Abstract
The otic placode, a specialized region of ectoderm, gives rise to components of the inner ear and shares many characteristics with the neural crest, including expression of the key transcription factor Sox10. Here, we show that in avian embryos, a highly conserved cranial neural crest enhancer, Sox10E2, also controls the onset of Sox10 expression in the otic placode. Interestingly, we show that different combinations of paralogous transcription factors (Sox8, Pea3 and cMyb versus Sox9, Ets1 and cMyb) are required to mediate Sox10E2 activity in the ear and neural crest, respectively. Mutating their binding motifs within Sox10E2 greatly reduces enhancer activity in the ear. Moreover, simultaneous knockdown of Sox8, Pea3 and cMyb eliminates not only the enhancer-driven reporter expression, but also the onset of endogenous Sox10 expression in the ear. Rescue experiments confirm that the specific combination of Myb together with Sox8 and Pea3 is responsible for the onset of Sox10 expression in the otic placode, as opposed to Myb plus Sox9 and Ets1 for neural crest Sox10 expression. Whereas SUMOylation of Sox8 is not required for the initial onset of Sox10 expression, it is necessary for later otic vesicle formation. This new role of Sox8, Pea3 and cMyb in controlling Sox10 expression via a common otic/neural crest enhancer suggests an evolutionarily conserved function for the combination of paralogous transcription factors in these tissues of distinct embryological origin.
Keywords: Ear, Cis-regulation, Sox10, Chick
INTRODUCTION
Placodes are defined as thickened regions of cranial ectoderm that delaminate or invaginate into the adjacent mesenchyme (Schlosser, 2008). Placodal cells contribute to the formation of the lens of the eye, sensory structures in the ear, auditory neurons and olfactory epithelium, as well as cranial ganglia. The best-studied cranial placode is the otic placode, which thickens from ectoderm adjacent to rhombomeres 5 and 6 (Hogan and Wright, 1992). In chicken embryos, the otic placode is first morphologically visible at Hamburger-Hamilton (HH) stages HH9-10, then invaginates and separates from the surface ectoderm to form the otic vesicle, and later undergoes complex morphogenetic rearrangements to generate components of the inner ear (Fekete and Wu, 2002), including the semicircular canals, cochlea and endolymphatic duct, as well as supporting cells and hair cells that form the sensory epithelium.
Like neural crest cells, the otic placode expresses members of the SoxE family of transcription factors, Sox8, Sox9 and Sox10. Whereas SoxE factors are known to play crucial roles in many aspects of neural crest development, from migration to differentiation, little is known about the transcriptional regulation of SoxE genes in the otic placode. It has been shown in Xenopus embryos that the SUMOylation state of SoxE proteins plays an important role in distinguishing their functions in neural crest and inner ear development (Taylor and Labonne, 2005). SoxE mutations, such as those observed in Wardenburg syndrome type IV, lead to defects in the ear (Dutton et al., 2009). These anomalies have been attributed to defects in formation of neural crest derivatives contributing to the ear, such as reduction in the number of melanocytes entering the ear (Bondurand et al., 2007) or glial cells of the spiral and vestibular ganglia (gVIII) (Evans and Noden, 2006). Recently, it was noted that Sox8, Sox9 and Sox10 are also expressed in the otic epithelium, suggesting that SoxE genes have a more direct role in the early development of the inner ear (Chiang et al., 2001; Dutton et al., 2001; O'Donnell et al., 2006; Betancur et al., 2010).
Although Sox10 serves as a useful marker for otic development, little is known about what controls its onset of expression in this region. To address this, we have taken advantage of the fact that the Sox10E2 enhancer, previously dissected for its regulatory activity in the cranial neural crest (Betancur et al., 2010), also drives reporter expression in the otic placode of the avian embryo. By probing for the upstream factors that regulate initial Sox10 expression in the otic region, we have uncovered intriguing similarities and differences between Sox10E2 activity in otic placode versus neural crest. Mutational analysis reveals that the binding motifs essential for the onset of Sox10E2-driven reporter expression in cranial neural crest are also required for the initial activity of the enhancer in the otic placode. Whereas the combination of Sox9, Ets1 and cMyb transcription factors activates Sox10E2 enhancer (Betancur et al., 2010), a different combination of paralogous transcriptional activators (Sox8 and Pea3) together with cMyb mediate the enhancing activity of Sox10E2 in the otic placode. Furthermore, we report that post-translational modification of Sox8 by SUMOylation is not required for the initial activation of Sox10 in the otic placode, but is necessary for the proper formation of otic vesicle. Our data suggest that, in the avian embryo, an evolutionarily conserved mechanism involving SoxE and Ets protein activity via a highly conserved amniote enhancer (Sox10E2) is responsible for the differential regulation of Sox10 onset of expression in otic placode and cranial neural crest cells.
MATERIALS AND METHODS
Ex ovo and in ovo electroporations
Chicken embryos were electroporated at HH4 following previously described procedures (Sauka-Spengler and Barembaum, 2008; Betancur et al., 2010). In morpholino-mediated knockdown experiments, unilateral ex ovo electroporations were performed.
Comparative genomic analyses and cloning of putative Sox10 regulatory regions
Highly conserved genomic regions were identified using ECR browser (http://rvista.dcode.org/). Binding motifs were predicted using Jaspar database (http://jaspar.genereg.net/cgi-bin/jaspar_db.pl) and P-Match program from Transfac database (http://www.gene-regulation.com/pub/programs.html) as previously described (Betancur et al., 2010). Putative regulatory regions were amplified with Expand High Fidelity Plus (Roche, Indianapolis, IN, USA) from chicken BAC DNA (BACPAC, Oakland, CA, USA) and cloned into the ptk-EGFP vector (Uchikawa et al., 2003). For details regarding ptk-Cherry and pCI H2B-RFP constructs generation, dissection and mutations of regulatory elements see Betancur et al. (Betancur et al., 2010).
Morpholinos
Morpholino-mediated knockdown experiments were performed by injecting translation-blocking, FITC-labeled morpholino antisense oligonucleotides (1 mM) in one half of the embryo at stages HH4-6 (right of the primitive streak) ex ovo. Morpholinos (MOs) used in this study were FITC tagged at the 3′ end. MOs were obtained from Gene Tools (Philomath, OR, USA) and their sequences are as follows: Control, 5′-ATGGCCTCGGAGCTGGAGAGCCTCA-3′; cMyb, 5′-ATGGCCGCGAGCTCCGCGTGCAGAT-3′; Pea3, 5′-CTGCTGGTCCACGTACCCCTTCATC-3′; Sox8, 5′-CTCCTCGGTCATGTTGAGCATTTGG-3′; Ets2, 5′-GTTTCTGATCGCAAATTCACTCATC-3′; Sox9, 5′-GGGTCTAGGAGATTCATGCGAGAAA-3′. In all cases, the morpholino was co-electroporated with enhancer constructs (2 μg/μl), control pCI H2B-RFP carrier DNA (1 μg/μl) or rescue cDNA constructs (2 μg/μl) for rescue experiments.
Over-expression constructs
Chick cMyb cDNA was amplified as previously described (Betancur et al., 2010). Chick Pea3, Xenopus Sox8 and Xenopus Sox9K61,365R (carrying two mutations for SUMO sites) cDNA were amplified from full length clones by PCR using the following primers (F, forward; R, reverse):
Pea3mut (used for rescue experiments, carrying mutations on the morpholino target site, in bold)
F, 5′-ATTACTCGAGGCAGGATGAAGGATTACGAAGGTTAGCAGGTGCCGTTCACTT;
R, 5′-TATTGATATCTGCAGAATTCGCCCTTGAATT.
Xenopus Sox8
F, 5′-ATTACTCGAGCCACCATGCTGAACATGAGTTCG;
R, 5′-TAATATCGATTTAAGGCCTTGTCAGGGT.
Xenopus Sox9K61,365R
F, 5′-ATTACTCGAGAATCTCTTGGATCCCTT;
R, 5′-TAATATCGATCTAGACTAGGGTCTTGTGAGCT.
Each construct cDNA was cloned into the pCI H2B-RFP overexpression vector using ClaI and XhoI or EcoRV.
Xenopus Sox8K230,346R was generated by a triple fragment fusion PCR.
