Abstract
Mediator is a multiprotein co-activator of RNA polymerase (Pol) II transcription. Mediator contains a conserved core that comprises the ‘head’ and ‘middle’ modules. We present here a structure–function analysis of the essential Med11/22 heterodimer, a part of the head module. Med11/22 forms a conserved four-helix bundle domain with C-terminal extensions, which bind the central head subunit Med17. A highly conserved patch on the bundle surface is required for stable transcription pre-initiation complex formation on a Pol II promoter in vitro and in vivo and may recruit the general transcription factor TFIIH. The bundle domain fold is also present in the Mediator middle module subcomplex Med7/21 and is predicted in the Mediator heterodimers Med2/3, Med4/9, Med10/14 and Med28/30. The bundle domain thus represents a common building block that has been multiplied and functionally diversified during Mediator evolution in eukaryotes.
INTRODUCTION
Transcription of eukaryotic protein-coding genes and several non-coding RNAs relies on Pol II, a set of general transcription factors (GTFs), namely TFIIB, D, E, F and H and co-activators such as the Mediator complex (1,2). While the GTFs are required for promoter recognition and opening, transcription start site selection and initial RNA synthesis, Mediator bridges between the general Pol II machinery and transcriptional activators at upstream DNA sites (3–6). Upon activator binding, Mediator undergoes conformational changes, which trigger its interaction with Pol II and promote pre-initiation complex (PIC) formation (7).
Mediator was purified from yeast (8,9), metazoans (10,11) and plants (12). Comparative genomics identified an ancient 17-subunit Mediator core that is conserved in eukaryotes (13,14). Mediator from the yeast Saccharomyces cerevisiae (Sc) is a 1.4 MDa multiprotein complex comprising 25 subunits, of which 11 are essential and 22 are at least partially conserved. Based on electron microscopy and biochemical analysis, mediator subunits were suggested to reside in four flexibly linked modules, the head, middle, tail and kinase module (15–17). The Sc head module comprises five essential subunits, Med6, Med8, Med11, Med17 (Srb4) and Med22 (Srb6) and two non-essential subunits, Med18 (Srb5) and Med20 (Srb2).
The head module is important for PIC assembly. It contacts the Pol II-TFIIF complex (18), the TATA binding protein (TBP) (16,19), TFIIB (16) and TFIIH (20). A temperature-sensitive mutation in the central head subunit Med17 (srb4-138) affects head module integrity, abolishes stimulation of basal transcription in vitro (18), prevents association of Pol II and GTFs with promoters in vivo (21,22) and causes a global shutdown of messenger RNA (mRNA) synthesis (23,24). Sc Med11 and Med22 bind and stabilize Med17 (16,18). A temperature-sensitive mutation in Med22 (srb6-107) causes a global decrease in mRNA synthesis, similar to srb4-138 (24). Point mutations in Med22 can act as extragenic suppressors of the srb4-138 phenotype (25), of the cold-sensitive phenotype of the Pol II C-terminal domain (CTD) truncation (26) and of the lethal CTD Ser2 phosphorylation site substitution mutation (27). Med11 interacts with the Rad3 subunit of TFIIH in a yeast two-hybrid assay and a point mutation in Med11 reduced promoter occupancy of the TFIIH kinase module (TFIIK) and consequently CTD Ser5 phosphorylation in vivo (20).
In this study, we show that Med11 and Med22 form a heterodimeric Med11/22 subcomplex. We describe a Med11 homologue in the yeast Schizosaccharomyces pombe (Sp) that had not been identified (28), strongly arguing that the Med11/22 subcomplex is conserved amongst eukaryotes. We further show that Med11/22 forms a four-helix bundle domain with similarity to the Med7/21 subcomplex in the middle module. We also demonstrate that the bundle is anchored to a C-terminal region of the central head subunit Med17 that appears to form an independently folded domain. We further identified a highly conserved surface patch on the bundle domain. This patch is required for Pol II transcription in vitro and in vivo and for stable PIC formation. Finally, we predict four additional heterodimeric four-helix bundles in Mediator, suggesting that this fold represents a common building block and providing insights into the evolution of Mediator.
MATERIALS AND METHODS
Preparation of recombinant Med11/22 variants
For recombinant expression of the heterodimeric Sc Med11/22 complex, the coding sequences were cloned as a bicistron (19) into a pET28b vector (Novagen) using NdeI/XhoI restriction sites resulting in a thrombin-cleavable N-terminal hexahistidine (6His) tag on Med22. Two point mutations were introduced to remove alternative bacterial start codons, nucleotide G132C in MED11 (no change in amino acid sequence) and nucleotide G81A in MED22 (amino acid change Met27Ile). Transformed Escherichia coli (E. coli) BL21 (DE3) RIL cells (Stratagene) were grown in LB medium at 37°C to an optical density at 600 nm (OD600) of 0.6. Expression was induced with 0.5 mM IPTG for 16 h at 18°C. Selenomethionine labeling was carried out as described (45,46). Cells were lysed by sonication in buffer A (20 mM Tris pH 8.0, 150 mM NaCl, 2 mM dithiothreitol) containing protease inhibitor cocktail (1 mM Leupetin, 2 mM Pepstatin A, 100 mM Phenylmethylsulfonyl fluoride, 280 mM Benzamidine). After centrifugation, the supernatant was loaded onto a 2 ml Ni-NTA column (Qiagen) equilibrated with buffer A. The column was washed with 10 column volumes (CVs) of buffer A containing 10 mM imidazole and with 10 CVs of buffer A containing 20 mM imidazole. The complex was eluted with buffer A containing 300 mM imidazole followed by overnight cleavage with thrombin while dialyzing against buffer B (20 mM Tris pH 8.0, 50 mM NaCl, 2 mM dithiothreitol). The proteins were further purified by anion exchange chromatography using a HiTrap Q HP column (GE Healthcare). The column was equilibrated with buffer B and the complex was eluted with a linear gradient of 25 CVs from 50 to 500 mM NaCl in buffer B. Subsequently, the sample was applied to a HiLoad Superdex-75 pg 26/60 size exclusion column (GE Healthcare) equilibrated with buffer A.
X-ray structure determination
For crystallization, the purified selenomethionine-labeled complex Med111-89/Med225-89 was concentrated to 10 mg/ml. Crystals were grown at 20°C in hanging drops over reservoirs containing 100 mM MES pH 6, 5.5% PEG 6000 and 100 mM MgCl2. Microseeding was performed to optimize crystals. Initial crystals were transferred into 100 µl reservoir solution and vortexed vigorously. 0.2 µl of the resulting microseeding solution was added to each 2 µl drop to nucleate crystal growth. Optimized crystals were harvested by gradually adding glycerol to a final concentration of 34% (v/v) and were subsequently flash-frozen in liquid nitrogen. Diffraction data was collected at 100 K on a PILATUS 6 M detector at the Swiss Light Source SLS, Villigen, Switzerland. Diffraction data were processed using XDS and XSCALE (47). The program SOLVE (48) identified 36 selenium sites in the asymmetric unit that were used for phasing. Solvent flattening, non-crystallographic symmetry averaging and initial model building were done with RESOLVE (48) and ARP/warp (49). The resulting electron density map allowed for manual building of most of Med11 and Med22 using COOT (50). The model was refined using conjugate gradient minimization in PHENIX (51). The asymmetric unit contained twelve Med11/22 heterodimers that deviated only slightly at the C–termini of the proteins. All structure figures depict chain A/B and were prepared using PyMOL (52).
Yeast strains and growth assays
Plasmids pRS316-MED11 and pRS316-MED22 were generated by cloning the respective open reading frame (ORF) plus 500 bp upstream and 300 bp downstream sequences into pRS316 (ATCC; URA3 marker) using SacII/ApaI restriction sites. Plasmids pRS315-MED11, pRS315-med1143–115, pRS315-med111–110, pRS315-med111–105, pRS315-med111–89, pRS315-med11-E17K/L24K, pRS315-med11-L73E/K80E, pRS315-med11-K80E/L84E, pRS315-med111–105-3HA, pRS315-med11-E17K/L24K-3HA, pRS315-MED22, pRS315-med2254–121, pRS315-med221–117, pRS315-med221–108 and pRS315-med221–89 were generated by cloning the respective ORF or mutant ORF plus 500 bp upstream and 300 bp downstream sequence into pRS315 (ATCC; LEU2 marker) using SacII/ApaI restriction sites. The heterozygous MED11/med11Δ and MED22/med22Δ Sc yeast strains were obtained from Euroscarf (Frankfurt, Germany) and transformed with pRS316-MED11 and pRS316-MED22 respectively. Diploids were sporulated and tetrads dissected on YPD plates. To assess functionality of mutants, pRS315 constructs were transformed into the respective shuffle strain. Equal amounts of freshly grown yeast cells in SC (-ura/-leu) were resuspended in water and 10-fold dilutions were spotted on 5-FOA and SC (-ura/-leu) plates. Viable mutant strains were streaked twice on 5-FOA plates and then on SC (-leu). Equal amounts of freshly grown yeast cells in SC (-leu) were resuspended in water, 10-fold dilutions were spotted on YPD plates and plates were subsequently incubated at 30°C and 37°C to assess fitness and temperature sensitivity. MED11 shuffle strains expressing C-terminal tandem affinity purification (TAP) tags on Kin28, Med7 and Rpb3 were generated by integrating a TAP-HIS3MX cassette in the respective genomic locus. The Sc wild-type strain BY4741 as well as med20Δ and med2Δ strains used for gene expression profiling were obtained from Open Biosystems. Correct gene disruptions and TAP-insertions were verified by PCR for all strains. Sp wild-type strain 972h- was used in this study. C-terminal TAP-tag on Med7 and C-terminal 3HA-tag on Med11 were introduced by integrating a TAP-KanMX4 cassette and a 3HA-ClonNat cassette in the respective genomic locus.
