Abstract
Quorum sensing (QS) is a cell-cell signaling mechanism that allows bacteria to monitor their population size and alter their behavior at high cell densities. Gram-negative bacteria use N-acylated L-homoserine lactones (AHLs) as their primary signals for QS. These signals are susceptible to lactone hydrolysis in biologically relevant media, and the ring-opened products are inactive QS signals. We have previously identified a range of non-native AHLs capable of strongly agonizing and antagonizing QS in Gram-negative bacteria. However, these abiotic AHLs are also prone to hydrolysis and inactivation and thereby have a relatively short time window for use (~12–48 h). Non-native QS modulators with reduced or no hydrolytic instability could have enhanced potencies and would be valuable as tools to study the mechanisms of QS in a range of environments (for example, on eukaryotic hosts). This study reports the design and synthesis of two libraries of new, non-hydrolyzable AHL mimics. The libraries were screened for QS modulatory activity using LasR, LuxR, and TraR bacterial reporter strains, and several new, abiotic agonists and antagonists of these receptors were identified.
Keywords: N-Acylated L-homoserine lactone, Gram-negative bacteria, Heterocycles, LuxR-type receptors, Pseudomonas aeruginosa, Quorum sensing
1. Introduction
Bacteria can monitor their population densities using a language of low molecular weight chemical signals in a process called quorum sensing (QS).1, 2 Depending on the environment, these signaling molecules accumulate in proportion to bacterial cell density. Once a critical population density is achieved,3 the bacteria can alter their gene expression levels in order to initiate behaviors that benefit the multicellular community.4–6 These behaviors are extremely diverse, and include biofilm formation, virulence factor production, root nodulation, swarming, antibiotic production, and bioluminescence.7–14 Many of these behaviors play important roles in mediating both pathogenic and symbiotic relationships with eukaryotic hosts.15–18 Notably, several of the most notorious human pathogens (e.g., Staphylococcus aureus and Pseudomonas aeruginosa) use QS to control virulence.19, 20 As such, there is significant interest in understanding the role and mechanisms of QS in pathogenesis with the goal of developing novel anti-infective strategies.21, 22
The use of abiotic small molecules23–26 and macromolecular probes27, 28 to modulate QS pathways has emerged as a viable strategy to study QS and control bacterial group behaviors with both spatial and temporal control. Most of these non-native molecules affect QS pathways by either intercepting or inactivating the native bacterial QS signal. Our laboratory has made recent contributions in this area through the design and synthesis of non-native small molecules that agonize and antagonize QS in Gram-negative bacteria.23, 29
Gram-negative bacteria largely use N-acylated L-homoserine lactones (AHLs) as their QS signals.6, 11, 12, 25, 30 In general, these signals are cell permeable and are generated by a synthase protein (a LuxI-type synthase). The AHLs are then sensed by a cognate, cytoplasmic receptor (a LuxR-type receptor) at sufficiently high intracellular AHL concentrations.31 The AHL:receptor complex typically dimerizes, binds specific DNA promoter sequences, and activates the transcription of genes that the bacteria utilize at high cell density.32 Over ~100 different LuxI/R-type QS circuits have been indentified in bacteria,7 and many species appear to utilize multiple LuxI/R-type circuits in tandem to control QS.33
With regard to the QS signals, there are ~25 known naturally occurring AHLs, and they only differ in the structure of their respective acyl groups.6 These groups are typically aliphatic (4–18 carbons) and have differing levels of oxidation at the 3-position.34, 35 LuxR-type proteins are generally highly selective for their native AHL, and the AHLs of other species can act as inhibitors.36 Therefore, acyl chain composition dictates species specificity for AHL signals.
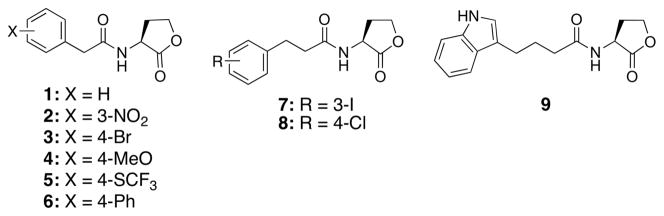
Over the past seven years, our laboratory has designed a range of AHLs that contain non-native acyl groups for use as chemical tools to study QS in Gram-negative bacteria.23 We have screened these compounds for activity in a range of species, including P. aeruginosa (for LasR and QscR),37, 38 Vibrio fischeri (LuxR),39 Agrobacterium tumefaciens (TraR),36 Pectobacterium carotovora (ExpR1/ExpR2),40 and Chromobacterium violaceum (CviR).41 Careful study of the active AHLs has revealed many structure-activity relationships (SARs) that dictate receptor selectivity for non-native AHL agonists and antagonists.42 Representative LuxR-type receptor agonists and antagonists reported by our laboratory are shown in Figure 1.
Figure 1.
Selected non-native AHLs reported by our laboratory that can agonize and antagonize LuxR-type receptors.
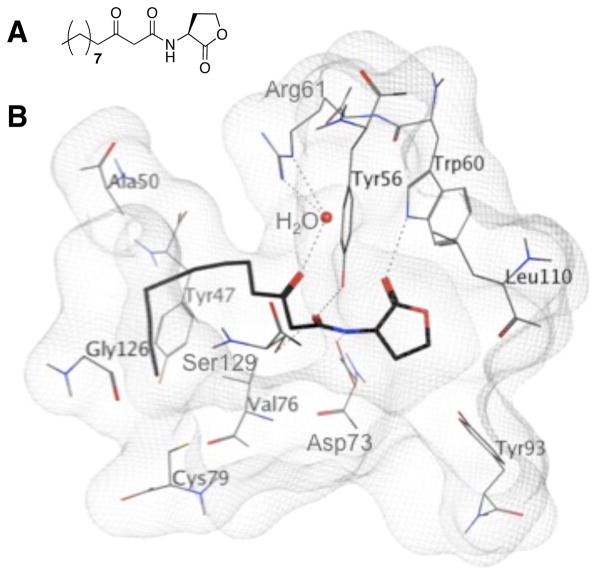
We currently seek to improve the potency of our lead AHL agonists and antagonists. It is well known that the AHL head group is prone to hydrolysis at pH values of 7 and above, and the hydrolyzed compound is inactive.43–45 For example, the native AHL for P. aeruginosa, N-(3-oxo)-dodecanoyl HL (OdDHL, Figure 2A), has a half-life of approximately two days in growth medium at 37 °C, while shorter chain AHLs hydrolyze in as little as six hours.46, 47 Our non-native AHLs share this same hydrolytic instability, and this can limit their application in biological experiments, particularly those over prolonged time periods.36 We reasoned that replacing the lactone head group in our lead AHLs with a non-hydrolyzable motif could engender heightened compound stability, and therefore, activity.
