Abstract
Most of the eukaryotic genome is transcribed, yielding a complex network of transcripts that includes tens of thousands of long noncoding RNAs with little or no protein-coding capacity. Although the vast majority of long noncoding RNAs have yet to be characterized thoroughly, many of these transcripts are unlikely to represent transcriptional “noise” as a significant number have been shown to exhibit cell type-specific expression, localization to subcellular compartments, and association with human diseases. Here, we highlight recent efforts that have identified a myriad of molecular functions for long noncoding RNAs. In some cases, it appears that simply the act of noncoding RNA transcription is sufficient to positively or negatively affect the expression of nearby genes. However, in many cases, the long noncoding RNAs themselves serve key regulatory roles that were assumed previously to be reserved for proteins, such as regulating the activity or localization of proteins and serving as organizational frameworks of subcellular structures. In addition, many long noncoding RNAs are processed to yield small RNAs or, conversely, modulate how other RNAs are processed. It is thus becoming increasingly clear that long noncoding RNAs can function via numerous paradigms and are key regulatory molecules in the cell.
Keywords: ncRNA, transcriptome, gene expression, transcriptional regulation, small RNAs, structural RNAs
The sequencing of the human genome provided quite a surprise to many when it was determined that there are only ∼20,000 protein-coding genes, representing <2% of the total genomic sequence (International Human Genome Sequencing Consortium 2004). Since other less complex eukaryotes like the nematode Caenorhabditis elegans have a very similar number of protein-coding genes, it quickly became clear that the developmental and physiological complexity of humans probably cannot be explained solely by the number of protein-coding genes. Alternative pre-mRNA splicing of protein-coding transcripts as well as post-translational modifications of proteins increase the diversity and functionality of the proteome, likely explaining part of this increased complexity. In addition, there has been an explosion of research addressing possible functional roles for the other 98% of the human genome that does not encode proteins. Rather unexpectedly, transcription is not limited to protein-coding regions, but is instead pervasive throughout the mammalian genome as demonstrated by large-scale cDNA cloning projects (Carninci et al. 2005; Katayama et al. 2005) and genomic tiling arrays (Bertone et al. 2004; Cheng et al. 2005; Birney et al. 2007; Kapranov et al. 2007a). In fact, >90% of the human genome is likely to be transcribed (Birney et al. 2007), yielding a complex network of overlapping transcripts that includes tens of thousands of long RNAs with little or no protein-coding capacity (for review, see Kapranov et al. 2007b).
There is still some debate as to whether this pervasive transcription represents largely useless transcription (transcriptional “noise”) (Wang et al. 2004; Struhl 2007; Ebisuya et al. 2008) or if these noncoding RNAs (ncRNAs) have functions that simply have not yet been identified (for review, see Mattick 2004). Considering that it has long been known that numerous noncoding transcripts—such as transfer RNAs, ribosomal RNAs, and spliceosomal RNAs—are critical components of many cellular machines, it seems highly likely that additional ncRNAs play key regulatory and functional roles. Supporting the biological relevance of these transcripts, multiple studies have shown that significant numbers of long ncRNAs are regulated during development (Blackshaw et al. 2004; Rinn et al. 2007; Dinger et al. 2008), exhibit cell type-specific expression (Ravasi et al. 2006; Mercer et al. 2008), localize to specific subcellular compartments (Hutchinson et al. 2007; Sone et al. 2007; Clemson et al. 2009; Sasaki et al. 2009; Sunwoo et al. 2009), and are associated with human diseases (for review, see Costa 2005; Szymanski et al. 2005; Prasanth and Spector 2007). In addition, evidence for evolutionary selection within some long ncRNAs has been found (Pollard et al. 2006; Pheasant and Mattick 2007; Ponjavic et al. 2007; Guttman et al. 2009).
In this review, we highlight recent studies that have revealed how long ncRNAs can function on the molecular level (Fig. 1). Although the functions of only a limited number of long noncoding transcripts have been identified, numerous paradigms are beginning to emerge. We first highlight examples in which simply the act of ncRNA transcription is sufficient to regulate the expression of nearby protein-coding genes. We then highlight how long ncRNAs themselves serve key regulatory roles, with a specific focus on functional paradigms. ncRNAs involved in dosage compensation in mammals or Drosophila will be mentioned only briefly, as these have been reviewed thoroughly elsewhere (Deng and Meller 2006; Masui and Heard 2006; Payer and Lee 2008).
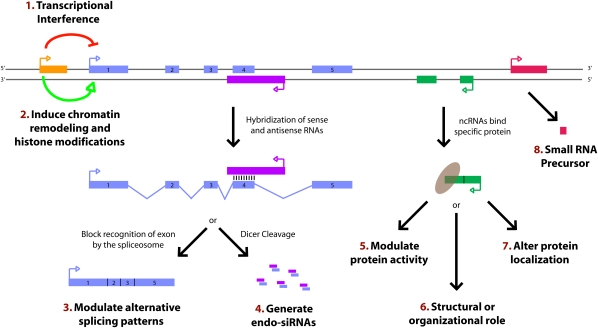
Figure 1.
Paradigms for how long ncRNAs function. Recent studies have identified a variety of regulatory paradigms for how long ncRNAs function, many of which are highlighted here. Transcription from an upstream noncoding promoter (orange) can negatively (1) or positively (2) affect expression of the downstream gene (blue) by inhibiting RNA polymerase II recruitment or inducing chromatin remodeling, respectively. (3) An antisense transcript (purple) is able to hybridize to the overlapping sense transcript (blue) and block recognition of the splice sites by the spliceosome, thus resulting in an alternatively spliced transcript. (4) Alternatively, hybridization of the sense and antisense transcripts can allow Dicer to generate endogenous siRNAs. By binding to specific protein partners, a noncoding transcript (green) can modulate the activity of the protein (5), serve as a structural component that allows a larger RNA–protein complex to form (6), or alter where the protein localizes in the cell (7). (8) Long ncRNAs (pink) can be processed to yield small RNAs, such as miRNAs, piRNAs, and other less well-characterized classes of small transcripts.
When the act of ncRNA transcription alone may be enough
The act of transcribing ncRNAs can have profound consequences on the ability of nearby genes to be expressed (Katayama et al. 2005). For example, transcription of a ncRNA across the promoter region of a downstream protein-coding gene can directly interfere with transcription factor binding, and thus prevent the protein-coding gene from being expressed (Martens et al. 2004). In fact, transcriptional interference mechanisms have been shown to regulate key developmental decisions, such as where homeotic (Hox) genes are expressed (Petruk et al. 2006) and whether Saccharomyces cerevisiae enters into meiosis (Hongay et al. 2006). Even if not directly interfering with a nearby promoter, transcription of ncRNAs can induce histone modifications that repress transcription initiation of overlapping protein-coding genes, as demonstrated at the yeast PHO84 (Camblong et al. 2007) and GAL1-10 gene clusters (Houseley et al. 2008). This is because transcriptional elongation causes histone marks to be added that prevent “spurious” transcription initiation from sites within the body of the transcript (Carrozza et al. 2005; Joshi and Struhl 2005; Keogh et al. 2005). Noncoding transcription has even been shown to induce the formation of heterochromatin at the p15 tumor suppressor gene locus that persisted after noncoding transcription was turned off, suggesting that the transient expression of ncRNAs can have long-lasting heritable effects on gene expression (Yu et al. 2008).
