Abstract
Pseudomonas aeruginosa is a ubiquitously distributed opportunistic pathogen that inhabits soil and water as well as animal-, human-, and plant-host-associated environments. The ubiquity would be attributed to its very versatile energy metabolism. P. aeruginosa has a highly branched respiratory chain terminated by multiple terminal oxidases and denitrification enzymes. Five terminal oxidases for aerobic respiration have been identified in the P. aeruginosa cells. Three of them, the cbb3-1 oxidase, the cbb3-2 oxidase, and the aa3 oxidase, are cytochrome c oxidases and the other two, the bo3 oxidase and the cyanide-insensitive oxidase, are quinol oxidases. Each oxidase has a specific affinity for oxygen, efficiency of energy coupling, and tolerance to various stresses such as cyanide and reactive nitrogen species. These terminal oxidases are used differentially according to the environmental conditions. P. aeruginosa also has a complete set of the denitrification enzymes that reduce nitrate to molecular nitrogen via nitrite, nitric oxide (NO), and nitrous oxide. These nitrogen oxides function as alternative electron acceptors and enable P. aeruginosa to grow under anaerobic conditions. One of the denitrification enzymes, NO reductase, is also expected to function for detoxification of NO produced by the host immune defense system. The control of the expression of these aerobic and anaerobic respiratory enzymes would contribute to the adaptation of P. aeruginosa to a wide range of environmental conditions including in the infected hosts. Characteristics of these respiratory enzymes and the regulatory system that controls the expression of the respiratory genes in the P. aeruginosa cells are overviewed in this article.
Keywords: respiration, terminal oxidase, denitrification, nitric oxide, Pseudomonas aeruginosa
Introduction
The opportunistic pathogen Pseudomonas aeruginosa has a remarkable ability to grow under a variety of environmental conditions, including soil and water as well as animal-, human-, and plant-host-associated environments. It is responsible for severe nosocomial infections in immunocompromised patients. In particular, it causes life-threatening chronic lung infection in patients with the inherited disease cystic fibrosis (CF; Lyczak et al., 2002). The genome of P. aeruginosa is relatively large (6.3 Mb) and carries a large number of genes for utilization of various carbon sources, energy metabolisms, and regulatory systems, which might contribute to the environmental adaptability of this bacterium (Stover et al., 2000). The main energy producing system of P. aeruginosa is respiration, which utilizes a proton motive force for ATP synthesis. In the case of eukaryotic respiration in mitochondria, the electron transfer pathway consists of four complexes, NADH dehydrogenase (complex I), succinate dehydrogenase (complex II), a cytochrome bc1 complex (complex III), and a cytochrome c oxidase (complex IV). Protons are pumped across the membrane during electron transfer through complexes I, III, and IV, producing the proton gradient. On the other hand, P. aeruginosa as well as many other bacterial species use a variety of electron donors and acceptors for respiration and therefore have far more complex and flexible electron transfer pathways. At least 17 respiratory dehydrogenases that are predicted to be responsible for feeding electrons from respiratory substrates into the quinone pool, including three types of NADH dehydrogenases and a succinate dehydrogenase, have been annotated in the genome of P. aeruginosa (Williams et al., 2007). P. aeruginosa has five terminal oxidases that catalyze the four-electron reduction of molecular oxygen to water (Matsushita et al., 1982, 1983; Fujiwara et al., 1992; Cunningham and Williams, 1995; Cunningham et al., 1997; Stover et al., 2000; Comolli and Donohue, 2002, 2004). Three of them are cytochrome c oxidases that receive electrons via the cytochrome bc1 complex and c-type cytochromes. The other two are quinol oxidases that receive electrons directly from ubiquinol (Figure 1). The respiratory chain is also branched to the denitrification enzymes that reduce nitrogen oxides. These alternative respiratory branches enable P. aeruginosa to grow under anaerobic conditions in the presence of nitrate or nitrite (Zumft, 1997). P. aeruginosa also has the ability to ferment arginine and pyruvate anaerobically. A fundamental understanding of the respiratory systems and the physiology of aerobic and anaerobic energy metabolism would be necessary for better comprehension of the ubiquity and pathogenicity of P. aeruginosa. Some excellent reviews on the aerobic and anaerobic respiration of P. aeruginosa are now available (Williams et al., 2007; Schobert and Jahn, 2010; Schobert and Tielen, 2010). This article will additionally focus on some recent information on the transcriptional regulation of the aerobic and anaerobic respiratory genes.
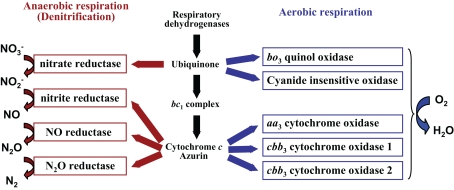
Figure 1.
Branched respiratory chain of P. aeruginosa. Under aerobic conditions, oxygen is utilized as a terminal electron acceptor and reduced to water by five terminal oxidases. Two quinol oxidases, the bo3 oxidase and the cyanide-insensitive oxidase, receive electrons directly from quinol. Three cytochrome c oxidases, the aa3 oxidase and the two cbb3 oxidases, receive electrons via the cytochrome bc1 complex and c-type cytochromes or a small blue-copper protein azurin. Under anaerobic conditions, electrons are transferred to nitrogen oxides via the denitrification enzymes.
Multiple Terminal Oxidases for Aerobic Respiration
Pseudomonas aeruginosa has five terminal oxidases for aerobic respiration (Figure 1; Matsushita et al., 1982, 1983; Fujiwara et al., 1992; Cunningham and Williams, 1995; Cunningham et al., 1997; Stover et al., 2000; Comolli and Donohue, 2002, 2004). Three of them, the cbb3-1 oxidase (Cbb3-1), the cbb3-2 oxidase (Cbb3-2), and the aa3 oxidase (Aa3), are cytochrome c oxidases. The other two, the cytochrome bo3 oxidase (Cyo) and the cyanide-insensitive oxidase (CIO), are quinol oxidases. These terminal oxidases are expected to have their specific affinity for oxygen, efficiency of proton-translocation, and resistance to various stresses such as cyanide and reactive nitrogen species. We have constructed five kinds of quadruple mutant strains, which lack four out of the five terminal oxidase gene clusters, and used them to characterize the features of each terminal oxidase (unpublished data). The Km values of Aa3, CIO, and Cyo for oxygen were high, whereas those of Cbb3-1 and Cbb3-2 were one order lower than those of the other three terminal oxidases, indicating that Aa3, CIO, and Cyo are low affinity enzymes and Cbb3-1 and Cbb3-2 are high affinity enzymes. The analysis of the proton-pumping activity (H+/O ratio) using the quadruple mutant strains showed that the electron transport complex terminated by Aa3 has the highest efficiency to create a proton gradient across the cell membrane, whereas that by CIO has the lowest efficiency. Carrying multiple terminal oxidases of such different characteristics and the differential use of them under different conditions must contribute to the ubiquity of P. aeruginosa in various environmental niches. Two redox-responsive transcriptional regulators, ANR (anaerobic regulation of arginine deiminase and nitrate reduction) and RoxSR, mainly regulate the expression of the terminal oxidase genes. ANR is a direct oxygen sensor and functions as a global regulator for anaerobic gene expression of P. aeruginosa (Zimmermann et al., 1991). RoxSR is a two-component transcriptional regulator consisting of the membrane-bound sensor kinase RoxS and the response regulator RoxR. RoxSR corresponds to PrrBA of Rhodobacter sphaeroides and RegBA of Rhodobacter capsulatus, which are the principal regulators controlling the expression of the genes for photosynthesis, carbon dioxide fixation, nitrogen fixation, and hydrogen metabolism, as well as numerous other functions in these purple photosynthetic bacteria (Dubbs and Tabita, 2004; Elsen et al., 2004; Eraso et al., 2008). The function and regulatory features of each terminal oxidase of P. aeruginosa are described below and in Figure 2.
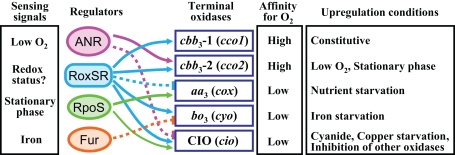
Figure 2.
Schematic model of the regulatory network controlling the multiple terminal oxidases in P. aeruginosa. The sensing signals for the regulators are shown in the left column. Affinity for oxygen and upregulation conditions of the terminal oxidases are shown in the right columns. Activation is indicated by arrows. Inhibition is indicated by bars with dotted lines.
The cytochrome cbb3 oxidases
The cbb3-type cytochrome c oxidase is phylogenetically the most distant member of the heme–copper oxidase superfamily and exclusively found in bacteria (Pitcher and Watmough, 2004). The X-ray structure of the enzyme from Pseudomonas stutzeri was reported recently (Buschmann et al., 2010). This type of enzyme is known to have very high affinity for oxygen and low proton-translocation efficiency. The Km values for oxygen were determined to be 7 and 40 nM for the enzymes from Bradyrhizobium japonicum and Campylobacter jejuni, respectively (Preisig et al., 1996; Jackson et al., 2007). The cbb3 oxidase is known to be induced under low oxygen conditions in many bacteria, such as Paracoccus denitrificans, R. sphaeroides, and R. capsulatus (Mouncey and Kaplan, 1998; Otten et al., 2001; Swem and Bauer, 2002). In the symbiotic nitrogen fixation bacterium B. japonicum, the cbb3 oxidase is essential for nitrogen fixation in hypoxic root nodules, in which the oxygen concentration is extremely low (∼20 nM; Preisig et al., 1993, 1996). The cbb3 oxidase is also known to be functioning in the obligately microaerophilic bacteria Helicobacter pylori and C. jejuni (Nagata et al., 1996; Jackson et al., 2007). From these observations, the cbb3 oxidase is recognized as the major player under the low oxygen environments.
The cbb3 oxidase is also known to have a repressive role in the PrrBA-dependent expression of the photosynthesis genes in R. sphaeroides (O'Gara et al., 1998; Oh and Kaplan, 1999, 2000). The photosynthesis genes are usually induced under low oxygen or anaerobic conditions in non-oxygenic photosynthetic bacteria. However, disruption of the cbb3 oxidase genes or inhibition of the cbb3 oxidase activity results in expression of the photosynthesis genes even under aerobic conditions in R. sphaeroides. It has been proposed that the cbb3 oxidase senses the electron flow through the respiratory chain and generates an inhibitory signal that prevents the activation of PrrBA (Oh et al., 2004).
The cbb3 oxidase is encoded by a tetracistronic operon ccoNOQP (fixNOQP). The ccoN (fixN) product is the catalytic subunit that contains the binuclear center consisting of a high spin heme b3 and CuB. The ccoO (fixO) and ccoP (fixP) genes encode transmembrane monoheme and diheme cytochrome c subunits, respectively. The gene product of ccoQ (fixQ) is thought to be required for stabilization of the core complex of the cbb3-type oxidases (Zufferey et al., 1996; Oh and Kaplan, 2002; Peters et al., 2008).
