Abstract
Algae biofuels may provide a viable alternative to fossil fuels; however, this technology must overcome a number of hurdles before it can compete in the fuel market and be broadly deployed. These challenges include strain identification and improvement, both in terms of oil productivity and crop protection, nutrient and resource allocation and use, and the production of co-products to improve the economics of the entire system. Although there is much excitement about the potential of algae biofuels, much work is still required in the field. In this article, we attempt to elucidate the major challenges to economic algal biofuels at scale, and improve the focus of the scientific community to address these challenges and move algal biofuels from promise to reality.
Importance & challenges of algal biofuels
The global economy requires fossil hydrocarbons to function, from producing plastics and fertilizers to providing the energy required for lighting, heating and transportation. With our increasing population and expanding economy, there will be increased fossil fuel use. As countries improve their gross domestic product per capita, data suggest that their fossil fuel use will increase, and competition for these limited resources will increase. In addition, there comes increasing atmospheric CO2 concentration, and the potential for significant greenhouse gas-mediated climate change [1], which now seems likely to affect all parts of the world. Finally, petroleum, which is partially derived from ancient algae deposits, is a limited resource that will eventually run out or become too expensive to recover [2–4]. These factors are driving the development of renewable energy sources that can supplant fossil fuels, and allow greater access to fuel resources for all nations, while greatly reducing carbon emissions into the atmosphere. A number of technologies have been examined as renewable energy sources and, although no single strategy is likely to provide a total solution, it seems possible that a combination of strategies can be employed that will substantially decrease our dependence on fossil fuels [4]. The challenge that remains is to develop renewable energy industries that operate sustainably and can be cost competitive with existing energy options.
Fossil fuels are used for the generation of electrical power, as well as liquid fuels. There are a variety of renewable or low atmospheric pollution technologies that can generate electrical power, including solar, wind, hydroelectric, geothermal and nuclear. However, renewable technologies to supplement or replace liquid fossil fuels are still in their early developmental stages. The International Energy Agency expects that biofuels will contribute 6% of total fuel use by 2030, but could expand significantly if undeveloped petroleum fields are not accessed or if substantial new fields are not identified (Figure 1). The most promising sustainable alternatives are almost exclusively categorized under the moniker ‘biofuels’. This term describes a diverse range of technologies that generate fuel with at least one component based on a biological system. The major technologies presently employed for biofuels begin with terrestrial plants and culminate with ethanol, whether this is corn starch to sugar to ethanol, or sugarcane sugars to ethanol. The regional success of some of these strategies is well noted; in particular, the sugarcane-to-ethanol production in Brazil [5]. To a lesser degree, oils from terrestrial plants – for example, soy and palm – are used to produce biodiesel. These strategies are functional at the small scale; however, as their use has increased, it is evident that they are not sustainable, owing to the enormous amount of agricultural land that would be required to supplant a significant fraction of petroleum using this strategy [6,7]. A number of hybrid strategies have been discussed or are currently being deployed. Examples of such strategies include conversion of cellulose to sugars for fermentation into fuel, and gasification of residual biomass into syngas that can then be used to produce liquid fuels [8]. Although each of these strategies is being used to produce fuels, they are insufficient to accommodate the global demand for liquid fuels.
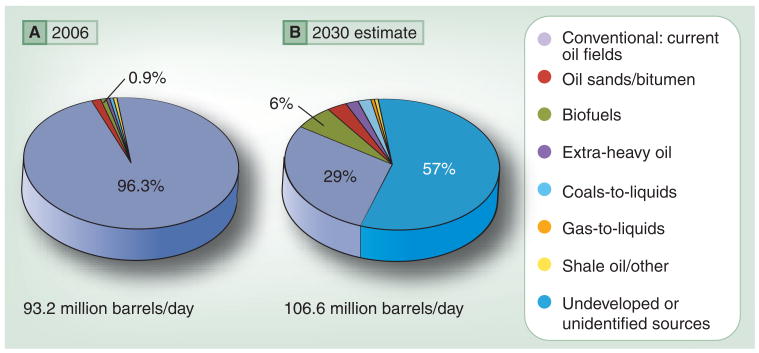
Figure 1. Previous and predicted global petroleum sources.
(A) Global liquid fuel use in 2006 was predominantly (96.3%) conventional petroleum, with slightly less than 1% being biofuels. (B) In 2030, the International Energy Agency estimates that 29% of liquid fuels will originate from current conventional oil sources, 57% will be from undeveloped or unidentified conventional oil sources and 6% will be biofuels [4]. The large gray area of undeveloped or unidentified sources provides ample and possibly necessary expansion for nonconventional sources.
In this article, we discuss the potential of a burgeoning alternative strategy: microalgae-produced liquid fuels. The high lipid content, high growth rate and ability to rapidly improve strains and produce co-products, without competing for arable land, make algae an exciting addition to the sustainable fuel portfolio. This article will focus on the requirements for establishing microalgae as an environmentally and economically viable platform, with an emphasis on combining fuel production with production of co-products, which we view as an essential strategy for the economic viability and, hence, broad adoption, of this potential fuel source (Figure 2).
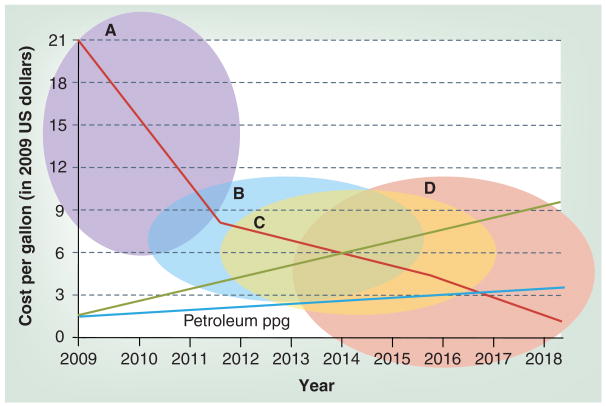
Figure 2. A combination of factors is expected to be required for algal fuels (red line) to become cost competitive with petroleum (green line: limited petroleum supply, resulting in increased costs; blue line: business as usual scenario).
These improvements will require years of research and cover (A) bioprospecting for high-oil-producing, low-input-requiring species; (B) engineering to improve growth, harvesting and nutrient recycling; (C) further strain improvement through breeding, selection and random mutagenesis; and (D) bioengineering to improve fuel traits, produce co-products and crop protection. Estimates given in this figure are for illustration purposes based on our best guesses. We believe that bioprospecting has high potential to identify a solid biofuel species in the next few years but subsequent improvement of that species, as well as solving engineering challenges to improve cost efficiency, will not occur as rapidly.
Present economic reality of liquid fuels
There are considerable challenges to making biofuels capable of competing with petroleum. Certainly, a premium price is warranted for clean fuels (fuels that have a 50% lower CO2 cradle-to-grave footprint than petroleum); however, estimated costs of a barrel of algae-based fuel using current technology is US$300–2600, compared with $40–80 (2009) for petroleum [9–12]. Although some estimates for a barrel of algae oil in specific regions reach as low as $84 [13]. The higher dollar estimates are more common and similar to our own estimates, and exclude algal oil from the current liquid fuel market. The challenges and strategies to tackle this economic discrepancy are discussed in this article.
Benefits of microalgal biofuels
Microalgae are a diverse group of single-celled organisms that have the potential to offer a variety of solutions for our liquid transportation fuel requirements through a number of avenues. Algal species grow in a wide range of aquatic environments, from freshwater through saturated saline. Algae efficiently use CO2, and are responsible for more than 40% of the global carbon fixation, with the majority of this productivity coming from marine microalgae [14,15]. Algae can produce biomass very rapidly, with some species doubling in as few as 6 h, and many exhibiting two doublings per day [16,17]. All algae have the capacity to produce energy-rich oils, and a number of microalgal species have been found to naturally accumulate high oil levels in total dry biomass [18]. For example, some Botryococcus spp. have been identified that have up to 50% of their dry mass stored as long-chain hydrocarbons [19]. With potentially millions of species, algal diversity gives researchers many options for identifying production strains and also provides sources for genetic information that can be used to improve these production strains. The microalgal species being investigated as potential biofuel crops originate from groups whose ancestral relationships are significantly broader than the most diverse land plants, providing a wealth of genetic diversity [20,21]. The groups most often considered when discussing microalgae are diatoms, green algae, golden brown, prymnesiophytes, eustigmatophytes and cyanobacteria [16], and members from all of these groups have been examined as potential fuel production strains. However, it should be noted that cyanobacteria are not algae but a class of photosynthetic bacteria.
Microalgae have additional advantages over terrestrial plants. Since they are single-celled organisms that duplicate by division, high-throughput technologies can be used to rapidly evolve strains. This can reduce processes that take years in crop plants, down to a few months in algae. Algae have a reduced impact on the environment compared with terrestrial sources of biomass used for biofuels [9]. They can be grown on land that would not be used for traditional agricultural, and are very efficient at removing nutrients from water. Thus, not only would production of algae biofuels minimize land use compared with biofuels produced from terrestrial plants but, in the process of culturing these microalgae, waste streams can be remediated. Potential waste streams include municipal wastewater to remove nitrates and phosphates before discharge, and flue gas of coal or other combustible-based power plants to capture sulfates and CO2 [22–24]. Algae production strains also have the potential to be bioengineered, allowing improvement of specific traits [25,26] and production of valuable co-products, which may allow algal biofuels to compete economically with petroleum. These characteristics make algae a platform with a high potential to produce cost-competitive biofuels.
Challenges for algal fuel commercialization
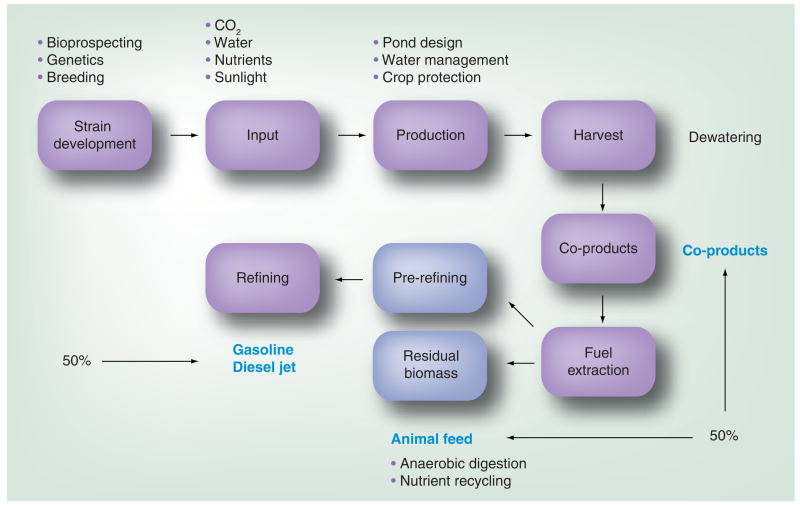
The high growth rates, reasonable growth densities and high oil contents have all been cited as reasons to invest significant capital to turn algae into biofuels. However, for algae to mature as an economically viable platform to offset petroleum and, consequently, mitigate CO2 release, there are a number of hurdles to overcome ranging from how and where to grow these algae, to improving oil extraction and fuel processing. The algal biofuels production chain is outlined in Figure 3 and shows that the major challenges include strain isolation, nutrient sourcing and utilization, production management, harvesting, coproduct development, fuel extraction, refining and residual biomass utilization.
Figure 3. Algal biofuels production chain.
Improved strains, as well as downstream efficiency, are integral aspects of the algae biofuel production strategy.
Making algal growth & harvesting more efficient
Improved engineering will make a significant impact on algae biofuel production. These improvements include efficient strategies for nutrient circulation and light exposure, and have been reviewed elsewhere [27,28]. In brief, there are significant challenges for engineers to either design photobioreactors (PBRs) that are cheap enough for large-scale deployment, or for engineers and biologists to combine forces to develop species that grow efficiently in low-cost open systems [29]. PBRs have advantages over open systems in that they can more easily maintain axenic cultures, and can maintain more controlled growth environments, which may lead to increases in productivity; however, contained systems are challenged by efficiencies in gas exchange and a requirement for supplemental cooling [28]. Despite the advantages of decreased contamination and increased productivity, it is unclear whether PBRs will ever become cost competitive with open pond systems. Regardless of the growth strategy employed, substantial improvements over current technologies for the growth, harvesting and extracting oil from algae need to be made, and coordinated efforts will be needed to couple engineering advances with improved production strains.
Improving oil extraction & downstream processing
Oil extraction is another challenge that is most easily addressed from the engineering side. There are three major strategies for extracting oil from algae: oil press/expeller, hexane extraction, and supercritical CO2 fluid extraction [30]. These technologies have all been successfully demonstrated but are relatively expensive, either in terms of equipment needed or energy required to extract the oil. Fortunately, all are amenable to engineering improvements. Once extracted, because crude algae oil is chemically similar to crude fossil fuel oil, the engineering challenges associated with algae oil conversion to usable liquid fuels are similar to those already well managed by petroleum companies, although improved catalysts will be required to improve gasoline production from bio-oil [31]. Because of these similarities, it seems reasonable to assume that collaborations between algae production companies and major oil companies are likely, since these companies have extensive experience maximizing downstream processing efficiencies.
Land use
Regardless of the growth strategy employed and efficiency of oil extraction, the scale of implementation that is required to replace a meaningful amount of fossil fuel is significant. In 2008, the USA alone required 19,497,950 barrels of oil per day [32]. For algae, or any other biofuel feedstock, to impact this number, significant acreage must be dedicated to production facilities, with estimates suggesting that 30 million acres will be required to meet US oil demand. Different models have been presented for how large-scale aquaculture can be achieved. Although both terrestrial strategies and marine strategies may be required, in this article we focus on the terrestrial aquaculture, since marine strategies are completely unknown at present and may require engineering significantly different from what is practiced today. The terrestrial models use land that is not presently used for food agriculture, and has minimal known environmental or other significant economic utility.
Water use
Water is potentially a major limiting factor in algal growth. Expansion of algal growth into nonarable land will require water; fortunately, many of these regions have substantial alkaline or saline water reservoirs beneath them, providing a significant source of nonpotable water that is suitable for growth of many algal species. Perhaps surprisingly, algae grown in open ponds have water requirements per unit area similar to that of cotton or wheat, but less than that of corn, to replenish the water lost in evaporation (for an overview of water requirements of terrestrial plants used in biofuel production see [33]). It is imperative when considering broad deployment of algae, to consider water use to avoid a future ‘water versus fuel’ debate. Although substantial alkaline reserves are available, water will remain a central issue for algae biofuels production and will need to be considered carefully as the industry expands.
