Abstract
Holoprosencephaly (HPE) is the most common disorder of human forebrain and facial development. Presently understood etiologies include both genetic and environmental factors, acting either alone, or more likely, in combination. The majority of patients without overt chromosomal abnormalities or recognizable associated syndromes have unidentified etiologies. A potential candidate gene, Twisted Gastrulation Homolog 1 (TWSG1), was previously suggested as a contributor to the complex genetics of human HPE based on (1) cytogenetic studies of patients with 18p deletions, (2) animal studies of TWSG1 deficient mice, and (3) the relationship of TWSG1 to bone morphogenetic protein (BMP) signaling, which modulates the primary pathway implicated in HPE, Sonic Hedgehog (SHH) signaling. Here we present the first analysis of a large cohort of patients with HPE for coding sequence variations in TWSG1. We also performed fine mapping of 18p for a subset of patients with partial 18p deletions. Surprisingly, minimal evidence for alterations of TWSG1 was found, suggesting that sequence alterations of TWSG1 are neither a common direct cause nor a frequent modifying factor for human HPE pathologies.
Keywords: Twisted Gastrulation Homolog 1, TWSG1, Tsg, holoprosencephaly, HPE, 18p, BMP
Introduction
Holoprosencephaly (HPE, OMIM 236100) occurs during the 3rd to 4th week after conception in humans and is the most common developmental disorder of the human forebrain [1]. HPE occurs with an estimated prevalence of 1/250 conceptuses [2] and 1/10,000 live births [3]. The most severe form of HPE is alobar HPE, in which there is a complete lack of separation of the cerebral hemispheres. Less severe forms are characterized by various degrees of cerebral separation, and include, in order of decreasing severity, semilobar, lobar, and middle interhemispheric variant (MIHV) HPE. Microform HPE describes patients with subtle midline facial abnormalities but without cerebral structural abnormalities by conventional neuroimaging techniques [4]. Craniofacial features range from cyclopia (one central eye) and a proboscis (nose-like appendage) to hypotelorism [5]. Clinical complications include neurological (primarily profound neurocognitive impairment and seizures) and endocrinological (primarily diabetes insipidus), as well as other medical problems [6].
Various genetic factors have been implicated in the underlying pathogenesis of HPE, including chromosomal abnormalities, such as trisomy 13 and triploidy [7], and intragenic mutations in and deletions of known HPE genes, such as Sonic Hedgehog (SHH) [5,8-11]. In addition to genetic disruptions, several environmental factors, such as gestational diabetes [12], and multiple malformation genetic syndromes, such as Smith-Lemli-Opitz Syndrome [13], are also associated with HPE. In the majority of patients without chromosomal abnormalities or syndromic forms of HPE (“non-chromosomal, non-syndromic HPE”), the etiology is not yet identifiable. Furthermore, the well-documented genotypic and phenotypic heterogeneity of HPE has supported the notion that the genetics of HPE are more complicated than previously thought [14]. For example, a specific mutation can be associated with a broad spectrum of phenotypes, most clearly demonstrated in large families that segregate an HPE-associated mutation [8,15].
Despite the apparent overall complexity of neural development, recent studies reveal that even among distantly related vertebrate species, there is a repeated, and relatively consistent, involvement and utilization of an integrated set of extracellular ligands that direct the basic patterning of the neural axis, including forebrain and spinal cord. The key extracellular ligands/factors include Hedgehogs, Fibroblast growth factors (FGFs), Wnts, TGF-βs (including bone morphogenetic proteins, or BMPs, and Nodals), and retinoic acid (see reviews [16-20]). Classic HPE, in particular, has been shown to be most closely associated with the SHH signaling pathway of the forebrain (reviewed in [21,22]). Some investigators propose that additional sub-types of HPE stem from disorders of dorsal midline brain establishment whose development requires intact BMP signals [23]. Given that substantial experimental evidence suggests cross-talk between multiple signaling centers as being key to understanding telencephalic development [24,25], our lab has recently undertaken a study of FGF8 signaling [26], and in the present study, the potential role of BMP signaling, in human HPE. Given the extensive redundancy of BMP ligands, antagonists, and receptors, we found it logistically difficult to study all of these factors simultaneously. Rather, we chose to begin our study with Twisted Gastrulation Homolog 1 (TWSG1, 18p11.22) as a surrogate candidate gene based on our considerations of the cytogenetic studies of patients with deletions of 18p and animal models demonstrating its role as a key modulator of BMP signaling.
The SHH signaling pathway is the primary pathway implicated in typical HPE. Mice which are either lacking a prechordal plate or deficient in Shh signaling demonstrate inadequate forebrain development and facial midline defects, including a proboscis-like extension and single optic vesicle [27]. Studies detailing the marker expression in mice deficient in Twsg1 have suggested that the forebrain defects are related to loss of signaling from forebrain organizing centers, including Fgf8 from the anterior neural ridge, and Shh from the prechordal plate [28]. Thus, these particular Twsg1 −/− murine models uncover mechanistic links with the same factors implicated in classical HPE dysmorphologies.
Several independent lines of evidence have suggested a role for TWSG1 in human HPE, including cytogenetic evidence, animal models, and the relationship of TWSG1 and BMP signaling, as well as our knowledge of the co-interactions with other pathways known to be associated with HPE. Twsg1 (called Tsg in non-mammals) is a highly conserved secreted protein with 24 cysteine residues and a hydrophobic signal sequence that are shared by human, mouse, zebrafish, and Xenopus Tsg homologs [29]. Twsg1 modulates the BMP signaling pathway with context-dependent inhibition [30,31] and enhancement [32,33]. Tsg functions as a fundamental component of the large macromolecular inhibitory complex that includes chordin-Tsg-BMP, and it prevents ligand binding to its cognate Type I receptors, BMPR1a or BMPR1b. This inactive complex can also lead to the re-release of active BMPs following the regulated cleavage of the chordin component by Tolloid [34-36]. Thus, the extracellular activities of BMPs are tightly regulated during vertebrate development, and they serve multiple distinct roles at several critical developmental time-points; the Tsg homologs are essential components in this regulation.
Recurrent observations of patients with HPE and 18p deletions led to the identification of a locus for HPE on 18p, termed HPE4 (OMIM #142946) [37], with further refinement to 18p11.3 [38]. Later, Transforming Growth Factor-Beta-Induced Factor (TGIF, OMIM 602630) was identified as a gene associated with HPE on 18p [39]. Tgif is expressed in the developing forebrain and craniofacial structures [39,40]. However, several independent labs have shown that targeted disruption of Tgif in mouse embryos results in normal brain patterning [40]. Moreover, gastrulation defects only occur when Tgif and a related gene, Tgif2, are both disrupted in mice [41]. In humans, no mutations in TGIF2 (OMIM 607294, 20q11.2-q12) have thus far been associated with HPE [42]. These apparent mouse vs. human differences have led to the research questions of the extent to which alterations in the TWSG1 gene might play a role in HPE pathogenesis and of how TWSG1 interacts with other genes on 18p.
Furthermore, successive studies of the prevalence of TGIF alterations in patients with human HPE have revealed that TGIF is altered in up to only ~1-2% of patients with non-chromosomal, non-syndromic HPE [5,9]. In addition, patients with both intragenic mutations in and deletions of TGIF fall along the milder end of the HPE spectrum (Keaton et al., unpublished results [43]). Finally, hemizygosity of TGIF does not necessarily result in HPE given that only ~10-15% of patients with 18p deletions have overt HPE [44]. Taken together, it has been suggested that an additional HPE gene is located on 18p, which may in turn modify the ultimate phenotypic expression of 18p deletions.
An additional important pathway in forebrain development involves BMPs, which are a subset of secreted cytokines within the TGF-β super-family of signaling molecules. BMPs must be attenuated during forebrain development given that they generally play negative roles in early stages of head formation wherein they favor epidermal fates at the expense of nervous tissue [45]. Ohkubo et al. [24] have demonstrated that a balance of Shh, Fgf8, and BMP signaling is likely required in order to generate patterning centers that can optimally coordinate the growth of the telencephalic and optic vesicles. Both increased BMP signaling, resulting in reduced Shh and Fgf8 expression, and decreased BMP signaling, with maintenance of Shh and Fgf8 expression, result in decreased proliferation and hypoplasia of the telencephalic and optic vesicles. Therefore, Tsg homologs that are context-dependent agonists or antagonists of BMP signaling emerge as attractive candidates for mutational analysis among patients with HPE.
