Abstract
Although patterns of somatic alterations have been reported for tumor genomes, little is known on how they compare with alterations present in non-tumor genomes. A comparison of the two would be crucial to better characterize the genetic alterations driving tumorigenesis. We sequenced the genomes of a lymphoblastoid (HCC1954BL) and a breast tumor (HCC1954) cell line derived from the same patient and compared the somatic alterations present in both. The lymphoblastoid genome presents a comparable number and similar spectrum of nucleotide substitutions to that found in the tumor genome. However, a significant difference in the ratio of non-synonymous to synonymous substitutions was observed between both genomes (P = 0.031). Protein–protein interaction analysis revealed that mutations in the tumor genome preferentially affect hub-genes (P = 0.0017) and are co-selected to present synergistic functions (P < 0.0001). KEGG analysis showed that in the tumor genome most mutated genes were organized into signaling pathways related to tumorigenesis. No such organization or synergy was observed in the lymphoblastoid genome. Our results indicate that endogenous mutagens and replication errors can generate the overall number of mutations required to drive tumorigenesis and that it is the combination rather than the frequency of mutations that is crucial to complete tumorigenic transformation.
INTRODUCTION
Somatic genetic alterations accumulate in the genome of all dividing cells as a result of DNA replication errors or exposure to mutagens. Some somatic alterations, known as driver mutations, confer a selective growth advantage to the cells and can cause cancer. Conversely, passenger alterations are biologically neutral and are expected to occur in normal genomes (1,2). Recent advances in sequencing technologies have allowed a comprehensive characterization of the genetic alterations occurring in individual tumors, including copy number variations (CNVs), chromosomal rearrangements, point mutations and small insertions and deletions (Indels) (3–11). However, the frequency and characteristics of the genetic alterations driving tumorigenesis are not currently well-defined.
Although the sequences of six-matched tumor and non-tumor genomes have been published (4–8,11), these studies were limited to the identification of somatic alterations present in the tumor genomes and there has been no attempt to define the somatic alterations present in the matched non-tumor genome. A comparison of the two would be crucial to better characterize the genetic changes that drive tumorigenesis, since we expect most, if not all, alterations present in the non-tumor genome to be passenger mutations.
Here, we have sequenced the genome of a breast tumor cell line (HCC1954) and a lymphoblastoid cell line (HCC1954BL) derived from the same patient and compared the genetic variations occurring in both. HCC1954 is an immortal, pseudo-tetraploid (12), publicly available tumor cell line derived from a hormone receptor negative, ERBB2 positive, primary breast tumor from a 61-year-old female patient. HCC1954BL is an Epstein-Barr virus (EBV)-transformed lymphoblastoid cell line derived from the same patient. Both cell lines received similar treatments in terms of the timing of establishment and in vitro propagation (36 passages) (13) and the sequencing data revealed both lines to be clonal, permitting the detection of somatic changes in both.
Using a combined sequencing approach, we were able to characterize chromosomal rearrangements and single nucleotide variations (SNVs) in the protein-coding regions of the HCC1954 and HCC1954BL genomes. By comparing the sets of rearrangements and SNVs present in both genomes, we were able to exclude those that are common to both genomes and correspond to inherited variations and to identify those that are exclusive to each genome and likely correspond to somatic alterations that have independently accumulated in the genomes of these cell lines during their lifespan. Thus, in the present work, in addition to the identification of somatic mutations present in the tumor genome, we have also identified those present in the matching lymphoblastoid genome and used a system biology approach to better characterize the functional differences between the set of altered genes present in both genomes.
We found that the HCC1954BL genome contains very few somatically acquired chromosomal rearrangements but, surprisingly, present a comparable number of somatic point mutations and a similar spectrum of nucleotide substitutions to that found in the HCC1954 genome. We also observed that, unlike in the HCC1954BL genome, non-synonymous mutations present in the HCC1954 genome were not randomly distributed and were co-selected to present synergistic tumor-promoting functions and to affect hub-genes in functional pathways related to tumorigenesis. Our results provide important insights into the normal mutational processes and into the functional implications of the accumulation of somatic mutations in a tumor and a matching lymphoblastoid genome.
MATERIALS AND METHODS
Cell lines and DNA extraction
HCC1954 and HCC1954BL cell lines were obtained from American Type Culture Collection (ATCC) and were maintained in RPMI medium containing 10% fetal bovine serum (FBS) and non-essential amino acids. Both cell lines received similar treatments in terms of the timing of establishment and in vitro propagation and present similar proliferation rates (data not shown). Both of them were received from ATCC at passage 25 after in vitro establishment and were maintained in culture until passage 36 when DNA was extracted for sequencing. DNA was isolated using the DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA, USA). Genomic DNA was treated with RNase Cocktail™ (Ambion Austin, TX, USA), followed by phenol–chloroform extraction and precipitation of the aqueous phase in 1/10 volume 3 M sodium acetate, pH5.2 and 100% ethanol.
Public genome data
The human reference genome sequence (NCBI build 36.1/hg18) was downloaded from the UCSC Genome Browser (http://genome.ucsc.edu). Alternative haplotype regions, including the immunoglobulin loci, were excluded from the reference sequence because of their highly polymorphic and rearranged structure. Human reference mRNA sequences (RefSeqs for coding mRNAs, ‘NM’ and non-coding mRNAs,‘NR’) were downloaded from NCBI (http://www.ncbi.nlm.nih.gov/RefSeq) and mapped to the human genome as previously described (14). Known SNPs were also downloaded from the UCSC Genome Browser (dbSNP version 130) and loaded into a MySQL database.
gDNA paired-end sequencing and mapping
A total of 5 µg of genomic DNA was randomly sheared using a Biorupter according to the manufacturer's instructions. The fragmented DNA was end-repaired using Klenow and T4 DNA polymerases and phosphorylated at the 5′-end with T4 polynucleotide kinase. A 3′ overhang was created using 3′–5′ exonuclease-deficient Klenow fragment and Illumina paired-end adaptor oligonucleotides were ligated to the created sticky ends. DNA fragments of ∼200 bp were size selected in 8% polyacrylamide gels and eluted from the gel overnight. Size-selected DNA was Polymerase Chain Reaction (PCR) amplified for 18 cycles to enrich for adapter-modified DNA fragments. A paired-end flow cell was prepared on the supplied cluster station according to the manufacturer's protocol. Clusters of PCR colonies were then sequenced on the Illumina GAII sequencing platform using the recommended protocols. Images from the instrument were processed to generate sequence files in FASTAQ format. Sequences were aligned to the human genome reference sequence (NCBI build 36.1/hg18) using Bowtie version 0.12.2 (15), allowing two mismatches in mapping. Duplicated read pairs with identical coordinates were merged, and only unambiguously mapped reads were used for the structural variation analysis.
