Abstract
The maintenance of organs and their regeneration in case of injury are crucial to the survival of all animals. High rates of tissue turnover and nearly unlimited regenerative capabilities make planarian flatworms an ideal system with which to investigate these important processes, yet little is known about the cell biology and anatomy of their organs. Here we focus on the planarian excretory system, which consists of internal protonephridial tubules. We find that these assemble into complex branching patterns with a stereotyped succession of cell types along their length. Organ regeneration is likely to originate from a precursor structure arising in the blastema, which undergoes extensive branching morphogenesis. In an RNAi screen of signaling molecules, we identified an EGF receptor (Smed-EGFR-5) as a crucial regulator of branching morphogenesis and maintenance. Overall, our characterization of the planarian protonephridial system establishes a new paradigm for regenerative organogenesis and provides a platform for exploring its functional and evolutionary homologies with vertebrate excretory systems.
Keywords: Planaria, Protonephridia, Branching morphogenesis
INTRODUCTION
Planarian flatworms have astonishing regenerative abilities (Reddien and Sánchez Alvarado, 2004). Arbitrary tissue fragments originating from almost any body plan position can regenerate into complete and perfectly proportioned animals. This ability is even more fascinating in face of the anatomical complexity of planarians. As members of the Lophotrochozoa, they contain a set of organ systems typically associated with higher animals, including a central nervous system (CNS; bi-lobed brain and ventral nerve cords), a muscle layer surrounding the body wall, a highly branched gut cavity, an excretory system (protonephridia), and complex arrays of sensory systems (e.g. chemo-, rheo- and photoreceptors). In order to restore the anatomical complexity of newly formed tissues, the regeneration of a complete animal from a random tissue fragment necessitates organogenesis on a massive scale. Regenerative organogenesis shares a number of problems with embryonic organogenesis (e.g. cell differentiation and morphogenesis), but raises further intriguing questions. Does organogenesis in regenerated tissues proceed de novo or by templated biogenesis from organ remnants? How is the regenerative response tuned to replace exactly the missing organ mass? How are functional and morphological integration between regenerated and pre-existing organ fragments achieved? Similar questions pertain to the limited, but medically important, regenerative abilities of vertebrate organs (e.g. the liver) (Pahlavan et al., 2006) and are generally not well understood.
In terms of organ regeneration, the planarian CNS has so far received the most attention (Cebria, 2007; Agata and Umesono, 2008). However, the multitude of cell types indicated by the rich and varied gene expression patterns and cell morphologies described to date make a mechanistic understanding of planarian brain regeneration a daunting endeavor (Collins et al., 2010; Nishimura et al., 2010; Nishimura et al., 2008; Nishimura et al., 2007; Cebria et al., 2002; Umesono et al., 1999). In search of a simpler structure to develop as a regenerative organogenesis model, we decided on the planarian excretory system, which consists of epithelial tubules that appear to end blindly in the mesenchyme. This feature defines the planarian excretory system as protonephridial, in contrast to metanephridial systems such as the vertebrate nephron, in which one terminus is located in an extracellular fluid compartment (Wilson and Webster, 1974). Protonephridia are found throughout the animal kingdom. Their evolutionary relationship with metanephridial systems such as the mammalian kidney remains a subject of intense debate and considerable interest due in great part to the limited suitability of traditional invertebrate model systems for studying kidney pathologies (Hyman, 1951; Wilson and Webster, 1974; Ruppert, 1994).
However, the anatomy of planarian protonephridia is not well understood. According to Hyman (Hyman, 1951), who remains the most comprehensive author on invertebrate anatomy, the planarian excretory system consists of anastomosing `main tubules' along the lateral body margins and their ciliated side branches. The lumen of the tubules is thought to be continuous with the outside via dorsally located nephridiopores. However, substantial disagreements in the underlying light microscopic observations from the late 19th and early 20th century reveal considerable uncertainty regarding this view (Chichkoff, 1892; Wilhelmi, 1906). On the ultrastructural level, electron microscopy studies described protonephridia as having the following components: (1) cylindrical cells located at the tip of the ciliated side branches, with narrowly apposed strands of cytoplasm forming a fenestrated barrel around a central bundle of cilia, the flickering movements of which gave rise to the term `flame cell' (McKanna, 1968a; Ishii, 1980a); (2) an initial ciliated tubule segment connected to the lumen of the flame cell barrel and purportedly composed of squamous epithelial cells; and (3) non-ciliated `main tubules' composed of a cuboidal epithelium (Pedersen, 1961; McKanna, 1968b; Ishii, 1980b).
Functional studies on protonephridia in planarians or other invertebrates are extremely scarce, but it is generally assumed that the concerted beating of the flame cell cilia bundle creates a pressure gradient to force tissue fluid across the fenestrations into the lumen of the tubule, where the lining epithelial cells modify the ultrafiltrate by absorption and secretion during its proximodistal passage and eventual release to the outside (Wilson and Webster, 1974). Likewise, knowledge regarding the molecular or functional identity of protonephridial cell types remains largely elusive (Finken-Eigen and Kunz, 1997; Pedersen, 1961; Skelly and Shoemaker, 2001).
We report here a systematic analysis of protonephridial structure and function with modern molecular biology tools. Our visualization of protonephridial architecture in planarians and the concomitant identification of specific markers for flame cells, proximal and distal tubule cells reveal a complex, branched epithelial organ consisting of multiple cell types. We found that protonephridia regenerate in a stereotypic sequence of events and we identified EGF signaling as a crucial regulator of protonephridial branching morphogenesis.
MATERIALS AND METHODS
Planarian maintenance
The CIW4 clonal line of Schmidtea mediterranea was maintained as described (Cebria and Newmark, 2005). One-week starved animals were used for all experiments.
Gene identification and cloning
All genes were cloned from an 8-day regeneration timecourse cDNA library prepared as described previously (Gurley et al., 2008). EGFR-5 was identified by performing BLAST analyses of the planarian genome against a panel of vertebrate and invertebrate EGFR sequences followed by reverse BLAST of the resulting hits against the human and Drosophila melanogaster genomes to ensure EGFR homology. The following primers (5′ to 3′) were used: DNAH-β3f, TAGCTGACCAAGAAGAAGAAGTGG; DNAH-β3r, CACAGACTTTAATGGA TCGACACC; CAVII-1f, TTATTTCTTGTCTCATCTCTTGATCTG; CAVII-1r, CAGGCACATGAAAATTGCAC; inx10f, ATGGTTCTTTCG GAATTCATAG; inx10r, AAATAAAATCATCTTTCAGTGGTAA AGTGGA; EGFR-5f-1, AGTGTGAACAACGATTAGGATG; and EGFR-5r-1, TCAGCAGGTTTCTCACATAC.
The 3′-end of the EGFR-5 sequence, which was exclusively used for the sequence analysis purposes of Fig. S2 in the supplementary material, was cloned with EGFR-5f-2 (TCTTTTACGGAATTGAG) and a poly(T) reverse primer.
