Abstract
The mechanism through which the large serine recombinases bind DNA is poorly understood. Alignments of ϕC31 integrase (Int) and its relatives indicate the presence of a conserved motif containing four cysteines resembling a zinc finger. Inductively coupled plasma–mass spectrometry (ICP–MS) confirmed that an Int monomer contains one atom of zinc. Pre-incubation of Int with ethylenediaminetetraacetic acid (EDTA) was detrimental for both recombination activity and DNA binding affinities but full activity could be restored by adding back Zn2+. Mutations in the cysteines and other highly conserved residues yielded proteins that were hypersensitive to proteases, suggesting that without zinc the domain is unfolded. Substitutions in the highly charged region between the conserved cysteines led to lowered DNA binding affinities while circular dichroism revealed that these variant Ints were not greatly affected in overall folding. Int was protected from inhibition by EDTA when DNA containing an attachment site was present suggesting that the zinc finger and the DNA are in close proximity. A truncated mutant of Int, hInt V371SUGA, lacking the putative zinc finger could bind DNA with low affinity. The data are consistent with there being at least two DNA binding motifs in Int one of which is the zinc finger-like motif.
INTRODUCTION
Temperate bacteriophages often encode an integrase (Int) that mediates site specific recombination between phage and host attachment sites, attP and attB, respectively, to produce the integrated prophage flanked by the recombinant sites, attL and attR. Excisive recombination between attL and attR regenerates the free circular phage genome containing an intact attP site and the attB site on the chromosome. Phage Ints are either tyrosine Ints and related to λ Int or they are serine Ints of which ϕC31 Int and Bxb1 gpInt are the best studied (1,2). The serine Ints and other large serine recombinases such as some transposases (e.g. TnpX from Tn4451) have N-terminal domains (NTDs) that are homologous in sequence, structure and function to the catalytic NTDs of the resolvase/invertases family of serine recombinases (1–6). The large serine recombinases however have much larger C-terminal domains (CTDs) of 300–500 amino acids compared with the small, ∼40 residue CTDs of the resolvase/invertases. The CTDs of the serine Ints are required for substrate recognition and for controlling the directionality of recombination i.e. integration versus excision (7–9).
φC31 Int requires only small (<50 bp) attP and attB sites for integration (10). AttP and attB share only 32% sequence identity and they each contain imperfect inverted repeats that flank the dinucleotide where the crossover occurs (11,12). AttL and attR are the products of reciprocal DNA exchange between attP and attB and therefore contain a half site of attP and a half site from attB. Excision (attL × attR recombination) requires Int but only occurs in the presence of the recombination directionality factor (RDF) or Xis, an accessory protein that controls the directionality of Int (T. Khaleel, E. Younger, A.R. McEwan, M.C.M. Smith, unpublished). The DNA substrates themselves also have a crucial role in controlling Int activity. In the absence of Xis, Int only recombines attP and attB and has no activity on any other pair of attachment sites (13). Moreover, without Xis, Int only brings attP and attB together to form a stable synaptic complex, an early stage in the recombination reaction pathway in which Int dimers are thought to bring two attachment sites together by the formation of Int tetramers (14). As no other pair of attachment sites will form stable synaptic complexes, it is clear that substrate binding is an allosteric event where attP, attB and attL/R elicit different conformational changes in Int that enable or disable protein–protein interactions and tetramerization (9,14,15).
Although the CTDs in ϕC31 and Bxb1 Ints have been shown to bind DNA there have been no reports on which motifs within these large domains might be making direct contacts with DNA. ϕC31 Int contains 605 amino acids and the CTD (residues 155–605) contains a number of recognizable motifs; the so-called ‘recombinase’ motif (pfam07508; amino acids ∼215–342), a putative zinc finger-like motif (Figure 1: amino acids 366–405), a putative coiled-coil domain (amino acids 445–524) known to be involved in oligomerization of Int and a region at the C-terminus rich in leucine, isoleucine and valine (1,9). Here we focus on the zinc finger-like motif. Generally zinc fingers are relatively small structural motifs that can mediate interactions with other macromolecules (16). The data presented here show that Int contains zinc and that zinc is essential for recombination activity and high-affinity DNA binding. Together with the properties of mutated Int proteins, the data suggest that the zinc finger-like motif coordinates zinc and is required for substrate recognition and high-affinity DNA binding.
Figure 1.
An alignment of the ϕC31 putative zinc binding motif with similar regions from a selection of related Ints. The alignment was performed with T-coffee (29) and then manipulated manually to align cysteine and glycine residues from related serine Ints with ϕC31 C405 and G377, respectively. ϕC31 Int coordinates are shown.
MATERIALS AND METHODS
Strains, cultures and plasmids
Escherichia coli DH5α [fhuA2 Δ(argF-lacZ)U169 phoA glnV44 Φ80 Δ(lacZ)M15 gyrA96 recA1 relA1 endA1 thi-1 hsdR17] and RapidTrans™ TAM1 [mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG] (Active motif) were used for DNA manipulations and in vivo recombination assays. Plasmid pARM010 was used for expression of an N-terminally 6× histidine-tagged Int derivative (hInt) and was constructed by ligating a PCR amplified DNA fragment (using pHS62 as the template and primers ARM4 and ARM5; Supplementary Table S1) cut with PciI/XhoI to pEHISTEV cut with NcoI/XhoI (13,17). Amino acid substitutions were introduced into pARM010, by site directed mutagenesis (SDM; Quickchange, Stratagene) with pHS62 as the template and primers as described in Supplementary Table S1 (8). The int alleles were sequenced for validation. Residues are numbered according to their position in wild-type Int (1–605) as expressed by pHS62 (13).
Plasmid pARM101 was used as a reporter for the in vivo activity assays and was constructed by exchanging the kanamycin resistance gene from pRT504 (14) with an apramycin resistance gene by recombineering in E. coli BW25113/pIJ790 (18). The DNA-encoding apramycin resistance was amplified by PCR using primers MS400 and MS401 (Supplementary Table S1) from the REDIRECT cassette encoded by pIJ773. Escherichia coli BL21 F− ompT gal dcm lon hsdSB(rB− mB−), λ(DE3), pLysS was used as a host for overexpression of Int and its derivatives.
