Abstract
This unit presents a specific and sensitive quantitative reverse-transcription PCR (RT-qPCR) method for measuring individual microRNAs (miRNAs) in tissue or cultured cells. MiRNAs are 17 – 24 nucleotides (nt) in length. Standard and quantitative PCR methods require a template that is at least twice the length of either of the specific forward or reverse primers, each typically ∼ 20 nt in length. Thus, the target minimum length is ≥ 40 nt, making miRNAs too short for standard RT-qPCR methods. In this assay, each of the RT-qPCR nucleic acid reagents, including the RT-primer, the forward and reverse PCR primers, and the hydrolysis probe, contain design features that, together, optimize miRNA specificity and assay sensitivity. The RT-primer contains a highly stable stem-loop structure that lengthens the target cDNA. The forward PCR primer adds additional length with nucleotides that optimize its melting temperature (Tm) and enhance assay specificity. The reverse primer disrupts the stem loop. Assay specificity is further optimized by placement of the probe over much of the original miRNA sequence, and the probe Tm is optimized by addition of a minor groove binding (MGB) moiety.
Keywords: miRNA, MIQE, RT-qPCR
Introduction
Real-time PCR is extensively used to quantify RNA. Standard procedures for conducting and publishing real-time PCR experiments have been recently codified in “The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments,” (Bustin et al., 2009). The method presented in this unit conforms with the recommended MIQE guidelines. The suggested MIQE nomenclature is used in this text (Table 1).
Table 1.
Recommended MIQE Nomenclature used in this Unit.
| MIQE term | Definition | Comment |
|---|---|---|
| qPCR | Quantitative Real-Time PCR | |
| RT-qPCR | Reverse-Transcription qPCR | |
| Reference gene | Gene used for normalization | Not to be called housekeeping gene |
| Hydrolysis probe | TaqMan probe | The chemistry can be applied outside of a single manufacturers' (Applied Biosystems, Inc.'s) purview |
| Cq | quantification cycle, the fractional PCR cycle used for quantification | Previously termed Ct (threshold cycle), Cp (crossing point), or TOP (take-off-point), coined by different manufacturers of real-time instruments |
| Run | A single PCR plate | |
| Reaction | A single well | |
| Biological replicates | Replicate samples within an experimental group | |
| Technical replicates | Replicate measures of a single sample |
Quantification of nucleic acids with qPCR and RT-qPCR is presented in depth in Unit 15.8. Increasing interest in the biological functions of small RNAs such as microRNAs (miRNAs) (see Units 26.1 and 26.4) warrant convenient methods for quantification of these small RNA species. However, miRNAs are too short to accommodate an adequate primer pair and probe for any level of specific amplification using standard methods. This unit describes a method for quantitative amplification of specific miRNAs (Chen et al., 2005) whereby the target cDNA is lengthened, and design specifics of the PCR forward primer and the hydrolysis probe combine to ensure specificity at great sensitivity. First strand cDNA synthesis with a highly stable stem-loop primer lengthens the target from its original ∼22 nt to >60 nt (Fig. 1). PCR amplification utilizes a forward primer that includes extra 5′ nts to adjust for an appropriate Tm, a universal reverse primer that is complementary to a sequence within the RT stem-loop primer, and a hydrolysis probe for detection and quantification of PCR products, enhanced for specificity by its conjugation to a minor groove binder (MGB) (Fig. 1). This approach is the basis for the miRNA RT-qPCR assays commercially available through Applied Biosystems, Inc. Applied Biosystems, Inc. has streamlined the process of designing specific assays and preparing those reagents (RT primer and PCR primers and probe) for investigators who provide newly discovered miRNA sequences. Advantages of this protocol over purchasing custom designed assays (nucleic acid reagents) from Applied Biosystems, Inc. include 1), standards for absolute quantification, and 2), reducing the costs of performing a large number of specific assays.
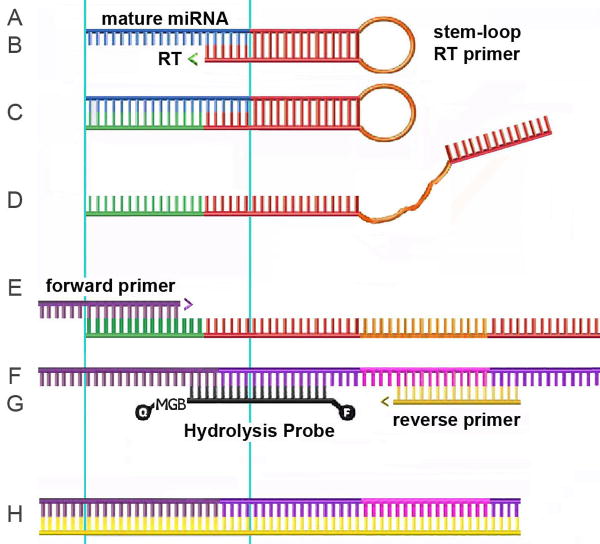
Figure 1.
Nucleic acid reagents used for and intermediate products generated in this method. A, Mature miRNA (blue). A-H, Light blue lines show the boundary of the mature miRNA sequence within the RT and qPCR reagent sequences; open arrowheads indicate directions of polymerization. B, The stem-loop primer 5′ 6 nt annealed with mature miRNA 3′ 6 nt; RT, reverse transcriptase. C and D, First strand cDNA, after polymerization, C, and heat denaturation, D. E, Forward primer with added 5′ nts. F, Second strand cDNA. G, Hydrolysis probe and reverse primer. H, PCR product defined by the 5′ termini of the forward and reverse primers.
This protocol incorporates absolute quantification (as in Unit 15.8.11), although relative quantification may also be adapted for this assay (Unit 15.8).
