Abstract
Fibronectin (FN) fragments found in chronic inflammatory diseases, including periodontal disease and arthritis may contribute to tissue destruction in part via induction of matrix metalloproteinases (MMPs). We previously showed that the 120-kDa FN fragment containing the central cell binding domain (120FN), dose-dependently induces MMP-1 (collagenase-1) in human periodontal ligament (PDL) cells, whereas intact FN did not elicit this response. Recently, we found that increase in MMP-1 expression is accompanied by a decreased osteoblastic phenotype in PDL cells. We hypothesized that 120FN inhibits osteoblastic differentiation of PDL cells by inducing MMP-1. The effects of increasing concentrations of the 120FN on MMP-1 expression and on osteoblastic markers was assessed in cultured PDL cells using Western blotting, qRT-PCR, and collagen degradation and alkaline phosphatase (AP) activity assays. The 120FN dose-dependently increased MMP-1 expression and activity concomitant with a decrease in AP activity. The increase in collagenase activity was largely attributed to increased MMP-1 expression. Concurrent with the decrease in AP activity, the 120FN reduced baseline and dexamethasone-induced gene expression of specific osteoblastic markers Runx2 and osteonectin, and diminished mineralized nodule formation. Finally, siRNA inhibition of 120FN-induced MMP-1 reduced collagenase expression and rescued the AP phenotype to baseline levels. These findings suggest that the disease-associated 120FN, in addition to having direct effects on tissue destruction by upregulating MMPs, could contribute to disease progression by impeding osteoblastic differentiation of osteogenic PDL cells, and consequently diminish bone regeneration.
Keywords: 120 kDa central cell binding domain of fibronectin, osteoblastic differentiation, periodontal ligament cells, MMP-1, mineralization
Introduction
Chronic inflammatory diseases such as periodontal disease are initiated by microorganisms, and perpetuated by systemic and local host responses to these agents resulting in the loss of bone. These microorganisms and byproducts of their action initiate an inflammatory process that enhances the expression of proteolytic enzymes, which lead to the dissolution of protein components into fragments. The proteolysis of matrix macromolecules generates bioactive fragments that in turn may accentuate the disease process. Fragments generated from the proteolysis of fibronectin (FN) are key contributors to this process.
FN is a large multifunctional glycoprotein present in the extracellular matrix (ECM) of many tissues including the periodontium. It contains multiple domains that bind collagen, heparin, fibrin and cells [1–4]. The regions between these domains are highly susceptible to proteolysis. Proteolysis, typically present at sites of inflammation, including periodontitis, arthritis, metastatic processes and injury leads to the generation of FN fragments [5–9]. These proteolytic fragments, which contain functional domains, mediate activities that are not induced by intact FN [10–13]. Thus, for example several FN fragments have been implicated in cartilage degradation in arthritis, and synovial fluid containing these fragments stimulates chondrocyte-mediated cartilage matrix degradation, presumably through induction of matrix metalloproteinases (MMPs) [13–15]. IN VITRO, these fragments also induce specific MMPs in synovial fibroblasts [10] fibrocartilaginous cells [14, 16] and periodontal ligament (PDL) cells [11].
Several FN fragments, including a 120-kDa fragment (120FN) containing the central cell-binding domain (CCBD), have been found in association with chronic inflammatory periodontal disease [7, 8, 17, 18]. The 120 FN, which is a byproduct of proteolysis of FN, has been found at concentrations of up to 1 μM in gingival crevicular fluid of patients with periodontitis and in synovial fluid of patients with arthritis [7, 9]. This FN fragment therefore is common to both these chronic inflammatory disorders, and may modulate disease pathogenesis or progression through various pathways including the induction of MMPs, including MMP-1 in PDL cells [11] and synoviocytes [9].
The basis by which the 120FN-induced MMPs contribute to disease progression and bone breakdown are not known, but may include their ability for direct degradation of the collagenous matrices and the facilitation of subsequent dissolution of the mineralized components of bone by osteoclasts [19]. Furthermore, on the basis of findings showing that increased levels of endogenous and exogenous collagenases inhibit osteoblastic differentiation of osteogenic cells including PDL cells [20–24], it is plausible that upregulation of collagenases by 120FN may delay or inhibit osteoblastic differentiation of PDL cells, thereby diminishing the pool of cells available for bone repair and regeneration. Although the 120FN induces the collagenolytic MMP-1 in PDL cells [11] and MMP-1 directly inhibits osteoblast differentiation of PDL cells [22, 24], there is currently no evidence that the 120FN-enhanced expression of endogenous MMP-1 undermines the osteoblastic differentiation of these cells. Therefore, the purpose of this study was to determine the direct effects of 120FN-induced MMP-1 in regulating the osteoblastic phenotype of PDL cells. PDL cells are a heterogenous population of multipotential cells that can undergo osteoblastic differentiation and mineralized nodule formation [25–28] and are essential to bone homeostasis IN VIVO. The findings of this study may provide insights into the mechanisms by which disease-associated FN fragments contribute to the perpetuation of bone loss or inhibition of bone regeneration in periodontitis and other inflammatory diseases.
Materials and Methods
Preparation and purification of the 120 kDa fibronectin fragment (120FN)
Chymotryptic digestion of fibronectin and subsequent purification was undertaken as previously described [29] with some modifications. Briefly human plasma fibronectin (Millipore, Billerica, MA) was digested with a-chymotrypsin (5U enzyme/mg FN, Sigma-Aldrich, St. Louis, MO) for 1 hour and 10 minutes at 37°C. The reaction was stopped by the addition of diphenyl carbomyl chloride (Sigma-Aldrich). The digested FN was subjected to gelatin-sepharose affinity chromatography (Gelatin-Sepharose™ 4B; GE Healthcare Bioscience Corp., Piscataway, NJ) to remove gelatin-binding fragments, and the subsequent flow through was chromatographed on heparin sepharose (Heparin-Sepharose™ 6 Fast Flow; Healthcare Bioscience Corp.) to remove heparin-binding fragments. The next flow through, containing primarily a 120 kDa fragment was pooled, dialyzed against water and concentrated using a 100 kDa molecular weight cut off filter (Amicon Ultra-100 filter device, Millipore, Billerica, MA). At each of these steps, the FN digests were characterized or the purity of the 120FN was analyzed by electrophoresis on 10% SDS PAGE gels followed by Coomassie blue staining. The final purified product was confirmed to be the 120 kDa central cell-binding domain (CCBD) of FN by Western blotting. The concentration of the 120FN was determined by Bradford microassay (Bio-Rad, Hercules, CA) according to manufacturer's instructions.
