Abstract
Circadian rhythms are regulated by a synchronized system of central and peripheral clocks. Here we show that a clock in the Drosophila fat body drives rhythmic expression of genes involved in metabolism, detoxification, the immune response and steroid hormone regulation. Some of these genes cycle even when the fat body clock is disrupted indicating they are regulated by exogenous factors. Food is an important stimulus as limiting food availability to a six-hour interval each day drives rhythmic expression of genes in the fat body. Restricting food to a time of day when consumption is typically low desynchronizes internal rhythms because it alters the phase of rhythmic gene expression in the fat body without affecting the brain clock. Flies maintained on this paradigm produce fewer eggs than those restricted to food at the normal time. These data suggest that desynchrony of endogenous rhythms, caused by aberrant feeding patterns, affects reproductive fitness.
INTRODUCTION
The circadian system controls many aspects of behavior and physiology, such as rest:activity or sleep/wake, body temperature, blood pressure, release of endocrine hormones and metabolic activity. Not surprisingly, these diverse rhythms are controlled by clocks located in different tissues. Thus, while a well-characterized clock in the central nervous system is sufficient to drive behavioral rhythms of rest:activity, clocks in peripheral tissues are required for the circadian control of many other processes, including metabolism (Allada and Chung, 2010; Glossop and Hardin, 2002; Green et al., 2008).
A major peripheral organ that contains its own clock is the mammalian liver. The liver clock incorporates signals from both the central clock and the environment to control a number of physiological processes including glucose, lipid and cholesterol metabolism as well as detoxification (Gachon et al., 2006; Lamia et al., 2008). In the fruit fly, Drosophila melanogaster, functions of the liver are served by an organ called the fat body. We recently identified a clock in the fat body and showed that this clock regulates feeding behavior and overall nutrient storage (Xu et al., 2008). However, like the liver, the fat body is a very complex organ that also functions in other physiological processes such as innate immunity, detoxification and reproduction (Lazareva et al., 2007; Leclerc and Reichhart, 2004). Whether these other processes are controlled by the clock in the fat body is not known; nor do we know how the fat body clock regulates metabolism. However, given that transcriptional regulation is a major component of clock output, the fat body clock likely exerts much of its control by driving rhythmic gene expression.
Typically, central and peripheral clocks are synchronized with each other and with environmental cycles (Maywood et al., 2007). However, restricted food availability can entrain clocks in metabolic tissues like liver, but not the brain, thereby leading to internal desynchrony (Damiola et al., 2000; Stokkan et al., 2001). This type of desynchrony is probably a common occurrence in modern society where people often eat at odd hours; however, the consequences are only starting to be realized (Arble et al., 2009). In general, little is known about the consequences of circadian disruption in animal systems. While experiments performed in cyanobacteria and plants have suggested that clocks provide a selective advantage to the organism (Johnson, 2001), such an effect of clocks on fitness has been difficult to demonstrate in animals. Only marginal effects on lifespan are observed in Drosophila that lack or have altered clocks (Klarsfeld and Rouyer, 1998), most likely because laboratory conditions fail to provide a competitive and selective environment. Another approach to assay the role of clocks in animal physiology and fitness would be to determine the effects of internal circadian desynchrony on organism function.
In this study, we used Drosophila to understand the control of physiology by peripheral oscillators. To this end, we performed microarray analysis on fat bodies throughout the circadian day to identify cycling transcripts controlled by the fat body clock. We then sought to determine the role of metabolic signals such as food availability in controlling the function of this peripheral clock and the central clock, using a restricted feeding paradigm. We report that a restricted feeding paradigm can drive rhythmic expression of circadian genes in the fat body, but does not affect expression of the same genes in the brain. As a result, a restricted feeding paradigm that provides food at the “wrong” time of day produces desynchrony between the clocks located in the fat body and the brain. Interestingly, such mistimed feeding affects the number of eggs produced, suggesting that synchrony of rhythms across tissues is important for overall organismal fitness.
RESULTS
Identification of cyclically expressed genes in the fat body
To identify circadian-regulated genes in the fat body of the adult fly, we performed high-resolution microarray analysis. We collected male, wildtype flies (UAS-DNCLK/+) every two hours over a 48 hour period in a light:dark cycle, dissected their fat bodies and isolated RNA that we then used to probe Affymetrix Drosophila microarrays (Figure 1). We used two statistical algorithms to identify cycling transcripts: COSOPT, which fits experimental data to cosine waves with varying phases and period lengths (Straume, 2004), and the Fisher's G-test, which uses Fourier transformations to screen for sinusoidal components (Wichert et al., 2004). From this analysis, 214 transcripts had p-values less than 0.05 for both algorithms and were thus considered rhythmic. To reduce false positives, the expression curves of each of these transcripts were screened manually by the following two criteria. To avoid less robust rhythms, we required at least a two-fold difference between peak and trough mRNA levels. Second, we required that the phase of expression be constant over the two-day experiment. These criteria whittled our list down to 137 fat body transcripts (Table 1).
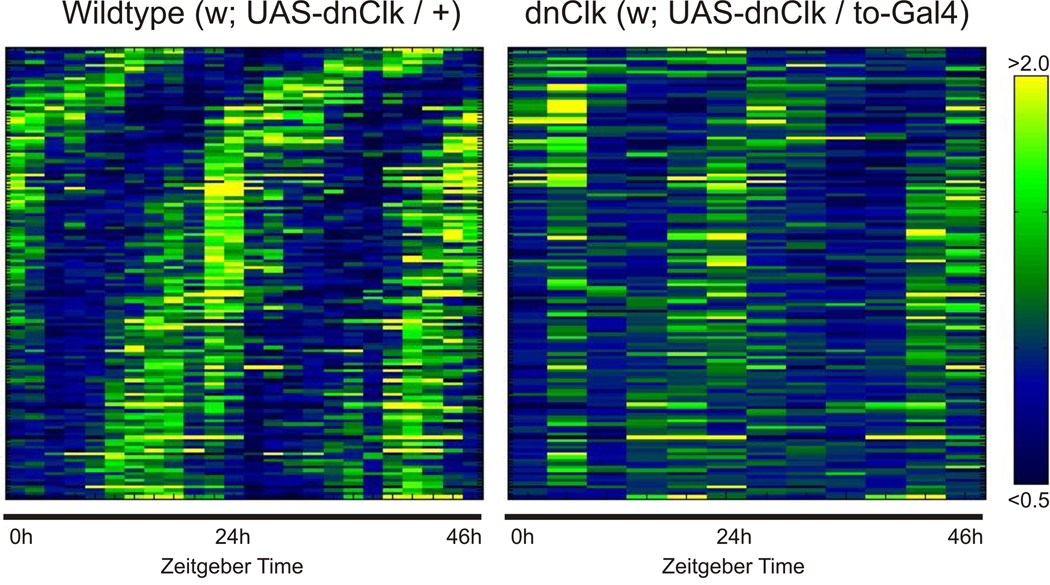
Figure 1. Rhythmic gene expression in the fat body is affected by expression of DN-CLK.
Microarray analysis was used to detect cycling transcripts in the fat bodies of wild type flies (left) and flies expressing DN-Clk flies (right). Individual transcripts were median normalized, sorted by phase, and plotted as a heatmap (yellow = high expression, blue = low expression).
Table 1.
Cyclically expressed genes in the Drosophila fat body. The 137 genes listed here were identified as described in the text. The “rhythm in DN-CLK” column indicates those whose expression profile was affected by disruption of the fat body clock.
| Gene | pMMC-β | FisherG-p | Molecular Function | rhythm in DNCLK fat body a |
Time of peak expression |
|---|---|---|---|---|---|
| Lipid metabolismb | |||||
| Sxe2 | 2.74E-04 | 4.68E-07 | phosphatidylserine-specific phospholipase A1 activity | R | ZT1-4 |
| CG31683 | 9.97E-03 | 4.98E-02 | phospholipase activity; phosphatidylcholine-sterol O-acyltransferase activity | AR | ZT17-20 |
| CG6432 | 1.57E-03 | 8.78E-04 | acetate-CoA ligase activity | AR | ZT13-16 |
| CG5077 | 4.20E-03 | 2.56E-03 | oxysterol binding | AR | ZT1-4 |
| CG31810 | 1.08E-02 | 3.19E-03 | oxidoreductase activity; steroid dehydrogenase activity | R | ZT1-4 |
| CG33110 | 1.82E-02 | 3.97E-03 | GNS1/SUR4 membrane protein, long chain fatty acid enlongase activity | AR | ZT9-12 |
| CG2781 | 2.71E-02 | 2.52E-02 | GNS1/SUR4 membrane protein, long chain fatty acid elongase activity | AR | ZT17-20 |
| CG16905 | 2.40E-03 | 4.53E-02 | GNS1/SUR4 membrane protein, long chain fatty acid elongase activity | AR | ZT17-20 |
| CG18609 /// | 3.43E-04 | 1.25E-06 | GNS1/SUR4 membrane protein, long chain fatty acid enlongase activity | R | ZT21-0 |
| DyakCG18609 | |||||
| CG9459 | 9.21E-04 | 8.93E-05 | GNS1/SUR4 membrane protein, long chain fatty acid enlongase activity | AR | ZT21-0 |
| CG9458 | 2.43E-03 | 4.38E-03 | GNS1/SUR4 membrane protein, long chain fatty acid enlongase activity | AR | ZT17-20 |
| CG9009 | 9.94E-03 | 5.28E-03 | long-chain-fatty-acid-CoA ligase activity | AR | ZT21-0 |
| CG7149 | 1.82E-03 | 2.45E-03 | diacylglycerol cholinephosphotransferase activity | AR | ZT5-8 |
| Nucleotide and nucleic acid metabolism | |||||
| CG5537 | 7.12E-03 | 5.07E-03 | uracil phosphoribosyltransferase activity | AR | ZT17-20 |
| CG5800 | 3.17E-02 | 2.43E-02 | RNA helicase activity; ATP-dependent helicase activity | AR | ZT21-0 |
| CG4038 | 7.70E-03 | 9.47E-03 | rRNA pseudouridylation guide activity; snoRNA binding | AR | ZT21-0 |
| Rrp46 | 4.12E-02 | 1.12E-02 | 3'-5'-exoribonuclease activity; RNA binding | AR | ZT21-0 |
| CG5728 | 2.27E-02 | 4.83E-02 | rRNA processing; regulation of alternative nuclear mRNA splicing, via spliceosome |
AR | ZT21-0 |
| CG14712 | 3.40E-03 | 1.11E-02 | aminoacyl-tRNA ligase activity | AR | ZT21-0 |
| Carbohydrate metabolism | |||||
| alpha-Man- | 7.67E-04 | 1.66E-05 | mannosyl-oligosaccharide 1,3-1,6-alpha-mannosidase activity | R | ZT21-0 |
| Gfat1 | 7.49E-03 | 2.27E-04 | glutamine-fructose-6-phosphate transaminase (isomerizing) activity | AR | ZT17-20 |
| Zw | 1.58E-03 | 8.48E-04 | oxidoreductase activity, glucose-6-phosphate dehydrogenase activity | AR | ZT9-12 |
