Abstract
DNA base-damage recognition in the base excision repair (BER) is a process operating on a wide variety of alkylated, oxidized and degraded bases. DNA glycosylases are the key enzymes which initiate the BER pathway by recognizing and excising the base damages guiding the damaged DNA through repair synthesis. We report here biochemical and structural evidence for the irreversible entrapment of DNA glycosylases by 5-hydroxy-5-methylhydantoin, an oxidized thymine lesion. The first crystal structure of a suicide complex between DNA glycosylase and unrepaired DNA has been solved. In this structure, the formamidopyrimidine-(Fapy) DNA glycosylase from Lactococcus lactis (LlFpg/LlMutM) is covalently bound to the hydantoin carbanucleoside-containing DNA. Coupling a structural approach by solving also the crystal structure of the non-covalent complex with site directed mutagenesis, this atypical suicide reaction mechanism was elucidated. It results from the nucleophilic attack of the catalytic N-terminal proline of LlFpg on the C5-carbon of the base moiety of the hydantoin lesion. The biological significance of this finding is discussed.
INTRODUCTION
The life of an organism and its durability from generation to generation depends on the stability of the genetic material contained in the DNA double helix. However, DNA is continuously exposed to harmful agents, from environmental and endogenous metabolic sources, which can modify its native structure (1). Among these agents, reactive oxygen species (ROS) cause numerous DNA base lesions that can interfere adversely with DNA metabolism (replication, transcription and recombination; 2,3). Thus, oxidized DNA base accumulation can lead to genomic instability resulting in cancer and age-related disorders (4). The deleterious effects of base damages are counteracted by specific DNA repair processes.
Oxidized base lesions are mainly repaired by base excision repair (BER; 5,6). The DNA glycosylases (monofunctional or bifunctional) initiate the BER pathway by recognizing and removing the base lesion thus leading to an abasic (AP) site in DNA. In the case of monofunctional enzymes, the resulting AP site is recognized by an AP endonuclease which cleaves the DNA backbone at the 5′ side of the AP site to generate a 5′-deoxyribose phosphate- (5′-dRp-) end (itself eliminated by the DNA polymerase 5′-dRp-lyase activity) and a free 3′-hydroxyl end for DNA polymerase repair synthesis. The bifunctional DNA glycosylase/AP lyase enzymes are responsible for the elimination of oxidized bases from DNA. In this case, the removal of the oxidized base (DNA glycosylase activity) is followed by the AP site cleavage at its 3′ side according to a β-elimination process (AP lyase activity) leading to a 3′-dRp, a substrate for AP endonucleases. In the course of both successive activities, DNA glycosylase/AP lyase enzymes remain covalently bound to DNA (imino–enzyme DNA complex, Figure1a). These enzymes have been classified as a function of their substrate specificities (oxidized purines or pyrimidines) and more recently, as a function of their 3D-folds (7,8). Most oxidized pyrimidines are excised by the bacterial Nth (EndoIII) and Nei (EndoVIII) enzymes and by their eukaryote functional homologues NTH1 and NEIL1, while oxidized purines are removed by the bacterial formamidopyrimidine-DNA glycosylase (Fpg or MutM) and its eukaryote homologue, the 8-oxoguanine-DNA glycosylase 1 (OGG1; 9).
Figure 1.
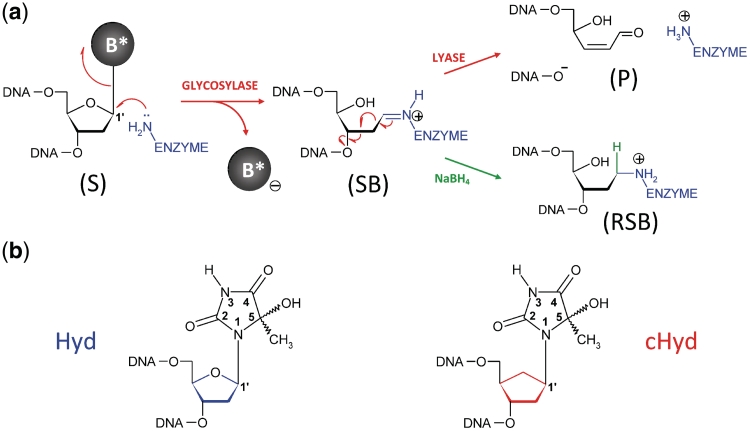
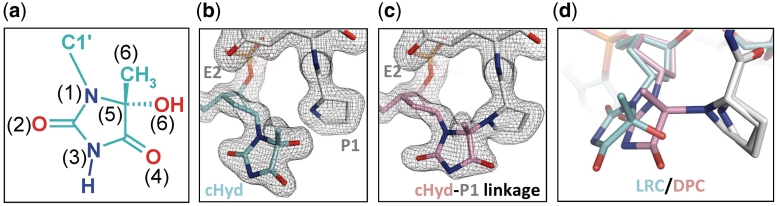
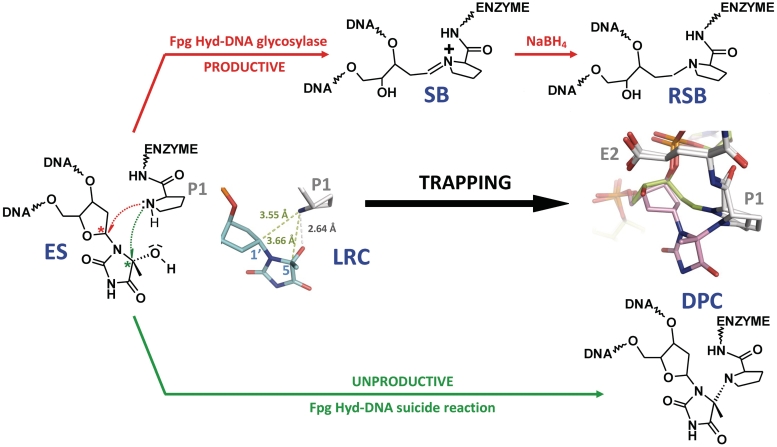
Repair of oxidized base by DNA glycosylase/AP lyases and structures of the nucleoside used in this study. (a) Glycosylase/lyase process. In the first reaction step, the base lesion (B*) contained in DNA (S, for substrate) was removed by the catalytic cleavage of the N-glycosydic bond (DNA glycosylase). An active site amino group (from an internal lysine or from the N-terminal amino group of the enzyme) was used as a nucleophile. This led to the formation of a covalent imino-enzyme DNA intermediate (SB, for Schiff base) between the C1′ of the damaged nucleoside and the catalytic amino group of the enzyme. In the second step of the reaction, the transient Schiff base intermediate underwent base-catalyzed β-elimination (AP lyase), resulting in strand scission at the C3′ side of the abasic site (P, for DNA cleavage product). In the case of Fpg, the β-elimination product underwent a δ-elimination process leading to the excision of the sugar and the formation of a one-nucleoside gap in DNA (Supplementary Figures S1 and S2). The formation of the Schiff base intermediate can be easily demonstrated by its irreversible stabilization in the presence of sodium borohydride (RSB, for reduced Schiff base). (b) Nucleoside and analogue used in the study. Preparation of 5-Hydroxy-5-methylhydantoin (Hyd, blue) and its carbanucleoside (cHyd, red) and their incorporation in synthetic oligonucleotides are described in the Supplementary Data.
During the past decade, our understanding of the general mechanism by which DNA glycosylases recognize base lesions has been greatly facilitated by combining expertise in synthetic chemistry of lesion- or analogue-containing short DNA duplexes and X-ray structures of DNA glycosylase bound to damaged DNA (8,10). From this point of view, Fpg is among the best documented enzymes (11–16). Crystal structures of Fpg/damaged DNA complexes reveal that through binding, the enzyme flips the damaged nucleoside [AP site, 8-oxo-7,8-dihydroguanine (8-oxoG), 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyG) and N7-benzyl-2,6-diamino-5-hydroxy-5-formamidopyrimidine (N7-benzyl-FapyG)] out of the DNA helix in an extrahelical binding pocket thus leading to the exposure of C1′ of the damage to the nucleophilic attack of the conserved N-terminal proline, P1, the catalytic residue responsible for the formation of the Schiff base (SB) (11–13,15) (Figure 1 and Supplementary Figure S1). Although Fpg has been identified as a DNA glycosylase/AP lyase specific for oxidized purines such as 8-oxo-7,8-dihydropurines (8-oxoG and 8-oxoA) (17,18) and imidazole-ring opened purines [FapyG and formamidopyrimidine derived from adenine (FapyA)] (11,12,19,20), some oxidized pyrimidines are also processed by the enzyme (21–23). This broad substrate specificity of Fpg seems to be shared by DNA glycosylases belonging to the helix-2 turns–helix (H2TH) structural superfamily (Fpg, Nei, NEIL1, etc.), whereas its eukaryotic functional homologue OGG1 [from the helix–hairpin–helix (HhH) superfamily] is rather specific for oxidized purines (8). In order to ascertain the Fpg substrate specificity limit and the functional and/or structural features determining an oxidized pyrimidine as a substrate for Fpg, we characterized the recognition and the excision of 5-hydroxy-5-methylhydantoin (Hyd) (Figure 1b) by Fpg from Lactococcus lactis (LlFpg or LlMutM). By studying the Hyd excision mechanism, we show that this lesion can entrap LlFpg, Nei and hNEIL1 but not Nth and OGG1 in an unproductive DNA–protein covalent (DPC) complex. We elucidated at the atomic level, the determinants of LlFpg binding to Hyd-DNA and the molecular mechanism of the suicide reaction by solving the crystal structures of LlFpg bound to Hyd-containing DNA in non-covalent and covalent binding modes.
