Abstract
Objective
Hypertension or high blood pressure is a strong correlate of diseases such as obesity and type 2 diabetes. We conducted a genome-wide linkage screen to identify susceptibility genes influencing systolic blood pressure (SBP) and diastolic blood pressure (DBP) in Mexican-Americans from the Veterans Administration Genetic Epidemiology Study (VAGES).
Methods
Using data from 1,089 individuals distributed across 266 families, we performed a multipoint linkage analysis to localize susceptibility loci for SBP and DBP by applying two models. In model 1, we added a sensible constant to the observed BP values in treated subjects [Tobin et al.; Stat Med 2005;24:2911–2935] to account for antihypertensive use (i.e. 15 and 10 mm Hg to SBP and DBP values, respectively). In model 2, we fixed values of 140 mm Hg for SBP and 90 mm Hg for DBP, if the treated values were less than the standard referenced treatment thresholds of 140/90 mm Hg for hypertensive status. However, if the observed treated BP values were found to be above these standard treatment thresholds, the actual observed treated BP values were retained in order not to reduce them by substitution of the treatment threshold values.
Results
The multipoint linkage analysis revealed strong linkage signals for SBP compared with DBP. The strongest evidence for linkage of SBP (model 1, LOD = 5.0; model 2, LOD = 3.6) was found on chromosome 6q14.1 near the marker D6S1031 (89 cM) in both models. In addition, some evidence for SBP linkage occurred on chromosomes 1q, 4p, and 16p. Most importantly, our major SBP linkage finding on chromosome 6q near marker D6S1031 was independently confirmed in a Caucasian population (LOD = 3.3). In summary, our study found evidence for a major locus on chromosome 6q influencing SBP levels in Mexican-Americans.
Key Words: Hypertension, Linkage, Antihypertensive medication, Genetic location, Heritability
Introduction
Hypertension or high blood pressure (HBP) is strongly associated with various diseases including obesity, type 2 diabetes (T2DM), end-stage renal disease, coronary artery and cerebrovascular diseases [1,2]. It is a component of the metabolic syndrome (MS), which predisposes to increased cardiovascular morbidity and mortality [3]. The increasing prevalence of HBP in the global populations has become a major public health issue. For example, the estimated overall prevalence of hypertension in the adult population of developed countries is ∼30% and has been predicted to rise to ∼60% in the next two decades [4,5]. In the US, 1 in 3 adults has HBP, and the age-standardized prevalence rate of hypertension increased from 24.4 to 28.9% in the US adult population within a period of ∼10 years [2,6]. Specifically, it increased from 23.3 to 27.4% in non-Hispanic whites, from 35.8 to 40.1% in non-Hispanic blacks, and from 25.0 to 27.1% in Mexican-Americans [6]. The total cost of HBP is estimated to be USD 69.4 billion in the year 2008 [2].
HBP is a strong correlate of various disease conditions including obesity and T2DM. In adults with diabetes, more than 75% have blood pressure (BP) levels ≥130/80 mm Hg or use antihypertensive medication [5]. The clustering of hypertension with diabetes tends to occur more likely in Hispanics than in blacks, whites, and other minority communities [7]. Also, more than 85% of those with MS have HBP or hypertension [3]. In addition, HBP is considered as an important risk factor for the development of chronic kidney disease (CKD) in the presence of obesity, MS, and microalbuminuria [3]. The increased prevalence of CKD from 1988–1994 to 1999–2004 in the US was partly explained by the increasing prevalence of diabetes and hypertension [8].
BP is a complex phenotype that is influenced by both genetic and environmental factors and their interactions [9,10]. The genetic evidence provided from studies using animal models, human families and twins suggested that genes account for 15–50% of the variation in BP levels [11,12,13,14]. In addition to the influences of age and gender, a number of environmental factors affect BP, including poor diet, lack of exercise, increased body weight, stress, smoking and alcohol intake [15]. Aside from the identification of causal mutations for various monogenic forms of familial hypertension [16,17], there have been continued efforts to localize susceptibility loci for BP or hypertension in the general population. Several genome-wide linkage screens have been performed to localize genes for BP or hypertension, which found evidence for suggestive or significant linkage signals on several chromosomal regions, e.g. 1q, 2p, 3p, 6q, 7q, 11q, 12q, 15q, 16q, 18q, and 19p [reviewed in [12], [13], [18,19,20]]. However, a consistent pattern of evidence for linkage of BP and hypertension across multiple studies was lacking. In addition, meta-analysis of genome-wide linkage screens for hypertension yielded no clear consensus on major chromosomal regions influencing hypertension or BP levels [21,22].
In recent years, several studies have used a genome-wide association study (GWAS) approach to localize susceptibility genes for BP or hypertension successfully (reviewed by Arora and Newton-Cheh [23], Hastie et al. [24], Rafiq et al. [25], and Ehret [26]). For example, two large GWASs, the Global Blood Pressure Genetics (Global BPgen) Consortium study [27] and the Cohorts for Heart and Aging Research in Genome Epidemiology (CHARGE) Consortium study [28], identified several significant common genetic variants/genes that influence variation in BP and hypertension (e.g. systolic BP, SBP: ATP2B1, CYP17A1, PLEKHA7, SH2B3; diastolic BP, DBP: ATP2B1, CACNB2, CSK-ULK3, SH2B3, TBX3-TBX5, ULK4, and hypertension: ATP2B1) [28]. However, it is recognized that the effect size of common variants identified by such GWASs is rather modest, and that these studies have explained only a small proportion of trait-specific heritabilities [25,26]. Therefore, the purpose of this study was to perform a genome-wide linkage screen to identify the susceptibility loci influencing BP in Mexican-American individuals from the Veterans Administration Genetic Epidemiology Study (VAGES), where the families were ascertained for T2DM.
