Abstract
BACKGROUND & AIMS
The hepatitis C virus (HCV) serine protease NS3/4A can cleave mitochondria-associated, anti-viral signaling protein (MAVS) and block retinoic acid-inducible gene I–mediated interferon (IFN) responses. Although this mechanism is thought to have an important role in HCV-mediated innate immunosuppression, its significance in viral persistence is not clear.
METHODS
We generated transgenic mice that express the HCV NS3/4A proteins specifically in the liver and challenged the animals with a recombinant vesicular stomatitis virus (VSV), a synthetic HCV genome, IFN-α, or IFN-β. We evaluated the effects of HCV serine protease on the innate immune responses and their interactions.
RESULTS
Expression of HCV NS3/4A resulted in cleavage of intrahepatic MAVS; challenge of transgenic mice with VSV or a synthetic HCV genome induced strong, type I IFN-mediated responses that were not significantly lower than those of control mice. Different challenge agents induced production of different ratios of IFN-α and -β, resulting in different autophagic responses and vesicular trafficking patterns of endoplasmic reticulum- and mitochondria-associated viral proteins. IFN-β promoted degradation of the viral proteins by the autolysosome. Variant isoforms of MAVS were associated with distinct, type I IFN-mediated autophagic responses; these responses have a role in trafficking of viral components to endosomal compartments that contain toll-like receptor -3.
CONCLUSIONS
IFN-β-mediates a distinct autophagic mechanism of anti-viral host defense. MAVS have an important role in type I IFN-induced autophagic trafficking of viral proteins.
Keywords: Autophagy, TLR3, liver disease, RIG-I
Hepatitis C virus (HCV) has infected an estimated 130 million people worldwide.1 Exposure to HCV typically leads to chronic liver disease, and it is the most common cause of cirrhosis, chronic liver failure and hepatocellular carcinoma. The immediate host response to acute HCV infection is triggered through cellular pattern recognition receptors (PRRs), such as the intra-cytoplasmic retinoic acid-inducible gene I (RIG-I) and the Toll-like receptor 3 (TLR3). These molecules signal through their respective adaptors, the mitochondria-associated anti-viral signaling protein (MAVS) (also called IPS-1, Cardif, and VISA) and the Toll/interleukin-1 receptor domain containing adaptor inducing IFN-β (TRIF), to activate the synthesis of IFN-β, IFN-α and proinflammatory cytokines. The secreted IFN-β and -α are recognized by a shared IFN-α/β receptor (IFNR). Their initial induction leads to the activation of an autocrine/paracrine positive feedback pathway, which results in the amplification of the response and induction of an antiviral state in the infected, as well as the neighboring bystander cells.2, 3
The mechanism of HCV persistence is complex and has often been linked to its ability to thwart the host immune response.4 Molecular studies have revealed several levels of host immune modulation by HCV.5 In one such mechanism, the HCV NS3/4A serine protease has been shown to interfere with the innate immune signaling pathways, through the specific cleavage of the adaptor proteins MAVS and TRIF.4 A signaling blockade imposed by this mechanism has been suggested to limit the level and diversity of IFN-stimulated gene (ISG) expression, leading to significant attenuation of host immunity.5–8 While the role of NS3/4A-mediated MAVS cleavage in HCV-mediated innate immune suppression has been largely substantiated in vitro and in vivo,9, 10 its significance in the clinical course of the disease remains unclear. TLR3 is upregulated during the ISG response in HCV-infected cells, and cleavage of TRIF has been demonstrated in vitro.4 However, the issue of how virus-derived dsRNA encounters TLR3, which is present on the cell surface or in intracellular endosomes, remains unresolved. In this context the cellular autophagy machinery has been suggested to play a role in delivering viral genetic material to endosomal TLRs.11 Autophagy is a cellular degradation pathway by which cytoplasmic constituents, including organelles, are directed to the lysosome. While autophagic pathways can regulate viral replication by influencing the anti-viral responses, many viruses have evolved to evade, subvert or exploit this innate response.12 HCV requires the autophagy machinery to initiate its replication.13 It induces an incomplete autophagic process at the surface of the ER and uses it as a membranous scaffold for RNA replication.14 Alternatively, the Atg5-Atg12 conjugate, a key regulator of autophagy, negatively regulates the type I IFN signaling by direct association with RIG-I and MAVS.15 Yet, it is not known if these two virus-related autophagic responses traverse.
HCV is seen to persist in infected patients, despite potent IFN induction and ISG expression.9, 10 Currently, the standard therapy with pegylated IFN-α and ribavirin is far from optimal; the sustained virological response is less than 50% among patients with genotype 1 HCV infection.4 However, high pre-treatment ISG levels in hepatitis C patients are more frequently associated with non-responsiveness to the IFN-α-based therapy,16, 17 which is rather perplexing, as an active IFN system is expected to potentiate the IFN-α treatment. To date, an understanding of the correlation between MAVS cleavage and innate immune response, as well as the IFN treatment outcome, has been largely impeded by the absence of robust small animal models suitable to address these issues in vivo.
To specifically delineate the role of the HCV NS3/4A-mediated innate immunocompromise in viral infection, we generated a novel transgenic mouse with hepatocyte-specific expression of NS3/4A proteins, and demonstrated that cleavage of MAVS by the HCV NS3/4A protease by itself does not impose an overwhelming blockade of the type I IFN response in the liver. Our results further show, for the first time, a differential effect of IFN-α and -β on HCV proteins in vivo due to the disparate upregulation of autophagic proteolysis and suggest an essential role of MAVS in the delivery of viral RNA to the endosomal compartments.
MATERIALS AND METHODS
Mice
Inducible NS3/4A transgenic mice were generated on the C57BL/6 × C3H background as detailed in Supplementary Materials and Methods and bred with Alb-Cre mice18 to generate mice with liver-specific, constitutive expression of HCV NS3/4A proteins. Double transgenic (NS3/4A × Alb-Cre) mice were euthanized around 6–8 weeks of age, and liver, spleen and lung tissues analyzed for constitutive expression of the NS3/4A transgene by western blot and immunohistochemistry. To determine the time-course induction of transgene expression in the NS3/4A single transgenic mice, a pCMVCre plasmid19 was hydrodynamically injected through the tail vein, and livers were harvested for western blot at 2-day intervals up to day 12. Mice were euthanized by CO2 asphyxiation. All experiments described were performed on 6- to 12-week-old animals, and control and transgenic groups were age- and sex-matched. All animal care and experimentation were performed according to the National Institute of Health Guidelines and with the approval of the Institutional Animal Care and Use Committee.
Challenge Studies
Double transgenic and littermate control mice were challenged with various inducers of type I IFN response, including a recombinant vesicular stomatitis virus, VSV-RFP (Indiana strain) (107 pfu); HCV genotype 1b genome complexed with a lipid-based in vivo transfection reagent (Altogen) (100 μg); poly(I:C) (Invivogen) (100 μg); poly(I:C)/LyoVec (Invivogen) (50 μg), a replication-deficient Adenovirus (AdLacZ, 2 × 109 pfu, ViraQuest); IFN-α (HyCult) (1,000 IU); and IFN-β (PBL InterferonSource) (10,000 IU). All challenge agents, except for the HCV genome, were delivered i.v. through the tail vein. The HCV genome was diluted in 5% glucose and hydrodynamically delivered. Mice were euthanized at 5 or 8 hours post-challenge, and the blood and livers were collected for analysis.