PDL cell culture
Primary human PDL cells were isolated from teeth extracted for orthodontic reasons or for therapeutic third molar extraction under IRB approval as previously described [11]. Cells were grown in a-MEM media supplemented with 10% fetal bovine serum, 1% penicillin / streptomycin and 1% fungizone. Cells from the third through sixth passage were used for all experiments. Cells were cultured in serum free media (a-MEM supplemented with 0.2% lactalbumin hydrolysate, LAH) and antibiotics to eliminate the complex effects of serum on cell responses. Cells were rinsed again after 6 hours and cultured in serum free medium with or without the 120FN (0.001–0.1μM) for various time points.
For transfection with MMP-1 siRNA, the PDL cells were plated at 30,000/cm2 in a-MEM with 10% FBS and incubated for 24 hours. The cells were transfected with 100 pmole/well of siRNA to MMP-1 or non-sense siRNA (Stealth TM RNAi Negative Control, Invitrogen Corp., Carlsbad, CA) using Lipofectamin2000 and Opti-MEM (Invitrogen). After 24 hours, the cells were washed and serum-free medium (with 0.2% LAH) without or with 120FN (0.1μM) was added. After 3 days of culture the cell-conditioned medium was collected for MMP assays. Cells were counted and lysed in distilled water for AP assay. Total RNA was extracted for quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) analysis for Runx2, osteonectin (ON) and MMP-1.
For mineralization experiments, cells were cultured in FN-depleted low serum (1%) medium containing ascorbic acid and β-glycerophosphate without or with 120FN (0.1μM). The medium was changed every third day. Serum FN was depleted by passing FBS through a heparin sepharose column [30, 31]. The experiments were terminated after 14 days, and the cells fixed for von Kossa staining [32].
FITC-collagen degradation assay
Total collagenase activity in conditioned medium was assessed by an FITC-collagen degradation assay as described previously [24]. A 96-well plate was coated with FITC-collagen (15μg/ml) (Chondrex, WA, USA) overnight at 4°C and washed twice with PBS. Cell conditioned medium (100μl) was added to the wells, and incubated at 37°C for 1 hour. The fluorescence intensity of degraded collagen products was determined with a microplate spectrofluorometer (Spectramax M2, Molecular Devices, Sunnyvale, CA, USA) with excitation at 494 nm and emission at 518 nm. As a reference, 100 μl of blank medium containing 3000 ng of bacterial collagenase was added to one set of wells for complete digestion of FITC-collagen. The data were converted to relative fluorescence units (RFU) of collagenase activity as described by the manufacturer and standardized to the total protein in the medium.
Substrate zymography
Gelatin substrate zymography was used to detect and characterize proteinase activity in PDL cell-conditioned medium. Cell-conditioned medium (20μl), was mixed with 4x sample buffer and electrophoretically resolved in 10% SDS-PAGE gels containing 2 mg/ml of gelatin (E1A grade, Biorad) as described previously [11]. Following electrophoresis, SDS was removed by washing the gels in 2.5% Triton X-100. The gels were placed in incubation buffer at 37°C for 24h, stained with 0.5% Coomassie blue and destained until proteinase bands were clearly visible.
Western immunoblotting
Western blots using standard methods were performed to determine the modulation of MMP-1 in conditioned medium by 120FN and MMP-1 siRNA or to characterize the purity of FN fragments. Equal volumes (20 μl) of conditioned medium were electrophoretically resolved on 10% SDS-PAGE gels, and the proteins transferred to PVDF membranes. Non-specific binding was blocked with 5% dry non-fat milk in tris-buffered saline with 0.1% Tween 20 (TBST) for one hour, and membranes were washed twice with TBST and incubated 3 h with rabbit anti-human MMP-1 or CCBD of FN primary antibodies (Chemicon International Inc.). After further washes with TBST, the membranes were incubated with a 1:1000 dilution of a peroxidase conjugated goat anti-rabbit antibody (Sigma-Aldrich) in TBST for 1 h. The membranes were washed again and the protein bands were visualized using enhanced chemiluminescence (Super Signal West Pico, Pierce, Rockford, IL).
Alkaline phosphatase activity assay
AP activity was assayed in cell lysates by enzymatic conversion of p-nitrophenylphosphate (p-NT) substrate to p-nitrophenol (Sigma-Aldrich) as described previously [32]. The amount of p-nitrophenol produced was measured spectrophotometrically at a wavelength of 410 nm, quantified against a standard curve in nM, and the results standardized by cell number per minute.
Quantitative reverse transcriptase-polymerase chain reaction
MMP-1, Runx2, ON, and GAPDH mRNA levels were assayed by qRT-PCR after 72h of 120FN treatment. A time course (18h, 36h and 72h) quantification for MMP-1 mRNA was also performed. Total RNA was extracted using RNeasy® Mini Kit (Qiagen Corp., Valencia, CA, USA) according to manufacturer’s instructions. Reverse transcription was performed on 50 ng total RNA in the initial mix with M-MLV reverse transcriptase according to the manufacturer's instructions (SuperScript™ III, Invitrogen). All primers and probes (GAPDH cat# Hs 99999905_m1; MMP-1 cat# Hs 00233958_m1, Runx2 cat# Hs 00231692_m1, ON cat# Hs 00277762_m1) were obtained commercially (TaqMan® Gene Expression Assay, Applied Biosystems, Foster City, CA) and RNAs amplified per manufacturer’s instructions (TaqMan® Universal PCR Master Mix, Applied Biosytems). Amplification was done under the following conditions: 50°C, 2 min; 95°C, 10 min; followed by 40 cycles of 94°C, 15 s and 60°C, 1 min. Data were analyzed using the ABI Prism 7500 SDS 1.2 Software (Applied Biosystems).
