Abstract
B-type natriuretic peptide (BNP) and its related peptides are biomarkers for the diagnosis of heart failure. Recent studies identified several O-glycosylation sites, including Thr-71, on human pro-BNP but the functional significance was unclear. In this study, we analyzed glycosylation and proteolytic processing of pro-BNP in cardiomyocytes. Human pro-BNP wild-type (WT) and mutants were expressed in HEK 293 cells and murine HL-1 cardiomyocytes. Pro-BNP and BNP were analyzed by immunoprecipitation and Western blotting. Glycosidases and glycosylation inhibitors were used to examine carbohydrates on pro-BNP. The effects of furin and corin expression on pro-BNP processing in cells also were examined. We found that in HEK 293 cells, recombinant pro-BNP contained significant amounts of O-glycans with terminal oligosialic acids. Mutation at Thr-71 reduced O-glycans on pro-BNP and increased pro-BNP processing. In HL-1 cardiomyocytes, residue Thr-71 contained little O-glycans, and pro-BNP WT and T71A mutant were processed similarly. In HEK 293 cells, pro-BNP was processed by furin. Mutations at Arg-73 and Arg-76, but not Lys-79, prevented pro-BNP processing. In HL-1 cardiomyocytes, which express furin and corin, single or double mutations at Arg-73, Arg-76 and Lys-79 did not prevent pro-BNP processing. Only when all these three residues were mutated, was pro-BNP processing completely blocked. Our data indicate that pro-BNP glycosylation in cardiomyocytes differed significantly from that in HEK 293 cells. In HEK 293 cells, furin cleaved pro-BNP at Arg-76 whereas in cardiomyocytes corin cleaved pro-BNP at multiple residues including Arg-73, Arg-76 and Lys-79.
Keywords: BNP, pro-BNP, corin, furin, glycosylation, cardiomyocytes
1. Introduction
B-type, or brain, natriuretic peptide (BNP) is a cardiac hormone that regulates blood pressure by promoting natriuresis, diauresis and vasodilation [1-3]. As a compensatory mechanism, BNP production is elevated in hypertrophic and failing hearts. Such a pathophysiologic response has been exploited to use BNP and its related peptides as biomarkers for the diagnosis of heart failure (HF) [4].
In cardiomyocytes, human BNP is made as a prepropeptide of 134 amino acids [5]. Removal of the 26-amino acid signal peptide generates pro-BNP of 108 amino acids, which is further cleaved to produce the C-terminal BNP 1-32 that is biologically active. To date, several proteases such as furin [6, 7] and corin [7-9] have been identified to process pro-BNP. Furin is a proprotein convertase in the Golgi of many cell types [10]. Corin is a cardiac protease that also activates atrial natriuretic peptide (ANP) [11-13]. In addition, dipeptidyl peptidase (DPP) IV has been shown to remove two N-terminal residues from BNP 1-32 to produce BNP 3-32 [14, 15].
In addition to proteolytic cleavage, pro-BNP undergoes other posttranslational modifications. Recent studies show that human plasma-derived pro-BNP and recombinant pro-BNP from mammalian cells contain significant amounts of O-glycans with terminal sialic acids [16-21]. T h e sialylated O-glycans protected pro-BNP from O-glycosidase digestion and stabilized pro-BNP in cell culture [17]. The O-glycans may also influence pro-BNP processing in cells. O-glycans on pro-BNP residue Thr-71 was reported to inhibit the propeptide processing in human embryonic kidney (HEK) 293 cells [21]. It was unknown if O-glycans have a similar inhibitory effect in cardiomyocytes.
Here we examined glycosylation and processing of pro-BNP in HEK 293 cells and cardiomyocytes. Our results showed that glycosylation and processing of pro-BNP in cardiomyocytes differed significantly from that in HEK 293 cells, and that in cardiomyocytes pro-BNP can be processed by corin at several different sites.
2. Materials and methods
2.1. Cell culture
HEK 293 cells were grown in 6-well plates in DMEM medium containing 10% fetal bovine serum (FBS). Murine atrial HL-1 cardiomyocytes from Dr. William Claycomb (Louisiana State University Medical Center) [22] were cultured in Claycomb medium (Sigma) with 10% FBS, 100 μM norepinephrine, and 4 mM L-glutamine. The cells were grown at 37°C in humidified incubators with 5% CO2 and 95% air.
2.2. Expression plasmids
Plasmids expressing human pro-BNP, corin and furin were reported previously [8, 23]. Plasmids expressing pro-BNP mutants T71A, R73A, R76A, K79A, R73A/R76A, R76A/K79A and R73A/R76A/K79A, in which residues Thr, Arg or Lys were replaced by Ala, were constructed by site-directed mutagenesis. All constructs were verified by DNA sequencing. Recombinant pro-BNPs encoded by these plasmids contained a V5 tag at their C-termini to facilitate protein detection.
