Abstract
Circularization of cellular mRNAs is a key event prior to translation initiation. We report that efficient translation of Saguaro cactus virus (SCV) requires a 3' translational enhancer (PTE) located partially in coding sequences. Unlike a similar PTE reported in the 3' UTR of Pea enation mosaic virus that does not engage in an RNA:RNA interaction (Wang Z. et al., J. Biol. Chem. 284, 14189–14202, 2009), the SCV PTE participates in long distance RNA:RNA interactions with hairpins located in the p26 ORF and in the 5' UTR of one subgenomic RNA. At least two additional RNA:RNA interactions are also present, one of which involves the p26 initiation codon. Similar PTE can be found in six additional carmoviruses that can putatively form long-distance interactions with 5' hairpins located in comparable positions.
Keywords: Cap-independent translation, long distance RNA:RNA interactions, Carmovirus, kissing loop interactions, 3' translational enhancers
Introduction
Translational control of gene expression is a key factor in controlling the proteome, ensuring the correct amassing of proteins in respon se to different stimuli with aberrancies leading to inappropriate cell proliferation (Ganoza et al., 2002). Since their discovery, plus-strand RNA viruses have been instrumental in elucidating mechanisms of translation. Their rapid life cycles, canonical and non -canonical termini and polycistronic genomes require highly sophisticated strategies for gene expression (Bushell and Sarnow, 2002; Thiebeauld et al., 2007). Many animal RNA viruses attract the translational machinery in a cap -independent process while suppressing host cap -dependent translation, resulting in severe crippling and death of host cells. Unlike most animal viruses, plant viruses require living cells to transit between cells, thus competition with the host for translation must involve processes that favor virus translation without adversely affecting host translation.
Most research on translation of cellular and viral RNAs concentrates on 5' UTR, where assembled pre-initiation complexes composed of 40S ribosomal subunit, met-tRNA and translation factors including multisubunit eIF4F recognize the 5' m 7GpppG-cap, and then scan in the 3' direction for the first initiation codon in a good context (Kozak, 1980, 1999; Marintchev and Wagner, 2004; Pestova et al., 2001; Preiss and Hentze, 2003). Animal plus-strand RNA viruses lacking 5' caps contain internal ribosome entry sites (IRESs) up stream of initiation codons. These large, structurally complex IRESs (300 to 1500 nt) use a variety of mechanisms to attract 40S subunits with a reduced need for translation factors and, in some cases, enhancement by trans-acting factors (Hellen and Sarnow, 2001; Isken et al., 2003; Martinez-Salas et al., 2008; Merrick, 2004).
Unlike our detailed knowledge of viral 5' UTR in translation, studies on 3' UTR have mainly concentrated on the function of the poly(A) tail, which promotes mRNA circularization and presumptive recycling of terminating ribosomes through interaction with poly(A) binding protein, which also binds to eIF4F (Gallie, 1991). The limited size of plant virus 5' UTRs (10 to 200 nt), however, caused investigators to search 3' UTRs for elements that enhance cap -independent translation (Dreher, 1999; Karetnikov et al., 2006; Karetnikov and Lehto, 2007; Miller and White, 2006). 3' UTR translational enhancers (3'TE) vary in sequence and secondary structure, and can be present in viruses containing or lacking a poly(A) tail (Kneller et al., 2006; Miller et al., 2007a; Miller et al., 2007b; Wang et al., 2010). Most 3'TE either encompass or are proximal to a hairpin with a loop sequence that forms a confirmed or postulated RNA:RNA bridge (minimal of 4 bp) with single-stranded complementary sequences near the 5' end (Miller and White, 2006). One plant virus element with an I-shaped structure (ISS) binds to binds to eIF4E and more tightly to eIF4F and participates in an RNA:RNA interaction with the 5' end allowing 40S subunits to enter and scan to the initiation codon (Nicholson et al., 2010). While other 3'TE are capable of binding to translation initiation factors, their mode of action remains elusive. 3'TE have also been identified in animal viruses such as flaviviruses and picornaviruses (Bradrick et al., 2006; Chiu et al., 2005; de Quinto et al., 2002; Dobrikova et al., 2003; Isken et al., 2004; Song et al., 2006; Weinlich et al., 2009).
We have recently discovered a novel translational enhancer in an internal location in the 3' UTR of the carmovirus, Turnip crinkle virus (TCV). This element, which is composed of three hairpins and two pseudoknots, folds into a tRNA -shaped 3D structure (TSS) that binds to the P-site of 60S ribosomal subunits and is part of a strong TE that also requires an adjacent hairpin and upstream poorly structured region for function (McCormack et al., 2008; Stupina et al., 2008). Of the 16 known carmoviruses, only the virus most closely related to TCV (Cardamine chlorotic fleck virus [CCFV]) has the combination of hairpins and pseudoknots necessary to adopt a TCV-like TSS (McCormack et al., 2008; Yuan et al., 2010).
For this report, putative translation elements in the 3' region of a different carmovirus were defined. We determined that Saguaro cactus virus (SCV) contains at least two important translation elements in its 3' UTR and adjacent upstream CP ORF: a Y-shaped structure also found in six additional carmoviruses and in the umbravirus Pea enation mosaic virus (PEMV; Wang et al., 2009) that is denoted the “PTE”, and a second, critical element near the 3' terminus. Unlike the PEMV PTE, the SCV PTE engages in long distance RNA:RNA interactions with 5'-proximal sequences in the genomic (g)RNA and smaller of the two subgenomic RNAs (sgRNA2) that are critical for efficient translation as assayed using reporter constructs in vivo.
Results
SCV contains a 3' PTE that spans the CP termination codon
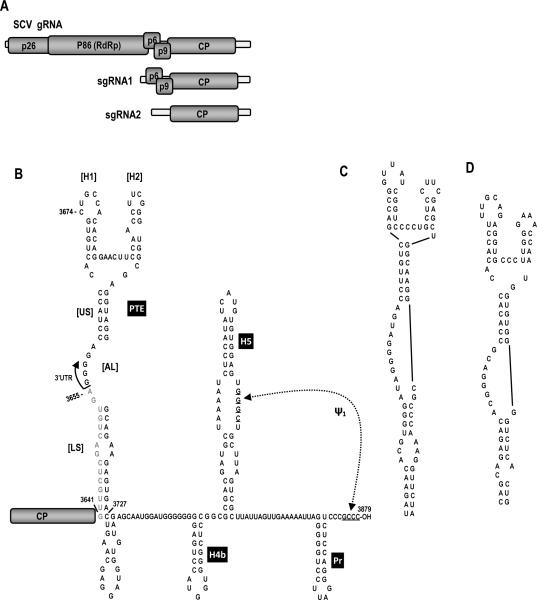
The genome organization and putative structure of the 3' UTR of SCV are shown in Fig. 1. SCV, as with most carmoviruses, encodes five proteins that are translated from the gRNA and two sgRNAs (Fig. 1A). The RNA structure in the SCV 3' region is based on computational predictions using mFold (Zuker, 2003) as well as phylogenetic comparisons with other members of the carmovirus genus. The 3' terminal hairpin, designated “Pr” according to TCV terminology, is similar to 3' terminal hairpins found in 15 of 16 carmoviruses (Yuan et al., 2010) and is the core promoter for transcription of complementary strands for the TCV satellite RNA, satC (Song and Simon, 1995). The 3' penultimate hairpin, H5, also found in 15 of 16 carmoviruses (Yuan et al., 2010), contains a large internal symmetrical loop with sequence identical to that of TCV. All carmoviruses that contain H5 are capable of forming a pseudoknot between the 3' side of their H5 large symmetrical loops and 3' terminal residues (Zhang et al., 2006b). Just upstream of H5 is hairpin H4b, and hairpins in similar locations are present in 14 of 16 carmoviruses (Yuan et al., 2010). Unlike TCV, SCV does not likely contain sequence capable of forming the stable pseudoknot that connects the apical loop of TCV H4b to sequence adjacent to the 3' base of H5. Additionally, SCV does not contain a hairp in just upstream of H4b known as H4a (McCormack et al., 2008). In TCV, H4a, H4b, the missing pseudoknot and an additional pseudoknot that links H4a to adjacent, upstream sequence, compose the TSS translational enhancer.
Fig. 1.