In situ hybridization and immunohistochemistry
Whole-mount and fluorescent in situ hybridizations were performed as previously described (Pearson et al., 2009). Following fluorescence or NBT/BCIP development, animals were incubated with anti-α-Tubulin antibody (1:300, NeoMarkers) or anti-acetylated-Tubulin antibody (1:500, Sigma) to detect ciliated sections of protonephridia. Primary antibodies were detected with Alexa Fluor-labeled anti-mouse secondary antibodies (1:500, Invitrogen). For documenting NBT/BCIP-developed whole-mount in situ specimens, animals were mounted in 80% glycerol and photographed using a Zeiss SteREO Lumar.V12 equipped with an AxioCam HRc camera. Whole-mount specimens stained with fluorescent markers were mounted in 2:1 benzyl benzoate:benzyl alcohol after dehydration in methanol and imaged on a Zeiss LSM510 live laser-scanning microscope. For sectioning, fluorescently stained whole-mount animals were dehydrated in a graded series of ethanol, incubated for ∼2 hours in 1:1 ethanol:Immuno-bed (Polysciences) and subsequently immersed in 100% Immuno-bed supplemented with catalyst according to the manufacturer's recommendations. Sections (10 μm) were collected on a Leica microtome equipped with a glass knife. Sections were mounted in Fluoromount-G (SouthernBiotech) and photographed using the Zeiss LSM510.
Histology
Specimens were prepared as follows. (1) Animals were fixed overnight at 4°C in 2.5% glutaraldehyde in 0.1 M sodium cacodylate, 1 mM CaCl2. (2) Animals were washed in wash buffer comprising 0.1 M sodium cacodylate supplemented with 1 mM CaCl2 and 1% sucrose (w/v) for 1 hour at room temperature (3-4 exchanges) and in distilled water for 1 hour at room temperature (3-4 exchanges). (3) Specimens were dehydrated in acetone at 30% (20 minutes), 50% (20 minutes), 70% (overnight), 90% (20 minutes, twice), and 100% (20 minutes, three times). (4) Specimens were embedded in epon-araldite (30% resin/acetone for 5 hours, 70% resin/acetone for 6 hours, 90% resin/acetone overnight, and fresh 100% resin for 8 hours, curing at 60°C for 2 days). (5) Thin sections (1 μm) were collected using an Ultracut UCT microtome (Leica), stained with Toluidine Blue, mounted in Cytoseal XYL (Richard-Allan Scientific), and photographed using a Zeiss Axiovert microscope. Quantifications were carried out independently by two observers.
Electron microscopy
Specimens were prepared using high-pressure freezing/freeze substitution as previously described (Pellettieri et al., 2010). Ultrathin (50 nm) sections were collected using an Ultracut UCT microtome. Transmission electron microscopy specimens were stained with 2.5% uranyl acetate for 4 minutes prior to imaging on a Hitachi H-7100 electron microscope equipped with a Gatan Orius CCD camera.
RNA interference (RNAi)
RNAi feedings were performed as described previously (Gurley et al., 2008; Rink et al., 2009). Six feedings at 2- to 3-day intervals were used in the unsuccessful attempt to elicit DNAH-β3(RNAi) and CAVII-1(RNAi) phenotypes. For inx10(RNAi) and EGFR-5(RNAi) experiments, animals were fed three times at 2- to 3-day intervals. For regeneration time series experiments, animals were amputated 3 days after the last feeding. For Fig. 5C and Fig. S5D,E in the supplementary material, RNAi-fed animals were additionally injected with EGFR-5 double-stranded (ds) RNA at 200 ng/μl 3 days after amputation. dsRNA was prepared using the MEGAscript RNAi kit (Ambion).
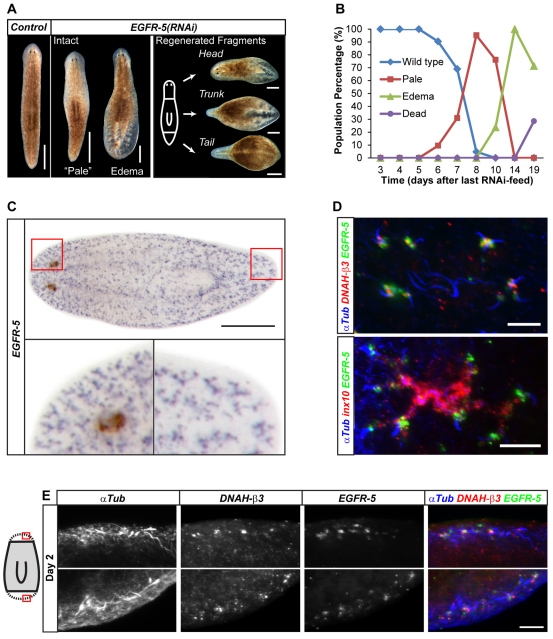
Fig. 5.
EGFR-5 is required for protonephridia function and is expressed in flame cells. (A) Gross morphological consequences of EGFR-5 knockdown in uncut planaria (left, 14 days after last feed) and regenerated fragments (right, 14 days after amputation). (B) Temporal succession of indicated phenotypes in a cohort of EGFR-5(RNAi)-fed animals (n=42). (C) Whole-mount in situ hybridization showing the expression pattern of EGFR-5 (NBT/BCIP development). The boxed regions are magnified beneath. (D) Fluorescent overlay of the EGFR-5 in situ pattern (green) with that of the indicated proximal markers. Images are maximum projections of confocal z-sections. (E) High-magnification views of EGFR-5 expression together with the indicated markers in early head (top row) and tail (bottom row) blastemas. Images are maximum projections of confocal z-sections. Scale bars: 500 μm in C and in A, intact; 300 μm in A, fragments; 50 μm in D; 20 μm in E.
RESULTS
Anatomy and ultrastructure of the Schmidtea mediterranea protonephridial system
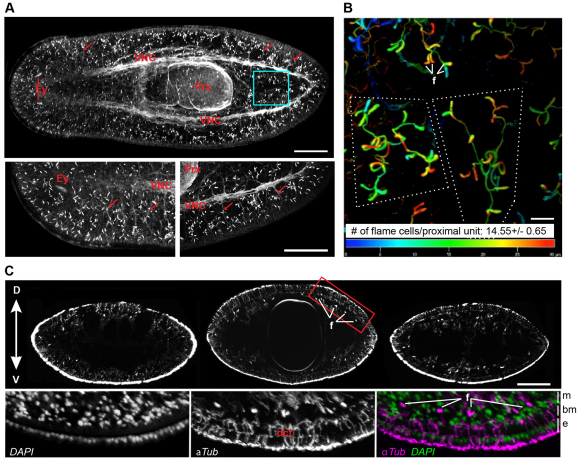
Sections of the planarian protonephridial system are known to be ciliated, and it has been reported that antibodies against tubulin, a major structural component of cilia, may label flame cells (Cebria and Newmark, 2005). We therefore used α-Tubulin staining of whole-mounted animals to gauge the general organization of protonephridia in planaria. Even though α-Tubulin staining also labeled other anatomical features (Fig. 1A,C), protonephridia were by far the brightest structures, allowing unambiguous tracing of their course through the tissue. The club-shaped cilia bundles of flame cells were readily apparent (Fig. 1A). Flame cells were highly abundant throughout the entire animal and appeared to be specifically aligned along the head margins (Fig. 1A, bottom left). At higher magnification, flame cells could be seen to connect to an α-Tubulin-positive network, which is likely to correspond to the ciliated tubule segments described in previous electron microscopy studies (Pedersen, 1961; McKanna, 1968b). Our whole-mount stains revealed a stereotypic organization of planarian protonephridia into tree-like units, whereby a common highly coiled `stem' splits into several thinner branches, each carrying one or two flame cells at its end. At least in the caudal regions of planarians, this arrangement results in a remarkably consistent number of 14 or 15 flame cells/unit (14.55±0.65), possibly indicating a stereotyped developmental sequence of protonephridia (Fig. 1B). Transverse sections showed protonephridial units to be entirely embedded in the mesenchyme and distributed without appreciable dorsoventral bias. Flame cells were mostly located immediately below the muscular layer that surrounds the planarian mesenchyme (Fig. 1C). The tubule stems usually faded out deeper into the mesenchyme, suggesting a transition into a non-ciliated tubule section undetectable by α-Tubulin staining. Although some protonephridial units were found deep in the mesenchyme (near CNS elements, between the two posterior gut branches and within the pharynx), the ciliated sections of the planarian protonephridia system appeared to mostly form a loose network around the surface of the mesenchyme (Fig. 1A,C).
