Abstract
Ecological specialisation concerns all species and underlies many major ecological and evolutionary patterns. Yet its status as a unifying concept is not always appreciated because of its similarity to concepts of the niche, the many levels of biological phenomena to which it applies, and the complexity of the mechanisms influencing it. The evolution of specialisation requires the coupling of constraints on adaptive evolution with covariation of genotype and environmental performance. This covariation itself depends upon organismal properties such as dispersal behaviour and life history and complexity in the environment stemming from factors such as species interactions and spatio-temporal heterogeneity in resources. Here, we develop a view on specialisation that integrates across the range of biological phenomena with the goal of developing a more predictive conceptual framework that specifically accounts for the importance of biotic complexity and coevolutionary events.
Keywords: Behaviour, coevolution, competition, constraints, environment, mutualism, niche, parasitism, predation, specialisation, species interactions
A NEED FOR CONCEPTUAL SYNTHESIS
There is renewed interest in how the ecological niche may evolve and how this affects population persistence and evolution. Identifying niche components that are labile to change either individually or as multi-trait complexes essentially amounts to understanding ecological specialisation. Despite the widespread interest that specialisation has generated going back to Darwin and the seminal synthesis by Futuyma & Moreno (1988), its importance as a broad unifying concept is not always appreciated. This is due to a lack of clarity in the terminology and in particular its similarity to concepts of the niche and local adaptation (see Glossary), its measurement, the many interacting factors influencing it (abiotic environment, genetic, individual, population and community), and the diverse biological phenomena to which it applies (physiological, functional, habitat, behavioural and taxonomic).
Ecological specialisation is the process of adaptation to a subset of possible environments (see Glossary). Specialisation underlies major patterns in the genesis, distribution and persistence of biological diversity. For example, the classic solution to the puzzle of what allows coexistence of competitors has been that specialisation on different resources (i.e. resource partitioning) reduces the strength of competition between species. This hypothesis has been remarkably successful in explaining patterns of diversity in animals, but less so for plants (Miller et al. 2005). Specialisation, however, plays equally critical roles in mechanisms advanced to explain coexistence of plant species, including specialisation on a non-resource environmental axis, such as temperature. Recent work suggests that interactions with specific enemies can play a primary role in local plant species coexistence (e.g. Mangan et al. 2010).
Given its broad biological relevance, ecological specialisation has been the subject of a number of recent reviews (e.g. Bolnick et al. 2003; Holt 2009; Ravigné et al. 2009; Devictor et al. 2010). However, despite the considerable attention received, our current knowledge of what specialisation entails, and the conditions that may favour its evolution, is incomplete. A substantial part of our knowledge comes from relatively simple situations in which resources exploited by focal organisms are abiotic or not evolving (Ravigné et al. 2009). However, considerable empirical work indicates that specialisation differs in predictable ways between different types of biotic interaction (Box 1). This important corpus of literature does not provide an overarching framework to understand the evolution of specialisation, owing mostly to the frequent omission of the impact of biotic complexity.
Recent insights building on decades of groundwork now enable a conceptual synthesis of the mechanisms underlying the evolution of ecological specialisation. Here, we review current knowledge, and develop a conceptual framework to promote our understanding of how specialisation may or may not evolve and how patterns in species interactions emerge. Synthesis of the literature points to two main interacting processes underlying specialisation: constraints on evolution and covariation of genotype with environmental performance. We further discuss the major role played by biotic effects and in particular multispecies interactions on the mode and complexity of specialisation.
FUNDAMENTALS OF SPECIALISATION
Specialisation occurs through adaptation to a restricted spectrum of environments and/or restriction in the availability of environments without evolutionary change (Bolnick et al. 2003; Devictor et al. 2010). In many ways, parallel to concepts of fundamental and realised niches, adaptation to a restricted range of environments is sometimes called ‘potential specificity’, while the observed use of these environments is termed ‘realised specificity’. Potential specificity is determined by evolutionary interactions between genotype and environment, whereas realised specificity reflects the impact of ecology, chance events and history on potential specificity (Bolnick et al. 2003; Devictor et al. 2010). Specialisation can also be viewed at different levels of biological organisation (e.g. among species in a community or among individuals within populations). As such, the processes of specialisation can involve divergence of a population on multiple types of environments, or the fixation of genotypes within a population on these same environment types. Bolnick et al. (2003) have emphasised that many apparently generalist species are in fact composed of a range of ecologically variable, individual specialists.
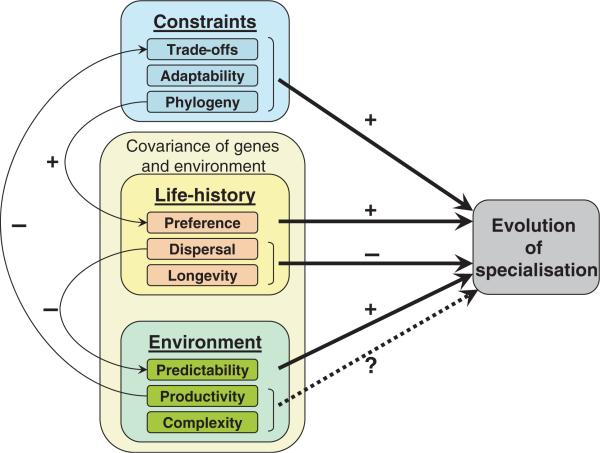
Adaptive specialisation is driven by constraints on performance across environments and covariance of genotype or species with these environments (Fig. 1). The covariance of genotypes or species with environment depends on behavioural and life history characteristics of the species and how they interact with the environment, and is reinforced when preference and performance are positively associated. The complexity of the environment itself, particularly the complexity of the biotic environment, can become a dominant factor driving the covariance of species with environment. We develop the individual elements of these processes in detail below.
Figure 1.
Conceptual model for the evolution of ecological specialisation. The two main forces affecting specialisation are fundamental biological constraints and the covariance between genes and environment. Generally, constraints tend to increase specialisation, although these can be mediated by environmental productivity, which can relax trade-offs. The effects of genotype × environment covariance depend on the component under consideration, how it is modified by constraints on individuals and populations, and interactions between life-history and environment. For example, preference for particular environments will increase the strength of genotype × environment correlations and thus favour increased specialisation; this may be further reinforced by phylogenetic constraints. By contrast, dispersal per se (i.e. uncoupled to preference or performance) will tend to decrease predictability and promote generalisation. All else being equal, variation in abiotic components of the environment will favour generalists, but the effects of community diversity are more difficult to characterise, especially when several species are involved in coevolutionary interactions.
CONSTRAINTS ON EVOLVABILITY
Theories of adaptive specialisation are based on differential adaptation to a subset of potentially encountered environments. Increased performance in some environments is generally assumed to be associated with decreased performance in others as a consequence of trade-offs or constraints (Kassen 2002). Constraints can result from limits of physiological performance, morphology or development. For example, the C4 photosynthetic pathways have greater water-use efficiency, but lower photosynthetic rates in cool, moist environments (Edwards et al. 2010). Such antagonistic pleiotropic effects generate unbreakable constraints, the existence of which is well established, having been studied in many fields of organismal biology (Laubichler & Maienschein 2009). However, the ubiquity of constraints is a matter of debate, with a recent comparative study suggesting weak trade-offs in locally adapted plant and animal species (Hereford 2009), and experimental demonstration of costs and fitness trade-offs for some advantageous phenotypes, such as host defence and pathogen infectivity, providing mixed support (Bingham & Agrawal 2010). Such inconsistencies, however, could reflect difficulties due to statistical power, measuring irrelevant traits (Phillips & Shine 2007), the multidimensionality of trade-offs and biotic interactions, and the fact that measuring specificity in such interactions is strongly context-dependent (i.e. the subset of genotypes actually evaluated – Box 2).