TAP
TAP were performed as described from 3 l of Sc culture (5 × 107 cells/ml) grown at 30°C in YPD medium with 2% glucose or from 2 l of Sp culture (OD600 of 6) grown at 32°C in YPD medium with 3% glucose (29,53). For small-scale co-immunoprecipitation in Sc 40 ml cultures (1 × 107 cells/ml) were grown. After centrifugation, the cells were resuspended in 1 ml TAP lysis buffer (50 mM Tris pH 7.5, 100 mM NaCl, 1.5 mM MgCl2, 0.15% NP-40, 0.5 mM DTT and protease inhibitor cocktail) and 1 ml silica-zirconia beads (Roth) was added. Lysis was done in a mixer mill (Retsch) at 4°C for 30 min at maximum speed. The cleared lysate was incubated with 25 μl IgG Sepharose 6 Fast Flow beads (GE Healthcare) at 4°C for 1 h on a turning wheel. After washing 5× with 1 ml TAP lysis buffer, the proteins were eluted by boiling beads in SDS–PAGE sample buffer.
Recombinant protein interaction assays
Plasmids encoding Sc 6His-Med22/Med11 variants were generated as described above. Sc Med17C (residues 377–687) was cloned into a pET21b vector (Novagen) using SalI/NotI restriction sites. A non-cleavable C-terminal 6His tag followed by a stop codon was introduced by PCR where applicable. The Sp His6-Med22/Med11 constructs were cloned as a bicistron into pET28b vector (Novagen) using NdeI/SacI restriction sites. The Sp Med17 was inserted into pCDF-Duet vector (Novagen) using NcoI/NotI restriction sites. For co-expression and co-purification the respective plasmids were co-transformed in E. coli BL21 (DE3) RIL cells (Stratagene) and grown in 5 ml LB medium at 37°C to an OD600 of 0.6. Expression was induced with 0.5 mM IPTG for 16 h at 18°C. Cells were harvested and resuspended in 1 ml lysis buffer (20 mM Tris pH 8.0, 150 mM NaCl, 2.5 mM MgCl2, 0.1 mM CaCl2, 10% glycerol, 1% Tween-20, 2 mg/ml lysozyme, 1 U/ml DNAse I, 2 mM DTT and protease inhibitor cocktail). Cell suspension was flash-frozen in liquid nitrogen then thawed and incubated for 1 h at 20°C while shaking. After centrifugation, the supernatant was incubated with 10 µl MagneHis beads (Promega) for 10 min at 4°C on a turning wheel. Beads were washed once with wash buffer (20 mM Tris pH 8.0, 5 mM DTT) containing 500 mM NaCl and twice with wash buffer containing 150 mM NaCl. Proteins were eluted with wash buffer containing 150 mM NaCl and 500 mM imidazole. Proteins were analyzed by SDS–PAGE and stained with coomassie blue.
In vitro assays with yeast nuclear extracts
Yeast nuclear extract preparation, in vitro transcription and immobilized template assay were done as described (41,54) with some changes. For yeast nuclear extract preparation 3 l of Sc were grown to 5 × 107 cells/ml in YPD with 2% glucose. Cells were harvested by centrifugation and the pellet was resuspended in 30 ml resuspension buffer (50 mM Tris pH 7.5, 20 mM EDTA, 30 mM DTT) and incubated at 30°C for 15 min. Afterwards, cells were centrifuged and resuspended in 20 ml YPD/S, 3 ml 2 M sorbitol, 3 ml resuspension buffer containing 18 mg zymolyase (Seikagaku) and protease inhibitor cocktail. Cells were incubated at 30°C for 15–60 min until ∼85% of cells were spheroplasted. A quantity of 100 ml of YPD/S was added and cells were centrifuged. Pellet was resuspended in 250 ml YPD/S and incubated at 30°C for 30 min. Afterwards, cells were washed twice with 200 ml ice-cold YPD/S and once with 200 ml 1 M ice-cold sorbitol. Cells were resuspended in 100 ml ice-cold buffer A (10 mM Tris pH 7.5, 18% polysucrose 400, 20 mM potassium acetate, 5 mM magnesium acetate, 1 mM EDTA, 0.5 mM spermidine, 0.15 mM spermine, 3 mM DTT and protease inhibitor cocktail) and lysed on ice in a Dounce glass homogenizer (Kontes) with a small pestle. Crude nuclei were isolated by centrifuging twice at 3800g for 8 min and twice for 5 min, transferring the supernatant each time to a new bottle. Afterwards, nuclei were centrifuged at 20 000g, washed once with 15 ml buffer B (100 mM Tris pH 8.0, 50 mM potassium acetate, 10 mM magnesium sulfate, 20% glycerol, 2 mM EDTA, 3 mM DTT and protease inhibitor cocktail) and flash-frozen in 15 ml fresh buffer B. The next day the nuclei were lysed by adding (NH4)2SO4 (pH 7.5) to a final concentration of 0.5 M and incubated at 4°C for 30 min on a turning wheel. Afterwards, nuclear lysate was centrifuged at 140 000g for 90 min at 4°C. Nuclear proteins in the supernatant were precipitated by addition of solid 0.35g (NH4)2SO4/ml and incubation at 4°C for 30 min on a turning wheel. After centrifugation, the nuclear proteins were resuspended in a small volume of buffer C (20 mM HEPES pH 7.6, 10 mM magnesium sulfate, 1 mM EGTA, 20% glycerol, 3 mM DTT and protease inhibitor cocktail) and dialyzed against buffer C containing 75 mM (NH4)2SO4. Nuclear extracts were flash frozen in liquid nitrogen and stored at −80°C.
Pol II in vitro transcription was driven from a wild-type HIS4 yeast promoter sequence (−428 to +24 respective to the start codon) inserted into pBluescript II KS+ plasmid using HindIII/BamHI restriction sites. The 25 µl reaction mixture contained 200 µg yeast nuclear extract, 200 ng of template plasmid, transcription buffer (10 mM HEPES pH 7.6, 50 mM potassium acetate, 0.5 mM EDTA, 2.5 mM magnesium acetate, 2.5 mM DTT), 192 µg of phosphocreatine, 0.2 µg of creatine phosphokinase, 10 U of RiboLock RNase inhibitor (Fermentas) and 100 µM nucleoside triphosphates (NTPs). For activated transcription, 200 ng of recombinant full-length Gcn4 was added. The reaction was incubated at 18°C for 60 min. RNA was isolated using the RNeasy MinElute kit (Qiagen). RNA was eluted from the column with 14 µl RNase-free water and in vitro transcripts were analyzed by primer extension. The 20 µl primer annealing reaction contained 12 µl eluate from RNeasy MinElute column, 0.125 pmol fluorescently labeled oligo (5′-Cy5-TTCACCAGTGAGACGGGCAACAGCCAAGCTC), 5 mM Tris pH 8.3, 75 mM KCl and 1 mM EDTA pH 8.0. After boiling the samples for 3 min at 95°C, primer was annealed for 45 min at 48°C. Afterwards, 40 µl reverse transcription mix [50 mM Tris pH 8.3, 75 mM KCl, 4.5 mM MgCl2, 25 mM DTT, 0.3 mM dNTPs, 12.5 U MuLV reverse transcriptase (Roche) and 1 µg actinomycin D] was added and the reaction was incubated for 30 min at 37°C. The resulting cDNA was ethanol precipitated and resuspended in 4 µl 0.04 mg/ml RNase A. After incubating 3 min at 18°C 4 µl loading dye were added (80% formamide, 25 mM EDTA, 1.5% bromophenolblue) and the samples were boiled for 1 min. Transcripts were separated on a 8% polyacrylamide/7 M urea TBE gel, scanned with a Typhoon 9400 and quantified with the ImageQuant software (GE Healthcare).