MATERIALS AND METHODS
Proteins and DNAs
All the biochemical and structural experiments presented in this study were carried out with Fpg from L. lactis (LlFpg). In the text, for reasons of convenience, the simplified annotation of ‘Fpg’ is generally used instead of LlFpg. To obtain ΔP1, P1G and E2Q mutant versions of Fpg, single mutations were achieved using the QuikChange® site-directed mutagenesis kit (Stratagene) with the pFlag-WT-LlFpg- plasmid as DNA template (12). The recombinant plasmids were then used to transform the Fpg defective E. coli strain BH540 (17). Overexpression and purification of the wild-type (wt) and mutant proteins were carried out as previously reported for the wt-LlFpg protein (12). The purified E. coli Endonuclease III (Nth) was a gift from Dr Serge Boiteux. The recombinant plasmids encoding Endonuclease VIII (Nei) and the human Nei-like protein 1 (hNEIL1) were a gift from Profs Susan Wallace and Sylvie Doublié. Overexpression of Nei and hNEIL1 in Escherichia coli and the protein purifications were carried out in the lab using procedures already described (24,25). The open reading frame of the wild-type yOGG1 was isolated by PCR using the plasmid pYSB160 as a template (26) and the oligonucleotide primers AAATGTCTTATAAATTCGG and CTCGAAGCTTCCCTAATCTATTTTTGCTTC. After EcoRI/HindIII digestion, the resulting PCR fragment was inserted by ligation into the linear EcoRI/HindIII pET30a expression vector (Novagen) and yOgg1 was overexpressed in the E. coli BL21(DE3) strain. After immobilizing by metal-affinity chromatography (IMAC) using Talon resin (Clontech), the homogeneous His-tagged protein was obtained after AcA54 (IBF-LKB) gel filtration chromatography and HS Poros perfusion chromatography (PerSeptiveBiosystem).
The protected phosphoramidites of Hyd (h) and cHyd (ch) (Figure 1b) were synthesized as described in the Supplementary Data, incorporated into oligonucleotides CTCTTT(h/ch)TTTCTCG (d2-Hyd and d2-cHyd, respectively, Supplementary Table S1), purified by MonoQ anion exchanger (Amersham Biosciences) and desalted by C-18 SEP-PAK cartridge (Waters). The G- or 8-oxoG-containing oligonucleotide in the same sequence was a gift from Dr Didier Gasparutto and the complementary strands GCGAGAAA(X)AAAGA (where X = A, T, C, G) were purchased from Eurogentech and also purified. Damage-containing strands were then annealed with their complementary strands to generate blunt-ended 14-mer DNA duplexes containing either G or 8-oxoG opposite C, Hyd opposite C or A and cHyd opposite X (where X = A, G, C or T). For the crystal structure analysis of Fpg bound to cHyd-containing DNA, we used, ‘d2-cHyd’ as complementary strands, GCGAGAAACAAAGA (INF1) and GAGAAACAAAGAGC (INF2). DNA duplexes [d2-cHyd:INF1] and [d2-cHyd:INF2] were used with LlFpg for the crystallization of the lesion recognition complex (LRC) and the DPC complex, respectively.
Enzyme assays
For DNA binding experiments (electrophoresis mobility shift assay, EMSA), DNA cleavage (glysosylase/lyase) and trapping assays, damaged strands were 5′-[32P]-labeled before annealing as previously described (15). Standard experimental conditions of EMSA and DNA cleavage assays have been already described (12,15). For the trapping assay (SDS–PAGE–TRAP), the 5′-[32P]-labeled Hyd- or cHyd-containing 14-mer DNA duplex (100 nM final concentration) was incubated with DNA glycosylase in 10 µl of 25 mM Hepes/NaOH, pH7.6, 125 mM NaCl, 1 mM Na2EDTA, 1.5 mM β-mercaptoethanol, 0.1 mM PMSF and 6.5% glycerol with or without 0.18 M NaBH4 for 20 min or 10 h, respectively. After incubation at 37°C, reactions were stopped by adding Laemmli sample buffer at pH 8.8 and heated at 85°C for 2 min. The resulting reaction mixtures were then analyzed by 10–16% discontinuous SDS–PAGE without stacking gel. After electrophoresis, gels were exposed for autoradiography, revealed using STORM-Imager and quantified using ImageQuant software.
Crystallizations of the cHyd/Fpg complexes, X-ray data collection and structure determination
The stock solutions of DNA/protein complexes (8 mg/ml) were prepared at 4°C by mixing the cHyd-containing 14-mer DNA duplex (where cHyd is opposite a cytosine, [cHyd:C] with 5′ and 3′ one overhanging bases [d2-cHyd:INF1] and [d2-cHyd:INF2], respectively) in 1.3 molar excess with the wild-type LlFpg. For the initial crystal screen exploration, we used three batches of cHyd-DNA, one containing d2-cHyd-1, the second d2-cHyd-2 or third, a mixture of both species (V/V) (Supplementary Table S1). Crystallization assays were performed by the hanging-drop vapor-diffusion method. At 20°C, crystals of the non-covalent lesion-recognition complex (LRC), suitable for X-ray analysis, were obtained in a few days by mixing the complex (with [d2-cHyd:INF1]) at 2.5 mg/ml with an equal volume of a well solution containing 0.1 M HEPES/NaOH, pH7.6 and 1.4 M sodium citrate. To obtain crystals of DPC, the stock solution of the complex (8 mg/ml, with [d2-cHyd:INF2]) was first incubated at 37°C for 1 h and then mixed with an equal volume of a well solution containing 0.1 M HEPES/NaOH, pH 7.6 and 1.6 M sodium citrate (pre-equilibrated at 37°C). Crystallization assays were then carried out at 37°C. After 3–4 days, small crystals appeared and were slowly grown for 1 month. Both the LRC and DPC crystals were directly flash-frozen in liquid nitrogen for X-ray data collection.
Diffraction data were collected at 100 K on beamline ID23-1 (ESRF, Grenoble), processed using XDS (27) and scaled in SCALA (28). The structures were solved by molecular replacement with PHASER (29) using only the protein of a previously published Fpg/DNA complex (PDB 1XC8) (11) as the search model. DNA molecules were built manually using COOT (30) and the complexes were then refined using simulated annealing procedures implemented into the PHENIX suite (31). The translation libration screw (TLS) refinement was used in the final cycles of the refinements. The loop residues 219–223 are missing in both final models. Calculations with MOLPROBITY (32) revealed that the percentages of amino acids in the most favored, allowed and disallowed regions of a Ramachandran plot are: 96.5%, 3.5%, 0.0% for LRC and 96.0%, 4.0%, 0.0% for DPC, respectively. We solved the crystal structures of Fpg bound to DNA duplex containing d2-cHyd-1, d2-cHyd-2 or a mixture of both epimeric species (Supplementary Table S1). The three structures are strictly identical. The best diffracting data set and structure refinement statistics were obtained with the epimeric mixture and are listed in Table 1. All structural descriptions in the text refer to this structure. Composite omit maps were calculated with PHENIX (31). Structure figures were generated with PyMOL (DeLano Scientific; http://www.pymol.org).
Table 1.
Data collection and refinement statistics
| LRC (293 K) | DPC (310 K) | |
|---|---|---|
| Data collection | ||
| Space group | P41212 | P41212 |
| Cell dimensions | ||
| a = b, c (Å) | 91.6, 142.8 | 91.7, 134.2 |
| Resolution (Å) | 48.0–1.8 (1.9–1.8)a | 48.1–1.8 (1.9–1.8)a |
| Rmerge | 0.057 (0.51) | 0.093 (0.48) |
| I/σI | 25.3 (4.5) | 11.4 (3.1) |
| Completeness (%) | 99.1 (96.5) | 99.9 (99.9) |
| Redundancy | 9.2 (9.0) | 5.0 (5.1) |
| Refinement | ||
| Resolution (Å) | 47.96–1.80 | 48.08–1.82 |
| No. reflections | 56 346 | 55 438 |
| Rwork/Rfree | 0.159/0.191 | 0.157/0.189 |
| No. of atoms | ||
| Protein | 2139 | 2162 |
| DNA | 566 | 565 |
| Zn | 1 | 1 |
| Glycerol | 6 | 6 |
| Water | 489 | 488 |
| B-factors (Å2) | ||
| Protein | 22.41 | 22.50 |
| DNA | 36.59 | 36.70 |
| Zn | 21.20 | 23.54 |
| Glycerol | 27.30 | 29.08 |
| Water | 40.78 | 37.49 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.008 | 0.022 |
| Bond angles (°) | 1.254 | 2.108 |
aValues in parentheses are for highest resolution shell.