Fig. 1.
Distribution of protonephridia. (A) Whole-mount α-Tubulin antibody staining. Magnified planarian head and tail regions are shown to the lower left and right, respectively. Flame cells appear as brightly stained club-shaped structures. Other anatomical features labeled by α-Tubulin staining are the ventral nerve cord (VNC), pharynx (Prx), eye cups (Ey) and peripheral nerves (arrows indicate examples). Images are maximum projections of confocal z-sections. The boxed region is magnified in B. (B) Depth-coded maximum projection of proximal units in the tail. Superficial structures appear in red, structures deep in the tissue as blue. Dotted lines outline single protonephridial units. f, examples of flame cells. (C) Transverse sections (top row) of α-Tubulin-stained whole-mount animals at the level of the photoreceptors (left), pharynx (center) and half-way between pharynx and tail tip (right). Flame cells (f) appear as bright dots. The boxed region is magnified beneath, showing nuclei (DAPI staining, left), α-Tubulin (center) and merge (right). e, surface epithelium; bm, basement membrane; m, mesenchyme; dct, non-protonephridial ductules of mucus-secreting cells. D, dorsal; V, ventral. Scale bars: 200 μm in A,C; 20 μm in B.
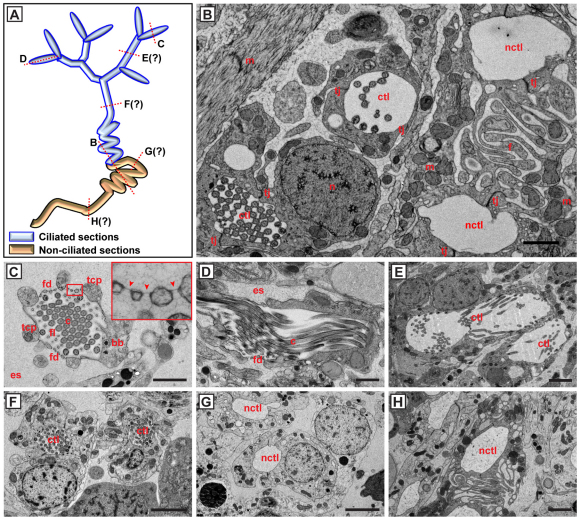
In parallel, we optimized high-pressure freezing methods for planarians, a method that can yield better tissue preservation for electron microscopy than traditional chemical fixatives (Fig. 2A-H) (Dernburg et al., 1998; Salvenmoser et al., 2010). A notable feature of high-pressure frozen specimens were large volumes of extracellular space between cells in the mesenchyme (Fig. 2B). The suggested loose organization is consistent with the almost instantaneous dissociation of the planarian mesenchyme upon removal of the epithelium (not shown). Readily identifiable ciliation and other criteria previously established in chemically fixed material (McKanna, 1968b; McKanna, 1968a; Ishii, 1980b; Ishii, 1980a) provided a set of morphological features for the identification of protonephridial structures. Flame cells were defined by the `filtration weir', which consists of closely apposed strands of cytoplasm surrounding a central cilia bundle, and by numerous microvilli between weir and cilia (Fig. 2C,D). The flame cells were often attached to a muscle fiber (not shown) and were always surrounded by a comparatively large volume of extracellular space (Fig. 2C), which in past studies using chemically fixed material appears to have occasionally been misinterpreted as `fixed parenchymal cells' (Pedersen, 1961; Ishii, 1980b). Cross-sections through ciliated tubules were much more frequent than through flame cells and tended to occur in clusters, consistent with the tortuous course of ciliated trunk and side branches (Fig. 1B). Interestingly, clusters of ciliated profiles were almost invariably accompanied by clusters of non-ciliated tubular cross-sections (Fig. 2B). Both types of lumen were formed by intercellular junctions between two cells, which in the case of non-ciliated tubules often showed dramatic folding of their cytoplasm, appearing as mitochondria-rich `loops' in cross-section (Fig. 2B,H). Owing to their spatial co-occurrence with ciliated profiles, the non-ciliated profiles are likely to correspond to a similarly sinusoidal continuation of the ciliated tubules. In both ciliated and non-ciliated tubule sections we observed morphological subtypes of the bounding cells (Fig. 2E-H), consistent with the functional differentiation of constituent cell types inferred previously [ciliated `ductule' and ciliated `collecting duct' (McKanna, 1968b); non-ciliated `trunk' and `distal tubule' (Ishii, 1980b); `transitional region' or `osmoregulatory duct' (McKanna, 1968b)]. Jointly, these experiments provide strong evidence to support the existence of multiple protonephridial cell types.
Fig. 2.
Ultrastructure of cell types in the planarian protonephridial system. (A) The approximate location of section planes. Question marks indicate uncertainty as to the exact position. (B) Overview image (high-pressure frozen specimen), showing ciliated tubule lumens (ctl) and non-ciliated tubule lumens (nctl). Lumens are formed by intercellular tight junctions (tj) between two tubule cells with laterally positioned nuclei (n). The cytoplasm of distal tubule cells is thrown into extensive folds (f), rich in mitochondria (m). (C) Coronal cross-section through a flame cell. The bundle of 9+2 cilia (c) is surrounded by a barrel of thick cytoplasmic processes (tcp), which support the filtration diaphragm (fd). Filaments (fl) inside the barrel have been interpreted as structural support. bb, basal body; es, extracellular space. Inset shows a magnification of the filtration diaphragm (arrowheads). (D) Longitudinal section through a flame cell. (E,F) Cross-sections through two types of ciliated proximal tubule lumen, bounded by squamous (E) or more cuboidal (F) cells. (G,H) Cross-sections through two types of non-ciliated distal tubule lumen, bounded by cuboidal (G) or extensively folded (H) cells. Scale bars: 1 μm in B-D; 2 μm in E-H.
Protonephridia are complex epithelial organs
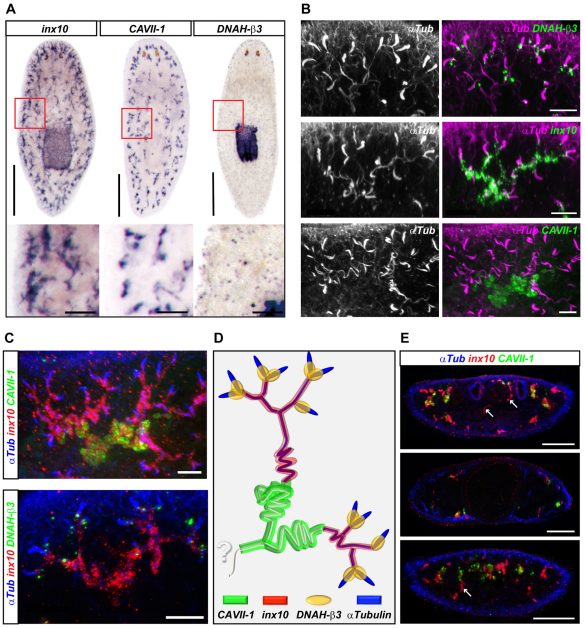
In order to identify molecular markers for the suspected diversity of protonephridial cell types, we screened whole-mount gene expression patterns for partial recapitulation of the α-Tubulin staining pattern (Fig. 3A). The gap junction gene Smed-innexin-10 (inx10) (Oviedo and Levin, 2007) and a carbonic anhydrase gene [H.14.9d or Smed-CAVII-1 (CAVII-1)] (Sánchez Alvarado et al., 2002), were both expressed in branching patterns, in which CAVII-1 branches appeared less complex and tended to terminate farther away from the body margins. The ciliary dynein heavy chain Smed-DNAH-β3 (DNAH-β3) was expressed in punctate foci that had a similarly uniform distribution as flame cells, yet with additional expression domains in the pharynx and along the body margins. All three genes indeed marked specific sections of protonephridia, as shown by colocalization with α-Tubulin immunostaining and double in situ hybridization experiments (Fig. 3B,C; see Movies 1 and 2 in the supplementary material).
