Abstract
Mitochondria play a key role in essential cellular functions. A deeper understanding of mitochondrial molecular processes is hampered by the difficulty of incorporating foreign nucleic acids into organelles. Mitochondria of most eukaryotic species import cytosolic tRNAs. Based on this natural process, we describe here a powerful shuttle system to internalize several types of RNAs into isolated mitochondria. We demonstrate that this tool is useful to investigate tRNA processing or mRNA editing in plant mitochondria. Furthermore, we show that the same strategy can be used to address both tRNA and mRNA to isolated mammalian mitochondria. We anticipate our novel approach to be the starting point for various studies on mitochondrial processes. Finally, our study provides new insights into the mechanism of RNA import into mitochondria.
INTRODUCTION
Mitochondria play important roles in diverse processes such as energy production, metabolic pathways, ageing and apoptosis. Although the mitochondrial (mt) genome only encodes a limited set of genes, their transcription, processing and translation is essential. Failure in any of these processes often lead to severe disorders such as cytoplasmic male sterility in plant (1) or pathological diseases in human (2). Our fundamental knowledge of mt gene expression and of the development of new therapeutic approaches is limited by important technical obstacles. In particular, the strategies available so far to manipulate the mt genetic system in vivo or in vitro are still scarce and difficult to handle (3).
In vivo, only the yeast Saccharomyces cerevisiae (4) and the unicellular alga Chlamydomonas reinhardtii (5) are amenable to stable mt transformation using a biolistic approach. Otherwise, no means currently exist to stably transform the mt DNA of higher eukaryotes. The few other strategies used in vivo and in vitro to study, manipulate and/or correct mt biogenesis rely on the import of macromolecules (proteins, DNA, RNA) into organelles (6,7). For example in vitro DNA uptake into isolated mitochondria has been obtained in various organisms through different strategies. Introduction and subsequent transcription of DNA has been established via electroporation on mammalian (8), trypanosomatid (9) and plant isolated mitochondria (10–12). Electroporation of mRNAs into isolated plant mitochondria has been described and has enabled the study of the processing of mRNA precursors (13). Direct DNA uptake and its subsequent expression have also been obtained with plant, yeast and mammalian mitochondria (14–17). All these approaches are promising but still present important drawbacks. It can be either the large amount of purified mitochondria required for experiment, the efficiency of internalization or expression of the nucleic acid and the difficulty in maintaining the integrity of mitochondria.
Natural import of RNA into mitochondria may represent another strategy. Cytosolic RNA species are imported into mitochondria of most eukaryotic cells and the vast majority of them are tRNAs (7,18). Two alternative approaches, exploiting a cryptic tRNA mt import system (19) or introducing an RNA import complex (20), have been successfully used to partially rescue mt myoclonic epilepsy and red ragged fibers (MERFF) dysfunction. The use of naturally imported RNA molecules other than tRNAs as vehicles to deliver sequences is also worthy of consideration. Another candidate is the 5S rRNA that is naturally imported into human mitochondria (21). Indeed, several 5S rRNA derivatives were obtained without losing their mt import capacity (22). The mt import of human RNase P and MRP RNAs has also been reported. Mt RNA targeting determinants present on these two RNAs were shown to allow their import in a polynucleotide phosphorylase-dependent manner (23). The challenge to incorporate foreign RNAs into mitochondria has just begun and the question whether mt import pathways can be diverted to address RNAs other than the naturally imported ones is open.
In the present study, we report a strategy based upon the use of a protein as a vector that enables the efficient introduction of various RNAs into isolated mitochondria. We show that, in vitro, the mammalian DiHydroFolateReductase (DHFR) is able to bind RNAs in a nonspecific manner. Then, we demonstrate that DHFR fused to a mitochondrial targeting sequence (MTS) can act as a carrier to introduce foreign RNAs into isolated plant mitochondria. We show that, once imported into the isolated organelle, a tRNA precursor is correctly processed leading to a mature tRNA. We then provide evidence that the unedited version of the full-length potato mt atp9 mRNA can be introduced into mitochondria and can be edited in organello. Finally, we demonstrate that the introduction of a foreign mRNA into isolated rat and human mitochondria can be obtained.
MATERIALS AND METHODS
DNA construction, plasmids and protein synthesis
The pSu9-DHFR (pDHFR) protein consists in the first 69 amino acids of subunit nine of the mt F0-ATPase (pSu9) of Neurospora crassa fused to the mouse dihydrofolate reductase (DHFR) (24). This construct was previously cloned into the pQE60 vector (gift from D. Rapaport, University of Tübingen, Germany). The pGFP consists in the first 69 amino acids of subunit nine of the mt F0-ATPase (pSu9) of Neurospora crassa fused to the green fluorescent protein (GFP) (25). This DNA construct was obtained using oligonucleotide-directed mutagenesis (15) and cloned into pQE60 vector. The GFP sequence was previously cloned into pQE60 vector and DHFR sequence is present on pQE40 vector. Thanks to the presence of a hexahistidinyl-tag, pDHFR, pGFP, DHFR and GFP expressed in Escherichia coli strain M15 were purified with Ni-NTA Superflow resin according to manufacturer’s recommendation (Qiagen, Valencia, CA, USA). The pDHFR and pGFP sequences were also cloned into the pCR®II-TOPO® vector in an orientation compatible with their transcription from the T7 promoter. (Invitrogen, Carlsbad, CA, USA). In vitro synthesis of pDHFR and pGFP proteins in the presence of [35S]-methionine (1000 Ci/mmol, Amersham) was carried out with the TNT® T7 Coupled Reticulocyte Lysate System (Promega) following the manufactor’s procedures.
In vitro transcription and labeling of synthetic RNAs
Clones corresponding to the Arabidopsis cytosolic tRNAAla and to the larch mt pre-tRNAHis are, respectively, described in refs. (26) and (15). The atp9 gene (27) was amplified with primers P5 and P6 using potato mt DNA as template. A fragment of the human mt genome (NC_012920) corresponding to the nad3 gene with its natural 100 flanking nucleotides was PCR-amplified with primers P7 and P8 (see Supplementary Table S1 for sequences). The DNA constructs were cloned into the pGEM-T-Easy vector (Promega, Madison, WI). The Riboprobe Kit (Promega, Madison WI, USA) was used to synthesize in vitro transcripts using either Sp6 or T7 polymerases under conditions previously described (25). To synthesize labeled in vitro transcripts, the conditions were identical except for the presence of [α-32P]UTP or of UTP Chromatide® Alexa Fluor® 488 (Molecular Probe, Eugene, OR).