RESULTS AND DISCUSSION
Evidence for Hyd-mediated stable DPC formation with Fpg
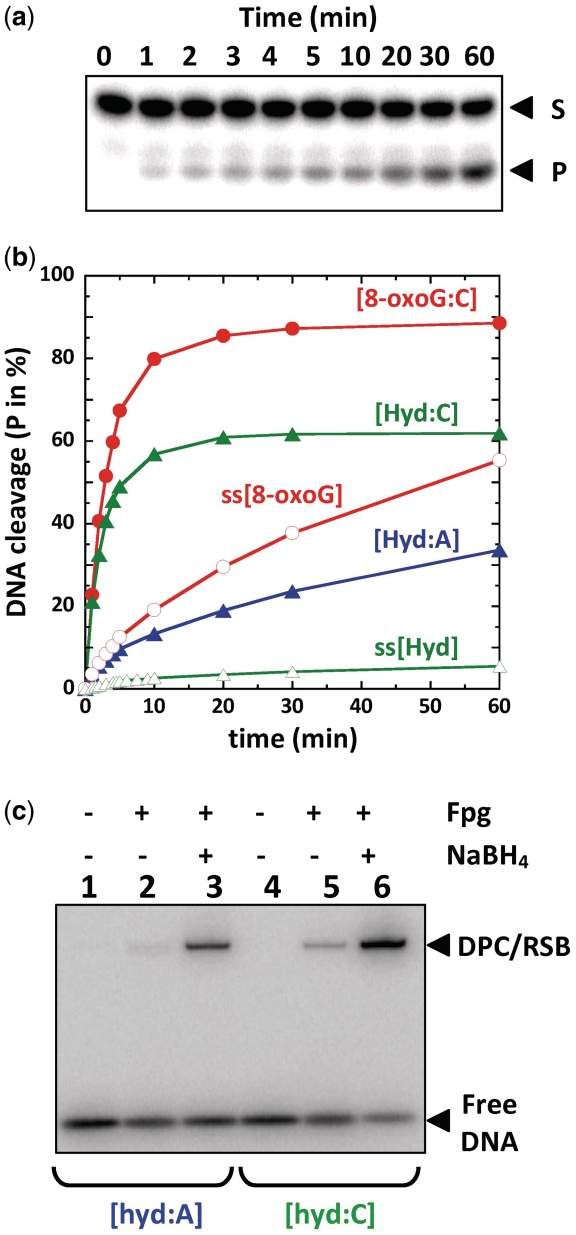
Among thymine oxidation and fragmentation products, Hyd is an abundant DNA lesion generated by hydroxyl radical-mediated oxidation, UV-A sensitized one-electron oxidations and ozone perepoxydation (Figure 1b) (33–35). Identified in both isolated and cellular DNA (36,37), this lesion constitutes either a replication block for some DNA polymerases (21) or can also be efficiently by-passed by low fidelity polymerases studied so far (38). The DNA replication interference studies thus revealed that Hyd lesions may be involved in mutagenesis and carcinogenesis processes if there is a failure in the cellular repair machineries. In previous works, Fpg, Nth, Nei, Ntg1, Ntg2 and NEIL1 DNA glycosylases were proposed to remove Hyd from DNA (21–23,39,40). With a view to study the substrate specificity of Fpg from L. lactis (LlFpg), single turnover experiments of the damaged base removal were carried out using the Hyd-containing DNA opposite A ([Hyd:A], see Supplementary Data Sup-1 for the chemical synthesis of Hyd-containing strand) and its canonical substrate 8-oxoG opposite C ([8-oxoG:C]) (17) as reference (both lesions being in the same DNA sequence context) (Figure 2a and b). Under our experimental conditions, 8-oxoG opposite C is >250 times better excised than Hyd opposite A (red and blue curves, respectively, in Figure 2b). Similarly, to lesion-containing single-stranded DNA (ss[8-oxoG] and ss[Hyd]), the [Hyd:A] duplex is a poor substrate for the LlFpg. This Fpg preference for lesion-containing double-stranded DNA was already observed for imidazole-ring opened purines (FapyG) (41). The E. coli Nth protein also displays this preference for Hyd-containing double-stranded DNA (green curves in Supplementary Figure S2a and b). Since Fpg is known for its susceptibility to the base opposite the lesion (17) and the error-prone effects of Hyd on DNA synthesis (38), we examined the excision of Hyd opposite a cytosine ([Hyd:C]) in the same sequence context (green curve and filled triangles in Figure 2b). Contrary to the [Hyd:A] duplex, Fpg excises Hyd opposite C with an efficiency similar to the excision of 8-oxoG opposite C and thus the [Hyd:C] duplex constitutes an excellent substrate for the enzyme. As already described for 8-oxoG and other oxidized purines (17,42,43), we found that an adenine opposite Hyd strongly affects its excision rate by Fpg (Hyd opposite A is 2000 × less efficiently excised than Hyd opposite C). Clearly, this is not the case for Nth which similarly excises Hyd opposite either A or C (Supplementary Figure S2b). The excision mechanism of Hyd was confirmed by trapping with NaBH4 the transient SB intermediate (reduced SB, reduced Schiffe base (RSB), green pathway in Figure 1a) formed between the enzyme and the DNA duplexes [Hyd:A] and [Hyd:C] (Figure 2c). The stable RSB was resolved by SDS–PAGE–TRAP (lanes 3 and 6, Figure 2c). Unexpectedly, without NaBH4, a DPC complex displaying a similar apparent molecular weight to that of RSB was also observed with both DNA probes (lanes 2 and 5, Figure 2c). As observed for the DNA glycosylase/lyase process (Figure 2b), the formation of both RSB and DPC was significantly stimulated when Hyd was opposite C. The formation of a NaBH4-independent DPC was not observed with Nth and Hyd-containing DNA duplex or with Fpg incubated with the [8-oxoG:C] duplex. Considering the glycosylase process (Figure 1a), we can propose that DPC did not result from a nucleophilic attack of the catalytic proline 1 (P1) on the C1′ carbon of the damaged nucleoside (Supplementary Figure S1).
Figure 2.
Classical and atypical processing of Hyd-containing DNA by Fpg (a) and (b) Single turnover Glycosylase/lyase assay of base damage-containing strand. 5′-[32P]-labeled double-stranded [X:Y] or single-stranded (ss[X]) oligonucleotides (100 pM) were incubated with Fpg at 5 nM (for [8-oxoG:C] and [Hyd:C]) or 1 µM (for [Hyd:A], ss[Hyd] and ss[8-oxoG]) as described in the ‘Materials and Methods’ section. In the time course, samples of the reaction mixture were analyzed by sequencing gel electrophoresis as previously described (15). Gels were then autoradiographed, (a) is an example done with the duplex [Hyd:A]) and the β−, δ-elimination product reaction product (P, see also Supplementary Figures S1 and S2) was quantified (b) for each experiment. (c) Formation of a stable DPC complex with Hyd-containing DNA duplexes visualized by SDS–PAGE–TRAP. Trapping assays with (+) or without (−) NaBH4 were carried out as described in ‘Materials and Methods’ section. The final Fpg concentration was 25 µM. A gel autoradiography is presented for the [Hyd:A] and [Hyd:C] DNA probes as indicated. DPC is for DNA-protein covalent complex NaBH4-independent and RSB for reduced Schiff base (Figure1a). Both DPC and RSB covalent complexes were not resolved by electrophoresis.
C1′ of Hyd is not the electrophile of the damaged nucleotide targeted by Fpg for DPC formation
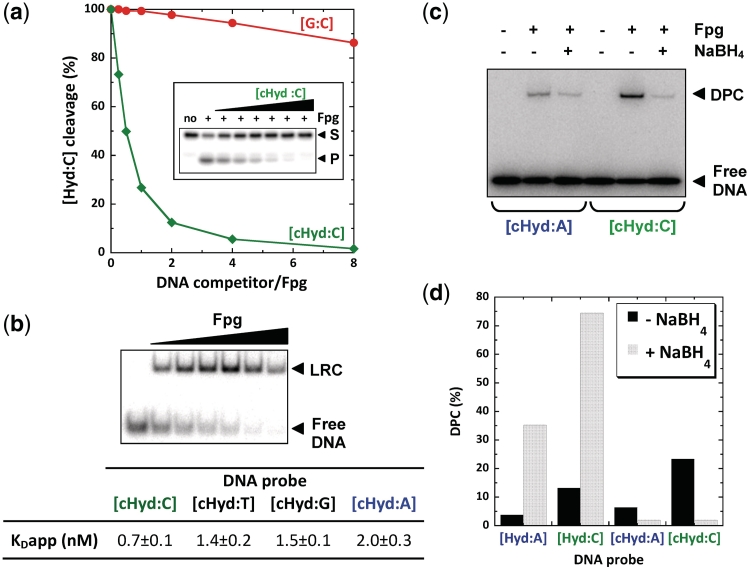
In order to exclude the possibility that a stable covalent bond was formed with the C1′ of Hyd due to an uncontrolled reduction in the absence of NaBH4 (Figure 1), DNA duplexes containing a carbanucleoside derivative of Hyd (cHyd; (Figure 1b, see Supplementary Data Sup-1 for its chemical synthesis) were tested. In this nucleoside analogue, a methylene group replaces the ring oxygen in the 2-deoxyribose moiety of Hyd; consequently, the glycosidic bond is stable to nucleophilic attack at C1′ by Fpg or Nth. Such nucleoside analogues have been extensively used for studying catalytic and DNA binding processes of DNA glycosylases and for analyzing structural determinants of these enzymes bound to damaged DNA bases (8,10). In previous work, we have used carbanucleosides of FapyG to solve the crystal structures of Fpg bound to FapyG-containing DNA (11,12). The carbanucleoside of 8-oxoG (c8-oxoG) was also used to characterize the interaction of Fpg with 8-oxoG-containing DNA and to elucidate its catalytic mechanism (44). Such chemically derivated nucleosides are considered as realistic substrate analogues for DNA glycosylases since their base pairing properties in duplex DNA are similar to those of the corresponding natural nucleosides (45,46). Molecular dynamic simulations of Fpg bound to FapyG-DNA in which the carbanucleoside has been replaced by a true 2-deoxyribose also provided evidence that carbanucleoside-containing lesions mimic well the corresponding natural lesions (47). Though, not a substrate for DNA glycosylases, DNA duplex containing cHyd opposite C ([cHyd:C]) constitutes a strong inhibitor of the Hyd-DNA glycosylase/AP lyase activity of Fpg when a normal DNA duplex [G:C] has little effect (Figure 3a). This immediately suggested that Fpg specifically interacts with DNA containing the cHyd analogue. To quantify this interaction, we determined the apparent dissociation constants of Fpg bound to cHyd opposite C, T, G or A by EMSA (Figure 3b). All DNA duplexes [cHyd:X] are high affinity ligands for the enzyme with KDapp values in a nanomolar range. However, Fpg displays an affinity 3-fold higher for cHyd opposite C than cHyd opposite A, whereas intermediate affinities were observed for cHyd opposite the other two bases. Similar binding affinities have been observed for 8-oxoG and FapyG (12,17). As we observed for the excision of Hyd by Fpg (Figure 2), the lesion recognition is also modulated by the nature of the base opposite cHyd with a preference for a C rather than an A. However, a strong substrate analogue interaction is not necessarily connected with an efficient excision of the damage. As an example, Nth binds the [cHyd:A] duplex less efficiently than does Fpg, while Hyd is better excised by Nth than by Fpg (Supplementary Figure S2). Based on the crystal structures of Fpg bound to oxidized purine-containing DNA (12,48), one can propose that a purine opposite the damage is associated with a steric hindrance inside the substrate binding pocket of the enzyme preventing the formation of a productive enzyme–substrate complex.