Figure 2.
A. The natural ligand for LasR in P. aeruginosa, OdDHL. B. View of OdDHL bound in the ligand binding pocket in LasR (from the reported X-ray crystal structure of the OdDHL:LasR N-terminal ligand binding domain complex).60 Hydrogen bonds are shown as dashed lines.
Others have previously examined lactone replacements in AHL analogs and uncovered a small set of compounds with moderate agonistic and antagonistic activities in LuxR-type proteins.43, 48–55 For example, the Suga group designed a 96-member library of OdDHL and N-butanoyl HL (BHL) analogs that contained a variety of heterocyclic and aromatic head groups, yet retained the native acyl groups, and evaluated the library for activity in both LasR and RhlR in P. aeruginosa.56, 57 Several active agonists and antagonists were uncovered, most notably analogs with phenolic and cyclohexanol head group replacements.58 Spring and coworkers have studied AHL analogs that contain cyclopentanone head groups, and shown that these compounds can strongly activate QS phenotypic responses in P. aeruginosa, Serratia ATCC39006, and P. carotovora.30, 50 In a related effort, Greenberg and co-workers have screened large, unbiased libraries of small molecules and identified several structurally unrelated, non-lactone compounds that can strongly modulate LasR.49, 59 Inspired by this recent work, we sought to examine the QS modulatory activity of chimeric compounds that contained (1) non-hydrolyzable head groups, and (2) our previously identified non-native acyl groups from lead AHLs.
Here we report the design and synthesis of a first- and second-generation library of AHL mimics containing non-lactone head groups. The first library (A) was designed to target LasR in P. aeruginosa. Active head groups from this library were then utilized for the design of chimeric ligands in a second-generation library (B). Both libraries were tested in bacterial reporter strains for activity in LasR, LuxR, and TraR. A set of new agonists and antagonists were found for these QS receptors. Several of these AHL mimics could be useful for biological experiments that require long time periods or elevated pH. Further, they also provide insights into which aspects of the lactone head group are necessary for LuxR-type receptor agonism and antagonism.
2. Results and Discussion
2.1. LasR Structural Considerations
We utilized OdDHL from P. aeruginosa as a design scaffold for the design of our first set of non-lactone derivatives (Library A). Similar to Suga’s earlier work (see above),56 we decided to retain the 3-oxo-dodecanoyl group from OdDHL in each library member, and simply replace the lactone with a non-native head group. In our choice of head groups, we scrutinized prior work that showed that a rigid aromatic or conjugated moiety adjacent to an aliphatic acyl chain could yield compounds active in LasR.43, 48–54, 61 We thus focused largely on aromatic head groups in our design of Library A. We also studied the X-ray crystal structure of the ligand-binding domain of LasR bound to OdDHL (Figure 2B) to ascertain molecular interactions essential for binding (making the assumption that non-lactone analogs could target the same ligand-binding site).60 For example, there is a water-mediated hydrogen bond between the 3-oxo group of OdDHL and the Arg61 guanidinium in the ligand-binding pocket. The acyl tail then packs into a hydrophobic cleft extending away from the lactone. We reasoned that these interactions would likely be maintained in Library A, with all members have 3-oxo dodecanoyl tails. A key hydrogen bond is also made between the OdDHL lactone carbonyl and Trp60 side chain NH. We sought, where possible, to retain and/or probe this hydrogen-bonding contact in Library A.
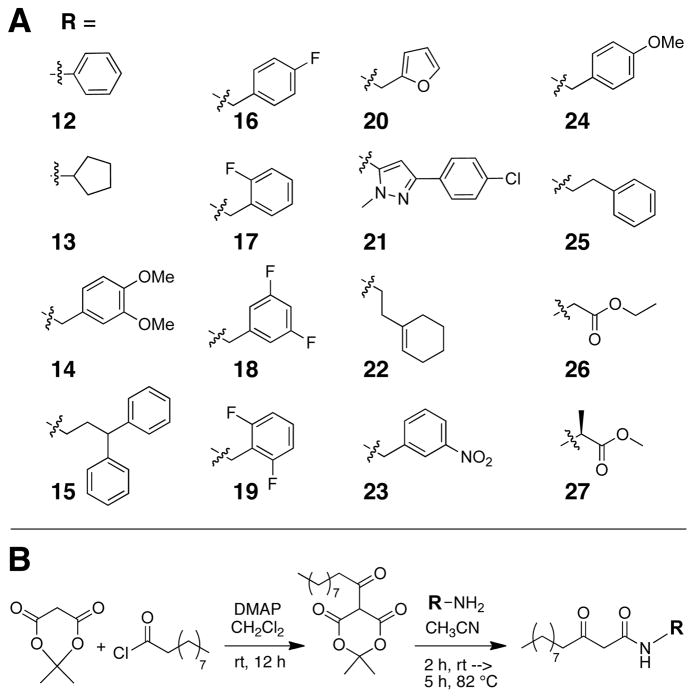
The heterocycles, carbocycles, and other head group mimics that we selected for incorporation into Library A are shown in Figure 3A. Fluorine substituted aromatic rings (16 and 17) were chosen as lactone carbonyl mimics to probe Trp60 interactions, due to fluorine’s ability to accept hydrogen bonds. Multiple fluorine substitutions were investigated in 18 and 19 to determine if Trp60 could hydrogen bond to multiple atoms given the correct spatial orientation. Non-hydrogen bond donors (in 14, 20, 21, and 24) were included to examine the effects of other electrostatic interactions. Library A also contained non-functionalized carbocycles (12, 13, 15, 22, and 25) to explore the necessity of the Trp60 binding interaction for LasR modulation.
Figure 3.
A. Non-lactone head groups (R) incorporated into Library A. B. Synthesis and overall structure of Library A members. DMAP = dimethyl aminopyridine. R is a novel head group shown in part A.