| Transportersb | |||||
| CG4797 | 2.68E-03 | 1.41E-03 | Major facilitator superfamily | AR | ZT13-16 |
| CG6126 | 1.61E-03 | 3.35E-03 | Major facilitator superfamily, organic cation transmembrane transporter activity | AR | ZT13-16 |
| CG6574 | 1.47E-02 | 1.31E-02 | Major facilitator superfamily, reduced folate carrier activity | AR | ZT21-0 |
| CG6231 | 2.59E-03 | 1.96E-03 | Major facilitator superfamily, secondary active organic cation transmembrane transporter activity |
AR | ZT13-16 |
| CG8389 | 1.35E-02 | 1.57E-02 | Major facilitator superfamily, secondary active monocarboxylate transmembrane transporter activity |
AR | ZT13-16 |
| CG8034 /// | 9.05E-03 | 6.60E-03 | Major facilitator superfamily | AR | ZT13-16 |
| DmirCG8034 | |||||
| CG1358 | 3.24E-02 | 1.24E-02 | Major facilitator superfamily | AR | ZT13-16 |
| CG6600 | 7.88E-03 | 9.16E-03 | Major facilitator superfamily (* when input CG6600 in Flybase, it turn out to be CG42269) |
AR | ZT13-16 |
| kar | 2.91E-02 | 7.20E-03 | Major facilitator superfamily, monocarboxylic acid transmembrane transporter activity |
AR | ZT13-16 |
| CG11147 | 6.87E-03 | 7.28E-03 | ATP-Binding Cassette (ABC) superfamily, ABC-2 type transporter, | AR | ZT17-20 |
| CG33970 | 3.60E-02 | 2.87E-02 | ATP-Binding Cassette (ABC) superfamily, ABC-2 type transporter | AR | ZT17-20 |
| Atet | 6.99E-03 | 3.88E-02 | ATP-Binding Cassette (ABC) superfamily, ABC-2 type transporter | AR | ZT17-20 |
| CG32792 | 1.93E-02 | 5.25E-03 | amiloride-sensitive sodium channel activity | AR | ZT21-0 |
| Esp | 7.61E-03 | 3.16E-03 | secondary active sulfate transmembrane transporter activity | AR | ZT21-0 |
| frc | 2.59E-02 | 4.79E-02 | pyrimidine nucleotide sugar transmembrane transporter activity | AR | ZT13-16 |
| Kinase | |||||
| CG10560 | 4.25E-02 | 3.19E-04 | protein kinase like, CHK_kinase-like | AR | ZT21-0 |
| CG31974 | 2.81E-03 | 1.93E-03 | protein kinase like, CHK_kinase-like | AR | ZT21-0 |
| CHKov2 | 1.62E-02 | 2.26E-02 | protein kinase like, CHK_kinase-like | AR | ZT21-0 |
| otk | 1.21E-03 | 5.46E-04 | transmembrane receptor protein tyrosine kinase activity | AR | ZT1-4 |
| CG3608 | 5.80E-03 | 4.93E-03 | protein kinase activity | AR | ZT17-20 |
| Phosphatase | |||||
| Ptp99A | 7.80E-03 | 6.29E-03 | transmembrane receptor protein tyrosine phosphatase activity | AR | ZT21-0 |
| CG32984 | 2.52E-02 | 1.28E-02 | catalytic activity; Alkaline-phosphatase-like | AR | ZT21-0 |
| Proteolysis | |||||
| CG31205 | 4.06E-04 | 5.43E-05 | serine-type endopeptidase activity | AR | ZT1-4 |
| asparagines- | 2.51E-03 | 3.38E-03 | asparagine synthase (glutamine-hydrolyzing) activity; serine-type | AR | ZT5-8 |
| synthetase | endopeptidase activity | ||||
| CG7220 | 4.31E-02 | 2.48E-02 | ubiquitin-protein ligase activity | AR | ZT13-16 |
| Usp36 | 1.03E-03 | 4.27E-03 | ubiquitin thiolesterase activity | AR | ZT21-0 |
| CG14218 | 3.07E-03 | 2.39E-02 | serine-type endopeptidase activity | AR | ZT21-0 |
| CG5639 | 1.25E-02 | 4.38E-02 | serine-type endopeptidase inhibitor activity | AR | ZT5-8 |
| CG7695 | 3.22E-02 | 1.24E-02 | Proteinase inhibitor I1, Kazal | AR | ZT5-8 |
| Oxidoreductase | |||||
| CG40486 | 4.61E-02 | 2.13E-05 | oxidoreductase activity | R | ZT13-16 |
| CG40485 | 5.08E-03 | 2.55E-02 | oxidoreductase activity | AR | ZT17-20 |
| CG4199 | 1.45E-02 | 1.49E-02 | oxidoreductase activity | AR | ZT13-16 |
| CG13833 | 6.67E-04 | 9.78E-08 | oxidoreductase activity | R | ZT21-0 |
| CG17562 | 8.82E-04 | 7.45E-10 | oxidoreductase activity | R | ZT1-4 |
| CG17560 | 2.78E-03 | 1.45E-08 | oxidoreductase activity | R | ZT1-4 |
| CG2065 | 2.87E-02 | 3.88E-02 | oxidoreductase activity | AR | ZT13-16 |
| CG10131 | 1.61E-02 | 2.84E-03 | oxidoreductase activity | AR | ZT9-12 |
| CG15629 | 1.20E-02 | 7.95E-04 | oxidoreductase activity | AR | ZT13-16 |
| CG1441 | 1.26E-02 | 9.57E-04 | reductase | AR | ZT21-0 |
| CG13091 | 7.26E-03 | 1.51E-03 | reductase | AR | ZT21-0 |
| Other metabolic enzymes | |||||
| CAH1 | 3.48E-02 | 2.66E-02 | carbonate dehydratase activity | R | ZT21-0 |
| Ahcy89E | 5.65E-03 | 1.77E-02 | adenosylhomocysteinase activity | AR | ZT13-16 |
| Jhe | 4.97E-03 | 5.47E-07 | juvenile-hormone esterase activity | AR | ZT21-0 |
| CG17323 | 3.05E-03 | 6.19E-08 | glucuronosyltransferase activity | R | ZT1-4 |
| CG13185 | 3.16E-03 | 7.84E-03 | ATPase activity | AR | ZT21-0 |
| alpha-Est10 | 4.27E-02 | 9.07E-03 | carboxylesterase activity | AR | ZT17-20 |
| CG13397 | 3.87E-02 | 2.02E-02 | alpha-N-acetylglucosaminidase activity | AR | ZT13-16 |
| Pde8 | 1.05E-02 | 3.60E-02 | 3',5'-cyclic-AMP phosphodiesterase activity; two-component response regulator activity |
AR | ZT21-0 |
| CG14934 | 1.68E-02 | 3.81E-02 | alpha-glucosidase activity; cation binding | R | ZT9-12 |
| e | 4.31E-03 | 9.42E-05 | beta-alanyl-dopamine synthase activity | R | ZT21-0 |
| CG13937 | 3.02E-02 | 1.02E-02 | HNK-1 sulfotransferase activity | AR | Zt17-20 |
| CG5156 | 3.61E-03 | 4.33E-05 | transferase activity, transferring acyl groups other than amino-acyl groups | R | ZT21-0 |
| Detoxification and other defense response | |||||
| Pli | 3.18E-03 | 3.06E-05 | Toll signaling pathway; signal transduction; immune response | AR | ZT17-20 |
| dro5 | 9.43E-04 | 1.18E-04 | molecular function is unknown | AR | ZT5-8 |
| CecC | 1.39E-02 | 4.33E-03 | Cecropin(potent antibacterial proteins) | AR | ZT21-0 |
| gk | 4.04E-03 | 8.93E-04 | molecular function is unknown | R | ZT21-0 |
| CG5846 | 6.44E-04 | 7.37E-05 | transcriptional factor, protein with the ankyrin repeat | AR | ZT17-20 |
| b6 | 1.49E-02 | 1.13E-02 | Concanavalin A-like lectin/glucanase, neuronal pentraxin receptor activity | AR | ZT1-4 |
| Cyp4d21 | 1.02E-03 | 1.14E-10 | electron carrier activity; monooxygenase activity | R | ZT17-20 |
| Cyp6a20 | 1.72E-03 | 5.38E-04 | electron carrier activity; monooxygenase activity | AR | ZT21-0 |
| Cyp304a1 | 5.68E-03 | 8.42E-03 | electron carrier activity; monooxygenase activity | AR | ZT17-20 |
| Cyp313a4 | 6.44E-03 | 3.54E-03 | electron carrier activity; monooxygenase activity | AR | ZT13-16 |
| Cyp6a21 | 3.25E-03 | 6.25E-04 | electron carrier activity; monooxygenase activity | R | ZT21-0 |
| Hormone binding | |||||
| CG1124 /// | 1.22E-03 | 5.08E-05 | odorant and hormone binding | AR | ZT5-8 |
| DyakCG1124 | |||||
| CG31189 | 2.11E-03 | 1.34E-04 | odorant and hormone binding | R | ZT21-0 |
| CG7079 | 2.07E-03 | 3.82E-04 | odorant and hormone binding | R | ZT21-0 |
| CG11852 /// | 2.25E-02 | 6.23E-03 | odorant and hormone binding | AR | ZT13-16 |
| DyakCG11852 | |||||
| Transcription | |||||
| vri | 1.18E-04 | 3.20E-09 | transcription regulator activity; transcription factor activity | R | ZT13-16 |
| tim | 4.79E-03 | 7.57E-08 | protein binding; protein heterodimerization activity | R | ZT13-16 |
| per | 6.86E-03 | 4.63E-08 | transcription corepressor activity | R | ZT13-16 |
| Clk | 1.90E-02 | 1.45E-03 | transcription factor activity | AR | ZT1-4 |
| sug | 1.74E-02 | 2.90E-02 | zinc ion binding; nucleic acid binding | AR | ZT13-16 |
| cry | 1.86E-03 | 2.56E-02 | G-protein coupled photoreceptor activity, transcription repressor activity; | R | ZT9-13 |
| Receptor and signal transduction | |||||
| RhoGAP93B | 5.13E-04 | 4.21E-05 | protein binding. | AR | ZT21-0 |
| AR-2 | 1.49E-03 | 1.43E-02 | allatostatin receptor activity; neuropeptide receptor activity; neuropeptide Y receptor activity |
AR | ZT21-0 |
| CG1657 /// | 5.63E-03 | 3.32E-02 | Rab guanyl-nucleotide exchange factor activity; GTPase activator activity; | AR | ZT17-20 |
| DmirCG1657 | sugar:hydrogen symporter activity | ||||
| CG14782 | 7.00E-03 | 4.05E-02 | guanyl-nucleotide exchange factor activity; zinc ion binding | AR | ZT13-16 |
| sas | 1.41E-02 | 1.71E-03 | receptor activity | R | ZT5-8 |
| Structural molecules | |||||
| Act5C | 8.77E-03 | 2.80E-04 | structural constituent of cytoskeleton | AR | ZT9-12 |
| Cpr100A | 1.05E-03 | 7.43E-05 | structural constituent of chitin-based cuticle | AR | ZT21-0 |
| Cpr67B | 1.20E-02 | 3.53E-04 | structural constituent of chitin-based cuticle | AR | ZT21-0 |
| CG15021 | 2.10E-02 | 7.98E-04 | Pistil-specific extensin-like protein | AR | ZT21-0 |
| CG31534 | 3.73E-03 | 2.09E-02 | zinc ion binding | AR | ZT17-20 |
| Gasp | 3.51E-03 | 3.78E-03 | structural constituent of peritrophic membrane | AR | ZT21-0 |
| Cell adhesion | |||||
| drpr | 1.28E-03 | 3.63E-03 | protein binding | AR | ZT13-16 |
| Fas1 | 3.05E-03 | 8.42E-03 | adhesion molecule binding | AR | ZT21-0 |
| Unknown biological function | |||||
| CG8964 | 1.30E-02 | 2.96E-02 | Immunoglobulin-like | R | ZT21-0 |
| CG11275 | 4.04E-02 | 1.27E-02 | BTB/POZ-like, Kelch related, protein binding | AR | ZT21-0 |
| CG7047 | 1.99E-02 | 5.12E-03 | Immunoglobulin/major histocompatibility complex, Arrestin-like | AR | ZT21-0 |
| slv | 4.37E-02 | 3.44E-03 | MtN3 and saliva related transmembrane protein, RAG1-activating protein | AR | ZT17-20 |
| CG34402 | 4.52E-03 | 8.69E-03 | CUB domain | AR | ZT1-4 |
| CG14696 /// | 8.27E-03 | 3.06E-02 | Arrestin-like | AR | ZT9-12 |
| DvarCG14696 | |||||
| CG9080 | 5.24E-04 | 1.48E-06 | molecular function is unknown | AR | ZT21-0 |
| CG31607 | 1.30E-03 | 5.26E-05 | molecular function is unknown | AR | ZT17-20 |
| CG40198 | 3.92E-03 | 4.99E-06 | molecular function is unknown | AR | ZT21-0 |
| CG11409 /// | 1.77E-02 | 4.14E-06 | molecular function is unknown | R | ZT9-12 |