Of course, not every gene becomes silenced due to the transcription of nearby ncRNAs. Continuous ncRNA transcription has, for example, been suggested to prevent the silencing of certain Hox genes by Polycomb group (PcG) proteins (Bender and Fitzgerald 2002; Hogga and Karch 2002; Rank et al. 2002; Schmitt et al. 2005). Additionally, transcription of long ncRNAs upstream of the Schizosaccharomyces pombe fbp1+ locus induces chromatin remodeling that is critical for transcriptional activation of the downstream protein-coding gene (Hirota et al. 2008). Interestingly, ncRNA transcription was found to initiate in a stepwise manner from multiple sites upstream of the fbp1+ promoter, causing chromatin opening to proceed progressively toward the mRNA transcription start site. The insertion of a transcriptional terminator within these ncRNAs prevents downstream chromatin remodeling, resulting in reduced recruitment of transcription factors to the fbp1+ promoter, and thus minimal induction of the mRNA. Similar stepwise remodeling of chromatin by ncRNAs has also been shown at the S. pombe ade6-M26 locus (Hirota and Ohta 2009).
A critical remaining question is whether chromatin remodeling occurs due to the act of ncRNA transcription, implying that the ncRNAs are simply nonfunctional by-products, or whether the ncRNAs themselves actively play a role; e.g., by recruiting chromatin remodeling or histone-modifying enzymes. At least at the yeast PHO5 locus, it appears to be the act of noncoding transcription rather than the ncRNA itself that contributes to chromatin plasticity and the ability of the protein-coding gene to be rapidly induced (Uhler et al. 2007). Originating from near the 3′ end of the PHO5 gene is an ∼2.4-kb antisense ncRNA that is rapidly degraded by the exosome, a phenomenon that has made detecting such unstable transcripts often difficult, except when RNA decay activities are depleted (Wyers et al. 2005; Davis and Ares 2006; Preker et al. 2008). Expression of the PHO5 ncRNA in trans had no effect on chromatin remodeling (Uhler et al. 2007). Therefore, rather than the unstable ncRNA itself playing a functional role at the PHO5 locus, it was argued that the actual act of ncRNA transcription affects the local rate of nucleosome exchange and/or turnover, allowing nucleosome eviction, and thus PHO5 transcription, to occur much more rapidly in response to phosphate starvation (Uhler et al. 2007). It should be noted that just because a transcript is degraded rapidly does not mean that it is nonfunctional, as unstable transcripts have, for example, been shown to repress transcription of the yeast Ty1 retrotransposon in trans (Berretta et al. 2008).
At the human DHFR locus, a long noncoding transcript originating from a region upstream of the major DHFR promoter acts to repress expression of the downstream protein-coding gene (Blume et al. 2003; Martianov et al. 2007). Interestingly, this ncRNA inhibits expression of DHFR both in cis and in trans by forming an RNA–DNA triplex structure with the DHFR promoter and directly interacting with TFIIB, which results in the disruption of the preinitiation complex at the DHFR promoter (Martianov et al. 2007). Therefore, depending on the gene locus, ncRNA transcription can have profound effects, both negatively and positively, on the ability of neighboring protein-coding genes to be expressed. In some cases, the act of transcription is sufficient to have functional consequences, but it is likely that many of the ncRNAs produced may play yet-to-be-identified active regulatory roles.
Long ncRNAs target proteins to specific genomic loci to affect transcription patterns
PcG proteins are known to bind and silence the expression of more than a thousand mammalian genes (Boyer et al. 2006; Bracken et al. 2006), yet how PcG proteins are recruited to these specific target sites in mammalian cells has been largely unclear. Several reports suggest that it likely may be long ncRNAs that target PcG proteins to specific genomic locations (Plath et al. 2003; Silva et al. 2003; Kohlmaier et al. 2004). Ezh2 (Enhancer of zeste homolog 2), a histone methyltransferase and member of Polycomb-repressive complex 2 (PRC2), was found to directly bind a 1.6-kb-long ncRNA known as RepA (Zhao et al. 2008). Interestingly, when transcription of a stably integrated ectopic RepA gene locus was induced, PcG proteins were recruited specifically to the gene locus, indicating that RepA is sufficient to recruit PRC2 to chromatin in vivo. The endogenous RepA ncRNA is transcribed from the Repeat A region of the Xist gene and has been proposed to play a key role in the early stages of mammalian X-chromosome inactivation (Zhao et al. 2008). Via a likely related mechanism, the ncRNA HOTAIR, derived from the HOXC locus, has been shown to interact with PcG proteins to regulate the HOXD locus in trans (Rinn et al. 2007).
Trithorax group (TrxG) proteins counteract the actions of PcG proteins to maintain active transcription states and, interestingly, also may be recruited to their target loci by long ncRNAs (Sanchez-Elsner et al. 2006). Certain ncRNAs from the Hox loci were shown to interact directly with the histone methyltransferase Ash1 in vitro and were proposed to target TrxG proteins to chromatin (Sanchez-Elsner et al. 2006). In fact, ectopic expression of these ncRNAs in trans was found to activate Hox gene expression in Drosophila S2 cells and wing imaginal discs (Sanchez-Elsner et al. 2006). However, another report did not observe a similar association between ectopic expression of these ncRNAs and transcriptional activation, and instead suggested that ncRNA transcription inhibits expression of the nearby Hox genes (Petruk et al. 2006, 2007; for review, see Lempradl and Ringrose 2008). Although there are still some discrepancies in the literature, it appears that certain ncRNAs play key roles in maintaining the active or inactive state of gene expression by modulating where PcG and TrxG proteins are targeted.
Numerous long ncRNAs, including Airn and Kcnq1ot1, are expressed from imprinted loci and have been suggested to function as key players in assuring that only one of the two parental alleles are expressed (for review, see Prasanth and Spector 2007; Peters and Robson 2008; Royo and Cavaille 2008). Recent work suggests that imprinted genes within a single cluster can unexpectedly be silenced by different mechanisms. In the mouse placenta, the ∼108-kb Airn ncRNA is required for the paternal-specific silencing in cis of a 400-kb region that includes the Slc22a3, Slc22a2, and Igf2r genes (Sleutels et al. 2002). Analogous to Xist, Airn is retained in the nucleus (Seidl et al. 2006) and appears to “coat” the imprinted locus on the paternal chromosome (Nagano et al. 2008). However, rather than uniformly localizing to the entire imprinted domain, Airn preferentially accumulates at the Slc22a3 promoter (Nagano et al. 2008). Air then interacts with the histone H3 Lys 9 methyltransferase G9a, leading to methylation and silencing of the paternal Slc22a3 promoter. Deleting G9a results in the loss of Slc22a3 imprinting, yet has no effect on Igf2r, which remains monoallelically expressed. Therefore, even though Airn is required for silencing both Slc22a3 and Igf2r on the paternal chromosome, it must do so by different mechanisms. It will be of great interest to tease apart the mechanisms by which ncRNAs are able to obtain such high functional specificity and recruit different proteins to different gene loci.
The imprinted ncRNA Kcnq1ot1 likewise accumulates nonuniformly along the Kcnq1 domain and interacts with G9a and PcG proteins (Pandey et al. 2008), as well as functions by transcriptional interference (Kanduri et al. 2006; Mohammad et al. 2008). Interestingly, certain genes in the Kcnq1 domain are only imprinted in the placenta, likely because the Kcnq1ot1 ncRNA interacts with G9a and PcG proteins in a lineage-specific manner, resulting in the establishment of repressive histone modifications on these genes only in certain cell types (Pandey et al. 2008). Specifically, an interaction between Kcnq1ot1 with G9a and PcG proteins was detected in the placenta, while no interaction was detected in the fetal liver.