Von Kossa staining
Cells were fixed in 100% ethanol, rehydrated in a graded ethanol series, washed, incubated in 5% silver nitrate for 1 hour at 37°C and exposed to bright light for 10 minutes. Mineralized nodules were examined under a microscope and quantified by video-densitometry [32]. A total of 6 different fields were counted for each well and mean ± SD of mineralized nodules per well is calculated.
Statistical analysis
The results were expressed as mean (+ SD) fold change in collagenase levels or activity or osteoblast marker gene levels over control baseline levels for a minimum of three experiments, each performed in triplicate. Data was analyzed by a two-way ANOVA followed by post-hoc Fisher’s PLSD test with a level of significance set at p < 0.05. The relationship between the fold change in AP activity and collagenase activity was determined by a Pearson’s correlation analysis.
Results
Purification of 120FN
Assay of the human plasma FN digests with a-chymotrypsin revealed a multitude of protein fragments (Fig. 1A), which when subjected to successive fractionation through gelatin-sepharose columns resulted in partial purification of 120 FN in addition to several other fragments (Fig. 1B). Subsequent elution through a heparin-sepharose column produced relatively purified 120FN and several smaller molecular weight fragments (Fig. 1C). Dialysis of this eluate using a 100 kDa centrifugation filter resulted in a purified 120FN (Fig. 1D), that was identified as the 120 kDa CCBD of FN (Fig. 1E) by Western blot using an antibody specific to this domain.
Figure 1.
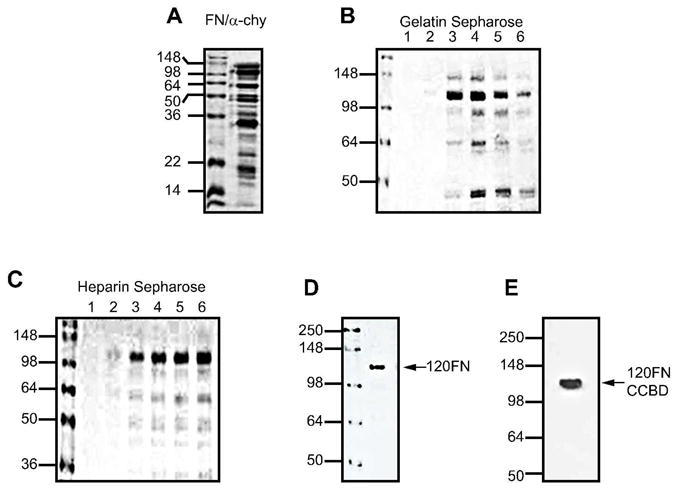
Purification scheme for the 120FN fragment. Human plasma FN was digested with a-chymotrypsin (A) and the 120 FN fragment containing the central cell binding-domain was collected in successive fractions (1 to 6) in flow through from gelatin-Sepharose (B) and heparin-Sepharose (C) column chromatography. The appropriate fractions were further purified by dialysis (D). For steps A to D, the samples were subjected to 10% SDS PAGE gel electrophoresis and stained by Coomassie blue. The identity of the purified fragment was further confirmed by Western blotting using an anti-central cell binding-domain (CCBD) antibody (E).
120FN dose- and time-dependently increases collagenase expression, while concomitantly decreasing AP activity in PDL cells
In order to determine the concentration of 120FN that produces maximal effects on MMP-1 expression and activity in PDL cells for use in subsequent experiments, we first performed 120FN dose-response experiments. The 120FN dose-dependently induced a 53/58-kDa gelatinolytic proteinase (Fig. 2A). This proteinase was identified as MMP-1 by Western blots, which also confirmed its dose-dependent induction by 120FN (Fig. 2B). Further confirmation of 120FN’s dose-dependent modulation of MMP-1 was provided by qRT-PCR, which showed a significant induction of MMP-1 mRNA at the two highest concentrations of the fragment (Fig. 2C). In contrast to the dose-dependent induction of MMP-1 protein and mRNA by 120FN, collagenase and AP activity were significantly modulated only at 0.1 μM concentration of 120FN (Figs. 2D and E). The contrasting findings of enhanced MMP-1 expression but no increase in collagenase activity at 0.01 μM 120FN may result from a concomitant but differential induction of MMP inhibitors at different concentrations of the 120FN and/or due to MMP-1 activation being modulated only at higher concentrations of 120FN [11].
Figure 2.