The SCV system. (A) Genome organization of SCV. SCV gRNA contains five ORFs. The viral replicase p86 is the ribosomal read -through product of p26. sgRNA1 is a bi-cistronic mRNA for expression of p6 and p9, which are movement proteins. sgRNA2 codes for the capsid protein (CP). (B) Putative secondary structure in the 3' UTR of SCV. The PTE is partially within the CP ORF (gray sequence). Pr, H5 and pseudoknot ψ1 are conserved in carmoviruses. Most carmoviruses also have H4b. H5 and H4b in TCV are part of the TCV TSS. US, upper stem; AL, asymmetric loop; LS, lower stem. (C) The similar PTE of PEMV, which is known to bind to translation initiation factor eIF4E (Wang et al., 2009). (D) The PTE of PMV, previously determined to be a 3'TE (Batten et al., 2006).
A phylogenetic search for conserved secondary structures in non-TSS carmoviruses led to the discovery that seven contain a topologically related Y-shaped structure (PTE) that is similar to a translational enhancer described for PEMV (Wang et al., 2009) and Panicum mosaic panicovirus (Batten et al., 2006; Fig. 1C). The SCV, PEMV and PMV PTE consist of two hairpins (H1 and H2), a supporting stem of 6 to 7 bp (US), a large asymmetric loop (AL) in the central region of the stem at the base of the hairpins that contains four consecutive guanylates flanked by adenylates, and a lower stem (LS) with a small symmetrical loop (Fig. 1B–D). The PEMV and PMV PTE reside entirely within their 3' UTRs while the SCV PTE spans the termination codon of the coat protein ORF.
Efficient translation of SCV reporter constructs requires UTRs and coding sequences from the 5' and 3' UTRs
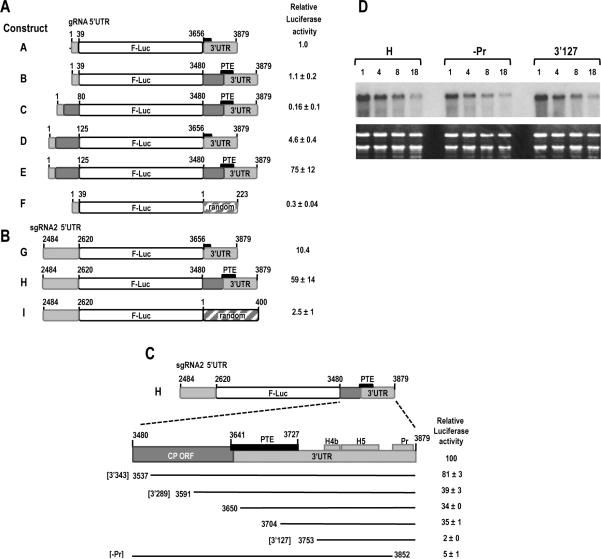
To determine if the SCV PTE is a translational enhancer and/ or to identify other possible translation elements in SCV, we generated a single luciferase (FLuc) reporter construct containing either viral or random 5' and 3' sequences and assayed for luciferase activity in Arabidopsis thaliana protoplasts co-transfected with a control luciferase (RLuc) construct. As shown in Fig. 2A, construct A containing the precise SCV 3' UTR (includes only a partial PTE) and precise gRNA 5' UTR (39 nt) was 3.3-fold more efficient at translation compared with a construct containing the gRNA 5' UTR and a random 3' sequence (compare constructs A and F). Extension of the SCV 3' sequence to contain upstream coding sequence (400 nt total) that includes the full-sized PTE, did not significantly improve translation directed by the gRNA 5' UTR (compare constructs A and B). Since this result suggested that additional 5' sequence might be required for functioning of the PTE (or other 3' element) as a translational enhancer, the 5' sequence was extended into the p26 ORF to position 80 or 125. While extension of 5' sequence to position 80 did not improve translation of the reporter construct containing the 3' 400 nt (Fig. 2A, construct C), extension to position 125 enhanced translation by 75-fold when the construct contained the extended 3' sequence and full-sized PTE (compare constructs A and E). Translation mediated by the gRNA 5' 125 nt was enhanced by a more modest 4-fold over the exact 5' UTR when the precise 3' UTR was present (compare constructs A and D). These results suggest that efficient translation mediated by gRNA 5' sequences requires coding sequences at both ends of the viral genome.
Fig. 2.
Identifying important translation sequences in SCV. (A) gRNA constructs contained either the gRNA 5' UTR (1–39 nt) or extended 5' sequence (1–80 or 1–125 nt) at the 5' end and either the exact 3' UTR (positions 3656–3879), extended 3' sequence (positions 3480–3879; 400 nt), or 223 nt of random sequence at the 3' end. Short black box denotes partial PTE element, with the larger black box labeled “PTE” denoting the full-sized PTE spanning the 3' UTR junction. Light grey regions are SCV UTR sequences and dark grey regions are SCV coding sequences. Transcripts were inoculated into A. thaliana protoplasts along with a control R-Luc construct and luciferase levels assayed 18 h later. Standard deviation in three replicate experiments is shown. (B) sgRNA2 constructs contained the exact 5' UTR (positions 2484–2620) at the 5' end and either the 3' UTR, 3' 400 nt, or 400 random nt at the 3' end. (C) Deletions were constructed in construct H, containing the 5' UTR of sgRNA2 and 3' 400 nt. Black lines represent remaining sequence. Identities of three fragments (3'343, 3'289 and -Pr) used in subsequent experiments are denoted. Positions of PTE, H4b, H5 and Pr are indicated. Standard deviation in three replicate experiments is shown. (D) Stability of parental construct H and deletion constructs –Pr and 3'127 in protoplasts. Transcribed RNAs were transfected into protoplasts and total RNA isolated at the times (in hours) indicated above each lane. RNA was subjected to RNA gel blot analysis using oligonucleotide probes complementary to the luciferase coding region.
To investigate requirements for translation of the smaller of the two sgRNAs (sgRNA2), the precise sgRNA2 5' UTR (137 nt) was incorporated into reporter constructs containing either the exact SCV 3' UTR, the SCV 3' 400 nt, or a random 400 nt sequence (Fig. 2B). When combined with the 400 nt random 3' sequence, the sgRNA2 5' UTR was 8-fold better at directing translation than the gRNA 5' UTR with the random 223 nt sequence (compare constructs I and F). In the presence of the precise SCV 3' UTR, the sgRN A2 5' UTR was 10.4-fold more efficient at translation compared with the gRNA 5' UTR (compare constructs G and A). Extending the SCV 3' UTR to include a total of 400 nt improved translation directed by the sgRNA2 5' UTR by nearly 6-fold. These results suggest that the sgRNA2 5' UTR is a stronger promoter of translation than the gRNA 5' UTR in the absence of SCV 3' sequences. As with the gRNA 5' region, efficient translation directed by the sgRNA2 5' UTR requires the 3' UTR and upstream coding sequences.
To begin delineating the SCV 3' sequences that enhance translation, deletions were generated in construct H, which contains the 5' UTR of sgRNA2 and the 3' 400 nt (positions 3480–3879) (Fig. 2C). Deleting from the 5' end of the 3' sequence to position 3537 resulted in a 19% drop in translation while deleting to position 3591 caused a 61% reduction. This reduced level of translation was essentially maintained when the deletions extended an additional 59 or 113 bases (to positions 3650 and 3704, respectively). Since these two latter deletions extended into the PTE without further reducing translation, a key translation element must either exist upstream of the PTE, or upstream sequences are required for proper PTE function. Deleting an additional 49 bases to position 3753 reduced translation to background levels suggesting that a critical element exists in the 3' region of the PTE or just downstream of the element. Deletion of only the Pr and short 3' terminal sequence also reduced translation to near background levels, suggesting that the 3' terminus is also critical for translation.
Since the Pr is a stable hairpin near the 3' terminus, the possibility existed that deletion of this hairpin affected the stability of the reporter RNA and thus indirectly impacted luciferase levels. To determine the effect of deleting the Pr hairpin on the stability of the RNA, construct H containing the sgRNA2 5' UTR and 3' 400 nt as well as the same construct without the Pr hairpin (-Pr) were transfected into protoplasts and RNA levels were assayed between 1 and 18 hours post-transfection. Also included in the assay was the construct containing the 3' 127 nt, which was also translated at a minimal level. No differences were found in the stability of the deletion-containing RNA transcripts compared with parental construct H transcripts (Fig. 2D). This result indicates that background levels of translation due to deletion of the Pr hairpin region or deletion through the PTE are due to absence of required element(s) and not reduced stability of the transcripts.
