Abstract
The aryl hydrocarbon receptor (AHR) and hypoxia inducible factors (HIFs) are transcription factors that control the adaptive response to toxicants, such as dioxins, and decreases in available oxygen, respectively. The AHR and HIFs utilize the same heterodimeric partner, the aryl hydrocarbon nuclear translocator (ARNT) for proper function. This requirement raises the possibility that cross-talk exists between these critical signaling systems. Single gene and reporter assays have yielded conflicting results regarding the nature of the competition for ARNT. Therefore, to determine the extent of cross-talk between the AHR and HIFs, a comprehensive analysis was performed using oligonucleotide arrays. The results identified 767 and 430 genes that are sensitive to cobalt chloride and 2,3,7,8-tetrachlorodibenzo-ρ-dioxin (TCDD) stimulation, respectively, with 308 and 176, respectively, exhibiting sensitivity to cross-talk. The overlap between these two sets consists of 33 unique genes including the classic target genes CYP1A1, carbonic anhydrase IX and those involved in lipid metabolism, and coagulation. Computational analysis of the regulatory region of these genes identified complex relationships between HIFs, AHR, their respective response elements as well as other DNA motifs, including the SRF, Sp-1, NF-kB, and AP-2 binding sites. These results suggest that HIF - AHR cross-talk is limited to genes with regulatory regions that contain specific motifs and architecture.
Introduction
The PAS (named for founding members; PER, ARNT, SIM) family of transcription factors act as sensors for various environmental stimuli, including hypoxia and specific classes of pollutants (1). As transcription factors, their principal reaction involves the modulation of gene expression that ultimately promotes an adaptive response to these stimuli. PAS transcription factors generally function as heterodimers that can have both cytosolic and nuclear components. The cytoplasmic component generally acts as a “sensor” for environmental stimuli and includes the aryl hydrocarbon receptor (AHR) and the alpha subunit of the hypoxia inducible factors (HIF1-3) (2,3). Once activated, these factors translocate to the nucleus and interact with the second class of the superfamily, the nuclear component, such as the aryl hydrocarbon nuclear translocator (ARNT, also known as HIF1β) (4,5). ARNT/HIF1β is the predominant binding partner for the AHR and HIF1α and therefore ARNT might act as a point of competition or cross-talk following activation of the AHR and HIF1α.
The AHR is a ligand activated transcription factor that responds to a broad range of planar aromatic hydrocarbons (6). Classic AHR ligands include environmental pollutants such as 2,3,7,8-tetrachlorodibenzo-ρ-dioxin (TCDD), naturally occurring compounds, such as indole-3-carbazole, and endogenous ligands such as tryptophan metabolites. In the absence of ligand, the cytosolic AHR is bound to the immunophilin-like protein, aryl hydrocarbon receptor-associated protein (ARA9) and a dimer of heat shock protein 90 (Hsp90) (7,8). Upon binding ligand, the AHR translocates to the nucleus where it heterodimerizes with ARNT. The transcriptionally active AHR:ARNT complex drives the expression of genes containing dioxin response elements (DREs, core sequence = GCGTG) such as the canonical AHR-responsive gene, cytochrome P450, P450 (9).
Hypoxia is defined as a decrease in available oxygen reaching the tissues of the body. The cellular response to hypoxia is a fine balance between adaptation and cell death and is primarily controlled by HIF1α (10). HIF1α is a cytosolic protein whose stability is regulated by a family of prolyl hydroxylases (11,12). These hydroxylases are oxygen dependent “sensors” for the hypoxia signaling cascade either directly or indirectly through changes in reactive oxygen species generated from complex III of the electron transport chain (13-16). In the presence of oxygen, HIF1α is hydroxylated and degraded via the Von Hippel Lindau tumor suppressor protein and the 26S proteosome pathway (17). In the absence of sufficient oxygen, the hydroxylase is inactive and the HIF1α protein becomes stabilized and translocates into the nucleus where it interacts with ARNT to drive the expression hypoxia response elements (HREs, core sequence = (G/A)CGTG) containing genes (18). Classic hypoxia inducible genes include vascular endothelial growth factor (VEGF), erythropoietin and most of the glycolytic enzyme genes (19,20).
The ability of ARNT to act as the heterodimeric partner for both HIF1α and AHR raises the possibility of cross-talk between these signaling cascades. In addition, the similarities between the HRE and DRE core sequences suggests HIF1:ARNT and AHR:ARNT might compete for the same regulatory sequence (21). Consequently, hypoxia:AHR cross-talk might have profound consequences on treatments aimed at hypoxic targets, such as tumors, and might influence the ability of the AHR to regulate the expression of various drug metabolizing enzymes. Previous in vitro and in vivo studies have shown varying degrees of competition between AHR and HIF signaling systems, presumably due to competition for ARNT or another critical cofactor (22-26). We hypothesized that this cross-talk may only apply to target genes which harbor select response element combinations that include but are not limited to DREs and HREs. Global gene expression analysis was performed in the human hepatoma HepB3 cells following exposure to TCDD, cobalt chloride (CoCl2) and their cotreatment. A set of genes responsive to cobalt or TCDD, with a subset influenced by co-treatment was identified. Computational analysis of the regulatory regions of a subset of these genes, influenced by individual treatment, identified motifs associated with TCDD or cobalt chloride modulated gene expression. The results suggest that cross-talk between the AHR and HIF1 signaling systems in Hep3B cells is not regulated by ARNT levels and is limited to a group of genes with specific promoter architecture.
Experimental Procedures
Cell Culture
Hep3B cells were maintained in αMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Hyclone, Logan, UT), 20 mM L-glutamine, 1 mM MEM non-essential amino acids, 100 mM Hepes (pH=7.4), 1,000 units/ml penicillin G and 1,000 μg/ml streptomycin sulfate (Invitrogen). Cells were approximately 70% confluent at the time of treatment. Cells were maintained at 37 °C, 5% CO2 and 21% O2 prior to treatment. 2,3,7,8-tetrachlorodibenzo-ρ-dioxin (TCDD, 10 nM) and cobalt chloride (100 μM) treatments were performed for 20 hours at 37°C, 5% CO2 and 21% O2. These doses for cobalt and TCDD were chosen for their ability to activate HIF1α and the AHR, respectively. Cobalt chloride has an established history as a hypoxic mimic and the chosen dose has a demonstrated ability to stabilize HIF1α and activate all classic HIF1α target genes, including VEGF and the glycolytic enzymes. The dose of TCDD was chosen for its ability to maximally activate the AHR (several times its Kd). Each treatment group was supplemented with dimethyl sulfoxide (DMSO) to a final concentration of 0.01%. Samples were compared to vehicle control (0.01% DMSO).