Animal studies in mouse, Xenopus tropicalis, zebrafish, and Drosophila have further demonstrated an association between Twsg1 and forebrain development. First, mice deficient in Twsg1 (Twsg1 −/−) show phenotypes that include HPE and cyclopia in a significant proportion of mice, most prominently against a C57BL/6 background [28]. Select Twsg1 −/− mice also have defects in eye development ranging from microphthalmia to lack of eye development [28], limited mandibular arch development or agnathia (absent mandible) [28,46], impaired foregut development and a rudimentary oropharynx that maintains communication with the nasopharynx [28], and salivary gland dysmorphogenesis [47]. Targeted disruption of Twsg1 in mice can also result in skeleto-lymphogenesis abnormalities [48], delayed ossification of cervical and upper thoracic regions, and truncated or discontinuous neural arches [28]. Next, X. tropicalis with reduced tsg due to morpholino oligonucleotides lack a forebrain [31]. Finally, Ross et al. [30] have shown that in zebrafish, blocking tsg1 function with morpholino oligonucleotides results in ventralization similar to chordin mutants, and that ectopic tsg1 mRNA has dorsalizing activity. They have further demonstrated in Drosophila that Tsg can co-operate with chordin to inhibit BMP signaling, resulting in ventralization.
Furthermore, mice that are double null for two BMP antagonists, chordin and noggin (Chrd−/−;Nog−/−), occasionally show HPE with a proboscis, cyclopia, and agnathia [49]. However, given that the double null mice have early lethality, Anderson et al. [50] studied mice with the genotype Chrd−/−;Nog+/−. They observed that these mice typically have three classes of defects, including a midline class (single nostril, proboscis, cyclopia, cleft palate), a truncation class, and a jaw class (lacking elements derived from the first branchial arch and frontonasal mass) [50, reviewed in 45].
Given the above evidence, we sought to test the role of TWSG1 in human HPE with a three-pronged approach. First, we performed fine mapping using of a subset of patients with 18p deletions (with known TGIF deletions) to better delineate whether TWSG1 was present or absent and to furthermore characterize the breakpoints. Second, given that both deletions and mutations affecting the same gene can result in HPE [22], we screened for sequence variations by performing high-throughput screening using High Resolution Melting (HRM) of nearly 350 patients with HPE, followed by direct DNA sequencing of HRM-detected variants. Third, we performed direct DNA sequencing on an additional cohort of patients with extra-cerebral phenotypic features that correlate with animal models. Here we present the first systematically documented study that examines a gene involved in BMP signaling in human HPE.
Materials and methods
Patient selection
All subjects provided informed consent for research participation in accordance with NHGRI IRB-approved protocols. Clinical data were collected in accordance with our NHGRI IRB-approved comprehensive clinical study on HPE.
The first arm of our study, in which we performed fine mapping of patients with 18p deletions, consisted of 10 patients with HPE and partial 18p deletions. For patient 10 with an unbalanced translocation 46,XX,der(18)t(6;18)(p24.1;p11.2)pat, resulting in a partial deletion of 18p and a partial duplication of 6p [38,51,52], DNA and lymphoblastoid cells were not available. As a surrogate marker for the patient, DNA and lymphoblastoid cells from the proband’s sibling, who had inherited a balanced translocation involving the same chromosomes, were used. The sample for patient 8 is available through the Coriell Institute for Medical Research, #GM18311. Patient 9 had an array comparative genomic hybridization (array CGH, aCGH) performed at an outside laboratory.
The second arm of our study, in which we performed high-throughput screening of TWSG1 using HRM (described below), consisted of 338 patients with HPE, 16 patients with possible HPE, and 6 unaffected relatives of probands. The entire clinical spectrum of HPE was represented in the cohort. In addition, 288 unrelated individuals were screened as controls, acquired as de-identified samples from Sigma-Aldrich Corporation and the European Collection of Cell Cultures (Human Random Control Panels 1, 3, and 4). HPE and control samples with melting profiles that deviated from wild type melting curves were directly sequenced.
The third arm of our study, in which we performed direct DNA sequencing of the TWSG1 coding region, consisted of 12 patients with features comparable to animal models of Twsg1 −/− [28]. Five of the patients had HPE, 2 of whom also had agnathia spectrum, 2 had vertebral abnormalities (butterfly vertebrae, C2-C3 vertebral fusion), and 1 had arhinia and microphthalmia. Seven of the patients had features of VACTERL association without HPE: 6 had definite and 1 had likely vertebral anomalies (including spina bifida occulta, hemivertebrae, butterfly vertebrae, dysplastic sacral vertebra, and/or congenital scoliosis), 5 had cardiac defects (including ventricular septal defect and congenital aortic stenosis with a bicuspid aortic valve), 5 had tracheo-esophageal fistulas, 3 had definite and 1 had likely renal anomalies (including renal hypoplasia, multicystic kidneys, and/or horseshoe kidneys), and 1 had radial limb anomalies (unilateral thumb hypoplasia and unilateral thumb aplasia). Sequencing was also performed on 2 patients with known TGIF variants to assess for a genetic epistasis event.
Finally, we also present additional clinical information gathered on 2 patients reported in the literature by Rosenfeld et al. [11], who do not have known HPE but who have partial 18p deletions involving the TWSG1 locus without involvement of TGIF.
Gene amplification
The sequence of the TWSG1 gene (NM_020648.5) was obtained from public database programs (http://www.ncbi.nlm.nih.gov) and the Bioinformatics site annotation provided by http://www.genome.ucsc.edu. Guidelines for the naming of the sequence variants conform to the recommendations of the human nomenclature committee (www.hgvs.org/mutnomen).
Oligonucleotide primers were designed using Oligo™ 6.8 for the four coding exons (exons 2-5) and exon/intron boundaries of TWSG1 (Table 1). The same primers were used for HRM, direct PCR amplification, and sequencing.
Table 1.
Primers for PCR amplification of the human TWSG1 gene, coding exons 2-5
| Exon | Forward | Reverse | Product size (bp) |
Tm* (°C) |
|---|---|---|---|---|
| 2 | 5′ CTG GGA GTT ACT GAT CAT CTT CTT 3′ |
5′ ATA AAC ATA ATG TTT CTG CTC TAC 3′ |
219 | 52 |
| 3 | 5′ ATT TGG AAT TTG AAG TTT AAC ATC 3′ |
5′ AGA AAT ATT AAG AAG TCT CCC TTG 3′ |
174 | 53 |
| 4 | 5′ CTA ACA AAA TCT TAT GAT ATT ACC 3′ |
5′ GGC GGA ATG GAA AAA GTC AAC T 3′ |
335 | 52 |
| 5 | 5′ TGC CCT GAA ATC TTA AAT TTT TGT 3′ |
5′ AAG TTG CTT TGG TTT GCA TTT GTC 3′ |
241 | 53 |
Annealing temperature for PCR amplification
HRM Roche LightCycler® 480
Each sample was amplified in a reaction volume of 7.5 μl, using 10 ng of DNA template, 3.75 μl of Roche High-Resolution Melting Master (containing FastStart Taq DNA polymerase, reaction buffer, dNTP mix, and HRM dye), 0.9 μl of magnesium chloride (Roche), 1.7 μl of molecular grade water (Cellgro), and 0.2 μM of each primer. All reactions were done using a Roche LightCycler® 480 (Roche, Ind.). PCR amplification parameters were: incubation at 95°C for 10 min, followed by 45 cycles of denaturation at 95°C for 10 sec, annealing 53°C for 15 sec, and extension at 72°C for 20 sec.
PCR assay for direct DNA sequencing
Each sample was amplified using a Roche system, in a reaction volume of 25 μl, using 25 ng of DNA template, 2.5 μl of 10X PCR buffer+MgCl2, 0.2 μl of FastStart Taq (5U/ μl), 0.2 mM of dNTP, 20.3 μl of molecular grade water (Cellgro), and 0.2 μM of each primer. All reactions were done using a PTC-225 thermocycler (MJ Research, Mass). PCR amplification parameters were: incubation at 95°C for 4 min, followed by 39 cycles of denaturation at 95°C for 30 sec, annealing at specific exon temperature (see Table 1) for 30 sec, and extension at 72°C for 1 min, with a final extension step of 72°C for 7 min.
High-Resolution DNA melting acquisition and analysis
High-Resolution DNA Melting (HRM) involves PCR in the presence of saturating DNA binding fluorescent dyes. Following PCR, DNA strands are separated using heat, and melting profiles and melting temperatures (Tm) are analyzed based on fluorescence data generated during DNA melting. HRM provides a closed-tube system, with reduced risk of contamination, decreased time for analysis, and with no additional processing necessary after PCR. The melting profile is primarily determined by GC content and amplicon length. HRM has previously been shown to detect single nucleotide polymorphisms (SNPs) in factor V (Leiden) and prothrombin, as well as to detect SNPs and mutations in other genes. HRM requires appropriate instrumentation, saturation dyes, and software for analysis (reviewed by Erali et al. [53]). In this candidate gene study, HRM was used to decrease the burden of sequencing by sequencing only samples with melting profiles that differed from wild type melting profiles.