Long ncRNAs modulate the activity of protein-binding partners
Many proteins bind to RNAs through a variety of RNA-binding motifs to modulate the processing, localization, and stability of the bound RNAs (for review, see Dreyfuss et al. 2002). Naturally, the converse is also true—RNAs can influence the activity and localization of the proteins they bind. For example, long ncRNAs can serve as key coactivators of proteins involved in transcriptional regulation. The ∼3.8-kb Evf-2 ncRNA, which is transcribed from an ultraconserved region, forms a complex with the homeodomain-containing protein Dlx2 (Feng et al. 2006). Using reporter-based assays, it was shown that Dlx2 acts a transcriptional enhancer only when the Evf-2 ncRNA is also present. Similarly, the ncRNA HSR1 (heat-shock RNA-1) forms a complex with HSF1 (heat-shock transcription factor 1), enabling the transcription factor to induce expression of heat-shock proteins during the cellular heat-shock response (Shamovsky et al. 2006), and an isoform of the ncRNA SRA (steroid receptor RNA activator) functions as a transcriptional coactivator of steroid receptors (Lanz et al. 1999). Conversely, noncoding transcripts derived from SINEs (short interspersed elements) bind to RNA polymerase II during heat shock to inhibit transcription of other mRNAs, such as actin (Allen et al. 2004; Espinoza et al. 2004; Mariner et al. 2008).
ncRNAs produced from the cyclin D1 (CCND1) promoter region have been shown recently to function as allosteric effectors of an RNA-binding protein known as TLS (Translocated in Liposarcoma) (Wang et al. 2008). These ncRNAs are variable in their lengths and of low abundance (generally less than two copies per cell), but are induced in response to DNA damage and remain bound to the chromatin in the CCND1 promoter region. Upon binding these ncRNAs, the TLS protein changes from an inactive to an active conformation, such that it binds and inhibits the enzymatic activities of the histone acetyltransferases CBP and p300, thus silencing CCND1 transcription.
Long ncRNAs also have been shown to modulate the activity of proteins by regulating their subcellular localization. The transcription factor NFAT (nuclear factor of activated T cells) localizes to the cytoplasm until calcium-dependent signals cause it to be imported into the nucleus, where it activates transcription of target genes (for review, see Hogan et al. 2003). One of the key regulators of NFAT trafficking happens to be a ncRNA known as NRON (noncoding repressor of NFAT) that is alternatively spliced (0.8–3.7 kb) (Willingham et al. 2005). By binding to members of the nucleocytoplasmic trafficking machinery, NRON specifically inhibits the nuclear accumulation of NFAT, but not that of other transcription factors such as p53 and NFκB that also translocate from the cytoplasm to nucleus.
Long ncRNAs as precursors for small RNAs
Recent genome-wide studies suggest that the function of a significant fraction of long unannotated transcripts may be to serve as precursors for small RNAs <200 nucleotides (nt) in length (Kapranov et al. 2007a; Fejes-Toth et al. 2009). In addition to microRNAs (miRNAs) that are usually generated via the sequential cleavage of long transcripts by Drosha and Dicer (Cai et al. 2004; Lee et al. 2004) and Piwi-interacting RNAs (piRNAs) that are likely generated via processing of a single long transcript (for review, see Aravin et al. 2007), there are many more small RNAs whose functions and mechanisms of biogenesis are less clear. For example, small RNAs have been found to cluster near the 5′ and 3′ ends of genes (Han et al. 2007; Kapranov et al. 2007a; Core et al. 2008; He et al. 2008; Preker et al. 2008; Seila et al. 2008; Fejes-Toth et al. 2009; Neil et al. 2009; Taft et al. 2009; Xu et al. 2009). Transfection of RNA mimetics to promoter-associated small RNAs (PASRs) were found to reduce expression of the overlapping mRNA promoter, indicating that these newly identified small RNAs impact gene expression (Fejes-Toth et al. 2009).
It now appears that many protein-coding mRNAs and long ncRNAs may be post-transcriptionally processed to yield many small RNAs that, curiously, have a 5′ cap structure (Fejes-Toth et al. 2009). Numerous small RNAs identified using next-generation sequencing technology were found to significantly overlap CAGE (cap analysis of gene expression) tags, which are thought to mark the 5′ ends of capped, long RNA transcripts (Fejes-Toth et al. 2009). Although many CAGE tags do mark transcription start sites, significant numbers were found in exonic regions and, in some cases, to even cross splice junctions, meaning they must have arisen from at least partially processed mRNAs. Therefore, it has been proposed that mature long transcripts (both protein-coding mRNAs and long ncRNAs) can be processed post-transcriptionally to yield small RNAs, which are then modified by the addition of a cap structure (Fig. 2A; Fejes-Toth et al. 2009).
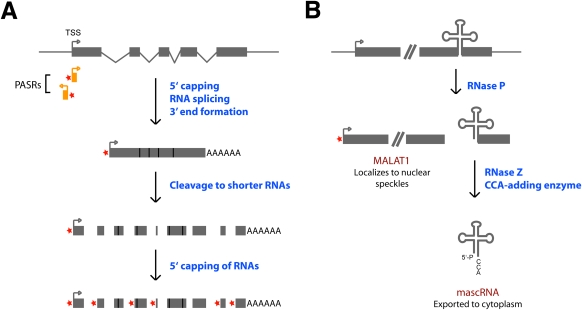
Figure 2.
Long ncRNAs are processed to yield small RNAs. (A) Many long processed transcripts can be post-transcriptionally cleaved to generate small RNAs (Fejes-Toth et al. 2009). A cap structure (denoted by a red star) is then added to the 5′ ends of many of these small RNAs. In addition, capped small RNAs, known as PASRs, map near the transcription start site (TSS) of many genes. (B) The nascent MALAT1 transcript is processed to yield two ncRNAs that localize to different subcellular compartments. Cleavage by RNase P simultaneously generates the 3′ end of the mature MALAT1 transcript and the 5′ end of mascRNA (Wilusz et al. 2008).
By processing a single long transcript into multiple smaller RNAs, each mature transcript can display a distinct subcellular localization and have a unique function. Recent work from our group provides a clear example of how a nascent transcript can be processed to yield two ncRNAs that localize to distinct subcellular compartments (Wilusz et al. 2008). MALAT1 (Metastasis-associated lung adenocarcinoma transcript 1), also known as NEAT2 (Hutchinson et al. 2007), is a long (∼7-kb) ncRNA that is misregulated in many human cancers (Ji et al. 2003; Lin et al. 2007) and was shown previously to be retained specifically in the nucleus in nuclear speckles (Hutchinson et al. 2007), domains that are thought to be involved in the assembly, modification, and/or storage of the pre-mRNA processing machinery (for review, see Lamond and Spector 2003). Upon probing for small RNAs mapping to the MALAT1 locus, we identified a highly conserved 61-nt tRNA-like small RNA (Wilusz et al. 2008). In stark contrast to the mature long MALAT1 transcript, the small RNA is localized exclusively to the cytoplasm, and thus we named it mascRNA, MALAT1-associated small cytoplasmic RNA.