Often the genetic basis underlying a given trade-off is unknown. Besides antagonistic pleiotropy, constraints can be generated by polygenic sources, such as epistatic interactions. However, these constraints may erode over time and therefore their role in promoting specialisation has been questioned (Joshi & Thompson 1995). Theory shows that specialisation under constraints may occur from either stabilising or directional selection on a character, and depend on, for example, the shape of the trade-off (e.g. Egas et al. 2004). Moreover, if the expression of adaptation costs is polygenic and these genes are not at equilibrium, then trade-offs are likely to be differentially expressed in time, thus decreasing selection for specialisation (Joshi & Thompson 1995). Clearly, work integrating across molecular, physiological and population levels, as made possible with recent technological advances (Box 3), is necessary to dissect the underlying genetics of trade-offs that result in specialisation and to develop a quantitative understanding of how these flow through to impacts on species and community ecology. Below, we evaluate the impacts of two types of constraints, those acting at the population level and those stemming from the phylogenetic history of the focal population.
Constraints at the population level
In addition to the trade-offs mediated directly by pleiotropic or genetic interactions at the individual level, specialisation could be affected through limitations on population evolvability, which will depend in part on the complexity of the genetic system of the trait under consideration. A recent theoretical study in multi-genic regulatory systems concluded that the main constraint on adaptability was the size of the gene network (Malcom 2011), with smaller networks being more adaptable (and thus promoting persistence in fluctuating environments).
At the most basic level, specialisation, like any other adaptive process, necessitates genetic variation within populations and hence may be limited by the rate of introduction of new alleles through mutation or gene flow, although high levels of migration can lead to a decrease in specialisation (Venail et al. 2008). For example, Kellermann et al. (2009) recently proposed that the lack of additive genetic variation in specialist Drosophila species might be responsible for their restricted geographical ranges. This is congruent with recent results suggesting that specialised taxa display less genetic and phenotypic variation than their generalist sister species (Kaci-Chaouch et al. 2008). But specialisation may also occur through the loss of either genetic variation or complexes of adaptive traits, for example through the accumulation of deleterious mutations due to relaxed selection associated with phenotypic plasticity (Snell-Rood et al. 2010), or in small populations that degrade performance in other environments (Kawecki 1994). The latter may occur ecologically when one environment increases in relative frequency relative to individual longevity, thus mitigating the impact of mutations with (slightly) deleterious effects in other environments and promoting specialisation (Jasmin & Kassen 2007). The potential importance of these latter processes in generating specificity in other biological situations requires further research.
Phylogenetic constraints
There is also a growing appreciation that a variety of constraints could have evolved at deeper nodes in species phylogenies and thus be inherited through evolutionary history (Diniz-Filho & Bini 2008). Phylogenetic conservatism constrains adaptability, due to the inheritance of traits conferring adaptation to ancestral environments – this renders some future mutations impossible due either to contingencies (e.g. developmental, metabolic), or because they would have deleterious effects, due for instance to pleiotropic effects associated with ancestral adaptations. For example, in Lamellodiscus parasitic flatworms, it was shown that 45% of observed host specificity is explained by phylogenetic constraints and 24% due to contemporary flatworm species exploiting environments (i.e. hosts) related to ancestral ones (Desdevises et al. 2002), meaning that only the remaining 30% of the observed specificity is explained by contemporary environmental conditions. Quantifying the proportion of current ecological traits of a species or population that is inherited through phylogenetic history is an important goal and requires special methods to accurately estimate the rate of trait change over time (Cooper et al. 2010).
COVARIATION OF GENOTYPE WITH ENVIRONMENT
Specialisation requires differential adaptation to a restricted set of environments and this is facilitated by positive covariation of specific genotypes with environments in which they tend to perform best. As a result, spatial and temporal aspects of environmental heterogeneity are likely to have opposing effects on local adaptation; the former will generally favour specialisation, while temporal variability will tend to promote generalisation (Kassen 2002; Abrams 2006a; Poisot et al. 2011a). Clearly though, the relevant scale of environmental variation (i.e. the spatio-temporal grain over which adaptation can occur) must be calibrated against life history and behavioural characters of the organism (e.g. dispersal ability, longevity). Similarly, the spatial and temporal scales of environmental variation can be modified or even driven by biotic interactions, which may either restrict (e.g. predators and parasites) or expand (e.g. symbiotic mutualists) the range of environments where persistence is possible. These aspects are covered in the following two sections.
Dispersal and life history
Coarse spatial environmental grain relative to individual movement will favour specialisation, since individuals will only experience a subset of environments encountered by the population, and populations only a subset of local and regional environments over which the species occurs (Levins 1968; Pandit et al. 2009). Widely dispersing organisms are more likely to have opportunities to expand their range (e.g. Davies & Pedersen 2008), although extremely strong trade-offs can lead to specialisation even under random dispersal (Levins 1968). Meta-analysis (Woolhouse & Gowtage-Sequeria 2005) indicates that pathogens expand their host range by switching to those they most frequently encounter, suggesting that frequent contacts between populations will (at least in the short-term) result in decreased specificity. Classic theory predicts that specialist genotypes are more likely to coexist when they experience a single environment throughout their life cycle (Levins 1968). Consistent with this prediction, sessile organisms or those with limited dispersal such as many plants, phytophagous mites and chewing lice, have locally adapted types coexisting at relatively small spatial scales (Reed & Hafner 1997).
While local dispersal can select for specialisation, the long-term persistence of specialists in a temporally variable environment depends upon the ability of the specialist to colonise new patches of its optimal environment, particularly when local extinctions occur. For example, recent empirical evidence (Brückmann et al. 2010) shows that decreasing habitat connectivity dramatically decreases the abundance of specialists (up to 69%) in both plants and butterflies. Alternatively, the recolonisation of new patches could be achieved via dispersal through time as enabled by the production of dormant structures (e.g. seeds in plants or ephippia in Daphnia spp.; Hairston & Kearns 2002). Such bet-hedging strategies have been shown to be important in the persistence of desert annuals, which specialise on good years (Venable 2007). The patterns emerging from these processes can nonetheless be obscured for organisms that display phenotypic plasticity or other mechanisms that increase environmental tolerance.
Behavioural selectivity
While fine-grained spatial and temporal heterogeneity in the environment and negative frequency dependent selection generally impede the evolution of specialisation, positive covariance of genotype with the environments in which it tends to perform best can be generated and reinforced by preferential movement or association. For example, Ravigné et al. (2009) used mathematical models to show that the joint evolution of habitat selection and preference increased the range of conditions allowing choosy specialists to coexist. Behavioural choice enables an organism to influence future individual performance and reduce energy wasted on sub-optimal environments. Environmental preference can become genetically linked with performance if the former leads to assortative mating, even in the presence of gene flow. An example of this can be seen in pea aphids (Acyrthosiphon pisum pisum), which vary in host preference – quantitative trait loci (QTLs) with antagonistic effects on performance on different hosts appear to be linked to QTLs that control habitat choice (Via & Hawthorne 2002). Abrams (2006b) showed how behavioural plasticity in host exploitation permitted more specialised types to persist in fluctuating environments, but enabled greater biotic complexity (i.e. the coexistence of specialist and generalist types). This feedback between behavioural traits (individual-level) and biotic complexity (community-level) will likely have complex consequences on specialisation and deserves further investigation.
A major unresolved issue is the apparent mismatch between preference and performance in some systems (Futuyma & Moreno 1988; Poore & Steinberg 1999), but not in others (e.g. Forister 2004). In some cases, such mismatches can be explained by the genetic basis of the traits involved, as demonstrated in the butterfly genus Papilio, where genes involved in preference and performance for host plant exploitation are independently transmitted (Thompson et al. 1990). A recent meta-analysis (Gripenberg et al. 2010) in insect–plant associations, however, suggests that the alignment of preferences and performances is more likely to be the rule rather than the exception, although this investigation needs to be expanded to include other biological systems. Mathematical analysis (Nosil et al. 2006) has shown that, all else being equal, covariance between preference and performance evolves to be higher in heterogeneous environments in which locally adapted populations migrate, than in homogeneous environments. This supports the idea that the evolution of specialisation can be driven by complex interactions between habitat structure and dispersal patterns.
ENVIRONMENTAL PREDICTABILITY AND SPECIES INTERACTIONS
Biotic and abiotic components of environments may themselves be altered by organisms, resulting in changed specialisation due to local adaptation or local extinction. Here, we identify four fundamental ways in which biological activity may qualitatively alter the covariation of genotypes with their favoured environment, thus either increasing or decreasing environmental predictability.