The LightCycler® 480 II system (Roche Applied Science, Indianapolis, Ind.) was used to identify putative sequence variants in TWSG1 in 354 samples from patients with HPE or possible HPE, 6 samples from relatives, and 288 control samples. 384-well plates were used, with SYBR Green I detection format. The samples were tested as four different amplicons, one for each of the four coding exons (exons 2-5). First, samples were pre-incubated at 95°C for 10 min, followed by PCR amplification (above). Melting curves were generated using pre-incubation at 95°C for 1 min, pre-hold at 40°C for 1 min, and increasing temperature from 65°C to 95°C at a programmed rate of 0.2°C/sec with fluorescence acquisition of 25 times per 1°C of temperature increase. Within the 384-well plates, either 8 or 16 water-containing blank samples were used as negative controls.
HRM curve analysis was performed according to the manufacturer’s recommendations using LightCycler® 480 Software release v.1.5.0 (Idaho Technology Inc., Idaho and Roche Applied Sciences, Ind.). Amplicons were normalized, and temperature-shifted fluorescence-over-temperature plots were identified. Samples with melting profiles that deviated from the wild type melting profiles were then column-purified and sequenced bi-directionally to confirm the presence of sequence changes.
DNA sequencing
Bidirectional DNA sequencing of TWSG1 was performed on samples with variant HRM melting profiles and on a selection of samples intended for direct sequencing only (9 probands and 1 sibling with 18p deletions, 7 patients with VACTERL association, 5 patients with phenotypic features similar to TWSG1 −/− mice, and 2 patients with known TGIF variants). For HRM variants, sequencing was performed on the PCR product amplified during the HRM analysis following column-purification; sequence variants were then confirmed following PCR re-amplification of the original DNA sample. Sequencing was performed at the DNA Sequencing Facility, National Institute of Neurological Disorders and Stroke (NINDS), NIH. DNA sequences were manually annotated and analyzed using Sequencher 4.7 (Gene Codes Corporation). The reference sequence used for TWSG1 was NM_020648.5 (assembly from March 2006, NCBI36/hg18).
Additional genes known to be associated with HPE were screened using research methods for the patients with 18p deletions and TWSG1 sequence variants. Sonic Hedgehog (SHH, NM_000193.2), ZIC2 (NM_007129.2), SIX3 (NM_005413.2), and TGIF (NM_003244.2) were sequenced using previously published methods with primers and conditions available on request [8,39,54,55]. An additional gene associated with HPE, GLI2, was also sequenced for patients 1-5 with 18p deletions, using previously published methods [56]. GLI2 is less frequently tested for HPE due to practical limitations given its large size. An additional candidate genetic element, the putative GLI2 promoter, was sequenced for all of the patients with 18p deletions, excluding patient 9, using oligonucleotide primers designed with Oligo™ 6.8 based on a published report of the sequence by Dennler et al. [57]. The following primers were used: (5′ TGTGGTGGCTGTGACTGCTGAGAG 3′; 5′ CACTTTCTTTCACAGTCCTGTGTC 3′; 329 bp, 55°C).
Evaluation of sequence changes
Publically available databases were used to analyze sequence alterations. Base pair changes were evaluated for conservation using EvoPrinterHD and Evodifferences Profile (http://evoprinter.ninds.nih.gov/) and UCSC Multiz Alignment and Conservation (http://genome.ucsc.edu/), and for potential splicing alteration using ESE Finder 3.0 (http://rulai.cshl.edu/cgi-bin/tools/ESE3/esefinder.cgi?process=home). The Database of Genomic Variants (http://projects.tcag.ca/variation/) was used to confirm documented SNPs. The single detected amino acid change was evaluated using PolyPhen (http://genetics.bwh.harvard.edu/pph/), Protein BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi), and Constraint-based Multiple Alignment Tool (COBALT, http://www.ncbi.nlm.nih.gov/tools/cobalt/cobalt.cgi).
Fluorescence in situ hybridization (FISH)
FISH was performed on 9 individuals, including 8 patients with HPE and 1 patient’s sibling. Due to the unavailability of a cell line for patient 10 with an unbalanced translocation of chromosomes 6 and 18, a cell line from the proband’s sibling with a balanced translocation of the same chromosomes was used as a surrogate marker for the patient. Bacterial artificial chromosomes (BACs) for TWSG1 (RP11-66J9) and TGIF (RP11-22C6) were selected using the UCSC Human Genome Browser (Mar. 2006, NCBI36/hg18). BACs were ordered from the clone library of the National Human Genome Research Institute (NHGRI) Physical Mapping Core and were isolated using a NucleoBond® AX 500 (Maxi) kit.
Metaphase preparation from EBV cell lines was performed by standard air-drying technique. FISH was performed with labeled DNA by Nick Translation technique, essentially as described by Dutra et al. [58]. On each slide, 200 ng of labeled probe was applied. Repeat sequences were blocked with Cot 1 (10X excess) competitor DNA. 10 ul of a hybridization mixture containing the labeled DNA in 50% formamide, 2x SSC, and 10% dextran sulfate were denatured at 75°C for 10 min and then incubated at 37°C for 30 min for pre-annealing. Slides were denatured and hybridized for at least 18 hours. Slides were counterstained with DAPI-Antifade.
Array CGH
Array CGH was performed on 8 probands with 18p deletions and 1 sibling with a balanced translocation, using SurePrint G3 Custom Microarrays with 180,000 (180K) probes distributed throughout the genome, with denser coverage over regions of clinical interest and chromosome X (Agilent Technologies, Inc.). Five-hundred ng of patient genomic DNA was labeled with Cyanine-5dUTP (Cy5) (Enzo Life Sciences, Inc.), and 500 ng of reference DNA (Human Genomic DNA, Promega) was labeled with Cyanine-3dUTP (Cy3) (Enzo Life Sciences, Inc.). The DNA samples were then blocked, hybridized onto the microarray slides, cleaned and scanned with a DNA Microarray Scanner with SureScan High-Resolution Technology (Agilent Technologies, Inc.) at 3 microns, double-pass, as recommended by the manufacturers. The array images were extracted using Feature Extraction Software Version 10.7.3.1 (Agilent Technologies, Inc.). The analysis was performed in Genomic Workbench Standard Edition Version 5.0.14 for aCGH (Agilent Technologies, Inc.).
Copy numbers were considered under the following parameters: loss of copy if 4 or more consecutive probes had a Log2 of the ratio of the normalized intensity of Cy5 over Cy3 equal to or less than −1, and gain of copy if 4 or more consecutive probes had a Log2 of the ratio of the normalized intensity of Cy5 over Cy3 equal to or greater than 0.58 [59].
Results
Fluorescence in situ hybridization (FISH)
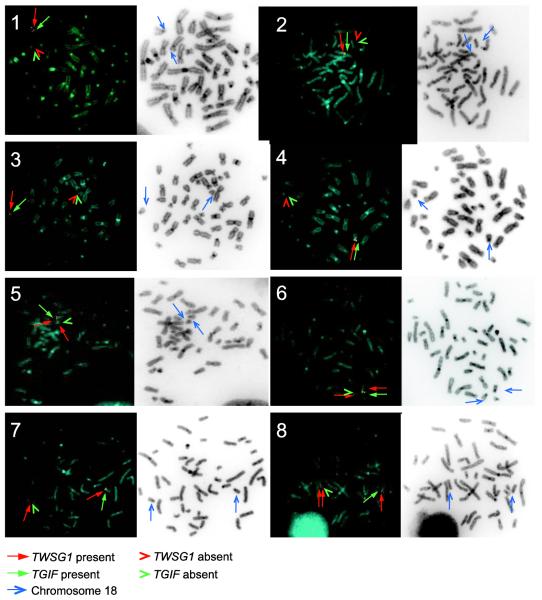
FISH was performed on 9 individuals with previously documented TGIF deletions in order to further characterize the presence or absence of TWSG1. The individuals studied included 8 probands with HPE and partial deletions of 18p, with or without duplications of 18p, and the sibling of a proband (patient 10) with an unbalanced translocation. FISH for the sibling showed that both TGIF and TWSG1 were part of the translocated segment of 18p on chromosome 6, thus implying that both genes were deleted in the proband.
Both TWSG1 and TGIF were deleted in 5 patients (4 severely affected and 1 mildly affected). TGIF alone was deleted in 3 patients (1 severely affected and 2 mildly affected). TGIF was deleted and TWSG1 duplicated in 1 severely affected patient. Of the 6 severely affected patients, 4 patients had additional cytogenetic abnormalities, including either an isochromosome 18q, partial deletion of Yp, partial duplication of 18p, or partial deletion of 6p. Table 2 and Figure 1.
Table 2.