The assay design necessitates a brief overview of miRNA biogenesis (Cullen, 2004). Mature miRNAs are transcribed within a long RNA that is termed the primary-miRNA (pri-miRNA). Pri-miRNA is processed in the nucleus by an RNase complex that generates an ∼ 60 nt imperfect stem-loop structure called pre-miRNA. Pre-miRNA is transported to the cytoplasm where it is cleaved and unwound by the RNase Dicer. Most pri-miRNAs yield one mature miRNA that derives from one of the strands of the stem, while the other strand and the loop are degraded; in some cases both stem strands are processed and retained as mature miRNAs. The fate of the pri- and pre-miRNA is degradation. However, in certain circumstances, pri-miRNA or pre-miRNA or both may be present in relative abundance (Cui, 2006). The first step in this assay is reverse transcription of the mature miRNA with a primer that anneals to only six nt of the 3′ end of the target. The stability of the stem-loop structure of the RT primer precludes its annealing to those 6 nt within the pri- or pre-miRNA, due to steric hinderence. However, if pre- and/or pri-miRNAs are present in great molar excess relative to the mature miRNA, some low level reverse transcription and subsequent qPCR of the embedded sequence may occur at a low level of efficiency. Even low levels of cross-amplification of these longer sequences could skew results. Thus, in these cases, it is important to separate very small RNAs from total RNA before performing the quantification. This protocol includes this as an option for preparing the sample RNA.
Assay design
The most important consideration for assay design is knowledge of the precise sequence of the miRNA of interest. Deep sequencing results have shown both 5′ and 3′ variations in newly discovered miRNAs, although this seems to be more pronounced with certain viruses, such as herpes simplex virus -1 and -2 (Umbach, 2008, Jurak, 2010). Shortened 5′ and/or 3′ ends may arise from degradation during processing of the sample. Lengthened ends can include post-processing modifications such as 3′ adenylation and uridylation (Ibrahim et al., 2010; Burroughs et al., 2010). Several considerations go into determining the precise sequence for which an assay will be designed. However, it is probable that the sequence represented in greatest abundance will be the correct one.
Strategic Planning: STEM-LOOP RT PRIMER and qPCR PRIMER and PROBE DESIGN (Figure 1)
1. Quantification Standard
This is the precise sequence of the miRNA of interest. Here, miR-H1, expressed by HSV-1 (Cui, 2006; Umbach, 2008; Jurak, 2010), will be used as an example.
The 20 nt RNA sequence of miR-H1 is the Quantification Standard:
5′-UGG AAG GAC GGG AAG UGG AA-3′
2. Stem-Loop (SL) RT primer
This combines 44 nt of the stem-loop sequence of Chen et al., 2005, 5′- GTC GTA TCC AGT GCA GGG TCC GAG GTA TTC GCA CTG GAT ACG AC -3′ with the complement of the six 3′ nt of the mature miRNA sequence (boxed)This is the stem-loop RT primer for miR-H1:
5′- GTC GTA TCC AGT GCA GGG TCC GAG GTA TTC GCA CTG GAT ACG AC

 -3′
-3′
3. Forward qPCR primer
Together the Forward qPCR Primer and the Probe should maximally cover the mature miRNA sequence. Take the first 12 to 17 nt of the 5′ end of the mature miRNA, and 3 to 7 additional 5′ nt selected for getting the Tm to 60°C. Start with the first 12 nt, as in this example below, so as to maximize probe coverage of the remaining miRNA sequence.
-
Take the first twelve 5′ nt from the mature miRNA sequence (transformed to DNA, boxed)
5′-
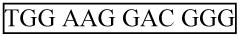 -3′
-3′
-
Add six nt to the 5′ end. The sequence of the six nt should be selected to confer upon the final oligo a Tm of 60 +/- 1 °C. An online calculator such as http:www.idtdna.com/analyzer/Applications/OligoAnalyzer can be useful. This is the miR-H1 Forward primer:
5′- CAC GCA
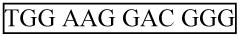 -3′
-3′
4. Reverse qPCR Primer
By using the same 44 nt stem-loop sequence for all RT primers, a universal primer can be derived from sequences within the stem-loop. We recommend this Reverse Primer:
5′-CCA GTG CAG GGT CCG AGG TA-3′
The reverse primer sequence reported by Chen et al., 2005, is: 5′- GTG CAG GGT CCG AGG T -3′. We found improved sensitivity with the longer primer.
5. Hydrolysis Probe
The length of this probe is tightly constrained, as >half the miRNA length is covered by the forward primer, The probe Tm should ideally be close to 70°C, and its length can be 12 to 17 nt. A minor groove binder (MGB), which is conjugated to the probe, is important for increasing the Tm of a shorter sequence. When designing this probe, it is helpful to envision the final forward and reverse PCR product strands.
-
Identify the location of the Universal Reverse qPCR Primer (lower case) in the 44 nt stem-loop sequence of Chen et al., 2005:
5′- GTC GTA Tcc agt gca ggg tcc gag gta TTC GCA CTG GAT ACG AC -3′
-
Add the 20 nt complement of the miRNA (boxed) to the 3′ end of the 44 nt stem loop sequence. This is the 62 nt first strand cDNA for miR-H1:
5′- GTC GTA Tcc agt gca ggg tcc gag gta TTC GCA CTG GAT ACG AC
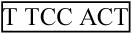
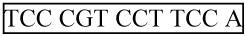 -3′
-3′
-
Determine the complement strand sequence. Add the six 5′ nt of the Forward qPCR Primer to the 5′ end and identify the Forward qPCR Primer in lower case. This is the 70 nt second strand cDNA of miR-H1:
5′- cac gca
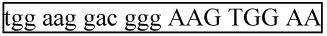 G TCG TAT CCA GTG CGA Ata cct cgg acc ctg cac tgg ATA CGA C -3′
G TCG TAT CCA GTG CGA Ata cct cgg acc ctg cac tgg ATA CGA C -3′
-
Finally, delete the nt 3′ of the Universal Reverse qPCR Primer sequence. This is the 63 nt forward strand of the final miR-H1 PCR product, and its complement is the reverse strand. Mature miR-H1 DNA is boxed, forward and reverse primers cover the italicized nucleotides, and the probe is in bold.