Participants and Methods
The Veterans Administration Genetic Epidemiology Study
The phenotypic and demographic information was collected from Mexican-American family members who were enrolled in the VAGES [29]. In the present study, genotypic and phenotypic data were available for 1,089 Mexican-Americans distributed across 266 mostly nuclear families. These individuals generated 2,002 relative pairs that were distributed across 9 relative pair categories (table 1). The VAGES families were ascertained on at least 2 sibs affected with T2DM and only 1 parent with T2DM. All family members, mostly from nuclear families, aged 18 years or above were invited to participate in the study. Our analysis also included 37 single and unrelated individuals with BP data available, since they could contribute to the evaluation of covariate effects. All participants signed a consent form approved by the Institutional Review Board, University of Texas Health Science Center, San Antonio, Tex., USA, after risks and procedures had been explained.
Table 1.
Distribution of relative pairs by category in the VAGES familiesa
| Relative pair | Relationship coefficientb | No. of pairs |
|---|---|---|
| Parent-offspring | 0.5000 | 504 |
| Sibs | 0.5000 | 1,168 |
| Grandparent-grandchild | 0.2500 | 21 |
| Avuncular | 0.2500 | 210 |
| Half-sibs | 0.2500 | 53 |
| Grand avuncular | 0.1250 | 6 |
| Half-avuncular | 0.1250 | .15 |
| First cousins | 0.1250 | 24 |
| First cousins, once removed | 0.0625 | 1 |
| Total | 2,002 | |
The total data used for the present study were obtained from 1,089 individuals distributed across 266 families.
The relationship coefficient is 2 × coefficient of kinship of two individuals.
Phenotype Data
The individuals of the present study were extensively phenotyped for various metabolic and anthropometric traits. In addition, demographic and medical history information was obtained on all examined participants. BP was measured three times using a standardized Colin Press-Mate (model BP-8800; Colin) automated cuff-inflation instrument, and an average of the last two SBP and DBP measures was used for genetic analysis. SBP and DBP values were available for 1,089 related plus 37 single and unrelated individuals to conduct the present study. The data related to antihypertensive treatment included all medications recorded for BP treatment, including angiotensin-converting enzyme or angiotensin receptor blocker. For the genetic analysis, BP values only within ±4 SDs from the mean were considered.
Genotype Data and Genetic Map
DNA was prepared from whole-blood samples (Gentra Systems, Minneapolis, Minn., USA). The Center for Inherited Disease Research (CIDR) performed a 10-cM genome scan using the DNA samples from the VAGES participants. For the present study, we used CIDR genotypic data on 385 highly polymorphic autosomal markers. Pedigree-validation and genotype-cleaning procedures, implemented in the PEDSYS [30] programs PREPREST and PRESWALK, respectively, were applied to these data. The pedigree-validation procedure utilizes the program PREST [31] to detect and resolve pedigree misspecifications. The genotype-cleaning procedure removes Mendelian inconsistencies and spurious recombinants by blanking genotypes which have been flagged by the program SimWalk2 [32] as probable genotyping errors. Overall, the blanking rate for errors was <0.22% of the total number of genotypes. The program INFER (PEDSYS) was used to infer unknown genotypes from the genotypes of relatives, when possible without ambiguity. As described previously [29], the genetic maps (map distance in Kosambi cM) used for this study were similar to the Marshfield map. The multipoint identical-by-descent matrices given a number of genetic markers (map distance in Haldane cM) were calculated using Markov chain Monte Carlo methods implemented in the program Loki [33,34]. To facilitate comparison with other studies, the locations of our linkage findings have been placed on the Marshfield genetic map.
Statistical Genetic Analysis
To estimate the heritability of BP and to test potential linkages between marker loci and BP, a variance components analytic approach was used [35,36,37]. This technique uses information from all possible biological relationships simultaneously in an attempt to disentangle the genetic architecture of a quantitative trait. Linkage of a genetic location for BP was tested using a general variance components method, which allows for marker locus-specific effects, residual genetic effects, random environmental effects, and covariate effects. Hypothesis testing of no linkage versus linkage was performed using the likelihood ratio test. LOD scores were obtained by converting the ln likelihood values into values of log to the base 10. A LOD score of 3.0 and above was considered as strong evidence in support of linkage. For the purpose of discussion, potential linkages are considered as those genetic locations across the genome with nominal p values of 0.01 or less (i.e. LOD scores ≥1.2).
The VAGES families were ascertained on T2DM probands; so, as a conservative approach, all genetic analyses were performed by correcting for the ascertainment by conditioning the likelihood for the family data on the phenotype (i.e. BP) of probands [38]. Since the VAGES families had more than 1 proband per family, we randomly chose one of the probands from each family to correct for the ascertainment bias since correcting for ascertainment bias in data sets ascertained on multiple affected probands can result in a loss of power to detect linkage [39,40]. To verify the findings from the multipoint linkage analyses, we determined trait-specific empirical p values based on the distribution of LOD scores observed under the hypothesis of no linkage. This null distribution was generated by repeatedly simulating a genetic marker, unlinked to the trait, which was then tested for linkage. We generated the empirical p values on the basis of LOD score distribution obtained from the 100,000 replicates. All genetic analyses were performed using the program SOLAR [36].