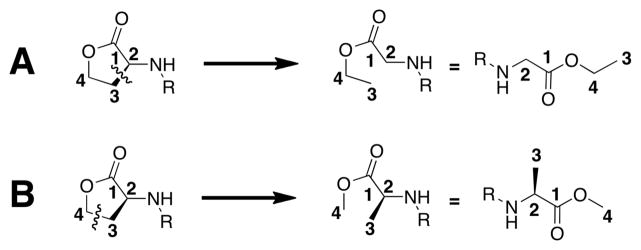
In addition to these cyclic head group replacements, we also incorporated glycine ethyl ester (26) and alanine methyl ester (27) as acyclic head groups into Library A (Figure 3A). These groups were selected to explore the effects of opening the lactone at bonds (typically) unavailable for cleavage in Nature; our design process is shown in Figure 4. The glycine ethyl ester analog was designed by conceptually breaking the bond between carbons 2 and 3. In turn, the alanine methyl ester analog originated from conceptually breaking the bond between carbons 3 and 4. Such non-native, alternatively “hydrolyzed” head groups have yet to be probed in AHL analogs, to our knowledge. We do note, however, these compounds will also be prone to hydrolysis, as they retain ester functionalities.62 We included them in this study nonetheless, as they represented novel head group replacements.
Figure 4.
Conceptual design of the unnatural, ring-opened forms of the lactone ring utilized in Library A. A. The glycine ethyl ester analog (26). B. The alanine methyl ester analog (27).
2.2. Synthesis of Library A
Library A was synthesized in parallel using standard solution-phase methods (Figure 3B). The head groups were incorporated as primary amines. A Meldrum’s acid derivative was used as a common dodecanoyl intermediate. Reaction of Meldrum’s acid with decanoyl chloride afforded the Meldrum’s acid derivative, which was subsequently refluxed with the desired head group amines to yield Library A. The 16 library members (12–27) were isolated in 35–70% yields with purities of 92–99% (See Supplementary Material).
2.3. Bacteriological Assays
Small molecules are typically screened for LuxR-type agonism or antagonism using bacterial reporter strains, and we used such cell-based assays in the current study.23 These bacterial reporter strains lack a functional LuxI-type synthase, yet retain a functional LuxR-type receptor. They typically contain a QS promoter fused to a reporter gene, and exogenous native AHL must be added to activate the system. Agonism or antagonism assays therefore can be performed by adding the compound alone or in competition with the native AHL ligand (at its EC50 value), respectively.
We utilized four bacterial reporter strains in this study to examine the activity of Library A (and eventually Library B) in LasR, LuxR, and TraR. Two strains were selected for the LasR screens: Escherichia coli DH5α (pJN105L + pSC11)63 and P. aeruginosa PA01 MW1 (pUM15).49 E. coli DH5α (pJN105L + pSC11) is a heterologous reporter strain containing one plasmid for the LasR gene and a second plasmid containing the promoter region for LasI fused to β-galactosidase (β-gal). LasR activity is read-out using a standard colorimetric assay with ortho-nitrophenyl-β-galactoside (ONPG) as the substrate for β-gal. The PA01 MW1 (pUM15) strain is a LasR reporter in P. aeruginosa that lacks a functional LasI and contains a plasmid with a LasR responsive promoter for Yellow Fluorescent Protein (YFP), which facilitates straightforward evaluation of LasR activity using fluorescence. Examining the library in both strains allowed us to study the effects of the AHL analogs on LasR in an isolated system (E. coli) and then in the presence of P. aeruginosa’s complex QS network (including RhlR and QscR) in the native PAO1 background. (We note that E. coli and P. aeruginosa have different compound uptake/efflux profiles, and this feature should be taken into account when comparing small molecule screening data between the two strains (see below)).
V. fischeri ESI 114 (Δ-LuxI)64 and A. tumefaciens WCF (pCF372)65 were used to examine the activity of library compounds in LuxR and TraR, respectively. The V. fischeri mutant strain lacks a functioning LuxI synthase, but retains its native lux operon, allowing a quantitative luminescent readout based on LuxR activity. Similarly, A. tumefaciens WCF (pCF372)65 lacks a functioning TraI, yet contains a plasmid with a TraR responsive promoter for the β-gal gene, thereby allowing for direct quantitation of TraR activity using absorbance.
We used our previously reported bacteriological assay protocols for small molecule screening, which allowed for comparisons to be made between the assay data reported here and our past work.42, 66 All compounds were screened at 10 μM in both agonism and antagonism assays. No effects on bacterial growth were observed over the time course of the reporter gene assays (4–16 h).
2.4. Biological Screening of Library A
We first evaluated Library A for LasR agonistic and antagonistic activities using the E. coli and P. aeruginosa reporter strains. The assay data are shown in Table 1. Several compounds were active in both strains, yet their relative activities were muted overall in the P. aeruginosa reporter relative to the E. coli reporter. We reason that this could be due to the P. aeruginosa strain containing competing LuxR-type receptors and/or the lowered cell permeability of P. aeruginosa relative to E. coli. P. aeruginosa is well known to have numerous efflux pumps that hamper cellular uptake of compounds.67 In general, sterically small and sparsely functionalized head groups appended to the 3-oxo-dodecanoyl group were the most active. We discuss trends in activity for the six most active compounds (12, 13, 17, 23, 24, and 26) below.
Table 1.
Antagonism and agonism assay data for Library A in LasR.[a]
| Compound | E. coli LasR | P. aeruginosa LasR | ||
|---|---|---|---|---|
| Antagonism [%] [b] | Agonism [%] [c] | Antagonism [%] [b] | Agonism [%] [c] | |
| 12 | 54 | 3 | 29 | 1 |
| 13 | −75 | 84 | −1 | 54 |
| 14 | 13 | 1 | 9 | 0 |
| 15 | 17 | 0 | 8 | 0 |
| 16 | 13 | 0 | 13 | 0 |
| 17 | 38 | 0 | 13 | 0 |
| 18 | 22 | 0 | 16 | 0 |
| 19 | 13 | 1 | 5 | 0 |
| 20 | 0 | 0 | 26 | 0 |
| 21 | 0 | 0 | 0 | 0 |
| 22 | 10 | 0 | −7 | 4 |
| 23 | 41 | 0 | 17 | 0 |
| 24 | −68 | 73 | 38 | 1 |
| 25 | −4 | 0 | 5 | 0 |
| 26 | −136 | 51 | 11 | 12 |
| 27 | −4 | 2 | 23 | 0 |
All synthetic compounds were screened at 10 μM. All assays were preformed in triplicate; error did not exceed ±10%. Positive controls were OdDHL at its EC50 value (in each strain) for antagonism assays, and at 100 times its EC50 value for agonism assays. Negative controls contained neither thiolactone nor natural AHL, and were subtracted from each sample to account for background. Negative inhibition values indicate that the compound activates at the tested concentration. Shaded rows highlight compounds of interest. See text for details of strains.
Antagonism assays were performed against OdDHL at its EC50 value in each strain: E. coli DH5α (pJN105L + pSC11) = 10 nM; P. aeruginosa PA01 MW1 (pUM15) = 1 μM.