Nutrient challenge
Algae require nutrients, light, water and a carbon source, most often CO2, for efficient growth. The major nutrients required by most algae include phosphorous, nitrogen, iron and sulfur. Often, the nutrient requirement necessary for algal growth is ignored, since algae are very efficient at sequestering these nutrients when present in their environment [34,35]. Changes in nutrient load and algal growth have been studies extensively in terms of eutrophication of lakes and coastal regions, but not as heavily in terms of productivity in large-scale aquaculture [36,37]. If terrestrial agriculture is a model for some of the challenges for algal aquaculture, then providing sufficient nutrients for large-scale algal growth is a significant challenge. Micro- and macro-nutrient supplements, or fertilizer, account for significant costs in the current terrestrial agriculture industry [38], and biofuels are not expected to be an exception. The use of fertilizers has been increasing globally. Unfortunately, many fertilizer components are generated from fossil fuels or mined and, as such, they are not renewable [39–42]. Algae, similar to plants, require sources of phosphorus, nitrogen and potassium, which are the major components of agricultural fertilizers, and large-scale aquaculture will impact these already limited supplies. In addition, optimal growth of many algal species requires chelated iron and sulfur.
Phosphorous makes up slightly less then 1% of total algal biomass and is required at approximately 0.03–0.06% in the medium to sustain algal growth. Fertilizers in the USA used for agriculture currently contain a less than optimal concentration of phosphate owing to limited supplies. Presently, less than 40 million tons of phosphate is mined from the USA annually, and the maximum phosphate production from this mining peaked in the late 1980s. If algal biofuels are to completely replace petroleum in the USA, an additional 53 million tons of phosphate must be acquired annually. This is a significant challenge, given that the total amount of phosphate in the USA is estimated to be approximately 2.8 billion tons. This leaves few options other then efficient recycling the phosphate back into the algae ponds or significantly increasing mining output, a prospect that would seem to provide a temporary solution at best.
Nitrogen, unlike phosphorous, is not limited in supply but is often a limiting macronutrient when it comes to plant and algae growth. Algae require nitrogen to be fixed into ammonia, nitrates and similar molecules, in order to be used as a nutrient source [43]. Some bacteria, such as rhizobia, have the ability to fix their own nitrogen and some form symbiotic relationships with terrestrial plants, providing the plants with this crucial nutrient to sustain protein and nucleic acid synthesis [42,44]. Some cyanobacteria also have the ability to fix nitrogen, while almost all algal species identified to date require an exogenous source of fixed nitrogen, and most prefer ammonia, as it is less energetically demanding than nitrate or nitrite [45]. Providing a cheap source of fixed nitrogen will be important for algae biofuel production, and the possibility of using nitrogen-fixing cyanobacteria to supply this nitrogen may help minimize these costs [46].
In the open oceans, iron is a major limiting nutrient for algal growth, as demonstrated by the induction of algal blooms by the addition of exogenous iron to open oceans [47]. Interestingly, the addition of iron to induce an algal bloom has been considered and tested as a strategy to sequester CO2 [47–49]. Biologically, iron is required for electron transport in all known photosynthetic organisms, including Chlamydomonas reinhardtii, and is typically found in iron-sulfur clusters in a variety of photosynthetic proteins [50]. Iron in its oxidized form is not optimal for uptake, and most algae prefer chelated iron. Fortunately, iron can be easily acquired and is more available than many of the other required nutrients.
Sulfur, in addition to its key role in the electron transport chain, is also required for protein synthesis and lipid metabolism. Sulfur deficiency has been shown to limit algal density and stunt growth [51]. Thus, it seems likely that sulfur will be important for optimal algal growth, and cost/benefit analysis will need to be considered to determine the optimal amount of sulfur to add to the media for the best economic return.
The acquisition of the aforementioned nutrients, as well as potassium and at least nine other micro- and macro-nutrients, should not be overlooked when considering the implications of scaling algal biofuel production to meaningful levels [52]. Many of the nutrients may be supplemented by combining nutrient-rich waste water or agricultural runoff with algal growth facilities, streamlining water remediation and optimizing economic fuel production. These strategies appear to be viable at some scale; however, alternative possibilities must also be developed. Ultimately, a combination of methods may be required, and perhaps a recycling of micro- and macro-nutrients will have to be developed for algae-based biofuels to reach a capacity that impacts present fossil fuel use. One of the most promising techniques for recycling nutrients in algal ponds is to use anaerobic digestion [53]. This bacterial process produces methane gas, while keeping the majority of the nutrients in a bacterial slurry that can be killed and the mix used for algal fertilizer. Methane gas is not currently a high-value commodity, but can help provide energy to operate algae farms, and cheap anaerobic digestion will preclude producing some types of higher value proteins in the algae. Therefore, a balance should be reached between efficient anaerobic digestion and high-value co-products, as shown in Figure 4.
Figure 4. To maximize algae biofuel sustainability, nutrients must be recycled.
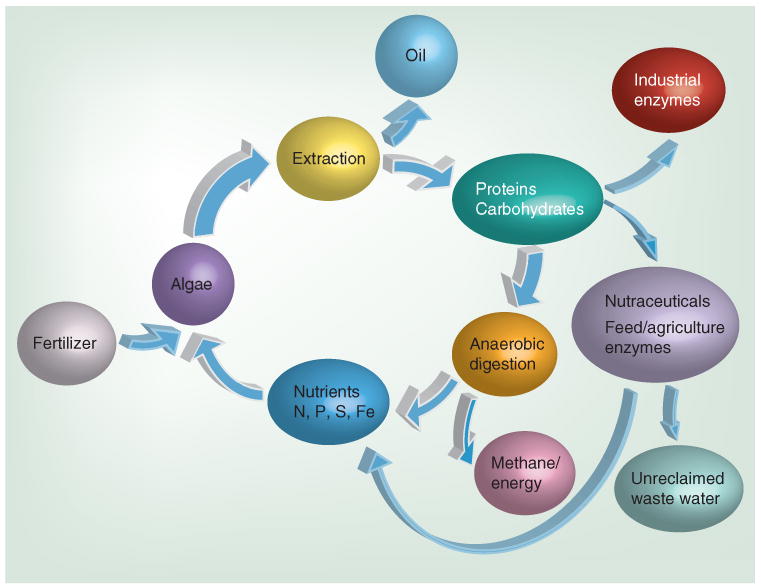
This is a model of how we expect nutrient utilization to occur as the field matures. Algae will be harvested and the oil will be extracted, the remaining biomass (carbohydrates/proteins) will either be recycled for nutrients through anaerobic digestion or similar means, producing methane gas and a nutrient-rich slurry, which can then be fed back into the algal pond, rather than exogenously produced fertilizers, or used to for high-value co-products, ranging from industrial enzymes, nutraceuticals or animal feed stocks. Some of these nutrients can be recycled through waste water, while others will be lost due to runoff.
Crop protection: minimizing algae death from biotic & abiotic factors
Much like terrestrial monocultures, large algal monocultures will be invaded by pests and pathogens, and therefore, crop protection is a major challenge to algal pond sustainability. Identifying strains resistant to pathogens, along with many other strategies, will need to be employed. These strategies, discussed later, may include engineering specific pest resistance into production species that have robust growth characteristics and significant lipid composition. Other approaches may include using multiple species, which may be sufficient to the slow spread of specific pests and minimize crop loss in large algal facilities.
Microalgal growth facilities can be an excellent habitat for a wide variety of undesirable guests. In most cases, these will be detrimental for algal growth by acting as competitors (other algae with low oil production or bacteria), parasites (virus, fungus or protozoans) or predators (protozoans, fungus or aquatic invertebrates [54–57]. Algae biofuel projects are considering both open and closed systems. These options have significantly different challenges. Closed systems, such as PBRs, have the potential to minimize contamination, but this comes at a high capital expense. Outdoor pond systems have lower initial capital costs, but historically these open pond systems have relied mainly on outcompeting contaminating organisms by using densely grown axenic (or nearly axenic) starter cultures [58,59]. This high-density inoculation allows algae populations to expand rapidly, minimizing end-product loss due to contamination. Unfortunately, this strategy might not be feasible for the extremely large culture volumes required for biofuel production, especially if continuous harvesting strategies are employed. Another solution to minimize contamination is to use microalgae that can grow under extreme conditions, which are not suitable for most of the potential contaminants. This would be the case with Dunaliella salina and Arthrospira, which can withstand up to 35% salinity and pH 10, respectively [59–61]. Unfortunately, not all production strains can survive in extreme conditions, and there is still the possibility for an extremophile contamination to arise [62].
Microalgae have developed morphological, behavioral and chemical mechanisms for defending themselves from pathogens and predators. Chemical defense is widely present in the ‘algae group’ against bacteria, fungus, protozoans, aquatic invertebrates, other algae and even viruses [63–65]. The majority of antibiotic extracts studied so far have been from marine macro- and micro-algae [65–67]; however, they are also present in many freshwater species [67–69]. Most antibiotics from microalgae have come from cyanobacteria, haptophytes, chrysophytes, diatoms, dinoflagellates and chlorophytes. The chemical nature of these substances is very diverse, including fatty acids, bromophenols, tanins, polysaccharides, alcohols, halogenated compounds, peptides, lipopeptides, alkaloids, amides, tertiary sulfoniums, and many other unique substances [65,66]. Some of these chemicals accumulate within cells so they only act after the algae is damaged or ingested. In other species, toxins are secreted into the media by the algae to avoid negative interactions [70]. The mode of action of toxins against predators can be further classified into acute toxicity, reduced fitness (growth reduction and/or reduced progeny), feeding inhibition and avoidance. Morphological and behavioral defense mechanisms also complement the chemical repertoire against algae grazers [57,70,71].
Given these natural defense mechanisms, it seems wise to take advantage of them, along with other strategies adapted from agriculture, to secure ‘crop’ protection for biofuel production. The simplest solution would be to pick a production strain with extremophile characteristics and a broad repertoire of antibiotic properties. However, this might not occur naturally in a single species, so the next simplest solution would be to coculture a set of microalgae that synergistically contribute to protect the entire crop. Additionally, a single species could be engineered to produce one or more of these algal antibiotics or other natural products [63,72,73]. However, these are mostly secondary metabolites that require several enzymes to be synthesized. As an alternative, antimicrobial peptides (AMPs) could be expressed from a single heterologous gene, as has been shown in the nucleus and chloroplast of plants [74,75]. Some of these molecules have been shown to be broad-spectrum antibacterial, antifungal or antiprotozoal agents for which pathogens have a limited capability to develop resistance [76,77]. Moreover, they can also be specifically designed and screened for a specific crop protection function [78].
Alternative bactericidal proteins (non-AMPs) have been expressed in algae. The Chlorella ellipsoidea nuclear genome was engineered to produce rabbit neutrophil peptide-1, which proved to be effective agent against human pathogens in vitro [79]. Transformed Chlamydomonas reindhartii chloroplast accumulated 5% total soluble protein (TSP) of the mammary-associated serum amyloid A3 peptide, but its antimicrobial activity was not tested [80,81]. Recently, expression of bovine lactoferricin was achieved in nuclear transformants of Nannochloropsis oculata. Algal extracts from strains expressing this protein were effective against Escherichia coli and Vibrio parahemolyticus [82].
The aforementioned proteins might be effective against microorganisms but aquatic invertebrates could still feed freely on these algae. A future solution might come from the expression of ‘insecticidal’ proteins, such as those with similar function to the ones from Bacillus thuringensis and Bacillus sphaericus. These proteins have already been expressed in plants and cyanobacteria, and shown to be detrimental towards aquatic insects, aquatic larvae and daphnids [73,83–85]. On the other hand, some aquatic invertebrates might be beneficial for algae, and could be used as a biological control strategy. Certain species have strict preference for prey other than algae, such as the heterotrophic protists [57,70]. For example, copepods have been shown to directly contribute to the blooming of the alga Phaeocystis (Haptophyceae) by selectively eating its protozoan predators [86].
Fundamentally, the challenges described can be overcome in algae, as they have in terrestrial crops, but this may require many years of basic research to understand algal/pathogen interactions that impact crop production. In addition, we will need to balance the cost of solutions relative to increase productivity and, hence. return on investment, and this analysis may prove difficult in the short term, as there are little fundamental data to base this analysis on.
Competition with petroleum: getting the price right
With current estimates of algal-based biofuels ranging from US$300–2600 per barrel based on current technology, technical hurdles need to be overcome to improve this price. Some of these improvements can come from improving growth strategies and engineering, as discussed previously, but improvements can also come from optimizing the use of the entire organism. Although the final price of a barrel of algae oil when production goes to large scale is difficult to extrapolate from the present small production facilities, system improvements will certainly bring costs down. Figure 5 illustrates our estimates of the relative impacts of technological improvements on the economic viability of algae biofuels. Most analysts do not predict full parity with petroleum in the near future. More likely, the initial selling point of algal fuels will be approximately twofold higher than petroleum, but the environmental costs will be substantially lower than our current strategy of depending on fossil fuels.
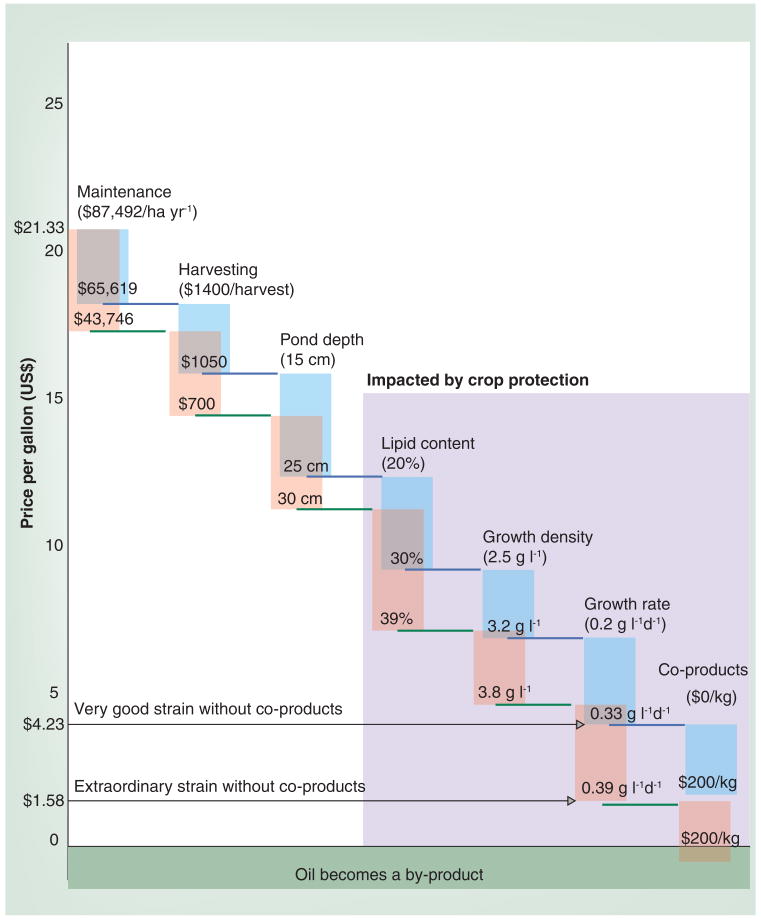
Figure 5. Additive effects of improvements on algal cost.