Two complete sets of the genes encoding the cbb3 cytochrome oxidases (Cbb3-1 and Cbb3-2) are tandemly clustered in the genome of P. aeruginosa (Stover et al., 2000). Cbb3-1 and Cbb3-2 are encoded by the ccoN1O1Q1P1 genes (PA1552–1554) and the ccoN2O2Q2P2 genes (PA1555–1557), respectively. The small ccoQ1 and ccoQ2 genes were not annotated in the genome previously (Stover et al., 2000). Tandem repeats of the genes for two cbb3-type cytochrome oxidases are also found in the genomes of other pseudomonads, such as Pseudomonas putida, Pseudomonas fluorescens, and P. stutzeri. In addition to the two complete sets of the cco gene clusters, two orphan ccoN-like genes (PA1856 and PA4133), which are highly similar to the main subunit genes of the cbb3 oxidases, have been identified in the P. aeruginosa genome (Stover et al., 2000). Both of these ccoN-like genes are followed by small genes (PA1855 and PA4134) that have similarity with the ccoQ genes. These extra ccoNQ-like gene clusters might have complementary functions for aerobic respiration.
The cco1 genes for Cbb3-1 were constitutively expressed at a high level even under high oxygen conditions and slightly downregulated at the stationary phase (Comolli and Donohue, 2004; Alvarez-Ortega and Harwood, 2007; Kawakami et al., 2010). Cbb3-1 seemed to be the dominant oxidase, especially under the normal aerobic growth conditions at exponential phase in LB medium. Disruption of the cco1 genes slightly affected the aerobic growth (Comolli and Donohue, 2004; Alvarez-Ortega and Harwood, 2007). Mutation of the cco1 genes also showed a small colony phenotype (Schurek et al., 2008). In contrast, the cco2 genes for Cbb3-2 were upregulated under low oxygen conditions or at the stationary phase. The expression level of Cbb3-2 surpassed that of Cbb3-1 under those induced conditions (Kawakami et al., 2010). ANR is involved in the transcriptional activation of the cco2 genes under low oxygen conditions (Comolli and Donohue, 2004; Kawakami et al., 2010). The induction of the cco2 genes at the stationary phase likely occurred because the oxygen concentration in the medium was depleted to the level that induces the activation by ANR. RoxSR is also involved in the induction of the cco2 genes (Kawakami et al., 2010). A similar RoxSR-dependent regulation was also reported in the case of cco1 (corresponding to cco2 of P. aeruginosa) in P. putida (Fernández-Piñar et al., 2008). A cco1 cco2 double mutant of P. aeruginosa showed very slow growth under a 2% oxygen condition and failed to grow under a 0.4% oxygen condition (Alvarez-Ortega and Harwood, 2007). These results indicate that Cbb3-1 plays a primary role in aerobic growth irrespective of oxygen concentration and that Cbb3-2 plays a compensatory or supplementary role under the oxygen-depleted conditions.
The cytochrome aa3 oxidase
The aa3-type cytochrome c oxidase is a major member of the heme–copper oxidase superfamily and is distributed in a wide range of bacteria. This type of heme–copper oxidase enzyme is closely related to the mitochondrial terminal oxidase and has a high proton-pumping activity. The aa3-type oxidases have low affinity for oxygen and usually play a dominant role under high oxygen conditions in many bacteria, such as P. denitrificans, B. japonicum, R. sphaeroides, and Bacillus subtilis (Bosma et al., 1987; Gabel and Maier, 1993; Flory and Donohue, 1997; Winstedt and von Wachenfeldt, 2000; Arai et al., 2008).
In P. aeruginosa, the aa3 oxidase (Aa3) is encoded by the coxBA-PA0107-coIII (coxC) gene cluster (PA0105–0108; Stover et al., 2000). The coxA, coxB, and coIII (coxC) genes encode subunits I, II, and III, respectively. Subunit I carries the heme a3−CuB binuclear catalytic center. Subunit II has a binding site of CuA, which is the electron transfer site with cytochrome c. PA0107 encodes a putative cytochrome c oxidase assembly protein involved in the insertion of copper into subunit I. The expression level of the cox genes is kept very low under normal laboratory growth conditions even at high oxygen tension, although these genes are slightly upregulated at the stationary phase in P. aeruginosa (Schuster et al., 2004; Alvarez-Ortega and Harwood, 2007). We recently found that the cox genes were significantly induced under starvation of carbon, nitrogen, or iron (Kawakami et al., 2010). The cox promoter was found to be dependent on a stationary phase sigma factor RpoS and repressed by RoxSR (Schuster et al., 2004; Kawakami et al., 2010). At the stationary phase, RpoS is highly expressed and activates the cox promoter, but the promoter is simultaneously repressed by RoxSR, which is expected to be active in the hypoxic high-cell-density stationary phase culture. Therefore, the expression level of the cox genes might be kept low under any nutrient-rich conditions. The cco1 cco2 cio cyo quadruple mutant, which is deficient in the four terminal oxidase gene clusters other than the cox gene cluster, was not able to grow under aerobic conditions. However, suppressor mutants that are able to grow aerobically emerged after aerobic incubation for several weeks (unpublished data). The cox structural genes had no mutation, but the roxS gene was inactivated and the RpoS-dependent promoter region of the cox genes was altered in the suppressor mutants, confirming the tight regulation of the cox genes by RoxSR and RpoS. The respiratory chain terminated by Aa3 has the highest ability to create a proton gradient across the membrane (the H+/O ratio is 6) among the five terminal oxidases of P. aeruginosa (unpublished data). Therefore, it is reasonable that Aa3 is utilized only under starvation conditions for efficient energy production. Utilization of Aa3 might be advantageous for proliferation in natural oligotrophic environments. However, if Aa3 were highly expressed under the nutrient-rich laboratory growth conditions, its high proton-pumping activity might cause an imbalance of energy and redox homeostasis in P. aeruginosa cells.
The bo3 quinol oxidase
The bo3-type quinol oxidase is also a member of the heme–copper oxidase superfamily. This type of the terminal oxidase is homologous to the aa3-type cytochrome oxidase but contains heme b and heme o. It lacks the CuA-binding site, which is involved in the interaction with cytochrome c, and receives electrons from ubiquinol. The bo3 oxidase of Escherichia coli is known to have a low affinity for oxygen and functions under high oxygen conditions (Cotter et al., 1990; D'Mello et al., 1995). It is encoded by the cyoABCDE genes in E. coli (Nakamura et al., 1997). The cyoA, cyoB, and cyoC genes encode the subunits corresponding to the subunits II, I, and III of the aa3-type cytochrome oxidase, respectively. The cyoD gene encodes subunit IV, which is proposed to assist the CuB-binding to subunit I during biosynthesis or assembly of the oxidase complex (Saiki et al., 1996). The cyoE encodes a protoheme IX farnesyltransferase (heme o synthetase), which is required for production of heme o from heme b (Saiki et al., 1992; Mogi et al., 1994).
The cyoABCDE genes (PA1317–1321) of the P. aeruginosa bo3 oxidase (Cyo) are highly homologous with the corresponding genes of E. coli. Cyo was identified as a quinol oxidase and shown to have a high Km value for oxygen by using the cco1 cco2 cox cio quadruple mutant (unpublished data). The H+/O ratio of Cyo was speculated to be 4, which was comparable to that of the bo3 oxidase from E. coli (Puustinen et al., 1989). The cyo genes were downregulated at the stationary phase or under the low oxygen conditions in P. aeruginosa cells (Alvarez-Ortega and Harwood, 2007; Kawakami et al., 2010). Considering that the affinity for oxygen is low, Cyo might function under high oxygen conditions as in the case of E. coli. However, because the expression level of the cyo genes was significantly lower than that of the cco1 genes without stresses, Cyo might make a minor contribution to the cell growth under normal laboratory growth conditions. The cyo genes were found to be significantly induced by iron starvation or in the presence of a nitric oxide (NO)-generating reagent, S-nitrosoglutathione (GSNO; Ochsner et al., 2002; Kawakami et al., 2010). Cyo is likely responsible for respiration under iron-limiting conditions and the expression of the cyo genes is regulated by the transcriptional regulator Fur (ferric uptake regulator), which is known to bind to DNA in the presence of Fe2+ and represses the iron-regulated promoters (Vasil and Ochsner, 1999; Vasil, 2007). Because the predicted number of iron atoms sequestered in the Cyo complex is lower than those of the other terminal oxidase complexes and the respiratory complex terminated by Cyo does not require the iron-containing cytochrome bc1 complex and soluble cytochrome c (Fujiwara et al., 1992; Thöny-Meyer, 1997), the demand for iron might be lower when Cyo is utilized. The cyo genes were upregulated by GSNO, but the role of Cyo in nitrosative stress resistance is not certain. Because Fur is known to lose its DNA-binding ability in the presence of NO (D'Autréaux et al., 2002), the upregulation by GSNO might be merely due to the inactivation of Fur by NO. An obligately aerobic bacterium, P. putida, also has five sets of terminal oxidases corresponding to those of P. aeruginosa (Ugidos et al., 2008). In P. putida, inactivation of the cyo genes relieves the catabolite repression of the phenol- or alkane-degradation genes (Petruschka et al., 2001; Dinamarca et al., 2002). The cyo mutation also leads to a significant change in the transcriptome profile, and the absence of Cyo in P. putida was compensated for by upregulation of CIO and one of the cbb3 oxidases corresponding to Cbb3-2 of P. aeruginosa (Morales et al., 2006). In contrast to P. aeruginosa, Cyo might make a major contribution to the aerobic growth of this obligately aerobic bacterium, although it remains to be investigated whether inactivation of Cyo has a significant influence in P. aeruginosa.
The cyanide-insensitive oxidase
The cyanide-insensitive quinol oxidase (CIO) has been found and described from P. aeruginosa and C. jejuni (Matsushita et al., 1983; Cunningham et al., 1997; Jackson et al., 2007). While the Cbb3-1, Cbb3-2, Aa3, and Cyo oxidases belong to the heme–copper oxidase superfamily, CIO is the only copper-free terminal oxidase in P. aeruginosa. It consists of two subunits encoded by the cioAB genes, which are highly homologous to the cydAB genes for the cytochrome bd-type quinol oxidases (Cunningham et al., 1997; Jünemann, 1997; Jackson et al., 2007). The bd oxidases have high affinity for oxygen and contain low-spin heme b558, high spin heme b595, and heme d (D'mello et al., 1996; Jünemann, 1997). E. coli has two bd oxidases (Bekker et al., 2009), one of which is known to be induced and predominant under oxygen-limited conditions (Rice and Hempfling, 1978; Cotter et al., 1990). In the free-living nitrogen-fixing bacterium Azotobacter vinelandii, the bd oxidase rapidly consumes oxygen and protects the oxygen-sensitive nitrogenase complex (Kelly et al., 1990; Poole and Hill, 1997). This phenomenon is called respiratory protection.