Treated BP Values and Models
The SBP and DBP data analyses were related to the same number of individuals (i.e. individuals with normal and treated BP values), but differed in the ways of considering the treated BP values for the genetic analyses. We used the approach by Tobin et al. [41] to more closely reflect the pretreatment BP values in our data. We called this method, as recommended by Cui et al. [42] and Tobin et al. [41], model 1, wherein a sensible constant was added to the observed BP values in treated subjects to account for antihypertensive use (i.e. 15 and 10 mm Hg to SBP and DBP values, respectively). In contrast to model 1, as a conservative approach, in model 2, we fixed values of 140 mm Hg for SBP and 90 mm Hg for DBP, if the treated values were less than the standard referenced treatment thresholds of 140/90 mm Hg for hypertensive status. However, if the observed treated BP values were found to be above these standard treatment thresholds, the actual observed treated BP values were retained in order not to reduce them by substitution of the treatment threshold values. The recommendations of the seventh Joint National Committee (JNC-7) were followed to define hypertension by setting the SBP and DBP values to 140/90 mm Hg [43]. The BP data were adjusted for covariate effects of age and sex terms and log-transformed body mass index (ln BMI) in both models.
Results
The clinical characteristics of 1,089 individuals with BP are reported in table 2. As can be seen, the mean age of the individuals was 50 years, 65% were females, 69% were affected with T2DM, and 37% of the individuals were found to be treated with anti-hypertensive medications. The mean BP values without and with adjustment for antihypertensive treatment are also reported in table 2. The coefficients of skewness and kurtosis for SBP in model 1 were 0.55 and 0.04, and in model 2 these values were 0.27 and 0.29. The same coefficients for DBP in model 1 were 0.10 and −0.06 and in model 2 −0.56 and −0.31.
Table 2.
Characteristics of 1,089 study subjects in 266 VAGES families
| Characteristics | |
|---|---|
| Subjects, na | 1,089 |
| Age, years | 49.9 ± 13.3 |
| Gender (% females) | 65 |
| Type 2 diabetes | 69 |
| Anti-hypertensive medicationb | 37 |
| Systolic blood pressure, mm Hg | 130.9 ± 19.4 |
| Model 1 systolic blood pressure, mm Hg | 136.2 ± 23.3 |
| Model 2 systolic blood pressure, mm Hg | 133.6 ± 19.1 |
| Diastolic blood pressure, mm Hg | 74.9 ± 11.0 |
| Model 1 diastolic blood pressure, mm Hg | 78.6 ± 12.6 |
| Model 2 diastolic blood pressure, mm Hg | 80.0 ± 11.8 |
| Body mass index | 33.2 ± 7.9 |
| ln body mass index | 3.5 ± 0.2 |
Figures are means ± SD or %, unless otherwise indicated.
ln = log transformed.
The analyses included 37 unrelated individuals in addition to the 1,089 subjects from the VAGES.
Antihypertensive treatment included all current medications including angiotensin-converting enzyme (ACE) or angiotensin receptor blocker (ARB).
Prior to conducting linkage analyses, we estimated the heritability (h2) to determine the genetic basis of BP (i.e. residuals), after accounting for the covariate effects of age and sex terms and ln BMI. Since in our linkage approach a multivariate normal distribution was assumed, we examined the distributional properties of the BP residuals on the basis of the coefficients of skewness and kurtosis. For SBP residuals, the coefficients of skewness and kurtosis after adjusting for the effects of age and sex terms and ln BMI were 0.52 and 0.67, and for DBP residuals, they were 0.10 and 0.02. Thus, the violation of nonnormality of the BP residuals in this study appears to be minor. In model 1, after accounting for the covariate effects, SBP and DBP phenotypes exhibited significant heritabilities (h2 = 38 and 34%, respectively; p < 0.0001). In model 2, the SBP and DBP phenotypes also exhibited significant heritabilities (h2 = 41 and 27%, respectively; p < 0.0001), after accounting for the covariate effects.
Multipoint Linkage Findings
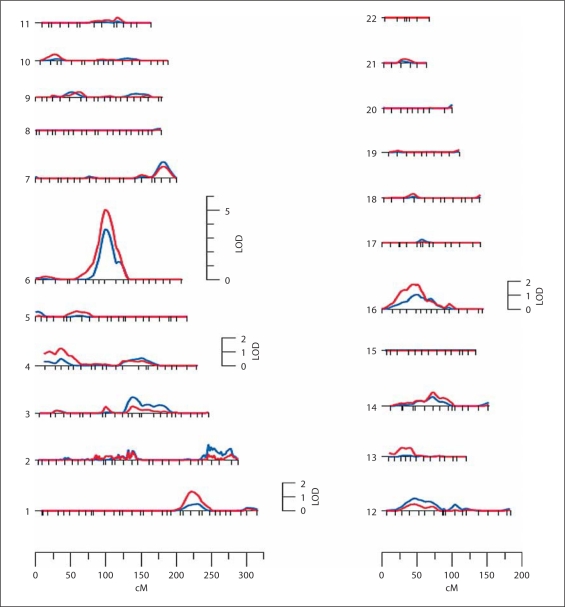
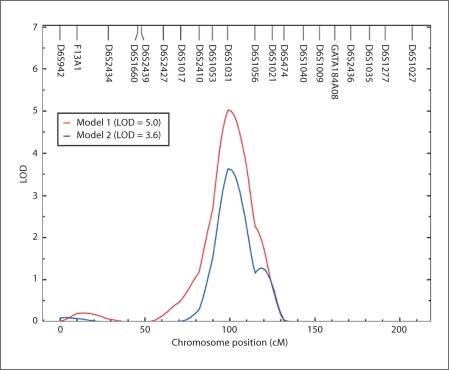
After determining that SBP and DBP were under significant additive genetic influence, we performed an autosomal genome-wide linkage analysis to identify quantitative trait loci for both SBP and DBP using the two models. We found strong and potential evidence for SBP linkage at various chromosomal regions (table 3), but none of the chromosomal regions showed potential evidence for linkage of DBP (data not shown). Therefore, only linkage results associated with SBP were discussed henceforth. Model 1 revealed strongest evidence for SBP linkage with a LOD score of 5.0 [empirical p = 1.0 × 10–5] on chromosome 6q14.1 at 89 cM from pter near marker D6S1031 (table 3, model 1; fig. 1 and 2). Three chromosomal regions showed potential evidence for SBP linkage: a genetic location between markers D16S3103 and D16S403 (LOD = 1.8) on chromosome 16p12.3–p12.1, a genetic location near marker D1S518 (LOD = 1.4) on chromosome 1q31.1, and a genetic location near marker D4S2639 (LOD = 1.2) on chromosome 4p15.31.