A long distance RNA:RNA interaction involving the 3' PTE is important for translation mediated by the 5' UTR of sgRNA2
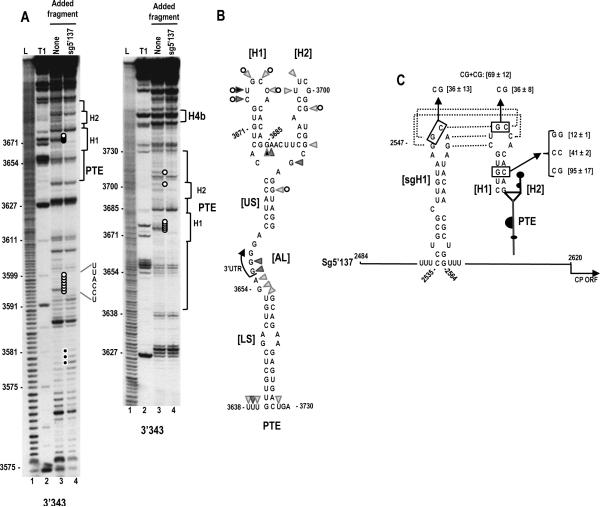
Although the deletion analysis was inconclusive as to the involvement of the PTE in translation, the striking similarity between the predicted SCV PTE structure and the PEMV/ PMV PTE translational enhancers suggested that the SCV PTE was a likely translation element. To first examine the validity of the predicted PTE secondary structure, in-line structure probing was used to evaluate the flexibility of PTE residues in a fragment containing the 3' 343 nt (3'343), which supported a high level of translation when paired with the sgRNA2 5' UTR (see Fig. 2C). In-line probing is based on self-cleavage of the RNA backbone, which requires in -line topology of the 2' hydroxyl, backbone phosphate and oxyanion leaving group. This topology only exists if the phosphate-sugar backbone can rotate about the C3'-O3' and O3'P bonds, which does not occur if the base is overly constricted by canonical/ non -canonical hydrogen bonding. Thus, the more flexible the nucleoside, the more backbone cleavage occurs. A typical in-line cleavage gel for the 3'343 fragment is shown in Fig. 3A, lane 3. The cleavage pattern in the vicinity of the PTE was consistent with the predicted structure, with all but two of the flexible residues confined to predicted loop regions (Fig. 3B). The lack of cleavages in the small symmetrical loop in the lower stem (LS) indicates that these residues are not flexible, which suggests that they may be constrained by non-Watson-Crick interactions across the stem.
Fig. 3.
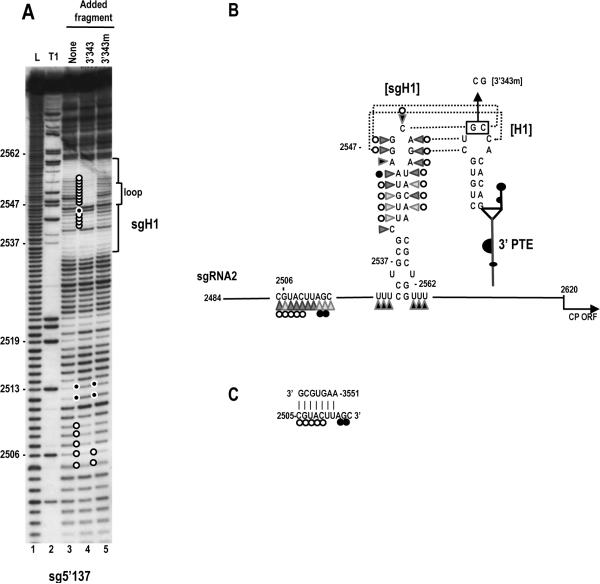
A kissing loop interaction exists between the 3' PTE and sgRNA2 5' UTR hairpin sgH1 that contributes to 3' translational enhancement. (A) In-line probing of fragment 3'343 in the absence and presence of the sgRNA2 5' UTR (5'137). The panel on the right is a longer run of the samples shown in the left panel. Fragment 3'343 was labeled at its 5' end and allowed to self cleave at 25°C for 14 h. Intensity levels of the bands in lanes 3 and 4 are proportional to the flexibility of the residues. L, alkaline-generated ladder; T1, RNase T1 digest of partially denatured RNA. Note loss of flexibility in the PTE H1 loop in the presence of fragment 5'137, which is consistent with base-pairing between the PTE H1 apical loop and PTE-complementary sequence in the sgH1 apical loop (see C). (B) Flexibility of residues in the 3' PTE region. Data is from lane 3 in (A). Darker triangles denote higher flexibility. Open circles reflect residues that lose flexibility in the presence of fragment 5'137. (C) Single and compensatory mutations generated in construct H that disrupt or reform the PTE H1 stem or the PTE RNA:RNA interaction. Levels of translation as a percentage of wt construct H are shown. Results are from three experiments with standard deviations given. Sequences that can putatively pair are connected by dotted lines.
To determine if the PTE is important for SCV translation, the 5' side PTE hairpin designated “H1” was disrupted by single mutations on both sides of the stem (Fig. 3C). This hairpin was chosen as its structure and loop sequence are conserved in most other putative carmovirus PTE (see Fig. 7A). Single alterations in the H1 stem of construct H, containing the sgRNA2 complete 5' UTR (sg5'137) and the 3' 400 nt, reduced translation to 41% or 12% of wt levels, while compensatory mutations that should reform the stem increased translation to 95% of wt levels (Fig. 3C). To determine if the loop of PTE H1 plays a role in translation, two bases were altered from GC to CG in construct H (Fig. 3C). These alterations reduced translation to 36% of wt. These results indicate that the SCV PTE H1 stem and loop contribute to translational enhancement by the 3' region of SCV.
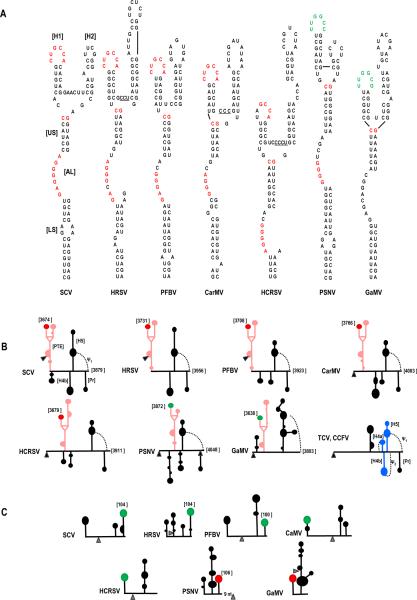
Fig. 7.
Comparison of PTE and PTE-interacting sequences in carmoviruses. (A) Sequences of PTE found in seven carmoviruses. Conserved sequences are colored alike. Note that the sequence in green contains the complement of the conserved GCCA sequence (UGGC). (B) Location of the carmovirus PTE in the 3' regions of the viral genomes. Black triangle denotes location of the stop codon for the CP ORFs. The PTE is in pink, the conserved H1 sequence CUGCCA is denoted by a red terminal loop and the conserved H1 sequence UUGGCC/ G is d enoted by a green terminal loop. Positions of the conserved sequences within the viral genome is given. Note that all viruses except GaMV have Pr, ψ1, H5 and H4b and none have ψ2. Secondary structures are based on mFold predictions and tertiary structures are based on visual observations. The structure of the 3' UTR of TCV and CCFV is provided for reference. (C) Location of PTE H1 complementary sequence near the 5' end of the gRNA. Note that all sequences are in apical loops of hairpins located either at the 5' terminus or positioned such that the loop sequence is 100 to 106 nt from the 5' end as denoted in the figure. The distance of these latter complementary sequences from the initiation codons (gray triangle) is highly variable. Secondary structures are based on mFold predictions. Sequences complementary to PTE H1 sequence CUGCCA is in red and UUGGCC/ G is in green. HRSV, Honeysuckle ringspot virus; PFBV, Pelargonium flower break virus; CarMV, Carnation mottle virus; PSNV; Pea stem necrosis virus; GaMV, Galinsoga mosaic virus.
One possible function for the PTE H1 in translation is through long-distance kissing loop interactions with sequences in the 5' regions of the gRNA and sgRNA2. A hairpin (sgH1) located at position 2535 in the center of the sgRNA2 5' UTR contains apical loop sequences capable of forming a 5 bp interaction with the PTE H1 apical loop (Fig. 3C). Converting the GC to CG in the sgH1 apical loop within construct H reduced translation to the same level as the GC alteration in the PTE H1 loop (36% of wt). When the sgH1 loop mutations were combined with the PTE H1 loop mutations, which were designed to be compensatory, translation was enhanced to 69% of wt (Fig. 3C).
