Abstract
Proteins of the activator protein-1 family are known to have roles in many physiological processes such as proliferation, apoptosis, and inflammation. However, their role in fat metabolism has yet to be defined in more detail. Here we study the impact of JunB deficiency on the metabolic state of mice. JunB knockout (JunB-KO) mice show markedly decreased weight gain, reduced fat mass, and a low survival rate compared with control mice. If fed a high-fat diet, the weight gain of JunB-KO mice is comparable to control mice and the survival rate improves dramatically. Along with normal expression of adipogenic marker genes in white adipose tissue (WAT) of JunB-KO mice, this suggests that adipogenesis per se is not affected by JunB deficiency. This is supported by in vitro data, because neither JunB-silenced 3T3-L1 cells nor mouse embryonic fibroblasts from JunB-KO mice show a change in adipogenic potential. Interestingly, the key enzymes of lipolysis, adipose triglyceride lipase and hormone-sensitive lipase, were significantly increased in WAT of fasted JunB-KO mice. Concomitantly, the ratio of plasma free fatty acids per gram fat mass was increased, suggesting an elevated lipolytic rate under fasting conditions. Furthermore, up-regulation of TNFα and reduced expression of perilipin indicate that this pathway is also involved in increased lipolytic rate in these mice. Additionally, JunB-KO mice are more insulin sensitive than controls and show up-regulation of lipogenic genes in skeletal muscle, indicating a shuttling of energy substrates from WAT to skeletal muscle. In summary, this study provides valuable insights into the impact of JunB deficiency on the metabolic state of mice.
JunB is a member of the activator protein-1 (AP-1) transcription factor family. Several AP-1 proteins, including Jun, Fos, and activating transcription factor family members dimerize to exert their actions (1). Among their effects, the most investigated is their capacity to modulate proliferation (for review see Refs. 1 and 2). In this regard, JunB has a dual role. It has been described as a growth-inhibiting protein that antagonizes the proliferative effects of c-Jun. It suppresses cell proliferation by direct transcriptional activation of the cell cycle suppressor p16INK4A (3) and by negatively regulating cyclin D1 (4). On the other hand, JunB has also a cell cycle-promoting role through activating the transcription of the cell cycle regulator cyclin A (5). However, AP-1 proteins not only act as oncogenes or tumor suppressors, but they are also involved in a broader spectrum of cellular events, such as apoptosis, differentiation, and inflammation (1, 6, 7). JunB and other AP-1 proteins are early responders to mitogenic stimuli, such as insulin, which up-regulates JunB in several model systems like human myotubes (8) and 3T3-L1 adipocytes (9). In vivo, JunB expression responds to plasma insulin levels in human skeletal muscle (SM) (10) and murine liver and adipose tissue (AT) (11) and is influenced by plasma glucose concentrations in mouse liver and AT (11, 12). All these facts highlight that JunB might have an important role in insulin-sensitive tissues, but its exact function remains unclear. Among the insulin-sensitive tissues, AT serves as an energy reservoir as well as an endocrine organ that secretes factors to maintain whole-body homeostasis. Until today, little was known about the influence of AP-1 proteins on adipocyte differentiation or fat cell metabolism. DeltaFosB, another AP-1 family member, inhibits adipogenesis both in vivo and in vitro (13). It down-regulates expression of adipogenic marker genes like PPARγ2 and C/EBPα by interfering with C/EBPβ (13). More recently, overexpression of JunB has been shown to block adipocytic differentiation in highly aggressive sarcomas and in 3T3-L1 cells, also through an interaction with C/EBPβ (14).
Given the scarce information on the effects of JunB in AT, despite that its up-regulation is found among the first responses of AT to nutrient signals such as insulin, the present paper addresses the role of JunB deficiency in AT metabolism. JunB knockout (JunB-KO) mice suffer from chronic myeloid leukemia-like disease (15, 16). It was reported that they are growth retarded and show reduced bone formation and longitudinal bone growth (17, 18). Here we show that the growth retardation of JunB-deficient mice correlates with a strongly reduced fat mass (and is also associated with high lethality). However, although expression levels of JunB have been shown to influence adipogenesis in vitro (14), we could not find any changes in the expression of adipogenic marker genes like C/EBPß, PPARγ, and C/EBPα, whereas the expression levels of adipose triglyceride lipase (ATGL) and hormone-sensitive lipase (HSL) were strongly increased in AT of JunB-KO mice. Additionally, we found TNFα expression significantly elevated in white AT (WAT) of JunB-KO mice, and concomitantly, perilipin expression was strongly reduced. From these results, we conclude that the reduced AT mass seems to be a consequence of increased lipolytic rate, most likely mediated by a combination of ATGL/HSL up-regulation and perilipin down-regulation.
Materials and Methods
Animal experiments
Mice carrying the floxed JunB allele (JunBfl/fl) were crossed to MORE-Cre mice to derive JunB-deficient mice (17). Only male mice were used for this study. Animals were kept on a 12-h light, 12-h dark cycle and on a standard laboratory chow diet (4.5% wt/wt fat, ssniff-Spezialdieaten GmbH, Soest, Germany). All animal procedures used were approved by the Austrian Bundesministerium für Wissenschaft und Forschung.
For the growth curves of wild-type (WT) and JunB-KO mice, all animals from several matings were weighed at the age of 5, 10, 15, 20, 40, 80, and 100 d after birth. For the survival rate, the percentage of surviving animals from several matings over a period of 12 wk was determined. The body mass composition (BMC) of WT and JunB-KO animals was measured at the age of 100 d using the minispec live mouse analyzer (7.5 MHz) from Bruker (Ettlingen, Germany). For feeding experiments, WT and JunB-KO mice at the age of 12 wk were put either on a chow diet or on a high-fat diet (HFD) containing 40% calories in fat (ssniff, Germany) for another 7 wk. Mice were weighed weekly after putting them on the respective diets. During these 7 wk, the survival rate of the mice was monitored in parallel. Additionally, during the last 3 wk of this feeding experiment, food intake of WT and JunB-KO mice on the chow diet was measured by weighing the food in each cage dispenser.
Analysis of plasma parameters
At the age of 3–4 months, blood was collected in the morning from fed or fasted animals (at least 10 h of nighttime fasting) by retroorbital bleeding. Plasma was produced (8000 × g at 4 C for 10 min) and immediately used to determine glucose (Sigma Chemical Co., St. Louis, MO), free fatty acids (FFA) (Wako Chemicals, Richmond, VA), and triglycerides (TG) (Infinity TG reagent; Thermo Electron, Noble Park North, Australia). Leptin and insulin were measured using ELISA following the manufacturers' instructions (mouse leptin ELISA kit from LINCO Research, St. Charles, MO, and rat insulin ELISA kit from Crystal Chem Inc., Downers Grove, IL). Plasma catecholamine (CA) levels were measured using the ClinRep Komplettkit for human plasma CA and reversed-phase HPLC electrochemical detection protocol described by Recipe Chemicals (Munich, Germany). In brief, the plasma volume was adjusted to 1 ml using water, 500 pg of internal standard was added, and CA was subsequently adsorbed to alumina, washed, and eluted according to the manufacturer's protocol. Forty microliters of the eluent was injected into the Recipe HPLC system consisting of an autosampler AS3000, an isocratic HPLC pump IP3000 (1 ml/min), a thermostat HT3000 (25 C), and the Clinrep CA column, coupled to the digital amperometric detector EC 3000 (electrode potential was set to 500 mV, sensitivity 10 nA). Data were analyzed using Clarity software and CA levels normalized according to the recovery of internal standard.
Glucose tolerance test (GTT) and insulin sensitivity test (IST)
Animals were subjected to tests at the age of 4 months. For the GTT, six WT and four JunB-KO mice were fasted for 6 h, and baseline blood samples (20 μl) were collected by tail bleeding (time zero). Subsequently, the mice received an ip injected bolus (1.7 g/kg) of 10% (wt/vol) d-glucose solution, and additional blood samples were drawn by tail bleeding after injection. Glucose was determined using the Glucometer from Roche (Roche Diagnostics Inc., Mannheim, Germany). For the IST, six mice from each group were fasted for 4 h, and afterward, 0.5 IU insulin per kilogram mouse was administered ip as a 0.1-IU/ml solution. Blood samples were collected again by tail bleeding at the time points indicated in the figures. The area under the curve (AUC) was calculated from both tests after correction for baseline blood glucose concentration.
