Abstract
The cocrystal structure of the PP7 bacteriophage coat protein in complex with its translational operator identifies a distinct mode of sequence-specific RNA recognition when compared to the well-characterized MS2 coat protein–RNA complex. The structure reveals the molecular basis of the PP7 coat protein’s ability to selectively bind its cognate RNA, and it demonstrates that the conserved β-sheet surface is a flexible architecture that can evolve to recognize diverse RNA hairpins.
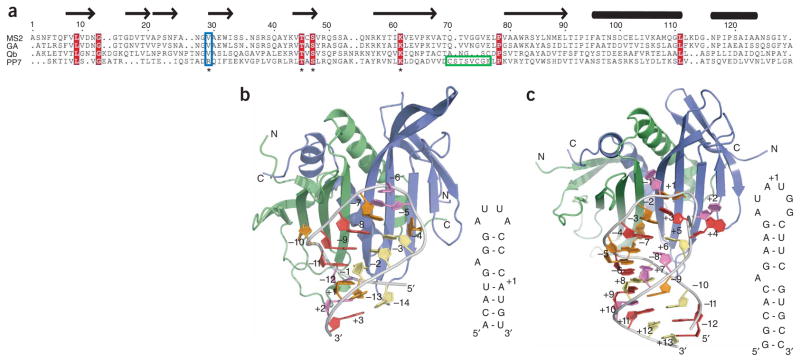
The inherent sequence diversity generated by RNA replication, which allows RNA viruses to evolve, must be tempered by the need for conservation of functional elements within the viral genome. The coat proteins of single-stranded (ss) RNA bacteriophages have both structural and regulatory roles in the viral life cycle, as they assemble into the mature capsid and regulate translation of the phage replicase through binding and sequestration of an RNA hairpin that contains the initiation site1. The coat protein and translational operator form a pair that must coevolve, so that mutation in one must be accompanied by compensatory mutations in the other. The Pseudomonas aeruginosa bacteriophage PP7 is a model system for exploring coevolution because of the extensive sequence divergence of both the PP7 coat protein and translational operator from other ssRNA bacteriophages2. The PP7 and MS2 coat proteins share only 15% sequence identity, and their cognate RNA hairpins differ in the position of the bulged adenosine as well as in the size and nucleotide composition of the loop (Fig. 1a). Biochemical experiments have shown that both coat proteins bind their own RNA hairpins with high affinity (Kd ~ 1 nM) and are able to discriminately bind in favor of their own RNA by ~1,000-fold3,4. In contrast to the PP7 coat protein, the Qβ coat protein also shares low sequence identity (21%) with the MS2 coat protein, yet it uses an RNA-binding mode similar to that of the MS2 coat protein5–7.
Figure 1.
Coat-protein sequence alignment and overview of the MS2 coat protein and PP7ΔFG complexes with RNA. (a) Alignment of four ssRNA bacteriophage coat proteins. (b,c) MS2 coat protein–RNA (2BU1) (b) and PP7ΔFG–RNA (c) complexes are shown as cartoons. In both structures the RNA hairpin (orange, adenine; red, guanine; violet, uridine; yellow, cytidine) binds across the extended β-sheet surface formed by the coat protein dimer (blue, green).
To investigate the structural basis of RNA recognition by the PP7 coat protein, we determined crystal structures of a truncated version of the protein that is deficient in capsid assembly (PP7ΔFG) both in the unbound form (1.6 Å resolution) and in complex with a 25-nt RNA hairpin (2.4 Å resolution) (Supplementary Table 1, Supplementary Fig. 1 and Supplementary Methods online). PP7ΔFG was generated by removing residues 67–75 and, notably, this truncation binds its translational operator with affinity similar to that of the full-length protein (Supplementary Fig. 2 online). The PP7ΔFG monomer adopts the topology characteristic of ssRNA bacteriophage coat proteins, with an N-terminal β hairpin, a five-stranded antiparallel β sheet and two C-terminal α helices (Supplementary Fig. 3a online)8–11. Antiparallel association of protomers in the dimer positions the last β strands adjacent to one another, resulting in a ten-stranded β sheet that comprises the RNA-binding surface. The interwoven packing of the α helices between each other and the N-terminal β hairpins further stabilizes the dimer, combining with the β sheet interface to bury ~3,000 Å 2 of solvent-accessible surface area. The PP7ΔFG dimer has almost identical free and bound structures (r.m.s. deviation < 0.9 Å for all Cα atoms) and is similar to that of the MS2 coat protein (r.m.s. deviation of < 2.3 Å for all Cα atoms) (Supplementary Fig. 3b).