Table 3.
Chromosomal regions potentially linked (LOD ≥1.2) to SBP using Tobin et al.'s method
| Nearest marker region | Distance from pter in the Marshfield map | Chromosomal location | Maximum LOD score |
|
|---|---|---|---|---|
| model 1a | model 2a | |||
| D1S518 | 202 | 01q31.1 | 1.4 | 0.4 |
| D4S2639 | 033 | 04p15.31 | 1.2 | 0.5 |
| D6S1031 | 089 | 06q14.1 | 5.0 | 3.6 |
| D16S3103–D16S403 | 032 – 44 | 16p12.3–p12.1 | 1.8 | 1.0 |
Please see text for a description of the models.
Fig. 1.
Summary of linkage findings of SBP in Mexican-Americans from the VAGES using model 1 (red line) and model 2 (blue line).
Fig. 2.
Multipoint linkage findings of SBP on chromosome 6q obtained from model 1 and model 2.
Using model 2, the evidence of linkage was observed for SBP on chromosome 6q near the same marker (D6S1031) witha LOD score of 3.6 (empirical p = 2.0 × 10–5) (table 3, model 2; fig. 1 and 2). As can be seen in table 3, there were no other regions identified by model 2 to be potentially linked with SBP. Thus, the maximum LOD score for SBP on chromosome 6q from the two models occurred at the same chromosomal location. The 1-LOD support intervals around the linkage peaks obtained from models 1 and 2 spanned a ∼23-cM-long region on chromosome 6q between markers D6S1053 (6q12, 80 cM) and D6S1056 (6q16.1, 103 cM) on the Marshfield map.
Discussion
In the present study, we used data on the Mexican-American population from the VAGES to identify susceptibility genes influencing variation in SBP and DBP traits. After finding that the heritability estimates derived from the two models were moderate and significant, we employed the variance components multipoint linkage technique to localize the susceptibility genes for SBP. Despite the differences in their magnitudes, the maximum LOD scores of 5.0 and 3.6 for SBP occurred at a genetic location on chromosome 6q near the marker D6S1031. In our linkage analyses, no attempt was made to correct for multiple testing in regard to the used correlated BP measures/models since they were not independent. For example, the phenotypic correlation between SBP and DBP in our study indicated that 39% of the variation is commonly shared by these correlated traits. Likewise, the commonly shared variation by the treatment-corrected SBP values by the two methods is 38% based on their phenotypic correlation.
Several other genome-wide linkage studies of BP found significant or potential linkages on chromosome 6q (table 4). As can be seen, we independently confirmed a significant linkage signal for SBP found in a European population from the National Heart, Lung and Blood Institute (NHLBI) Family Heart Study, which is a multi-center, population-based study, consisting of 2,959 individuals from 500 white families [44]. This study performed a linkage analysis using different methods on how to account for antihypertensive use in treated individuals. Briefly, one of their methods included individuals who were taking antihypertensive medication and fixed their treated BP values to 140/90 mm Hg. Using these data, a significant linkage signal (LOD = 3.3) for SBP values was found on chromosome 6q near the same marker as reported in the present study (D6S1031) [44]. In another study, a potential linkage finding (LOD = 2.5) for DBP was reported at the same 6q chromosomal region in Dutch families with familial combined hyperlipidemia [45]. Thus, the BP susceptibility loci in these studies mapped to the same marker (D6S1031) region on chromosome 6q14.1 as reported in the present study. In the San Antonio Family Heart Study, another Mexican-American family study in San Antonio without any overlap with the VAGES, a potential linkage signal (LOD = 2.2) for SBP was observed ∼10 cM away from our linkage peak [46]. As shown in table 4, some other studies observed BP linkage signals on chromosome 6q, but which were closer to the qter. For example, Krushkal et al. [47], Adeyemo et al. [13] and Caulfield et al. [20] implicated the 6q23–q27 chromosomal region as influencing variation in SBP and/or essential hypertension.
Table 4.
Correspondence of our SBP linkage finding with other studies on chromosome 6q
| Population | Trait | Chromosomal location | Marker region implicated | Position cM (Marshfield map) | LOD score | Reference |
|---|---|---|---|---|---|---|
| Mexican-Americans | SBP | 6q12 | D6S1053 | 080 | 2.2 | Rutherford et al., 2007 [46] |
| Dutch families | DBP | 6q14.1 | D6S1031 | 089 | 2.5 | Allayee et al. 2001 [45] |
| White families | SBP | 6q14.1 | D6S1031 | 089 | 3.3 | Hunt et al., 2002 [44] |
| Present study | SBP | 6q14.1 | D6S1031 | 089 | 3.6 | Puppala et al. |
| White families (Americans) | SBP | 6q23.3 – 6q24.2 | D6S1009-D6S1003 | 138–144 | 0.0001a | Krushkal et al., 1999 [47] |
| Nigerian families | SBP | 6q26 | D6S1277 | 173 | 2.9 | Adeyemo et al., 2005 [13] |
| White sibling pairs (Europeans) | essential hypertension | 6q27 | D6S281 | 190 | 3.2 | Caulfield et al., 2003 [20] |
SBP = Systolic blood pressure; DBP = diastolic blood pressure.
p value.