Protein expression and purification
Int, hInt and Int variants including the 6 × histidine tagged CTD fragment (hCTD), were purified as described previously (8,14). Western blots were performed on proteins taken from three stages of the expression and purification procedure of hInt mutants i.e. whole cell extracts, the crude cell lysates and a fraction following Ni2+ affinity chromatography. Aliquots were then loaded onto a 4–12% PAGE gel, blotted onto nylon membranes and probed with a rabbit polyclonal antibody to Int followed by a secondary horse raddish peroxidise (HRP) conjugate goat anti-rabbit Ig antibody or an anti-His tag antibody-HRP conjugate (Qiagen). Detection was performed using an enhanced chemiluminescent (ECL) substrate (Pierce). A Superose-6 and a Superdex S-200 column were used for analytical size exclusion chromatography (SEC) on an AKTA FPLC (GE Healthcare) pre-equilibrated with GF buffer (20 mM Tris–HCl pH 8.0, 500 mM NaCl, 10 mM β-mercaptoethanol). The apparent molecular weights were determined by comparison of peaks obtained at A280 with those obtained with molecular weight standards (Bio-Rad).
Recombination assays
In vivo recombination assays were performed by the introduction of a plasmid expressing int into E. coli DH5α (pARM101) and selection on agar containing IPTG, X-gal, apramycin and kanamycin (9,14). A plasmid expressing wild-type Int gives 100% white colonies in this assay. Subculture from blue colonies on the same medium was used to detect any white colonies that might segregate and would thus be indicative of very low levels of Int activity.
In vitro recombination assays were performed as described previously except that modifications were made to buffers (19). The effect of EDTA upon Int activity was assayed by supplementing RxE buffer [10 mM Tris–PO4, 100 mM NaCl, 5 mM DTT, 5 mM spermidine, 4.5% glycerol and 0.5 mg/ml bovine serum albumin (BSA)] with EDTA as indicated. Int (final concentration 1 µM) was added to the modified recombination buffer and pre-incubated (30°C, 1 h) prior to the addition of pRT702 and pRT600 containing attP and attB, respectively (final concentrations ∼3 nM). Reactions were further incubated (30°C, 2 h) and the reaction products were then cut with HindIII and separated by electrophoresis (0.8% agarose/TBE gel). In vitro assays to measure the protection against EDTA by att sites were performed by pre-incubation (1 h, 30°C) of Int in RS buffer (10 mM Tris–PO4, 100 mM Na2SO4, 5 mM DTT, 5 mM spermidine, 4.5% glycerol and 0.5 mg/ml BSA) with or without 2 mM EDTA and with or without linear 50-mer att sites, made from annealed DNA oligonucleotides, at a 2:1 molar ratio of Int monomer: 50-mer att site (400 nM Int and 200 nM att DNA). Recombination was then assayed as above by addition of a partner att site located on a supercoiled plasmid and in reactions that did not contain an att site during pre-incubation, a 50-mer att site. Reaction mixtures were digested with SacI and separated by agarose gel electrophoresis to detect recombinant products.
In vitro recombination was also assayed using the reporter plasmid pRT508 that contains attP and attB flanking lacZ (9). Approximately 0.15 nM pRT508 was used in an in vitro recombination assay; the reaction mixture was introduced into DH5α and the proportion of blue versus white colonies growing on agar containing ampicillin, IPTG and XGal indicated the recombination activity.
In vitro DNA binding assays
In vitro DNA binding affinities were determined as described previously using radiolabelled annealed oligonucleotides as probes in electrophoretic mobility shift assays (EMSAs) (8). For standard affinity assays 1.0 ng labelled probe in binding buffer (20 mM Tris–HCl, pH 8.0, 50 mM KCl, 5% glycerol, 70 μg/ml sonicated salmon sperm DNA, 0.5 mg/ml BSA) and Int (final concentrations of 0–1333 nM) were mixed and incubated (30°C, 1 h). Reactions were loaded onto a 4% non-denaturing polyacrylamide gel, dried on filter paper and the radioactive bands detected using a Fuji FLA-3000 phophorimager. To study the effects of EDTA and zinc on DNA binding, binding buffer was replaced with RS buffer supplemented with 70 μg/ml of sonicated salmon sperm DNA. Int (final concentrations 0–1333 nM) was pre-incubated (30°C, 30 min) with or without EDTA in modified RS buffer prior to addition of ZnSO4 (or other metal salt, final concentrations 0.5 mM) and 1 ng labelled probe. Reactions were then incubated (30°C, 1 h) and the complexes separated and detected as for the standard assay.
ICP–MS
Protein-bound sulphur and zinc were assayed by coupling SEC to ICP–MS. Sample (5–40 μl) of ∼18 µM Int was injected onto a HiTrap SM 5 ml desalting column (SEC) and eluted using buffer [10 mM Tris–HCl (normapur, VWR), 50 mM KCl (analR, BDH) pH 7.40] and a flow rate of 0.7 ml/min. The mobile phase pumped through the SEC column by a high performance liquid chromatography (HPLC) system (Accela, Thermo Scientific) was coupled to the Meinhard-nebuliser of an Element-II ICP–MS (Thermo Scientific) via a T-piece which allowed the introduction of 74Ge as internal standard. Data were recorded at medium resolution with a dwell time per isotope ratio of 10 ms/20 peaks (250 ms total/isotope). No evidence of interference between the 64Zn+ and 32S2+ signals was observed. To determine sulphur to zinc ratios 32S, 64Zn isotopes were measured alongside 74Ge (as internal standard, BDH). For calibration stock solutions of 100 mM methionine (Sigma, 98%), 10 mM ZnSO4 (1000 p.p.m., BDH) and 10 mM EDTA (normalpur, VWR) were prepared (and further dilutions as required) as standards. To verify sensitivity and accuracy of the Zn/S ratios carbonic anhydrase from bovine erythrocytes (2500 U/mg protein, Sigma) and superoxide dismutase from bovine erythrocytes (7000 U/mg protein Sigma) were used. Quantification of isotope was determined via integrated peak area.