RNA import assay into isolated mitochondria
Highly purified mitochondria were isolated from potato tubers as described previously (28). Import assays were carried out using tRNA import conditions essentially as described in ref. (26). Briefly, RNA was incubated with isolated mitochondria at 25°C for 20 min. Upon incubation, the reaction was treated with RNases to remove non imported RNA followed by RNA isolation. When indicated, purified pDHFR or pGFP (35 or 70 fmol) was added to the import medium. When necessary, upon RNA import into isolated mitochondria, mitoplasts were generated before performing the RNase treatment. To do so, an osmotic shock was used to break the outer mt membrane and obtain mitoplasts. Practically, upon import, mitochondria were centrifuged 5 min at 10 000g, resuspended in 10 mM potassium phosphate pH 7.5 (1 ml for 0.4 mg of proteins), kept on ice for 45 s and centrifuged again before resuspension in washing buffer [see isolation of mitochondria, (28)]. Quality and integrity of mitoplasts were classically checked by western blots or by exogenously added cytochrome c reduction test (14,29). Analysis of the RNA samples recovered upon import into isolated mitochondria depends on the RNA substrate used. Radioactively labeled tRNA or tRNA precursor samples were separated by electrophoresis through 7 M urea–15% (w/v) polyacrylamide gels. After electrophoresis, the gels were dried and either submitted to autoradiography or either exposed to a phosphorimager plate, allowing quantification with a Fla-7000 phosphorimager using the Image Gauge software (Fujifilm). Radioactively labeled mRNA samples were separated on agarose-formaldehyde gels. After electrophoresis, the RNAs were transferred from gel to nylon hybond N+ membrane using manufacturer’s instruction and the membrane was then either submitted to autoradiography or exposed to a phosphorimager plate as described above. Non-radioactive RNA samples were analyzed using RT–PCR or cRT–PCR as described in ref. (15). For RNA import assay into isolated human or rat mitochondria, mitochondria were first isolated according to ref. (16). An RNA import assays were conducted essentially as for RNA import into isolated potato mitochondria, but with the import medium described for protein import in ref. (30). Mitoplasts were generated and their integrity checked according to ref. (16). Protocol for preparation of yeast W303 mitochondria was the same as described in ref. (31). Nucleic acids import conditions were inspired from ref. (32) in the presence of 35 fmol purified pDHFR. To obtain mitoplasts after incubation, the protocol described by Glick was used (33). Briefly, mitochondria were resuspended in a KH2PO4/K2HPO4 pH 7.5 10 mM solution (1 ml for 0.4 mg of proteins) and kept on ice for 15 min. Osmolarity was reestablished before performing the RNase treatment by resuspending mitoplasts in purification buffer.
Protein import into isolated mitochondria
Import of 35S-labeled fusion protein pDHFR and pGFP was performed as previously described (34).
Microscopic observations
In vitro import of fluorescent tRNA into potato isolated mitochondria was observed using a Zeiss LSM 510 confocal microscope. The Alexa Fluor 488 UTP incorporated in tRNA molecules fluoresces at 561 nm when excited at 488 nm. When indicated, mitochondria were labeled with the vital fluorochrome MitoTrackerTM Orange (Molecular Probes) (final concentration of 0.5 pM in import buffer). Images were treated with the LSM 510 version 2.8 program and analyzed using Image J software.
RESULTS
Incorporation of tRNA into plant mitochondria using a protein shuttle system
In yeast and human, respectively, tRNALys and 5S rRNA mt import requires cytosolic factors. These factors play the role of carriers and allow the translocation of RNAs across the protein import pathway (22,35). Conversely, in plants, we previously showed that tRNAs can be translocated with a low efficiency into isolated mitochondria through the voltage-dependent anion channel (VDAC) and without the assistance of a cytosolic extract (26). However interestingly, we showed that although the pathways used in vitro for tRNA and protein mt import are different in higher plants, two major components of the protein import machinery (TOM20 and TOM40) are required for tRNA mt import (25). This led us to speculate that, even though tRNAs can enter plant mitochondria without any added factors, this import might be facilitated in the presence of an appropriate co-import factor.