Agonism assays were normalized to the positive control (OdDHL) in each strain.
The unsubstituted, phenyl (aniline) analog 12 was the strongest LasR antagonist identified in the E. coli LasR strain, with the meta-nitro benzyl (23) and ortho-fluoro benzyl (17) analogs being the second and third most active LasR antagonists, respectively. Additional fluoro substituents (18 and 19) or a para-fluoro substituent (16) yielded less active compounds. The antagonistic activities of 12, 17, and 23 corroborated earlier findings by Greenberg, Suga, and Kim demonstrating that related aromatic analogs are LasR antagonists (albeit in alternate strains for Suga and Kim).49, 56, 68 As aniline derivative 12 was the most active overall, this finding does question the proposed critical nature of the AHL lactone carbonyl for LasR binding with Trp60 (assuming these analogs target the same site via their antagonism mechanism). In turn, the relatively high activity of aromatic analogs overall suggests that π−π stacking or hydrophobic interactions could play a role in LasR binding.
Cyclopentyl analog 13 was found to be a strong LasR agonist in both reporter strains. Again, this high level of activity suggests that either the hydrogen bond to the native AHL lactone carbonyl is not as crucial as originally thought, or that 13 binds LasR in an alternative manner. We note that Ishida and co-workers have reported a cyclopentyl compound similar to 13, yet containing a simple (non-3-oxo) decanoyl tail;51 interestingly, they found that this derivative was a LasR antagonist in a P. aeruginosa strain, as opposed to an agonist like 13. These opposite activities highlight the importance of the 3-oxo group and chain length in LasR modulatory activity.
The para-methoxy benzyl analog (24) displays conflicting activities between the two reporter strains: it is a strong LasR agonist in the E. coli reporter, but a weak LasR antagonist in the P. aeruginosa reporter. These data suggest that 24 may not solely target LasR in the P. aeruginosa strain. The differing activities of 24 relative to its meta-nitro analog 23 (see above) suggest that subtle steric and electronic changes to the benzyl head group can have significant effects on ligand activity in LasR, at least in this E. coli reporter strain. Benzyl analog 24 represents an excellent candidate for further testing in additional heterologous reporter strains containing QscR and RhlR, which also play a role in regulating the overall P. aeruginosa QS response.69, 70 We note, however, that we cannot discount compound permeability differences between the strains also contributing to these disparate activity profiles (see above).
The “ring opened” lactone analogs 26 and 27 provided some interesting activity trends. First, only the glycine ethyl ester analog (26) showed appreciable agonistic activity in LasR, indicating that the ring opening in 27 strongly demoted compound activity relative to “ring closed” OdDHL, while the ring opening in 26 had less of an effect relative to OdDHL. Second, while 26 was a moderate agonist of LasR in E. coli, it was only minimally active in the P. aeruginosa strain. This trend is somewhat similar to that observed for para-methoxy benzyl analog 24 above, so analogous logic could apply for its possible mode of action.
Library A was also tested for activity in LuxR and TraR using the reporter strains introduced above. However, compound activities were low to modest in these species (See Supplementary Material for full data). These low activities were perhaps to be expected, however, as Library A was designed to target LasR in P. aeruginosa, and reinforces prior work demonstrating that the length of the AHL acyl tail, even in non-lactone derivatives, can give ligands receptor selectivity.36
2.5. Design of Library B
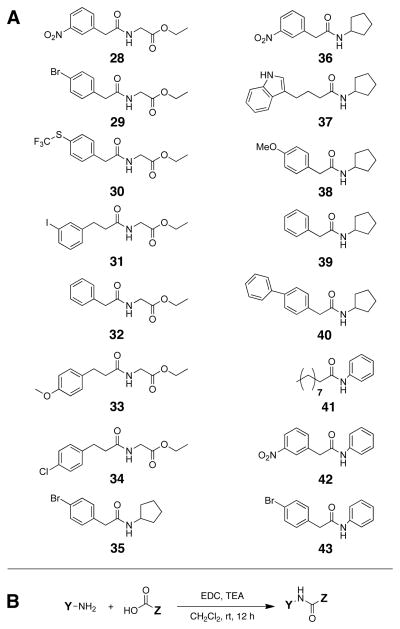
Library B was designed around the most active head groups uncovered in Library A (aniline, cyclopropyl amine, and glycine ethyl ester in 12, 13 and 26, respectively; Figure 5A). To these head groups, we appended several acyl tails that have shown a range of activities when incorporated in our previously reported AHL analogs (see Figure 1).36, 39, 42 We note that many of these earlier AHL analogs were found to be active in LuxR, TraR, and/or LasR. For example, the para-bromo and para-phenyl AHLs (3 and 6) are strong antagonists of both LuxR and TraR, the meta-iodo phenylpropanoyl HL (7) is a strong antagonist of both LasR and LuxR, and indole 9 targets LasR selectively. The receptor selectivity profiles of AHLs 3, 6, 7, and 9 prompted us to use their acyl groups in Library B to examine if such selectivity trends would be retained in chimeric AHL mimics. Likewise, the divergent activity of 2 (a strong LuxR agonist and a strong LasR antagonist) made the meta-NO2 phenylacetanoyl group an attractive choice for studying contrasting receptor modulatory activity. We also included the acyl tail from para-methoxy phenylacetanoyl HL (4), a more moderate LasR antagonist, in order to investigate whether this moderate activity profile would also be maintained with alternate head groups. The unfunctionalized phenylacetanoyl tail (from 1), which is inactive in most LuxR-type receptors when appended to a native lactone head group, was included to test if this inactivity would be maintained in Library B. Lastly, the para-chloro phenylpropanoyl group from AHL (8), which is a strong LuxR, LasR, and TraR antagonist, was selected to probe the ability of non-lactone AHL mimics to maintain such multi-receptor, or more “broad-spectrum,” activities.
Figure 5.
A. Structures of compounds in Library B. B. Synthesis of Library B. Y represents the lead heterocycles identified from Library B. Z represents the acyl tails from several of our previously reported non-native AHLs. EDC = 1-ethyl-3-(3-dimethyl aminopropyl) carbodiimide (EDC). TEA = triethyl amine
2.6. Synthesis of Library B
Library B was synthesized in solution via standard carbodiimide-mediated amide bond coupling reactions (EDC) between the three head group amines and the respective carboxylic acids (Figure 5B). The 16 library members (28–43) were isolated in 60–95% yields with purities of 89–99% (See Supplementary Material).