Figure 3.
The carbanucleoside of Hyd (cHyd) is a suicide inhibitor for Fpg (a) Inhibition of the Fpg Hyd-DNA Glycosylase/lyase activity by cHyd-containing DNA duplex inhibition was carried out in standard conditions (see ‘Materials and Methods’ section) in the presence of the radio-labeled undamaged DNA [G:C] (red curve) or [cHyd:C] (green curve). (b) Apparent dissociation constants extracted from EMSA titration experiments. The autoradiography of a titration experiment of [cHyd:C] DNA duplex by increasing Fpg concentrations is presented. Under the chosen incubation conditions (20 min at 4°C), only the non-covalent lesion recognition complex (LRC) is formed. The apparent dissociation constants (KDapp) were determined for [cHyd:X] DNA probes where X = A, G, C or T (table). (c) Stable DNA-protein covalent complex (DPC) with cHyd-containing DNA duplexes. SDS-PAGE-TRAP was carried out as described in Figure2d using [cHyd:A] and [cHyd:C] as DNA probes. (d) Comparative analysis of trapping assays performed with Hyd and cHyd Quantifications were extracted from the SDS-PAGE-TRAP experiments of Figures 2c and 3c.
Considering all these observations, the cHyd analogue-containing DNA constitutes an ideal ligand to test the possibility of trapping Fpg in a stable DPC complex different from the imino enzyme–DNA intermediate (SB) trapped by NaBH4 (RSB) (Figures 1a and 2c). Such an experiment is presented in Figure 3c. Unambiguously, Fpg is entrapped by DNA duplex containing cHyd opposite A or C and thus the trapping does not result from an artifactual NaBH4-independent reduction. Although not a catalytic process (the enzyme is modified after the suicide reaction), the DPC formation is time- and protein concentration-dependent (Supplementary Figure S3a). Since that was observed for the glycosylase activity and cHyd-DNA binding, the DPC formation is more effective when the analogue is opposite a C and contained in DNA duplex rather than single-stranded DNA (Supplementary Figure S3b and c). Thus, the suppression of the electrophilicity of C1′ in the carbanucleoside cHyd did not prevent the DPC formation indicating that the structural nature of DPC is significantly different from the RSB (Figures 2c and 3c). The DPC formation seems to correspond to an irreversible and abortive reaction because it is not associated with DNA cleavage. In addition, cHyd is more efficient than Hyd in promoting DPC formation (Figure 3d and Supplementary Figure S3c) suggesting a competition between productive and unproductive reactions with the natural substrate Hyd (Figure2c). From this point of view, cHyd can be considered as a suicide inhibitor for Fpg.
The 3D structures of Fpg bound to Hyd reveal that the catalytic P1 of the enzyme is involved in the suicide reaction
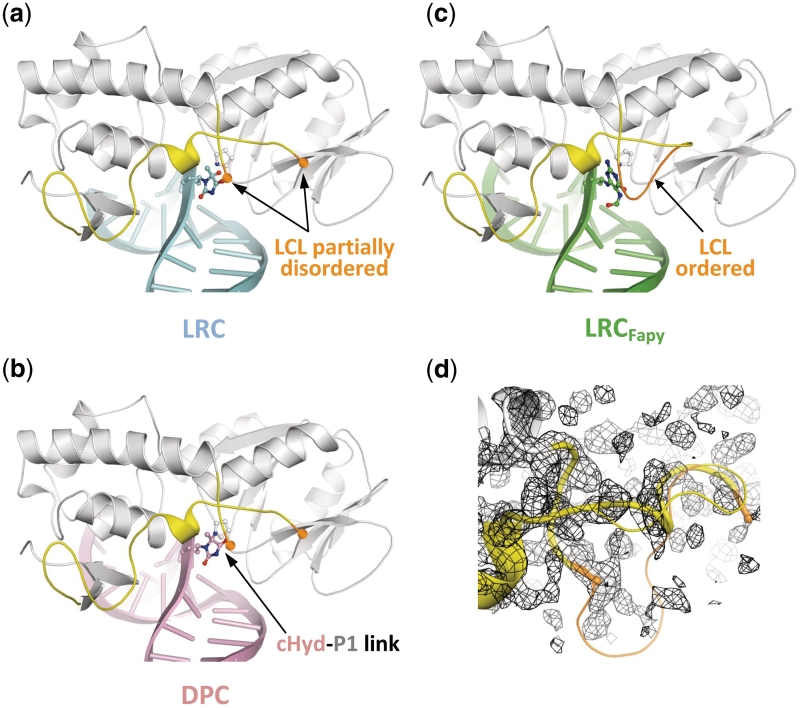
To unravel the molecular mechanism of the enzyme entrapment by Hyd and to elucidate the precise nature of DPC, we solved the crystal structure of Fpg from L. lactis bound to a 14-mer DNA duplex [cHyd/C]. Exploiting the temperature sensitivity of the DPC formation (Supplementary Figure S4a), we obtained crystals of DPC and of the non-covalent complex (referred to lesion recognition complex, LRC, representing the enzyme–substrate complex before catalysis) (Supplementary Figure S4b also see ‘Materials and Methods’ section). Both complexes crystallized in the space group P41212. The structures of LRC and DPC were solved by molecular replacement using the crystal structure of Fpg bound to FapyG-containing DNA as starting model (11) and refined to 1.8 Å resolution (Table 1). Both crystal structures are very similar to each other and also to the crystal structure of Fpg/cFapyG-DNA complex (Figure 4a, b and c). Only residues 219–223 of the flexible part of the loop αF-β9 [also named the lesion-capping loop (LCL)] are missing in the electron density maps of the present structures testifying to the dynamic behavior of LCL (12) in protein/DNA complexes (Figure 4d). The RMSD between both structures is 0.322 Å and only 0.101 Å on protein Cα if we exclude DNA in the calculation. RMSD differences essentially depended on the ends of DNA duplexes we used for crystallization (see ‘Materials and Methods’ section). As previously observed with the abasic (AP) site, 8-oxoG and FapyG residues (11–15), through DNA binding Fpg induced a strong torsion of the DNA helical axis centered on the damaged nucleoside. Protein residues involved in the recognition of the DNA backbone are identical to those identified previously (15). In both structures, the electron density maps in the vicinity of the active sites of the enzyme are perfectly resolved and show the damaged nucleoside cHyd in an extrahelical ‘syn’ conformation (Figure 5b and Supplementary Table S5). Similar to 8-oxoG, FapyG or dihydrouracil (DHU) and as expected for an efficient Fpg substrate, the Hyd base moiety in LRC is positioned by the enzyme inside the substrate binding site (Supplementary Figure S5 and upper panels in Figure 6). In such a localization, the C1′ of the damaged nucleoside is positioned at 3.55 Å from the catalytic amine of P1 and thus, exquisitely exposed to its nucleophilic attack.
Figure 4.
Overview of the LRC and DPC structures. (a, b and c) Overall structures of Fpg Hyd recognition complex (LRC, blue), Hyd-DNA Fpg covalent complex (DPC, pink) and Fpg FapyG recognition complex (LRCFapy, green, pdbid:1XC8). The ribbon representation of Fpg is in grey and the lesion-capping loop (LCL) is highlighted in yellow. In the structure of LCRFapy, the highly flexible part of LCL is ordered and highlighted in orange. (d) Close-up views of LCL. The partially disordered LCL (thick cylinder) from LRC is superimposed with the full LCL from LRCFapy (thin cylinder).
Figure 5.
Close-up views of Fpg active site region in both LRC and DPC complex (a) Hyd lesion atom nomenclature. (b and c) View showing σA-weighted 2Fo − Fc simulated annealing omit maps contoured at 1.3 SD for LRC and DPC complexes, respectively. Elements of protein (gray) and DNA (blue or pink) are represented in stick models with oxygen, nitrogen and phosphate atoms colored in red, dark blue and orange, respectively. (d) Superimposition of both LRC and DPC crystal structures.
Figure 6.
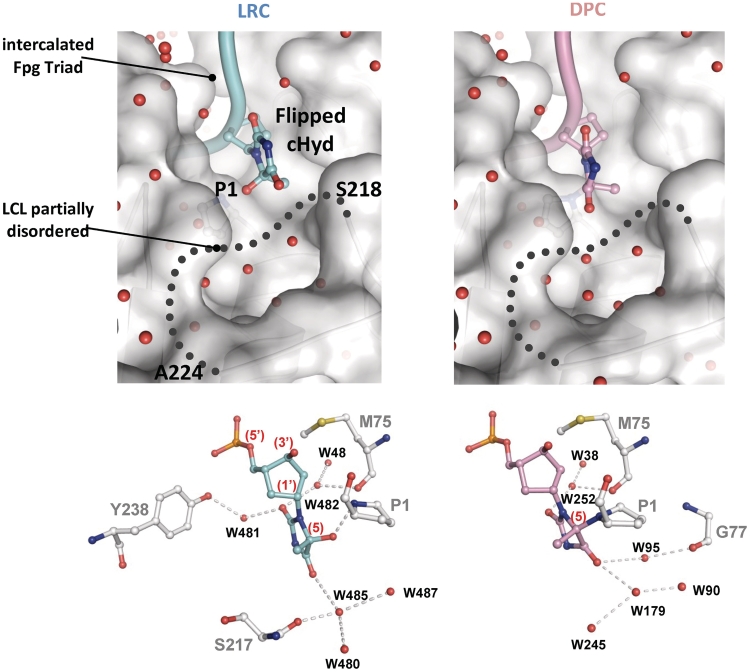
Protein–DNA contacts inside the base lesion recognition pocket of Fpg. Upper panels show focus views of the extruded damaged nucleoside, free and covalently bound (LRC and DPC, respectively) inside the substrate binding pocket of Fpg. The solvent accessible surface area and the ribbon drawing of the protein are indicated in grey. The side chain of P1 is also indicated. The damaged strand of DNA is represented by a cylinder and the lesion by its ball-and-stick model (in pale blue and pink for LRC and DPC, respectively). The missing part of LCL (residues 218–224) is represented by a gray dotted line. Small red spheres are for structural water molecules. Lower panels show the interaction network (grey dashed lines) between the enzyme and the damaged nucleobase. The Fpg binding residues are in grey and water molecule-mediated interactions are represented by red balls. N, O, S and P atoms are represented by blue, red, yellow, and orange balls, respectively.