2.3. Transfection, immunoprecipitation and Western blotting
Plasmids were transfected into HEK 293 and HL-1 cells using FuGENE (Roche Diagnostics) or Lipofectamine 2000 (Invitrogen) reagents. Conditioned medium from the transfected cells was collected and recombinant proteins were immunoprecipitated by an anti-V5 antibody. The cells were washed with a buffer and lysed in a solution containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% Nonidet P-40 (v/v), and a protease inhibitor cocktail (1:100 dilution, Sigma). Pro-BNP and BNP in the conditioned medium and cell lysate were analyzed by Western blotting using an anti-V5 antibody (Invitrogen), as described previously [24]. To quantify pro-BNP processing, the optical density of bands on X-ray films was measured densitometrically, and the percentage of pro-BNP to BNP conversion was calculated using computer software, as described previously [25].
2.4. Inhibition of O-glycosylatioin in cultured cells
To inhibit O-glycosylation on pro-BNP, we used benzyl 2-acetamido-2-deoxy-α-D-galactopyranoside (Ben-gal) (Sigma), which inhibits UDP-GlcNAc:GalNAc-β1,3-N-acetylglucosaminyl-transferase activity [26]. HEK 293 and HL-1 cells expressing recombinant pro-BNP were grown in the presence of Ben-gal (0.7 mmol/L for HEK 293 cells and 4 mmol/L for HL-1 cells) or vehicle control (DMSO) at 37°C overnight. The conditioned medium was collected and pro-BNP and BNP were analyzed by immunoprecipitation and Western blotting.
2.5. Glycosidase digestion
Glycosidases including PNGase F from Chryseobacterium meningosepticum, O-glycosidase from Streptococcus pneumonia and α(2-3,6,8,9) neuraminidase (also called sialidase A) from Arthrobacter ureafaciens (Prozyme) were used to analyze the carbohydrate contents on pro-BNP. Additional sialidases from Streptococcus pneumoniae, Clostridium perfringens, and Vibrio cholerae (Prozyme) that cleave α(2-3)-, α(2-3,6)- or α(2-3,6,8)-linked sialic acids, respectively, were used to predict sialic acid structures on pro-BNP. The conditioned medium containing pro-BNP from transfected HEK 293 and HL-1 cells was treated with glycosidases, either individually or in combination, at 37°C for 3 h. Proteins were analyzed by Western blotting using an anti-V5-HRP antibody.
2.6. Statistical analysis
Statistical analysis was done using Student's t-test. Data were presented as means ± S.D. A p value of <0.05 was considered to be statistically significant.
3. Results
3.1. Human pro-BNP WT and T71A mutant expressed in HEK 293 and HL-1 cells
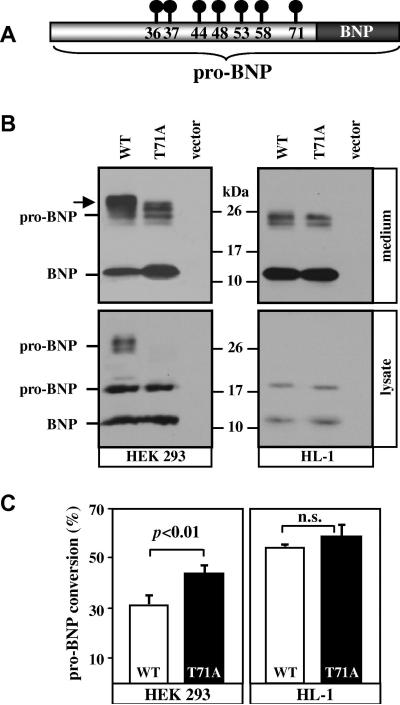
Previous studies identified several O-glycosylation sites, including Thr-71, in human recombinant pro-BNP from HEK 293 cells (Fig. 1A). To examine the importance of pro-BNP residue Thr-71 in glycosylation in cardiomyocytes, we expressed pro-BNP WT and T71A mutant in HEK 293 and HL-1 cells. In the conditioned medium from HEK 293 cells (Fig. 1B, top left), WT pro-BNP and BNP with a C-terminal V5 tag were detected on Western blots as ~26- and ~12-kDa bands, respectively. In the cell lysate (Fig. 1B, bottom left), two bands of pro-BNP, representing glycosylated (~26 kDa) and non-glycosylated (~20 kDa) forms, and a band of BNP (~12 kDa) were detected. Pro-BNP T71A from HEK 293 cell conditioned medium appeared smaller than pro-BNP WT in molecular mass (Fig. 1B, arrow in top left), indicating that the mutation prevented glycosylation at Thr-71. In HEK 293 cell lysate (Fig. 1B, bottom left), the non-glycosylated pro-BNP and mature BNP, but not the glycosylated pro-BNP, were detected in samples with pro-BNP T71A, indicating that most of the glycosylated pro-BNP T71A was secreted rapidly from the cells.