Although the cioAB genes for CIO are highly homologous to the cydAB genes for the bd oxidase, CIOs from P. aeruginosa and C. jejuni lack the spectral features for heme b595 and heme d (Matsushita et al., 1983; Cunningham and Williams, 1995; Cunningham et al., 1997; Jackson et al., 2007). It had been considered that the hemes of the bd oxidase are replaced by other unknown redox centers in CIO. However, recent analysis using the membrane vesicle of a CIO-overproducing Gluconobacter oxydans revealed that CIO carries all of the hemes b558, b595, and d. The absence of the spectroscopic properties was predicted to be caused by the unique ligand-binding properties of CIO (Mogi et al., 2009). A distinctive feature that could be used easily to discriminate CIO from the bd oxidase is that the conserved sequence of the periplasmic loop (Q-loop) that contains the putative quinol oxidizing site is significantly shorter in CioA than in CydA (Cunningham et al., 1997). CIO is known to have higher resistance to cyanide than Cyo (Mogi et al., 2009). In contrast to the finding that the bd oxidases have high affinity for oxygen, CIO of C. jejuni was reported to have low affinity for oxygen, with a Km value of 0.8 μM (Jackson et al., 2007). It has been recognized that CIO of P. aeruginosa has high affinity for oxygen because the cco1 cco2 cio triple mutant, which lacks Cbb3-1, Cbb3-2, and CIO, could not grow microaerobically under a 2% O2 concentration (Alvarez-Ortega and Harwood, 2007). However, our recent analysis using the cco1 cco2 cox cyo quadruple mutant revealed that the Km value of CIO of P. aeruginosa for oxygen is significantly higher than those of Cbb3-1 and Cbb3-2 and comparable to those of Aa3 and Cyo, indicating that CIO of P. aeruginosa is a low affinity enzyme like CIO of C. jejuni (unpublished data).
Pseudomonas aeruginosa produces a respiratory chain inhibitor, hydrogen cyanide, at concentrations up to 300 μM under low oxygen conditions. The concentrations are high enough to inhibit the activity of the heme–copper oxidases. Therefore, CIO is believed to have a role as an electron sink under cyanogenic conditions (Cunningham and Williams, 1995; Cunningham et al., 1997; Blumer and Haas, 2000). The reason why the cco1 cco2 cio triple mutant could not grow under the 2% O2 condition (Alvarez-Ortega and Harwood, 2007) was probably because of the inhibition of Aa3 and Cyo by the endogenous cyanide. Cyanide is reported to be the primary toxic factor responsible for the paralytic killing of Caenorhabditis elegans by P. aeruginosa (Gallagher and Manoil, 2001). CIO appeared to be required for this pathogenicity because the mutant of the cioAB genes (PA3929–3930) had lower killing activity against the nematoda (Zlosnik et al., 2006).
The cioAB genes are upregulated at stationary phase or at very low ambient oxygen concentration (0.4–0.5%) in P. aeruginosa (Cooper et al., 2003; Alvarez-Ortega and Harwood, 2007; Kawakami et al., 2010). Respiratory chain inhibitors, cyanide and sodium nitroprusside (SNP), significantly induce the cio genes (Comolli and Donohue, 2002; Cooper et al., 2003; Kawakami et al., 2010). The cio genes are also induced by copper starvation or disruption of the senC gene (PA0114), which encodes a putative copper chaperone for the heme–copper oxidases (Frangipani et al., 2008; Frangipani and Haas, 2009). Since CIO is the only non-heme–copper oxidase in P. aeruginosa, CIO must be important for respiration under copper-restricted conditions. The cio genes are positively regulated by RoxSR and disruption of the cco1 genes for Cbb3-1 causes upregulation of the cio promoter (Comolli and Donohue, 2004). Regulation of the RoxSR activity by the electron flow through the cbb3 oxidase might be operative in P. aeruginosa as in the case of PrrBA in R. sphaeroides (Oh and Kaplan, 2000). The expression of CIO is negatively regulated by ANR and an anr mutant has been shown to express a high level of CIO, especially under low oxygen concentrations (Cooper et al., 2003). It seems likely that ANR prevents the overproduction of CIO when it is not necessary. The stationary phase sigma factor RpoS also activates the expression of the cio genes, although its contribution is minor when ANR or RoxSR is operative (Kawakami et al., 2010). The regulation of the cio genes in P. aeruginosa is thus complicated, but CIO seems to be controlled so as to be expressed when the other terminal oxidases of the heme–copper superfamily are not functioning.
Regulatory factors controlling the multiple terminal oxidases
Two redox-responsive transcriptional regulators, ANR and RoxSR, and a stationary phase sigma factor RpoS play dominant roles in the control of the multiple terminal oxidases in P. aeruginosa (Figure 2). ANR functions as a global regulator for anaerobic gene expression in response to oxygen depletion (Galimand et al., 1991; Sawers, 1991; Zimmermann et al., 1991). Approximately 170 transcription units are predicted to belong to the ANR regulon (Trunk et al., 2010). ANR is an analog of E. coli FNR (fumarate nitrate reductase regulator), which senses intracellular oxygen levels by an oxygen-sensitive [4Fe–4S]2+ cluster bound to N-terminal Cys residues (Kiley and Beinert, 1998; Unden et al., 2002).
RoxSR is an analog of PrrBA/RegBA of purple photosynthetic bacteria, which activates expression of the photosynthesis genes under low oxygen conditions (Dubbs and Tabita, 2004; Elsen et al., 2004; Eraso et al., 2008). It has been proposed that PrrBA of R. sphaeroides receives an inhibitory signal from the electron flow through the cbb3 oxidase because the PrrBA-dependent genes are upregulated even under aerobic conditions when the cbb3 oxidase activity is blocked by inhibitors or mutations (O'Gara et al., 1998; Oh and Kaplan, 1999, 2000; Kim et al., 2007). The activity of RegBA of R. capsulatus and Rhodospirillum rubrum is proposed to be controlled by the redox status of ubiquinones (Grammel and Ghosh, 2008; Wu and Bauer, 2010). The sensing signal of RoxSR of P. aeruginosa is not certain at present. Mutation of the cco1 genes for Cbb3-1 caused upregulation of the RoxSR-dependent expression of CIO in P. aeruginosa PAK, suggesting the possibility that RoxSR senses the electron flow through Cbb3-1 (Comolli and Donohue, 2004). However, because Cbb3-1 is the dominant terminal oxidase under normal aerobic growth conditions, deletion of Cbb3-1 might greatly influence the redox status of the ubiquinone pool. In the case of P. putida, in which Cyo might make a major contribution under aerobic conditions, the absence of Cyo causes upregulation of CIO as well as a significant change in the transcriptome profile (Morales et al., 2006). In any case, the activity of RoxSR of Pseudomonas species is expected to be controlled, directly or indirectly, by the redox status of the respiratory chain.
ANR activates the expression of Cbb3-2, which is a high affinity enzyme and predicted to be dominant under low oxygen conditions (Ray and Williams, 1997; Comolli and Donohue, 2004). ANR represses the expression of CIO (Cunningham et al., 1997; Cooper et al., 2003; Comolli and Donohue, 2004), which was recently found to be a low affinity enzyme (unpublished data). Another low affinity enzyme, Cyo, is probably repressed in an indirect manner by ANR (Kawakami et al., 2010), which is in contrast to the direct repression of Cyo by ANR in P. putida (Ugidos et al., 2008). RoxSR regulates the expression of all five terminal oxidases, though whether directly or indirectly is not certain (Comolli and Donohue, 2002; Kawakami et al., 2010). Aa3 is repressed and the other four terminal oxidases are activated by RoxSR. Some other genes related to respiratory function, such as hemB and nuoAL, are under the control of RoxSR, indicating that RoxSR plays an extensive role in the regulation of respiration in P. aeruginosa (Kawakami et al., 2010). Cbb3-2, Aa3, and CIO are upregulated at the stationary phase. RpoS plays a significant role in the expression of Aa3, but make a minor contribution to the regulation of CIO and no contribution to that of Cbb3-2 (Cooper et al., 2003; Schuster et al., 2004; Kawakami et al., 2010). RpoS is known to be necessary for survival under carbon starvation in P. aeruginosa (Jørgensen et al., 1999; Suh et al., 1999). Therefore, the induction of Aa3 under nutrient starvation conditions might be mainly the effect of the function of RpoS.
Regulation of the terminal oxidases not only by peripheral oxygen tension by ANR but also by the redox status of the respiratory chain by RoxSR might be a sophisticated mechanism for fine tuning of multiple enzymes with different characteristics, because the redox status of the respiratory components is significantly affected by nutritional conditions and respiratory stressors as well as oxygen availability. Availability of nitrogen oxides that act as alternative electron acceptors for anaerobic respiration might also influence the redox status of the respiratory chain under low oxygen conditions.
Microaerobic physiology of P. aeruginosa
The cbb3 oxidases are dominantly expressed and function even under the aerobic conditions in P. aeruginosa. This feature is unique to P. aeruginosa, because the cbb3 oxidases are known to have very high affinity for oxygen and are usually repressed under high oxygen conditions in other bacterial species (Preisig et al., 1996; Mouncey and Kaplan, 1998; Otten et al., 2001; Swem and Bauer, 2002; Jackson et al., 2007). Our preliminary experiment revealed that both of the two cbb3 oxidases of P. aeruginosa also have high affinity for oxygen (unpublished data). The low affinity enzymes are highly induced only under starvation or stressed conditions (Kawakami et al., 2010). This is in contrast to the case of the non-pathogenic obligately aerobic bacterium P. putida, which is phylogenetically close to P. aeruginosa and has the same set of five terminal oxidases, but in which Cyo might make a major contribution under aerobic conditions (Morales et al., 2006; Fernández-Piñar et al., 2008; Ugidos et al., 2008).
The dominant expression and function of the high affinity enzymes might indicate that the P. aeruginosa cells are maintained in a microaerobic state even under aerobic conditions. P. aeruginosa actively produces a microaerobic environment even at high aeration rates by reducing the transfer rate of oxygen from the gas phase into the liquid phase (Sabra et al., 2002). This phenomenon was predicted to be due to the production of biosurfactants such as rhamnolipid. P. aeruginosa strains infected in the CF airway show the mucoid phenotype by overproduction of the exopolysaccharide alginate, which is known to act as a physical barrier to oxygen entering the cell. Even the non-mucoid strain PAO1 forms a mucoid polysaccharide capsule, which might consist primarily of alginate, on the cell surface as a response to oxidative stress (Sabra et al., 2002). Alginate is also reported to restrict the diffusion of oxygen (Hassett, 1996). A nitrogen-fixing bacterium, A. vinelandii, which is closely related to Pseudomonas, is also known to produce alginate as a barrier to protect oxygen-sensitive nitrogenase from oxygen (Sabra et al., 2000).
The microaerobic physiology of P. aeruginosa might be significantly related to its pathogenesis. During pulmonary infection, P. aeruginosa is exposed to reactive oxygen species produced by the host immune cells. The physical blockage of oxygen transfer by production of the polysaccharide layer and reduction of the oxygen diffusion rate would be advantageous for P. aeruginosa to survive under such oxidative stress conditions. Moreover, release of virulence factors, such as elastase and pyocyanin, into the culture supernatant of P. aeruginosa has been reported to be enhanced by the microaerobic culture conditions (Sabra et al., 2002).
Anaerobic Energy Metabolism
Pseudomonas aeruginosa had been considered as an obligately aerobic bacterium previously, but it is now recognized to be highly adapted to anaerobic conditions. Because the P. aeruginosa-infected mucus in the CF airway is depleted of oxygen, the anaerobic physiology of P. aeruginosa is believed to be important for its pathogenesis (Yoon et al., 2002; Schobert and Jahn, 2010; Schobert and Tielen, 2010). In the absence of oxygen, P. aeruginosa can grow by dissimilatory nitrate respiration by using nitrogen oxides as alternative terminal electron acceptors of the respiratory chain. This process is called denitrification because soluble nitrate and nitrite are reduced to and released as gaseous nitrous oxide (N2O) or dinitrogen (N2; Zumft, 1997). This pathway is ecologically important because it is one of the very few routes for generating atmospheric N2. Complete denitrification consists of four sequential steps to reduce nitrate to N2 via nitrite, nitric oxide (NO), and N2O. Each step of the pathway is catalyzed by individual metalloenzymes, i.e., nitrate reductase, nitrite reductase, NO reductase, and N2O reductase (Figure 3).