Unambiguously and in contrast to LRC, the electron density between the C5 of the cHyd base moiety and the nitrogen atom of the N-terminal proline (P1) of Fpg is continuous in DPC, indicating the presence of a covalent link between the protein and the damaged DNA as expected by biochemical analysis (compare Figure 5b and c). Surprisingly, the length of the covalent link (1.64 Å) is slightly longer than that generally expected (1.47 Å) for a C-N linkage. This lengthening is concomitant with a slight deformation of the hydantoin ring moiety which appears slightly non-planar in DPC in contrast to LRC. This deformation probably results from a slight shortening of some cHyd-ring bonds. This geometrical phenomenon is possibly connected to the presumed n→σ* orbital interaction for the bonds in the axial C5 epimer of the hydantoin ring moiety (the lone pair-lone pair repulsion within C–N is expected to enhance the n→σ* donation) and is classically described as the anomeric effect (49). The local superimposition of DPC and LRC structures indicates that the covalent link results in bringing the cHyd base moiety closer to P1 (without significantly changing the torsion angles of the cyclopentane ring moiety; Supplementary Table S5). Although P1 undergoes some changes in its ring torsion angles it remains at about the same position in both structures (Figure 5d). Thus, instead of attacking the C1′ of the damaged nucleoside (Figure 1a), P1 performs a nucleophilic attack on the epimeric C5 carbon of the cHyd base moiety. Indubitably, the suicide reaction leads to an unproductive complex as expected by biochemical experiments with Hyd- and cHyd-containing DNA (Figures 2 and 3).
The atypical binding mode of Hyd by Fpg is responsible for the enzyme entrapment
Contrary to Fpg bound to 8-oxoG, FapyG or 5,6-dihydrouracil, the residues 219–223 of LCL were not visible in the electron density maps of LRC and DPC structures (Figure 4d). In previous works, we defined two extreme conformations for LCL: (i) a closed conformation in which the highly flexible part of the loop is ordered and where its peptidic backbone is directly involved in the binding of the extrahelical damaged nucleobase (the O6 group for 8-oxoG, the O6 formyl group of FapyG and the O4 group for DHU) and (ii) an open conformation in which LCL is partly disordered (with AP site and N7-Benzyl-FapyG) (12,15). In the case of Hyd, the O4 group is not in a position which enables its stable interaction with LCL. Interestingly, mutations in LCL, which alter its conformation near the residues involved in lesion recognition, completely abrogate the Fpg 8-oxoG-DNA glycosylase activity without affecting its AP lyase activity (50). The ‘open’ conformation of LCL in the LRC structure was unexpected since Hyd opposite C is an efficient substrate similar to 8-oxoG opposite C (Figure 2). The LCL ‘open’ conformation is similar to those observed in wild-type Fpg bound to AP site (14,15,23,48) or to N7-benzyl-FapyG (12) and in the deltaP1-Fpg mutant bound to FapyG (11). In the present state of knowledge, it therefore appears difficult to extract a general rule for the precise role of LCL in the Fpg base excision process. On the basis of our present structures, we can propose that, contrary to 8-oxoG and FapyG, the closed LCL conformation is probably not required for the recognition and removal of extrahelical Hyd.
Another intriguing point about Hyd recognition concerns the interaction network established between the enzyme and the extrahelical Hyd nucleoside inside the catalytic site. Compared to the recognition of 8-oxoG, FapyG or DHU, few interactions are observed in the LRC structure though the base moiety of the damage completely enters inside the substrate binding site (left upper panel in Figure 6 and Supplementary Figure S5). The recognition of Hyd rests on only one direct hydrogen bond established between the amine position of the N-terminal P1 and the hydroxyl group at the C5 position of Hyd. Three water molecule-mediated interactions are also found and involve the hydroxyl group of Y238, the carbonyl backbone of the intercalated M75 for the recognition of the O2 position of Hyd and the hydroxyl group of S217 (one residue of LCL) for the recognition of the O4 position of Hyd. For comparison, all the hydrogen donors and acceptors of the FapyG base moiety are contacted by the enzyme (eight direct interactions and seven interactions mediated by water molecules; 12). Therefore, the correct positioning of the extrahelical Hyd inside the substrate binding site results, first, from the flip-out of the damaged nucleoside (numerous interactions with the DNA backbone especially those bordering the damage) and, second, essentially rests on the direct hydrogen interaction P1-Hyd. In addition, the peculiar Hyd binding mode we observed in LRC is also significantly different from that of DHU (a fully reduced pyrimidine base lesion) which is achieved by six direct interactions neutralizing the hydrogen donors and acceptors of the base moiety (Supplementary Figure S6) (13). However, all protein residues contacting Hyd are strictly conserved in the Fpg family testifying to a relative specificity of the recognition. S217 (equivalent to S219 in Fpg from Bacillus stearothermophilus) is also involved in the recognition of 8-oxoG, FapyG, N7-benzyl-FapyG and DHU, whereas Y238 and M75 are recruited only for the recognition of Fapy derivatives.
The use by Fpg of the catalytic P1 as a substrate binding residue was also unexpected for an efficiently metabolized substrate. Indeed, P1 is free of interaction in the crystal structures of Fpg bound to 8-oxoG, FapyG-, DHU- and AP site-containing DNA. However, an exception can be made for the recognition of the bulky lesion, Bz-FapyG, in which P1 directly interacts with the O8 formyl group of the purine opened imidazole-ring (12). Compared with the recognition of FapyG, we proposed that this interaction is unfavorable for an efficient excision of Bz-FapyG by the enzyme. In the present case, the P1-Hyd interaction results in keeping P1 in the immediate vicinity of the electrophilic C5 atom of Hyd. Therefore, in LRC, P1 is exquisitely positioned to perform a nucleophilic attack on C5 of the damaged nucleobase which results in the formation of the suicide complex DPC (see the proposed reaction mechanism in ‘Conclusions’ section). The most significant difference between LRC and DPC is the P1-Hyd covalent link. As in LRC, Hyd remains extrahelical in DPC and has lost the interactions with Y238 and S217. A new interaction mediated by water molecules involves G77 of Fpg, a residue also recruited for the recognition of Bz-FapyG (12). By comparing the structures of LRC and DPC, the atypical binding mode of Hyd in LRC provides a satisfactory explanation for the formation of DPC.
Structural and functional insights into the molecular mechanism of Hyd-mediated DPC
In both LRC and DPC structures, electron density in the substrate binding pocket of Fpg is plainly visible and interpretable for the atoms of Hyd (Figure 5); although the Hyd-DNA used in crystallization consisted of a [5R,5S]-mixture of the damage (see ‘Materials and Methods’ section and Supplementary Data), Hyd appears to bear only the R-configurated C5 in our structures, thus suggesting stereochemical selectivity in the binding and suicide reaction. Considering the probable stereochemical heterogeneity of the cHyd-containing DNA in solution (see the preliminary epimerization study in Supplementary Data) and the binding experiments (Figure 3b), the apparent configuration retention can result from at least three possibilities: (i) under the biochemical and crystallization conditions we used, the 5R-diastereomer was the more abundant species in solution (passive process) or (ii) Fpg efficiently binds only to the 5R-diastereomer according to an extruded binding mode and thus causes a 5S* to 5R* epimerization shift in solution or (iii) Fpg binds both epimers and performed the 5R-configuration retention inside the binding pocket by an unknown mechanism (the latter two possibilities corresponding to an active process mediated by Fpg). The stereochemical selection of the 5R-configuration is clearly supported by both, the strong interaction between the ammonium group of P1 and the hydroxyl group of Hyd in LRC and the covalent link P1-Hyd in DPC (see above). Such an interaction in LRC cannot mature with C5-OH of Hyd harboring the 5S-configuration. The atypical binding mode of Hyd involving P1 as a residue contacting the damage is probably directly connected to the stereoselectivity of the suicide reaction.
Biochemical experiments with the natural Hyd indicate that Hyd can be both, a true substrate and a suicide substrate whereas cHyd just behaves as a suicide substrate with a significant preference for a C opposite the damage (Figures 2 and 3). Comparison between Hyd and cHyd allowed us to obviously demonstrate clearly that the suicide reaction is different from the DNA glycosylase process. Interestingly, X-ray structures of Fpg bound to cHyd-DNA unambiguously demonstrate that the N-terminal P1 is the nucleophile involved in both reactions (Figure 7). According to this result, the suicide reaction corresponds to condensation associated with the loss of a water molecule. The structure of LRC supports the possibility of achieving both reactions with the catalytic P1. Indeed, the nitrogen group of P1 is exquisitely positioned to perform either a nucleophilic attack on C1′ (DNA glycosylase activity) or on C5 (suicide reaction) of Hyd (Figure 7). From the chemical point of view, the direct attack of C5 by P1 appears however unlikely considering that in the LRC crystal structure the observed angle of attack of the amine nucleophile is highly unfavorable for C5-OH bond cleavage. Thus, we can propose a reaction mechanism in which the 5R-OH group of Hyd can be involved in the activation of P1 through proton transfer (Supplementary Figure S7). In such a mechanism, the extruded Hyd acts as a co-factor of the enzyme by activating P1 through its C5-OH group. According to this mechanism, the P1 nucleophile and the Hyd OH-leaving group are activated in one step. This leads to a planar transition state (an iminium cation in equilibrium with a carbocation) which then undergoes the P1 nucleophilic attack either on C1′ (DNA glycosylase) or on C5-Hyd (suicide reaction) (Supplementary Figure S7). In the case of the suicide reaction, only the 5R-DPC is formed because the P1 nucleophilic attack on C5 comes from one site dictated by the precise architecture of the Fpg substrate binding pocket. The involvement of P1 in the suicide reaction supported by X-ray structures (Figure 7) was confirmed by a site directed-mutagenesis study of the known Fpg catalytic residues (Figure 8a) (12,14,15,51). As expected, the substitution or deletion of P1 (P1G and ΔP1) significantly affects the suicide reaction. Likewise, the DNA glycosylase inactive mutant E2Q is also affected in the suicide reaction. The position of E2 in the present structures as compared to that of the structures of Fpg bound to the AP site or FapyG is identical. Thus, the precise role played by E2 remains to be elucidated but its involvement in both Hyd-DNA glycosylase and Hyd-mediated suicide reaction is clearly demonstrated.