Silencing of JunB using short hairpin RNA (shRNA) lentivirus particles
One control nontargeting shRNA lentivirus and four shRNA lentiviruses directed against JunB were purchased from Sigma (MISSION shRNA lentiviral particles NM_008416). Gene silencing in 3T3-L1 cells was performed as previously described elsewhere (19).
Cell culture and differentiation analysis
3T3-L1 cells were grown and induced to differentiate as previously described elsewhere (19). Mouse embryonic fibroblasts (MEF) from WT and JunB-KO animals were harvested from intercrosses between heterozygous JunB-KO mice as described elsewhere (20). MEF were grown in culture medium (αMEM, 10% fetal bovine serum, 2 mm l-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin; all from Invitrogen/Life Technologies, Inc., Carlsbad, CA) at 37 C and 5% CO2. Medium was refreshed every third day, and cells were subcultivated (at a 3:1 rate) before they reached confluence. At passage one, cells (two million/ml per tube) were frozen in liquid nitrogen in medium with a supplementation of 10% dimethylsulfoxide (Sigma). Differentiation experiments were performed at passage three as follows: a standard MDI mix (1 μm dexamethasone, 0.5 mm isobutylmethylxanthine, and 5 μg/ml insulin) was supplemented in the culture medium, along with 1 μm rosiglitazone (Alexis, Biochemicals, Lausanne, Switzerland), to 2-d postconfluent cells. After 3 d, medium was replaced by growth medium supplemented with 1 μg/ml insulin.
Lipid content of the differentiated cells was determined by oil red O staining (21) and by TG content analysis. Briefly, 3T3-L1 cells were washed with PBS, collected in 0.5 ml PBS per well, and sonicated twice for 20 sec on ice. TG content was measured using Infinity TG reagent (Thermo Electron). Values were corrected by protein content measurement using the bicinchoninic acid reagent (Pierce Biotechnology, Rockford, IL).
Gene expression analysis
Murine tissues such as SM and epididymal WAT were harvested after overnight fasting and washed with PBS, and RNA was isolated using Trizol according to the manufacturer's protocol guidelines. For cell culture experiments, cells were washed with PBS and harvested using the RNA isolation kit from Macherey-Nagel (Düren, Germany). Gene expression was assessed by real-time RT-PCR using an ABI Prism 7700 sequence detector system using SYBR Green PCR master mix (Applied Biosystems, Foster City, CA). Gene expression was normalized using TFIIß as reference gene. Relative mRNA expression levels were calculated using averaged ΔΔCt values for each biological replicate (as implemented in Ref. 22). The following primer sequences were used for RT-PCR: TFIIß forward GTCACATGTCCGAATCATCCA, and reverse, TCAATAACTCGGTCCCCTACAA; ATGL forward, GTCCTTCACCATCCGCTTGTT, and reverse, CTCTTGGCCCTCATCACCAG; PPARγ forward, CACAATGCCATCAGGTTTGG, and reverse, CAGCTTCTCCTTCTCGGCCT; C/EBPα forward, ATCTGCGAGCACGAGACGTC, and reverse, TGTCGGCTGTGCTGGAAGA; C/EBPß forward, GGACTTGATGCAATCCGGA, and reverse, AACCCCGCAGGAACATCTTTA; aP2 forward, CGACAGGAAGGTGAAGAGCATC, and reverse, ACCACCAGCTTGTCACCATCT; HSL forward, GCAAGATCAAAGCCTCAGCG, and reverse, GCCATATTGTCTTCTGCGAGTGT; Glut4 forward, CTGTCGCTGGTTTCTCCAACT, and reverse, GCATCCGCAACATACTGGAA; LPL forward, CTCAGATGCCCTACAAAGTGTTCC, and reverse, TCTCGAAGGCCTGGTTGTGT; FAS forward, GCTGTAGCACACATCCTAGGCA, and reverse, TCGTGTTCTCGTTCCAGGATC; SCD1 forward, ATCGCCTCTGGAGCCACAC, and reverse, ACACGTCATTCTGGAACGCC; JunB forward, GGCTTTGCGGACGGTTT, and reverse, GGCGTCACGTGGTTCATCT; perilipin forward, GGTACACTATGTGCCGCTTCC, and reverse, CTTTGCGCTCCGCCTCT; TNFα forward, CATCTTCTCAAAATTCGAGTGACAA, and reverse, TGGGAGTAGACAAGGTACAACCC; ACL forward, TCCTACAAAGAGGTGGCAGAACT, and reverse, GGCTTGAACCCCTTCTGGAT; CD36 forward, GGCCAAGCTATTGCGACAT, and reverse, CAGATCCGAACACAGCGTAGA; CGI-58 forward, TGGTGTCCCACATCTACATCA, and reverse, CAGCGTCCATATTCTGTTTCCA.
Western blot analysis
WAT of WT and JunB-KO mice was harvested after o/n fasting. The tissues were homogenized in buffer containing 0.25 m sucrose, 1 mm EDTA (pH 7), 1 mm EDTA, and 1× protease inhibitor cocktail. The homogenates were centrifuged at 1000 × g at 4 C for 20 min. The liquid intermediates between lipid layer and pellet were transferred to a new tube and mixed 1:1 with lysis buffer [50 mm Tris-HCl (pH 6.8), 10% glycerol, 2.5% sodium dodecyl sulfate, 1× protease inhibitor cocktail, 1 mm phenylmethylsulfonyl fluoride]. Control [nontargeting control construct (ntc)] and JunB-silenced 3T3-L1 cells (sil1 and sil2) were harvested for protein analysis by scraping into lysis buffer after two washes with ice-cold PBS. The protein suspensions were incubated at 90 C for 10 min. Thereafter, benzonase (Merck, Darmstadt, Germany) was added followed by incubation at room temperature for 1 h. Protein concentration was determined with the bicinchoninic acid protein assay kit from Pierce according to the manufacturer's instructions. Each lane of a 10% Bis-Tris gel (NuPAGE; Invitrogen) was loaded with 20 μg sample. After electrophoresis, gels were blotted to nitrocellulose membranes (Invitrogen). Blots were blocked and incubated with the following antibodies: anti-JunB (Santa Cruz Biotechnology, Santa Cruz, CA; diluted 1:300), anti-ATGL (1:1000; Cayman Chemical, Ann Arbor, MI), and anti-β-actin (1:25,000; Sigma). For chemiluminescent detection, a horseradish peroxidase-conjugated secondary antibody was used (antimouse 1:5000 for JunB, antimouse 1:2000 for ß-actin, and antirabbit 1:5000 for ATGL; Pierce), and enhanced chemiluminescence component (Pierce) served as substrate. Developed films were scanned with HP Scanjet at 600 dpi.
Statistical analysis
If not otherwise stated, results are mean values (± sd) from at least three independent experiments. Statistical significance was determined using the two-tailed Student′s t test.