In our model, our starting strain, with characteristics in brackets, has a break-even price of US$21.33. Significant improvements in all factors (blue) or exceptional improvements in all factors (pink) have break-even prices of US$4.23 or US$1.58 before co-products are considered. Our model consists of seven major factors: 1) annual maintenance costs: this includes personnel, land taxes, fertilizer costs, upkeep, water and power; 2) harvesting costs: this cost is incurred every time algae reaches harvesting stage which is a function of growth rate, and maximum growth density (it includes costs for water extraction, oil extraction and oil transport from harvest site to sales destination); 3) pond depth: this is how deep the ponds can be made while the algae still maintain optimal growth rates. The remaining four are characteristics of the algae; 4) lipid content; 5) growth rate; 6) maximum growth density; 7) marketable co-products produced by the algae. Based on our analysis, improvements in all seven factors are required for algae to come close to competitive with petroleum. Maximizing the last four characteristics require good crop protection, stressing the importance of developments in this field before the large capital investment required to build full scale algae biofuel farms. It is important to note this economic model does not include the initial capital costs to build the initial farms.
From the bench to the pond: strategies to make algae biofuels viable
Taking algae from a potential biofuel producer to large-scale production will be dictated by a combination of factors that include economic viability, and the perceived value of CO2 mitigation by this technology. In addition, early successes in both business and academic environments will promote funding of the research necessary to optimize and validate this technology. To get a sense of the potential progress required, we generated a simplified economic analysis using a Scenedesmus spp., which produced 0.21 g/l/day of biomass, with a lipid content of 21% [18]. In our model, petroleum-based oil would have to cost approximately $710 per barrel for this organism to be economically viable at these growth and lipid accumulation rates using existing production technologies. One surprising result from this model is the importance of growth density in comparison to growth rate. Although growth rate is important for overall productivity, our model predicts that higher growth densities improve economic viability more rapidly than a proportional increase in growth rate, since the expense of harvesting and fuel extraction outweighs the capital expense of building a larger facility to get the same overall total production. If we assume that, with thorough characterization of extant species, we can identify a species that has a growth rate of 0.3 g/l/day and 40% oil content, then the resulting price of algae oil would be approximately $310 per barrel. This number could potentially be improved with breeding and selection or molecular genetics to further optimize the production strain, but there is no guarantee that a strain with these characteristics will be identified owing to the complex interaction between growth rates and oil accumulation. Similarly, there is no way to validate that our model is correct, since a number of costs implicit in the model are estimates based on previous work of others [9–12], and extrapolated from traditional agriculture data [39]. In the following sections, we discuss the major strategies to overcome this price gap.
Bioprospecting: utilizing natural diversity to increase productivity
Algae are an extremely diverse group that contains many thousands of known species, and potentially hundreds of thousands. The great diversity of algal species provides a wide range of starting strains for fuel production. This presents an incredible opportunity, but also a significant challenge. Characterizing species for application in industrial processes requires substantial effort. To move a species into an applied pipeline after initial species identification, significant physiological, biochemical and genetic characterization must occur. This characterization includes establishing optimal growth conditions (i.e., temperature, nutrient levels, salinity and pH), growth characterization (i.e., rate of growth and final culture density), and analysis of metabolite accumulation (i.e., lipid composition and accumulation). In parallel, functional genomics (genomics, proteomics and metabolomics) can provide insight into metabolic pathways present in these species and provide a foundation for future metabolic engineering. The US government-sponsored Aquatic Species Program (ASP) provided initial characterization of a few hundred species, from the 1970s through to the mid-1990s [16]. This work has continued in a number of laboratories around the world, but remains an area of biology that is largely unexplored. General descriptions of the few algal classes in which one or more species has been characterized now follow.
Diatoms
This diverse group, with more than 100,000 estimated species, is currently one of the most prolific primary marine producers. Furthermore, it is believed that some oil deposits originated from diatom biomass [87]. Diatoms are distinguished by their ornate bipartite shells, which are composed primarily of polymerized silicates. In silica-limited environments, diatoms have been found to accumulate lipids; however, growth is reduced [88]. Although this class represents a relatively untapped pool of biodiversity for biofuels, only a limited amount of research has been done to understand how these algae accumulate lipids, while the majority of the work has focused on understanding how diatoms generate their shells. In addition, diatom species have been identified that generate a number of interesting biomolecules that have human health benefits or commercial application, such as omega-3 fatty acids [89,90]. Breeding approaches, as well as using forward genetics to better understand their basic biology, has been hampered by the fact that they are diploid during vegetative growth, unlike many other algal groups. Although other diploid species have become foundational model organisms (e.g. Arabidopsis, Drosophila and mice), the diatom sexual lifecycle is poorly understood, making them recalcitrant to traditional forward genetic exploration. Whole-genome sequences have been released for two species, Thalassiosira pseudonana and Phaeodactylum tricornutum, and recent work has shown stable nuclear transformation of these two species and a few other diatoms [91–94]. The sequenced genomes, in conjunction with nuclear transformation, may allow reverse genetic approaches, such as siRNAs, to dissect the molecular pathways regulating diatom metabolism, allowing us to improve their potential as biofuel-generating organisms and our understanding of their basic biology [95].
Chlorophyceae & trebouxiophyceae
These green algae are the most heavily studied algal groups, primarily owing to the establishment of the chlorophyceae, C. reinhardtii, as an algal model organism. One reason for this focus on green algae is their shared common ancestry with vascular plants. The extant green algae consist of approximately 8000 species, and member species can be found throughout freshwater and marine habitats, and a few species (e.g., D. salina) have been shown to survive in high/saturated saline solutions. Green algae have been used extensively in aquaculture, mainly for the production of secondary metabolites, such as β-carotene and astaxanthin. The genomes of a number of green algae have been sequenced, and molecular tools are in place to transform both the chloroplast and nuclear genome of C. reinhardtii [96–98]. Current work is underway to transform plastids of additional species in our laboratory, as well as others, while nuclear transformation has been achieved in a number of other green algae species, such as members of the Chlorella and volvox genuses [99,100]. In terms of oil production, of the published algal species, members of the Scenedesmus genus have been identified as potential oil-producing species, with both rapid growth, as well as relatively high lipid content [18,101,102]. In addition, a number of species have been identified that produce interesting metabolites, such as Botryococcus spp. that produce the triterpenoid botryoccenes, a potential fuel molecule that requires minimal refining [103].
Cyanobacteria
Cyanobacteria were initially examined for biofuel production and found to accumulate relatively low levels of lipids [16]; however, recently, there has been renewed interest in cyanobacteria as a potential biofuel source because of the ability of some species to grow in extreme environments, and the potential to rapidly engineer these species. Although lipid production in cyanobacteria may never reach that of the eukaryotic algae, the ability to easily manipulate the cyanobacterial genome may allow them to be engineered to produce other biofuel precusor molecules. Cyanobacteria may also be able to be manipulated to secrete biofuel molecules. Another potential advantage of some cyanobacteria is their ability to fix nitrogen. Although growing two or more species as a consortia has not been carefully modeled to date, it is possible that a nitrogen-fixing cyanobacteria, cocultured with a lipid-rich algae, may form a synergistic relationship, allowing the total biomass density to expand above current estimates, while minimizing the cost of adding nitrates to the system. In fact, recently, a diatom species has been characterized, which has endosymbiosed a nitrogen-fixing cyanobacterium, suggesting that such relationships are indeed beneficial to at least one of the two species [104,105].
Through further study of these diverse classes, we may be able to identify new species with improved growth characteristics, improved fuel production or natural co-products, which can improve the viability of algal biofuels. In addition, genomic data from these species can be used as sources of new genes and, combined with screening for natural products, may illuminate new metabolic pathways, which may be useful for engineering other species.
Growth of algal strains
A key characteristic for determining the viability of newly identified algal species as potential biofuel strains is their growth characteristics. Growth rates and maximum growth densities must be characterized in terms of real-world growth conditions rather than laboratory conditions. These conditions can be simulated to some degree in the laboratory by using appropriate media, mixing condition and light regimes; however, production estimates to date, which are primarily based on laboratory conditions in enclosed photobioreactors, will need to be recalculated for industrially scalable systems before they can be considered meaningful for biofuel production, whether they are open ponds or advanced photobioreactors. While useful knowledge can be gained from these types of investigations, in order for true economic viability to be assessed, and for algae to move forward from the bench to the pond, all aspects of algal physiology must be assessed under real-world conditions, rather than the theoretically achievable results under idealized conditions.
Optimizing the growth of algae in open ponds is a key component of reaching economic viability, and remains a major challenge for the industry. Identifying species that grow well under these conditions is a focal point of research in many laboratories. Algae can grow in a wide variety of temperatures, with growth being limited primarily by nutrient availability and light. Light provides the energy for carbon fixation, and is converted to chemical energy through photosynthesis, providing the building blocks for biofuel production. Algae growth rates are often limited by light penetration into the ponds from both self-shading and light absorption by the water, and these constraints are major determining factors of pond depth. The identification of production species that have adapted to harvest shorter wavelengths of light, which have greater water depth penetration, may allow algal pond depth to be increased beyond the current 15–30 cm [106]. This will decrease the total land required for algal biofuel production.
To date, algal growth in open pond systems have been able to produce 0.06–0.231 g/l/day of biomass [16,106,107] and nearly 3 g/l/day in bioreactors [108]. These numbers may improve through genetics and breeding, as has been accomplished in crop species, as demonstrated by Montsanto and Pioneer's successes in improving the yields of maize or improvements in engineering for photobioreactors. Direct molecular genetic modification may also aid in improving growth characteristics. The failure to change metabolism owing to environmentally adverse conditions may be disadvantageous for survival in a competitive environment; however, impeding these pathways may allow for more consistent yields in partially controlled environments [109], such as those being considered for algal biofuel production. In addition, a number of engineering strategies have been examined to increase both light and nutrient utilization, which is discussed elsewhere [110]. At least one algal company has explored eliminating the light component entirely, growing algae nonphotosynthetically, using a reduced carbon source (Solazyme). In these conditions, algal biomass can accumulate more rapidly and reach higher densities, since they avoid issues with shading and have a readily available carbon source. However, since the vast majority of reduced carbon sources are derived from photosynthesis, this strategy simply displaces the need for photosynthesis to another plant, while adding the burden of shipping the reduced carbon source to the site of algal fermentation. In addition, adding a reduced carbon source to algae cultures will increase the presence of unwanted pests, potentially making the scale-up to the volumes required for significant biofuel production problematic.
Breeding & classical genetics to improve & identify traits
Artificial selection of desired traits in agricultural plants has probably been occurring since the dawn of agriculture. The two traditional strategies have been identification of traits of interest from the naturally occurring diversity of the crop species, and selectively combining these traits through interbreeding. A clear success of this strategy is the diversity of Brassica oleracea cultivars from broccoli to cabbage. Directed and successful breeding of some species of microalgae is possible, but little is known about the sexual lifecycle of the vast majority of species. This is partly owing to the enormous diversity in the algae groups, from the diploid diatoms to the haploid chlorophyceae, such that processes occurring in one algae species are not necessarily applicable to other species. However, perhaps more limiting is the general lack of information on the entire lifecycle of many algae, mainly owing to their small size and aqueous lifecycle. There is potential to improve algae for biofuels by developing the tools for selective breeding and using these to move traits that have an impact on biofuels between closely related species, or to improve specific strains of one species. These breeding strategies have an advantage, in that polygenic traits can be moved between strains; however, currently mutagenesis and molecular genetics are at the forefront of algal strain improvement.
'Omics of algal biofuels
In the past 10 years, biological sciences have seen an explosion of strategies that examine entire classes of molecules from a whole organism or cell type, collectively describe as 'omes, including genomes, proteomes, transcriptomes, lipidomes and others. Technologies used to study 'omes strive to analyze entire molecule classes, rather than by piecemeal. The relative merits and weaknesses of these strategies can be debated; however, these 'omic fields will play a vital role when characterizing new species, as researchers expand beyond classical model organisms. These technologies have been especially valuable in unicellular organisms. Broad application in the natural product field is already occurring to identify operons that may encode enzymes to produce new valuable products through comparative genomics. As organism complexity increases, the challenges of 'omic strategies increase. Fortunately, microalgae are amenable to many of the 'omic techniques developed for bacteria and single-celled eukaryotes, such as S. cerevisae. The technologies to apply 'omics to micro-algae are constantly increasing, and we now describe some of the broad applicability of these technologies to improve algal biofuels.
Sorting through the broad range of species being considered for algal biofuels requires a concerted effort to evaluate a number of traits. The primary traits of interest are growth rates, growth density, lipid accumulation, resistance to predators and harvestability. Many of these are probably multigene traits, and species with sequenced genomes will offer significant advantages to those without. We can use comparative genomics to understand key aspects of the basic biology of algae, and whole-genome sequences significantly improve the ease of bioengineering, from the traditional breeding and mapping of genes that control a trait, to knock-out or -down genes that affect pathways of interest. Genomes, when well annotated, not only provide knowledge of metabolic pathways and enzymes that are present in the system, but also help in identification of elements that will be required for regulating gene expression, including promoters and regulatory elements, both of which aid in improved metabolic engineering. Although genome sequencing was once a rate-limiting factor, advances in sequencing technologies have significantly reduced the time and cost of sequencing entire genomes, making high-throughput genome sequencing feasible. To date, a number of algal genomes have been sequenced, or are in process of being sequenced and these numbers are expected to expand dramatically as potential biofuel strains are identified from environmental isolates.