Modification of the physical environment
In some sense, the simplest situation is one in which the physical environment is modified, or constructed, to create locally favourable conditions for the organism that increase predictability. Ecosystem engineering and niche construction describe situations in which organisms create favourable environments used by other species and themselves respectively (e.g. earthworms, beavers). Niche construction for example, by increasing correlation of genotype with environment, can then enhance specialisation with respect to the created environment.
The inclusion of species interactions (e.g. predator-prey, competition, host-symbiont) adds a layer of complexity to predictions about specialisation, given the potential for both ecological and evolutionary knock-on effects and feedbacks, context-dependent selection and the nature of the interaction (e.g. competition, parasitism, predation or mutualism). We discuss these situations next.
Altered access to favourable environments by competition and enemy-victim associations
The predictability of access to different environments can be reduced through competitive pre-emption by other species (Fig. 2). This preemption could be the result of competition with evolved specialists in other species. Given that selection to reduce intraguild competition is fundamental to the process of specialisation, one might expect that greater competition would facilitate specialisation. However, phylogenetic analyses do not indicate that lineages become more specialised over macroevolutionary time, which suggests that competition can favour the emergence of generalists as well (Johnson et al. 2009), perhaps as a result of increased environmental variance due to temporal heterogeneity in competition.
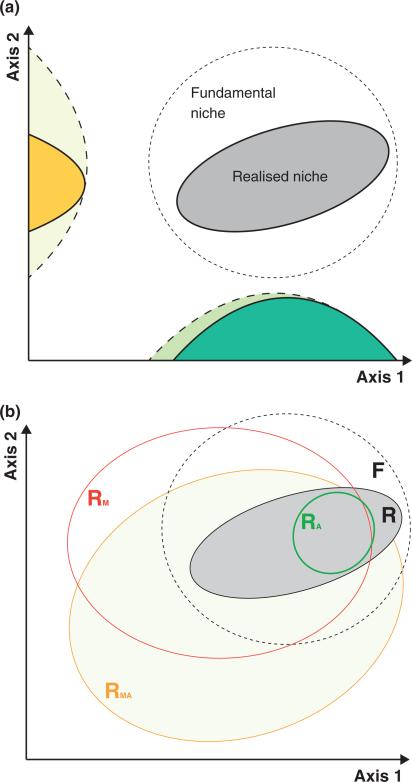
Figure 2.
Links between specificity and niche space, and the relevance of biotic interactions. (a) While the niche is classically defined as the intersection of tolerances over multiple environmental axes, specificity is the breadth of tolerance on each axis (Futuyma & Moreno 1988). Integrating specificity over all environmental axes defines the niche; thus fundamental and realised niches are, respectively, associated with potential and realised specificity [coloured areas correspond to hypothetical relative frequencies in tolerances or performances (dashed line = fundamental; solid line = realised) for each independent environmental axis]. How specialisation and niche evolution will relate depends on the correlations and pleiotropic effects linking niche axes. (b) The traditional definition of the fundamental niche (F) is that it represents the total multi-dimensional ecological space in which a species could persist. The realised niche (R) is the ecological space in which a species actually persists and is at least partly dependent on biotic interactions with other species. Biotic interactions have the potential to change the presence of a focal species along one or more of the axes that define F. For example, associations with antagonists (predators, pathogens or competitors) may further constrain the realised niche (RA), while facultative mutualists may increase the potential for a species to expand or shift its realised (and even fundamental) niche along one or more axes depending on the nature of interactions with other species in the community (RM,A and RM).
Interactions with antagonists could similarly alter associations of genotype and environment, through reduced fitness in habitats frequented by antagonists. Examples include behavioural shifts to avoid enemies or reduce disease risk (e.g. primate social groups are thought to shift home territories to minimise exposure to parasites that build up in the environment), local extinction-recolonisation dynamics within existing (long-standing) host–pathogen interactions, or more widespread extinction and reduction of host ranges by emerging pathogens (e.g. chytridiomycosis in amphibians; Tasmanian devil facial tumour). Alternatively, the potential for introduced pathogens to fundamentally change ecosystem structure through impacts on the distribution of key native hosts could also alter conditions favouring specialisation in invaded communities. For example, there are an increasing number of cases where exotic fungal pathogens have significantly altered forest ecosystems (Loo 2008); we still know far too little about the longer term ecological and evolutionary consequences of such invasions.
Currently, it is difficult to make general predictions as to whether the outcome of such interactions should lead to increased or reduced specialisation and well-characterised empirical examples are scarce. In some situations, disruption of correlations of genotype with their optimal environment will favour generality as suggested by a theoretical model demonstrating that phytoplankton can avoid marine virus predation by evenly exploiting several nutrients (Menge et al. 2011). In other cases, the result may be greater specialisation (e.g. where the victim is restricted to a narrower range of environments in which enemies cannot persist). For example, an empirical study demonstrated that the generalist caterpillar Grammia geneura altered its pattern of food preference in the presence of a parasitoid (Singer et al. 2004); in this case, specialisation on a moderately toxic plant of low nutritive value conferred protection against the enemy.
Niche expansion through the benefits conferred by mutualisms
In contrast to antagonistic interactions, mutualists (particularly microbial symbionts) can expand host tolerance to environments (Fig. 2) and thereby increase generalism (e.g. mycorrhizal fungi increasing plant access to soil resources). In effect, these symbiotic associations alleviate constraints (i.e. trade-offs) that might otherwise form the foundations of resource specialisation. For example, invasive plants may perform better in new environments if they can benefit from the presence of mutualists similar in their function or strategy to those existing in the plant's natural range (Richardson et al. 2000). However, associations with symbionts themselves have costs, and these costs may ultimately affect the evolutionary dynamics of their hosts (Bever et al. 2010). Finally, recent empirical evidence indicates that interactions with mutualists can lead hosts to specialise in the same niche space, resulting in mutualism-induced competition (Elias et al. 2008).
Coevolutionary interactions
The nature of an ecological interaction (mutualistic or antagonistic) may result in positive or negative changes in the density, the distribution and the quality of species-as-environments. For example, theoretical work shows that if a predator has a sufficient impact on its prey populations, then the former can be selected to either increase or decrease specialisation (e.g. Abrams 2006a). The processes underlying specialisation in coevolutionary associations may differ from those where only one species evolves. Below, we review literature pertaining to two broad classes of coevolving interactions: mutualisms and enemy–victim associations.
Mutualism
Although antagonistic associations and in particular, parasitism, are often regarded as producing high levels of specialisation, recent empirical evidence shows that in insect sister groups, coevolution can lead mutualist taxa to evolve higher levels of specialisation than their antagonistic counterparts (Kawakita et al. 2010). While common symbiotic mutualists [e.g. N2-fixing bacteria (Thrall et al. 2008)] may have low specificity of association, they can have high specificity of impacts, in many ways similar to the issue of potential and realised specificity (Devictor et al. 2010). Specificity of association can contribute to the evolutionary stability of mutualisms if there is a positive association between specificity and the effectiveness of mutualism, as has been found for example in nematode-bacterium mutualisms (Chapuis et al. 2009). Preferential allocation to the most beneficial symbionts after association, as has been shown in mycorrhizal fungi (Bever et al. 2009) and rhizobia (Kiers et al. 2003), can also reinforce the mutualism. These coevolutionary dynamics could generate positive frequency dependence, which can reinforce specificity and lead to codivergence (Machado et al. 2005; Elias et al. 2008).
Reinforcing coevolution could also yield three-way specialisation in the context of host–symbiont environment interactions, in which the host genotype performs best when matched with the specialised symbiont in a particular environment. Some evidence of such co-adaptation has been found in acacia-rhizobia associations along a salinity gradient (Thrall et al. 2008). In a more dramatic illustration of mutualist mediation of resource use, plant tolerance to high temperatures has been shown to be conferred by a mutualistic endophytic fungus that itself requires infection with a virus to persist in these environments (Marquez et al. 2007). In this case, specialisation across a physical gradient requires mutualism with the fungal endophyte on the one hand and affinity of the fungus for the virus on the other hand. Recent evidence indicates that parasitoids may evolve faster when faced with hosts employing symbiont-induced defences (Dion et al. 2011), to maintain pace with on-going coevolution between the host and its defensive symbiont.