RNA Extraction
Following treatment, duplicate cell samples were removed from the tissue culture dish by trypsinization and washed in PBS (4 °C). RNA was extracted by homogenization (Polytron, Kinematica, Lucerne, Switzerland) in TRIzol reagent (GIBCO/BRL, Gaithersburg MD) (added to cell pellet) at maximum speed for 90-120 s. Following a 5 min., room temperature incubation, a 1/5 vol of chloroform was added, agitated, and subjected to centrifugation at 12,000 × g for 15 min. The aqueous phase was removed, and the RNA was precipitated upon the addition of a half vol of isopropanol. The RNA was furthered purified with the RNAeasy Total RNA isolation kit (Qiagen, Valencia, CA) according to manufacturer’s specifications. Finally the purified total RNA was eluted in 10 μL of diethylpyrocarbonate (DEPC) treated H2O, and quantity and integrity were characterized using a DU640 UV spectrophotometer (Beckman, Fullerton, CA) and a Bioanalyzer 2100 (Agilent, Palo Alto, CA).
RNA Labeling
Five micrograms of total RNA from two separate biological replicates were used to make first strand cDNA using the Superscript Choice system (Gibco/BRL) and a T7 promotor/oligo-dT primer (Gibco/BRL). Second strand cDNA was also made with the Superscript Choice system (Invitrogen). The resulting cDNA was subjected to phenol:chloroform purification, ammonium acetate precipitation, and used as a template to make biotinylated amplified antisense cRNA using T7 RNA polymerase (Enzo kit, Affymetrix, Santa Clara, CA). Twenty micrograms cRNA was fragmented to a range of 20 to 100 bases in length using fragmentation buffer (200 mM Tris-acetate, pH 8.1, 500 mM KOAc, 150 mM MgOAc) and heating for 35 min at 94 °C. The quality of cRNA and size distribution of fragmented cRNA were examined by both agarose and polyacrylamide gel electrophoresis.
Hybridization
20 μg of cRNA was hybridized to a U95A version 1 gene chip (Affymetrix) with 1X 4-morpholineethanesulfonic acid (MES) hybridization buffer using standard protocols outlined in the Gene Chip® Expression Analysis Technical Manual (Affymetrix). Hybridization was conducted in a GeneChip Hybridization Oven for 16 hours at 45 °C. Following hybridization, the arrays were washed on a Genechip Fluidics Station 400 according to manufacturer’s instructions (Affymetrix). The arrays were scanned using a Hewlett Packard 2500A Gene Array Scanner and the raw images were visually inspected for defects, proper grid alignment, and converted into CEL files using the MAS5 Software Suite (Affymetrix). Finally, quality of cRNA was assessed by examining 3′/5′ ratios for GAPDH oligonucleotides present on the arrays.
Data Analysis
Background subtraction and single intensity measure for each transcript was derived from multiple probe sets by means of the GCRMA algorithm using the ‘full model tag’ in R (http://www.r-project.org). The GCRMA algorithm was chosen for its performance in reporting low and high level expression over other methods, as well as its dynamic range for single probe sets (27-29). Differentially expressed genes that are statistically significant were determined by analysis of variance (ANOVA). Fold change calculations were performed in Excel on data that was median-scaled to a global intensity target value of 100. For each treatment vs. control condition, genes that changed were assigned based on a P value of <0.05 and a fold change value of > 1.5.
Quantitative Real-Time PCR (qRT-PCR) Analysis
Changes in gene expression were observed by microarray analyses and verified by real-time PCR performed on an Applied Biosystems Prism 7000 Sequence detection System (Foster City, CA) as previously described (30). Analysis was performed on triplicate samples that were treated in an identical way as those used in the GeneChip experiments. Briefly, cDNA was synthesized from total RNA (1 μg per sample per treatment, n=6) in a reverse transcriptase reaction in 20 μl of 1X First Strand Synthesis buffer (Invitrogen) containing 1 μg oligo (5′-T21VN-3′), 0.2 mM dNTPs, 10 mM DTT, and 200 IU of Superscript II reverse transcriptase (Invitrogen). The reaction mixture was incubated at 42 °C for 60 min, and stopped by incubation at 75 °C for 15 min. Amplification of cDNA (1/20) was performed using SYBR® Green PCR buffer (1x AmpliTaqTM Gold PCR Buffer, 0.025 U/ml AmpliTaqTM Gold (Perkin-Elmer, Wellesley, MA), 0.2 mM dNTPs, 1 ng/μl 6-carboxy-X-rhodamine, 1:40,000 diluted SYBR® Green Dye and 3% DMSO) and 0.1 μM primers. The thermal cycling parameters were 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. Prior to analyzing the samples, standard curves of purified, target-specific amplicons were created. Briefly, gene specific oligonucleotides were used to PCR amplify the gene product from a pooled sample of prepared cDNA, the concentration of the amplicons was determined by UV spectophotometery and a standard curve was created (100 to 100 million copies). The mRNA expression for each gene was determined by comparison to its respective standard curve. This measurement was controlled for RNA quality, quantity, and RT efficiency by normalizing it to the expression level of the β-Actin gene. Each primer set produced a single product as determined by melt-curve analysis and amplicons were of the appropriate size, as analyzed by agarose gel electrophoresis. Statistical significance was determined using normalized fold changes and ANOVA. Primers were designed using the web-based application Primer3 (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi) biasing towards the 3′ end of the transcript to maximize the likelihood of giving a gene specific product. The settings used in Primer3 were 125 base-pair amplicon, 20mers, 60 °C melting temperatures and all other as defaults. Primer sequences were analyzed by BLAST. Gene names, accession numbers, and forward and reverse primer sequences are listed in Table 1.
Table 1.