As mentioned previously, the possibility of finding biofuel production strains that combine rapid growth rates, high lipid yields and an ability to grow to relatively high densities while ideally having good crop protection and harvestability characteristics is low. However, upon identifying strains that have one or more of these characteristics, advanced genomic approaches should help elucidate the pathways that allow these traits to persist. By gaining an increased understanding of how extant species maintain these distinct traits, researchers will more rapidly discern whether a new species has potential for development as a biofuel production strain. Although groups are assembling significant genomic data because of the high efficiency and relatively ‘low’ cost of full-genome sequencing, there is a disconnection between sequence generation and production of useful genetic information, including correlation of biological function with a specific gene or cluster of genes. Fortunately, high-throughput sequencing strategies can be adapted to generate transcriptome data, either by aligning cDNA sequence data to an assembled genome or through de novo transcriptome assembly [111,112]. This is a good first step in understanding gene expression in production species; however, an emphasis must be placed on correlating gene expression with phenotype in these species.
Certainly, approaches can be used to rapidly identify genes that correlate with high lipid content, but these are only possible with high-throughput sequencing. In order for these approaches to yield results, basic characterization of species must also occur. For biofuels, the basic traits mentioned should be considered, and independent groups from the academic and the commercial sector will need to continue to define the most desirable characteristics. As these traits are identified, there is potential to use modified quantitative trait loci (QTL) strategies, and apply these across species to identify orthologs that regulate similar traits in other species [113]. These strategies, and other technologies, may expedite the identification of genes that have the potential to improve biofuel viability and the engineering of production strains.
Bioengineering: improving traits through synthetic biology
Identification of an ideal, unmodified biofuel organism that fits into the established infrastructure for harvesting, extraction and purification, and is economically viable, is a possibility; however, a much more likely scenario is the identification of a variety of species that each have one or a few of these desirable traits. These traits, when engineered into a single strain, may be sufficient to result in an economically viable production strain. In addition to strain improvements in fuel production, using genes identified from other algae species may allow for improved expression of heterologous proteins, which either have high value as a protein coproduct or enzymatically produce a high-value coproduct. Both of these strategies are being investigated to improve the economics of algal biofuels.
Transformation technologies
To date, only a few algal species have been successfully genetically manipulated. These modifications have come in the form of induced random mutagenesis to identify new genes that play important roles in processes of interest. Additional mutagenic strategies include insertional mutagenesis of the nuclear genome, allowing the isolation of tagged mutant genes, although there is some debate on whether this strategy has strong preference for specific genomic regions [114]. Heterologous gene expression has also been used as a means to modify biological function, and the nuclear genomes of a number of algae have been transformed, with a variety of reporter genes, as well as drug-resistance genes; however, extensive analysis of transgene expression has only been performed in Chlamydomonas. In Chlamydomonas, researchers have encountered problems with transgene silencing [115]. Strategies to mitigate nuclear silencing have been attempted, including identifying strains that have mutations in the silencing pathways [115,116]. In addition to the nuclear genome, the plastid genome of Chlamydomonas has been successfully transformed and has become a consistent method for heterologous protein expression [117,118]. Although C. reinhardtii may not be a good biofuel production species, the technologies established in this species have potential for application in other algal species. We will discuss some of the successes and failures in algal transformation and their implications in biofuels.
The development of heterologous gene expression tools in algae have followed a path similar that that used in other research organisms. A common strategy to verify that a phenotype of interest is caused by a mutation in a specific gene is to insert a second, wild-type copy of the gene into the host genome to test whether this transgene can complement the mutant phenotype. This strategy is especially valuable in organisms that are haploid during the majority of their lifecycle, such as the chlorophyte alga, because it allows researchers to determine whether loss-of-function or gain-of-function mutations cause the observed phenotype. Early successes in nuclear transformation of C. reinhardtii started in the late 1980s [119,120], with high-efficiency transformation of the nuclear genome first described in 1990 [121,122]. These strategies were quickly adopted for gene identification of loss-of-function mutations by transforming large cosmid, yeast or bacterial artificial chromosomes (YACs or BACs) containing pieces of the C. reinhardtii genome into mutant C. reinhardtii backgrounds [123–125], allowing researchers to clone genes based on transcomplementation. Nuclear gene expression for transcomplementation has been used extensively in C. reinhardtii for gene identification with relatively good success; however, algal nuclear transformation has also been used for heterologous gene expression.
The goal of heterologous gene expression was originally to select transformants that had taken up foreign DNA and separate them from those that did not. Initially, the potential for this approach was determined by complementing a photosynthetically deficient mutant with the endogenous OEE-1 gene [122], since heterologous marker genes had shown little success. As genome information became more available, it became clear that the C. reinhardtii nuclear genome contained significant GC codon bias, suggesting failure of nuclear heterologous gene expression maybe owing to incorrect codon usage of the inserted gene, although this bias does not preclude heterologous gene expression [126]. However, improving codon optimization did improve selectable marker expression and, thus, improve their use [127]. However, despite the improvement in codon optimization, it was found that C. reinhardtii can and often does efficiently silence or downregulate nonrequired heterologous genes when expressed at high levels, perhaps as a defense against viral infection.
Overcoming this nuclear gene silencing in C. reinhardtii has been an ongoing challenge. A number of mutants have been identified that have decreased gene silencing capabilities, often through the disruption of the small RNA pathways [115]. In addition, strategies that couple heterologous gene expression to expression of genes that provide a significant advantage to the algae have also found significant success. Interestingly, not all algae appear to show this aggressive silencing of heterologous genes. The diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana have both shown stable integration and expression of marker genes, including genes that do not necessarily impart a significant advantage (e.g., GFP); however, extensive data on the expression of heterologous genes in most algae, including these diatoms, is still being generated. For a list of microalgal species that have been transformed, please see Table 1. Improving our ability to manipulate the nuclear genome of many of these species in a stable and reproducible fashion will greatly facilitate our ability to produce biofuels and bioproducts.
Table 1.
Transformed microalgae species.
| Species/variety | Class | Method | Ref. |
|---|---|---|---|
| Nuclear transformation | |||
| Chlamydomonas reinhardtii | Chlorophyceae | Bombardment, si carbide whisker, electroporation, glass bead, grobacterium | [122,128–132] |
| Dunaliella salina | Chlorophyceae | electroporation, bombardment, glass beads | [133–136] |
| Gonium pectorale | Chlorophyceae | Bombardment | [99] |
| Haematococcus pluvialis | Chlorophyceae | Bombardment (transient) | [137] |
| Volvox carteri | Chlorophyceae | Bombardment | [138] |
| Cyclotella cryptica | Diatom | Bombardment | [92] |
| Cylindrotheca fusiformis | Diatom | Bombardment | [139] |
| Navicula saprophila | Diatom | Bombardment | [92] |
| Phaeodactylum tricornutum | Diatom | Bombardment | [91,94] |
| Thalassiosira weissflogii | Diatom | Bombardment | [94] |
| Thalassiosira pseudonana | Diatom | Bombardment | [93] |
| Amphidinium spp. | Dinoflagellate | Si carbide whisker | [140] |
| Symbiodinium microadriaticum | Dinoflagellate | Si carbide whisker | [140] |
| Chlorella ellipsoidea | Trebouxiophyceae | Bombardment protoplast, transformation, electroporation | [141,142] |
| Chlorella saccharophila | Trebouxiophyceae | Electroporation | [143] |
| Chlorella vulgaris | Trebouxiophyceae | Protoplast transformation | [144] |
| Chloroplast transformation | |||
| Euglena gracilis | Euglenaceae (kingdom: Excavata) | Bombardment (transient) | [145] |
| Chlamydomonas reinhardtii | Chlorophyceae | Bombardment | [129] |
| Porphyridium spp. | Porphyridiophyceae | Bombardment | [146] |
| Phaeodactylum tricornutum | Diatom | Bombardment (none yet) | [147] |
Stable chloroplast transformation was first accomplished in the green algae C. reinhartdii in 1989 when Boynton and co-workers restored the photosynthetic capacity of a chloroplast mutant by cell bombardment with high-velocity microprojectiles coated with the wild-type gene [129]. In contrast to nuclear transformation, chloroplast transformation occurs through homologous recombination rather than random integration and does not show gene silencing. As shown in Table 1, more then 20 different algae have now been transformed in either the nucleus or plastid, with all major classes having at least one report of foreign gene expression.
The first attempts at expression of recombinant proteins in the chloroplast of C. reinhardtii involved the bacterial neomycin phosphotransferase [148] and β-glucuronidase genes [149], both driven by C. reinhardtii chloroplast promoters. These studies showed stable accumulation of recombinant mRNAs, but no protein accumulation could be detected. Using chloroplast promoters and 5′ untranslated regions (UTRs) to drive the expression of the bacterial aadA gene, encoding aminoglycoside adenine transferase (AAD), stable accumulation of a foreign protein in transgenic chloroplasts was first reported in 1991 as the ability of the transformed cells to resist spectinomycin treatment and by an enzymatic assay of AAD activity [150]. The first identification of foreign protein expression using western blot analysis was shown in 1999 for the bacterial B(β)-glucuronidase (GUS) reporter protein, driven by either the rbcL, psbA or atpA promoters and 5′ UTRs [151]. Similarly, expression of Renilla luciferase protein was also shown by western blot analysis in 1999 [152]. Although protein accumulation in each of these initial studies appeared to be very low, probably less than 0.01% of soluble protein, the recovery of enzymatic activity of the recombinant proteins in all cases indicated that properly folded foreign proteins could be expressed in chloroplasts of C. reinhardtii. More recently, as our understanding of chloroplast gene regulation has improved, reports have shown recombinant proteins accumulating as high as 10–20% of total soluble protein [80,153]. However, there is significantly more to be learned, since different lines that have identical plastid insertions can have quite disparate protein accumulation. This suggests that second site mutations (probably nuclear) may be induced during the chloroplast transformation event.
Although protein expression in algae has been improved over the last 20 years, application of these technologies outside of the laboratory has been limited. The growing interest in biofuels, has renewed interest in improving heterologous protein expression for real-world applications, with a goal for heterologous protein expression of increased lipid production. This was attempted by simply overexpressing the acetyl-CoA carboxylase (ACC) in the diatom Cyclotella cryptica to improve lipid content by increasing the first enzyme in the lipid biogenesis pathway. Unfortunately, this initial experiment did not yield increased lipid production [16]. As the complexities of lipid regulation have been further elucidated, the possibilities for modifying lipid metabolism have increased [154]. These modifications may increase lipid content, as well as generate easily processed lipids for fuels. Another major recent focus in biofuels is lipid secretion. Cyanobacteria have been engineered to secrete as much as 133 mg/l/day [155]. The ability to secrete lipids may improve efficiency in harvesting and nutrient utilization, as well as provide some protection from contamination. Similar strategies have been proposed for diatoms and other eukaryotic algae but have yet to be implemented to our knowledge. Although lipid quantity and secretion have seen limited success, improvements in lipid composition and content in higher plants have been attempted. For the most part, these improvements have been undertaken to modify the carbon chain length and degrees of saturation to produce neutraceuticals, such as omega-3 and omega-6 fatty acids [89,156,157]. These successful experiments suggest that improving lipid production in algae is a tenable option to improve biofuel charactieristics.
Improving fuel molecules
Algae can be harnessed to produce a number of molecules that can be used for fuels. The current trend is to use the fatty acids that are naturally produced in algae, since many species produce these lipids at a substantial percentage of their total mass. In-depth discussions of algal lipid production have been reviewed elsewhere [158–160]. In brief, for fatty acid production, acetyl CoA is converted to malonyl CoA through the activity of acetyl CoA carboxylase (ACCase). Fatty acid synthase (FAS) then transfers the malonyl moiety from the malonyl CoA to an acyl carrier protein (ACP), generating malonyl-ACP. Subsequent condensation steps catalyzed by FAS proteins elongate the growing acyl chain, while elongases and desaturases further modify the fatty acids. Most algae species accumulate fatty acids with a range in length of 16–20 carbons, with no to as many as five sites of unsaturation. Although the fatty acid composition of different algae species is distinct, once extracted, its chemical profile is not expected to be substantially different from the chemical mixes currently found in sweet light crude oil. The enzymes involved in oil production may be able to be manipulated, thus increasing the oil production of these species, and the resulting oil can be used after being transesterified as diesel or undergo carbon cracking to produce biogasoline or biojet fuels [161]. To maximize fuel production and minimize costs, knowing the exact lipid content in terms of carbon chain length is important, so that chemical modification can be optimized. Lipid composition can be determined through lipid extraction, fractionation and subsequent mass spectrometry analysis. Different methods of lipid extraction from hexane extraction to pressing strategies extract lipids with different efficiencies and can yield different lipid profiles and returns. It is important that the extraction strategies be optimized depending on downstream industrial applications.
Although harvesting endogenous lipids, or even improved lipids through bioengineering is the most likely strategy for biofuel production, algae can produce a number of other potential fuel molecules. Much discussion has surrounded the potential of developing a hydrogen economy. A number of algal species can be induced to produce hydrogen gas using the reducing potential of photosynthesis in a reaction catalyzed by a hydrogenase [162]; however, both the movement towards a hydrogen economy and the ability to engineer algal species to produce substantial amounts of hydrogen in a regulated manner have moved forward at a slower rate than once predicted. Hydrogen production in algae has been reviewed elsewhere [162]. In addition to hydrogen, algae produce hexose sugars, which can be fermented to produce ethanol, butanol and potentially longer chain hydrocarbons. Although this strategy has not been extensively studied, the high photosynthetic efficiency of algae coupled with high biomass per area and the low cellulose content of many species makes algae a candidate that could be considered for subsequent fermentation.
Although both hydrogen- and sugar-based biofuel production could potentially impact the use of petroleum fuels, algae have the potential to produce a number of secondary metabolites that have characteristics much closer to existing petroleum fuels. The most promising of these are the terpenes, which offer a potential new fuel source outside of fatty acids that is compatible with our existing fuel framework. Terpenes are polymers of isoprene C5H8. If produced in high quantities, hemiterpenes (single isoprene units) and monoterpenes (C10H16) could function in current gasoline engines with only minor engine modifications [163]. Longer isoprene chains could be used for biodiesel or cracked into biogasoline or jet fuels. To date, terpene accumulation for fuels has not been a large focus of algal biology, since many terpenes and terpene derivatives have properties that make them more valuable for other uses, such as flavor, fragrances and antibiotics [164,165]. However, if algae can be engineered to produce higher levels of certain terpenes, then these molecules present an alternative to fatty acids as an algal biofuel.
