Abstract
Background & Aims
RAC1 is a GTPase that has an evolutionarily conserved role in coordinating immune defenses, from plants to mammals. Chronic inflammatory bowel diseases (IBD) are associated with dysregulation of immune defenses. We studied the role of RAC1 in IBD using human genetic and functional studies and animal models of colitis.
Methods
We used a candidate gene approach to HapMap-Tag single nucleotide polymorphisms (SNPs) in a discovery cohort; findings were confirmed in 2 additional cohorts. RAC1 mRNA expression was examined from peripheral blood cells of patients. Colitis was induced in mice with conditional disruption of Rac1 in phagocytes by administration of dextran sulphate sodium (DSS).
Results
We observed a genetic association between RAC1 with ulcerative colitis (UC) in a discovery cohort, 2 independent replication cohorts, and in combined analysis for the SNPs rs10951982 (Pcombined UC = 3.3 × 10–8, odds ratio [OR]=1.43 [1.26–1.63]) and rs4720672 (Pcombined UC=4.7 × 10–6, OR=1.36 [1.19–1.58]). Patients with IBD who had the rs10951982 risk allele had increased expression of RAC1, compared to those without this allele. Conditional disruption of Rac1 in macrophage and neutrophils of mice protected them against DSS-induced colitis.
Conclusion
Studies of human tissue samples and knockout mice demonstrated a role for the GTPase RAC1 in the development of UC; increased expression of RAC1 was associated with susceptibility to colitis.
Keywords: innate immunity, Crohn's disease, CD, Rac-1 knockout
Introduction
IBD is a chronic relapsing and remitting disease that affects 1 in 250 individuals of European ancestry. Although the exact etiology of IBD is unknown, it is speculated that IBD occurs in genetically susceptible individuals as a result of dysregulated immune response to gut flora after exposure to an as yet unidentified environmental stimulus1. Genetic association studies have identified innate immunity as a critical component in the development of IBD2–8. However, these studies have identified only 23% of the susceptibility determinants for CD9 and 16% for UC10.
RAC1 belongs to the Ras superfamily of GTP-binding proteins that act as “molecular switches” and influence a number of key cellular functions critical for innate immunity11 including NOD2 and TLR2 regulation12, leukocyte chemotaxis13, barrier defence14, and bacterial killing15. RAC1 function is also disrupted by multiple bacterial cytotoxins16. Furthermore, RAC1 is a target of the commonly used IBD treatment azathioprine, the effects of which are thought to be mediated through T cell apoptosis17 and altered phagocyte chemotaxis to sites of intestinal inflammation18. The importance of RAC1 in innate immunity as seen through its conservation from plants19 to mammals led to our detailed exploration of the RAC1 gene involvement in IBD.
Materials and Methods
SNP Analysis and Genotyping
International HapMap project20 (http://www.hapmap.org) Caucasian (CEU) Phase II data were used to select tag SNPs (MAF > 1%) that span the RAC1 gene and flanking regions through the “Tagger” software program21. Twelve tag SNPs covering RAC1 region (Chromosome 7, 6386318 to 6411760) were captured with r2 > 0.8 (1 SNP was removed during QC because the MAF was < 1%; See Supplemental Methods). Genotype analysis of samples was done using the Illumina® Goldengate Custom Chip genotyping system (Toronto discovery) and Taqman (North America Replication and Scotland Validation) at The Centre for Applied Genomics, Hospital for Sick Children, Toronto and the University of Edinburgh.
Subjects (for Quality Control, and Population Stratification please see Supplemental Methods)
All subjects in this study were of European descent by self-reporting of ethnic heritage. All probands had a confirmed diagnosis of IBD and fulfilled standard diagnostic criteria. Phenotypic characterization was based on the Montreal classification22. Perianal disease included only those CD patients with perianal abscess and/or fistulae. Study subject phenotypic information and DNA samples were obtained with institutional review ethics board approval for IBD genetic studies at the Hospital for Sick Children and Mount Sinai Hospital in Toronto. Replication cohorts had REB approval for genetic and phenotypic studies at the individual institutions. Written informed consent was obtained from all participants.
The discovery cohort included patients recruited from the Hospital for Sick Children and Mt Sinai Hospital in Toronto with local and NIDDK control individuals. The first replication cohort consisted of 1836 Caucasian individuals from North American including 443 CD and 477 UC patients, and 916 controls (NIDDK patients recruited from Chicago and Pittsburgh with North American control individuals obtained from the Centre for Applied Genomics). The second cohort consisted of 2449 individuals exclusively recruited from Scotland including 691 CD and 615 UC patients, and 1143 controls. All patients and controls individuals were non-related Caucasian individuals. Only two RAC1 SNPs were genotyped in the replication cohorts and both had call rates greater than 98% in both cohorts. Part of these cohorts have been used in previous GWAS including the all the NIDDK patients in the North American replication5,23 and 374 individual from Scotland in the Pediatric IBD GWAS24. None of the replication cohort control individuals were genotyped in previous IBD GWAS.
Preliminary Analysis
HAPLOVIEW25 was used to obtain LD patterns, obtaining descriptive statistics and summaries of the SNPs. PLINK version 1.0626 was used to test for Hardy-Weinberg equilibrium (HWE) for each marker based on Pearson's chi-square test. Descriptive statistics of demographic variables were generated using SAS version 9.2 (SAS institute, Cary, NC). The Wilcoxon Rank Sum Test and Chi-square test were used to identify differences in demographic variables between subgroups.
Association Analysis
The analysis was applied in three stages. In stage one, association analyses were applied to detect the associations with the 11 RAC1 SNPs (1 SNP was removed during QC because the MAF was < 1%; See Supplemental Methods) and IBD, CD, and UC vs. healthy controls (HC). Logistic regression models were applied for the additive genetic model, and Pearson chi-square tests were applied for dominant and recessive genetic models. Although we used an additive genetic model for primary analysis27 we also explored dominant and recessive genetic models for sensitive analysis. Throughout the report the p-values are the dominant genetic model p-value. Odds ratios (OR) and 95% confidence intervals (CI) were estimated for the disease compared to the control group. The association, adjusting for selected principal component vectors from the EIGENSTRAT analysis, was tested using conditional logistic regression (SAS v9.2, Cary, NC).
In stage two, the 2 RAC1 SNPs identified from the discovery cohort were genotyped in a replication cohort (North America) and independent validation cohort (Scotland). Independent analysis was applied on the replication cohort and validation cohort. Combined effect estimates from all three IBD cohorts were estimated using a logistic regression model. All P-values are two-sided.
