Abstract
Objective
The low prevalence of ovarian cancer demands both high sensitivity (>75%) and specificity (99.6%) to achieve a positive predictive value of 10% for successful early detection. Utilizing a two stage strategy where serum marker(s) prompt the performance of transvaginal sonography (TVS) in a limited number (2%) of women could reduce the requisite specificity for serum markers to 98%. We have attempted to improve sensitivity by combining CA125 with proteomic markers.
Methods
Sera from 41 patients with early stage (I/II) and 51 with late stage (III/IV) epithelial ovarian cancer, 40 with benign disease and 99 healthy individuals, were analyzed to measure 7 proteins [Apolipoprotein A1 (Apo-A1), truncated transthyretin (TT), transferrin, hepcidin, ß-2-microglobulin (ß2M), Connective Tissue Activating Protein III (CTAPIII), and Inter-alpha-trypsin inhibitor heavy chain 4 (ITIH4)]. Statistical models were fit by logistic regression, followed by optimization of factors retained in the models determined by optimizing the Akaike Information Criterion. A validation set included 136 stage I ovarian cancers, 140 benign pelvic masses and 174 healthy controls.
Results
In a training set analysis, the 3 most effective biomarkers (Apo-A1, TT and CTAPIII) exhibited 54% sensitivity at 98% specificity, CA125 alone produced 68% sensitivity and the combination increased sensitivity to 88%. In a validation set, the marker panel plus CA125 produced a sensitivity of 84% at 98% specificity (P= 0.015, McNemar's test).
Conclusion
Combining a panel of proteomic markers with CA125 could provide a first step in a sequential two-stage strategy with TVS for early detection of ovarian cancer.
INTRODUCTION
Over the last two decades, improvements in cytoreductive surgery and combination chemotherapy have increased median survival for patients with ovarian cancer, but have not affected the rate of cure. Long term survival depends critically on the stage of disease at diagnosis. Advanced epithelial ovarian cancer in stage III or IV has a poor prognosis with long-term survival obtained in less than 30% of patients [1]. When cancer is limited to the ovaries in stage I, up to 90% of patients can be cured with currently available therapy. However, only 20% of ovarian cancers are currently diagnosed in stage I. Given the prevalence of ovarian cancer in the general population (1 in 2,500 for sporadic ovarian cancer in the postmenopausal population at greatest risk), a screening strategy must have a sensitivity for early stage disease of at least 75% and a specificity of 99.6% to achieve a positive predictive value of 10%, i.e, 10 operations for each case of ovarian cancer detected.
Attempts to detect ovarian cancer at an early stage have utilized serum biomarkers, alone or in combination with transvaginal sonography. Of the serum biomarkers, CA125 has been studied most extensively [2,3]. CA125 is a mucin encoded by the MUC16 gene expressed in 80% of epithelial ovarian cancers [4]. CA125 is shed from ovarian cancer cells and levels can rise exponentially 10 to 21 months before diagnosis, but detects only 50%- 60% of early stage disease. Although CA125 has a specificity of 99% in the postmenopausal population, it does not achieve the required 99.6%. In a premenopausal population, the specificity is further compromised by the presence of endometriosis, adenomyosis and retrograde menstruation that can elevate antigen levels.
Studies have suggested that adequate specificity can be attained to screen a post-menopausal population at average risk using a two step strategy, where rising values of serum marker(s) prompt transvaginal sonography (TVS) in a small fraction of women. An algorithm has been developed to identify significant changes from year to year [5]. In the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS) in the United Kingdom, 50,078 women have been screened with annual determinations of CA125 using this approach [6]. During the first two years of study, CA125 followed by TVS produced a sensitivity of 89.5%, a specificity of 99.8% and a positive predictive value of 35%. In the United States, a similar screening trial has involved 3,252 postmenopausal women followed annually with CA125 using the same algorithm coordinated by the Ovarian SPORE at M.D. Anderson Cancer Center [7]. Eight operations were performed and 5 ovarian cancers detected – three invasive and two borderline – all in early stage (I/II). Overall, the positive predictive value for the entire screen was 37%, consistent with the outcome in the U.K.
The two stage strategy will be limited, as 20% of ovarian cancers do not express CA125; thus, multiple markers may be required to optimize sensitivity. In one recent study, 96 putative biomarkers were assayed in sera from healthy postmenopausal individuals and from women with early stage (I/II) and late stage (III/IV) ovarian cancer using a multiplex platform [8]. A panel of 4 biomarkers – CA125, HE4, CEA and sVCAM1 – produced a sensitivity of 86% for early stage (I/II) and 95% for late stage (III/IV) at 98% specificity, the value required for cost-effective referral to TVS. Consequently, it appears that multiple biomarkers can improve upon the sensitivity of CA125 alone, while maintaining requisite specificity.