Identifying natural traits to improve the economics of production strains
In addition to improved lipid production, other strain-specific improvements are being considered, including crop protection, salt tolerance, growth at high pH, improved nutrient utilization and traits that lead to more efficient harvesting, such as flocculation. The economic impact from these improvements will allow for decrease operational costs. For example, flocculation will allow improved harvesting by making it easier to concentrate algae, decreasing the cost of water extraction. A variety of algae have been characterized for their fouling of marine equipment. Some of the qualities that cause fouling include rapid growth on specific substrates. Further studies of these attributes may lead to improved flocculation and extraction, by understanding how and why these species grow and adhere to these specific substrates. These data may allow these traits to be exploited, so that algae will aggregate after a specific induction event or to a specific surface.
In general, the literature on algal strain improvements is minimal; however, extrapolation from successes in other system may provide a blueprint for some of these improvements. One such improvement is better salt tolerance conferred through expression of glycinebetaine and polyamines in terrestrial plants [166,167]. Despite the potential value of these improvements, their real value must be determined experimentally in each species.
Improved economics through the production of natural co-products
The extraction and sale of natural or engineered co-products along with algae oil could positively impact the economics of algae-based biofuels. If the infrastructure for algae fuel production is in place, expansion of these facilities to include protein or other coproduct purification can be added at a fraction of total cost. The postprocessing residue from algae oil extraction consists primarily of proteins and carbohydrates. Conventional use of these by-products might include anaerobic digestion to generate methane gas [163], combustion for energy production or, perhaps, use as animal feed, although algae are not presently sold as animal feed outside of the aquaculture industry. The high protein content of most microalgae and their amino acid composition makes them suitable for human and animal nutrition. The cyanobacteria Arthrospira (i.e., Spirulina) has a 60–71% dry-weight protein content, and is widely used as a food supplement for humans, cattle, poultry, aquarium fish, ornamental birds and horses [168]. Algae biomass is also an essential source of nutrients for fish, mollusk and shrimp in the aquaculture industry. The most popular algae genera are Tetraselmis, Nannochloropsis, Isochrysis, Pavlova, Navicula, Nitzschia, Chaetoceros, Skeletonema, Phaeodactylum and Thalassiosira [169,170]. Chlorella is also regarded as an excellent nutrient source for humans but it also produces a high valuable molecule, β-1,3-glucan. This polysaccharide is a recognized immunostimulator, a free radical scavenger and a reducer of blood lipids [171].
Given their diverse nature, microalgae can produce a wide variety of nutrients and secondary metabolites that are beneficial for human or animals. Valuable current or potential co-products include carotenoids, and long-chain polyunsaturated fatty acids (LCPUFAs). Microalgae can also produce a wide variety of useful carotenoids, such as lutein, zeaxanthin, lycopene, bixin, β-carotene and astaxanthin. However, commercial production is mainly confined to the latter two [172–174]. β-carotene is produced by the marine algae D. salina, which can accumulate up to 14% of its dry weight as this pigment under stress conditions [175]. This carotenoid is an orange pigment, widely used as a natural food colorant. It is also a strong antioxidant and a precursor of vitamin A [176,177]. The main producer of astaxanthin is the freshwater algae, Haematococcus pluvialis, which can accumulate up to 4% of its dry weight as this pigment [178]. Astaxanthin is a red pigment, mainly used as a feed additive for coloring salmon, carp, red seabream, shrimp and chickens. It is also used as a food supplement for humans, given that it is an extraordinary antioxidant [179,180].
Microalgae can also synthesize LCPUFAs, including omega-3 and omega-6. These are essential for humans and marine animals but they are only available in a very limited selection of foods [181,182]. Docosahexaenoic acid (DHA) is an omega-3 fatty acid commercially produced by Crypthecodinium and Schizochytrium for infant formulas and aquaculture feeds. Algae can also efficiently produce other important LCPUFAs but are currently not the main commercial source of these fatty acids. These LCPUFAs include eicosapentanoic acid (produced by Nannochloropsis, Phaeodactylum and Nitzschia), arachidonic acid (produced by Porphyridium) and γ-linoleic acid (produced by Arthrospira) [183].
Additional minor commercial products from microalgae are phycobiliproteins, used as food and research dyes (Arthrospira and Porphyridium) [184,185], extracts for cosmetics (Nannochloropsis and Dunaliella) [186], and stable isotope biomolecules used for research (Phaeodactylum and Arthrospira) [187].
Microalgae can synthesize many other unique molecules with commercial potential, such as toxins, vitamins, antibiotics, sterols, lectins, mycosporine-like amino acids, halogenated compounds and polyketides. In some instances, the expression of molecules that improve crop protection may also have pharmaceutical value. For further reading see [183,188–192].
These natural co-products have potential to provide a bridge while the economics of algal biofuels improve. Early on, owing to the large market for fuels and companies establishing niches, it is most likely that diverse coproduct-producing strains will be used, rather than an optimal single strain. In addition, many of these co-products will be coextracted with the lipids using current strategies, decreasing their value as a coproduct. Improving extraction techniques or dedicating a percentage of the algal crop to these higher value products depending on demand (e.g., with terrestrial agriculture) may further close the economic gap between petroleum and algae biofuels (Table 2).
Table 2.
Potential algae co-products.
| Product | Price (US$) | Market estimation (US$ × 106/year) |
|---|---|---|
| Biomass for humans | 50 per kg | 1250–3800 |
| Aquaculture feed | 70 per l; >160 per kg | ∼700 |
| β-carotene | 300–3000 per kg | >280 |
| Astaxanthin | 2,500–10,000 per kg | 150–200 |
| Polyunsaturated fatty acid | 60 per g | ∼1530 |
| Phycobiliproteins | 3.25–17 per mg | 12–50 |
| Radiolabeled metabolites | 60–38,000 per g | 5–13 |
Byproducts economics
Co-products derived from the protein fraction of algae
An alternative coproduct strategy is to genetically engineer algae to produce higher value proteins, specifically industrial enzymes and animal feed supplements. These co-products would enhance the value of the protein residual from biofuel production strains. The current market values for recombinant proteins range between US$5 and $10,000 per kg. Although the higher value end of this spectrum consists of small niche markets, there are many industrial enzymes that can be produced, with values ranging between $25 and $50 per kg, and for which large markets currently exist [25]. With sufficient expression and economic purification of these protein co-products, the price gap between fossil fuels and algae-based biofuels can be minimized.
Future perspective
We have discussed strategies to make algae-based fuels costs competitive with petroleum. Bioprospecting is of importance to identify algal species that have desired traits (e.g. high lipid content, growth rates, growth densities and/or the presence of valuable co-products), while growing on low-cost media. Despite the potential of this strategy, the most likely scenario is that bioprospecting will not identify species that are cost competitive with petroleum, and subsequent genetic engineering and breeding will be required to bring these strains to economic viability. The range of potential for engineering algae is just beginning to be realized, from improving lipid biogenesis and improving crop protection, to producing valuable enzyme or protein co-products. No sustainable technology is without its challenges but blind promotion of those technologies without honest consideration of the long-term implications may lead to the acceptance of strategies whose long-term consequences outweigh their short-term benefits. We have presented what we view as the most important current and upcoming challenges of algae biofuels but, as with any new industry, the more we learn the more we realize that challenges exist that we had not foreseen. Even given these uncertainties, we believe that fuel production from algae can be cost competitive and widely scalable and deployable in the next 7–10 years, but only if we continue to expand our understanding of these amazing organisms as we expand our ability to engineer them for the specific task of developing a new energy industry.
Acknowledgments
Stephen Mayfield is a founder of and has a financial interest in Sapphire Energy an algal biofuel company, but this work should not be considered to reflect the views of Sapphire Energy. This report was supported by a grant from the US Air Force #FA9550-09-1-0336. Special thanks to Michael Burkart and Evan Stephens for their comments.
Key terms
- Fertilizer
The major components of fertilizers are derived from nonrenewable sources, from phosphorus mines, to ammonia generated through petroleum cracking.
- Commodity
Commercialization of biofuels requires that the biofuel is interchangeable and competitive with current fuels on the market. As such, its price is subject to supply and demand of the competing fuels and, as such, is a commodity.
- Crop protection
Broad deployment of monocultured algae will require protection from both biotic and abiotic factors that allow maximal sustainable growth. Crop protection is a critical focus of western agriculture and will be crucial for aqua culture for biofuels.
- Sustainability
Initial lifecycle analyses of biofuels are often incomplete, as demonstrated with corn to ethanol; however, sustainability should also be applied to business plans and whether both the direct and indirect costs associated with the process of interest are truly covered by the revenue generated by the company.
- Hydrocarbon cracking
The majority of petroleum is not immediately useable, and the various carbon chains found therein require reformation through oil refining, including catalytic cracking of longer-chain hydrocarbons into shorter-chain hydrocarbons.
Footnotes
Financial & competing interests disclosure: The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪ ▪ of considerable
- 1.Parry ML, Intergovernmental Panel on Climate Change, Working Group II, World Meteorological Organization, United Nations Environment Programme . Summary for Policymakers. In: Parry ML, Canziani OF, Palutikof JP, van der Linden PJ, Hanson CE, editors. Climate Change 2007: Impacts, Adaptation and Vulnerability Contribution of Working Group II to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge University Press; Cambridge, UK: 2007. [Google Scholar]
- 2.Dyni JR. Scientific Investigations Report 2005-5294. US Geological Survey; VA, USA: 2006. Geology and resources of some world oil-shale deposits. [Google Scholar]
- 3.Schindler J, Zittel W. Crude Oil – The Supply Outlook. Vol. 102. Energy Watch Group; Ottobrunn, Germany: 2008. [Google Scholar]
- 4.Energy Information Administration. International Energy Outlook. Vol. 284. EIA; DC, USA: 2009. [Google Scholar]
- 5.Nass LL, Pereira PAA, Ellis D. Biofuels in Brazil: an overview. Crop Sci. 2007;47:2228–2237. [Google Scholar]
- 6.Fargione J, Hill J, Tilman D, Polasky S, Hawthorne P. Land clearing and the biofuel carbon debt. Science. 2008;319(5867):1235–1238. doi: 10.1126/science.1152747. [DOI] [PubMed] [Google Scholar]; ▪ Promotes looking at the impact of biofuels total lifecycle analysis, and redefines what should be looked at in that analysis.
- 7.Searchinger T, Heimlich R, Houghton RA, et al. Use of U.S. croplands for biofuels increases greenhouse gases through emissions from land-use change. Science. 2008;319(5867):1238–1240. doi: 10.1126/science.1151861. [DOI] [PubMed] [Google Scholar]; ▪ Promotes looking at the impact of biofuels total lifecycle analysis and redefines what should be looked at in that analysis.
- 8.Huber GW, Iborra S, Corma A. Synthesis of transportation fuels from biomass: chemistry, catalysts, and engineering. Chem Rev. 2006;106(9):4044–4098. doi: 10.1021/cr068360d. [DOI] [PubMed] [Google Scholar]; ▪ Excellent review of the chemistry behind conversion of biomass to useable fuels.
- 9.Dismukes GC, Carrieri D, Bennette N, Ananyev GM, Posewitz MC. Aquatic phototrophs: efficient alternatives to land-based crops for biofuels. Curr Opin Biotechnol. 2008;19(3):235–240. doi: 10.1016/j.copbio.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 10.Borowitzka MA. Algal biotechnology products and processes – matching science and economics. J Applied Phycology. 1992;4:267–279. [Google Scholar]
- 11.Chisti Y. Biodiesel from microalgae. Biotechnol Adv. 2007;25(3):294–306. doi: 10.1016/j.biotechadv.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 12.Alabi AO, Tampier M, Bibeau E. Microalgae Technogies and Processes for Biofuels/Bioenergy Production in British Columbia. The BC Innovation Council; BC, Canada: 2009. [Google Scholar]
- 13.Huntley ME, Redalje DG. CO2 mitigation and renewable oil from photosynthetic microbes: a new appraisal. Mitigat Adapt Strategies Global Change. 2006;12:573–608. [Google Scholar]
- 14.Falkowski PG, Barber RT, Smetacek VV. Biogeochemical controls and feedbacks on ocean primary production. Science. 1998;281(5374):200–207. doi: 10.1126/science.281.5374.200. [DOI] [PubMed] [Google Scholar]
- 15.Parker MS, Mock T, Armbrust EV. Genomic insights into marine microalgae. Annu Rev Genet. 2008;42:619–645. doi: 10.1146/annurev.genet.42.110807.091417. [DOI] [PubMed] [Google Scholar]
- 16.Sheehan J, Dunahay T, Benemann J, Roessler P. A Look Back at the U S Department of Energy's Aquatic Species Program – Biodiesel from Algae. Vol. 328. National Renewable Energy Laboratory; CO, USA: 1998. [Google Scholar]; ▪ ▪ Outlines some of the early successes and challenges faced by groups currently addressing algae biofuels.
- 17.Huesemann MH, Hausmann TS, Bartha R, Aksoy M, Weissman JC, Benemann JR. Biomass productivities in wild type and pigment mutant of Cyclotella sp. (diatom) Appl Biochem Biotechnol. 2009;157(3):507–526. doi: 10.1007/s12010-008-8298-9. [DOI] [PubMed] [Google Scholar]
- 18.Rodolfi L, Chini Zittelli G, Bassi N, et al. Microalgae for oil: strain selection, induction of lipid synthesis and outdoor mass cultivation in a low-cost photobioreactor. Biotechnol Bioeng. 2009;102(1):100–112. doi: 10.1002/bit.22033. [DOI] [PubMed] [Google Scholar]; ▪ Strain selection is of major importance for commercial biofuel production. The authors provide a solid foundation for comparing potential producitivity of multiple species. This area is under-represented in biofuels literature and the authors should be commended.