Pol II immobilized template assays were done on a linear yeast HIS4 promoter. Templates were amplified by PCR from the template plasmid described above (forward primer: 5′-biotin-TAATGCAGCTGGCACGACAGG; reverse primer: 5′-GCCGCTCTAGCTGCATTAATG) and purified with PCR purification kit (Qiagen) and phenol–chloroform extraction. A quantity of 200 µg of magnetic streptavidin beads (Dynabeads M-280, Invitrogen) were used per reaction carrying ∼2.5 pmol of template. Coupled beads were blocked for 15 min at 20°C with transcription buffer containing 60 mg/ml casein and 5 mg/ml polyvinylpyrrolidone and subsequently for 15 min at 20°C with transcription buffer containing 0.5 mM biotin. After washing, beads were resuspended in 20 µl transcription buffer, 5 pmol recombinant full-length Gcn4 were added and incubated for 10 min at 20°C while shaking. Afterwards, beads were washed twice and again resuspended in 20 µl transcription buffer. Immobilized template reaction of 100 µl contained 20 µl beads, 1 mg yeast nuclear extract, 5 µg competitor DNA (HaeIII digested genomic E. coli DNA), transcription buffer, 768 µg of phosphocreatine, 0.8 µg of creatine phosphokinase, 0.05% NP-40. Assembly was done for 1 h at 4°C while shaking followed by washing twice with transcription buffer containing 2 µg/ml competitor DNA, twice with transcription buffer containing 500 mM potassium acetate and twice with transcription buffer. Proteins were eluted by boiling beads in SDS-loading dye and subsequently analyzed by SDS–PAGE and western blot. Antibodies against Med2 (Santa Cruz), Rpb3 (Neoclone), Rpb11 (Neoclone), Srb4 (gift of the Hahn laboratory), TAF4 (Abcam), TFIIB (Abcam) were used in this study.
Gene expression profiling
All experiments were performed in YPD medium with 2% glucose. For microarray analysis, at least two independent colonies were used for inoculation and overnight cultures were diluted in fresh medium to 1 × 106 cells/ml (25 ml cultures, 160 rpm shaking incubator, 30°C). Cells were harvested in early log-phase (1 × 107 cells/ml) by centrifugation. Total RNA was prepared after cell lysis using a mixer mill (Retsch) and subsequent purification using the RNAeasy kit (Qiagen). Total RNA was prepared with the RiboPureYeast Kit (Ambion) following the manufacturer’s instructions. All following steps were conducted according to the Affymetrix GeneChip 3′IVT Kit protocol. Briefly, one-cycle cDNA synthesis was performed with 250–300 ng of total RNA. In vitro reverse transcription labeling was carried out for 16 h. The fragmented samples were hybridized for 16 h on Yeast Genome 2.0 expression arrays (Affymetrix), washed and stained using a Fluidics 450 station and scanned on an Affymetrix GeneArray scanner 3000 7 G. Data analysis was performed using R/Bioconductor (55). Sp probes were filtered out prior to normalization with the GCRMA algorithm (56). Linear model fitting and multiple testing correction using an empirical Bayes approach was performed using the LIMMA package (57). Differentially expressed genes were defined as having an adjusted P < 0.05 and an estimated fold change >2.0 (calculated as the fold change of the average expression in the triplicate measurements). Hierarchical clustering was calculated using TIGR MeV application (58). Gene ontology (GO) analysis (38) of differentially expressed genes was performed to identify overrepresented biological processes using the GO term finder web tool (http://go.princeton.edu/cgi-bin/GOTermFinder).
Chromatin immunoprecipitation
All chromatin immunoprecipitation experiments were performed in biological duplicates in YPD medium with 2% glucose as previously described in detail (59,60). Since the Med11/22 mutant strains displayed severe growth defects and larger cell sizes compared to wild-type, cells were counted instead of measuring optical density. To minimize the risk of acquiring a rescue mutation and ensure biological significance of our observations, the used biological duplicates were already separated before shuffling out the respective rescue plasmid and several rounds of selection. Phenotype and growth was monitored closely at each step. Overnight cultures were diluted in fresh medium to 1 × 106 cells/ml (40 ml cultures, 160 rpm shaking incubator, 30°C) and grown to early log-phase (1 × 107 cells/ml) before formaldehyde cross-linking. Input and immunoprecipitated samples were assayed by quantitative real-time PCR to assess occupancy of proteins at three different promoters. Primer pairs directed against the promoter of the highly transcribed ILV5 gene (ACCCAGTATTTTCCCTTTCC; TTGTCTATATGTTTTTGTCTTGC), the housekeeping gene ADH1 (TTTCCTTCCTTCATTCACGCACA; TCAAGTAACTGGAAGGAAGGCCGTA), the glucose-repressed GAL1 gene (GGGTAATTAATCAGCGAAGC; GGTTATGCAGCTTTTCCATT) as well as against a heterochromatic control region of chromosome V (TGCGTACAAAAAGTGTCAAGAGATT; ATGCGCAAGAAGGTGCCTAT) were used. All primer pairs had PCR efficiencies in the range of 95–100%. Total of 25 µl PCR reactions contained 1 μl DNA template, 2 μl of 10 μM primer pairs and 12.5 μl iTaq SYBR Green Supermix (Bio-Rad). Quantitative PCR was performed on a Bio-Rad CFX96 Real-Time System (Bio-Rad Laboratories, Inc.) using a 3 min denaturing step at 95°C, followed by 49 cycles of 30 s at 95°C, 30 s at 61°C and 15 s at 72°C. Threshold cycle (Ct) values were determined using the Ct determination mode ‘Regression’ of Bio-Rad CFX Manager software package (Version 1.1). Fold enrichment over heterochromatic control region was determined and calculated as described (59).
RESULTS
Revised Med11 N-terminus
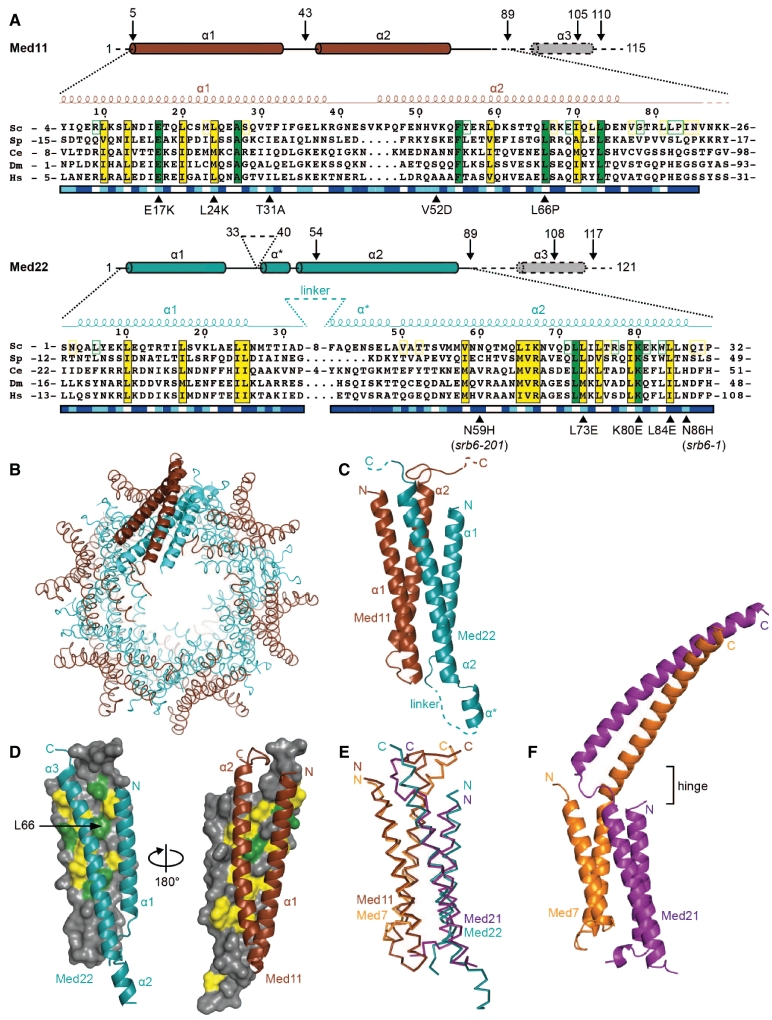
Based on published interaction data, we assumed that Sc Med11 and Med22 form a stable heterodimer. Indeed, the two subunits could be co-expressed in recombinant form (19,29) in E. coli and subsequently co-purified. When we analyzed recombinant Med11/22 and endogenous purified Mediator by SDS–PAGE, we noticed a discrepancy in the electrophoretic mobility of Med11 (data not shown). Endogenous Med11 appeared smaller than the variant expressed recombinantly. A comparison of the annotated MED11 ORF with experimentally determined transcription start sites derived from cDNA sequencing of full-length 5′-capped mRNAs (30) revealed that the start codon of MED11 was incorrectly assigned in the databases. The real initiator methionine corresponds to residue 17 of the original annotation; thus, the real protein is smaller than previously thought. We corrected the amino acid numbering and used the new numbering throughout.