Fig. 1. Expression of pro-BNP and T71A mutant in HEK 293 and HL-1 cells.
(A) Schematic presentation of pro-BNP and BNP. Reported O-glycosylated residues are indicated by circle-and-line symbols. (B) Plasmids expressing pro-BNP WT and T71A mutant or a control vector were transfected into HEK 293 (left) or HL-1 (right) cells. Pro-BNP and BNP in the conditioned medium (top) and cell lysate (bottom) were analyzed by Western blotting. An arrow (top left) indicates the difference in molecular mass between pro-BNP WT and T71A. Data were representative of 4 independent experiments. (C) Quantitative analysis of pro-BNP processing. Percentage of pro-BNP processing was calculated by densitometric analysis of Western blots. Data are mean ± S.D. from 4 independent experiments. n.s., not statistically significant.
In HL-1 cells (Fig 1B, right), pro-BNP and BNP bands of similar sizes from WT and T71A mutant were detected in the conditioned medium (~24 kDa for pro-BNP and ~12 kDa for BNP) and cell lysate (~20 kDa for pro-BNP and ~12 kDa for BNP), suggesting that glycosylation at Thr-71 may not be significant in cardiomyocytes.
By densitometric analysis of the Western blots, the percentage of pro-BNP to BNP conversion for WT was much less than T71A mutant in HEK 293 cells (Fig. 1C, left), but similar to that of T71A mutant in HL-1 cells (Fig. 1C, right).
3.2. Glycosylation on pro-BNP T71A in HEK 293 and HL-1 cells
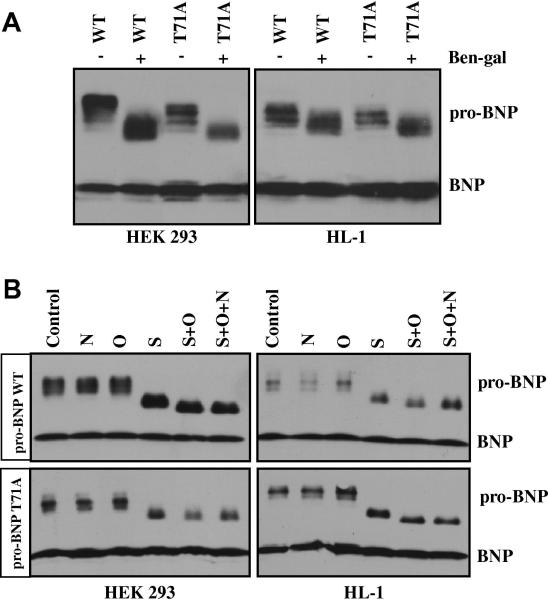
To examine O-glycosylation on pro-BNP, we treated HEK 293 and HL-1 cells expressing pro-BNP WT and T71A mutant with an O-glycosylation inhibitor, Ben-gal. The presence of Ben-gal in the culture medium reduced the apparent molecular mass of pro-BNP WT and T71A in HEK 293 (Fig. 2A, left) and HL-1 (Fig. 2A, right) cells, which was consistent with the previous finding of O-glycosylation in pro-BNP and indicated that mutation at Thr-71 did not prevent O-glycosylation at other pro-BNP residues.
Fig. 2. O-glycosylation in pro-BNP WT and T71A.
(A) HEK 293 (left) and HL-1 (right) cells expressing pro-BNP WT or T71A mutant were cultured with (+) or without (-) Ben-gal for 24 h. Pro-BNP and BNP in the conditioned medium were analyzed by Western blotting. Data were representative for at least three independent experiments. (B) Pro-BNP WT (top) and T71A mutant (bottom) from HEK 293 (left) or HL-1 (right) cells were digested with PNGase F (F), O-glycosidase (O), and sialidase A (S), individually or in combination, and analyzed by Western blotting. Data were representative for at least three independent experiments.
3.3. Treatment of pro-BNP T71A with glycosidases
To verify this result, we incubated pro-BNP WT and T71A mutant from HEK 293 and HL-1 cells with PNGase F, O-glycosidase, and sialidase A, individually or in combination. As shown in Fig. 2B, treatment of PNGase F or O-glycosidase alone did not alter the apparent molecular mass of pro-BNP WT (top) and T71A (bottom). When pro-BNPs were treated with sialidase A, alone or with O-glycosidase, the apparent molecular mass was reduced, indicating that sialylated O-glycans were present on pro-BNP WT and T71A mutant. PNGase F in combination with sialidase A and O-glycosidase showed no further reduction of the apparent mass of pro-BNP WT and T71A mutant, confirming that pro-BNP WT and T71A mutant contained significant amounts of sialylated O-glycans but little N-glycans.