There are no known major candidate genes for BP in our 6q chromosomal region of interest. The ∼23-cM-long (∼30 Mb) 1-LOD support interval region around the SBP linkage peak (89 cM, 76 Mb) on chromosome 6q is flanked by markers D6S1053 (6q12; 80 cM, 65 Mb) and D6S1056 (6q16.1; 103 cM, 94 Mb) on the Marshfield map, and it contains approximately 180 genes (NCBI Build 37). As noted earlier, numerous BP susceptibility variants/genes have been identified by recently conducted GWASs. However, none of the reported BP/hypertension GWA findings occurred in the 1-LOD support interval around the linkage peak found in our study (reviewed by Arora and Newton-Cheh [23], Hastie et al. [24], Rafiq et al. [25], and Ehret [26]). As in the case of other GWASs of complex diseases, the concordance of linkage and GWASs of BP/hypertension is far from perfect. The GWASs, generally using a case-control design involving unrelated individuals, have been shown to be successful in associating common variation with BP susceptibility. In contrast to linkage studies, the chromosomal regions of interest identified by GWASs involve small confidence regions. However, the issues of missing heritability, rare variants, and synthetic associations need to be addressed. These concerns highlight the potential role of rare variants which are undetectable with the use of the current GWAS-related technologies that focus on common variants [48,49]. In this context, it should be noted that large family-based studies have a high likelihood of detecting the rare variants, given that multiple individuals in large families can be associated with such sequence variants.
To verify whether any BP data were found in the recent GWASs to be modestly associated in our 6q chromosomal region of interest, a survey of the overall association findings from these studies was performed. As reported mostly in their supplementary materials, modest associations related to 1 or more BP traits (i.e. SBP, DBP and hypertension) that occurred in the 1-LOD support interval were found by 6 studies (Levy et al. [50], WTCCC [51], Org et al. [52], Yang et al. [53], Adeyemo et al. [54], Hiura et al. [55]). For example, an association (trend p = 9.4 × 10–5) between hypertension and rs276699 (76.5 Mb) was found in a British population [51]. In a Japanese population, the best modest associations were found between SBP and rs9353859 (p = 5.0 × 10–4 at 92.3 Mb) and DBP and rs1482567 (p = 1.8 × 10–5 at 72.7 Mb), respectively [55]. In the Framingham Heart Study data, a modest association was observed between DBP and rs2509458 (p = 6.9 × 10–6 at 88.7 Mb [50]).
Interestingly, SNP × SNP interaction tests in a GWAS of young-onset hypertension in the Han Chinese population of Taiwan revealed a significant pair-wise interaction between rs1886985 on chromosome 6q14.1 (76.7 Mb) and rs6129969 on chromosome 20q12 (40.5 Mb) associated with young-onset hypertension [53]. Three types of interaction tests were performed, where the p values ranged from 1.9 × 10–4 to 7.6 × 10–9. This finding was validated in the second-stage analysis. Given that rs1886985 represents the gene IMPG1 (which encodes interphotoreceptor matrix proteoglycan 1) region, these authors performed a preliminary gene expression analysis of IMPG1 and concluded that its expression may be involved in the modification of blood vessel structure, in turn affecting the activity and stability of proteins and signaling molecules within the matrix [53].
To address the issue of the potential influence of T2DM at the identified SBP susceptibility locus on 6q14.1 in the VAGES family data enriched for T2DM due to the ascertainment scheme, we performed 2 additional analyses. We found no evidence for linkage of T2DM (LOD = 0) at the location where we found strong evidence for linkage of SBP (at 89 cM on chromosome 6q14.1). We also conducted bivariate linkage analyses of T2DM and SBP at the same genetic location using models 1 and 2, and found no evidence for shared major gene influences (model 1, p = 0.37 and model 2, p = 0.62) on T2DM and SBP. These observations reveal that the major SBP locus found in this study is not likely to be a T2DM susceptibility locus, and that it influences variation in SBP without any confounding effects on T2DM. Recently, we reported suggestive evidence for linkage of plasma triglyceride levels (LOD = 2.2) on chromosome 6q in the same population as reported in the present study [28]. However, the chromosomal region of interest reported in the present study is approximately 98 cM far from that reported by our previous study [28], thus suggesting that these 2 genetic regions on chromosome 6q are unlikely to be the same.
In summary, we performed genome-wide linkage analyses to map susceptibility genes for SBP and DBP in Mexican-Americans, and we found strong evidence for a major susceptibility gene influencing SBP on chromosome 6q14.1. Most importantly, our significant SBP linkage finding independently confirmed an earlier report of a major susceptibility locus at the same location on chromosome 6q14.1 by the NHLBI Family Heart Study relating to a European population. Potential linkages and modest genome-wide associations for BP/hypertension were also found in our 6q chromosomal region of interest. Although our findings correspond well with those reported in a Caucasian population, it is possible that the present linkage finding may be related to T2DM-associated hypertension, given that our VAGES families were enriched with T2DM individuals. Our study also provided some evidence of linkage for SBP on chromosomes 1q, 4p and 16p. In consideration of the major and potential linkage findings together with the modest associations for BP in the same 6q chromosomal region by several studies involving ethnically diverse populations, region-specific genomic surveys at a finer level appear to be urgently needed for identifying the putative functional variants that influence variation in SBP. Such refined gene discovery efforts on chromosome 6q may ultimately help to pave the way for prevention and treatment of coronary artery and cerebrovascular disease complications.