Med11/22 structure solution
We prepared the correct full-length recombinant Med11/22 heterodimer in pure recombinant form. In contrast to the initial preparation, this complex crystallized. The crystals could, however, not be refined, likely due to flexibility in the poorly conserved C-termini of both subunits. We, therefore, tested various truncated protein variants for their solubility and crystallization behavior and found that a variant comprising the highly conserved core of Med11/22 (Med115–89/Med221–89, Figure 1) resulted in crystals diffracting up to a resolution of 2.1 Å. The X-ray structure was solved by selenomethionine labeling and multi-wavelength anomalous diffraction and was refined to a free Rfactor of 20.8% (Table 1). The crystals contained an unusual sphere-like arrangement of 12 heterodimers in the asymmetric unit (Figure 1B). Disordered in the crystals was only a non-conserved Med22 linker (residues 33–40) between helix α1 and the non-conserved helix α*. Deletion of this linker in yeast had no phenotype (Supplementary Figure S1).
Figure 1.
Structure of Med11/22 Mediator subcomplex. (A) Multiple sequence alignment of the conserved core of Med11 and Med22 from Saccharomyces cerevisiae (Sc), Schizosaccharomyces pombe (Sp), Caenorhabditis elegans (Ce), Drosophila melanogaster (Dm) and Homo sapiens (Hs). The corrected numbering of Sc Med11 is used. Invariant and conserved residues are highlighted in green and yellow, respectively. Additionally, residues that are invariant or conserved among the yeast family Saccharomycotinae (Sc, Candida glabrata, Candida albicans, Ashbya gossypii, Kluyveromyce lactis and Debaryomyces hansenii) are highlighted with green and yellow frames on the Sc sequence, respectively. Surface accessibility is indicated below the sequences (blue, high; cyan, intermediate; white, buried). Secondary structure elements of the conserved structural core are shown above the sequences (spirals, α-helices; lines, ordered but without secondary structure; dashed lines, disordered in crystal construct). In addition schematic views of the full proteins are shown. Consensus secondary structure predictions (61–63) for the C-termini of Sc Med11 and Med22 are indicated with dashed lines and gray filling. All truncations relevant for this study are indicated with arrows and the residue number. Previously reported as well as mutations generated in this study are marked with filled triangles. Sequence alignments were done with MUSCLE (64) and figures were prepared with ESPript (65). (B) Ribbon-model representation of the 12 heterodimers within the asymmetric unit. Med11 and Med22 are depicted in brown and cyan, respectively. (C) Ribbon-model representation of the Med11/22 crystal structure. Secondary structure elements are labeled according to (A). The linker between helices α1 and α* of Med22 (residues 33–40) was disordered. C-termini of Med11 and Med22 had to be removed from crystallization construct and are therefore lacking in the structure. (D) Dimer interface conservation. On the left, Med11 is shown in surface representation together with a ribbon model of Med22. The view is related to (C) by a 90° rotation around a vertical axis. On the right, Med22 is shown in surface representation together with a ribbon model of Med11. The two views are related by a 180° rotation around a vertical axis. (E) Superimposition of Cα traces of the four-helix bundle folds of Med11/22 (brown and cyan; this study) and Med7C/21 [orange and purple; (31)]. (F) Ribbon-model representation of the complete Med7C/21 structure (pdb accession code 1YKE). The reported flexible hinge region is indicated.
Table 1.
Data collection and refinement statistics
| Peak | Inflection | Remote | |
|---|---|---|---|
| Data collection | |||
| Space group | P 21212 | ||
| Cell parameters (Å) | |||
| a | 148.5 | ||
| b | 173.8 | ||
| c | 101.6 | ||
| Wavelength (Å) | 0.9796 | 0.9797 | 0.9720 |
| Resolution range (Å)a | 100–2.05 (2.10–2.05) | 100–2.2 (2.26–2.20) | 100–2.3 (2.36–2.30) |
| Completeness (%) | 98.9 (99.1) | 98.9 (99.1) | 98.9 (98.7) |
| Unique reflections | 315 134 (23 359) | 256 151 (19 022) | 224 540 (16 639) |
| Redundancy | 2.9 (3.0) | 2.9 (2.9) | 2.9 (2.8) |
| Rsym (%) | 7.2 (57.8) | 7.1 (52.6) | 7.6 (55.9) |
| <I>/<σI> | 10.8 (2.0) | 11.5 (2.2) | 11.5 (2.2) |
| Refinement | |||
| Number of residues | 1891 | ||
| Number of non-hydrogen atoms | 16 483 | ||
| Number of solvent molecules | 964 | ||
| RMS bond deviation (Å) | 0.008 | ||
| RMS angle deviation (°) | 0.968 | ||
| Ramachandran plot (preferred/allowed) | 99.3/0.7 | ||
| Rcryst (%) | 17.0 | ||
| Rfree (%)b | 20.8 |
aThe numbers in parenthesis correspond to the highest resolution shell
b5% of the data were set aside for free Rfactor calculation.
A helix bundle building block in Mediator
Med11/22 forms an antiparallel four-helix bundle (Figure 1C). This bundle fold is required in vivo since deletion of helix α1 in Med11 (med1143–115) was lethal in yeast and deletion of α1 and α* of Med22 (med2254–121) caused a growth defect (Figure 2). Unexpectedly, the bundle fold resembles the previously reported structure of the heterodimeric Med7C/21 subcomplex of the middle module (31). The Med11/22 and Med7C/21 structures show a root mean square deviation of 3.1 Å over 121 Cα atoms (Figure 1E). By combining results from HHPred (32) and secondary structure predictions, we detected a total of six possible heterodimeric four-helix bundles in Mediator. Nine out of 17 Mediator core subunits and two metazoan-specific subunits could participate in bundle formation (Figure 3 and Supplementary Table S1). Heterodimer formation has been experimentally verified for Med11/22 in the head (this study), Med4/9 (33) and Med7/21 (31) in the middle and Med2/3 in the tail (34). Additional bundles are apparently present in the Med10/14 heterodimer in the middle module (33) and in a metazoan-specific Med28/30 subcomplex. These results establish the four-helix bundle fold as a common building block of Mediator that is apparently found in all modules except for the dissociable kinase module.
Figure 2.
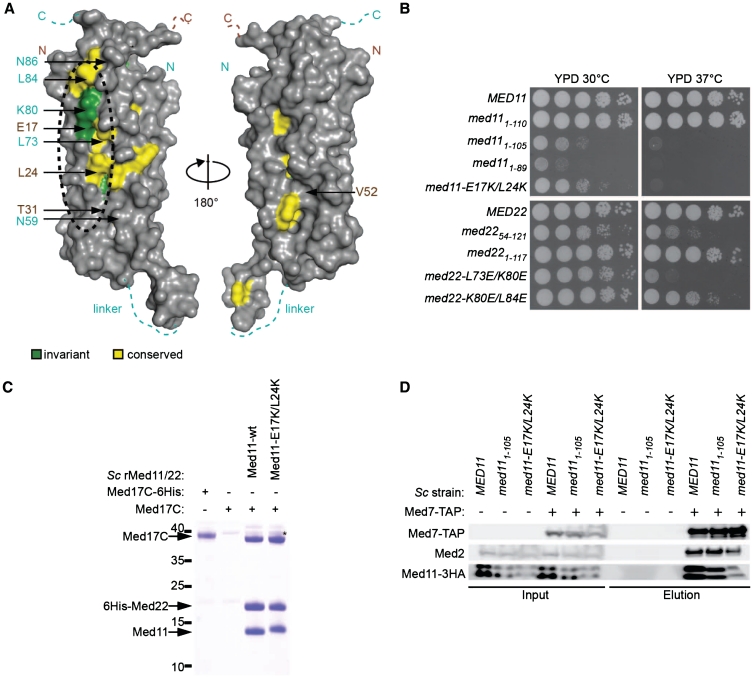
C-terminal extensions of Med11/22 are essential for viability. Yeast complementation assays. MED11 and MED22 constructs including 500 bp upstream of the start codon and 300 bp downstream of the stop codon were cloned into a pRS315 plasmid (LEU2) and transformed into the respective yeast shuffle strains. On 5FOA plates the URA3 shuffle plasmid encoding the respective full-length gene is shuffled out. Yeast cells lacking either the N- or C-terminus of Med11 or the C-terminus of Med22 are inviable. Cells lacking the N-terminal helix of Med22 or the last 10 amino acids of Med11 display a slow growth phenotype.