For: 5′- cac gca
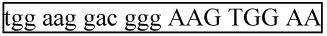 G TCG TAT CCA GTG CGA Ata cct cgg acc ctg cac tgg-3′
G TCG TAT CCA GTG CGA Ata cct cgg acc ctg cac tgg-3′Rev: 5′- cca gtg cag ggt ccg agg taT TCG CAC TGG ATA CGA C

 tgc gtg -3′
tgc gtg -3′
-
The probe (bold, in Reverse PCR strand) spans the mature miRNA-stem-loop sequence junction, does not overlap the forward primer, and maximizes length to increase its Tm. It is synthesized with a 5′ FAM moiety, and 3′ quencher and a 3′ minor-groove binder (MGB) that raises its Tm and confers higher specificity. This is the miR-H1 probe:
5′- (FAM) TGG ATA CGA C
 -3′ (MGB)(Quencher)
-3′ (MGB)(Quencher)
Materials
Fresh or frozen tissue (5 to 50 μg wet weight) and/or 106 -107 cultured cells
Nuclease-free water
Nuclease-free TE
mirVana miRNA Isolation Kit (Applied Biosystems, Inc. AM1560) (also see Units 4.2, 26.4, and 26.7)
3M sodium acetate, molecular biology grade
100% ethanol, molecular biology grade
NanoDrop® ND-1000 or ND-2000 (ThermoScientific) or comparable spectrophotometer flashPAGE Fractionator System (Applied Biosystems, Inc.) (optional); components: (1) the flashPAGE Fractionator unit, (2) pre-cast gels, (3) loading and running buffers, and (4) (optional) flashPAGE Clean-up Kit for rapid concentration of fractionated samples
Working stocks of assay nucleic acid reagents (see Reagents and Solutions):
Quantification Standards at 2.3×1010 molecules/μL
5× stocks (5.0 nM) of stem loop primers
20× stocks of combined Forward Primer, Reverse Primer, and Probe
TaqMan MicroRNA RT Kit (Applied Biosystems, Inc. 4366596), contents: 10X RT buffer, dNTP mix w/dTTP (100 mM total), RNase Inhibitor (20U/μL), and Multiscribe (MuLV) RT
Labnet Miniplate Spinner mps 1000 (Labnet International)
96 well assay plates and sealing film for reverse transcriptions
Standard thermal cycler (optional) with 96-well platform (optional)
TaqMan Universal PCR Master Mix, No AmpErase UNG (Applied Biosystems, Inc. 4324018)
Real time thermal cycler, e.g. Step-One Plus (Applied Biosystems, Inc.)
Optical 96-well plates and optical sealing film specific for the instrument
Microsoft Excel (or alternative spreadsheet)
STEM-LOOP RT-qPCR for miRNAs
First, design, prepare, and test the reagents (Figure 2), then apply the assay to experimental samples. This protocol describes a plate layout in which two assays are run together on a set of 12 samples (Figure 3). In this example, a reference miRNA (miR-A) is assayed along with the miRNA of interest (miR-B).
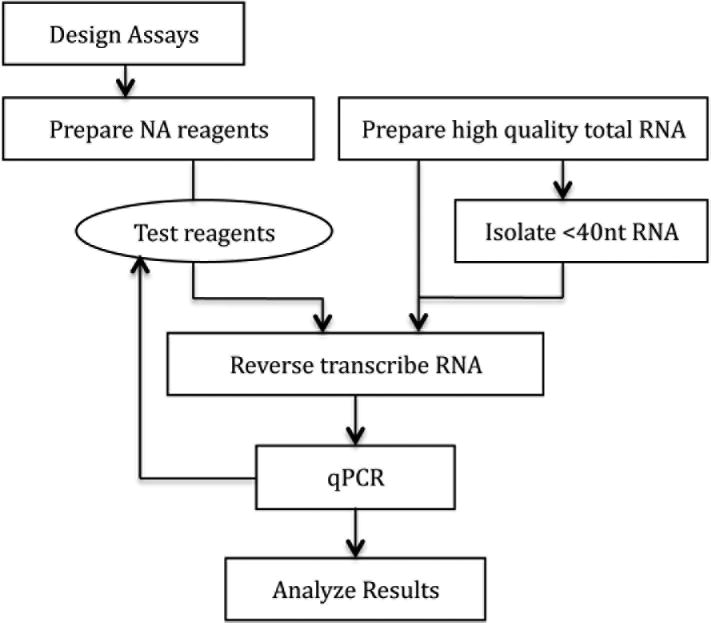
Figure 2.
Flowchart. Once the reagents have been designed, received, and prepared, it is advisable to test them before assaying experimental samples.
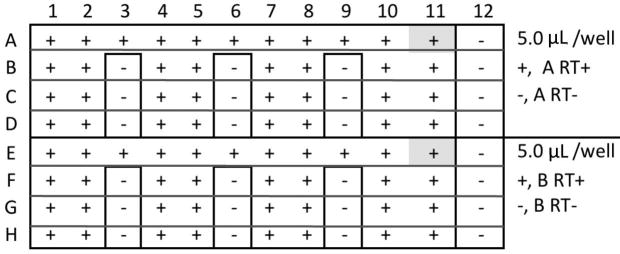
Figure 3.
Layout of 96-well plate for two assays, A (Reference miRNA) and B (new miRNA) and 12 samples, S1 – S12. Negative control wells are shaded. “8,” “7,” etc., log10 number of standard miRNA molecules per reaction. +, -, presence or absence or RT in RT reaction. NTC, no template control; water replaces RNA in the RT reaction. In this example, the reference miRNA, miR-A, is abundant so the standard dilutions used are between 108 and 104 copies per reaction, in duplicate. The second highest standard dilution is also assayed in the absence of RT, (“7”-). The new miRNA, miR-B, is less abundant so the standard dilutions used are between 106 and 102 copies per reaction; the second highest standard dilution is also assayed in the absence of RT, (“5”-). Twelve samples, S1 – S12, are assayed for both miRNAs. These should include a mock control and several biological replicates from each experimental group. Technical replicates are assayed in adjacent wells, with a single, adjacent, RT- negative control for every sample in every assay.