Statistical Analysis
For statistical analyses, the 2-tailed student t test was used. Analysis of variance was performed by using ANOVA. P values < 0.05 were considered significant. P values > 0.05 were considered not significant.
Additional Methods
Procedures described in Supplementary Materials and Methods include those for VSV passage and titration; in vitro transcription of HCV genome; qRT-PCR for relative quantitation of IFN-β and ISG56 mRNA levels and transgene transcription analysis; qPCR for AdLacZ copy numbers; immunostaining and fluorescent microscopy; western blot; ELISA; immunoprecipitation; and isolation of cellular vesicles.
RESULTS
Characterization of Transgenic Mice
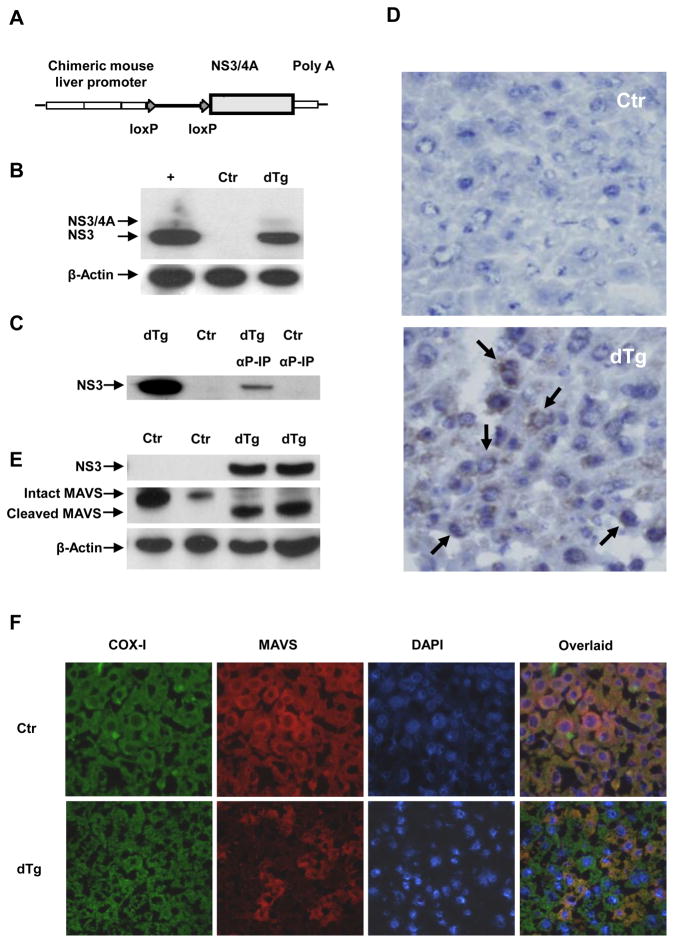
To delineate the effect of HCV NS3/4A on the innate immune responses, we generated Cre/loxP-inducible, NS3/4A transgenic mice by using the pLIVE-NS3/4A liver-specific construct (Fig. 1A). To render the mice capable of expressing NS3/4A constitutively, we bred the transgenic founders with an ALB-Cre partner that has liver-specific expression of Cre-recombinase.18 Double transgenic mice (NS3/4A × Alb-Cre) from 6 founder lines were analyzed for transgene expression. Of the 6 lines screened, only one line showed detectable transgene expression (Fig. 1B–1D). Both the NS3/4A fusion protein and the cleaved NS3 (the major species) were detected in western blots (Fig. 1B). Cleavage of the NS3/4A junction indicated that the NS3 serine protease was functionally active. NS3 expression was further confirmed by immunoprecipitation in the double transgenic livers (Fig. 1C). Immunohistochemical analyses demonstrated the perinuclear distribution of NS3 (Fig. 1D), consistent with its ER-centric subcellular localization20 in 88.8 % (± 2.4 % SD) of the hepatocytes, although the levels of expression varied among cells. However, we were unable to study NS3 by immunofluorescence in greater detail due to the high background and autofluorescence of the liver tissues.
Figure 1.
Cre/loxP recombination-mediated conditional transgenic mice with liver-specific expression of HCV NS3/4A were generated. (A) Schematic representation of the pLive-NS3/4A construct. The HCV genotype 1b NS3/4A genomic region was cloned under the control of the mouse alpha fetoprotein enhancer II and the mouse minimal albumin promoter. (B) Detection of NS3 and NS3/4A protein expression in double transgenic (NS3/4A × Alb-Cre) (dTg) mice by western blot. The lane on the far left (+) is the positive control derived from 2–3 C+ cells. (C) Detection of NS3 protein by immunoprecipitation with an HCV serine protease-specific antiserum. (D) Detection of NS3 in double transgenic liver by immunohistochemistry (40 × magnification). Arrows point to NS3 positively-stained cells. (E) Detection of intrahepatic MAVS by western blot (with pooled anti-MAVS antibodies, 4983, Cell Signaling plus AT107, Alexis). (F) Detection of intrahepatic MAVS by immunofluorescence. Micrographs (40 × magnification) of control and dTg mice. Shown are individual and merged images of sections stained with DAPI and anti-MAVS (4983, Cell Signaling) and anti-COX- I (A6403, Invitrogen).
Since HCV NS3/4A serine protease is known to cleave murine MAVS,6, 8, 21 we examined the NS3/4A-expressing mouse livers for MAVS cleavage. Indeed, both intact and cleaved MAVS (80% and 73% cleaved) were detected by western blotting in the double transgenic livers, but only intact MAVS was present in the control ones (Fig. 1E). Immunofluorescent staining with an antibody against intact MAVS showed that MAVS was detectable in the cytosol of most hepatocytes and co-localized with the mitochondrial marker COX-I in the livers of the control mice (Fig. 1F). However, in the livers of double transgenic mice, the intact MAVS was only detectable in 21.2 % (± 4.9 % SD) of the hepatocytes. In addition, the liver-specificity and Cre-mediated inducibility of NS3/4A in these mice make this animal model versatile for a variety of experimental settings (Supplementary Fig. 1).