| DmirCG11409 | |||||
| CG30121 | 1.46E-02 | 6.59E-05 | molecular function is unknown | AR | ZT21-0 |
| CG32801 | 7.74E-03 | 2.32E-04 | molecular function is unknown | AR | ZT21-0 |
| CG7224 /// | 1.22E-02 | 4.29E-04 | molecular function is unknown | AR | ZT17-20 |
| DyakCG7224 | |||||
| CG15210 | 7.20E-03 | 5.35E-04 | molecular function is unknown | AR | ZT13-16 |
| CG5073 | 1.82E-03 | 7.17E-04 | molecular function is unknown | AR | ZT21-0 |
| CG33998 | 2.37E-02 | 1.75E-03 | molecular function is unknown | AR | ZT21-0 |
| CG31157 | 1.13E-03 | 2.79E-03 | molecular function is unknown | AR | ZT21-0 |
| CG32694 | 4.70E-03 | 4.47E-03 | molecular function is unknown | AR | ZT21-0 |
| CG9972 | 9.22E-03 | 4.47E-03 | molecular function is unknown | AR | ZT5-8 |
| CG32512 | 3.02E-03 | 1.05E-02 | molecular function is unknown | AR | ZT13-16 |
| CG30033 | 7.76E-04 | 1.16E-02 | molecular function is unknown | R | ZT21-0 |
| CG13694 | 2.04E-03 | 1.19E-02 | molecular function is unknown | AR | ZT17-20 |
| Vago | 2.40E-02 | 1.74E-02 | molecular function is unknown | AR | ZT13-16 |
| CG1311 | 3.51E-02 | 2.25E-02 | molecular function is unknown | AR | ZT21-0 |
| CG31344 | 2.05E-02 | 2.93E-02 | molecular function is unknown | AR | ZT21-0 |
| CG9542 | 1.04E-02 | 3.21E-02 | molecular function is unknown | AR | ZT9-12 |
| CG7330 | 9.32E-04 | 3.32E-02 | molecular function is unknown | AR | ZT1-4 |
| CG30343 | 3.25E-03 | 8.90E-03 | molecular function is unknown | AR | ZT17-20 |
| CG3631 | 9.83E-03 | 2.50E-02 | molecular function is unknown | AR | ZT17-20 |
Fat body where the clock is disrupted by expression of a dominant negative CLK protein
The transporter and GNS1/SUR4 families of genes include molecules listed as such on flybase although not identified by DAVID (Supplemental Table 2)
Based on their peak time of expression, these rhythmic genes were divided into six groups. 40% of these genes peaked in the late night (ZT 21 to ZT 0, where ZT 0 or Zeitgeber Time 0=lights on and ZT 12=lights off). Conversely, only nine genes peaked during the mid-late day (ZT 5 to ZT 12) (Supplemental Figure 1 and Table 1). As discussed below, we were able to identify specific classes of genes that were enriched among the cycling transcripts. We also verified the cycling of seven candidate genes by quantitative PCR and selected some of these for further analysis (see section below on restricted feeding).
Regulation of cyclically-expressed transcripts in flies with a disrupted fat body clock
As discussed earlier, circadian clocks exist in both central clock neurons of the brain and various peripheral tissues including the fat body (Glossop and Hardin, 2002; Xu et al., 2008); therefore, rhythmic expression of genes in the fat body can be controlled by either the central brain clock, or autonomously by the local fat body clock. To determine the importance of the fat body clock in this rhythmic gene expression, we performed microarray analysis on flies in which we disrupted the fat body clock through expression of a dominant negative CLK (DN-CLK) protein by the fat body-specific takeout (to)-Gal4 driver (Dauwalder et al., 2002; Tanoue et al., 2004). Figure 1 shows a heatmap representation of these data. In the wild type samples, there are two clear peaks and troughs of expression over the course of the two-day experiment. In contrast, the transcripts from DN-CLK flies show either severely dampened oscillations or are arrhythmic. We then compared the expression profiles of the 137 transcripts, identified above, in wild type and DN-CLK fat bodies by two-way ANOVA. As the DN-CLK fat bodies were collected at four-hour intervals, we used a four-hour resolution for the ANOVA. Rhythmic expression of 81 transcripts was affected by disruption of the fat body clock, indicating that the cycling of most mRNAs is controlled by the local fat body clock (Table 1). This is generally consistent with recently published data for the mammalian liver (Kornmann et al., 2007) although the percentage of genes affected by the local clock was lower in our experiments (60%) than in those of Kornmann et al. In addition to rhythmicity, overall levels of several rhythmic genes were either increased or decreased by disruption of the fat body clock (Supplemental Table 1).
Rhythmic genes are involved in different functions of the fat body
As a first step towards understanding how circadian transcription controls fat body physiology, we analyzed cycling transcripts in the fat body with DAVID (Huang da et al., 2009) to identify enriched functional categories. As expected, the known clock genes are enriched (classified in Supplemental Table 2 as genes involved in locomotor rhythm), as well as several other classes of genes, some of which are described below:
Lipid and carbohydrate metabolism
Our analysis identified two large groups of metabolic genes. The first group includes 15 transporter genes, of which nine belong to the Major Facilitator Superfamily (MFS), and three are ATP-Binding Cassette (ABC) Superfamily genes. The other group of metabolic genes is implicated in lipid metabolism; this includes the six GNS1/SUR4 membrane proteins, which have long-chain fatty acid elongase activity, and one long-chain-fatty-acid-CoA ligase, CG9009. Both these groups of genes show peak expression at night. Eight of the nine MFS genes of the transporter group as well as all three ABC genes peak during the early to mid night. None of the transporters peak during the daytime. Likewise, five of the six GNS1/SUR4 genes, as well as CG9009, peak at night, specifically during the mid-late nighttime (ZT17-ZT0). Such clustering of the expression of genes with similar function may determine the phases of the physiological phenomena they underlie. Not only do the rhythmic GNS1/SUR4 family genes have a similar phase, four of them - CG18609, CG9459, CG9458, and CG16905 - are expressed exclusively in the heart and fat body (from the FlyAtlas website). Interestingly, three of these genes are in close proximity on chromosome 3R, which may represent a recent gene duplication event or be indicative of co-regulated transcription.
Immune response and detoxification
The fat body is important for detoxification and immune functions in Drosophila. Since both these functions display a circadian rhythm (Krishnan et al., 2008; Lee and Edery, 2008), the fat body clock most likely drives these rhythms. Consistent with this idea, we identified several rhythmically expressed immune and detoxification genes, including five cytochrome P450 genes, eleven oxidoreductase genes and some genes involved in ubiquitin-related protein degradation pathways (Table1).
Reproduction
Drosophila reproduction involves multiple, sequential steps, from an elaborate courtship ritual to copulation to fertilization and finally egg-laying. While a circadian rhythm of actual courtship or mating has not been described, proximity between the male and female appears to be highest at night (Fujii et al., 2007). In addition, egg laying occurs in a rhythmic fashion (Allemand, 1976; T. Manjunatha et al., 2008). Moreover, levels of Juvenile Hormome (JH), a key regulator of reproduction, oscillate in the haemolymph of the honey bee (M et al., 2001) and a putative JH binding protein, takeout (to), is expressed rhythmically in Drosophila (Sarov-Blat et al., 2000). Our data show that four other members of the to superfamily (CG7079, CG31189, CG1124, and CG11852) are expressed rhythmically in the fat body. Moreover, we find that Juvenile hormone esterase (Jhe), which breaks down JH, is expressed rhythmically. The cycling of these genes could occur in response to feedback by cyclically expressed JH, but the results still indicate an interaction between the fat body clock and the rhythmic regulation of JH. Although most accounts of JH function are limited to analysis of females, our data obtained from male flies suggest that regulated expression of JH may also be important in males.
Besides JH, sexually dimorphic cuticular pheromones are important hormones for reproduction, in particular for courtship behavior (Dickson, 2008). In Drosophila, the expression and emission of these sex pheromones is clock regulated, but the central clock in the ventral lateral neurons is dispensable for this rhythm (Krupp et al., 2008), suggesting the involvement of peripheral clocks. Sex pheromones are typically produced in oenocytes, which may have been included in our fat body preparations since it is difficult to cleanly dissect out adult fat bodies. However, it is also possible that some key enzymes for sex pheromone synthesis come from the fat body. The synthesis of sex pheromones includes three major steps: First, desaturase1 (desat1) transforms palmitic acid to palmitoleic acid; then elongases with chain-length specificity lengthen the carbon chain of palmitoleic acid to 23 and 25 carbons in males and 27 and 29 carbons in females; finally, the very long chain fatty acid is decarboxylated to form hydrocarbons. There are at least 19 potential long chain fatty acid elongase genes in Drosophila (Chertemps et al., 2007), six of which are on our list of rhythmic genes (Table 1). This result, along with the finding that desat1 is a clock controlled gene (Krupp et al., 2008), strongly supports a role of peripheral clocks in regulating the rhythmic expression of sex pheromones.