Prepare RNA samples: total RNA and <40nt RNA
1. Prepare high quality total RNA. Verify the RNA quality and measure its quantity; depending on the relative abundance of the miRNA of interest, start with 0.2 to 4.0 μg total RNA in up to 40 μL for flashPAGE isolation of <40 nt RNA (described below).
Having tested several RNA isolation methods, we found that mirVana miRNA Isolation Kit (Applied Biosystems, Inc.) generated the highest quality and most consistent results, possibly because this kit was specifically developed to maximize recovery of small RNAs. Quality may be assessed by running a test sample on a Northern blot (Unit 4.9) and quantity may be measured with a spectrophotometer, an Agilent Analyzer, or a NanoDrop Spectrophotometer (Units A3D and A3J). It is not generally necessary to DNase the total RNA or <40 nt fraction for miRNA RT qPCR when prepared with the mirVana miRNA Isolation Kit.
2. (optional) Isolate <40nt small RNAs from total RNA. There are two methods by which this can be done: gel isolation (Unit 26.4, steps 3 to 11), or flashPAGE Fractionator System per manufacturer's protocol (Applied Biosystems, Inc.). The <40 nt fraction can be cleaned up with a commercial clean-up product (flashPAGE Clean-up Kit, Applied Biosystems, Inc.) and eluted with 30 μL elution buffer. Alternatively, bring the volume to 90 μL, add 10 μL 3M sodium acetate, add 400 μL 100% ethanol, mix well, leave at -20°C overnight or longer, and recover the <40 nt RNA with centrifugation at 20,000 × g for 45 min at 4°C, followed by a wash with 80% ethanol. The RNA pellet may not be visible. However, if it is quite large, then wash the pellet again with 70% ethanol. Resuspend in 50 ul RNase-free TE. The <40 nt RNA can be quantified (CPMB Appendices A.3D and/or A3J). Quantification of an appropriate reference miRNA will demonstrate the relative recovery.
Total RNA is the starting material for samples in which most of the cells express the miRNA of interest at levels greater than the pri-miRNA or pre-miRNA from which it is derived. If this is not the case, <40 nt RNA can be isolated from total RNA with gel electrophoresis. Additionally, flashPAGE can be used to gain sensitivity The flashPAGE method is relatively inexpensive, and requires no radioactivity, so it is the method of choice presented here. The flashPAGE Clean-up Kit is more convenient for fewer samples, and the overnight precipitation is more convenient when working with a larger number of samples. Reserve “before” and “after” samples to assess recovery using a miRNA RT-qPCR assay of an abundant reference miRNA such as let7a.
Dilute miRNA quantification standards
3. Make 1/10 serial dilutions of Std A and Std B (separately) in nuclease-free TE.
These can also be diluted in an RNA solution that does not contain the miRNA of interest to verify specificity.
Keep working stock of 2.3×1010 molecule/μL labeled “10” on dry ice when at the bench. On ice, set up 10 small tubes containing 27 μL nuclease-free water and labeled “9” through “1.” Quickly thaw the concentrated standard labeled “10,” vortex, spin briefly, and transfer 3μL to the first tube labeled “9.” Mix well. Transfer 3μL of that to the next tube, “8,” mix well, and so on, for both standards.
If the miRNA of interest is abundant enough to detect readily on a Northern blot, use standards at 107to 103molecules per reaction, otherwise use 105to 101.
Prepare and load RT reaction master mixes
4. Label 4 screw-cap tubes, A+, A-, B+, B-. Make 4 RT master mixes, an RT+ and an RT- for each miR-A and miR-B, (Table 2).
Table 2.
RT master mixes.
| 48 wells, n= | 35 | 13 | A+ | A- | B+ | B- |
|---|---|---|---|---|---|---|
| + RT | - RT | + RT batch | - RT batch | + RT batch | - RT batch | |
| Each (μL) |
Each (μL) |
× 40 (μL) |
× 20 (μL) |
× 40 (μL) |
× 20 (μL) |
|
| 100 mM dNTPs | 0.075 | 0.075 | 3.0 | 1.5 | 3.0 | 1.5 |
| RT | 0.5 | 0 | 20 | --- | 20 | --- |
| 10× Buffer | 0.75 | 0.75 | 30.0 | 15.0 | 30.0 | 15.0 |
| RNaseInhibitor | 0.095 | 0.095 | 3.8 | 1.9 | 3.8 | 1.9 |
| H2O | 2.08 | 2.58 | 103.2 | 51.6 | 103.2 | 51.6 |
| 5× A RT Primer | 1.5 | 1.5 | 60.0 | 30.0 | --- | --- |
| 5× B RT Primer | 1.5 | 1.5 | --- | --- | 60.0 | 30.0 |
| total Vol | 5 | 5 | 200 | 100 | 200 | 100 |
Do not allow the stem loop primers to warm up above room temperature to insure the stability of the stem loop structure.
5. Load 96-well RT reaction plate with the four RT+/- reagents and “non template control” (NTC). (Figure 4). Seal a 96-well reaction plate with transparent sealing tape.
Figure 4.
RT plate for two assays, A and B, and 12 Samples, +/- RT. NTC, A11 and E11, shaded.
RT-: Draw lines around the wells that require RT- reaction mix. Cut on lines with a razor blade and remove the sealing tape with forceps. Aliquot 5.0 μL A- (RT-) reaction mix into each RT- well of Rows A through D; Aliquot 5.0 μL B- (RT-) reaction mix into each RT- well of Rows E through H. Replace the bits of sealing tape to cover the RT- wells.
RT+: Remove the main part of the sealing tape. Aliquot 5.0 μL A+ reaction mix into all remaining wells of Rows A through D; Aliquot 5.0 μL B+ reaction mix into all remaining wells of Rows E through H.