Exome capture, sequencing and mapping
A total of 34.1 Mb of the human genome corresponding to approximately 180 000 coding exons (28.4 Mb) and adjacent intergenic and intronic regions (5.7 Mb) was captured using the Nimblegen Sequence Capture 2.1 M Human Exome array v1.0. Briefly, genomic DNA samples were fragmented, ligated to adapters and hybridized to the exome capture array. Unbound fragments were washed away and the target-enriched DNA was eluted. Enriched samples were amplified by Ligation-Mediated PCR (LM-PCR). The adapters used during enrichment were designed for direct integration into the workflow of the 454 GS FLX instrument, eliminating the library construction process. We continued the protocol from the DNA library quantification and emulsion PCR according to manufacturer's protocol. Sequencing was performed using the 454 GS FLX Titanium platform. Sequences were aligned to the human genome reference sequence (NCBI build 36.1/hg 18) using BLAT program (parameters: noTrimA –tileSize = 12 –minScore = 200 −minIdentity = 92 –out = pslx) (16). Duplicated reads with identical coordinates were merged, and only unambiguously mapped reads were used for SNV calling and mutation detection.
Data deposition and availability
Quality scores and FASTA sequences generated for HCC1954 and HCC1954BL were uploaded to the Short Read Archive under the accession numbers ERA010917 and ERA011762 and are publicly available.
Structural variation analysis
Illumina paired-end reads that failed to align to the human genome reference sequence in the expected orientation or distance were used in the structural variation analysis after removing those that mapped to highly repetitive regions within 1 Mb of a centromeric or telomeric region. Reads for which the two ends aligned to within the expected distance, but with one of the two ends in the incorrect orientation were also excluded from the analysis because these reads are likely to be artifacts generated by mispriming of the Illumina sequencing oligonucleotide or intramolecular rearrangements generated during library amplification. Structural variants were called from Bowtie alignments of genomic paired-end sequences, requiring at least five independent read pairs in HCC1954 and no read pairs representing the rearrangement in HCC1954BL and vice versa. Interchromosomal rearrangements were called when each read from the same pair was mapped uniquely, but in distinct chromosomes. Intrachromosomal deletions were called when the read-pairs mapped in an expected order and orientation, but in a distance greater than the expected (average + 4*SD). Intrachromosomal tandem duplications were called when read-pairs mapped in an expected orientation, but in an unexpected order and distance. Intrachromosomal inversions were called when read-pairs mapped in an expected order, but in an unexpected orientation and distance (for a review, see Ref. 17). Structural variants represented by read pairs that mapped to within 500 bp of a previously identified copy number polymorphism were removed from the final list of somatic chromosomal rearrangements.
SNVs and point mutation analysis
SNVs and somatic point mutations were independently called for each cell line from BLAT alignments of capture sequences to the human reference genome. SNVs supported by at least three reads with base quality ≥20 were called for the HCC1954 and HCC1954BL genomes. To assess the sensitivity of our SNV detection strategy, we have compared our SNV calls to SNV calls extracted from genotyping array data available for both cell lines (GEO: GSE12019 and GSE13373), as previously described (5). SNVs common to both genomes and/or already described in dbSNP were excluded from the somatic point mutation analysis, since they likely correspond to inherited sequence variants. Somatic point mutations were then identified, requiring at least three high-quality independent reads (≥3 reads with Phred score ≥20 and ≥1 read with Phred score ≥30) reporting the variant in HCC1954 and no reads reporting the variant in HCC1954BL and vice versa. Variant reads were required to represent at least 20% of the total number of reads covering the variant genomic position to filter somatic mutations that might have eventually arisen during in vitro propagation of both cell lines and present in a small subpopulation of the cells. A depth of at least 5× was required to assure sufficient coverage in both cell lines. We also excluded from the point mutation analysis false mutation calls residing in regions of loss-of-heterozygosity (LOH) in either of the cell lines. Briefly, the zygosity of approximately 250 000 known SNPs represented in the Affymetrix SNP array was determined for both cell lines using hybridization data available in public databases (GEO: GSE12019 and GSE13373). LOH regions (where SNPs were heterozygous in the normal but homozygous in the tumor and vice versa) were identified using Hidden Markov Model algorithms as previously described (7,8). Results were manually inspected and exome sequence data were used to confirm the SNP array analysis and to increase resolution in regions of low-SNP density represented in the Affymetrix SNP array.
Ratio of non-synonymous to synonymous substitutions
Monte Carlo simulations were used to determine if the ratio of non-synonymous to synonymous substitutions (dN/dS) observed for HCC1954 and HCC1954BL were significantly different from that expected by chance (null hypothesis). In these simulations, values expected by change were obtained from 1000 random sets of 64 mutations for HCC1954 and 30 mutations for HCC1954BL occurring in the coding region of known human RefSeq genes. To calculate if the difference between the dN/dS ratios observed for HCC1954 and HCC1954BL was significant, we performed a χ2 test using the HCC1954BL dN/dS values as expected values for HCC1954.
PCR and Sanger sequencing validation
Primers for validation were designed to target regions immediately flanking point mutations using Primers3 software (http://frodo.wi.mit.ed/primer3/). PCR amplification was performed on HCC1954 and HCC1954BL genomic DNA using Taq Platinum Hi-Fidelity Polymerase (Invitrogene), following standard protocols. PCR product purity and size were assessed on 2% agarose gels stained with GelRed (Biotium). Sanger sequencing was performed using an ABI3130 Capillary DNA Analyzer. Sequence trace files were manually analyzed for point mutations. All confirmations were performed using both HCC1954 and HCC1954BL DNA to determine, if the variants were somatic or germline. Validated non-synonymous mutations were classified as non-conservative if they result in changes in amino acid charge and polarity. Functional proteins domains were determined using Pfam and HMMER package (E < 0.001). A Perl-script was used to cross information on the position of non-synonymous mutations and functional domains. Evolutionary conserved amino acid residues were obtained from UCSC Genome Browser Conservation Track (http://genome.ucsc.edu). Amino acid conservation between 10 different species, Pan troglodytes (Chimp), Macaca mulatta (Rhesus), Mus musculus (mouse), Rattus norvegicus (rat), Canis lupus familiaris (Dog), Loxodonta africana (elephant), Monodelphis domestica (Opossum), Gallus gallus (chicken) and Takifugu rubripes (fugu), was manually determined.