However, symbiotic mutualists and hosts do not always coadapt, as is reflected in the imperfect patterns seen in studies of phylogenetic codivergence (Machado et al. 2005) and as might be predicted, the non-correspondence of host and symbiont fitness. For example, laboratory manipulations have demonstrated the degradation of mutualism due to poor correspondence between symbiont fitness and benefit to host in plant-mycorrhizal fungal interactions (Bever 2002). The negative frequency dependent dynamics generated from potential non-correspondence of host and symbiont fitnesses are similar to that found in host pathogen coevolution and illustrate the complexities generated when biological organisms are the environment.
Enemy–victim
When an organism itself is the environment, there is the possibility of at least transitory passive specialisation, such as would be the case when a victim species (e.g. a host or prey) evolves resistance to a non-evolving enemy species (e.g. a pathogen or predator), thereby restricting the latter's host range. More generally, there can be active coevolution that increases or decreases covariation of genotypes with their optimal biotic environments. For example, given the strong trade-offs that defensive genes are likely to encompass (Strauss et al. 2002), we might expect increased victim-specificity to evolve; this may partly depend on the genetic architecture of resistance (e.g. major genes vs. more quantitative situations) as well as life-history. Some of the clearest examples of specificity come from microbial pathogens. Plant-pathogen interactions, for example, range from generalist to specialist associations and recent study has demonstrated genetically controlled variation in host specificity (Barrett et al. 2009).
Considerable research on microbial systems has addressed how specialists and generalists may coexist. When only exploiters evolve, resource evenness (i.e. greater diversity, be it biotic or abiotic) promotes generalisation, but does not promote increased coexistence between specialists and generalists (Kassen 2002). When both antagonists coevolve, however, theory predicts the reciprocal selective pressures to exploit and resist can create tremendous genotypic diversity (Yoder & Nuismer 2010). Empirical study indicates that this will favour not only the emergence of generalist exploiters, but also their coexistence with highly specialised species (e.g. Coberly et al. 2009). The increased coexistence observed in coevolutionary antagonistic interactions may be due to the diversification of exploitation and resistance strategies, which persist at least transiently until they are either fixed or go extinct.
There is considerable empirical evidence in host–pathogen systems that host phylogenetic structure is an important determinant of host-switches, with pathogens more likely to acquire new hosts that are evolutionarily related to their original host. For example, infectious diseases of primates are more often shared between closely related hosts (Davies & Pedersen 2008). Conversely, prey or host species may be able to defend themselves more efficiently against related exploiters (i.e. that share common exploitation mechanisms), be they related due to common ancestry or by the result of convergent evolution, which will obscure the dynamics of specificity in coevolving systems. Interestingly, experimental inoculation studies with plant floral smuts suggest that both host and pathogen phylogenies are important predictors of the potential for host shifts (Vienne et al. 2009). Diversification in biotic resources will likewise alter specificity patterns. A resource speciation event can trigger exploiter speciation, which may lead to the emergence of multiple specialists instead of a single generalist. In this context, specificity is the outcome of a resource-driven evolutionary event (Benkman 2003).
PRODUCTIVITY AND COMMUNITY DIVERSITY
In the previous sections, we discussed several mechanisms by which species interact with and change both abiotic and biotic elements of their environments. These mechanisms may variously promote or disrupt the potential for specialisation to emerge and persist, depending on their influence on correlations between genotype and environment. In the next section, we go beyond species interactions and focus on the interplay between two key axes that have the potential to further alter ecological and evolutionary predictions regarding specialisation: community diversity and environmental productivity.
For given strengths of trade-offs and environmental heterogeneity, specialists may be threatened by extinction at lower bounds of habitat productivity, unless they are able to disperse to patches with reduced levels of competition (thereby reducing specialisation). As a result, dispersal may be expected to decrease and the level of specialisation to increase with productivity. This process is thought to contribute to the correlation of species richness with latitude that is observed in many taxa (e.g. Hillebrand & Matthiessen 2009). Expectations for changes in levels of specialisation within antagonistic coevolving systems with increasing total productivity are less clear. Thus, higher environmental productivity could increase the frequency of multiple infections thereby exacerbating competition for hosts that could favour specialisation in enemies (Thrall et al. 2007). Alternatively, increasing productivity could increase contact with alternative hosts, thereby reducing correlations between symbiont genotypes and their favoured environments, and selecting for increased generalism (Thrall et al. 2007). Consistent with this latter expectation, phages exploit a greater range of putative bacterial genotypes in high compared to low resource environments (Poisot et al. 2011b). In addition, recent theoretical work has shown a strong impact of resource dynamics on the range of pathogen specialisation achieved through coevolution (Poisot et al. 2011a), with consequences for epidemiological features of the pathogen community. By contrast, in certain symbiotic mutualisms, increasing productivity may result in reduced allocation to nutritional symbionts and hence declining specificities due to limited long-term viability of small specialist populations.
Empirical work shows that specialised herbivores may avoid consumption by predators, but that there is limited support for the role of enemy-free space in fostering resource specialisation (Berdegue et al. 1996). Work over the past few decades has shown that specialisation in herbivores can also be partly explained by host plant chemistry (Ode 2006). However, both enemy avoidance and the chemicals involved in specialisation can interact, as shown by the gastropod Costaciella oceliffera that specialises on a toxic plant which reduces the fitness of its predators (Hay et al. 1990). The frog-eating Floodplain death adder (Acanthophis praelongus) displays the ability to selectively delay ingestion of killed frogs depending on their toxicity (Phillips & Shine 2007). In this case, diversification of prey chemical defences did not prompt predator specialisation through evolution of anti-toxic compounds tailored to each frog species. These results are congruent with the view that the observed specificity of an organism is contingent, not only upon chemical coevolution (i.e. physiological constraints) and the community matrix within which it exists (Thompson 1988), but also on behavioural responses. Specialisation will likely jointly evolve with community diversity and the nature of this dynamic will likely depend upon environmental quality. A recent conceptual model (Thrall et al. 2007) hypothesises that as community diversity increases, pathogen specificity is predicted to decline, whereas mutualist specificity will increase. To our knowledge, empirical data do not exist to test this expectation, nor do we know the extent to which antagonists and mutualists exert reciprocal selective pressures on one another and how their specificities may co-evolve in complex communities and across productivity gradients. Further theoretical and empirical work on the potential for such feedbacks is necessary prior to the generation of a cohesive theoretical framework that would generate predictions across landscapes.
CONCLUDING REMARKS
Recent theory, empirical data and experiments indicate that a small number of fundamental ecological and evolutionary mechanisms are responsible for the genesis and maintenance of specialisation and correlated traits that reinforce and protect it. Here, we have focused on individuals, populations and species interactions, and as such have not considered in detail the many other manifestations of specialisation, including sexual selection, adaptive radiation, epidemiology, species invasions, species ranges, extinctions and community structure. This highlights the fact that specialisation is a property of any biological function with different levels or types of expression, and any scale, from molecules, to individuals, through to populations, species, communities and ecosystems. We suggest that the same ecological and evolutionary processes that mould ecological specialisation in species interactions and approaches to its quantification (Box 2), will generalise to (or have analogues with) other forms of biological specialisation.
There are clearly considerable gaps in our understanding of ecological specialisation, spanning areas ranging from population ecology to macro-evolution (Box 4). For example, why do antagonistic and mutualistic interactions exhibit different distributions of specialisation (Box 1)? We believe that the most fruitful research will aim to understand the complex ecological and evolutionary feedbacks that drive the evolution of specialisation and the persistence of specialised species in a broader community context, and the integration of expectations when both productivity and community complexity can vary. Essential to these approaches will be a better understanding of the joint roles of variation in both abiotic and biotic environments and the genetic interactions arbitrating individual and population adaptation (Box 3). While considerable theory has examined the role of spatial heterogeneity in species interactions, relatively few attempts have been made to explore the community dynamics of evolving (or non-evolving) specialists and generalists in complex landscapes, and how these may in turn further drive environmental predictability and the genotype by environment correlations that underpin specialisation. Such integrative approaches would have clear relevance for understanding and managing biological diversity.