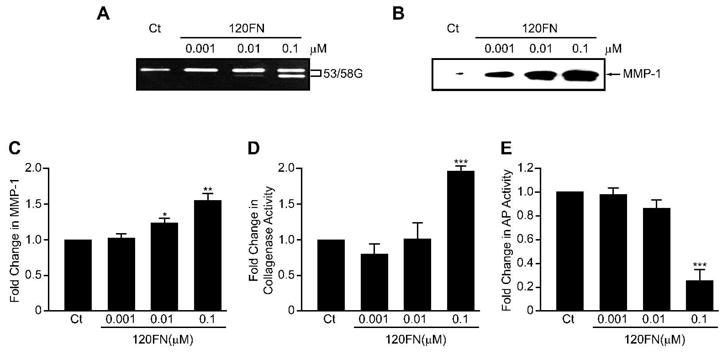
The 120FN dose-dependently induces MMP-1 in PDL cells. Early passage PDL cells were cultured in serum-free medium in the absence or presence of 120FN (0.001–0.1 μM) for 3 days. Substrate zymography on cell-conditioned media demonstrated a dose-dependent increases in a 53/58 gelatinolytic proteinase (53/58G) (A), which was identified as MMP-1 and its dose-dependent induction confirmed on Western blot (B). Extracted RNA was assayed by qRT-PCR for MMP-1 gene expression (C), cell-conditioned media was assayed for collagenase activity by FITC-collagen degradation assay (D), and cell lysates were assayed for AP activity (E). Data are shown as mean (+ SD) fold change relative to control baseline levels (Ct) for three experiments performed in triplicate. (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
Because 0.1 μM 120FN produced significant increases in MMP-1 expression and collagenase activity, we next utilized this concentration of 120FN to determine its time-dependent induction of collagenase and its potential modulation of AP activity in PDL cells. Cells incubated with 0.1 μM 120FN exhibited increasing expression of MMP-1 protein levels (Fig. 3A and B), gene levels (Fig. 3C) and collagenase activity (Fig. 3D) over time. This increase in collagenase activity and MMP-1 mRNA was significant (p < 0.05) at 36 and 72 hours of culture, relative to control untreated cells and to levels at the beginning of the experiment. In contrast, while control cells demonstrated increasing levels of AP activity at 36 and 72 hours of culture, cells cultured with 120FN maintained the AP phenotype at control levels over time (Fig. 3E), This finding suggests that the induction of MMP-1 by 120FN inhibits the time-dependent increases in AP expression in PDL cells. A strong negative correlation was observed between collagenase and AP activities (p < 0.001, r = −0.725). Since treatment with 0.1 μM 120FN produced statistically significant effects on collagenase and AP activities at 72 hours, we used this concentration of the fragment and this time-point for subsequent experiments.
Figure 3.
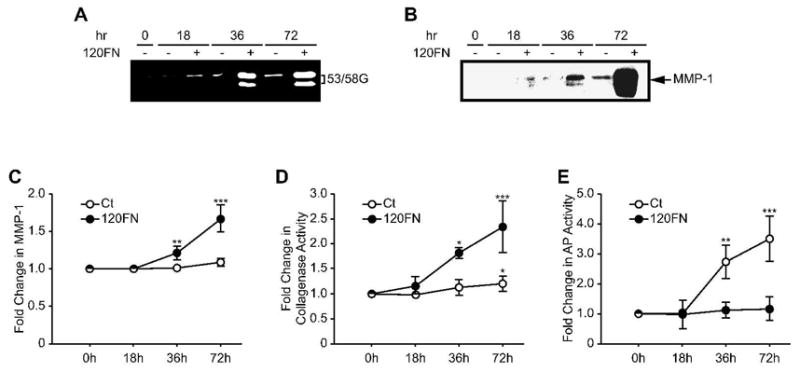
The 120FN induces collagenase activity and concomitantly decreases AP activity in PDL cells. Early passage PDL cells were cultured in serum-free medium in the absence or presence of 120FN (0.1 μM) for various time points. Cell-conditioned medium assayed by substrate zymography demonstrated a time-dependent 120FN-mediated induction of 53/58 gelatinolytic proteinase (53/58G) (A), identified as MMP-1 by Western blotting (B). This was accompanied by a time-dependent induction of MMP-1 gene expression assayed by qRT-PCR (C) and collagenase activity analyzed by FITC-collagen degradation assay on cell-conditioned medium (D). Assays on cell extract revealed that relative to control untreated cells, 120FN inhibits the time-dependent increase in AP activity (E). Data are shown as mean (+ SD) fold change relative to control baseline levels (Ct) for three experiments performed in triplicate. (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
The induction of MMP-1 by 120FN is accompanied by inhibition of the expression of osteoblast-specific markers
To further examine the role of 120FN in modulating osteoblast differentiation in PDL cells, we assessed its effects on osteoblast-specific markers, Runx2 and ON by qRT-PCR as well as AP activity. Cells cultured in pro-osteogenic dexamethasone were used as positive controls. As demonstrated in previous experiments and studies, 0.1 μM 120FN enhanced, while dexamethasone repressed collagenase activity (Fig. 4A) and MMP-1 gene expression (Fig. 4B) [24]. Dexamethasone’s repression of collagenase was accompanied by a significant increase in AP activity (130%), Runx2 (40%) and ON (20%) gene expression relative to control cells (Fig. 4C, D and E). In contrast, the induction of collagenase activity and MMP-1 gene expression by 120FN was accompanied by significant suppression of AP activity (70%), as well as Runx2 (23%), and ON (58%) gene expression relative to untreated cells.
Figure 4.