2.7. Biological Screening of Library B
Library B was screened for LasR antagonism and agonism using the same reporter strains and protocols as for Library A. The results of these screens are shown in Table 2. In general, similar to the LasR screening data for Library A, the E. coli LasR strain yielded compounds with heightened activities relative to the P. aeruginosa LasR strain. The most active compound indentified was the meta-nitro phenylacetanoyl glycine methyl ester (28), which displayed a novel activity profile (see below).
Table 2.
Antagonism and agonism assay data for Library B in LasR.[a]
| Compound | E. coli LasR | P. aeruginosa LasR | ||
|---|---|---|---|---|
| Antagonism [%] | Agonism [%] | Antagonism [%] | Agonism [%] | |
| 28 | 41 | 93 | −21 | 0 |
| 29 | 33 | 0 | −15 | 0 |
| 30 | 33 | 0 | −27 | 0 |
| 31 | 38 | 0 | −17 | 0 |
| 32 | 38 | 0 | −10 | 0 |
| 33 | 36 | 0 | −6 | 0 |
| 34 | 38 | 0 | −5 | 0 |
| 35 | 11 | 0 | −11 | 0 |
| 36 | 29 | 0 | −5 | 0 |
| 37 | 20 | 0 | −10 | 2 |
| 38 | 24 | 6 | −1 | 0 |
| 39 | 11 | 0 | 1 | 0 |
| 40 | 4 | 0 | −26 | 1 |
| 41 | 20 | 3 | 5 | 0 |
| 42 | 12 | 0 | 0 | 1 |
| 43 | 20 | 3 | 12 | 0 |
See footnotes [a–c] in Table 1.
Interestingly, many Library B compounds exhibited slight agonistic activities in the antagonism assay in the P. aeruginosa LasR strain, yet failed to agonize LasR in the agonism assay in this same strain (i.e., in the absence of OdDHL). The meta-nitro and para-thioether glycine ethyl ester chimeras (28 and 30), and the biphenyl cyclopentyl chimera (40) showed greater than 20% agonistic activities only in the LasR antagonism assay. We have recently seen this activity trend in the P. aeruginosa LasR reporter for another class of non-lactone AHL mimics (i.e., thiolactones).71 Our current hypothesis is that such non-native ligands are potentially capable of forming heterodimers of LuxR-type proteins bound to the native AHL and the non-native ligand (both present in the competitive antagonism assay) that are more active than homodimeric complexes formed from native AHL alone. In turn, these non-native ligands cannot form active homodimers of the same receptors, and thus are inactive in the agonism assay. While ongoing work in our laboratory is directed at more fully understanding this potential mode of action, analogs 28, 30, and 40 exhibit activity profiles in LasR that are suggestive that they could operate via such a cooperative agonism mechanism.
The majority of the glycine ethyl ester derivatives in Library B (28–33) were moderate LasR antagonists (30–40%) in the E. coli reporter strain (Table 2). However, the meta-nitro chimera 28 was also an excellent LasR agonist in this strain (when tested at the same concentration in the absence of OdDHL), activating to 93% of the level of OdDHL. One possible explanation for such a divergent activity profile is the following: homodimers of LasR complexed with either 28 or OdDHL are can agonize the system, yet the heterodimeric complex of LasR:28/LasR:ODdHL is inactive. An antagonistic effect would therefore be observed only when the natural ligand is present. We have named compounds that can behave in this manner putative “bimodal binders” because of their dual antagonistic and agonistic properties, and expound on this pathway further in a related recent study.71 Additional studies are certainly required to fully understand the mechanisms by which 28 elicits its conflicting activities in LasR, and are ongoing.
Cyclopentyl derivatives 35–40 and aniline derivatives 41–43 were only weak antagonists of the E. coli LasR reporter strain and largely inactive in the P. aeruginosa LasR reporter strain (Table 2). The only exception was the biphenyl cyclopentyl chimera 40, which activated LasR only in the antagonism assay and thus exhibited a potential cooperative agonistic profile (see above). Together with the screening data for Library A in LasR, these results suggest that the native OdDHL acyl chain generally engenders higher ligand activities in LasR relative to non-native acyl groups when appended to the non-native head groups in Library B.
We next screened Library B in LuxR and TraR reporter strains (see above) for both antagonistic and agonistic activities. The screening data are shown in Table 3. Four moderate to strong LuxR modulators were indentified: 30, 31, 33, and 36. However, TraR was largely unresponsive to Library B (data not shown). This is perhaps expected, as TraR is believed to have a much more rigid ligand binding pocket relative to other LuxR-type proteins72, 73 and is less tolerant in general to non-native AHL analogs.66
Table 3.
Antagonism and agonism assay data for Library B in LuxR.[a]
| Compound | V. fischeri LuxR | |
|---|---|---|
| Antagonism [%] [b] | Agonism [%] [c] | |
| 28 | 19 | 0 |
| 29 | 14 | 0 |
| 30 | 43 | 0 |
| 31 | 89 | 0 |
| 32 | 15 | 0 |
| 33 | 40 | 0 |
| 34 | 14 | 0 |
| 35 | 35 | 0 |
| 36 | 62 | 33 |
| 37 | 25 | 4 |
| 38 | 18 | 3 |
| 39 | 19 | 0 |
| 40 | 17 | 5 |
| 41 | 2 | 7 |
| 42 | 0 | 0 |
| 43 | 1 | 0 |
See footnote [a] in Table 1.
Antagonism assays were performed against OHHL at its the EC50 value in the V. fischeri ESI 114 (Δ-LuxI) strain = 2 μM.
Agonism assays were normalized to the positive control (100 μM OHHL).
The most active LuxR modulator indentified in Library B was the meta-iodo phenylpropanoyl glycine ethyl ester derivative 31, which was a very strong (89%) LuxR antagonist. Notably, the AHL analog of 31, AHL 7 (Figure 1), displays analogous inhibitory activity in LuxR,66 suggesting that antagonism is largely dependent on acyl group structure, as opposed to head group structure, for these two compounds. Aryl glycine ethyl ester derivates 30 and 33 were also LuxR antagonists, yet were 2-fold less active (~40%).
The meta-nitro cyclopentyl derivative 36 was the only LuxR modulator in Library B not based on glycine ethyl ester (Table 3). Cyclopentyl analog 36 was a 62% LuxR antagonist, yet was also capable of agonizing LuxR by 33%. We note that the AHL analog (3) of 36 is a highly potent agonist in LuxR.39 The dual activity of 36 in LuxR is reminiscent of the meta-nitro chimera 28 in LasR (see above), and suggests that 36 may behave as a bimodal binder with LuxR, forming inactive heterodimers of LuxR when the native ligand (OHHL) is present, yet active homodimers alone.