Primer information for qRTPCR
| Descriptions | Gene Abbrev. |
REF Seq | Amplicon Size |
Forward | Reverse |
|---|---|---|---|---|---|
| adrenomedullin | ADM | NM_001124 | 127 | ggtgcagaagaatccgagtg | acgccgtgagaaatcagttt |
| BCL2/adenovirus E1B 19kDa interacting protein 3-like | BNIP3L | NM_004331 | 121 | agcatgacccaacctaccac | tcttcaggccaaaaagggta |
| chemokine (C-C motif) ligand 20 | CCL20 | NM_004591 | 124 | gtggcttttctggaatggaa | caagtccagtgaggcacaaa |
| cyclin-dependent kinase inhibitor 1C | CDKN1C | NM_000076 | 129 | agagatcagcgcctgagaag | tgggctctaaattggctcac |
| glycoprotein hormones, alpha polypeptide | CGA | NM_000735 | 129 | cccactccactaaggtccaa | cgtgtggttctccactttga |
| ceruloplasmin (ferroxidase) | CP | NM_000096 | 128 | cccatggggtacaaacagag | agcccatggaatacaagcag |
| cytochrome P450, family 1, subfamily A, polypeptide 1 | Cyp1A1 | NM_000499 | 122 | cttccgacactcttccttcg | ggttgatctgccactggttt |
| cytochrome P450, family 1, subfamily B, polypeptide 1 | Cyp1B1 | NM_000104 | 119 | atggcctcatcaacaaggac | gagccaggatggagatgaag |
| dual specificity phosphatase 5 | DUSP5 | NM_004419 | 133 | cctgctaaaactgggatgga | ctatctcactggggcagcat |
| fibrinogen alpha chain | FGA | NM_000508 | 140 | agccgatcatgaaggaacac | tactggatcccggtagcttg |
| fibrinogen beta chain | FGB | NM_005141 | 126 | gatgggagaaaacaggacca | catccaccaccgtcttcttt |
| potassium inwardly-rectifying channel, subfamily J | KCNJ8 | NM_004982 | 138 | gatcatctgccacgtgattg | gttcgtgcttgtgtggtgat |
| lysyl oxidase | LOX | NM_002317 | 133 | gcacacacacagggattgag | ccaggtagctggggtttaca |
| lumican | LUM | NM_002345 | 124 | atcagcaacatccctgatga | caaccagggatgacacattg |
| metallothionein 1X | MT1X | NM_005952 | 124 | gcaaatgcaaagagtgcaaa | cagcagctgcacttgtctga |
| v-myc myelocytomatosis viral oncogene homolog | MYC | NM_002467 | 122 | ccgaggagaatgtcaagagg | ggccttttcattgttttcca |
| phosphoenolpyruvate carboxykinase 2 | PCK2 | NM_004563 | 137 | gggtgctagactggatctgc | gagggagaacagctgagtgg |
| SMAD, mothers against DPP homolog 5 | SMAD5 | NM_001001419 | 120 | atctcaggtctcccagagca | tgcagaagaaatgccttcaa |
| UDP glucuronosyltransferase 1 | UGT1A1 | NM_007120 | 133 | gtgcctttatcacccatgct | tccagctcccttagtctcca |
| beta-Actin | ACTB | NM_001101 | 116 | ctcttccagccttccttcct | agcactgtgttggcgtacag |
Computational Scanning for DREs and HREs
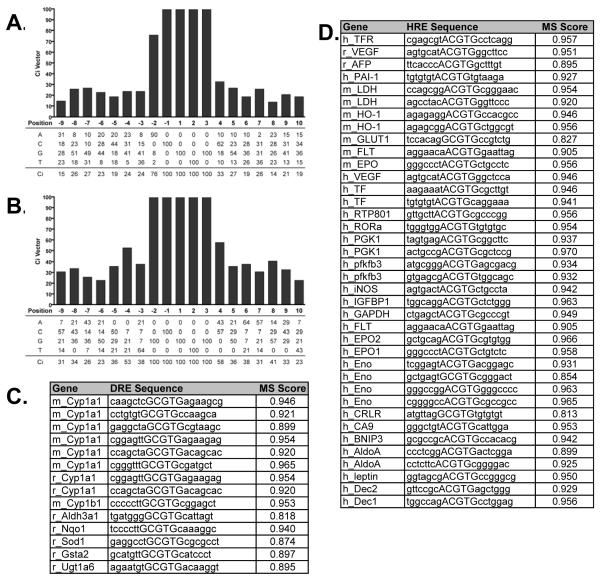
Computational identification and matrix similarity (MS) scores (31) of putative DREs and HREs were performed using 19 base-pair position weight matrices (PWMs) (Figure 1A and 1B) as previously developed by Sun et. al. (32). The PWMs were constructed using the sequences of reported functional response elements that were positive in either electrophoretic mobility shift of transient transfection assays. In total, 14 DREs (Figure 1C) and 38 HREs (Figure 1D) were used in the development of the PWMs. Each matrix consists of the 5 base-pair core HRE or DRE, (G/A)CGTG (Figure 1A) and GCGTG (Figure 1B) respectively, and the adjacent variant 7 base-pair flanking sequences of the bona fide functional response elements. The sequences of the response elements were subsequently scanned using the PWMs to determine the threshold MS scores (0.818 for DREs and 0.813 for HREs) (32). The MS scores for the 14 functional DREs and 38 HREs are listed in Figure 1C and 1D respectively. Furthermore, the consensus index (Ci) vector was calculated for both the HRE and DRE PWMs, where the Ci vectors represents the conservation of the individual nucleotide positions in the matrices (31). A complete list of information regarding the sequences used in creating the PWMs can be found in Supplemental Table 1 and Sun et. al. 2004.
Figure 1.
HRE and DRE position weight matrices and the sequences used in their construction
Position weight matrices were created as described in materials and methods. The HRE (A) and DRE (B) PWMs are displayed relative to the “G” of the second half of the binding site. The genes and sequences used for the creation of the DRE (C) and HRE (D) PWMs and their respective MS scores are listed. Sequence information regarding HREs used in construction of PWM can be found in Supplemental Table 1.
The genomic sequences (−5,000 base-pair to the transcriptional start site (TSS) and the 5′ untranslated region (UTR)) of the 33 RefSeq genes were extracted from the University of California, Santa Cruz (UCSC) Genome Browser (http://www.genome.ucsc.edu) (Build #35) and scanned for exact matches to the DRE and HRE core sequences on both the positive and negative strands using the PWMs. The MS score for each match was computed and those with scores greater than the threshold values are expected to have both a greater probability of possessing a measurable binding affinity and presumable biological relevance.
Identification of Over-Represented Short Sequence Motifs in Gene Regulatory Regions
Gene regulatory regions, defined as starting −5,000 kilobase relative to the transcription start site through the 5′ UTR, were obtained from the UCSC Genome Browser for all known genes assigned a mature RefSeq mRNA accession. These sequences were stored in the Gene Regulatory Subsystem of our toxicogenomic database, dbZach (http://dbzach.fst.msu.edu, (33)), to facilitate further analysis. All 5–10 nucleotide short sequence motifs were identified using a sliding window method (34). An empirical Bayes implementation of the Wilcoxon’s Rank Sum Test, similar to those used for microarray analysis (35,36), was used to identify 5-10 nucleotide motifs that are over-represented in one population compared to another. This method computes a posterior probability, which represents the likelihood of that result occurring (e.g., a posterior probability of 0.90 means there is a 90% probability the result is true). The Transfac database (37) was queried to annotate the motifs, and identify potential binding proteins. Absolute numbers of individual motifs that were determined to be over-represented in the cobalt or TCDD treatment groups were then analyzed by hierarchical clustering (unweighted pair-group method using arithmetic averages using correlation distances, http://gepas.bioinfo.cnio.es/cgi-bin/cluster (38)). These values were clustered with the GeneChip expression data found in Figure 4.
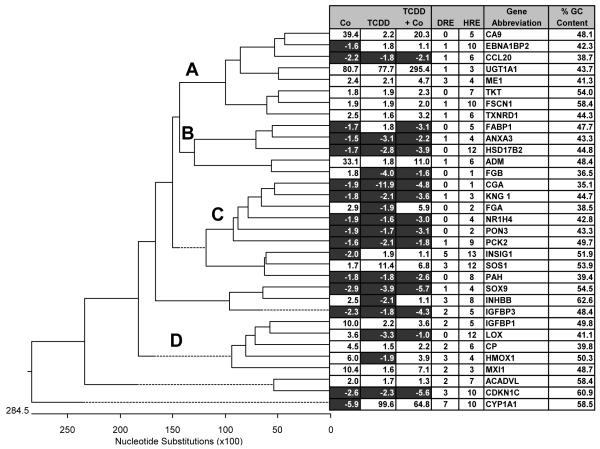
Figure 4.