Fig. 3.
Molecular anatomy of protonephridia. (A) Whole-mount in situ hybridization showing the expression patterns of the indicated marker genes (NBT/BCIP development). Anterior to the top. The boxed regions are magnified beneath. (B) Fluorescent overlay of indicated in situ patterns (green, right) with anti-α-Tubulin staining (pink and left). Images are maximum projections of confocal z-sections. (C) Fluorescent overlay of double in situ patterns (red and green) with anti-α-Tubulin staining (blue). Images are maximum projections of confocal z-sections. (D) Protonephridial molecular anatomy. See text for details. Question mark indicates unknown continuation of distal tubules. (E) Cross-sections of double-labeled whole-mount planaria (red, inx10; green, CAVII-1) overlaid with anti-α-Tubulin staining (blue). Sections were taken at the level of the photoreceptors (top), pharynx (center) and half-way between pharynx and tail tip (bottom). Arrows indicate examples of inx10-positive transverse tubules. Scale bars: 500 μm in A, top; 100 μm in A, bottom; 20 μm in B,C; 200 μm in E.
Together, these markers permit the following molecular description of protonephridial anatomy (Fig. 3D). DNAH-β3-positive flame cells connect to inx10-expressing ciliated tubules, which transition into CAVII-1-positive tubules. The tightly coiled CAVII-1-positive tubules are no longer ciliated, and thus are very likely to correspond to the non-ciliated tubule profiles seen by electron microscopy (Fig. 2B,G,H) (McKanna, 1968b; Ishii, 1980b). CAVII-1-positive segments often connected two neighboring proximal units, continuing the convergence trend of many proximal elements into fewer and fewer distal structures. Based on our molecular markers and previous efforts to clarify the confusing nomenclature (Ishii, 1980b), we refer to the ciliated and inx10-expressing segments as `proximal tubules' and to the non-ciliated CAVII-1-expressing segments as `distal tubules'. Unlike the uniformly distributed proximal units, distal tubules showed a clear bias toward the dorsal side in transverse sections (Fig. 3E). The connection to ventral proximal units was maintained via long inx10-positive tubule segments transgressing the mesenchyme (Fig. 3E). However, distal tubules still appeared to terminate abruptly in the mesenchyme. Hence, markers for yet more distal segments remain to be discovered. Similarly, we were unable to define the
connection of protonephridia to the outside with the available reagents. A historical study on non-specifically stained sections (Wilhelmi, 1906) is usually quoted as evidence for the termination of protonephridia at dorsally located nephridiopores (Hyman, 1951), but alternatives, such as drainage into the gut, remain possible.
Altogether, our ultrastructural and molecular marker analyses indicate that planarian protonephridia constitute a complex epithelial organ system that consists of multiple cell types organized into an intricate branching pattern.
Protonephridia regeneration
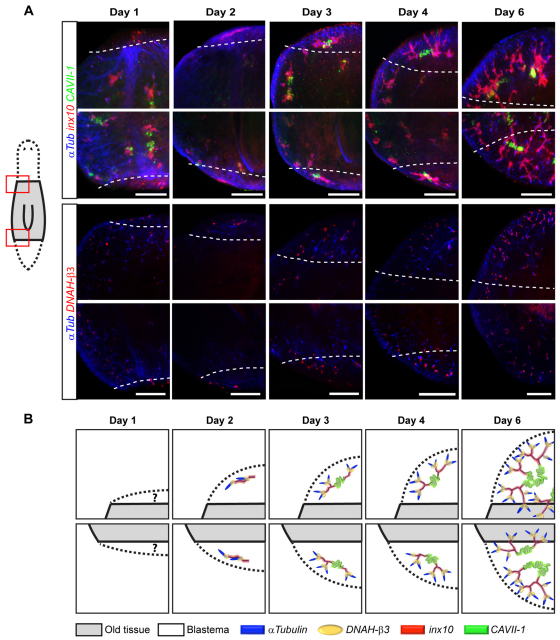
Having characterized markers for protonephridial cell types, we next explored how the cellular and morphological complexity of these organs is restored in the course of regeneration. Using multicolor in situ experiments at defined time points after amputation, we examined the temporal sequence and morphology of marker expression in head and tail blastemas (Fig. 4A,B). At 1 day post-amputation, the flame cell marker (DNAH-β3) and the proximal marker (inx10) produced diffuse and grainy signals at the wound margin. The small volume of new tissue at this early time point did not allow unambiguous distinction between old and new tissues and the variability in the signal observed between different animals could indicate background staining. However, 2 days after amputation, the proximal marker was prominently expressed in a rod-shaped structure embedded within the blastema, which was also associated with punctate flame cell marker expression. Even in high-magnification confocal z-stacks, we could not detect proximal marker-positive connections between this rod and protonephridia in the old tissue. Moreover, the morphology and temporal appearance of the structure were highly stereotyped, invariably occurring as a size-matched pair on either side of the midline in both head and tail blastema. Interestingly, the temporal snapshots of the regeneration timecourse experiments suggested that this structure might be the precursor of all protonephridia regeneration in the new tissue, which is why we refer to it as the proto-tubule.
Fig. 4.
Protonephridia regeneration. (A) Magnified view of left anterior (rows 1 and 3) and left posterior (rows 2 and 4) blastema of regenerating planaria trunk fragments at the indicated time points after amputation. Dashed lines demarcate the boundary between old and new tissue, as inferred from autofluorescence in the infra-red channel (not shown). In addition to anti-α-Tubulin antibody staining (blue), the top two rows were hybridized with the proximal marker probe (inx10, red) and the distal marker probe (CAVII-1, green), and the bottom two rows were hybridized with the flame cell probe (DNAH-β3, red). Images are maximum projections of confocal z-sections. Scale bars: 100 μm. (B) Graphic representation of the regeneration sequence.
The distal marker CAVII-1 was first expressed on the third day after amputation, its initial expression domain invariably bisecting the proto-tubule. Beginning on day 3, the inx10-positive proximal segments underwent extensive branching morphogenesis. Branching first became evident on day 3 (coincident with dispersal of the DNAH-β3 signal) and branch elongation towards the blastema margins was especially prominent on day 4. Even though branching appeared to be slightly delayed in tail blastemas as compared with head blastemas, protonephridia morphology in both cases became practically indistinguishable from that of uncut animals by day 6 after amputation, suggesting that organ regeneration, as assessed with the current set of markers, was complete by this time point.
Overall, the highly stereotyped regeneration of protonephridia from a precursor structure argues in favor of de novo organogenesis in regenerating tissues.