Figure 3.
A heterodimeric four-helix bundle building block in Mediator. Schematic depiction of Mediator subunits predicted to share the heterodimeric bundle fold. α-helices from crystal structures or from predictions (61–63) are indicated as boxes drawn to scale. Helix α1, α2 and C-terminal helical extensions are colored in black, gray and white, respectively. Check marks indicate experimentally confirmed heterodimers (co-expression and co-purification). Structural homology to the published Med7C/21 structure was predicted with HHPred (http://toolkit.lmb.uni-muenchen.de/hhpred) for all listed subunits except the highly divergent Sc subunits Med2 and Med3. The P-value and score for the HHPred searches are given. When the human protein sequence was used for the HHPred search, the P-values are marked with an asterisk.
Essential C-terminal helices extend from the Med11/22 helix bundle
In the structure of Med7C/21 (31), the four-helix bundle is connected to C-terminal helical extensions from both subunits that form a coiled-coil (Figure 1F). The C-terminal extensions of Med11 and Med22 are not present in the Med11/22 structure, but are predicted to each contain an additional helix (Figure 1A and Supplementary Figure S2). However, in contrast to Med7C/21, the C-terminal helices are not predicted to form a coiled-coil and are connected to the bundle through longer, probably flexible linkers. To investigate the importance of the predicted Med11/22 C-terminal helices in vivo, we generated several Med11/22 truncation variants (Figure 1A) and tested them in yeast complementation assays (Figure 2). Removal of either helix (med111–89 or med221–89) or even partial truncation of the Med22 C-terminal helix (med221–108) was lethal under standard conditions. Thus, the apparently flexible and helical C-terminal extensions from the Med11/22 bundle are required for normal cell growth.
Med11/22 extensions bind a Med17 C-terminal domain
We next asked whether the C-terminal extensions anchor the Med11/22 bundle within the head module by an interaction with Med17, the architectural head subunit. We could indeed detect in vitro binding of Med11/22 to a soluble C-terminal domain of Med17 (Med17C, residues 377–687) (Figure 4A, lanes 1 and 3). To map the Med17 binding determinant in Med11/22, we co-expressed and co-purified truncated Med11/22 variants with Med17C in E. coli. Recombinant Med11/22 heterodimers containing Med1143–115 could not be co-purified, whereas Med111–89, Med111–105, Med2254–121 and Med221–89 all formed stable complexes (Figure 4A). Truncation of either predicted C-terminal helix (Figure 4A, lanes 6, 7, 10–12) prevented co-purification of Med17C with Med11/22, while truncation of the Med22 N-terminus (Figure 4A, lane 4) or C-terminal truncations that did not affect the C-terminal helices (Figure 4A, lanes 5 and 9) had no effect. We conclude that the C-terminal helices of Med11/22 anchor the four-helix bundle to Med17C and that the C-terminus of Med22 is absolutely required for this interaction in vitro and in vivo. This is in accordance with yeast two-hybrid results showing that the Sc Med11–Med17 interaction is lost upon C-terminal truncation of Med11 (20). The consistency of these in vitro binding data with the above in vivo phenotyping further suggests that Med11/22 anchoring to Med17C is essential for cellular growth.
Figure 4.
C-terminal extensions of Med11/22 bind a Med17 C-terminal domain. (A) Co-expression in E. coli and co-purification of Sc a C-terminal domain of Med17C (residues 377–687) with Sc 6His-Med22/Med11 constructs using nickel magnetic beads. A co-purifying contaminant is marked with an asterisk. (B) Co-immunoprecipitation of the putative Sp Med11-3HA with Sp Med7-TAP from Sp lysate using IgG agarose beads. Med7 was eluted natively using tobacco etch virus protease. Med7 was detected in the input using peroxidase/anti-peroxidase antibody complex. Med11 was detected in both input and eluate using anti-HA antibody. (C) Co-expression in E. coli and co-purification of Sp Med17 (lanes 1–3) and Sp Med17C (residues 257–545; lanes 4–6) with Sp His6-Med22/Med11 constructs using nickel magnetic beads.
A conserved Med17C/11/22 Mediator subcomplex
The helix bundle of Med11/22 appears to be conserved among eukaryotes, because amino acid residues that constitute the Med11–Med22 interface are conserved (Figure 1D). However, a Med11 homolog has thus far not been found in Sp (28). We could however predict a remote Med11 homology in the Sp protein SPAC644.10 (35). Indeed, Sp SPAC644.10 could be co-immunoprecipitated with Med7, suggesting it is a Mediator subunit (Figure 4B). Amplification and sequencing of the SPAC644.10 cDNA clone revealed the absence of an annotated intron and enabled co-expression and co-purification of recombinant Sp SPAC644.10 with Sp Med22 (Figure 4). This establishes SPAC644.10 as the Sp Med11 homolog and confirms the conservation of the Med11/22 bundle. To investigate whether the conservation extends to the anchoring of Med11/22 on Med17C, we tested whether a trimeric Sp Med17/11/22 subcomplex could be obtained after subunit co-expression in E. coli and this was indeed achieved (Figure 4C, lane 1). We mapped a soluble C-terminal domain of Sp Med17 (Med17C, residues 257–545) that was sufficient for Med11/22 binding (Figure 4C, lane 4). Truncation of the C-terminus of Sp Med22 did not affect Med11/22 heterodimer formation, but abolished Med17 binding (Figure 4C, lane 3 and 6). Truncation of the C-terminus of Sp Med11 (Med111–91) had almost no effect (Figure 4C, lane 2 and 5). Thus, the interaction of the C-terminal extension from the Med11/22 bundle with Med17C is conserved between the distantly related yeast species Sp and Sc and the Med17C/11/22 subcomplex is therefore a conserved architectural unit of the Mediator head.
A highly conserved interaction patch on Med11/22
Despite the importance of the head module for PIC formation, only few contacts between PIC components and head module subunits have been reported. Thus, we wanted to identify potential interaction surfaces on Med11/22 that could account for contacts to PIC components. Plotting sequence conservation (Figure 5A) and electrostatic surface charge (Supplementary Figure S3) onto the Med11/22 structure revealed a large, highly conserved surface patch with exposed hydrophobic residues on one side of the bundle domain. Several known Med11/22 mutations (20,25,26), namely srb6-201 (med22-N59H), srb6-1 (med22-N86H), med11-T31A (original annotation: med11-T47A), are located around this patch (Figure 5A). Mutations reported to affect interaction of Med11 with Med22 or Med17 (20,36), namely med11-V52D (original annotation: med11-V68D), med11-G92S (original annotation: med11-G108S) and med11-L66P (original annotation: med11-L82P), are however located distant from this patch or in the hydrophobic core (Figure 5A and Figure 1D). This suggests that the conserved surface patch is involved in the interaction with components of the PIC.
Figure 5.
A conserved interaction patch on Med11/22. (A) Surface conservation of the Med11/22 four-helix bundle. Invariant and conserved residues from yeast to human are highlighted in green and yellow, respectively. Residues of Med11 (brown) and Med22 (cyan) targeted by mutagenesis in this study and in previous reports are indicated with arrows. The orientation is identical to Figure 1C. The two views are related by a 180° rotation around the vertical axis. (B) Spot dilutions of yeast strains carrying structure-based mutations on the conserved surface patch and viable truncations (Figure 2) on YPD plates at 30°C and 37°C. (C) Co-expression in E. coli and co-purification of Sc Med17C with Sc 6His-Med22/Med11 and Sc 6His-Med22/Med11-E17K/L24K using nickel magnetic beads. (D) Co-immunoprecipitation of Sc Med11-3HA (Mediator head module) and Sc Med2 (Mediator tail module) with Sc Med7-TAP (Mediator middle module) from wild-type, med111–105 and med11-E17K/L24K yeast strains.