Fusion 1:
F, 5′-TACAGCTCCTGGGCAACGTG;
K230R_R, 5′-TGGTGAAGATCAGTCCTAGGGGTGGTTGGAGG.
Fusion 2:
K230R_ F, 5′-CCTCCAACCACCCCTAGGACTGATCTTCACCA;
K346R_R, 5′-CTCAGCTGCTCTGTCCTGATGTGGGGTCTCT.
Fusion 3:
K346R_F, 5′-AGAGACCCCACATCAGGACAGAGCAGCTGAG;
R, 5′-GCTTCGGCCAGTAACGTTAG.
The final product was digested with SphI/ClaI and cloned into pCI XSox8 H2B-RFP. This created a Sox8 with two SUMOylation sites mutated: lysine 230 to arginine and lysine 346 to arginine.
Each of the over-expression constructs or combinations were electroporated in embryos at approximately HH4-6.
In situ hybridization
Whole-mount in situ hybridization was performed using a procedure previously described (Wilkinson, 1992). Probes were prepared from chicken EST clones obtained from ARK genomics (Scotland, UK) and Geneservice (Cambridge, UK), with the exception of Sox10 and Sox8 probes, which were prepared from full-length cDNA constructs.
Microscopy and immunohistochemistry
The electroporated embryos were collected at stages HH9-15, fixed in 4% paraformaldehyde overnight and then washed three times in PBS at room temperature. Whole embryos were imaged using a Zeiss axioscope2 Plus fluorescence microscope and then cryosectioned as previously described (Betancur et al., 2010). Sections of embryos treated with morpholinos were subsequently washed and immunostained with anti-Pax2 primary antibody (1:1000; Zymed, CA, USA) followed by Alexa Fluor594-conjugated donkey anti-rabbit secondary antibody (1:1000; Molecular Probes, OR, USA). Sections were coverslipped and imaged using the same imaging procedure described for the whole mounts.
RESULTS
Three core Sox10 enhancers, Sox10L8, Sox10E1 and Sox10E2, have regulatory activity in the forming chick otic placode and vesicle
As a first step in examining how expression of SoxE genes is developmentally regulated in the ear, we focused on dissecting conserved elements in the vicinity of the coding region of chick Sox10. This SoxE family member is expressed throughout development of both the ear and the neural crest (Cheng et al., 2000). Conserved genomic regions in the vicinity of Sox10 were tested for regulatory activity by electroporating the whole epiblast of chicken embryos at stage HH4. Three fragments, denoted Sox10E2, Sox10E1 and Sox10L8, exhibited regulatory activity in the ear and neural crest, as previously described (Betancur et al., 2010) (see Fig. S1 in the supplementary material).
Embryos electroporated with a construct containing the Sox10E2 enhancer (2.1 kb downstream of Sox10 coding sequence) first exhibited strong EGFP reporter expression at stage HH9+, coincident with the onset of endogenous Sox10 expression in the forming otic placode (Fig. 1A,A′). Sox10E2-driven reporter activity persisted through stage 16 (data not shown), whereas the other two active enhancers, Sox10E1 (situated 1.1 kb downstream) and Sox10L8 (6.1 kb upstream) only drove reporter expression later, at the otic vesicle stage (HH12-13) (Fig. 1B-E′). Thus, only Sox10E2 can mediate the early reporter expression that coincides with the first appearance of endogenous Sox10 transcripts in the otic placode. Interestingly, the 264-bp Sox10E2 core regulatory region has also been shown to be essential for the initial activation of Sox10 expression in the cranial neural crest (Betancur et al., 2010).
Fig. 1.
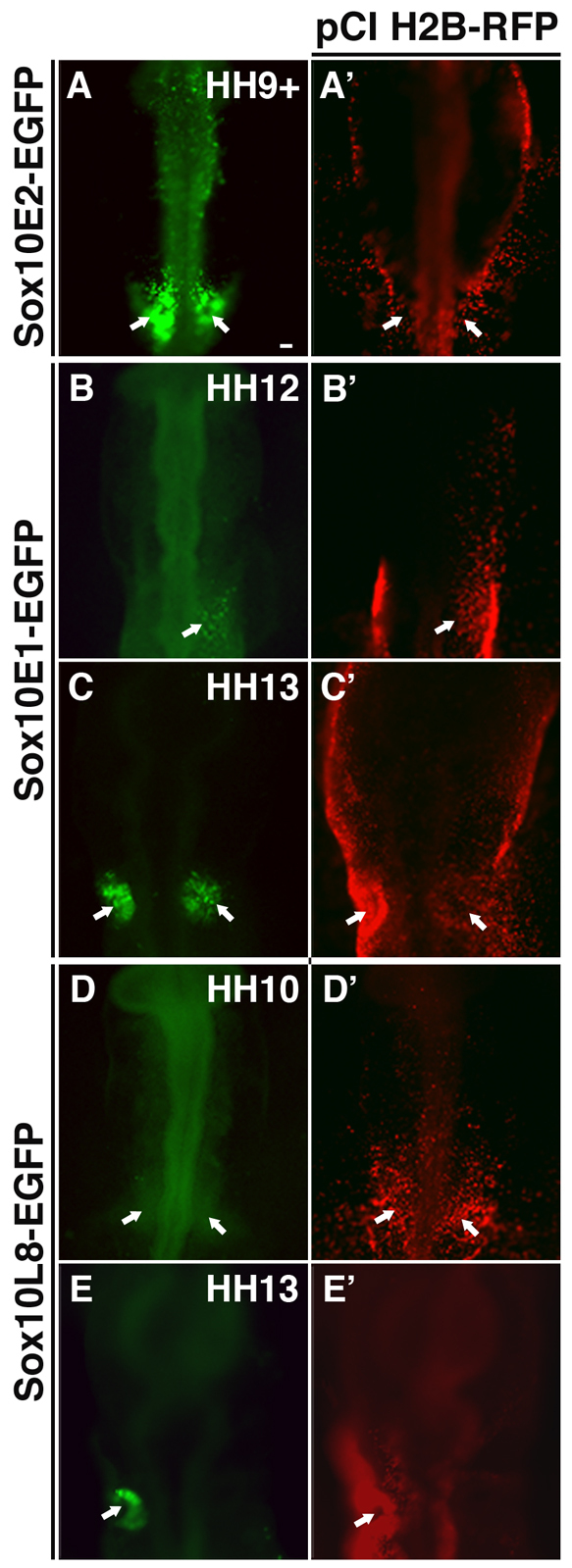
Distinct EGFP reporter activity driven by the Sox10 enhancers Sox10E2, Sox10E1 and Sox10L8 is observed at different stages of otic development. (A-E′) EGFP reporter expression is activated by the Sox10 enhancer Sox10E2 as the otic placode becomes distinguishable at stage HH9+ in chicken embryos (A, arrows). By contrast, EGFP reporter expression under the control of enhancers Sox10E1 (B,C) and Sox10L8 (D,E) only appears later as the otic vesicle begins to invaginate (arrows). Panels A'-E' show the same embryos as in A-E, respectively, and show near ubiquitous expression of the co-electroporated tracer pCI H2B-RFP. Scale bar: 50 μm.
Binding motifs for SoxE, Myb and Ets transcription factors are required for strong enhancing activity of Sox10E2 in the developing otic placode
To identify core elements responsible for otic activity of the Sox10E2 enhancer, we dissected the 264-bp fragment probing an auxiliary, flanking region at the 5′ end, as well as a median 160-bp amniotic evolutionarily conserved region (AECR) (Fig. 2A). Regulatory activity of each subfragment was tested by electroporating versions with selected mutations at stage HH4. Initial dissections showed that a 138-bp fragment containing the 3′ end portion of the 160-bp AECR was sufficient to activate weak, tissue-specific reporter expression (Fig. 2A, E2.1; n=6). The 5′ terminal auxiliary region failed to elicit regulatory activity (Fig. 2A, E2.2; n=4), whereas the 160-bp AECR yielded weak EGFP expression (Fig. 2A, E2.3; n=6). These results suggest that the minimal binding motifs required for tissue-specific activity of the enhancer in the otic placode are contained in the 3′ end terminal half of Sox10E2. For optimal enhancing activity, there appear to be many dispersed functional binding motifs within Sox10E2, some of which are located in the 5′ terminus. This experimental information, together with bioinformatic analysis, was used to identify putative transcription factor binding motifs to predict potential inputs important for otic placode formation.