Circular dichroism spectroscopy
Circular dichroism (CD) Spectra of the protein solutions were obtained using a Jasco J-810 spectropolarimeter. Far UV CD measurements (185–260 nm) were collected in quartz cells of 0.02 cm path length using the following parameters: temp 25°C; scan speed 50 nm/min; bandwidth 1.0 nm; response 0.5 s and data pitch 0.2 nm. Int and derivatives were exchanged into a buffer compatible with CD measurements [25 mM Tris–PO4 (pH 7.5), 200 mM Na2SO4] with a desalting column (5 ml HiTrap, GE Healthcare). Chloride ions present in the normal purification/storage buffer absorb strongly below 200 nm which would have affected the reliability of the data. The activity of the protein in RS buffer containing sodium sulphate instead of sodium chloride was tested and showed no loss of activity.
Protein concentrations were determined using a spectrophotometer at 280 nm using an estimated extinction coefficient, ε280, for Int, hint and hCTD of 78 380, 85 830 and 79 870 M−1 cm−1, respectively. In all measurements, the CD spectrum of the buffer was subtracted from each protein spectrum. To obtain final CD spectra, five CD scans were accumulated and averaged for each sample. The spectra were corrected for protein concentration and cell path length before being analysed by DICHROWEB (20,21) an online server which hosts the various algorithms used to estimate protein secondary structures. Near UV CD measurements (250–320 nm) were obtained in a 0.5 cm quartz cuvette using the following parameters: temp 25°C; scan speed 10 nm/min; bandwidth 1.0 nm; response 2 s and data pitch 0.2 nm.
RESULTS
The CTDs of the large serine recombinases contain a conserved motif that resembles a zinc finger (Figure 1), but does not conform to known classes (16). Four conserved cysteines present in most large serine recombinases could coordinate a zinc atom and, if so, this motif is likely to be a new class of zinc finger.
φC31 Int contains zinc
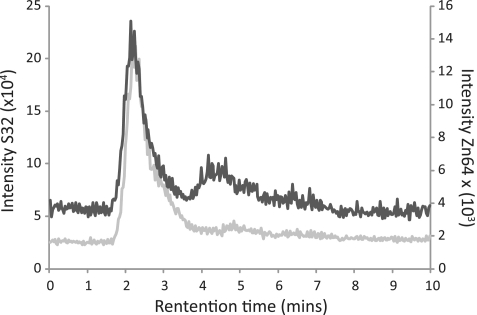
To determine whether zinc is present, Int was analysed by HPLC–ICP–MS. Concomitant analysis of sulphur and zinc showed that the Int containing fraction eluting from the HPLC column contained zinc (Figure 2). No other metals were detected by ICP–MS in Int. Known amounts of the S and Zn were used as standards and peak integration was used to obtain a ratio of sulphur to zinc. At an injection volume of 40 µl a reproducible ratio of 15:1 S:Zn was obtained or just under one atom of zinc bound to one monomer of Int (Table 1). The discrepancy from the expected ratio of 17:1 may be explained by zinc attrition in the column or the result of contamination with sulphur. Zinc was also present in the isolated large CTD of Int, hCTD, containing amino acid 155–605 fused to an N-terminal histidine tag (8) (Table 1), but was undetectable in a truncated Int, hIntV371SUGA containing a stop codon within the zinc finger-like motif (data not shown). These data localize the zinc binding motif to within the CTD.
Figure 2.
Quantitation of zinc (light grey) and sulphur (dark grey) in ϕC31 Int eluting from SEC and elements detected simultaneously by ICP-MS.
Table 1.
ICP–MS analysis of sulphur and zinc in Int and hCTD
| Injection volume (µl) | Sulphur (µmol/ml) | Zinc (µmol/ml) | S/Zn |
|---|---|---|---|
| Integrase | |||
| 5 | 0.0666 | 0.00461 | 14.5 |
| 10 | 0.0645 | 0.00426 | 15.1 |
| 20 | 0.108 | 0.00646 | 16.6 |
| 40 | 0.121 | 0.00791 | 15.3 |
| Mean (SD) | 15.4 (0.9) | ||
| hCTD | |||
| 5 | 0.00237 | 0.000157 | 15.3 |
| 10 | 0.00381 | 0.000236 | 15.1 |
| 20 | 0.00515 | 0.000315 | 16.6 |
| Mean (SD) | 15.9 (0.5) |
Zinc binding by Int is specific and essential for recombination activity
To assess the biological importance of metal binding by the zinc finger-like motif, Int was treated with metal chelators or a cysteine modifier and used in an activity assay. Int was pre-incubated with increasing concentrations of EDTA prior to the addition of substrates attP and attB and further incubation to allow recombination to occur. Pre-incubation with EDTA had an inhibitory effect on the integration reaction, even at the lowest EDTA concentration (Figure 3, top panel, lanes 1–6). Int activity was sometimes reduced after pre-incubation, even in the absence of added chelator (e.g. Figure 3, top panel, lanes 1 and 7). It was possible, however, to completely reconstitute Int activity by addition of ZnSO4 (final concentration 0.5 mM) at all concentrations of EDTA tested, including the control sample that contained no EDTA (Figure 3, middle panel, lanes 13–18). This restoration of full activity suggests that Int can lose zinc, either by leaching in the absence of EDTA or due to the chelation of zinc by EDTA (Note that addition of zinc to Int without pre-incubation had no effect on activity). Addition of exogenous zinc after treatment with chelator restored Int activity, suggesting that zinc is rapidly rebound by Int and is sufficiently stable, even when the chelator is in excess, to drive recombination.
Figure 3.
ϕC31 Int is inactivated by pre-incubation with EDTA. Int (1 µM) was pre-incubated with increasing concentrations of EDTA prior to the addition of substrates (pRT600 and pRT702) to assay integration (top panel, lanes 1–6). At the same time as adding substrates metal salts were added [final concentration 0.5 mM: NiSO4 (top panel, lanes 7–12), MgSO4, (top panel, lanes 13–18), CaSO4 (middle panel, lanes 1–6), FeSO4 (middle panel, lanes 7–11), ZnSO4 (middle panels, lanes 13–18), MnSO4 (bottom panel, lanes 1–6), CuSO4 (bottom panel, lanes 7–12) or CoSO4 (bottom panel, lanes 13–18)]. Recombination reactions were incubated (30°C, 1 h) and recombinant products were detected by restriction with HindIII and separation by agarose gel electrophoresis. The substrates pRT702 (2491 bp) or pRT600 (3035 bp) are each cut once by HindIII and the product is a co-integrate (pRT600702) that is cleaved by HindIII to yield 5435 and 91 bp bands. M are molecular weight markers (NEB, 1 kb ladder). Only ZnSO4 could restore full integration activity.