In stage 3, the confirmed RAC1 SNPs were further explored. Haplotype analysis was applied on this region on multiple marker blocks. Imputation of an additional 73 ungenotyped SNPs in the RAC1 region was performed using released Phase II/III CEU HapMap data (BEAGLE). The imputed SNPs were analyzed on the IBD phenotypes using an additive genetic model.
Subgroup Analysis
In addition to comparing IBD, CD, and UC to HC, we applied subgroup analysis to evaluate the genetic effect on the disease risk of the IBD sub-population according to the Montreal Classification system. The sub-population comparisons were applied for each of the genetic markers on ileal only (L1), colonic only (L2), ileo-colonic (L3), ileal any (L1/L3), colon only (L2 plus UC), colon any (L2/L3 plus UC), perianal, and early onset IBD patients (diagnosis age < 19) vs. HC. Different genetic models were used to test single marker associations between each of the subgroups. The analyses were applied for the discovery cohort, North American replication cohort, Scottish replication cohort, and the pooled samples separately.
Animal Experimentation
Rac1-KO mice were generated, bred and genotyped (mixed Sv129 black 6 and Balb/c background) as previously described28. Mice used in these experiments were between 8 and 12 wks old, and all control mice were littermates of the Rac1-KO mice. Rac1-KO mice exhibited no obvious phenotype. Mice were housed in a pathogen free environment and fed standard diet with free access to water. All experiments were approved by the Animal Care Ethics panel at the University of Toronto.
DSS Model
Dextran sodium sulphate (DSS) experiments were performed as previously described29. For the Rac1-KO experiments 12 KO and 18 WT mice were treated with 5% DSS in their drinking water for 7 days. Mice were monitored daily for weight loss, stool consistency, rectal bleeding and general appearance. For further DSS, Cytokine, and MPO methods please see Supplemental Methods.
Results
Genetic Studies
Candidate Gene Approach
We first examined the role of RAC1 in IBD using a candidate gene approach. After strict quality control (QC; 1 SNP was removed during QC because the MAF was < 1%; See Supplemental Methods) measures, 11 RAC1 tag SNPs (Supplemental Figure 1) were successfully genotyped in the discovery cohort consisting of 2049 subjects (656 CD, 544 UC, and 849 controls; Supplementary Table 1A). The rs10951982 RAC1 SNP was significantly associated with IBD after Bonferroni correction threshold for 11 SNPs examined for IBD, CD and UC (α = 1.5 × 10−3; Table 1 shows the association for discovery SNPs based on the minor allele). rs10951982 is not located in a known regulatory element or in strong LD (r2 > 0.25) with any HapMap SNP located in other genes on Chromosome 7.
Table 1.
Genetic Association in Discovery Cohort.
| rs# | Position | MAF Controls | PIBD | OR (95% CI) | PCD | OR (95% CI) | PUC | OR (95% CI) |
|---|---|---|---|---|---|---|---|---|
| rs836488 | 6,386,318 | 0.0366032 | 1.1 × 10−1 | 1.33 (0.93–1.87) | 5.4 × 10−1 | 1.13 (0.75–1.71) | 4.0 × 10−2 | 1.51 (1.02–1.74 |
| rs10499343 | 6,386,519 | 0.122743 | 1.1 × 10−1 | 0.98 (0.81–1.19) | 2.5 × 10−1 | 0.86 (0.67–1.11) | 1.5 × 10−1 | 1.10 (0.92–1.11) |
| rs10951982 | 6,389,081 | 0.217911 | 1.3 × 10−3 | 0.78 (0.67–0.91) | 4.0 × 10−3 | 0.73 (0.59–0.96) | 1.0 × 10−2 | 0.75 (0.63–0.96) |
| rs10238136 | 6,389,871 | 0.034163 | 2.2 × 10−1 | 0.81 (0.57–1.14 | 3.5 × 10−1 | 0.82 (0.54–1.23) | 3.6 × 10−1 | 0.81 (0.52–1.23) |
| rs702484 | 6,398,458 | 0.275012 | 1.1 × 10−1 | 0.89 (0.77–1.02) | 8.1 × 10−1 | 0.83 (0.67–1.02) | 2.7 × 10−1 | 0.88 (0.74–1.02) |
| rs836547 | 6,405,443 | 0.0961445 | 8.7 × 10−1 | 1.01 (0.82–1.25) | 8.7 × 10−2 | 0.96 (0.73–1.25) | 7.8 × 10−1 | 1.04 (0.80–1.25) |
| rs2303364 | 6,408,213 | 0.0471883 | 4.6 × 10−1 | 1.11 (0.82–1.51) | 76 × 10−1 | 0.97 (0.67–1.39) | 4.6 × 10−1 | 1.30 (0.89–1.39) |
| rs4720672 | 6,410,364 | 0.155198 | 1.1 × 10−2 | 0.76 (0.63–0.94) | 2.5 × 10−2 | 0.77 (0.61–0.96) | 4.0 × 10−2 | 0.78 (0.65–0.96) |
| rs836554 | 6,411,760 | 0.229136 | 5.1 × 10−1 | 0.95 (0.82–1.10) | 1.3 × 10−1 | 0.85 (0.69–1.05) | 4.5 × 10−1 | 1.08 (0.88–1.05) |
| rs3813517 | 6,415,057 | 0.0258663 | 9.9 × 10−2 | 1.00 (0.67–1.48) | 8.9 × 10−1 | 0.96 (0.64–1.55) | 7.1 × 10−1 | 1.09 (0.66–1.55) |
| rs836559 | 6,415,288 | 0.435334 | 1.8 × 10−2 | 0.85 (0.75–0.97) | 1.5 × 10−3 | 0.70 (0.56–0.87) | 1.0 × 10−1 | 0.82 (0.79–0.87) |
MAF – minor allelic frequency. P-vale and OR based on Dominant genetic model and MAF.