Unlike tRNAs that are transcribed by RNA polymerase III, mascRNA is generated via processing of the MALAT1 nascent transcript (Fig. 2B; Wilusz et al. 2008). RNase P recognizes the tRNA-like structure in the nascent RNA polymerase II transcript and then cleaves to simultaneously generate the 3′ end of the mature MALAT1 transcript and the 5′ end of mascRNA. Additional enzymes involved in tRNA biogenesis, including RNase Z and the CCA-adding enzyme, then further process the small RNA prior to its export to the cytoplasm. While MALAT1 is very stable, mascRNA is fairly rapidly degraded—so not only do the two ncRNAs localize to separate subcellular locations, they also have vastly different half-lives and presumably distinct functions. MEN β, a >20-kb ncRNA that is retained in the nucleus in paraspeckles, is processed at its 3′ end by a similar mechanism (Sunwoo et al. 2009). Based on genome-wide studies (Kapranov et al. 2007a; Fejes-Toth et al. 2009), it is likely that many genetic loci generate multiple ncRNA transcripts, which localize to different subcellular compartments and may not fit into already known and characterized classes of ncRNAs.
The RNAi machinery has well-characterized roles in the generation of miRNAs and siRNAs that regulate gene expression post-transcriptionally (for review, see Lee et al. 2006; Jaskiewicz and Filipowicz 2008). However, a recent report implicates these enzymes in the processing of long transcripts to small RNAs that likely do not function as miRNAs (Ganesan and Rao 2008). In mice, a 2.4-kb unspliced, polyadenylated nuclear-retained ncRNA known as mrhl is processed by Drosha to yield an 80-nt small RNA (Ganesan and Rao 2008). Interestingly, the 80-nt transcript is not further processed by Dicer in vivo, probably because it is retained in the nucleus in association with chromatin.
Last, there is emerging evidence that Xist and Tsix, two long ncRNAs that regulate mammalian X-chromosome inactivation, may also be processed to yield small RNAs (Ogawa et al. 2008). Developmentally regulated small RNAs between 25 and 42 nt in length map to the Xist and Tsix loci. Because Xist and Tsix form a dsRNA duplex in vivo and expression of the small RNAs is diminished when Dicer is deleted, it was suggested that processing of dsRNA generates these small RNAs (Ogawa et al. 2008). However, Dicer is not currently known to cleave RNAs to small transcripts in this size range (25–42 nt), suggesting that Dicer may play an indirect role in the biogenesis of these small RNAs and that Xist and Tsix are processed via other mechanisms. In addition, two recent reports suggest that X inactivation does not depend on Dicer (Nesterova et al. 2008; Kanellopoulou et al. 2009). Although miRNAs and piRNAs have received the most attention of late, it is becoming increasingly clear that long RNA transcripts are processed to yield many other classes of small RNAs with likely very different and unique functions.
Long ncRNAs affect the processing of other RNAs
Long ncRNAs can be processed to yield small RNAs, but they can also affect how other transcripts are processed; for example, by modulating their ability to be cut into small RNAs or changing their pre-mRNA splicing patterns. The 800-nt spliced and polyadenylated C. elegans rncs-1 (RNA noncoding, starvation up-regulated) ncRNA inhibits the production of small RNAs from other transcripts in trans (Hellwig and Bass 2008). Despite containing an almost perfectly double-stranded helix of ∼300 base pairs, this transcript is not a Dicer substrate because branched structures flanking the central double-stranded helix inhibit its processing. Instead, rncs-1 functions in trans to inhibit Dicer activity. Upon overexpressing or deleting rncs-1 in vivo, the expression levels of certain siRNAs were found to decrease or increase, respectively, with a corresponding change in the mRNA levels of their gene targets (Hellwig and Bass 2008). Therefore, it has been proposed that rncs-1 binds to Dicer or accessory dsRNA-binding proteins to compete with other dsRNAs involved in gene silencing.
Certain long ncRNAs are likely able to base-pair with small RNAs to modulate their activities. For example, by interacting with miRNAs, long noncoding transcripts could competitively inhibit the ability of miRNAs to interact with their mRNA targets, analogous to how artificial miRNA sponges function (Ebert et al. 2007). This target mimicry mechanism is used by the long ncRNA IPS1 (INDUCED BY PHOSPHATE STARVATION 1) in Arabidopsis thaliana (Franco-Zorrilla et al. 2007). The ∼550-nt IPS1 ncRNA is poorly evolutionarily conserved except for a short 23-nt motif that is highly complementary to miR-399, a miRNA that is induced in response to phosphate starvation, although the base-pairing is interrupted by a mismatched loop at the expected miRNA cleavage site. This interruption in base-pairing causes the IPS1 ncRNA to be noncleavable and instead allows IPS1 to sequester miR-399, resulting in increased expression of miR-399 target genes (Franco-Zorrilla et al. 2007).
Several recent reports indicate that RNA transcripts derived from pseudogenes can surprisingly cause mRNAs from the functional protein-coding copy of the gene to be processed to small RNAs (Tam et al. 2008; Watanabe et al. 2008). This is because long antisense transcripts produced from pseudogenes are able to hybridize to their corresponding spliced mRNAs, resulting in the formation of dsRNAs that are cleaved by Dicer to endogenous siRNAs (endo-siRNAs). The coding mRNA is thus consumed to generate endo-siRNAs that may direct RISC (RNA-induced silencing complex) to cleave additional copies of the mRNA transcript, resulting in further down-regulation of the protein-coding gene (Tam et al. 2008; Watanabe et al. 2008). Thus, these results show that pseudogenes are not simply nonfunctional elements that eventually will be lost, but instead are key regulators of gene expression when transcribed as long ncRNAs.
Like pseudogenes, some natural antisense transcripts (NATs) are able to hybridize to overlapping genes and generate endo-siRNAs (Czech et al. 2008; Ghildiyal et al. 2008; Okamura et al. 2008; Watanabe et al. 2008). In addition, there are numerous examples of NATs modulating the alternative splicing patterns of their overlapping genes (Krystal et al. 1990; Munroe and Lazar 1991; Yan et al. 2005), such as at the Zeb2/Sip1 gene locus. Zeb2/Sip1 is a transcriptional repressor of E-cadherin whose expression is tightly regulated during epithelial–mesenchymal transition (EMT) (Beltran et al. 2008). Translation of the Zeb2/Sip1 protein requires an internal ribosome entry site (IRES); however, in epithelial cells, the region containing the IRES is spliced out of the mature mRNA. Upon EMT, a NAT is produced that is complementary to the 5′ splice site of this intron, thus blocking the spliceosome from removing the IRES from the mature mRNA and enabling expression of the Zeb2/Sip1 protein (Beltran et al. 2008).