Box 1 Patterns of specialisation across different biotic interactions.
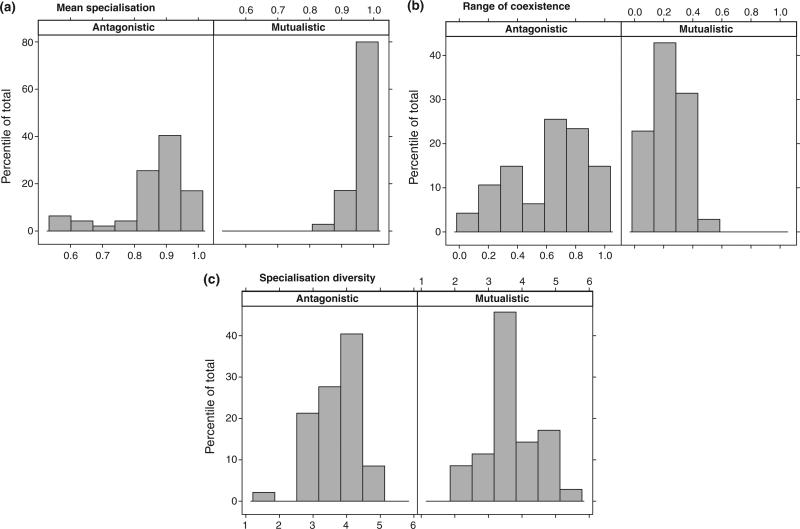
The biological and ecological mechanisms involved in different types of biotic interactions can give rise to various patterns of organisation (Fortuna et al. 2010). The costs of interacting with multiples species and the benefits received from the interaction can alter the number of links and their distributions over evolutionary time (Jordano et al. 2003). To date most research has focused on the distribution of links in mutualistic networks (e.g. Bascompte et al. 2003), but data on other types of interaction exist. To test if patterns of species specialisation differ between antagonistic and mutualistic interactions, we compiled 82 bipartite networks available on the IWDB database (http://www.nceas.ucsb.edu/interactionweb/ obtained in January 2011) and calculated specialisation of the upper trophic level using PDI (see Poisot et al. 2011b and Box 2). For the distribution of specialisation in each network, we calculated mean specialisation, the range of coexistence (i.e. the difference between maximal and minimal specialisation values) and specialisation diversity (as measured by the Shannon–Wiener index).
The data were analysed for mutualistic (n = 35) and antagonistic (n = 47) webs and differences in the means were statistically evaluated using Kruskal–Wallis tests. We found that antagonistic webs were on average less specialised (Fig. 3a, 0.85 vs. 0.92, P < 10–3) and permitted greater coexistence between species with different levels of specialisation (Fig. 3b, 0.61 vs. 0.23, P < 10–5). The diversity of degrees of specialisation did not differ between webs (Fig. 3c, P = 0.65). This simple analysis suggests that different mechanisms (and/or similar mechanisms acting with different intensities) may shape specialisation patterns in both antagonistic and mutualistic interactions. An objective of future research is to untangle the relative roles of constraints, costs and benefits associated with the interaction (or avoidance thereof in the case of antagonistic systems) in specialisation patterns across biotic interactions.
Box 2 Conceptual and quantitative issues in the measurement of species specificity.
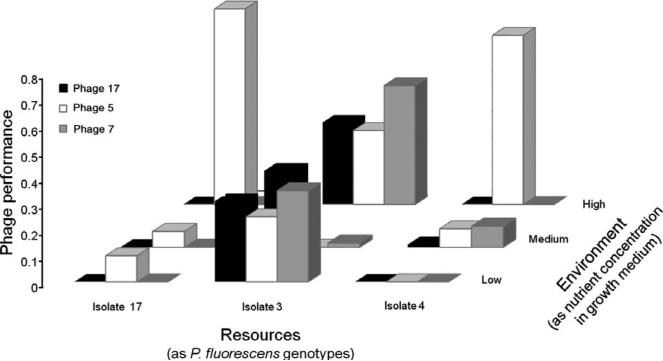
While recent reviews on specialisation have focused on the definition of specificity and its measurement at individual (Bolnick 2002) and community (Devictor et al. 2010) levels, no review to our knowledge has addressed its measurement at the population level, despite the evolutionary relevance of population level processes. Generating methodologies for measuring specificity has proven conceptually challenging. Specificity can be defined as the breadth occupied on each niche axis (Futuyma & Moreno 1988; Fig. 2a, main text). As is shown in Fig. 4, some species can simultaneously be a generalist on one axis and a specialist on another (e.g. phage 17 shows the same performance on bacterial isolate 3 across environments, but displays different performances across bacterial isolates within a single habitat). In this example, measuring the same trait solely across habitats would therefore result in incomplete or biased estimates of specificity. Similarly, measuring specificity using only the number of virulent loci may fail to account for the genetic structure of host resistance.
Moreover, there is uncertainty about how specificity should be measured. Blüthgen et al. (2006) propose the sampling-robust measure d’ for specificity estimates at the species level. Its wide applicability is, however, limited because of: (1) the heuristic process involved in its normalisation, (2) the impossibility to use continuous data and (3) non-independence with regard to the performances of other species in the community. Recently, Poisot et al. (2011b) proposed the Paired Differences Index, which employs continuous performance data. However, as of present no formal attempt has been made to compare the relative suitabilities of different specificity measures.
Box 3 Genomic approaches to investigating specificity in evolutionary and coevolutionary interactions.
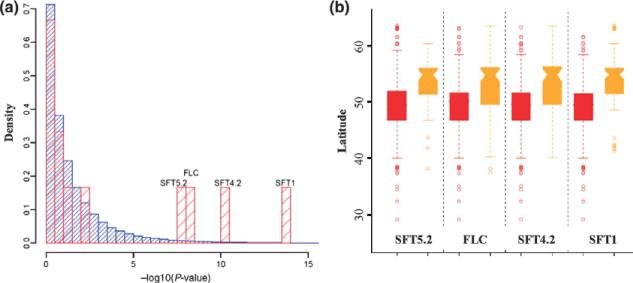
Evolutionary changes in specialisation of populations, species or communities can be studied at the genomic level. A possible approach to achieve this is by identifying among-individual or among-species genomic differences in patterns of linkage or natural selection and associating those with the likely responsible environmental agents. For instance, allelic variation at QTLs associated with flowering time in Arabidopsis thaliana was shown to follow a latitudinal distribution [Fig. 5 (Li et al. 2010)]. The QTLs directly controlled performance across different environments, as maximum seed yield was observed at an optimal flowering time, unique both geographically and temporally.
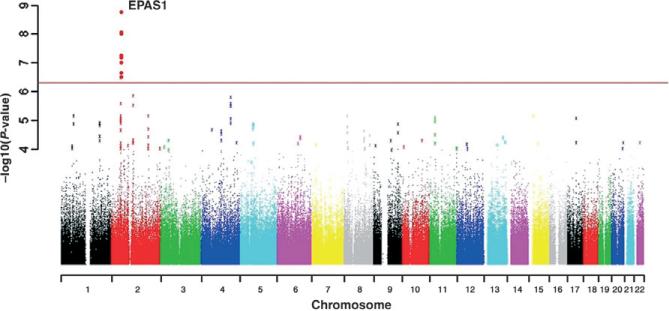
As another example, the identification of fixed mutations in EPAS1, a gene that controls haemoglobin production, in human populations of Tibet could be associated with the ability to settle in high altitude, hypoxic environments (Fig. 6, from Beall et al. 2010). In this case, the mutant alleles would correspond to the evolution of a generalist, as Tibetans have evolved to be able to maintain a constant red blood cell concentration across different altitude ranges. They do not suffer known fitness trade-offs associated with the mutation.
Finally, studies of DNA sequence variation in parasites of plants and animals have shed light on the genetic basis of pathogenicity. For example, in plant parasites such as oomycetes and rust fungi, genes responsible for host range and host specificity typically exhibit very high rates of non-synonymous vs. synonymous mutations (i.e. diversifying selection; Soanes & Talbot 2008). Further applications of association studies could include studies of species abundance rather than DNA polymorphism (e.g. by assessing the composition of microbial species communities across ecological gradients using microbial DNA arrays; Brodie et al. 2007). There are an increasing number of initiatives that aim to address the functional genetics of local adaptation. The integration of molecular and population level approaches show promise for yielding insights into the genetic nature of trade-offs and the resulting changes in specialisation.