Restricted feeding drives rhythmic expression of genes in the fat body but not in the brain
Our data indicate that genes involved in many different processes are expressed rhythmically under the control of the fat body clock. While some of these genes are expressed with the same phase, others have different phases, which may be optimally suited to their respective function. To address the importance of maintaining appropriately phased expression of the different functional classes of genes, we sought to identify a method by which these phases could be decoupled. Reasoning that the expression of metabolic genes might be responsive to food, we maintained flies on a restricted feeding paradigm whereby they were provided food for only six hours a day. Because in Drosophila, not only the central clock, but also clocks in various peripheral tissues can detect light, and light may be a stronger Zeitgeber (entraining stimulus) than any other environmental cue (Oishi et al., 2004), we conducted these experiments in constant darkness (DD). As reported previously (Xu et al., 2008), Drosophila normally consume more food during the late nighttime and early daytime, so we chose two intervals, CT9-CT15, and CT21-CT3, the first corresponding to the trough, and the second to the peak of the Drosophila feeding rhythm (Xu et al., 2008). Flies were maintained on one or the other of these paradigms for six days in DD, and assayed for rhythmic gene expression. Controls were given constant access to food and simply moved from one food vial to another at the onset and offset of restricted feeding.
As in adult Drosophila heads (Yang and Sehgal, 2001), we found that the circadian expression of clock genes in the Drosophila fat body dampens in extended constant darkness. After six days in DD, neither tim nor per mRNA cycling was detected in the Drosophila fat body under ad libitum feeding conditions. However, when food was restricted to CT9-CT15, a time when flies normally eat little, rhythmic expression of both genes was observed but with a peak at CT4-8, which is about 8–12 hours advanced compared to the phase entrained by light:dark (LD) cycles (Fig 2).
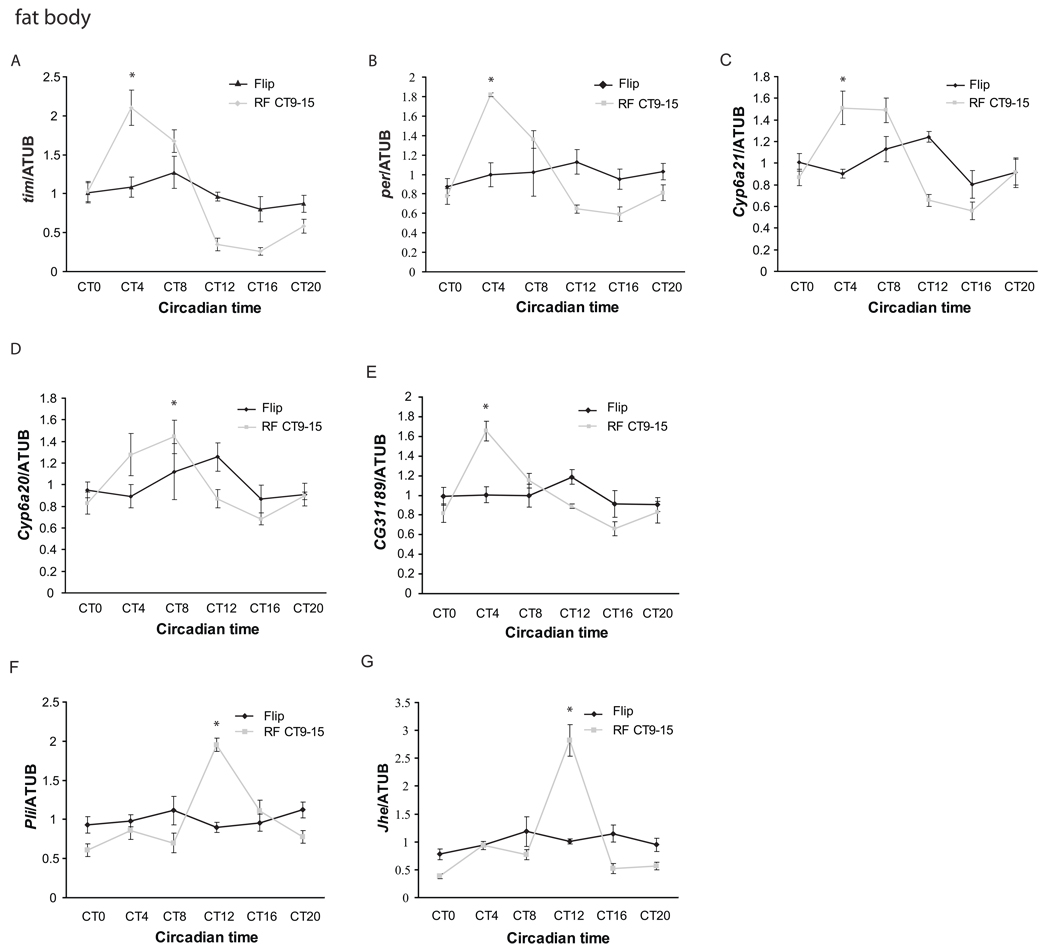
Figure 2. Restricted feeding between CT9 and CT15 drives rhythmic expression of clock genes and clock-regulated genes in the Drosophila fat body.
Quantitative PCR was performed to assay expression of seven cyclically expressed genes at different times of day in the fat body (A–G) of flies maintained on ad lib food (red) or restricted to food between CT9 and CT15 (black). Flies were transferred to a 1% agar vial from a normal food vial on day 1 in DD. Restricted feeding was started on day 2 and continued until day 6 at which point the flies were collected. Of the seven genes assayed - tim, per, cyp6a21, cyp6a20, CG31189, Jhe, and pli –all except per and CG31189 were also assayed in the brain. Expression values are normalized to α-tubulin. Each experiment was repeated at least three times, and the error bars represent S.E.M. Asterisks denote the time of peak expression of genes (p<0.05 compared to the trough point by two-tailed Student’s t-test with unequal variance) in flies that underwent restricted feeding. The cycling was also independently verified by ANOVA.
Restricted feeding (RF) between CT9-15 (CT9-15) had varied effects on the other clock-controlled genes we examined in the fat body. We selected these genes from among those whose cycling in LD we had confirmed by qPCR (see above), and they include one takeout family gene CG31189, two cytochrome P450 genes, cyp6a20 and cyp6a21, a possible immune response gene pli, and the gene expressing Juvenile hormone esterase (Jhe). As for tim and per, none of these genes showed a clear rhythmic pattern on the sixth day in DD under ad libitum conditions. Under RF CT9-15, rhythmic expression of CG31189, cyp6a21, and cyp6a20 was restored, with peaks shifted to CT4-8 (Fig 2C–E). The other two genes, pli and Jhe, were also expressed rhythmically in RF CT9-15, but they displayed sharp peaks right at the time of feeding (Fig 2F–G), which raises the possibility that their transcription is stimulated directly by food. Food-induced rhythmic expression of fat body genes did not persist when flies were transferred to ad lib conditions (Supplemental Figure 2).
To determine if RF affects central clock function, we assayed expression of the same genes in the brain. As in the fat body, on the 6th day in DD, we did not detect rhythmic expression of any of these genes in brain when food was constantly available (expression of Jhe and CG31189 was too low for reliable qPCR analysis, most likely because these are expressed at low levels in the brain). However, in contrast to the effect on the fat body, RF did not drive rhythmic expression of these genes in the brain (Fig 3). This result is consistent with the finding in mice that restricted feeding can change the phase of clocks in peripheral tissues like the liver but not the central clock in the SCN (Damiola et al., 2000; Stokkan et al., 2001). We also examined the effect of RF on rest:activity behavior and found that the phase was affected such that the animals were more active just before the time of food availability (Supplemental Figure 3). However, remnants of the old phase were evident as bouts of activity even while the flies were feeding. Moreover, when the flies were transferred back from RF to ad lib conditions, they reverted to the phase of activity seen prior to RF and also in controls maintained constantly on ad lib food. Based upon these factors, we believe the heightened pre-food activity during RF represents a response to starvation rather than entrained anticipation of food. Starvation typically induces a foraging response that manifests as increased activity in these assays.
Figure 3. Restricted feeding does not drive rhythmic gene expression in the brain.
As in Figure 2, flies were maintained on ad lib food (red) or restricted to food between CT9 and CT15 (black) from days 2–6 of DD. Quantitative RT-PCR was performed to assay expression of five genes expressed cyclically in the fat body (see Figure 2) at different times of day (A–D). Expression values are normalized to α-tubulin. Each experiment was performed at least twice. Error bars represent standard deviation.