Bonferroni correction threshold: for 11 SNP examined in 3 IBD (CD, UC and IBD); α = 1.5 × 10−3
Genetic Replication
To confirm this association, we examined rs10951982 and rs4720672 in two independent cohorts. The replication cohort was comprised of 1836 Caucasian subjects including 443 CD and 477 UC patients, and 916 controls recruited from North America and a second validation cohort from Scotland comprised of 2449 Caucasian subjects including 691 CD and 615 UC patients, and 1143 controls (Supplementary Table 1B and 1C). The signal for IBD and UC was replicated in both cohorts with similar ORs and minor allelic frequency (Table 2). Further exploration of genetic models showed that the signal was strongest in the dominant model (reported here and Supplemental Table 3). Combined analysis (1790 CD and 1636 UC patients, and 2908 controls) showed that both SNPs were significantly associated with IBD, CD, and UC (Pcombined IBD = 4.2 × 10−7, OR = 1.29 (1.17–1.44), Pcombined CD = 3.7 × 10−3, OR = 1.19 (1.06–1.34), and Pcombined UC = 3.3 × 10−8, OR = 1.43 (1.26–1.63) for rs10951982; and Pcombined IBD = 7.7 × 10−7, OR = 1.34 (1.17–1.47), Pcombined CD = 3.7 × 10−4, OR = 1.26 (1.11–1.44), and Pcombined UC = 4.7 × 10−6, OR= 1.36 (1.19–1.58) for rs4720672).
Table 2.
Discovery, replication, and pooled analysis showing association between RAC1 SNPs and IBD, CD, and UC.
| SNP | Position | Cohort | Number of Individuals (Risk Allele Frequency) | Number of Patients (Risk Allele Frequency) | P-value | OR (95% CI) |
|---|---|---|---|---|---|---|
| rs10951982 | 6389081 | IBD | ||||
| Toronto | 849 (0.78) | 1200 (0.80) | 1.3 × 10−3 | 1.34 (1.12–1.62) | ||
| North America | 916 (0.76) | 918 (0.79) | 4.0 × 10−3 | 1.31 (1.08–1.62) | ||
| Scotland | 1106 (0.74) | 1288 (0.75) | 2.0 × 10−2 | 1.21 (1.03–1.44) | ||
| Combined | 2871 (0.75) | 3346 (0.79) | 4.2 × 10−7 | 1.29 (1.17–1.44) | ||
| CD | ||||||
| Toronto | 656 (0.80) | 4.0 × 10−3 | 1.36 (1.09−1.69) | |||
| North America | 441 (0.77) | 1.9 × 10−1 | 1.16 (1.08–1.47) | |||
| Scotland | 650 (0.75) | 4.7 × 10−1 | 1.07 (1.11–1.34) | |||
| Combined | 1747 (0.77) | 3.7 × 10−3 | 1.19 (1.06–1.34) | |||
| UC | ||||||
| Toronto | 544 (0.81) | 1.4 × 10−2 | 1.34 (1.09–1.63) | |||
| North America | 477 (0.80) | 8.1 × 10−4 | 1.49 (1.17–1.89) | |||
| Scotland | 578 (0.78) | 4.0 × 10−2 | 1.43 (1.75–0.87) | |||
| Combined | 1599 (0.80) | 3.3 × 10−8 | 1.43 (1.26–1.63) | |||
|
| ||||||
| rs4720672 | 6410364 | IBD | ||||
| Toronto | 849 (0.83) | 1200 (0.86) | 1.1 × 10−2 | 1.28 (1.06–1.56) | ||
| North America | 916 (0.82) | 918 (0.85) | 5.5 × 10−3 | 1.33 (1.08–1.62) | ||
| Scotland | 1106 (0.80) | 1288 (0.83) | 5.0 × 10−3 | 1.28 (1.08–1.53) | ||
| Combined | 2871 (0.82) | 3346 (0.85) | 7.7 × 10−7 | 1.34 (1.17–1.47) | ||
| CD | ||||||
| Toronto | 656 (0.86) | 2.6 × 10−2 | 1.29 (1.03–1.62) | |||
| North America | 441 (0.84) | 5.0 × 10−2 | 1.28 (1.00–1.63) | |||
| Scotland | 650 (0.82) | 9.0 × 10−2 | 1.14 (1.031.44) | |||
| Combined | 1747 (0.84) | 3.7 × 10−4 | 1.26 (1.11–1.44) | |||
| UC | ||||||
| Toronto | 544 (0.86) | 4.9 × 10−2 | 1.26 (1.03–1.62) | |||
| North America | 477 (0.86) | 1.0 × 10−2 | 1.36 (1.07–1.75) | |||
| Scotland | 578 (0.85) | 3.0 × 10−3 | 1.41 (1.12–1.75) | |||
| Combined | 1599 (0.85) | 4.7 × 10−6 | 1.36 (1.19–1.58) | |||
As rs10951982 and rs4720672 are in LD (r2 = 0.64, controls), we applied haplotype analysis30. The haplotype of this two-SNP block is significantly associated with IBD, CD, and UC (PIBD = 9.1 × 10−3, PCD = 5.0 × 10−2, and PUC = 3.1 × 10−2, haplotype omnibus test). Further haplotype analysis on different RAC1 SNP blocks did not show significant associations (data not shown).
Genotype-Phenotype Analysis
As the initial analysis indicated that the genetic signal was strongest in IBD and UC, the role of these SNPs was examined by disease location using the Montreal Classification22 (Table 3). We analyzed “any” colonic IBD (combined UC, colonic only CD (L2), and ileo-colonic CD (L3)) and found significant association with both SNPs in the combined analysis (Pcombined = 7.8 × 10−8, OR= 1.33 (1.20–1.49) for rs10951982; and Pcombined = 1.6 × 10−7, OR= 1.35 (1.20–1.51) for rs4720672). These SNP remained significant after Bonferroni correction (Bonferroni correction threshold: for 11 SNP examined in 8 IBD sub-phenotypes; α = 5.6 × 10−4). Further significant association was found with colonic only IBD (combined UC and colonic only CD (L2)) and early onset IBD (diagnosis < 19 years of age).
Table 3.
Genotype phenotype analysis RAC1 SNPs in the combined analysis.