An epidermal growth factor receptor is required for protonephridial function
In order to identify components of the signaling network orchestrating protonephridia differentiation and morphogenesis, we performed an RNAi screen of a candidate library comprising ∼400 planarian homologs of conserved signaling pathway components (J.C.R. and K. A. Gurley, unpublished). Previous studies have reported massive bloating of animals fed RNAi against cilia components (Reddien et al., 2005; Rink et al., 2009) or against the proximal marker gene inx10 (Oviedo and Levin, 2007). In osmotic shock experiments and accompanying histological sections, we observed that such bloating is in fact caused by severe edema formation upon functional impairment of the ciliated and inx10-expressing proximal tubule (see Fig. S1 in the supplementary material). Edema formation also provided a readily apparent screening phenotype for potential protonephridia genes.
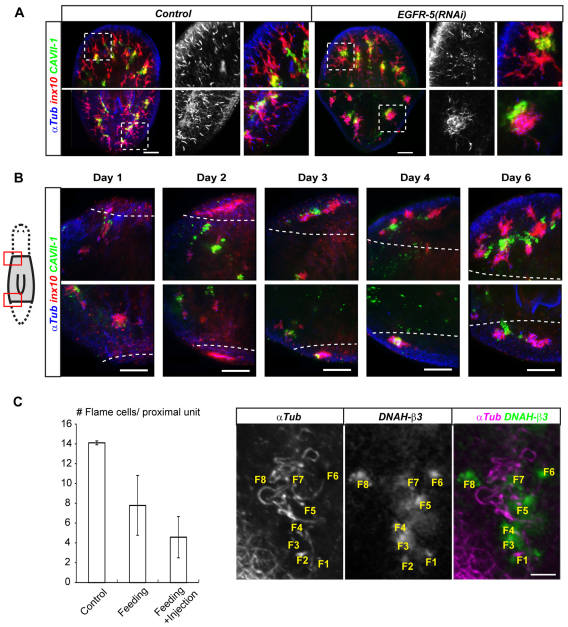
Our screen identified the epidermal growth factor receptor (EGFR) homolog Smed-EGFR-5 (EGFR-5), which, when knocked down, led to edema formation in intact and regenerating animals, similar to inx10(RNAi) (Fig. 5A; see Fig. S2 in the supplementary material). Even though planarians have five EGFR family members (J.C.R., unpublished), which represents an unusual expansion of this gene family amongst invertebrates (Stein and Staros, 2006), only RNAi of EGFR-5 led to edema formation. The first indication of a phenotype in uncut EGFR-5(RNAi) animals was an apparent depigmentation in the anterior half of the animal (`pale') beginning at day 5 after the last RNAi feed (Fig. 5B). Within a further 3 days, pale animals progressed to tail edema formation as in inx10(RNAi) animals. Starting at day 14 after the last RNAi feed, lesions became apparent, which progressed to eventual lysis and death of all EGFR-5(RNAi) animals.
Consistent with a direct role in protonephridial function, EGFR-5 was expressed seemingly exclusively in Y-shaped branches that were reminiscent of proximal protonephridia segments (Fig. 5C). Multicolor in situ hybridizations confirmed co-expression with the proximal marker inx10 and revealed particularly high expression levels in DNAH-β3-positive flame cells (Fig. 5D; see Movie 3 in the supplementary material). Consistently, EGFR-5 expression was already detectable, along with the flame cell marker, by 48 hours of regeneration, at the proto-tubule stage (Fig. 5E).
Taken together, these data identify EGFR-5 not only as a second marker for flame cells and the adjacent terminal proximal branches, but also as a molecule with an important role in maintaining the functional integrity of the proximal segment of protonephridia.
EGFR-5 is required for flame cell maintenance and branching morphology
In order to understand how EGFR-5 might influence protonephridia function, we examined protonephridia regeneration in EGFR-5(RNAi) animals. A first survey of regenerated heads and tails 14 days after amputation demonstrated that all three cell type markers were present, indicating that the respective cell types had differentiated (see Fig. S3A in the supplementary material). However, the expression patterns, especially those of the proximal markers, were severely disturbed. Multicolor confocal imaging experiments (Fig. 6A) demonstrated abnormal thickening of proximal branches and misdirected branch extension towards the posterior in head fragments. In tail fragments, however, the few remaining proximal segments were coiled into tight balls (Fig. 6A).
Fig. 6.
EGFR-5(RNAi) phenotype ontogeny in regeneration. (A) Late stage morphological defects of regenerated protonephridia in EGFR-5(RNAi) animals as compared with control planaria at 14 days post-amputation. Head (top row) and tail (bottom row) of representative animals are shown; magnifications show the boxed region from the respective overview images. Monochrome shows α-Tubulin staining alone. Images are maximum projections of confocal z-sections. (B) Magnified view of left anterior (top row) and left posterior (bottom row) blastema of regenerating trunk fragments of EGFR-5(RNAi) animals at the indicated time points after amputation. Refer to Fig. 4 for control. Dashed lines demarcate the boundary between old and new tissue, as inferred from autofluorescence in the infra-red channel (not shown). α-Tubulin antibody staining (blue) was combined with the proximal marker probe (inx10, red) and the distal marker probe (CAVII-1, green). Images are maximum projections of confocal z-sections. (C) Flame cell quantification in 14-day EGFR-5(RNAi) regenerates having received either the standard RNAi dosage used throughout this study (Feeding) or an additional injection of EGFR-5 dsRNA on the third day of regeneration (Feeding + Injection). Three proximal units in six animals were scored for each time point. Error bars represent s.e.m. To the right is shown an illustration of the flame cell counting procedure using the indicated markers. For greater sensitivity, the flame cell marker DNAH-β3 was developed with the non-fluorescent substrate NBT/BCIP. Flame cells (F, numbered) were defined as the spatial coincidence of an NBT/BCIP focus with a terminal tubule segment in image z-stacks. The images shown are maximum projections of a z-stack; the NBT/BCIP brightfield image was brightness-inverted and pseudo-colored green. Scale bars: 100 μm.
Regeneration timecourse experiments provided insights into the ontogenesis of these defects (Fig. 6B; see Fig. S3B in the supplementary material; refer to Fig. 4A for control): The proto-tubule appeared to form normally in regenerating EGFR5(RNAi) animals, but severe branching and branch extension defects became apparent from day 3 onwards. The sprouting and peripherally directed extension of proximal branches from the proto-tubule were severely inhibited, such that by day 6, when control animals had regenerated the complete proximal arborizations, EGFR-5(RNAi) animals displayed only short, partly posteriorly misoriented inx10-positive bundles in the head, but hardly any signs of branch extension in tails.
A further abnormality of protonephridia regenerated under EGFR-5(RNAi) was that of less prominent flame cell cilia bundles in the α-Tubulin channel (Fig. 6A). Also, the flame cell marker expression pattern was affected (see Fig. S3A,B in the supplementary material), suggesting possible defects in flame cell specification or maintenance. Indeed, quantification of flame cell numbers in EGFR-5(RNAi) animals 14 days after amputation (Fig. 6C) revealed a decrease to an average of eight flame cells/proximal unit. An additional injection of EGFR-5 dsRNA on the third day of regeneration, administered in order to boost the lessening knockdown efficiency (see Fig. S3C in the supplementary material), resulted in a further decrease to about five flame cells/proximal unit. Thus, the regeneration of protonephridia under EGFR-5(RNAi) caused both morphological defects in proximal arborizations and a reduced number of flame cells per proximal unit.