Alignment and sequence analysis of the promoters and 5′ UTR of core genes
5000 bp of upstream sequences and the 5′ UTRs were aligned by ClustalW and compared to expression patterns from oligonucleotide chips. The promoters were also analyzed for HREs and DREs using a PWM (See methods). Finally, the GC content was also calculated. Up-regulated genes are shown in white and down-regulated genes are shown in dark gray. Four distinct clusters are also noted (A-D).
Results
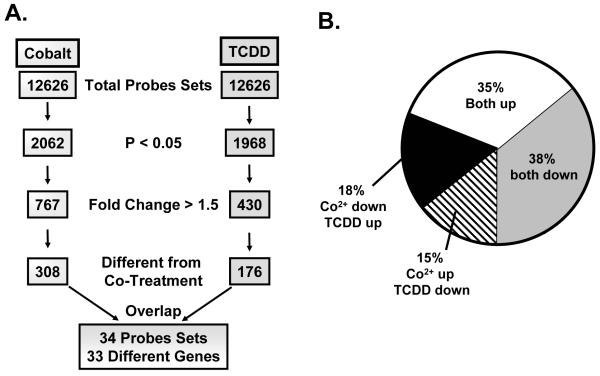
Identification of genes potentially susceptible to cross-talk between the AHR and HIF signaling cascades was performed on a high density oligonucleotide array. Hep3B cells were treated with DMSO (0.01%, vehicle control), cobalt chloride (100 mM), TCDD (10 nM) or cobalt chloride and TCDD (100 mM and 10 nM respectively) for 20 hours and global gene expression patterns were analyzed. The procedure to identify these genes is outlined in Figure 2A. Briefly, significant (p < 0.05) gene expression changes following a single treatment were identified and further pared using a 1.5 fold cutoff leaving 767 and 430 probe sets for cobalt and TCDD treatment, respectively. These genes were then analyzed for significant differences between single treatment and co-treatment as determined by paired student’s t-test. 308 and 176 probe sets were identified with these criteria for cobalt and TCDD treated gene groups respectively. These two groups constitute the genes that can be influenced by cross-talk from either single treatment. Of these, 34 probe sets (33 genes) were represented in both lists (Figure 2A). These 33 genes represent a group of targets that are affected by both single treatments (i.e. AHR and HIF responsive) and whose expression is modulated by cross-talk.
Figure 2.
Analysis of genomic data and comparison of 33 target genes.
The genomic data was analyzed in a four step process (A). First, genes were screened for significance (p<0.05). Second, these genes were filtered for those that displayed greater than 1.5 fold change. Third, the list was analyzed for those that were significantly altered (p<0.05) when single treatment was compared to co-treatment. Finally, the cobalt and TCDD lists were compared for overlap. (B) Direction of expression changes was analyzed for the 34 probe sets identified in the screen. A complete list of cobalt and TCDD influenced genes can be found in supplemental data (Suppl. Tables 5 and 6).
Since the analysis was performed with no bias towards direction of change, it is possible that some of the changes would be in the same direction or in opposite directions. Analysis showed that a majority of these genes displayed the “same” direction changes, either both up or both down. Only 33% of the genes exhibited an opposite pattern of expression (Figure 2B). These results suggest a more complex interaction between the AHR and HIF signaling cascades and identify genes where this interaction may lead to antagonism (i.e. less than additive), or synergy (i.e. more than additive).
The list of 33 genes include prototypical dioxin and hypoxia responsive genes, notably P450 and heme oxygenase-1. These genes exhibit a classic cross-talk expression pattern. For example, P450 is induced approximately 100-fold in the presence of TCDD, while it is repressed almost 6-fold after cobalt treatment. Following co-treatment, gene expression is only induced 65-fold (Table 2). A similar pattern is seen for heme oxygenase-1, where it is induced by cobalt, repressed by TCDD, and only partially induced by co-treatment. This type of competition is not the only type of regulation observed. SOX-9 and CDKN1C displayed clear evidence of additive regulation in which individual treatments modify expression in the same direction and co-treatment yields an additive expression value (Table 2). In addition, UGT1A1 displays a level of synergy between treatments. These results suggest that simple regulation or competition for ARNT or other cellular factors cannot fully explain all of the expression changes seen in the genomic screen.
Table 2.
Microarray Fold Changes and information for 33 core cross-talk genes
| Fold Change | ||||||
|---|---|---|---|---|---|---|
| Probe Set | Co | TCDD | TCDD+Co | REFSeq | Descriptions | Abbreviation |
| 1025_g_at | −5.9 | 99.6 | 64.8 | NM_000499 | cytochrome P450, family 1, subfamily A | CYP1A1 |
| 33436_at | −2.9 | −3.9 | −5.7 | NM_000346 | SRY (sex determining region Y)-box 9 | SOX9 |
| 1787_at | −2.6 | −2.3 | −5.6 | NM_000076 | cyclin-dependent kinase inhibitor 1C | CDKN1C |
| 37319_at | −2.3 | −1.8 | −4.3 | NM_000598 | insulin-like growth factor binding protein 3 | IGFBP3 |
| 39545_at | −2.2 | −2.3 | −3.9 | NM_000076 | cyclin-dependent kinase inhibitor 1C | CDKN1C |
| 40385_at | −2.2 | −1.8 | −2.1 | NM_004591 | chemokine (C-C motif) ligand 20 | CCL20 |
| 35303_at | −2.0 | 1.9 | 1.1 | NM_005542 | insulin induced gene 1 | INSIG1 |
| 38519_at | −1.9 | −1.6 | −3.0 | NM_005123 | nuclear receptor subfamily 1, group H, member 4 | NR1H4 |
| 39352_at | −1.9 | −11.9 | −4.8 | NM_000735 | glycoprotein hormones, alpha polypeptide | CGA |
| 41424_at | −1.9 | −1.7 | −3.1 | NM_000940 | paraxonase 3 | PON3 |
| 33701_at | −1.8 | −1.8 | −2.6 | NM_000277 | phenylalanine hydroxylase | PAH |
| 37235_g_at | −1.8 | −2.1 | −3.6 | NM_000893 | kininogen 1 | KNG 1 |
| 38586_at | −1.7 | 1.8 | −3.1 | NM_001443 | fatty acid binding protein 1 | FABP1 |
| 38178_at | −1.7 | −2.8 | −3.9 | NM_002153 | hydroxysteroid (17-beta) dehydrogenase 2 | HSD17B2 |
| 36135_at | −1.6 | 1.8 | 1.1 | NM_006824 | EBNA1 binding protein 2 | EBNA1BP2 |
| 37188_at | −1.6 | −2.1 | −1.8 | NM_004563 | phosphoenolpyruvate carboxykinase 2 | PCK2 |
| 31792_at | −1.5 | −3.1 | −2.2 | NM_005139 | annexin A3 | ANXA3 |
| 33260_at | 1.7 | 11.4 | 6.8 | NM_005633 | son of sevenless homolog 1 | SOS1 |