Proteomic methods have been utilized to search for new biomarkers that can detect early stage ovarian cancer [9]. Our group has evaluated 7 proteomic biomarkers that were previously identified with SELDI-TOF-MS to distinguish malignant from benign pelvic masses.
MATERIALS AND METHODS
Human specimens
Using IRB approved protocols and having obtained informed consent, blood was drawn pre-operatively from patients undergoing surgery. Benign or malignant pelvic masses were diagnosed and staged (if malignant) by board certified gynecologic oncologists. Normal samples were drawn from healthy postmenopausal women during office visits to participate in a screening trial to detect early stage ovarian cancer [7]. Serum was kept on ice, separated promptly on the same day and stored at -80°C until assay. A training set from the MDACC sample bank consisted of available sera from 41 patients with early stage ovarian cancer (22 stage I and 19 stage II), 51 with late stage (III/IV) ovarian cancer, 40 with benign disease and 99 healthy individuals (Table 1A). The histology of early stage samples was 39% serous, 10% mucinous (muc), 20% endometrioid and 31% mixed (endo, serous, muc). Late stage histotypes were 73% serous, 10% endometrioid and 17% mixed. A blinded test set contained serum samples from the GOG consisting of 136 patients with stage I ovarian cancers, 140 patients with benign pelvic masses and 50 healthy controls. The histology of GOG cancer samples was 18% serous, 24% mucinus, 42% endometrioid, 15% clear cell and 1% unknown. This set was supplemented with 124 healthy controls (also blinded) from the MDACC sample bank (Table 1B).
Table 1.
Number of patient serum samples analyzed per group. A) Samples from the MD Anderson Cancer Center (MDACC) tissue bank used in the training set analysis. B) Samples from the GOG used in the blinded test set analysis. The GOG set was supplemented with normal samples from MDACC.
| A) Training Set |
B) Validation Set |
|||||
|---|---|---|---|---|---|---|
| Age | Age | |||||
| Sample # | Median | Range | Sample # | Median | Range | |
| Stage I | 22 | 56 | 20-87 | 136 | 54 | 15-87 |
| Stage II | 19 | 60 | 41-88 | ~ | ~ | ~ |
| Stage III | 41 | 59 | 31-85 | ~ | ~ | ~ |
| Stage IV | 10 | 59 | 32-83 | ~ | ~ | ~ |
| Benign | 40 | 48 | 26-80 | 140 | 50 | 19-85 |
| Normal (MDA) | 99 | 62 | 51-77 | 124 | 61 | 51-77 |
| Normal (GOG) | 0 | ~ | ~ | 50 | 44 | 25-65 |
Biomarker assays
CA125II measurements were performed at MDACC with a Roche immunoassay. SELDI-TOF-MS protocols were followed to measure a truncated form of transthyretin (TT), apolipoprotein A1 (Apo-A1), transferrin, hepcidin, ß-2-microglobulin (ß2M), Connective Tissue Activating Peptide III (CTAPIII), and a cleavage fragment of inter-alpha-trypsin inhibitor heavy chain 4 (ITIH4). A Biomek 2000 and Tecan robot were used for liquid handling of samples and reagents. Three types of ProteinChip arrays and four assay conditions were used to analyze the seven proteins. ITIH4, HepC and ß2 microglobulin were captured on copper chelated IMAC50 arrays (HIB protocol); CTAPIII and transferrin on copper chelated IMAC50 arrays (CTAP, Tr protocol ); ApoA1 on hydrophobic retention H50 arrays and TT on anionic exchange Q10 arrays. E. coli celllysate (10mg/mL) was used as an exogenous complex protein matrix and was added to the CTAPIII, TT and ApoA1 specimen diluent buffers prior to the dilution of the specimen. This provided consistent analyte binding to the ProteinChip arrays and also acted as an internal standard for ion intensity normalization. Detailed array preparationis provided in the supplemental materials.
Data management
Data was collected using ProteinChip Data Manager Software and a mass reader (PCS Series 4000, Enterprise Edition, mass spectrometer). The average mass for each spectrum peak analyzed was: ITIH4 at 3273da, hepcidin at 2790da, B2M at 11731da, CTAPIII at 9289da, transferrin at 79908da, ApoA1 at 28080da and the average of two TT peaks of 13841da (unmodified), and 13880da (cysteinylated).