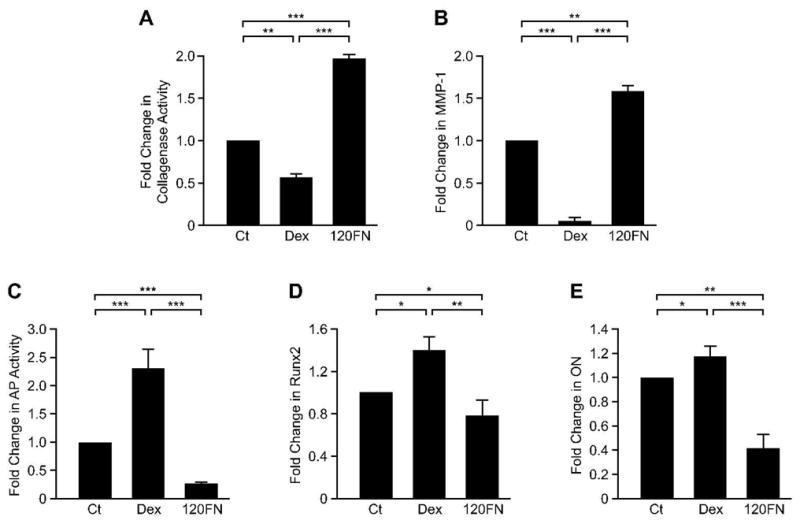
The 120FN inhibits the expression of osteoblastic markers in PDL cells concomitant to induction of MMP-1. PDL cells were cultured in serum-free medium in the absence or presence of 0.1μM 120FN. Cells cultured with the pro-osteogenic dexamethasone (Dex) were used as positive controls. Cell conditioned medium was assayed for collagenase activity (A), cell lysates analyzed for AP activity (C), and total RNA was extracted and subjected to qRT-PCR for MMP-1 (B), Runx2 (D) and ON (E). Data are shown as mean (+ SD) fold change relative to control (Ct) for three independent experiments, each performed in triplicate. (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
120FN-mediated suppression of alkaline phosphatase activity is specifically mediated through MMP-1
Evidence that the 120FN inhibits osteoblastic differentiation by inducing MMP-1 expression was sought by specifically inhibiting MMP-1 with siRNA in the presence and absence of this fragment. Western blotting confirmed successful silencing of MMP-1 with siRNA and its induction by 120FN (Fig. 5A). Furthermore, the induction of MMP-1 by 120FN was largely suppressed in the presence of MMP-1 siRNA. These effects of 120FN and MMP-1 siRNA on collagenase activity mimicked that of their effects on MMP-1 expression (Fig. 5B). The changes in collagenase levels and activity were accompanied by an inverse regulation of AP activity (Fig. 5C). Specifically, 120FN’s induction of MMP-1 was paralleled by a decrease in the AP phenotype of the cells, while the suppression of constitutive MMP-1 expression was accompanied by increased AP activity. Finally, the suppression of 120FN-induced MMP-1 by siRNA rescued the AP phenotype similar to that in control untreated PDL cells. This finding confirms the contribution of 120FN-enhanced MMP-1 in modulating AP in PDL cells.
Figure 5.
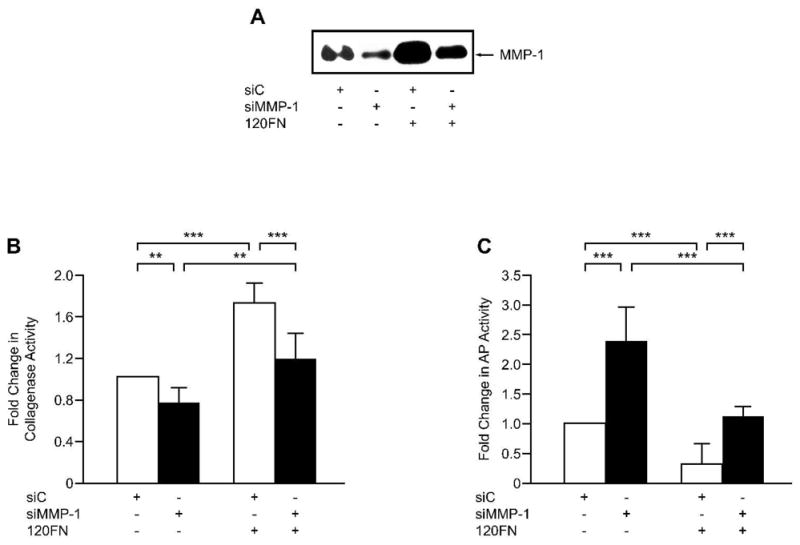
The inhibition of AP phenotype by the 120FN is mediated through its induction of MMP-1. Early passage PDL cells were transfected with 100 pmole of siRNA to MMP-1 (siMMP-1) or scrambled control siRNA (siC) and maintained in serum-free medium for 3days. The cell-conditioned medium was assayed by Western blot for MMP-1 (A) and collagenase activity (B), and the cell extracts assayed for AP activity (C). Data are shown as mean (+ SD) fold change relative to control for three independent experiments performed in triplicate. (*, p < 0.05; **, p < 0.01; ***, p < 0.001).
The 120FN decreases mineralized nodule formation by PDL cells under osteogenic and mineralizing conditions
Finally, we examined the effects of 120FN on the deposition of mineralized matrix by PDL cells in the absence and presence of pro-osteogenic and mineralizing conditions. As expected and as shown previously [32, 33] mineralized nodule formation was significantly enhanced (p < 0.001) in cells treated with β-glycerophosphate and ascorbic acid relative to cells cultured in control media (Fig. 6A and B). β-glycerophosphate and ascorbic acid produced significantly greater mineralized nodule formation even in cells cultured in the presence of 120FN relative to control cells. However, under osteogenic/mineralizing conditions the presence of the 120FN resulted in a significant (p < 0.001) reduction in mineralized nodule formation relative to cultures in which the fragment was not added.
Figure 6.
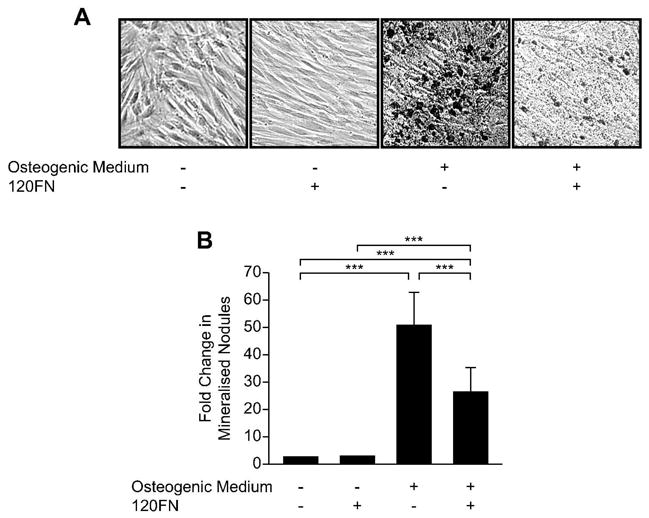
The 120FN inhibits mineralized nodule formation in PDL cells. PDL cells were cultured in osteogenic/mineralizing FN-free serum medium without or with 120FN (0.1 μM) for 14 days. The cells and matrix were subjected to von Kossa staining (A). For quantification, mineralized nodules were counted for a total of six different fields per well (B). Data are shown as mean (+ SD) fold change relative to control baseline levels for three experiments performed in triplicate (***, p < 0.001).