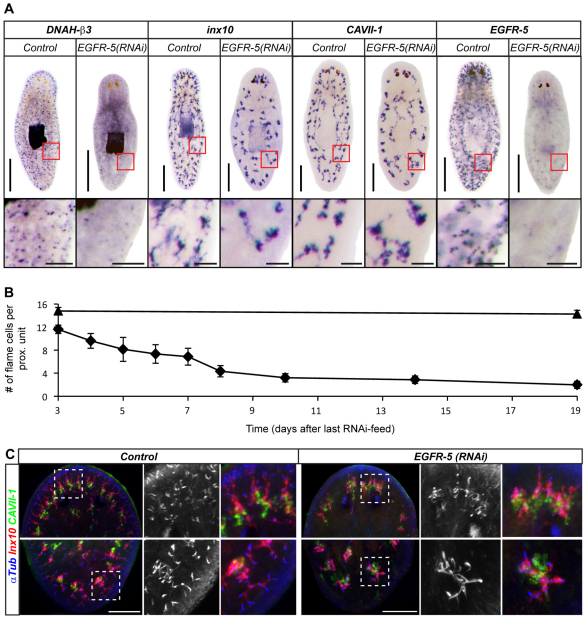
The same spectrum of phenotypes was also observed in non-regenerating EGFR-5(RNAi) animals. Expression of the flame cell marker was severely reduced 14 days after the last RNAi feed (Fig. 7A). A quantification of flame cell numbers revealed a rapid decline in response to EGFR-5(RNAi), reducing their numbers to two flame cells/proximal unit around day 14 (Fig. 7B). Such loss of flame cells is likely to explain the edema formation and eventual lysis in EGFR-5(RNAi) animals. In addition, proximal arborizations were also severely affected under EGFR-5(RNAi) (Fig. 7C). However, in contrast to the disorganized and misdirected branching patterns observed in regenerating animals (Fig. 6A), proximal arborizations were severely shortened to a few, short coils, especially in caudal regions. Interestingly, the gradual loss of flame cells (Fig. 7B) was paralleled by a collapse of proximal arborizations (see Fig. S4 in the supplementary material).
Fig. 7.
EGFR-5(RNAi) phenotype ontogeny in intact animals. (A) Planarian protonephridial marker gene expression in control and EGFR-5(RNAi) animals 14 days after last RNAi feed. Each pair was developed under identical conditions. The boxed regions are magnified beneath. (B) Timecourse quantification of average flame cell numbers per proximal unit. Diamonds, EGFR-5(RNAi) animals; triangles, controls. Flame cells were counted in confocal z-stacks of α-Tubulin/DNAH-β3 double-labeled whole-mount animals. Three proximal units in five animals were scored for each time point. Error bars represent s.e.m. (C) Morphological defects of protonephridia in EGFR-5(RNAi) animals as compared with control 14-days after the last RNAi feed. Head (top row) and tail (bottom row) of representative animals are shown; magnifications show the boxed region from the respective overview images. Monochrome shows α-Tubulin staining alone. Images are maximum projections of confocal z-sections. Scale bars: 500 μm in A, top; 100 μm in C and in A, bottom.
Our data thus indicate that EGFR-5 is required both for flame cell maintenance and for guiding branch extension of protonephridia. The close association between the two phenotypes further suggests that they might be mechanistically linked.
DISCUSSION
Our results establish the planarian protonephridia as a bona fide epithelial organ system. A variety of cell types assemble into complex tubular arbors with stereotypic proximodistal organization, in which abundant proximal elements converge into fewer and fewer distal structures. Such complexity at the architectural and morphological level place planarian protonephridia on a par with other branched organ systems such as trachea in insects and lungs, mammary glands, blood vessels and the kidney in vertebrates (Lu and Werb, 2008; Beyenbach et al., 2010; Costantini and Kopan, 2010). All of the above examples rely critically on branching morphogenesis to establish the specific morphology necessary for proper organ function. It is, therefore, not too surprising that our screen of signaling components required for protonephridia-mediated tissue fluid homeostasis identified a gene with functions in branching morphogenesis.
The first phenotype resulting from EGFR-5(RNAi) comprised dramatic effects on the branching pattern of protonephridia. In non-regenerating RNAi-fed animals, the fanned-out organization of proximal branches collapsed until only short scrawl-like structures remained (Fig. 7C). In regenerating RNAi animals, by contrast, we observed misorientation of the branches in the head and dramatic coiling of proximal tubules into tight balls in the tail region (Fig. 6A). Timecourse experiments provided insights into the ontogeny of these seemingly disparate phenotypes. In intact animals, the collapse of proximal arborizations correlated with the loss of flame cells (see below). In regenerating animals, the misdirected branch extension phenotypes appeared to develop in two phases. Up until regeneration day 6, EGFR-5(RNAi) animals displayed markedly reduced branching and branch extension in comparison to control animals (Fig. 6B, Fig. 4A). In order to generate the striking coils in tails or disorganized branch patterns in heads of 14-day regenerates (Fig. 6A), a burst of misguided proximal branch extension must occur between days 6 and 14. The reason for the distinct outcomes in heads versus tails involves differential RNAi sensitivities: both misguided branch extension and coil formation were part of a phenotypic series, but, for unknown reasons, tail tissues responded more strongly to a given dose of RNAi than head tissues (see Fig. S5A-C in the supplementary material). This effect might also contribute to edema formation preferentially in tails of EGFR-5(RNAi) animals (Fig. 5A). Moreover, we noted that the switch in branch extension capabilities around regeneration day 6 coincided with a lessening knockdown efficiency of EGFR-5 (see Fig. S3C in the supplementary material). The misdirected branch extension between day 6 and 14 is therefore likely to involve inappropriate levels or timing of EGFR-5 expression, whereas the early inhibition of branch extension correlated with low EGFR-5 levels. Similarly, the collapse and shortening of proximal branches observed in non-regenerating EGFR-5(RNAi) animals occurred under low and sustained knockdown of EGFR-5 (Fig. 7A). Hence, a shared feature between the intact and regeneration phenotypes is a requirement for EGFR-5 in causing and maintaining branch extension.
Second, RNAi-mediated knockdown of EGFR-5 caused a loss of flame cells, the cell type at the tip of protonephridial branches. These cells are thought to represent the entry point of interstitial fluid into the protonephridial system by means of ultrafiltration, analogous to the role of the glomerulus in the human kidney (Pedersen, 1961). Hence, the dramatic edema formation in EGFR-5(RNAi) animals is likely to result primarily from a loss of flame cells, rather than from the altered branching morphology of protonephridia. In intact animals, the number of flame cells per proximal unit declined rapidly in response to EGFR-5(RNAi), from 14-15 cells/unit at the onset of RNAi feeding to only 2 cells/unit at the lysis stage (Fig. 7B). In regenerating animals, we could also measure an RNAi dose-dependent decrease in flame cells/proximal unit 14 days post-amputation (Fig. 6C). These data establish a definite role of EGFR-5 in maintaining flame cells (and possibly also the adjacent proximal branches). Whether EGFR-5 is also required for flame cell differentiation during regeneration and/or homeostasis is currently difficult to ascertain. Flame cell marker expression was detectable early on in regenerating EGFR-5(RNAi) animals (see Fig. S3B in the supplementary material), but the tools to determine whether their numbers are normal and whether or not the differentiation process is affected are presently lacking. Moreover, the seemingly regeneration-induced decrease in EGFR-5 RNAi efficiency (see Fig. S3C in the supplementary material) represents a second experimental obstacle to addressing early roles of EGFR-5 in flame cell differentiation.
Regardless of the specific role of EGFR-5 in flame cells, the strong correlation between flame cell phenotypes and proximal branching morphology remains an important observation. In intact animals, we observed a gradual collapse of arborizations concomitant with the loss of flame cells. In regenerating animals, the severity of the branching defects at day 14 post-amputation correlated with the number of flame cells (Fig. 6C; see Fig. S5D,E in the supplementary material). Hence, a mechanistic link between flame cells and the establishment and maintenance of proximal branching patterns seems likely.