3. Conclusion
We have designed and synthesized two focused libraries of non-lactone AHL mimics and evaluated their activities as agonists and antagonists of LasR, LuxR, and TraR. This study was motivated by our interest in enhancing the hydrolytic stability of such autoinducer mimics in order to potentially increase their stability and potency for use as chemical tools to study QS. Sixteen novel non-hydrolyzable head groups were explored in the first-generation library (A), and three head groups (aniline, cyclopentyl amine, and glycine ethyl ester) yielded the most potent LasR modulators when derivatized with the OdDHL acyl tail (i.e., in analogs 12, 13, and 26, respectively). These compounds were largely selective for LasR over LuxR and TraR, which reinforces previous work that demonstrated that acyl chain structure is critical for receptor selectivity, even if the native lactone head group is removed.42
We next designed a 16-member second-generation library (B) of chimeric molecules composed of the three lead head groups from Library A appended to the acyl groups of a subset of our previously reported non-native AHLs. Several potent modulators were uncovered, including the glycine ethyl ester analogs 28 and 31, which were a strong LasR agonist and LuxR antagonist, respectively. It was surprising that these novel ring-opened, achiral ligands were the most active in Library B, and motivates further study of acyclic lactone head group replacements in autoinducer analogs.
Additional work will be required to further improve the potency of these lead compounds and to fully understand their mechanisms of action, particularly with regard to potential cooperative agonism and bimodal binding pathways.71 Nevertheless, the results of this study suggest that the lactone head group is not required in small molecule modulators of LuxR-type proteins, and corroborate several previous reports of active non-lactone ligands. With regard to LasR, our data for certain AHL mimics suggest that the lactone hydrogen bond to Trp60 in LasR may not be essential for agonistic or antagonistic activity, assuming that these compounds target the OdDHL binding site. We of course cannot discount the possibility that alternate hydrogen-bond acceptors in selected AHL mimics could replace the native AHL lactone carbonyl (e.g., esters in the glycine ethyl ester derivatives) in this interaction.
The new QS agonists and antagonists reported here could have value as mechanistic probes to study QS. For example, certain non-hydrolyzable analogs may be useful for controlled-release studies or for experiments examining QS responses in relation to ligand diffusibility and population density over time spans ranging from several days to weeks.74 Further studies are warranted beforehand to fully characterize their interactions with LuxR-type receptors, and such experiments are ongoing in our laboratory.
4. Experimental Section
4.1. Compound Synthesis
All chemical reagents were purchased from commercial sources (Alfa-Aesar, Aldrich, and Acros) and used without further purification. Solvents were purchased from commercial sources (Aldrich and J.T. Baker) and used as obtained, with the exception of dichloromethane (CH2Cl2), which was distilled over calcium hydride immediately prior to use.
The Meldrum’s acid intermediate used in the synthesis of Library A was generated by dissolving Meldrum’s acid (0.5 g, 3.5 mmol) in CH2Cl2 (50 mL) under nitrogen in an ice bath (Figure 3B). DMAP (0.86 g, 7 mmol) was added to the mixture and allowed to completely dissolve. Dodecanoyl chloride (0.72 mL, 3.5 mmol) was then added slowly over an hour. The reaction mixture was allowed to stir on ice for an additional hour, and then was allowed to come to room temperature (rt) and stir overnight. The reaction mixture was washed 2x with 2M HCl and 1x with brine. The organic layers were isolated, dried over MgSO4, filtered, and concentrated to yield the crude Meldrum’s acid intermediate that was used immediately in the next amide-coupling step.
Library A were synthesized by stirring the desired amine head group (0.16 mmol) with the Meldrum’s acid intermediate from above (48 mg, 0.16 mmol) in acetonitrile (15 mL) at rt under nitrogen for 2 h (Figure 3B). The reaction mixture was then refluxed for an additional 5 h to ensure complete reaction. The solvent was removed in vacuo, and the reaction residue was redissolved in ethyl acetate and washed 1x each with saturated NaHCO3, 1M NaHSO4, and brine. The organic layers were combined, dried over MgSO4, filtered, and concentrated to yield the Library A members. As needed, members of Library A were further purified by silica gel column chromatography (25% EtOAc/Hex).
The glycine ethyl ester head group in 26 was synthesized as previously reported,75 and commercially available alanine methyl ester was utilized to synthesize 27. Library B was synthesized using EDC couplings and purified as previously reported.37, 57
4.2. Bacterial Assays
Bacterial reporter gene assays for E. coli DH5α (pJN105L + pSC11), V. fischeri ESI 114 (Δ-LuxI), and A. tumefaciens WCF (pCF372) were conducted as previously described.36 Reporter gene assays for P. aeruginosa PA01 MW1 (pUM15) were modified from reported procedure.49 See Supplementary Material for additional details.
Supplementary Material
Acknowledgments
Financial support for this work was provided by the NIH (AI063326), Greater Milwaukee Foundation Shaw Scientist Program, Burroughs Welcome Fund, Camille & Henry Dreyfus Foundation, Research Corporation, and Johnson & Johnson. C.E.M. was supported in part through a DOD (Air Force Office of Scientific Research) National Defense Science and Engineering Graduate (NDSEG) Fellowship (32 CFR 168a). We gratefully acknowledge Professors Peter Greenberg (University of Washington), Stephen Winans (Cornell University), and Edward Ruby (UW–Madison) for donations of bacterial strains and advice on their manipulation. We also thank Karla Camancho for her assistance in purifying members of Library A.