Figure 7.
Molecular mechanism of the suicide reaction compared to that of the DNA glycosylase catalytic reaction. The crystal structure of the lesion recognition complex (LRC) between Fpg and cHyd-DNA duplex can be considered as the non-covalent (enzyme/substrate) recognition complex (ES) formed between Fpg and Hyd-DNA duplex. The structural view indicates the hydrogen bond between P1 and the damaged base (gray dashed line) and the distances between the enzyme nucleophile and both electrophiles of the damaged nucleoside (pale green dashed lines). After its activation (ammonium deprotonation, Supplementary Figure S1), the amino group of the N-terminal proline (P1) of Fpg can act as a nucleophile to attack either C1′ (red pathway) or the 5R-epimer conformation of C5 (green pathway) of the nucleoside. The Hyd-DNA glycosylase activity of the enzyme leads to the imino-enzyme DNA intermediate (SB) (associated with the removal of the base damage) which can be intercepted by its irreversible NaBH4 reduction (reduced Schiff base, RSB). The nucleophilic attack at the C5 position of the damaged base leads to the unproductive DPC complex (see Supplementary Figure S7 for a proposed reaction mechanism). A superposition view of the crystal structures of the borohydride trapped complex (RSB, in green, pdbid: 1K82) and DPC (in pink, this work) is shown.
Figure 8.
Trapping assays with Fpg mutants and other DNA glycosylases (a) The [cHyd:C]-trapping assays were carried out as previously described with no enzyme (−), the wild-type LlFpg enzyme (WT) and single-mutation P1G, ΔP1 and E2Q versions of LlFpg. (b) Similar comparative assays were performed by incubating the [cHyd:C] and [cHyd:A] DNA probes (as indicated) with no enzyme (lanes 1), Fpg (lanes 2), Nei (lanes 3) or hNEIL1 (lanes 4).
Since an active amine (an internal lysine or the N-terminal residue) is used as nucleophile by the bifunctional DNA glycosylases (8), we then examined the possibility of entrapping other DNA glycosylases by Hyd. Interestingly, and whatever the experimental conditions we used, Nth and yOGG1 escape from the entrapment by Hyd, whereas Hyd opposite A or C is a very efficient substrate for Nth (Supplementary Figure S2) and is not processed by yOGG1 when paired with the four normal nucleobases (22). In the absence of knowledge of the crystal structure of Nth bound to the oxidized pyrimidine-containing DNA, we are unable to explain this fact. However, we can propose that Nth (and OGG1) has selected a substrate binding mode significantly different from that of Fpg which prevents its entrapment by Hyd. In contrast to Nth and OGG1, it was relatively easy to entrap the E. coli Nei protein and its human functional homologue, the Nei-like protein 1 (hNEIL1), both bifunctional DNA glycosylases specific for oxidized pyrimidines (Figure 8b). The apparent molecular weights of DPCs observed with the three enzymes are compatible with those of the free enzymes (∼30, ∼31 and 44.7 kDa for Nei, Fpg and hNEIL1, respectively). The molecular basis of Nei and hNEIL1 entrapment remains to be elucidated. However, considering that both enzymes are members of the same structural superfamily of Fpg, we can propose that their N-terminal catalytic P1 are also involved in the suicide reaction mediated by these enzymes (23). In conclusion, Hyd-containing DNA potentially constitutes a molecular trap for several bifunctional DNA glycosylases in Prokaryotes and Eukaryotes from the H2TH structural superfamily (Fpg, Nei, NEIL1, NEIL2, etc.) whatever their canonical substrate specificity (8). On the contrary, the bifunctional DNA glycosylases belonging to the HhH structural superfamily such as Nth and OGG1 were not entrapped by Hyd.
CONCLUSION
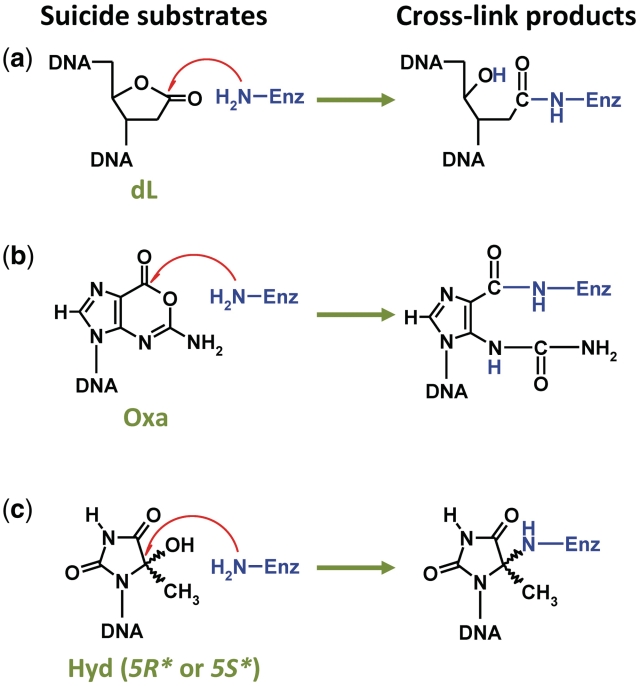
Although involved in genome stability, repair pathways might potentiate intrinsic problems for the cell in peculiar situations, as observed with the DPC formed when DNA glycosylase/AP lyase enzymes encounter a Hyd residue in DNA. This results in transforming a simple base lesion into a more complex bulky lesion. In this work, we deciphered at the molecular and atomic levels, the cross-linking reaction happening between Hyd and Fpg. We showed that the catalytic amine of the enzyme (P1) is directly responsible for the Fpg entrapment by Hyd. Nevertheless, the unproductive reaction we describe here is not very efficient and the currently available data indicate that evolution has disfavored a rapid reaction that generates stable DPC. However, under strong oxidative stress, Hyd might be formed at such a rate that its complete excision by the bacterial Nth or its eukaryote homologue (NTH1) is compromised, thus causing accumulation of DPC involving DNA glycosylases such as Fpg and Nei (NEIL1 and NEIL2 in eukaryotes). Two scenarios can be considered: (i) DPC compromises repair by hiding the lesion from the DNA glycosylases able to correctly excise the damaged base and by ‘hijacking’ other DNA glycosylases from their natural substrates in DNA and (ii) the deleterious effect of DPC is by-passed by constituting a cell signal which permits the recruitment of other DNA repair systems at the target site under oxidative stress; both scenarios not being exclusive. The ‘derailment’ of repair enzymes (first scenario) has already been proposed for other DPC sources such as the covalent trapping of Polβ and other AP lyases by 2-deoxyribonolactone (dL) and oxanine (Oxa) (Figure 9) (52–54), the unproductive and stable non-covalent complex between a DNA glycosylase and a bulky lesion (12), the covalent entrapment of topoisomerase resulting from an incomplete reaction at single-stranded cleavage sites (55) and various chemical and physical agents-induced DPCs (56). On the other hand, the second scenario has been proposed to prevent dL-induced DPC formation by a rapid set up of the long-patch BER pathway (57). Since major DPCs or unproductive repair enzyme/DNA lesion complexes could represent typical bulky/helix distorting adducts (this is the case of Fpg entrapped by Hyd), which can be considered as replication blocks, they are expected to be substrates for the nucleotide excision repair (NER) and homologous recombination (HR) machineries (56,58). Likewise, it has recently been proposed that the unproductive 5′,8-cyclo-2′-deoxyadenosine recognition complex with the DNA glycosylase/AP lyase NEIL1 (also observed with Fpg; 59) is necessary for its NER repair (60). However, the biological consequences of Hyd-mediated DPC remain to be elucidated and the entrapment of other DNA glycosylases such as Nei and NEIL proteins must be improved.
Figure 9.
Three molecular traps for bifunctional DNA glycosylases. Covalent trapping by (a) 2-deoxyribonolactone- (dL) (53), (b) Oxanine- (Oxa) (54) and (c) 5-hydroxy-5-methylhydantoin-containing DNA (Hyd or cHyd, this work).