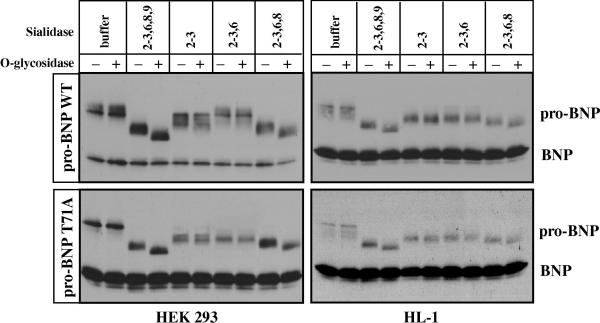
To gain insights into sialic acid structures on pro-BNP, we treated pro-BNP WT and T71A mutant with sialidases that favor specific linkages. When pro-BNP WT and T71A from HEK 293 and HL-1 cells were treated with sialidases that cleave α(2-3,6,8,9)- and α(2-3,6,8)-linkages, the apparent molecular mass of pro-BNP WT and T71A mutant was significantly reduced to a similar degree (Fig. 3). Treatment of sialidase that cleaves α(2-3)-linked sialic acids caused a smaller reduction in pro-BNP molecular mass, which was similar to the effect of sialidase that cleaves α(2-3,6)-linkages, indicating that most sialic acids on pro-BNP existed in the forms of either α(2-8)- or α(2-3)-linked moieties.
Fig. 3. Pro-BNP WT and T71A mutant digested with different sialidases.
Pro-BNP WT (top) and T71A mutant (bottom) from HEK 293 (left) or HL-1 (right) cells were digested with O-glycosidase with (+) or without (-) sialidases that favor specific linkages. Pro-BNP and BNP were analyzed by Western blotting. Data were representative for at least three independent experiments.
3.4. Processing of pro-BNP WT and T71A in HEK 293 and HL-1 cells
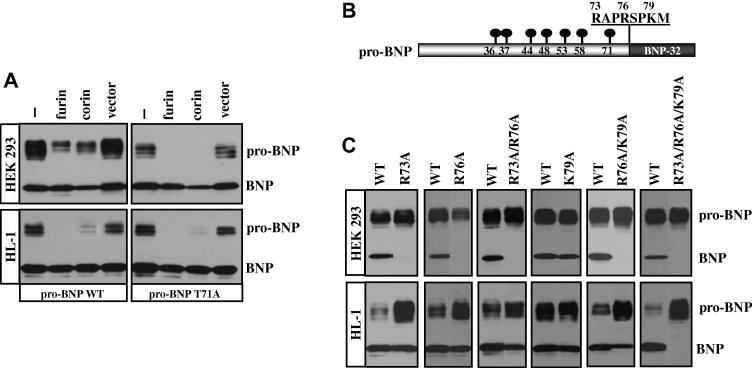
We examined the processing of pro-BNP WT and T71A mutant. When pro-BNP WT was expressed in HEK 293 or HL-1 cells, a significant portion of pro-BNP in the conditioned medium remained unprocessed (Fig. 4A, left). When recombinant furin or corin was co-expressed in these cells, pro-BNP processing was mostly completed in HL-1 cells (Fig. 4A, bottom left) but there were still significant portions of unprocessed pro-BNP in HEK 293 cells (Fig. 4A, top left). Similarly, significant portions of unprocessed pro-BNP T71A were present in the conditioned medium from HEK 293 and HL-1 cells (Fig. 4A, right). However, when recombinant furin or corin was co-expressed in these cells, pro-BNP T71A processing was mostly completed. The results showed that O-glycosylation at Thr-71 inhibited pro-BNP processing in HEK 293 cells, and that Thr-71 residue, which was not O-glycosylated in HL-1 cells, had little effect on pro-BNP processing in these cardiomyocytes.
Fig. 4. Processing of pro-BNP WT and mutants in HEK 293 and HL-1 cells.
(A) Pro-BNP WT (left) and T71A mutant (right) were expressed in HEK 293 (top) and HL-1 (bottom) cells that were either parental or co-transfected with plasmids expressing furin or corin, or a control vector. Pro-BNP and BNP in the conditioned medium were analyzed by Western blotting. (B) Schematic presentation of human pro-BNP with O-glycosylated residues and the cleavage site. (C) Pro-BNP WT and mutants with single, double or triple mutations were expressed in HEK 293 (top) or HL-1 (bottom) cells. Pro-BNP and BNP in the conditioned medium were analyzed by Western blotting. Data were representative for at least three independent experiments.