To investigate the functional importance of this patch we generated structure-based surface mutations. Strains carrying double point mutations of adjacent surface residues (med11-E17K/L24K, med22-L73E/K80E and med22-K80E/L84E) exhibited various degrees of temperature-sensitivity (Figure 5B). The med11-E17K/L24K strain exhibited a strong growth defect at all temperatures, similar to med111–105. However, unlike the Med11 truncation, the patch mutation did not affect anchoring of Med11/22 to Med17C in vitro (Figure 5C). Since Med11/22 is essential for Med17 stability (18) we purified Mediator from wild-type, med111–105 and med11-E17K/L24K yeast strains to test complex integrity. These mutations did apparently not affect Mediator architecture since Med7-TAP (middle module) co-immunoprecipitated subunits Med2 (tail module) and Med11 (head module) (Figure 5D and Supplementary Figure S4). Thus, the phenotype observed for the Med11 mutants does not appear to be the result of impaired Mediator integrity, but seems to be a direct effect of impaired Med11/22 interactions with other factors.
Med11/22 is a functionally distinct submodule
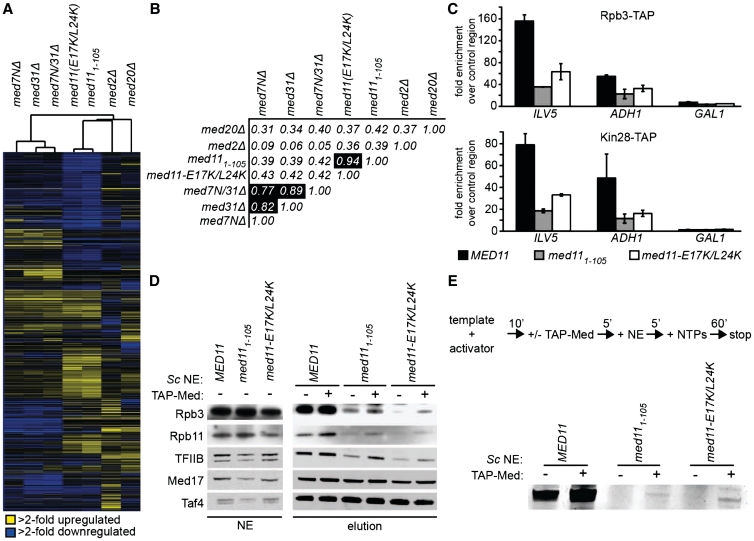
To test whether mutations in Med11/22 affect transcription in vivo, we performed genome-wide gene-expression profiling of the med111–105 and med11-E17K/L24K yeast strains (Figure 6A). The MED11 mutants form a distinct cluster with a Pearson correlation coefficient of 0.94, correlating only moderately with med20Δ (head module), med2Δ (tail module) and with mutations affecting the previously described functional submodule Med7N/31 (middle module) (37) (Figure 6B and Supplementary Table SII). In total, 604 and 450 genes were significantly changed (fold expression change >2-fold; P < 0.05) in med111–105 and med11-E17K/L24K, respectively (Supplementary Figure S5A, Supplementary Table SII). A total of 401 genes were significantly changed in both mutants, with 160 genes up-regulated and 241 genes downregulated. The number of significantly changed genes in both MED11 mutant strains is higher than in any other Mediator mutant tested (Supplementary Figure S5B). The effect on gene expression is rather pleiotropic without any significant enrichment for specific GO terms (38). This indicates a distinct function of the Med11/22 subcomplex in gene regulation and a more global role in contrast to the more gene-specific roles of previously reported non-essential Mediator submodules (29,37,39).
Figure 6.
Med11/22 is a functional Mediator submodule in vitro and in vivo. (A) Hierarchical cluster diagram (Pearson correlation) of genes exhibiting significantly altered mRNA levels (>2-fold, vertical axis) for different Mediator mutant strains (horizontal axis). Changes in mRNA levels compared to the wild-type strain are depicted in yellow (up), blue (down) or black (no change). (B) Pearson correlation matrix for expression profiles of different Mediator mutants (1, very high correlation; 0, no correlation; −1, very high anti-correlation). (C) Chromatin immunoprecipitation of Rpb3-TAP (Pol II) and Kin28-TAP (TFIIH kinase module) in wild-type, med111–105 and med11-E17K/L24K strains grown to early exponential phase in YPD medium containing 2% glucose. Fold enrichment over a heterochromatic control region is shown for the yeast promoters of a highly expressed gene (ILV5), a housekeeping gene (ADH1) and a glucose-repressed gene (GAL1). (D) In vitro PIC assembly of wild-type, med111–105 and med11-E17K/L24K nuclear extracts (NE) on the immobilized HIS4 yeast promoter. PIC formation of mutant extracts was partially rescued by adding 2 pmol tandem-affinity purified Mediator complex (TAP-Med) to the nuclear extracts prior to PIC assembly. Presence of Pol II (Rpb3 & Rpb11), TFIIB, Mediator (Med17) and TFIID (Taf4) was tested by western Blot. (E) In vitro transcription assay with wild-type, med111–105 and med11-E17K/L24K nuclear extracts (NE) on the HIS4 yeast promoter. Order of addition is shown on top. Transcription was partially rescued by adding 2 pmol tandem-affinity purified Mediator complex (TAP-Med).
The Med11/22 surface patch functions in PIC stabilization
Since Med11 was previously reported to stabilize the TFIIK subcomplex of TFIIH and, to some extent, Pol II at promoters (20), we performed chromatin immunoprecipitations of Kin28, the kinase subunit of TFIIK and the Pol II subunit Rpb3 in wild-type, med111–105 and med11-E17K/L24K strains (Figure 6C). In the mutant strains, we observed decreased occupancies for both Kin28 and Rpb3 at active promoters, indicating a defect in stable PIC formation. To test whether the decrease is a direct effect of impaired Med11/22 function or an indirect effect of global deregulation of gene expression, we performed in vitro immobilized template assays with yeast nuclear extracts (Figure 6D). PICs were assembled on an immobilized HIS4 yeast promoter in a Gcn4-dependent manner, washed and subsequently analyzed by western blot. Consistent with the in vivo results, both mutant extracts displayed a decreased occupancy of Pol II after washing. As expected, the occupancy of TFIIB, interacting directly with Pol II, is also decreased. Mediator and TFIID, which are recruited directly by the transcriptional activator Gcn4 (40), remain unaffected. Consistently, nuclear extracts from the MED11 mutant strains were inactive in in vitro transcription assays (Figure 6E). Since addition of purified wild-type Mediator to the mutant extracts partially rescued the recruitment of the basal machinery and consequently also transcriptional activity, the observed defects can be directly linked to Mediator function. These results show that the conserved surface patch on the Med11/22 bundle is required for stable PIC formation.
DISCUSSION
To understand the regulatory mechanisms of eukaryotic transcription, a detailed structural and functional dissection of the involved multiprotein complexes is required. In this study, we extend our previous structure–function analysis of the general transcription co-activator Mediator. We combine structural biology, yeast genetics, biochemistry and gene-expression profiling, to define and functionally characterize the Mediator subcomplex Med11/22, to provide a more detailed understanding of Mediator head module architecture and to obtain insights into Mediator conservation and evolution. We report the structure of a conserved four-helix bundle domain in Med11/22 that puts reported mutations (20,36) into a molecular framework and identifies a highly conserved surface patch that is required for a distinct widespread function of the Med11/22 subcomplex.
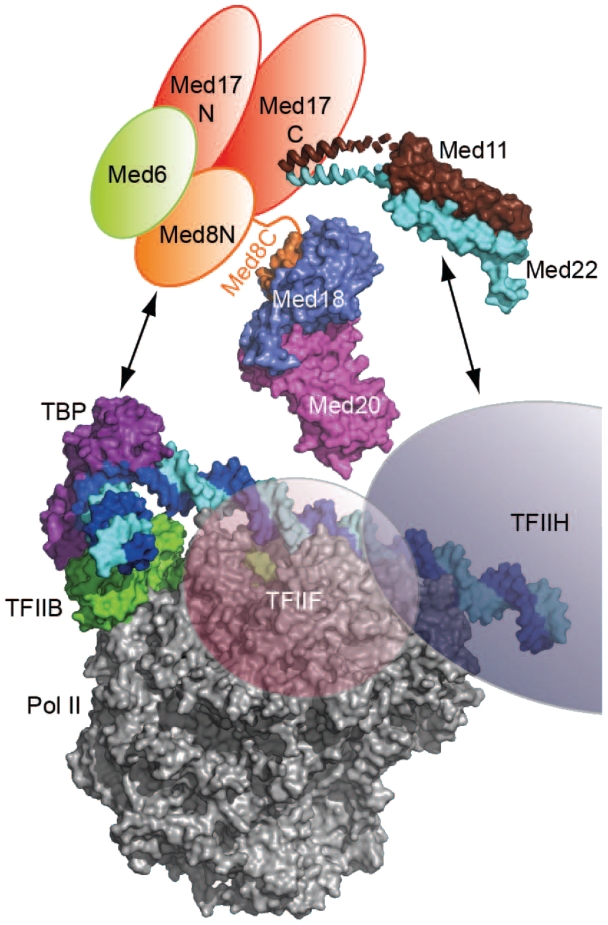
We suggest that the Med11/22 surface patch mediates an essential interaction with TFIIH, because the previously described temperature-sensitive mutation med11-T31A (20) locates near the patch. This mutation was shown to decrease interaction of Med11 with the TFIIH subunit Rad3 in yeast two-hybrid assays and promoter occupancy of the TFIIH kinase module TFIIK. In contrast to the mutation med11-T31A, which was discovered genetically, the mutation med11-E17K/L24K reported here was identified based on structural data and had a strong general growth defect that allowed us to conduct functional studies under standard growth conditions. Chromatin immunoprecipitation showed a decrease of TFIIK and Pol II occupancies at active promoters in vivo, indicating a defect in PIC formation. Yeast nuclear extracts from the mutant strain were defective in stable PIC formation and inactive in transcription in vitro. The defects result from impairing the function of the Med11/22 submodule and not the head module per se, since, in contrast to the reported med17 mutant srb4-138 (41), activator-mediated recruitment of Mediator to the promoter was not affected. Thus the Med11/22 submodule specifically functions in promoting stable PIC formation. We propose that the conserved submodular architecture of the Mediator head enables multiple transient interactions with PIC components, including TBP (16) and TFIIH (20) (Figure 7), thereby stabilizing the PIC and facilitating open complex formation and initial RNA synthesis.