Results
JunB-KO mice show growth retardation, lower survival rate, and reduced fat mass compared with WT littermates
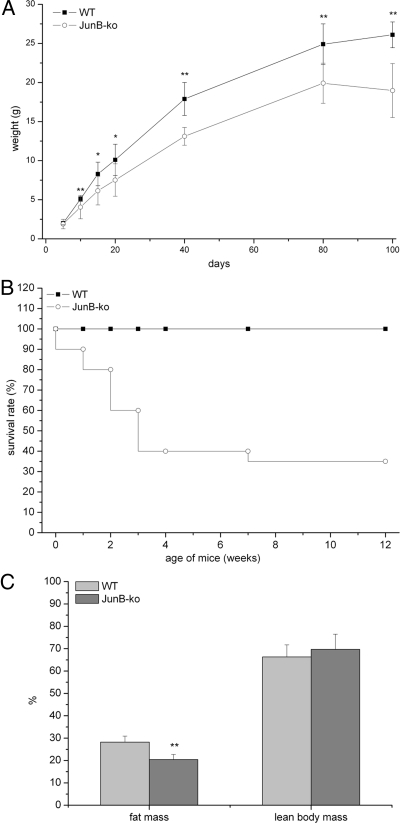
To assess the consequences of JunB deficiency on body weight, heterozygous JunB-KO mice were mated, and the offspring were weighed during the suckling period every fifth day and after weaning at the age of 40, 80, and 100 d. JunB-KO mice appeared normal immediately after birth compared with their WT littermates. During suckling, JunB-KO mice gained significantly less weight than their WT littermates, and at the age of 3 months, JunB-KO mice weighed approximately 25% less than their WT littermates (Fig. 1A). JunB-KO mice were born with Mendelian frequency. However, their survival rate dropped under 50% during weaning, and only 35% of the JunB-KO mice reached the age of 12 wk (Fig. 1B). Heterozygous JunB-KO mice on the other hand could not be distinguished from their WT littermates, because they had the same weight and survival rate as WT controls. Comparing the BMC of 3- to 4-month-old JunB-KO mice with the BMC of WT controls revealed a significant reduction of fat mass in JunB-KO mice (19% fat mass compared with 28% in WT mice), whereas lean body mass was not significantly altered (Fig. 1C). When compared with WT mice, liver and brown AT of JunB-KO mice showed the same percent weight, appearance, and TG content (data not shown). To evaluate whether JunB-KO mice gained less weight due to a reduced food intake, we measured the daily food intake over a period of 3 wk. JunB-KO mice even showed a slight but not significant increase in food intake compared with WT animals (0.19 ± 0.3 and 0.16 ± 0.2 g diet/g mouse · d in JunB-KO and control mice, respectively). Thus, the altered fat to body weight ratio in JunB-deficient animals is independent of their food intake.
Fig. 1.
JunB-KO mice show strong growth retardation and increased death rate. A, Growth curve of JunB-KO (○) mice and WT littermates (■) (n = 10). Weight (grams) was measured at the age of 5, 10, 15, 20, 40, 80, and 100 d after birth. Statistical significance was determined using the two-tailed Student's t test. *, P < 0.05; **, P < 0.01. B, Survival rate of JunB-KO mice and WT littermates (n = 20, at the beginning of the experiment) on a chow diet from birth until the age of 12 wk. C, JunB-KO mice have strongly reduced fat mass, whereas lean body mass (percent body weight) was not changed in comparison with WT controls. BMC of JunB-KO mice and WT littermates (n = 7) was measured at the age of 100 d using the minispec live mouse analyzer (7.5 MHz) from Bruker. Statistical significance was determined using the two-tailed Student's t test. **, P < 0.01.
Modulation of plasma parameters due to JunB deficiency
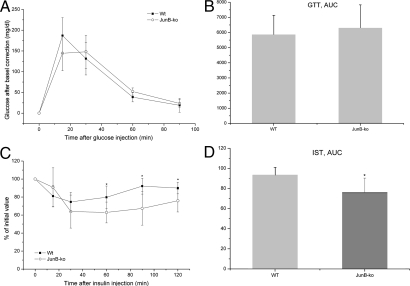
Table 1 contains the plasma parameters measured in mice at the age of 3–4 months in fed and/or overnight fasted state. In the fed state, we found significantly elevated TG levels in JunB-deficient mice, whereas no changes in TG levels could be found in the fasted state (Table 1). For FFA levels, we could measure a trend of reduction in plasma of JunB-KO mice compared with their WT controls in fasted and fed states (Table 1). Interestingly, JunB-deficient mice showed significantly reduced fasting glucose levels in comparison with their WT littermates. Also, plasma insulin levels were significantly reduced in the fasted state in JunB-KO mice in comparison with their WT littermates, whereas there was no significant difference in insulin in the fed state (Table 1). Concomitant lower glucose and insulin levels during fasting suggest increased insulin sensitivity in the JunB-KO mice. To address this, we subsequently performed GTT and IST in these mice. Figure 2A shows that, after baseline-correction, the glucose profile in JunB-KO mice is similar to that of the WT mice. Concomitantly, the AUC (Fig. 2B) shows that JunB-KO mice have the same glucose tolerance as WT littermates. Instead, the clearance of plasma glucose observed after a single ip injection of insulin was significantly more pronounced in JunB-deficient mice than in WT animals as depicted in the IST after baseline correction (Fig. 2C) and in the AUC that is significantly lower in JunB-KO mice (Fig 2D), indicating that JunB-KO mice are more insulin sensitive than WT controls. Furthermore, we investigated whether JunB-KO mice have reduced leptin secretion. Table 1 shows that secreted leptin levels were also significantly reduced in JunB-KO mice. However, calculated leptin to WAT mass ratio was not changed in JunB-KO mice in comparison with their WT littermates (Table 1), suggesting that JunB-deficient mice produce less leptin due to reduced AT mass. However, as shown above, the food intake was not significantly elevated in JunB-KO mice. Finally, we measured catecholamine levels of JunB-KO mice and WT controls and found no significant differences (Table 1).
Table 1.
Plasma parameters of 3- to 4-month-old JunB-KO and WT mice
| Fed state |
Fasted state |
|||
|---|---|---|---|---|
| WT | JunB-KO | WT | JunB-KO | |
| TG (mmol/liter) | 0.77 ± 0.1 | 1.04 ± 0.2b | 1.24 ± 0.28 | 1.27 ± 0.29 |
| FFA (mmol/liter) | 0.60 ± 0.1 | 0.55 ± 0.2 | 1.05 ± 0.16 | 0.84 ± 0.19 |
| Glucose (mg/dl) | 64.6 ± 9.93 | 47.3 ± 6.62a | ||
| Insulin (ng/ml) | 0.84 ± 0.158 | 0.99 ± 0.352 | 0.52 ± 0.075 | 0.45 ± 0.038a |
| Leptin (ng/ml) | 3.45 ± 0.69 | 2.14 ± 0.31b | ||
| Leptin (ng/ml · g fat mass) | 0.48 ± 0.10 | 0.55 ± 0.08 | ||
| Epinephrine (pg/ml) | 219.9 ± 32 | 231.9 ± 7 | ||
| Norepinephrine (pg/ml) | 453.1 ± 148 | 317.5 ± 55 | ||
Blood samples from JunB-KO mice and their WT littermates were taken either in fed state (at 0900 h) or after overnight fasting. Plasma parameters were analyzed as described in Materials and Methods. Statistical significance was determined using the two-tailed Student's t test.
P < 0.05.
P < 0.01.
Fig. 2.
JunB-KO mice have elevated insulin sensitivity. A, For the GTT, six WT (■) and four JunB-KO (○) mice at the age of 4 months were fasted for 6 h. Baseline blood samples were then collected by tail bleeding (time zero). Subsequently, the mice received an ip injection of glucose, and additional blood samples were drawn by tail bleeding at 15, 30, 60, and 90 min after injection. Glucose was determined using the Glucometer from Roche. The curve for the GTT was baseline corrected. Statistical significance was determined using the two-tailed Student's t test. B, To examine glucose tolerance of WT (■) and JunB-KO (○) mice, the glucose AUC was calculated after correction for baseline blood glucose concentrations. C, For the IST, six mice from each group were fasted for 4 h, and afterward, 0.5 IU insulin/kg mouse was administered ip. Blood samples were collected by tail bleeding at the time points indicated in the figure. Glucose was determined using the Glucometer from Roche. The curve for the IST was baseline corrected. Statistical significance was determined using the two-tailed Student's t test. *, P < 0.05. D, The insulin sensitivity, the decrease of plasma glucose after injection of insulin, was determined by the glucose AUC calculated after correction for baseline blood glucose concentration. Statistical significance was determined using the two-tailed Student's t test. *, P < 0.05.
HFD ameliorates JunB-KO phenotype and survival rate
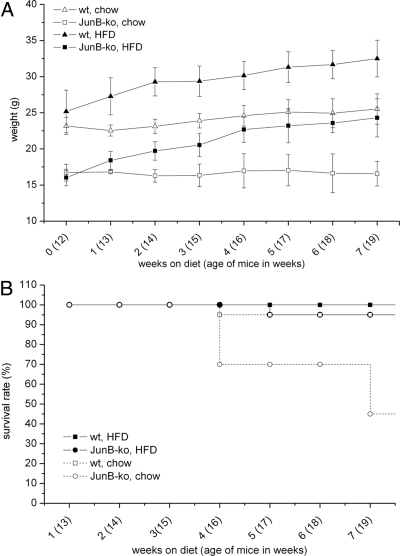
To delineate whether JunB-KO mice are also impaired in their ability to gain weight upon a high-caloric diet, 12-wk-old JunB-deficient mice and their WT littermates were put on a HFD for 7 wk or continued on chow diet for the same time. Figure 3A shows that under a chow diet, JunB-KO mice did not increase body weight like WT mice did (as observed for the first 100 d in Fig. 1A). Interestingly, on a HFD, JunB-deficient mice showed nearly the same weight gain as their WT littermates (Fig. 3A).