Statistical analysis
Method A
Statistical analysis was based on peak intensity after filtering, baseline correction, and normalization of each spectrum to average ion current. Using this normalized peak intensity data from the training samples, logistic regression models were built using six different training sets (normal vs stage I (N v I); normal vs stage I and II (N v E); normal vs all stages, (N v C); normal and benign vs stage I (NB v I); normal and benign vs stage I and II (NB v E); normal and benign vs all stages (NB v C)). This was followed by a stepwise optimization of factors retained in the models determined by utilizing the Akaike Information Criterion to eliminate redundant variables. We refit models and ROC curves from 1000 bootstrap samples of the training data in order to compute empirical p-values to test the hypothesis that the ROC curve is better than (i.e., closer to the upper left-hand corner) the performance of CA125 alone with a fixed cutoff. Each logistic regression model uses the normalized peak intensities of the proteins to calculate a posterior probability that these values arose from a cancer sample. In order to produce the ROC curves, we varied the threshold that this posterior probability must exceed in order to call a sample cancer rather than normal. In order to get a binary prediction, we used the threshold that corresponded to 98% specificity for detecting early stage ovarian cancer on the training data. These optimized models using three proteins were used to analyze the blinded GOG test sample set.
Method B
To reduce the potential of model overfitting resulting in poor generalization performance on independent data, we chose to utilize a simple linear prediction model derived using linear discriminant analysis (LDA) with bootstrap. The actual training sample set included only stage I ovarian cancer cases and healthy controls from the training set (N=121). Derivation of models used the seven biomarkers only and seven biomarkers plus CA125. Briefly, in each bootstrap run, the training set was split 60%/40% with randomization into two subsets. The larger subset was used to derive an LDA classifier and then tested on the two subsets. If the areas-under-curve in receiver-operating-characteristic (ROC) curve analysis were greater than 0.85 for both subsets and the sensitivities at a fixed 98% specificity were greater than 70% for both subsets, the corresponding LDA would be saved. The bootstrap procedure was repeated until a fixed number of “qualified” LDAs had been reached. A final linear prediction model was determined by averaging the saved LDAs.
RESULTS
Three proteomic markers enhance the sensitivity of CA125 for detecting early stage ovarian cancer
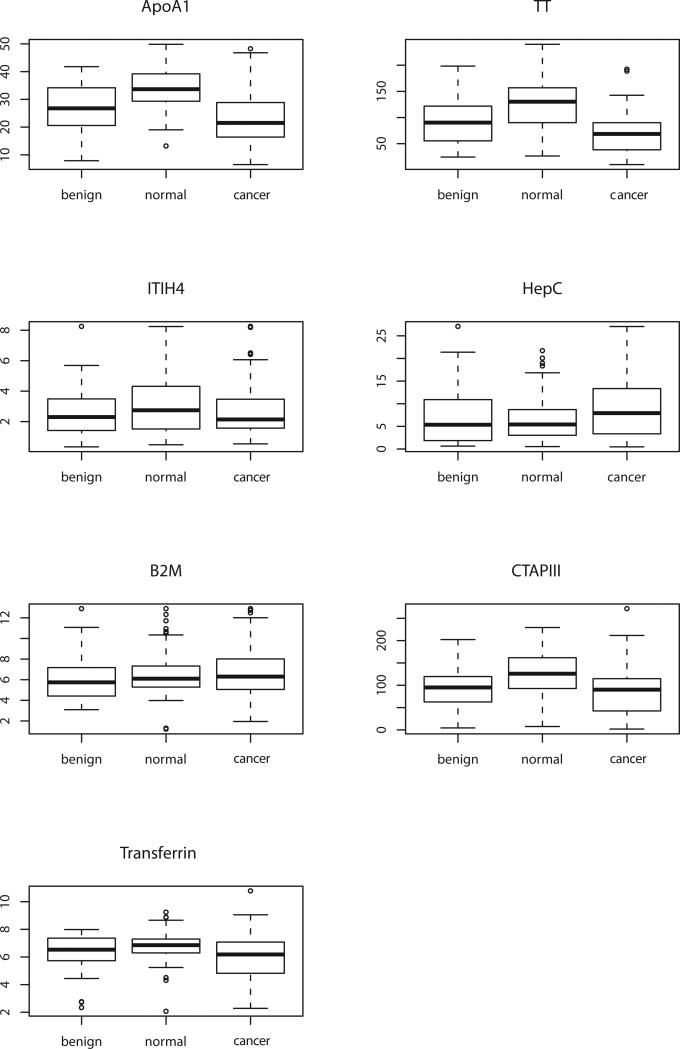
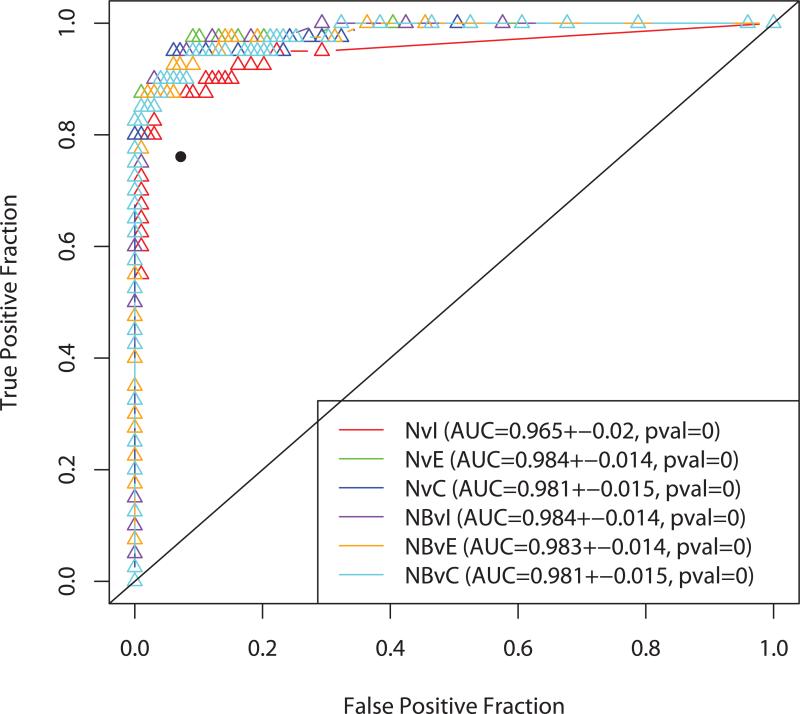