Interestingly, branching morphogenesis generally depends on specialized cells at the tubule tips (Lu and Werb, 2008). Tip cells specified by, and responding to, FGF signaling guide the extension and morphogenesis of Drosophila tracheal tubules (Ghabrial and Krasnow, 2006). Vertebrate kidney development relies on tip cells specified via differential RET receptor signaling to guide branch outgrowth from the ureteric bud (Chi et al., 2009), and vertebrate blood vessel development represents yet another example in which migratory tip cells act as the `motor' for elongation and positioning of a tubular network (Hellstrom et al., 2007; Siekmann and Lawson, 2007). By analogy, and in light of our data, flame cells might act as tip cells in protonephridia morphogenesis, besides their roles in organ physiology. Furthermore, EGFR-5 might fulfil the widespread requirement for receptor tyrosine kinase (RTK) signaling in specifying and guiding the `tip motor' (Andrew and Ewald, 2010). The collapse of proximal arborizations concomitant with the loss of flame cells in intact EGFR-5(RNAi) animals (see Fig. S4 in the supplementary material) indicates a persisting tip cell function, which might reflect the anchoring of tubule ends to the muscular layer via the prominent flame cell filopodia that are visible in electron micrographs (McKanna, 1968a; Ishii, 1980a).
Altogether, the branching morphogenesis of planarian protonephridia revealed by our studies represents a fascinating example of biological pattern formation on several levels (Fig. 1A,B). First, individual protonephridial units tile in a non-overlapping manner. Second, within units, branches extend in a spatially efficient manner. Third, proximal units contain a remarkably consistent number of flame cells (14.55±0.65), amounting to 14 or 15 flame cells per proximal unit in caudal regions. The mechanistic basis of these patterns, in particular the hypothesis that limiting quantities of an EGFR-5 ligand might be involved, represents an interesting area for future exploration. In addition, the striking food supply-dependent variations in planarian body size raise questions regarding the scaling of protonephridial capacity with animal size. The animals used in this study ranged between 0.8 and 2.5 mm in length; hence, it appears likely that the number of flame cells/unit remains constant at 14 or 15 irrespective of animal size. The consequence of capacity adjustments via the addition or removal of entire protonephridial units also seems in agreement with the mechanism of protonephridial regeneration in forming tissues.
Our data strongly suggest that protonephridia regenerate de novo, rather than by growth and extension from pre-existing units. A proximal marker-expressing structure, which we called the proto-tubule, appears to initiate organ regeneration. The rod-shaped proto-tubule emerged at ∼36 hours, embedded within the blastema (Fig. 4A). Besides the fact that we could not detect connections to pre-existing protonephridia, the remarkable consistency in the timing of appearance, position, size and symmetry between right and left blastema halves strongly suggest de novo formation of the proto-tubule. We cannot exclude a contribution of pre-existing protonephridia to the final organ complement in the new tissue, for example by dynamic reorientation of proximal branches near the blastema boundary. However, the temporal snapshots of our timecourse experiments suggest that the majority of protonephridia in the new tissue originate from the morphogenetic remodeling of the proto-tubule. One interesting aspect of this remodeling is the invariable bisection of the proto-tubule by distal marker-expressing cells on day 3 (Fig. 4A), which is likely to represent the ontogenetic basis for the convergence of two proximal units into one distal unit (Fig. 3).
Besides the mechanisms driving branching and branch extension, the origin of the proto-tubule raises further fascinating problems. In analogy with epithelial tube formation in other systems (Lubarsky and Krasnow, 2003), invagination from the overlying epithelium or condensation of blastema cells are plausible mechanisms. The intermittent ciliation within 48-hour proto-tubules (Fig. 5E) might indicate a focal mode of lumen formation, which is typical of solid precursor structures (Dong et al., 2009). Moreover, a study in the polyclad flatworm Imogine mcgrathi suggests embryonic protonephridia formation from mesodermal cells (Younossi-Hartenstein and Hartenstein, 2000), which is why we currently tend to favor a condensation mechanism. The relatively poor preservation of cell boundaries by our in situ hybridization protocol is one of the current obstacles in further addressing the cell biology of such a fascinating example of non-embryonic organogenesis. The same problem also precluded us from tracing protonephridia to their site of termination.
Altogether, our ultrastructural, molecular and functional dissection of planarian protonephridia define a novel experimental paradigm for studying the various processes involved in the assembly, morphogenesis and maintenance of an epithelial organ, as well as its evolution. The cellular complexity of the planarian protonephridia revealed by our work suggests that studies of this organ system will not only complement those of other molecularly tractable, yet highly derived, excretory systems (e.g. the single excretory cell of C. elegans, or the uncoupling between ultrafiltration and absorption/secretion in the Malpighian tubules of Drosophila), but also might help to elucidate the functional and evolutionary relationships that define the invertebrate and vertebrate excretory systems.
Supplementary Material
Acknowledgments
We thank members of the A.S.A. laboratory for critical reading of the manuscript and Nina Strömgren Allen for helpful discussions of electron micrographs. A.S.A. is a Howard Hughes Medical Institute Investigator. This work was also supported by NIH NIGMS R37 GM57260 to A.S.A. Deposited in PMC for release after 6 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.066852/-/DC1
These authors contributed equally to this work
References
- Agata K., Umesono Y. (2008). Brain regeneration from pluripotent stem cells in planarian. Philos. Trans. R. Soc. Lond. B Biol. Sci. 363, 2071-2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew D. J., Ewald A. J. (2010). Morphogenesis of epithelial tubes: Insights into tube formation, elongation, and elaboration. Dev. Biol. 341, 34-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyenbach K. W., Skaer H., Dow J. A. (2010). The developmental, molecular, and transport biology of Malpighian tubules. Annu. Rev. Entomol. 55, 351-374 [DOI] [PubMed] [Google Scholar]
- Cebria F. (2007). Regenerating the central nervous system: how easy for planarians! Dev. Genes Evol. 217, 733-748 [DOI] [PubMed] [Google Scholar]
- Cebria F., Newmark P. A. (2005). Planarian homologs of netrin and netrin receptor are required for proper regeneration of the central nervous system and the maintenance of nervous system architecture. Development 132, 3691-3703 [DOI] [PubMed] [Google Scholar]
- Cebria F., Kudome T., Nakazawa M., Mineta K., Ikeo K., Gojobori T., Agata K. (2002). The expression of neural-specific genes reveals the structural and molecular complexity of the planarian central nervous system. Mech. Dev. 116, 199-204 [DOI] [PubMed] [Google Scholar]