To provide additional support for this long-distance interaction, in-line probing was repeated for fragment 3'343 in the presence of the sgRNA2 5' UTR (fragment sg5'137). A direct RNA:RNA interaction between fragments should be discernable by loss of residue flexibility in the PTE H1 loop (since intermolecular base-pairing would significantly constrain residue flexibility). Addition of fragment sg5'137 to fragment 3'343 followed by in-line probing reduced the level of cleavage for all residues in the loop of PTE H1 (Fig. 3A right, lane 4, five white adjacent circles). In addition, residues in the stem of PTE H2 and the upper stem (US) that were slightly flexible in the in -line cleavage pattern of the PTE alone were no longer flexible in the presence of sg5'137 (Fig. 3A, right, single white circles). Interestingly, additional alterations in the cleavage pattern were found upstream of the PTE (between positions 3578 to 3599) when the two fragments were combined. The reasons for these additional cleavage alterations are not known, but may represent another RNA:RNA interaction.
We next examined the effect of the PTE on the cleavage pattern of sgH1 in fragment sg5'137 (Fig. 4). In-line probing of sg5'137 produced a cleavage pattern that was consistent with the predicted sgH1 structure (Fig. 4A, lane 3). Residues in the apical loop and upper stem region generated higher levels of cleavage indicating substantial flexibility, while residues in the remainder of the stem were susceptible to low levels of cleavage. The bulged cytidylate at position 2540 was significantly more flexible than adjacent base-paired stem residues. However, the symmetrical bulge in the lower stem contained minimally flexible residues, suggesting that the bulged uridylates are constrained by hydrogen bonding, possibly with each other across the stem.
Fig. 4.
Effect of the PTE kissing loop interaction on the structure of the sgRNA2 5' UTR in the vicinity of sgH1. (A) In-line probing of the sgRNA2 5' UTR (5'137) in the absence and presence of fragment 3'343 and 3'343 with mutations disrupting the RNA:RNA interaction (3'343m). The 5'137 fragment was labeled at its 5' end and allowed to self cleave at 25°C for 14 h in the absence and presence of 3'343 or 3'343m. L, alkaline-generated ladder; T1, RNase T1 digest of partially denatured RNA. Open and closed circles denote residues that lose or gain flexibility, respectively, in the presence of wt 3'343 or 3'343m. Note enhanced stability of the residues in the sgH1 stem and loop when 5'137 and wt 3'343 are combined, which is consistent with base-pairing between sgH1 and PTE. Additional changes are also found in an upstream region. (B) Flexibility of residues in the sgH1 region of the sgRNA2 5' UTR. Data is from (A). Darker triangles denote higher flexibility. Open and closed circles denote residues that lose or gain flexibility, respectively, in the presence of wt 3'343. Sequences that can putatively pair are connected by dotted lines. The mutations in 3'343m that disrupt the RNA:RNA interaction are shown. (C) Possible pairing between sequence at positions 3551–3557 in the 3' region of SCV and sg5'137. Open and closed circles are as designated in (B).
Two regions of sg5'137 were strongly affected by addition of fragment 3'343 prior to cleavage: nearly the entire sgH1 hairpin, and a small region upstream of the hairpin between positions 2505 and 2513 (Fig. 4A, lane 4). Nearly every residue in the sgH1 stem that were previously susceptible to low but detectable cleavage, now exhibited background cleavage levels, indicating that the interaction between sg5'137 and 3'343 significantly reduces the flexibility of residues in the sgH1 stem. In addition, cleavages in sgH1 apical loop residues predicted to pair with the 3'PTE were reduced to background levels (Fig. 4A, lane 4, white circles). Only two residues in the upper portion of sgH1 were unchanged: the remaining unpaired adenylate at position 2546 in the apical loop, and the bulged cytidylate at position 2540 in the mid -stem region. This result is consistent with an RNA:RNA interaction between the 3' PTE H1 loop and the apical loop of sgH1, which supports the comp ensatory mutagenesis analysis (Fig. 3C).
Residues in positions 2505 to 2509 also exhibited reduced flexibility in the presence of 3'343 in all three replicates of this experiment, while residues in positions 2512 and 2513 displayed enhanced flexibility. This suggests either the existence of an additional RNA:RNA interaction or that the interaction between sgH1 and the PTE H1 generates an upstream conformational shift that alters the flexibility of these residues. To determine if the RNA:RNA interaction between sgH1 and PTE H1 is necessary for the upstream cleavage pattern alterations between positions 2505 and 2513, the in -line cleavage profile was determined for sg5'137 in the presence of fragment 3'343m, which contains a two base alteration in PTE H1 (GC to CG) that disrupts the RNA:RN A interaction. Our reasoning was that if the altered flexibility of residues 2505–2513 is unrelated to the PTE RNA:RNA interaction, then the cleavage pattern of these residues should be unaffected when this PTE interaction is prevented. As shown in Fig. 4A lane 5, addition of 3'343m to labeled sg5'137 eliminates the cleavage pattern differences in sgH1 due to the PTE interaction. The cleavage pattern between residues 2505 and 2513, however, was not similarly restored to the wt pattern yet differed from the cleavage pattern found in the presence of wt 3'343. Residues in positions 2507 to 2509 no longer exhibited the reduced flexibility found in the presence of wt 3'343 while residues in positions 2505, 2506 and 2512 and 2513 retained their altered flexibility. This result suggests that while the PTE:sgH1 interaction contributes to the changed cleavage pattern between positions 2505 and 2513, these changes are not due to a conformational shift in the region that results from this RNA:RNA interaction. Rath er, it is likely that a second RNA:RNA interaction exists in this upstream region whose formation is partially dependent on the downstream PTE:sgH1 interaction. A search of the 3' 343 nt region for sequence complementary to positions 2505 to 2509 revealed one possibility between residues 3551 and 3557 (Fig. 4C). This possible interaction is currently being investigated.
A long distance RNA:RNA interaction involving the PTE and a p26 ORF hairpin is important of the gRNA
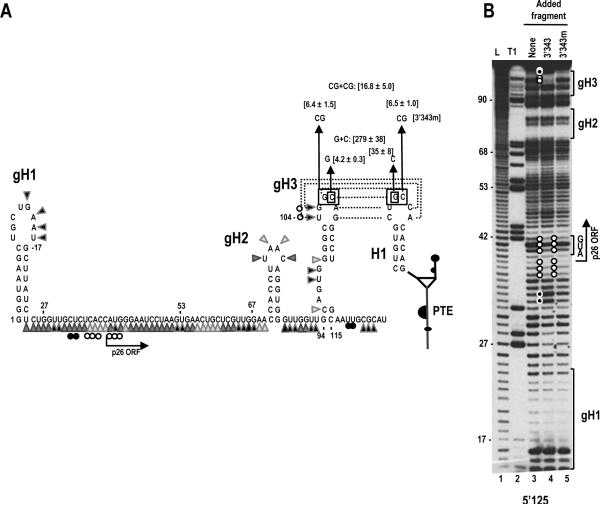
A putative hairpin (gH3) located 94 nt from the 5' end of the gRNA in the p26 ORF contains a 6 nt apical loop that is fully complementary to the 6 nt loop of YSS H1 (Fig. 5A). Hairpin gH3 is located within the portion of the 5' 125 nt fragment that substantially enhances translation of the reporter construct in the presence of the 3' 400 nt (Fig. 2A). To investigate if a long distance interaction takes place between YSS H1 and gH3, two base changes (GC to CG) were engineered into the gH3 apical loop that are complementary to the two base changes previously constructed in YSS H1. Mutations were generated in luciferase construct E, containing the 5' 125 nt of the gRNA and the 3' 400 nt (Fig. 2A). Each of the alterations caused nearly identical decreases in luciferase levels, to 6.4 or 6.5% of wt construct E. Unexpectedly, the combined presumptive compensatory mutations did not substantially restore luciferase levels, reaching only 16.8% of wt levels. Since the possibility existed that this particular RNA:RNA interaction requires sequence specificity, or that the two base alteration disrupted the structure of gH3, the experiment was repeated with mutations in only one of the two adjacent residues in the apical loops of gH3 and YSS H1 (Fig. 5A). Altering only the guanylate in the PTE H1 apical loop to a cytidylate reduced translation of construct E to 35% of wt, while changing the cytidylate at position 107 in the apical loop of gH3 to a guanylate had an effect similar to the double mutation, with levels reduced to 4.2% of wt. Combining the two mutations enhanced translation to 279% of wt (Fig. 5A). Altogether, these results support a long-distance RNA:RNA interaction between gH3 and PTE H1 that requires more sequence specificity in the interacting residues than needed for the PTE/ sgH1 interaction described above.