| 38789_at | 1.8 | 1.9 | 2.3 | NM_001064 | transketolase | TKT |
| 37019_at | 1.8 | −4.0 | −1.6 | NM_005141 | fibrinogen beta chain | FGB |
| 39070_at | 1.9 | 1.9 | 2.0 | NM_003088 | fascin homolog 1 | FSCN1 |
| 38376_at | 2.0 | 1.7 | 1.3 | NM_000018 | acyl-Coenzyme A dehydrogenase | ACADVL |
| 31824_at | 2.4 | 2.1 | 4.7 | NM_002395 | malic enzyme 1 | ME1 |
| 39425_at | 2.5 | 1.6 | 3.2 | NM_003330 | thioredoxin reductase 1 | TXNRD1 |
| 38545_at | 2.5 | −2.1 | 1.1 | NM_002193 | inhibin, beta B | INHBB |
| 38825_at | 2.9 | −1.9 | 5.9 | NM_000508 | fibrinogen alpha chain | FGA |
| 38637_at | 3.6 | −3.3 | −1.0 | NM_002317 | lysyl oxidase | LOX |
| 39008_at | 4.5 | 1.5 | 2.2 | NM_000096 | ceruloplasmin | CP |
| 33802_at | 6.0 | −1.9 | 3.9 | NM_002133 | heme oxygenase (decycling) 1 | HMOX1 |
| 1232_s_at | 10.0 | 2.2 | 3.6 | NM_000596 | insulin-like growth factor binding protein 1 | IGFBP1 |
| 39072_at | 10.4 | 1.6 | 7.1 | NM_005962 | MAX interactor 1 | MXI1 |
| 34777_at | 33.1 | 1.8 | 11.0 | NM_001124 | adrenomedullin | ADM |
| 40309_at | 39.4 | 2.2 | 20.3 | NM_001216 | carbonic anhydrase IX | CA9 |
| 32392_s_at | 80.7 | 77.7 | 295.4 | NM_007120 | UDP glucuronosyltransferase 1 | UGT1A1 |
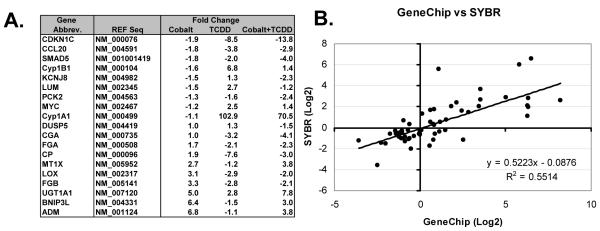
The expression patterns of nineteen different genes, including 11 from Table 2, were verified by qRTPCR. These genes were a mixture of classic and novel cobalt- or TCDD-inducible genes identified in the individual treatment groups (Supplemental Data, Table 3 and 4) and cross-talk analysis (Table 2). In most cases, these results verified those of the Gene Chip data (Figure 3). 49 of the 57 gene expression changes (19 different genes under 3 different treatments) were verified by qRTPCR (Figure 3B). Interestingly, in the gene chip data, fibrinogen alpha subunit (FGA) was up-regulated upon co-treatment, while the fibrinogen beta subunit (FGB) was down-regulated under similar conditions (Table 2). qRTPCR results showed that these two subunits were expressed in a similar pattern under each treatment condition (Figure 3A). Since, different pools of RNA were used for the qRTPCR and microarray experiments, cell line or culture differences can not be ruled out as an explanation for this difference.
Figure 3.
qRTPCR verification of expression changes for selected cross-talk genes.
qRTPCR was performed on 19 genes using b-Actin for normalization. Gene abbreviations, accession numbers used in primer design, and fold changes relative to vehicle control are listed (A). A direct comparison between qRTPCR and Gene Chip results was performed and analyzed by linear regression (B).
One possible explanation for the different expression patterns exhibited by the 33 genes (Table 2) that display cross-talk regulation is promoter context of response elements. To initially characterize differences between these promoters, genomic sequences 5000 base-pair upstream of the transcription start codon and the 5′ UTR were analyzed for homology, HREs, DREs, and GC content. The promoter and 5′ UTRs from the 33 genes were aligned using the ClustalW algorithm. Interestingly, the sequences clustered based on their expression patterns with four clusters containing 4 or more sequences (A-D, Figure 4). Group A was predominantly up-regulated in both treatments to an equal level and several were additive. Group B was primarily inhibited and showed a tendency for competition where the co-treatment group was somewhere between the two individual expression levels. Group C was predominantly down-regulated in both treatments, and Group D showed higher expression in the cobalt treatment compared to that of TCDD (Figure 4). This pattern was not unsystematic or due to some inherent base pair composition found within the promoters and/or 5′ UTR of the genes analyzed, since randomization (http://www.cellbiol.com/cgi-bin/randomizer/sequence_randomizer.html) of the promoter sequence yielded no consistent pattern with relation to expression. In addition, random sequences exhibited no relation to the alignment seen in Figure 4 (data not shown). These results suggest that motifs within the analyzed regions contained the regulatory information that influenced the cross-talk expression patterns observed.
Differences in the regulation patterns of the various genes could also be explained by the number of hypoxia or dioxin response elements located within the regulatory region. Comparison of these motif sequences to HRE and DRE PWMs suggests that DREs were better represented in Group D and this bias did not correlate with an increase in GC content (Figure 4). The analysis did identify more HREs than DREs, primarily due to the fact that the HRE PWM was only exclusive at 4 positions, compared to 5 positions in the DRE (Figure 1 and 4). Given the redundancy within the core sequences of the DREs and HREs, it is also possible that several sequences could register as both DREs and HREs. To determine the level of overlap, each DRE and HRE was further scored using the opposing PWM. For example, each DRE listed in figure 4 was scored with the HRE PWM and a similar cut-off (the MS score had to be higher than the lowest HRE used to create PWM) was applied. The results show that 40% of the DREs also scored positive as HREs while only 14% of the HREs scored positive as DREs (Supplemental data Tables 8 and 9). However, there was no relationship between HRE-DRE overlaps and the type of cross-talk observed. Finally, it is possible that base pairing relationships, G/C vs. A/T, may bias the association of a promoter into one group or another. Based on GC content there was no correlation between base-pairing relationships and gene expression patterns or groupings, suggesting that other information within the promoter regulated expression (Figure 4).
