of heat shock regulatory elements. Mol. Cell. Biol. 1988;8:3761–3769. doi: 10.1128/mcb.8.9.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarge KD, Murphy SP, Morimoto RI. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol. Cell. Biol. 1993;13:1392–1407. doi: 10.1128/mcb.13.3.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liang D, Benko Z, Agbottah E, Bukrinsky M, Zhao RY. Anti-vpr activities of heat shock protein 27. Mol. Med. 2007;13:229–239. doi: 10.2119/2007-00004.Liang. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahmad N, Venkatesan S. Nef protein of HIV-1 is a transcriptional repressor of HIV-1 LTR. Science. 1988;241:1481–1485. doi: 10.1126/science.3262235. [DOI] [PubMed] [Google Scholar]
- 45.Witte V, Laffert B, Rosorius O, Lischka P, Blume K, Galler G, Stilper A, Willbold D, d'Aloja P, Sixt M, et al. HIV-1 Nef mimics an integrin receptor signal that recruits the polycomb group protein Eed to the plasma membrane. Mol. Cell. 2004;13:179–190. doi: 10.1016/s1097-2765(04)00004-8. [DOI] [PubMed] [Google Scholar]
- 46.Xie Y, Chen C, Stevenson MA, Auron PE, Calderwood SK. Heat shock factor 1 represses transcription of the IL-1beta gene through physical interaction with the nuclear factor of interleukin 6. J. Biol. Chem. 2002;277:11802–11810. doi: 10.1074/jbc.M109296200. [DOI] [PubMed] [Google Scholar]
- 47.Ignatenko NA, Gerner EW. Regulation of the HIV1 long terminal repeat by mutant heat shock factor. Exp. Cell Res. 2003;288:1–8. doi: 10.1016/s0014-4827(03)00186-1. [DOI] [PubMed] [Google Scholar]
- 48.Inouye S, Fujimoto M, Nakamura T, Takaki E, Hayashida N, Hai T, Nakai A. Heat shock transcription factor 1 opens chromatin structure of interleukin-6 promoter to facilitate binding of an activator or a repressor. J. Biol. Chem. 2007;282:33210–33217. doi: 10.1074/jbc.M704471200. [DOI] [PubMed] [Google Scholar]
- 49.Di Cesare S, Poccia F, Mastino A, Colizzi V. Surface expressed heat-shock proteins by stressed or human immunodeficiency virus (HIV)-infected lymphoid cells represent the target for antibody-dependent cellular cytotoxicity. Immunology. 1992;76:341–343. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.