Fig. 5.
A kissing loop interaction between the 3' PTE and gRNA p26 ORF coding sequences contributes to 3' translational enhancement. (A) Putative structure of the 5' 125 nt of SCV. The secondary structure shown is based on mFold predictions and results of in-line probing (see B). The potential pairing between hairpin gH3 and the PTE H1 is shown with dotted lines connecting the paired residues. Flexibility of residues in the 5' 125 nt region is from lane 3 in (B). Darker triangles denote higher flexibility. Open and closed circles reflect residues that lose or gain flexibility, respectively in the presence of fragment 3'343. Single and compensatory mutations generated in construct E that disrupt or reform the PTE RNA:RNA interaction are shown. Levels of translation as a percentage of wt construct E are given. Results are from three experiments with standard deviations given. (B) In-line probing of labeled fragment 5'125 in the absence and presence of fragments 3'343 and 3'343m. L, alkaline-generated ladder; T1, RNase T1 digest of partially denatured RNA. Note that two gH3 loop residues lose flexibility and two residues near the base of gH3 show enhanced flexibility in the presence of fragment 3'343 only (lane 4, top), which is consistent with base-pairing between the apical loops of PTE H1 and gH3 (see A). Also note the presence of a second region of altered flexibility in the vicinity of the p26 initiation codon, which is retained in the absence of the PTE-mediated RNA:RNA interaction (lane 5) suggesting the presence of a second RNA:RNA interaction.
To determine if the PTE/ gH3 RNA:RNA interaction is detectable by in-line probing, cleavage profiles were determined for the gRNA 5' 125 nt fragment (5'125) in the presence and absence of fragment 3'343. As with fragment sg5'137, a direct RNA:RNA interaction between gRNA gH3 and PTE H1 should be discernable by the loss of flexibility of residues in the 5'125 gH3 apical loop. The cleavage pattern of 5'125 was consistent with the presence of three hairpins: a 5' terminal hairpin (gH1) and two hairpins in the p26 coding region, including gH3 (Fig. 5B lane 3). When fragment 3'343 was added prior to the cleavage reaction, the cleavage pattern of 5'125 was significantly altered in three regions: 1) the apical loop of gH3, where the highly flexible residues at positions 104 and 105 lost their flexibility, which is consistent with the predicted RNA:RNA interaction with PTE H1; 2) two residues at the 3' base of gH3, whose flexibility was substantially enhanced; and an upstream region (positions 32–42) that includes the initiation codon (Fig. 5A and B).
To determine if the cleavage differences in all three locations were dependent on maintenance of the PTE/ gH3 interaction, in-line probing of 5'125 was repeated in the presence of fragment 3'343m, which contains a two base alteration in the PTE H1 apical loop. As shown in Fig 5B lane 5, the cleavage differences in the gH3 apical loop, the downstream adjacent region and the two residues at positions 32 and 33 were restored to their wt pattern, indicating that structure changes in these regions require the PTE/ gH3 interaction. In contrast, the reduced flexibility of residues in positions 36 to 42 remained despite the loss of the PTE/ gH3 interaction. This result strongly suggests the existence of a second RNA:RNA interaction that includes the initiation codon.
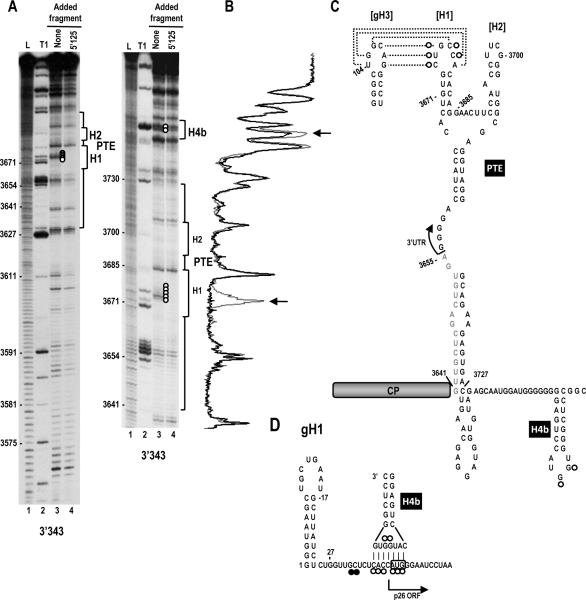
In-line probing was next conducted for fragment 3'343 in the presence and absence of fragment 5'125 (Fig. 6). Addition of fragment 5'125 to 3'343 before cleavage significantly reduced the flexibility of all residues in the PTE H1 apical loop (Fig. 6A, five adjacent open circles), consistent the RNA:RNA interaction between PTE H1 and gRNA gH3. We also noticed very reproducible differences in the cleavage pattern in the apical loop of H 4b when the 3' and 5' fragments were combined. The strong cleavages in the two guanylates in the H4b loop were reduced by approximately 50% in multiple independent experiments. In contrast, these H4b residues were never reduced in intensity when combined with fragment sg5'137 (see Fig. 3A, right). Interestingly, the sequence of the lower stem and apical loop of H 4b (5'CAUGGUG) is fully complementary to sequence at the initiation codon of the p26 ORF (5'CACCAUG) (Fig. 6D). The validity of this putative additional RNA:RNA interaction is currently being investigated.
Fig. 6.
Two long-distance interactions occur between the 5' end of the SCV genomic RNA and the 3' region. (A) In-line probing of fragment 3'343 in the absence and presence of the gRNA fragment 5'125. The panel on the right is a longer run of the samples shown in the left panel. L, alkaline-generated ladder; T1, RNase T1 digest of partially denatured RNA. Note loss of flexibility in the PTE H1 loop (lane 4, five consecutive white circles) in the presence of fragment 5'125, which is consistent with base-pairing between the PTE H1 apical loop and PTE-complementary sequence in hairpin gH3 (see C). Cleavages of two residues in the H4b apical loop were also reproducibly reduced in intensity in the presence of 5'125 but not 5'137 (see Fig. 3A). (B) Densitometry tracing of the right side autoradiograph in A. Gray tracing is from lane 3 and black tracing is from lane 4. Arrows denote positions where the cleavage pattern differs between the two lanes. (C) Location of cleavage pattern differences between lanes 3 and 4 in A. Open circles reflect residues that lose flexibility in the presence of fragment 5'125. (D) Possible pairing between the apical loop of H4b and the region of reduced flexibility in the 5'125 fragment in the presence of 3'343. The start of the p26 ORF is indicated. The initiation codon is boxed.
Discussion
Long-distance protein-mediated or RNA:RNA interactions allow for the circularization of an RNA and for elements to function in locations distal to where they reside in the genome (Edgil and Harris, 2006; Liu et al., 2009; Miller and White, 2006; Villordo et al., 2010). For the carmovirus TCV, ribosome-binding translation elements that overlap replication elements are found in the 3' UTR, allowing for a conformational rearrangement of local RNA structures mediated by RdRp binding that coordinates the switch between replication and translation (Stupina et al., 2008; Yuan et al., 2009). Repositioning of 3' proximal translation elements to the 5' end in TCV has been proposed to be facilitated by a bridge mediated by a single ribosome binding simultaneously to two distal elements (Stupina et al., 2011), and not a more common long-distance RNA:RNA interaction found in several small plus-strand RNA plant viruses engaged in cap-independent translation (Miller and White, 2006; Pettit Kneller et al., 2006).
To determine what aspects of translation in TCV are present in other carmoviruses, we conducted a phylogenetic search for conserved RNA structures in other carmoviruses. This led to the identification of the PTE-type element, which is present in seven carmoviruses. Comparison of the seven PTE structures reveals several conserved features (Fig. 7A). All PTE have H1 hairpins containing a stem of 4 to 6 bp (5/ 7 with 5 bp) and a 6 nt loop. The loop sequences are highly conserved with four PTE (including SCV) containing the sequence “CUGCCA” and two containing the near complement, “UUGGCC/ G”. The apical loop of the PTE H 1 of PMV, a member of the same virus family (Tombusviridae), contains the similar sequence “UUGCAG”. This conservation in sequence and structure likely reflects the important role of H 1 in participating in the long distance RNA:RNA interactions, and may explain why two base alterations in the kissing-loop interaction between PTE H1 and sgH1 were poorly compensatory (Fig. 4).