Acknowledgements
We thank Marcel J. Fourcaudot and Lenore M. Rodriguez for their excellent technical assistance. We thank our nurses, James King, John Kincaid, Rose Kaminski-Graham, and Norma Diaz, for their excellent care of the patients throughout the study. This work was supported by a Veterans Administration Epidemiologic grant. This study was also supported in part by grants from the National Institutes of Health (DK42273, DK47482, DK53889, DK70746, and MH59490), and by an American Heart Association National Scientist Development Grant. We thank the Center for Inherited Disease Research (CIDR) for providing a genome scan using the VAGES data. We thank the participants of the VAGES and are grateful for their participation and cooperation.
References
- 1.Fields LE, Burt VL, Cutler JA, Hughes J, Roccella EJ, Sorlie P. The burden of adult hypertension in the United States 1999 to 2000: a rising tide. Hypertension. 2004;44:398–404. doi: 10.1161/01.HYP.0000142248.54761.56. [DOI] [PubMed] [Google Scholar]
- 2.Rosamond W, Flegal K, Furie K, Go A, Greenlund K, Haase N, Hailpern SM, Ho M, Howard V, Kissela B, Kittner S, Lloyd-Jones D, McDermott M, Meigs J, Moy C, Nichol G, O'Donnell C, Roger V, Sorlie P, Steinberger J, Thom T, Wilson M, Hong Y, American Heart Association Statistics Committee and Stroke Statistics Subcommittee Heart disease and stroke statistics – 2008 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2008;117:e25–e146. doi: 10.1161/CIRCULATIONAHA.107.187998. [DOI] [PubMed] [Google Scholar]
- 3.Franklin SS. Hypertension in the metabolic syndrome. Metab Syndr Relat Disord. 2006;4:287–298. doi: 10.1089/met.2006.4.287. [DOI] [PubMed] [Google Scholar]
- 4.Kearney PM, Whelton M, Reynolds K, Muntner P, Whelton PK, He J. Global burden of hypertension: analysis of worldwide data. Lancet. 2005;365:217–223. doi: 10.1016/S0140-6736(05)17741-1. [DOI] [PubMed] [Google Scholar]
- 5.Bakris GL, Sowers JR. ASH Position Paper: treatment of hypertension in patients with diabetes – an update. J Clin Hypertens (Greenwich) 2008;10:707–713. doi: 10.1111/j.1751-7176.2008.00012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cutler JA, Sorlie PD, Wolz M, Thom T, Fields LE, Roccella EJ. Trends in hypertension prevalence, awareness, treatment, and control rates in United States adults between 1988–1994 and 1999–2004. Hypertension. 2008;52:818–827. doi: 10.1161/HYPERTENSIONAHA.108.113357. [DOI] [PubMed] [Google Scholar]
- 7.Hypertension in America: a national reading. Am J Manag Care. 2005;11:S383–S385. [PubMed] [Google Scholar]
- 8.Coresh J, Selvin E, Stevens LA, Manzi J, Kusek JW, Eggers P, Van Lente F, Levey AS. Prevalence of chronic kidney disease in the United States. JAMA. 2007;298:2038–2047. doi: 10.1001/jama.298.17.2038. [DOI] [PubMed] [Google Scholar]
- 9.Lifton RP. Molecular genetics of human blood pressure variation. Science. 1996;272:676–680. doi: 10.1126/science.272.5262.676. [DOI] [PubMed] [Google Scholar]
- 10.Hamet P, Pausova Z, Adarichev V, Adaricheva K, Tremblay J. Hypertension: genes and environment. J Hypertens. 1998;16:397–418. doi: 10.1097/00004872-199816040-00001. [DOI] [PubMed] [Google Scholar]
- 11.Hsueh WC, Mitchell BD, Schneider JL, Wagner MJ, Bell CJ, Nanthakumar E, Shuldiner AR. QTL influencing blood pressure maps to the region of PPH1 on chromosome 2q31–34 in Old Order Amish. Circulation. 2000;101:2810–2816. doi: 10.1161/01.cir.101.24.2810. [DOI] [PubMed] [Google Scholar]
- 12.Samani NJ. Genome scans for hypertension and blood pressure regulation. Am J Hypertens. 2003;16:167–171. doi: 10.1016/s0895-7061(02)03244-2. [DOI] [PubMed] [Google Scholar]
- 13.Adeyemo A, Luke A, Wu X, Cooper RS, Kan D, Omotade O, Zhu X. Genetic effects on blood pressure localized to chromosomes 6 and 7. J Hypertens. 2005;23:1367–1373. doi: 10.1097/01.hjh.0000173519.06353.8b. [DOI] [PubMed] [Google Scholar]
- 14.Padmanabhan S, Melander O, Hastie C, Menni C, Delles C, Connell JM, Dominiczak AF. Hypertension and genome-wide association studies: combining high fidelity phenotyping and hypercontrols. J Hypertens. 2008;26:1275–1281. doi: 10.1097/HJH.0b013e3282ff634f. [DOI] [PubMed] [Google Scholar]