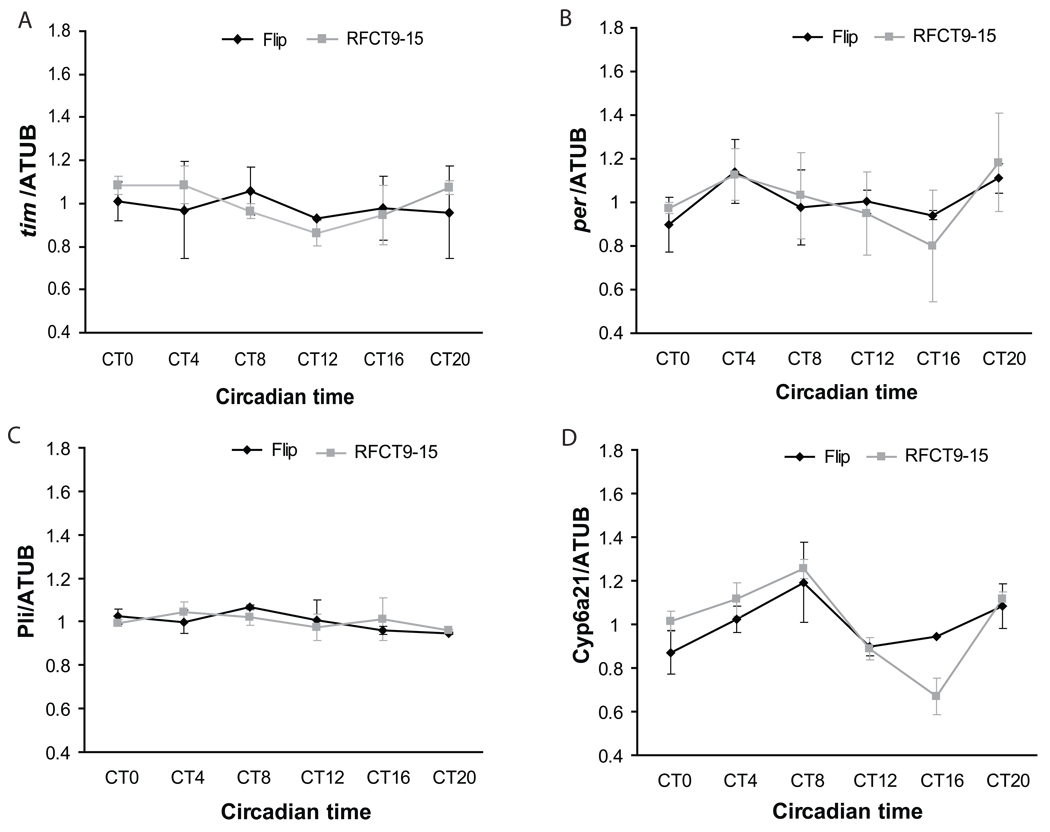
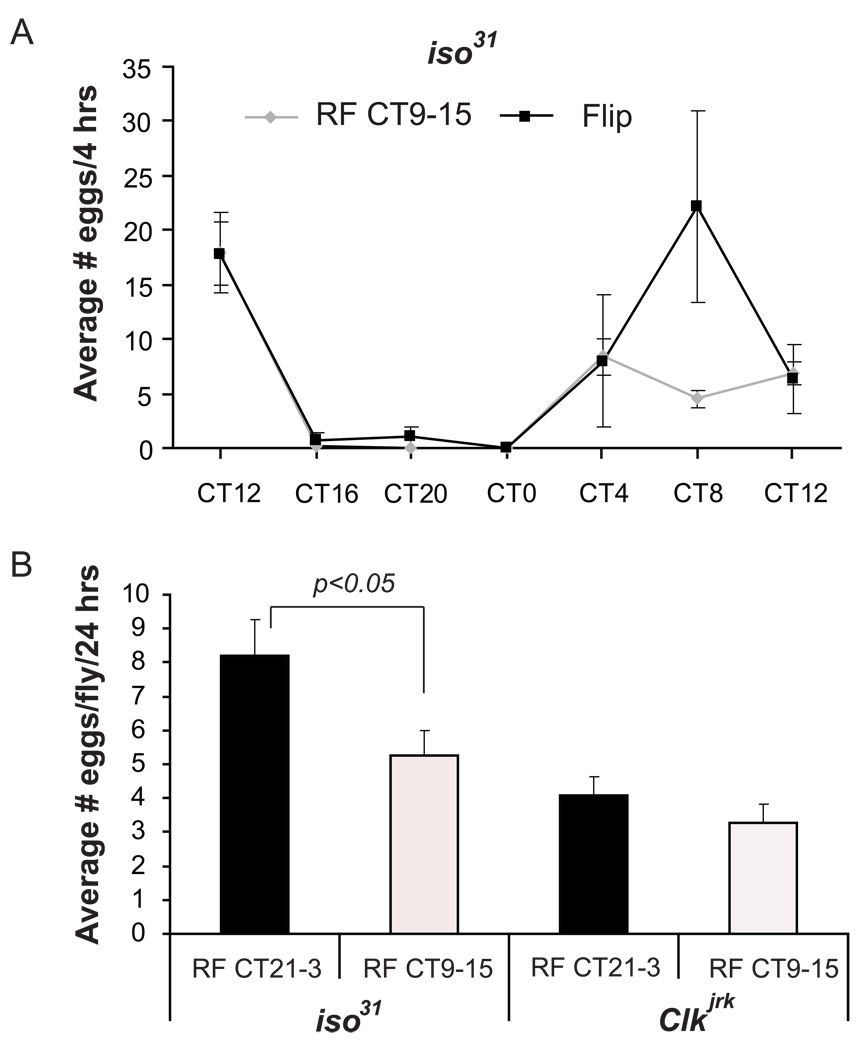
Given that food can drive the cycling of fat body genes, why is rhythmic expression of clock genes in the fat body undetectable after six days in DD? The most likely possibility is that the feeding rhythm does not persist for such an extended period of time in DD. We previously reported that even by the third day in DD it dampens considerably (Xu et al., 2008), which may account for the dampened molecular rhythms. However, restricting feeding to times when flies normally eat should restore the normal rhythmic pattern of gene expression. Indeed, we found that RF CT21-CT3, the normal peak time of feeding, restored rhythmic expression of tim with its normal phase (Supplemental Fig 4A). We also tested several other genes and obtained similar results (Supplemental Fig 4B–D).
Together these data demonstrate that an RF protocol will drive rhythmic expression of circadian genes in the fat body, but not in the brain. The timing of RF can restore the normal LD phase or shift rhythmic gene expression to a new one.
Restricted feeding affects the fat body clock through both Clk dependent and Clk independent pathways
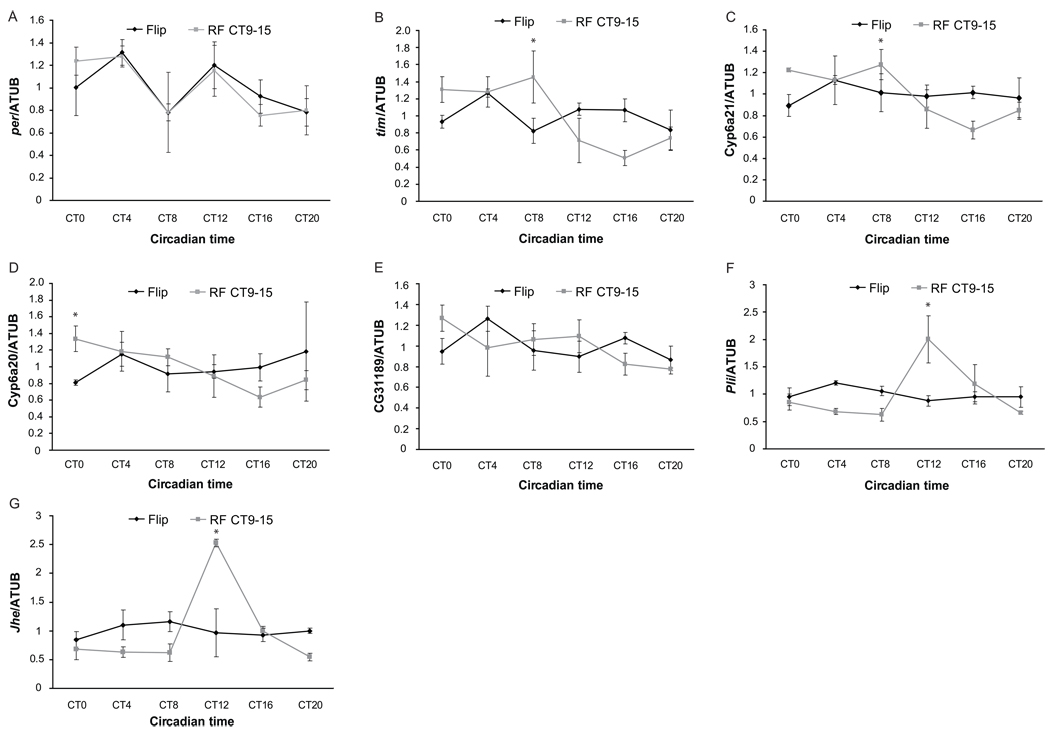
To determine if the effect of feeding on rhythmic gene expression in the fat body is through the circadian clock, we subjected per01 flies, which lack the central clock gene period (per), to a RF paradigm. Surprisingly, these flies were also driven by RF to display rhythmic expression of candidate fat body genes, suggesting that feeding can drive the cycling of circadian genes (Supplemental Fig 5). We also tested the effect of RF in flies lacking the core clock activator, Clock (Clk). RF of ClkJrk flies could only partially restore rhythms, as cyclic gene expression in the ClkJrk fat body was much weaker than in wild type flies (Fig 4A–E). This was true for both clock genes and output genes. There were exceptions: rhythms in pli and Jhe expression were strong in the ClkJrk fat body (Fig 4F and G). These data support the idea that expression of pli and Jhe is driven entirely by feeding and suggest that feeding promotes rhythmic gene expression in the fat body through two pathways. For some clock controlled genes, like pli and Jhe, RF acts solely through a clock-independent pathway; for clock genes such as tim and other output genes, the effect of RF is mediated by Clk-dependent and Clk-independent pathways, so the effect is much weaker in the absence of Clk.
Figure 4. The effect of restricted feeding on the rhythm and phase of cycling genes is mediated by both Clk-dependent and Clk-independent pathways.
Quantitative PCR analysis in fat bodies of control ClkJrk flies (black) and ClkJrk flies fed between CT9 and CT15 (red). These experiments were repeated at least twice, and error bars represent the S.E.M. In ClkJrk flies, restricted feeding has less of an effect on the cyclic expression of some genes such as (A) per, (B) tim, (C) cyp6a21, (D) cyp6a20, and (E) CG31189, but can drive the cycling of pli (F) and Jhe (G) in a similar manner as in WT flies. Error bars represent standard deviation.
Circadian misalignment caused by restricted feeding affects fly egg production
While being one of the fly’s major metabolic organs, the fat body also contributes to reproduction in females by synthesizing lipids that are loaded into the unfertilized egg to provide energy for the developing embryo (Bownes, 1994). Due to this important reproductive function of the fat body, and the fact that many cycling genes in the fat body function in juvenile hormone and sex pheromone biosynthesis (Supplemental Table 2), we hypothesized that the fat body clock regulates rhythms of behaviors related to reproduction, such as the rhythm of egg-laying. To test this idea, we first examined the timing of egg deposition, a previously described rhythmic behavior in Drosophila (Allemand, 1976; T. Manjunatha et al., 2008).