NTC: Immediately after loading the plate with RT+ and RT- reaction mixes, load 2.5 μL RNA solution that does not contain the miRNA of interest into the two NTC wells, A11 and E11 (Fig. 4, shaded).
6. Remove all sealing film and replace with a fresh sheet. Leave on ice.
Load the RT plate with standard dilutions and experimental sample RNAs
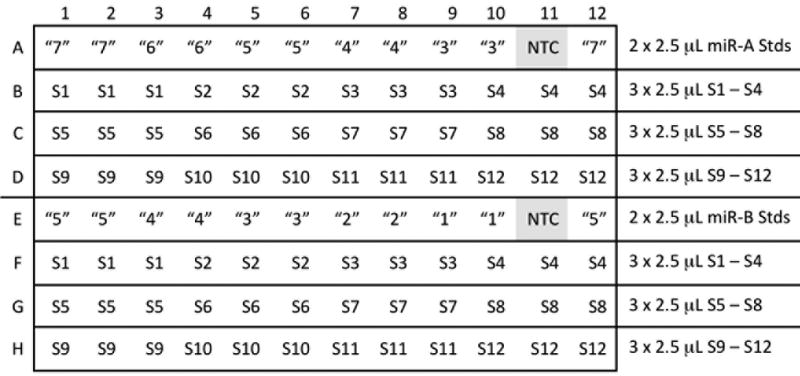
7. Load 96-well RT reaction plate with 2.5 μL standard dilutions as in Fig. 5. Underline Rows A, D, and E on the new sealing tape; cut with blade. Remove sealing tape from row A. Aliquot 2.5 μL miR-A standard “4” into A10 and A9, then “5” into A8 and A7, “6” into A6 and A5, “7” into A4 and A3, and “8” into A2 and A1. Aliquot 2.5 μL “7” into A12. Replace the strip of sealing tape. Remove sealing tape from row E. Aliquot 2.5 μL miR-B standard “2” into E10 and E9, then “3” into E8 and E7, “4” into E6 and E5, “5” into E4 and E3, and “6” into E2 and E1. Aliquot 2.5 μL “5” into E12. Replace the strip of sealing tape.
Figure 5.
Standard and Sample loading.
8. Load 96-well RT reaction plate with 2.5 μL experimental RNA samples as in (Figure 5). Underline Rows B, C, F, and G on the sealing tape; cut with blade. Remove sealing tape from rows B and F. Aliquot 2.5 μL sample #1 (S1) into B1, B2, B3, F1, F2, and F3. Aliquot 2.5 μL S2 into B4, B5, B6, F4, F5, and F6. Continue for all samples. Seal tightly with sealing film. Centrifuge plate briefly in Mini Plate Spinner.
Note: Each assay requires 3 × 2.5 μL = 7.5 μL of sample; to run a panel of assays, bring the volume of sample to (n+1) assays× 7.5 μL; include appropriate reference genes in the count. Example: to assay A, B, C, and D miRNAs, and E, F, and G reference genes (7 assays), bring the Sample volume to (7+1) ×7.5μL = 60 μL.
Reverse transcription
9. Run RT plate on a 96-well PCR platform, with sequential incubations at 16°C for 30 min, 42°C for 30 min, and 85°C for 5 min. Cool the plate and centrifuge plate briefly in Mini Plate Spinner.
qPCR
10. Make one PCR reaction mix per assay, A and B, (Table 3). Each 20X, the mixture of Forward and Reverse primers and Hydrolysis probe, is specific to each assay. Aliquot 18.7 μL reaction mix A into each well of Rows A, B, C and D of a PCR plate (optical grade, specific for the real time instrument). Aliquot 18.7 μL reaction mix B into each well of Rows E, F, G, and H.
Table 3. qPCR Reaction mixes.
| Reaction Mix n=48 |
ea: (μL) |
A (μL) |
B (μL) |
|---|---|---|---|
| 2× Universal | 10 | 500 | 500 |
| H2O | 7.67 | 388 | 388 |
| A 20× | 1 | 50 | --- |
| B 20× | 1 | --- | 50 |
|
| |||
| F v | 18.67 | 938 | 938 |
11. Transfer 1.3 μL RT product from the 96-well RT plate to the 96-well PCR plate. Seal with optical-grade film. Centrifuge plate briefly in Mini Plate Spinner.
12. Run real-time PCR at standard conditions:
| 1 cycle | 10.0 min | 95°C |
| 40 cycles | 15 sec | 95°C |
| 1.0 min | 60° C |
Including a single initial cycle of 2 min at 50°C makes no difference to the outcome.
Data Analysis
See also Unit 15.8, steps 10 to 14, which refer to absolute quantification.
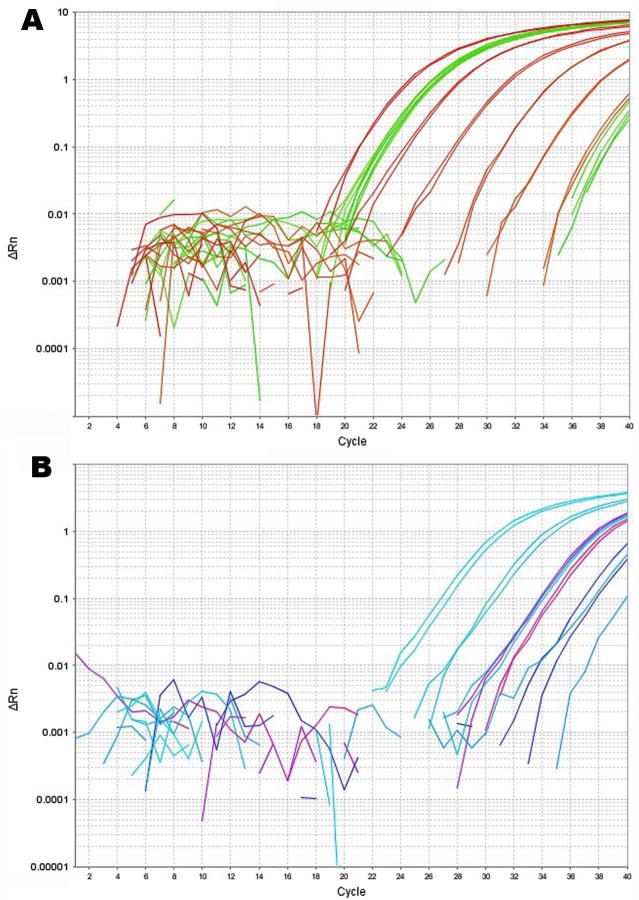
13. View the amplification plots of the standard reactions to ensure that the assay is performing correctly (Fig. 6). Each 10-fold dilution should have a ∼3.3 cycle difference. The samples should all be contained within the high and low ends of the respective standard curve.