| Phenotype (Montreal Classification) | # Patients | SNP | P | OR (95% CI) | SNP | P | OR (95% CI) |
|---|---|---|---|---|---|---|---|
| Heal “only” CD (LI) | 476 | rsl0951982 | 1.0 × 10−1 | 1.19 (1.03–1.47) | rs4720672 | 1.8 × 10−1 | 1.16 (1.07–1.44) |
| Heal “any” (L1/L3) | 1260 | 3.6 × 10−3 | 1.23 (1.07–1.40) | 1.9 × 10−3 | 1.26 (1.08–1.47) | ||
| Colonic “only” CD (L2) | 513 | 1.0 × 10−1 | 1.17 (1.03–1.44) | 1.0 × 10−2 | 1.31, (1.06–1.61) | ||
| Ileo-colonic CD (L3) | 784 | 6.1 × 10−3 | 1.36 (1.06–1.49) | 1.5 × −4 | 1.33 (1.12–1.61) | ||
| Colon “only” IBD (UC/L2 CD) | 2112 | 1.5 × 10−7 | 1.36 (1.21–1.53) | 1.3 × 10−6 | 1.35 (1.55–1.53) | ||
| Colon “any” IBD (UC/L2/L3 CD) | 2896 | 7.8 × 10−8 | 1.33 (1.20–1.49) | 1.6 × 10−7 | 1.35 (1.20–1.51) | ||
| Early onset IBD | 1132 | 2.9 × 10−6 | 1.38 (1.20–1.61) | 2.0 × 10−5 | 1.33 (1.19–1.61) | ||
| CD with perianal disease (p) | 446 | 6.0 × 10−3 | 1.35 (1.08–1.66) | 1.6 × 10−3 | 1.44 (1.14–1.85) | ||
P-value based on combined analysis. Early onset IBD (diagnosis > 19 years of age). Bonferroni correction threshold: for 11 SNP examined in 8 IBD sub-phenotypes; α = 5.6 × 10−4
Imputation Analysis
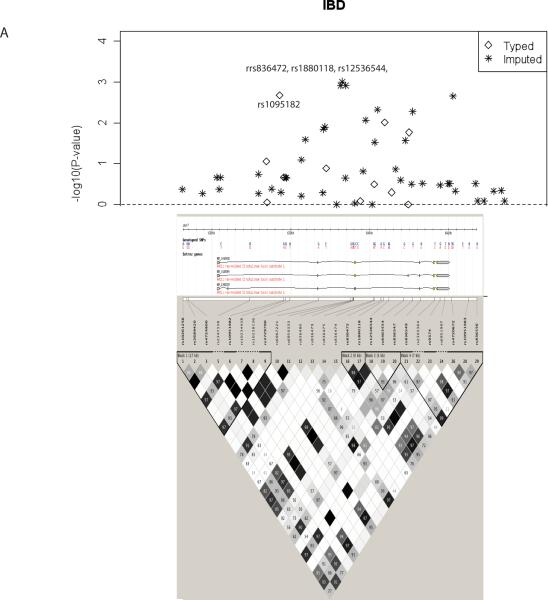
We next imputed 73 SNPs in the RAC1 region not initially genotyped using released Phase II/III CEU HapMap data to identify any additional significant SNPs in our discovery cohort. Imputation analysis revealed multiple HapMap SNPs with nominal significant levels of association (Figure 1A for IBD and Supplemental Figure 2 for UC and CD). Three SNPs located in intron 3 of RAC1 in strong LD (r2 > 0.80) with rs10951982 showed the strongest association with IBD and UC (for IBD: rs836472, P = 1.2 × 10−3, OR = 1.33; rs1880118, P = 9.8 × 10−4, OR = 1.33; and rs12536544, P = 1.2 × 10−3, OR = 1.33). Together these data implicate this region of RAC1 as harboring the causal variant; however, the imputed SNPs are based on in silico analysis and further sequencing in this region is required to determine the causal variant(s).
Figure 1. RAC1 Imputation Analysis.
Regional blots plot of the negative decadic logarithm of the P-values of SNPs in the RAC1 region including both genotyped (diamond) and imputation (*) SNPs, based on the 2049 individuals from the discovery cohort. In IBD the strongest signal comes from 3 SNPs (rs836472, rs1880118, rs12536544) located at 6401429 to 6402426 of Chromosome 7 that are in strong LD (r2 > 0.80) with rs10951982. RAC1 gene is depicted below LD plot with genotyed SNP in bold and r2 SNP map.
RAC1 Expression
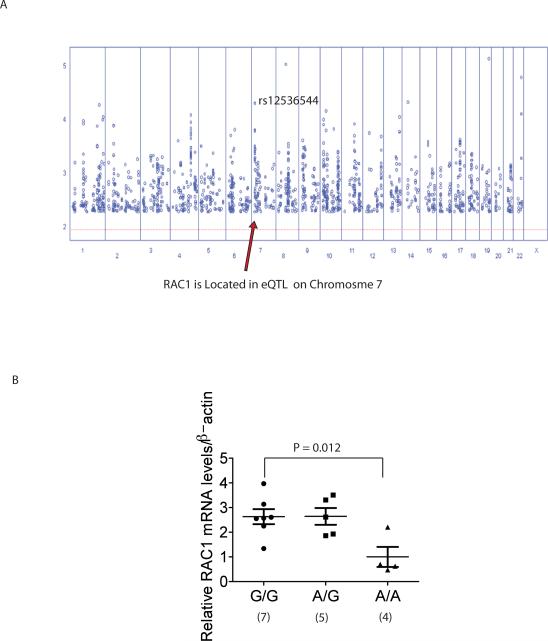
Our study indicates that RAC1 rs10951982 risk allele increases the susceptibility to the development of colonic IBD. A previous study demonstrated that SNPs in the RAC1 gene region are in an expression quantitative trait loci (eQTLs) in lymphoblastoid cell lines (Figure 2A)31. The area of maximal LOD score (3.3 – 4.3) on chromosome 7 occurs in the RAC1 gene region with several SNPs (rs37279790, rs836472, rs1880118, rs12536544, and rs10951983) in strong linkage disequilibrium to rs10951982 (r2 of 0.82 to 1.0). The additive effect for the major allele corresponded to a cis-acting regulatory effect with an increase in standard deviation units for RAC131 (Figure 2A and Supplemental Table 4). Furthermore, mRNA from freshly isolated peripheral blood cells (PBCs) in 16 IBD patients with colonic disease showed increased RAC1 expression based on genotype (Figure 2B). Carriage of the risk `G' allele resulted in increased RAC1 expression (comparison of normalized expression for GG (7 patients) / AG (5 patients) to AA (4 patients) using non-parametric comparison and Wilcoxon exact test P = 0.008; and AA vs GG Wilcoxon exact test P = 0.012). These results indicate that carriage of the risk allele of rs10951982 results in increased RAC1 expression. There were no differences in RAC1 splicing based on genotype in these patients (data not shown). Further expression analysis is required to determine the effect of these SNPs on RAC1 expression, splicing, and mRNA stability in both healthy individuals and IBD patients.
Figure 2. RAC1 Expression Based on Genotype.
(A) Plot of LOD score (y-axis) and SNP position of each chromosome (x-axis). The eQTL maximal LOD score on chromosome 7 is the RAC1 SNP (rs12536544) that is in strong disequilibrium to rs10951982 (r2 of 0.82 to 1.0).
(B) RAC1 expression from mRNA isolated from PBCs from 16 IBD patients with colonic disease based on rs10951982 genotype. `G' allele is the risk allele. Expression was normalized to β-actin expression. Wilcoxon exact test.