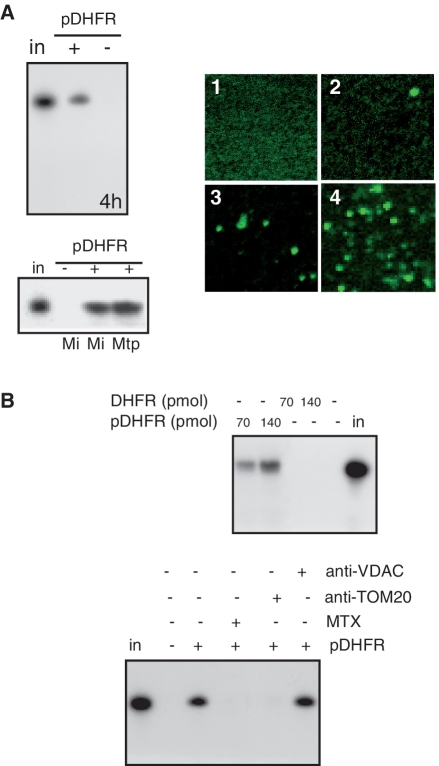
To test this hypothesis, the protein pDHFR was chosen as a candidate carrier. It consists on a chimeric protein made by the first 69 amino acids of subunit nine of the mt F0-ATPase of Neurospora crassa fused to the mouse DHFR. This chimeric protein was chosen as a co-import vector for two major reasons. First, it is efficiently targeted to yeast (36), plant and human mitochondria (Supplementary Figure S1a and S1c). Second, DHFR is a RNA binding protein able to bind its own mRNA in vivo (37) and we provide evidence that DHFR is also able to bind other RNAs nonspecifically in vitro (Supplementary Figure S2). As a first approach to validate our hypothesis, in vitro import assay of plant cytosolic tRNAAla into isolated potato mitochondria was performed as previously described (26), in the presence or not of pDHFR. Upon incubation, the reaction was treated with RNases to remove non imported RNA, followed by RNA isolation and gel analysis. As shown in Figure 1A (and Supplementary Figure S3a), when pDHFR is added to the import medium, tRNA mt import efficiency is considerably increased and up to 10% of the tRNA input can be retrieved into the organelle. In the absence of mitochondria or when mitochondria are treated with Triton X-100 prior to the RNAse treatment, no signal is detected (Supplementary Figure S4a and S4b), thus demonstrating that the detection of non-degraded RNA is not merely due to a protection by DHFR. Furthermore, DHFR, without pSu9 targeting sequence is not able to promote tRNA import into mitochondria (Figure 1B). Similarly, pGFP, a chimeric protein not able to bind RNAs (25) cannot promote RNA uptake (Supplementary Figure S3b) even though the protein can be imported into isolated mitochondria (Supplementary Figure S1b). An additional control experiment was the addition of a folate analogue, methotrexate, prior to the RNA import assay. By interacting with DHFR, methotrexate inhibits its import into mitochondria (38). We show here that addition of methotrexate also inhibits tRNA import (Figure 1C). The amount of incorporated RNA remained stable when mitochondria were first submitted to an osmotic shock to break the outer membranes after the import step and before the treatment with RNases. This indicates that the tRNA transcript has likely reached the mt matrix (Figure 1A). Using an independent technique with a fluorescent tRNA transcript, the effect of pDHFR on tRNA mt targeting and/or import can be directly observed by confocal microscopy (Figure 1A and Supplementary Figure S4c). In the absence of pDHFR, very little fluorescence is associated with mitochondria. By contrast, most mitochondria incubated for 20 min with pDHFR shows fluorescence, again indicating that pDHFR facilitates the targeting of the RNA to mitochondria. To assess the quality of the purification step, mitochondria were stained with the mt marker, mitotracker (Supplementary Figure S4). Finally, while in the absence of protein factors, tRNAs are translocated through the outer mt membrane via VDAC (25), in the presence of pDHFR, no inhibition of tRNA mt import by VDAC antibodies is observed. This demonstrates that this channel is not associated with the RNA import process when pDHFR is added to the import medium. In contrast, the use of antibodies against TOM20, the major receptor of the protein translocase complex in the outer mt membrane still inhibits the internalization of the RNA into mitochondria (Figure 1B). Altogether, our data suggest that tRNAs can be co-imported with pDHFR into isolated plant mitochondria.
Figure 1.
pDHFR increases tRNA import into isolated potato mitochondria. (A) On the left: 32P-labeled in vitro-transcribed tRNAAla was incubated with isolated potato mitochondria (25) in the absence (−) or presence (+) of 35 pmol of pDHFR. Following standard import conditions, RNase treatment was performed either on mitochondria (Mi) or on mitoplasts (Mtp). RNAs were fractionated on a denaturing polyacrylamide gel. Equivalent loading was checked by ethidium bromide staining prior to autoradiography visualization (4 h exposure). On the right: visualization under confocal microscope of Alexa Fluor-labeled in vitro transcribed tRNAAla incubated with isolated potato mitochondria in the absence (2) or presence (3 and 4) of 35 pmol of pDHFR. Visualization was performed after 5 min (3) or 25 min (2 and 4) of incubation. Alexa Fluor-labeled in vitro transcribed tRNAAla in import medium without mitochondria was used as a control (1). (B) 32P-labeled in vitro-transcribed tRNAAla was incubated with isolated potato mitochondria in the absence (−) or presence (70 and 140 pmol) of pDHFR or DHFR. Methotrexate (MTX, 50 nM), or antibodies against VDAC or TOM20 (25) were added to the standard import mixture and incubated for 10 min before adding the labeled tRNAAla. in: 10% of input RNA (2 fmol).
Processing of larch mt tRNAHis precursor into potato mitochondria
As the interaction between pDHFR and RNAs is not specific in vitro, we then wanted to test whether longer RNAs other than tRNAs can be imported into isolated potato mitochondria using the pDHFR as a shuttle. We chose a 250-nt-long precursor RNA of the larch mt tRNAHis. Larch mt tRNAHis contains three cytidine to uridine editing sites that are required for its correct processing (39) (Figure 2A). Both the radiolabeled edited or unedited forms of the transcript were incubated with potato mitochondria in the presence or not of pDHFR. Upon incubation, the edited form of the tRNAHis precursor is successfully incorporated into isolated mitochondria and processed in a mature-size tRNA. However, no RNA product is visible when the non-edited RNA precursor is incubated with isolated mitochondria (Figure 2B). From the incubation of the RNA with isolated mitochondria to the extraction of mt RNA, there is a time span of ∼80 min. The non-edited RNA, not recognized by mt processing enzymes, is a naked molecule that should be directly and rapidly submitted to degradation by the RNases involved in quality control and surveillance in mitochondria. This is in agreement with our previous data showing that only the edited form of the tRNA precursor is processed in organello while the non-edited RNA precursor is degraded (15).
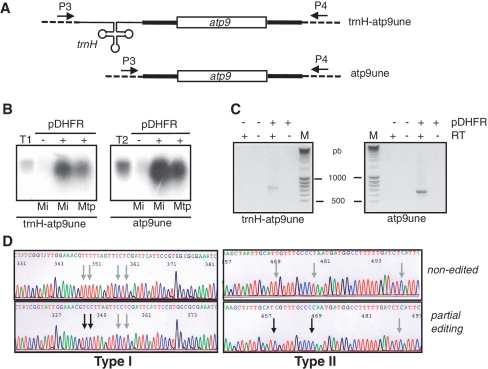
Figure 2.