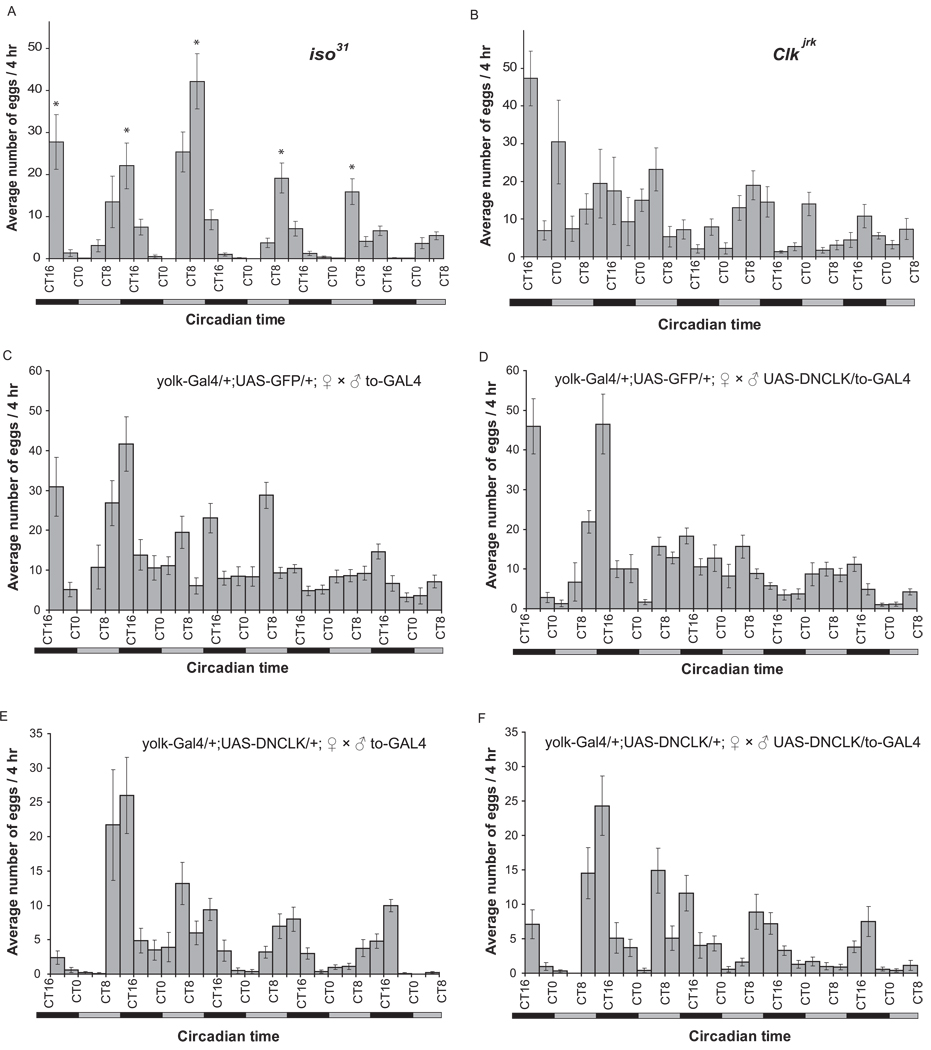
Consistent with previous reports (Allemand, 1976; T. Manjunatha et al., 2008), we found that wild type flies exhibit a strong egg-laying rhythm in constant darkness with a period slightly less than 24 hours and a peak at CT8-CT16 (Fig 5A). To confirm that this behavior is under circadian control, we also measured the egg-laying rhythm in two clock mutants, per0 and ClkJrk. and assessed the resulting rhythms statistically using JTK_Cycle {Hughes, 2010 #1. per0 flies maintain an egg laying rhythm with an inconsistent period length and ClkJrk mutants lack an egg laying rhythm (Fig 5B and data not shown), supporting the idea that the timing of egg laying in Drosophila is under clock control. To assess the contribution of the fat body clock to the control of the egg laying rhythm, we measured egg deposition in females expressing DN-Clk specifically in the fat body under the control of yolk-Gal4. We found that although the total number of eggs laid by females with an ablated fat body clock is lower than in wild type flies (and the eggs are infertile), the egg laying rhythm is unaffected by ablation of the fat body clock (Fig 5C and 5E). Similar results were observed following matings where either just the male (Fig 5D) or both the male and female had a disrupted fat body clock (Fig 5F), thereby also excluding a role for the male fat body clock in regulating this rhythm. Together, these data suggest that while egg-laying behavior is controlled by the circadian system, clocks in both the male and female fat body are dispensable for regulating this rhythm.
Figure 5. The egg-laying rhythm does not require clocks in the male or the female fat body.
Flies were entrained to LD for 3 days, and from the first day in DD, the egg-laying profiles of (A) iso31 flies, (B) ClkJrk flies (C) control female flies mated with control males, (D) control female flies mated with male flies lacking a fat body clock, (E) female flies lacking a fat body clock mated with control males, and (F) female flies lacking a fat body clock mated with male flies also lacking a fat body clock, were recorded for 5 consecutive days. For (A) and (B), the experiments were done twice with 10 vials, of 10 flies each, for each genotype per experiment. The final data for each time point represent the average of 20 recorded values, and the data are presented as mean±SEM. For (C)–(F), the data for each time point were from an average of 10 recorded values for each genotype, and the data are presented as mean±SEM. The data were analyzed for a rhythm using JTK_Cycle. A ~24 hour rhythm was detected in all samples except (B) and (C). Since transgenes present in C are also present in some of the other conditions, lack of a rhythm in this control condition probably arises from a spurious data point.
Because a number of rhythmic genes involved in reproduction are controlled by the fat body clock while egg laying behavior itself is not, it is possible that the fat body clock is synchronized with clocks elsewhere to control egg production and deposition. Since restricted feeding at CT9-15 drives rhythmic expression of circadian genes in the fat body, but not in all tissues, it likely leads to desynchrony among these different clocks and could affect the egg-laying rhythm. To test this hypothesis, we maintained flies on the six-day CT9-15 restricted feeding paradigm described above, and then measured the egg-laying rhythm. If the egg-laying rhythm is controlled by a clock resistant to restricted feeding, such treatment should not affect the egg-laying rhythm measured on the first day of ad lib feeding following the RF protocol. This was indeed the case. We found that the egg-laying patterns of wild type flies were similar in the handled control and the RF group in the first 24-hour period after restricted feeding treatment (Fig 6A). Collectively, these results suggest that restricted feeding does not affect the egg laying rhythm or the clock that controls it.
Figure 6. The effect of restricted feeding on the timing and number of eggs laid.
Flies were maintained on a restricted feeding paradigm for six days, and then transferred to ad lib food. (A) On the first day after RF, the egg-laying rhythm was measured, as described in Figure 4, in wildtype iso31 flies maintained on RF CT9-15. (B) The total number of eggs laid on the first day following RF was measured in wildtype and ClkJrk flies maintained on RF CT9-15 or RF CT21-3. Iso31 flies fed between CT9 and CT15 lay fewer eggs than those fed between CT21 and CT3. This difference is not seen in ClkJrk flies. The experiments were performed twice with 10 vials for each genotype per experiment. For (B) since eggs were collected over a 24-hour period, only five flies were housed per vial. Since the survival rate was different from vial to vial, the data are plotted per fly, with the error bars indicating variability across vials. Statistical significance was determined by two-tailed Student’s t-test with unequal variance. Error bars represent S.E.M for parts A and B.
Although restricted feeding does not affect the timing of egg deposition, we asked if the daily time of feeding could affect the animal’s reproductive capacity. As noted above, RF21-3 is in concordance with the endogenous feeding rhythm; also, it drives rhythmic gene expression with the same phase as the LD cycle (Supplemental Figure 4). On the other hand, RF9-15 drives feeding and metabolic gene expression with a phase that defies the endogenous rhythm. We compared the effects of these two regimes by placing wild type and ClkJrk flies on each of the two restricted feeding paradigms and counting the total number of eggs laid per fly during the first 24-hour period following RF. We found that flies fed at the “right” time (CT21-3) laid more eggs than those fed at the “wrong” time (CT9-15) (Fig 6B), indicating that circadian desynchrony caused by feeding at the “wrong” time of day leads to a defect in overall reproductive capacity. To confirm that this defect was due to a problem in the circadian system, we subjected ClkJrk flies to the same treatment. While these flies lay fewer eggs overall {Beaver, 2002 #1262}, they laid a similar number of eggs regardless of whether they were fed between CT9-15 or CT21-3 (Fig 6B). These data support the idea that the altered time of feeding interferes with endogenous circadian function.
Since changes in caloric intake can affect egg production, we also measured food consumption in flies maintained on the two different RF paradigms. The amount of food consumed by the two groups was equivalent indicating that differences in caloric intake do not account for the differential number of eggs laid (Supplemental Figure 6). Moreover, ClkJrk flies consumed the same amount of food as controls, even though they lay fewer eggs, supporting the idea that egg production under these conditions is largely independent of food intake (Supplemental Figure 6).
DISCUSSION
We report here that genes involved in major physiological processes such as metabolism, steroid hormone regulation, detoxification and the immune response are expressed with a circadian rhythm in the fly fat body. Most genes expressed cyclically in the fat body show peak expression at ZT17-24, although some peak at ZT5-12. Interestingly, genes of the Major Facilitator Superfamily, which encode small molecule transporters, show highest expression in the early evening (ZT13-16). Since this is the time when feeding usually decreases, the transporters may serve to transport molecules for nutrient storage. Egg-laying, which is regulated by steroid hormones and may utilize lipids (these are deposited in the egg), also peaks in the early evening (CT12-16) (Fig 5). Although the egg-laying rhythm does not require the fat body clock, appropriately timed expression of hormones and nutrients in the fat body may contribute to overall egg production. Interestingly, genes involved in lipid metabolism and sex pheromone synthesis, such as GNS1/SUR4 membrane proteins which have long chain fatty acid elongase activity, peak at ZT17-24, after stored lipids have been utilized during egg-laying. High expression of these genes may be required to replenish lipid stores and regulate the circulating pheromone balance in the body. On the other hand, genes involved in the breakdown of steroid hormones, such as Jhe, peak in the late night/early morning (ZT21-0) (Table 1), when egg laying is low and feeding is initiated. The phasing of gene expression in this fashion may allow for temporal separation of physiological processes.
Several groups have used microarrays to identify circadian regulated genes in the Drosophila head and body (Ceriani et al., 2002; Claridge-Chang et al., 2001; Lin et al., 2002; McDonald and Rosbash, 2001; Ueda et al., 2002). We compared our data with the results of these groups and found several genes in common. For instance, the detoxification genes, Cyp4d21and Cyp6a21, were detected in several studies, and Cyp6a20 was also identified by McDonald and Rosbash (McDonald and Rosbash, 2001). Other examples include the takeout (to) family genes and MFS transporters. McDonald and Rosbash reported rhythmic expression in fly heads of six takeout family members, 2 of which were also identified by our microarray (McDonald and Rosbash, 2001). Similarly, two of the MSF transporter genes we identified were reported as circadian genes in fly heads by Claridge-Chang et al (Claridge-Chang et al., 2001). Since fat bodies are also found in fly heads, and all three of these groups of genes have fat body-relevant functions, it is possible that the rhythmic expression of these genes in the head derives from head fat body cells.
There are also some differences between our microarray results and the previous reports. One of these is in the time of peak expression of cycling genes. Ceriani et al found that most genes fell into two categories that peaked at ZT8 and ZT20 respectively (Ceriani et al., 2002). Likewise, Hashimoto reported two major populations with peaks at around ZT10 and ZT20 (Ueda et al., 2002). Claridge-Chang et al found that the peaks were more evenly distributed throughout the day, with slightly fewer genes peaking between ZT4-8(19) and ZT8-12(17) than at other time periods (Claridge-Chang et al., 2001). Our data in this respect are most like those of Claridge-Chang. Different experimental designs and different backgrounds could account for these variations, but a major factor is probably the source of the tissue. All other laboratories used RNA from either Drosophila heads or bodies, both of which contain many different tissues. We used a fat body-enriched preparation that contained few other cell types so our phase distribution is relatively specific for the phase of rhythmic functions in the fat body. While heads contain fat bodies, the cycling, and perhaps even the expression, of genes in the fat body may have been masked in the other studies by signals from other parts of the head. These findings also suggest that although the phase of clock gene expression in different tissues is relatively similar, the rhythm of output genes may be quite different based upon the physiological function of the tissue.