KEGG and protein–protein interactions analysis
The list of genes carrying non-synonymous mutations validated by Sanger sequencing for each cell line was uploaded to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database for the pathway enrichment analysis. Genes carrying non-synonymous mutations were used since these mutations are more likely to have a functional impact when compared to synonymous mutations. Monte Carlo simulations were performed in which 1000 random sets of 45 genes and 12 genes were evaluated regarding KEGG pathway enrichment to address whether the obtained results were significantly different (P < 0.05) from those expected by chance. Simulations were performed using all known coding genes and KEGG pathways (200 annotated pathways). For the protein–protein interactions (PPI) analysis, data from the following databases were merged: MINT (December 2007 version), BIOGRID (2.0.37), INTACT (January 2008 version), HPRD (September 2007 version), BIND (May 2006 version) and DIP (January 2008 version). These databases include both high-throughput experiments and low-throughput ones curated from the literature. When a field was present for the technique used to discover the interaction, those registers with an entry that referred to one of the several mass spectrometry-based methods were excluded to avoid including indirect interactions. Exclusively functional interactions, for example, numerous exclusively genetic interactions present in BIOGRID, were also excluded. Only genes carrying non-synonymous mutations validated by Sanger sequencing were used in the PPI analysis. Monte Carlo simulations were performed in which 10 000 random sets of 25 and 8 genes with known PPI were evaluated regarding the degree of interaction and the number of common interaction partners among mutated proteins to address whether the obtained results were significantly different (P < 0.05) from those expected by chance.
RESULTS
Sequencing strategy and genome coverage
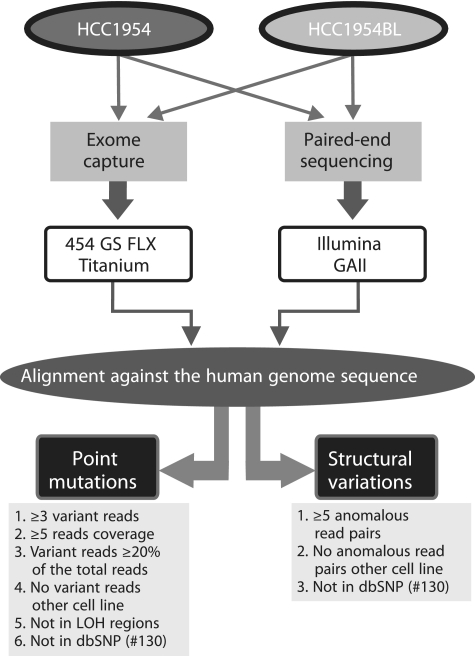
A combined sequencing strategy was used to characterize chromosomal rearrangements and point mutations in protein-coding regions of the HCC1954 and HCC1954BL genomes (Figure 1). Approximately 381 million and 347 million 35–75 bp paired-end reads from 200 bp DNA inserts were generated for HCC1954 and HCC1954BL, respectively, using an Illumina GAII sequencing platform (Figure 1 and Table 1). For HCC1954, ∼254 million paired-end reads were unambiguously mapped to the reference genome, which based on the average insert size of ∼200 bp (Supplementary Figure S1) generated 8× physical coverage. Similar numbers were generated for HCC1954BL (∼237 million mapped paired-end reads and 8× physical coverage). Paired-end reads that aligned discordantly with respect to each other on the reference genome were then used to detect chromosomal rearrangements in the HCC1954 and HCC1954BL genomes (Figure 1).
Figure 1.
Sequencing strategy. Outline of the sequencing strategy and bioinformatics algorithms used for the identification of point mutations and structural chromosomal rearrangements in the HCC1954 and HCC1954BL genomes.
Table 1.
Summary of sequence generation and mapping to the reference human genome sequence for the HCC1954 and HCC1954BL cell lines
| HCC1954 |
HCC1954BL |
|||
|---|---|---|---|---|
| Capture sequencing | Paired-end sequencing | Capture sequencing | Paired-end sequencing | |
| Total number of reads | 5 996 389 | 381 274 888 | 6 265 250 | 347 891 568 |
| Mapped reads | 5 212 428 | 254 326 859 | 5 106 763 | 237 886 727 |
| Percentage of mapped reads | 86.9 | 66.7 | 81.5 | 68.4 |
| Total number of nucleotides | 3 143 589 263 | 19 392 752 128 | 3 252 428 887 | 15 693 171 704 |
| Mapped nucleotides | 2 257 027 363 | 13 432 965 012 | 2 175 120 803 | 11 166 288 816 |
| Percentage of mapped nucleotides | 71.8 | 69.3 | 66.7 | 71.1 |
In addition, the NimbleGen Sequence Capture Human Exome Array was used to capture 34.1 Mb of the human genome corresponding to approximately 180 000 coding exons (28.4 Mb) and adjacent regions (5.7 Mb). Approximately, 2.3 Gb of unambiguously mapped sequences were generated for each cell line and over 32% of the bases mapped to the targeted regions (Figure 1 and Table 1). The average fold-coverage was 22.8 for HCC1954 and 21.7 for HCC1954BL (Supplementary Figure S2). Captured sequences mapped to the reference genome were then used for the detection of SNVs and somatic point mutations in protein-coding exons and adjacent regions from both genomes (Figure 1). For HCC1954, 95.2% of targeted bases were covered at least once and 92% met our criteria for SNV calling. Similar numbers were obtained for HCC1954BL (95.7% of targeted bases covered at least once and 93.8% met our criteria for variant calling).
Chromosomal rearrangements
We used a paired-end sequencing strategy to identify structural chromosome variations present in the HCC1954 and HCC1954BL genomes (18). Paired-end reads that aligned discordantly with respect to each other, on the reference human genome, were identified for HCC1954 (76 407 paired-end reads) and HCC1954BL (55 967 paired-end reads). From this set of aligned reads, we excluded those that precisely duplicated other sequences derived from the same library, those that could not be unumbiguosly mapped to the human genome, and those that mapped within 1 Mb of a telomeric or centromeric sequence gap. Structural variants were called from the remaining paired-end sequences, requiring at least five independent read pairs exclusively reporting the variant in one of the cell lines. Structural variants represented by read pairs that mapped to within 500 bp of a known copy number polymorphism were removed from the final list of somatic chromosomal rearrangements (Figure 1).