Given that it is not the absolute number of HREs or DREs within the regulatory region or their basic composition that dictates expression, promoter analysis for other motifs that might correlate with the expression pattern in Figure 4 were also examined. A computational word search was performed to identify over-represented motifs/elements 5000 base-pair upstream of the start codon and in the 5′ UTR. The 33 genes from Figure 4 were not sufficient to perform a word analysis; therefore, the analysis was conducted on a non-redundant set of 65 active genes from each of the cobalt and TCDD treatment groups for which gene regulatory sequences. The lists of over-represented motifs in cobalt and TCDD groups were combined, and the frequencies of each motif occurrences were calculated for each gene. 27 different words, corresponding to 15 different known transcription response elements, included Sp1, serum response elements (SRE) and NF-kB sites, were over-represented in the cobalt treated group. Conversely, 9 different words, corresponding to 6 different elements were over-represented in the TCDD treated group. These included a c/EBP-a, GREs and two types of GATA response elements (Figure 5).
Figure 5.
Promoter analysis for over-represented motifs and correlation with expression
The promoters of 65 different genes from the cobalt and TCDD treatment groups were analyzed for motifs that were over-represented as described in material and methods. Information about the motif, its corresponding response element as determined by TRANSFAC, and the statistical values are represented for the Cobalt group (A) and the TCDD group (B) as defined in materials in methods. The total number of response elements listed in A and B (including HREs and DREs) were then identified in the regulatory regions of the 33 genes from Fig. 4 and correlated with expression data via hierarchical clustering. A dendrogram correlating the GeneChip expression data (Co2+, TCDD and TCDD+Co2+) with the various response elements is displayed (C). Complete table of motifs identified in the analysis and the table used to create the dendrogram can be found in supplemental data (Tables 2-4).
The regulatory regions of the 33 genes identified in Figure 4 were analyzed for the over-represented response elements and the correlation between expression patterns from the microarray and the frequency of occurrence of these 23 major response elements (15 from cobalt treated group, 6 from the TCDD group and HREs and DREs) using hierarchical clustering. All of the expression data clustered together with the SRE (Figure 5C). A second cluster showed strong correlation with the expression data which included EGR-1, Sp-1, DRE, MyoD, NF-kB AP-2 and HRE elements. These results suggest that the expression patterns in Figure 4 are well correlated with the number of SREs in the respective promoters and to a lesser extent to a group of 7 other response elements, including the HREs and DREs.
Discussion
Understanding the nature and extent of the cross-talk between the hypoxia and AHR signaling cascades is important for determining the role of each signaling cascade in directly modulating the function of the other. The results presented here suggest a gene-specific cross-talk. In some instances the cross-talk was shown to be additive (e.g. IGFBP3, UGT1A1, and CDKN1C) while in others it was competitive (e.g. P450, IHBB, and LOX). To establish a possible mechanism for the selective nature of the cross-talk, a small portion of the putative regulatory region of a subset of these genes was analyzed for HREs and DREs. There was no apparent correlation between the number of these types of response elements and the nature of the cross-talk observed. Computational analysis of the regulatory regions proximal to the Transcription Starting Site (TSS) identified over-represented motifs within these regions suggesting that AHR-HIF1 cross-talk can not be explained completely by competition for ARNT and most likely involves other cofactors, response elements and promoter context.
Differences in expression patterns for those genes affected by TCDD and cobalt cross-talk suggest that a variety of mechanisms are involved. If the cross-talk was solely dependent upon competition for ARNT evidence of interactions would be wide spread and would not include additive responses. In contrast only 33 genes exhibited an interaction, with approximately 30% displayed additive tendencies, including ME1, SRY, CDKN1C and NR1H4. This pattern suggests that ARNT is not limiting and alternative mechanisms are involved. These mechanisms probably involve competition and/or synergy between other co-activators/co-repressors or other signaling systems that are being influenced and the cross-talk is a result of secondary signaling. For example, TCDD might inhibit the expression of a transcription factor necessary for cobalt-induced expression of another TCDD target gene. Alternatively, cobalt exposure might activate a signal that leads to post-translational modification of the AHR, thus altering its activity. Comparable interactions have been described between steroid receptors and other signaling cascades ((39) and references within). Finally, these alternative pathways likely involve other cascades that influence the ability of AHR and HIF1 to alter expression patterns. The predominance of SREs present in the regulatory region of genes exhibiting cross-talk suggests that one of these alternative inputs is the serum response cascade.
Presence of the SRE motif correlates with the expression patterns elicited by various treatments, suggesting a regulatory role in gene expression. Surprisingly, the number of SREs across the 33 genes in Table 2 is more predictive of cross-talk than the number of HREs or DREs (Figure 4). The 33 cross-talk genes were identified after meeting two criteria: Their expression was influenced by each individual treatment and this expression was further altered upon co-treatment. The original hypothesis that cross-talk was due to ARNT competition meant that this two step process would identify the subset of genes most prone to cross-talk. Given the different types of interactions (eg. additive and competitive) simple competition for ARNT does not satisfactorily explain cross-talk in Hep3B cells and these requirements might have biased the results to select for other transcription factor pathways, such as the SRE.
The serum response factor (SRF) binds the SRE and regulates the expression of a variety of genes including plasminogen activator inhibitor 1 (PAI-1), early growth response factor -1 (EGR-1) and glucose transporter-1 (Glut-1) (40-42). Interestingly, each of these genes is a target for hypoxia-mediated transcription. The over-representation of SREs in the cobalt treated group suggests that HIF1 and SRF signaling pathways may converge upon a select group of genes and the correlation of SREs with the set of cross-talk genes from Table 2 might suggest that these genes are important for the response to these two signaling pathways.
Genes exhibiting cross-talk fall into a variety of ontological categories, with several distinct processes and functions being over-represented, including blood coagulation, cell proliferation, fatty acid oxidation, metal binding and oxidoreductases (Supplemental Data, Table 7) suggesting HIF-AHR interactions may alter these endogenous processes. For example, hypoxia influences the disposition of a variety of drugs through multiple mechanisms, including altering the expression of phase 1 enzymes, including several oxidoreductases and a patient who has been pre-exposed to AHR ligands might be further compromised in their ability to dispose of drugs (43). The putative role of cross-talk might be most evident during development of the liver and heart. Mice lacking functional AHR have aberrant liver development and are prone to hypertension and cardiac hypertrophy (44). Liver development is abnormal in AHR null mice as a result of decreased peripheral profusion (44) possibly due to an imbalance in hypoxia-induced fibrin expression that ultimately influences angiogenesis. Over-production of fibrin and its subsequent deposition into the local circulation might explain the excess hematopoeitic cells in the AHR -/- liver and the subsequent increase in peripheral resistance (2). Similarily, cardiac hypertrophy in AHR -/- mice appears correlated with endothelin-1, angiotensin II, and HIF1α (45-47). Therefore, AHR-HIF1a interactions may adversely affect heart and liver development following signaling cross-talk.