Animal Studies: Rac1 Conditional Knockout Mice Studies
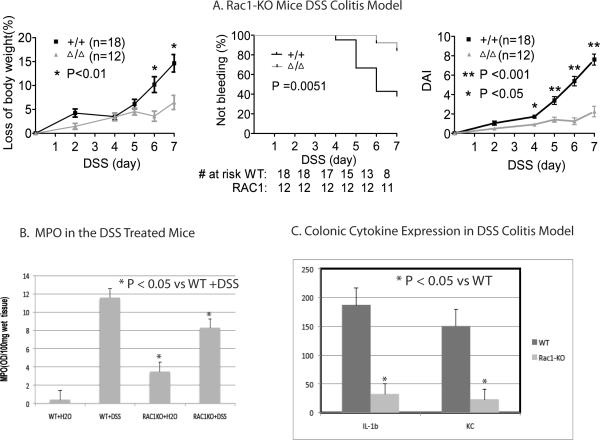
As Rac1 knockouts are embryonic lethal in mice32, we next examined the neutrophil and macrophage conditional Rac1 knockout (Rac1-KO) mice28. These mice had no obvious colonic phenotype with normal bowel histology although they did show increased MPO activity (Figure 3B). As shown in Figure 3A, the Rac1-KO mice were protected from developing DSS-induced colitis. Rac1-KO mice show decreased weight loss after 6 days (P < 0.01; Wilcoxon rank sum test). WT mice began bleeding after day 4 with 10/18 mice bleeding on day 7. In comparison, only 1/12 Rac1-KO was bleeding on day 7 (P = 0.0051; log rank test = 7.3, hazard ratio 4.6, 95% CI 1.5–14). WT mice had significantly worse DAI scores starting on day 4 (P < 0.05 on day 4, and P < 0.001 on days 5, 6, and 7 Wilcoxon rank sum test). Comparison of the distal colon on day seven of DSS treatment showed reduced histological scores with fewer ulcers in the distal colonic in the Rac1-KO group (P=0.008 analyzed as a dichotomous variable with a cutoff of 0 – 2 or > 3 ulcers using Fisher's exact test; Supplemental Figure 3). Neutrophil migration into the colon was reduced in the Rac1-KO mice compared to WT as assessed by MPO activity (Figure 3C. P < 0.01; Wilcoxon rank sum test). Cytokine profiles for colonic lystates from Rac1-KO mice treated with DSS showed significantly reduced level of the proinflammatory cytokine IL-1β and the neutrophil chemokine KC compared to WT mice (Figure 3C; P < 0.05; Mann Whitney test Two Tailed). Furthermore, there was impaired up-regulation of proinflammatory cytokines including IL-1β, IL-12 and TNFα in response to DSS induced colitis as compared to WT (P < 0.05, Mann Whitney test Two Tailed; Supplemental Figure 4). Splenocytes isolated from DSS-treated Rac1-KO mice and stimulated with lipopolysaccharide (LPS) also showed a similar decrease in production of inflammatory cytokines (Supplemental Figure 5) further demonstrating the reduced inflammation in these KO mice.
Figure 3. Impact of impairing Rac1 function in DSS induced colitis in mice.
(A) Rac1-KO. Left Panel graphs changes in weight with DSS treatment. Middle Panel shows changes in rectal bleeding. # at risk indicates number of mice not bleeding at each time point. Right Panel shows Disease Activity Index. (Error bars represent SEM).
(B) Colonic MPO Activity. Colonic neutrophil infiltration was measured by MPO activity. MPO activity was assessed in the distal colon of WT mice (WT + water), Rac1-KO mice (Rac1-KO + water), and Rac1-KO mice treated with 5% DSS (Rac1-KO + DSS). (Wilcoxon rank sum test; Error bars represent SEM).
(C) Colonic Cytokine Assay. Cytokine assays performed using MESO scale discovery mouse TH1/TH2 9-plex assay kit. (Mann Whitney test Two Tailed; Error bars represent SEM).
Discussion
Recent IBD GWAS have identified innate immunity as a critical component in the pathogenesis of IBD2–8. The identification of RAC1's association with IBD further strengthens this link with innate immunity. The number of IBD susceptibility genes has increased dramatically due to meta-analysis of recent GWAS and the signal for RAC1 has not been reported to date. However, in the 1st International IBD Genetics Consortium (IIBDGC) meta-analysis for UC, SNPs in strong LD (r2 > 0.80; and identified in our imputation analysis Figure 1A) to our reported SNP (rs10951982) showed strong association to UC (rs12536544, Pmeta = 8.80 × 10−5; rs836472, Pmeta = 2.60 × 10−5; rs1880118, Pmeta = 6.3 × 10−4; and rs10951983, UC Pmeta = 1.2 × 10−4)10. Although these SNPs have not yet been reported due to the stringent criteria for replication in that UC meta-analysis10, these meta-analysis results strongly implicate RAC1 as a novel UC susceptibility gene. Furthermore, the RAC1 SNP rs12536544 genotyped in the meta-analysis was at the maximum eQTL for RAC1 as shown in Figure 2A (P = 10−7) indicating that the risk allele for these SNPs increased RAC1 expression. Interestingly, this novel genetic association is also found with the colonic IBD (UC and colonic CD) phenotype, a phenotype that has yet to be explored in IBD GWAS.
We observed that reduction or loss of Rac1 expression through conditional neutrophil/macrophage KO resulted in protection from the development of DSS induced colitis. We chose the DSS model as it has recently been shown to be a reasonable model of UC33. This protection from DSS induced colitis was accompanied by a reduction in MPO, an indirect marker of neutrophil colonic infiltration, and a reduction in the colonic proinflammatory cytokine IL-1β and the neutrophil chemokine KC. We have previously shown that loss of Rac1 in these mice results in profound defects in inflammatory recruitment and migration to chemotactic stimuli28. The down-regulation of the neutrophil chemokine KC would also result in decreased neutrophil migration and reduced inflammation into the intestine34. Therefore, it is possible that the protection from DSS induced colitis in these mice is due to delayed neutrophil infiltration as observed when neutrophil recruitment is impaired through CXCR2 blocking antibodies35. This is also consistent with other animal models of inflammation where loss of Rac1 function leads to decreased neutrophil recruitment leading to attenuation of inflammation28, 36, 37.