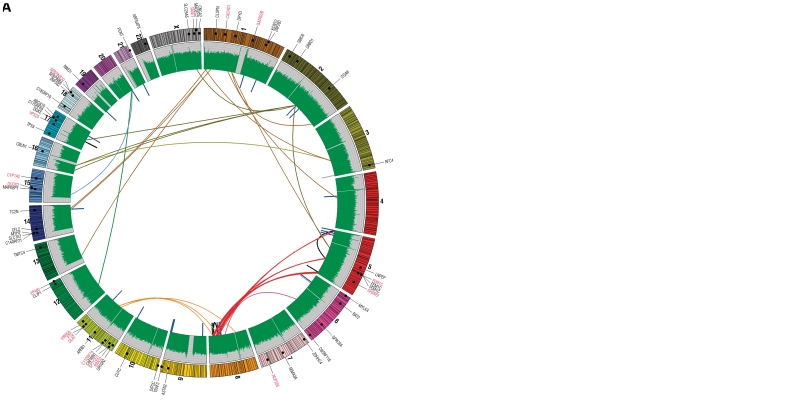
A total of 94 structural rearrangements were detected in HCC1954 including 49 interchromosomal events, 30 deletions, 11 inversions and 4 duplications (Table 2 and Figure 2). Of the 49 interchromosomal events, 38 affected genic regions and 22 had already been described for HCC1954 (10,19). In contrast, no interchromosomal rearrangements were detected in the HCC1954BL genome and all four intrachromosomal rearrangements, including two deletions and two inversions, were located in intergenic or intronic regions (Table 2 and Figure 2).
Table 2.
Somatic point mutations and structural variations in the HCC1954 and HCC1954BL genomes
| Somatic variations | HCC1954 | HCC1954BL |
|---|---|---|
| N (%) | N (%) | |
| Point mutations | 274 (100) | 173 (100) |
| Coding | 64 (23.36) | 30 (17.3) |
| Nonsense | 2 (0.73) | 3 (1.7) |
| Missense | 45 (16.42) | 15 (8.7) |
| Synonymous | 17 (6.20) | 12 (6.9) |
| Non-coding | 14 (5.11) | 15 (8.7) |
| UTR | 13 (4.74) | 13 (7.5) |
| ncRNA | 1 (0.36) | 2 (1.2) |
| miRNA | 0 (0) | 0 (0) |
| Intronic | 179 (65.33) | 114 (65.9) |
| Splice site | 0 (0) | 0 (0) |
| Other intronic | 179 (65.33) | 114 (65.9) |
| Intergenic | 17 (6.20) | 14 (8.1) |
| Structural variations | 94 (100) | 4 (100) |
| Interchromosomal | 49 (52.1) | 0 (0) |
| Intrachromosomal | 45 (47.9) | 4 (100) |
| Deletions | 30 (31.9) | 2 (50.0) |
| Inversions | 11 (11.7) | 2 (50.0) |
| Duplications | 4 (4.3) | 0 (0) |
UTR = untranslated region, ncRNA = non-coding RNA.
Figure 2.
Circos plot representing somatic point mutations and structural variations in the (A) HCC1954 and (B) HCC1954BL genomes. Chromosome representations are shown around the outer ring and are oriented in a clockwise direction. Other tracks contain (from outside to inside) point mutations as dots (non-synonymous labeled in back and synonymous labeled in red), physical coverage of the genome by paired-end reads in green, interchromosomal rearrangements represented by colored lines linking two chromosomes (different colors representing interchromosomal rearrangements are determined by the first chromosome in the circos in the clockwise direction starting with chromosome 1), intrachromosomal deletions as blue lines, inversions as black lines and duplications as gray lines.
Variant calling and somatic point mutations
Captured sequences (on-target and off-target) mapped to the reference human genome were used for variant calling and point mutation detection in coding regions and adjacent sequences. A total of 82 355 and 83 474 SNVs supported by at least three reads with base quality ≥20 were called for the HCC1954 and HCC1954BL genomes, respectively (Table 3). As expected, most of the SNVs were common to both genomes and the majority (92%) of these inherited variants has already been described in dbSNP. The rate of novel variant discovery (8%) for this individual is consistent with other published whole human genome sequences (20–24).
Table 3.
Single nucleotide variations identified in the HCC1954 and HCC1954BL genomes
| HCC1954 | HCC1954BL | |
|---|---|---|
| N (%) in dbSNP | N (%) in dbSNP | |
| Substitutions | 82 355 (92.68) | 83 474 (93.60) |
| Coding | 11 717 (90.92) | 12 373 (93.84) |
| Intronic | 60 314 (92.53) | 61 428 (93.77) |
| UTR | 3419 (92.57) | 3570 (94.04) |
| ncRNA | 256 (96.87) | 260 (96.92) |
| Intergenic | 6649 (91.84) | 5843 (90.86) |
| Indels | 689 (52.10) | 587 (52.81) |
| Coding | 38 (50.00) | 31 (51.61) |
| Intronic | 595 (52.43) | 506 (54.15) |
| UTR | 30 (46.66) | 26 (42.30) |
| ncRNA | 1 (100.00) | 1 (0.00) |
| Intergenic | 25 (52.00) | 23 (39.13) |
UTR = untranslated region, ncRNA = non-coding RNA
To assess the coverage depth and sensitivity of our SNV calling strategy, we compared our SNV calls to SNV calls extracted from genotyping array data available for both cell lines. (GEO: GSE12019 and GSE13373) as previously described (5). In total, 93.7% and 97.8% of the heterozygous array calls located within the captured regions were sequenced at least once for HCC1954 and HCC1954BL, respectively, and that 80.8% and 83.3% of the heterozygous array calls for HCC1954 and HCC1954BL, respectively, were correctly identified by our sequencing strategy and SNV calling criteria. The difference in the SNV calling efficiency observed between both cell lines is not statistically significant (P = 0.69, χ2 = 0.16, df = 1) and together these results demonstrated that both genomes were sufficiently and equally covered for SNV calling and mutation detection.
To identify SNVs that are specific to each cell line and likely to correspond to somatic point mutations occurring in each genome, we excluded those already described as a known SNP in dbSNP and established a set of stringent filtering criteria (Figure 1). Briefly, variants had to be represented by at least three high-quality reads from one cell line and no reads from the other. Variant reads were required to correspond to at least 20% of the total number of reads covering the variant position to eliminate point mutations that might have eventually arisen during in vitro culturing and a depth of at least 5× for the variant base was also required for each cell line to assure sufficient coverage in both cell lines. Variants for HCC1954BL were further filtered to remove those residing in regions of LOH in HCC1954 (see ‘Materials and Methods’ section).
A total of 274 point mutations were predicted in the HCC1954 genome of which 64 (23.4%) occurred in protein-coding regions, 14 (5.1%) in non-coding regions, 179 (65.3%) in intronic regions and 17 (6.2%) in intergenic regions (Figure 2 and Table 2). Of the 64 point mutations occurring in coding regions, 47 (73.4%) were predicted to cause amino acid changes (non-synonymous), including 45 that were missense and two that were nonsense.