Figure 6.
Amplification plots from one qPCR run for two assays, A, Reference miRNA (here, let7a), and B, miRNA of interest (here, HSV-1 miR-H4-3p). A, Standard curve, NTC, and standard miRNA RT- all from Row A, all in red, and three experimental samples in duplicate with their respective RT- controls all from Row B, all in green. Technical replicates and biological replicates from each of two experimental groups were constant for the reference miRNA. Non-specific amplifications arose in the experimental sample negative controls (far right, green) but not in the standard miRNA negative control. B, Standard curve, NTC, and standard miRNA RT- all from Row E, all in light blue, and three samples in duplicate with their respective RT- controls from Row F (blue, pink, and purple). Non-specific amplifications were absent in both the experimental sample negative controls and in the standard miRNA negative control, but at this low level, the duplicates were compromised. Technical replicates performed well above 103 molecules/reaction.
14. Transfer the results (Export Results) e.g. via a flash drive to a computer that has Excel installed. Open the results in Excel. Save original data in a protected folder. Create a copy to perform the data analyses.
All real time platforms have their pre-installed instrument software for collection and reporting fluorescence during amplification, and the preinstalled software can perform varying degrees of analysis of the results depending on the platform. In addition, there are several calculation and statistical applications available (see Logan, 2006, for in depth discussion of this topic, and http://bioinformatics.gene-quantification.info). The company's technical support representative should be consulted. The following calculations performed manually allow one to maintain greater vigilance in monitoring the validity of one's biological results.
15. Plot the Cq (also referred to as Ct, see Table 1) vs log 10 molecules per reaction for each assay A and B. The “7,” “6,” “5,” etc. designation indicates the log 10 number of molecules per reaction. Apply the linear regression formula y = mx + b, where y = Cq, m = slope, x = the quantity of the unknown sample in log molecules per reaction, and b = intercept. Also determine the correlation coefficient, R2, in Excel. Calculate amplification efficiency, E, which is given as E =10∧(-1/slope).
Maintain a record of the date, slope, intercept, R2, and E for each assay, and compare these parameters with each new run to ensure that the standards and other reagents are performing properly.
16. Interpolate Cqs with the regression curve specific to the assay, which yields log 10 molecules per reaction, and average these values per sample. If the instrument software has already calculated these values, compare with the values you obtain. If there are no discrepancies, the values obtained using the instrument software can be accepted. If there are discrepancies, contact the representative of the real-time instrument company. It is good practice to periodically make this comparison.
17. Divide the results (average log miR-B molecules per reaction), by the reference gene results (average log miR-A molecules per reaction) for each sample. This will normalize for differences between samples in recovery of both total RNA and the <40nt RNA.
These are log-transformed values. To divide log-transformed numbers, subtract the denominator, D, from the numerator, N: N/D = logN-logD. D is the reference miRNA, miR-A, and N is the test miRNA, miR-B. Log (miR-B / miR-A) = log miR–B - log miR-A. For example, say log miR-A = 7.25 and log miR-B = 4.40, log (miR-B / miR-A) = 4.4 – 7.25 = -2.85; 10-285= 0.014.
To calculate copies per cell, per unit tissue, or per unit total RNA, experimentally determine the average copy number of the reference miRNA, miR-A, in the cell, tissue, or RNA of interest, then multiply this factor by (copies miR-B/copies miR-A).
Reagents and Solutions
Working stocks
1. Quantification Standard: Order RNA, 100 μM, gel- or HPLC-purified from a preferred vendor, such as Integrated DNA Technologies (IDT). Verify the concentration. Dilute to 2.3×1010 molec/μL. Store in multiple aliquots at -80°C.
100 μM = 100 × 10-12 moles / μL.
100 × 10-12 moles / μL × Avogadro's Number 6.023 × 1023 molecules/mole = 602.3 × 1011 molecules/μL.
-
Dilute 1/200 to 0.5 μM = 3 × 1011 molecules/μL:
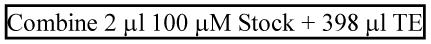
-
Dilute 1/13 to 2.3 × 1010 molecules/μL [(2.31 × 1010) / (3 × 1011) = 1/ 13]:
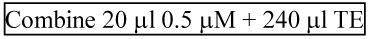
2.5μL of 1/10 serially diluted standard are reverse-transcribed in a total volume of 7.5 μL. Of that, 1.3μL are transferred to a PCR plate for amplification. Thus, (1.3μL × 2.5μL / 7.5μL)× 2.3 × 10nmolecules/μL= 1 × 10nmolecules/reaction.
The concentration of each nucleic acid reagent, even from the most reputable supplier, should be checked on a spectrophotometer prior to making working stock solutions.
2. SL RT primer: Order 50 nt DNA oligonucleotide, 100 μM, gel- or HPLC-purified (IDT is recommended). Verify its concentration. Re-fold loop RT-primers as follows:
Transfer to new tube, then overlay with 100 μL molecular-biology grade mineral oil. Heat to 95°C for 10m. Reduce heat slowly to 75°C. Hold temp at 75, 68, 65 and 62°C for an hour each, hold at 60°C for several more hours. Transfer primer from under oil to new tube. This step should never need repeating if the primer is never heated above room temperature. Make a 5× working stock of 5.0 nM. Store both the 100 μM and the working stocks at -20°C.