Using the carbanucleoside cHyd, we demonstrated that Hyd-mediated DPC formation corresponds to a suicide and unproductive reaction and we probed at the atomic level the structures of LRC and DPC. Therefore, cHyd behaves as a DNA glycosylase inhibitor of Fpg, Nei and NEIL1 proteins (suicide process) and hOGG1 and Nth (unproductive non-covalent process). If we consider that agents which target DNA repair enzymes would act to overcome tumor resistance to chemotherapy and radiotherapy (many of these agents exerting their effects through their ability to damage DNA), the development of drugs based on the direct inhibition of DNA repair enzymes is pertinent in anticancer therapy. This concept is under investigation for several DNA repair targets such as BER enzymes (61). Therefore, inhibitors of the Poly(ADP)ribose polymerase (a BER DNA repair enzyme), currently tested in the pre-clinical phase I, enhance the cytotoxic effects of ionizing radiation and DNA-damaging chemotherapy agents (62). More recently, signal interfering DNA (siDNA) technology proposed to use short modified DNA molecules [mimicking double-strand breaks (DSB)], to inhibit the DSB repair processes in cancer cells in association with radiotherapy or chemotherapy (63,64). Similarly, one can imagine that cHyd-containing oligonucleotides delivered to cancer cells will reduce the BER capacity by DNA glycosylase/AP lyase covalent trapping. Impairing the oxidized base removal thus potentiates the efficacy of anticancer drugs and radiotherapy. From this point of view, dL- and Oxa-containing DNA are also good candidates for this approach (Figure 9). The structural and functional determinants of DPC revealed by X-ray structures provide a starting point for the rational design and synthesis of more efficient cHyd-derivatives. The future perspective of designing novel inhibitors rests on a better understanding of inhibition at the atomic level. With this aim in view, the crystal structures of Hyd-DPC formed with Nei structural family proteins are under investigation in our labs.
ACCESSION NUMBERS
The atomic coordinates of LRC and DPC have been deposited in the Protein Data Bank under the accession codes 2XZF and 2XZU, respectively.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Association pour la Recherche contre le cancer (ARC, France); La ligue contre le cancer Inter-régionale (Committee of Indre and Cher, France); Electricité de France (EDF); the Centre national de la recherche scientifique (CNRS, INSB, PEPS program 2009); the Volkswagen foundation; Deutsche Forschungsgemeinschaft (SFB 646); the Alexander von Humboldt Foundation for a postdoctoral fellowship to M.A.I. Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are greatly indebted to the ESRF staff (ESRF ID23-1 beam line, Grenoble, France) for their help in diffraction data collection, to Barbara Steigenberger for her excellent technical assistance, to Susan Wallace and Sylvie Doublié for the generous gift of the expression plasmid encoding the E. coli Nei and hNEIL1 proteins, to Serge Boiteux and Vincent Aucagne for a critical reading of the article and to Elizabeth Rowley-Jolivet for the English proofreading of the article.
REFERENCES
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Cadet J, Douki T, Gasparutto D, Ravanat JL. Oxidative damage to DNA: formation, measurement and biochemical features. Mutat. Res. 2003;531:5–23. doi: 10.1016/j.mrfmmm.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Cadet J, Berger M, Douki T, Ravanat JL. Oxidative damage to DNA: formation, measurement, and biological significance. Rev. Physiol. Biochem. Pharmacol. 1997;131:1–87. doi: 10.1007/3-540-61992-5_5. [DOI] [PubMed] [Google Scholar]
- 4.Maynard S, Schurman SH, Harboe C, de Souza-Pinto NC, Bohr VA. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis. 2009;30:2–10. doi: 10.1093/carcin/bgn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dizdaroglu M. Base-excision repair of oxidative DNA damage by DNA glycosylases. Mutat. Res. 2005;591:45–59. doi: 10.1016/j.mrfmmm.2005.01.033. [DOI] [PubMed] [Google Scholar]
- 6.Xu G, Herzig M, Rotrekl V, Walter CA. Base excision repair, aging and health span. Mech. Ageing Dev. 2008;129:366–382. doi: 10.1016/j.mad.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hazra TK, Das A, Das S, Choudhury S, Kow YW, Roy R. Oxidative DNA damage repair in mammalian cells: a new perspective. DNA Repair. 2007;6:470–480. doi: 10.1016/j.dnarep.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalhus B, Laerdahl JK, Backe PH, Bjoras M. DNA base repair–recognition and initiation of catalysis. FEMS Microbiol. Rev. 2009;33:1044–1078. doi: 10.1111/j.1574-6976.2009.00188.x. [DOI] [PubMed] [Google Scholar]
- 9.van der Kemp PA, Thomas D, Barbey R, de Oliveira R, Boiteux S. Cloning and expression in Escherichia coli of the OGG1 gene of Saccharomyces cerevisiae, which codes for a DNA glycosylase that excises 7,8-dihydro-8-oxoguanine and 2,6-diamino-4-hydroxy-5-N-methylformamidopyrimidine. Proc. Natl Acad. Sci. USA. 1996;93:5197–5202. doi: 10.1073/pnas.93.11.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scharer OD. Chemistry and biology of DNA repair. Angew. Chem. Int. Ed. Engl. 2003;42:2946–2974. doi: 10.1002/anie.200200523. [DOI] [PubMed] [Google Scholar]
- 11.Coste F, Ober M, Carell T, Boiteux S, Zelwer C, Castaing B. Structural basis for the recognition of the FapydG lesion (2,6-diamino-4-hydroxy-5-formamidopyrimidine) by formamidopyrimidine-DNA glycosylase. J. Biol. Chem. 2004;279:44074–44083. doi: 10.1074/jbc.M405928200. [DOI] [PubMed] [Google Scholar]
- 12.Coste F, Ober M, Le Bihan YV, Izquierdo MA, Hervouet N, Mueller H, Carell T, Castaing B. Bacterial base excision repair enzyme Fpg recognizes bulky N7-substituted-FapydG lesion via unproductive binding mode. Chem. Biol. 2008;15:706–717. doi: 10.1016/j.chembiol.2008.05.014. [DOI] [PubMed] [Google Scholar]
- 13.Fromme JC, Verdine GL. DNA lesion recognition by the bacterial repair enzyme MutM. J. Biol. Chem. 2003;278:51543–51548. doi: 10.1074/jbc.M307768200. [DOI] [PubMed] [Google Scholar]
- 14.Pereira de Jesus K, Serre L, Zelwer C, Castaing B. Structural insights into abasic site for Fpg specific binding and catalysis: comparative high-resolution crystallographic studies of Fpg bound to various models of abasic site analogues-containing DNA. Nucleic Acids Res. 2005;33:5936–5944. doi: 10.1093/nar/gki879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Serre L, Pereira de Jesus K, Boiteux S, Zelwer C, Castaing B. Crystal structure of the Lactococcus lactis formamidopyrimidine-DNA glycosylase bound to an abasic site analogue-containing DNA. EMBO J. 2002;21:2854–2865. doi: 10.1093/emboj/cdf304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilboa R, Zharkov DO, Golan G, Fernandes AS, Gerchman SE, Matz E, Kycia JH, Grollman AP, Shoham G. Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA. J. Biol. Chem. 2002;277:19811–19816. doi: 10.1074/jbc.M202058200. [DOI] [PubMed] [Google Scholar]
- 17.Castaing B, Geiger A, Seliger H, Nehls P, Laval J, Zelwer C, Boiteux S. Cleavage and binding of a DNA fragment containing a single 8-oxoguanine by wild type and mutant FPG proteins. Nucleic Acids Res. 1993;21:2899–2905. doi: 10.1093/nar/21.12.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tchou J, Bodepudi V, Shibutani S, Antoshechkin I, Miller J, Grollman AP, Johnson F. Substrate specificity of Fpg protein. Recognition and cleavage of oxidatively damaged DNA. J. Biol. Chem. 1994;269:15318–15324. [PubMed] [Google Scholar]
- 19.Chetsanga CJ, Lindahl T. Release of 7-methylguanine residues whose imidazole rings have been opened from damaged DNA by a DNA glycosylase from Escherichia coli. Nucleic Acids Res. 1979;6:3673–3684. doi: 10.1093/nar/6.11.3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dizdaroglu M, Kirkali G, Jaruga P. Formamidopyrimidines in DNA: mechanisms of formation, repair, and biological effects. Free Radic. Biol. Med. 2008;45:1610–1621. doi: 10.1016/j.freeradbiomed.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 21.Gasparutto D, Ait-Abbas M, Jaquinod M, Boiteux S, Cadet J. Repair and coding properties of 5-hydroxy-5-methylhydantoin nucleosides inserted into DNA oligomers. Chem. Res. Toxicol. 2000;13:575–584. doi: 10.1021/tx000005+. [DOI] [PubMed] [Google Scholar]
- 22.Gasparutto D, Muller E, Boiteux S, Cadet J. Excision of the oxidatively formed 5-hydroxyhydantoin and 5-hydroxy-5-methylhydantoin pyrimidine lesions by Escherichia coli and Saccharomyces cerevisiae DNA N-glycosylases. Biochim. Biophys. Acta. 2009;1790:16–24. doi: 10.1016/j.bbagen.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Zharkov DO, Shoham G, Grollman AP. Structural characterization of the Fpg family of DNA glycosylases. DNA Repair. 2003;2:839–862. doi: 10.1016/s1568-7864(03)00084-3. [DOI] [PubMed] [Google Scholar]
- 24.Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair. 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 25.Jiang D, Hatahet Z, Blaisdell JO, Melamede RJ, Wallace SS. Escherichia coli endonuclease VIII: cloning, sequencing, and overexpression of the nei structural gene and characterization of nei and nei nth mutants. J. Bacteriol. 1997;179:3773–3782. doi: 10.1128/jb.179.11.3773-3782.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Girard PM, Boiteux S. Repair of oxidized DNA bases in the yeast Saccharomyces cerevisiae. Biochimie. 1997;79:559–566. doi: 10.1016/s0300-9084(97)82004-4. [DOI] [PubMed] [Google Scholar]