The ability of other cations (Mn2+, Cu2+, Co2+, Mg2+, Ca2+, Fe2+, Ni2+) to restore activity was tested but only Ca2+ and Mg2+ gave any observable recovery of activity after EDTA treatment (Figure 3). ICP–MS tests on all the salt solutions showed that they all contained small amounts of zinc that in the case of the calcium and magnesium salts may have restored some Int activity without poisoning. Overall the data show that Int specifically requires zinc for activity. In a time course of inhibition of Int activity by EDTA a 20 min pre-incubation was sufficient to abolish activity (Supplementary Figure S1). Despite several attempts we were not able to purify the apo-Int after treatment with EDTA. EDTA treated Int was used in SEC–ICP–MS analysis and although the signal was low due to poor protein solubility, no zinc was detected in the fractions containing Int (data not shown). The cysteine modifying compound iodoacetate also strongly inhibited the activity of Int, consistent with cysteines having an important role in recombination activity (Supplementary Figure S2). Taken together these data indicate that Int monomers coordinate a zinc atom, probably via the conserved cysteines, and that zinc is essential for recombination activity.
Zinc is required for high-affinity DNA binding by Int
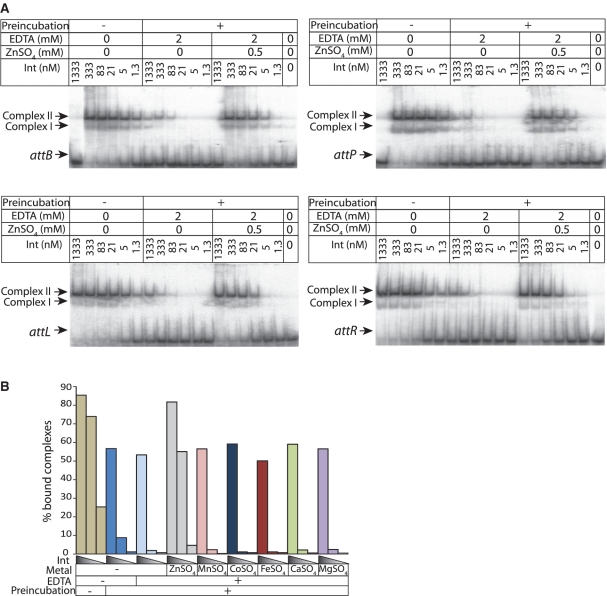
A possible role for the zinc finger-like motif in Int is in substrate recognition and DNA binding. We, therefore, tested the effect of EDTA on Int DNA binding activity in EMSA experiments (Figure 4A). Pre-incubation of Int in the presence of 2 mM EDTA led to greatly reduced affinities (50% binding of substrates was ≥1333 nM) for all four attachment sites. Affinities could be partly restored on addition of 0.5 mM ZnSO4 (50% binding of probe was between 21 and 83 nM compared to 5 to 21 nM for the non-EDTA treated, non-pre-incubated Int control). Pre-incubation in the absence of EDTA also partly inhibited Int binding activity but this too could be restored by addition of ZnSO4 (Figure 4B). Other metal ions were tested for their ability to restore DNA binding after pre-incubation and but only zinc ions could restore full activity to Int pre-incubated with EDTA (Figure 4B). The EDTA sensitive binding activity of Int is therefore dependent specifically on zinc ions.
Figure 4.
High-affinity DNA binding by Int requires ZnSO4. (A) The relative affinities of Int for DNA substrates (in modified RS buffer; see ‘Material and Methods’ section) without pre-incubation, with pre-incubation in the presence of EDTA and with pre-incubation with EDTA followed by addition of ZnSO4 were measured. Pre-incubation was performed in RS buffer or RS buffer supplemented with 2 mM EDTA (30 min) and followed by addition of radiolabelled probe and either 0 or 0.5 mM ZnSO4. The binding reactions were incubated (1 h, 30°C) then subjected to non-denaturing PAGE and the radiolabel detected as described previously (8). The arrows indicate free att site, Int:att site complex I (I) and int:att site complex II (II). (B) The % bound complexes at three different Int concentrations (1333, 83 and 5 nM) to an attB probe obtained without pre-incubation (tan), pre-incubation without EDTA (mid blue), pre-incubation with EDTA (light blue) and pre-incubation with EDTA and addition of ZnSO4 (grey), MnSO4 (pink), CoSO4 (dark blue), FeSO4 (red), CaSO4 (green), MgSO4 (mauve) to a final concentration 0.5 mM were calculated using AIDA software after phosphorimaging. Only addition of ZnSO4 could restore high-affinity DNA binding.
These data indicate that EDTA inhibits DNA binding by Int, presumably through chelating zinc that is lost from Int during pre-incubation. However, even at EDTA concentrations that completely inhibit Int recombination activity, Int can still bind the attachment sites although with greatly reduced affinities (Figure 4A). Binding by Int to its substrates is, therefore, comprised of EDTA resistant and EDTA sensitive binding. The EDTA sensitivity implies that high-affinity DNA binding requires zinc.
Cysteines and other conserved residues in the zinc finger-like motif are required for protein stability
Four conserved cysteines, C366, C368, C387 and C405, could coordinate the zinc atom and substitution of these residues might enable purification of an apo-protein (Figure 1). These residues, and two other conserved amino acids, Y385 and M372, were substituted in hInt, an N-terminally His-tagged derivative of Int. All of the mutated proteins hIntC366A, hIntC368A, hIntM372A, hIntM372I, hIntY385F, hIntC387A, hIntC405A and a double mutant, hIntC366A, C368A, gave rise to the same breakdown products (∼44 and ∼22 kDa) after expression in E. coli (Supplementary Figure S3 and data not shown). Western blot analysis and peptide mass fingerprinting confirmed that the ∼44 kDa band was an hInt fragment containing the His-tag and the NTD but truncated at the zinc finger-like motif (data not shown). Coordination of zinc during expression is, therefore, vital for correct folding of the protein and without this the unfolded protein is liable to attack from E. coli proteases. Purified wild-type Int treated with EDTA does not breakdown indicating that the protein is stable in the absence of proteases (data not shown). These data indicate the importance of these residues to the stable folding of this motif. Unsurprisingly these mutated proteins showed no recombination activity in an in vivo recombination assay (data not shown).