3.5. Pro-BNP processing sites in HEK 293 and HL-1 cells
Previous studies showed that both furin and corin cleaved pro-BNP. To examine the cleavage sites of furin- and corin-mediated pro-BNP processing, we tested pro-BNP mutants with altered residues at Arg-73, Arg-76 and Lys-79 around the cleavage site (Fig. 4B). In HEK 293 cells, which express furin but not corin, mutations at Arg-73, Arg-76 or Arg-73/Arg-76 prevented pro-BNP processing (Fig. 4C, top). In contrast, mutation at Lys-79 alone did not alter pro-BNP processing (Fig. 4C, top). Only when mutation at Lys-79 was combined with mutations at Arg-76 or Arg-73/Arg-76, was pro-BNP processing prevented. The results showed that Arg-73 and Arg-76, but not Lys-79, were important for furin-mediated pro-BNP processing in HEK 293 cells.
In HL-1 cells, in contrast, mutations at Arg-73 and Arg-76, alone or in combination, only slightly reduced pro-BNP processing (Fig. 4C, bottom). Nor did mutation at Lys-79, alone or in combination with Arg-76 mutation, prevent pro-BNP processing. Only when residues Arg-73, Arg-76 and Lys-79 were all mutated, was pro-BNP processing prevented (Fig. 4C, bottom), indicating that in HL-1 cells, which express both furin and corin, pro-BNP was processed at multiple sites including Arg-73, Arg-76, and Lys-79.
4. Discussion
ANP, BNP and C-type natriuretic peptide (CNP) are natriuretic peptide family members, which are made as inactive proforms and activated by proteolytic enzymes. Recent studies have identified significant amounts of O-glycans in human pro-BNP and this posttranslational modification appears to be unique for pro-BNP because no carbohydrates were detected in pro-ANP or pro-CNP [17]. In general, carbohydrates participate in protein trafficking, enzyme activation, and protein-protein interactions [25, 27, 28]. The function of O-glycans on pro-BNP is not fully understood. We showed that O-glycans increased pro-BNP stability in culture [17]. Others reported that O-glycans at residue Thr-71 markedly inhibited recombinant pro-BNP processing in HEK 293 cells [21]. This finding is intriguing, raising an inevitable question of how pro-BNP is possibly processed if similar O-glycosylation occurs in cardiomyocytes.
To address this question, we analyzed pro-BNP WT and T71A mutant in HEK 293 cells and HL-1 cardiomyocytes. We found that pro-BNP WT and T71A mutant in HEK 293 and HL-1 cells contained O-glycans with terminal sialic acids via α(2-3) and/or α(2-8) linkages (Figs. 2 and 3). The extent of pro-BNP O-glycosylation in cardiomyocytes, however, was less than that in HEK 293 cells. Consistent with the previous report of O-glycans on residue Thr-71 [20, 21], T71A mutation reduced pro-BNP glycosylation in HEK 293 cells (Fig. 1A). In these cells, pro-BNP WT, compared to T71A mutant, was more resistant to endogenous furin- and recombinant furin- or corin-mediated processing (Figs. 1C and 4A), supporting a role of O-glycans on Thr-71 in inhibiting pro-BNP processing.
Unlike in HEK 293 cells, residue Thr-71 contained little O-glycans when pro-BNP was expressed in HL-1 cardiomyocytes, as indicated by similar migration bands of pro-BNP and BNP from WT and T71A mutant on Western blots (Fig. 1B). Consistently, processing of pro-BNP WT and T71A mutant was similar in HL-1 cells, with or without furin or corin co-expression (Figs. 1C and 4A). Thus, our results indicate that O-glycosylation at pro-BNP residue Thr-71 identified in HEK 293 cells may not occur in cardiomyocytes. It is likely, therefore, that the function of O-glycans on pro-BNP may be involved primarily in peptide stability and/or extracellular distribution rather than inhibiting pro-BNP processing in cardiomyocytes.
Previously, we and others reported that furin and corin processed pro-BNP [6-9]. The sequence-specificity of furin- and corin-mediated pro-BNP processing is not well defined. Using non-glycosylated pro-BNP made in E. coli, Semenov et al. showed that furin cleaved pro-BNP at Arg-76, generating BNP 1-32, whereas corin cleaved pro-BNP at Lys-79, generating BNP 4-32 [7]. More recently, corin was shown to be shed from the cell surface and soluble corin was detected in human plasma [29-34]. Ichiki et al. reported that pro-BNP was processed by plasma soluble corin to produce BNP 1-32 and BNP 3-32 [32]. Because BNP 3-32 was derived from the cleavage between Pro-78↓Lys-79, it is unlikely that this fragment was cleaved directly by corin, which as a trypsin-like enzyme, favors basic residues [35]. Most likely, BNP 1-32 was produced first by plasma soluble corin and in turn trimmed N-terminally by DPP IV to BNP 3-32 [14, 15].