Levels of the seven biomarkers (Apo-A1, TT, transferrin, hepcidin, ß2M, CTAPIII and ITIH4) were measured in a training set including 41 patients with stage I/II epithelial ovarian cancer, 40 with benign disease and 99 healthyindividuals (Figure 1). Only 4 of the 7 biomarkers (Apo-A1, TT, CTAPIII and transferrin) distinguished patients with early stage ovarian cancer from healthy individuals using both one way analysis of variance and a Wilcoxon test. In each case, levels in sera from ovarian cancer patients were lower than in healthy controls. Although four of the seven markers (TT, ApoA1, ITIH4, and transferrin) were significantly different in normal versus stage I disease, three biomarkers (TT, CTAPIII and Apo-A1) were sufficient for maximum separation between both normal (or normal and benign) and early stage (I - II) or all stages (I – IV) of disease. We calculated the sensitivity of the models, estimated from the normal controls and early cancer samples in the training data at 98% specificity (Table 2). CA125 (>35 U/mL) alone distinguished early stage (I-II) patients from healthy women with 68% sensitivity and the three biomarkers alone (Apo-A1, TT and CTAPIII) exhibited 54% sensitivity. However, when CA125 was combined with the proteomic biomarkers (Apo-A1, TT and CTAPIII) the combination reached a sensitivity of 88% (Table 2). The ROC curves were essentially equivalent for all six models where CA125 alone (dichotomized at 35 U/ml) had about the same performance as all models just using the serum markers. Figure 2 contains ROC curves for the three proteomic markers + CA125 and shows that adding the markers to CA125 has improved sensitivity.
Figure 1.
Box plots for levels of seven proteomic biomarkers in sera from women with benign pelvic masses, healthy individuals and patients with early stage epithelial ovarian cancer. One-way analysis of variance indicates that levels of Apo-A1 (P<0.00001), TT (P<0.00001), CTAPIII (P<0.0001), transferrin (P=0.00015) and hepcidin (P=0.00172) are significantly lower in ovarian cancer patients than in controls. With Wilcoxon analysis, only Apo-A1, TT, CTAPIII and transferrin were significant (P<0.05).
Table 2.
Sensitivity at 98% specificity of models using proteomic markers with or without CA125- Model A analysis. N v I marker panel consists of TT, ApoA1, ITIH4 and transferrin. All other models, marker panel consists of TT, ApoA1 and CTAPIII. Models were fit using the training set data for normal samples (N) or normal and benign samples (NB) as controls and either stage I (I), early cancer (E; stages I and II), or all cancer samples (C) as cases.
| Model | With CA125 | Without CA125 |
|---|---|---|
| NvI | 0.80 | 0.51 |
| NvE | 0.88 | 0.54 |
| NvC | 0.85 | 0.39 |
| NBvI | 0.85 | 0.49 |
| NBvE | 0.85 | 0.59 |
| NBvC | 0.85 | 0.34 |
Figure 2.
Plots of the ROC curves for models trained on six subsets of the MD Anderson data. These six models did use CA125. The isolated black point is the performance of CA125 Alone. The p value was computed by applying the Wilcoxon-Mann-Whitney test to the AUC value (bootstrap empirical p-value = 0.038).