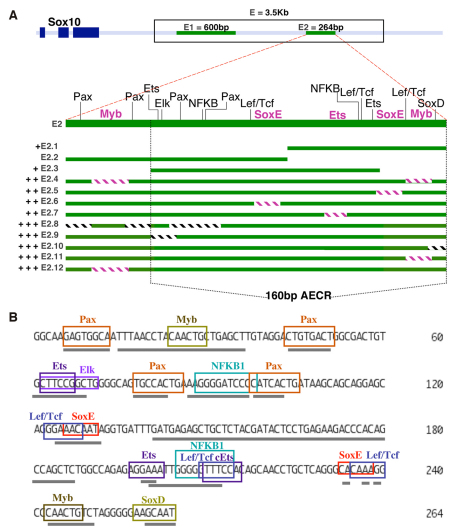
Fig. 2.
Analysis of reporter expression driven by different versions of enhancer Sox10E2 in the otic placode. (A) Diagram showing enhancers Sox10E1 and Sox10E2 located downstream of the dark blue squares representing Sox10 exons. Modified versions of Sox10E2 were created by dissecting the 264-bp fragment into smaller fragments, cutting within or around the 160-bp amniotic evolutionary conserved region (AECR; area between black dotted lines), or by mutating computationally located putative binding motifs (blocks of hatched lines). E2.8-E2.12 represent subfragments of Sox10E2 that showed strong regulatory activity in the otic placode. Sox10 E2.1 and E2.3-E2.7 subfragments displayed weak enhancing activity and E2.2 had no regulatory activity. +, ++ and +++ represent low, medium and high levels of observed reporter expression, respectively. Blocks of lavender hatched lines represent identified functional binding motifs. (B) Binding motifs for transcription factors identified computationally are depicted within the Sox10E2 genomic sequence. Gray underlines mark the specific nucleotide sequences that were mutated to test whether the individual or combination of binding motifs were relevant for the function of the Sox10E2 regulatory region.
Detailed mutational analysis showed that mutating two Myb (Fig. 2A, E2.4; Fig. 2B and Fig. 3B,B′; n=7) and either SoxE (Fig. 2A, E2.5-E2.6; Fig. 2B and Fig. 3C,C′; n=13) or one Ets (Fig. 2A, E2.7; Fig. 2B and Fig. 3D,D′; n=8) binding motifs significantly decreased EGFP reporter activity in the otic placode. By contrast, mutation of other sites, such as Pax, NFKB (Fig. 2A, E2.8; Fig. 2B and Fig. 3E,E′; n=13), Elk/Ets (Fig. 2A, E2.9; Fig. 2B and data not shown; n=7) or SoxD (Fig. 2A, E2.10; Fig. 2B and data not shown; n=10) within the Sox10E2 did not alter levels of EGFP expression, compared with those observed when reporter was driven by the intact enhancer (Fig. 2A, E2; Fig. 2B and Fig. 3A,A′; n=12). Dissection and mutational results demonstrated that a minimal fragment containing a single SoxE and Ets site is sufficient to activate weak reporter expression (Fig. 2A, E2.1 or E2.3). The addition of another SoxE and a single Myb functional site leads to recovery of strong EGFP expression (Fig. 2A, E2.11 and E2.12).
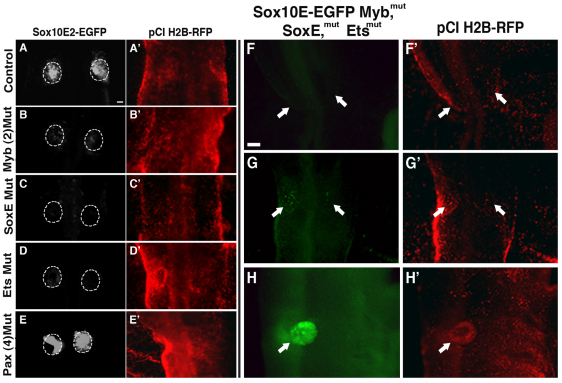
Fig. 3.
SoxE, Ets and Myb binding motifs are required for the early enhancing activity of module Sox10E2 in the otic placode. (A-E) EGFP reporter expression is greatly decreased in the otic placode of HH11-HH12 chicken embryos after mutating both Myb sites (B), and either SoxE (C) or Ets (D) binding motifs within enhancer Sox10E2 compared with control (A). By contrast, no significant change in reporter expression is observed when mutating four Pax binding motifs simultaneously (E). Dotted outline demarcates the otic placode area. Scale bar: 50 μm. (F-H) Furthermore, a 3.5-kb Sox10E genomic fragment bearing mutations in SoxE, Ets and Myb binding motifs within the Sox10E2 enhancer region, but intact Sox10E1 enhancer, completely abolishes EGFP expression at HH9+ when endogenous Sox10 expression is first observed in the forming otic placode (F, arrows). Weak and scattered EGFP activity reappears around HH12-HH13 (G, arrows) when the otic vesicle begins to take shape and is back to normal levels around HH15 (H, arrow). Scale bar: 100 μm. A′-H′ correspond to A-H respectively and show efficient expression of the co-electroporated tracer pCI H2B-RFP.
Interestingly, the simultaneous mutation of all five functional binding motifs (two Myb, one SoxE and two Ets) within the 3.5-kb genomic fragment that also includes a late otic enhancer Sox10E1 (Fig. 2A, E) completely eliminated reporter expression at stage HH9+ (Fig. 3F,F′). The results show that these binding motifs are essential for the early regulation/onset of Sox10 expression in the otic placode. At around stage HH12-13, scattered EGFP signal began to reappear (Fig. 3G,G′) and reporter accumulated by stage HH15 in the already formed otic vesicle (Fig. 3H,H′). This recovery of reporter expression is most likely to be mediated by the Sox10E1 element included in this construct, which appears to play a later maintenance role.
Knockdown of cMyb, Pea3 or Sox8, but not Ets2 or Sox9, reduces EGFP reporter expression by acting on Sox10E2
Armed with the finding that Ets, Myb and SoxE DNA binding sites are required for optimal reporter expression, we next examined candidate transcription factors that might bind to these sites. The expression of several transcription factors, including Sox8 and the Ets family member Pea3, preceded that of Sox10 and persisted during otic placode formation in chicken embryos (Fig. 4A-F′). By contrast, expression of Sox9 followed that of Sox10 in the chick ear (McKeown et al., 2005; Bagheri-Fam et al., 2006; Lunn et al., 2007). We confirmed by in situ hybridization that cMyb is present within the otic territory at stage HH9 and remains so at HH11 (Fig. 4G-I′), at the time when Sox10 transcripts can be detected within the placode. Similarly, Ets2 transcripts were found in the forming otic region, at HH7, just prior to Sox10 onset and persisted at stage HH9+ (data not shown). The expression data suggest cMyb, Pea3, Ets2, Sox8 and/or Sox9 as candidates for controlling initial, or perhaps maintaining, Sox10 expression in the otic placode via the Sox10E2 regulatory element.
Fig. 4.