Antiviral Responses in the NS3/4A-Expressing Mice
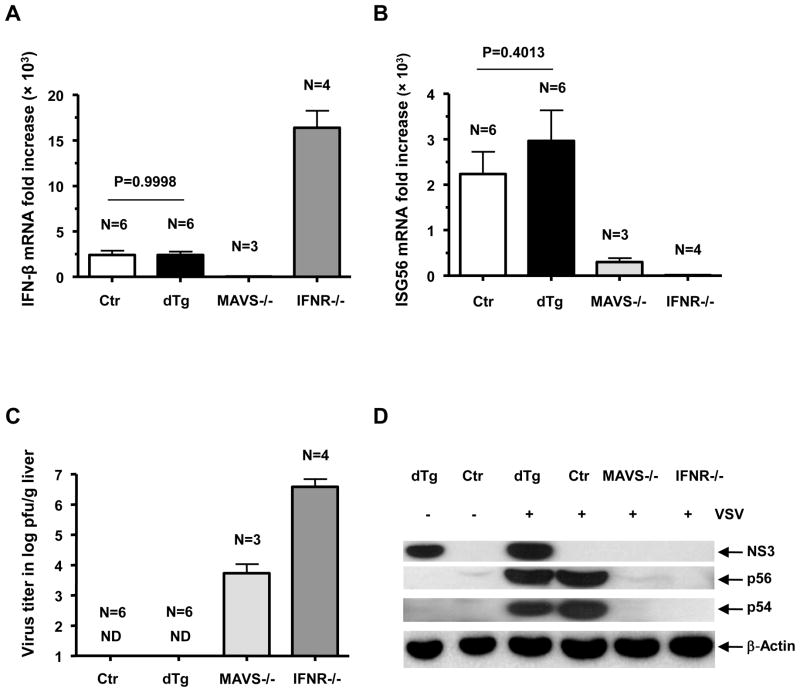
Cleavage of MAVS by the NS3/4A protease has been suggested as a pivotal mechanism by which HCV blunts the IFN response mediated through the RIG-I signaling pathway.6, 8 We, therefore, tested the NS3/4A transgenic mouse’s ability to fend off an RNA virus infection. By using a recombinant VSV (107 pfu), known to elicit a rapid type I IFN response through RIG-I, we challenged the double transgenic and littermate control mice and euthanized them at 5 hours post-injection. Surprisingly, IFN-β gene expression and the downstream ISG response were induced at a comparable level in both double transgenic and control mice (Fig. 2A, B and D). Both groups of animals cleared the virus promptly (Fig. 2C). In comparison, IFNR−/− and MAVS−/− mice showed a severe compromise in their antiviral response, as evidenced by their viral presence at 5 hours post-challenge. Earlier, it was suggested that plasmacytoid dendritic cells in MAVS−/− mice can produce IFN-α through the TLR7 pathway independent of RIG-I activation.21, 22 This mechanism may be responsible for the reduced VSV titers despite the absence of a notable IFN-β response in the MAVS−/− livers (Fig. 2A and C).5
Figure 2.
Intrahepatic type I IFN response at 5 hours post-i.v. challenge with 107 pfu of recombinant VSV. qRT-PCR detection of (A) IFN-β mRNA and (B) ISG56 mRNA (fold increase). (C) Virus titer (log pfu/g) in the liver at 5 hours post-challenge. (D) Gene expression following VSV challenge, detected by western blot. ND, Not detectable. Error bars represent standard error of mean (SEM)
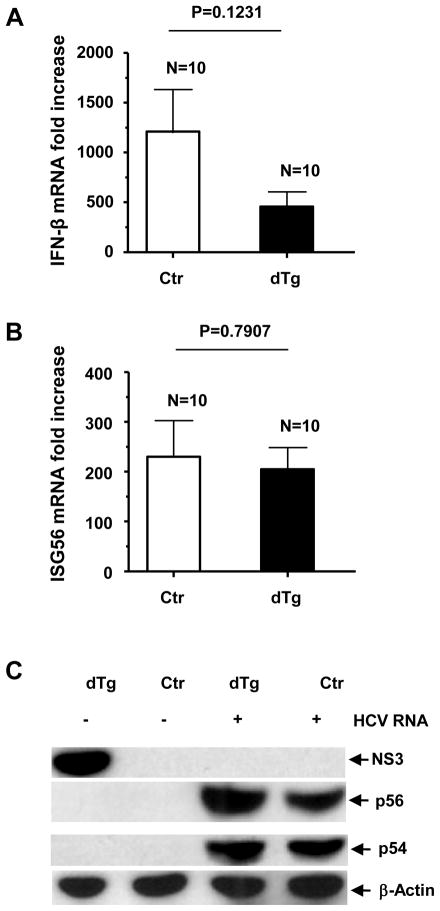
The 3′ untranslated region (UTR) of the HCV genome contains the HCV-specific, pathogen-associated molecular pattern substrate of RIG-I.23 To further define the role of NS3/4A in antiviral host defense, we hydrodynamically delivered HCV genomic RNA to the double transgenic mice and littermate controls. The HCV RNA challenge resulted in a potent induction of IFN-β and ISG responses (Fig. 3A–C) in the transgene-expressing mice, which were not significantly lower than those in the controls. Surprisingly, we found that the NS3 protein became undetectable after the HCV RNA challenge (Fig. 3C). To test whether the observed sequestration of NS3 was linked to activation of the RIG-I signaling pathway in the double transgenic mice, we challenged the animals with several additional RIG-I-independent (AdLacZ and polyI:C) and -dependent (polyI:C/LyoVec) IFN inducers. Following the i.v. injections, both groups of animals exhibited comparable IFN responses, and the NS3 protein became undetectable in the double transgenic mice (Supplementary Figs. 2–4).
Figure 3.
Intrahepatic type I IFN response at 8 hours following hydrodynamic injection of 100 μg synthetic HCV RNA genome. The livers were harvested following systemic PBS perfusion to remove contaminating blood. qRT-PCR detection of (A) IFN-β mRNA and (B) ISG56 mRNA (fold increase). (C) Gene expression following HCV-N challenge, detected by western blot. Error bars represent SEM.