3. Forward and Universal Reverse qPCR primers: Order DNA oligonucleotides, 100 μM, gel- or HPLC-purified (IDT is recommended). Verify their concentration. Store at -20°C.
4. MGB Hydrolysis Probe: Order 100 μM DNA oligonucleotide with 5′-FAM and 3′-MGB modifications, e.g. 5′- (FAM) TGG ATA CGA CTT CCA CT -3′ (MGB)(Quencher), available from Applied Biosystems, Inc.
5. Combine Forward and Reverse primers and Hydrolysis probe to make a 20X PCR Reaction Reagent for final concentrations of 1.5, 0.7, and 0.8 μM, respectively.
| 1× fc (μM) | 20× (μM) | Combine (μL) | |
| 100 μM Specific Forward primer | 1.5 | 30.0 | 30 |
| 100 μM Reverse primer | 0.7 | 14.0 | 14 |
| 100 μM Specific Probe | 0.8 | 16.0 | 16 |
| H2O | 40 | ||
| 20×; store at -20°C | 100 |
Commentary
Background
The topic of miRNA is covered in depth in Chapter 26. The topic of qPCR (real time PCR, see Table 1) is covered in depth in Unit 15.8; however it is important to become familiar with the MIQE guidelines as well (Bustin, 2009). The assay detailed here is one of many tools that can be used to study miRNA biology. Methods for detection of microRNAs include cloning, deep sequencing, northern, in situ, and solution hybridization, microarray analysis, and direct or nondirect RT-qPCR. Cloning followed by sequencing or, more recently, deep sequencing, have led the field in discovery of miRNAs, with deep sequencing revealing many less abundant microRNAs. The number of “hits” per unique sequence in deep sequencing is relatively quantitative. Cloning miRNAs is considered labor intensive, and deep sequencing may be prohibitively expensive, but with shared resources more institutions are making it available to investigators. Hybridization-based gene expression analysis often take days of processing time, can be quantitative but with a limited dynamic range (1 to 2 log), and may require 1-10 μg of total RNA per sample. These remain the least expensive of the methods in start up and operating costs. Microarray analysis and RT-qPCR for miRNAs with known sequences offer many advantages for gene expression analysis including sensitivity to the single cell level, high specificity, and high throughput. Both require costly instrumentation, which is becoming more commonly available. There are a few different RT-qPCR methods available and companies that support them. Direct RT-qPCR means that total RNA (or <40 nt RNA) is directly reverse transcribed then amplified by qPCR. Nondirect RT-PCR means that the miRNA of interest is polyadenylated prior to reverse transcription (for example, NCode™ miRNA RT-qPCR System from Invitrogen). We elected to explore the former, direct RT-qPCR with the stem-loop RT primer, when it was first published, for the following reasons. Compared to the nondirect approach, 1) there is less handling of RNA and therefore less opportunity for contamination or RNA degradation, 2) one less enzymatic step (each enzymatic step can introduce variation, and can reduce recovery), 3) the stem-loop method discriminates between mature and pre- or pri-miRNA by 100-fold and 4), the stem-loop method discriminates between single base mismatches by 100-fold (Chen, 2005). NCode™ miRNA RT-qPCR System (Invitrogen) also has a miRNA amplification system/kit that generates senseRNA that reportedly preserves the relative abundance of different miRNAs; the stem-loop method has not seemed to need this additional amplification of miRNA in order to quantitatively detect low abundance miRNAs within complex mixtures. The costs of the primers are competitive while the cost of the probe may seem high. Compared to providing Applied Biosystems with a sequence and commissioning an assay made for that sequence, designing, procuring, and mixing one's own nucleic acid reagents as presented here, represents a savings. Direct comparison between this RT-qPCR method, northern blot hybridization, and relative number of reads from deep sequencing can show quantitative discrepancies, especially where the genomic sequence is GC-rich. Discrepancies in the quantity of a given miRNA measured by different methods may arise from technical (e.g. purification quality), chemical (e.g. relative combined efficiencies of each of a series of enzymatic steps, with regard to RT-qPCR and deep sequencing), and biological variations (e.g. contained within the sequence itself). This RT-qPCR method is based on a synthetic miRNA standard and is sensitive to single nt substitutions.
This protocol was adapted from TaqMan™ MicroRNA RT Kit (Applied Biosystems, Inc.). The Applied Biosystems, Inc. literature does not contain primer and probe concentrations. We initially tested the primers and probe at the concentrations reported by Chen, et al., 2005: 50 nM RT Primer (final concentration), 0.2 μM Probe, 1.5 μM Forward primer, and 0.7 μM Reverse primer. Upon optimization of several assays, we found that a lower concentration of RT primer (1 nM final concentration) gave greatly reduced non-specific amplifications while improving sensitivity. Also, a slightly higher concentration of hydrolysis probe (0.8 μM) gave more consistent results at lower concentrations. The differences may be more applicable to rare miRNAs, and less important with more abundant species.
In this protocol we used a 96-well plate format. The protocol could be modified to work on a 384 well plate.