The ability of HIF1α and AHR to influence each other’s transcriptional activity on a genome scale appears to involve multiple factors and our results suggest that HREs, DREs and promoter context, play a role in mediating this cross-talk. It is also likely that promoter context and other transcription factors play a major role in determining gene expression behavior following hypoxia and/or TCDD treatment. In fact, the direction and magnitude of change, following exposure to either hypoxia or TCDD, when compared to hypoxia and TCDD, is probably more dependent upon additional transcription factors than previously suspected. Therefore, understanding the role of cross-talk between hypoxia and the AHR will require a more thorough characterization of HIFs and the AHR interactions with other transcription factors and their respective ability to influence basic transcriptional machinery.
Supplementary Material
Acknowledgment
Special thanks to Melinda Kochenderfer for her help in preparing this manuscript. J.J.L. and T.R.Z. are partially supported by the Michigan Agriculture Experimental Station. This work was supported by funds from the National Institute of Health (NIH) R01-ES12186.
Glossary
- AHR
aryl hydrocarbon receptor
- HIF1-3
hypoxia inducible factors
- ARNT
aryl hydrocarbon nuclear translocator
- TCDD
2,3,7,8-tetrachlorodibenzo-ρ-dioxin
- ARA9
aryl hydrocarbon receptor-associated protein
- DREs
dioxin response elements
- P-450
cytochrome P450
- HREs
hypoxia response elements
- DMSO
dimethyl sulfoxide
- qRT-PCR
Quantitative Real-Time PCR
- MS
matrix similarity
- PWMs
position weight matrices
- Ci
consensus index
References
- (1).Gu Y-Z, Hogenesch J, Bradfield C. The PAS superfamily: Sensors of environmental and developmental signals. Academic Press; 2000. pp. 519–561. [DOI] [PubMed] [Google Scholar]
- (2).Schmidt JV, Carver LA, Bradfield CA. Molecular characterization of the murine Ahr gene. Organization, promoter analysis, and chromosomal assignment. J. Biol. Chem. 1993;268:22203–22209. [PubMed] [Google Scholar]
- (3).Semenza GL, Agani F, Booth G, Forsythe J, Iyer N, Jiang BH, Leung S, Roe R, Wiener C, Yu A. Structural and functional analysis of hypoxia-inducible factor 1. Kidney Int. 1997;51:553–555. doi: 10.1038/ki.1997.77. [DOI] [PubMed] [Google Scholar]
- (4).Reyes H, Reisz-Porszasz S, Hankinson O. Identification of the Ah receptor nuclear translocator protein (Arnt) as a component of the DNA binding form of the Ah receptor. Science. 1992;256:1193–1195. doi: 10.1126/science.256.5060.1193. [DOI] [PubMed] [Google Scholar]
- (5).Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Bradfield CA, Poland A. A competitive binding assay for 2,3,7,8-tetrachlorodibenzo-p-dioxin and related ligands of the Ah receptor. Mol Pharmacol. 1988;34:682–688. [PubMed] [Google Scholar]
- (7).Carver LA, Bradfield CA. Ligand dependent interaction of the Ah receptor with a novel immunophilin homolog in vivo. J. Biol. Chem. 1997;272:11452–11456. doi: 10.1074/jbc.272.17.11452. [DOI] [PubMed] [Google Scholar]
- (8).Ma Q, Whitlock JP. A novel cytoplasmic protein that interacts with the Ah receptor, contains tetratricopeptide repeat motifs, and augments the transcriptional response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. J. Biol. Chem. 1997;272:8878–8884. [PubMed] [Google Scholar]
- (9).Whitlock JP, Denison MS, Fisher JM, Shen ES. Induction of hepatic cytochrome P450 gene expression by 2,3,7,8-tetrachlorodibenzo-p-dioxin. Mol. Biol. Med. 1989;6:169–178. [PubMed] [Google Scholar]
- (10).Wang GL, Semenza GL. Characterization of hypoxia-inducible factor 1 and regulation of DNA binding activity by hypoxia. J Biol. Chem. 1993;268:21513–21518. [PubMed] [Google Scholar]
- (11).Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- (12).Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O’Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, Tian YM, Masson N, Hamilton DL, Jaakkola P, Barstead R, Hodgkin J, Maxwell PH, Pugh CW, Schofield CJ, Ratcliffe PJ. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- (13).Berra E, Benizri E, Ginouves A, Volmat V, Roux D, Pouyssegur J. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1alpha in normoxia. EMBO J. 2003;22:4082–4090. doi: 10.1093/emboj/cdg392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Brunelle JK, Bell EL, Quesada NM, Vercauteren K, Tiranti V, Zeviani M, Scarpulla RC, Chandel NS. Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metab. 2005;1:409–414. doi: 10.1016/j.cmet.2005.05.002. [DOI] [PubMed] [Google Scholar]
- (15).Guzy RD, Hoyos B, Robin E, Chen H, Liu L, Mansfield KD, Simon MC, Hammerling U, Schumacker PT. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 2005;1:401–408. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- (16).Mansfield KD, Guzy RD, Pan Y, Young RM, Cash TP, Schumacker PT, Simon MC. Mitochondrial dysfunction resulting from loss of cytochrome c impairs cellular oxygen sensing and hypoxic HIF-alpha activation. Cell. Metab. 2005;1:393–399. doi: 10.1016/j.cmet.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Mole DR, Maxwell PH, Pugh CW, Ratcliffe PJ. Regulation of HIF by the von Hippel-Lindau tumour suppressor: implications for cellular oxygen sensing. IUBMB Life. 2001;52:43–47. doi: 10.1080/15216540252774757. [DOI] [PubMed] [Google Scholar]
- (18).Semenza GL, Jiang BH, Leung SW, Passantino R, Concordet JP, Maire P, Giallongo A. Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J Biol. Chem. 1996;271:32529–32537. doi: 10.1074/jbc.271.51.32529. [DOI] [PubMed] [Google Scholar]
- (19).Minchenko A, Salceda S, Bauer T, Caro J. Hypoxia regulatory elements of the human vascular endothelial growth factor gene. Cell. Mol. Biol. Res. 1994;40:35–39. [PubMed] [Google Scholar]
- (20).Semenza GL, Roth PH, Fang HM, Wang GL. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J Biol. Chem. 1994;269:23757–23763. [PubMed] [Google Scholar]
- (21).Hogenesch JB, Chan WC, Jackiw VH, Brown RC, Gu YZ, Pray-Grant M, Perdew GH, Bradfield CA. Characterization of a subset of the basic-helix-loop-helix-PAS superfamily that interact with components of the dioxin signaling pathway. J Biol Chem. 1997;272:8581–8593. doi: 10.1074/jbc.272.13.8581. [DOI] [PubMed] [Google Scholar]
- (22).Chan WK, Yao G, Gu YZ, Bradfield CA. Cross-talk between the aryl hydrocarbon receptor and hypoxia inducible factor signaling pathways. Demonstration of competition and compensation. J. Biol. Chem. 1999;274:12115–12123. doi: 10.1074/jbc.274.17.12115. [DOI] [PubMed] [Google Scholar]
- (23).Gassmann M, Kvietikova I, Rolfs A, Wenger RH. Oxygen- and dioxin-regulated gene expression in mouse hepatoma cells. Kidney Int. 1997;51:567–574. doi: 10.1038/ki.1997.81. [DOI] [PubMed] [Google Scholar]
- (24).Gradin K, McGuire J, Wenger RH, Kvietikova I, Whitelaw ML, Toftgard R, Tora L, Gassmann M, Poellinger L. Functional interference between hypoxia and dioxin signal transduction pathways: competition for recruitment of the Arnt transcription factor. Mol. Cell. Biol. 1996;16:5221–5231. doi: 10.1128/mcb.16.10.5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Nie M, Blankenship AL, Giesy JP. Interactions between aryl hydrocarbon receptor (AhR) and hypoxia signaling pathways. Environ. Toxicol. Pharmacol. 2001;10:17–27. doi: 10.1016/s1382-6689(01)00065-5. [DOI] [PubMed] [Google Scholar]
- (26).Pollenz RS, Davarinos NA, Shearer TP. Analysis of aryl hydrocarbon receptor-mediated signaling during physiological hypoxia reveals lack of competition for the aryl hydrocarbon nuclear translocator transcription factor. Mol. Pharmacol. 1999;56:1127–1137. doi: 10.1124/mol.56.6.1127. [DOI] [PubMed] [Google Scholar]
- (27).Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Ihaka R, Gentlemen R. R: A language for data analysis and graphics. J. Comp. Graph. Stats. 1996;5:299–314. [Google Scholar]
- (29).Wu Z, Irizarry RA, Gentlemen R, Murillo FM, Spencer F. A model based background adjustment for oligonucleotide expression arrays. 2004 http://www.bioinformatica.unito.it/downloads/complexity/gcpaper.pdf:1-29.