The same set of criteria was used to predict mutations present in the HCC1954BL genome. A total of 173 point mutations were predicted for HCC1954BL, of which 30 (17.3%) occurred in protein-coding regions, 15 (8.7%) in non-coding regions, 114 (65.9%) in intronic regions and 14 (8.1%) in intergenic regions (Figure 2 and Table 2). Of the 30 point mutations occurring in coding regions, 18 (60%) were non-synonymous, including 15 that were missense and 3 that were nonsense.
Non-synonymous substitutions and mutation spectrum
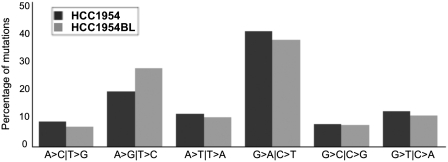
Since some of the mutations present in the HCC1954 genome could have arisen during normal breast development as a result of the normal mutational process, we next sought to determine if the set of mutations occurring in the HCC1954 genome is enriched for driver mutations and/or occur in genes that are functionally related to the tumorigenesis. To address this issue, we first analyzed the ratio of non-synonymous to synonymous substitutions (dNs/dS) in the protein-coding regions of both genomes. The dNs/dS ratio is commonly used to estimate the degree of selection on non-synonymous changes, assuming that most synonymous mutations are biologically neutral. This ratio for HCC1954 is 2.8 (P = 0.37, Monte Carlo simulation) and for HCC1954BL is 1.5 (P = 0.18, Monte Carlo simulation), not significantly different from that expected by chance. However, it is notable that the difference in the non-synonymous/synonymous ratio between the cell lines is statistically significant (P = 0.031; χ2 = 4.68; df = 1), indicating that non-synonymous mutations are more frequent in HCC1954 than in HCC1954BL. We also investigated the type of nucleotide changes present in HCC1954 and HCC1954BL genomes. Both genomes presented a similar spectrum of nucleotide substitutions, with a predominance of transitions, which change a purine to purine (A↔G) or pyrimidine to pyrimidine (C↔T) (Figure 3).
Figure 3.
Spectrum of nucleotide substitutions in the HCC1954 and HCC1954BL genomes. Frequency of point mutations in each of the six possible nucleotide substitution classes (A > C|T > G, A > G|T > C, A > T|T > A, G > A|C > T, G > C|C > G, G > T|C > A) observed in the HCC1954 (blue) and HCC1954BL (orange) genomes.
Sanger validation and KEGG analysis
To better characterize the genetic changes that drive tumorigenesis, we validated by capillary sequencing all non-synonymous point mutations present in the HCC1954 and HCC1954BL genomes. Of the 47 non-synonymous mutations present in the HCC1954 genome, 33 (70.2%) have already been described in the literature and 12 out of the 14 (85.7%) novel non-synonymous mutations were confirmed by capillary sequencing (Supplementary Table S1). Of the 18 non-synonymous point mutations predicted for HCC1954BL, 12 (66.6%) were also detected by capillary sequencing (Supplementary Table S2). Of the 45 non-synonymous mutations identified for HCC1954, 29 (64.4%) result in non-conservative amino acid changes, 19 (42.2%) occur within functional protein domains and 42 (93.3%) occur in evolutionary conserved amino acids residues. For HCC1954BL 8 (66.6%) of the 12 non-synonymous mutations result in non-conservative amino acid changes, 6 (50%) occur within functional protein domains and 11 (91.66%) occur in evolutionary conserved amino acids residues.
We next compared the sets of genes with validated non-synonymous mutations present in the two genomes. First, we determined whether these sets were enriched for specific signaling pathways related to tumorigenesis. According to KEGG (25), the set of mutated genes in HCC1954 was significantly enriched for genes related to apoptosis (P = 0.017, Monte Carlo Simulation), MAPK signaling (P = 0.023, Monte Carlo Simulation), axon guidance and cell migration (P = 0.033, Monte Carlo Simulation) and main pathways altered in cancer (P = 0.037, Monte Carlo Simulation). In contrast, no enrichment for any specific pathway related to cancer was observed for the set of genes mutated in HCC1954BL (Table 4).
Table 4.
KEGG pathway analysis for genes with validated non-synonymous mutations present in the HCC1954 and HCC1954BL genomes
| KEGG ID | KEGG annotation | Number of genes in the pathway | Gene Name | P-value |
|---|---|---|---|---|
| HCC1954 | ||||
| hsa05222 | Small cell lung cancer | 3 | ITGA6 TP53 TRAF2 | 0.0003 |
| hsa05410 | Hypertrophic cardiomyopathy | 2 | ITGA6 MYH7 | 0.0167 |
| hsa04210 | Apoptosis | 2 | TP53 TRAF2 | 0.0169 |
| hsa05414 | Dilated cardiomyopathy | 2 | ITGA6 MYH7 | 0.0191 |
| hsa04010 | MAPK signaling pathway | 3 | ARRB1 TP53 TRAF2 | 0.0237 |
| hsa00770 | Pantothenate and CoA biosynthesis | 1 | DPYD | 0.0325 |
| hsa04360 | Axon guidance | 2 | CFL2 SEMA3A | 0.0335 |
| hsa04614 | Renin-angiotensin system | 1 | LNPEP | 0.0372 |
| hsa05200 | Pathways in cancer | 3 | ITGA6 TP53 TRAF2 | 0.0375 |
| HCC1954BL | ||||
| hsa03440 | Homologous recombination | 1 | EME1 | 0.0234 |
| hsa00310 | Lysine degradation | 1 | SETD2 | 0.0382 |
| hsa04740 | Olfactory transduction | 2 | OR51E2 OR2D2 | 0.0421 |
Protein–protein Interaction and functional networks
We also examined known PPI to investigate the organization of mutated genes into functional networks. We first determined for each genome the percentage of genes with validated non-synonymous mutations that had at least one known protein interaction with any other protein. Similar percentages were obtained for both the HCC1954 (55.5%, 25/45) and HCC1954BL (66.7%, 8/12) cell lines, indicating that there is no difference in terms of representation of the mutated genes for each cell line in this PPI database (P = 0.729, χ2 = 0.12, df = 1).