Long ncRNAs serve as structural RNAs
The mammalian cell nucleus is not only compartmentalized such that it is separate from the cytoplasm, but also such that it contains many membraneless compartments that serve specialized functions (for review, see Spector 2001, 2006). How many of these subcellular bodies form and are maintained is unclear, but recent work suggests that long ncRNAs may, in some cases, serve as key structural components. Within the nucleus, a number of RNA-binding proteins, including paraspeckle protein component 1 (PSPC1, also known as PSP1α), NONO (also known as p54/nrb), and the 68-kDa subunit of cleavage factor Im, as well as a nuclear-retained mRNA (CTN-RNA), localize to paraspeckles (Fox et al. 2002, 2005; Dettwiler et al. 2004; Prasanth et al. 2005). Although the exact function of paraspeckles is unclear, they have been suggested to function as storage sites for nuclear-retained RNAs (Prasanth et al. 2005). Interestingly, RNase A treatment disrupts the structural integrity of paraspeckles (Fox et al. 2005; Prasanth et al. 2005), suggesting that RNA(s) may be a critical component of these nuclear structures. Three recent reports, including one from our group, have identified the MEN ɛ/β long ncRNAs as these critical RNA components of paraspeckles (Clemson et al. 2009; Sasaki et al. 2009; Sunwoo et al. 2009). Transcribed from the same RNA polymerase II promoter, MEN ɛ (also known as NEAT1 [Hutchinson et al. 2007]) and MEN β differ in the location of their 3′ ends, but both are retained in the nucleus in paraspeckles (Sasaki et al. 2009; Sunwoo et al. 2009). Unlike depleting CTN-RNA expression (Prasanth et al. 2005), depletion of MEN ɛ/β results in the disruption of paraspeckles, arguing that these long ncRNAs are required for paraspeckle establishment and maintenance (Fig. 3; Clemson et al. 2009; Sasaki et al. 2009; Sunwoo et al. 2009).
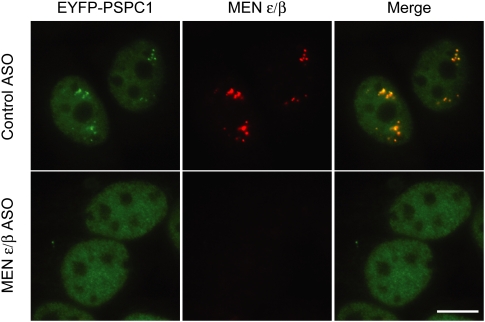
Figure 3.
MEN ɛ/β ncRNAs are essential for the structural integrity of paraspeckles. The MEN ɛ/β ncRNAs localize to paraspeckles in HeLa cells as shown by the colocalization of an RNA FISH probe with PSPC1, a known protein component of paraspeckles, fused to EYFP. Upon treating the cells with an antisense oligonucleotide (ASO) complementary to MEN ɛ/β to deplete expression of the ncRNAs, paraspeckles are disrupted. In contrast, a control ASO has no effect on paraspeckle integrity. Bar, 10 μm.
RNA has also been found to serve a structural role in the organization and maintenance of the cellular cytoskeleton as well as the mitotic spindle. In Xenopus oocytes, the proper organization of the cytokeratin cytoskeleton is dependent on two RNAs, the Xlsirts ncRNA and the VegT mRNA, which are integrated within the cytoskeleton (Kloc et al. 2005, 2007). Depletion of either transcript using antisense oligonucleotides disrupts the cytokeratin network, but not the actin cytoskeleton. Interestingly, although VegT is a protein-coding mRNA, blocking its translation had no effect on the cytokeratin network (Heasman et al. 2001; Kloc et al. 2005), arguing that the RNA itself is functioning to maintain the cytoskeleton. Likewise, a large number of RNAs, particularly ribosomal RNAs as well as a number of uncharacterized transcripts, have been found to associate with the mitotic spindle (Blower et al. 2005). RNase A treatment disrupts spindle assembly and causes the spindle to collapse, although treatment with translation inhibitors have no effect, arguing that these RNAs play a translation-independent role in spindle assembly in M phase (Blower et al. 2005).
Considering the great variety of mRNA localization patterns that are observed during early Drosophila embryogenesis (Lecuyer et al. 2007), it is tempting to speculate that many more RNAs (especially ncRNAs) may have structural and organization roles in the cell. For example, Xist (Zhang et al. 2007) and Kcnq1ot1 (Mohammad et al. 2008) both cause silenced chromatin to localize to the perinucleolar region during S phase of the cell cycle.
Not so fast—short ORFs can be translated
Many transcripts are classified as noncoding on the basis of not having ORFs longer than 50–100 amino acids. However, the Drosophila tarsal-less (tal) gene provides a telling example of the importance of validating these sorts of bioinformatic predictions. The tal gene expresses a 1.5-kb transcript that contains only ORFs of <50 amino acids and was, therefore, originally classified as a long ncRNA. However, several 33-nt ORFs within the tal gene are actually translated into 11-amino-acid-long peptides that control key tissue morphogenesis and pattern formation events during Drosophila development (Galindo et al. 2007; Kondo et al. 2007; Pueyo and Couso 2008). Due to practical and statistical reasons, ORFs as short as these in tal are generally systematically eliminated from gene annotations, but clearly need to be considered when addressing the function of an unannotated transcript. Additionally, some ncRNA genes, such as SRA (Steroid receptor RNA activator), appear to yield multiple RNA isoforms, some of which can be translated (for review, see Leygue 2007), thus allowing a gene to have functions carried out by both RNA and protein.
Perspectives
It is quickly becoming clear that long ncRNAs can have numerous molecular functions, including modulating transcriptional patterns, regulating protein activities, serving structural or organizational roles, altering RNA processing events, and serving as precursors to small RNAs (Fig. 1). Although only a very small portion of known long ncRNAs has been thoroughly characterized to date, future work will likely identify many more transcripts that fit into these and other functional paradigms. In addition, future work will further address the question of whether the act of ncRNA transcription itself is enough to have functional consequences, or if many of the resulting ncRNAs actually have functions in cis that cannot be recapitulated by ectopic expression in trans.
A major current challenge is to understand how the molecular functions of these long ncRNAs affect the organism. For example, long ncRNAs have been implicated in numerous developmental events (for review, see Amaral and Mattick 2008), such as the formation of photoreceptors in the developing retina (Young et al. 2005) and the regulation of cell survival and cell cycle progression during mammary gland development (Ginger et al. 2006). The generation of knockout animal models will likely reveal many insights and definitively show that many long ncRNAs are not transcriptional “noise,” but are instead required for normal development.
Numerous long ncRNAs are misregulated in various diseases, especially cancer (for review, see Costa 2005; Prasanth and Spector 2007), and some have been found to be very sensitive and specific markers of tumors, such as DD3 (also known as PCA3) in prostate tumors (de Kok et al. 2002). However, the mechanisms by which these transcripts may affect tumor initiation and/or progression are currently unknown. Long ncRNAs thus remain a relatively unexplored area in disease research, which may allow us to identify new therapeutic targets. Recent work on Alzheimer's disease has identified a ncRNA antisense to the β-secretase (BACE1) gene, which generates amyloid β (Aβ), that may aid in driving the disease (Faghihi et al. 2008). The ∼2-kb ncRNA is induced in response to numerous cell stressors, including serum starvation and Aβ peptides and, unfortunately, increases the stability of the BACE1 mRNA, thus leading to even more Aβ peptides and the deleterious feed-forward cycle of disease progression. However, treatment with siRNAs against the ncRNA reduces the levels of Aβ peptides, suggesting that this noncoding transcript may serve as an attractive drug target candidate for Alzheimer's disease.
Although it has been classically assumed that most genetic information is expressed as and transacted by proteins, it is now clear that transcription is pervasive throughout the eukaryotic genome, yielding many functional ncRNAs with key regulatory roles. Many surprises have surfaced over the past few years, and it is certain that future research will provide many more unexpected insights into the functions of ncRNAs.
Acknowledgments
We thank members of the Spector laboratory for many helpful discussions and comments. Research in the Spector laboratory is supported by NIH/GM42694, 5P01CA013106-38, and NIH/EY18244. J.E.W. is supported by a Beckman Graduate Studentship at the Watson School of Biological Sciences.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1800909.