Many of the genes expressed cyclically in the fat body are controlled by the fat body clock, as evidenced by loss of their cycling in flies where the fat body clock is specifically ablated. Others are under the control of extrinsic factors as they continue to cycle in the absence of a fat body clock. While the fat body clock may contribute to the rhythmic expression of these genes, it does not appear to be essential. The extrinsic factors may be clocks elsewhere, or even environmental or metabolic cues. For instance, expression of two of the genes we characterized in detail here is acutely induced by feeding. It is possible that many other genes will fall into this category. A small minority of genes is expressed cyclically only when the fat body clock is ablated suggesting that the clock in the fat body typically serves to dampen the cycling of these genes. The reason why this may be important is unclear; it could help to maintain adequate levels of critical proteins.
We also studied the entrainment of the fat body clock by environmental cues. Since the fat body is an important nutrient sensor and metabolic organ for Drosophila, we examined the effect of changes in nutrient levels. By placing flies on a restricted feeding paradigm, we drove the cycling of clock genes and clock-controlled genes in the fat body with a phase that was dictated by the timing of feeding. The clock in the brain was not shifted by the same treatment, indicating that this effect is likely tissue-autonomous with no involvement of the central clock in the brain. A similar situation occurs in mammals where circadian genes in the liver are affected by restricted feeding while the central clock in the suprachiasmatic nucleus is not (Damiola et al., 2000; Stokkan et al., 2001).
We report that the effect of the restricted feeding paradigm does not require the major clock protein, PER (Supplemental Figure 4), suggesting that the main circadian transcription/translation loop is dispensable in this process. This is consistent with the data of Vollmers et al, who found that restricted feeding can restore rhythmic expression of hundreds of gene in oscillator-deficient Cry1−/−;Cry2−/− mice (Vollmers et al., 2009). Like PER in Drosophila, the CRY proteins in mammals are the major transcriptional repressors in the mammalian clock; thus, loss of the circadian repressors does not affect the response to RF. However, we found that the effect of RF on the fat body clock is significantly weakened when the major transcriptional factor, CLK, is missing, indicating that CLK contributes to rhythmic gene expression in the fat body in response to restricted feeding. In addition, given that some effects of RF persist in the absence of CLK, a second pathway, which is totally independent of major clock genes, is clearly involved.
One factor that could mediate the effects of RF is the stress response caused by food deprivation. Stress responses can trigger the expression of various genes, including the major metabolic genes in the fat body, to adjust the body to a hostile environment (Ekengren et al., 2001; Liu et al.). It is likely that these metabolic genes are under multiple controls. For example, when food resources are ample, they could be under the control of the fat body clock and/or the central clock in the brain; under emergency situations, such as food scarcity, the stress response pathway may directly regulate expression of these genes. This stress response pathway may also shift the phase of clocks in organs crucial for nutrient metabolism, such as the fat body, to adapt to this environmental change. Under chronic conditions of limited food availability, the entrained clock in the fat body may take over from the stress response pathway to regulate target genes.
Regardless of the precise mechanisms by which RF drives rhythmic gene expression, we suggest that even under conditions of ad lib food availability rhythmic feeding contributes to the cycling of gene expression in the fat body. The rhythm of feeding under these conditions is driven by clocks that are entrained by light:dark cycles. Since all other rhythms, such as rest:activity, are also entrained by LD cycles, there is general synchrony among the various rhythmic processes. In constant darkness, rhythmic rest:activity persists, as does the cycling of clock gene expression in specific brain neurons (Yang and Sehgal, 2001), but the cycling of clock genes in other tissues dampens. Although we have not assayed the feeding rhythm after seven days in DD, the dampening evident at three days suggests that this too would be difficult to detect. Under RF conditions, the brain is unaffected and while the rest:activity cycle shows a change (discussed above), this most likely reflects a response to starvation rather than entrainment to RF (Supplemental Figure 3). We note that altered behavior in mammals exposed to RF may also result from clock-independent mechanisms (Storch and Weitz, 2009). However, RF affects gene expression in metabolic tissues, in case of Drosophila producing a robust rhythm in the fat body. This could reflect initiation of a new rhythm or enhancement of an underlying one. In either event, it appears that when this rhythm maintains the same phase relationship to the brain cycle as seen in LD, deleterious consequences of RF are minimized. In contrast, reproductive fitness is compromised when flies eat at the "wrong" time of the day. While differences in caloric intake can alter egg production, we show that food consumption is the same under both RF conditions analyzed. It is likely that following 18 hours of starvation, the homeostatic need for food overcomes any circadian regulation.
Functional clocks have been identified in the majority of organs throughout the body and it is hypothesized that proper synchrony of these clocks is essential for physiological homeostasis. While it was originally thought that the Drosophila central clock and/or light control the phases of the peripheral clocks, we provide evidence that a peripheral clock in the fat body can be driven independently of the central clock by restricted feeding. Since nutritional cues will affect the fat body/liver clock while leaving the central clock intact, altering an animal’s feeding time leads to desynchrony, the significance of which is just beginning to be elucidated. A recent study demonstrated that mistimed feeding in rats leads to obesity, suggesting that circadian desynchrony has severe consequences for metabolism (Arble et al., 2009). We show here that mistimed feeding in flies leads to decreased reproductive fitness. Lack of such a decrease in ClkJrk flies suggests that circadian clocks are required for this effect. On the other hand, since RF can drive rhythmic gene expression in fat bodies of clockless flies (per0 mutants), it is possible that the desynchrony is not between circadian clocks, but rather between rhythmic gene expression in different tissues. Regardless, these data suggest a link between synchronized rhythmic gene expression across tissues, metabolism and overall fitness. Clearly, this has tremendous implications in modern day society where people often have aberrant eating schedules and, as a result, likely have internal circadian desynchrony.
EXPERIMENTAL PROCEDURES
RNA preparation and microarray
Newly eclosed male flies were entrained for 5 days to 12h light:12h dark cycles (LD) at 25°C. On the 6th day, wild type flies (w;UAS-DNCLK/+;) were collected every 2 hr for 2 consecutive days, for a total of 24 time points. Flies with a disrupted fat body clock (w;UAS-DNCLK/takeout-Gal4) were collected every 4 hr for a total of 12 time points. Abdominal fat bodies were dissected as described previously (DiAngelo and Birnbaum, 2009). For each time point, fat bodies from 10 fly abdomens were pooled for RNA preparation. Total RNA was extracted from all fat body samples using Trizol reagent (Invitrogen), and sent to the Penn Microarray Facility; cDNA synthesis, biotin labeling of cRNA, and hybridization of the Affymetrix Drosophila Genome 2.0 chips were performed by the Penn Microarray Facility as recommended by the manufacturer (Affymetrix). Cel to text file condensation was performed using the MAS 5.0 algorithm in the Microarray Suite v.4 (Affymetrix) and visualized using the GeneSpring (Silicon Genetics) software package. Detection of cycling transcripts as well as estimation of period length and phase in microarray data was performed as previously described (Hughes et al., 2009). Briefly, COSOPT (Straume, 2004) and Fisher’s G-test (Wichert et al., 2004) were used to estimate the likelihood that a given transcript was cycling (p-values) and the false-discovery rate (q-value) was estimated using the method described by Storey et al. (Storey and Tibshirani, 2003). No expression cutoff was used. To generate heatmaps (Figure 1) signal intensities for all 136 cycling transcripts were median normalized and imported into MATLAB 7.10.0 (MathWorks) using custom scripts (available on demand). DAVID analysis was performed as previously described (Huang da et al., 2009). All .cel files have been deposited into GEO (Accession number: GSE24503) and are freely available. ANOVAs were performed using custom-built scripts in R (available on demand) using a p-value cutoff of < 0.05.
Primer Design and Real-Time Quantitative PCR
For each time point, abdominal fat bodies or brains from 20 flies were collected for RNA preparation. Total RNA was extracted using Trizol reagent (Invitrogen). cDNA was synthesized using a high capacity cDNA reverse transcription kit (Applied Biosystems). The levels of mRNA for different genes were measured by using SYBR-GREEN PCR Mix (Applied Biosystems) in an ABI 7000 Sequence Detection System (Applied BioSystems). The following primers were designed using ABI PrimerExpress software- for α-tubulin mRNA: 5’ primer, cgt ctg gac cac aag ttc ga; 3’ primer, cct cca tac cct cac caa cgt;; for tim mRNA: 5’ primer, tgg ctg cac tga tgg act tg; 3’ primer, ccc agc gat tgc att gg; for per mRNA: 5’ primer, cgt caa tcc atg gtc ccg; 3’ primer, cct gaa aga cgc gat ggt g; for CG31189 mRNA: 5’ primer, cgt tgg tgg agt aca gaa atg act; 3’ primer, atc caa cca tat gat cca tcg at; for pli mRNA: 5’ primer, tgc gct acg gtg aac tag tga ta; 3’ primer, gtc gtc gac cgc gat ctc; for Jhe mRNA: 5’ primer, cac gga ggc gaa ggt gat a; 3’ primer, gaa ttc ctg ggt ttt cca aac tt; for cyp6a20 mRNA: 5’ primer, gaa aca aca taa tgg gaa att gga t; 3’ primer, gct tcc tca tgg tct cat caa tg; for cyp6a21 mRNA: 5’ primer, gtt gta tcg gaa acc ctt cga tt; 3’ primer, aac ctc ata gtc ctc cag gca tt. The level of α-tubulin mRNA was used as a control for the total RNA content in each sample. The values for other RNAs were normalized to those of α-tubulin.
Restricted Feeding
Newly eclosed male flies were entrained in LD for at least three days, and then transferred to constant darkness (DD). For flies undergoing restricted feeding (RF), on the first day in DD, flies were switched from normal food (cornmeal/yeast/molasses/agar) to 1% agar; starting the following day, these flies were fed normal food for a specific six hour period each day and maintained in a 1% agar vial the rest of the time. Control flies were simply flipped to a new vial containing normal food during the RF period and flipped back at the end of the RF. Both control and RF treated flies were collected by rapid freezing on dry ice on the 6th day of the RF process. Fat body RNA samples were collected for QPCR analysis.
Evaluation of Egg-laying
Ten newly eclosed female and five newly eclosed male flies were transferred to a normal food vial and entrained in LD for at least three days before transfer to DD. For the egg laying rhythm, which was monitored from the first day in DD, flies were transferred to a new food vial every four hours for five consecutive days. Eggs laid in every vial were counted and recorded as the number of egg laid during the 4-hour period. For each genotype, at least 12 independent vials were used to get statistically reliable results. To determine the effect of RF on total egg laying output, two groups of flies were entrained in LD for three days and then transferred to DD at ZT0 and ZT12 respectively. Three hours after transfer to DD, flies were transferred to 1% agar vials, and for the next 7 days, the flies transferred to DD at ZT12 were fed only from CT9 to CT15, while the other group was fed from CT21 to CT3. On the 7th day, after feeding, instead of transferring back to 1% agar vials, flies were kept in the same food vial for another 18 hours. Eggs laid in these vials provided the total number of eggs laid during the first 24-hour period after RF treatment. For each group, at least 12 independent vials were used to get statistically reliable results. Statistical tests for rhythmicity (Figure 4) were performed using JTK_Cycle as previously described (Hughes et al., 2010) with a period-length window of 20–28 hours.