The kissing loop interaction between SCV PTE H1 and sgH1 caused a significant reduction in flexibility in the sgH1 stem. This reduced flexibility could be caused by enhanced stabilization of the hairpin stem, or it could result from conversion of an initial kissing loop interaction into a more stable extended du plex by putative pairing of the entire PTE H1 hairpin with the sgH1 stem (cUGUGCUGCCacACAG and cUUGUaaGGCAGaUACAa; putative paired bases are capitalized). This latter possibility would be similar to the two-step process for dimerization of HCV (Shetty et al., 2010) and HIV (Laughrea and Jette, 1994; Skripkin et al., 1994), which require viral-encoded proteins to achieve the more stable complex (Muriaux et al., 1996; Muriaux et al., 1995; Shetty et al., 2010). While this is an intriguing possibility, it sh ould be noted that this extended pairing is not discernable for gH3, and that none of the other carmovirus putative interactions have this extended pairing capability.
The SCV PTE joins a growing list of 3' TEs that can pair with 5' elements to bridge the ends of gRNA and sgRNA (Fabian and White, 2004, 2006; Guo et al., 2001; Karetnikov et al., 2006; Karetnikov and Lehto, 2008; Miller and White, 2006; Nicholson et al., 2010; Treder et al., 2008; Xu and White, 2009). These elements fold into diverse 2D conformations including Y-shaped structures (Fabian and White, 2004, 2006; Xu and White, 2009) and I-shaped structures (Nicholson et al., 2010). The Y-shaped structures, conserved among all viruses in the Tombusvirus genus (Xu and White, 2009), share some similar features with the carmoviral PTE including a branched stem upper structure, and the 5' side hairpin participating in the RNA:RNA interaction. The sequences engaged in the kissing loop interactions are also similar in length and composition, consisting of between 5–7 bases and including the conserved sequence “CCA”. The 5' gRNA sequences that interact with the Tombusvirus 3'TE are also located in 5' terminal elements, similar to the location of the presumptive interacting sequences for some of the carmoviral PTE.
The PEMV PTE H1, which is not involved in any discernable RNA:RNA interaction (Wang et al., 2009), contains sequence unrelated to the other PTE. Interestingly, the necessity for specific sequences in the H1 loop may differ dep ending on the location of the 5' complementary sequence. Compensatory alterations involving two adjacent bases restored nearly 70% of wt translation when assaying the interaction between the sgRNA2 sgH1 and the PTE, while the identical compensatory altera tions with the gRNA gH3 only restored 16% of wt translation levels. Single base compensatory alterations of the latter interaction produced substantially more translation than the wt interaction. We currently have no explanation for the enhanced translation of this mutant interaction.
The H2 hairpins of the PTE are highly variable in sequence, length, and presence of interior bulges. The upper stem (US) of the PTE ranges from 6 to 8 bp, and all (including PMV and PEMV) have a C:G pair that closes the top of the stem. The lower stems (LS) of the PTEs are variable in length and sequence, with the majority containing small symmetric or asymmetric loops (Fig. 7A). The PTE large asymmetrical loop (AL) contains between 6 and 8 nt, with nearly all accommodating at least three consecutive guanylates. These guanylate residues are required for eIF4E binding to the PEMV PTE in vitro and in vivo (Wang et al., 2009). The sequence between H1 and H2 in the PEMV PTE (CCCU) was also important for eIF4E binding and was proposed to participate in a pseudoknot with sequences in the asymmetric loop, although direct evidence for this interaction is lacking. While PMV and 3/ 7 carmovirus PTE contain sequence similar or identical to this conserved sequence between H1 and H2 (Fig. 7A, underlined residues), this is not true for the remaining carmoviruses including SCV. A pseudoknot between the sequence connecting H1 and H2 and the asymmetric loop is also improbable for SCV (three possible paired bases would include two G:U pairs) and not discernable for two other carmoviruses, with only one or no bases between the two hairpins. Whether SCV interacts with eIF4E or other translation initiation factors is currently being investigated.
A previous report on translation of carmovirus Hibiscus chlorotic ringspot virus (HCRSV) identified a 6-nt sequence as important for translation (Koh et al., 2002). This sequence, GGGCAG, lies within the large asymmetric loop of the newly identified HCRSV PTE element (Fig. 7A), which is critical for function of the PEMV PTE. The HCRSV report also concluded that a nearby hairpin was not important for translation. The alterations generated in this hairpin, now known to be H2 in its PTE, do not affect the structure of the H2 lower stem and thus are unlikely to have significantly affected the structure of the HCRSV PTE. It should also be noted that results presented in this report (Koh et al., 2002) relied on the premise that the 3' translational enhancer does not influence translation from the 5' end of the viral genome, a faulty premise given the current results on translation of SCV.
The location of PTE in the 3' region of these viruses is variable (Fig. 7B). Three viruses (SCV, PFBV, HRSV) have PTE in id entical locations that commence in the CP coding region and terminate in the 3' UTR, while the PTE of the remaining four viruses (as well as PEMV and PMV) reside wholly within their 3' UTR. The structure of the RNA between the PTE and the downstream conserved H5 hairpin is also variable as predicted by mFold (Zuker, 2003). Interestingly, while 5/ 7 carmoviruses with PTE have an H4b-type hairpin just upstream of H5, none of these viruses appear capable of forming a stable pseud oknot (ψ2) between the H4b apical loop and sequence 3' of H5. In contrast, all non-PTE carmoviruses with H4b contain sequence capable of forming ψ2. This suggests that the H4b loop sequences in PTE carmoviruses are available for an alternative function(s). For SCV, this alterative function may be an RNA:RNA interaction with sequence that includes the p26 initiation codon. This possibility is based on finding that the H4b apical loop and adjacent stem sequence can potentially form 7 bp with sequence in the vicinity of the p26 initiation codon. In addition, both the H4b loop and sequence in the vicinity of this initiation codon lose flexibility when 5' and 3' fragments are combined, even when the PTE/ gH3 interaction is eliminated (Fig. 5).
Our results also suggest the possibility of at least one additional RNA:RNA interaction involving sequence between positions 2505 and 2509 in the 5' UTR of sgRNA2 (Fig. 4A). Reduced flexibility of some of the residues in this location remained when the PTE H1 was combined with a mutant sg5'137 fragment that eliminates the RNA:RNA interaction (3'343m). This suggests that altered flexibility of these residues is partly, but not fully, dependent on the PTE H1/ sgH1 interaction. The sequence in the 3' region with the most extensive complementarity resides between positions 3551 and 3557 (Fig. 4C). This putative interaction is being investigated.
All carmoviral PTE contain putative long-distance interacting sequences in apical loops of hairpins located in both the 5' region of the gRNA (Fig. 7C) and the sgRNA2 5' UTR. In the gRNA, putative interacting sequences were found in two locations: either in a 5' terminal hairpin (CaMV, GaMV, HCRSV [and also PMV]), or between 100 and 106 nt from the 5' end (SCV, HRSV, PFBV, PSNV). For viruses with short 5' UTR, the PTE H1 complementary sequence that is distal to the 5' end resides in the coding region of the 5' proximal ORF. For PSNV, which has a more extensive 5' UTR, the sequence is still within the 5' UTR. This suggests that the 5' sequence that interacts with PTE H1 is not randomly placed, but rather exists in a particular location necessary to fulfill its function in translation. For sgRNA2, the interacting sequences are all located in apical loops of hairpins between 74 and 90 nt upstream of the CP initiation codon, which is centrally positioned within the sgRNA2 5' UTRs of carmoviruses whose sgRNA2 transcription start sites are known. Four of the seven carmoviruses harboring known or presumptive PTE can also putatively form kissing-loop interactions with a small hairpin located just upstream from their sgRNA1 initiation codons (not shown). The sgRNA1 5' UTRs that have been mapped are of similar limited size as the sgRNA1 5' UTR of SCV (26 nt). SCV does not contain any obvious PTE-interacting sequence within its sgRNA1 5' UTR or nearby within the coding sequence. How or whether the PTE influences translation of this viral mRNA is not known.
The conservation of structure and location of sequences with the capacity to interact with carmoviral PTE raises the question of why the PEMV PTE does not engage in a similar long-distance interaction. We have recently found that a second, partial PTE is positioned adjacent to the PEMV PTE and this partial PTE does engage in a long-distance RNA:RNA interaction in a similar fashion as that described here for SCV (F. Gao and A.E. Simon, in preparation). The PEMV PTE therefore, differs from the SCV PTE by having its RNA:RNA interaction relocated to a nearby element. Further investigations of the negative and/ or positive properties of retaining the long-distance interaction ability within the longer PTE will expand our understanding of this novel 3' translation element.