Differential Effect of IFN-α and -β on NS3
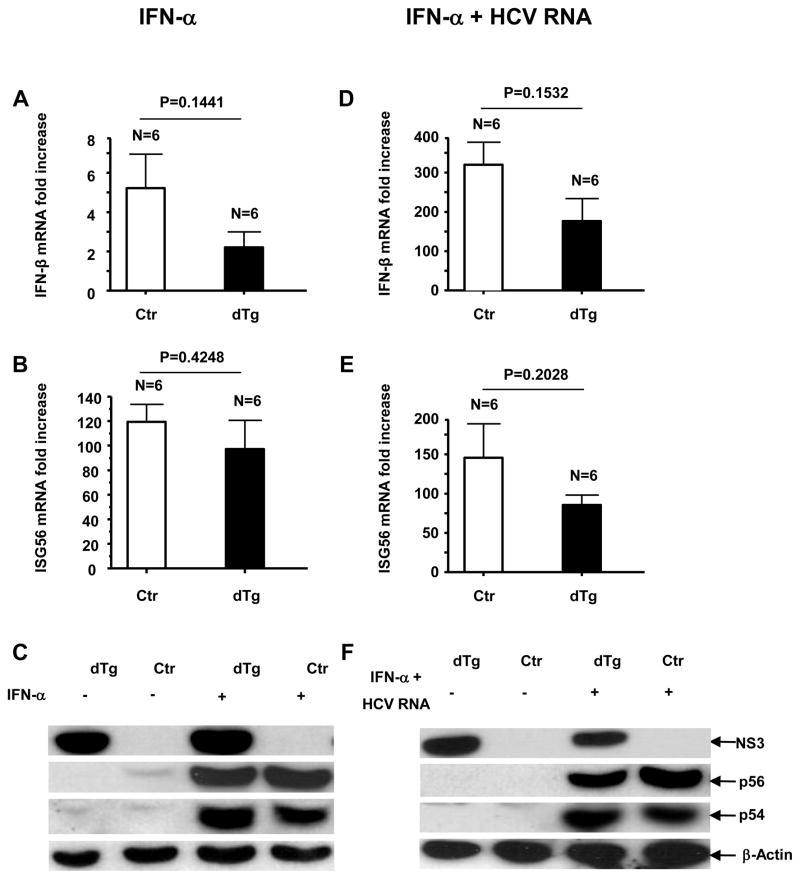
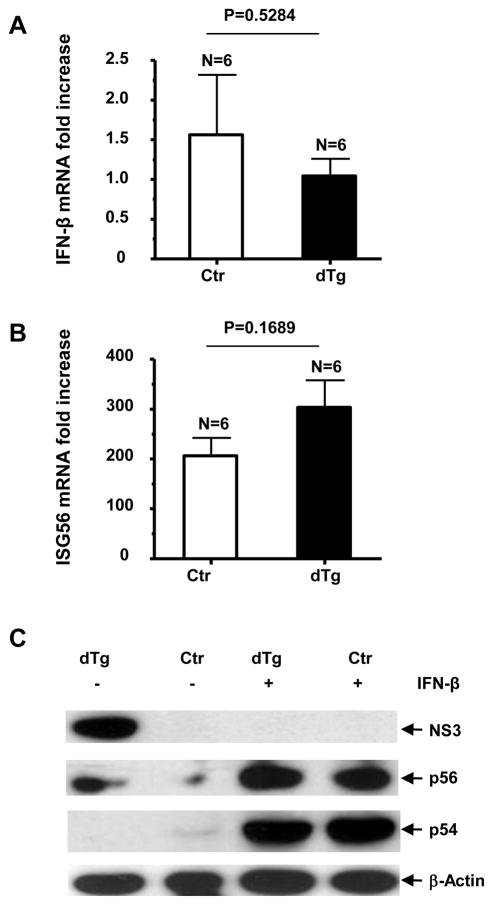
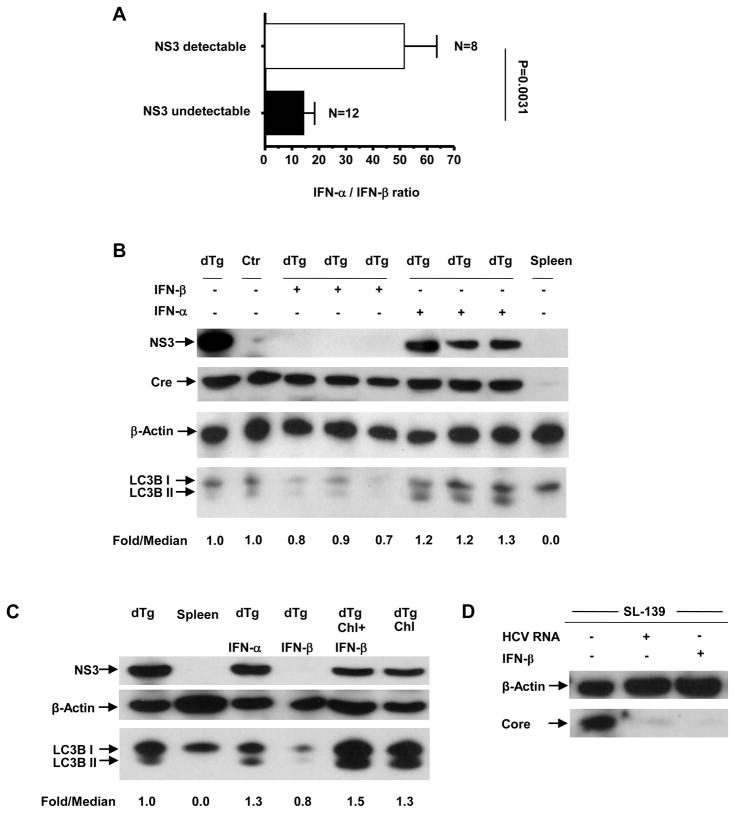
To ascertain if the NS3 sequestration following the HCV genome challenge was caused by the type I IFN response, we injected double transgenic and control mice with recombinant IFNs. IFN-α injection elicited a marked ISG response, but limited IFN-β induction, in both groups of mice (Fig. 4A–C). Interestingly, the NS3 protein was detectable in the double transgenic mice from both the IFN-α-treated and untreated group (Fig. 4C). We subsequently challenged double transgenic mice with HCV RNA plus IFN-α by hydrodynamic injection and found that the NS3 persisted despite the presence of HCV RNA (Fig. 4F), indicating that IFN-α prevented the NS3-suppressive effect of HCV RNA. Moreover, these mice showed a lower induction of IFN-β (Fig. 4D), as compared to those challenged with HCV RNA alone (Fig. 3A). To determine whether the repression of the NS3 protein following HCV RNA challenge was due to a higher relative IFN-β response, we injected NS3-expressing and non-expressing mice with recombinant IFN-β. IFN-β elicited a substantial ISG response in both groups of mice (Fig. 5B and C) and the repression of the NS3 protein in the double transgenic mice (Fig. 5C). We then examined the association between the relative ratios of induced IFN-α to IFN-β and NS3 repression in vivo by analyzing the serum levels of IFN-α and -β by ELISA. A significant correlation was found between a relatively strong induction of IFN-β and intrahepatic repression of the HCV protease, regardless of challenging agents (Fig. 6A and Supplementary Table 1). We further confirmed the absence of detectable NS3 protein in these mice by immunoprecipitation and immunohistochemistry (data not shown).
Figure 4.
Detection of intrahepatic (A) IFN-β mRNA fold increase and (B) ISG56 mRNA (fold increase) by qRT-PCR, 5 hours post-i.v. injection with 1000 IU IFN-α. (C) Gene expression following IFN-α injection, detected by western blot. (D–F) Intrahepatic type I IFN response in NS3-expressing mice at 8 hours following the hydrodynamic injection of 100 μg synthetic HCV RNA genome plus 1000 IU IFN-α. The livers were harvested following systemic PBS perfusion to remove contaminating blood. qRT-PCR detection of (D) IFN-β mRNA and (E) ISG56 mRNA (fold increase). (F) Gene expression following HCV RNA plus IFN-α challenge, detected by western blot. Error bars represent SEM.
Figure 5.
Detection of intrahepatic (A) IFN-β mRNA fold increase and (B) ISG56 mRNA fold increase by qRT-PCR, 5 hours post-i.v. injection with 10,000 IU IFN-β. (C) Gene expression following IFN-β injection, detected by western blot. Error bars represent SEM.
Figure 6.