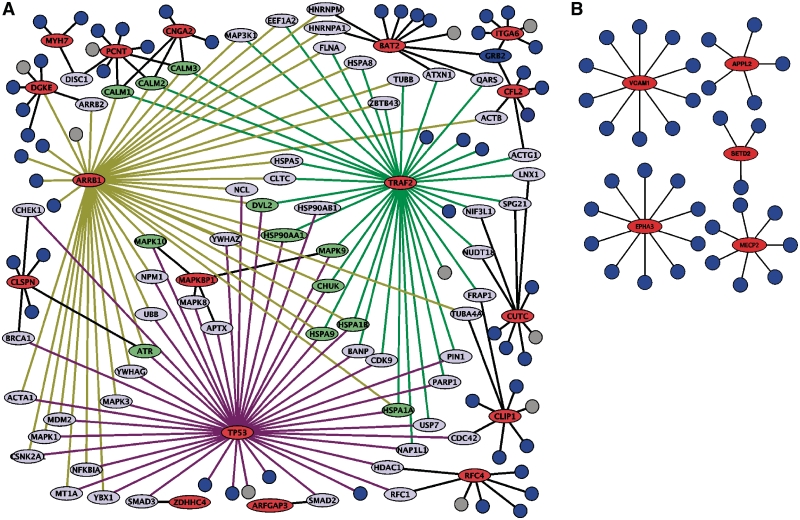
We then analyzed the average number of interactions for each mutated protein because proteins with larger numbers of interactions have been suggested to serve as essential hubs of molecular pathways (26). Proteins mutated in HCC1954 interact with a higher average number of partners than proteins mutated in HCC1954BL (avg 33.2 versus 5.1 proteins, Figure 4). To exclude the possibility that the higher degree of interaction observed for HCC1954 was due to the larger number of mutated proteins in this cell line, a Monte Carlo simulation was performed in which 10 000 random sets of 25 proteins with known PPI were evaluated regarding their degree of interaction. Only 17 out of the 10 000 simulated sets had a higher average degree than that observed for mutated proteins in HCC1954 (P = 0.0017, Monte Carlo simulation), indicating that the number of interactions observed for proteins mutated in HCC1954 was significantly different from that expected by chance. The same strategy showed that the average degree of interaction for proteins mutated in HCC1954BL was similar to that expected by chance (P = 0.875, Monte Carlo Simulation). To confirm that the differences in the degree of interaction observed between both cell lines were not influenced by the smaller number of mutated proteins in HCC1954BL, a Monte Carlo simulation was again performed in which 1000 random sets of 5 proteins from HCC1954 carrying non-synonymous mutations and presenting known PPI were evaluated regarding the average number of interactions. The number obtained is higher than that obtained for the five mutated proteins in HCC1954BL (34.7 versus 5.1) and again different from that expected by chance (P = 0.001, Monte Carlo simulation).
Figure 4.
Protein–protein interactions networks for mutated genes in HCC1954 (A) and HCC1954BL (B). Proteins with validated non-synonymous mutations are represented as red circles and each line represents a confident interaction. Interaction partners with mutated genes are represented in green if they interact with three mutated proteins or in light blue if they interact with two mutated proteins.
Interestingly, genes related to apoptosis (TP53, TRAF2, SLC25A5), MAPK signaling (TP53, ARRB1, TRAF2), cell adhesion (ITGA6), cytoskeleton organization (PCNT, CLIP1) and cell cycle (RFC4, PCNT) were among the HCC1954 mutated proteins displaying a higher number of interactions (≥10 interactions, Figure 4). To evaluate if the higher degree of interaction observed for HCC1954 was just a consequence of the proteins mutated in this cell line belonging to molecular pathways with higher connectivity, we selected all proteins from all KEGG pathways containing at least one mutated protein in HCC1954 and HCC1954BL (e.g. these sets include all proteins from MAPK pathway for HCC1954 and all proteins from Lysine degradation pathway for HCC1954BL) and calculated the average number of interactions for both sets of proteins. The set for HCC1954 contained 2311 proteins with an average of 18.4 interactions per protein. The set for HCC1954BL contained 395 proteins with an average of 22.1 interactions per protein. These values are not significantly different from each other (P = 0.0921, t-test = 1.16875; df = 503.76), indicating that the higher degree of interaction observed for HCC1954 is not a direct consequence of the mutated proteins belonging to pathways with higher connectivity.
We also looked for common interaction partners among the mutated proteins because mutated proteins with common partners would probably have synergistic tumor-promoting functions (27). Approximately 68% (17/25) of the mutated proteins in HCC1954 shared at least one common partner, as opposed to none of the mutated proteins in HCC1954BL (Figure 4). Simulations to correct for the difference in the number of mutated proteins in both genomes, revealed that the number of mutated proteins sharing a common partner was significantly different from that expected by chance for HCC1954 (P < 0.0001, Monte Carlo simulation) but not for HCC1954BL (P = 0.855, Monte Carlo simulation). Again, to confirm that the differences in the number of common interaction partners observed between both cell lines were not influenced by the smaller number of mutated proteins in HCC1954BL, a Monte Carlo simulation was again performed in which 1000 random sets of 5 proteins from HCC1954 carrying non-synonymous mutations and presenting known PPI were evaluated regarding the average number of common interaction partners. The number obtained is higher than that obtained for the five mutated proteins in HCC1954BL (3.3 versus 0) and again different from that expected by chance (P = 0.0245, Monte Carlo simulation).
A total of 64 common partners were identified for proteins mutated in HCC1954, of which 51, 10 and 3 interact with 2, 3 and 4 mutated proteins in HCC1954, respectively. Again, the number of common partners observed for HCC1954 was significantly different from that expected by chance (P < 0.0001, Monte Carlo simulation). Key cancer genes such as BRCA1, CDC42, CHECK1, MDM2, MAP3K1/3 and SMAD2/3 were among the 64 common interaction partners (Figure 4).
Finally, we also investigated the organization of mutated proteins into functional networks in other tumor genomes recently sequenced. Similar patterns of synergy were observed for the set of genes carrying non-synonymous point mutations in melanoma, glioblastoma, lung and breast-tumor genomes (Table 5). Although the average number of interactions for mutated proteins varied significantly between the different tumors analyzed (8.1–32.5 interactions), the percentage of mutated genes with common partners was similar among all tumors analyzed (varying from 41% to 66%) and different from that expected by chance (Table 5). Among the five tumors analyzed, the estrogen receptor-positive metastatic lobular breast cancer was the one presenting the most similar functional organization to HCC1954 with an average of 32.5 interactions for each mutated protein, 44% of the mutated proteins with common partners and 28 common partners among the mutated proteins. All these values are significantly different from that expected by chance (P = 0.0034, P = 0.0001 and P = 0.0013, respectively, Monte Carlo Simulations).
Table 5.