- 27.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 1993;26:795–800. [Google Scholar]
- 28.Evans PR. Jnt CCP4/ESFEACBM Newsl. Protein Crystallogr. 1997;33:22–24. [Google Scholar]
- 29.McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 32.Davis IW, Murray LW, Richardson JS, Richardson DC. MOLPROBITY: structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 2004;32:W615–619. doi: 10.1093/nar/gkh398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Téoule R, Cadet J. Radiation-induced degradation of the base component in DNA and related substances. In: Hüttermann J, Kohnlein W, Téoule R, Bertinchamps AJ, editors. Effects of ionizing radiation on DNA. Berlin: Spring; 1978. pp. 171–203. [Google Scholar]
- 34.von Sonntag C, Schuchmann HP. The radiolysis of pyrimidines in aqueous solutions: an updating review. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1986;49:1–34. doi: 10.1080/09553008514552201. [DOI] [PubMed] [Google Scholar]
- 35.Girault I, Molko D, Cadet J. Ozonolysis of thymidine: isolation and identification of the main oxidation products. Free Radic Res. 1994;20:315–325. doi: 10.3109/10715769409145631. [DOI] [PubMed] [Google Scholar]
- 36.Kasprzak KS, Jaruga P, Zastawny TH, North SL, Riggs CW, Olinski R, Dizdaroglu M. Oxidative DNA base damage and its repair in kidneys and livers of nickel(II)-treated male F344 rats. Carcinogenesis. 1997;18:271–277. doi: 10.1093/carcin/18.2.271. [DOI] [PubMed] [Google Scholar]
- 37.Senturker S, Karahalil B, Inal M, Yilmaz H, Muslumanoglu H, Gedikoglu G, Dizdaroglu M. Oxidative DNA base damage and antioxidant enzyme levels in childhood acute lymphoblastic leukemia. FEBS Lett. 1997;416:286–290. doi: 10.1016/s0014-5793(97)01226-x. [DOI] [PubMed] [Google Scholar]
- 38.d'Abbadie M, Hofreiter M, Vaisman A, Loakes D, Gasparutto D, Cadet J, Woodgate R, Paabo S, Holliger P. Molecular breeding of polymerases for amplification of ancient DNA. Nat. Biotechnol. 2007;25:939–943. doi: 10.1038/nbt1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaruga P, Birincioglu M, Rosenquist TA, Dizdaroglu M. Mouse NEIL1 protein is specific for excision of 2,6-diamino-4-hydroxy-5-formamidopyrimidine and 4,6-diamino-5-formamidopyrimidine from oxidatively damaged DNA. Biochemistry. 2004;43:15909–15914. doi: 10.1021/bi048162l. [DOI] [PubMed] [Google Scholar]
- 40.Senturker S, Auffret van der Kemp P, You HJ, Doetsch PW, Dizdaroglu M, Boiteux S. Substrate specificities of the ntg1 and ntg2 proteins of Saccharomyces cerevisiae for oxidized DNA bases are not identical. Nucleic Acids Res. 1998;26:5270–5276. doi: 10.1093/nar/26.23.5270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boiteux S, O'Connor TR, Lederer F, Gouyette A, Laval J. Homogeneous Escherichia coli FPG protein. A DNA glycosylase which excises imidazole ring-opened purines and nicks DNA at apurinic/apyrimidinic sites. J. Biol. Chem. 1990;265:3916–3922. [PubMed] [Google Scholar]
- 42.Guo Y, Bandaru V, Jaruga P, Zhao X, Burrows CJ, Iwai S, Dizdaroglu M, Bond JP, Wallace SS. The oxidative DNA glycosylases of Mycobacterium tuberculosis exhibit different substrate preferences from their Escherichia coli counterparts. DNA Repair. 9:177–190. doi: 10.1016/j.dnarep.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hazra TK, Muller JG, Manuel RC, Burrows CJ, Lloyd RS, Mitra S. Repair of hydantoins, one electron oxidation product of 8-oxoguanine, by DNA glycosylases of Escherichia coli. Nucleic Acids Res. 2001;29:1967–1974. doi: 10.1093/nar/29.9.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zharkov DO, Rieger RA, Iden CR, Grollman AP. NH2-terminal proline acts as a nucleophile in the glycosylase/AP-lyase reaction catalyzed by Escherichia coli formamidopyrimidine-DNA glycosylase (Fpg) protein. J. Biol. Chem. 1997;272:5335–5341. doi: 10.1074/jbc.272.8.5335. [DOI] [PubMed] [Google Scholar]
- 45.Ober M, Linne U, Gierlich J, Carell T. The two main DNA lesions 8-Oxo-7,8-dihydroguanine and 2,6-diamino-5-formamido-4-hydroxypyrimidine exhibit strongly different pairing properties. Angew. Chem. Int. Ed. Engl. 2003;42:4947–4951. doi: 10.1002/anie.200351287. [DOI] [PubMed] [Google Scholar]
- 46.Ober M, Muller H, Pieck C, Gierlich J, Carell T. Base pairing and replicative processing of the formamidopyrimidine-dG DNA lesion. J. Am. Chem. Soc. 2005;127:18143–18149. doi: 10.1021/ja0549188. [DOI] [PubMed] [Google Scholar]
- 47.Song K, Kelso C, de los Santos C, Grollman AP, Simmerling C. Molecular simulations reveal a common binding mode for glycosylase binding of oxidatively damaged DNA lesions. J. Am. Chem. Soc. 2007;129:14536–14537. doi: 10.1021/ja075128w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fromme JC, Verdine GL. Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM. Nat. Struct. Biol. 2002;9:544–552. doi: 10.1038/nsb809. [DOI] [PubMed] [Google Scholar]
- 49.Juaristi E, Cuevas G. Recent studies of the Anomeric effect. Tetrahedron. 1992;48:5019–5087. [Google Scholar]
- 50.Qi Y, Spong MC, Nam K, Karplus M, Verdine GL. Entrapment and structure of an extrahelical guanine attempting to enter the active site of a bacterial DNA glycosylase, MutM. J. Biol. Chem. 2009;285:1468–1478. doi: 10.1074/jbc.M109.069799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lavrukhin OV, Lloyd RS. Involvement of phylogenetically conserved acidic amino acid residues in catalysis by an oxidative DNA damage enzyme formamidopyrimidine glycosylase. Biochemistry. 2000;39:15266–15271. doi: 10.1021/bi001587x. [DOI] [PubMed] [Google Scholar]
- 52.Sung JS, Demple B. Analysis of base excision DNA repair of the oxidative lesion 2-deoxyribonolactone and the formation of DNA-protein cross-links. Methods Enzymol. 2006;408:48–64. doi: 10.1016/S0076-6879(06)08004-9. [DOI] [PubMed] [Google Scholar]
- 53.Nakano T, Terato H, Asagoshi K, Masaoka A, Mukuta M, Ohyama Y, Suzuki T, Makino K, Ide H. DNA-protein cross-link formation mediated by oxanine. A novel genotoxic mechanism of nitric oxide-induced DNA damage. J. Biol. Chem. 2003;278:25264–25272. doi: 10.1074/jbc.M212847200. [DOI] [PubMed] [Google Scholar]
- 54.Kroeger KM, Hashimoto M, Kow YW, Greenberg MM. Cross-linking of 2-deoxyribonolactone and its beta-elimination product by base excision repair enzymes. Biochemistry. 2003;42:2449–2455. doi: 10.1021/bi027168c. [DOI] [PubMed] [Google Scholar]
- 55.Lebedeva N, Auffret van der Kemp P, Bjornsti MA, Lavrik O, Boiteux S. Trapping of DNA topoisomerase I on nick-containing DNA in cell free extracts of Saccharomyces cerevisiae. DNA Repair. 2006;5:799–809. doi: 10.1016/j.dnarep.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 56.Barker S, Weinfeld M, Murray D. DNA-protein crosslinks: their induction, repair, and biological consequences. Mutat. Res. 2005;589:111–135. doi: 10.1016/j.mrrev.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 57.Sung JS, DeMott MS, Demple B. Long-patch base excision DNA repair of 2-deoxyribonolactone prevents the formation of DNA-protein cross-links with DNA polymerase beta. J. Biol. Chem. 2005;280:39095–39103. doi: 10.1074/jbc.M506480200. [DOI] [PubMed] [Google Scholar]
- 58.Nakano T, Katafuchi A, Matsubara M, Terato H, Tsuboi T, Masuda T, Tatsumoto T, Pack SP, Makino K, Croteau DL, et al. Homologous recombination but not nucleotide excision repair plays a pivotal role in tolerance of DNA-protein cross-links in mammalian cells. J. Biol. Chem. 2009;284:27065–27076. doi: 10.1074/jbc.M109.019174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Muller E, Gasparutto D, Castaing B, Favier A, Cadet J. Recognition of cyclonucleoside lesions by the Lactococcus lactis FPG protein. Nucleos. Nucleot. Nucleic Acids. 2003;22:1563–1565. doi: 10.1081/NCN-120023034. [DOI] [PubMed] [Google Scholar]
- 60.Jaruga P, Xiao Y, Vartanian V, Lloyd RS, Dizdaroglu M. Evidence for the involvement of DNA repair enzyme NEIL1 in nucleotide excision repair of (5′R)- and (5′S)-8,5′-cyclo-2′-deoxyadenosines. Biochemistry. 2010;49:1053–1055. doi: 10.1021/bi902161f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pallis AG, Karamouzis MV. DNA repair pathways and their implication in cancer treatment. Cancer Metastasis Rev. 2010;29:677–685. doi: 10.1007/s10555-010-9258-8. [DOI] [PubMed] [Google Scholar]
- 62.Drew Y, Plummer R. PARP inhibitors in cancer therapy: two modes of attack on the cancer cell widening the clinical applications. Drug. Resist. Updat. 2009;12:153–156. doi: 10.1016/j.drup.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 63.Quanz M, Berthault N, Roulin C, Roy M, Herbette A, Agrario C, Alberti C, Josserand V, Coll JL, Sastre-Garau X, et al. Small-molecule drugs mimicking DNA damage: a new strategy for sensitizing tumors to radiotherapy. Clin. Cancer Res. 2009;15:1308–1316. doi: 10.1158/1078-0432.CCR-08-2108. [DOI] [PubMed] [Google Scholar]
- 64.Quanz M, Chassoux D, Berthault N, Agrario C, Sun JS, Dutreix M. Hyperactivation of DNA-PK by double-strand break mimicking molecules disorganizes DNA damage response. PLoS ONE. 2009;4:e6298. doi: 10.1371/journal.pone.0006298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.