Although the causal variant has not been established in this study, expression datasets22 and our patient's expression studies demonstrates that the RAC1 risk allele identified here increases RAC1 expression. These human expression experiments are complemented by our animal model experiments that show that decreased Rac1 activity or loss Rac1 expression leads to protection from the development of DSS induced colitis. The cytokine profile observed from colonic lysates from DSS treated animals with conditional macrophage/neutrophil Rac1-KO demonstrated defective proinflammatory production. Overall, these results suggest that carriage of the RAC1 risk allele results in higher expression of RAC1 leading to increased neutrophil recruitment into the colon and subsequent increased proinflammatory cytokine expression in colonic. Along with RAC1 dependent T-cell apoptosis17, one of the proposed mechanisms for the medication commonly used to maintain IBD patients in remission, azathioprine, is through inhibition of neutrophil trafficking into the colon18. Therefore it is intriguing to speculate that patients carrying the RAC1 risk allele have higher levels of RAC1 expression with increased neutrophil migration into the colon and that these patients may benefit from treatment with azathioprine.
RAC1 is expressed in most cell types and has divergent and critical role in numerous cellular pathways. The functional studies presented here focus on its role in the neutrophil-macrophage cell lineage. It is also plausible that the association of RAC1 with IBD is mediated through an alternate pathway not examined here. The divergent functions of RAC1 in innate immunity, barrier defense, and T cell apoptosis makes further study of this protein important to our understanding of IBD.
Supplementary Material
Acknowledgements
We wish to acknowledge the work of Karoline Fielder at the Hospital for Sick Children, Joanne Stempak at Mt Sinai and Dr Elaine Nimmo, Dr Richard Russell and Hazel Drummond in Edinburgh. AMM is supported by a transition award from the Crohn's and Colitis Foundation of Canada (CCFC)/ Canadian Association of Gastroenterology (CAG)/ Canadian Institute for Health Research (CIHR), a Canadian Child Health Clinician Scientist Program (Strategic Training Initiatives in Health Research Program – CIHR) award and an Early Researcher Award from the Ontario Ministry of Research and Innovation. JB is supported by a CCFC/CAG summer studentship. TW is supported by CCFC and AstraZenca Partnered fellowships from the CAG/CIHR. PMS is a recipient of Canada Research Chair in Gastrointestinal Disease. David C Wilson is the holder of a Medical Research Council Patient Cohorts Research Initiative award (G0800675). Financial assistance was also provided by the Wellcome Trust Programme Grant (072789/Z/03/Z), Action Medical Research, the Chief Scientist Office of the Scottish Government Health Department, andthe GI/Nutrition Research Fund, Child Life and Health, University of Edinburgh. MSS is supported by the Gale and Graham Wright Research Chair in Digestive Diseases at Mount Sinai Hospital and funding from CCFC and NIDDK (DK-06-504). John H. Brumell, PhD, holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. Funding was provided by a CIHR operating grant (MOP97756) to AMM and JHB.
AMM is supported by a transition award from the Crohn's and Colitis Foundation of Canada (CCFC)/ Canadian Association of Gastroenterology (CAG)/ Canadian Institute for Health Research (CIHR), a Canadian Child Health Clinician Scientist Program (Strategic Training Initiatives in Health Research Program – CIHR) award and an Early Researcher Award from the Ontario Ministry of Research and Innovation. JB is supported by a CCFC/CAG summer studentship. PMS is a recipient of Canada Research Chair in Gastrointestinal Disease. David C Wilson is the holder of a Medical Research Council Patient Cohorts Research Initiative award (G0800675). Financial assistance was also provided by the Wellcome Trust Programme Grant (072789/Z/03/Z), Action Medical Research, the Chief Scientist Office of the Scottish Government Health Department, and the GI/Nutrition Research Fund, Child Life and Health, University of Edinburgh. MSS is supported by the Gale and Graham Wright Research Chair in Digestive Diseases at Mount Sinai Hospital and funding from CCFC and NIDDK (DK-06-504). John H. Brumell, PhD, holds an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund. Funding was provided by a CIHR operating grant (MOP97756) to AMM and JHB.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflict of interest exists.
References
- 1.Abraham C, Cho JH. Inflammatory Bowel Disease. N Engl J Med. 2009;361:2066–2078. doi: 10.1056/NEJMra0804647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith AM, Rahman FZ, Hayee B, Graham SJ, Marks DJ, Sewell GW, Palmer CD, Wilde J, Foxwell BM, Gloger IS, Sweeting T, Marsh M, Walker AP, Bloom SL, Segal AW. Disordered macrophage cytokine secretion underlies impaired acute inflammation and bacterial clearance in Crohn's disease. J Exp Med. 2009;206:1883–97. doi: 10.1084/jem.20091233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, Britton H, Moran T, Karaliuskas R, Duerr RH, Achkar JP, Brant SR, Bayless TM, Kirschner BS, Hanauer SB, Nunez G, Cho JH. A frameshift mutation in NOD2 associated with susceptibility to Crohn's disease. Nature. 2001;411:603–6. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 4.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, Huse K, Albrecht M, Mayr G, De La Vega FM, Briggs J, Gunther S, Prescott NJ, Onnie CM, Hasler R, Sipos B, Folsch UR, Lengauer T, Platzer M, Mathew CG, Krawczak M, Schreiber S. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 5.