(A) Correlation of IFN-α/β ratio and the intrahepatic steady-state NS3. The IFN-α and -β levels were analyzed by ELISA (pg/ml). The NS3 detectable group included 8 mice, 6 challenged with recombinant VSV and 2 challenged with poly(I:C). The NS3 undetectable group included 12 mice, 3 challenged with HCV RNA, 4 challenged with poly(I:C), 2 challenged with poly(I:C)/LyoVec and 3 challenged with AdLacZ. Error bars represent SEM. (B) Detection of autophagy marker LC3B by western blot post-IFN-α and IFN-β challenge. LC3B-II/LC3B-I fold/median ratios were calculated after normalizing each sample band to the respective β-actin band. Detection of Cre by western blot. (C) Correlation of intrahepatic NS3 degradation with the level of autophagic flux following injection of 0.08 mg/kg of chloroquine alone, as well as coupled with 10,000 IU of IFN-β by western blot. LC3B-II/LC3B-I, fold/median ratios were calculated after normalizing each sample band to the respective β-actin band. (D) Detection of HCV core protein by western blot in the liver of a transgenic mouse line (SL-139) following hydrodynamic challenge with HCV RNA and i.v. challenge with IFN-β.
IFN-β-Mediated Degradation of NS3
To examine whether the suppression of NS3 was due to regulation of the transgene expression, we first tested the levels of NS3 mRNA in the double transgenic mouse livers with qRT-PCR following challenge with IFN-α and -β, but found no significant alterations in the transcript levels (Supplementary Fig. 5A). Since NS3/4A has a half-life of approximately 26 hours in cell culture,20 we reasoned that the repression of NS3 was not due to a block of de novo protein synthesis, but rather to its rapid degradation. Type I IFN can stimulate the generation of immunoproteasomes in chimpanzee livers as early as 2 weeks following HCV infection.24 To test whether NS3 degradation in our double transgenic mice resulted from an accelerated protein turnover through this immunoproteasomal pathway, we injected the double transgenic mice i.v. with a proteasomal inhibitor, bortezomib (1 mg/kg),25 1 hour before IFN-β challenge. Western blot analyses demonstrated no detectable NS3, even in the presence of proteasomal blockade (Supplementary Fig. 5B), thereby ruling out the possibility of immunoproteasome-mediated NS3 degradation. We further showed that there was no differential induction of ISG15 by IFN-α and -β by western blot, and thus excluded the possible involvement of ISGylation in this NS3 repression (Supplementary Fig. 5C).
The sequestration of subcellular components such as ER and mitochondria for autophagic degradation in hepatocytes has been well documented.26 Since HCV NS3/4A co-localizes with these organelles, we examined LC3B, a functional reporter of autophagy, in the liver lysates of the IFN-challenged mice. LC3B is cleaved at its C terminus to form LC3B-I. Upon induction of autophagy, LC3B-I is covalently conjugated to phosphatidylethanolamine to form LC3B-II. LC3B-II is localized on both the luminal and cytosolic surfaces of autophagosomes. While the LC3B-II on the luminal surface is degraded in the autolysosome, that on the cytosolic surface is delipidated and recycled.27 Western blot analysis showed that IFN-α challenge resulted in increased LC3B-II levels in the double transgenic mice (Fig. 6B), a finding which suggests the accumulation of LC3B-II and upregulation of autophagy. On the other hand, IFN-β challenge resulted in a decrease in both LC3B-I and -II levels (Fig. 6B). As the accelerated degradation of the luminal LC3B-II would result in an overall deficit of the cellular LC3B pool, the reduced LC3B-I and -II levels were indicative of an upregulation of autophagy, as well as an enhanced autolysosomal proteolysis. A differential induction of autophagy by IFN-α and -β pointed to the possible involvement of this mechanism in the degradation of NS3.
To ascertain if the enhancement of autophagic proteolysis by IFN-β was indeed responsible for the repression of NS3, we injected mice i.p. with the autophagolysosome inhibitor chloroquine (0.08 mg/kg) 1 hour prior to IFN-β challenge. Chloroquine inhibits lysosomal degradation of autophagosomes by raising lysosomal pH to suppress the activity of lysosomal acid hydrolases.28 As expected, the blockage of autophagic proteolysis prevented the IFN-β-elicited repression of NS3 in the double transgenic liver tissues (Fig. 6C). Collectively, these results suggested that an accelerated autophagic proteolysis triggered by an IFN-β response resulted in the rapid degradation of NS3. To verify that the IFN-β-mediated degradation of NS3 was not due to its ectopic expression, we examined the cytosolic protein, Cre, in IFN-α- and -β-treated, double transgenic liver tissues by western blot. Interestingly, the Cre protein was consistently detectable in these livers following IFN-β challenge (Fig. 6B). In contrast, when an HCV structural transgenic mouse line (SL-139)29 was challenged with either HCV RNA or IFN-β, HCV core protein was repressed (Fig. 6D). These results indicated that only ER and/or mitochondria-associated proteins were targeted by the IFN-induced autophagic pathways in mouse liver.
Differential sequestration of NS3 by type I IFNs
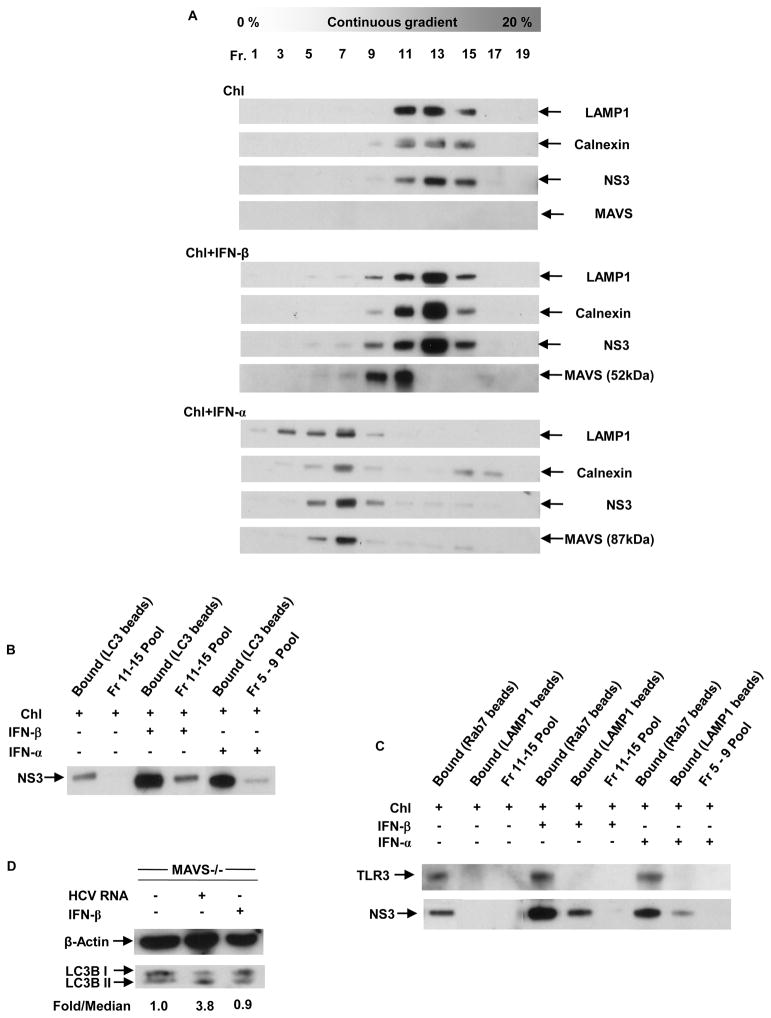