Fig. 3.
JunB-KO mice recover from their phenotype under HFD. The 12-wk-old mice were fed either a standard chow or a HFD for 7 more weeks. A, Growth curve of JunB-KO mice and WT littermates (n ≥ 6). Weight (grams) was measured weekly over a period of 7 wk in JunB-KO mice (□, on chow diet; ■, on HFD) and WT littermates (▵, on chow diet; ▴, on HFD). B, Survival rate of JunB-KO mice and WT littermates (n = 20, at the beginning of the experiment). Survival rate of the mice was monitored weekly over a period of 7 wk in mice on control diet (□, WT mice; ○, JunB-KO) and HFD (■, WT mice; ●, JunB-KO).
We also evaluated whether JunB-deficient mice on a HFD show an increased survival rate. For that, we monitored the percentage of survival of WT and JunB-KO mice during the 7 wk on the HFD. As depicted in Fig. 3B, only 5% of the JunB-KO mice died on the HFD, whereas 55% of the JunB-KO mice died on the chow diet. These data indicate that the capacity to gain weight is retained in JunB-KO mice, which strongly reverses the early death of these mice.
JunB is not required for adipogenic differentiation of 3T3-L1 cells and MEF
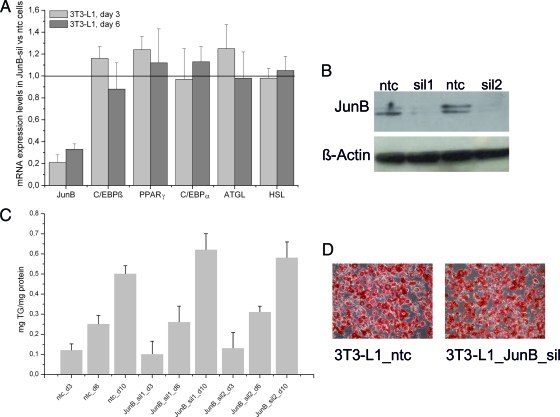
Recently, it has been published that overexpression of JunB blocks adipocytic differentiation in highly aggressive sarcomas and in 3T3-L1 cells through an interaction with C/EBPß (14). To investigate whether JunB deficiency has a direct influence on fat cell differentiation or on the expression of lipolytic enzymes in vitro, we stably silenced JunB in 3T3-L1 cells using lentiviral shRNA. Silencing efficiency of knockdown lentiviruses harboring a shRNA construct targeted against JunB was determined by quantitative RT-PCR and protein analysis and compared with cells infected with a ntc. For two shRNA constructs, JunB gene expression was significantly reduced in JunB-silenced cells compared with ntc cells on mRNA (Fig. 4A) and protein level (Fig. 4B). After adding the standard differentiation cocktail (MDI) to ntc and JunB-silenced cells, we measured the TG content on d 3, 6, and 10. As depicted in Fig. 4C, TG content was not significantly influenced by JunB silencing, which is also shown by oil red O staining (Fig. 4D). In addition, adipogenic marker genes like C/EBPβ, C/EBPα, and PPARγ as well as the expression of ATGL and HSL were unchanged in JunB-silenced cells compared with ntc (Fig. 4A). To employ a more physiological model, we isolated MEF from the offspring of JunB+/− matings and subjected them to adipogenic differentiation. MEF from JunB-deficient embryos were indistinguishable from WT MEF in terms of TG accumulation as well as expression of adipogenic and lipolytic marker genes (data not shown). These results propose that JunB does not directly regulate the expression of adipocytic and lipolytic genes, at least in vitro.
Fig. 4.
Expression of JunB is not required for 3T3-L1 differentiation. 3T3-L1 cells were infected with the MISSION TRC lentiviral particles coding for JunB shRNA (two independent replicates) or a nontargeting shRNA as control (ntc), selected for puromycin resistance, expanded as a mixed population, and induced to differentiate using MDI cocktail. A, JunB silencing has no impact on the expression of genes important for differentiation and lipolysis in differentiating 3T3-L1 cells. Expression of C/EBPß, PPARγ, C/EBPα, ATGL, and HSL in JunB-silenced 3T3-L1 cells was compared with control cells (ntc) at d 3 and 6. B, Western blot analysis was performed to confirm silencing of JunB on protein level (two independently silenced 3T3-L1 cells named sil1 and sil2). β-Actin served as loading control. C, TG accumulation in 3T3-L1 cells is unchanged upon silencing of JunB (JunB_sil1 and JunB_sil2) in comparison with ntc. TG content of the cell lysates was measured on d 3, 6, and 10 after induction of differentiation and normalized to protein content. D, Oil red O staining of control and JunB-silenced 3T3-L1 cells on d 10 of differentiation.
ATGL and HSL expression as well as lipolytic rate are increased in JunB-KO mice
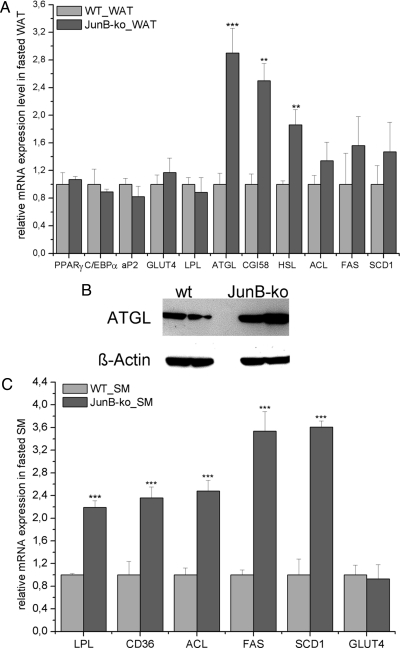
To investigate whether the marked reduction of fat mass in JunB-KO mice is caused by defective adipogenesis, we analyzed the expression of key genes of adipocyte differentiation. No changes in the expression of PPARγ, C/EBPα, and C/EBPß were detected (Fig. 5A). Hence, we conclude that the reduced AT mass in JunB-KO mice is not a result of reduced adipogenic potential. Next, we investigated whether JunB-KO mice might counteract their reduced WAT mass by increasing the expression of genes for FA and glucose uptake or for lipogenesis. We found neither expression of lipoprotein lipase (LPL), a protein important for FA supply to WAT, nor expression of Glut4, the main transporter involved in glucose uptake into WAT, changed in fasted (Fig. 5A) or fed (data not shown) JunB-KO mice compared with WT controls. Additionally, lipogenic genes like ATP-citrate lyase (ACL), FA synthase (FAS) and steroyl-CoA-desaturase 1 (SCD1) were not changed in WAT of JunB-KO mice (Fig. 5A). Next, we analyzed whether the reduction of AT mass in JunB-KO mice might be due to increased lipolysis. Interestingly, we found ATGL, the main lipase responsible for intracellular TG hydrolysis, to be strongly up-regulated on mRNA and protein levels in WAT of fasted JunB-KO mice (Fig. 5, A and B, respectively). Furthermore, comparative gene identification-58 (CGI-58), an important activator of ATGL lipolysis, is significantly increased in WAT of JunB-KO mice (Fig. 5A). In addition, HSL mRNA expression was significantly increased (Fig. 5A). Finally, the plasma FFA levels in relation to total adipose mass (23, 24) of fasted JunB-deficient mice are 1.5-fold elevated compared with WT animals (0.216 ± 0.077 mmol/liter · g fat mass in JunB-KO mice compared with 0.146 ± 0.028 mmol/liter · g fat mass in WT animals; P value = 0.065). This suggests an increase in the lipolytic rate, which might be a response to counteract the reduced energy substrate serum levels (FFA and glucose; Table 1) and the resulting reduced energy supply for the periphery.