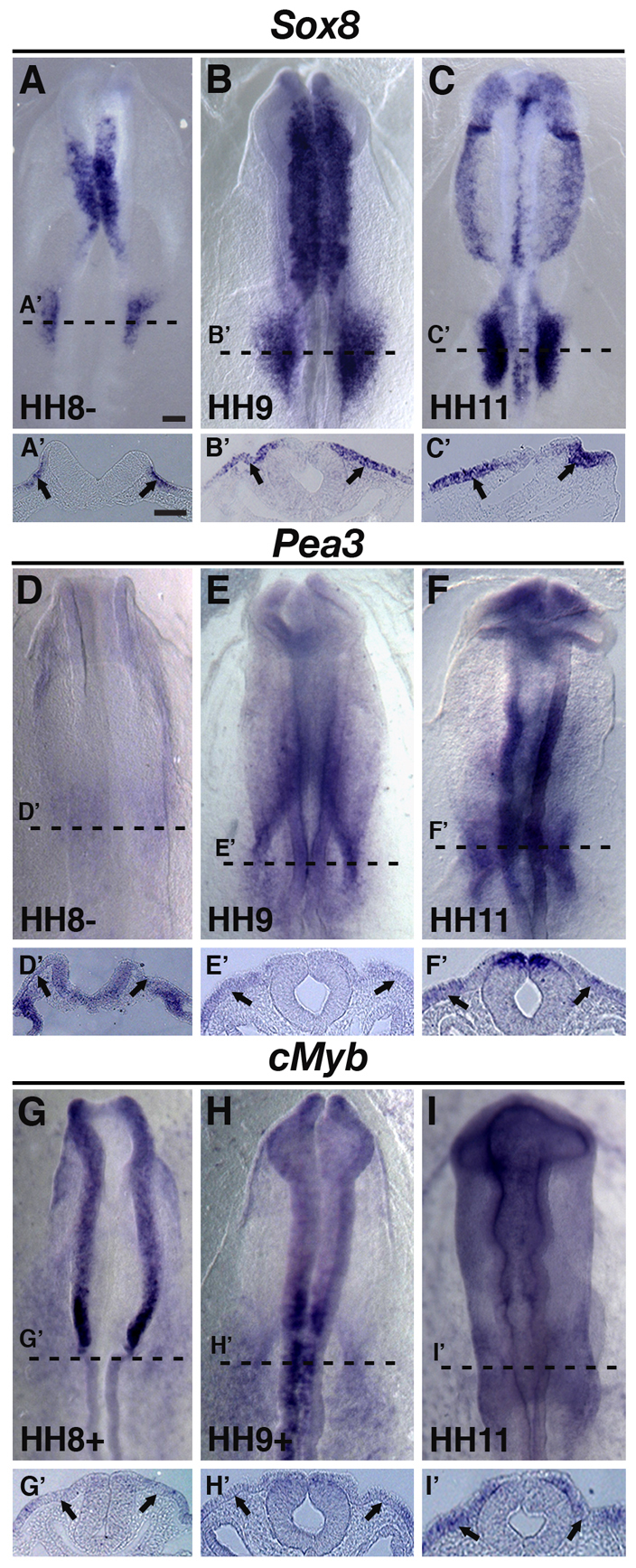
Sox8, Pea3 and cMyb are expressed in the presumptive otic area prior to placode formation and Sox10 expression. (A-F′) By HH8, Sox8 and Pea3 transcripts are observed in the presumptive otic placode region (A,A',D,D' arrows). At HH9+ and HH11, Sox8 and Pea3 continue to be expressed in the developing otic placode (B-C',E-F', arrows). (G-I′) Endogenous cMyb is also observed at HH9-HH11 in the presumptive otic and forming placode region (arrows). A′-I′ show cross-sectional views of A-I, respectively, taken at the level shown by the dashed line. Scale bars: 100 μm for A-I; 50 μm for A'-I'.
To test this, we designed morpholino antisense oligonucleotides to knock down each factor specifically and examined the subsequent effects on Sox10E2 regulatory activity. Because Sox10 is expressed prior to Sox9 in the otic placode, the prediction was that knocking down Sox9 would not affect Sox10 onset of expression in the otic placode. Morpholino-mediated knockdown of proteins, expression of which precedes that of Sox10, such as cMyb (Fig. 5D-D′; n=6), Pea3 (Fig. 5E-E′; n=7) or Sox8 (Fig. 5F-F′; n=8), caused a significant reduction of Sox10E2-driven reporter expression, when compared with embryos treated with control morpholino (Fig. 5A-A′; n=5). Interestingly, a more dramatic effect on reporter expression was observed when cMyb, Pea3 and Sox8 were knocked down simultaneously (Fig. 5G-G′; n=6). By contrast, little or no effect was noted with morpholino against either Sox9 (Fig. 5B-B′; n=6) or Ets2 (Fig. 5C-C′; n=10). These results are consistent with a scenario in which cMyb, Pea3 or/and Sox8 regulate initial Sox10 expression in the otic placode, through their corresponding functional SoxE, Ets and Myb binding motifs situated within the Sox10E2 regulatory region.
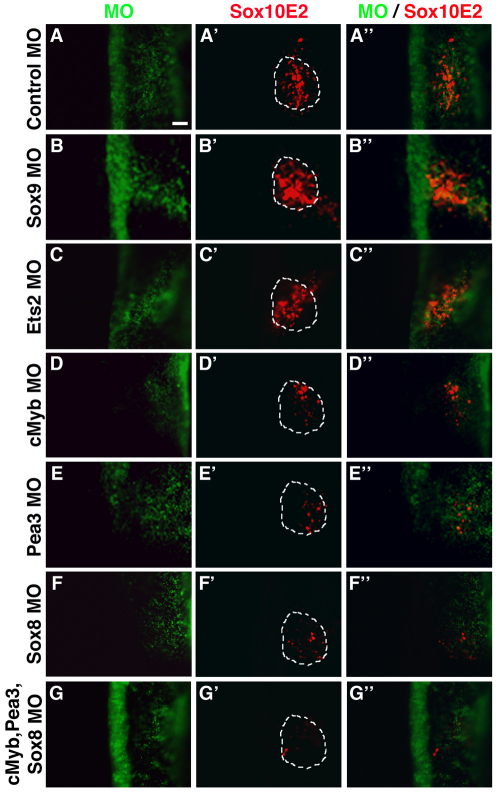
Fig. 5.
cMyb, Pea3 or Sox8 morpholino-mediated knockdown dramatically reduces Sox10E2 regulatory activity in the otic placode. (A-A′) FITC-labeled morpholino control (green) does not affect Sox10E2-driven Cherry reporter expression (red) in the otic placode of HH11-HH12 chicken embryos. (B-C′) Similarly, Sox9 (B-B′) or Ets2 (C-C′) morpholinos do not affect Sox10E2 regulatory activity. (D-F′) By contrast, morpholinos against either cMyb (D-D′), Pea3 (E-E′) or Sox8 (F-F′), dramatically reduce Cherry expression driven by the Sox10E2 enhancer. (G-G′) Cherry expression is almost entirely abolished when cMyb, Pea3 and Sox8 are knocked-down simultaneously. Embryos were electroporated on the right side only. Dotted outline demarcates the otic area. Scale bar: 50 μm.
Endogenous Sox10 expression is reduced in the otic placode when either cMyb, Pea3 or Sox8 are knocked down
Because Sox10E2-driven reporter expression was reduced in the presence of morpholinos against cMyb, Sox8 or Pea3, we investigated next whether these perturbations have an effect on endogenous Sox10 expression. Knocking down cMyb (Fig. 6B,B′; n=6), Pea3 (Fig. 6C,C′; n=6) or Sox8 (Fig. 6D,D′; n=6) significantly reduced Sox10 expression in the otic placode region of embryos analyzed at stage HH11. By contrast, no effect on Sox10 expression in rhombomere (R) 4 was observed in the same embryos when knocking down Pea3 (Fig. 6C′) and only a slight reduction when using morpholino against either cMyb or Sox8 (Fig. 6B′,D′). This is not surprising as both of these genes are expressed by pre-migratory neural crest cells. Furthermore, we demonstrated previously that cMyb is directly involved in the early regulation of Sox10 expression in cranial neural crest cells (Betancur et al., 2010). Embryos electroporated with control morpholino (Fig. 6A-A′; n=8) or Ets2 morpholino (data not shown) showed no change in endogenous Sox10 expression in the otic placode and at the R4 level. To check that loss of Sox10 expression within the otic placode was not an indirect effect due to change in cell fate, we used the preplacodal and otic marker, Pax2, to immunostain embryos treated with morpholino following knockdown of these upstream regulators. We observed no significant change in Pax2 expression after Pea3 morpholino electroporation, for instance, when compared with the contralateral side or in embryos treated with control morpholino (see Fig. S2 in the supplementary material; data not shown).