To determine the sequence-specificity of furin- and corin-mediated pro-BNP processing, we tested a series of pro-BNP mutants around the cleavage site. In HEK 293 cells expressing furin but not corin, mutations at Arg-73 and Arg-76, either alone or in combination, prevented pro-BNP processing (Fig. 4C, top). In contrast, mutation at Lys-79 had little effect on pro-BNP processing (Fig. 4C, top), indicating that furin cleaved pro-BNP at Arg-76, consistent with the furin recognition consensus sequence, RXXR↓, where the cleavage at the P1 Arg requires another basic residue at the P4 position [10].
Interestingly, in HL-1 cells expressing both furin and corin, single mutations at Arg-73, Arg-76, or Lys-79 did not prevent pro-BNP processing. Moreover, neither double mutations at Arg-73/Arg-76 nor Arg-76/Lys-79 prevented pro-BNP processing. Only when residues Arg-73, Arg-76 and Lys-79 were all mutated, was pro-BNP processing blocked (Fig. 4C, bottom). These data indicate that in HL-1 cells corin cleaved pro-BNP at multiple sites, including Arg-73, Arg-76 and Lys-79. Previous studies have shown that HL-1 cells retained all structural and functional characteristics of adult cardiomyocytes [22]. Our results suggest that in hearts corin may cleave pro-BNP at different sites. Consistent with this hypothesis, BNP fragments with different N-termini have been found in human plasma, including BNPs 1-32, 3-32 and 4-32 [36]. In principle, BNP 1-32 may be from corin- or furin-mediated cleavage, whereas BNP 4-32 may come from corin-mediated cleavage. BNP 3-32 is most likely from DPP IV-mediated cleavage of BNP 1-32 [14, 15]. Thus, our results provide new insights into the biochemical basis of pro-BNP processing. Studies have shown that corin expression is up-regulated in hypertrophic and failing hearts [37, 38]. High levels of plasma BNPs 1-32, 3-32 and 4-32 are found in patients with HF [36]. Further studies are needed to determine if the cleavage of pro-BNP by corin is regulated in the heart under physiological and pathological conditions.
Acknowledgments
We thank Dr. William Claycomb for HL-1 cells. This work was supported in part by grants from the Bakken Heart-Brain Institute, the National Institutes of Health (HL089298, HL089298-S1, HD064634), the National Natural Science Foundation of China (31070716), and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
Abbreviations
- ANP
atrial natriuretic peptide
- BNP
B-type or brain natriuretic peptide
- CNP
C-type natriuretic peptide
- HEK
human embryonic kidney
- HF
heart failure
- NT
N-terminal
- FBS
fetal bovine serum
- Ben-gal
Benzyl 2-acetamido-2-deoxy-α-D-galactopyranoside
References
- 1.Daniels LB, Maisel AS, Natriuretic peptides J. Am Coll Cardiol. 2007;50:2357–2368. doi: 10.1016/j.jacc.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 2.Goetze JP. Biochemistry of pro-B-type natriuretic peptide-derived peptides: the endocrine heart revisited. Clin Chem. 2004;50:1503–1510. doi: 10.1373/clinchem.2004.034272. [DOI] [PubMed] [Google Scholar]
- 3.Mair J. Biochemistry of B-type natriuretic peptide--where are we now? Clin Chem Lab Med. 2008;46:1507–1514. doi: 10.1515/CCLM.2008.295. [DOI] [PubMed] [Google Scholar]
- 4.Maisel AS, Krishnaswamy P, Nowak RM, McCord J, Hollander JE, Duc P, Omland T, Storrow AB, Abraham WT, Wu AH, Clopton P, Steg PG, Westheim A, Knudsen CW, Perez A, Kazanegra R, Herrmann HC, McCullough PA. Rapid measurement of B-type natriuretic peptide in the emergency diagnosis of heart failure. N Engl J Med. 2002;347:161–167. doi: 10.1056/NEJMoa020233. [DOI] [PubMed] [Google Scholar]
- 5.Wu Q, Xu-Cai YO, Chen S, Wang W. Corin: new insights into the natriuretic peptide system. Kidney Int. 2009;75:142–146. doi: 10.1038/ki.2008.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sawada Y, Suda M, Yokoyama H, Kanda T, Sakamaki T, Tanaka S, Nagai R, Abe S, Takeuchi T. Stretch-induced hypertrophic growth of cardiocytes and processing of brain-type natriuretic peptide are controlled by proprotein-processing endoprotease furin. J Biol Chem. 1997;272:20545–20554. doi: 10.1074/jbc.272.33.20545. [DOI] [PubMed] [Google Scholar]