FISH and aCGH results for patients with holoprosencephaly and partial 18p deletions
| Patient | Previous cytogenetic descriptions |
aCGH results (chromosome 18)° |
FISH results (TWSG1 - RP11-66J9, TGIF - RP11-22C6) |
HPE severity |
Other clinical feature s |
Sex | References ◆ |
|---|---|---|---|---|---|---|---|
| 1 | 46,XY,idic(18q) | arr 18p11.32p11(108, 819- 15,370,683)x1; arr 18q11.1q23(16,78 3,849- 76,113,817)x3 |
ish del(18)(p11.22p11.31)(R P11-66J9−,RP11-22C6−) |
Severe | Cyclopi a |
M | Overhauser et al., 1995 [38] |
| 2 | 46XX,del(18)(p11.1) | arr 18p11.32p11.21(3 8,826- 15,155,151)x1 |
ish del(18)(p11.22p11.31)(R P11-66J9−,RP11-22C6−) |
Mild (normal brain) |
DD, SMCI |
F | Overhauser et al., 1995 [38]; Aughton et al., 1991 [65] |
| 3 | 46,XX,del(18)(p11) | arr 18p11.32p11.21(1 08,151- 15,155,151)x1 |
ish del(18)(p11.22p11.31)(R P11-66J9−,RP11-22C6−) |
Severe (alobar) |
Unkno wn |
F | Overhauser et al., 1995 [38] |
| 4 | 45,X,dic(Y;18)(q;p11.2) | arr 18p11.32p11.21(5 9,754- 13,639,732)x1 |
ish del(18)(p11.22p11.31)(R P11-66J9−,RP11-22C6−) |
Severe | Cyclopi a |
M | Overhauser et al., 1995 [38]; Münke et al., 1988 [66] |
| 5 | 46,XY,r(18)(p11.31q23 ) |
arr 18p11.32p11.32(5 9,813- 4,012,544)x1; arr 18q12.1(26,114,4 73-26,261,205)x1 |
ish 18p11.22(RP11- 66J9x2),del(18)(p11.31p 11.31)(RP11-22C6−) |
Mild (normal brain) |
DD, SMCI |
M | Overhauser et al., 1995 [38] |
| 6 | Submicroscopic TGIF deletion |
arr 18p11.32p11.31(5 9,754- 5,216,552)x1 |
ish 18p11.22(RP11- 66J9x2),del(18)(p11.31p 11.31)(RP11-22C6−) |
Likely severe |
Optic hypopl asia, cleft lip, DI, epileps y |
M | Bendavid et al., 2006 [10]; Keaton et al., unpublished results [43] |
| 7 | Submicroscopic TGIF deletion |
arr 18p11.31(3,184,0 26-4,960,239)x1 |
ish 18p11.22(RP11- 66J9x2),del(18)(p11.31p 11.31)(RP11-22C6−) |
Mild (normal brain) |
SMCI, absent labial frenulu m, pyrifor m apertur e stenosi s, normal develo pment |
M | Keaton et al., unpublished results [43] |
| 8 | 46,XX,der(18)dup(p11. 1p11.3)del(p11.3) |
arr 18p11.32p11.31(4 ,316- 3,638,271)x1; arr 18p11.31p11.21(3 ,638,330- 15,155,151)x3 |
ish dup(18)(p11.22p11.22)( RP11- 66J9x3),del(p11.31p11.3 1)(RP11-22C6−) |
Likely severe |
Cleft lip/ palate, single nare, DI, epileps y |
F | Bendavid et al., 2006 [10] |
| 9 | N/A | arr 18p11.32p11.22 (1,543- 9,719,894)x1; arr 18p11.22p11.21 (9,720,723- 15,391,751)x3 de novo |
N/A | Mild | SMCI, pyrifor m apertur e stenosi s, epibulb ar lipoder moid, hearing loss |
M | This report |
| 10* | 46,XX,der(18)t(6;18)(p 24.1;p11.21)pat |
N/A | N/A | Severe | Agnathi a |
F | Overhauser et al., 1995 [38]; Krassikoff and Sekhon, 1989 [51] |
Fine mapping of the minimal critical region of 18p was performed for patients with 18p deletions of various sizes, using FISH and aCGH. °aCGH results are only given as they apply to 18p, and they were not replicated by an independent method, other than the FISH studies.
Cytogenetic descriptions and clinical features previously published as noted; clinical features for patients 8 and 9 have not previously been reported.
A sample from patient 10 was not available for testing; as a surrogate, the patient’s clinically normal brother with 46,XY,t(6;18)(p24.1;p11.21) was tested; on aCGH, he had no clinically significant copy number variations on 18p, and FISH showed ish t(6;18)(RP11-22C6+, RP11-66J9+; RP11-22C6−, RP11-66J9−). DD – developmental delay; DI – diabetes insipidus; N/A – not applicable; SMCI – single maxillary central incisor.
Fig. 1.
FISH was used to confirm the presence or absence of TWSG1 and TGIF, a gene known to be associated with HPE, for 8 patients with holoprosencephaly (HPE) and deletions of 18p. Red arrow indicates TWSG1 present, red arrowhead indicates TWSG1 absent, green arrow indicates TGIF present, and green arrowhead indicates TGIF absent. Patient identification indicated by numbers. The HPE types of patients 1, 3, 4, 6, and 8 are severe or likely severe. A sample from patient 10 was not available for testing; as a surrogate, the patient’s clinically normal brother with 46,XY,t(6;18)(p24.1;p11.21) was tested; FISH showed ish t(6;18)(RP11-22C6+, RP11-66J9+; RP11-22C6−, RP11-66J9−) (not shown).
Array CGH
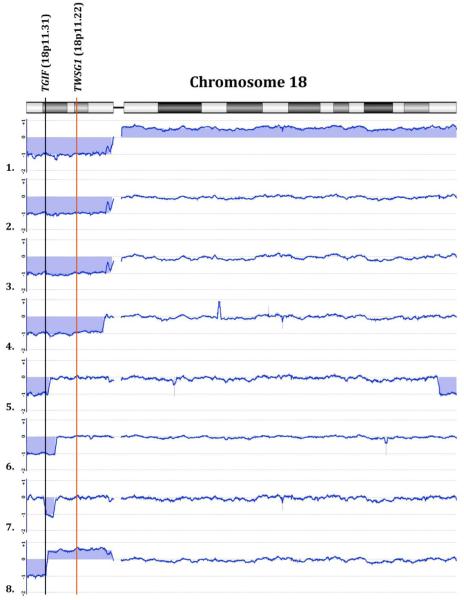
Using array CGH technology, we confirmed the FISH results for TGIF and TWSG1 of the 9 individuals with partial deletions of 18p (discussed above) and refined the estimation of breakpoints. The sibling with a balanced translocation had no copy number variants on 18p known to be clinically significant. In addition, our analysis includes a previously unreported mildly affected patient with an aCGH performed at an outside testing center that showed deletion of TGIF and TWSG1. Table 2 and Figure 2.
Fig. 2.
Array CGH was applied to better characterize the breakpoints on 18p for 8 patients with holoprosencephaly (HPE) and deletions of 18p. A schematic of chromosome 18 is represented, with the aCGH results of chromosome 18 for each patient. Differences in copy number of regions of the chromosome are shaded in blue. For each aCGH figure, a difference in the negative direction of the y-axis indicates a deletion, and a difference in the positive direction of the y-axis indicates a duplication. Three patients (2, 3, 8) share the same proximal breakpoint near the centromere (chr18:15,155,151). Patient identification indicated by numbers. The HPE types of patients 1, 3, 4, 6, and 8 are severe or likely severe.
High Resolution Melting and DNA sequencing
HRM was used to screen 338 patients with HPE, 16 patients with possible HPE, and 6 unaffected relatives, in addition to 288 control samples. Each sample was run as four amplicons, one for each coding exon. Of the 1,440 amplicons from patients with HPE, possible HPE, or unaffected relatives, 124 amplicons (8.6%) overall were sequenced due to variant melting profiles, including 24 (6.7%) for exon 2, 41 (11.4%) for exon 3, 37 (10.3%) for exon 4, and 22 (6.1%) for exon 5. Of the 1,152 amplicons from controls, 46 amplicons (4.0%) were sequenced due to variant melting profiles, including 10 (3.5%) for exon 2, 9 (3.1%) for exon 3, 16 (5.6%) for exon 4, and 11 (3.8%) for exon 5. There were 24/2,592 amplicon reactions (0.9%), including control and patient samples, that did not amplify with HRM, which were then directly amplified with PCR and sequenced.