Discussion
We previously demonstrated a strong negative relationship between repression of MMP-1 expression and activity by dexamethasone and induction of several osteoblastic markers including AP, osteocalcin and ON in PDL cells [24, 32]. We further demonstrated an inverse relationship between collagenase and AP activities using both chemical inhibitors of MMPs and MMP-1 siRNA [22, 24]. In long-term cultures, the dexamethasone-mediated decrease in collagenase expression and enhanced osteoblastic markers was accompanied by a significant increase in mineralized nodule formation in dexamethasone treated versus control cells [32]. Conversely, overexpression or addition of exogenous MMP-1 results in an inhibition of most osteoblast markers in osteogenic PDL cells [22]. Together these studies suggest an important contribution of collagenolytic MMPs in regulating osteoblastic differentiation of PDL cells.
The findings of the current study extend these previous observations [22, 24] by showing that osteoblastic differentiation can be inhibited even when endogenous MMP-1 expression is secondarily enhanced by bioactive agents such as 120FN that are commonly found in association with chronic inflammatory conditions. Our findings show a consistent inverse relationship between the disease-associated 120FN-induced MMP-1 levels and osteoblastic markers in PDL cells, and demonstrate that 120FN inhibits osteoblastic differentiation and mineralization of PDL cells specifically by inducing MMP-1 expression in these cells. In addition to these effects of 120FN on osteoblastogenesis, these findings may also offer insights into the basis for previous observations for the repression of osteoblastic differentiation by proinflammatory cytokines such as by IL-1β [21]. In these studies, it was noted that the treatment of PDL cells with IL-1β suppressed the expression of osteoblast markers even under osteogenic conditions. Although this inhibition of osteoblastic differentiation was accompanied by increases in collagenase levels, a direct cause-and-effect relationship between IL-1β-induced collagenase and osteoblast differentiation was not determined. Our studies provide the first evidence of a link between exogenously stimulated MMP-1 expression and repression of osteoblastic phenotype in osteogenic cells.
Although MMPs induced by pro-inflammatory cytokines have been found in association with inflammatory disorders including periodontal disease and arthritis and implicated in tissue destruction, our study provides the first evidence of a different mechanism by which these agents and MMPs may exacerbate tissue degeneration. An inhibition of osteoblastic differentiation by FN fragments and other collagenase inducing agents could eventually lead to a diminished pool of osteoblasts and a subsequent decrease in bone formation and reparative capacity of the tissue. Taken together these data suggest that FN fragments may perpetuate periodontal disease by increasing net bone loss through induction of specific MMPs, which in turn leads to disturbances in the osteogenic capacity of the cells. Thus, the current data suggest a novel mechanism for bone remodeling mediated by disease-associated FN fragments, and outline factors important in maintaining equilibrium between bone formation and bone resorption in preserving healthy periodontal tissues. Identification of such regulatory pathways provides new insights into the pathogenesis of periodontal disease and chronic inflammation, which may point to potential avenues for therapeutic intervention.
The MMP-1 inductive capacity of the 120FN resides in peptide V, an 8 amino acid sequence in region III-14 of FN, which lacks the RGD sequence located in region III-10 of FN [10]. Therefore, it is highly likely that osteoblast differentiation of PDL cells is indirectly regulated by peptide V. Although the basis by which collagenase mediates the inhibition of osteoblast differentiation are not currently known, it may involve one or more possible mechanisms. These include the MMP-mediated changes to the stable collagenous matrix [20, 21, 24], which may modify the cues provided to the cells including those from altered collagen-integrin interactions [34-39]. Additionally, MMPs may modulate changes in the bioavailability of matrix bound bioactive agents through the release of matrix bound growth factors into solution or activate latent bioactive agents [40-42]. Of these mechanisms, there is substantial evidence that the collagen-initiated signals responsible for osteoblast differentiation are mediated via integrins. These studies have implicated α2 and β1-containing integrins in modulating osteoblast differentiation [35, 36, 38, 39]. In context of our findings on the effects of 120FN-induced MMP-1 in modulating osteoblastic differentiation, it is conceivable that degradation of collagen disrupts this collagen-integrin signaling leading to delay or attenuation of osteoblastic differentiation. In contrast, inhibition of MMP-1 may allow faster and more substantial accumulation of collagen [22] leading to earlier and enhanced differentiation of osteogenic cells into osteoblasts via this collagen-integrin cascade.
Acknowledgments
This work was supported by NIH-R01-DE16671 to SK and NIH-R01-DE013725 to YLK.