Protein–protein interaction analysis for genes with non-synonymous mutations in other solid tumors
| References | Tumor type | Number of genes with non-synonymous mutations | Number of mutated genes with PPI information (%) | Average number of interactions for mutated genes (P-value) | Number of mutated genes with common partner (%) (P-value) | Number of common partners (P-value) |
|---|---|---|---|---|---|---|
| Pleasance et al. (8) | Lung | 90 | 50 (56) | 11.6 (0.2692) | 33 (66) (0.0001) | 42 (0.0870) |
| Pleasance et al. (7) | Melanoma | 188 | 100 (53) | 8.3 (0.8344) | 69 (69) (0.0001) | 103 (0.3130) |
| Ding et al. (4) | Breast basal | 29 | 17 (59) | 8.1 (0.2210) | 7 (41) (0.0001) | 7 (0.0132) |
| Shah et al. (9) | Breast lobular | 32 | 16 (50) | 32.5 (0.0034) | 7 (44) (0.0001) | 28 (0.0011) |
| Clark et al. (3) | GBM | 110 | 40 (36) | 12.9 (0.7269) | 18 (45) (0.0001) | 13 (0.1896) |
| Galante et al. (this study) | Breast HCC1954 | 45 | 25 (56) | 33.2 (0.0017) | 17 (68) (0.0001) | 64 (0.0001) |
DISCUSSION
In this study, we were able to identify, for the first time to our knowledge, the somatically acquired genetic alterations present in the genome of a lymphoblastoid and a tumor cell derived from the same individual. By also characterizing the somatic mutations present in the lymphoblastoid genome, we were able to show how a non-tumor somatic tissue evolves over the same timescale and under similar environmental conditions when compared to a tumor genome, providing important insights into the normal mutational processes and into the functional implications of the accumulation of somatic alterations in a tumor and a matching lymphoblastoid genome.
A highly complex pattern of chromosomal rearrangements was exclusively observed in the tumor genome with most of these rearrangements affecting genic regions. In agreement with these observations, in a previous study in which exome and transcriptome data from HCC1954 and HCC1954BL were combined to identify genes with LOH and allele-specific expression (ASE), large LOH regions, harboring genes with ASE and known tumor suppressor characteristics, were only detected in the tumor genome (28). Together these observations support the concept that chromosomal instability is a key feature of human cancer and a driving force of tumorigenesis (29).
Interestingly, the number of somatically acquired point mutations and the spectrum of nucleotide substitutions found in the lymphoblastoid genome were comparable to that present in the tumor genome. It has been proposed that normal point mutation rates are insufficient to account for all point mutations observed in tumors and that tumor cells must acquire a mutator phenotype, which increases the accumulation of point mutations in the tumor genome (30). The number of point mutations present in both genomes analyzed is in agreement with the estimated spontaneous mutation rate of normal human cells (∼2.10–10/bp per cell division) (31) and the difference in the total number of point mutations observed for the HCC1954 and HCC1954BL genomes (274/173 = 1.58) is probably due to the higher DNA content of the tumor cell line rather than to the existence of a mutator phenotype (30). Both cell lines also presented a similar spectrum of nucleotide substitutions with a predominance of transitions. A similar frequency of G > A|C > T transitions was observed for other recently reported breast-tumor genomes (4,9) and a comparison to the spectrum of nucleotide substitutions reported for a melanoma and a lung cancer cell line indicates that neither genome under study had signs of the influence of external mutagens such as tobacco or ultraviolet light (7,8). Together these results indicate that the influence of endogenous mutagens and replication errors are sufficient to generate the overall number of point mutations required to drive tumorigenesis and that tumor cells do not necessarily need to acquire a mutator phenotype to increase the accumulation of point mutations in their genomes.
Although, we cannot completely discard the possibility that some of the somatic point mutations detected in HCC1954 and HCC1954BL genomes result from in vitro culturing and EBV transformation (in the case of HCC1954BL), we do not expect these mutations to be representative or to influence our observations and overall conclusions. Both cell lines received similar treatments in terms of the timing of establishment and in vitro propagation and have not been maintained in culture for a long period (36 passages), reducing the probability of introducing mutations during the culturing process. We have also set up very stringent criteria for somatic point mutation detection to filter most, if not all, non-clonal mutations eventually introduced during in vitro culturing of these cell lines (see ‘Materials and Methods’ section). Moreover, it has been recently demonstrated that clonal point mutations rarely arise during in vitro or in vivo experimental growth of tumor cells (32). Jones et al. have analyzed 289 mutations present in 18 cell lines or xenografts, each derived from a different primary tumor. Over 99% of these mutations were also present in the primary tumors (29). Finally, several studies have demonstrated a very strong correlation when DNA from EBV-transformed lymphoblastoid cell lines was compared to DNA from the corresponding blood samples (33), indicating that EBV transformation does not substantially alters the mutation rate and genetic stability of transformed cells. Indeed EBV-transformed lymphoblastoid cell lines have been widely used as source of DNA in genetic screening studies and have also been used as source of normal DNA in whole genome studies on the occurrence of somatic mutations in tumor genomes (7,8).
We also do not expect our observations and conclusions to be significantly affected by sequencing errors. Since one of our criteria for point mutation detection was to have at least 20% of the reads reporting the mutated allele, this represents an average of four reads considering the average exome coverage of 22×. If we use a false-positive error rate of ∼2% for each position per read for the 454 sequencing platform (34), our error rate per position would be 16 per 100 Mb. Since the exome captured region comprises ∼31 Mb, the expected number of false positive mutations per genome is between 4 and 5 mutations. Nevertheless, it is important to emphasize that we took a conservative approach and used for all downstream analysis (KEGG and PPI) only those mutations that were further validated by Sanger sequencing.
Significant functional differences were observed between the set of genes mutated in both cell lines, indicating that mutations in the tumor genome are not randomly distributed. We showed that non-synonymous point mutations are more frequently found in the tumor genome and that they preferentially affect hub-genes in molecular pathways related to tumorigenesis. Moreover, by looking for common interaction partners among mutated proteins, we demonstrated, for the the first time, that mutations in the tumor genome are co-selected to present synergistic tumor-promoting functions. This observation was further extended to other individual tumor genomes sequenced. Similar patterns of synergy were observed for the set of genes carrying non-synonymous point mutations in melanoma, glioblastoma, lung and breast-tumor genomes.
The functional differences observed between the sets of genes mutated in the lymphoblastoid and tumor genomes raise questions regarding the number and strength of the driver genetic alterations required for tumorigenesis. If a tumor cell were to require just a small number of ‘strong’ driver alterations, we would not expect to see the striking functional association of the genes mutated in the tumor because the majority of the mutations would be passengers. Our results thus support the model in which the tumor genome has a few ‘strong’ driver mutations and several ‘weak’ driver mutations that act in a synergistic fashion to disrupt molecular pathways related to tumorigenesis (35). Although, this model has been proposed in the literature for some time, to our knowledge, this is the first time the existence of strong and weak driver mutations is evidenced by comparing genome-wide mutation data generated from tumor and non-tumor tissues derived from the same individual. We would expect these ‘weak’ drivers to be infrequently mutated and insufficient to form tumors in the absence of the strong drivers. Distinguishing ‘weak’ drivers from passenger mutations will require a systems biology approach to dissect the individual roles of mutated genes and examine their interactions within the networks and pathways related to tumorigenesis. Moreover, this approach will require the analysis of large numbers of normal genome sequences to identify the permutations of mutated genes that do not result in tumorigenesis.