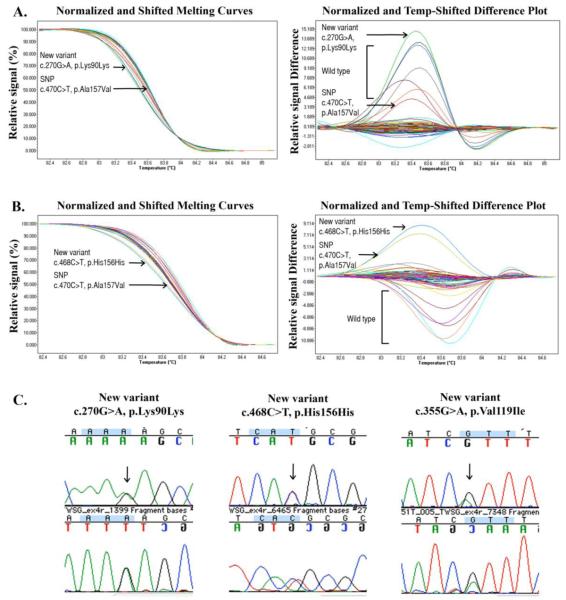
Of the 170 total HRM variant samples sequenced, 21 samples (12.4%) had sequence variants, which were comprised of 5 different sequence changes. Two of the sequence variants were previously undocumented synonymous coding sequence variants identified in patients and not in controls. The first variant (c.270G>A, p.Lys90Lys) was identified in a 10-year-old female with lobar HPE, distortion of the midbrain and falx and partial agenesis of the corpus callosum, whose facial anomalies included a cleft lip and palate. The second (c.468C>T, p.His156His) was identified in a 44-year old Caucasian female with lobar HPE, bilateral retinal colobamata, hypertelorism, macroglossia, esotropia, severe cognitive impairment and behavioral problems, balance problems, primary amenorrhea, and a cardiac malformation consisting of a partial atrioventricular (AV) canal defect. Figure 3.
Fig. 3.
High Resolution Melting (HRM) profiles of patients with sequence variants in TWSG1 and accompanying chromatograms. Temperature (°C) is represented on the x-axis, and relative signal (%) or relative signal difference is represented on the y-axis. Each curve represents a different patient sample. The curves clustering around the x-axis are presumably wild type. Following direct sequencing of HRM variants, two previously undocumented synonymous changes were identified in 2 patients with HPE, both in exon 4, which were not present in controls. Sequencing of the remaining HRM variants revealed wild type sequences. Normalized and Shifted Melting Curves (left) and Normalized and Temp-Shifted Difference Plot (right) are shown. Two different cohorts of patients were tested in (a) and (b). (a) Sequencing of the green curve revealed c.270G>A, p.Lys90Lys. Sequencing of the red curve revealed a known SNP (c.470C>T, p.Ala157Val, rs34595349). (b) Sequencing of the blue curve revealed c.468C>T, p.His156His. Sequencing of the yellow curve revealed the same known SNP as above. (c) Chromatograms illustrating the previously undocumented variants. The third chromatogram was a non-synonymous variant (c.355G>A, p.Val119Ile) identified in patient 8, not found in controls, which was detected through direct sequencing of TWSG1 for patients with 18p deletions.
Two previously documented SNPs were confirmed in both patient and control samples, including c.470C>T, p.Ala157Val (rs34595349) in 4 controls and 6 patients, and c.558G>A, p.Glu186Glu (rs72948180) in 2 controls and 4 patients. A previously undocumented SNP was identified in 2 controls and 1 patient (c.144G>A, p.Pro48Pro).
Direct DNA sequencing - Twisted Gastrulation Homolog 1 (TWSG1)
First, direct DNA sequencing of TWSG1 was performed on the 9 individuals tested with FISH, including 8 probands with HPE and 18p deletions, and 1 sibling of a proband with an 18p deletion for whom DNA was not available. A previously undocumented non-synonymous variant (c.355G>A, p.Val119Ile) was identified in patient 8 and was not observed in HRM controls. The patient had a partial 18p deletion/duplication, which involved deletion of TGIF and duplication of TWSG1. Inheritance could not be determined. The female patient had severe HPE identified on prenatal ultrasound consistent with alobar HPE, with a single large ventricle and absent cortical sulci and gyri, hydrocephalus ex vacuo and a small cerebellum. In addition, she had a midline cleft lip and palate, severe bitemporal narrowing, depressed supraorbital areas, and a sloping forehead. At 2 months of age, clinical characteristics included tonic-clonic seizures, failure to thrive, likely partial diabetes insipidus, and hypothalamic dysfunction. She died at 9 months of age secondary to a cardiopulmonary arrest.
In addition, direct DNA sequencing of TWSG1 was performed on 12 patients with features comparable to animal models of Twsg1 −/− [28] or with VACTERL association and prominent vertebral and foregut anomalies, and on 2 patients with known TGIF variants. No TWSG1 sequence variations were identified among this group of patients.
Additional genes
Screening through research methods of the four genes most commonly implicated in HPE (SHH, ZIC2, SIX3, and TGIF) revealed no pathogenic variants in the patients with 18p deletions or in the patients with previously undocumented TWSG1 variants. No pathogenic sequence variants in GLI2 were identified in the 5 patients with 18p deletions who were additionally screened for GLI2 variants (exons 2-14). One previously undocumented variant in the GLI2 promoter (g.121,549,834C>T), of unknown significance, was observed in patients 4 and 6.
Analysis of sequence variants
The non-synonymous variant (c.355G>A, p.Val119Ile) found in patient 8 with an 18p deletion and duplication showed high conservation at the nucleotide and amino acid level. Using EvoPrinterHD and the Evodifferences Profile, the base showed conservation in Chimpanzee, Rhesus-Monkey, Marmoset, Dog, Horse, Cow, Armadillo, and Mouse. Using COBALT, the amino acid demonstrated conservation in Rhesus Monkey, Horse, Dog, Chicken, Xenopus laevis, and Drosophila. The base change was not predicted to alter splicing (ESE Finder), and the amino acid change was predicted to have a benign effect on protein function (PolyPhen) consistent with similar biochemical properties of the two amino acids.
Of the two synonymous variants found in patients with HPE and not in controls, one (c.270G>A, p.Lys90Lys) showed high conservation and was conserved in the same EvoPrinter species noted above. It was not predicted to alter splicing. The other synonymous variant (c.468C>T, p.His156His) had mild conservation and was predicted to have mild effects on splicing.
Clinical descriptions of patients with TWSG1 deletions only
Two patients were identified through a study by Rosenfeld et al. [11] as having 18p deletions based on aCGH (Signature Genomic Laboratories, Seattle, WA, USA) that involve TWSG1 but not TGIF. Upon collection of further clinical data from the authors and a referring clinician, HPE was found to be absent in the first patient (by brain MRI) and could not be confirmed or ruled out in the second patient. The first patient is a 25-year-old female initially evaluated for chronic pancreatitis. Related to this issue, no mutations were identified in CFTR, PRSS1, and SPINK1. A brain MRI performed at 13 years of age was normal. She has macrocephaly (head circumference 59 cm, >98th centile), irregular helical contour with attached lobules, borderline low-set ears, a broad neck, mildly short fourth metacarpals, and cubitus valgus. Neurological exam reveals that she is a toe walker. She has a measured IQ of 97, but has learning disabilities, particularly with math and spelling. Medical history additionally includes hypothyroidism, type 2 diabetes mellitus, depression, and obesity (BMI 39.4 kg/m2). There is no family history of seizures or learning problems. An oligo array, SignatureChip OS (Signature Genomic Laboratories, Spokane, WA), showed a de novo 3.2 Mb deletion at chr18:8,562,765-11,724,781 (hg18).
The second patient is a 7-year-old female with unspecified dysmorphic features and a seizure disorder. No brain imaging was documented. Two different BAC arrays, Signature V4.0 and Signature MarkerChip, showed deletion of TWSG1 and neighboring genes on 18p11.22, confirmed to be de novo. Distal probes on v4 and three pericentric probes were not deleted.
Discussion
Our studies represent, to our knowledge, the first comprehensive attempt to inventory the sequence variations detectable in the human TWSG1 gene in both control individuals and patients with holoprosencephaly spectrum disorders. Surprisingly, we find that there is very limited sequence variation of the coding region of the TWSG1 gene in either group. Our study sample of individuals with HPE is of sufficient size to identify likely pathologically significant mutations in a novel candidate gene provided that they are present in aggregate among at least 1% of patients. Several genes associated with HPE, including TGIF and GLI2, are nearly at this lower limit threshold of sensitivity, while others are even less commonly implicated, including PTCH, DISP1, and FGF8, making definitive genetic molecular analysis quite complicated [22]. Furthermore, detecting simple sequence variants does not necessarily correlate with important functional changes in the altered protein. Consequently, a layer of functional genomics is required to authenticate most sequence changes.
Our data provide compelling evidence that coding region variations of the TWSG1 gene are not a common determinant or co-morbid genetic change that might directly account for HPE or its variability in clinical manifestations. It does not rule out, however, that non-coding genetic variations in the presently poorly understood regulatory elements of TWSG1 or related genes are prevalent or potentially important. Furthermore, based on our analysis, we cannot exclude TWSG1 as a major factor in the etiology of HPE in early embryos. In animal studies, deficiencies of TWSG1 are proven to cause HPE as characterized in early embryonic mouse phenotypes. However, our study samples are primarily from live-born infants, and thus we may be missing cases of early prenatal lethality related to TWSG1. An additional limitation to our approach was to use pre-screening by HRM to focus on cases of interest. While it is generally agreed that the sensitivity of HRM is high [60,61], it is not uniformly successful in identifying all classes of genetic variation. As discussed by Erali et al. [53], melting analysis may be complicated by the type of base change, the presence of a homozygous variant, or the presence of common non-disease causing variants.