Footnotes
DISCLOSURES: NONE
References
- 1.Barkalow FJ, Schwarzbauer JE. Localization of the major heparin-binding site in fibronectin. J Biol Chem. 1991;266:7812–7818. [PubMed] [Google Scholar]
- 2.Hahn LH, Yamada KM. Isolation and biological characterization of active fragments of the adhesive glycoprotein fibronectin. Cell. 1979;18:1043–1051. doi: 10.1016/0092-8674(79)90217-4. [DOI] [PubMed] [Google Scholar]
- 3.Hynes RO, Yamada KM. Fibronectins: multifunctional modular glycoproteins. J Cell Biol. 1982;95:369–377. doi: 10.1083/jcb.95.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pierschbacher MD, Ruoslahti E. Variants of the cell recognition site of fibronectin that retain attachment-promoting activity. Proc Natl Acad Sci U S A. 1984;81:5985–5988. doi: 10.1073/pnas.81.19.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clemmensen I, Andersen RB. Different molecular forms of fibronectin in rheumatoid synovial fluid. Arthritis Rheum. 1982;25:25–31. doi: 10.1002/art.1780250104. [DOI] [PubMed] [Google Scholar]
- 6.Griffiths AM, Herbert KE, Perrett D, Scott DL. Fragmented fibronectin and other synovial fluid proteins in chronic arthritis: their relation to immune complexes. Clin Chim Acta. 1989;184:133–146. doi: 10.1016/0009-8981(89)90283-0. [DOI] [PubMed] [Google Scholar]
- 7.Huynh QN, Wang S, Tafolla E, Gansky SA, Kapila S, Armitage GC, Kapila YL. Specific fibronectin fragments as markers of periodontal disease status. J Periodontol. 2002;73:1101–1110. doi: 10.1902/jop.2002.73.10.1101. [DOI] [PubMed] [Google Scholar]
- 8.Talonpoika J, Heino J, Larjava H, Hakkinen L, Paunio K. Gingival crevicular fluid fibronectin degradation in periodontal health and disease. Scand J Dent Res. 1989;97:415–421. doi: 10.1111/j.1600-0722.1989.tb01455.x. [DOI] [PubMed] [Google Scholar]
- 9.Xie DL, Meyers R, Homandberg GA. Fibronectin fragments in osteoarthritic synovial fluid. J Rheumatol. 1992;19:1448–1452. [PubMed] [Google Scholar]
- 10.Huhtala P, Humphries MJ, McCarthy JB, Tremble PM, Werb Z, Damsky CH. Cooperative signaling by alpha 5 beta 1 and alpha 4 beta 1 integrins regulates metalloproteinase gene expression in fibroblasts adhering to fibronectin. J Cell Biol. 1995;129:867–879. doi: 10.1083/jcb.129.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kapila YL, Kapila S, Johnson PW. Fibronectin and fibronectin fragments modulate the expression of proteinases and proteinase inhibitors in human periodontal ligament cells. Matrix Biol. 1996;15:251–261. doi: 10.1016/s0945-053x(96)90116-x. [DOI] [PubMed] [Google Scholar]
- 12.Werb Z, Tremble PM, Behrendtsen O, Crowley E, Damsky CH. Signal transduction through the fibronectin receptor induces collagenase and stromelysin gene expression. J Cell Biol. 1989;109:877–889. doi: 10.1083/jcb.109.2.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie DL, Hui F, Meyers R, Homandberg GA. Cartilage chondrolysis by fibronectin fragments is associated with release of several proteinases: stromelysin plays a major role in chondrolysis. Arch Biochem Biophys. 1994;311:205–212. doi: 10.1006/abbi.1994.1228. [DOI] [PubMed] [Google Scholar]
- 14.Homandberg GA, Meyers R, Xie DL. Fibronectin fragments cause chondrolysis of bovine articular cartilage slices in culture. J Biol Chem. 1992;267:3597–3604. [PubMed] [Google Scholar]
- 15.Homandberg GA, Hui F. Arg-Gly-Asp-Ser peptide analogs suppress cartilage chondrolytic activities of integrin-binding and nonbinding fibronectin fragments. Arch Biochem Biophys. 1994;310:40–48. doi: 10.1006/abbi.1994.1137. [DOI] [PubMed] [Google Scholar]
- 16.Hu B, Kapila YL, Buddhikot M, Shiga M, Kapila S. Coordinate induction of collagenase-1, stromelysin-1 and urokinase plasminogen activator (uPA) by the 120-kDa cell-binding fibronectin fragment in fibrocartilaginous cells: uPA contributes to activation of procollagenase-1. Matrix Biol. 2000;19:657–669. doi: 10.1016/s0945-053x(00)00114-1. [DOI] [PubMed] [Google Scholar]
- 17.Lopatin DE, Caffesse ER, Bye FL, Caffesse RG. Concentrations of fibronectin in the sera and crevicular fluid in various stages of periodontal disease. J Clin Periodontol. 1989;16:359–364. doi: 10.1111/j.1600-051x.1989.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 18.Talonpoika JT, Soderling E, Paunio K. Characterization of fibronectin and fibrin(ogen) fragments in gingival crevicular fluid. Scand J Dent Res. 1993;101:26–32. doi: 10.1111/j.1600-0722.1993.tb01641.x. [DOI] [PubMed] [Google Scholar]
- 19.Chambers TJ, Darby JA, Fuller K. Mammalian collagenase predisposes bone surfaces to osteoclastic resorption. Cell Tissue Res. 1985;241:671–675. doi: 10.1007/BF00214590. [DOI] [PubMed] [Google Scholar]
- 20.Franceschi RT, Iyer BS, Cui Y. Effects of ascorbic acid on collagen matrix formation and osteoblast differentiation in murine MC3T3-E1 cells. J Bone Miner Res. 1994;9:843–854. doi: 10.1002/jbmr.5650090610. [DOI] [PubMed] [Google Scholar]
- 21.Chien HH, Lin WL, Cho MI. Interleukin-1beta-induced release of matrix proteins into culture media causes inhibition of mineralization of nodules formed by periodontal ligament cells in vitro. Calcif Tissue Int. 1999;64:402–413. doi: 10.1007/pl00005822. [DOI] [PubMed] [Google Scholar]
- 22.Hayami T, Kapila YL, Kapila S. MMP-1 (collagenase-1) and MMP-13 (collagenase-3) differentially regulate markers of osteoblastic differentiation in osteogenic cells. Matrix Biol. 2008;27:682–692. doi: 10.1016/j.matbio.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takiguchi T, Kobayashi M, Suzuki R, Yamaguchi A, Isatsu K, Nishihara T, Nagumo M, Hasegawa K. Recombinant human bone morphogenetic protein-2 stimulates osteoblast differentiation and suppresses matrix metalloproteinase-1 production in human bone cells isolated from mandibulae. J Periodontal Res. 1998;33:476–485. doi: 10.1111/j.1600-0765.1998.tb02347.x. [DOI] [PubMed] [Google Scholar]