- 19.Kojima E, Zhang K. Growth and hydrocarbon production of microalga Botryococcus braunii in bubble column photobioreactors. J Biosci Bioeng. 1999;87(6):811–815. doi: 10.1016/s1389-1723(99)80158-3. [DOI] [PubMed] [Google Scholar]
- 20.Deschamps P, Moreira D. Signal conflicts in the phylogeny of the primary photosynthetic eukaryotes. Mol Biol Evol. 2009;26(12):2745–2753. doi: 10.1093/molbev/msp189. [DOI] [PubMed] [Google Scholar]
- 21.Reeb VC, Peglar MT, Yoon HS, et al. Interrelationships of chromalveolates within a broadly sampled tree of photosynthetic protists. Mol Phylogenet Evol. 2009;53(1):202–211. doi: 10.1016/j.ympev.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 22.He P, Xu S, Zhang H, et al. Bioremediation efficiency in the removal of dissolved inorganic nutrients by the red seaweed, Porphyra yezoensis, cultivated in the open sea. Water Res. 2008;42(4–5):1281–1289. doi: 10.1016/j.watres.2007.09.023. [DOI] [PubMed] [Google Scholar]
- 23.Fierro S, Sanchez-Saavedra Mdel P, Copalcua C. Nitrate and phosphate removal by chitosan immobilized Scenedesmus. Bioresour Technol. 2008;99(5):1274–1279. doi: 10.1016/j.biortech.2007.02.043. [DOI] [PubMed] [Google Scholar]
- 24.Douskova I, Doucha J, Livansky K, et al. Simultaneous flue gas bioremediation and reduction of microalgal biomass production costs. Appl Microbiol Biotechnol. 2009;82(1):179–185. doi: 10.1007/s00253-008-1811-9. [DOI] [PubMed] [Google Scholar]
- 25.Rosenberg JN, Oyler GA, Wilkinson L, Betenbaugh MJ. A green light for engineered algae: redirecting metabolism to fuel a biotechnology revolution. Curr Opin Biotechnol. 2008;19(5):430–436. doi: 10.1016/j.copbio.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 26.Zaslavskaia LA, Lippmeier JC, Shih C, Ehrhardt D, Grossman AR, Apt KE. Trophic conversion of an obligate photoautotrophic organism through metabolic engineering. Science. 2001;292(5524):2073–2075. doi: 10.1126/science.160015. [DOI] [PubMed] [Google Scholar]
- 27.Lehr F, Posten C. Closed photo-bioreactors as tools for biofuel production. Curr Opin Biotechnol. 2009;20(3):280–285. doi: 10.1016/j.copbio.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 28.Chisti Y. Biodiesel from microalgae beats bioethanol. Trends Biotechnol. 2008;26(3):126–131. doi: 10.1016/j.tibtech.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 29.Borowitzka MA. Commercial production of microalgae: ponds, tanks, tubes and fermenters. J Biotechnology. 1999;70:313–321. [Google Scholar]
- 30.Krichnavaruk S, Shotipruk A, Goto M, Pavasant P. Supercritical carbon dioxide extraction of astaxanthin from Haematococcus pluvialis with vegetable oils as co-solvent. Bioresour Technol. 2008;99(13):5556–5560. doi: 10.1016/j.biortech.2007.10.049. [DOI] [PubMed] [Google Scholar]
- 31.Maher KD, Bressler DC. Pyrolysis of triglyceride materials for the production of renewable fuels and chemicals. Bioresour Technol. 2007;98(12):2351–2368. doi: 10.1016/j.biortech.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 32.US Energy Information Administration. United States Energy Profile. EIA; DC, USA: 2010. [Google Scholar]
- 33.Gerbens-Leenes W, Hoekstra AY, Van Der Meer TH. The water footprint of bioenergy. Proc Natl Acad Sci USA. 2009;106(25):10219–10223. doi: 10.1073/pnas.0812619106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marchetti A, Parker MS, Moccia LP, et al. Ferritin is used for iron storage in bloom-forming marine pennate diatoms. Nature. 2009;457(7228):467–470. doi: 10.1038/nature07539. [DOI] [PubMed] [Google Scholar]
- 35.Ruiz-Marin A, Mendoza-Espinosa LG, Stephenson T. Growth and nutrient removal in free and immobilized green algae in batch and semi-continuous cultures treating real wastewater. Bioresour Technol. 2009;101(1):58–64. doi: 10.1016/j.biortech.2009.02.076. [DOI] [PubMed] [Google Scholar]
- 36.Conley DJ, Paerl HW, Howarth RW, et al. Ecology. Controlling eutrophication: nitrogen and phosphorus. Science. 2009;323(5917):1014–1015. doi: 10.1126/science.1167755. [DOI] [PubMed] [Google Scholar]
- 37.Liu D, Keesing JK, Xing Q, Shi P. World's largest macroalgal bloom caused by expansion of seaweed aquaculture in China. Mar Pollut Bull. 2009;58(6):888–895. doi: 10.1016/j.marpolbul.2009.01.013. [DOI] [PubMed] [Google Scholar]
- 38.US Department of Agriculture. Farm Production Expenditures 2008 Summary. Vol. 80. US Department of Agriculture; DC, USA: 2009. [Google Scholar]
- 39.Byerlee D, De Janvry A, World Bank . Agriculture for Development. World Bank; DC, USA: 2007. [Google Scholar]
- 40.Vaccari DA. Phosphorus: a looming crisis. Sci Am. 2009;300(6):54–59. doi: 10.1038/scientificamerican0609-54. [DOI] [PubMed] [Google Scholar]
- 41.Funderberg E. Why are nitrogen prices so high. Pasture Range. 2009;5 [Google Scholar]
- 42.Vance CP. Symbiotic nitrogen fixation and phosphorus acquisition. Plant nutrition in a world of declining renewable resources. Plant Physiol. 2001;127(2):390–397. [PMC free article] [PubMed] [Google Scholar]
- 43.Ryther JH, Dunstan WM. Nitrogen, phosphorus, and eutrophication in the coastal marine environment. Science. 1971;171(3975):1008–1013. doi: 10.1126/science.171.3975.1008. [DOI] [PubMed] [Google Scholar]
- 44.Long SR. Rhizobium–legume nodulation: life together in the underground. Cell. 1989;56(2):203–214. doi: 10.1016/0092-8674(89)90893-3. [DOI] [PubMed] [Google Scholar]
- 45.Inokuchi R, Kuma Ki, Miyata T, Okada M. Nitrogen-assimilating enzymes in land plants and algae: phylogenic and physiological perspectives. Physiol Plant. 2002;116(1):1–11. doi: 10.1034/j.1399-3054.2002.1160101.x. [DOI] [PubMed] [Google Scholar]
- 46.Berman-Frank I, Lundgren P, Falkowski P. Nitrogen fixation and photosynthetic oxygen evolution in cyanobacteria. Res Microbiol. 2003;154(3):157–164. doi: 10.1016/S0923-2508(03)00029-9. [DOI] [PubMed] [Google Scholar]
- 47.Coale KH, Johnson KS, Chavez FP, et al. Southern ocean iron enrichment experiment: carbon cycling in high- and low-si waters. Science. 2004;304(5669):408–414. doi: 10.1126/science.1089778. [DOI] [PubMed] [Google Scholar]
- 48.Boyd PW, Law CS, Wong CS, et al. The decline and fate of an iron-induced subarctic phytoplankton bloom. Nature. 2004;428(6982):549–553. doi: 10.1038/nature02437. [DOI] [PubMed] [Google Scholar]
- 49.Coale KH, Johnson KS, Fitzwater SE, et al. A massive phytoplankton bloom induced by an ecosystem-scale iron fertilization experiment in the equatorial pacific ocean. Nature. 1996;383(6600):495–501. doi: 10.1038/383495a0. [DOI] [PubMed] [Google Scholar]
- 50.Godman J, Balk J. Genome analysis of chlamydomonas reinhardtii reveals the existence of multiple, compartmentalized iron-sulfur protein assembly machineries of different evolutionary origins. Genetics. 2008;179(1):59–68. doi: 10.1534/genetics.107.086033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yildiz FH, Davies JP, Grossman AC. Characterization of sulfate transport in Chlamydomonas reinhardtii during sulfur-limited and sulfur-sufficient growth. Plant Physiol. 1994;104:981–987. doi: 10.1104/pp.104.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maathuis FJ. Physiological functions of mineral macronutrients. Curr Opin Plant Biol. 2009;12(3):250–258. doi: 10.1016/j.pbi.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 53.Sialve B, Bernet N, Bernard O. Anaerobic digestion of microalgae as a necessary step to make microalgal biodiesel sustainable. Biotechnol Advances. 2009;27(4):409–416. doi: 10.1016/j.biotechadv.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 54.Brussaard CPD. Viral control of phytoplankton populations – a review. J Eurkaryotic Microbiol. 2004;51(2):125–138. doi: 10.1111/j.1550-7408.2004.tb00537.x. [DOI] [PubMed] [Google Scholar]
- 55.Mayali X, Azam F. Algicidal bacteria in the sea and their impact on algal blooms. J Eurkaryotic Microbiol. 2004;51(2):139–144. doi: 10.1111/j.1550-7408.2004.tb00538.x. [DOI] [PubMed] [Google Scholar]
- 56.Park M, Yih W, Coats DW. Parasites and phytoplankton, with special emphasis on dinoflagellate infections. J Eurkaryotic Microbiol. 2004;51(2):145–155. doi: 10.1111/j.1550-7408.2004.tb00539.x. [DOI] [PubMed] [Google Scholar]
- 57.Tillmann U. Interactions between planktonic microalgae and protozoan grazers. J Eurkaryotic Microbiol. 2004;51(2):156–168. doi: 10.1111/j.1550-7408.2004.tb00540.x. [DOI] [PubMed] [Google Scholar]
- 58.Lebeau T, Robert JM. Diatom cultivation and biotechnologically relevant products. Part I. Cultivation at various scales. Appl Microbiol Biotechnol. 2003;60(6):612–623. doi: 10.1007/s00253-002-1176-4. [DOI] [PubMed] [Google Scholar]
- 59.Matsudo MC, Bezerra RP, Sato S, Perego P, Converti A, Carvalho JCM. Repeat fed-batch cultivation of Arthrospira (spirulina) patensis using urea as nitrogen source. Biochem Engineer J. 2008;43(1):52–57. [Google Scholar]
- 60.Ginzburg M, Ginzburg BZ. Interrelationships of light, temperature, sodium chloride and carbon source in growth of halotolerant and halophilic strains of Dunaliella. Eur J Phycology. 1981;16(3):313–324. [Google Scholar]
- 61.Dubinsky Z, Rotem J. Relations between algal populations and the pH of their media. Oecologia. 1974;16:53–69. doi: 10.1007/BF00345087. [DOI] [PubMed] [Google Scholar]
- 62.Borowitzka MA. Culturing microalgae in outdoor ponds. In: Andersen RA, editor. Algal Culturing Techniques. Vol. 218. Academic Press; NY, USA: 2005. [Google Scholar]
- 63.Kulik MM. The potential for using cyanobacteria (blue-green algae) and algae in the biological control of plant pathogenic bacteria and fungi. Eur J Plant Pathol. 1995;101(6):585–599. [Google Scholar]
- 64.Thyrhaug R, Larsen A, Thingstad Tf, Bratbak G. Stable coexistence in marine algal host–virus systems. Marine Ecol Progress Series. 2003;254:27–35. [Google Scholar]
- 65.Bhadury P, Wright PC. Exploitation of marine algae: biogenic compounds for potential antifouling applications. Planta. 2004;219(4):561–578. doi: 10.1007/s00425-004-1307-5. [DOI] [PubMed] [Google Scholar]
- 66.Pesando D. Antibacterial and antifungal activities of marine algae. Intro Applied Phycol. 1990:3–26. [Google Scholar]
- 67.Gupta AB, Shrivastava GC. On antibiotic properties of some fresh water algae. Hydrobiologia. 1965;25(1):285–288. [Google Scholar]
- 68.Chu CY, Liao WR, Huang R, Lin LP. Haemagglutinating and antibiotic activities of freshwater microalgae. World J Microbiol Biotechnol. 2004;20(8):817–825. [Google Scholar]
- 69.Santoyo S, Rodríguez-Meizoso I, Cifuentes A, et al. Green processes based on the extraction with pressurized fluids to obtain potent antimicrobials from Haematococcus pluvialis microalgae. LWT Food Science Technol. 2009;42(7):1213–1218. [Google Scholar]
- 70.Wolfe GV. The chemical defense ecology of marine unicellular plankton: constraints, mechanisms, and impacts. Biological Bull. 2000;198(2):225. doi: 10.2307/1542526. [DOI] [PubMed] [Google Scholar]
- 71.Pohnert G. Diatom/copepod interactions in plankton: the indirect chemical defense of unicellular algae. Chem Bio Chem. 2005;6(6):946–959. doi: 10.1002/cbic.200400348. [DOI] [PubMed] [Google Scholar]
- 72.Copping LG, Duke SO. Natural products that have been used commercially as crop protection agents. Pest Management Sci. 2007;63(6):524–554. doi: 10.1002/ps.1378. [DOI] [PubMed] [Google Scholar]
- 73.Berry JP, Gantar M, Perez MH, Berry G, Noriega FG. Cyanobacterial toxins as allelochemicals with potential applications as algaecides, herbicides and insecticides. Mar Drugs. 2008;6:117–146. doi: 10.3390/md20080007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Degray G, Rajasekaran K, Smith F, Sanford J, Daniell H. Expression of an antimicrobial peptide via the chloroplast genome to control phytopathogenic bacteria and fungi. Plant Physiol. 2001;127(3):852. [PMC free article] [PubMed] [Google Scholar]
- 75.Oard SV, Enright FM. Expression of the antimicrobial peptides in plants to control phytopathogenic bacteria and fungi. Plant Cell Reports. 2006;25(6):561–572. doi: 10.1007/s00299-005-0102-5. [DOI] [PubMed] [Google Scholar]
- 76.Montesinos E. Antimicrobial peptides and plant disease control. FEMS Microbiology Lett. 2007;270(1):1–11. doi: 10.1111/j.1574-6968.2007.00683.x. [DOI] [PubMed] [Google Scholar]