Fig. 5.
Changes in gene/protein expression in WAT and SM of fasted JunB-KO mice. JunB-KO mice and their WT controls (n ≥ 3) were fasted overnight, and afterward, tissues were collected and RNA isolated for gene expression analysis, assessed by real-time PCR. Gene expression was normalized to TFIIß, and relative mRNA expression levels were calculated using averaged ΔΔCt. For each gene, values are relative to the WT animals. For Western blot analysis, protein was isolated from WAT of JunB-KO mice and their WT controls (n = 2). ß-Actin served as loading control. Statistical significance was determined using the two-tailed Student's t test. **, P < 0.01; ***, P < 0.001. A, mRNA expression levels in WAT. ATGL, CGI-58, and HSL mRNA expression were significantly increased in WAT of JunB-deficient mice. B, Protein expression levels of ATGL were analyzed by Western blot as described in the Materials and Methods. ATGL expression was strongly increased in WAT of JunB-KO mice. C, mRNA expression levels in SM. LPL, CD36, and some lipogenic genes were strongly increased in SM of JunB-deficient mice.
In line with this, LPL and CD36 expression are highly up-regulated in SM of fasted JunB-deficient mice (Fig. 5C), probably to supply the SM with sufficient amounts of FA to satisfy the demand for ß-oxidation. Also, lipogenic genes like ACL, FAS, and SCD1 were highly up-regulated in the SM of fasted JunB-KO mice, as depicted in Fig. 5C. From these data, we conclude that JunB-KO mice increase TG hydrolysis in WAT to supply the periphery with energy. Additionally, because LPL and CD36 are increased in SM of JunB-KO, we hypothesize that JunB-KO mice try to maintain SM energy supply at the expense of WAT.
TNFα expression is strongly increased in JunB-KO mice
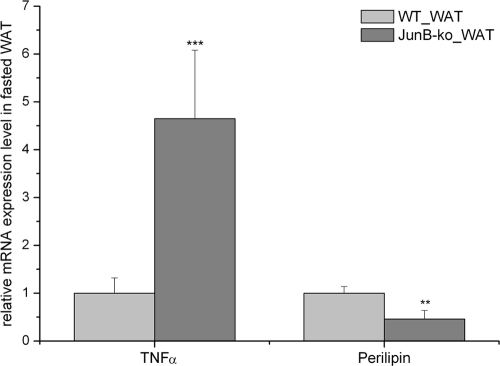
Our cell culture experiments showed that JunB knockdown does not regulate genes involved in adipogenesis and lipolysis per se. However, JunB has been shown to be involved in the control of TNFα shedding and expression (6, 25), and many studies have implicated TNFα in the regulation of lipolysis (26–30). We found TNFα mRNA expression to be significantly increased in WAT of JunB-deficient mice (Fig. 6). Furthermore, expression of perilipin, a lipid droplet surrounding protein that restricts lipolysis, was strongly reduced in these mice (Fig. 6). From these results, we propose that an increase in TNFα expression may contribute to the increased lipolytic rate in JunB-deficient mice via down-regulation of perilipin.
Fig. 6.
Expression of TNFα and perilipin in WAT of fasted JunB-KO mice. JunB-KO mice and their WT controls (n ≥ 3) were fasted overnight, and afterward, tissues were collected and RNA isolated for gene expression analysis, assessed by real-time PCR. Gene expression was normalized to TFIIß, and relative mRNA expression levels were calculated using averaged ΔΔCt. For each gene, values are relative to the WT animals. Statistical significance was determined using the 2-tailed Student's t test. **, P < 0.01; ***, P < 0.001.
Discussion
In this study, we have explored the effects of JunB deficiency on AT metabolism in mice. Lack of JunB in the embryo is lethal, but strategies have been developed to obtain viable JunB-deficient mice (17, 16). We found that these JunB-deficient animals have strongly reduced weight gain reflected by massively reduced AT mass, leading to a death rate of approximately 70% within the first 4 months after birth. The fact that these mice have strongly reduced AT mass could be due either to a failure to acquire fat or to a progressive loss of fat while the animals are growing. To analyze whether JunB is involved in the acquisition of fat stores, we examined whether JunB is involved in adipogenesis. In fact, it has been previously shown that JunB overexpression reduces adipocytic differentiation through an interaction with C/EBPß (14). However, our results show that JunB silencing in 3T3-L1 changed neither lipid accumulation nor expression of adipogenic genes. Furthermore, the in vitro adipogenic capacity of primary cells (MEF) obtained from JunB-KO mice was indistinguishable from WT MEF. Additionally, we measured the gene expression of adipocytic key genes like PPARγ, C/EBPα, and C/EBPβ in AT of JunB-KO mice and found no differences in comparison with WT littermates. Finally, we investigated the effects of an energy-rich diet in JunB-deficient mice. On a HFD, the survival rate of JunB-KO mice could be significantly improved, possibly by preventing further weight loss. These results show that JunB-KO animals can properly acquire fat, and that the capacity of adipogenesis is still retained in these mice. All these data suggest that JunB deficiency has no influence on adipogenesis in these mice.
Interestingly, gene expression analysis of WAT of fasted JunB-KO mice showed an increased HSL expression and strong up-regulation of ATGL and CGI-58 expression. Additionally, the plasma FFA to WAT mass ratio of fasted JunB-KO mice is elevated, suggesting increased lipolysis in agreement with the increased expression of lipolytic genes. At least in human fat cells and for HSL, a strong positive correlation between lipolytic activity and mRNA expression has been shown (31). Therefore, AT reduction in JunB-KO mice could be due to an increased lipolytic rate leading to decreased fat accretion. Whether this possible increased lipolysis is the cause or the consequence of the metabolic derangements in JunB-KO mice remains unclear. However, ATGL and HSL are highly up-regulated in WAT of fasted JunB-KO mice, whereas expression of both is not changed in JunB-silenced 3T3-L1 cells, suggesting no direct regulation of ATGL and HSL via JunB. Thus, the increase of ATGL and HSL might be due to a systemic effect/factor in JunB-KO mice, like the significantly decreased insulin levels. Insulin has been shown to be an important hormonal regulator of HSL and ATGL expression and activity (32–34).
JunB-KO mice develop a chronic myeloid leukemia-like disease later in life (17), but we report here that a phenotype resembling cachexia develops early postnatally in these mice, before the onset of leukemia. Cachexia is recognized as a severe wasting syndrome associated with cancer or other nonmalignant conditions (35). However, JunB-KO mice reflect only some hallmarks of cachexia while lacking others. For instance, cachexia is characterized by loss of muscle mass with or without loss of fat mass (36), but lean body mass of JunB-KO mice is not different from WT mice. Furthermore, insulin resistance and anorexia are often associated with cachexia (35), which cannot be recapitulated in JunB-KO mice (they are more insulin sensitive than WT mice, and their food intake is equal to WT mice), although increased lipolysis in cancer cachexia has also been shown to occur independently of malnutrition (24, 36, 37). We also found that JunB-KO mice on a HFD can normalize their weight to WT levels within 4–5 wk. The topic of nutritional reversion of cachexia is also highly controversial. For cancer cachexia, it has been shown that neither oral nor iv nutritional supplementation can reverse the weight loss in cachectic subjects (38). On the other hand, HFD have previously been shown to ameliorate cachexia symptoms in other noncancerous animal models (38, 39). Finally, strongly arguing in favor of a cachexia-resembling phenotype of JunB-KO mice, their decreased body (and fat) weight might be caused by increased transcriptional up-regulation of the lipolytic pathway in WAT. Increased lipolysis in cancer cachexia was previously attributed to up-regulation of HSL expression (23, 24), whereas ATGL expression was not changed (23). AT loss and increased lipolysis have also been shown to be associated with noncancer cachexia (40–43); however, to our knowledge, neither HSL nor ATGL have been directly linked to non-cancer-associated cachexia. Thus, our data suggest that also up-regulation of ATGL, along with HSL, could be involved in the formation of a cachectic phenotype.