Processing of larch tRNAHis precursor upon RNA uptake using the pDHFR protein shuttle into isolated potato mitochondria. (A) Schematic representation of the larch mt tRNA precursor transcript identified in (39). The −1 and +1 positions sensitive to plant mt RNase P cleavage (42) are indicated. Primers P1 and P2 used for circularized RT–PCR (cRT–PCR) analysis are indicated (see Supplementary Table S1 for sequences). (B) 32P-labeled in vitro-transcribed larch mt edited (ptrnH-ed) or unedited (ptrnH-uned) precursor was incubated for 30 min with isolated potato mitochondria under standard conditions in the absence (−) or presence (+) of 35 pmol of pDHFR. Positions of the precursor and mature larch mt tRNAHis are indicated by arrows. Schemes of these RNAs are on the left side of the polyacrylamide gel used to analyze the processing products generated after import. in: input edited (2.5 fmol) or non-edited (5 fmol) RNA. T, 32P-labeled in vitro-transcribed tRNAAla as a size marker. (C) Schematic representation of the import and processing of non-radioactive larch mt tRNA precursor into isolated potato mitochondria. Upon RNA uptake and processing, RNA molecules were amplified by cRT–PCR using primers P1 and P2 and in the absence (−) or presence (+) of reverse transcriptase and analyzed on an agarose gel (the negative image of the ethidium-stained gel is presented). M, migration of a DNA ladder. (D) The 5′ and 3′ termini of 14 clones determined by the sequencing analysis of the PCR product seen in (C) are presented. Nucleotides present in the mature tRNA are written in blue and nucleotides corresponding to unprocessed 5′- or 3′-termini are in black. The RNase P potential cleavage sites are indicated as −1 and +1 positions. The extra CCA sequence found in five clones is written in red and bold.
To confirm the uptake and processing of the edited tRNAHis precursor into mitochondria, an mt RNA import assay was performed with the non-radiolabeled transcript. Following import, mt RNAs were extracted and further analyzed by circularized RT–PCR (cRT–PCR). A specific reverse transcriptase-dependent product of ∼80 nt was amplified (Figure 2C) and cloned. Out of 14 sequences (Figure 2D), four sequences (clones 10–13) correspond to fully processed tRNAHis, each harboring the non-encoded CCA sequence at the 3′-end and a 5′ extremity with the additional G residue (called G−1), a hallmark of tRNAHis molecules (40). These processing steps are carried out by the mt processing enzymes, RNase P, RNase Z and tRNA nucleotidyl-transferase located in the matrix. This demonstrates that the tRNA precursor transcript was imported in vitro into the matrix compartment of isolated potato mitochondria. Furthermore, it was previously reported that the 5′-end cleavage achieved by RNase P precedes 3′-end processing by RNase Z (41). Although most of our data are consistent with this, one sequence (clone 1) shows a tRNA with a fully processed 3′-end but a non-mature 5′-end. This indicates that cleavage by RNase Z and the tRNA nucleotidyl-transferase activity can precede cleavage by RNase P at the 5′-end. So far, based on in vitro analysis, it was assumed that the 3′-end maturation occurs via a precise endonucleolytic cut immediately after the discriminator nucleotide at position 73 (41). Here, in 9 sequences, the 3′-endonucleolytic cut is observed from 1 to 6 nts downstream the discriminator nucleotide. Thus, to subsequently remove these remaining nucleotides, an exonuclease not identified so far is very likely required. This is reminiscent to the situation found in prokaryotes (41), but not described in plant mitochondria to date. Finally, in the clones analyzed here, two types of 5′-termini are observed: either the tRNA ends at G−1 (in 10 sequences) or ends at G+1 (in four sequences). This is in full agreement with the fact that the extra G−1 residue is generated via two pathways in plant mitochondria: either the G−1 is genome-encoded and retained during maturation or it is post-transcriptionally added by a tRNAHis guanylyl transferase activity (42). As a whole, the possibility of incorporating a 250-nt-long exogenous RNA into isolated mitochondria provides new insights about the processing of plant mt tRNAs in organello and validates the efficiency of the pDHFR as a shuttle system.
Incorporation of the full-length atp9 mRNA into potato mitochondria
As described above, the presence of pDHFR allowed the efficient incorporation of tRNAs or tRNA precursors into isolated mitochondria. We then investigated the capacity of the protein shuttle to drive full-length mRNA into isolated potato mitochondria. The potato mt atp9 mRNA was chosen to test this hypothesis (43). So far, the two transcripts imported into mitochondria using pDHFR contain a tRNA sequence. To verify whether the protein shuttle developed here is dependent or not on a tRNA structure, the non-edited atp9 mRNA was fused or not to the edited version of larch mt tRNAHis precursor RNA (Figure 3A and Supplementary Figure S5) and both radiolabeled RNA substrates were used for RNA import assay. As shown on Figure 3B and C, the 815-nt-long trnH-atp9 RNA and the atp9 mRNA (651-nt long) are imported into isolated mitochondria only when pDHFR is added to the import medium. Furthermore, when mitoplasts are generated after the RNA import step, more than 50% of the incorporated RNA is resistant to RNase treatment showing that the mRNA transcript reached the mt matrix (Figure 3B). Compared to tRNA uptake, where 10% of the input is classically retrieved into mitochondria, we were not able to incorporate >5% of mRNA input into the organelle. Although no measurement of Kd was performed, we observed similar RNA binding capacity of the various RNA substrates we tested using Northwestern experiments (e.g. Supplementary Figure S2b). These results suggest that the import efficiency is limited by the size of the RNA molecule rather than by the binding capacity to DHFR. Finally, the editing status of atp9 mRNA was analyzed. Following the internalization of the non-radiolabeled unedited form of atp9 mRNA, RT–PCR was performed on mt RNAs and the 651-nt-long specific reverse transcriptase-dependent amplicon (Figure 3C) was cloned and sequenced. Out of 15 sequenced clones, six were found partially edited (Figure 3D). As the editing machinery is located in the mt matrix, this further supports the data presented above. Altogether, this shows that the mt import system developed here does not depend on a tRNA structure and allows the efficient incorporation of full-length mRNA into the organelle.
Figure 3.