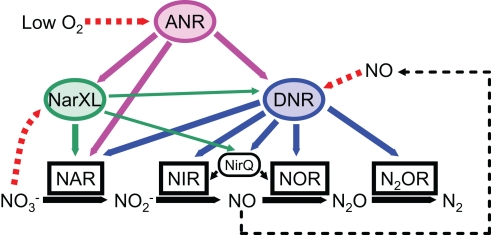
Figure 3.
Schematic model of the regulatory network controlling the denitrification genes in P. aeruginosa. ANR activates the expression of DNR under anaerobic or low oxygen conditions. DNR activates the expression of all denitrification genes in response to nitric oxide. A two-component nitrate sensing regulator, NarXL is required for expression of the nar genes encoding nitrate reductase. Both ANR and DNR can activate the nar gene expression. NirQ is predicted to be involved in the fine tuning of the activities of nitrite reductase and nitric oxide reductase. NAR, NIR, NOR, and N2OR indicate nitrate reductase, nitrite reductase, nitric oxide reductase, and nitrous oxide reductase, respectively.
Denitrification enzymes and genes
Nitrate reductase catalyzes the first step of denitrification, reduction of nitrate to nitrite. Three types of nitrate reductases, Nar, Nap, and Nas, which are localized to the cytoplasmic membrane, periplasm, and cytoplasm, respectively, are encoded in the genome of P. aeruginosa (Berks et al., 1995; Stover et al., 2000). They all contain a molybdopterin guanine dinucleotide cofactor. The membrane-bound Nar is the enzyme responsible for anaerobic nitrate respiration of P. aeruginosa in CF sputum (Palmer et al., 2007). Nar is encoded in the narK1K2GHJI gene cluster (PA3872–3877). NarG, NarH, and NarI are the structural subunits of the enzyme and NarJ is required for the assembly of the functional enzyme (Philippot and Højberg, 1999). The narK1 and narK2 genes encode putative nitrate/nitrite transporters homologous to each other, but only narK2 has been reported to be required for denitrifying growth (Sharma et al., 2006). The narXL genes (PA3878–3879) encoding a two-component transcriptional regulator, NarXL, which is required for the nitrate-responsive expression of the narK1K2GHJI operon, is located upstream from the narK1 gene (Schreiber et al., 2007). Reduction of nitrate by Nar is coupled to quinol oxidation and consumes two protons from the cytoplasm, thereby contributing to creation of a proton gradient across the membrane (Zumft, 1997).
The periplasmic enzyme Nap and related proteins are encoded in the napEFDABC gene cluster (PA1172–1177). Nap is a quinol oxidase, but nitrate reduction in the periplasm by Nap does not contribute to generation of the proton gradient. The physiological function of Nap in P. aeruginosa is not certain at present. In R. sphaeroides, Nap is regulated by both nitrate and electron supply and predicted to be involved in redox balancing by using nitrate as an ancillary oxidant to dissipate excess reductant (Gavira et al., 2002). The cytoplasmic enzyme Nas is exclusively involved in nitrate assimilation. The nasC gene (PA1779) for Nas is clustered with the nirBD genes (PA1780–1781) encoding assimilatory nitrite reductase, which catalyzes the reduction of nitrite to ammonium (Berks et al., 1995).
Nitrite reductase catalyzes the second step of denitrification, reduction of nitrite to NO. Two types of dissimilatory nitrite reductases, the copper-containing type and the cytochrome cd1-type, have been reported so far and P. aeruginosa has the latter type (Zumft, 1997). Both types are located in the periplasm. The cytochrome cd1 nitrite reductase consists of two identical subunits, each containing a covalently attached heme c and a non-covalently bound heme d1, which is a prosthetic group unique to this type of nitrite reductase (Silvestrini et al., 1994). In P. aeruginosa, nitrite reductase is encoded in the nirSMCFDLGHJEN gene cluster (PA0509–0519). nirS is the structural gene for the enzyme (Silvestrini et al., 1989). nirM encodes cytochrome c-551 and nirC encodes another monoheme cytochrome c, and both these cytochromes mediate electron transfer from the cytochrome bc1 complex to nitrite reductase (Arai et al., 1990; Nordling et al., 1990; Hasegawa et al., 2001, 2003). The nirFDLGHJE genes are necessary for the biosynthesis of heme d1 (Kawasaki et al., 1995, 1997). nirN encodes a c-type cytochrome, which is similar to nirS, but its function is not certain (Hasegawa et al., 2001).
Reduction of NO to N2O is catalyzed by NO reductase (Hendriks et al., 2000; Zumft, 2005). Two types of bacterial NO reductases, designated cNOR and qNOR, have been characterized. cNOR is a membrane-bound cytochrome bc complex and receives electrons from soluble cytochrome c. qNOR lacks the cytochrome c component and receives electrons from quinol. P. aeruginosa and many denitrification bacteria have cNOR. qNOR has been found in bacteria and cyanobacteria such as Ralstonia eutropha, Neisseria gonorrhoeae, and Synechocystis sp. PCC6803 (Cramm et al., 1999; Householder et al., 2000; Büsch et al., 2002). Bacterial NO reductase shows similarity to the main subunit of cytochrome c oxidases of the heme–copper superfamily and is predicted to be the ancestor of oxygen-based respiratory enzymes (Saraste, 1994; Saraste and Castresana, 1994; Zumft, 2005). NO reductases have no proton-pumping activity, but cNOR contributes to generation of the proton gradient across the membrane because the electrons for its reaction are supplied through the cytochrome bc1 complex via soluble cytochrome c. The NO reductase of P. aeruginosa has been purified and characterized and its crystal structure has been reported recently (Kumita et al., 2004; Hino et al., 2010). NO reductase is encoded in the norCBD operon (PA0523–0525), which is clustered with the nir genes for nitrite reductase in the genome of P. aeruginosa (Arai et al., 1995a). norC and norB encode cytochrome c and cytochrome b subunits of the enzyme, respectively. norD encodes a soluble protein predicted to be required for production of the active enzyme. NorB carries the binuclear catalytic center consisting of heme b3 and non-heme FeB. NorC mediates electron transfer from soluble cytochrome c to NorB (Zumft, 2005). NO reductase functions not only for anaerobic energy conservation as a respiratory enzyme, but also for detoxification of exogenous NO. The machinery for detoxification of NO and its derivative reactive nitrogen species is necessary because the infected P. aeruginosa cells are subjected to nitrosative stress by the attack of the host immune system. The NO reductase-deficient mutant of P. aeruginosa shows a reduced survival rate in NO-producing macrophages (Kakishima et al., 2007). P. aeruginosa has another NO-detoxification enzyme, flavohemoglobin, which is encoded by the fhp gene (PA2664; Arai et al., 2005). Flavohemoglobin is known to exhibit NO dioxygenase activity under aerobic conditions and NO reductase activity under anaerobic conditions (Poole and Hughes, 2000). However, flavohemoglobin of P. aeruginosa is necessary for detoxification of NO under aerobic conditions but does not support denitrifying growth by complementing the function of NO reductase under anaerobic conditions (Arai et al., 2005). The fhp gene is regulated by the NO-responsive transcriptional regulator FhpR and transcribed as an operon with the following two genes, ppyR (PA2663) and nnrS (PA2662), which encode putative membrane proteins. Disruption of these ppyR and nnrS genes has no effect on the aerobic tolerance to reactive nitrogen species, but the nnrS mutant shows very poor growth under anaerobic denitrification conditions (unpublished data). The ppyR gene has been reported to have a role in the formation of biofilm (Attila et al., 2008).
The nirQOP operon (PA0520–0522) is located between the structural genes for nitrite reductase (nirS) and NO reductase (norCB; Arai et al., 1994, 1996, 1998). The activities of nitrite reductase and NO reductase should be coordinately regulated in order to avoid accumulation of highly cytotoxic intermediate, NO. The function of NirQ is predicted to be the fine tuning of the expression and activation of nitrite reductase and NO reductase (Jüngst and Zumft, 1992; Arai et al., 1996). The nirO and nirP genes encode transmembrane proteins. NirO has sequence similarity to subunit III of cytochrome c oxidases. The function of the nirOP genes is not certain, but it has been proposed that these genes are involved in efficient energy conservation under anaerobic conditions (Arai et al., 1996, 1998).
The final step of the denitrification pathway, reduction of N2O to N2, is catalyzed by N2O reductase. This enzyme has been intensively studied in P. stutzeri and P. denitrificans (Zumft, 1997; Zumft and Kroneck, 2007) and has also been purified and characterized from P. aeruginosa (SooHoo and Hollocher, 1991). N2O reductase is a periplasmic enzyme and predicted to receive electrons from the cytochrome bc1 complex via soluble cytochrome c or pseudoazurin. It has two types of copper centers, the mixed-valent dinuclear CuA species at the electron entry site and the tetranuclear CuZ center at the catalytically active site (Zumft and Kroneck, 2007). The CuA site of N2O reductase shows similarity to the corresponding electron entry site in subunit II of cytochrome oxidases of the heme–copper superfamily, suggesting an evolutionary relationship between the two enzymes for anaerobic and aerobic respiration (Saraste, 1994; Saraste and Castresana, 1994). In P. aeruginosa, N2O reductase is encoded in the nosRZDFYL operon (PA3391–3396; Arai et al., 2003). The structural gene for the enzyme is nosZ. The nosDFY gene products are thought to be involved in the processing and insertion of copper into the enzyme. The nosL gene product is proposed to be an outer membrane disulfide isomerase. nosR encodes a membrane protein of unknown function. Many denitrifying bacteria can grow on N2O as the only electron acceptor under anaerobic conditions. However, P. aeruginosa cannot grow on exogenous N2O as the only electron acceptor, although it can utilize endogenous N2O for the generation of energy for growth during denitrification. This is probably because the nosR promoter is regulated by NO and exogenous N2O does not induce the nos genes (SooHoo and Hollocher, 1990; Arai et al., 2003).
Transcriptional regulation of the denitrification genes
Denitrification enzymes are induced under anaerobic or low oxygen conditions in the presence of nitrate or nitrite (Arai et al., 1991a,b; Figure 3). Two transcriptional regulators, ANR and DNR (dissimilatory nitrate respiration regulator), are required for full expression of all denitrification genes (Arai et al., 1994, 1995b, 1997, 1999, 2003; Ye et al., 1995; Schreiber et al., 2007). NarXL regulates some of the denitrification promoters, such as narK1, nirQ, and dnr (Schreiber et al., 2007). Denitrification is also regulated by quorum-sensing signal molecules (Yoon et al., 2002; Toyofuku et al., 2007, 2008).