- 77.Rivas L, Luque-Ortega, Andreu D. Amphibian antimicrobial peptides and protozoa: Lessons from parasites. BBA Biomembranes. 2009;1788(8):1570–1581. doi: 10.1016/j.bbamem.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 78.Marcos JF, Muñoz A, Pérez-Payá E, Misra S, López-García B. Identification and rational design of novel antimicrobial peptides for plant protection. Ann Rev Phytopathol. 2008;46:273. doi: 10.1146/annurev.phyto.121307.094843. [DOI] [PubMed] [Google Scholar]
- 79.Chen Y, Wang Y, Sun Y, Zhang L, Li W. Highly efficient expression of rabbit neutrophil peptide-1 gene in chlorella ellipsoidea cells. Curr Genet. 2001;39(5):365–370. doi: 10.1007/s002940100205. [DOI] [PubMed] [Google Scholar]
- 80.Manuell AL, Beligni MV, Elder JH, et al. Robust expression of a bioactive mammalian protein in chlamydomonas chloroplast. Plant Biotechnol J. 2007;5(3):402–412. doi: 10.1111/j.1467-7652.2007.00249.x. [DOI] [PubMed] [Google Scholar]
- 81.Molenaar AJ, Harris DP, Rajan GH, et al. The acute-phase protein serum amyloid a3 is expressed in the bovine mammary gland and plays a role in host defence. Biomarkers. 2009;14(1):26–37. doi: 10.1080/13547500902730714. [DOI] [PubMed] [Google Scholar]
- 82.Li SS, Tsai HJ. Transgenic microalgae as a non-antibiotic bactericide producer to defend against bacterial pathogen infection in the fish digestive tract. Fish Shellfish Immunol. 2009;26(2):316–325. doi: 10.1016/j.fsi.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 83.Kuci Ska J, Lonc E, Rydzanicz K. Transgenic bioinsecticides inimical to parasites, but imical to environment. Wiadomo Ci Parazytologiczne. 2003;49(1):11. [PubMed] [Google Scholar]
- 84.Rosi-Marshall Ej, Tank Jl, Royer Tv, et al. Toxins in transgenic crop byproducts may affect headwater stream ecosystems. Proc Natl Acad Sci USA. 2007;104(41):16204. doi: 10.1073/pnas.0707177104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bøhn T, Primicerio R, Hessen DO, Traavik T. Reduced fitness of Daphnia magna fed a bt-transgenic maize variety. Arch Environment Contamin Toxicol. 2008;55(4):584–592. doi: 10.1007/s00244-008-9150-5. [DOI] [PubMed] [Google Scholar]
- 86.Hansen FC, Reckermann M, Klein Breteler WCM, Riegman R. Phaeocystis blooming enhanced by copepod predation on protozoa: evidence from incubation experiments. Marine Ecol Progress Series. 1993;102:51–51. [Google Scholar]
- 87.Armbrust EV. The life of diatoms in the World's oceans. Nature. 2009;459(7244):185–192. doi: 10.1038/nature08057. [DOI] [PubMed] [Google Scholar]
- 88.Roessler PG. Changes in the activities of various lipid and carbohydrate biosynthetic enzymes in the diatom Cyclotella cryptica in response to silicon deficiency. Arch Biochem Biophys. 1988;267(2):521–528. doi: 10.1016/0003-9861(88)90059-8. [DOI] [PubMed] [Google Scholar]
- 89.Petrie JR, Shrestha P, Mansour MP, Nichols PD, Liu Q, Singh SP. Metabolic engineering of omega-3 long-chain polyunsaturated fatty acids in plants using an acyl-CoA δ6-desaturase with omega3-preference from the marine microalga micromonas pusilla. Metab Eng. 2009;12(3):233–240. doi: 10.1016/j.ymben.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 90.Napier JA, Beaudoin F, Michaelson LV, Sayanova O. The production of long chain polyunsaturated fatty acids in transgenic plants by reverse-engineering. Biochimie. 2004;86(11):785–792. doi: 10.1016/j.biochi.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 91.Apt KE, Kroth-Pancic PG, Grossman AR. Stable nuclear transformation of the diatom Phaeodactylum tricornutum. Mol Gen Genet. 1996;252(5):572–579. doi: 10.1007/BF02172403. [DOI] [PubMed] [Google Scholar]
- 92.Dunahay TG, Jarvis EE, Roessler PG. Genetic transformation of the diatoms Cyclotella cryptica and Navicula saprophila. J Phycol. 1995;31(6):1004–1012. [Google Scholar]
- 93.Poulsen N, Kroger N. A new molecular tool for transgenic diatoms: control of mrna and protein biosynthesis by an inducible promoter-terminator cassette. FEBS J. 2005;272(13):3413–3423. doi: 10.1111/j.1742-4658.2005.04760.x. [DOI] [PubMed] [Google Scholar]
- 94.Falciatore A, Casotti R, Leblanc C, Abrescia C, Bowler C. Transformation of nonselectable reporter genes in marine diatoms. Mar Biotechnol (NY) 1999;1(3):239–251. doi: 10.1007/pl00011773. [DOI] [PubMed] [Google Scholar]
- 95.De Riso V, Raniello R, Maumus F, Rogato A, Bowler C, Falciatore A. Gene silencing in the marine diatom Phaeodactylum tricornutum. Nucleic Acids Res. 2009;37(14):e96. doi: 10.1093/nar/gkp448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Boynton JE, Gillham NW. Genetics and transformation of mitochondria in the green alga Chlamydomonas. Methods Enzymol. 1996;264:279–296. doi: 10.1016/s0076-6879(96)64027-0. [DOI] [PubMed] [Google Scholar]
- 97.Boynton JE, Gillham NW. Chloroplast transformation in Chlamydomonas. Methods Enzymol. 1993;217:510–536. doi: 10.1016/0076-6879(93)17087-l. [DOI] [PubMed] [Google Scholar]
- 98.Kindle KL, Schnell RA, Fernandez E, Lefebvre PA. Stable nuclear transformation of Chlamydomonas using the Chlamydomonas gene for nitrate reductase. J Cell Biol. 1989;109(6 Pt 1):2589–2601. doi: 10.1083/jcb.109.6.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lerche K, Hallmann A. Stable nuclear transformation of Gonium pectorale. BMC Biotechnol. 2009;9:64. doi: 10.1186/1472-6750-9-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hawkins RL, Nakamura M. Expression of human growth hormone by the eukaryotic alga, Chlorella. Curr Microbiol. 1999;38(6):335–341. doi: 10.1007/pl00006813. [DOI] [PubMed] [Google Scholar]
- 101.Ahlgren G, Lundstedt L, Brett M, Forsberg C. Lipid composition and food quality of some freshwater phytoplankton for cladoceran zooplankters. J Plankton Res. 1990;12(4):809–818. [Google Scholar]
- 102.Xin L, Hong-Ying H, Jia Y. Lipid accumulation and nutrient removal properties of a newly isolated freshwater microalga, Scenedesmus sp. LX1, growing in secondary effluent. N Biotechnol. 2009;27(1):59–63. doi: 10.1016/j.nbt.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 103.Metzger P, Largeau C. Botryococcus braunii: a rich source for hydrocarbons and related ether lipids. Appl Microbiol Biotechnol. 2005;66(5):486–496. doi: 10.1007/s00253-004-1779-z. [DOI] [PubMed] [Google Scholar]
- 104.Kneip C, Voss C, Lockhart PJ, Maier UG. The cyanobacterial endosymbiont of the unicellular algae rhopalodia gibba shows reductive genome evolution. BMC Evol Biol. 2008;8:30. doi: 10.1186/1471-2148-8-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Carpenter EJ, Janson S. Intracellular cyanobacterial symbionts in the marine diatom Climacodium frauenfeldianum (bacillariophyceae) J Phycol. 2000;36(3):540–544. doi: 10.1046/j.1529-8817.2000.99163.x. [DOI] [PubMed] [Google Scholar]
- 106.Moheimani NR, Borowitzka MA. Limits to productivity of the alga Pleurochrysis carterae (haptophyta) grown in outdoor raceway ponds. Biotechnol Bioeng. 2007;96(1):27–36. doi: 10.1002/bit.21169. [DOI] [PubMed] [Google Scholar]
- 107.Blanco AM, Moreno J, Del Campo JA, Rivas J, Guerrero MG. Outdoor cultivation of lutein-rich cells of Muriellopsis sp. in open ponds. Appl Microbiol Biotechnol. 2007;73(6):1259–1266. doi: 10.1007/s00253-006-0598-9. [DOI] [PubMed] [Google Scholar]
- 108.Doucha J, Livansky K. Productivity, CO2/O2 exchange and hydraulics in outdoor open high density microalgal (Chlorella sp.) photobioreactors operated in a middle and southern european climate. J Applied Phycology. 2006;18(6):811–826. [Google Scholar]
- 109.Tao Y, Ferrer Jl, Ljung K, et al. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell. 2008;133(1):164–176. doi: 10.1016/j.cell.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Greenwell HC, Laurens LM, Shields RJ, Lovitt RW, Flynn KJ. Placing microalgae on the biofuels priority list: a review of the technological challenges. J R Soc Interface. 2009 doi: 10.1098/rsif.2009.0322. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chaisson Mj, Brinza D, Pevzner PA. De novo fragment assembly with short mate-paired reads: does the read length matter? Genome Res. 2009;19(2):336–346. doi: 10.1101/gr.079053.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lister R, Gregory BD, Ecker JR. Next is now: new technologies for sequencing of genomes, transcriptomes, and beyond. Curr Opin Plant Biol. 2009;12(2):107–118. doi: 10.1016/j.pbi.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Borevitz JO, Maloof JN, Lutes J, et al. Quantitative trait loci controlling light and hormone response in two accessions of Arabidopsis thaliana. Genetics. 2002;160(2):683–696. doi: 10.1093/genetics/160.2.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Galvan A, Gonzalez-Ballester D, Fernandez E. Insertional mutagenesis as a tool to study genes/functions in Chlamydomonas. Adv Exp Med Biol. 2007;616:77–89. doi: 10.1007/978-0-387-75532-8_7. [DOI] [PubMed] [Google Scholar]
- 115.Jeong Br, Wu-Scharf D, Zhang C, Cerutti H. Suppressors of transcriptional transgenic silencing in Chlamydomonas are sensitive to DNA-damaging agents and reactivate transposable elements. Proc Natl Acad Sci USA. 2002;99(2):1076–1081. doi: 10.1073/pnas.022392999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Casas-Mollano JA, Van Dijk K, Eisenhart J, Cerutti H. SET3P monomethylates histone H3 on lysine 9 and is required for the silencing of tandemly repeated transgenes in Chlamydomonas. Nucleic Acids Res. 2007;35(3):939–950. doi: 10.1093/nar/gkl1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mayfield SP, Manuell AL, Chen S, et al. Chlamydomonas reinhardtii chloroplasts as protein factories. Curr Opin Biotechnol. 2007;18(2):126–133. doi: 10.1016/j.copbio.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 118.Purton S. Tools and techniques for chloroplast transformation of Chlamydomonas. Adv Exp Med Biol. 2007;616:34–45. doi: 10.1007/978-0-387-75532-8_4. [DOI] [PubMed] [Google Scholar]
- 119.Bingham SE, Cox JC, Strem MD. Expression of foreign DNA in Chlamydomonas reinhardtii. FEMS Microbiol Lett. 1989;53(1–2):77–81. doi: 10.1016/0378-1097(89)90369-8. [DOI] [PubMed] [Google Scholar]
- 120.Debuchy R, Purton S, Rochaix JD. The argininosuccinate lyase gene of Chlamydomonas reinhardtii: an important tool for nuclear transformation and for correlating the genetic and molecular maps of the Arg7 locus. EMBO J. 1989;8(10):2803–2809. doi: 10.1002/j.1460-2075.1989.tb08426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kindle KL. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 1990;87(3):1228–1232. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mayfield SP, Kindle KL. Stable nuclear transformation of Chlamydomonas reinhardtii by using a C. reinhardtii gene as the selectable marker. Proc Natl Acad Sci USA. 1990;87(6):2087–2091. doi: 10.1073/pnas.87.6.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Purton S, Rochaix JD. Complementation of a Chlamydomonas reinhardtii mutant using a genomic cosmid library. Plant Mol Biol. 1994;24(3):533–537. doi: 10.1007/BF00024121. [DOI] [PubMed] [Google Scholar]
- 124.Zhang H, Herman PL, Weeks DP. Gene isolation through genomic complementation using an indexed library of Chlamydomonas reinhardtii DNA. Plant Mol Biol. 1994;24(4):663–672. doi: 10.1007/BF00023562. [DOI] [PubMed] [Google Scholar]
- 125.Vashishtha M, Segil G, Hall JL. Direct complementation of Chlamydomonas mutants with amplified yac DNA. Genomics. 1996;36(3):459–467. doi: 10.1006/geno.1996.0491. [DOI] [PubMed] [Google Scholar]
- 126.Hall LM, Taylor KB, Jones DD. Expression of a foreign gene in Chlamydomonas reinhardtii. Gene. 1993;124(1):75–81. doi: 10.1016/0378-1119(93)90763-s. [DOI] [PubMed] [Google Scholar]
- 127.Fuhrmann M, Oertel W, Hegemann P. A synthetic gene coding for the green fluorescent protein (GFP) is a versatile reporter in Chlamydomonas reinhardtii. Plant J. 1999;19(3):353–361. doi: 10.1046/j.1365-313x.1999.00526.x. [DOI] [PubMed] [Google Scholar]
- 128.Rochaix JD, Van Dillewijn J. Transformation of the green alga Chlamydomonas reinhardii with yeast DNA. Nature. 1982;296(5852):70–72. doi: 10.1038/296070a0. [DOI] [PubMed] [Google Scholar]
- 129.Boynton JE, Gillham NW, Harris EH, et al. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science. 1988;240(4858):1534–1538. doi: 10.1126/science.2897716. [DOI] [PubMed] [Google Scholar]; ▪ First pubication describing particle bombardment for algal chloroplast transformation.