- (30).Vengellur A, Woods BG, Ryan HE, Johnson RS, LaPres JJ. Gene expression profiling of the hypoxia signaling pathway in hypoxia inducible factor 1 null mouse embryonic fibroblasts. Gene Expression. 2003;11:181–197. doi: 10.3727/000000003108749062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–84. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Sun YV, Boverhof DR, Burgoon LD, Fielden MR, Zacharewski TR. Comparative analysis of dioxin response elements in human, mouse and rat genomic sequences. Nucleic Acids Res. 2004;32:4512–23. doi: 10.1093/nar/gkh782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Burgoon LD, Boutros PC, Dere E, Zacharewski TR. dbZach: A MIAME-Compliant Toxicogenomic Supportive Relational Database. Toxicol. Sci. 2006;90:558–568. doi: 10.1093/toxsci/kfj097. [DOI] [PubMed] [Google Scholar]
- (34).Tajima F. Determination of window size for analyzing DNA sequences. J. Mol. Evol. 1991;33:470–3. doi: 10.1007/BF02103140. [DOI] [PubMed] [Google Scholar]
- (35).Eckel JE, Gennings C, Chinchilli VM, Burgoon LD, Zacharewski TR. Empirical bayes gene screening tool for time-course or dose-response microarray data. J. Biopharm. Stat. 2004;14:647–670. doi: 10.1081/BIP-200025656. [DOI] [PubMed] [Google Scholar]
- (36).Efron B, Tibshirani R. Empirical bayes methods and false discovery rates for microarrays. Genet. Epidemiol. 2002;23:70–86. doi: 10.1002/gepi.1124. [DOI] [PubMed] [Google Scholar]
- (37).Wingender E. Transfac, Transpath and Cytomer as starting points for an ontology of regulatory networks. In Silico Biol. 2004;4:55–61. [PubMed] [Google Scholar]
- (38).Herrero J, Al-Shahrour F, Diaz-Uriarte R, Mateos A, Vaquerizas JM, Santoyo J, Dopazo J. GEPAS: a web-based resource for microarray gene expression data analysis. Nucleic Acids Res. 2003;31:3461–3467. doi: 10.1093/nar/gkg591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Wang L, Zhang X, Farrar WL, Yang X. Transcriptional crosstalk between nuclear receptors and cytokine signal transduction pathways in immunity. Cell. Mol. Immunol. 2004;1:416–24. [PubMed] [Google Scholar]
- (40).Chang H, Shyu KG, Lin S, Tsai SC, Wang BW, Liu YC, Sung YL, Lee CC. The plasminogen activator inhibitor-1 gene is induced by cell adhesion through the MEK/ERK pathway. J. Biomed. Sci. 2003;10:738–745. doi: 10.1159/000073961. [DOI] [PubMed] [Google Scholar]
- (41).Ebert BL, Firth JD, Ratcliffe PJ. Hypoxia and mitochondrial inhibitors regulate expression of glucose transporter-1 via distinct cis-acting sequences. J. Biol. Chem. 1995;270:29083–29089. doi: 10.1074/jbc.270.49.29083. [DOI] [PubMed] [Google Scholar]
- (42).Yan S-F, Lu J, Zou YS, Soh-Won J, Cohen DM, Buttrick PM, Cooper DR, Steinberg SF, Mackman N, Pinsky DJ, Stern DM. Hypoxia-associated Induction of Early Growth Response-1 Gene Expression. J Biol. Chem. 1999;274:15030–15040. doi: 10.1074/jbc.274.21.15030. [DOI] [PubMed] [Google Scholar]
- (43).Fradette C, Du Souich P. Effect of hypoxia on cytochrome P450 activity and expression. Curr Drug Metab. 2004;5:257–71. doi: 10.2174/1389200043335577. [DOI] [PubMed] [Google Scholar]
- (44).Harstad EB, Guite CA, Thomae TL, Bradfield CA. Liver deformation in Ahr-null mice: evidence for aberrant hepatic perfusion in early development. Mol Pharmacol. 2006;69:1534–1541. doi: 10.1124/mol.105.020107. [DOI] [PubMed] [Google Scholar]
- (45).Lund AK, Goens MB, Kanagy NL, Walker MK. Cardiac hypertrophy in aryl hydrocarbon receptor null mice is correlated with elevated angiotensin II, endothelin-1, and mean arterial blood pressure. Toxicol. Appl. Pharmacol. 2003;193:177–187. doi: 10.1016/j.taap.2003.08.008. [DOI] [PubMed] [Google Scholar]
- (46).Lund AK, Goens MB, Nunez BA, Walker M. Characterizing the role of endothelin-1 in the progression of cardiac hypertrophy in aryl hydrocarbon receptor (AhR) null mice. Toxicol. Appl. Pharmacol. 2006;212:127–135. doi: 10.1016/j.taap.2005.07.005. [DOI] [PubMed] [Google Scholar]
- (47).Thackaberry EA, Gabaldon DM, Walker MK, Smith SM. Aryl hydrocarbon receptor null mice develop cardiac hypertrophy and increased hypoxia-inducible factor-1alpha in the absence of cardiac hypoxia. Cardiovasc. Toxicol. 2002;2:263–274. doi: 10.1385/ct:2:4:263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.