Material and Methods
Generation of Constructs
Single-luciferase reporter construct T7-Fluc was the progenitor for the translation constructs. The 5' end of this construct contains a T7 promoter, followed by BamH1 and SacI restriction sites into which 5' genomic or subgenomic fragments, generated by PCR, were inserted. SCV 3' fragments were introduced downstream of the Fluc coding region using PmlI and SspI restriction sites. Oligonucleotide-mediated site-directed mutagenesis was used to generate point mutations. All constructs were verified by sequencing.
Protoplast preparation, inoculations and RNA gel blots
Luciferase reporter constructs were linearized by Ssp1I and subjected to transcription in-vitro using T7 RNA polymerase. Protoplasts were generated from stationary callus cultures of Arabidopsis thaliana ecotype Co1-0, using cellulase and pectinase to remove the cell wall as previously described (Zhang et al., 2006a). Thirty micrograms of uncapped transcripts were inoculated into 5×106 protoplasts using 50% poly-ethylene glycol along with 10 μg of uncapped transcripts containing a Renilla luciferase (Rluc) reporter as an internal control. Protoplasts were harvested 18 h later, cells lysed using 1× passive lysis buffer (Promega) and luciferase activ ity measured using a TD 20/ 20 luminometer (Turner Designs). For RNA stability assays, cells were harvested at between 1 and 18 h post-transfection and total RNA extracted using RNA extraction buffer (50 mM Tri-HCl pH 7.5, 5 mM EDTA, 100 mM NaCl, 1% SDS). RNA was subjected to electrophoresis through 1% agarose gels and then transferred to nitrocullose membranes. Luciferase reporter RNA was detected using a combination of four different 32P-labeled oligonucleotides designed to be complementary to the luciferase coding region.
In-line probing
RNA fragments subjected to in-line cleavage were generated by PCR. A T7 RNA polymerase promoter was incorporated at the 5' end of each fragment during PCR. In-vitro transcribed RNAs were purified by agarose gel electrophoresis, dephosphorylated using Antarctic phosphatase (NEB), labeled at the 5' end by using γ32P-ATP and T4 poly nucleotide kinase (NEB) and then re-purified through 5% polyacrylamide gels. Labeled fragments were denatured at 75°C and slow cooled to room temperature. In-line cleavage reactions were performed at 25°C by incubating 5 pmol of labeled RNAs in in-line cleavage buffer [50 mM Tris-HCl (pH 8.5), 20 mM MgCl2] for 14 h. For competition assays, 10-fold molar excess of unlabelled fragments (50 pmol) were combined with the labeled fragment (5 pmol) and the combined fragments subjected to in-line cleavage as described above. RNA cleavage ladders were produced by incubating 5 pmol of 5' end-labeled RNA in a solution containing 1 μg yeast tRNA, 50 mM NaHCO3/ Na2CO3 (pH 9.2), and 1 mM EDTA for 5 min at 95°C. To generate RNase T1 digest ladders, 5 pmol of denatured 5' end-labeled RNAs were incubated in 1 μg yeast tRNA, 20 mM sodium citrate (pH 5.0), 1 mM EDTA, 7 M urea, and 1 U RNase T1 (Ambion) for 2 min at 25°C. All reactions were ethanol precipitated and resuspended in acrylamide gel loading buffer II (Ambion). Before electrophoresis, fragments were heated at 95°C for 2 min, and then snap cooled on ice. Electrophoresis was performed through 8 M urea, 8% or 10% denaturing polyacrylamide gels followed by autoradiography. At least two independent in-line probing assays were performed for each fragment.
Acknowledgements
This work was supported by a grant from the U.S. Public Health Service (GM 061515-05A2/ G120CD) to A.E.S. M. C. was supported by NIH Institutional Training Grant 2T32AI051967-06A1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Batten JS, Desvoyes B, Yamamura Y, Scholthof KBG. A translational enhancer element on the 3'-proximal end of the Panicum mosaic virus genome. FEBS Let. 2006;580:2591–2597. doi: 10.1016/j.febslet.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Bradrick SS, Walters RW, Gromeier M. The hepatitis C virus 3'-untranslated region or a poly(A) tract promote efficient translation subsequent to the initiation phase. Nucl. Acids Res. 2006;34:1293–1303. doi: 10.1093/nar/gkl019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushell M, Sarnow P. Hijacking the translation apparatus by RNA viruses. J. Cell Biol. 2002;158:395–399. doi: 10.1083/jcb.200205044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu WW, Kinney RM, Dreher TW. Control of translation by the 5'- and 3'-terminal regions of the dengue virus genome. J. Virol. 2005;79:8303–8315. doi: 10.1128/JVI.79.13.8303-8315.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Quinto SL, Saiz M, de la Morena D, Sobrino F, Martinez-Salas E. IRES-driven translation is stimulated separately by the FMDV 3'-NCR and poly(A) sequences. Nucl. Acids Res. 2002;30:4398–4405. doi: 10.1093/nar/gkf569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrikova E, Florez P, Bradrick S, Gromeier M. Activity of a type 1 picor navirus internal ribosomal entry site is determined by sequences within the 3' nontranslated region. Proc. Natl. Acad. Sci. USA. 2003;100:15125–15130. doi: 10.1073/pnas.2436464100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher TW. Functions of the 3'-untranslated regions of positive strand RNA viral genomes. Ann. Rev. Phytopathol. 1999;37:151–174. doi: 10.1146/annurev.phyto.37.1.151. [DOI] [PubMed] [Google Scholar]
- Edgil D, Harris E. End -to-end communication in the modulation of translation by mammalian RNA viruses. Virus Res. 2006;119:43–51. doi: 10.1016/j.virusres.2005.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian MR, White KA. 5'-3' RNA-RNA interaction facilitates cap - and poly(A) tail-independent translation of tomato bushy stunt virus mRNA - A potential common mechanism for Tombusviridae. J. Biol. Chem. 2004;279:28862–28872. doi: 10.1074/jbc.M401272200. [DOI] [PubMed] [Google Scholar]
- Fabian MR, White KA. Analysis of a 3'-translation enhancer in a tombusvirus: A dynamic model for RNA-RNA interactions of mRNA termini. RNA. 2006;12:1304–1314. doi: 10.1261/rna.69506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR. The CAP and poly(A) tail function synergistically to regulate messenger -RNA translational efficiency. Genes Dev. 1991;5:2108–2116. doi: 10.1101/gad.5.11.2108. [DOI] [PubMed] [Google Scholar]
- Ganoza MC, Kiel MC, Aoki H. Evolutionary conservation of reactions in translation. Microbiol. Mol. Biol. Rev. 2002;66:460–485. doi: 10.1128/MMBR.66.3.460-485.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Allen EM, Miller WA. Base-pairing between untranslated regions facilitates translation of uncapped, nonpolyadenylated viral RNA. Mol. Cell. 2001;7:1103–1109. doi: 10.1016/s1097-2765(01)00252-0. [DOI] [PubMed] [Google Scholar]
- Hellen CUT, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- Isken O, Grassmann CW, Sarisky RT, Kann M, Zhang S, Grosse F, Kao PN, Behrens SE. Members of the NF90/ NFAR protein group are involved in the life cycle of a positive-strand RNA virus. EMBO J. 2003;22:5655–5665. doi: 10.1093/emboj/cdg562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken O, Grassmann CW, Yu HY, Behrens SE. Complex signals in the genomic 3' nontranslated region of bovine viral diarrhea virus coordinate translation and replication of the viral RNA. RNA. 2004;10:1637–1652. doi: 10.1261/rna.7290904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karetnikov A, Keranen M, Lehto K. Role of the RNA2 3' non-translated region of Blackcurrant reversion nepovirus in translational regulation. Virology. 2006;354:178–191. doi: 10.1016/j.virol.2006.06.028. [DOI] [PubMed] [Google Scholar]
- Karetnikov A, Lehto K. The RNA2 5' leader of Blackcurrant reversion virus mediates efficient in vivo translation through an internal ribosomal entry site mechanism. J. Gen. Virol. 2007;88:286–297. doi: 10.1099/vir.0.82307-0. [DOI] [PubMed] [Google Scholar]
- Karetnikov A, Lehto K. Translation mechanisms involving long-distance base pairing interactions between the 5' and 3' non-translated regions and internal ribosomal entry are conserved for both genomic RNAs of Blackcurrant reversion nepovirus. Virology. 2008;371:292–308. doi: 10.1016/j.virol.2007.10.003. [DOI] [PubMed] [Google Scholar]