The dnr gene (PA0527) encoding DNR is clustered with the nir–nor genes for nitrite reductase and NO reductase. Both ANR and DNR belong to the CRP/FNR superfamily of transcriptional regulators and activate a synthetic promoter that has a consensus FNR-binding motif (TTGAT----ATCAA; Hasegawa et al., 1998). However, the promoters of the denitrification genes, which have sequences similar to the FNR-binding motif, are activated only by DNR, but not by ANR. Expression of DNR is under the control of ANR. Thus, the ANR-mediated anaerobic induction of the denitrification genes is an indirect event that occurs by way of DNR (Arai et al., 1997). The hemN, hemF, and narK1 promoters are known to be recognized by both ANR and DNR (Rompf et al., 1998; Schreiber et al., 2007). It is not certain how ANR and DNR distinguish their target promoters. Transcriptome analysis of the ANR and DNR regulons revealed that DNR specifically regulates the denitrification genes (Trunk et al., 2010). DNR specifically senses NO and does not respond to CO (Arai et al., 2003). Heme is required for the in vivo activity of DNR (Castiglione et al., 2009). The crystal structures of the apo-forms of full-length and truncated DNR have been solved recently (Giardina et al., 2008, 2009, 2011). The structural analyses showed that DNR undergoes a very large conformation rearrangement on activation.
Anaerobic fermentation
Pseudomonas aeruginosa is able to grow in the absence of oxygen and nitrogen oxides, although very slowly, by using arginine as an energy source in rich medium (Shoesmith and Sherris, 1960; Vander Wauven et al., 1984). Anaerobic degradation of arginine to ornithine through the arginine deiminase (ADI) pathway is a non-redox process but produces 1 mol of ATP per arginine by a substrate-level phosphorylation. The ADI pathway enzymes are encoded in the arcDABC operon (PA5170–5173; Lüthi et al., 1990). The arcABC genes encode arginine deiminase, catabolic ornithine carbamoyltransferase, and carbamate kinase, respectively. arcD encodes an arginine-ornithine antiporter. The arc genes are induced by ANR in response to oxygen depletion (Gamper et al., 1991). An arginine-responsive regulator, ArgR, which is involved in the regulation of many arginine metabolic pathways, enhances the arcD promoter activity (Lu et al., 1999). The expression of the arc genes is partially repressed by NarXL in the presence of nitrate, indicating that the more energetically efficient denitrification is preferred over arginine fermentation (Benkert et al., 2008).
Pyruvate fermentation allows long-term anaerobic survival of P. aeruginosa under stationary conditions, although it does not sustain significant anaerobic growth (Eschbach et al., 2004). Acetate kinase and phosphotransacetylase encoded by the ackA-pta operon (PA0835–0836) and NADH-dependent lactate dehydrogenase encoded by ldhA (PA0927) are responsible for pyruvate fermentation. ATP is produced by the acetate kinase activity. Lactate dehydrogenase functions for reoxidation of NADH produced by the pyruvate dehydrogenase activity. ANR and the integration host factor (IHF) are required for the expression of the ackA-pts operon (Eschbach et al., 2004).
Role of pyocyanin in anaerobic survival
Pseudomonas aeruginosa produces a small redox-active phenazine compound, pyocyanin. Pyocyanin generates reactive oxygen species and acts as an antibiotic agent in the soil or a virulence factor during infection. Recently, Newman's group proposed that pyocyanin has additional roles for maintenance of redox homeostasis and control of multicellular behavior (Price-Whelan et al., 2007; Dietrich et al., 2008; Ramos et al., 2010; Wang et al., 2010; Figure 4). Pyocyanin decreases the intracellular NADH levels under energy starvation conditions, indicating that the reoxidation of NADH could be coupled to the reduction of pyocyanin. Because pyocyanin is auto-oxidized by oxygen, it could function as an extracellular electron shuttle. When the P. aeruginosa cells form a biofilm or colony, a steep oxygen gradient is present in the cell community. The cells at the bottom are limited for oxygen but pyocyanin could serve as an alternative electron acceptor. The reduced pyocyanin could be reoxidized after diffusion to the oxygen-rich surface. Thus, pyocyanin contributes to the redox homeostasis and survival of the cells in the deeper anaerobic niches. Further experimental evidence would be required to certify this hypothesis. Pyocyanin is also reported to stimulate pyruvate excretion by decreasing the carbon flux through the central metabolic pathway in P. aeruginosa PA14 at the late stationary phase (Price-Whelan et al., 2007). The pyruvate secreted by the cells at the aerobic niches, such as the surfaces of biofilms, might support the survival of the cells at anaerobic niches of the community as a substrate of anaerobic pyruvate fermentation.
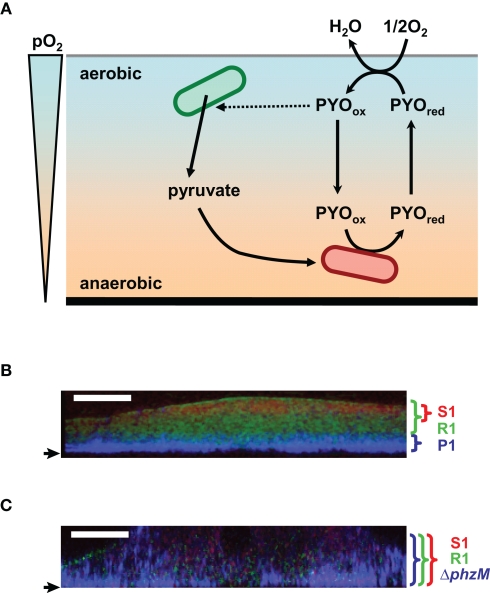
Figure 4.
Hypothetical model of the role of pyocyanin in anaerobic survival in biofilm and construction of the multilayered structure of the multispecies biofilm. (A) Pyocyanin is predicted to act as an electron acceptor for the anaerobic cells and shuttle electrons between anaerobic and aerobic niches. Pyocyanin stimulates excretion of pyruvate. The secreted pyruvate is expected to be utilized for the anaerobic pyruvate fermentation. PYOred and PYOox indicate the reduced- and oxidized-forms of pyocyanin, respectively. pO2 indicates partial oxygen pressure. (B,C) FISH images of three-species biofilms in vertical sections. A pyocyanin-overproducing P. aeruginosa strains P1 (P1) and its pyocyanin-non-producing derivative (ΔphzM) appear blue with a Cy3-labeled probe, A pyocyanin-resistant Raoultella strain (R1) and a pyocyanin-sensitive Brevibacillus strain (S1) appear green and red with the FITC- and Cy5-labeled probes, respectively. Strain P1 forms a multilayered biofilm with strains R1 and S1 (B). An intermingled biofilm is formed when strain ΔphzM is used (C). Small arrows indicate the bottom of biofilms. White bars indicate 50 μm.
Another example of the relationship between pyocyanin and biofilm lifestyle has been reported in a mixed-species biofilm (Narisawa et al., 2008; Figures 4B,C). The pyocyanin-overproducing P. aeruginosa strain P1 forms a multilayered multispecies biofilm when cocultured with a pyocyanin-resistant Raoultella strain R1 and a pyocyanin-sensitive Brevibacillus strain S1. The layer of strain P1 at the bottom was covered with the layers of resistant and sensitive strains. The sensitive strain was separated from strain P1 by the layer of the resistant strain. This multilayered structure is expected to be advantageous for strain P1 for protection from antibiotics, grazing, desiccation, and many other stresses, although the bottom of the biofilm is disadvantageous for oxygen acquisition. A pyocyanin non-producing derivative of strain P1 forms an intermingled multispecies biofilm with the pyocyanin-resistant and -sensitive strains. P. aeruginosa PAO1, which poorly produces pyocyanin under anaerobic conditions, does not construct the multilayered biofilm. These results indicate that pyocyanin has the ability to construct the organized layered structure in the multispecies biofilm. Pyocyanin might also serve as an electron acceptor for the anaerobic preservation of the P. aeruginosa cells at the oxygen-depleted bottom of the multilayered biofilm.
Conclusions and Future Perspectives
The respiratory chain of P. aeruginosa is highly branched and terminated either by multiple terminal oxidases of different characteristics or by the denitrification enzymes that reduce nitrogen oxides. This versatile respiratory function as well as fermentative energy generating systems contribute to the ubiquitous distribution and persistence of P. aeruginosa in various environments under both aerobic and anaerobic conditions. One of the characteristic features of the respiratory chain of P. aeruginosa is that high affinity terminal oxidases of the cbb3-type are dominant even under aerobic conditions. P. aeruginosa seems to actively produce microaerobic environments by itself. The microaerobic and anaerobic physiology of P. aeruginosa is significantly related to its pathogenicity. All terminal oxidases for aerobic respiration are directly or indirectly regulated by RoxSR, which is predicted to sense the redox status of the respiratory chain either by the redox status of the ubiquinone pool or by the electron flow through the terminal oxidases. Because ubiquinone is located at the pivotal point of the divergent electron transport chain of both aerobic and anaerobic respiration (Figure 1), its redox status must be important for the traffic control of the electron flow. The oxygen sensing global regulator ANR directly regulates a subset of the terminal oxidases and indirectly induces all denitrification enzymes. The NO-sensing regulator DNR directly regulates the expression of all denitrification enzymes. Thus, the respiratory function of P. aeruginosa is efficiently controlled by oxygen, NO, and the redox status of the respiratory chain.
In recent years, bacteria-specific energy metabolism has been garnering increasing attention as a therapeutic target (Hurdle et al., 2011). Most of the classical antibiotics target the bioprocesses of actively growing bacteria, such as biosynthesis of proteins, DNA, and peptidoglycan. However, these antibiotics are not effective for eradicating persistent infections, in which most of the bacterial cells are under slow-growing and/or non-growing conditions. Because maintenance of the cellular energy and redox homeostasis is necessary even for the non-growing cells to maintain viability, inhibition of the energy metabolism is expected to be effective to kill the persisting cells. The problem is the versatility of the bacterial respiratory chains when considering the respiratory components as drug targets. Actually, inactivation of a subset of the multiple terminal oxidases of P. aeruginosa has no or only minor effect. In contrast to the rich variety of the enzymes of energy metabolism, the regulatory machinery for the energy metabolism is less variable, with many bacterial species sharing common regulatory systems. Therefore, the regulatory systems might be worth considering as a possible drug target.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
I thank N. Narisawa, T. Kawakami, M. Kuroki, T. Osamura, and K. Yamamoto for providing figures and unpublished data and for their critical reading of the manuscript.