Footnotes
Compound characterization data, biological assay protocols, and additional screening data.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Camilli A, Bassler BL. Science. 2006;311:1113. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Platt TG, Fuqua C. Trends Microbiol. 2010;18:383. doi: 10.1016/j.tim.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boedicker JQ, Vincent ME, Ismagilov RF. Angew Chem Int Edit. 2009;48:5908. doi: 10.1002/anie.200901550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ng WL, Bassler BL. Annu Rev Genet. 2009;43:197. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bassler BL, Losick R. Cell. 2006;125:237. doi: 10.1016/j.cell.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 6.Fuqua C, Greenberg EP. Nat Rev Mol Cell. 2002;3:685. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- 7.Dickschat JS. Nat Prod Rep. 2010;27:343. doi: 10.1039/b804469b. [DOI] [PubMed] [Google Scholar]
- 8.Dobretsov S, Teplitski M, Paul V. Biofouling. 2009;25:413. doi: 10.1080/08927010902853516. [DOI] [PubMed] [Google Scholar]
- 9.Novick RP, Geisinger E. Annu Rev Genet. 2008;42:541. doi: 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 10.Barnard AML, Bowden SD, Burr T, Coulthurst SJ, Monson RE, Salmond GPC. Philos T Roy Soc B. 2007;362:1165. doi: 10.1098/rstb.2007.2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nyholm SV, Mcfall-Ngai MJ. Nat Rev Microbiol. 2004;2:632. doi: 10.1038/nrmicro957. [DOI] [PubMed] [Google Scholar]
- 12.Wisniewski-Dye F, Downie JA. Antonie Van Leeuwenhoek. 2002;81:397. doi: 10.1023/a:1020501104051. [DOI] [PubMed] [Google Scholar]
- 13.Kuipers OP, de Ruyter PGGA, Kleerebezem M, de Vos WM. J Biotechnol. 1998;64:15. [Google Scholar]
- 14.Winans SC. Mol Microbiol. 1991;5:2345. doi: 10.1111/j.1365-2958.1991.tb02080.x. [DOI] [PubMed] [Google Scholar]
- 15.Pacheco AR, Sperandio V. Curr Opin Microbiol. 2009;12:192. doi: 10.1016/j.mib.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parsek MR, Greenberg EP. Proc Natl Acad Sci U S A. 2000;97:8789. doi: 10.1073/pnas.97.16.8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu J, Winans SC. Proc Natl Acad Sci U S A. 1999;96:4832. doi: 10.1073/pnas.96.9.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Kievit TR. Environ Microbiol. 2009;11:279. doi: 10.1111/j.1462-2920.2008.01792.x. [DOI] [PubMed] [Google Scholar]
- 19.Smith RS, Iglewski BH. Curr Opin Microbiol. 2003;6:56. doi: 10.1016/s1369-5274(03)00008-0. [DOI] [PubMed] [Google Scholar]
- 20.George EA, Muir TW. Chem Bio Chem. 2007;8:847. doi: 10.1002/cbic.200700023. [DOI] [PubMed] [Google Scholar]
- 21.Smith RS, Iglewski BH. J Clin Invest. 2003;112:1460. doi: 10.1172/JCI20364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rasko DA, Sperandio V. Nat Rev Drug Discov. 2010;9:117. doi: 10.1038/nrd3013. [DOI] [PubMed] [Google Scholar]
- 23.Geske GD, O’Neill JC, Blackwell HE. Chem Soc Rev. 2008;37:1432. doi: 10.1039/b703021p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amara N, Mashiach R, Amar D, Krief P, Spieser SAH, Bottomley MJ, Aharoni A, Meijler MM. J Am Chem Soc. 2009;131:10610. doi: 10.1021/ja903292v. [DOI] [PubMed] [Google Scholar]
- 25.Galloway WRJD, Hodgkinson JT, Bowden SD, Welch M, Spring DR. Chem Rev. 2011;111:28. doi: 10.1021/cr100109t. [DOI] [PubMed] [Google Scholar]
- 26.Stevens AM, Queneau Y, Soulere L, von Bodman S, Doutheau A. Chem Rev. 2011;111:4. doi: 10.1021/cr100064s. [DOI] [PubMed] [Google Scholar]
- 27.Kaufmann GF, Sartorio R, Lee SH, Mee JM, Altobell LJ, Kujawa DP, Jeffries E, Clapham B, Meijler MM, Janda KD. J Am Chem Soc. 2006;128:2802. doi: 10.1021/ja0578698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Amara N, Krom BP, Kaufmann GF, Meijler MM. Chem Rev. 2011;111:195. doi: 10.1021/cr100101c. [DOI] [PubMed] [Google Scholar]
- 29.Mattmann ME, Blackwell HE. J Org Chem. 2010;75:6737. doi: 10.1021/jo101237e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Welch M, Dutton JM, Glansdorp FG, Thomas GL, Smith DS, Coulthurst SJ, Barnard AML, Salmond GPC, Spring DR. Bioorg Med Chem Lett. 2005;15:4235. doi: 10.1016/j.bmcl.2005.06.066. [DOI] [PubMed] [Google Scholar]
- 31.Eberhard A, Burlingame AL, Eberhard C, Kenyon GL, Nealson KH, Oppenheimer NJ. Biochemistry (Mosc) 1981;20:2444. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- 32.Schuster M, Greenberg EP. Chemical Communication among Bacteria. Washington, DC: ASM Press; 2008. [Google Scholar]
- 33.Patankar AV, Gonzalez JE. FEMS Microbiol Rev. 2009;33:739. doi: 10.1111/j.1574-6976.2009.00163.x. [DOI] [PubMed] [Google Scholar]
- 34.Schaefer AL, Greenberg EP, Oliver CM, Oda Y, Huang JJ, Bittan-Banin G, Peres CM, Schmidt S, Juhaszova K, Sufrin JR, Harwood CS. Nature. 2008;454:595. doi: 10.1038/nature07088. [DOI] [PubMed] [Google Scholar]
- 35.Churchill MEA, Chen LL. Chem Rev. 2011;111:68. doi: 10.1021/cr1000817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geske GD, O’Neill JC, Miller DM, Mattmann ME, Blackwell HE. J Am Chem Soc. 2007;129:13613. doi: 10.1021/ja074135h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geske GD, Wezeman RJ, Siegel AP, Blackwell HE. J Am Chem Soc. 2005;127:12762. doi: 10.1021/ja0530321. [DOI] [PubMed] [Google Scholar]