Though IFN-β could induce degradation of NS3 through the upregulation of autophagy, it was still not known whether this mechanism involved the direct degradation of the viral proteins in autolysosomes or the removal of an essential host-derived stabilization factor. To address this issue, we isolated the vesicular organelles from chloroquine pre-injected normal livers, as well as from IFN-α- and β-challenged, double transgenic livers by iodixanol density gradient centrifugation. Every alternate fraction of the gradient was analyzed by western blotting for NS3, the late-endosome/lysosome marker LAMP1, the ER marker calnexin and MAVS (Fig. 7A). While all proteins analyzed lined up in similar fractions in individual livers, they were present in relatively heavier fractions in the case of normal and IFN-β-treated livers as compared to those from IFN-α-treated livers. This suggested a differential sequestration of NS3 by the two type I IFNs. To confirm the autophagic sequestration of NS3, we isolated the autophagosomes from the NS3-positive fractions by immunoprecipitation with an anti-LC3 antibody. Immunoblot analyses revealed that NS3 was indeed sequestered in autophagosomes and that both type I IFNs upregulated the autophagic sequestration of NS3 (Fig. 7B). To determine autophagosome-lysosome fusion, we first immunoprecipitated the NS3-positive fractions with an antibody against the GTPase Rab7, followed by immunoprecipitation with anti-LAMP1 (Fig. 7C). Since LAMP1 is common to late-endosomes and lysosomes, the Rab7 pre-cleanup was carried out to exclude the late-endosome and/or amphisomes (fused autophagosomes and endosomes) from these fractions. This sequential pull-down revealed that only the IFN-β-induced autophagosomes were being directed to the lysosome for degradation. More importantly, the presence of IFN-α-induced vesicles in relatively lighter fractions suggests that these vesicles could be trafficked away from the lysosomes, as lysosomal vesicles are known to behave as dense organelles following ultracentrifugation on a variety of density gradients.30
Figure 7.
(A) Vesicular organelle fractionation on iodixanol gradient. Light mitochondrial pellets from liver homogenates of chloroquine pre-injected normal and IFN-α- and β-challenged, dTg mice were mixed with 20% iodixanol, and 20 fractions were recovered by gradient ultracentrifugation. The distribution profiles of LAMP1, calnexin, NS3 and MAVS were examined by western blot analysis of equal volume aliquots of the indicated fractions. (B) Sequestration of NS3 in autophagosomes was examined by LC3B pull-down. Aliquots (50-μl) of all NS3-positive iodixanol fractions from individual livers were pooled and immunoprecipitated with an anti-LC3B antibody, followed by immunoblot detection with anti-NS3. (C) Fusion of NS3-containing vesicles with lysosomes was examined by sequential Rab7 and LAMP1 pull-down. Aliquots (50-μl) of all NS3-positive iodixanol fractions from individual livers were pooled and immunoprecipitated first with an anti-Rab7 antibody, followed by an anti-LAMP1 antibody and immunoblot detection with anti-NS3 and anti-TLR3. (D) Detection of autophagy marker LC3B in MAVS−/− mice by western blot following hydrodynamic challenge with HCV RNA and i.v. challenge with IFN-β. LC3B-II/LC3B-I fold/median ratios were calculated after normalizing each sample band to the respective β-actin band.
Role of MAVS in type I IFN-induced autophagic response
Interestingly, different isoforms of MAVS were detected in the NS3-positive vesicular fractions from IFN-α- and -β-challenged livers (Fig. 7A). While only the lower molecular weight isoform of MAVS was seen in the fractions from an IFN-β-treated liver, the NS3-positive fractions from the IFN-α-treated livers showed the presence of only the higher molecular weight isoform of MAVS. No MAVS was detected in the absence of IFNs. To examine the role of MAVS in the IFN-induced autophagic response, we analyzed the LC3B in MAVS−/− livers post-challenge with HCV RNA and IFN-β. HCV RNA induced the accumulation of LC3B-II, suggesting the upregulation of autophagy. On the other hand, IFN-β challenge did not cause any change in the level of cellular autophagy in the absence of MAVS (Fig. 7D). As HCV RNA is known to induce incomplete autophagy at the surface of the ER,14 these results indicate that the type I IFNs induce an independent autophagic response at the surface of the mitochondria, which involves different isoforms of MAVS. Additionally, the IFN-induced autophagic response steers the cellular autophagosomes (including those induced by viral RNA) towards the endosomal compartments (Fig. 7C). Thus, these results implicate a possible role of MAVS in delivering viral components to TLR3-containing endosomes, which possibly results in a second wave of TLR3-mediated IFN-β response. Since the murine homologue of TRIF lacks the HCV NS3/4A cleavage site,7 a second wave of TLR3 signaling would explain the absence of significant compromise of the IFN-β response in the double transgenic mice upon viral RNA challenge, in spite of NS3-4A-mediated MAVS cleavage.
DISCUSSION
Through co-evolution with its host, HCV has developed several strategies to escape from the host’s immune system.4, 5 Convincing evidence has demonstrated that HCV NS3/4A serine protease can cleave the RIG-I signaling adaptor MAVS and compromise type I IFN and ISG responses in virus-infected cells.6, 8, 10, 21, 22 However, the precise role of this mechanism in intra-hepatic immune regulation remains unclear. In this study, we generated a transgenic mouse with liver-specific expression of the HCV NS3/4A and used this model to examine the effect of the viral protease on antiviral responses in vivo. We found no evidence of patent immunosuppression or a delay of viral clearance despite NS3/4A-mediated MAVS cleavage in a considerable cell population in the liver (Fig. 1). The absence of demonstrable immunosuppression cannot be attributed to the lack of a need for effective innate immune responses to clear VSV, since the virus persisted in the livers of MAVS−/− mice at 5 hours post-infection (Fig. 2). More interestingly, IFNR−/− animals that produced high levels of IFN-β without detectable ISG responses also failed to clear VSV (Fig. 2). These data demonstrated that an initial IFN-β induction is insufficient to clear VSV, as IFN-β effector functions are entirely dependent on the autocrine/paracrine positive feedback loop.