- 130.Dunahay TG. Transformation of Chlamydomonas reinhardtii with silicon carbide whiskers. Biotechniques. 1993;15(3):452–455. 457–458, 460. [PubMed] [Google Scholar]
- 131.Brown LE, Sprecher SL, Keller LR. Introduction of exogenous DNA into Chlamydomonas reinhardtii by electroporation. Mol Cell Biol. 1991;11(4):2328–2332. doi: 10.1128/mcb.11.4.2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rajam MV, Kumar SV. Green alga (Chlamydomonas reinhardtii) Methods Mol Biol. 2006;344:421–433. doi: 10.1385/1-59745-131-2:421. [DOI] [PubMed] [Google Scholar]
- 133.Geng DG, Wang YQ, Wang P, Li WB, Sun YR. Stable expression of hepatitis B surface antigen gene in Dunaliella salina (chlorophyta) J Applied Phycology. 2003;15(6):451–456. [Google Scholar]
- 134.Sun Y, Yang ZY, Gao XS, Li QY, Zhang QQ, Xu ZK. Expression of foreign genes in Dunaliella by electroporation. Mol Biotechnol. 2005;30(3):185–192. doi: 10.1385/MB:30:3:185. [DOI] [PubMed] [Google Scholar]
- 135.Feng SY, Xue LX, Liu HT, Lu PJ. Improvement of efficiency of genetic transformation for Dunaliella salina by glass beads method. Mol Biol Rep. 2009;36(6):1433–1439. doi: 10.1007/s11033-008-9333-1. [DOI] [PubMed] [Google Scholar]
- 136.Tan CP, Qin S, Zhang Q, Jiang P, Zhao FQ. Establishment of a micro-particle bombardment transformation system for Dunaliella salina. J Microbiol. 2005;43(4):361–365. [PubMed] [Google Scholar]
- 137.Teng CY, Qin S, Liu JG, Yu DZ, Liang CW, Tseng CK. Transient expression of lacz in bombarded unicellular green alga Haematococcus pluvialis. J Applied Phycology. 2002;14(6):497–500. [Google Scholar]
- 138.Hallmann A, Rappel A. Genetic engineering of the multicellular green alga Volvox: a modified and multiplied bacterial antibiotic resistance gene as a dominant selectable marker. Plant J. 1999;17(1):99–109. doi: 10.1046/j.1365-313x.1999.00342.x. [DOI] [PubMed] [Google Scholar]
- 139.Fischer H, Robl I, Sumper M, Kroger N. Targeting and covalent modification of cell wall and membrane proteins heterologously expressed in the diatom Cylindrotheca fusiformis (bacillariophyceae) J Phycol. 1999;35(1):113–120. [Google Scholar]
- 140.Ten Lohuis MR, Miller DJ. Genetic transformation of dinoflagellates (Amphidinium and Symbiodinium): expression of gus in microalgae using heterologous promoter constructs. Plant J. 1998;13(3):427–435. [Google Scholar]
- 141.Jarvis EE, Brown LM. Transient expression of firefly luciferase in protoplasts of the green-alga Chlorella ellipsoidea. Curr Genet. 1991;19(4):317–321. [Google Scholar]
- 142.Dawson HN, Burlingame R, Cannons AC. Stable transformation of Chlorella: Rescue of nitrate reductase-deficient mutants with the nitrate reductase gene. Curr Microbiol. 1997;35(6):356–362. doi: 10.1007/s002849900268. [DOI] [PubMed] [Google Scholar]
- 143.Maruyama M, Horakova I, Honda H, Xing XH, Shiragami N, Unno H. Introduction of foreign DNA into Chlorella saccharophila by electroporation. Biotechnol Techniques. 1994;8(11):821–826. [Google Scholar]
- 144.Hawkins Rl, Nakamura M. Expression of human growth hormone by the eukaryotic alga, Chlorella. Curr Microbiol. 1999;38(6):335–341. doi: 10.1007/pl00006813. [DOI] [PubMed] [Google Scholar]
- 145.Doetsch NA, Favreau MR, Kuscuoglu N, Thompson MD, Hallick RD. Chloroplast transformation in Euglena gracilis: splicing of a group III twintron transcribed from a transgenic psbk operon. Curr Genet. 2001;39(1):49–60. doi: 10.1007/s002940000174. [DOI] [PubMed] [Google Scholar]
- 146.Lapidot M, Raveh D, Sivan A, Arad SM, Shapira M. Stable chloroplast transformation of the unicellular red alga Porphyridium species. Plant Physiol. 2002;129(1):7–12. doi: 10.1104/pp.011023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Materna AC. Development of molecular tools in the diatom Phaedactylum tricornutum. Fachbereich Biologie. 2006;126 [Google Scholar]
- 148.Blowers AD, Bogorad L, Shark KB, Sanford JC. Studies on Chlamydomonas chloroplast transformation: foreign DNA can be stably maintained in the chromosome. Plant Cell. 1989;1(1):123–132. doi: 10.1105/tpc.1.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Blowers AD, Ellmore GS, Klein U, Bogorad L. Transcriptional analysis of endogenous and foreign genes in chloroplast transformants of Chlamydomonas. Plant Cell. 1990;2(11):1059–1070. doi: 10.1105/tpc.2.11.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Goldschmidt-Clermont M. Transgenic expression of aminoglycoside adenine transferase in the chloroplast: a selectable marker of site-directed transformation of Chlamydomonas. Nucleic Acids Res. 1991;19(15):4083–4089. doi: 10.1093/nar/19.15.4083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ishikura K, Takaoka Y, Kato K, Sekine M, Yoshida K, Shinmyo A. Expression of a foreign gene in Chlamydomonas reinhardtii chloroplast. J Biosci Bioeng. 1999;87(3):307–314. doi: 10.1016/s1389-1723(99)80037-1. [DOI] [PubMed] [Google Scholar]
- 152.Minko I, Holloway SP, Nikaido S, et al. Renilla luciferase as a vital reporter for chloroplast gene expression in Chlamydomonas. Mol Gen Genet. 1999;262(3):421–425. doi: 10.1007/s004380051101. [DOI] [PubMed] [Google Scholar]
- 153.Surzycki R, Greenham K, Kitayama K, et al. Factors effecting expression of vaccines in microalgae. Biologicals. 2009;37(3):133–138. doi: 10.1016/j.biologicals.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 154.Weselake RJ, Taylor DC, Rahman MH, et al. Increasing the flow of carbon into seed oil. Biotechnol Adv. 2009;27(6):866–878. doi: 10.1016/j.biotechadv.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 155.Liu X, Brune D, Vermaas W, Curtiss R., 3rd Production and secretion of fatty acids in genetically engineered cyanobacteria. Proc Natl Acad Sci USA. 2009 doi: 10.1073/pnas.1001946107. Epub ahead of print. [DOI] [PubMed] [Google Scholar]; ▪ Lipid secretion by algae is an exciting area for future research.
- 156.Damude HG, Kinney AJ. Engineering oilseed plants for a sustainable, land-based source of long chain polyunsaturated fatty acids. Lipids. 2007;42(3):179–185. doi: 10.1007/s11745-007-3049-1. [DOI] [PubMed] [Google Scholar]
- 157.Wu G, Truksa M, Datla N, et al. Stepwise engineering to produce high yields of very long-chain polyunsaturated fatty acids in plants. Nat Biotechnol. 2005;23(8):1013–1017. doi: 10.1038/nbt1107. [DOI] [PubMed] [Google Scholar]
- 158.Uttaro AD. Biosynthesis of polyunsaturated fatty acids in lower eukaryotes. IUBMB Life. 2006;58(10):563–571. doi: 10.1080/15216540600920899. [DOI] [PubMed] [Google Scholar]
- 159.Guschina IA, Harwood JL. Lipids and lipid metabolism in eukaryotic algae. Prog Lipid Res. 2006;45(2):160–186. doi: 10.1016/j.plipres.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 160.Ratledge C. Fatty acid biosynthesis in microorganisms being used for single cell oil production. Biochimie. 2004;86(11):807–815. doi: 10.1016/j.biochi.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 161.Benson TJ, Hernandez R, White MG, et al. Heterogenous cracking of an unsaturated fatty acid and reaction intermediates on H+ZSM-5 catalyst. Clean. 2008;36(8):652–656. [Google Scholar]
- 162.Melis A, Happe T. Hydrogen production. Green algae as a source of energy. Plant Physiol. 2001;127(3):740–748. [PMC free article] [PubMed] [Google Scholar]
- 163.Sialve B, Bernet N, Bernard O. Anaerobic digestion of microalgae as a necessary step to make microalgal biodiesel sustainable. Biotechnol Adv. 2009;27(4):409–416. doi: 10.1016/j.biotechadv.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 164.Connolly JD, Hill RA. Triterpenoids. Nat Prod Rep. 2008;27(1):79–132. doi: 10.1039/b808530g. [DOI] [PubMed] [Google Scholar]
- 165.Kirby J, Keasling JD. Biosynthesis of plant isoprenoids: perspectives for microbial engineering. Annu Rev Plant Biol. 2009;60:335–355. doi: 10.1146/annurev.arplant.043008.091955. [DOI] [PubMed] [Google Scholar]
- 166.Kalamaki MS, Alexandrou D, Lazari D, et al. Over-expression of a tomato N-acetyl-l-glutamate synthase gene (SLNAGS1) in Arabidopsis thaliana results in high ornithine levels and increased tolerance in salt and drought stresses. J Exp Bot. 2009;60(6):1859–1871. doi: 10.1093/jxb/erp072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Yang X, Liang Z, Wen X, Lu C. Genetic engineering of the biosynthesis of glycinebetaine leads to increased tolerance of photosynthesis to salt stress in transgenic tobacco plants. Plant Mol Biol. 2008;66(1–2):73–86. doi: 10.1007/s11103-007-9253-9. [DOI] [PubMed] [Google Scholar]
- 168.Becker W. Microalgae in human and animal nutrition. In: Richmond A, editor. Handbook of Microalgal Culture: Biotechnology & Applied Phycology. Blackwell; Oxford, UK: 2003. pp. 312–351. [Google Scholar]
- 169.Borowitzka MA. Microalgae for aquaculture: opportunities and constraints. J Appl Phycol. 1997;9(5):393–401. [Google Scholar]
- 170.Brown MR. Nutritional value and use of microalgae in aquaculture. Avances en nutrición acuícola VI Memorias del VI Simposium Internacional de Nutrición Acuícola. 2003 [Google Scholar]
- 171.Iwamoto H. Industrial production of microalgal cell-mass and secondary products – major industrial species – chlorella. In: Richmond A, editor. Handbook of Microalgal Culture: Biotechnology & Applied Phycology. Blackwell; Oxford, UK: 2003. pp. 255–263. [Google Scholar]
- 172.Vilchez C, Garbayo I, Lobato MV, Vega JM. Microalgae-mediated chemicals production and wastes removal. Enzyme Microbial Technol. 1997;20(8):562–572. [Google Scholar]
- 173.Del Campo JA, Moreno J, Rodr Guez H, Angeles Vargas M, Rivas J, Guerrero MG. Carotenoid content of chlorophycean microalgae: factors determining lutein accumulation in Muriellopsis sp. (chlorophyta) J Biotechnol. 2000;76(1):51–59. doi: 10.1016/s0168-1656(99)00178-9. [DOI] [PubMed] [Google Scholar]
- 174.Jin E, Polle Jew, Lee H, Hyun S, Chang M. Xanthophylls in microalgae: from biosynthesis to biotechnological mass production and application. J Microbiol Biotechnol. 2003;13(2):165–174. [Google Scholar]
- 175.Ye ZW, Jiang JG, Wu GH. Biosynthesis and regulation of carotenoids in Dunaliella: progresses and prospects. Biotechnol Advances. 2008;26(4):352–360. doi: 10.1016/j.biotechadv.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 176.Ben-Amotz A. Handbook of Microalgal Culture: Biotechnology & Applied Phycology. Blackwell Science; Oxford, UK: 2004. Industrial production of microalgal cell-mass and secondary products – major industrial species: Dunaliella; pp. 273–280. [Google Scholar]
- 177.Raja R, Hemaiswarya S, Rengasamy R. Exploitation of Dunaliella for β-carotene production. Applied Microbiol Biotechnol. 2007;74(3):517. doi: 10.1007/s00253-006-0777-8. [DOI] [PubMed] [Google Scholar]
- 178.Eonseon J, Lee CG, Polle JEW. Secondary carotenoid accumulation in Haematococcus (chlorophyceae): biosynthesis, regulation, and biotechnology. J Microbiol Biotechnol. 2006;16(6):821–831. [Google Scholar]
- 179.Guerin M, Huntley ME, Olaizola M. Haematococcus astaxanthin: applications for human health and nutrition. Trends Biotechnol. 2003;21(5):210. doi: 10.1016/S0167-7799(03)00078-7. [DOI] [PubMed] [Google Scholar]
- 180.Cysewski GR, Lorenz T. Industrial production of microalgal cell-mass and secondary products. Major industrial species – Haematococcus. In: Richmond A, editor. Handbook of Microalgal Culture. Blackwell; Oxford, UK: 2004. pp. 281–288. [Google Scholar]
- 181.Ratledge C. Fatty acid biosynthesis in microorganisms being used for single cell oil production. Biochimie. 2004;86(11):807–815. doi: 10.1016/j.biochi.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 182.Ward Op, Singh A. Omega-3/6 fatty acids: alternative sources of production. Process Biochem. 2005;40(12):3627–3652. [Google Scholar]
- 183.Spolaore P, Joannis-Cassan C, Duran E, Isambert A. Commercial applications of microalgae. J Biosci Bioengin. 2006;101(2):87. doi: 10.1263/jbb.101.87. [DOI] [PubMed] [Google Scholar]
- 184.Prasanna R, Sood A, Suresh A, Nayak S, Kaushik BD. Potentials and applications of algal pigments in biology and industry. Acta Botanica Hungarica. 2007;49(1):131–156. [Google Scholar]
- 185.Sekar S, Chandramohan M. Phycobiliproteins as a commodity: trends in applied research, patents and commercialization. J Applied Phycology. 2008;20(2):113–136. [Google Scholar]
- 186.Stolz P, Obermayer B. Manufacturing microalgae for skin care. Cosmetics Toiletries. 2005;120(3):99–106. [Google Scholar]
- 187.Acien Fernandez FG, Fernandez Sevilla JM, Egorova-Zachernyuk TA, Molina Grima E. Cost-effective production of 13C, 15N stable isotope-labelled biomass from phototrophic microalgae for various biotechnological applications. Biomol Engineer. 2005;22(5–6):193–200. doi: 10.1016/j.bioeng.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 188.Pulz O, Gross W. Valuable products from biotechnology of microalgae. Applied Microbiol Biotechnol. 2004;65(6):635. doi: 10.1007/s00253-004-1647-x. [DOI] [PubMed] [Google Scholar]
- 189.Singh S, Kate BN, Banerjee UC. Bioactive compounds from cyanobacteria and microalgae: an overview. Critical Rev Biotechnol. 2005;25(3):73–95. doi: 10.1080/07388550500248498. [DOI] [PubMed] [Google Scholar]
- 190.Cardozo KHM, Guaratini T, Barros MP, et al. Metabolites from algae with economical impact. Comparative Biochem Physiology (Pt C) 2007;146(1–2):60–78. doi: 10.1016/j.cbpc.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 191.Hallmann A. Algal transgenics and biotechnology. Transgenic Plant J. 2007;1:81–98. [Google Scholar]
- 192.Raja R, Hemaiswarya S, Kumar NA, Sridhar S, Rengasamy R. A perspective on the biotechnological potential of microalgae. Critical Rev Microbiol. 2008;34(2):77. doi: 10.1080/10408410802086783. [DOI] [PubMed] [Google Scholar]