- 7.Semenov AG, Tamm NN, Seferian KR, Postnikov AB, Karpova NS, Serebryanaya DV, Koshkina EV, Krasnoselsky MI, Katrukha AG. Processing of pro-B-type natriuretic peptide: furin and corin as candidate convertases. Clin Chem. 2010;56:1166–1176. doi: 10.1373/clinchem.2010.143883. [DOI] [PubMed] [Google Scholar]
- 8.Wang W, Liao X, Fukuda K, Knappe S, Wu F, Dries DL, Qin J, Wu Q. Corin variant associated with hypertension and cardiac hypertrophy exhibits impaired zymogen activation and natriuretic peptide processing activity. Circ Res. 2008;103:502–508. doi: 10.1161/CIRCRESAHA.108.177352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan W, Wu F, Morser J, Wu Q. Corin, a transmembrane cardiac serine protease, acts as a pro-atrial natriuretic peptide-converting enzyme. Proc Natl Acad Sci U.S.A. 2000;97:8525–8529. doi: 10.1073/pnas.150149097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Steiner DF, Smeekens SP, Ohagi S, Chan SJ. The new enzymology of precursor processing endoproteases. J Biol Chem. 1992;267:23435–23438. [PubMed] [Google Scholar]
- 11.Pan J, Hinzmann B, Yan W, Wu F, Morser J, Wu Q. Genomic structures of the human and murine corin genes and functional GATA elements in their promoters. J Biol Chem. 2002;277:38390–38398. doi: 10.1074/jbc.M205686200. [DOI] [PubMed] [Google Scholar]
- 12.Wu Q. The serine protease corin in cardiovascular biology and disease. Front Biosci. 2007;12:4179–4190. doi: 10.2741/2379. [DOI] [PubMed] [Google Scholar]
- 13.Yan W, Sheng N, Seto M, Morser J, Wu Q. Corin, a mosaic transmembrane serine protease encoded by a novel cDNA from human heart. J Biol Chem. 1999;274:14926–14935. doi: 10.1074/jbc.274.21.14926. [DOI] [PubMed] [Google Scholar]
- 14.Boerrigter G, Costello-Boerrigter LC, Harty GJ, Lapp H, Burnett JC., Jr. Des-serine proline brain natriuretic peptide 3-32 in cardiorenal regulation. Am J Physiol Regul Integr Comp Physiol. 2007;292:R897–901. doi: 10.1152/ajpregu.00569.2006. [DOI] [PubMed] [Google Scholar]
- 15.Brandt I, Lambeir AM, Ketelslegers JM, Vanderheyden M, Scharpe S, De Meester I. Dipeptidyl-peptidase IV converts intact B-type natriuretic peptide into its des-SerPro form. Clin Chem. 2006;52:82–87. doi: 10.1373/clinchem.2005.057638. [DOI] [PubMed] [Google Scholar]
- 16.Crimmins DL, Kao JL. A glycosylated form of the human cardiac hormone pro B-type natriuretic peptide is an intrinsically unstructured monomeric protein. Arch Biochem Biophys. 2008;475:36–41. doi: 10.1016/j.abb.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang J, Pristera N, Wang W, Zhang X, Wu Q. Effect of sialylated O-glycans in pro-brain natriuretic peptide stability. Clin Chem. 2010;56:959–966. doi: 10.1373/clinchem.2009.140558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang F, O'Rear J, Schellenberger U, Tai L, Lasecki M, Schreiner GF, Apple FS, Maisel AS, Pollitt NS, Protter AA. Evidence for functional heterogeneity of circulating B-type natriuretic peptide. J Am Coll Cardiol. 2007;49:1071–1078. doi: 10.1016/j.jacc.2006.10.063. [DOI] [PubMed] [Google Scholar]
- 19.Mair J. Clinical significance of pro-B-type natriuretic peptide glycosylation and processing. Clin Chem. 2009;55:394–397. doi: 10.1373/clinchem.2008.119271. [DOI] [PubMed] [Google Scholar]
- 20.Schellenberger U, O'Rear J, Guzzetta A, Jue RA, Protter AA, Pollitt NS. The precursor to B-type natriuretic peptide is an O-linked glycoprotein. Arch Biochem Biophys. 2006;451:160–166. doi: 10.1016/j.abb.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 21.Semenov AG, Postnikov AB, Tamm NN, Seferian KR, Karpova NS, Bloshchitsyna MN, Koshkina EV, Krasnoselsky MI, Serebryanaya DV, Katrukha AG. Processing of pro-brain natriuretic peptide is suppressed by O-glycosylation in the region close to the cleavage site. Clin Chem. 2009;55:489–498. doi: 10.1373/clinchem.2008.113373. [DOI] [PubMed] [Google Scholar]