Estimates of the proportion of infected hepatocytes vary widely in hepatitis C patients, ranging from less than 10% to almost 50%.31 The intrahepatic distribution of HCV RNA was seen to be focal and correlated with the IFN-β induction.32 However, due to involvement of the IFN-positive feedback loop in the neighboring and non-parenchymal cells, ISG induction has often been seen across the liver.9, 32, 33 During the course of this study, we observed that the NS3/4A-expressing mice, when challenged, showed a robust ISG induction, which was not necessarily proportional to their relative IFN-β induction (Figs. 2–5). Therefore, the activation of the IFN-positive feedback pathway could partially compensate for NS3-mediated MAVS cleavage in the livers of these animals. These data are consistent with the observation that HCV infection results in various levels of nuclear translocation of IRF-3 and a heightened antiviral state in immortalized human hepatocytes.34 In patients, MAVS cleavage and suppression of IFN response in the infected cells do not seem to correlate with the ability of the virus to establish chronic infection.17
Although IFN-α and -β share many physicochemical and pharmacological properties, they differ in their regulation and biological effects during viral infections.35, 36 IFN-β is induced mainly through immediate early kinetics by activation of the constitutively expressed IRF-3 protein. On the other hand, most members of the IFN-α family are induced via IRF-7 and upregulated through the IFN-positive feedback loop.5 Thus, potent expression of IFN-α occurs only after the primary induction of IFN-β in virus-infected cells, and the immunomodulatory activities of IFN-β are dependent on its upregulation of IFN-α, pointing to a synergistic effect of the two. In this study, we found that the two type I IFNs mediate a disparate sequestration and trafficking of ER and/or mitochondria-associated viral proteins through an autophagic mechanism in vivo. Interestingly, while IFN-β favored autolysomal degradation of the sequestered proteins, IFN-α steered (Fig. 4F) the vesicular compartments away from the lysosomes. More importantly, the two isoforms of MAVS may play a role in these differential type I IFN-induced autophagic responses. This mechanism could be highly relevant in HCV infection, as the virus life cycle is closely associated with these organelles and involves vesicular compartmentalization of viral components.37 Collectively, the current data support the view that a balance between the IFN-α and -β levels is crucial for efficient anti-HCV host defense, as these two IFNs can play distinctive roles in virus clearance. Interestingly, in a study involving 5 chimpanzees infected with HCV, 3 spontaneous resolvers all mounted robust IFN-α and -β responses in the liver up to 28 weeks post-infection.24
Owing to the levels and percentages of NS3 expression in the hepatocytes of our double transgenic mice, some MAVS molecules perhaps were not cleaved in the liver (Fig. 1). Earlier, MAVS has been shown to interact with the signaling molecules, TRIF and TRAF6, and these interactions are critical for TLR3-mediated type I IFN and NFκB signaling.39 It has also been suggested that while TLR3 plays an important role in systemic responses to RNA viral infection, the first wave of type I IFN production through cell-autonomous recognition of viral infection is driven by the RIG-I-MAVS pathway.22, 38 Due to the absence of an HCV-permissive cell line containing a functional TLR3-dependent pathway, it has been difficult to evaluate the role of TLR3 in restricting HCV infection. Our results lead us to suggest that the IFN-induced autophagy can facilitate the trafficking of viral components to the endosomal compartments. This mechanism may allow for a TLR3-mediated IFN-β response. Since mouse TRIF is not susceptible to HCV NS3/4A cleavage,7 this TLR3 signaling would explain the absence of a significant compromise of the IFN-β response in the double transgenic mice upon viral RNA challenge, despite the efficient cleavage of MAVS. On the other hand, in the case of human cells, the simultaneous cleavage of MAVS and TRIF by the HCV protease is likely to modulate at least the IFN-β level within the virus infected cell, if not the global ISG response in the liver. This is likely to result in a specific compromise of the anti-HCV host defense as the cellular PRRs have access to HCV RNA only within the milieu of the infected cell. Conversely, in a case like the hepatitis A virus, which can also mediate both MAVS and TRIF cleavages,4 the viral RNA released at the time of an infectious burst can engage TLR3 on the surface of uninfected cells in the tissue leading to activation of the IFN-β signaling response. Since the type I IFN-mediated autophagic responses may be altered in hepatoma cell lines,36 investigation of HCV replication in physiologic host cells is required to understand the impact of these phenomena in the course of HCV infection.
In summary, by using our HCV NS3/4A transgenic mouse model, we have demonstrated that MAVS cleavage alone does not compromise global antiviral response in the liver and that MAVS plays a role in type I IFN-mediated autophagic responses. More importantly, our data bring to light a unique IFN-β-mediated mechanism of anti-viral host defense. Further studies are underway to delineate the role of this mechanism in hepatitis C persistence and pathogenesis.
Supplementary Material
Acknowledgments
Grant Support: This work was supported by National Institutes of Health Grant AI069142 (to J.S.) and AI043003 (to L.S.). M.M.D. is a James W. McLaughlin Postdoctoral Fellow and a Hans Popper Postdoctoral Fellow of the American Liver Foundation.
We thank C. Rice for the HCV antiserum, Z. Chen for the MAVS−/− mice, J. Yan for assistance with the animal experiments, B. Gelman for Abs and technical suggestions, S. Lemon for the pHCV-N plasmid and helpful discussion, A. Brasier and M. Yi for their critical comments, and M. Susman for her editorial help on the manuscript.
Abbreviations used in this paper
- AdLacZ
recombinant Adenovirus LacZ
- COX I
cytochrome c oxidase subunit I
- dTg
double transgenic
- ER
endoplasmic reticulum
- HCV
hepatitis C virus
- IFN
interferon
- ISG
IFN-stimulated gene
- IFNR
IFN receptor
- IFNR−/−
IFN-α/β receptor knockout
- LC3B
microtubule-associated protein I light chain 3B
- RIG-I
retinoic acid-inducible gene I
- MAVS
mitochondria-associated anti-viral signaling protein
- PAMP
pathogen-associated molecular pattern
- TLR3
Toll-like receptor 3
- TRIF
Toll/interleukin-1 receptor domain containing adaptor inducing IFN-β
- UTR
untranslated region
- VSV
vesicular stomatitis virus
Footnotes
Disclosures: The authors disclose no conflicts.