Using Method B, for the training samples (ovarian cancer and healthy control only), the multivariate models using seven biomarkers alone had a sensitivity of 63.4% CI (52.8-73.2%) at the fixed specificity of 98% CI (92.9-99.8%) and an ROC area-under-curve (AUC) of 0.93 CI (0.89-0.97). When CA125 was included in the multivariate model, at the same specificity, the sensitivity reached 86.0% CI (77.3-92.3%) and the ROC AUC improved to 0.97 CI (0.94-1.00).
CA125 and three proteomic biomarkers provide 84% sensitivity at 98% specificity for detecting stage I ovarian cancer in an independent validation set
The six models that were generated during analysis of the training sample set were applied to the blinded GOG data using Statistical Method A. The N v I model predicted substantially fewer cancers and more normals than CA125 alone (Table 3). This suggested that it was more conservative than CA125 and would have lower sensitivity with higher specificity. Because our cutoff was designed primarily to achieve the same specificity while improving the sensitivity, this observation strongly suggested that the N v I model focused on the wrong features. However, while there were slight differences in the other 5 models, the agreement between models in calling a sample normal or cancer was very high (predictions were identical for greater than 95% of unknown samples) and identified 50 to 60 more cancers than CA125 alone (Table 3). Since we wanted to increase sensitivity, we looked at the agreement between the five models considered to do so by counting, for each sample, the number of models that predicted that it was a cancer sample. In a histogram plotting agreement from 0 - 5 on the x axis, if the count is 0, then all models agree the sample is normal. If the result is 5, all models agree the sample is from a woman with cancer. This perfect agreement was achieved for 429 samples (230 called normal and 199 called cancer). Because the five models agreed on which markers to include and they agreed on the predictions for more than 95% of the validation samples, we chose to use the N v E model because 1) it only used healthy women and early stage (I or II) ovarian cancer samples to train the model, 2) the threshold for the posterior probability cutoff in this model was close to the 50% level, and is therefore easier to explain and 3) the performance on the training data predicted cancers in all stages even though it was only trained on the early stage cancers.
Table 3.
Comparison between model predictions incorporating proteomic markers and predictons based on the level of CA125.
| CA125 predictions | CA125 predictions | ||||
|---|---|---|---|---|---|
| N v I | Cancer | Normal | NB v I | Cancer | Normal |
| Cancer | 94 | 7 | Cancer | 150 | 60 |
| Normal | 62 | 286 | Normal | 6 | 233 |
| N v E | NB v E | ||||
| Cancer | 152 | 63 | Cancer | 151 | 58 |
| Normal | 4 | 230 | Normal | 5 | 235 |
| N v C | NB v C | ||||
| Cancer | 156 | 50 | Cancer | 156 | 55 |
| Normal | 0 | 243 | Normal | 0 | 238 |
Upon breaking the code, the results of the N v E model run on blinded GOG samples revealed that CA125 (>35U/ml) alone exhibited a sensitivity of 79% for predicting stage I cancer (Table 4A). Adding the proteomic markers to CA125 resulted in an increase in sensitivity to 88% at 95% specificity (Table 4A) and 84% sensitivity at 98% specificity (P= 0.015, McNemar's test) (Table 4B).
Table 4.
A. Performance of CA125 and proteomic markers on the Stage I Validation Set using Method A and 3 markers (TT, ApoA1, CTAPIII) or method B with 7 Biomarkers (TT, Apo-A1, CTAPIII, transferrin (Tfr), hepcidin, ß2M, and ITIH4). B. Model performance with Specificity set at 98% (p= 0.015).
| A. | |||
|---|---|---|---|
| Model | Cutoff | Sensitivity | Specificity |
| CA125 | 35 IU | 78.7% (107/136) | 95.4% (166/174) |
| Method A Markers Only | 0.896 | 56.6% (77/136) | 99.4% (173/174) |
| Method A Markers + CA125 | 0.672 | 88.2% (120/136) | 95.4% (166/174) |
| Method B Markers Only | 0.781 | 64.0% (87/136) | 97.1% (169/174) |
| Method B Markers+CA125 | 1.11 | 86.8% (118/136) | 93.7% (163/174) |
| B. | |||
|---|---|---|---|
| Model | Cutoff | Sensitivity | Significance (1-tailed p) |
| CA125 | 48 IU | 72.1% (98/136) | |
| Method A Markers + CA125 | 0.85 | 83.8% (114/136) | 0.0008 |
| Method B Markers+CA125 | 1.236 | 81.6% (111/136) | 0.0235 |
Using Statistical Method B, CA125 alone provided 72% (63.7-79.4%) sensitivity at 98% specificity (Table 4B). The 7 biomarkers alone provided 64% (52.5-68.9%) sensitivity and the combination 82% (74.1-87.7%) sensitivity. When ROC curves were drawn, CA125 yielded an AUC of 0.93 (0.90-0.96%), the 7 biomarkers 0.92 (0.89-0.95%) and the combination 0.97 (0.96-0.99%). The improvement in ROC AUC by adding the seven biomarkers to CA125 is statistically significant (p<0.05) over that of CA125 alone.