Critical Parameters/Trouble shooting
It is important to start with and demonstrate high-quality RNA and test the nucleic acid reagents prior to committing any experimental samples to the assay. The most critical parameter is the re-folding of the stem-loop into its stable structure. When this primer is subjected to high temperatures, some melting may occur. If it is incompletely folded, the reverse transcription of non-specific cDNA may occur. Where the miRNA of interest is rare, non-specific cDNA could exceed the specific cDNA and potentially contribute to non-specific PCR amplification. Given the combined specificities of the forward primer and probe, non-specific PCR amplification should not contribute to the fluorescent signal. However, unscheduled amplifications could interfere with the robustness of the assay by, for example, competing for and depleting reaction components. The experimental samples typically contain a complex mixture of RNA; some of this is likely to be reverse transcribed by the incompletely folded sequences of the stem loop primer, generating non-specific amplification targets. First test the assay on the synthetic RNA standard. If there is no amplification or if there are abnormal amplification results, be sure to evaluate each step of the procedure, and perform routine troubleshooting as presented in Unit 15.8. In particular, ensure that your reagents, consumables, and techniques are RNase-free, and confirm that each reagent has the correct sequence. If the assay appears to be working well but does not appear to be specific to 103 molecules/reaction and/or generates amplification plots with the no template controls, re-fold the RT primer and try again. Further optimization of the limit of sensitivity may not be of additional value if you are measuring microRNA that is relatively abundant. Finally, it may be possible that the precise ends of the miRNA of interest are heterogenous and may contain non-templated nucleotides. The precise 3′end sequence determines the stem loop RT primer and the probe sequences, and the precise 5′ end determines the forward PCR primer. Cloning may be necessary to determine the precise sequence.
Anticipated Results
This protocol was developed for simultaneous testing of up to 12 samples that include biological replicates of several experimental groups including a biological negative control (i.e. a sample lacking the miRNA of interest), with each sample tested in duplicate (technical replicates) and in the absence of reverse transcriptase (RT negative controls), in a run that also contains quantification standards and negative PCR controls, such that the set of twelve samples are simultaneously assayed for two different miRNAs. For example, one may assay a reference miRNA and a test miRNA for all twelve samples in the same run. When this is repeated for the testing of additional samples, the on-plate standard curves also serve to monitor the assay performance and additionally act as plate-to-plate normalization controls.
Time Considerations
After designing the nucleic acid reagents, obtaining them can take up to several days. One hold-up can be in ensuring good quality control of some small RNA sequences (i.e. the quantification standards). The stem-loop primer can be re-folded to a stable structure overnight in a standard thermo-cycler. Once the working stocks are prepared and aliquots properly stored, it is important to test the assay on the quantification standard before committing experimental samples to the assay. A simple dilution series can be made in small test tubes with and without RT, reverse transcribed, run on the real time instrument. This is to establish the dynamic range of the particular assay, and ensure that the reagents are working as expected. Preparation of the working stocks and testing the assay can be performed in one to two days.
Preparation of the experimental samples is likely to take longer. They can be accumulated as total RNA or <40nt RNA and stored at -70 to -80°C. The MirVana kit allows processing of as many as twelve samples at one time. FlashPAGE isolation takes ∼12 minutes per sample for the electrophoresis and is followed by purifying the group of samples together. Thus, allow two days or more for sample preparation. Once the samples and reagents are assembled, a single plate containing two assays and twelve samples can easily be run in one day. This method can also be adapted to a 384-well format. The set-up of the RT step is the time-limiting step. Using a 384-well format would ideally be performed robotically. Alternatively, four separate RT plates can be set-up and reverse transcribed and can be stored at -20°C. When four plates are ready, they can be transferred to a single 384-well plate for qPCR. Although this may seem like a higher-throughput option, the actual set-up time is not changed, and without robotic assistance it may be overwhelming and potentially compromise one's experimental samples.
Running one 96-well plate with two assays per plate per day permits keeping abreast of the data analysis and allows for more continuous monitoring of the assay performance.
Acknowledgments
This work was performed in the laboratory of Dr. Don Coen. The author warmly thanks Dr. Coen for his generous support. The author also warmly thanks Ramey Packer, Seamus McCarron, Jean Pesola, and Igor Jurak for their critical reading of this manuscript.
Footnotes
Internet Resources
http://www.idtdna.com/analyzer/Applications/OligoAnalyzer/
When one types in an oligo sequence, this internet tool returns the complement sequence, length, GC content, melting temperature, molecular weight, molecular extinction coefficient, nmol/OD260, and μg/OD260. This resource is very user-friendly.
http://bioinformatics.gene-quantification.info
This resource offers bioinformatical, biostatical as well as multi-dimensional expression software tools, which are well described in Logan, 2009, Chapter 5, Data Analysis Software, by Michael W. Pfaffl, Jo Vandesompele, and Mikael Kubista.
Literature Cited
- Burroughs AM, Ando Y, de Hoon MJL, Tomaru Y, Nishibu T, Ukekawa R, Funakoshi T, Kurokawa T, Suzuki H, Hayashizaki Y, Daubl CO. A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Res. 2010;20:1398–1410. doi: 10.1101/gr.106054.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clinical Chemestry. 2009;55:1–12. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Realtime quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui C, Griffiths A, Li G, Silva L, Kramer MF, Gaasterland T, Wang XJ, Coen DM. Prediction and identification of herpes simplex virus 1-encoded microRNAs. JVirol. 2006;80:5499–5508. doi: 10.1128/JVI.00200-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen B. Transcription and processing of human microRNA precursors. Molec Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Ibrahim F, Rymarquis LA, Kim EJ, Becker J, Balassa E, Green PJ, Cerutti H. Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc Natl Acad Sci USA. 2010;107:3906–11. doi: 10.1073/pnas.0912632107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurak I, Kramer MF, Mellor JC, van Lint AL, Roth FP, Knipe DM, Coen DM. Numerous conserved and divergent microRNAs expressed by herpes simplex viruses 1 and 2. JVirol. 2010;84:4659–4672. doi: 10.1128/JVI.02725-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan J, Edwards K, Saunders N. Real-time PCR: Current Technology and Applications. Norfolk, UK: Caister Academic Press; 2009. [Google Scholar]
- Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454:780–783. doi: 10.1038/nature07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Key References
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry. 2009;55:1–12. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]; This seminal paper provides sensible and comprehensive guidelines for setting up, conducting, analyzing, and reporting the results of real-time PCR experiments
- Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, Lao KQ, Livak KJ, Guegler KJ. Realtime quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper describes the stem-loop approach and demonstrates the specificity the stem loop primer (and coupled PCR reagents) for the mature miRNA and for its precise sequence