The global organization of all six PP7ΔFG–RNA complexes in the asymmetric unit resembles that of the MS2 coat protein–RNA complex, despite the differences in RNA targets (Fig. 1b,c). The RNA hairpin binds across the PP7ΔFG dimer interface and buries ~950 Å 2 of solvent-accessible surface area. The RNA stem comprises five base pairs below and four base pairs above the bulged adenosine (A(−7)) and adopts the standard A-form helical conformation. All of the ribose sugars in the hairpin have C3′-endo sugar puckers except for four (A(−7), A+1, G+3, G+4) that adopt C2′-endo conformations.
In the RNA loop, three of the six bases (A(−2), A+1 and G+3) form a purine stack that continues the base stacking of the helical stem (Fig. 2a). The foundation of this stack is formed by A–2, which stacks directly upon the G(−3)–C+5 base pair at the top of the stem. The RNA backbone kinks at the U+2 phosphate and positions the G+3 base in the middle of the purine stack between the A(−2) and A+1 bases. The U(−1) base is flipped out and away from the purine stack, and its conformation is stabilized by interactions with the PP7ΔFG dimer. Both U+2 and G+4 extend away from the RNA loop as well, but their role in stabilizing the complex is unclear because they do not form equivalent interactions in the six complexes within the asymmetric unit. In complexes that do not make crystal contacts to this region, the electron density for the U+2 and G+4 bases is weak, suggesting that these nucleotides do not play a prominent role in binding. The structure of the RNA loop is consistent with previous SELEX experiments that found that U(−1), A+1 and G+3 were conserved and that the nucleotide in position −2 must be a purine12.
Figure 2.
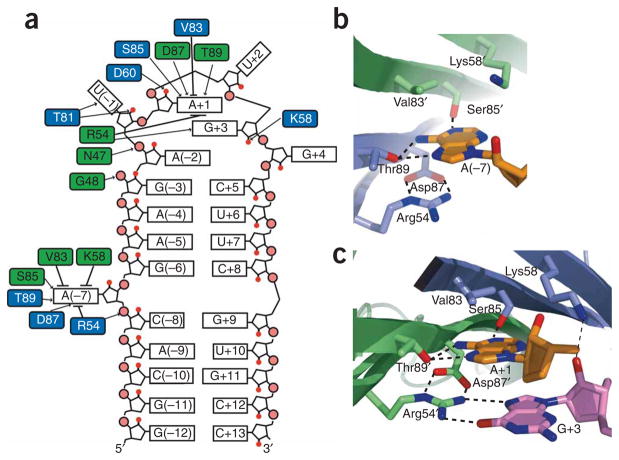
RNA-protein interface. (a) Schematic representation of PP7ΔFG interactions with the RNA hairpin. Black arrows, hydrogen bonds; black lines, van der Waals and stacking interactions. (b,c) The A(−7) (b) and A+1 (c) recognition pockets are shown, with the adenosine nucleotides (orange) and PP7ΔFG residues (blue, green) as sticks. Potential hydrogen bonds are shown as dashed lines.
The bulged adenosine (A(−7)) and the adenosine that forms the top level of the purine stack (A+1) are recognized by nearly identical interactions made by symmetrically positioned pockets on the PP7ΔFG dimer (Fig. 2a–c). The binding pockets are located along the dimer interface and are formed by six residues (Arg54, Lys58, Val83, Ser85, Asp87 and Thr89), with each protomer contributing three residues. Because many of the interactions are redundant, recognition of A(−7) will be described in detail and only the differences found in the A+1 binding pocket will be highlighted. For clarity, the prime symbol denotes residues located on the opposite protomer of the PP7ΔFG dimer.
The upper surface of the A(−7) binding pocket is formed by the hydrophobic side chain of Val83′ and the aliphatic portion of the Lys58′ side chain, which make van der Waals contacts with the base (Fig. 2b,c). In the A+1 pocket, the side chain amine of Lys58 hydrogen bonds to the 2′OH of G+3 and likely stabilizes its C2′-endo sugar pucker (Fig. 2c). Asp87, Thr89 and Ser85′ form the middle level of the pocket and make sequence-specific contacts to the adenine base. The side chain OH of Thr89 is within hydrogen-bonding distance of both the adenine N7 and N6 exocyclic amines. The backbone carbonyl of Asp87 is also within hydrogen-bonding distance of the N6 exocyclic amine. The third RNA-protein interaction at this level is a hydrogen bond between the OH of Ser85′ and adenine N1. Arg54, whose guanidinium group makes a cation-π stacking interaction with the adenine base, forms the base of the binding pocket. The position of the Arg54 side chain is buttressed by hydrogen bonds to the side chain of Asp87. In the A+1 pocket, the Arg54′ guanidinium group makes two hydrogen bonds to O6 and N7 of G+3 to specifically recognize the guanine base. (Fig. 2c). The importance of Lys58 and Arg54 in RNA recognition is supported by experiments that demonstrate that mutation of these residues leads to severe repression defects4.