The detailed analysis of the non-random cytogenetic rearrangements that led to the original descriptions of the HPE loci, including HPE4 on 18p, were the first indications of a single genetic event (albeit with the duplication or deletion of numerous genes) being both necessary and sufficient to produce HPE phenotypes. Once subsequent mutational analysis of genes within the molecularly defined minimal critical regions was applied, we, and others, were able to identify at least one gene per region that was an independent target of mutation in unrelated HPE families. However, as we have noted above (see Introduction), there has always been the possibility that additional genes in cis, or at completely different chromosomal locations, might also be responsible for the ultimate clinical manifestations. Our re-analysis of the 18p deletion cases leads us to conclude that the strongest predictor of HPE manifestations is the status of the TGIF gene and not that of TWSG1. Further analysis of patients with HPE and 18p deletions involving TGIF are reviewed in Rosenfeld et al. [11] and Keaton et al., unpublished results [43]. Although more cases need to be studied, we cannot establish a direct link between the deletion or mutation of TWSG1 and overt HPE malformations. Hence, the absence of frequent non-synonymous coding region changes, or prima facia deleterious changes in the gene, supports the hypothesis of a benign effect of haploinsufficiency for TWSG1 with respect to HPE-related pathologies.
The exquisite regulation of BMP signaling in which Tsg homologs participate underlies a fundamental aspect of vertebrate dorsal-ventral patterning that is highly conserved throughout phylogeny (see De Robertis [36]). Severe genetic alterations in non-redundant factors are likely to be causes of early embryonic lethality in humans. In contrast, mice tolerate numerous deleterious alleles in the biochemically similar BMP antagonists chordin and noggin before HPE pathologies become prevalent [50, reviewed in 45]. Therefore, at least in principal, it seems likely that subtle genetic variation in the BMP signaling pathway has many unexplored opportunities to affect mutations in more classically understood HPE-related pathologies. One case in point is the requirement for BMP signaling in dorsal midline development of the cortical hem and choroid plexus, and its proposed association with the MIHV of HPE (reviewed in [23]). Our sample set includes only 3 confirmed cases of this uncommon HPE variant, and therefore we cannot rule in or exclude a role of BMP signaling in its causation. However, we would note that pathologically relevant mutations in several HPE genes can be identified in this sub-type of MIHV HPE (Lacbawan et al. [62], Keaton et al., unpublished results [43], and Muenke lab, unpublished results), and it is no longer considered by us as an exclusive pathological aberration of dorsally expressed genes, such as Zic2. Rather, we now appreciate that Zic2 has both an early function during gastrulation [63] and a late role following neurulation that can account for the significant range of phenotypes encountered in HPE patients.
Given the multiple roles for BMP signaling in a variety of developmental contexts, it is worth proposing that mutational analysis of TWSG1 should continue to be considered in additional phenotypes beyond those examined in this study. Recently, Zakin et al. [64] reported a genetic interaction between Bmp7 and Tsg in the mouse resulting in sirenomelia. Similarly, the growth of the mandibular arch and salivary glands are influenced by Bmp signaling and its modulation by Tsg in animal models [46,47]. Therefore, our approach to the genetic analysis of the human TWSG1 gene may prove useful in additional patient groups with specific phenotypes related to disturbances in BMP signaling.
In practical molecular diagnostic terms, while it is challenging to identify even a single significant change in a patient with HPE, it is even rarer to identify two potentially co-morbid interacting mutations. Although such combinations of alterations have been previously reported as anecdotal cases, the plausibility of there being an established genetic interaction between them has been largely neglected or unproven. Therefore, our attempt to ascertain the mutational status of the other HPE genes in the 18p cases was entirely reasonable despite the negative findings, but should not be construed as disproving the “multiple-hit” hypothesis of HPE [14]. Rather, one must conclude that outside of the most common HPE-associated genes (SHH, ZIC2, and SIX3), as well as TGIF and GLI2, it is difficult to detect additional significant genetic alterations, at least in the genes tested thus far. It remains interesting that no additional co-morbid mutations were detected in these 18p deletion cases, though additional genes deleted from 18p may also be contributing to the HPE pathogenesis. Nevertheless, given the strong likelihood that HPE pathologies result from a synergistic effect of genetic variation in several different genes, instead of following classical recessive genetic patterns extrapolated from animal models, it remains important that the described interactions between BMP signaling and more classical HPE-related disease mechanisms be explored further by additional studies. As genomic technologies involving Next-Generation Sequencing become more inexpensive and practical in the clinical and research settings, the benefits of the basic biological description of genetic interactions between BMP signaling and related pathways will be able to be better data-mined for evidence of salient interactions.
Acknowledgments
We appreciate the many participating families who have enabled physicians and researchers to advance our understanding of holoprosencephaly. The authors thank the following groups and individuals: Carter Centers for Brain Research in Holoprosencephaly and Related Malformations; Howard Hughes Medical Institute – National Institutes of Health Research Scholars Program; Settara Chandrasekharappa, Ph.D. (NHGRI Genomics Core, Cancer Genetics Branch) for assistance with BAC selection; James W. Nagle and Debbie Kauffman (NINDS DNA Sequencing Facility) for assistance with DNA sequencing; Adam Woolfe, Ph.D. and Laura Elnitski, Ph.D. (NHGRI, Genome Technology Branch) for assistance with the evaluation of splicing variants; Karin Gaudenz (The Stowers Institute, University of Kansas Medical Center) and Lawrence Marsh (The University of California at Irvine) for earlier TWSG1 studies; Ward Odenwald (NINDS) for assistance with EvoPrinter; and Nan Zhou, Bob Long, and Manu S. Raam (NHGRI, Medical Genetics Branch) for lab preparation.
This research was supported by the Division of Intramural Research, National Human Genome Research Institute, National Institutes of Health, Department of Health and Human Services, USA.
Funding received from the National Institutes of Health (NIH).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Lipinski RJ, Godin EA, O’leary-Moore SK, Parnell SE, Sulik KK. Genesis of teratogen-induced holoprosencephaly in mice. Am. J. Med. Genet. C Semin. Med. Genet. 2010;154C:29–42. doi: 10.1002/ajmg.c.30239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Shiota K, Yamada S. Early pathogenesis of holoprosencephaly. Am. J. Med. Genet. C Semin. Med. Genet. 2010;154C:22–28. doi: 10.1002/ajmg.c.30248. [DOI] [PubMed] [Google Scholar]
- [3].Orioli IM, Castilla EE. Epidemiology of holoprosencephaly: Prevalence and risk factors. Am. J. Med. Genet. C Semin. Med. Genet. 2010;154C:13–21. doi: 10.1002/ajmg.c.30233. [DOI] [PubMed] [Google Scholar]
- [4].Hahn JS, Barnes PD. Neuroimaging advances in holoprosencephaly: Refining the spectrum of the midline malformation. Am. J. Med. Genet. C Semin. Med. Genet. 2010;154C:120–132. doi: 10.1002/ajmg.c.30238. [DOI] [PubMed] [Google Scholar]