ACCESSION NUMBERS
SRA, ERA010917, ERA011762.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Ludwig Institute for Cancer Research; The Conrad N Hilton Foundation; Conselho Nacional de Desenvolvimento Científico e Tecnológico—CNPq; Fogarty International Center [D43TW007015]—National Institutes of Health (to P.A.F.G., S.J.S., L.O.). Funding for open access charge: Ludwig Institute for Cancer Research.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors acknowledge Daniel Ohara and Jose Eduardo Kroll for technical support.
REFERENCES
- 1.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat. Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark MJ, Homer N, O'Connor BD, Chen Z, Eskin A, Lee H, Merriman B, Nelson SF. U87MG decoded: the genomic sequence of a cytogenetically aberrant human cancer cell line. PLoS Genet. 2010;6:e1000832. doi: 10.1371/journal.pgen.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordonez GR, Bignell GR, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pleasance ED, Stephens PJ, O'Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, Delaney A, Gelmon K, Guliany R, Senz J, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 10.Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee W, Jiang Z, Liu J, Haverty PM, Guan Y, Stinson J, Yue P, Zhang Y, Pant KP, Bhatt D, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465:473–477. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- 12.Bignell GR, Santarius T, Pole JC, Butler AP, Perry J, Pleasance E, Greenman C, Menzies A, Taylor S, Edkins S, et al. Architectures of somatic genomic rearrangement in human cancer amplicons at sequence-level resolution. Genome Res. 2007;17:1296–1303. doi: 10.1101/gr.6522707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gazdar AF, Kurvari V, Virmani A, Gollahon L, Sakaguchi M, Westerfield M, Kodagoda D, Stasny V, Cunningham HT, Wistuba II, et al. Characterization of paired tumor and non-tumor cell lines established from patients with breast cancer. Int. J. Cancer. 1998;78:766–774. doi: 10.1002/(sici)1097-0215(19981209)78:6<766::aid-ijc15>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 14.Galante PA, Vidal DO, de Souza JE, Camargo AA, de Souza SJ. Sense-antisense pairs in mammals: functional and evolutionary considerations. Genome Biol. 2007;8:R40. doi: 10.1186/gb-2007-8-3-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Medvedev P, Stanciu M, Brudno M. Computational methods for discovering structural variation with next-generation sequencing. Nat. Meth. 2009;6:S13–S20. doi: 10.1038/nmeth.1374. [DOI] [PubMed] [Google Scholar]
- 18.Campbell PJ, Stephens PJ, Pleasance ED, O'Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat. Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhao Q, Caballero OL, Levy S, Stevenson BJ, Iseli C, de Souza SJ, Galante PA, Busam D, Leversha MA, Chadalavada K, et al. Transcriptome-guided characterization of genomic rearrangements in a breast cancer cell line. Proc. Natl Acad. Sci. USA. 2009;106:1886–1891. doi: 10.1073/pnas.0812945106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahn SM, Kim TH, Lee S, Kim D, Ghang H, Kim DS, Kim BC, Kim SY, Kim WY, Kim C, et al. The first Korean genome sequence and analysis: full genome sequencing for a socio-ethnic group. Genome Res. 2009;19:1622–1629. doi: 10.1101/gr.092197.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schuster SC, Miller W, Ratan A, Tomsho LP, Giardine B, Kasson LR, Harris RS, Petersen DC, Zhao F, Qi J, et al. Complete Khoisan and Bantu genomes from southern Africa. Nature. 2010;463:943–947. doi: 10.1038/nature08795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J, Li J, Zhang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen YJ, Makhijani V, Roth GT, et al. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–876. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 25.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jonsson PF, Bates PA. Global topological features of cancer proteins in the human interactome. Bioinformatics. 2006;22:2291–2297. doi: 10.1093/bioinformatics/btl390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bredel M, Scholtens DM, Harsh GR, Bredel C, Chandler JP, Renfrow JJ, Yadav AK, Vogel H, Scheck AC, Tibshirani R, et al. A network model of a cooperative genetic landscape in brain tumors. JAMA. 2009;302:261–275. doi: 10.1001/jama.2009.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Q, Kirkness EF, Caballero OL, Galante PA, Parmigiani RB, Edsall L, Kuan S, Ye Z, Levy S, Vasconcelos AT, et al. Systematic detection of putative tumor suppressor genes through the combined use of exome and transcriptome sequencing. Genome Biol. 2011;11:R114. doi: 10.1186/gb-2010-11-11-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Michor F, Iwasa Y, Vogelstein B, Lengauer C, Nowak MA. Can chromosomal instability initiate tumorigenesis? Sem. Cancer Biol. 2005;15:43–49. doi: 10.1016/j.semcancer.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 30.Bielas JH, Loeb KR, Rubin BP, True LD, Loeb LA. Human cancers express a mutator phenotype. Proc. Natl Acad. Sci. USA. 2006;103:18238–18242. doi: 10.1073/pnas.0607057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Albertini RJ, Nicklas JA, O'Neill JP, Robison SH. In vivo somatic mutations in humans: measurement and analysis. Annu. Rev. Genet. 1990;24:305–326. doi: 10.1146/annurev.ge.24.120190.001513. [DOI] [PubMed] [Google Scholar]
- 32.Jones S, Chen WD, Parmigiani G, Diehl F, Beerenwinkel N, Antal T, Traulsen A, Nowak MA, Siegel C, Velculescu VE, et al. Comparative lesion sequencing provides insights into tumor evolution. Proc. Natl Acad. Sci. USA. 2008;105:4283–4288. doi: 10.1073/pnas.0712345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sie L, Loong S, Tan EK. Utility of lymphoblastoid cell lines. J. Neurosci. Res. 2009;87:1953–1959. doi: 10.1002/jnr.22000. [DOI] [PubMed] [Google Scholar]
- 34.Harismendy O, Ng PC, Strausberg RL, Wang X, Stockwell TB, Beeson KY, Schork NJ, Murray SS, Topol EJ, Levy S, et al. Evaluation of next generation sequencing platforms for population targeted sequencing studies. Genome Biol. 2009;10:R32. doi: 10.1186/gb-2009-10-3-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.