Substitutions in the zinc finger-like motif affect DNA binding affinities
Amino acid substitutions were generated between S374 and I381 (Table 2). These residues were chosen as they are in a putative loop between two zinc coordinating cysteines. In addition they flank a partially conserved glycine, a residue that allows for sharp turns in the peptide backbone (Figure 1). The polar and charged amino acids could potentially make contacts with the DNA. The mutated proteins were purified, tested for in vitro recombination activity and DNA binding activity (Table 2 and Figure 5). While almost all the mutated proteins had changes in affinity compared with wild-type Int, the most severely affected proteins for integration activity were those with substitutions in four consecutive residues S374 to G377.
Table 2.
Integration activities and DNA binding affinities of Int mutants
| Plasmid | Int variant | In vivo activity: % white/+ or – in subculture | In vitro activity: transformation assay (% white) | Int concentrations (nM) for 50% binding of probe |
|||
|---|---|---|---|---|---|---|---|
| attB | attP | attL | attR | ||||
| pHS62 | Int | 100/+ | 91 | 21–83 | 21–83 | 21 | 21 |
| pARM010 | hINT | 100/+ | 87 | 21–83 | 21–83 | 21 | 21 |
| pADD002 | hIntS374A | 42/+ | 78 | 83 | 21–83 | 5–21 | 5–21 |
| pARM024 | hIntS374L | 0/− | 2 | >1333 | 1333 | 1333 | 330 |
| pARM070 | hIntK375A | 2/+ | 20 | 333 | 83–333 | 83–333 | 83–333 |
| pARM103 | hIntK375Q | 0/− | <1 | 333 | 83 | 21–83 | 21 |
| pARM104 | hIntK375E | 0/− | <1 | 1333 | 83 | 83 | 21 |
| pARM071 | hIntR376A | 1/+ | <1 | 1333 | 83 | 83 | 21 |
| pARM105 | hIntR376E | 0/− | <1 | >1333 | >1333 | >1333 | >1333 |
| pARM096 | hIntR376P | 0/− | <1 | >1333 | 333 | 21 | 21 |
| pADD001 | hIntG377L | 61/+ | 78 | 21–83 | 21–83 | 21–83 | 21 |
| pADD010 | hIntG377A | 100/+ | 84 | 21–83 | 21–83 | 21–83 | 21–83 |
| pARM020 | hIntG377D | 40/+ | 58 | 21–83 | 21 | 5–21 | 5–21 |
| pARM021 | hIntG377F | 4/+ | <1 | 83–333 | 83–333 | 83–333 | 83 |
| pARM022 | hIntG377K | 0/− | 1 | 1333 | 21–83 | 83 | 21–83 |
| PARM072 | hIntE378A | 100/+ | 94 | 333 | 83–333 | 83–333 | 83–333 |
| pARM073 | hIntE379A | 100/+ | 93 | 83–333 | 83–333 | 83–333 | 83–333 |
| pARM074 | hIntS380A | 100/+ | 95 | 333 | 83–333 | 83–333 | 83–333 |
| pARM107 | hIntS380D | 32/+ | 29 | 21 | 21 | 5 | 5 |
| pARM108 | hIntS380N | 12/+ | 70 | 5–21 | 5 | 5 | 1.3 |
| pARM109 | hIntS380T | 100/+ | 94 | 1.3–5 | 5 | 1.3–5 | 1.3 |
| pARM100 | hIntI381M | 100/+ | 95 | ND | ND | ND | ND |
| pARM106 | hIntV371SUGA | 0/− | <1 | >1333 | 333–1331 | >1333 | >1333 |
Bold indicates a lower affinity for a substrate than the wild-type Int. ‘ND’ indicates that the data were not determined.
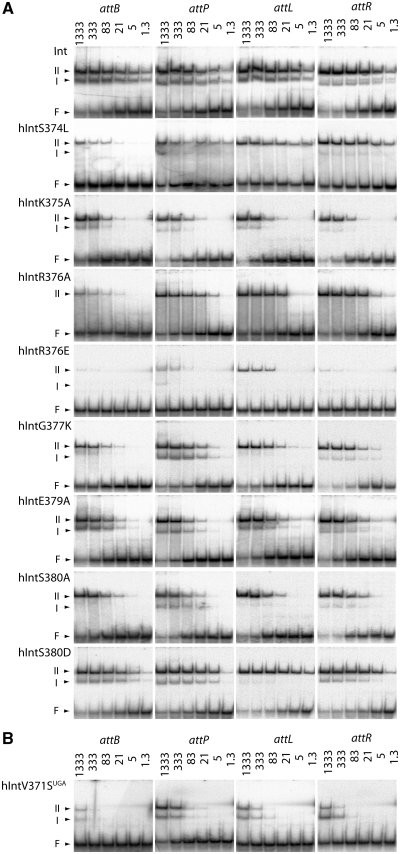
Figure 5.
Substrate binding by Int and mutated derivatives. (A) Int and hInt derivatives containing single residue substitutions were incubated at the final concentrations indicated (nM) with labelled annealed oligonucleotides containing attB, attP, attL and attR sequences in binding buffer (see ‘Materials and Methods’ section) and the bound complexes separated by non-denaturing gel electrophoresis and the label detected by phosphorimaging. The arrows are as described in Figure 4. (B) Substrate binding (as in A) by the truncated hInt derivative, hIntV371UGA.