References
- Allen TA, Von Kaenel S, Goodrich JA, Kugel JF. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat Struct Mol Biol. 2004;11:816–821. doi: 10.1038/nsmb813. [DOI] [PubMed] [Google Scholar]
- Amaral PP, Mattick JS. Noncoding RNA in development. Mamm Genome. 2008;19:454–492. doi: 10.1007/s00335-008-9136-7. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J. The Piwi–piRNA pathway provides an adaptive defense in the transposon arms race. Science. 2007;318:761–764. doi: 10.1126/science.1146484. [DOI] [PubMed] [Google Scholar]
- Beltran M, Puig I, Pena C, Garcia JM, Alvarez AB, Pena R, Bonilla F, de Herreros AG. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial–mesenchymal transition. Genes & Dev. 2008;22:756–769. doi: 10.1101/gad.455708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender W, Fitzgerald DP. Transcription activates repressed domains in the Drosophila bithorax complex. Development. 2002;129:4923–4930. doi: 10.1242/dev.129.21.4923. [DOI] [PubMed] [Google Scholar]
- Berretta J, Pinskaya M, Morillon A. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes & Dev. 2008;22:615–626. doi: 10.1101/gad.458008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S, et al. Global identification of human transcribed sequences with genome tiling arrays. Science. 2004;306:2242–2246. doi: 10.1126/science.1103388. [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshaw S, Harpavat S, Trimarchi J, Cai L, Huang H, Kuo WP, Weber G, Lee K, Fraioli RE, Cho SH, et al. Genomic analysis of mouse retinal development. PLoS Biol. 2004;2:E247. doi: 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blower MD, Nachury M, Heald R, Weis K. A Rae1-containing ribonucleoprotein complex is required for mitotic spindle assembly. Cell. 2005;121:223–234. doi: 10.1016/j.cell.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Blume SW, Meng Z, Shrestha K, Snyder RC, Emanuel PD. The 5′-untranslated RNA of the human dhfr minor transcript alters transcription pre-initiation complex assembly at the major (core) promoter. J Cell Biochem. 2003;88:165–180. doi: 10.1002/jcb.10326. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes & Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell. 2007;131:706–717. doi: 10.1016/j.cell.2007.09.014. [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa FF. Non-coding RNAs: New players in eukaryotic biology. Gene. 2005;357:83–94. doi: 10.1016/j.gene.2005.06.019. [DOI] [PubMed] [Google Scholar]
- Czech B, Malone CD, Zhou R, Stark A, Schlingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, et al. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis CA, Ares M., Jr Accumulation of unstable promoter-associated transcripts upon loss of the nuclear exosome subunit Rrp6p in Saccharomyces cerevisiae. Proc Natl Acad Sci. 2006;103:3262–3267. doi: 10.1073/pnas.0507783103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kok JB, Verhaegh GW, Roelofs RW, Hessels D, Kiemeney LA, Aalders TW, Swinkels DW, Schalken JA. DD3(PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Res. 2002;62:2695–2698. [PubMed] [Google Scholar]
- Deng X, Meller VH. Non-coding RNA in fly dosage compensation. Trends Biochem Sci. 2006;31:526–532. doi: 10.1016/j.tibs.2006.07.007. [DOI] [PubMed] [Google Scholar]
- Dettwiler S, Aringhieri C, Cardinale S, Keller W, Barabino SM. Distinct sequence motifs within the 68-kDa subunit of cleavage factor Im mediate RNA binding, protein-protein interactions, and subcellular localization. J Biol Chem. 2004;279:35788–35797. doi: 10.1074/jbc.M403927200. [DOI] [PubMed] [Google Scholar]
- Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008;18:1433–1445. doi: 10.1101/gr.078378.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721–726. doi: 10.1038/nmeth1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebisuya M, Yamamoto T, Nakajima M, Nishida E. Ripples from neighbouring transcription. Nat Cell Biol. 2008;10:1106–1113. doi: 10.1038/ncb1771. [DOI] [PubMed] [Google Scholar]
- Espinoza CA, Allen TA, Hieb AR, Kugel JF, Goodrich JA. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat Struct Mol Biol. 2004;11:822–829. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- Faghihi MA, Modarresi F, Khalil AM, Wood DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G, 3rd, Kenny PJ, Wahlestedt C. Expression of a noncoding RNA is elevated in Alzheimer's disease and drives rapid feed-forward regulation of β-secretase. Nat Med. 2008;14:723–730. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fejes-Toth K, Sotirova V, Sachidanandam R, Assaf G, Hannon GJ, Kapranov P, Foissac S, Willingham AT, Duttagupta R, Dumais E, et al. Post-transcriptional processing generates a diversity of 5′-modified long and short RNAs. Nature. 2009;457:1028–1032. doi: 10.1038/nature07759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Bi C, Clark BS, Mady R, Shah P, Kohtz JD. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes & Dev. 2006;20:1470–1484. doi: 10.1101/gad.1416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox AH, Lam YW, Leung AK, Lyon CE, Andersen J, Mann M, Lamond AI. Paraspeckles: A novel nuclear domain. Curr Biol. 2002;12:13–25. doi: 10.1016/s0960-9822(01)00632-7. [DOI] [PubMed] [Google Scholar]
- Fox AH, Bond CS, Lamond AI. P54nrb forms a heterodimer with PSP1 that localizes to paraspeckles in an RNA-dependent manner. Mol Biol Cell. 2005;16:5304–5315. doi: 10.1091/mbc.E05-06-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet. 2007;39:1033–1037. doi: 10.1038/ng2079. [DOI] [PubMed] [Google Scholar]
- Galindo MI, Pueyo JI, Fouix S, Bishop SA, Couso JP. Peptides encoded by short ORFs control development and define a new eukaryotic gene family. PLoS Biol. 2007;5:e106. doi: 10.1371/journal.pbio.0050106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan G, Rao SM. A novel noncoding RNA processed by Drosha is restricted to nucleus in mouse. RNA. 2008;14:1399–1410. doi: 10.1261/rna.838308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Seitz H, Horwich MD, Li C, Du T, Lee S, Xu J, Kittler EL, Zapp ML, Weng Z, et al. Endogenous siRNAs derived from transposons and mRNAs in Drosophila somatic cells. Science. 2008;320:1077–1081. doi: 10.1126/science.1157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginger MR, Shore AN, Contreras A, Rijnkels M, Miller J, Gonzalez-Rimbau MF, Rosen JM. A noncoding RNA is a potential marker of cell fate during mammary gland development. Proc Natl Acad Sci. 2006;103:5781–5786. doi: 10.1073/pnas.0600745103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Kim D, Morris KV. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc Natl Acad Sci. 2007;104:12422–12427. doi: 10.1073/pnas.0701635104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman J, Wessely O, Langland R, Craig EJ, Kessler DS. Vegetal localization of maternal mRNAs is disrupted by VegT depletion. Dev Biol. 2001;240:377–386. doi: 10.1006/dbio.2001.0495. [DOI] [PubMed] [Google Scholar]