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, Shugart YY, Griffiths AM, Targan SR, Ippoliti AF, Bernard EJ, Mei L, Nicolae DL, Regueiro M, Schumm LP, Steinhart AH, Rotter JI, Duerr RH, Cho JH, Daly MJ, Brant SR. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Villani AC, Lemire M, Fortin G, Louis E, Silverberg MS, Collette C, Baba N, Libioulle C, Belaiche J, Bitton A, Gaudet D, Cohen A, Langelier D, Fortin PR, Wither JE, Sarfati M, Rutgeerts P, Rioux JD, Vermeire S, Hudson TJ, Franchimont D. Common variants in the NLRP3 region contribute to Crohn's disease susceptibility. Nat Genet. 2009;41:71–6. doi: 10.1038/ng285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M, Binder V, Finkel Y, Cortot A, Modigliani R, Laurent-Puig P, Gower-Rousseau C, Macry J, Colombel JF, Sahbatou M, Thomas G. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 8.Parkes M, Barrett JC, Prescott NJ, Tremelling M, Anderson CA, Fisher SA, Roberts RG, Nimmo ER, Cummings FR, Soars D, Drummond H, Lees CW, Khawaja SA, Bagnall R, Burke DA, Todhunter CE, Ahmad T, Onnie CM, McArdle W, Strachan D, Bethel G, Bryan C, Lewis CM, Deloukas P, Forbes A, Sanderson J, Jewell DP, Satsangi J, Mansfield JC, Cardon L, Mathew CG. Sequence variants in the autophagy gene IRGM and multiple other replicating loci contribute to Crohn's disease susceptibility. Nat Genet. 2007;39:830–2. doi: 10.1038/ng2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franke A, McGovern DP, Barrett JC, Wang K, Radford-Smith GL, Ahmad T, Lees CW, Balschun T, Lee J, Roberts R, Anderson CA, Bis JC, Bumpstead S, Ellinghaus D, Festen EM, Georges M, Green T, Haritunians T, Jostins L, Latiano A, Mathew CG, Montgomery GW, Prescott NJ, Raychaudhuri S, Rotter JI, Schumm P, Sharma Y, Simms LA, Taylor KD, Whiteman D, Wijmenga C, Baldassano RN, Barclay M, Bayless TM, Brand S, Buning C, Cohen A, Colombel JF, Cottone M, Stronati L, Denson T, De Vos M, D'Inca R, Dubinsky M, Edwards C, Florin T, Franchimont D, Gearry R, Glas J, Van Gossum A, Guthery SL, Halfvarson J, Verspaget HW, Hugot JP, Karban A, Laukens D, Lawrance I, Lemann M, Levine A, Libioulle C, Louis E, Mowat C, Newman W, Panes J, Phillips A, Proctor DD, Regueiro M, Russell R, Rutgeerts P, Sanderson J, Sans M, Seibold F, Steinhart AH, Stokkers PC, Torkvist L, Kullak-Ublick G, Wilson D, Walters T, Targan SR, Brant SR, Rioux JD, D'Amato M, Weersma RK, Kugathasan S, Griffiths AM, Mansfield JC, Vermeire S, Duerr RH, Silverberg MS, Satsangi J, Schreiber S, Cho JH, Annese V, Hakonarson H, Daly MJ, Parkes M. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 42:1118–25. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anderson CA, Boucher G, Lees CW, Franke A, D'Amato M, Taylor KD, Lee JC, Goyette P, Imielinski M, Latiano A, Lagace C, Scott R, Amininejad L, Bumpstead S, Baidoo L, Baldassano RN, Barclay M, Bayless TM, Brand S, Buning C, Colombel JF, Denson LA, De Vos M, Dubinsky M, Edwards C, Ellinghaus D, Fehrmann RS, Floyd JA, Florin T, Franchimont D, Franke L, Georges M, Glas J, Glazer NL, Guthery SL, Haritunians T, Hayward NK, Hugot JP, Jobin G, Laukens D, Lawrance I, Lemann M, Levine A, Libioulle C, Louis E, McGovern DP, Milla M, Montgomery GW, Morley KI, Mowat C, Ng A, Newman W, Ophoff RA, Papi L, Palmieri O, Peyrin-Biroulet L, Panes J, Phillips A, Prescott NJ, Proctor DD, Roberts R, Russell R, Rutgeerts P, Sanderson J, Sans M, Schumm P, Seibold F, Sharma Y, Simms LA, Seielstad M, Steinhart AH, Targan SR, van den Berg LH, Vatn M, Verspaget H, Walters T, Wijmenga C, Wilson DC, Westra HJ, Xavier RJ, Zhao ZZ, Ponsioen CY, Andersen V, Torkvist L, Gazouli M, Anagnou NP, Karlsen TH, Kupcinskas L, Sventoraityte J, Mansfield JC, Kugathasan S, Silverberg MS, Halfvarson J, Rotter JI, Mathew CG, Griffiths AM, Gearry R, Ahmad T, Brant SR, Chamaillard M, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 43:246–52. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bokoch GM. Regulation of innate immunity by Rho GTPases. Trends Cell Biol. 2005;15:163–71. doi: 10.1016/j.tcb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 12.Eitel J, Krull M, Hocke AC, N'Guessan PD, Zahlten J, Schmeck B, Slevogt H, Hippenstiel S, Suttorp N, Opitz B. Beta-PIX and Rac1 GTPase mediate trafficking and negative regulation of NOD2. J Immunol. 2008;181:2664–71. doi: 10.4049/jimmunol.181.4.2664. [DOI] [PubMed] [Google Scholar]
- 13.Zhang H, Sun C, Glogauer M, Bokoch GM. Human neutrophils coordinate chemotaxis by differential activation of Rac1 and Rac2. J Immunol. 2009;183:2718–28. doi: 10.4049/jimmunol.0900849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noren NK, Niessen CM, Gumbiner BM, Burridge K. Cadherin engagement regulates Rho family GTPases. J Biol Chem. 2001;276:33305–8. doi: 10.1074/jbc.C100306200. [DOI] [PubMed] [Google Scholar]
- 15.Koh AL, Sun CX, Zhu F, Glogauer M. The role of Rac1 and Rac2 in bacterial killing. Cell Immunol. 2005;235:92–7. doi: 10.1016/j.cellimm.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 16.Aktories K, Barbieri JT. Bacterial cytotoxins: targeting eukaryotic switches. Nat Rev Microbiol. 2005;3:397–410. doi: 10.1038/nrmicro1150. [DOI] [PubMed] [Google Scholar]
- 17.Tiede I, Fritz G, Strand S, Poppe D, Dvorsky R, Strand D, Lehr HA, Wirtz S, Becker C, Atreya R, Mudter J, Hildner K, Bartsch B, Holtmann M, Blumberg R, Walczak H, Iven H, Galle PR, Ahmadian MR, Neurath MF. CD28-dependent Rac1 activation is the molecular target of azathioprine in primary human CD4+ T lymphocytes. J Clin Invest. 2003;111:1133–45. doi: 10.1172/JCI16432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murthy SN, Anania T, Clearfield HR. Azathioprine reduces extravasation and neutrophil trafficking in immune complex-mediated inflammation in the rat colon. Agents Actions. 1991;34:244–6. doi: 10.1007/BF01993293. [DOI] [PubMed] [Google Scholar]