- [5].Solomon BD, Mercier S, Vélez JI, Pineda-Alvarez DE, Wyllie A, Zhou N, Dubourg C, David V, Odent S, Roessler E, Muenke M. Analysis of genotype-phenotype correlations in human holoprosencephaly. Am. J. Med. Genet. C Semin. Med. Genet. 2010;154C:133–141. doi: 10.1002/ajmg.c.30240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Levey EB, Stashinko E, Clegg NJ, Delgado MR. Management of children with holoprosencephaly. Am. J. Med. Genet. C Semin. Med. Genet. 2010;154C:183–190. doi: 10.1002/ajmg.c.30254. [DOI] [PubMed] [Google Scholar]
- [7].Solomon BD, Rosenbaum KN, Meck JM, Muenke M. Holoprosencephaly due to numeric chromosome abnormalities. Am. J. Med. Genet. C Semin. Med. Genet. 2010;154C:146–148. doi: 10.1002/ajmg.c.30232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Roessler E, Belloni E, Gaudenz K, Jay P, Berta P, Scherer SW, Tsui LC, Muenke M. Mutations in the human Sonic Hedgehog gene cause holoprosencephaly. Nat. Genet. 1996;14:357–360. doi: 10.1038/ng1196-357. [DOI] [PubMed] [Google Scholar]
- [9].Dubourg C, Lazaro L, Pasquier L, Bendavid C, Blayau M, Le Duff F, Durou MR, Odent S, David V. Molecular screening of SHH, ZIC2, SIX3, and TGIF genes in patients with features of holoprosencephaly spectrum: Mutation review and genotype-phenotype correlations. Hum. Mutat. 2004;24:43–51. doi: 10.1002/humu.20056. [DOI] [PubMed] [Google Scholar]
- [10].Bendavid C, Dubourg C, Gicquel I, Pasquier L, Saugier-Veber P, Durou MR, Jaillard S, Frebourg T, Haddad BR, Henry C, Odent S, David V. Molecular evaluation of foetuses with holoprosencephaly shows high incidence of microdeletions in the HPE genes. Hum. Genet. 2006;119:1–8. doi: 10.1007/s00439-005-0097-6. [DOI] [PubMed] [Google Scholar]
- [11].Rosenfeld JA, Ballif BC, Martin DM, Aylsworth AS, Bejjani BA, Torchia BS, Shaffer LG. Clinical characterization of individuals with deletions of genes in holoprosencephaly pathways by aCGH refines the phenotypic spectrum of HPE. Hum. Genet. 2010 doi: 10.1007/s00439-009-0778-7. epub ahead of print. [DOI] [PubMed] [Google Scholar]
- [12].Miller EA, Rasmussen SA, Siega-Riz AM, Frias JL, Honein MA. National Birth Defects Prevention Study, Risk factors for non-syndromic holoprosencephaly in the National Birth Defects Prevention Study. Am. J. Med. Genet. C Semin. Med. Genet. 2010;154C:62–72. doi: 10.1002/ajmg.c.30244. [DOI] [PubMed] [Google Scholar]
- [13].Weaver DD, Solomon BD, Akin-Samson K, Kelley RI, Muenke M. Cyclopia (synophthalmia) in Smith-Lemli-Opitz Syndrome: First reported case and consideration of mechanism. Am. J. Med. Genet. C Semin. Med. Genet. 2010;154C:142–145. doi: 10.1002/ajmg.c.30241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ming JE, Muenke M. Multiple hits during early embryonic development: digenic diseases and holoprosencephaly. Am. J. Hum. Genet. 2002;71:1017–1032. doi: 10.1086/344412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Solomon BD, Lacbawan F, Jain M, Domené S, Roessler E, Moore C, Dobyns WB, Muenke M. A novel SIX3 mutation segregates with holoprosencephaly in a large family. Am. J. Med. Genet. A. 2009;149A:919–925. doi: 10.1002/ajmg.a.32813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wilson SW, Houart C. Early steps in the development of the forebrain. Dev Cell. 2004;6:167–181. doi: 10.1016/s1534-5807(04)00027-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Liu A, Niswander LA. Bone morphogenetic protein signalling and vertebrate nervous system development. Nat. Rev. Neurosci. 2005;6:945–954. doi: 10.1038/nrn1805. [DOI] [PubMed] [Google Scholar]
- [18].Monuki ES. The morphogen signaling network in forebrain development and holoprosencephaly. J. Neuropathol. Exp. Neurol. 2007;66:566–575. doi: 10.1097/nen.0b013e3180986e1b. [DOI] [PubMed] [Google Scholar]
- [19].Krauss RS. Holoprosencephaly: new models, new insights. Expert Rev Mol Med. 2007;9:1–17. doi: 10.1017/S1462399407000440. [DOI] [PubMed] [Google Scholar]
- [20].Schachter KA, Krauss RS. Murine models of holoprosencephaly. Curr. Top. Dev. Biol. 2008;84:139–170. doi: 10.1016/S0070-2153(08)00603-0. [DOI] [PubMed] [Google Scholar]
- [21].Lupo G, Harris WA, Lewis KE. Mechanisms of ventral patterning in the vertebrate nervous system. Nat. Rev. Neurosci. 2006;7:103–114. doi: 10.1038/nrn1843. [DOI] [PubMed] [Google Scholar]
- [22].Roessler E, Muenke M. The molecular genetics of holoprosencephaly. Am. J. Med. Genet. C Semin. Med. Genet. 2010;154C:52–61. doi: 10.1002/ajmg.c.30236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Fernandes M, Hébert JM. The ups and downs of holoprosencephaly: dorsal versus ventral patterning forces. Clin. Genet. 2008;73:413–423. doi: 10.1111/j.1399-0004.2008.00994.x. [DOI] [PubMed] [Google Scholar]
- [24].Ohkubo Y, Chiang C, Rubenstein JL. Coordinate regulation and synergistic actions of BMP4, SHH and FGF8 in the rostral prosencephalon regulate morphogenesis of the telencephalic and optic vesicles. Neuroscience. 2002;111:1–17. doi: 10.1016/s0306-4522(01)00616-9. [DOI] [PubMed] [Google Scholar]
- [25].Storm EE, Garel S, Borello U, Hebert JM, Martinez SM, McConnell SK, Martin GR, Rubenstein JL. Dose-dependent functions of Fgf8 in regulating telencephalic patterning centers. Development. 2006;133:1831–1844. doi: 10.1242/dev.02324. [DOI] [PubMed] [Google Scholar]
- [26].Arauz RF, Solomon BD, Pineda-Alvarez DE, Parsons JA, Roessler E, Muenke M. A hypomorphic allele in the FGF8 gene contributes to holoprosencephaly and is allelic to gonadotropin-releasing hormone deficiency in humans. Mol. Syndromol. 2010;1:59–66. doi: 10.1159/000302285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chiang C, Litingtung Y, Lee E, Young KE, Corden JL, Westphal H, Beachy PA. Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature. 1996;383:407–413. doi: 10.1038/383407a0. [DOI] [PubMed] [Google Scholar]
- [28].Petryk A, Anderson RM, Jarcho MP, Leaf I, Carlson CS, Klingensmith J, Shawlot W, O’Connor MB. The mammalian twisted gastrulation gene functions in foregut and craniofacial development. Dev. Biol. 2004;267:374–386. doi: 10.1016/j.ydbio.2003.11.015. [DOI] [PubMed] [Google Scholar]
- [29].Graf D, Timmons PM, Hitchins M, Episkopou V, Moore G, Ito T, Fujiyama A, Fisher AG, Merkenschlager M. Evolutionary conservation, developmental expression, and genomic mapping of mammalian Twisted gastrulation. Mamm. Genome. 2001;12:554–560. doi: 10.1007/s0033501-0005-x. [DOI] [PubMed] [Google Scholar]
- [30].Ross JJ, Shimmi O, Vilmos P, Petryk A, Kim H, Gaudenz K, Hermanson S, Ekker SC, O’Connor MB, Marsh JL. Twisted gastrulation is a conserved extracellular BMP antagonist. Nature. 2001;410:479–483. doi: 10.1038/35068578. [DOI] [PubMed] [Google Scholar]
- [31].Wills A, Harland RM, Khokha MK. Twisted gastrulation is required for forebrain specification and cooperates with Chordin to inhibit BMP signaling during X. tropicalis gastrulation. Dev. Biol. 2006;289:166–178. doi: 10.1016/j.ydbio.2005.10.022. [DOI] [PubMed] [Google Scholar]
- [32].Little SC, Mullens MC. Twisted gastrulation promotes BMP signaling in zebrafish dorsal-ventral axial patterning. Development. 2004;131:5825–5835. doi: 10.1242/dev.01464. [DOI] [PubMed] [Google Scholar]
- [33].Oelgeschläger M, Larraín J, Geissert D, De Robertis EM. The evolutionarily conserved BMP-binding protein Twisted gastrulation promotes BMP signalling. Nature. 2000;405:757–763. doi: 10.1038/35015500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Larraín J, Oelgeschläger M, Ketpura NI, Reversade B, Zakin L, De Robertis EM. Proteolytic cleavage of chordin as a switch for the dual activities of twisted gastrulation in BMP signaling. Development. 2001;128:4439–4447. doi: 10.1242/dev.128.22.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Zakin L, De Robertis EM. Extracellular regulation of BMP signaling. Cur Biol. 2010;20:R89–92. doi: 10.1016/j.cub.2009.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].De Robertis EM. Spemann’s organizer and the self-regulation of embryonic fields. Mech. Dev. 2009;126:925–941. doi: 10.1016/j.mod.2009.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Frézal J, Schinzel A. Report of the committee on clinical disorders, chromosome aberrations and uniparental disomy. Cytogenet. Cell Genet. 1991;58:986–1052. [Google Scholar]
- [38].Overhauser J, Mitchell HF, Zackai EH, Tick DB, Rojas K, Muenke M. Physical mapping of the holoprosencephaly critical region in 18p11.3. Am. J. Hum. Genet. 1995;57:1080–1085. [PMC free article] [PubMed] [Google Scholar]