Two types of DNA binding affinity defects were observed with this set of Int variants; those that had reduced affinities for all the att sites (hIntS374L, hIntK375A, hIntR376E, hIntG377F, hIntE378A, hIntE379A and hIntS380A) and those whose affinities to attB were affected more than to other att sites (for e.g. hIntK375Q, hIntK375E, hIntR376A and hIntG377K; Table 2 and Figure 5). In some proteins (hIntK375E, hIntR376A, hIntG377K), attL binding also appeared to be more affected that attP or attR binding. attB and attL both contain a B half site that has been shown previously to have a greater contribution than the B′ half site to the activity of the attB site (15). One substitution, hIntR376E, abolished nearly all DNA binding (Table 2 and Figure 5). Thus an intact zinc finger-like motif is required for high-affinity DNA binding and may have a role in distinguishing between an attB and an attP site.
CD spectroscopy and SEC to evaluate secondary structure of binding defective mutants
The DNA binding defects in the Int variants could be due to loss of direct contacts between protein and DNA or due to gross structural changes that indirectly perturb DNA binding affinity. Int variants with some of the more severe binding defects were, therefore, analysed by CD and SEC to try to detect structural changes that might be correlated with poor DNA binding (Figure 6; Supplementary Tables S1 and S2).
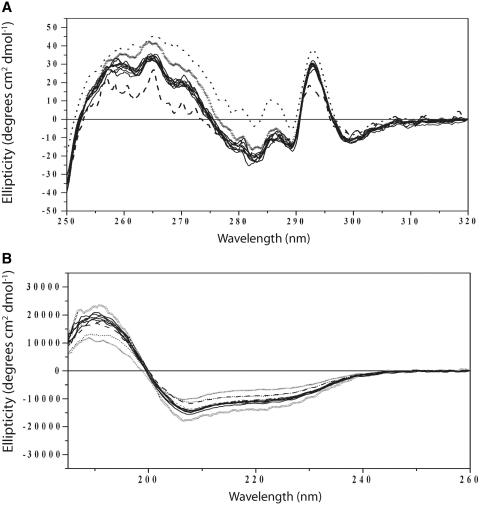
Figure 6.
CD of ϕC31 Int and derivatives. Superimposed spectra of all the protein analysed are shown (panel A; near UV, panel B; far UV). The outliers in the near UV are hCTD (dotted line) and the wild-type Int (dashed line). All the remaining spectra in the near UV superimpose with the hInt protein. In the far UV the outliers are hCTD (crosses) and hIntS380D (circles); hInt and Int are the dashed and dotted lines, respectively. In the far UV spectra, all the hInt derivatives with an amino acid substitution therefore have slightly enhanced ellipticity compared with the hInt.
The near UV CD spectra showed that the environments of the aromatic residues in wild-type Int and most of the variant proteins were essentially the same and strongly implied that there were no gross structural changes in the variant proteins (Figure 6A). The proteins with the most deviant fingerprints in the near UV region were hCTD (expected as this protein has lost the entire NTD) and hIntK375Q, an Int variant that is recombinationally inactive and defective in attB binding but able to bind attP, attL and attR with near normal affinities (Table 2). Most of the Int mutants showed a slight increase in the intensity of their ellipticity values in the far UV region which implies (assuming accurate protein estimations) that the amino acid substitutions in all these mutants have all undergone small, similar changes to overall secondary structural conformation, such as stiffening or removal of structural constraints that affect the tertiary conformation (Supplementary Table S2). The major outliers in the far UV were hCTD and hIntS380D, a partially active protein with very high binding affinities to all the attachment sites (Figure 6B, Table 2). Overall the differences between the secondary structures of the Int mutants compared with hInt or Int were small. In particular, there was little change in the estimated proportions of turn or disorder although the mutant proteins appeared reproducibly to have marginally more predicted alpha helix compared with β-sheet (Supplementary Table S2). Crucially none of the observed differences in CD spectra could be correlated with changes in DNA binding affinities or recombination activities.
Analysis of the Int variants by SEC indicated no changes in oligomeric structures; all the Int proteins with amino acid substitutions behaved as dimers in solution (Supplementary Table S3).
A truncated Int lacking an intact zinc finger-like motif has reduced binding affinities
A by product of the site-directed mutagenesis experiments was an int allele encoding a V371S substitution followed by a stop codon (hIntV371SUGA). This protein, truncated within the zinc finger-like motif, binds the att sites with much lower affinities than hInt but still generates the two complexes (I and II) typically observed when Int binds its substrates (Figure 5B). The residual binding activity by hIntV371SUGA shows that there is another, as yet uncharacterized, DNA recognition and binding motif in Int. Binding to the substrates by hIntV371SUGA resembled binding by EDTA-treated Int as Int–DNA complexes were only observed at the highest Int concentrations. Thus an intact zinc finger-like motif is required for high-affinity DNA binding.
Inhibition of Int activity by EDTA can be prevented by the presence of attachment sites
If the zinc finger is indeed a DNA recognition and binding motif and comes into close contact with the attachment sites, we hypothesized that Int in complex with the att sites should be less sensitive to EDTA than Int in free solution.
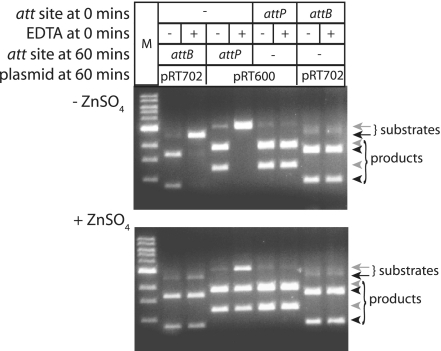
Annealed oligonucleotides containing the sequences of attP or attB were used to attain stoichiometric ratios of att site 50-mer to a dimer of Int. Int was pre-incubated in the presence or absence of the 50-mer att site and in the presence or absence of EDTA for 1 h. Recombination activity was then assayed by addition of a partner att site encoded on a supercoiled plasmid and, in those reactions that did not contain an att site at the start of pre-incubation, a 50-mer att site (Figure 7). Recombination yielded linearized plasmid which could be further digested to give restriction fragments diagnostic of recombination products. As before, pre-incubation of Int with EDTA inhibited attP × attB recombination and activity could be mostly restored by addition of ZnSO4. However, if the attP or attB 50-mers were added at the start of pre-incubatin with EDTA, no inhibition of recombination activity was observed suggesting that these substrates could protect Int from the zinc chelating effects of EDTA (Figure 7).
Figure 7.