Editing of atp9 non-edited mRNA upon RNA uptake using the pDHFR protein shuttle into isolated potato mitochondria. (A) Schematic representation of the atp9 RNA used for mt import. Larch mt edited tRNAHis (trnH) precursor RNA (cloverleaf and thin lines) is fused or not to the full-length potato mt atp9 mRNA (open box: coding sequence; bold lines: 5′- and 3′-UTR). The nucleotide sequence is presented in Supplementary Figure S5. Primers P3 and P4 used for RT–PCR analysis and indicated with arrows (see Supplemental Table S1 for sequences) are located on the sequence corresponding to the pGEM-T Easy region (dashed lines) remaining upon cloning and transcription. (B) 32P-labeled RNAs corresponding to either the non-edited atp9 version of the trnH-atp9 (trnH-atp9une) or of the atp9 (atp9une) were incubated in the absence (−) or presence (+) of pDHFR. Upon incubation, nucleic acids were extracted from mitochondria (Mi) or from mitoplasts (Mtp) post-treated with RNases. T1 and T2, corresponding input RNAs (1 and 2.5 fmol, respectively). (C) Similar experiments to those depicted in (B) but with non-radioactive RNAs. Imported RNAs were then analyzed by RT–PCR using the primers P3/P4, specific to exogenous transcripts and in the absence (−) or presence (+) of reverse transcriptase. Negative images of ethidium-stained gels are presented. Fragment sizes were determined with a DNA ladder (M). (D) DNA sequence of sites partially edited upon import of atp9 mRNA non-edited version into isolated potato mitochondria. The non-edited version is compared to partially edited sequences (partial editing) obtained upon RNA uptake. Non-edited sites are indicated with gray arrows, the four edited sites with black arrows. Two types (type I and II) of partially edited sequences were found. Among the 15 sequences analyzed, two are of type I, four of type II and the other are non-edited.
Import of foreign RNAs into isolated mitochondria of other organisms
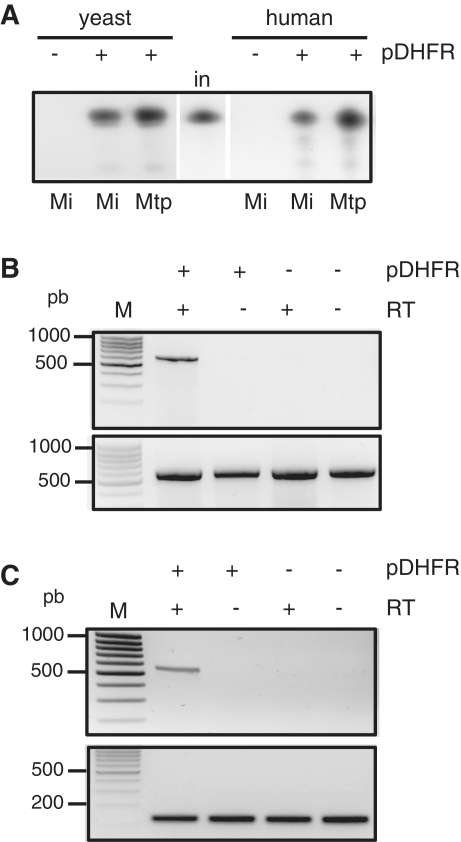
The yeast tRNALys and the human 5S rRNA specifically interact with the precursor of yeast mt lysyl-tRNA synthetase and the human mt thiosulfate sulfurtransferase (rhodanese) respectively. Then these RNAs are co-imported with the proteins across the mt protein import channel (22,35). In both cases, the interaction and import depend on specific import signals. We therefore wondered whether the pDHFR shuttle allow the incorporation of RNAs into isolated yeast or mammalian mitochondria, independently on their sequence and/or structure. Plant cytosolic tRNAAla is not naturally present in yeast or human mitochondria and do not contain any of the RNA import motifs identified so far. Mitochondria isolated from yeast or human cells were incubated with radiolabelled tRNAAla transcript in the presence or not of pDHFR. As shown on Figure 4A, plant tRNAAla is not imported in vitro into mitochondria of yeast or human cells in the absence of any added factor but is efficiently incorporated into mitochondria (up to 5% of the input) when pDHFR is present in the import medium. This result shows that the mt import system based on the pDHFR carrier can be extended to organisms other than plants. Yeast mitochondria can be transformed while mammalian mitochondria cannot. Hence, we then focused our study on mammalian cells. We thus tested the capability to address the potato mt atp9 mRNA into isolated human mitochondria. Upon import assay with mitochondria followed by RNase post-treatment of the organelles, total mt RNAs were extracted. The atp9 mRNA was detected by RT–PCR only when the protein shuttle was added to the import medium (Figure 4B). Finally in a similar way, a 630-nt-long transcript containing the entire human mt nad3 mRNA was successfully imported into isolated rat mitochondria using the same approach (Figure 4C).
Figure 4.
RNA import into isolated yeast and mammalian mitochondria using the pDHFR shuttle. (A) 32P-labeled in vitro-transcribed plant tRNAAla was incubated with isolated yeast or human mitochondria in the absence (−) or presence (+) of 35 pmol of pDHFR (pDHFR). Following standard import conditions, RNase treatment was performed either on mitochondria (Mi) or on mitoplasts (Mtp). RNAs were fractionated on a denaturing polyacrylamide gel. Equivalent loading was checked by ethidium bromide staining prior to autoradiography visualization. in: 10% of input RNA (2 fmol). (B) Non-radioactive potato atp9 mRNA transcript was incubated with human mitochondria in the absence (−) or presence (+) of pDHFR. Upon incubation, nucleic acids were extracted from mitochondria post-treated with RNases. Upper panel: imported RNAs were analyzed by RT–PCR using the primers P3/P4 specific to exogenous transcripts (see Figure 3A) and in the absence (−) or presence (+) of reverse transcriptase. Lower panel: as a control, the region corresponding to the nad3 gene of the human mt genome was PCR amplified using the primers P7/P8. Negative images of ethidium-stained gels are presented. Fragment sizes were determined with a DNA ladder (M). (C) Non-radioactive human nad3 mRNA transcript was incubated with rat mitochondria in the absence (−) or presence (+) of pDHFR. Upon incubation, nucleic acids were extracted from mitochondria post-treated with RNases. Upper panel: imported RNAs were analyzed by RT–PCR using the primers P7/P8 specific to human nad3 mRNA and in the absence (−) or presence (+) of reverse transcriptase. Lower panel: as a control, a region corresponding to the trnR gene of the rat mt genome was PCR amplified using primers P9/P10. Negative images of ethidium-stained gels are presented. Fragment sizes were determined with a DNA ladder (M).