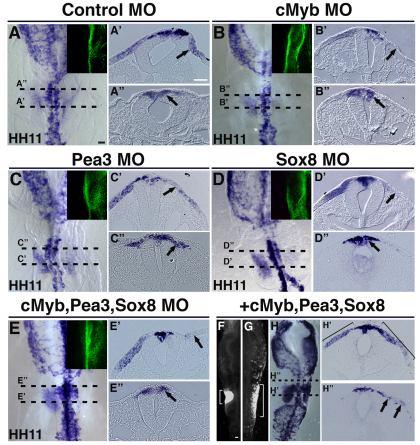
Fig. 6.
cMyb, Pea3 and Sox8 regulate the onset of Sox10 expression in the otic placode. (A-A′) FITC-labeled control morpholino does not affect Sox10 expression in the otic placode at HH11 (arrows). (B-E′) By contrast, cMyb (B-B′, arrows) or Pea3 (C-C′, arrows) morpholino perturbations strongly decrease Sox10 expression at HH11. Similarly, knockdown of Sox8 reduces Sox10 expression at HH11 (D-D′, arrows) in the otic region. Furthermore, depletion of endogenous Sox10 expression in the otic placode is observed when cMyb, Pea3 and Sox8 are knocked down simultaneously (E-E′, arrows). A′-E′ show transverse sections showing the effects of knock-down of cMyb, Pea3, Sox8 or all three factors together on Sox10 expression (arrows) at the level of rhombomere 4. Green panels (A-E, right corner of each image) show the specific FITC-labeled morpholino localization on the right side of the embryo for each of the morpholino treatments. (F-H′) Simultaneous over-expression of cMyb, Pea3 and Sox8 causes an expansion of Sox10E2-EGFP reporter expression (G, bracket) near the otic placode area when compared with embryos electroporated with Sox10E2 EGFP only (F, bracket). Furthermore, an expansion of endogenous Sox10 expression in the otic placode (H,H' bracket) and ectopic expression at rhombomere 3 level (H,H′, arrows) are observed on the cMyb-Pea3-Sox8-treated right side of the embryo when compared with the untreated contralateral left side. Scale bar: 50 μm.
Whereas single knockdown of each candidate factor individually reduced early Sox10 expression, simultaneous inactivation of all three upstream regulators, cMyb, Pea3 and Sox8, nearly completely eliminated endogenous Sox10 expression in the ear at stage HH11 (Fig. 6E,E′; n=5) without affecting gene expression in neural crest cells (Fig. 6E′) and resulted in reduced expression in the otic vesicle at stage 13 (data not shown). These results concur with mutational analysis of Myb, Ets and SoxE binding sites in a larger genomic fragment containing both Sox10E2 and Sox10E1 enhancers. The later recovery of endogenous Sox10 expression is likely to be due to the regulatory activity of other enhancers found in Sox10 genomic locus, including Sox10E1. We often noted a thinning of the ectoderm layer after morpholino knockdowns that affect Sox10 expression. This is probably a secondary effect on the expression of other genes, probably downstream of Sox10, that are required for the normal formation of the otic placode.
Over-expression and rescue using Pea3, cMyb and Sox8
In contrast to the knockdown of these factors, over-expressing cMyb, Pea3 and Sox8 together with Sox10E2 EGFP resulted in expansion of both the reporter signal (Fig. 6G; n=10) and the endogenous Sox10 domain (Fig. 6H,H′; right side) in the vicinity of the ear when compared with `Sox10E2 EGFP only' electroporated embryos (Fig. 6F; n=10) or the control unelectroporated left side (Fig. 6H,H′). Interestingly, ectopic Sox10 expression was also noticed in the ectoderm adjacent to rhombomeres 3 and 4 (Fig. 6H′; right side).
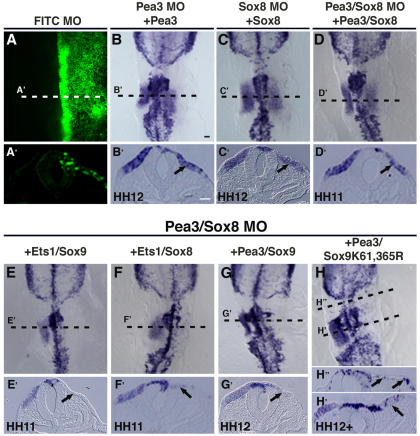
To test further the specificity of action of Pea3 and Sox8 morpholinos and to determine whether this particular set of paralogs is required to activate Sox10 expression in the otic placode region, we performed rescue experiments in which embryos were co-electroporated with morpholinos in combination with one or more rescue construct. Co-electroporation of Pea3 morpholino with Pea3 expression construct rescues the defect in Sox10 expression within the otic region (Fig. 7B,B′; n=5/7). Similarly, Sox8 morpholino-mediated loss of Sox10 was rescued by a Sox8 (Xenopus Sox8) expression construct (Fig. 7C,C′; n=6/7). Furthermore, the double Pea3-Sox8 morpholino knockdown phenotype was rescued when combined with expression of Pea3-Sox8 cDNA constructs (Fig. 7D,D′; n=5/5). By contrast, the paralogous genes Ets1 and Sox9, expressed in the neural crest, failed to rescue Pea3-Sox8 knockdown. For example, neither Ets1 plus Sox9, Ets1 plus Sox8, nor Pea3 plus Sox9 were able to rescue the Sox10 phenotype (Fig. 7E,G′; n=9/9). Previous studies have shown that a SUMO site-mutated form of SoxE (XSox9K61,365R), dramatically increases Sox10 expression in neural crest precursors and also can rescue neural crest defects in Sox10-depleted embryos (Taylor and Labonne, 2005). Given this, we tested whether the expression of this Sox9 SUMO-mutated form (XSox9K61,365R) was capable of rescuing Sox10-depleted expression in the otic placode, caused by Pea3-Sox8 protein knockdown and, hence, replace Sox8 activity. The results showed that, like the wild-type Sox9, the SUMO-mutated form, in combination with Pea3, is unable to recover Sox10 expression in the otic region (Fig. 7H,H′). Interestingly, it does induce ectopic Sox10 expression in the adjacent ectoderm (Fig. 7H,H′), similar to the effect of combined over-expression of cMyb, Pea3 and Sox8 (Fig. 6H,H′). Lastly, to test whether SUMOylation of Sox8 is required for Sox10 transcriptional activation in the otic placode, we generated a Xenopus Sox8 SUMO-mutated version, by replacing lysines 230 and 346 with arginine residues (XSox8K230,345R). We find that the Sox8 SUMO-mutated construct, alone or in combination with Pea3, is capable of rescuing initial Sox10 expression, after Sox8 morpholino- or Sox8-Pea3-morpholino-mediated knockdown. We also observed ectopic Sox10 expression in the adjacent ectoderm (Fig. 8A,B; n=14/14). However, the normal formation of the otic vesicle at later stages was not rescued (Fig. 8C,D; n=4/4).
Fig. 7.
Pea3 and Sox8 specifically regulate Sox10 expression during early otic placode development. (A-D′) Effects on Sox10 expression in the otic placode can be rescued by co-electroporating Pea3, Sox8 or combined morpholinos with corresponding Pea3, Sox8 or expression constructs. Panels A and A′ demonstrate that the FITC morpholino for each of the treatments is localized specifically on the right side of the embryo. (E-H′) The combined expression of either Ets1 and Sox9 (E,E′), Ets1 and Sox8 (F,F′), or Pea3 and Sox9 (G,G′) failed to rescue the observed reduction of Sox10 expression in the otic placode caused by the Pea3 and Sox8 morpholino treatment. Expressing a SUMO site mutated form of SoxE (XSox9K61,365R) together with Pea3 also failed to rescue Sox10 expression in the otic placode (H,H′). However, this combination of factors was able to induce ectopic Sox10 expression in the ectoderm at rhombomeres 3 and 4 (H,H′). Scale bars: 50 μm.
Fig. 8.