- 38.Mattmann ME, Shipway PM, Heth NJ, Blackwell HE. Chem Bio Chem. 2011;12:942. doi: 10.1002/cbic.201000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Geske GD, O’Neill JC, Blackwell HE. ACS Chem Biol. 2007;2:315. doi: 10.1021/cb700036x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palmer AG, Streng E, Jewell KA, Blackwell HE. Chem Bio Chem. 2011;12:138. doi: 10.1002/cbic.201000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Praneenararat T, Geske GD, Blackwell HE. Org Lett. 2009;11:4600. doi: 10.1021/ol901871y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geske GD, Mattmann ME, Blackwell HE. Bioorg Med Chem Lett. 2008;18:5978. doi: 10.1016/j.bmcl.2008.07.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schaefer AL, Hanzelka BL, Eberhard A, Greenberg EP. JBacteriol. 1996;178:2897. doi: 10.1128/jb.178.10.2897-2901.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eberhard A, Widrig CA, MaBath P, Schineller JB. Arch Microbiol. 1986;146:35. doi: 10.1007/BF00690155. [DOI] [PubMed] [Google Scholar]
- 45.Byers JT, Salmond CLGPC, Welch M. J Bacteriol. 2002;184:1163. doi: 10.1128/jb.184.4.1163-1171.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yates EA, Philipp B, Buckley C, Atkinson S, Chhabra SR, Sockett RE, Goldner M, Dessaux Y, Camara M, Smith H, Williams P. Infect Immun. 2002;70:5635. doi: 10.1128/IAI.70.10.5635-5646.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Glansdorp FG, Thomas GL, Lee JK, Dutton JM, Salmond GPC, Welch M, Spring DR. Org Biomol Chem. 2004;2:3329. doi: 10.1039/B412802H. [DOI] [PubMed] [Google Scholar]
- 48.Passador L, Tucker KD, Guertin KR, Journet MP, Kende AS, Iglewski BH. J Bacteriol. 1996;178:5995. doi: 10.1128/jb.178.20.5995-6000.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muh U, Schuster M, Heim R, Singh A, Olson E, Greenberg EP. Antimicrob Agents Chemother. 2006;50:3674. doi: 10.1128/AAC.00665-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee LYW, Hupfield T, Nicholson RL, Hodgkinson JT, Su X, Thomas GL, Salmond PC, Welch M, Spring DR. Molecular Bio Systems. 2008;4:505. doi: 10.1039/b801563e. [DOI] [PubMed] [Google Scholar]
- 51.Ishida T, Ikeda T, Takiguchi N, Kuroda A, Ohtake H, Kato J. Appl Environ Microbiol. 2007;73:3183. doi: 10.1128/AEM.02233-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim C, Kim J, Park HY, Park HJ, Lee JH, Kim CK, Yoon J. Appl Microbiol Biotechnol. 2008;80:37. doi: 10.1007/s00253-008-1474-6. [DOI] [PubMed] [Google Scholar]
- 53.Estephane J, Dauvergne J, Soulere L, Reverchon S, Queneau Y, Doutheau A. Bioorg Med Chem Lett. 2008;18:4321. doi: 10.1016/j.bmcl.2008.06.090. [DOI] [PubMed] [Google Scholar]
- 54.Hjelmgaard T, Persson T, Rasmussen TB, Givskov M, Nielsen J. Bioorg Med Chem. 2003;11:3261. doi: 10.1016/s0968-0896(03)00295-5. [DOI] [PubMed] [Google Scholar]
- 55.Suga H, Smith KM. Curr Opin Chem Biol. 2003;7:586. doi: 10.1016/j.cbpa.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 56.Smith KM, Bu Y, Suga H. Chem Biol. 2003;10:563. doi: 10.1016/s1074-5521(03)00107-8. [DOI] [PubMed] [Google Scholar]
- 57.Smith KM, Bu YG, Suga H. Chem Biol. 2003;10:81. doi: 10.1016/s1074-5521(03)00002-4. [DOI] [PubMed] [Google Scholar]
- 58.Jog GJ, Igarashi J, Suga H. Chem Biol. 2006;13:123. doi: 10.1016/j.chembiol.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 59.Muh U, Hare BL, Duerkop BA, Schuster M, Hanzelka BL, Heim R, Olson ER, Greenberg EP. Proc Natl Acad Sci U S A. 2006;103:16948. doi: 10.1073/pnas.0608348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bottomley MJ, Muraglia E, Bazzo R, Carfi A. J Biol Chem. 2007;282:13592. doi: 10.1074/jbc.M700556200. [DOI] [PubMed] [Google Scholar]
- 61.Suga H, Smith KM. Curr Opin Chem Biol. 2003;7:586. doi: 10.1016/j.cbpa.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 62.Katseres NS, Reading DW, Shayya L, DiCesare JC, Purser GH. Biochem Biophys Res Commun. 2009;386:363. doi: 10.1016/j.bbrc.2009.06.037. [DOI] [PubMed] [Google Scholar]
- 63.Lee JH, Lequette Y, Greenberg EP. Mol Microbiol. 2006;59:602. doi: 10.1111/j.1365-2958.2005.04960.x. [DOI] [PubMed] [Google Scholar]
- 64.Lupp C, Urbanowski M, Greenberg EP, Ruby EG. Mol Microbiol. 2003;50:319. doi: 10.1046/j.1365-2958.2003.t01-1-03585.x. [DOI] [PubMed] [Google Scholar]
- 65.Zhu J, Beaber JW, More MI, Fuqua C, Eberhard A, Winans SC. J Bacteriol. 1998;180:5398. doi: 10.1128/jb.180.20.5398-5405.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Geske GD, Neill OJC, Miller DM, Wezeman RJ, Mattmann ME, Lin Q, Blackwell HE. Chem Bio Chem. 2008;9:389. doi: 10.1002/cbic.200700551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Maseda H, Sawada I, Saito K, Uchiyama H, Nakae T, Nomura N. Antimicrob Agents Chemother. 2004;48:1320. doi: 10.1128/AAC.48.4.1320-1328.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kim C, Kim J, Park HY, Lee JH, Park HJ, Kim CK, Yoon J. Appl Microbiol Biotechnol. 2009;83:1095. doi: 10.1007/s00253-009-1954-3. [DOI] [PubMed] [Google Scholar]
- 69.Chugani SA, Whiteley M, Lee KM, D’Argenio D, Manoil C, Greenberg EP. Proc Natl Acad Sci U S A. 2001;98:2752. doi: 10.1073/pnas.051624298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Brint JM, Ohman DE. J Bacteriol. 1995;177:7155. doi: 10.1128/jb.177.24.7155-7163.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.McInnis CE, Blackwell HE. Bioorg Med Chem. 2011;XX:XXX. doi: 10.1016/j.bmc.2011.06.071. Ms. Ref. No.: BMC-D-11-00637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vannini A, Volpari C, Gargioli C, Muraglia E, Cortese R, De Francesco R, Neddermann P, Di Marco S. EMBO J. 2002;21:4393. doi: 10.1093/emboj/cdf459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang RG, Pappas T, Brace JL, Miller PC, Oulmassov T, Molyneauz JM, Anderson JC, Bashkin JK, Winans SC, Joachimiak A. Nature. 2002;417:971. doi: 10.1038/nature00833. [DOI] [PubMed] [Google Scholar]
- 74.Breitbach AS, Broderick AH, Jewell CM, Gunasekaran S, Lin Q, Lynn DM, Blackwell HE. Chem Commun. 2010;47:370. doi: 10.1039/c0cc02316g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Braibante MEF, Braibante HTS, Morel AF, Costa CC, Lima MG. J Brazil Chem Soc. 2006;17:184. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.