- 22.Claycomb WC, Lanson NA, Jr., Stallworth BS, Egeland DB, Delcarpio JB, Bahinski A, Izzo NJ., Jr. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci U S A. 1998;95:2979–2984. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu C, Wu F, Pan J, Morser J, Wu Q. Furin-mediated processing of pro-C-type natriuretic peptide. J Biol Chem. 2003;278:25847–25852. doi: 10.1074/jbc.M301223200. [DOI] [PubMed] [Google Scholar]
- 24.Knappe S, Wu F, Madlansacay MR, Wu Q. Identification of domain structures in the propeptide of corin essential for the processing of proatrial natriuretic peptide. J Biol Chem. 2004;279:34464–34471. doi: 10.1074/jbc.M405041200. [DOI] [PubMed] [Google Scholar]
- 25.Liao X, Wang W, Chen S, Wu Q. Role of glycosylation in corin zymogen activation. J Biol Chem. 2007;282:27728–27735. doi: 10.1074/jbc.M703687200. [DOI] [PubMed] [Google Scholar]
- 26.Kuan SF, Byrd JC, Basbaum C, Kim YS. Inhibition of mucin glycosylation by aryl-N- acetyl-alpha-galactosaminides in human colon cancer cells. J Biol Chem. 1989;264:19271–19277. [PubMed] [Google Scholar]
- 27.Eklund EA, Freeze HH. Essentials of glycosylation. Semin Pediatr Neurol. 2005;12:134–143. doi: 10.1016/j.spen.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 28.Varki A. Biological roles of oligosaccharides: all of the theories are correct. Glycobiology. 1993;3:97–130. doi: 10.1093/glycob/3.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong N, Chen S, Yang J, He L, Liu P, Zhen D, Li L, Zhou Y, Ruan C, Plow E, Wu Q. Plasma soluble corin in patients with heart failure. Circ Heart Fail. 2010;3:207–211. doi: 10.1161/CIRCHEARTFAILURE.109.903849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong N, Dong J, Liu P, Xu L, Shi S, Wu Q. Effects of anticoagulants on human plasma soluble corin levels measured by ELISA. Clin Chim Acta. 2010;411:1998–2003. doi: 10.1016/j.cca.2010.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ibebuogu UN, Gladysheva IP, Houng AK, Reed GL. Decompensated heart failure is associated with reduced corin levels and decreased cleavage of pro-atrial natriuretic Peptide. Circ Heart Fail. 2011;4:114–120. doi: 10.1161/CIRCHEARTFAILURE.109.895581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ichiki T, Huntley BK, Heublein DM, Sandberg SM, McKie PM, Martin FL, Jougasaki M, Burnett JC., Jr. Corin is present in the normal human heart, kidney, and blood, with pro-B-type natriuretic peptide processing in the circulation. Clin Chem. 2011;57:40–47. doi: 10.1373/clinchem.2010.153908. [DOI] [PubMed] [Google Scholar]
- 33.Jiang J, Wu S, Wang W, Chen S, Peng J, Zhang X, Wu Q. Ectodomain shedding and autocleavage of the cardiac membrane protease corin. J Biol Chem. 2011;286:10066–10072. doi: 10.1074/jbc.M110.185082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peleg A, Jaffe AS, Hasin Y. Enzyme-linked immunoabsorbent assay for detection of human serine protease corin in blood. Clin Chim Acta. 2009;409:85–89. doi: 10.1016/j.cca.2009.09.004. [DOI] [PubMed] [Google Scholar]
- 35.Knappe S, Wu F, Masikat MR, Morser J, Wu Q. Functional analysis of the transmembrane domain and activation cleavage of human corin. J Biol Chem. 2003;278:52363–52370. doi: 10.1074/jbc.M309991200. [DOI] [PubMed] [Google Scholar]
- 36.Niederkofler EE, Kiernan UA, O'Rear J, Menon S, Saghir S, Protter AA, Nelson RW, Schellenberger U. Detection of endogenous B-type natriuretic peptide at very low concentrations in patients with heart failure. Circ Heart Fail. 2008;1:258–264. doi: 10.1161/CIRCHEARTFAILURE.108.790774. [DOI] [PubMed] [Google Scholar]
- 37.Chen S, Sen S, Young D, Wang W, Moravec CS, Wu Q. Protease corin expression and activity in failing hearts. Am J Physiol Heart Circ Physiol. 2010;299:H1687–1692. doi: 10.1152/ajpheart.00399.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tran KL, Lu X, Lei M, Feng Q, Wu Q. Upregulation of corin gene expression in hypertrophic cardiomyocytes and failing myocardium. Am J Physiol Heart Circ Physiol. 2004;287:H1625–1631. doi: 10.1152/ajpheart.00298.2004. [DOI] [PubMed] [Google Scholar]