- Hellwig S, Bass BL. A starvation-induced noncoding RNA modulates expression of Dicer-regulated genes. Proc Natl Acad Sci. 2008;105:12897–12902. doi: 10.1073/pnas.0805118105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K, Ohta K. Cascade transcription of mRNA-type long non-coding RNAs (mlonRNAs) and local chromatin remodeling. Epigenetics. 2009;4:5–7. doi: 10.4161/epi.4.1.7353. [DOI] [PubMed] [Google Scholar]
- Hirota K, Miyoshi T, Kugou K, Hoffman CS, Shibata T, Ohta K. Stepwise chromatin remodelling by a cascade of transcription initiation of non-coding RNAs. Nature. 2008;456:130–134. doi: 10.1038/nature07348. [DOI] [PubMed] [Google Scholar]
- Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes & Dev. 2003;17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- Hogga I, Karch F. Transcription through the iab-7 cis-regulatory domain of the bithorax complex interferes with maintenance of Polycomb-mediated silencing. Development. 2002;129:4915–4922. doi: 10.1242/dev.129.21.4915. [DOI] [PubMed] [Google Scholar]
- Hongay CF, Grisafi PL, Galitski T, Fink GR. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell. 2006;127:735–745. doi: 10.1016/j.cell.2006.09.038. [DOI] [PubMed] [Google Scholar]
- Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell. 2008;32:685–695. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- Jaskiewicz L, Filipowicz W. Role of Dicer in posttranscriptional RNA silencing. Curr Top Microbiol Immunol. 2008;320:77–97. doi: 10.1007/978-3-540-75157-1_4. [DOI] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- Joshi AA, Struhl K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell. 2005;20:971–978. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Kanduri C, Thakur N, Pandey RR. The length of the transcript encoded from the Kcnq1ot1 antisense promoter determines the degree of silencing. EMBO J. 2006;25:2096–2106. doi: 10.1038/sj.emboj.7601090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Dimitrov SD, Chen X, Colin C, Plath K, Livingston DM. X chromosome inactivation in the absence of Dicer. Proc Natl Acad Sci. 2009;106:1122–1127. doi: 10.1073/pnas.0812210106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007a;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007b;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, et al. Cotranscriptional Set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell. 2005;123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- Kloc M, Wilk K, Vargas D, Shirato Y, Bilinski S, Etkin LD. Potential structural role of non-coding and coding RNAs in the organization of the cytoskeleton at the vegetal cortex of Xenopus oocytes. Development. 2005;132:3445–3457. doi: 10.1242/dev.01919. [DOI] [PubMed] [Google Scholar]
- Kloc M, Bilinski S, Dougherty MT. Organization of cytokeratin cytoskeleton and germ plasm in the vegetal cortex of Xenopus laevis oocytes depends on coding and non-coding RNAs: Three-dimensional and ultrastructural analysis. Exp Cell Res. 2007;313:1639–1651. doi: 10.1016/j.yexcr.2007.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlmaier A, Savarese F, Lachner M, Martens J, Jenuwein T, Wutz A. A chromosomal memory triggered by Xist regulates histone methylation in X inactivation. PLoS Biol. 2004;2:E171. doi: 10.1371/journal.pbio.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Hashimoto Y, Kato K, Inagaki S, Hayashi S, Kageyama Y. Small peptide regulators of actin-based cell morphogenesis encoded by a polycistronic mRNA. Nat Cell Biol. 2007;9:660–665. doi: 10.1038/ncb1595. [DOI] [PubMed] [Google Scholar]
- Krystal GW, Armstrong BC, Battey JF. N-myc mRNA forms an RNA–RNA duplex with endogenous antisense transcripts. Mol Cell Biol. 1990;10:4180–4191. doi: 10.1128/mcb.10.8.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamond AI, Spector DL. Nuclear speckles: A model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, Tsai MJ, O'Malley BW. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell. 1999;97:17–27. doi: 10.1016/s0092-8674(00)80711-4. [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 2007;131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Han J, Yeom KH, Jin H, Kim VN. Drosha in primary microRNA processing. Cold Spring Harb Symp Quant Biol. 2006;71:51–57. doi: 10.1101/sqb.2006.71.041. [DOI] [PubMed] [Google Scholar]
- Lempradl A, Ringrose L. How does noncoding transcription regulate Hox genes? Bioessays. 2008;30:110–121. doi: 10.1002/bies.20704. [DOI] [PubMed] [Google Scholar]
- Leygue E. Steroid receptor RNA activator (SRA1): Unusual bifaceted gene products with suspected relevance to breast cancer. Nucl Recept Signal. 2007;5:e006. doi: 10.1621/nrs.05006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Maeda S, Liu C, Karin M, Edgington TS. A large noncoding RNA is a marker for murine hepatocellular carcinomas and a spectrum of human carcinomas. Oncogene. 2007;26:851–858. doi: 10.1038/sj.onc.1209846. [DOI] [PubMed] [Google Scholar]
- Mariner PD, Walters RD, Espinoza CA, Drullinger LF, Wagner SD, Kugel JF, Goodrich JA. Human Alu RNA is a modular transacting repressor of mRNA transcription during heat shock. Mol Cell. 2008;29:499–509. doi: 10.1016/j.molcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- Martens JA, Laprade L, Winston F. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature. 2004;429:571–574. doi: 10.1038/nature02538. [DOI] [PubMed] [Google Scholar]
- Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–670. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- Masui O, Heard E. RNA and protein actors in X-chromosome inactivation. Cold Spring Harb Symp Quant Biol. 2006;71:419–428. doi: 10.1101/sqb.2006.71.058. [DOI] [PubMed] [Google Scholar]
- Mattick JS. RNA regulation: A new genetics? Nat Rev Genet. 2004;5:316–323. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Sunkin SM, Mehler MF, Mattick JS. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci. 2008;105:716–721. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad F, Pandey RR, Nagano T, Chakalova L, Mondal T, Fraser P, Kanduri C. Kcnq1ot1/Lit1 noncoding RNA mediates transcriptional silencing by targeting to the perinucleolar region. Mol Cell Biol. 2008;28:3713–3728. doi: 10.1128/MCB.02263-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munroe SH, Lazar MA. Inhibition of c-erbA mRNA splicing by a naturally occurring antisense RNA. J Biol Chem. 1991;266:22083–22086. [PubMed] [Google Scholar]
- Nagano T, Mitchell JA, Sanz LA, Pauler FM, Ferguson-Smith AC, Feil R, Fraser P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science. 2008;322:1717–1720. doi: 10.1126/science.1163802. [DOI] [PubMed] [Google Scholar]
- Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- Nesterova TB, Popova BC, Cobb BS, Norton S, Senner CE, Tang YA, Spruce T, Rodriguez TA, Sado T, Merkenschlager M, et al. Dicer regulates Xist promoter methylation in ES cells indirectly through transcriptional control of Dnmt3a. Epigenetics Chromatin. 2008;1:2. doi: 10.1186/1756-8935-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y, Sun BK, Lee JT. Intersection of the RNA interference and X-inactivation pathways. Science. 2008;320:1336–1341. doi: 10.1126/science.1157676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Chung WJ, Ruby JG, Guo H, Bartel DP, Lai EC. The Drosophila hairpin RNA pathway generates endogenous short interfering RNAs. Nature. 2008;453:803–806. doi: 10.1038/nature07015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- Payer B, Lee JT. X chromosome dosage compensation: How mammals keep the balance. Annu Rev Genet. 2008;42:733–772. doi: 10.1146/annurev.genet.42.110807.091711. [DOI] [PubMed] [Google Scholar]