Inhibition of Int by EDTA can be prevented by addition of one of the recombination substrates. Int (400 nM) was incubated for 60 min with or without EDTA and with or without a protecting 50-mer att site (200 nM). Int activity was then measured by addition of a partner att site encoded on a plasmid (attP; pRT702, attB; pRT600) and in reactions that contained no att site during pre-incubation, a 50-mer att site. The recombination reactions were incubated (30°C, 1 h) and the products of recombination were analysed after digestion with ScaI. The substrates pRT702 (2491 bp; black arrow) or pRT600 (3035 bp; grey arrow) are each cut once by ScaI but if they have undergone recombination with their partner att sites contained on the 50-mers, the linear products are cleaved to give 1671 and 869 bp (pRT702; black arrowheads) and 1887 and 1200 bp (pRT600; grey arrowheads). M are molecular weight markers. The lower panel shows that inhibition by EDTA could be prevented if ZnSO4 (final concentration 0.5 mM) is added at the start of pre-incubation.
No protection was observed when a non-specific oligonucleotide or if poly dI-dC were added to the pre-incubation mix (data not shown). These data show that Int specifically bound to its attachments sites is protected from the inhibitory activity of EDTA suggesting that the zinc finger-like motif is in direct contact with the DNA.
DISCUSSION
Zinc fingers are a large family of small protein domains with diverse sequence, size and range of functions. Mostly zinc fingers are structural domains involved in interactions with other molecules, particularly nucleic acids and proteins (16,22). Zinc binding can often be predicted from sequence as typical zinc binding is through interactions with cysteine, histidine, glutamic acid or aspartic acid side chains, but most frequently through tetra-coordination via cysteines (16). Here we present the evidence for a putative zinc finger in ϕC31 Int. An Int monomer contains a Zn2+ atom and both recombinase and DNA binding activities of Int were sensitive to EDTA. Mutant Ints containing substitutions in the cysteines that are thought to be coordinating the Zn2+ atom are unstable indicating that this motif requires zinc for structural integrity.
Evidence is also presented that the putative zinc finger is directly involved in recognition and binding to the attachment sites. Amino acid substitutions can have dramatic effects on DNA binding affinity by zinc fingers (23,24). Similarly amino acid substitutions in the zinc finger-like motif in ϕC31 Int also reduced the DNA binding affinities to variable extents, the most severe of which could be due to an ionic clash (hIntR376E) and appears to abolish DNA binding to a greater extent than either treatment of Int with EDTA or truncation of Int at V371 (hIntV371UGA). Mutant Ints with lesser reductions in affinity could be due to loss of contacts (substitutions with alanine) or steric blocks (e.g. G377K, G377F; Table 2) (25,26). We showed that the att sites could protect Int against inhibition by EDTA; this is probably because the DNA and the zinc finger-like motif are in close proximity and the DNA prevents EDTA from accessing the Zn2+. High-affinity DNA binding by Int is EDTA sensitive but that there is also an EDTA resistant, low affinity activity. Indeed the truncated protein, hIntV371SUGA, indicates that Int has at least one other DNA binding region. In Bxb1 gpInt a similar construct to ϕC31 hIntV371SUGA also binds DNA with low affinity and substitutions in both Bxb1 gpInt and TnpX in regions equivalent to the truncated CTD in hIntV371SUGA affect DNA binding (7,27). Despite this strong evidence for direct contact by the zinc finger-like motif to DNA, we cannot yet rule out an indirect effect in which this motif influences either this second DNA binding region or an as yet unidentified DNA binding motif elsewhere in the protein.
In the absence of Xis Int forms a synaptic complex only when attP and attB are substrates indicating that binding to these substrates evokes different Int conformations as a result of different protein:DNA recognition events (1,7,10). A fundamental question to address given that attP and attB are such different sequences is whether Int has two DNA binding regions; one for P type arms and one for B arms. This seems unlikely given that mutants described here can be generally reduced in all att site binding. However, many of the Int variants were more affected in attB than to attL/R and attP, indicating that the interaction with attB, and possibly B type arms, might involve a different set of contacts to those made with attP. Moreover recombination activity is more likely to be lost by amino acid substitutions in S374 to G377 than in E378 to I381, although DNA binding can be affected by substitutions throughout the zinc finger-like motif (Table 2, Figure 5). Thus contacts made to the attB and attP sites via S374, K375 and R376 could be critical for inducing conformation changes required for activation of recombination. In our experiments we noted a distinct reduction in the amount of complex I formed in the EMSA experiments with hIntS374L, hIntR376A and hIntG377K compared with wild-type Int. Int binds to all the substrates to generate two complexes (I and II) that we have previously interpreted as containing either a monomer or a dimer of Int, respectively (10). Possibly the dimer complex is more stable in some of the Int variants than in the wild-type Int complexes and that destabilizing the dimer interface may therefore be one of the changes that occurs to generate the active synaptic tetramer.
In this article, we have demonstrated for the first time the importance of zinc in a phage-encoded Int that is likely a part of a DNA binding motif. The serine Ints are widely used as tools for genome engineering and gene therapy, [for example, see Ref. (28)]. The mechanism through which these recombinases recognize their DNA substrates and activate recombination is of interest for rational engineering and improvement of recombination efficiency in heterologous environments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Biotechnology and Biological Sciences Research Council (BBSRC), UK (project BB/H001212/1); Funding for research: BBSRC (projects B19375, BB/H001212/1 and BB/F011687/1) and the University of Aberdeen.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Dr Nigel Grindley for useful discussions and suggestions for experiments and to Mrs Amanda Davidson and Dr Paul Rowley who generated some of the initial mutants. The authors also thank the University of Aberdeen Proteomics facility for the preparation and analysis of protein samples.