DISCUSSION
In this study, a combination of Apo-A1, TT, CTAPIII and CA125 achieved a sensitivity of up to 84% at a specificity of 98% to distinguish patients with early stage ovarian cancer from healthy individuals (Method A). No greater sensitivity was achieved using all 7 biomarkers and other analytical techniques (Method B). The sensitivity and specificity of this panel are comparable to results with a four biomarker panel selected from 96 candidate antigens measured by immunoassays with multiplex techniques [8]. The seven biomarkers initially evaluated in the present study (TT, Apo-A1, ß2M, Tfr, hepcidin, CTAPIII and ITIH4) have been used to distinguish benign from malignant pelvic masses [10]. Immunoassays for CA125 and four of these biomarkers (TT, Apo-A1, ß2M and transferrin) constitute the OVA1 test, recently approved by the United States Food and Drug Administration to triage patients with malignant pelvic masses for surgery by gynecologic oncologists. Immunoassays for three of the proteomic biomarkers (Apo-A1, TT and ITIH4) in combination with CA125 improved upon thesensitivity of CA125 alone for detecting early stage disease [11]. An independent study validated the utility of Apo-A1 and TT for distinguishing patients with ovarian cancer of all stages from patients with benign ovarian masses or gastrointestinal disease. A model that included age, CA125, Apo-A1 and 6 forms of post-translationally modified TT produced a sensitivity of 79% with a specificity of 94% [12]. In the present study, use of SELDI analysis, rather than immunoassay, and the substitution of CTAPIII for ITIH4increased sensitivity for early stage disease in the validation set to 84% at a higher specificity of 98%.
CTAPIII is a member of the β thromboglobulin family of proteins associated with platelet α-granules [13]. CTAPIII can be cleaved by cathepsin G to produce the biologically active chemokine β-thromboglobulin neutrophil-activating peptide 2 (NAP-2) which mediates chemotaxis, adherence and degranulation of neutrophils. One report suggests that CTAPIII can be found in normal cervical epithelial cells and expression is lost during malignant transformation [14]. In the present study, CTAPIII levels were decreased in early stage ovarian cancer. Similar observations have been made in pediatric patients with acute lymphoblastic leukemia [15]. The mechanism(s) responsible for dysregulation of CTAPIII remains unknown, but in the case of ovarian cancer it seems likely that decreased levels relate to the host response.
Down regulation of Apo-A1 and TT are also likely to be regulated by a hostresponse to cancer. In addition to the studies described above, decreased serum Apo-A1 levels have been observed in ovarian cancer patients in multiple reports [16,17]. Synthesized in the liver, TT has been used as a sensitive marker for malnutrition [18]. As a “visceral” protein, TT synthesis is downregulated when acute phase reactants are produced in response to acute or chronic illness. TT has not been detected by immunohistochemistry in ovarian cancers; serum levels decline with increasing severity of disease suggesting that TT may be negatively regulated by inflammation in ovarian cancer patients [18]. In contrast to CTAPIII, Apo-A1 and TT, CA125 is expressed and shed directly by ovarian cancers and elevated levels have been observed in 50-60% of patients with early stage disease [19]. In the present study, CA125 was elevated (>35 U/mL) in 68-79% of cases, a higher fraction than reported in the literature. As the training set was obtained at M.D. Anderson Cancer Center and the validation set from the Gynecologic Oncology Group, gynecologic oncologists operated on the patients in both groups. Consequently, inadequate staging is not likely to be the cause of a greater fraction of positive CA125 values.
While the improved sensitivity observed with a combination of CA125 and the three proteomic markers is encouraging, a critical question is whether the four biomarker panel can detect small volumes of pre-clinical disease prior to conventional diagnosis. Recent studies suggest that high grade cancers with typical ovarian histotypes can arise from the fallopian tube, endometriosis or the peritoneal cavity itself. Kurman and colleagues have described two classes of ovarian cancer: Type I tumors of low grade with Ras and Raf mutations that generally present in early stage; and Type II tumors of high grade with p53 mutations that metastasize early and present in an advanced stage [20]. The challenge is to detect Type II cancers before they have metastasized. In prophylactic oophorectomies in patients with germ line BRCA1/2 mutations, small cancers have been detected with p53 mutations. As p53 mutation has correlated with advanced stage, it has been assumed that very small tumors metastasize. While this is possible, the natural history of hereditary and sporadic ovarian cancers has not been well documented. If however, detecting Type II ovarian cancer at an early stage proves difficult, identifying women with small volume disease is a reasonable alternative, as these are the patients who benefit most from chemotherapy. Detection of type II cancers in a high risk population has been particularly difficult and use of multiple markers might improve sensitivity.