References
- Alvarez-Ortega C., Harwood C. S. (2007). Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol. Microbiol. 65, 153–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai H., Hayashi M., Kuroi A., Ishii M., Igarashi Y. (2005). Transcriptional regulation of the flavohemoglobin gene for aerobic nitric oxide detoxification by the second nitric oxide-responsive regulator of Pseudomonas aeruginosa. J. Bacteriol. 187, 3960–3968 10.1128/JB.187.12.3960-3968.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai H., Igarashi Y., Kodama T. (1991a). Anaerobically induced expression of the nitrite reductase cytochrome c-551 operon from Pseudomonas aeruginosa. FEBS Lett. 280, 351–353 10.1016/0014-5793(91)80329-2 [DOI] [PubMed] [Google Scholar]
- Arai H., Igarashi Y., Kodama T. (1991b). Nitrite activates the transcription of the Pseudomonas aeruginosa nitrite reductase and cytochrome c-551 operon under anaerobic conditions. FEBS Lett. 288, 227–228 10.1016/0014-5793(91)81040-F [DOI] [PubMed] [Google Scholar]
- Arai H., Igarashi Y., Kodama T. (1994). Structure and ANR-dependent transcription of the nir genes for denitrification from Pseudomonas aeruginosa. Biosci. Biotechnol. Biochem. 58, 1286–1291 [DOI] [PubMed] [Google Scholar]
- Arai H., Igarashi Y., Kodama T. (1995a). The structural genes for nitric oxide reductase from Pseudomonas aeruginosa. Biochim. Biophys. Acta 1261, 279–284 10.1016/0167-4781(95)00018-C [DOI] [PubMed] [Google Scholar]
- Arai H., Igarashi Y., Kodama T. (1995b). Expression of the nir and nor genes for denitrification of Pseudomonas aeruginosa requires a novel CRP/FNR-related transcriptional regulator, DNR, in addition to ANR. FEBS Lett. 371, 73–76 10.1016/0014-5793(95)00885-D [DOI] [PubMed] [Google Scholar]
- Arai H., Kawasaki S., Igarashi Y., Kodama T. (1996). “Arrangement and transcriptional regulation of the denitrification genes in Pseudomonas aeruginosa,” in Molecular Biology of Pseudomonads, eds Nakazawa T., Furukawa K., Haas D., Silver S. (Washington DC: ASM Press; ), 298–308 [Google Scholar]
- Arai H., Kodama T., Igarashi Y. (1997). Cascade regulation of the two CRP/FNR-related transcriptional regulators (ANR and DNR) and the denitrification enzymes in Pseudomonas aeruginosa. Mol. Microbiol. 25, 1141–1148 [DOI] [PubMed] [Google Scholar]
- Arai H., Kodama T., Igarashi Y. (1998). The role of the nirQOP genes in energy conservation during anaerobic growth of Pseudomonas aeruginosa. Biosci. Biotechnol. Biochem. 62, 1995–1999 [DOI] [PubMed] [Google Scholar]
- Arai H., Kodama T., Igarashi Y. (1999). Effect of nitrogen oxides on expression of the nir and nor genes for denitrification in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 170, 19–24 [DOI] [PubMed] [Google Scholar]
- Arai H., Mizutani M., Igarashi Y. (2003). Transcriptional regulation of the nos genes for nitrous oxide reductase in Pseudomonas aeruginosa. Microbiology 149, 29–36 10.1099/mic.0.25936-0 [DOI] [PubMed] [Google Scholar]
- Arai H., Roh J. H., Kaplan S. (2008). Transcriptome dynamics during the transition from anaerobic photosynthesis to aerobic respiration in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 190, 286–299 10.1128/JB.01375-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai H., Sanbongi Y., Igarashi Y., Kodama T. (1990). Cloning and sequencing of the gene encoding cytochrome c -551 from Pseudomonas aeruginosa. FEBS Lett. 261, 196–198 10.1016/0014-5793(90)80669-A [DOI] [PubMed] [Google Scholar]
- Attila C., Ueda A., Wood T. K. (2008). PA2663 (PpyR) increases biofilm formation in Pseudomonas aeruginosa PAO1 through the psl operon and stimulates virulence and quorum-sensing phenotypes. Appl. Microbiol. Biotechnol. 78, 293–307 [DOI] [PubMed] [Google Scholar]
- Bekker M., de Vries S., Ter Beek A., Hellingwerf K. J., Teixeira de Mattos M. J. (2009). Respiration of Escherichia coli can be fully uncoupled via the nonelectrogenic terminal cytochrome bd-II oxidase. J. Bacteriol. 191, 5510–5517 10.1128/JB.00562-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benkert B., Quäck N., Schreiber K., Jaensch L., Jahn D., Schobert M. (2008). Nitrate-responsive NarX-NarL represses arginine-mediated induction of the Pseudomonas aeruginosa arginine fermentation arcDABC operon. Microbiology 154, 3053–3060 10.1099/mic.0.2008/018929-0 [DOI] [PubMed] [Google Scholar]
- Berks B. C., Ferguson S. J., Moir J. W. B., Richardson D. J. (1995). Enzymes and associated electron transport systems that catalyse the respiratory reduction of nitrogen oxides and oxyanions. Biochim. Biophys. Acta 1232, 97–173 [DOI] [PubMed] [Google Scholar]
- Blumer C., Haas D. (2000). Mechanism, regulation, and ecological role of bacterial cyanide biosynthesis. Arch. Microbiol. 173, 170–177 [DOI] [PubMed] [Google Scholar]
- Bosma G., Braster M., Stouthamer A. H., van Verseveld H. W. (1987). Isolation and characterization of ubiquinol oxidase complexes from Paracoccus denitrificans cells cultured under various limiting growth conditions in the chemostat. Eur. J. Biochem. 165, 657–663 [DOI] [PubMed] [Google Scholar]
- Büsch A., Friedrich B., Cramm R. (2002). Characterization of the norB gene, encoding nitric oxide reductase, in the nondenitrifying cyanobacterium Synechocystis sp. strain PCC6803. Appl. Environ. Microbiol. 68, 668–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschmann S., Warkentin E., Xie H., Langer J. D., Ermler U., Michel H. (2010). The structure of cbb3 cytochrome oxidase provides insights into proton pumping. Science 329, 327–330 10.1126/science.1187303 [DOI] [PubMed] [Google Scholar]
- Castiglione N., Rinaldo S., Giardina G., Cutruzzolà F. (2009). The transcription factor DNR from Pseudomonas aeruginosa specifically requires nitric oxide and haem for the activation of a target promoter in Escherichia coli. Microbiology 155, 2838–2844 10.1099/mic.0.028027-0 [DOI] [PubMed] [Google Scholar]
- Comolli J. C., Donohue T. J. (2002). Pseudomonas aeruginosa RoxR, a response regulator related to Rhodobacter sphaeroides PrrA, activates expression of the cyanide-insensitive terminal oxidase. Mol. Microbiol. 45, 755–768 [DOI] [PubMed] [Google Scholar]
- Comolli J. C., Donohue T. J. (2004). Differences in two Pseudomonas aeruginosa cbb3 cytochrome oxidases. Mol. Microbiol. 51, 1193–1203 [DOI] [PubMed] [Google Scholar]
- Cooper M., Tavankar G. R., Williams H. D. (2003). Regulation of expression of the cyanide-insensitive terminal oxidase in Pseudomonas aeruginosa. Microbiology 149, 1275–1284 10.1099/mic.0.26017-0 [DOI] [PubMed] [Google Scholar]
- Cotter P. A., Chepuri V., Geniss R. B., Gunsalus R. P. (1990). Cytochrome o (cyoABCDE) and d (cydAB) oxidase gene expression in Escherichia coli is regulated by oxygen, pH, and the fnr gene product. J. Bacteriol. 172, 6333–6338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cramm R., Pohlmann A., Friedrich B. (1999). Purification and characterization of the single-component nitric oxide reductase from Ralstonia eutropha H16. FEBS Lett. 460, 6–10 10.1016/S0014-5793(99)01315-0 [DOI] [PubMed] [Google Scholar]
- Cunningham L., Pitt M., Williams H. D. (1997). The cioAB genes from Pseudomonas aeruginosa code for a novel cyanide-insensitive terminal oxidase related to the cytochrome bd quinol oxidases. Mol. Microbiol. 24, 579–591 [DOI] [PubMed] [Google Scholar]
- Cunningham L., Williams H. D. (1995). Isolation and characterization of mutants defective in the cyanide-insensitive respiratory pathway of Pseudomonas aeruginosa. J. Bacteriol. 177, 432–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Autréaux B., Touati D., Bersch B., Latour J.-M., Michaud-Soret I. (2002). Direct inhibition by nitric oxide of the transcriptional ferric uptake regulation protein via nitrosylation of the iron. Proc. Natl. Acad. Sci. U.S.A. 99, 16619–16624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich L. E. P., Teal T. K., Price-Whelan A., Newman D. K. (2008). Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science 321, 1203–1206 10.1126/science.1160619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinamarca M. A., Ruiz-Manzano A., Rojo F. (2002). Inactivation of cytochrome o ubiquinol oxidase relieves catabolic repression of the Pseudomonas putida GPo1 alkane degradation pathway. J. Bacteriol. 184, 3785–3793 10.1128/JB.184.14.3785-3793.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Mello R., Hill S., Poole R. K. (1995). The oxygen affinity of cytochrome bo’ in Escherichia coli determined by the deoxygenation of oxyleghemoglobin and oxymyoglobin: Km values for oxygen are in the submicromolar range. J. Bacteriol. 177, 867–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'mello R., Hill S., Poole R. K. (1996). The cytochrome bd quinol oxidase in Escherichia coli has an extremely high oxygen affinity and two oxygen-binding haems: implications for regulation of activity in vivo by oxygen inhibition. Microbiology 142, 755–763 10.1099/00221287-142-4-755 [DOI] [PubMed] [Google Scholar]
- Dubbs J. M., Tabita F. R. (2004). Regulators of nonsulfur purple phototrophic bacteria and the interactive control of CO2 assimilation, nitrogen fixation, hydrogen metabolism and energy generation. FEMS Microbiol. Rev. 28, 353–376 [DOI] [PubMed] [Google Scholar]
- Elsen S., Swem L. R., Swem D. L., Bauer C. E. (2004). RegB/RegA, a highly conserved redox-responding global two-component regulatory system. Microbiol. Mol. Biol. Rev. 68, 263–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eraso J. M., Roh J. H., Zeng X., Callister S. J., Lipton M. S., Kaplan S. (2008). Role of the global transcriptional regulator PrrA in Rhodobacter sphaeroides 2.4.1: combined transcriptome and proteome analysis. J. Bacteriol. 190, 4831–4848 10.1128/JB.00301-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eschbach M., Schreiber K., Trunk K., Buer J., Jahn D., Schobert M. (2004). Long-term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. J. Bacteriol. 186, 4596–4604 10.1128/JB.186.14.4596-4604.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Piñar R., Ramos J. L., Rodríguez-Herva J. J., Espinosa-Urgel M. (2008). A two-component regulatory system integrates redox state and population density sensing in Pseudomonas putida. J. Bacteriol. 190, 7666–7674 10.1128/JB.00868-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory J. E., Donohue T. J. (1997). Transcriptional control of several aerobically induced cytochrome structural genes in Rhodobacter sphaeroides. Microbiology 143, 3101–3110 10.1099/00221287-143-10-3101 [DOI] [PubMed] [Google Scholar]