- 15.Mullins LJ, Bailey MA, Mullins JJ. Hypertension, kidney, and transgenics: a fresh perspective. Physiol Rev. 2006;86:709–746. doi: 10.1152/physrev.00016.2005. [DOI] [PubMed] [Google Scholar]
- 16.Lifton RP. Genetic dissection of human blood pressure variation: common pathways from rare phenotypes. Harvey Lect. 2004–2005;100:71–101. [PubMed] [Google Scholar]
- 17.O'Shaughnessy KM. Dissecting complex traits: recent advances in hypertension genomics. Genome Med. 2009;28:1. doi: 10.1186/gm43. 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mein CA, Caulfield MJ, Dobson RJ, Munroe PB. Genetics of essential hypertension. Hum Mol Genet. 2004;13:R169–R175. doi: 10.1093/hmg/ddh078. [DOI] [PubMed] [Google Scholar]
- 19.Binder A. A review of the genetics of essential hypertension. Curr Opin Cardiol. 2007;22:176–184. doi: 10.1097/HCO.0b013e3280d357f9. [DOI] [PubMed] [Google Scholar]
- 20.Caulfield M, Munroe P, Pembroke J, Samani N, Dominiczak A, Brown M, Benjamin N, Webster J, Ratcliffe P, O'Shea S, Papp J, Taylor E, Dobson R, Knight J, Newhouse S, Hooper J, Lee W, Brain N, Clayton D, Lathrop GM, Farrall M, Connell J, MRC British Genetics of Hypertension Study Genome-wide mapping of human loci for essential hypertension. Lancet. 2003;361:2118–2123. doi: 10.1016/S0140-6736(03)13722-1. [DOI] [PubMed] [Google Scholar]
- 21.Province MA, Kardia SL, Ranade K, Rao DC, Thiel BA, Cooper RS, Risch N, Turner ST, Cox DR, Hunt SC, Weder AB, Boerwinkle E. A meta-analysis of genome-wide linkage scans for hypertension: the National Heart, Lung and Blood Institute Family Blood Pressure Program. Am J Hypertens. 2003;16:144–147. doi: 10.1016/s0895-7061(02)03248-x. [DOI] [PubMed] [Google Scholar]
- 22.Liu W, Zhao W, Chase GA. Genome scan meta-analysis for hypertension. Am J Hypertens. 2004;17:1100–1106. doi: 10.1016/j.amjhyper.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 23.Arora P, Newton-Cheh C. Blood pressure and human genetic variation in the general population. Curr Opin Cardiol. 2010 doi: 10.1097/HCO.0b013e3283383e2c. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hastie CE, Padmanabhan S, Dominiczak AE. Genome-wide association studies of hypertension: light at the end of the tunnel. Int J Hypertens. 2010;2010:509–581. doi: 10.4061/2010/509581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rafiq S, Anand S, Roberts R. Genome-wide association studies of hypertension: have they been fruitful? J Cardiovasc Transl Res. 2010;3:189–196. doi: 10.1007/s12265-010-9183-9. [DOI] [PubMed] [Google Scholar]
- 26.Ehret GB. Genome-wide association studies: contribution of genomics to understanding blood pressure and essential hypertension. Curr Hypertens Rep. 2010;12:17–25. doi: 10.1007/s11906-009-0086-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newton-Cheh C, Cook NR, VanDenburgh M, Rimm EB, Ridker PM, Albert CM. A common variant at 9p21 is associated with sudden and arrhythmic cardiac death. Circulation. 2009;120:2062–2068. doi: 10.1161/CIRCULATIONAHA.109.879049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, Glazer NL, Morrison AC, Johnson AD, Aspelund T, Aulchenko Y, Lumley T, Köttgen A, Vasan RS, Rivadeneira F, Eiriksdottir G, Guo X, Arking DE, Mitchell GF, Mattace-Raso FU, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet. 2009;41:677–687. doi: 10.1038/ng.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coletta DK, Schneider J, Hu SL, Dyer TD, Puppala S, Farook VS, Arya R, Lehman DM, Blangero J, Defronzo RA, Duggirala R, Jenkinson CP. Genome-wide linkage scan for genes influencing plasma triglyceride levels in the Veterans Administration Genetic Epidemiology Study. Diabetes. 2009;58:279–284. doi: 10.2337/db08-0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dyke B: PEDSYS: a pedigree data management system. PGI. Tech rep no. 2, Population Genetics Laboratory, Department of Genetics, Southwest Foundation for Biomedical Research, San Antonio, TX (1999).