TNFα have among others been shown to elevate lipolysis in WAT (26, 27, 44, 45, 47). Recently, Guinea-Viniegra et al. (6) reported that Jun proteins control TNFα shedding and epidermal inflammation. They demonstrated that the loss of Jun proteins in the epidermis evokes a TNFα-dependent cytokine cascade, inducing severe skin inflammation and perinatal death of newborns by depletion of glycogen and lipid reservoirs, and describe this as a cachectic phenotype (6). We found TNFα expression highly up-regulated in WAT of fasted JunB-KO mice. Additionally, we found the expression of perilipin, a lipid droplet-coating and lipolysis-restricting protein, to be strongly reduced in WAT of JunB-KO mice. TNFα has been described to activate lipolysis via down-regulation and phosphorylation of perilipin (26–30). Thus, we propose that elevated TNFα expression locally increases the lipolytic rate in WAT of JunB-deficient mice by down-regulation of perilipin. Notably, higher expression levels of ATGL and HSL, along with TNFα, in WAT of JunB-KO mice constitute an inconsistency with previous reports that show reduced ATGL and reduced or unchanged HSL expression upon increased TNFα levels (48–52). This discrepancy could stem from the fact that the majority of these reports are performed in vitro (49, 50, 52). The few studies that measure the effect of TNFα on ATGL and HSL expression in vivo do so in healthy mice with intact AT (51), which is not the case in JunB-KO mice that seem to be in a permanent catabolic state. Additionally, the repressive effect of TNFα on the expression of many adipogenic genes is most likely mediated by a down-regulation of PPARγ (49, 51, 53), of which both HSL and ATGL are direct target genes (54, 55). In AT of JunB-KO mice, however, PPARγ expression is comparable to WT control mice. Interestingly, perilipin (also a PPARγ target gene) (56) is down-regulated in AT of JunB-KO mice. A recent publication showed that perilipin mRNA regulation by TNFα depends strongly on nuclear factor-κB signaling, which was not the case for ATGL mRNA (47). Hence, it is possible that TNFα affects transcription of individual genes via separate pathways (i.e. nuclear factor-κB or PPARγ signaling). Also, TNFα treatment was shown to lead to a dose-dependent up-regulation of PPARγ and its targets (57), indicating that TNFα concentration and timing (acute vs. chronically changed levels) are of major importance and define the context of TNFα's effect on lipolytic genes. Finally, we find significantly reduced circulating insulin in fasted JunB-KO mice. Because insulin is known as a pivotal lipolytic hormone (32–34), its lower level in JunB-KO mice might facilitate up-regulation of ATGL and HSL.
Increased TNFα-induced lipolysis has been linked to insulin resistance, at least in part due to the increased FA levels in plasma, which accumulate in other nonadipose tissues (58). In fact, in JunB-KO mice, the consumed energy is not adequately stored in WAT, and these mice show increased plasma TG levels in the fed condition. LPL and Glut4 gene expression were not modified in WAT, suggesting that JunB-KO mice show no tendency to counteract AT loss. Thus, we tested glucose tolerance and insulin sensitivity. We found that JunB-KO mice and their WT littermates have the same tolerance to glucose, and therefore, the pancreatic insulin secretion seems not to be altered. However, the IST showed that there is ameliorated insulin sensitivity in JunB-KO mice, which might in turn lead to the lower glucose and insulin levels displayed by these mice. This result is somewhat surprising because most of the more severely lipodystrophic mouse models are insulin resistant (59–61). However, there are also mouse models with reduced AT mass that develop neither a fatty liver nor insulin resistance (62). The increased insulin sensitivity could be an effect of the low AT mass, much like the reduction of AT mass in obesity has been related to amelioration of insulin sensitivity in part due to reduced FA levels (63, 64). In agreement, a model with a nonsecretable TNFα transgenically expressed in an adipocyte-specific manner shows local adipose insulin resistance but does not develop systemic insulin resistance (46). These mice have lower AT mass and no changes in Glut4 expression in WAT, as observed in the JunB-KO mice. In addition, the SM mRNA expression level of Glut4 was not modified, as would have been expected if the JunB-KO animals suffered from a systemic TNFα-induced insulin resistance. Instead, we found the expression of lipogenic genes up-regulated in SM, presumably to counteract reduced ß-oxidation which could occur as a result of the lower serum FA levels in fasted JunB-KO mice. Furthermore, LPL expression in SM was also up-regulated in JunB-KO mice. During fasting, this could be a mechanism to shuffle more FA from hydrolyzed lipoprotein-associated TG to the SM to provide more energy when serum FA and glucose levels are low. In addition, we have not observed fatty liver symptoms in JunB-KO mice, which also indicate that FA derived from TG are not stored ectopically.
In summary, we show here that JunB-deficient mice have strongly reduced AT mass and that enzymes of lipolysis (ATGL and HSL) were significantly increased in WAT of these mice as well as the ratio of plasma FFA per gram fat mass, suggesting an increased lipolysis. However, our data indicate that JunB deficiency neither interferes with adipogenesis nor directly influences lipolytic gene expression. Furthermore, we propose that TNFα up-regulation in WAT of JunB-KO mice, and concomitantly reduced perilipin levels present a possible underlying mechanism as well as a link between increased lipolytic rate and JunB deficiency.
Acknowledgments
We thank Florian Stoeger and Claudia Neuhold for technical assistance. We are very thankful to Erwin F. Wagner for providing the JunBfl/fl and MORE-Cre mice.
This work was supported by the Austrian Ministry for Science and Research, GEN-AU project GOLD-Genomics of Lipid-Associated Disorders (FFG 820979 C3), and the Austrian Science Fund SFB, Project Lipotoxicity (F 30 01).
Current address for M.P.: Department of Biochemistry and Biotechnology, Universitat Rovira i Virgili, 43007 Tarragona, Spain.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- AP-1
- Activator protein 1
- AT
- adipose tissue
- ATGL
- adipose triglyceride lipase
- AUC
- area under the curve
- BMC
- body mass composition
- CA
- catecholamine
- CGI-58
- comparative gene identification-58
- FFA
- free fatty acid
- GTT
- glucose tolerance test
- IST
- insulin sensitivity test
- JunB-KO
- JunB knockout
- LPL
- lipoprotein lipase
- MEF
- mouse embryonic fibroblasts
- ntc
- nontargeting control construct
- shRNA
- short hairpin RNA
- SM
- skeletal muscle
- TG
- triglyceride
- WAT
- white AT.