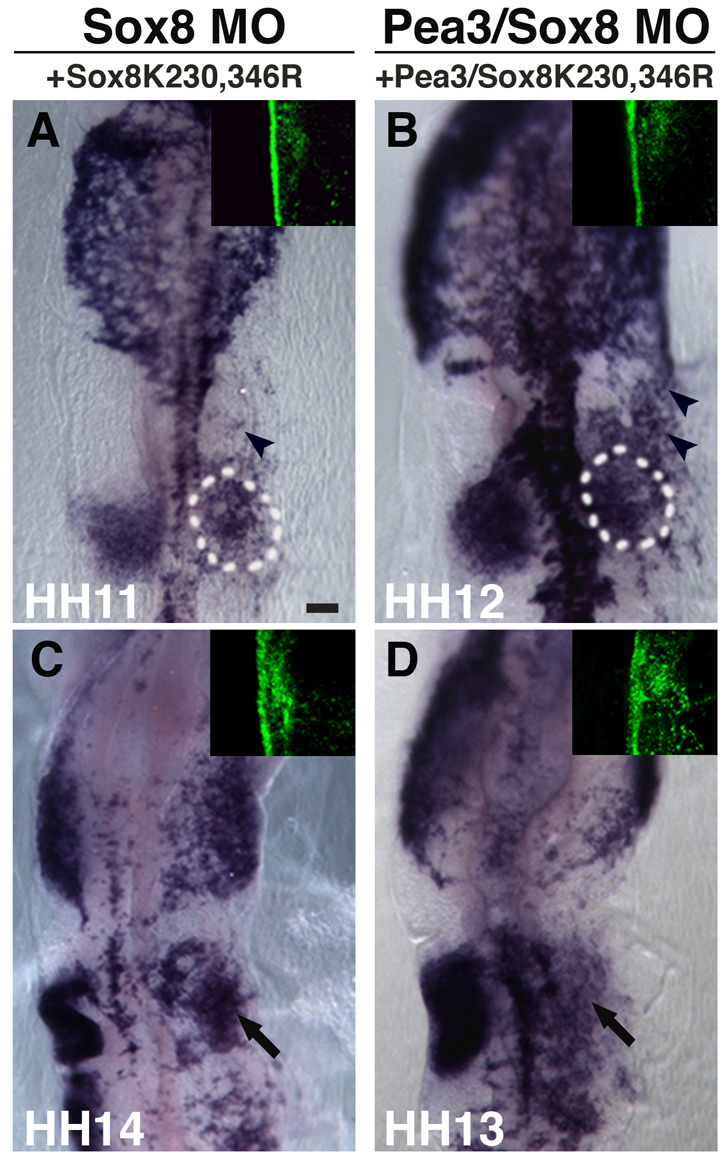
SUMOylation of Sox8 is not necessary for the initial Sox10 expression in the otic region. (A) At stages HH11-HH12, Sox10 expression in the otic placode is recovered when rescuing by co-electroporating Sox8 morpholino and the construct expressing the Sox8 SUMO-mutated form (XSox8K230,346R). (B) A similar result is observed when rescuing Sox8 and Pea3 morpholino-treated embryos with Sox8 SUMO-mutated (XSox8K230,346R) and Pea3 expression constructs combined. (C,D) However, at stages HH13-HH14, Sox8 expression of SUMO mutated form alone (C) or in combination with Pea3 (D) fails to rescue later defect in otic vesicle formation (arrows) caused by Sox8 or Sox8-Pea3 knockdown, respectively. Arrowheads in panels A and B indicate Sox10 ectopic expression. Green panels (right corner of each image) show the specific FITC-labeled morpholino localization on the right side of the embryo for each of the morpholino treatments. Scale bar: 100 μm.
DISCUSSION
A Sox10 enhancer controls gene expression in the forming otic placode
In the chicken embryo, we find that three Sox10 genomic fragments, Sox10L8, Sox10E1 and Sox10E2 have regulatory activity in the developing otic region. In particular, onset of the 264-bp Sox10E2 activity correlates with endogenous Sox10 expression in the forming otic placode, whereas the activities of the other two regulatory regions start some ten hours later. This suggests that the Sox10E2 enhancer is responsible for initiation of Sox10 expression in the ear, whereas Sox10L8 and Sox10E1, and probably other unidentified regulatory regions, are recruited to maintain Sox10 expression during later ear development. Interestingly, we have shown that this same enhancer, Sox10E2, is necessary for initial Sox10 expression in the nascent cranial neural crest (Betancur et al., 2010), albeit with different, paralogous inputs. Thus, the same Sox10 regulatory region functions to mediate Sox10 transcriptional activation in two different cell populations (the cranial crest cells and placode cells), just as each of the populations becomes morphologically recognizable.
cMyb, in combination with Sox8 and Pea3, activates Sox10 gene expression in the otic placode
In the otic placode, our findings suggest that the onset of Sox10 expression via the Sox10E2 element is mediated by binding of the transcription factor cMyb, in combination with Sox8 and Pea3, demonstrating a novel function for all of these regulators. Consistent with this possibility, cMyb, Sox8, Pea3 and Ets2 all are expressed prior to Sox10 in the chick otic placode, contrary to the paralogous genes Ets1 and Sox9, expression of which appears after Sox10. Although Ets2 can bind to and activate Pea3 consensus sites (Buttice and Kurkinen, 1993), our morpholino-mediated inactivation of cMyb, Sox8 and Pea3 but not Ets2 results in reduction of Sox10E2-driven reporter expression in the otic placode. This suggests that cMyb, Sox8 and Pea3 can individually control and converge on the Sox10E2 regulatory region and all are necessary to initiate its reporter expression in the placode cells.
Mutational analysis revealed that SoxE, Ets and either one of two Myb binding motifs, previously found to be necessary for the expression of Sox10E2-driven reporter in neural crest cells, are also essential for the strong activation of Sox10E2 in the otic placode. However, some differences were noted. Individual mutations decreased reporter signal intensity in the ear rather than completely abolishing it, as was the case observed in cranial neural crest. Rather, the cluster of two SoxE, one Ets and either Myb site was sufficient to drive strong tissue-specific, Sox10-like expression in the otic placode. Furthermore, simultaneous mutation of all sites within a larger genomic context delayed reporter onset until stage HH12-13, demonstrating that the synergistic activity of this cluster of binding motifs is necessary for initiation of Sox10E2 enhancer regulatory activity in the placode cells. Within this mutated context, it is perhaps Sox10E1 that regulates gene expression within the invaginating placode at later stages (HH12-13).
Whereas onset of endogenous Sox10 expression in neural crest cells requires each of the Sox10 activators (cMyb, Sox9 and Ets1) individually, elimination of a single factor controlling the expression in the otic placode (cMyb, Sox8 or Pea3) reduces, but fails to abolish, either exogenous Sox10E2 reporter activity or the onset of endogenous Sox10 expression. However, simultaneous depletion of the three factors completely abolishes Sox10 expression during otic placode development (HH9-HH12). Later, Sox10 expression appears to recover, when the otic vesicle begins to take shape. This is most likely to be mediated by different inputs on other Sox10 cis-regulatory regions, including Sox10E1 and Sox10L8 described here.
It is interesting to note that the same binding sites appear to be activated by different transcription factors in a tissue-specific manner. It is known that transcription factor binding sites can often be recognized by multiple transcription factors. For example, Ets2 has been shown to bind in vitro to a Pea3 motif located in the human stromelysin proximal regulatory region (Buttice and Kurkinen, 1993). Ets1 and Pea3 can both recognize the same Ets binding motifs (Fisher et al., 1991). Moreover, in embryonal carcinoma cells, both Ets1 and Pea3 can regulate expression of the TGF-beta receptor gene (TbetaR-II) through the same enhancer (Kopp et al., 2004). Similarly, there is evidence that Sox9 and Sox10 can share the same binding motifs within other Sox10 regulatory regions in mouse (Werner et al., 2007), making it likely that Sox8 utilizes the same site.
SoxE group genes play a conserved role in neural crest cells and otic placode during evolution
Previous studies have shown that Sox8, Sox9 and Sox10 genes, belonging to the SoxE family of transcription factors, are expressed by both neural crest and otic placode cells across different vertebrate taxa. However, the timing of onset of expression for different SoxE paralogs in the two populations varies across species. For instance, whereas in Xenopus neural crest progenitors, Sox8 expression is followed by Sox9 and Sox10, in chicken neural crest, Sox9 is the first SoxE factor to appear closely followed by Sox8 and Sox10 (Hong and Saint-Jeannet, 2005). Conversely, in the otic region of the chicken embryo, Sox9 is expressed only later, in the formed vesicle, whereas the early forming territory is marked first by Sox8 and then by Sox10 expression (McKeown et al., 2005).