Box 4 Key issues for future research.
Ecological drivers and community structure
Does specialisation within natural communities increase or decrease ecological stability?
How do complex abiotic and biotic environmental landscapes influence specialisation?
How does the level of trophic complexity within a community influence the evolution of specialisation?
At what trophic levels is specialisation most likely to emerge?
Evolutionary processes
Are abiotic or biotic factors more likely to drive specialisation over short to medium timescales?
To what extent do trade-offs vs. mutation accumulation through genetic drift drive specialisation?
Life-history characteristics and behaviour
Is the evolution of specialisation more likely for behaviourally plastic species than locally dispersing ones?
What life-history features (e.g. longevity, dispersal, life-cycle complexity and diet) favour the evolution of specificity?
Does life-history distinguish generalists from specialists within phylogenetic clades?
Evolutionary implications of specialisation in biotic interactions
Are specialised pathogens more virulent than generalists, and likewise, are specialised mutualists generally more beneficial?
Do specialised hosts preferentially associate with specialised symbionts, and generalist hosts with generalist symbionts?
Do coevolutionary dynamics result in increased specialisation with increasing environmental productivity?
Macro-evolutionary patterns
Is there a trend towards greater specialisation in phylogenetic clades?
Do molecular patterns of selection differ between generalist and specialist genomes (e.g. for the latter, divergence might be greater in more localised and specific regions of genomes)?
How does specialisation relate to the diversification potential of a species, and should we expect to find generalist clades to be more species-rich than specialists ones?
Figure 3.
(a) Mean specialisation, (b) range of coexistence and (c) diversity in the levels of specialisation in 82 bipartite networks for antagonistic and mutualistic interactions. Antagonistic networks are less specialised on average, but exhibit a wider range of degree of specialisation. There is no significant difference between specialisation diversity between the two types of interactions.
Figure 4.
Performances of three bacteriophage isolates on three bacterial host isolates (P. fluorescens) as measured in environments of increasing productivity. This figure illustrates how specificity estimates for one axis may depend on other environmental axes. Ideally, specificity for one activity should be measured by controlling for other influential environmental variables. Data from Poisot et al. (2011b).
Figure 5.
(a) Histogram of the P-values for correlations between SNP alleles and latitude. Histogram in blue represents the genome-wide distribution (172243 SNPs with MAF > 10%), and red represents the 12 candidate QTL SNPs. (b) Latitudinal distribution of the alleles at the four QTL (Col allele in red and non-Col allele in orange). From Li et al. (2010).
Figure 6.
A genome-wide allelic differentiation scan that compares Tibetan residents at 3 200–3 500 m in Yunnan Province, China with HapMap Han samples. Eight SNPs near one another and EPAS1 have genome-wide significance. The horizontal axis is the genomic position, with colours indicating chromosomes. The vertical axis is the negative log of SNP-by-SNP P-values generated from the Yunnan Tibetan vs. HapMap Han comparison. The red line indicates the threshold for genome-wide significance used. From Beall et al. (2010).
ACKNOWLEDGEMENTS
We thank V. Devictor for discussions on the semantics of specialisation and three referees for their insightful comments. We are grateful to the curators and contributors of the IWDB database. This study was funded by ‘EvolStress’ (ANR-09-BLAN-099-01), ‘EvoRange’ (2009-PEXT-011-01) and ‘CoMute’ (ANR-06-BLAN-0164-01) to MEH, National Institutes of Health (NIH grant 5RO1 GM074265-01A2) and a CSIRO Newton Turner Career Award to PHT, and NSF-DEB-0919434, NSFDEB-1050237 and NIH-5 R01 GM092660 to JDB. We thank E. McCauley and the National Center for Ecological Analysis and Synthesis, Santa Barbara, for hosting our working group. This is contribution 2011-065 of the Institut des Sciences de l’Evolution de Montpellier. Due to space limitations many relevant publications could not be cited, particularly in cases where topical reviews are available.
GLOSSARY
- Coevolution
Reciprocal evolution in each of two or more species resulting from their interactions
- Constraints
Properties of the genotype or phenotype that result in an organism achieving different fitness in different environments
- Environment vs. habitat
Environment is one or more dimensions of the habitat affecting organism condition
- Fundamental niche
The ensemble of abiotic environments in which a population can persist without external immigration, when not limited by habitat size or biotic interactions
- Generalisation
Ecological and evolutionary processes that result in persistence in an increased range of environments or lessened skew in performance over exploited environments
- Local adaptation
Higher average performance of a population in a local habitat compared with away habitats
- Niche
The set of environments suitable for population persistence. Specialisation contrasts with the niche in that the former refers to the breadth of the latter or to its component environmental axes
- Niche conservatism vs. niche evolution
Niche conservatism is the tendency for species to maintain ancestral traits defining their niche, whereas niche evolution is the lability of traits, permitting either shifting or extension of the fundamental niche
- Performance
Quantitative traits correlated with relative fitness
- Realised niche
Limitations to, or extension of, the fundamental niche resulting from species interactions, chance events and history
- Specialist, generalist
A specialist adapts to or persists in a narrower range of environments or habitats than a generalist
- Specialisation
The process by which an organism adapts to an increasingly narrow subset of its possible environments and persists in an increasingly narrow range of habitats
- Specificity
Displaying differential adaptation to a subset of suitable environments. Whereas specialisation refers to the process or tendency of change, specificity is the state of adaptation to environments at any given time or place
- Tolerance
Level of fitness compensation in a stressful environment
- Trade-off
Change in a phenotypic trait resulting in the change in one or more other traits, due to pleiotropic (individual constraint) or epistatic (selection) effects
REFERENCES
- Abrams PA. Adaptive change in the resource-exploitation traits of a generalist consumer: the evolution and coexistence of generalists and specialists. Evolution. 2006a;60:427–439. [PubMed] [Google Scholar]
- Abrams PA. The prerequisites for and likelihood of generalist-specialist coexistence. Am. Nat. 2006b;167:329–342. doi: 10.1086/499382. [DOI] [PubMed] [Google Scholar]
- Barrett LG, Kniskern JM, Bodenhausen N, Zhang W, Bergelson J. Continua of specificity and virulence in plant host–pathogen interactions: causes and consequences. New Phytol. 2009;183:513–529. doi: 10.1111/j.1469-8137.2009.02927.x. [DOI] [PubMed] [Google Scholar]
- Bascompte J, Jordano P, Melián CJ, Olesen JM. The nested assembly of plant–animal mutualistic networks. Proc. Natl. Acad. Sci. USA. 2003;100:9383–9387. doi: 10.1073/pnas.1633576100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall CM, Cavalleri GL, Deng L, Elston RC, Gao Y, Knight J, et al. Natural selection on EPAS1 (HIF2α) associated with low hemoglobin concentration in Tibetan highlanders. Proc. Natl. Acad. Sci. USA. 2010;107:11459–11464. doi: 10.1073/pnas.1002443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkman CW. Divergent selection drives the adaptive radiation of crossbills. Evolution. 2003;57:1176–1181. doi: 10.1111/j.0014-3820.2003.tb00326.x. [DOI] [PubMed] [Google Scholar]