- Kneller ELP, Rakotondrafara AM, Miller WA. Cap-independent translation of plant viral RNAs. Virus Res. 2006;119:63–75. doi: 10.1016/j.virusres.2005.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh DC-Y, Liu DX, Wong S-M. A six-nucleotide segment within the 3' untranslated region of Hibiscus chlorotic ringspot virus plays an essential role in translational enhancement. J. Virol. 2002;76:1144–1153. doi: 10.1128/JVI.76.3.1144-1153.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Evaluation of the scanning model for initiation of protein -synthesis in eukaryotes. Cell. 1980;22:7–8. doi: 10.1016/0092-8674(80)90148-8. [DOI] [PubMed] [Google Scholar]
- Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- Laughrea M, Jette L. A 19-nucleotide sequence upstream of the 5' major splice donor is part of the dimerization domain of human -immunodeficiency virus-1 genomic RNA. Biochemistry. 1994;33:13464–13474. doi: 10.1021/bi00249a035. [DOI] [PubMed] [Google Scholar]
- Liu Y, Wimmer E, Paul AV. Cis-acting RNA elements in human and animal plus-strand RNA viruses. Biochim. Biophys. Acta. 2009;1789:495–517. doi: 10.1016/j.bbagrm.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marintchev A, Wagner G. Translation initiation: structures, mechanisms and evolution. Quart. Rev. Biophys. 2004;37:197–284. doi: 10.1017/S0033583505004026. [DOI] [PubMed] [Google Scholar]
- Martinez-Salas E, Pacheco A, Serrano P, Fernandez N. New insights into internal ribosome entry site elements relevant for viral gene expression. J. Gen. Virol. 2008;89:611–626. doi: 10.1099/vir.0.83426-0. [DOI] [PubMed] [Google Scholar]
- McCormack JC, Yuan XF, Yingling YG, Kasprzak W, Zamora RE, Shapiro BA, Simon AE. Structural domains within the 3' untranslated region of Turnip crinkle virus. J. Virol. 2008;82:8706–8720. doi: 10.1128/JVI.00416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick WC. Cap-dependent and cap-independent translation in eukaryotic systems. Gene. 2004;332:1–11. doi: 10.1016/j.gene.2004.02.051. [DOI] [PubMed] [Google Scholar]
- Miller W, Wang Z, Treder K, Kraft J. Cap-independent translation via diverse elements in the 3' untranslated regions of plant viral RNAs. Phytopathology. 2007a;97:S77–S77. doi: 10.1042/BST0351629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WA, Wang Z, Treder K. The amazing diversity of cap -independent translation elements in the 3'-untranslated regions of plant viral RNAs. Biochemi. Soc. Trans. 2007b;35:1629–1633. doi: 10.1042/BST0351629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WA, White KA. Long-distance RNA-RNA interactions in plant virus gene expression and replication. Ann. Rev. Phytopath. 2006;44:447–467. doi: 10.1146/annurev.phyto.44.070505.143353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriaux D, Fosse P, Paoletti J. A kissing complex together with a stable dimer is involved in the HIV-1(Lai) RNA dimerization process in vitro. Biochemistry. 1996;35:5075–5082. doi: 10.1021/bi952822s. [DOI] [PubMed] [Google Scholar]
- Muriaux D, Girard PM, Bonnetmathoniere B, Paoletti J. Dimerization of HIV-1(LAI) RNA at low inonic strength. An autocomplementary sequence in the 59 leader region is evidenced by an antisense oligonucleotide. J. Biol. Chem. 1995;270:8209–8216. doi: 10.1074/jbc.270.14.8209. [DOI] [PubMed] [Google Scholar]
- Nicholson BL, Wu BD, Chevtchenko I, White KA. Tombusvirus recruitment of host translational machinery via the 3' UTR. RNA. 2010;16:1402–1419. doi: 10.1261/rna.2135210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG, Lomakin IB, Pilipenko EV, Shatsky IN, Agol VI, Hellen CUT. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. USA. 2001;98:7029–7036. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettit Kneller EL, Rakotondrafara AM, Miller WA. Cap-independent translation of plant viral RNAs. Virus Res. 2006;119:63–75. doi: 10.1016/j.virusres.2005.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss T, Hentze MW. Starting the protein synthesis machine: eukaryotic translation initiation. Bioessays. 2003;25:1201–1211. doi: 10.1002/bies.10362. [DOI] [PubMed] [Google Scholar]
- Shetty S, Kim S, Shimakami T, Lemon SM, Mihailescu MR. Hepatitis C virus genomic RNA dimerization is mediated via a kissing complex intermediate. RNA. 2010;16:913–925. doi: 10.1261/rna.1960410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skripkin E, Paillart JC, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human immunodeficiency-virus type-1 RNA dimerization in vitro. Proc. Natl. Acad. Sci. USA. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CZ, Simon AE. Requirement of a 3'-terminal stem-loop in in vitro transcription by an RNA-dependent RNA-polymerase. J. Mol. Biol. 1995;254:6–14. doi: 10.1006/jmbi.1995.0594. [DOI] [PubMed] [Google Scholar]
- Song YT, Friebe P, Tzima E, Junemann C, Bartenschlager R, Niepmann M. The hepatitis C virus RNA 3'-untranslated region strongly enhances translation directed by the internal ribosome entry site. J. Virol. 2006;80:11579–11588. doi: 10.1128/JVI.00675-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupina VA, Meskauskas A, McCormack JC, Yingling YG, Shapiro BA, Dinman JD, Simon AE. The 3' proximal translational enhancer of Turnip crinkle virus binds to 60S ribosomal subunits. RNA. 2008;14:2379–2393. doi: 10.1261/rna.1227808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupina VA, Yuan X, Meskauskas A, Dinman JD, Simon AE. Ribosome binding to a 5' translational enhancer is altered in the presence of the 3'UTR in cap -independent translation of Turnip crinkle virus. J. Virol. 2011;85:4638–4653. doi: 10.1128/JVI.00005-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiebeauld O, Pooggin M, Ryabova L. Alternative translation strategies in plant viruses. Plant Viruses. 2007;1:20. [Google Scholar]
- Treder K, Kneller ELP, Allen EM, Wang ZH, Browning KS, Miller WA. The 3' cap-independent translation element of Barley yellow dwarf virus binds eIF4F via the eIF4G subunit to initiate translation. RNA. 2008;14:134–147. doi: 10.1261/rna.777308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villordo SM, Alvarez DE, Gamarnik AV. A balance between circular and linear forms of the dengue virus genome is crucial for viral replication. RNA. 2010;16:2325–2335. doi: 10.1261/rna.2120410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kraft JJ, Hui AY, Miller WA. Structural plasticity of Barley yellow dwarf virus-like cap-independent translation elements in four genera of plant viral RNAs. Virology. 2010;402:177–186. doi: 10.1016/j.virol.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZH, Treder K, Miller WA. Structure of a Viral Cap-independent Translation Element That Functions via High Affinity Binding to the eIF4E Subunit of eIF4F. J. Biol. Chem. 2009;284:14189–14202. doi: 10.1074/jbc.M808841200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinlich S, Huttelmaier S, Schierhorn A, Behrens SE, Ostareck-Lederer A, Ostareck DH. IGF2BP1 enhances HCV IRES-mediated translation initiation via the 3' UTR. RNA. 2009;15:1528–1542. doi: 10.1261/rna.1578409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, White KA. RNA-Based regulation of transcription and translation of Aureusvirus subgenomic mRNA1. J. Virol. 2009;83:10096–10105. doi: 10.1128/JVI.00376-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan XF, Shi KR, Meskauskas A, Simon AE. The 3' end of Turnip crinkle virus contains a highly interactive structure including a translational enhancer that is disrupted by binding to the RNA-dependent RNA polymerase. RNA. 2009;15:1849–1864. doi: 10.1261/rna.1708709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan XF, Shi KR, Young MYL, Simon AE. The terminal loop of a 3' proximal hairpin plays a critical role in replication and the structure of the 3' region of Turnip crinkle virus. Virology. 2010;402:271–280. doi: 10.1016/j.virol.2010.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JC, Zhang GH, Guo R, Shapiro BA, Simon AE. A pseudoknot in a preactive form of a viral RNA is part of a structural switch activating minus -strand synthesis. J. Virol. 2006a;80:9181–9191. doi: 10.1128/JVI.00295-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JC, Zhang GH, McCormack JC, Simon AE. Evolution of virus-derived sequences for high-level replication of a subviral RNA. Virology. 2006b;351:476–488. doi: 10.1016/j.virol.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucl. Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]