References
- 1. Shaulian E, Karin M. 2002. AP-1 as a regulator of cell life and death. Nat Cell Biol 4:E131–E136 [DOI] [PubMed] [Google Scholar]
- 2. Shaulian E. 2010. AP-1—the Jun proteins: oncogenes or tumor suppressors in disguise? Cell Signal 22:894–899 [DOI] [PubMed] [Google Scholar]
- 3. Passegué E, Wagner EF. 2000. JunB suppresses cell proliferation by transcriptional activation of p16(INK4a) expression. EMBO J 19:2969–2979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bakiri L, Lallemand D, Bossy-Wetzel E, Yaniv M. 2000. Cell cycle-dependent variations in c-Jun and JunB phosphorylation: a role in the control of cyclin D1 expression. EMBO J 19:2056–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Andrecht S, Kolbus A, Hartenstein B, Angel P, Schorpp-Kistner M. 2002. Cell cycle promoting activity of JunB through cyclin A activation. J Biol Chem 277:35961–35968 [DOI] [PubMed] [Google Scholar]
- 6. Guinea-Viniegra J, Zenz R, Scheuch H, Hnisz D, Holcmann M, Bakiri L, Schonthaler HB, Sibilia M, Wagner EF. 2009. TNFα shedding and epidermal inflammation are controlled by Jun proteins. Genes Dev 23:2663–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yogev O, Goldberg R, Anzi S, Yogev O, Shaulian E. 2010. Jun proteins are starvation-regulated inhibitors of autophagy. Cancer Res 70:2318–2327 [DOI] [PubMed] [Google Scholar]
- 8. Hansen L, Gaster M, Oakeley EJ, Brusgaard K, Damsgaard Nielsen EM, Beck-Nielsen H, Pedersen O, Hemmings BA. 2004. Expression profiling of insulin action in human myotubes: induction of inflammatory and pro-angiogenic pathways in relationship with glycogen synthesis and type 2 diabetes. Biochem Biophys Res Commun 323:685–695 [DOI] [PubMed] [Google Scholar]
- 9. Sartipy P, Loskutoff DJ. 2003. Expression profiling identifies genes that continue to respond to insulin in adipocytes made insulin-resistant by treatment with tumor necrosis factor-α. J Biol Chem 278:52298–52306 [DOI] [PubMed] [Google Scholar]
- 10. Coletta DK, Balas B, Chavez AO, Baig M, Abdul-Ghani M, Kashyap SR, Folli F, Tripathy D, Mandarino LJ, Cornell JE, Defronzo RA, Jenkinson CP. 2008. Effect of acute physiological hyperinsulinemia on gene expression in human skeletal muscle in vivo. Am J Physiol Endocrinol Metab 294:E910–E917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mizuno TM, Funabashi T, Kleopoulos SP, Mobbs CV. 2004. Specific preservation of biosynthetic responses to insulin in adipose tissue may contribute to hyperleptinemia in insulin-resistant obese mice. J Nutr 134:1045–1050 [DOI] [PubMed] [Google Scholar]
- 12. Hoene M, Franken H, Fritsche L, Lehmann R, Pohl AK, Häring HU, Zell A, Schleicher ED, Weigert C. 2010. Activation of the mitogen-activated protein kinase (MAPK) signalling pathway in the liver of mice is related to plasma glucose levels after acute exercise. Diabetologia 53:1131–1141 [DOI] [PubMed] [Google Scholar]
- 13. Sabatakos G, Sims NA, Chen J, Aoki K, Kelz MB, Amling M, Bouali Y, Mukhopadhyay K, Ford K, Nestler EJ, Baron R. 2000. Overexpression of DeltaFosB transcription factor(s) increases bone formation and inhibits adipogenesis. Nat Med 6:985–990 [DOI] [PubMed] [Google Scholar]
- 14. Mariani O, Brennetot C, Coindre JM, Gruel N, Ganem C, Delattre O, Stern MH, Aurias A. 2007. JUN oncogene amplification and overexpression block adipocytic differentiation in highly aggressive sarcomas. Cancer Cell 11:361–374 [DOI] [PubMed] [Google Scholar]
- 15. Passegué E, Jochum W, Schorpp-Kistner M, Möhle-Steinlein U, Wagner EF. 2001. Chronic myeloid leukemia with increased granulocyte progenitors in mice lacking junB expression in the myeloid lineage. Cell 104:21–32 [DOI] [PubMed] [Google Scholar]
- 16. Passegué E, Wagner EF, Weissman IL. 2004. JunB deficiency leads to a myeloproliferative disorder arising from hematopoietic stem cells. Cell 119:431–443 [DOI] [PubMed] [Google Scholar]
- 17. Kenner L, Hoebertz A, Beil T, Keon N, Karreth F, Eferl R, Scheuch H, Szremska A, Amling M, Schorpp-Kistner M, Angel P, Wagner EF. 2004. Mice lacking JunB are osteopenic due to cell-autonomous osteoblast and osteoclast defects. J Cell Biol 164:613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hess J, Hartenstein B, Teurich S, Schmidt D, Schorpp-Kistner M, Angel P. 2003. Defective endochondral ossification in mice with strongly compromised expression of JunB. J Cell Sci 116:4587–4596 [DOI] [PubMed] [Google Scholar]
- 19. Bogner-Strauss JG, Prokesch A, Sanchez-Cabo F, Rieder D, Hackl H, Duszka K, Krogsdam A, Di Camillo B, Walenta E, Klatzer A, Lass A, Pinent M, Wong WC, Eisenhaber F, Trajanoski Z. 2010. Reconstruction of gene association network reveals a transmembrane protein required for adipogenesis and targeted by PPARγ. Cell Mol Life Sci 67:4049–4064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Todaro GJ, Green H. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol 17:299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Soukas A, Socci ND, Saatkamp BD, Novelli S, Friedman JM. 2001. Distinct transcriptional profiles of adipogenesis in vivo and in vitro. J Biol Chem 276:34167–34174 [DOI] [PubMed] [Google Scholar]
- 22. Pabinger S, Thallinger GG, Snajder R, Eichhorn H, Rader R, Trajanoski Z. 2009. QPCR: application for real-time PCR data management and analysis. BMC Bioinformatics 10:268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Agustsson T, Rydén M, Hoffstedt J, van Harmelen V, Dicker A, Laurencikiene J, Isaksson B, Permert J, Arner P. 2007. Mechanism of increased lipolysis in cancer cachexia. Cancer Res 67:5531–5537 [DOI] [PubMed] [Google Scholar]
- 24. Rydén M, Agustsson T, Laurencikiene J, Britton T, Sjölin E, Isaksson B, Permert J, Arner P. 2008. Lipolysis—not inflammation, cell death, or lipogenesis—is involved in adipose tissue loss in cancer cachexia. Cancer 113:1695–1704 [DOI] [PubMed] [Google Scholar]
- 25. Zenz R, Eferl R, Kenner L, Florin L, Hummerich L, Mehic D, Scheuch H, Angel P, Tschachler E, Wagner EF. 2005. Psoriasis-like skin disease and arthritis caused by inducible epidermal deletion of Jun proteins. Nature 437:369–375 [DOI] [PubMed] [Google Scholar]
- 26. Zhang HH, Halbleib M, Ahmad F, Manganiello VC, Greenberg AS. 2002. Tumor necrosis factor-α stimulates lipolysis in differentiated human adipocytes through activation of extracellular signal-related kinase and elevation of intracellular cAMP. Diabetes 51:2929–2935 [DOI] [PubMed] [Google Scholar]
- 27. Rydén M, Arvidsson E, Blomqvist L, Perbeck L, Dicker A, Arner P. 2004. Targets for TNF-α-induced lipolysis in human adipocytes. Biochem Biophys Res Commun 318:168–175 [DOI] [PubMed] [Google Scholar]
- 28. Souza SC, de Vargas LM, Yamamoto MT, Lien P, Franciosa MD, Moss LG, Greenberg AS. 1998. Overexpression of perilipin A and B blocks the ability of tumor necrosis factor α to increase lipolysis in 3T3-L1 adipocytes. J Biol Chem 273:24665–24669 [DOI] [PubMed] [Google Scholar]
- 29. Bézaire V, Mairal A, Anesia R, Lefort C, Langin D. 2009. Chronic TNFα and cAMP pre-treatment of human adipocytes alter HSL, ATGL and perilipin to regulate basal and stimulated lipolysis. FEBS Lett 583:3045–3049 [DOI] [PubMed] [Google Scholar]
- 30. Ryden M, Dicker A, van Harmelen V, Hauner H, Brunnberg M, Perbeck L, Lonnqvist F, Arner P. 2002. Mapping of early signaling events in tumor necrosis factor-α-mediated lipolysis in human fat cells. J Biol Chem 277:1085–1091 [DOI] [PubMed] [Google Scholar]
- 31. Large V, Arner P, Reynisdottir S, Grober J, Van Harmelen V, Holm C, Langin D. 1998. Hormone-sensitive lipase expression and activity in relation to lipolysis in human fat cells. J Lipid Res 39:1688–1695 [PubMed] [Google Scholar]
- 32. Holm C, Osterlund T, Laurell H, Contreras JA. 2000. Molecular mechanisms regulating hormone-sensitive lipase and lipolysis. Annu Rev Nutr 20:365–393 [DOI] [PubMed] [Google Scholar]