Author Contributions: M.M.D., T.C. and J.S. conceived the study, M.M.D., B.G. and A.A.K. performed experiments, M.M.D., R.A.D., L.S., T.C. and J.S. analyzed the data and M.M.D., T.C. and J.S. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alter MJ. Epidemiology of hepatitis C virus infection. World J Gastroenterol. 2007;13:2436–2441. doi: 10.3748/wjg.v13.i17.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horner SM, Gale M., Jr Intracellular innate immune cascades and interferon defenses that control hepatitis C virus. J Interferon Cytokine Res. 2009;29:489–498. doi: 10.1089/jir.2009.0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaitin DA, Roisman LC, Jaks E, et al. Inquiring into the differential action of interferons (IFNs): an IFN-alpha2 mutant with enhanced affinity to IFNAR1 is functionally similar to IFN-beta. Mol Cell Biol. 2006;26:1888–1897. doi: 10.1128/MCB.26.5.1888-1897.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lemon SM. Induction and evasion of innate antiviral responses by hepatitis C virus. J Biol Chem. 2010;285:22741–22747. doi: 10.1074/jbc.R109.099556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Foy E, Li K, Sumpter R, et al. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc Natl Acad Sci USA. 2005;102:2986–2991. doi: 10.1073/pnas.0408707102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li K, Foy E, Ferreon JC, et al. Immune evasion by hepatitis C virus NS3/4A protease-mediated cleavage of the Toll-like receptor 3 adaptor protein TRIF. Proc Natl Acad Sci USA. 2005;102:2992–2997. doi: 10.1073/pnas.0408824102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meylan E, Curran J, Hofmann K, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 8.Lau DT, Fish PM, Sinha M, et al. Interferon regulatory factor-3 activation, hepatic interferon-stimulated gene expression, and immune cell infiltration in hepatitis C virus patients. Hepatology. 2008;47:799–809. doi: 10.1002/hep.22076. [DOI] [PubMed] [Google Scholar]
- 9.Bellecave P, Sarasin-Filipowicz M, Donze O, et al. Cleavage of mitochondrial antiviral signaling protein in the liver of patients with chronic hepatitis C correlates with a reduced activation of the endogenous interferon system. Hepatology. 2010;51:1127–1136. doi: 10.1002/hep.23426. [DOI] [PubMed] [Google Scholar]
- 10.Levine B, Deretic V. Unveiling the roles of autophagy in innate and adaptive immunity. Nat Rev Immunol. 2007;7:767–777. doi: 10.1038/nri2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dreux M, Chisari FV. Viruses and the autophagy machinery. Cell Cycle. 2010;9:1295–1307. doi: 10.4161/cc.9.7.11109. [DOI] [PubMed] [Google Scholar]
- 12.Dreux M, Gastaminza P, Wieland SF, Chisari FV. The autophagy machinery is required to initiate hepatitis C virus replication. Proc Natl Acad Sci USA. 2009;106:14046–14051. doi: 10.1073/pnas.0907344106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sir D, Chen WL, Choi J, et al. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology. 2008;48:1054–1061. doi: 10.1002/hep.22464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jounai N, Takeshita F, Kobiyama K, et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci USA. 2007;104:14050–14055. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Honda M, Sakai A, Yamashita T, et al. Hepatic ISG expression is associated with genetic variation in interleukin 28B and the outcome of IFN therapy for chronic hepatitis C. Gastroenterology. 2010;139:499–509. doi: 10.1053/j.gastro.2010.04.049. [DOI] [PubMed] [Google Scholar]
- 16.Sarasin-Filipowicz M, Oakeley EJ, Duong FH, et al. Interferon signaling and treatment outcome in chronic hepatitis C. Proc Natl Acad Sci USA. 2008;105:7034–7039. doi: 10.1073/pnas.0707882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Postic C, Shiota M, Niswender KD, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic beta cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274:305–315. doi: 10.1074/jbc.274.1.305. [DOI] [PubMed] [Google Scholar]
- 18.Le Y, Gagneten S, Tombaccini D, et al. Nuclear targeting determinants of the phage P1 cre DNA recombinase. Nucleic Acids Res. 1999;27:4703–4709. doi: 10.1093/nar/27.24.4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolk B, Sansonno D, Krausslich HG, et al. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline-regulated cell lines. J Virol. 2000;74:2293–2304. doi: 10.1128/jvi.74.5.2293-2304.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li XD, Sun L, Seth RB, et al. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci USA. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Q, Sun L, Liu H-H, et al. The specific and essential role of MAVS in antiviral Innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 22.Carney DS, Gale JM. HCV regulation of host defense. In: Tan S-T, editor. Hepatitis C virus: Genomes and molecular biology. Norfolk, UK: Horizon Biosciences; 2006. pp. 375–398. [PubMed] [Google Scholar]
- 23.Saito T, Owen DM, Jiang F, et al. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454:523–527. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shin EC, Seifert U, Kato T, et al. Virus-induced type I IFN stimulates generation of immunoproteasomes at the site of infection. J Clin Invest. 2006;116:3006–3014. doi: 10.1172/JCI29832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williamson MJ, Silva MD, Terkelsen J, et al. The relationship among tumor architecture, pharmacokinetics, pharmacodynamics, and efficacy of bortezomib in mouse xenograft models. Mol Cancer Ther. 2009;8:3234–3243. doi: 10.1158/1535-7163.MCT-09-0239. [DOI] [PubMed] [Google Scholar]
- 26.Yin XM, Ding WX, Gao W. Autophagy in the liver. Hepatology. 2008;47:1773–1785. doi: 10.1002/hep.22146. [DOI] [PubMed] [Google Scholar]
- 27.Rubinsztein DC, Cuervo AM, Ravikumar B, et al. In search of an “autophagomometer”. Autophagy. 2009;5:585–589. doi: 10.4161/auto.5.5.8823. [DOI] [PubMed] [Google Scholar]
- 28.Jiang M, Liu K, Luo J, Dong Z. Autophagy is a renoprotective mechanism during in vitro hypoxia and in vivo ischemia-reperfusion injury. Am J Pathol. 2010;176:1181–1192. doi: 10.2353/ajpath.2010.090594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korenaga M, Wang T, Li Y, et al. Hepatitis C virus core protein inhibits mitochondrial electron transport and increases reactive oxygen species (ROS) production. J Biol Chem. 2005;280:37481–37488. doi: 10.1074/jbc.M506412200. [DOI] [PubMed] [Google Scholar]
- 30.Luzio JP, Pryor PR, Bright NA. Lysosomes: fusion and function. Nat Rev Mol Cell Biol. 2007;8:622–632. doi: 10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- 31.Bigger CB, Brasky KM, Lanford RE. DNA microarray analysis of chimpanzee liver during acute resolving hepatitis C virus infection. J Virol. 2001;75:7059–7066. doi: 10.1128/JVI.75.15.7059-7066.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stiffler JD, Nguyen M, Sohn JA, et al. Focal distribution of hepatitis C virus RNA in infected livers. PLoS One. 2009;4:e6661. doi: 10.1371/journal.pone.0006661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacQuillan GC, Mamotte C, Reed WD, et al. Upregulation of endogenous intrahepatic interferon stimulated genes during chronic hepatitis C virus infection. J Med Virol. 2003;70:219–227. doi: 10.1002/jmv.10381. [DOI] [PubMed] [Google Scholar]
- 34.Kanda T, Steele R, Ray R, Ray RB. Hepatitis C virus infection induces the beta interferon signaling pathway in immortalized human hepatocytes. J Virol. 2007;81:12375–12381. doi: 10.1128/JVI.01695-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Der SD, Zhou A, Williams BRG, Silverman RH. Identification of genes differentially regulated by interferon α, β, or γ using oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsumoto K, Okano J, Murawaki Y. Differential effects of interferon alpha-2b and beta on the signaling pathways in human liver cancer cells. J Gastroenterol. 2005;40:722–732. doi: 10.1007/s00535-005-1616-x. [DOI] [PubMed] [Google Scholar]
- 37.Targett-Adams P, Boulant S, McLauchlan J. Visualization of double-stranded RNA in cells supporting hepatitis C virus RNA replication. J Virol. 2008;82:2182–2195. doi: 10.1128/JVI.01565-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu LG, Wang YY, Han KJ, et al. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.