- Frangipani E., Haas D. (2009). Copper acquisition by the SenC protein regulates aerobic respiration in Pseudomonas aeruginosa PAO1. FEMS Microbiol. Lett. 298, 234–240 [DOI] [PubMed] [Google Scholar]
- Frangipani E., Slaveykova V. I., Reimmann C., Haas D. (2008). Adaptation of aerobically growing Pseudomonas aeruginosa to copper starvation. J. Bacteriol. 190, 6706–6717 10.1128/JB.00450-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara T., Fukumori Y., Yamanaka T. (1992). A novel terminal oxidase, cytochrome baa3 purified from aerobically grown Pseudomonas aeruginosa : it shows a clear difference between resting state and pulsed state. J. Biochem. 112, 290–298 [DOI] [PubMed] [Google Scholar]
- Gabel C., Maier R. J. (1993). Oxygen-dependent transcriptional regulation of cytochrome aa3 in Bradyrhizobium japonicum. J. Bacteriol. 175, 128–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galimand M., Gamper M., Zimmermann A., Haas D. (1991). Positive FNR-like control of anaerobic arginine degradation and nitrate respiration in Pseudomonas aeruginosa. J. Bacteriol. 173, 1598–1606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher L. A., Manoil C. (2001). Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J. Bacteriol. 183, 6207–6214 10.1128/JB.183.21.6207-6214.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamper M., Zimmermann A., Haas D. (1991). Anaerobic regulation of transcription initiation in the arcDABC operon of Pseudomonas aeruginosa. J. Bacteriol. 173, 4742–4250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavira M., Roldán M. D., Castillo F., Moreno-Vivián C. (2002). Regulation of nap gene expression and periplasmic nitrate reductase activity in the phototrophic bacterium Rhodobacter sphaeroides DSM158. J. Bacteriol. 184, 1693–1702 10.1128/JB.184.6.1693-1702.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giardina G., Castiglione N., Caruso M., Cutruzzolà F., Rinaldo S. (2011). The Pseudomonas aeruginosa DNR transcription factor: light and shade of nitric oxide-sensing mechanisms. Biochem. Soc. Trans. 39, 294–298 [DOI] [PubMed] [Google Scholar]
- Giardina G., Rinaldo S., Castiglione N., Caruso M., Cutruzzolà F. (2009). A dramatic conformational rearrangement is necessary for the activation of DNR from Pseudomonas aeruginosa. Crystal structure of wild-type DNR. Proteins 77, 174–180 10.1002/prot.22428 [DOI] [PubMed] [Google Scholar]
- Giardina G., Rinaldo S., Johnson K. A., Di Matteo A., Brunori M., Cutruzzolà F. (2008). NO sensing in Pseudomonas aeruginosa : structure of the transcriptional regulator DNR. J. Mol. Biol. 378, 1002–1015 [DOI] [PubMed] [Google Scholar]
- Grammel H., Ghosh R. (2008). Redox-state dynamics of ubiquinone-10 imply cooperative regulation of photosynthetic membrane expression in Rhodospirillum rubrum. J. Bacteriol. 190, 4912–4921 10.1128/JB.00423-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa N., Arai H., Igarashi Y. (1998). Activation of a consensus FNR-dependent promoter by DNR of Pseudomonas aeruginosa in response to nitrite. FEMS Microbiol. Lett. 166, 213–217 [DOI] [PubMed] [Google Scholar]
- Hasegawa N., Arai H., Igarashi Y. (2001). Two c-type cytochromes, NirM and NirC, encoded in the nir gene cluster of Pseudomonas aeruginosa act as electron donors for nitrite reductase. Biochem. Biophys. Res. Commun. 288, 1223–1230 [DOI] [PubMed] [Google Scholar]
- Hasegawa N., Arai H., Igarashi Y. (2003). Need for cytochrome bc1 complex for dissimilatory nitrite reduction of Pseudomonas aeruginosa. Biosci. Biotechnol. Biochem. 67, 121–126 [DOI] [PubMed] [Google Scholar]
- Hassett D. J. (1996). Anaerobic production of alginate by Pseudomonas aeruginosa : alginate restricts diffusion of oxygen. J. Bacteriol. 178, 7322–7325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendriks J., Oubrie A., Castresana J., Urbani A., Gemeinhardt S., Saraste M. (2000). Nitric oxide reductases in bacteria. Biochim. Biophys. Acta 1459, 266–273 [DOI] [PubMed] [Google Scholar]
- Hino T., Matsumoto Y., Nagano S., Sugimoto H., Fukumori Y., Murata T., Iwata S., Shiro Y. (2010). Structural basis of biological N2O generation by bacterial nitric oxide reductase. Science 330, 1666–1670 10.1126/science.1195591 [DOI] [PubMed] [Google Scholar]
- Householder T. C., Fozo E. M., Cardinale J. A., Clark V. L. (2000). Gonococcal nitric oxide reductase is encoded by a single gene, norB, which is required for anaerobic growth and is induced by nitric oxide. Infect. Immun. 68, 5241–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurdle J. G., O'Neill A. J., Chopra I., Lee R. E. (2011). Targeting bacterial membrane function: an underexploited mechanism for treating persistent infections. Nat. Rev. Microbiol. 9, 62–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R. J., Elvers K. T., Lee L. J., Gidley M. D., Wainwright L. M., Lightfoot J., Park S. F., Poole R. K. (2007). Oxygen reactivity of both respiratory oxidases in Campylobacter jejuni : the cydAB genes encode a cyanide-resistant, low-affinity oxidase that is not of the cytochrome bd type. J. Bacteriol. 189, 1604–1615 10.1128/JB.00897-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen F., Bally M., Chapon-Herve V., Michel G., Lazdunski A., Williams P., Stewart G. S. A. B. (1999). RpoS-dependent stress tolerance in Pseudomonas aeruginosa. Microbiology 145, 835–844 10.1099/13500872-145-4-835 [DOI] [PubMed] [Google Scholar]
- Jünemann S. (1997). Cytochrome bd terminal oxidase. Biochim. Biophys. Acta 1321, 107–127 [DOI] [PubMed] [Google Scholar]
- Jüngst A., Zumft W. G. (1992). Interdependence of respiratory NO reduction and nitrite reduction revealed by mutagenesis of nirQ, a novel gene in the denitrification gene cluster of Pseudomonas stutzeri. FEBS Lett. 314, 308–314 10.1016/0014-5793(92)81495-8 [DOI] [PubMed] [Google Scholar]
- Kakishima K., Shiratsuchi A., Taoka A., Nakanishi Y., Fukumori Y. (2007). Participation of nitric oxide reductase in survival of Pseudomonas aeruginosa in LPS-activated macrophages. Biochem. Biophys. Res. Commun. 355, 587–591 [DOI] [PubMed] [Google Scholar]
- Kawakami T., Kuroki M., Ishii M., Igarashi Y., Arai H. (2010). Differential expression of multiple terminal oxidases for aerobic respiration in Pseudomonas aeruginosa. Environ. Microbiol. 12, 1399–1412 [DOI] [PubMed] [Google Scholar]
- Kawasaki S., Arai H., Igarashi Y., Kodama T. (1995). Sequencing and characterization of the downstream region of the genes encoding nitrite reductase and cytochrome c-551 (nirSM) from Pseudomonas aeruginosa : identification of the gene necessary for biosynthesis of heme d1. Gene 167, 87–91 10.1016/0378-1119(95)00641-9 [DOI] [PubMed] [Google Scholar]
- Kawasaki S., Arai H., Kodama T., Igarashi Y. (1997). Gene cluster for dissimilatory nitrite reductase (nir) from Pseudomonas aeruginosa : sequencing and identification of a locus for heme d1 biosynthesis. J. Bacteriol. 179, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly M. J. S., Poole R. K., Yates M. G., Kennedy C. (1990). Cloning and mutagenesis of genes encoding the cytochrome bd terminal oxidase complex in Azotobacter vinelandii : mutants deficient in the cytochrome d complex are unable to fix nitrogen in air. J. Bacteriol. 172, 6010–6019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiley P. J., Beinert H. (1998). Oxygen sensing by the global regulator, FNR: the role of the iron-sulfur cluster. FEMS Microbiol. Rev. 22, 341–352 [DOI] [PubMed] [Google Scholar]
- Kim Y.-J., Ko I.-J., Lee J.-M., Kang H.-Y., Kim Y. M., Kaplan S., Oh J.-I. (2007). Dominant role of the cbb3 oxidase in regulation of photosynthesis gene expression through the PrrBA system in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 189, 5617–5625 10.1128/JB.00443-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumita H., Matsuura K., Hino T., Takahashi S., Hori H., Fukumori Y., Morishima I., Shiro Y. (2004). NO reduction by nitric-oxide reductase from denitrifying bacterium Pseudomonas aeruginosa : characterization of reaction intermediates that appear in the single turnover cycle. J. Biol. Chem. 279, 55247–55254 [DOI] [PubMed] [Google Scholar]
- Lu C. D., Winteler H., Abdelal A., Haas D. (1999). The ArgR regulatory protein, a helper to the anaerobic regulator ANR during transcriptional activation of the arcD promoter in Pseudomonas aeruginosa. J. Bacteriol. 181, 2459–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüthi E., Baur H., Gamper M., Brunner F., Villeval D., Mercenier A., Haas D. (1990). The arc operon for anaerobic arginine catabolism in Pseudomonas aeruginosa contains an additional gene, arcD, encoding a membrane protein. Gene 87, 37–43 10.1016/0378-1119(90)90493-B [DOI] [PubMed] [Google Scholar]
- Lyczak J. B., Cannon C. L., Pier G. B. (2002). Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15, 194–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita K., Shinagawa E., Adachi O., Ameyama M. (1982). o-type cytochrome oxidase in the membrane of aerobically grown Pseudomonas aeruginosa. FEBS Lett. 139, 255–258 10.1016/0014-5793(82)80864-8 [DOI] [PubMed] [Google Scholar]
- Matsushita K., Yamada M., Shinagawa E., Adachi O., Ameyama M. (1983). Membrane-bound respiratory chain of Pseudomonas aeruginosa grown aerobically. A KCN-insensitive alternate oxidase chain and its energetics. J. Biochem. 93, 1137–1144 [DOI] [PubMed] [Google Scholar]
- Mogi T., Ano Y., Nakatsuka T., Toyama H., Muroi A., Miyoshi H., Migita C. T., Ui H., Shiomi K., Omura S., Kita K., Matsushita K. (2009). Biochemical and spectroscopic properties of cyanide-insensitive quinol oxidase from Gluconobacter oxydans. J. Biochem. 146, 263–271 10.1093/jb/mvp067 [DOI] [PubMed] [Google Scholar]
- Mogi T., Saiki K., Anraku Y. (1994). Biosynthesis and functional role of haem O and haem A. Mol. Microbiol. 14, 391–398 [DOI] [PubMed] [Google Scholar]
- Morales G., Ugidos A., Rojo F. (2006). Inactivation of the Pseudomonas putida cytochrome o ubiquinol oxidase leads to a significant change in the transcriptome and to increased expression of the CIO and cbb3-1 terminal oxidases. Environ. Microbiol. 8, 1764–1774 [DOI] [PubMed] [Google Scholar]
- Mouncey N. J., Kaplan S. (1998). Oxygen regulation of the ccoN gene encoding a component of the cbb3 oxidase in Rhodobacter sphaeroides 2.4.1T: involvement of the FnrL protein. J. Bacteriol. 180, 2228–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagata K., Tsukita S., Tamura T., Sone N. (1996). A cb-type cytochrome-c oxidase terminates the respiratory chain in Helicobacter pylori. Microbiology 142, 1757–1763 10.1099/13500872-142-7-1757 [DOI] [PubMed] [Google Scholar]