- Berdegue M, Trumble JT, Hare JD, Redak RA. Is it enemy-free space? The evidence for terrestrial insects and freshwater arthropods Ecol. Entomol. 1996;21:203–217. [Google Scholar]
- Bever JD. Negative feedback within a mutualism: host-specific growth of mycorrhizal fungi reduces plant benefit. Proc. Biol. Sci. 2002;269:2595–2601. doi: 10.1098/rspb.2002.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bever JD, Richardson SC, Lawrence BM, Holmes J, Watson M. Preferential allocation to beneficial symbiont with spatial structure maintains mycorrhizal mutualism. Ecol. Lett. 2009;12:13–21. doi: 10.1111/j.1461-0248.2008.01254.x. [DOI] [PubMed] [Google Scholar]
- Bever JD, Dickie IA, Facelli E, Facelli JM, Klironomos J, Moora M, et al. Rooting theories of plant community ecology in microbial interactions. Trends Ecol. Evol. 2010;8:468–478. doi: 10.1016/j.tree.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingham RA, Agrawal AA. Specificity and trade-offs in the induced plant defence of common milkweed Asclepias syriaca to two lepidopteran herbivores. J. Ecol. 2010;98:1014–1022. [Google Scholar]
- Blüthgen N, Menzel F, Blüthgen N. Measuring specialization in species interaction networks. BMC Ecol. 2006;6:9–14. doi: 10.1186/1472-6785-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolnick DI. Measuring individual-level resource specialization. Ecology. 2002;69:18–29. [Google Scholar]
- Bolnick DI, Svanbäck R, Fordyce JA, Yang LH, Davis JM, Hulsey CD, et al. The ecology of individuals: incidence and implications of individual specialization. Am. Nat. 2003;161:1–28. doi: 10.1086/343878. [DOI] [PubMed] [Google Scholar]
- Brodie EL, DeSantis TZ, Parker JPM, Zubietta IX, Piceno YM, Andersen GL. Urban aerosols harbor diverse and dynamic bacterial populations. Proc. Natl. Acad. Sci. USA. 2007;104:299–304. doi: 10.1073/pnas.0608255104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brückmann SV, Krauss J, Steffan-Dewenter I. Butterfly and plant specialists suffer from reduced connectivity in fragmented landscapes. J. Appl. Ecol. 2010;47:799–809. [Google Scholar]
- Chapuis E, Emelianoff V, Paulmier V, Le Brun N, Pagès S, Sicard M, et al. Manifold aspects of specificity in a nematode-bacterium mutualism. J. Evol. Biol. 2009;22:2104–2117. doi: 10.1111/j.1420-9101.2009.01829.x. [DOI] [PubMed] [Google Scholar]
- Coberly LC, Wei W, Sampson KY, Millstein J, Wichman HA, Krone SM. Space, time, and host evolution facilitate coexistence of competing bacteriophages: theory and experiment. Am. Nat. 2009;173:E121–E138. doi: 10.1086/597226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper N, Jetz W, Freckleton RP. Phylogenetic comparative approaches for studying niche conservatism. J. Evol. Biol. 2010;23:2529–2539. doi: 10.1111/j.1420-9101.2010.02144.x. [DOI] [PubMed] [Google Scholar]
- Davies TJ, Pedersen AB. Phylogeny and geography predict pathogen community similarity in wild primates and humans. Proc. Biol. Sci. 2008;275:1695–1701. doi: 10.1098/rspb.2008.0284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desdevises Y, Morand S, Legendre P. Evolution and determinants of host specificity in the genus Lamellodiscus (Monogenea). Biol. J. Linn. Soc. 2002;77:431–443. [Google Scholar]
- Devictor V, Clavel J, Julliard R, Lavergne S, Mouillot D, Thuiller W, et al. Defining and measuring ecological specialization. J. Appl. Ecol. 2010;47:15–25. [Google Scholar]
- Diniz-Filho JAF, Bini LM. Macroecology, global change and the shadow of forgotten ancestors. Glob. Ecol. Biogeo. 2008;17:11–17. [Google Scholar]
- Dion E, Zélé F, Simon J-C, Christin PA, Outreman Y. Rapid evolution of parasitoids when faced with the symbiont-mediated resistance of their hosts. J. Evol. Biol. 2011;24:741–750. doi: 10.1111/j.1420-9101.2010.02207.x. [DOI] [PubMed] [Google Scholar]
- Edwards EJ, Osborne CP, Strömberg CAE, Smith SA, Bond WJ, et al. The origins of C4 grasslands: integrating evolutionary and ecosystem science. Science. 2010;328:587–591. doi: 10.1126/science.1177216. [DOI] [PubMed] [Google Scholar]
- Egas M, Dieckmann U, Sabelis MW. Evolution restricts the coexistence of specialists and generalists: the role of trade-off structure. Am. Nat. 2004;163:518–531. doi: 10.1086/382599. [DOI] [PubMed] [Google Scholar]
- Elias M, Gompert Z, Jiggins C, Willmott K. Mutualistic interactions drive ecological niche convergence in a diverse butterfly community. PLoS Biol. 2008;6:e300. doi: 10.1371/journal.pbio.0060300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forister ML. Oviposition preference and larval performance within a diverging lineage of lycaenid butterflies. Ecol. Entomol. 2004;29:264–272. [Google Scholar]
- Fortuna MA, Stouffer DB, Olesen JM, Jordano P, Mouillot D, Krasnov B, et al. Nestedness versus modularity in ecological networks: two sides of the same coin? J. Anim. Ecol. 2010;78:811–817. doi: 10.1111/j.1365-2656.2010.01688.x. [DOI] [PubMed] [Google Scholar]
- Futuyma DJ, Moreno G. The evolution of ecological specialization. Ann. Rev. Ecol. Syst. 1988;19:207–233. [Google Scholar]
- Gripenberg S, Mayhew PJ, Parnell M, Roslin T. A meta-analysis of preference-performance relationships in phytophagous insects. Ecol. Lett. 2010;13:383–393. doi: 10.1111/j.1461-0248.2009.01433.x. [DOI] [PubMed] [Google Scholar]
- Hairston NG, Kearns CM. Temporal dispersal: ecological and evolutionary aspects of zooplankton egg banks and the role of sediment mixing. Int. Comp. Biol. 2002;42:481–491. doi: 10.1093/icb/42.3.481. [DOI] [PubMed] [Google Scholar]
- Hay ME, Duffy JE, Paul VJ, Renaud PE, Fenical W. Specialist herbivores reduce their susceptibility to predation by feeding on the chemically defended seaweed Avrainvillea longicaulis. Limnol. Oceano. 1990;35:1734–1743. [Google Scholar]
- Hereford J. A quantitative survey of local adaptation and fitness trade-offs. Am. Nat. 2009;173:579–588. doi: 10.1086/597611. [DOI] [PubMed] [Google Scholar]
- Hillebrand H, Matthiessen B. Biodiversity in a complex world: consolidation and progress in functional biodiversity research. Ecol. Lett. 2009;12:1405–1419. doi: 10.1111/j.1461-0248.2009.01388.x. [DOI] [PubMed] [Google Scholar]
- Holt RD. Bringing the Hutchinsonian niche into the 21st century: ecological and evolutionary perspectives. Proc. Natl. Acad. Sci. USA. 2009;106(Suppl. 2):19659–19665. doi: 10.1073/pnas.0905137106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasmin J-N, Kassen R. On the experimental evolution of specialization and diversity in heterogeneous environments. Ecol. Lett. 2007;10:272–281. doi: 10.1111/j.1461-0248.2007.01021.x. [DOI] [PubMed] [Google Scholar]
- Johnson KP, Malenke JR, Clayton DH. Competition promotes the evolution of host generalists in obligate parasites. Proc. Biol. Sci. 2009;276:3921–3926. doi: 10.1098/rspb.2009.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordano P, Bascompte J, Olesen JM. Invariant properties in coevolutionary networks of plant-animal interactions. Ecol. Lett. 2003;6:69–81. [Google Scholar]
- Joshi A, Thompson JN. Trade-offs and the evolution of host specialization. Evol. Ecol. 1995;9:82–92. [Google Scholar]
- Kaci-Chaouch T, Verneau O, Desdevises Y. Host specificity is linked to intraspecific variability in the genus Lamellodiscus (Monogenea). Parasitology. 2008;135:607–616. doi: 10.1017/S003118200800437X. [DOI] [PubMed] [Google Scholar]
- Kassen R. The experimental evolution of specialists, generalists, and the maintenance of diversity. J. Evol. Biol. 2002;15:173–190. [Google Scholar]