The PP7ΔFG–RNA complex is further stabilized by several hydrogen bonds and electrostatic interactions outside of the adenine recognition pockets. The backbone amide of Gly48′ and the side chain of Asn47′ make hydrogen bonds to phosphate oxygens of A(−2) and U(−1), respectively. The U(−1) nucleotide is extended away from the loop into a pocket formed by Thr51′, Ala52′, Val91′ and Thr81. The backbone amide and side chain OH of Thr81 make hydrogen bonds to the O2 and 2′-OH of U(−1). Another potential hydrogen bond exists between the side chain carboxylic acid of Asp60 and the A+1 O2′, which may stabilize its C2′-endo sugar pucker. A similar interaction is observed in the MS2 coat protein–RNA complex, where Glu63 is hydrogen bonded to the U(−5) 2′ OH13. Crystal-packing differences result in the side chain of Arg24 stacking with the guanine base of G+4 and contacting either its O4 or phosphate oxygens. Weak electron density and alternate conformations of the Arg24 side chain suggest that this interaction does not contribute substantially to the overall affinity of complex. There are also several positively charged residues (Arg24′, Arg39, Arg45′) that may participate in favorable electrostatic interactions with the phosphate backbone of the RNA.
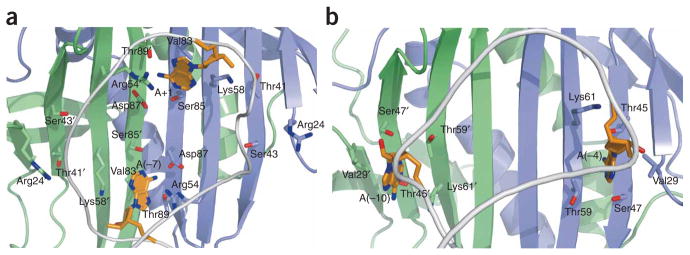
Although the PP7 and MS2 coat proteins share similar protein scaffolds, their RNA-binding surfaces have evolved to specifically recognize distinct RNA hairpins. The most notable difference between the two structures is the location of the adenine-recognition pockets, which are important components of binding for both coat proteins12,14,15. In the PP7 coat protein, the pockets are aligned along the dimer axis and are formed by residues from both protomers (Fig. 3a). These pockets are rotated ~90° with respect to the dimer axis in the MS2 coat protein and consequently are composed of residues from only one protomer (Fig. 3b). In the MS2 coat protein, four residues (Val29, Thr45, Ser47, Lys61; MS2 numbering) that are highly conserved in ssRNA bacteriophage coat proteins form the A(−4) and A(−10) binding pockets (Figs. 1a and 3b)15. Because of the different orientation of the binding pockets in the PP7 and MS2 coat proteins, these conserved residues have distinct functions. Of the four residues, only the conserved lysine (Lys61 in MS2 and Lys58 in PP7) is involved in RNA binding in both complexes, albeit via different interactions (Fig. 3a,b). In the PP7 coat protein, the aliphatic portion of the Lys58 side chain combines with Val83 to form the top wall of the pocket, whereas in the MS2 coat protein, Lys61 forms the bottom wall. In the MS2 coat protein, Thr45 and Ser47 line the A(−4) and A(−10) binding pockets and recognize the adenine base; however, Ser85 and Thr89, not Thr41 and Ser43, perform the analogous role in the PP7 coat protein. Consistent with the interactions found in the PP7ΔFG cocrystal structure, mutagenesis of Thr41 and Ser43 demonstrated that these residues do not contribute to RNA binding4. The hydrophobic Val29 side chain in the MS2 coat protein forms the top surface of the binding pockets, but in the PP7 coat protein this residue is replaced by an arginine (Arg24). This substitution to a much larger polar side chain is unique among phage coat proteins to PP7 and may be a key determinant in preventing interaction with the MS2 hairpin.
Figure 3.
Orientation of adenine recognition pockets. (a,b) The position of the adenine recognition pockets is rotated approximately 90° with respect to the dimer axis between the PP7ΔFG (a) and MS2 (b) coat proteins. Cartoon ribbon shows the position of the phosphate backbone (gray) and bound adenosines are shown as sticks (orange).
The PP7 and MS2 coat proteins represent two distinct solutions to the problem of sequence-specific recognition of an asymmetric RNA hairpin by a symmetric binding surface. The coevolution of the PP7 coat protein and its translational operator has resulted in the formation of adenine recognition pockets whose orientation relative to the dimer axis differ considerably from those of the MS2 coat protein. This dramatic rearrangement explains the inability of the two coat proteins to bind the other’s RNA target. The extended β-sheet surface revealed by the PP7ΔFG-RNA structure is a flexible scaffold capable of recognizing diverse RNAs with high specificity. By engineering coat proteins with altered RNA target specificities, it will be possible to expand the experimental applications of ssRNA bacteriophage coat proteins in RNA affinity purification, tethering and fluorescence labeling of RNAs for live cell imaging16–19.