- 33. Kershaw EE, Hamm JK, Verhagen LA, Peroni O, Katic M, Flier JS. 2006. Adipose triglyceride lipase: function, regulation by insulin, and comparison with adiponutrin. Diabetes 55:148–157 [PMC free article] [PubMed] [Google Scholar]
- 34. Lafontan M, Langin D. 2009. Lipolysis and lipid mobilization in human adipose tissue. Prog Lipid Res 48:275–297 [DOI] [PubMed] [Google Scholar]
- 35. Rydén M, Arner P. 2007. Fat loss in cachexia: is there a role for adipocyte lipolysis? Clin Nutr 26:1–6 [DOI] [PubMed] [Google Scholar]
- 36. Bosaeus I, Daneryd P, Svanberg E, Lundholm K. 2001. Dietary intake and resting energy expenditure in relation to weight loss in unselected cancer patients. Int J Cancer 93:380–383 [DOI] [PubMed] [Google Scholar]
- 37. Shaw JH, Wolfe RR. 1987. Fatty acid and glycerol kinetics in septic patients and in patients with gastrointestinal cancer. The response to glucose infusion and parenteral feeding. Ann Surg 205:368–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gonçalves N, Roncon-Albuquerque R, Jr, Oliveira M, Quina-Rodrigues C, Lourenço AP, Leite-Moreira AF. 2010. A high-calorie diet attenuates cachexia and adipose tissue inflammation in monocrotaline-induced pulmonary hypertensive rats. Rev Port Cardiol 29:391–400 [PubMed] [Google Scholar]
- 39. Kim HJ, Vaziri ND, Norris K, An WS, Quiroz Y, Rodriguez-Iturbe B. 2010. High-calorie diet with moderate protein restriction prevents cachexia and ameliorates oxidative stress, inflammation and proteinuria in experimental chronic kidney disease. Clin Exp Nephrol 14:536–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Anker SD, Ponikowski PP, Clark AL, Leyva F, Rauchhaus M, Kemp M, Teixeira MM, Hellewell PG, Hooper J, Poole-Wilson PA, Coats AJ. 1999. Cytokines and neurohormones relating to body composition alterations in the wasting syndrome of chronic heart failure. Eur Heart J 20:683–693 [DOI] [PubMed] [Google Scholar]
- 41. Wilson DO, Rogers RM, Wright EC, Anthonisen NR. 1989. Body weight in chronic obstructive pulmonary disease. The National Institutes of Health Intermittent Positive-Pressure Breathing Trial. Am Rev Respir Dis 139:1435–1438 [DOI] [PubMed] [Google Scholar]
- 42. Jakobsson P, Jorfeldt L, von Schenck H. 1995. Fat metabolism and its response to infusion of insulin and glucose in patients with advanced chronic obstructive pulmonary disease. Clin Physiol 15:319–329 [DOI] [PubMed] [Google Scholar]
- 43. Heijligenberg R, Romijn JA, Klein S, Endert E, Sauerwein HP. 1997. Lipolytic sensitivity to catecholamines in patients with human immunodeficiency virus infection. Am J Clin Nutr 66:633–638 [DOI] [PubMed] [Google Scholar]
- 44. Rahn Landström T, Mei J, Karlsson M, Manganiello V, Degerman E. 2000. Down-regulation of cyclic-nucleotide phosphodiesterase 3B in 3T3-L1 adipocytes induced by tumour necrosis factor α and cAMP. Biochem J 346(Pt 2):337–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gasic S, Tian B, Green A. 1999. Tumor necrosis factor α stimulates lipolysis in adipocytes by decreasing Gi protein concentrations. J Biol Chem 274:6770–6775 [DOI] [PubMed] [Google Scholar]
- 46. Xu H, Hirosumi J, Uysal KT, Guler AD, Hotamisligil GS. 2002. Exclusive action of transmembrane TNF α in adipose tissue leads to reduced adipose mass and local but not systemic insulin resistance. Endocrinology 143:1502–1511 [DOI] [PubMed] [Google Scholar]
- 47. Laurencikiene J, van Harmelen V, Arvidsson Nordström E, Dicker A, Blomqvist L, Näslund E, Langin D, Arner P, Rydén M. 2007. NF-κB is important for TNF-α-induced lipolysis in human adipocytes. J Lipid Res 48:1069–1077 [DOI] [PubMed] [Google Scholar]
- 48. Green A, Dobias SB, Walters DJ, Brasier AR. 1994. Tumor necrosis factor increases the rate of lipolysis in primary cultures of adipocytes without altering levels of hormone-sensitive lipase. Endocrinology 134:2581–2588 [DOI] [PubMed] [Google Scholar]
- 49. Kim JY, Tillison K, Lee JH, Rearick DA, Smas CM. 2006. The adipose tissue triglyceride lipase ATGL/PNPLA2 is downregulated by insulin and TNF-α in 3T3-L1 adipocytes and is a target for transactivation by PPARγ. Am J Physiol Endocrinol Metab 291:E115–E127 [DOI] [PubMed] [Google Scholar]
- 50. Kralisch S, Klein J, Lossner U, Bluher M, Paschke R, Stumvoll M, Fasshauer M. 2005. Isoproterenol, TNFα, and insulin downregulate adipose triglyceride lipase in 3T3-L1 adipocytes. Mol Cell Endocrinol 240:43–49 [DOI] [PubMed] [Google Scholar]
- 51. Li L, Yang G, Shi S, Yang M, Liu H, Boden G. 2009. The adipose triglyceride lipase, adiponectin and visfatin are downregulated by tumor necrosis factor-α (TNF-α) in vivo. Cytokine 45:12–19 [DOI] [PubMed] [Google Scholar]
- 52. Sumida M, Sekiya K, Okuda H, Tanaka Y, Shiosaka T. 1990. Inhibitory effect of tumor necrosis factor on gene expression of hormone sensitive lipase in 3T3-L1 adipocytes. J Biochem 107:1–2 [DOI] [PubMed] [Google Scholar]
- 53. Guilherme A, Tesz GJ, Guntur KV, Czech MP. 2009. Tumor necrosis factor-α induces caspase-mediated cleavage of peroxisome proliferator-activated receptor γ in adipocytes. J Biol Chem 284:17082–17091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Deng T, Shan S, Li PP, Shen ZF, Lu XP, Cheng J, Ning ZQ. 2006. Peroxisome proliferator-activated receptor-γ transcriptionally up-regulates hormone-sensitive lipase via the involvement of specificity protein-1. Endocrinology 147:875–884 [DOI] [PubMed] [Google Scholar]
- 55. Kershaw EE, Schupp M, Guan HP, Gardner NP, Lazar MA, Flier JS. 2007. PPARγ regulates adipose triglyceride lipase in adipocytes in vitro and in vivo. Am J Physiol Endocrinol Metab 293:E1736–E1745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Dalen KT, Schoonjans K, Ulven SM, Weedon-Fekjaer MS, Bentzen TG, Koutnikova H, Auwerx J, Nebb HI. 2004. Adipose tissue expression of the lipid droplet-associating proteins S3–12 and perilipin is controlled by peroxisome proliferator-activated receptor-γ. Diabetes 53:1243–1252 [DOI] [PubMed] [Google Scholar]
- 57. Zhao S, Dong S. 2008. Effect of tumor necrosis factor α on cholesterol efflux in adipocytes. Clin Chim Acta 389:67–71 [DOI] [PubMed] [Google Scholar]
- 58. Cawthorn WP, Sethi JK. 2008. TNF-α and adipocyte biology. FEBS Lett 582:117–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Duan SZ, Ivashchenko CY, Whitesall SE, D'Alecy LG, Duquaine DC, Brosius FC, 3rd, Gonzalez FJ, Vinson C, Pierre MA, Milstone DS, Mortensen RM. 2007. Hypotension, lipodystrophy, and insulin resistance in generalized PPARγ-deficient mice rescued from embryonic lethality. J Clin Invest 117:812–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Moitra J, Mason MM, Olive M, Krylov D, Gavrilova O, Marcus-Samuels B, Feigenbaum L, Lee E, Aoyama T, Eckhaus M, Reitman ML, Vinson C. 1998. Life without white fat: a transgenic mouse. Genes Dev 12:3168–3181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shimomura I, Hammer RE, Richardson JA, Ikemoto S, Bashmakov Y, Goldstein JL, Brown MS. 1998. Insulin resistance and diabetes mellitus in transgenic mice expressing nuclear SREBP-1c in adipose tissue: model for congenital generalized lipodystrophy. Genes Dev 12:3182–3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Reue K, Phan J. 2006. Metabolic consequences of lipodystrophy in mouse models. Curr Opin Clin Nutr Metab Care 9:436–441 [DOI] [PubMed] [Google Scholar]
- 63. Frayn KN, Tan GD, Karpe F. 2007. Adipose tissue: a key target for diabetes pathophysiology and treatment? Horm Metab Res 39:739–742 [DOI] [PubMed] [Google Scholar]
- 64. Karpe F, Tan GD. 2005. Adipose tissue function in the insulin-resistance syndrome. Biochem Soc Trans 33:1045–1048 [DOI] [PubMed] [Google Scholar]