- 31.McPeek MS, Sun L. Statistical tests for detection of misspecified relationships by use of genome-screen data. Am J Hum Genet. 2000;66:1076–1094. doi: 10.1086/302800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sobel E, Papp JC, Lange K. Detection and integration of genotyping errors in statistical genetics. Am J Hum Genet. 2002;70:496–508. doi: 10.1086/338920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heath SC. Markov chain Monte Carlo segregation and linkage analysis for oligogenic models. Am J Hum Genet. 1997;61:748–760. doi: 10.1086/515506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heath SC, Snow GL, Thompson EA, Tseng C, Wijsman EM. MCMC segregation and linkage analysis. Genet Epidemiol. 1997;14:1011–1015. doi: 10.1002/(SICI)1098-2272(1997)14:6<1011::AID-GEPI75>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 35.Amos CI. Robust variance components approach for assessing genetic linkage in pedigrees. Am J Hum Genet. 1994;54:535–543. [PMC free article] [PubMed] [Google Scholar]
- 36.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62:1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hopper J, Matthews J. Extensions to multivariate normal models for pedigree analysis. Ann Hum Genet. 1982;46:373–383. doi: 10.1111/j.1469-1809.1982.tb01588.x. [DOI] [PubMed] [Google Scholar]
- 38.Boehnke M, Lange K. Ascertainment and goodness of fit of variance component models for pedigree data. Prog Clin Biol Res. 1984;147:173–192. [PubMed] [Google Scholar]
- 39.Duggirala R, Almasy L, Blangero J, Jenkinson CP, Arya R, DeFronzo RA, Stern MP, O'Connell P, American Diabetes Association GENNID Study Group Further evidence for type 2 diabetes susceptibility locus on chromosome 11q. Genet Epidemiol. 2003;24:240–242. doi: 10.1002/gepi.10233. [DOI] [PubMed] [Google Scholar]
- 40.Comuzzie AG, Williams JT. Correcting for ascertainment bias in the COGA data set. Genet Epidemiol. 1999;17(suppl 1):S109–S114. doi: 10.1002/gepi.1370170719. [DOI] [PubMed] [Google Scholar]
- 41.Tobin MD, Sheehan NA, Scurrah KJ, Burton PR. Adjusting for treatment effects in studies of quantitative traits: antihypertensive therapy and systolic blood pressure. Stat Med. 2005;24:2911–2935. doi: 10.1002/sim.2165. [DOI] [PubMed] [Google Scholar]
- 42.Cui JS, Hopper JL, Harrap SB. Antihypertensive treatments obscure familial contributions to blood pressure variation. Hypertension. 2003;41:207–210. doi: 10.1161/01.hyp.0000044938.94050.e3. [DOI] [PubMed] [Google Scholar]
- 43.Chobanian AV, Bakris GL, Black HR, Cushman WC, Green LA, Izzo JL, Jr, Jones DW, Materson BJ, Oparil S, Wright JT, Jr, Roccella EJ, et al. The Seventh Report of the Joint National Committee on Prevention, Detection, Evaluation, and Treatment of High Blood Pressure: the JNC 7 report. JAMA. 2003;289:2560–2572. doi: 10.1001/jama.289.19.2560. [DOI] [PubMed] [Google Scholar]
- 44.Hunt SC, Ellison RC, Atwood LD, Pankow JS, Province MA, Leppert MF. Genome scans for blood pressure and hypertension: the National Heart, Lung, and Blood Institute Family Heart Study. Hypertension. 2002;40:1–6. doi: 10.1161/01.hyp.0000022660.28915.b1. [DOI] [PubMed] [Google Scholar]
- 45.Allayee H, de Bruin TW, Michelle Dominguez K, Cheng LS, Ipp E, Cantor RM, Krass KL, Keulen ET, Aouizerat BE, Lusis AJ, Rotter JI. Genome scan for blood pressure in Dutch dyslipidemic families reveals linkage to a locus on chromosome 4p. Hypertension. 2001;38:773–778. doi: 10.1161/hy1001.092617. [DOI] [PubMed] [Google Scholar]
- 46.Rutherford S, Cai G, Lopez-Alvarenga JC, Kent JW, Voruganti VS, Proffitt JM, Curran JE, Johnson MP, Dyer TD, Jowett JB, Bastarrachea RA, Atwood LD, Goring HH, Maccluer JW, Moses EK, Blangero J, Comuzzie AG, Cole SA. A chromosome 11q quantitative-trait locus influences change of blood-pressure measurements over time in Mexican Americans of the San Antonio Family Heart Study. Am J Hum Genet. 2007;81:744–755. doi: 10.1086/521151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krushkal J, Ferrell R, Mockrin SC, Turner ST, Sing CF, Boerwinkle E. Genome-wide linkage analyses of systolic blood pressure using highly discordant siblings. Circulation. 1999;99:1407–1410. doi: 10.1161/01.cir.99.11.1407. [DOI] [PubMed] [Google Scholar]
- 48.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Levy D, Larson MG, Benjamin EJ, Newton-Cheh C, Wang TJ, Hwang SJ, Vasan RS, Mitchell GF. Framingham Heart Study 100K Project: genome-wide associations for blood pressure and arterial stiffness. BMC Med Genet. 2007;8(suppl 1):S3. doi: 10.1186/1471-2350-8-S1-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Org E, Eyheramendy S, Juhanson P, Gieger C, Lichtner P, Klopp N, Veldre G, Döring A, Viigimaa M, Sõber S, Tomberg K, Eckstein G, KORA, Kelgo P, Rebane T, Shaw-Hawkins S, Howard P, Onipinla A, Dobson RJ, Newhouse SJ, Brown M, Dominiczak A, Connell J, Samani N, Farrall M, BRIGHT, Caulfield MJ, Munroe PB, Illig T, Wichmann HE, Meitinger T, Laan M. Genome-wide scan identifies CDH13 as a novel susceptibility locus contributing to blood pressure determination in two European populations. Hum Mol Genet. 2009;18:2288–2296. doi: 10.1093/hmg/ddp135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang HC, Liang YJ, Wu YL, Chung CM, Chiang KM, Ho HY, Ting CT, Lin TH, Sheu SH, Tsai WC, Chen JH, Leu HB, Yin WH, Chiu TY, Chen CI, Fann CS, Wu JY, Lin TN, Lin SJ, Chen YT, Chen JW, Pan WH. Genome-wide association study of young-onset hypertension in the Han Chinese population of Taiwan. PLoS One. 2009;4:e5459. doi: 10.1371/journal.pone.0005459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adeyemo A, Gerry N, Chen G, Herbert A, Doumatey A, Huang H, Zhou J, Lashley K, Chen Y, Christman M, Rotimi C. A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet. 2009;5:e1000564. doi: 10.1371/journal.pgen.1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hiura Y, Tabara Y, Kokubo Y, Okamura T, Miki T, Tomoike H, Iwai N. A genome-wide association study of hypertension-related phenotypes in a Japanese population. Circ J. 2010;74:2353–2359. doi: 10.1253/circj.cj-10-0353. [DOI] [PubMed] [Google Scholar]