In Xenopus, Sox9 has been reported to be involved in the invagination of the otic, based on the observation that Sox9 morphant placodes do not attach to the neural tube, fail to invaginate and undergo apoptosis (Barrionuevo et al., 2008). In zebrafish, sox9b deletion mutants show slightly smaller ears whereas double mutants for sox9a and sox9b lack or have vestigial otic vesicles, owing to a failure in the otic placode induction (Liu et al., 2003; Yan et al., 2005). The differences in Sox9 loss-of-function effects observed in zebrafish versus Xenopus might be due to differences in otic formation between the two species. In zebrafish, rather than invagination, the formation of the otic cup involves cavitation. Similar to Sox9, zebrafish sox10 mutants display subtle vesicle shape defects. At later stages, the size of the vesicle is smaller than normal, partially owing to cell death (Dutton et al., 2009). A role for Sox9 and Sox10 in cell survival and maintenance of multipotency in the otic placode is not surprising given its previously demonstrated similar functions in neural crest cells (Kim et al., 2003; Cheung et al., 2005). Just as in the neural crest, cross-regulatory interactions among SoxE genes appear to play a role during ear development. For instance, de-repression of sox9 is observed in zebrafish sox10 mutants, whereas maintenance of sox10 expression throughout otic epithelia development depends upon SoxE function. Mutating Sox10 and simultaneously knocking down sox9a and sox9b is sufficient to greatly disrupt sox10 expression (Dutton et al., 2009). Nevertheless, these gene perturbations do not completely eliminate sox10 expression, suggesting that other factors, such as Sox8, also might be involved in the regulation of sox10.
Morpholino-mediated knockdown experiments show that Pea3, Sox8 and cMyb are essential for Sox10 expression in the developing ear. As expected, we find that expression of cognate proteins can rescue the morpholino-mediated loss-of-function phenotypes. However, their paralogs were unable to do so, indicating that neural crest-versus placode-specific binding of SoxE or Ets family members to the corresponding motifs within Sox10E2 might be mediated by additional co-factors. Post-translational modifications of SoxE proteins have been shown to affect their association with specific partners in the otic vesicle versus neural crest (Taylor and Labonne, 2005). Accordingly, we tested the ability of SUMO-mutated versions of Sox9 (XSox9K61,365R) and Sox8 (XSox8K230,346R) to rescue of Sox10 expression in the chicken otic placode after Sox8 or Sox8-Pea3 morpholino (MO) knockdown. Neither wild-type Sox9 nor SUMO-mutated Sox9 in combination with Pea3, were able to rescue the knockdown morphant phenotype induced by loss of Pea3 and Sox8 in the otic placode area. This is consistent with the possibility that paralogous genes, deployed in different territories, also acquire different transactivation partners. It remains possible, however, that the lack of Sox10 transcript and proper otic vesicle formation in XSox9K61,365R rescues is an indirect consequence of XSox9K61,365R mis-expression that depletes other early otic placode genes, such as Pax8, as previously reported (Taylor and Labonne, 2005). By contrast, expression of Sox8 completely rescues the Sox8 morpholino-mediated phenotypes whereas the Sox8 SUMO-mutated version (XSox8K230,346R) rescues only initial expression of Sox10 in the otic placode but not the later defect in otic vesicle formation. The latter result is in line with a previous study showing that expression of a SUMO-mutated version of Sox9 leads to failure of formation of the otic vesicle, but can rescue Sox10 gene expression in the neural crest (Taylor and Labonne, 2005). Importantly, as SUMOylation is known to interfere with binding of transcription factors to their direct downstream targets (Girard and Goossens, 2006), the fact that Sox8 SUMOylation is not necessary for initial expression of Sox10 agrees with its role as a direct activator of Sox10.
Our experiments reveal that the simultaneous ectopic mis-expression of cMyb, Pea3 and Sox8 is sufficient to induce both the ectopic expression of EGFP reporter driven by enhancer Sox10E2 and Sox10 mRNA in the ectoderm, adjacent to the hindbrain in a region of ectoderm competent to respond to otic induction (Groves and Bronner-Fraser, 2000). Similarly, expression of SoxE SUMO-mutated protein alone or in combination with Pea3 induced ectopic Sox10 expression in this ectoderm. This might indicate that in a fairly naïve area where neural crest cells or otic placode signature genes are not normally present, either Sox8 or Sox9 plus Pea3 are sufficient to induce Sox10 expression, possibly by binding the corresponding motifs within Sox10E2.
Common neural crest and otic placode evolutionarily conserved enhancers
Despite different paralog usage across species, the shared expression of SoxE genes in both neural crest and otic placodes is a common feature of vertebrates. We find that it is even conserved in basal vertebrates, such as lamprey (Sauka-Spengler and Bronner-Fraser, 2008). Intriguingly, onset of Sox10 expression is regulated by one common neural crest and otic placode enhancer. Similarly, other Sox10 enhancers characterized in mouse display activity in both cell populations (Werner et al., 2007), as does a human Sox9 regulatory element (Bagheri-Fam et al., 2006). Bagheri-Fam and colleagues showed in a mouse transgenic line that a human Sox9 enhancer (E3) is capable of regulating weak initial expression of Sox9 in cranial neural crest cells and otic placode as early as E8.5, when initial endogenous Sox9 expression is first observed in these two distinct regions. Furthermore, they identified potential binding motifs in silico within E3 for factors that play central roles during the development of the cranial neural crest and inner ear. However, the functionality of these candidate binding motifs and their roles remain to be investigated in the neural crest and otic placode populations. In the case of amniote neural crest and otic development, it appears that the Sox10 and Sox9 enhancers have retained their ability to respond in two different cellular environments with little or no rearrangement at the cis-regulatory level.
A possibility is that the neuroepithelial layer, containing neural crest and placode progeny, co-opted similar gene batteries that reflect a common regulatory code (set of transcriptional regulators) and then each population evolved independently as influenced by their environments, which in turn created differential expression of paralog genes. For instance, in the most basal chordate, the cephalochordate amphioxus, most neural plate border specifiers are found in the neural plate border whereas most neural crest specifiers, with the exception of Snail that is found in the neural tube, are expressed in the underlying mesoderm (Meulemans and Bronner-Fraser, 2005; Meulemans and Bronner-Fraser, 2007; Baker, 2008). It is possible that a group of genes forming the `neural crest specifier module' was recruited by some cells in the neuroepithelial domain (future neural crest), whereas SoxE genes were co-opted simultaneously by presumptive neural crest and otic placode cells found in the same domain.
It is important to note that, although the Sox10E2 neural crest and otic placode enhancer is highly conserved across amniotes, it has not yet been found in anamniotes. In fact, such regions might be difficult to find owing to evolutionary distance, such that many non-essential regions might have been added or lost, whereas essential motifs might be rearranged to such an extent that it is difficult to identify conserved regions. Conversely, although organization and type of binding motifs for associated upstream regulators might be conserved, the overall sequence conservation may be poor.
In summary, the present results show that the Sox10E2 regulatory region is a dynamic enhancer, in which similar binding motifs are employed for regulation of Sox10 in two different populations of cells: the cranial neural crest and otic placode. Although SoxE, Ets or both Myb binding motifs enhance the activity of Sox10E2 in placodal cells, each motif is not completely necessary for some basal regulatory activity to occur. Importantly, we find that the combined action of transcription factors cMyb, Sox8 and Pea3 is required to initiate Sox10 gene expression, thereby uncovering a novel regulatory function for these factors in otic placode development. Intriguingly, different family members of the SoxE and Ets family mediate initiation of Sox10 expression in the neural crest by binding to the same motifs within Sox10E2. Thus, the function of a regulatory region has been conserved to activate gene expression in two different cell populations, whereas its capacity to respond to the same set of upstream gene activators has changed. This might be due to a combination of gene duplication/divergence and changes in the cis-regulatory machinery that control the timing and spatial expression of these upstream factors, creating a different activating code for the Sox10 enhancer that is unique to each cell population.