- [39].Gripp KW, Wotton D, Edwards MC, Roessler E, Ades L, Meinecke P, Richieri-Costa A, Zackai EH, Massagué J, Muenke M, Elledge SJ. Mutations in TGIF cause holoprosencephaly and link NODAL signalling to human neural axis determination. Nat. Genet. 2000;25:205–208. doi: 10.1038/76074. [DOI] [PubMed] [Google Scholar]
- [40].Jin JZ, Gu S, McKinney P, Ding J. Expression and functional analysis of Tgif during mouse midline development. Dev. Dyn. 2006;235:547–553. doi: 10.1002/dvdy.20642. [DOI] [PubMed] [Google Scholar]
- [41].Powers SE, Taniguchi K, Yen W, Melhuish TA, Shen J, Walsh CA, Sutherland AE, Wotton D. Tgif1 and Tgif2 regulate Nodal signaling and are required for gastrulation. Development. 2010;137:249–259. doi: 10.1242/dev.040782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].El-Jaick KB, Powers SE, Bartholin L, Myers KR, Hahn J, Orioli IM, Ouspenskaia M, Lacbawan F, Roessler E, Wotton D, Muenke M. Functional analysis of mutations in TGIF associated with holoprosencephaly. Mol. Genet. Metab. 2007;90:97–111. doi: 10.1016/j.ymgme.2006.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Keaton A, Solomon BD, Kauvar EF, El -Jaick KB, Gropman AL, Zafer Y, Meck JM, Bale SJ, Grange DK, Haddad BR, Gowans GC, Clegg NJ, Delgado MR, Hahn JS, Pineda-Alvarez DE, Lacbawan F, Vélez JI, Roessler E, Muenke M. Mutations in TGIF in human holoprosencephaly: correlation between genotype and phenotype. doi: 10.1159/000328203. Unpublished results. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Turleau C. Monosomy 18p. Orphanet J. Rare Dis. 2008;3:4. doi: 10.1186/1750-1172-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Klingensmith J, Matsui M, Yang YP, Anderson RM. Roles of bone morphogenetic protein signaling and its antagonism in holoprosencephaly. Am. J. Med. Genet. C Semin. Med. Genet. 2010;154C:43–51. doi: 10.1002/ajmg.c.30256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].MacKenzie B, Wolff R, Lowe N, Billington CJ, Jr, Peterson A, Schmidt B, Graf D, Mina M, Gopalakrishnan R, Petryk A. Twisted gastrulation limits apoptosis in the distal region of the mandibular arch in mice. Dev. Biol. 2009;328:13–23. doi: 10.1016/j.ydbio.2008.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Melnick M, Petryk A, Abichaker G, Witcher D, Person AD, Jaskoll T. Embryonic salivary gland dysmorphogenesis in Twisted gastrulation deficient mice. Arch. Oral Biol. 2006;51:433–438. doi: 10.1016/j.archoralbio.2005.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Nosaka T, Morita S, Kitamura H, Nakajima H, Shibata F, Morikawa Y, Kataoka Y, Ebihara Y, Kawashima T, Itoh T, Ozaki K, Senba E, Tsuji K, Makishima F, Yoshida N, Kitamura T. Mammalian twisted gastrulation is essential for skeleto-lymphogenesis. Mol. Cell. Biol. 2003;23:2969–2980. doi: 10.1128/MCB.23.8.2969-2980.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Bachiller D, Klingensmith J, Kemp C, Belo JA, Anderson RM, May SR, McMahon JA, McMahon AP, Harland RM, Rossant J, De Robertis EM. The organizer factors Chordin and Noggin are required for mouse forebrain development. Nature. 2000;403:658–661. doi: 10.1038/35001072. [DOI] [PubMed] [Google Scholar]
- [50].Anderson RM, Lawrence AR, Stottmann RW, Bachiller D, Klingensmith J. Chordin and noggin promote organizing centers of forebrain development in the mouse. Development. 2002;129:4975–4987. doi: 10.1242/dev.129.21.4975. [DOI] [PubMed] [Google Scholar]
- [51].Krassikoff N, Sekhon GS. Familial agnathia-holoprosencephaly caused by an inherited unbalanced translocation and not autosomal recessive inheritance. Am J Med Genet. 1989;34:255–257. doi: 10.1002/ajmg.1320340227. [DOI] [PubMed] [Google Scholar]
- [52].Pauli RM, Pettersen JC, Arya S, Gilbert EF. Familial agnathia-holoprosencephaly. Am. J. Med. Genet. 1983;14:677–698. doi: 10.1002/ajmg.1320140411. [DOI] [PubMed] [Google Scholar]
- [53].Erali M, Voelkerding KV, Wittwer CT. High resolution melting applications for clinical laboratory medicine. Exp. Mol. Pathol. 2008;85:50–58. doi: 10.1016/j.yexmp.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Brown SA, Warburton D, Brown LY, Yu CY, Roeder ER, Stengel-Rutkowski S, Hennekam RC, Muenke M. Holoprosencephaly due to mutations in ZIC2, a homologue of Drosophila odd-paired. Nat. Genet. 1998;20:180–183. doi: 10.1038/2484. [DOI] [PubMed] [Google Scholar]
- [55].Wallis DE, Roessler E, Hehr U, Nanni L, Wiltshire T, Richieri-Costa A, Gillessen-Kaesbach G, Zackai EH, Rommens J, Muenke M. Mutations in the homeodomain of the human SIX3 gene cause holoprosencephaly. Nat. Genet. 1999;22:196–198. doi: 10.1038/9718. [DOI] [PubMed] [Google Scholar]
- [56].Roessler E, Du YZ, Mullor JL, Casas E, Allen WP, Gillessen-Kaesbach G, Roeder ER, Ming JE, Ruiz i Altaba A, Muenke M. Loss-of-function mutations in the human GLI2 gene are associated with pituitary anomalies and holoprosencephaly-like features. Proc. Natl. Acad. Sci. USA. 2003;100:13424–13429. doi: 10.1073/pnas.2235734100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Dennler S, André J, Verrecchia F, Mauviel A. Cloning of the human GLI2 Promoter: transcriptional activation by transforming growth factor-beta via SMAD3/beta-catenin cooperation. J. Biol. Chem. 2009;284:31523–31531. doi: 10.1074/jbc.M109.059964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Dutra AS, Mignot E, Puck JM. Gene localization and syntenic mapping by FISH in the dog. Cytogenet. Cell. Genet. 1996;74:113–117. doi: 10.1159/000134395. [DOI] [PubMed] [Google Scholar]
- [59].Vermeesch JR, Melotte C, Froyen G, Van Vooren S, Dutta B, Maas N, Vermeulen S, Menten B, Speleman F, De Moor B, Van Hummelen P, Marynen P, Fryns JP, Devriendt K. Molecular karyotyping: array CGH quality criteria for constitutional genetic diagnosis. J. Histochem. Cytochem. 2005;53:413–422. doi: 10.1369/jhc.4A6436.2005. [DOI] [PubMed] [Google Scholar]
- [60].Reed GH, Wittwer CT. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin. Chem. 2004;50:1748–1754. doi: 10.1373/clinchem.2003.029751. [DOI] [PubMed] [Google Scholar]
- [61].Audrezet MP, Dabricot A, Le Marechal C, Ferec C. Validation of high-resolution DNA melting analysis for mutation scanning of the Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) gene. J. Mol. Diagn. 2008;10:424–434. doi: 10.2353/jmoldx.2008.080056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Lacbawan F, Solomon BD, Roessler E, El-Jaick K, Domené S, Vélez JI, Zhou N, Hadley D, Balog JZ, Long R, Fryer A, Smith W, Omar S, McLean SD, Clarkson K, Lichty A, Clegg NJ, Delgado MR, Levey E, Stashinko E, Potocki L, Vanallen MI, Clayton-Smith J, Donnai D, Bianchi DW, Juliusson PB, Njølstad PR, Brunner HG, Carey JC, Hehr U, Müsebeck J, Wieacker PF, Postra A, Hennekam RC, van den Boogaard MJ, van Haeringen A, Paulussen A, Herbergs J, Schrander-Stumpel CT, Janecke AR, Chitayat D, Hahn J, McDonald-McGinn DM, Zackai EH, Dobyns WB, Muenke M. Clinical spectrum of SIX3-associated mutations in holoprosencephaly: correlation between genotype, phenotype and function. J. Med. Genet. 2009;46:389–398. doi: 10.1136/jmg.2008.063818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Warr N, Powles-Glover N, Chappell A, Robson J, Norris D, Arkell RM. Zic2-associated holoprosencephaly is caused by a transient defect in the organizer region during gastrulation. Hum. Mol. Genet. 2008;17:2986–2996. doi: 10.1093/hmg/ddn197. [DOI] [PubMed] [Google Scholar]
- [64].Zakin L, Reversade B, Kuroda H, Lyons KM, De Robertis EM. Sirenomelia in Bmp7 and Tsg compound mutant mice: requirment for Bmp signaling in the development of ventral posterior mesoderm. Development. 2005;132:2489–2499. doi: 10.1242/dev.01822. [DOI] [PubMed] [Google Scholar]
- [65].Aughton DJ, AlSaadi AA, Transue DJ. Single maxillary central incisor in a girl with del(18p) syndrome. J. Med. Genet. 1991;28:530–532. doi: 10.1136/jmg.28.8.530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Münke M, Page DC, Brown LG, Armson BA, Zackai EH, Mennuti MT, Emanuel BS. Molecular detection of a Yp/18 translocation in a 45,X holoprosencephalic male. Hum. Genet. 1988;80:219–223. doi: 10.1007/BF01790089. [DOI] [PubMed] [Google Scholar]