Supplementary Material
HIGHLIGHTS.
Eighty percent of women diagnosed with ovarian cancer already have late stage disease because the symptoms are common and are often associated with a wide variety of underlying causes.
If discovered in stage I, 90% of ovarian cancers can be treated successfully. To date, there are no suitable biomarkers for screening women at normal risk to detect early stage ovarian cancer.
A single CA125 measurement has been shown to be useful in determinations of recurrence but is not suitable for an early detection strategy.
This study tests a panel of protein markers found to be significantly different in the serum of normal subjects and women with benign gynecologic disease or ovarian cancer.
Finding new biomarkers or a combination of markers plus CA125 is essential for the development of a diagnostic assay that can detect curable stage I disease.
Acknowledgments
GRANT SUPPORT This work was supported by funds from the M.D. Anderson SPORE in Ovarian CancerNCI P50 CA83639, the Bioinformatics Shared Resources of the M.D. Anderson CCSGNCI P30 CA16672 and philanthropic support from Golfers Against Cancer, the Tracey JoWilson Foundation and the Mossy Foundation.
Footnotes
CONFLICT OF INTEREST Robert C Bast receives royalties for the discovery of CA125 and serves on the advisory board for Fujurebio Diagnostics, Vermillion, Inc and Illumina, Inc. Zhen Zhang received funding through a sponsored research agreement between JHCI and Vermillion, Inc and has IP associated with discovery of biomarkers discussed in manuscript. Eric Fung is an employee of Vermillion, Inc and is a shareholder. Christine Yip is an employee of Vermillion, Inc. and is a shareholder.
LITERATURE CITED
- 1.Berek JS, Friedlander ML, Bast RC., Jr . Ovarian Cancer. In: Hong WK, Bast RC Jr, Hait W, Kufe D, Pollock RE, Weichselbaum RR, Holland JF, Frei E III, editors. Holland-Frei Cancer Medicine. 8th Edition PMPH-USA; Shelton, CT: 2010. pp. 1344–1375. [Google Scholar]
- 2.Bast RC, Jr, Klug TL, St. John E, Jenison E, Niloff JM, Lazarus H, Berkowitz RS, Leavitt T, Griffiths CT, Parker L, Zurawski VR, Jr, Knapp RC. A radioimmunoassay using a monoclonal antibody to monitor the course of epithelial ovarian cancer. New Engl J Med. 1983;309:883–887. doi: 10.1056/NEJM198310133091503. [DOI] [PubMed] [Google Scholar]
- 3.Moore RG, McLaughlan S, Bast RC., Jr Current state of biomarker development for clinical application in epithelial ovarian cancer. Gynecol Oncol. 2010;116:240–5. doi: 10.1016/j.ygyno.2009.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosen D, Wang L, Atkinson JN, Yu Y, Lu K, Diamandis EP, Hellstrom I, Mok SC, Liu J, Bast RC., Jr Potential markers that complement expression of CA 125 in epithelial ovarian cancer. Gynecol Oncol. 2005;99:267–77. doi: 10.1016/j.ygyno.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 5.Skates SJ, Xu FJ, Yu YH, Sjovall K, Einhorn N, Chang Y, et al. Toward an optimal algorithm for ovarian cancer screening with longitudinal tumor markers. Cancer. 1995;76(10 Suppl):2004–2010. doi: 10.1002/1097-0142(19951115)76:10+<2004::aid-cncr2820761317>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 6.Menon U, Gentry-Maharaj A, Hallett R, Ryan A, Burnell M, Sharma A, et al. Sensitivity and specificity of multimodal and ultrasound screening for ovarian cancer, and stage distribution of detected cancers: results of the prevalence screen of the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS). Lancet Oncol. 2009;10(4):327–340. doi: 10.1016/S1470-2045(09)70026-9. [DOI] [PubMed] [Google Scholar]
- 7.Lu KH, Skates S, Bevers T, Adeyinka O, Newland W, Moore R, Leeds L, Harris L, Harris S, Fritsche H, Bast RC., Jr A prospective U.S. ovarian cancer screening study using the risk of ovarian cancer algorithm (ROCA). J Clin Oncol. 2010;28:15s. (suppl; abstr 5003) [Google Scholar]