- 19.Nakashima A, Chen L, Thao NP, Fujiwara M, Wong HL, Kuwano M, Umemura K, Shirasu K, Kawasaki T, Shimamoto K. RACK1 functions in rice innate immunity by interacting with the Rac1 immune complex. Plant Cell. 2008;20:2265–79. doi: 10.1105/tpc.107.054395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The International HapMap Project Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 21.de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–23. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- 22.Silverberg MS, Satsangi J, Ahmad T, Arnott ID, Bernstein CN, Brant SR, Caprilli R, Colombel JF, Gasche C, Geboes K, Jewell DP, Karban A, Loftus EV, Jr, Pena AS, Riddell RH, Sachar DB, Schreiber S, Steinhart AH, Targan SR, Vermeire S, Warren BF. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: Report of a Working Party of the 2005 Montreal World Congress of Gastroenterology. Can J Gastroenterol. 2005;19(Suppl A):5–36. doi: 10.1155/2005/269076. [DOI] [PubMed] [Google Scholar]
- 23.Silverberg MS, Cho JH, Rioux JD, McGovern DP, Wu J, Annese V, Achkar JP, Goyette P, Scott R, Xu W, Barmada MM, Klei L, Daly MJ, Abraham C, Bayless TM, Bossa F, Griffiths AM, Ippoliti AF, Lahaie RG, Latiano A, Pare P, Proctor DD, Regueiro MD, Steinhart AH, Targan SR, Schumm LP, Kistner EO, Lee AT, Gregersen PK, Rotter JI, Brant SR, Taylor KD, Roeder K, Duerr RH. Ulcerative colitis-risk loci on chromosomes 1p36 and 12q15 found by genome-wide association study. Nat Genet. 2009;41:216–20. doi: 10.1038/ng.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Imielinski M, Baldassano RN, Griffiths A, Russell RK, Annese V, Dubinsky M, Kugathasan S, Bradfield JP, Walters TD, Sleiman P, Kim CE, Muise A, Wang K, Glessner JT, Saeed S, Zhang H, Frackelton EC, Hou C, Flory JH, Otieno G, Chiavacci RM, Grundmeier R, Castro M, Latiano A, Dallapiccola B, Stempak J, Abrams DJ, Taylor K, McGovern D, Silber G, Wrobel I, Quiros A, Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, Brant SR, Silverberg MS, Taylor KD, Barmuda MM, Bitton A, Dassopoulos T, Datta LW, Green T, Griffiths AM, Kistner EO, Murtha MT, Regueiro MD, Rotter JI, Schumm LP, Steinhart AH, Targan SR, Xavier RJ, Libioulle C, Sandor C, Lathrop M, Belaiche J, Dewit O, Gut I, Heath S, Laukens D, Mni M, Rutgeerts P, Van Gossum A, Zelenika D, Franchimont D, Hugot JP, de Vos M, Vermeire S, Louis E, Cardon LR, Anderson CA, Drummond H, Nimmo E, Ahmad T, Prescott NJ, Onnie CM, Fisher SA, Marchini J, Ghori J, Bumpstead S, Gwillam R, Tremelling M, Delukas P, Mansfield J, Jewell D, Satsangi J, Mathew CG, Parkes M, Georges M, Daly MJ, Heyman MB, Ferry GD, Kirschner B, Lee J, Essers J, Grand R, Stephens M, et al. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat Genet. 2009;41:1335–40. doi: 10.1038/ng.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 26.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freidlin B, Zheng G, Li Z, Gastwirth JL. Trend tests for case-control studies of genetic markers: power, sample size and robustness. Hum Hered. 2002;53:146–52. doi: 10.1159/000064976. [DOI] [PubMed] [Google Scholar]
- 28.Glogauer M, Marchal CC, Zhu F, Worku A, Clausen BE, Foerster I, Marks P, Downey GP, Dinauer M, Kwiatkowski DJ. Rac1 deletion in mouse neutrophils has selective effects on neutrophil functions. J Immunol. 2003;170:5652–7. doi: 10.4049/jimmunol.170.11.5652. [DOI] [PubMed] [Google Scholar]
- 29.Muise AM, Walters T, Wine E, Griffiths AM, Turner D, Duerr RH, Regueiro MD, Ngan BY, Xu W, Sherman PM, Silverberg MS, Rotin D. Protein-tyrosine phosphatase sigma is associated with ulcerative colitis. Curr Biol. 2007;17:1212–8. doi: 10.1016/j.cub.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 30.Purcell S, Daly MJ, Sham PC. WHAP: haplotype-based association analysis. Bioinformatics. 2007;23:255–6. doi: 10.1093/bioinformatics/btl580. [DOI] [PubMed] [Google Scholar]
- 31.Dixon AL, Liang L, Moffatt MF, Chen W, Heath S, Wong KC, Taylor J, Burnett E, Gut I, Farrall M, Lathrop GM, Abecasis GR, Cookson WO. A genome-wide association study of global gene expression. Nat Genet. 2007;39:1202–7. doi: 10.1038/ng2109. [DOI] [PubMed] [Google Scholar]
- 32.Sugihara K, Nakatsuji N, Nakamura K, Nakao K, Hashimoto R, Otani H, Sakagami H, Kondo H, Nozawa S, Aiba A, Katsuki M. Rac1 is required for the formation of three germ layers during gastrulation. Oncogene. 1998;17:3427–33. doi: 10.1038/sj.onc.1202595. [DOI] [PubMed] [Google Scholar]
- 33.Yan Y, Kolachala V, Dalmasso G, Nguyen H, Laroui H, Sitaraman SV, Merlin D. Temporal and spatial analysis of clinical and molecular parameters in dextran sodium sulfate induced colitis. PLoS One. 2009;4:e6073. doi: 10.1371/journal.pone.0006073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song F, Ito K, Denning TL, Kuninger D, Papaconstantinou J, Gourley W, Klimpel G, Balish E, Hokanson J, Ernst PB. Expression of the neutrophil chemokine KC in the colon of mice with enterocolitis and by intestinal epithelial cell lines: effects of flora and proinflammatory cytokines. J Immunol. 1999;162:2275–80. [PubMed] [Google Scholar]
- 35.Farooq SM, Stillie R, Svensson M, Svanborg C, Strieter RM, Stadnyk AW. Therapeutic effect of blocking CXCR2 on neutrophil recruitment and dextran sodium sulfate-induced colitis. J Pharmacol Exp Ther. 2009;329:123–9. doi: 10.1124/jpet.108.145862. [DOI] [PubMed] [Google Scholar]
- 36.Filippi MD, Szczur K, Harris CE, Berclaz PY. Rho GTPase Rac1 is critical for neutrophil migration into the lung. Blood. 2007;109:1257–64. doi: 10.1182/blood-2006-04-017731. [DOI] [PubMed] [Google Scholar]
- 37.Binker MG, Binker-Cosen AA, Gaisano HY, Cosen-Binker LI. Inhibition of Rac1 decreases the severity of pancreatitis and pancreatitis-associated lung injury in mice. Exp Physiol. 2008;93:1091–103. doi: 10.1113/expphysiol.2008.043141. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.