DISCUSSION
Altogether, the data provided in our article show that the pDHFR protein can be used as an efficient shuttle to carry RNA into mitochondria. Our data present a simple and straightforward system to incorporate any RNA of interest into isolated mitochondria. One of the first reports of nucleic acid import through the mt membranes also involved the protein import machinery. In that case, the C-terminus of an mt precursor protein was covalently linked to a short oligonucleotide, allowing its incorporation into yeast mitochondria (44). This strategy is limited by the fact that the nucleic acid is still attached to the protein once in mitochondria and no experiment showing the functionality of the DNA was provided. Electroporation has also been successfully used to incorporate nucleic acids into isolated human or plant mitochondria and the functionality of the incorporated DNA or RNA was demonstrated (8,45,46). Yet, a major drawback in using electroporation is that most mitochondria lose their integrity, thus requiring large amounts of organelles. Here, our non-damaging method uses ten times less material and represents a valuable alternative for the in organello studies of mt RNA biogenesis. Interestingly, the natural competence of yeast or human mitochondria to import RNAs involve carrier proteins such as pre-LysRS or rhodanese that specifically drive tRNALys or 5S rRNA into mitochondria using the protein import machinery (22,35). In both cases, RNA derivatives could be introduced into mitochondria in order to study the sequence and structure requirements of these imported RNAs. Using pDHFR as an mt RNA vector shows no constraints in term of sequence or structure motifs required for the RNA to be imported, because the protein likely has the capacity to bind any RNA in vitro. Importantly, RNAs of up to 800 nt in size were successfully imported into mitochondria showing that the shuttle system is not restricted to short RNAs. Whether a size limitation in the mt import capacity exists will need to be further examined. This system can be used with mitochondria isolated from various organisms where mt transformation is still not possible, such as mitochondria of higher plants or human. Mitochondrial processing events such as cis- or trans-splicing or editing of mRNA in plant mitochondria or processing of overlapping transcripts in mammalian mitochondria could be studied in more details using this in vitro system. Furthermore, as mostly nothing is known on mt translation in plant and human mitochondria, the possibility to address full-length mRNA into mitochondria opens the door to their potential translation and thus to the study of this important process. Finally, a similar in vivo approach could be designed. Using as a protein shuttle an RNA binding protein recognizing a specific motif is required. For instance, an interesting candidate is the coat protein of the MS2 phage that is able to specifically recognize a stem loop motif. This recognition is at the basis of the ‘green RNA technology’ (47). If the MS2 protein fused to an mt targeting sequence and the RNA of interest fused to the stem loop recognition motif are both expressed in transgenic cells, then we can expect the targeting of the RNA into mitochondria. Thus, our approach enlarges the set of tools for studying mt genetics.
Although this system is artificial, it raises interesting questions on the natural RNA mt import mechanisms. We previously showed in vitro that tRNAs can enter plant mitochondria at a low level without any added protein factors (26). Strikingly, the data presented here support the idea that, as in yeast and human, plant can import RNA thanks to a protein carrier and components of the protein import machinery in vitro. Understanding whether plant mitochondria use carrier proteins and whether the two options (with and without a carrier protein) occur in vivo will require further studies that are beyond the scope of this article. Finally it is usually admitted that RNA mt import has a polyphyletic origin with various independent mechanisms (7,48). Components of the protein import machinery are involved in yeast, human and in plant mt RNA import. Carrier proteins were found for yeast and human RNA import and our data suggest that plants may also have the possibility to use carrier proteins for the natural import of tRNAs into mitochondria. Although such aspects were not found so far in protozoans, we can wonder whether, contrarily to what is generally believed, there is a common ancestral process at the origin of the present-day RNA mt import and as now hypothesized in recent reviews (18), there might be a strong connection between protein and RNA mt import.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Supported by the Centre National de la Recherche Scientifique, the University of Strasbourg, the Agence National pour la Recherche Scientifique (ANR-09-BLAN-0240-01); Fondi di Ateneo dell’Universita degli studi di Bari. French Ministère Délégué à l’Enseignement Supérieur et à la Recherche, fellowship (to F.S.); Università degli Studi di Bari ‘Aldo Moro’, 2-year research grant (to A.P.). Funding for open access charges: IBMP, CNRS.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank D. Rapaport for the gift of pDHFR construct; M. Messmer and H. Lange for critical reading of the manuscript.