- Kawakita A, Okamoto T, Goto R, Kato M. Mutualism favours higher host specificity than does antagonism in plant-herbivore interaction. Proc. Biol. Sci. 2010;277:2765–2774. doi: 10.1098/rspb.2010.0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawecki TJ. Accumulation of deleterious mutations and the evolutionary cost of being a generalist. Am. Nat. 1994;144:833–838. [Google Scholar]
- Kellermann V, van Heerwaarden B, Sgrò CM, Hoffmann AA. Fundamental evolutionary limits in ecological traits drive Drosophila species distributions. Science. 2009;325:1244–1246. doi: 10.1126/science.1175443. [DOI] [PubMed] [Google Scholar]
- Kiers ET, Rosseau RA, West SA, Denison RF. Host sanctions and the legume-rhizobium mutualism. Nature. 2003;425:78–81. doi: 10.1038/nature01931. [DOI] [PubMed] [Google Scholar]
- Laubichler MD, Maienschein J. Form and Function in Developmental Evolution. Cambridge University Press; Cambridge: 2009. [Google Scholar]
- Levins R. Evolution in Changing Environments. Princeton University; New Jersey: 1968. [Google Scholar]
- Li Y, Huang Y, Bergelson J, Nordborg M, Borevitz JO. Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2010;107:211499–221204. doi: 10.1073/pnas.1007431107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo JA. Ecological impacts of non-indigenous invasive fungi as forest pathogens. Biol. Inv. 2008;11:81–96. [Google Scholar]
- Machado CA, Robbins N, Gilbert MTP, Herre EA. Critical review of host specificity and its coevolutionary implications in the fig/fig-wasp mutualism. Proc. Natl. Acad. Sci. USA. 2005;102(Suppl. 1):6558–6565. doi: 10.1073/pnas.0501840102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malcom JW. Smaller gene networks permit longer persistence in fast-changing environments. PLoS ONE. 2011;6:e14747. doi: 10.1371/journal.pone.0014747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S, Schnitzer S, Herre E, Mack KML, Valencia MC, Sanchez EI, et al. Negative plant-soil feedback predicts tree-species relative abundance in a tropical forest. Nature. 2010;466:752–755. doi: 10.1038/nature09273. [DOI] [PubMed] [Google Scholar]
- Marquez LM, Redman RS, Rodriguez RJ, Roossinck MJ. A virus in a fungus in a plant: three-way symbiosis required for thermal tolerance. Science. 2007;315:513. doi: 10.1126/science.1136237. [DOI] [PubMed] [Google Scholar]
- Menge DNL, Ballantyne F, Weitz JS. Dynamics of nutrient uptake strategies: lessons from the tortoise and the hare. Theor. Ecol. 2011;1:163–177. doi: 10.1007/s12080-010-0110-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller TE, Burns JH, Munguia P, Walters EL, Kneitel JM, Richards PM, et al. A critical review of twenty years use of the resource-ratio theory. Am. Nat. 2005;165:439–448. doi: 10.1086/428681. [DOI] [PubMed] [Google Scholar]
- Nosil P, Crespi BJ, Sandoval CP, Kirkpatrick M. Migration and the genetic covariance between habitat preference and performance. Am. Nat. 2006;167:E66–E78. doi: 10.1086/499383. [DOI] [PubMed] [Google Scholar]
- Ode PJ. Plant chemistry and natural enemy fitness: effects on herbivore and natural enemy interactions. Ann. Rev. Entomol. 2006;51:163–185. doi: 10.1146/annurev.ento.51.110104.151110. [DOI] [PubMed] [Google Scholar]
- Pandit SN, Kolasa J, Cottenie K. Contrasts between habitat generalists and specialists: an empirical extension to the basic metacommunity framework. Ecology. 2009;90:2253–2262. doi: 10.1890/08-0851.1. [DOI] [PubMed] [Google Scholar]
- Phillips B, Shine R. When dinner is dangerous: toxic frogs elicit species-specific responses from a generalist snake predator. Am. Nat. 2007;170:936–942. doi: 10.1086/522845. [DOI] [PubMed] [Google Scholar]
- Poisot T, Thrall PH, Hochberg ME. Trophic network structure emerges through antagonistic coevolution in temporally varying environments. Proc. Biol. Sci. 2011a doi: 10.1098/rspb.2011.0826. DOI: 10.1098/rspb.2011.0826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poisot T, Lepennetier G, Martinez E, Ramsayer J, Hochberg ME. Resource availability affects the structure of a natural bacteria-bacteriophage community. Biol. Lett. 2011b;7:201–204. doi: 10.1098/rsbl.2010.0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poore AGB, Steinberg PD. Preference–performance relationships and effects of host plant choice in an herbivorous marine amphipod. Ecol. Monogr. 1999;69:443–464. [Google Scholar]
- Ravigné V, Dieckmann U, Olivieri I. Live where you thrive: joint evolution of habitat choice and local adaptation facilitates specialization and promotes diversity. Am. Nat. 2009;174:141–169. doi: 10.1086/605369. [DOI] [PubMed] [Google Scholar]
- Reed DL, Hafner MS. Host specificity of chewing lice on pocket gophers: a potential mechanism for cospeciation. J. Mammal. 1997;78:655–660. [Google Scholar]
- Richardson DM, Allsopp N, D'antonio CM, Milton SJ, Rejmanek M. Plant invasions – the role of mutualisms. Biol. Rev. 2000;75:65–93. doi: 10.1017/s0006323199005435. [DOI] [PubMed] [Google Scholar]
- Singer MC, Carriere Y, Theuring C, Hartmann T. Disentangling food quality from resistance against parasitoids: diet choice by a generalist caterpillar. Am. Nat. 2004;164:423–429. doi: 10.1086/423152. [DOI] [PubMed] [Google Scholar]
- Snell-Rood EC, Van Dyken JD, Cruickshank T, Wade MJ, Moczek AP. Toward a population genetic framework of developmental evolution: the costs, limits, and consequences of phenotypic plasticity. Bioessays. 2010;32:71–81. doi: 10.1002/bies.200900132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soanes DM, Talbot NJ. Moving targets: rapid evolution of oomycete effectors. Trends Microbiol. 2008;16:507–510. doi: 10.1016/j.tim.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Strauss SY, Rudgers JA, Lau JA, Irwin RE. Direct and ecological costs of resistance to herbivory. Trends Ecol. Evol. 2002;17:278–285. [Google Scholar]
- Thompson JN. Coevolution and alternative hypotheses on insect/plant interactions. Ecology. 1988;69:893–895. [Google Scholar]
- Thompson JN, Wehling W, Podolsky R. Evolutionary genetics of host use in swallowtail butterflies. Nature. 1990;344:148–150. [Google Scholar]
- Thrall PH, Hochberg ME, Burdon JJ, Bever JD. Coevolution of symbiotic mutualists and parasites in a community context. Trends Ecol. Evol. 2007;22:120–126. doi: 10.1016/j.tree.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Thrall PH, Bever JD, Slattery JF. Rhizobial mediation of Acacia adaptation to soil salinity: evidence of underlying trade-offs and tests of expected patterns. J. Ecol. 2008;96:746–755. [Google Scholar]
- Venable DL. Bet hedging in a guild of desert annuals. Ecology. 2007;88:1086–1090. doi: 10.1890/06-1495. [DOI] [PubMed] [Google Scholar]
- Venail PA, MacLean RC, Bouvier T, Brockhurst MA, Hochberg ME, Mouquet N. Diversity and productivity peak at intermediate dispersal rate in evolving metacommunities. Nature. 2008;452:210–213. doi: 10.1038/nature06554. [DOI] [PubMed] [Google Scholar]
- Via S, Hawthorne DJ. The genetic architecture of ecological specialization: correlated gene effects on host use and habitat choice in pea aphids. Am. Nat. 2002;159:76–88.. doi: 10.1086/338374. [DOI] [PubMed] [Google Scholar]
- Vienne DM, de, Hood ME, Giraud T. Phylogenetic determinants of potential host shifts in fungal pathogens. J. Evol. Biol. 2009;22:2532–2541. doi: 10.1111/j.1420-9101.2009.01878.x. [DOI] [PubMed] [Google Scholar]
- Woolhouse ME, Gowtage-Sequeria S. Host range and emerging and reemerging pathogens. Emerg. Infect. Dis. 2005;11:1842–1848. doi: 10.3201/eid1112.050997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JB, Nuismer SL. When does coevolution promote diversification? Am. Nat. 2010;176:802–817. doi: 10.1086/657048. [DOI] [PubMed] [Google Scholar]