Supplementary Material
Acknowledgments
This work was supported by the US National Institutes of Health (grants AR-41480 and EB-002060 to R.H.S and National Research Service Award institutional training grant (5T32HL007675-19) support to J.A.C.) and the Albert Einstein Cancer Center. The authors wish to thank the staff at the National Synchrotron Light Source X29a beamline for assistance with data collection, the staff at the Argonne Advanced Photon Source Structural GenomiX Collaborative Access Team beamline for express crystallography data collection and G. Arenas, M. Hennig, U. Meier, S. Nguyen and S. Ryder for helpful discussions.
Footnotes
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
AUTHOR CONTRIBUTIONS
J.A.C. designed and performed the experiments. Y.P. assisted with crystallography. J.A.C., Y.P., S.C.A. and R.H.S. wrote and discussed the manuscript.
Note: Supplementary information is available on the Nature Structural & Molecular Biology website.
Accession codes. Protein Data Bank: Coordinates have been deposited with accession codes 2QUD (PP7ΔFG) and 2QUX (PP7ΔFG–RNA complex).
References
- 1.Kozak M, Nathans D. Bacteriol Rev. 1972;36:109–134. doi: 10.1128/br.36.1.109-134.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olsthoorn RC, Garde G, Dayhuff T, Atkins JF, Van Duin J. Virology. 1995;206:611–625. doi: 10.1016/s0042-6822(95)80078-6. [DOI] [PubMed] [Google Scholar]
- 3.Carey J, Cameron V, de Haseth PL, Uhlenbeck OC. Biochemistry. 1983;22:2601–2610. doi: 10.1021/bi00280a002. [DOI] [PubMed] [Google Scholar]
- 4.Lim F, Downey TP, Peabody DS. J Biol Chem. 2001;276:22507–22513. doi: 10.1074/jbc.M102411200. [DOI] [PubMed] [Google Scholar]
- 5.Spingola M, Peabody DS. Nucleic Acids Res. 1997;25:2808–2815. doi: 10.1093/nar/25.14.2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim F, Spingola M, Peabody DS. J Biol Chem. 1996;271:31839–31845. doi: 10.1074/jbc.271.50.31839. [DOI] [PubMed] [Google Scholar]
- 7.Horn WT, et al. Structure. 2006;14:487–495. doi: 10.1016/j.str.2005.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Valegard K, Liljas L, Fridborg K, Unge T. Nature. 1990;345:36–41. doi: 10.1038/345036a0. [DOI] [PubMed] [Google Scholar]
- 9.Ni CZ, et al. Structure. 1995;3:255–263. doi: 10.1016/S0969-2126(01)00156-3. [DOI] [PubMed] [Google Scholar]
- 10.Golmohammadi R, Fridborg K, Bundule M, Valegard K, Liljas L. Structure. 1996;4:543–554. doi: 10.1016/s0969-2126(96)00060-3. [DOI] [PubMed] [Google Scholar]
- 11.Tars K, Bundule M, Fridborg K, Liljas L. J Mol Biol. 1997;271:759–773. doi: 10.1006/jmbi.1997.1214. [DOI] [PubMed] [Google Scholar]
- 12.Lim F, Peabody DS. Nucleic Acids Res. 2002;30:4138–4144. doi: 10.1093/nar/gkf552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valegard K, et al. J Mol Biol. 1997;270:724–738. doi: 10.1006/jmbi.1997.1144. [DOI] [PubMed] [Google Scholar]
- 14.Wu HN, Uhlenbeck OC. Biochemistry. 1987;26:8221–8227. doi: 10.1021/bi00399a030. [DOI] [PubMed] [Google Scholar]
- 15.Valegard K, Murray JB, Stockley PG, Stonehouse NJ, Liljas L. Nature. 1994;371:623–626. doi: 10.1038/371623a0. [DOI] [PubMed] [Google Scholar]
- 16.Bardwell VJ, Wickens M. Nucleic Acids Res. 1990;18:6587–6594. doi: 10.1093/nar/18.22.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hogg JR, Collins K. RNA. 2007;13:868–880. doi: 10.1261/rna.565207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lykke-Andersen J, Shu MD, Steitz JA. Cell. 2000;103:1121–1131. doi: 10.1016/s0092-8674(00)00214-2. [DOI] [PubMed] [Google Scholar]
- 19.Bertrand E, et al. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.