- Chi X., Michos O., Shakya R., Riccio P., Enomoto H., Licht J. D., Asai N., Takahashi M., Ohgami N., Kato M., et al. (2009). Ret-dependent cell rearrangements in the Wolffian duct epithelium initiate ureteric bud morphogenesis. Dev. Cell 17, 199-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chichkoff G. D. (1892). Recherches sur les Dendrocoeles d'eau douce (Triclades). Arch. Biol. (Liege) 12, 435-568 [Google Scholar]
- Collins J. J., 3rd, Hou X., Romanova E. V., Lambrus B. G., Miller C. M., Saberi A., Sweedler J. V., Newmark P. A. (2010). Genome-wide analyses reveal a role for peptide hormones in planarian germline development. PLoS Biol. 8, e1000509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini F., Kopan R. (2010). Patterning a complex organ: branching morphogenesis and nephron segmentation in kidney development. Dev. Cell 18, 698-712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg A. F., McDonald K., Moulder G., Barstead R., Dresser M., Villeneuve A. M. (1998). Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94, 387-398 [DOI] [PubMed] [Google Scholar]
- Dong B., Horie T., Denker E., Kusakabe T., Tsuda M., Smith W. C., Jiang D. (2009). Tube formation by complex cellular processes in Ciona intestinalis notochord. Dev. Biol. 330, 237-249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finken-Eigen M., Kunz W. (1997). Schistosoma mansoni: gene structure and localization of a homologue to cysteine protease ER 60. Exp. Parasitol. 86, 1-7 [DOI] [PubMed] [Google Scholar]
- Ghabrial A. S., Krasnow M. A. (2006). Social interactions among epithelial cells during tracheal branching morphogenesis. Nature 441, 746-749 [DOI] [PubMed] [Google Scholar]
- Gurley K., Rink J., Sánchez Alvarado A. (2008). Beta-catenin defines head versus tail identity during planarian regeneration and homeostasis. Science 319, 323-327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellstrom M., Phng L. K., Hofmann J. J., Wallgard E., Coultas L., Lindblom P., Alva J., Nilsson A. K., Karlsson L., Gaiano N., et al. (2007). Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature 445, 776-780 [DOI] [PubMed] [Google Scholar]
- Hyman L. H. (1951). The Invertebrates: Platyhelminthes and Rhynchocoela. II. New York: McGraw-Hill; [Google Scholar]
- Ishii S. (1980a). The ultrastructure of the protonephridial flame cell of the freshwater planarian Bdellocephala brunnea. Cell Tissue Res. 206, 441-449 [DOI] [PubMed] [Google Scholar]
- Ishii S. (1980b). The ultrastructure of the protonephridial tubules of the freshwater planarian Bdellocephala brunnea. Cell Tissue Res. 206, 451-458 [DOI] [PubMed] [Google Scholar]
- Lu P., Werb Z. (2008). Patterning mechanisms of branched organs. Science 322, 1506-1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubarsky B., Krasnow M. A. (2003). Tube morphogenesis: making and shaping biological tubes. Cell 112, 19-28 [DOI] [PubMed] [Google Scholar]
- McKanna J. A. (1968a). Fine structure of the protonephridial system in Planaria. I. Flame cells. Z. Zellforsch. Mikrosk. Anat. 92, 509-523 [DOI] [PubMed] [Google Scholar]
- McKanna J. A. (1968b). Fine structure of the protonephridial system in Planaria. II. Ductules, collecting ducts, and osmoregulatory cells. Z. Zellforsch. Mikrosk. Anat. 92, 524-535 [DOI] [PubMed] [Google Scholar]
- Nishimura K., Kitamura Y., Inoue T., Umesono Y., Sano S., Yoshimoto K., Inden M., Takata K., Taniguchi T., Shimohama S., et al. (2007). Reconstruction of dopaminergic neural network and locomotion function in planarian regenerates. Dev. Neurobiol. 67, 1059-1078 [DOI] [PubMed] [Google Scholar]
- Nishimura K., Kitamura Y., Umesono Y., Takeuchi K., Takata K., Taniguchi T., Agata K. (2008). Identification of glutamic acid decarboxylase gene and distribution of GABAergic nervous system in the planarian Dugesia japonica. Neuroscience 153, 1103-1114 [DOI] [PubMed] [Google Scholar]
- Nishimura K., Kitamura Y., Taniguchi T., Agata K. (2010). Analysis of motor function modulated by cholinergic neurons in planarian Dugesia japonica. Neuroscience 168, 18-30 [DOI] [PubMed] [Google Scholar]
- Oviedo N. J., Levin M. (2007). smedinx-11 is a planarian stem cell gap junction gene required for regeneration and homeostasis. Development 134, 3121-3131 [DOI] [PubMed] [Google Scholar]
- Pahlavan P. S., Feldmann R. E., Jr, Zavos C., Kountouras J. (2006). Prometheus' challenge: molecular, cellular and systemic aspects of liver regeneration. J. Surg. Res. 134, 238-251 [DOI] [PubMed] [Google Scholar]
- Pearson B. J., Eisenhoffer G. T., Gurley K. A., Rink J. C., Miller D. E., Sánchez Alvarado A. (2009). Formaldehyde-based whole-mount in situ hybridization method for planarians. Dev. Dyn. 238, 443-450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K. J. (1961). Some obserfations on the fine structure of planarian protonephridia and gastrodermal phagocytes. Z. Zellforsch. 53, 609-628 [Google Scholar]
- Pellettieri J., Fitzgerald P., Watanabe S., Mancuso J., Green D. R., Sánchez Alvarado A. (2010). Cell death and tissue remodeling in planarian regeneration. Dev. Biol. 338, 76-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddien P., Sánchez Alvarado A. (2004). Fundamentals of planarian regeneration. Annu. Rev. Cell Dev. Biol. 20, 725-757 [DOI] [PubMed] [Google Scholar]
- Reddien P. W., Bermange A. L., Murfitt K. J., Jennings J. R., Sánchez Alvarado A. (2005). Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria. Dev. Cell 8, 635-649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J. C., Gurley K. A., Elliott S. A., Sánchez Alvarado A. (2009). Planarian Hh signaling regulates regeneration polarity and links Hh pathway evolution to cilia. Science 326, 1406-1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruppert E. (1994). Evolutionary origin of the vertebrate nephron. Am. Zool. 34, 542-553 [Google Scholar]
- Salvenmoser W., Egger B., Achatz J. G., Ladurner P., Hess M. W. (2010). Electron microscopy of flatworms: standard and cryo-preparation methods. Methods Cell Biol. 96, 307-330 [DOI] [PubMed] [Google Scholar]
- Sánchez Alvarado A., Newmark P. A., Robb S. M., Juste R. (2002). The Schmidtea mediterranea database as a molecular resource for studying platyhelminthes, stem cells and regeneration. Development 129, 5659-5665 [DOI] [PubMed] [Google Scholar]
- Siekmann A. F., Lawson N. D. (2007). Notch signalling limits angiogenic cell behaviour in developing zebrafish arteries. Nature 445, 781-784 [DOI] [PubMed] [Google Scholar]
- Skelly P. J., Shoemaker C. B. (2001). Schistosoma mansoni proteases Sm31 (cathepsin B) and Sm32 (legumain) are expressed in the cecum and protonephridia of cercariae. J. Parasitol. 87, 1218-1221 [DOI] [PubMed] [Google Scholar]
- Stein R. A., Staros J. V. (2006). Insights into the evolution of the ErbB receptor family and their ligands from sequence analysis. BMC Evol. Biol. 6, 79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesono Y., Watanabe K., Agata K. (1999). Distinct structural domains in the planarian brain defined by the expression of evolutionarily conserved homeobox genes. Dev. Genes Evol. 209, 31-39 [DOI] [PubMed] [Google Scholar]
- Wilhelmi J. (1906). Untersuchungen ueber die Exkretionsorgane der Suesswassertricladen. Ztschr. Wiss. Zool. 80, 544-575 [Google Scholar]
- Wilson R. A., Webster L. A. (1974). Protonephridia. Biol. Rev. Camb. Philos. Soc. 49, 127-160 [DOI] [PubMed] [Google Scholar]
- Younossi-Hartenstein A., Hartenstein V. (2000). The embryonic development of the polyclad flatworm Imogine mcgrathi. Dev. Genes Evol. 210, 383-398 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.