- 24.Hayami T, Zhang Q, Kapila Y, Kapila S. Dexamethasone's enhancement of osteoblastic markers in human periodontal ligament cells is associated with inhibition of collagenase expression. Bone. 2007;40:93–104. doi: 10.1016/j.bone.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 25.Seo BM, Miura M, Gronthos S, Bartold PM, Batouli S, Brahim J, Young M, Robey PG, Wang CY, Shi S. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet. 2004;364:149–155. 26. doi: 10.1016/S0140-6736(04)16627-0. [DOI] [PubMed] [Google Scholar]
- Chen SC, Marino V, Gronthos S, Bartold PM. Location of putative stem cells in human periodontal ligament. J Periodontal Res. 2006;41:547–553. doi: 10.1111/j.1600-0765.2006.00904.x. [DOI] [PubMed] [Google Scholar]
- 27.Nagatomo K, Komaki M, Sekiya I, Sakaguchi Y, Noguchi K, Oda S, Muneta T, Ishikawa I. Stem cell properties of human periodontal ligament cells. J Periodontal Res. 2006;41:303–310. doi: 10.1111/j.1600-0765.2006.00870.x. [DOI] [PubMed] [Google Scholar]
- 28.Xu J, Wang W, Kapila Y, Lotz J, Kapila S. Multiple differentiation capacity of STRO-1+/CD146+ PDL mesenchymal progenitor cells. Stem Cells Dev. 2009;18:487–496. doi: 10.1089/scd.2008.0113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruoslahti E, Hayman EG, Engvall E, Cothran WC, Butler WT. Alignment of biologically active domains in the fibronectin molecule. J Biol Chem. 1981;256:7277–7281. [PubMed] [Google Scholar]
- 30.Ruoslahti E, Hayman EG, Pierschbacher M, Engvall E. Fibronectin: purification, immunochemical properties, and biological activities. Methods Enzymol. 1982;82(Pt A):803–831. doi: 10.1016/0076-6879(82)82103-4. [DOI] [PubMed] [Google Scholar]
- 31.Underwood PA, Bean PA, Mitchell SM, Whitelock JM. Specific affinity depletion of cell adhesion molecules and growth factors from serum. J Immunol Methods. 2001;247:217–224. doi: 10.1016/s0022-1759(00)00327-6. [DOI] [PubMed] [Google Scholar]
- 32.Shiga M, Kapila YL, Zhang Q, Hayami T, Kapila S. Ascorbic acid induces collagenase-1 in human periodontal ligament cells but not in MC3T3-E1 osteoblast-like cells: potential association between collagenase expression and changes in alkaline phosphatase phenotype. J Bone Miner Res. 2003;18:67–77. doi: 10.1359/jbmr.2003.18.1.67. [DOI] [PubMed] [Google Scholar]
- 33.Aronow MA, Gerstenfeld LC, Owen TA, Tassinari MS, Stein GS, Lian JB. Factors that promote progressive development of the osteoblast phenotype in cultured fetal rat calvaria cells. J Cell Physiol. 1990;143:213–221. doi: 10.1002/jcp.1041430203. [DOI] [PubMed] [Google Scholar]
- 34.Cheng SL, Lai CF, Blystone SD, Avioli LV. Bone mineralization and osteoblast differentiation are negatively modulated by integrin alpha(v)beta3. J Bone Miner Res. 2001;16:277–288. doi: 10.1359/jbmr.2001.16.2.277. [DOI] [PubMed] [Google Scholar]
- 35.Jikko A, Harris SE, Chen D, Mendrick DL, Damsky CH. Collagen integrin receptors regulate early osteoblast differentiation induced by BMP-2. J Bone Miner Res. 1999;14:1075–1083. doi: 10.1359/jbmr.1999.14.7.1075. [DOI] [PubMed] [Google Scholar]
- 36.Mizuno M, Fujisawa R, Kuboki Y. Type I collagen-induced osteoblastic differentiation of bone-marrow cells mediated by collagen-alpha2beta1 integrin interaction. J Cell Physiol. 2000;184:207–213. doi: 10.1002/1097-4652(200008)184:2<207::AID-JCP8>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 37.Schneider GB, Zaharias R, Stanford C. Osteoblast integrin adhesion and signaling regulate mineralization. J Dent Res. 2001;80:1540–1544. doi: 10.1177/00220345010800061201. [DOI] [PubMed] [Google Scholar]
- 38.Xiao G, Wang D, Benson MD, Karsenty G, Franceschi RT. Role of the alpha2-integrin in osteoblast-specific gene expression and activation of the Osf2 transcription factor. J Biol Chem. 1998;273:32988–32994. doi: 10.1074/jbc.273.49.32988. [DOI] [PubMed] [Google Scholar]
- 39.Xiao G, Gopalakrishnan R, Jiang D, Reith E, Benson MD, Franceschi RT. Bone morphogenetic proteins, extracellular matrix, and mitogen-activated protein kinase signaling pathways are required for osteoblast-specific gene expression and differentiation in MC3T3-E1 cells. J Bone Miner Res. 2002;17:101–110. doi: 10.1359/jbmr.2002.17.1.101. [DOI] [PubMed] [Google Scholar]
- 40.Karsdal MA, Larsen L, Engsig MT, Lou H, Ferreras M, Lochter A, Delaisse JM, Foged NT. Matrix metalloproteinase-dependent activation of latent transforming growth factor-beta controls the conversion of osteoblasts into osteocytes by blocking osteoblast apoptosis. J Biol Chem. 2002;277:44061–44067. doi: 10.1074/jbc.M207205200. [DOI] [PubMed] [Google Scholar]
- 41.Lee S, Jilani SM, Nikolova GV, Carpizo D, Iruela-Arispe ML. Processing of VEGF-A by matrix metalloproteinases regulates bioavailability and vascular patterning in tumors. J Cell Biol. 2005;169:681–691. doi: 10.1083/jcb.200409115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakamura M, Miyamoto S, Maeda H, Ishii G, Hasebe T, Chiba T, Asaka M, Ochiai A. Matrix metalloproteinase-7 degrades all insulin-like growth factor binding proteins and facilitates insulin-like growth factor bioavailability. Biochem Biophys Res Commun. 2005;333:1011–1016. doi: 10.1016/j.bbrc.2005.06.010. [DOI] [PubMed] [Google Scholar]