- Peters J, Robson JE. Imprinted noncoding RNAs. Mamm Genome. 2008;19:493–502. doi: 10.1007/s00335-008-9139-4. [DOI] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Riley KM, Hodgson J, Schweisguth F, Hirose S, Jaynes JB, Brock HW, Mazo A. Transcription of bxd noncoding RNAs promoted by trithorax represses Ubx in cis by transcriptional interference. Cell. 2006;127:1209–1221. doi: 10.1016/j.cell.2006.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Brock HW, Mazo A. A model for initiation of mosaic HOX gene expression patterns by non-coding RNAs in early embryos. RNA Biol. 2007;4:1–6. doi: 10.4161/rna.4.1.4300. [DOI] [PubMed] [Google Scholar]
- Pheasant M, Mattick JS. Raising the estimate of functional human sequences. Genome Res. 2007;17:1245–1253. doi: 10.1101/gr.6406307. [DOI] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- Pollard KS, Salama SR, Lambert N, Lambot MA, Coppens S, Pedersen JS, Katzman S, King B, Onodera C, Siepel A, et al. An RNA gene expressed during cortical development evolved rapidly in humans. Nature. 2006;443:167–172. doi: 10.1038/nature05113. [DOI] [PubMed] [Google Scholar]
- Ponjavic J, Ponting CP, Lunter G. Functionality or transcriptional noise? Evidence for selection within long noncoding RNAs. Genome Res. 2007;17:556–565. doi: 10.1101/gr.6036807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth KV, Spector DL. Eukaryotic regulatory RNAs: An answer to the ‘genome complexity’ conundrum. Genes & Dev. 2007;21:11–42. doi: 10.1101/gad.1484207. [DOI] [PubMed] [Google Scholar]
- Prasanth KV, Prasanth SG, Xuan Z, Hearn S, Freier SM, Bennett CF, Zhang MQ, Spector DL. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- Pueyo JI, Couso JP. The 11-aminoacid long Tarsal-less peptides trigger a cell signal in Drosophila leg development. Dev Biol. 2008;324:192–201. doi: 10.1016/j.ydbio.2008.08.025. [DOI] [PubMed] [Google Scholar]
- Rank G, Prestel M, Paro R. Transcription through intergenic chromosomal memory elements of the Drosophila bithorax complex correlates with an epigenetic switch. Mol Cell Biol. 2002;22:8026–8034. doi: 10.1128/MCB.22.22.8026-8034.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravasi T, Suzuki H, Pang KC, Katayama S, Furuno M, Okunishi R, Fukuda S, Ru K, Frith MC, Gongora MM, et al. Experimental validation of the regulated expression of large numbers of non-coding RNAs from the mouse genome. Genome Res. 2006;16:11–19. doi: 10.1101/gr.4200206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo H, Cavaille J. Non-coding RNAs in imprinted gene clusters. Biol Cell. 2008;100:149–166. doi: 10.1042/BC20070126. [DOI] [PubMed] [Google Scholar]
- Sanchez-Elsner T, Gou D, Kremmer E, Sauer F. Noncoding RNAs of trithorax response elements recruit Drosophila Ash1 to Ultrabithorax. Science. 2006;311:1118–1123. doi: 10.1126/science.1117705. [DOI] [PubMed] [Google Scholar]
- Sasaki YTF, Ideue T, Sano M, Mituyama T, Hirose T. MEN ɛ/β noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt S, Prestel M, Paro R. Intergenic transcription through a polycomb group response element counteracts silencing. Genes & Dev. 2005;19:697–708. doi: 10.1101/gad.326205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidl CI, Stricker SH, Barlow DP. The imprinted Air ncRNA is an atypical RNAPII transcript that evades splicing and escapes nuclear export. EMBO J. 2006;25:3565–3575. doi: 10.1038/sj.emboj.7601245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamovsky I, Ivannikov M, Kandel ES, Gershon D, Nudler E. RNA-mediated response to heat shock in mammalian cells. Nature. 2006;440:556–560. doi: 10.1038/nature04518. [DOI] [PubMed] [Google Scholar]
- Silva J, Mak W, Zvetkova I, Appanah R, Nesterova TB, Webster Z, Peters AH, Jenuwein T, Otte AP, Brockdorff N. Establishment of histone H3 methylation on the inactive X chromosome requires transient recruitment of Eed–Enx1 polycomb group complexes. Dev Cell. 2003;4:481–495. doi: 10.1016/s1534-5807(03)00068-6. [DOI] [PubMed] [Google Scholar]
- Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J Cell Sci. 2007;120:2498–2506. doi: 10.1242/jcs.009357. [DOI] [PubMed] [Google Scholar]
- Spector DL. Nuclear domains. J Cell Sci. 2001;114:2891–2893. doi: 10.1242/jcs.114.16.2891. [DOI] [PubMed] [Google Scholar]
- Spector DL. SnapShot: Cellular bodies. Cell. 2006;127:1071. doi: 10.1016/j.cell.2006.11.026. [DOI] [PubMed] [Google Scholar]
- Struhl K. Transcriptional noise and the fidelity of initiation by RNA polymerase II. Nat Struct Mol Biol. 2007;14:103–105. doi: 10.1038/nsmb0207-103. [DOI] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, Spector DL. MEN ɛ/β nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski M, Barciszewska MZ, Erdmann VA, Barciszewski J. A new frontier for molecular medicine: Noncoding RNAs. Biochim Biophys Acta. 2005;1756:65–75. doi: 10.1016/j.bbcan.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, Lassmann T, Forrest AR, Grimmond SM, Schroder K, et al. Tiny RNAs associated with transcription start sites in animals. Nat Genet. 2009;41:572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature. 2008;453:534–538. doi: 10.1038/nature06904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhler JP, Hertel C, Svejstrup JQ. A role for noncoding transcription in activation of the yeast PHO5 gene. Proc Natl Acad Sci. 2007;104:8011–8016. doi: 10.1073/pnas.0702431104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhang J, Zheng H, Li J, Liu D, Li H, Samudrala R, Yu J, Wong GK. Mouse transcriptome: Neutral evolution of ‘non-coding’ complementary DNAs. Nature. 2004;431:757. doi: 10.1038/nature03016. [DOI] [PubMed] [Google Scholar]
- Wang X, Arai S, Song X, Reichart D, Du K, Pascual G, Tempst P, Rosenfeld MG, Glass CK, Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature. 2008;453:539–543. doi: 10.1038/nature06908. [DOI] [PubMed] [Google Scholar]
- Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- Wilusz JE, Freier SM, Spector DL. 3′ End processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008;135:919–932. doi: 10.1016/j.cell.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan MD, Hong CC, Lai GM, Cheng AL, Lin YW, Chuang SE. Identification and characterization of a novel gene Saf transcribed from the opposite strand of Fas. Hum Mol Genet. 2005;14:1465–1474. doi: 10.1093/hmg/ddi156. [DOI] [PubMed] [Google Scholar]
- Young TL, Matsuda T, Cepko CL. The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol. 2005;15:501–512. doi: 10.1016/j.cub.2005.02.027. [DOI] [PubMed] [Google Scholar]
- Yu W, Gius D, Onyango P, Muldoon-Jacobs K, Karp J, Feinberg AP, Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LF, Huynh KD, Lee JT. Perinucleolar targeting of the inactive X during S phase: Evidence for a role in the maintenance of silencing. Cell. 2007;129:693–706. doi: 10.1016/j.cell.2007.03.036. [DOI] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]