REFERENCES
- 1.Havey MJ. The use of cytoplasmic male sterility for hybrid seed production. In: Daniell H, Chase C, editors. Molecular Biology and Biotechnology of Plant Organelles. Dordrecht: Springer; 2004. pp. 623–634. [Google Scholar]
- 2.Wallace DC. Mitochondrial DNA mutations in disease and aging. Environ. Mol. Mutagen. 2010;51:440–450. doi: 10.1002/em.20586. [DOI] [PubMed] [Google Scholar]
- 3.Kyriakouli DS, Boesch P, Taylor RW, Lightowlers RN. Progress and prospects: gene therapy for mitochondrial DNA disease. Gene Ther. 2008;15:1017–1023. doi: 10.1038/gt.2008.91. [DOI] [PubMed] [Google Scholar]
- 4.Fox TD, Sanford JC, McMullin TW. Plasmids can stably transform yeast mitochondria lacking endogenous mtDNA. Proc. Natl Acad. Sci. USA. 1988;85:7288–7292. doi: 10.1073/pnas.85.19.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Remacle C, Cardol P, Coosemans N, Gaisne M, Bonnefoy N. High-efficiency biolistic transformation of Chlamydomonas mitochondria can be used to insert mutations in complex I genes. Proc. Natl Acad. Sci. USA. 2006;103:4771–4776. doi: 10.1073/pnas.0509501103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rustin PH, Jacobs HT, Dietrich A, Lightowlers RN, Tarassov I, Corral-Debrinski IM. Targeting allotopic material to the mitochondrial compartment: new tools for better understanding mitochondrial physiology and prospect for therapy. Med. Sci. 2007;23:519–525. doi: 10.1051/medsci/2007235519. [DOI] [PubMed] [Google Scholar]
- 7.Sieber F, Duchêne AM, Maréchal-Drouard L. Mitochondrial RNA import: from diversity of natural mechanisms to potential applications. Int. Rev. Cell Mol. Biol. 2011;287:145–190. doi: 10.1016/B978-0-12-386043-9.00004-9. [DOI] [PubMed] [Google Scholar]
- 8.Collombet JM, Wheeler VC, Vogel F, Coutelle C. Introduction of plasmid DNA into isolated mitochondria by electroporation. A novel approach toward gene correction for mitochondrial disorders. J. Biol. Chem. 1997;272:5342–5347. doi: 10.1074/jbc.272.8.5342. [DOI] [PubMed] [Google Scholar]
- 9.Estévez AM, Thiemann OH, Alfonzo JD, Simpson L. T7 RNA polymerase-driven transcription in mitochondria of Leishmania tarentolae and Trypanosoma brucei. Mol. Biochem. Parasitol. 1999;103:251–259. doi: 10.1016/s0166-6851(99)00139-5. [DOI] [PubMed] [Google Scholar]
- 10.Castandet B, Choury D, Bégu D, Jordana X, Araya A. Intron RNA editing is essential for splicing in plant mitochondria. Nucleic Acids Res. 2010;38:7112–7120. doi: 10.1093/nar/gkq591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Farre JC, Leon G, Jordana X, Araya A. Cis Recognition elements in plant mitochondrion RNA editing. Mol. Cell Biol. 2001;21:6731–6737. doi: 10.1128/MCB.21.20.6731-6737.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Staudinger M, Bolle N, Kempken F. Mitochondrial electroporation and in organello RNA editing of chimeric atp6 transcripts. Mol. Genet. Genomics. 2005;273:130–136. doi: 10.1007/s00438-005-1117-x. [DOI] [PubMed] [Google Scholar]
- 13.Hinrichsen I, Bolle N, Paun L, Kempken F. RNA processing in plant mitochondria is independent of transcription. Plant Mol. Biol. 2009;70:663–668. doi: 10.1007/s11103-009-9498-6. [DOI] [PubMed] [Google Scholar]
- 14.Koulintchenko M, Konstantinov Y, Dietrich A. Plant mitochondria actively import DNA via the permeability transition pore complex. EMBO J. 2003;22:1245–1254. doi: 10.1093/emboj/cdg128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Placido A, Gagliardi D, Gallerani R, Grienenberger JM, Maréchal-Drouard L. Fate of a larch unedited tRNA precursor expressed in potato mitochondria. J. Biol. Chem. 2005;280:33573–33579. doi: 10.1074/jbc.M505269200. [DOI] [PubMed] [Google Scholar]
- 16.Koulintchenko M, Temperley RJ, Mason PA, Dietrich A, Lightowlers RN. Natural competence of mammalian mitochondria allows the molecular investigation of mitochondrial gene expression. Hum. Mol. Genet. 2006;15:143–154. doi: 10.1093/hmg/ddi435. [DOI] [PubMed] [Google Scholar]
- 17.Weber-Lotfi F, Ibrahim N, Boesch P, Cosset A, Konstantinov Y, Lightowlers N, Dietrich RNA. Developing a genetic approach to investigate the mechanism of mitochondrial competence for DNA import. Biochim. Biophys. Acta. 2009;1787:320–327. doi: 10.1016/j.bbabio.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lithgow T, Schneider A. Evolution of macromolecular import pathways in mitochondria, hydrogenosomes and mitosomes. Philos. Trans. R. Soc. Lond., B, Biol Sci. 2010;365:799–817. doi: 10.1098/rstb.2009.0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolesnikova OA, Entelis NS, Jacquin-Becker C, Goltzene F, Chrzanowska-Lightowlers ZM, Lightowlers RN, Martin RP, Tarassov I. Nuclear DNA-encoded tRNAs targeted into mitochondria can rescue a mitochondrial DNA mutation associated with the MERRF syndrome in cultured human cells. Hum. Mol. Genet. 2004;13:2519–2534. doi: 10.1093/hmg/ddh267. [DOI] [PubMed] [Google Scholar]
- 20.Mahata B, Mukherjee S, Mishra S, Bandyopadhyay A, Adhya S. Functional delivery of a cytosolic tRNA into mutant mitochondria of human cells. Science. 2006;314:471–474. doi: 10.1126/science.1129754. [DOI] [PubMed] [Google Scholar]