- 8.Yurkovetsky Z, Lomakin A, Skates S, Menon U, Marrangoni A, Velikokhatnaya L, Nolen B, Winans M, Modugno F, Marks J, Godwin A, Gorelik E, Badgwell D, Bast RC, Jr, Lokshin A. Development of a multimarker assay for early detection of ovarian cancer. J Clin Oncol. 2010;28:2159–66. doi: 10.1200/JCO.2008.19.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim G, Minig L, Kohn EC. Proteomic profiling in ovarian cancer. Int J Gynecol Cancer. 2009;19:S2–S6. doi: 10.1111/IGC.0b013e3181c03929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hogdall E, Fung ET, Zhang Z, Engelholm SA, Petri AL, Risum S, Lundvall L, Nedergaard L, Pedersen AT, Hartwell D, Yip C, Wang Z, Meng XY, Thurasiraman V, Lomas L, Chan DW, Hogdall C. Prospective independent validation of a marker panel for distinguishing malignant from benign pelvic masses.. Gynecol Oncol; 39th Annual Meeting of the Society of Gynecologic Oncologists; 2008. Abstract. [Google Scholar]
- 11.Zhang Z, Bast RC, Jr, Yu Y, Li J, Sokoll LJ, Rai AJ, Rosenzweig JM, Cameron B, Wang YY, Meng XY, Berchuck A, Van Haaften-Day C, Hacker NF, de Bruijn HW, van der Zee AG, Jacobs IJ, Fung ET, Chan DW. Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Res. 2004;64:5882–90. doi: 10.1158/0008-5472.CAN-04-0746. [DOI] [PubMed] [Google Scholar]
- 12.Moore LE, Fung ET, McGuire M, Rabkin CC, Molinaro A, Wang Z, Zhang F, Wang J, Yip C, Meng X-Y, Pfeiffer RM. Evaluation of apolipoprotein A1 and posttranslationally modified forms of transthyretin as biomarkers for ovarian cancer detection in an independent study population. Cancer Epidemiol Biomarkers Prev. 2006;15:1641–46. doi: 10.1158/1055-9965.EPI-05-0980. [DOI] [PubMed] [Google Scholar]
- 13.Brandt E, Petersen F, Ludwig A, Ehlert JE, Bock L, Flad H-D. The b-thromboglobulins and platelet factor 4: blood platelet-derived CXC chemokines with divergent roles in early neutrophil regulation. J. Leukoc. Biol. 2000;67:471–478. doi: 10.1002/jlb.67.4.471. [DOI] [PubMed] [Google Scholar]
- 14.Grisaru D, Vlodavsky I, Prus D, Levavi H, Lessing JB, Eldor A, Friedmann Y. Connective Tissue Activating Peptide III expression disappears progressively with increased dysplasia in human cervical epithelium. Gynecol Oncol. 2000;79:23–27. doi: 10.1006/gyno.2000.5915. [DOI] [PubMed] [Google Scholar]
- 15.Shi L, Zhang J, Wu P, Feng K, Li J, Xie Z, Xue P, Cai T, Cui Z, Chen X, Hou J, Zhang J, Yang F. Discovery and identification of potential biomarkers of pediatric Acute Lymphoblastic Leukemia. Proteome Science. 2009;7:7. doi: 10.1186/1477-5956-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuesel AC, Kroft T, Prefontaine M, Smith IC. Lipoprotein(a) and CA125 levels in the plasma of patients with benign and malignant ovarian disease. Int J Cancer. 1992;52:341–6. doi: 10.1002/ijc.2910520302. [DOI] [PubMed] [Google Scholar]
- 17.Kozak KR, Su F, Whitelegge JP, Faull K, Reddy S, Farias-Eisner R. Characterization of serum biomarkers for detection of early stage ovarian cancer. Proteomics. 2005;5:4589–96. doi: 10.1002/pmic.200500093. [DOI] [PubMed] [Google Scholar]
- 18.Gericke B, Raila1 J, Sehouli J, Haebel S, Könsgen D, Mustea A, Schweigert FJ. Microheterogeneity of transthyretin in serum and ascitic fluid of ovarian cancer patients. BMC Cancer. 2005;5:133. doi: 10.1186/1471-2407-5-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobs I, Bast RC., Jr The CA 125 tumour-associated antigen: A review of the literature. Hum Reprod. 1989;4:1–12. doi: 10.1093/oxfordjournals.humrep.a136832. [DOI] [PubMed] [Google Scholar]
- 20.Kurman RJ, Visvanathan K, Roden R, Wu TC, Shih I-E. Early detection and treatment of ovarian cancer: shifting from early stage to minimal volume of disease based on a new model of carcinogenesis. Am J Obstet Gynecol. 2008;198:351–356. doi: 10.1016/j.ajog.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.