Highlights.
The Drosophila fat body clock controls rhythmic expression of genes involved in many processes
Restricted feeding can drive rhythmic gene expression in the fat body, but not in the brain
Restricted feeding can drive rhythmic gene expression in fat bodies of clockless flies
Mistimed feeding desynchronizes rhythms across tissues and impairs reproductive fitness
Supplementary Material
Acknowledgements
We thank members of our laboratories for their input. MATLAB scripts were generously provided by Laura Hughes. This work was funded in part by 1R01NS048471, 1R56NS048471 and P01 AG017628. JBH is supported by P50 MH074924 (to Joseph S. Takahashi), 1R01NS054794 from NINDS, and 1R01HL097800 from NHBLI. JRD was supported by training grant T32-AG000255. AS is an Investigator of the HHMI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allada R, Chung BY. Circadian organization of behavior and physiology in Drosophila. Annu Rev Physiol. 2010;72:605–624. doi: 10.1146/annurev-physiol-021909-135815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allemand R. Influence of light condition modification on the circadian rhythm of vitellogenesis and ovulation in Drosophila melanogaster. J Insect Physiol. 1976;22:1075–1080. doi: 10.1016/0022-1910(76)90116-5. [DOI] [PubMed] [Google Scholar]
- Arble DM, Bass J, Laposky AD, Vitaterna MH, Turek FW. Circadian timing of food intake contributes to weight gain. Obesity (Silver Spring) 2009;17:2100–2102. doi: 10.1038/oby.2009.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bownes M. The regulation of the yolk protein genes, a family of sex differentiation genes in Drosophila melanogaster. Bioessays. 1994;16:745–752. doi: 10.1002/bies.950161009. [DOI] [PubMed] [Google Scholar]
- Ceriani MF, Hogenesch JB, Yanovsky M, Panda S, Straume M, Kay SA. Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J Neurosci. 2002;22:9305–9319. doi: 10.1523/JNEUROSCI.22-21-09305.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertemps T, Duportets L, Labeur C, Ueda R, Takahashi K, Saigo K, Wicker-Thomas C. A female-biased expressed elongase involved in long-chain hydrocarbon biosynthesis and courtship behavior in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2007;104:4273–4278. doi: 10.1073/pnas.0608142104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claridge-Chang A, Wijnen H, Naef F, Boothroyd C, Rajewsky N, Young MW. Circadian regulation of gene expression systems in the Drosophila head. Neuron. 2001;32:657–671. doi: 10.1016/s0896-6273(01)00515-3. [DOI] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes & Development. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauwalder B, Tsujimoto S, Moss J, Mattox W. The Drosophila takeout gene is regulated by the somatic sex-determination pathway and affects male courtship behavior. Genes & Development. 2002;16:2879–2892. doi: 10.1101/gad.1010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiAngelo JR, Birnbaum MJ. Regulation of fat cell mass by insulin in Drosophila melanogaster. Mol Cell Biol. 2009;29:6341–6352. doi: 10.1128/MCB.00675-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson BJ. Wired for sex: the neurobiology of Drosophila mating decisions. Science. 2008;322:904–909. doi: 10.1126/science.1159276. [DOI] [PubMed] [Google Scholar]
- Ekengren S, Tryselius Y, Dushay MS, Liu G, Steiner H, Hultmark D. A humoral stress response in Drosophila. Curr Biol. 2001;11:1479. doi: 10.1016/s0960-9822(01)00452-3. [DOI] [PubMed] [Google Scholar]
- Fujii S, Krishnan P, Hardin P, Amrein H. Nocturnal male sex drive in Drosophila. Curr Biol. 2007;17:244–251. doi: 10.1016/j.cub.2006.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachon F, Olela FF, Schaad O, Descombes P, Schibler U. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006;4:25–36. doi: 10.1016/j.cmet.2006.04.015. [DOI] [PubMed] [Google Scholar]
- Glossop NR, Hardin PE. Central and peripheral circadian oscillator mechanisms in flies and mammals. J Cell Sci. 2002;115:3369–3377. doi: 10.1242/jcs.115.17.3369. [DOI] [PubMed] [Google Scholar]
- Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Zheng X, Yang J, Imamichi T, Stephens R, Lempicki RA. Extracting biological meaning from large gene lists with DAVID. Chapter 13. Curr Protoc Bioinformatics. 2009 doi: 10.1002/0471250953.bi1311s27. Unit 13 11. [DOI] [PubMed] [Google Scholar]
- Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, Baggs JE, Panda S, Hogenesch JB. Harmonics of circadian gene transcription in mammals. PLoS Genet. 2009;5:e1000442. doi: 10.1371/journal.pgen.1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CH. Endogenous timekeepers in photosynthetic organisms. Annual Review of Physiology. 2001;63:695–728. doi: 10.1146/annurev.physiol.63.1.695. [DOI] [PubMed] [Google Scholar]
- Klarsfeld A, Rouyer F. Effect of circadian rhythms and LD periodicity on the lifespan of Drosophila melanogaster. Journal of Biological Rhythms. 1998;13:471–478. doi: 10.1177/074873098129000309. [DOI] [PubMed] [Google Scholar]
- Kornmann B, Schaad O, Bujard H, Takahashi JS, Sch1ibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan N, Davis AJ, Giebultowicz JM. Circadian regulation of response to oxidative stress in Drosophila melanogaster. Biochem Biophys Res Commun. 2008;374:299–303. doi: 10.1016/j.bbrc.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupp JJ, Kent C, Billeter JC, Azanchi R, So AK, Schonfeld JA, Smith BP, Lucas C, Levine JD. Social experience modifies pheromone expression and mating behavior in male Drosophila melanogaster. Curr Biol. 2008;18:1373–1383. doi: 10.1016/j.cub.2008.07.089. [DOI] [PubMed] [Google Scholar]
- Lamia KA, Storch KF, Weitz CJ. Physiological significance of a peripheral tissue circadian clock. Proc Natl Acad Sci U S A. 2008;105:15172–15177. doi: 10.1073/pnas.0806717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazareva AA, Roman G, Mattox W, Hardin PE, Dauwalder B. A role for the adult fat body in Drosophila male courtship behavior. PLoS Genet. 2007;3:e16. doi: 10.1371/journal.pgen.0030016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclerc V, Reichhart JM. The immune response of Drosophila melanogaster. Immunol Rev. 2004;198:59–71. doi: 10.1111/j.0105-2896.2004.0130.x. [DOI] [PubMed] [Google Scholar]
- Lee JE, Edery I. Circadian regulation in the ability of Drosophila to combat pathogenic infections. Curr Biol. 2008;18:195–199. doi: 10.1016/j.cub.2007.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Han M, Shimada B, Wang L, Gibler TM, Amarakone A, Awad TA, Stormo GD, Van Gelder RN, Taghert PH. Influence of the period-dependent circadian clock on diurnal, circadian, and aperiodic gene expression in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2002;99:9562–9567. doi: 10.1073/pnas.132269699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhou S, Ma L, Tian L, Wang S, Sheng Z, Jiang RJ, Bendena WG, Li S. Transcriptional regulation of the insulin signaling pathway genes by starvation and 20-hydroxyecdysone in the Bombyx fat body. J Insect Physiol. doi: 10.1016/j.jinsphys.2010.02.011. [DOI] [PubMed] [Google Scholar]
- M.Elekonich M, Schulz DJ, Bloch G, Robinson GE. Juvenile hormone levels in honey bee (Apis mellifera L.) foragers: foraging experience and diurnal variation. J Insect Physiol. 2001;47:1119–1125. doi: 10.1016/s0022-1910(01)00090-7. [DOI] [PubMed] [Google Scholar]
- Manjunatha T, Hari Dass S, Sharma VK. Egg-laying rhythm in Drosophila melanogaster. J Genet. 2008;87:495–504. doi: 10.1007/s12041-008-0072-9. [DOI] [PubMed] [Google Scholar]
- Maywood ES, O'Neill JS, Chesham JE, Hastings MH. Minireview: The circadian clockwork of the suprachiasmatic nuclei--analysis of a cellular oscillator that drives endocrine rhythms. Endocrinology. 2007;148:5624–5634. doi: 10.1210/en.2007-0660. [DOI] [PubMed] [Google Scholar]
- McDonald MJ, Rosbash M. Microarray analysis and organization of circadian gene expression in Drosophila. Cell. 2001;107:567–578. doi: 10.1016/s0092-8674(01)00545-1. [DOI] [PubMed] [Google Scholar]
- Oishi K, Shiota M, Sakamoto K, Kasamatsu M, Ishida N. Feeding is not a more potent Zeitgeber than the light-dark cycle in Drosophila. Neuroreport. 2004;15:739–743. doi: 10.1097/00001756-200403220-00034. [DOI] [PubMed] [Google Scholar]
- Sarov-Blat L, So WV, Liu L, Rosbash M. The Drosophila takeout gene is a novel molecular link between circadian rhythms and feeding behavior. Cell. 2000;101:647–656. doi: 10.1016/s0092-8674(00)80876-4. [DOI] [PubMed] [Google Scholar]
- Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- Storch KF, Weitz CJ. Daily rhythms of food-anticipatory behavioral activity do not require the known circadian clock. Proc Natl Acad Sci U S A. 2009;106:6808–6813. doi: 10.1073/pnas.0902063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straume M. DNA microarray time series analysis: automated statistical assessment of circadian rhythms in gene expression patterning. Methods Enzymol. 2004;383:149–166. doi: 10.1016/S0076-6879(04)83007-6. [DOI] [PubMed] [Google Scholar]
- Tanoue S, Krishnan P, Krishnan B, Dryer SE, Hardin PE. Circadian clocks in antennal neurons are necessary and sufficient for olfaction rhythms in Drosophila. Curr Biol. 2004;14:638–649. doi: 10.1016/j.cub.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Ueda HR, Matsumoto A, Kawamura M, Iino M, Tanimura T, Hashimoto S. Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. Journal of Biological Chemistry. 2002;277:14048–14052. doi: 10.1074/jbc.C100765200. [DOI] [PubMed] [Google Scholar]
- Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci U S A. 2009;106:21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichert S, Fokianos K, Strimmer K. Identifying periodically expressed transcripts in microarray time series data. Bioinformatics. 2004;20:5–20. doi: 10.1093/bioinformatics/btg364. [DOI] [PubMed] [Google Scholar]
- Xu K, Zheng X, Sehgal A. Regulation of feeding and metabolism by neuronal and peripheral clocks in Drosophila. Cell Metab. 2008;8:289–300. doi: 10.1016/j.cmet.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Sehgal A. Role of molecular oscillations in generating behavioral rhythms in Drosophila. Neuron. 2001;29:453–467. doi: 10.1016/s0896-6273(01)00218-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.