Supplementary Material
Acknowledgments
We thank H. Kondoh for the pTK-EGFP reporter construct; Y. C. Cheng for full length Sox10; V. Lee for full length cSox8; C. LaBonne for XSox9K61R,365R; J.-P. Saint-Jeannet for full length XSox8; M. Dvorak for the cMyb antibody; M. Jones and J. Tan for technical help; M. Barembaum for full length cPea3; and C. Baker for helpful discussions. This project was supported by NIH grants HD037105 and DE16459. Deposited in PMC for release after 12 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.057836/-/DC1
References
- Bagheri-Fam S., Barrionuevo F., Dohrmann U., Gunther T., Schule R., Kemler R., Mallo M., Kanzler B., Scherer G. (2006). Long-range upstream and downstream enhancers control distinct subsets of the complex spatiotemporal Sox9 expression pattern. Dev. Biol. 291, 382-397 [DOI] [PubMed] [Google Scholar]
- Baker C. V. (2008). The evolution and elaboration of vertebrate neural crest cells. Curr. Opin. Genet. Dev. 18, 536-543 [DOI] [PubMed] [Google Scholar]
- Barrionuevo F., Naumann A., Bagheri-Fam S., Speth V., Taketo M. M., Scherer G., Neubuser A. (2008). Sox9 is required for invagination of the otic placode in mice. Dev. Biol. 317, 213-224 [DOI] [PubMed] [Google Scholar]
- Betancur P. A., Sauka-Spengler T., Bronner-Fraser M. (2010). A key regulatory enhancer for cranial neural crest: genomic code for Sox10. Proc. Natl. Acad. Sci. USA 107, 3570-3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondurand N., Dastot-Le Moal F., Stanchina L., Collot N., Baral V., Marlin S., Attie-Bitach T., Giurgea I., Skopinski L., Reardon W., et al. (2007). Deletions at the SOX10 gene locus cause Waardenburg syndrome types 2 and 4. Am. J. Hum. Genet. 81, 1169-1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttice G., Kurkinen M. (1993). A polyomavirus enhancer A-binding protein-3 site and Ets-2 protein have a major role in the 12-O-tetradecanoylphorbol-13-acetate response of the human stromelysin gene. J. Biol. Chem. 268, 7196-7204 [PubMed] [Google Scholar]
- Cheng Y., Cheung M., Abu-Elmagd M. M., Orme A., Scotting P. J. (2000). Chick sox10, a transcription factor expressed in both early neural crest cells and central nervous system. Brain Res. Dev. Brain Res. 121, 233-241 [DOI] [PubMed] [Google Scholar]
- Cheung M., Chaboissier M. C., Mynett A., Hirst E., Schedl A., Briscoe J. (2005). The transcriptional control of trunk neural crest induction, survival, and delamination. Dev. Cell 8, 179-192 [DOI] [PubMed] [Google Scholar]
- Chiang E. F., Pai C. I., Wyatt M., Yan Y. L., Postlethwait J., Chung B. (2001). Two sox9 genes on duplicated zebrafish chromosomes: expression of similar transcription activators in distinct sites. Dev. Biol. 231, 149-163 [DOI] [PubMed] [Google Scholar]
- Dutton K., Abbas L., Spencer J., Brannon C., Mowbray C., Nikaido M., Kelsh R. N., Whitfield T. T. (2009). A zebrafish model for Waardenburg syndrome type IV reveals diverse roles for Sox10 in the otic vesicle. Dis. Model. Mech. 2, 68-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutton K. A., Pauliny A., Lopes S. S., Elworthy S., Carney T. J., Rauch J., Geisler R., Haffter P., Kelsh R. N. (2001). Zebrafish colourless encodes sox10 and specifies non-ectomesenchymal neural crest fates. Development 128, 4113-4125 [DOI] [PubMed] [Google Scholar]
- Evans D. J., Noden D. M. (2006). Spatial relations between avian craniofacial neural crest and paraxial mesoderm cells. Dev. Dyn. 235, 1310-1325 [DOI] [PubMed] [Google Scholar]
- Fekete D. M., Wu D. K. (2002). Revisiting cell fate specification in the inner ear. Curr. Opin. Neurobiol. 12, 35-42 [DOI] [PubMed] [Google Scholar]
- Fisher R. J., Mavrothalassitis G., Kondoh A., Papas T. S. (1991). High-affinity DNA-protein interactions of the cellular ETS1 protein: the determination of the ETS binding motif. Oncogene 6, 2249-2254 [PubMed] [Google Scholar]
- Girard M., Goossens M. (2006). Sumoylation of the SOX10 transcription factor regulates its transcriptional activity. FEBS Lett. 580, 1635-1641 [DOI] [PubMed] [Google Scholar]
- Groves A. K., Bronner-Fraser M. (2000). Competence, specification and commitment in otic placode induction. Development 127, 3489-3499 [DOI] [PubMed] [Google Scholar]
- Hogan B., Wright C. (1992). Developmental biology-the making of the ear. Nature 355, 494-495 [DOI] [PubMed] [Google Scholar]
- Hong C. S., Saint-Jeannet J. P. (2005). Sox proteins and neural crest development. Semin. Cell Dev. Biol. 16, 694-703 [DOI] [PubMed] [Google Scholar]
- Kim J., Lo L., Dormand E., Anderson D. J. (2003). SOX10 maintains multipotency and inhibits neuronal differentiation of neural crest stem cells. Neuron 38, 17-31 [DOI] [PubMed] [Google Scholar]
- Kopp J. L., Wilder P. J., Desler M., Kim J. H., Hou J., Nowling T., Rizzino A. (2004). Unique and selective effects of five Ets family members, Elf3, Ets1, Ets2, PEA3, and PU.1, on the promoter of the type II transforming growth factor-beta receptor gene. J. Biol. Chem. 279, 19407-19420 [DOI] [PubMed] [Google Scholar]
- Liu D., Chu H., Maves L., Yan Y. L., Morcos P. A., Postlethwait J. H., Westerfield M. (2003). Fgf3 and Fgf8 dependent and independent transcription factors are required for otic placode specification. Development 130, 2213-2224 [DOI] [PubMed] [Google Scholar]
- Lunn J. S., Fishwick K. J., Halley P. A., Storey K. G. (2007). A spatial and temporal map of FGF/Erk1/2 activity and response repertoires in the early chick embryo. Dev. Biol. 302, 536-552 [DOI] [PubMed] [Google Scholar]
- McKeown S. J., Lee V. M., Bronner-Fraser M., Newgreen D. F., Farlie P. G. (2005). Sox10 overexpression induces neural crest-like cells from all dorsoventral levels of the neural tube but inhibits differentiation. Dev. Dyn. 233, 430-444 [DOI] [PubMed] [Google Scholar]
- Meulemans D., Bronner-Fraser M. (2005). Central role of gene cooption in neural crest evolution. J. Exp. Zool. B Mol. Dev. Evol. 304, 298-303 [DOI] [PubMed] [Google Scholar]
- Meulemans D., Bronner-Fraser M. (2007). Insights from amphioxus into the evolution of vertebrate cartilage. PLoS ONE 2, e787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell M., Hong C. S., Huang X., Delnicki R. J., Saint-Jeannet J. P. (2006). Functional analysis of Sox8 during neural crest development in Xenopus. Development 133, 3817-3826 [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T., Barembaum M. (2008). Chapter 12 gain- and loss-of-function approaches in the chick embryo. Methods Cell Biol. 87, 237-256 [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T., Bronner-Fraser M. (2008). Insights from a sea lamprey into the evolution of neural crest gene regulatory network. Biol. Bull. 214, 303-314 [DOI] [PubMed] [Google Scholar]
- Schlosser G. (2008). Do vertebrate neural crest and cranial placodes have a common evolutionary origin? BioEssays 30, 659-672 [DOI] [PubMed] [Google Scholar]
- Taylor K. M., Labonne C. (2005). SoxE factors function equivalently during neural crest and inner ear development and their activity is regulated by SUMOylation. Dev. Cell 9, 593-603 [DOI] [PubMed] [Google Scholar]
- Uchikawa M., Ishida Y., Takemoto T., Kamachi Y., Kondoh H. (2003). Functional analysis of chicken Sox2 enhancers highlights an array of diverse regulatory elements that are conserved in mammals. Dev. Cell 4, 509-519 [DOI] [PubMed] [Google Scholar]
- Werner T., Hammer A., Wahlbuhl M., Bosl M. R., Wegner M. (2007). Multiple conserved regulatory elements with overlapping functions determine Sox10 expression in mouse embryogenesis. Nucleic Acids Res. 35, 6526-6538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson D. G. (1992). Whole mount in situ hybridisation of vertebrate embryos. In In situ Hybridization: A Practical Approach (ed. Wilkinson D. G.), pp. 75-83 Oxford: IRL Press; [Google Scholar]
- Yan Y. L., Willoughby J., Liu D., Crump J. G., Wilson C., Miller C. T., Singer A., Kimmel C., Westerfield M., Postlethwait J. H. (2005). A pair of Sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development 132, 1069-1083 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.