REFERENCES
- 1.Smith MCM, Brown WR, McEwan AR, Rowley PA. Site-specific recombination by ϕC31 integrase and other large serine recombinases. Biochem. Soc. Trans. 2010;38:388–394. doi: 10.1042/BST0380388. [DOI] [PubMed] [Google Scholar]
- 2.Grindley NDF, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Ann. Rev. Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 3.Smith MCM, Thorpe HM. Diversity in the serine recombinases. Mol. Microbiol. 2002;44:299–307. doi: 10.1046/j.1365-2958.2002.02891.x. [DOI] [PubMed] [Google Scholar]
- 4.Yuan P, Gupta K, Van Duyne GD. Tetrameric structure of a serine integrase catalytic domain. Structure. 2008;16:1275–1286. doi: 10.1016/j.str.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 5.Sanderson MR, Freemont PS, Rice PA, Goldman A, Hatfull GF, Grindley NDF, Steitz TA. The crystal structure of the catalytic domain of the site-specific recombination enzyme gamma delta resolvase at 2.7 A resolution. Cell. 1990;63:1323–1329. doi: 10.1016/0092-8674(90)90427-g. [DOI] [PubMed] [Google Scholar]
- 6.Ghosh P, Kim AI, Hatfull GF. The orientation of mycobacteriophage Bxb1 integration is solely dependent on the central dinucleotide of attP and attB. Mol. Cell. 2003;12:1101–1111. doi: 10.1016/s1097-2765(03)00444-1. [DOI] [PubMed] [Google Scholar]
- 7.Ghosh P, Pannunzio NR, Hatfull GF. Synapsis in phage Bxb1 integration: selection mechanism for the correct pair of recombination sites. J. Mol. Biol. 2005;349:331–348. doi: 10.1016/j.jmb.2005.03.043. [DOI] [PubMed] [Google Scholar]
- 8.McEwan AR, Rowley PA, Smith MCM. DNA binding and synapsis by the large C-terminal domain of ϕC31 integrase. Nucleic Acids Res. 2009;37:4764–4773. doi: 10.1093/nar/gkp485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rowley PA, Smith MCA, Younger E, Smith MCM. A motif in the C-terminal domain of ϕC31 integrase controls the directionality of recombination. Nucleic Acids Res. 2008;36:3879–3891. doi: 10.1093/nar/gkn269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thorpe HM, Wilson SE, Smith MCM. Control of directionality in the site-specific recombination system of the Streptomyces phage ϕC31. Mol. Microbiol. 2000;38:232–241. doi: 10.1046/j.1365-2958.2000.02142.x. [DOI] [PubMed] [Google Scholar]
- 11.Kuhstoss S, Rao RN. Analysis of the integration function of the streptomycete bacteriophage ϕC31. J. Mol. Biol. 1991;222:897–908. doi: 10.1016/0022-2836(91)90584-s. [DOI] [PubMed] [Google Scholar]
- 12.Combes P, Till R, Bee S, Smith MCM. The streptomyces genome contains multiple pseudo-attB sites for the ϕC31-encoded site-specific recombination system. J. Bacteriol. 2002;184:5746–5752. doi: 10.1128/JB.184.20.5746-5752.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thorpe HM, Smith MCM. In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc. Natl Acad. Sci. USA. 1998;95:5505–5510. doi: 10.1073/pnas.95.10.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith MCA, Till R, Brady K, Soultanas P, Thorpe H, Smith MCM. Synapsis and DNA cleavage in ϕC31 integrase-mediated site-specific recombination. Nucleic Acids Res. 2004;32:2607–2617. doi: 10.1093/nar/gkh538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta M, Till R, Smith MCM. Sequences in attB that affect the ability of ϕC31 integrase to synapse and to activate DNA cleavage. Nucleic Acids Res. 2007;35:3407–3419. doi: 10.1093/nar/gkm206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krishna SS, Majumdar I, Grishin NV. Structural classification of zinc fingers: survey and summary. Nucleic Acids Res. 2003;31:532–550. doi: 10.1093/nar/gkg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu H, Naismith JH. A simple and efficient expression and purification system using two newly constructed vectors. Protein expression and purification. 2009;63:102–111. doi: 10.1016/j.pep.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gust B, Kieser T, Chater KF. REDIRECT Technology: PCR-targeting System in Streptomyces coelicolor. John Innes Centre, Norwich; 2002. [Google Scholar]
- 19.Smith MCA, Till R, Smith MCM. Switching the polarity of a bacteriophage integration system. Mol. Microbiol. 2004;51:1719–1728. doi: 10.1111/j.1365-2958.2003.03942.x. [DOI] [PubMed] [Google Scholar]
- 20.Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–W673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitmore L, Wallace BA. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 22.Gamsjaeger R, Liew CK, Loughlin FE, Crossley M, Mackay JP. Sticky fingers: zinc-fingers as protein-recognition motifs. TIBS. 2007;32:63–70. doi: 10.1016/j.tibs.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 23.Nardelli J, Gibson T, Charnay P. Zinc finger-DNA recognition: analysis of base specificity by site-directed mutagenesis. Nucleic Acids Res. 1992;20:4137–4144. doi: 10.1093/nar/20.16.4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clouaire T, Roussigne M, Ecochard V, Mathe C, Amalric F, Girard JP. The THAP domain of THAP1 is a large C2CH module with zinc-dependent sequence-specific DNA-binding activity. Proc. Natl Acad. Sci. USA. 2005;102:6907–6912. doi: 10.1073/pnas.0406882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luscombe NM, Laskowski RA, Thornton JM. Amino acid-base interactions: a three-dimensional analysis of protein-DNA interactions at an atomic level. Nucleic Acids Res. 2001;29:2860–2874. doi: 10.1093/nar/29.13.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rohs R, Jin X, West SM, Joshi R, Honig B, Mann RS. Origins of specificity in protein-DNA recognition. Ann. Rev. Biochem. 2010;79:233–269. doi: 10.1146/annurev-biochem-060408-091030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adams V, Lucet IS, Tynan FE, Chiarezza M, Howarth PM, Kim J, Rossjohn J, Lyras D, Rood JI. Two distinct regions of the large serine recombinase TnpX are required for DNA binding and biological function. Mol. Microbiol. 2006;60:591–601. doi: 10.1111/j.1365-2958.2006.05120.x. [DOI] [PubMed] [Google Scholar]
- 28.Calos MP. The ϕC31 integrase system for gene therapy. Curr. Gene Therapy. 2006;6:633–645. doi: 10.2174/156652306779010642. [DOI] [PubMed] [Google Scholar]
- 29.Poirot O, O'Toole E, Notredame C. Tcoffee@igs: a web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res. 2003; 31:3503–3506. doi: 10.1093/nar/gkg522. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.