- 21.Magalhaes PJ, Andreu AL, Schon EA. Evidence for the presence of 5S rRNA in mammalian mitochondria. Mol. Biol. Cell. 1998;9:2375–2382. doi: 10.1091/mbc.9.9.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smirnov A, Comte C, Mager-Heckel A-M, Addis V, Krasheninnikov IA, Martin RP, Entelis NS, Tarassov I. Mitochondrial enzyme rhodanese is essential for 5S ribosomal RNA import into human mitochondria. J. Biol. Chem. 2010;285:30792–30803. doi: 10.1074/jbc.M110.151183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang G, Chen HW, Oktay Y, Zhang J, Allen EL, Smith GM, Fan KC, Hong JS, French SW, McCaffery JM, et al. PNPASE regulates RNA import into mitochondria. Cell. 2010;142:456–467. doi: 10.1016/j.cell.2010.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rapaport D, Neupert W. Biogenesis of Tom40, core component of the TOM complex of mitochondria. J. Cell Biol. 1999;146:321–331. doi: 10.1083/jcb.146.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salinas T, Duchêne AM, Delage L, Nilsson S, Glaser E, Zaepfel M, Maréchal-Drouard L. The voltage-dependent anion channel, a major component of the tRNA import machinery in plant mitochondria. Proc. Natl Acad. Sci. USA. 2006;103:18362–18367. doi: 10.1073/pnas.0606449103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delage L, Dietrich A, Cosset A, Maréchal-Drouard L. In vitro import of a nuclearly encoded tRNA into mitochondria of Solanum tuberosum. Mol. Cell. Biol. 2003;23:4000–4012. doi: 10.1128/MCB.23.11.4000-4012.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dell'Orto P, Moenne A, Graves PV, Jordana X. The potato mitochondrial ATP synthase subunit 9: gene structure, RNA editing and partial protein sequence. Plant Sci. 1993;88:45–53. [Google Scholar]
- 28.Fey J, Vermel M, Grienenberger J, Maréchal-Drouard L, Gualberto JM. Characterization of a plant mitochondrial active chromosome. FEBS Lett. 1999;458:124–128. doi: 10.1016/s0014-5793(99)01140-0. [DOI] [PubMed] [Google Scholar]
- 29.Douce R, Christensen EL, Bonner WD. Preparation of intact plant mitochondria. Biochim. Biophys. Acta. 1972;275:148–160. doi: 10.1016/0005-2728(72)90035-7. [DOI] [PubMed] [Google Scholar]
- 30.Messmer M, Florentz C, Schwenzer H, Scheper GC, van der Knaap S, Maréchal-Drouard M, Sissler LM. A human pathology-related mutation prevents import of an aminoacyl-tRNA synthetase into mitochondria. Biochem. J. 2011;433:441–446. doi: 10.1042/BJ20101902. [DOI] [PubMed] [Google Scholar]
- 31.Daum G, Bohni P, Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxydase are located in the intermembrane space of mitochondria. J. Biol. Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- 32.Tarassov I, Entelis N, Martin RP. Mitochondrial import of a cytoplasmic lysine-tRNA in yeast is mediated by cooperation of cytoplasmic and mitochondrial lysyl-tRNA synthetases. EMBO J. 1995;14:3461–3471. doi: 10.1002/j.1460-2075.1995.tb07352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glick BS. Pathways and energetics of mitochondrial protein import in Saccharomyces cerevisiae. Meth. Enzymol. 1995;260:224–231. doi: 10.1016/0076-6879(95)60140-6. [DOI] [PubMed] [Google Scholar]
- 34.Duchêne AM, Giritch A, Hoffmann B, Cognat V, Lancelin D, Peeters NM, Zaepfel M, Maréchal-Drouard L, Small ID. Dual targeting is the rule for organellar aminoacyl-tRNA synthetases in Arabidopsis thaliana. Proc Natl Acad. Sci. USA. 2005;102:16484–16489. doi: 10.1073/pnas.0504682102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tarassov I, Kamenski P, Kolesnikova O, Karicheva O, Martin RP, Krasheninnikov IA, Entelis N. Import of nuclear DNA-encoded RNAs into mitochondria and mitochondrial translation. Cell Cycle. 2007;6:2473–2477. doi: 10.4161/cc.6.20.4783. [DOI] [PubMed] [Google Scholar]
- 36.Rapaport D, Neupert W, Lill R. Mitochondrial protein import. Tom40 plays a major role in targeting and translocation of preproteins by forming a specific binding site for the presequence. J. Biol. Chem. 1997;272:18725–18731. doi: 10.1074/jbc.272.30.18725. [DOI] [PubMed] [Google Scholar]
- 37.Tai N, Ding Y, Schmitz JC, Chu E. Identification of critical amino acid residues on human dihydrofolate reductase protein that mediate RNA recognition. Nucleic Acids Res. 2002;30:4481–4488. doi: 10.1093/nar/gkf562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eilers M, Schatz G. Binding of a specific ligand inhibits import of a purified precursor protein into mitochondria. Nature. 1986;322:228–232. doi: 10.1038/322228a0. [DOI] [PubMed] [Google Scholar]
- 39.Maréchal-Drouard L, Kumar R, Remacle C, Small I. RNA editing of larch mitochondrial tRNA(His) precursors is a prerequisite for processing. Nucleic Acids Res. 1996;24:3229–3234. doi: 10.1093/nar/24.16.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marck C, Grosjean H. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA. 2002;8:1189–1232. doi: 10.1017/s1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morl M, Marchfelder A. The final cut. The importance of tRNA 3'-processing. EMBO Rep. 2001;2:17–20. doi: 10.1093/embo-reports/kve006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Placido A, Sieber F, Gobert A, Gallerani R, Giegé P, Maréchal-Drouard L. Plant mitochondria use two pathways for the biogenesis of tRNAHis. Nucleic Acids Res. 2010;38:7711–7717. doi: 10.1093/nar/gkq646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perrin R, Meyer EH, Zaepfel M, Kim YJ, Mache R, Grienenberger JM, Gualberto JM, Gagliardi D. Two exoribonucleases act sequentially to process mature 3'-ends of atp9 mRNAs in Arabidopsis mitochondria. J. Biol. Chem. 2004;279:25440–25446. doi: 10.1074/jbc.M401182200. [DOI] [PubMed] [Google Scholar]
- 44.Vestweber D, Schatz G. DNA-protein conjugates can enter mitochondria via the protein import pathway. Nature. 1989;338:170–172. doi: 10.1038/338170a0. [DOI] [PubMed] [Google Scholar]
- 45.Yoon YG, Koob MD. Efficient cloning and engineering of entire mitochondrial genomes in Escherichia coli and transfer to transcriptionally active mitochondria. Nucleic Acids Res. 2003;31:1407–1415. doi: 10.1093/nar/gkg228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Farre JC, Araya A. Gene expression in isolated plant mitochondria: high fidelity of transcription, splicing and editing of a transgene product in electroporated organelles. Nucleic Acids Res. 2001;29:2484–2491. doi: 10.1093/nar/29.12.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beach DL, Salmon ED, Bloom K. Localization and anchoring of mRNA in budding yeast. Curr. Biol. 1999;9:569–578. doi: 10.1016/s0960-9822(99)80260-7. [DOI] [PubMed] [Google Scholar]
- 48.Alfonzo JD, Söll D. Mitochondrial tRNA import-the challenge to understand has just begun. Biol. Chem. 2009;390:717–722. doi: 10.1515/BC.2009.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.