Figure 7.
Submodular architecture of the Mediator head and relative size of the Pol II PIC. Crystal structures of the essential Med11/22 four-helix bundle (this article), the previously described non-essential Med8C/18/20 subcomplex (29) and a molecular model of the Pol II–TBP–TFIIB–DNA promoter closed complex (66) are drawn to scale. Locations of the general factors TFIIF and H, as determined by biochemical probing (67,68), are indicated by semi-transparent ellipsoids. Mediator head interactions with PIC components, namely Med8/18/20-TBP (19) and Med11/22–TFIIH (20), are indicated by arrows.
Another intriguing observation is the presence of up to six related helix bundle folds, to which about one-half of the Mediator core subunits contribute. The presence of related bundle domains in different Mediator modules elucidates the evolutionary origin of Mediator. Large protein complexes with high functional modularity might generally have evolved through duplication of genes-encoding dimers (42). For example, the 15-subunit TFIID complex contains five heterodimeric subcomplexes interacting through a common histone-fold domain (43,44). Subsequent divergent evolution of paralogous subunits could then generate asymmetry and diversification of protein interactions for functional specialization. Rapid evolution of Mediator by dimer duplication and diversification befits the finding that Mediator is only present in eukaryotes although the general transcription machineries are conserved between eukaryotes and archaea.
ACCESSION NUMBERS
The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID code: 3R84). Microarray data were submitted to the ArrayExpress database (http://www.ebi.ac.uk/microarray) under accession number E-MEXP-3150.
SUPPLEMENTARY Data
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft (SFB646, TR5, FOR1068, NIM); EU grant 3D Repertoire (contract no. LSHG-CT-2005-512028); European Molecular Biology Organization; Boehringer Ingelheim Fonds (to M.S.); Elite Network of Bavaria (to M.S.); LMUinnovativ project Bioimaging Network (BIN) (to P.C.); LMUexcellent research professorship (to P.C.). Funding for open access charge: Deutsche Forschungsgemeinschaft.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Heidi Feldmann and the members of the Cramer laboratory for help and discussions. We are particularly grateful to Kerstin Maier and Tobias Koschubs for help with the gene expression profiling and Andreas Mayer for help with ChIP assays. We thank the members of the crystallization facility at the MPI for Biochemistry (Martinsried) and the Hahn laboratory for the kind gift of yeast strains, plasmids and antibodies. Part of this work was performed at the Swiss Light Source (SLS) at the Paul Scherrer Institute (Villigen, Switzerland).
REFERENCES
- 1.Sikorski TW, Buratowski S. The basal initiation machinery: beyond the general transcription factors. Curr. Opin. Cell Biol. 2009;21:344–351. doi: 10.1016/j.ceb.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 3.Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 2010;11:761–772. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjorklund S, Gustafsson C. The yeast Mediator complex and its regulation. Trends Biochem. Sci. 2005;30:240–244. doi: 10.1016/j.tibs.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 5.Kornberg R. Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 2005;30:235–239. doi: 10.1016/j.tibs.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 6.Naar A, Lemon B, Tjian R. Transcriptional coactivator complexes. Annu. Rev. Biochem. 2001;70:475–501. doi: 10.1146/annurev.biochem.70.1.475. [DOI] [PubMed] [Google Scholar]
- 7.Meyer KD, Lin S-C, Bernecky C, Gao Y, Taatjes DJ. p53 activates transcription by directing structural shifts in Mediator. Nat. Struct. Mol. Biol. 2010;17:753–760. doi: 10.1038/nsmb.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flanagan P, Kelleher R, Sayre M, Tschochner H, Kornberg R. A mediator required for activation of RNA polymerase II transcription in vitro. Nature. 1991;350:436–438. doi: 10.1038/350436a0. [DOI] [PubMed] [Google Scholar]
- 9.Kelleher R, Flanagan P, Kornberg R. A novel mediator between activator proteins and the RNA polymerase II transcription apparatus. Cell. 1990;61:1209–1215. doi: 10.1016/0092-8674(90)90685-8. [DOI] [PubMed] [Google Scholar]
- 10.Park JM, Werner J, Kim JM, Lis JT, Kim YJ. Mediator, not holoenzyme, is directly recruited to the heat shock promoter by HSF upon heat shock. Mol. Cell. 2001;8:9–19. doi: 10.1016/s1097-2765(01)00296-9. [DOI] [PubMed] [Google Scholar]
- 11.Fondell JD, Ge H, Roeder RG. Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl Acad. Sci. USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bäckström S, Elfving N, Nilsson R, Wingsle G, Björklund S. Purification of a plant Mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol. Cell. 2007;26:717–729. doi: 10.1016/j.molcel.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 13.Bourbon H-M. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 2008;36:3993–4008. doi: 10.1093/nar/gkn349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boube M, Joulia L, Cribbs D, Bourbon H-M. Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell. 2002;110:143–151. doi: 10.1016/s0092-8674(02)00830-9. [DOI] [PubMed] [Google Scholar]
- 15.Cai G, Imasaki T, Yamada K, Cardelli F, Takagi Y, Asturias FJ. Mediator head module structure and functional interactions. Nat. Struct. Mol. Biol. 2010;17:273–279. doi: 10.1038/nsmb.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang J, Kim S, Hwang M, Han S, Lee Y, Kim Y. The structural and functional organization of the yeast mediator complex. J. Biol. Chem. 2001;276:42003–42010. doi: 10.1074/jbc.M105961200. [DOI] [PubMed] [Google Scholar]
- 17.Asturias F, Jiang Y, Myers L, Gustafsson C, Kornberg R. Conserved structures of mediator and RNA polymerase II holoenzyme. Science. 1999;283:985–987. doi: 10.1126/science.283.5404.985. [DOI] [PubMed] [Google Scholar]
- 18.Takagi Y, Calero G, Komori H, Brown JA, Ehrensberger AH, Hudmon A, Asturias F, Kornberg RD. Head module control of mediator interactions. Mol. Cell. 2006;23:355–364. doi: 10.1016/j.molcel.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 19.Larivière L, Geiger S, Hoeppner S, Röther S, Strässer K, Cramer P. Structure and TBP binding of the Mediator head subcomplex Med8-Med18-Med20. Nat. Struct. Mol. Biol. 2006;13:895–901. doi: 10.1038/nsmb1143. [DOI] [PubMed] [Google Scholar]
- 20.Esnault C, Ghavi-Helm Y, Brun S, Soutourina J, Van Berkum N, Boschiero C, Holstege F, Werner M. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol. Cell. 2008;31:337–346. doi: 10.1016/j.molcel.2008.06.021. [DOI] [PubMed] [Google Scholar]
- 21.Bhaumik SR, Raha T, Aiello DP, Green MR. In vivo target of a transcriptional activator revealed by fluorescence resonance energy transfer. Genes Dev. 2004;18:333–343. doi: 10.1101/gad.1148404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ansari S, He Q, Morse R. Mediator complex association with constitutively transcribed genes in yeast. Proc. Natl Acad. Sci. USA. 2009;106:16734–16739. doi: 10.1073/pnas.0905103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holstege F, Jennings E, Wyrick J, Lee T, Hengartner C, Green M, Golub T, Lander E, Young R. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 24.Thompson C, Young R. General requirement for RNA polymerase II holoenzymes in vivo. Proc. Natl Acad. Sci. USA. 1995;92:4587–4590. doi: 10.1073/pnas.92.10.4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee T, Wyrick J, Koh S, Jennings E, Gadbois E, Young R. Interplay of positive and negative regulators in transcription initiation by RNA polymerase II holoenzyme. Mol. Cell. Biol. 1998;18:4455–4462. doi: 10.1128/mcb.18.8.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson C, Koleske A, Chao D, Young R. A multisubunit complex associated with the RNA polymerase II CTD and TATA-binding protein in yeast. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- 27.Yuryev A, Corden JL. Suppression analysis reveals a functional difference between the serines in positions two and five in the consensus sequence of the C-terminal domain of yeast RNA polymerase II. Genetics. 1996;143:661–671. doi: 10.1093/genetics/143.2.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spahr H, Samuelsen C, Baraznenok V, Ernest I, Huylebroeck D, Remacle J, Samuelsson T, Kieselbach T, Holmberg S, Gustafsson C. Analysis of Schizosaccharomyces pombe mediator reveals a set of essential subunits conserved between yeast and metazoan cells. Proc. Natl Acad. Sci. USA. 2001;98:11985–11990. doi: 10.1073/pnas.211253898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larivière L, Seizl M, van Wageningen S, Röther S, van de Pasch L, Feldmann H, Strässer K, Hahn S, Holstege FCP, Cramer P. Structure-system correlation identifies a gene regulatory Mediator submodule. Genes Dev. 2008;22:872–877. doi: 10.1101/gad.465108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miura F, Kawaguchi N, Sese J, Toyoda A, Hattori M, Morishita S, Ito T. A large-scale full-length cDNA analysis to explore the budding yeast transcriptome. Proc. Natl Acad. Sci. USA. 2006;103:17846–17851. doi: 10.1073/pnas.0605645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baumli S, Hoeppner S, Cramer P. A conserved mediator hinge revealed in the structure of the MED7.MED21 (Med7.Srb7) heterodimer. J. Biol. Chem. 2005;280:18171–18178. doi: 10.1074/jbc.M413466200. [DOI] [PubMed] [Google Scholar]
- 32.Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koschubs T, Lorenzen K, Baumli S, Sandström S, Heck AJR, Cramer P. Preparation and topology of the Mediator middle module. Nucleic Acids Res. 2010;38:3186–3195. doi: 10.1093/nar/gkq029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beve J, Hu G, Myers L, Balciunas D, Werngren O, Hultenby K, Wibom R, Ronne H, Gustafsson C. The structural and functional role of Med5 in the yeast Mediator tail module. J. Biol. Chem. 2005;280:41366–41372. doi: 10.1074/jbc.M511181200. [DOI] [PubMed] [Google Scholar]
- 35.Wood V, Gwilliam R, Rajandream M-A, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 36.Han S, Lee Y, Gim B, Ryu G, Park S, Lane W, Kim Y. Activator-specific requirement of yeast mediator proteins for RNA polymerase II transcriptional activation. Mol. Cell. Biol. 1999;19:979–988. doi: 10.1128/mcb.19.2.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koschubs T, Seizl M, Larivière L, Kurth F, Baumli S, Martin DE, Cramer P. Identification, structure, and functional requirement of the Mediator submodule Med7N/31. EMBO J. 2009;28:69–80. doi: 10.1038/emboj.2008.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. GO::TermFinder–open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van de Peppel J, Kettelarij N, van Bakel H, Kockelkorn T, van Leenen D, Holstege F. Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol. Cell. 2005;19:511–522. doi: 10.1016/j.molcel.2005.06.033. [DOI] [PubMed] [Google Scholar]
- 40.Herbig E, Warfield L, Fish L, Fishburn J, Knutson B, Moorefield B, Pacheco D, Hahn S. Mechanism of Mediator recruitment by tandem Gcn4 activation domains and three Gal11 activator-binding domains. Mol. Cell. Biol. 2010;30:2376. doi: 10.1128/MCB.01046-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ranish JA, Yudkovsky N, Hahn S. Intermediates in formation and activity of the RNA polymerase II preinitiation complex: holoenzyme recruitment and a postrecruitment role for the TATA box and TFIIB. Genes Dev. 1999;13:49–63. doi: 10.1101/gad.13.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pereira-Leal JB, Levy ED, Kamp C, Teichmann SA. Evolution of protein complexes by duplication of homomeric interactions. Genome Biol. 2007;8:R51. doi: 10.1186/gb-2007-8-4-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gangloff YG, Romier C, Thuault S, Werten S, Davidson I. The histone fold is a key structural motif of transcription factor TFIID. Trends Biochem. Sci. 2001;26:250–257. doi: 10.1016/s0968-0004(00)01741-2. [DOI] [PubMed] [Google Scholar]
- 44.Leurent C, Sanders S, Ruhlmann C, Mallouh V, Weil PA, Kirschner DB, Tora L, Schultz P. Mapping histone fold TAFs within yeast TFIID. EMBO J. 2002;21:3424–3433. doi: 10.1093/emboj/cdf342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meinhart A, Blobel J, Cramer P. An extended winged helix domain in general transcription factor E/IIE alpha. J. Biol. Chem. 2003;278:48267–48274. doi: 10.1074/jbc.M307874200. [DOI] [PubMed] [Google Scholar]
- 46.Budisa N, Steipe B, Demange P, Eckerskorn C, Kellermann J, Huber R. High-level biosynthetic substitution of methionine in proteins by its analogs 2-aminohexanoic acid, selenomethionine, telluromethionine and ethionine in Escherichia coli. Eur. J. Biochem. 1995;230:788–796. doi: 10.1111/j.1432-1033.1995.tb20622.x. [DOI] [PubMed] [Google Scholar]
- 47.Kabsch W. Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 1993;26:795–800. [Google Scholar]
- 48.Terwilliger T, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. D. Biol. Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perrakis A, Harkiolaki M, Wilson KS, Lamzin VS. ARP/wARP and molecular replacement. Acta Crystallogr. D. 2001;57:1445–1450. doi: 10.1107/s0907444901014007. [DOI] [PubMed] [Google Scholar]
- 50.Emsley P, Cowtan K. Coot: Model-Building Tools for Molecular Graphics. Acta Crystallogr. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.McCoy A, Grosse-Kunstleve R, Adams P, Winn M, Storoni L, Read R. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.DeLano W. DeLano Scientific. CA, USA: San Carlos; 2002. [DOI] [PubMed] [Google Scholar]
- 53.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 54.Ranish J, Hahn S. The yeast general transcription factor TFIIA is composed of two polypeptide subunits. J. Biol. Chem. 1991;266:19320–19327. [PubMed] [Google Scholar]
- 55.Gentleman R, Carey V, Bates D, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu Z, Irizarry R, Gentleman R, Martinez-Murillo F, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J. Am. Stat. Assoc. 2004;99:909–917. [Google Scholar]
- 57.Smyth G. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 58.Saeed A, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 59.Aparicio O, Geisberg JV, Sekinger E, Yang A, Moqtaderi Z, Struhl K. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr. Protoc. Mol. Biol. 2005 doi: 10.1002/0471142727.mb2103s69. Chapter 21, Unit 21.23. [DOI] [PubMed] [Google Scholar]
- 60.Mayer A, Lidschreiber M, Siebert M, Leike K, Söding J, Cramer P. Uniform transitions of the general RNA polymerase II transcription complex. Nat. Struct. Mol. Biol. 2010;17:1272–1278. doi: 10.1038/nsmb.1903. [DOI] [PubMed] [Google Scholar]
- 61.Cuff JA, Barton GJ. Evaluation and improvement of multiple sequence methods for protein secondary structure prediction. Proteins. 1999;34:508–519. doi: 10.1002/(sici)1097-0134(19990301)34:4<508::aid-prot10>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 62.Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 63.Ouali M, King RD. Cascaded multiple classifiers for secondary structure prediction. Protein Sci. 2000;9:1162–1176. doi: 10.1110/ps.9.6.1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Edgar R. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gouet P, Courcelle E, Stuart D, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 66.Kostrewa D, Zeller ME, Armache K-J, Seizl M, Leike K, Thomm M, Cramer P. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature. 2009;462:323–330. doi: 10.1038/nature08548. [DOI] [PubMed] [Google Scholar]
- 67.Chen H-T, Warfield L, Hahn S. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat. Struct. Mol. Biol. 2007;14:696–703. doi: 10.1038/nsmb1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim TK, Ebright RH, Reinberg D. Mechanism of ATP-dependent promoter melting by transcription factor IIH. Science. 2000;288:1418–1422. doi: 10.1126/science.288.5470.1418. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.