- Nakamura H., Saiki K., Mogi T., Anraku Y. (1997). Assignment and functional roles of the cyoABCDE gene products required for the Escherichia coli bo-type quinol oxidase. J. Biochem. 122, 415–421 [DOI] [PubMed] [Google Scholar]
- Narisawa N., Haruta S., Arai H., Ishii M., Igarashi Y. (2008). Coexistence of antibiotic-producing and antibiotic-sensitive bacteria in biofilms is mediated by resistant bacteria. Appl. Environ. Microbiol. 74, 3887–3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordling M., Young S., Karlsson B. G., Lundberg L. G. (1990). The structural gene for cytochrome c551 from Pseudomonas aeruginosa. The nucleotide sequence shows a location downstream of the nitrite reductase gene. FEBS Lett. 259, 230–232 10.1016/0014-5793(90)80015-B [DOI] [PubMed] [Google Scholar]
- Ochsner U. A., Wilderman P. J., Vasil A. I., Vasil M. L. (2002). GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa : identification of novel pyoverdine biosynthesis genes. Mol. Microbiol. 45, 1277–1287 [DOI] [PubMed] [Google Scholar]
- O'Gara J. P., Eraso J. M., Kaplan S. (1998). A redox-responsive pathway for aerobic regulation of photosynthesis gene expression in Rhodobacter sphaeroides 2.4.1. J. Bacteriol. 180, 4044–4050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J. I., Kaplan S. (1999). The cbb3 terminal oxidase of Rhodobacter sphaeroides 2.4.1: structural and functional implications for the regulation of spectral complex formation. Biochemistry 38, 2688–2696 10.1021/bi9825100 [DOI] [PubMed] [Google Scholar]
- Oh J. I., Kaplan S. (2000). Redox signaling: globalization of gene expression. EMBO J. 19, 4237–4247 10.1093/emboj/19.16.4237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J. I., Kaplan S. (2002). Oxygen adaptation – The role of the CcoQ subunit of the cbb3 cytochrome c oxidase of Rhodobacter sphaeroides 2.4.1. J. Biol. Chem. 277, 16220–16228 [DOI] [PubMed] [Google Scholar]
- Oh J. I., Ko I. J., Kaplan S. (2004). Reconstitution of the Rhodobacter sphaeroides cbb3-PrrBA signal transduction pathway in vitro. Biochemistry 43, 7915–7923 10.1021/bi0496440 [DOI] [PubMed] [Google Scholar]
- Otten M. F., Stork D. M., Reijnders W. N., Westerhoff H. V., Van Spanning R. J. M. (2001). Regulation of expression of terminal oxidases in Paracoccus denitrificans. Eur. J. Biochem. 268, 2486–2497 [DOI] [PubMed] [Google Scholar]
- Palmer K. L., Brown S. A., Whiteley M. (2007). Membrane-bound nitrate reductase is required for anaerobic growth in cystic fibrosis sputum. J. Bacteriol. 189, 4449–4455 10.1128/JB.00162-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A., Kulajta C., Pawlik G., Daldal F., Koch H.-G. (2008). Stability of the cbb3-type cytochrome oxidase requires specific CcoQ–CcoP interactions. J. Bacteriol. 190, 5576–5586 10.1128/JB.00534-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruschka L., Burchhardt G., Müller C., Weihe C., Herrmann H. (2001). The cyo operon of Pseudomonas putida is involved in catabolite repression of phenol degradation. Mol. Genet. Genomics 266, 199–206 [DOI] [PubMed] [Google Scholar]
- Philippot L., Højberg O. (1999). Dissimilatory nitrate reductases in bacteria. Biochim. Biophys. Acta 1446, 1–23 [DOI] [PubMed] [Google Scholar]
- Pitcher R. S., Watmough N. J. (2004). The bacterial cytochrome cbb3 oxidases. Biochim. Biophys. Acta 1655, 388–399 [DOI] [PubMed] [Google Scholar]
- Poole R. K., Hill S. (1997). Respiratory protection of nitrogenase activity in Azotobacter vinelandii – roles of the terminal oxidases. Biosci. Rep. 17, 303–317 [DOI] [PubMed] [Google Scholar]
- Poole R. K., Hughes M. N. (2000). New functions for the ancient globin family: bacterial responses to nitric oxide and nitrosative stress. Mol. Microbiol. 36, 775–783 [DOI] [PubMed] [Google Scholar]
- Preisig O., Anthamatten D., Hennecke H. (1993). Genes for a microaerobically induced oxidase complex in Bradyrhizobium japonicum are essential for a nitrogen-fixing endosymbiosis. Proc. Natl. Acad. Sci. U.S.A. 90, 3309–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preisig O., Zufferey R., Thöny-Meyer L., Appleby C. A., Hennecke H. (1996). A high-affinity cbb3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum. J. Bacteriol. 178, 1532–1538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price-Whelan A., Dietrich L. E. P., Newman D. K. (2007). Pyocyanin alters redox homeostasis and carbon flux through central metabolic pathways in Pseudomonas aeruginosa PA14. J. Bacteriol. 189, 6372–6381 10.1128/JB.00505-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puustinen A., Finel M., Virkki M., Wikström M. (1989). Cytochrome o (bo) is a proton pump in Paracoccus denitrificans and Escherichia coli. FEBS Lett. 249, 163–167 10.1016/0014-5793(89)80616-7 [DOI] [PubMed] [Google Scholar]
- Ramos I., Dietrich L. E. P., Price-Whelan A., Newman D. K. (2010). Phenazines affect biofilm formation by Pseudomonas aeruginosa in similar ways at various scales. Res. Microbiol. 161, 187–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A., Williams H. D. (1997). The effects of mutation of the anr gene on the aerobic respiratory chain of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 156, 227–232 [DOI] [PubMed] [Google Scholar]
- Rice C. W., Hempfling W. P. (1978). Oxygen-limited continuous culture and respiratory energy conservation in Escherichia coli. J. Bacteriol. 134, 115–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompf A., Hungerer C., Hoffman T., Lindenmeyer M., Römling U., Groβ U., Doss M. O., Arai H., Igarashi Y., Jahn D. (1998). Regulation of Pseudomonas aeruginosa hemF and hemN by the dual action of the redox response regulators Anr and Dnr. Mol. Microbiol. 29, 985–997 [DOI] [PubMed] [Google Scholar]
- Sabra W., Kim E.-J., Zeng A.-P. (2002). Physiological responses of Pseudomonas aeruginosa PAO1 to oxidative stress in controlled microaerobic and aerobic cultures. Microbiology 148, 3195–3202 [DOI] [PubMed] [Google Scholar]
- Sabra W., Zeng A.-P., Lünsdorf H., Deckwer W.-D. (2000). Effect of oxygen on formation and structure of Azotobacter vinelandii alginate and its role in protecting nitrogenase. Appl. Environ. Microbiol. 66, 4037–4044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki K., Mogi T., Anraku Y. (1992). Heme O biosynthesis in Escherichia coli : the cyoE gene in the cytochrome bo operon encodes a protoheme IX farnesyltransferase. Biochem. Biophys. Res. Commun. 189, 1491–1497 [DOI] [PubMed] [Google Scholar]
- Saiki K., Nakamura H., Mogi T., Anraku Y. (1996). Probing a role of subunit IV of the Escherichia coli bo-type ubiquinol oxidase by deletion and cross-linking analyses. J. Biol. Chem. 271, 15336–15340 [DOI] [PubMed] [Google Scholar]
- Saraste M. (1994). Structure and evolution of cytochrome oxidase. Antonie Van Leeuwenhoek 65, 285–287 10.1007/BF00872214 [DOI] [PubMed] [Google Scholar]
- Saraste M., Castresana J. (1994). Cytochrome oxidase evolved by tinkering with denitrification enzymes. FEBS Lett. 341, 1–4 10.1016/0014-5793(94)80228-9 [DOI] [PubMed] [Google Scholar]
- Sawers R. G. (1991). Identification and molecular characterization of a transcriptional regulator from Pseudomonas aeruginosa PAO1 exhibiting structural and functional similarity to the FNR protein of Escherichia coli. Mol. Microbiol. 5, 1469–1481 [DOI] [PubMed] [Google Scholar]
- Schobert M., Jahn D. (2010). Anaerobic physiology of Pseudomonas aeruginosa in the cystic fibrosis lung. Int. J. Med. Microbiol. 300, 549–556 [DOI] [PubMed] [Google Scholar]
- Schobert M., Tielen P. (2010). Contribution of oxygen-limiting conditions to persistent infection of Pseudomonas aeruginosa. Future Microbiol. 5, 603–621 10.2217/fmb.10.16 [DOI] [PubMed] [Google Scholar]
- Schreiber K., Krieger R., Benkert B., Eschbach M., Arai H., Schobert M., Jahn D. (2007). The anaerobic regulatory network required for Pseudomonas aeruginosa nitrate respiration. J. Bacteriol. 189, 4310–4314 10.1128/JB.00240-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurek K. N., Marr A. K., Taylor P. K., Wiegand I., Semenec L., Khaira B. K., Hancock R. E. W. (2008). Novel genetic determinants of low-level aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 52, 4213–4219 10.1128/AAC.00507-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster M., Hawkins A. C., Harwood C. S., Greenberg E. P. (2004). The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol. Microbiol. 51, 973–985 [DOI] [PubMed] [Google Scholar]
- Sharma V., Noriega C. E., Rowe J. J. (2006). Involvement of NarK1 and NarK2 proteins in transport of nitrate and nitrite in the denitrifying bacterium Pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol. 72, 695–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoesmith J. H., Sherris J. C. (1960). Studies on the mechanism of arginine-activated motility in a Pseudomonas strain. J. Gen. Microbiol. 22, 10–24 10.1099/00221287-22-1-10 [DOI] [PubMed] [Google Scholar]
- Silvestrini M. C., Falcinelli S., Ciabatti I., Cutruzzolà F., Brunori M. (1994). Pseudomonas aeruginosa nitrite reductase (or cytochrome oxidase): an overview. Biochimie 76, 641–654 10.1016/0300-9084(94)90141-4 [DOI] [PubMed] [Google Scholar]
- Silvestrini M. C., Galeotti C. L., Gervais M., Schininá E., Barra D., Bossa F., Brunori M. (1989). Nitrite reductase from Pseudomonas aeruginosa : sequence of the gene and the protein. FEBS Lett. 254, 33–38 10.1016/0014-5793(89)81004-X [DOI] [PubMed] [Google Scholar]
- SooHoo C. K., Hollocher T. C. (1990). Loss of nitrous oxide reductase in Pseudomonas aeruginosa cultured under N2O as determined by rocket immunoelectrophoresis. Appl. Environ. Microbiol. 56, 3591–3592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SooHoo C. K., Hollocher T. C. (1991). Purification and characterization of nitrous oxide reductase from Pseudomonas aeruginosa strain P2. J. Biol. Chem. 266, 2203–2209 [PubMed] [Google Scholar]
- Stover C. K., Pham X. Q., Erwin A. L., Mizoguchi S. D., Warrener P., Hickey M. J., Brinkman F. S. L., Hufnagle W. O., Kowalik D. J., Lagrou M., Garber R. L., Goltry L., Tolentino E., Westbrock-Wadman S., Yuan Y., Brody L. L., Coulter S. N., Folger K. R., Kas A., Larbig K., Lim R., Smith K., Spencer D., Wong G. K.-S., Wu Z., Paulsen I. T., Reizer J., Saier M. H., Hancock R. E. W., Lory S., Olson M. V. (2000). Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406, 959–964 10.1038/35023079 [DOI] [PubMed] [Google Scholar]
- Suh S.-J., Silo-Suh L., Woods D. E., Hassett D. J., West S. E. H., Ohman D. E. (1999). Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J. Bacteriol. 181, 3890–3897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swem D. L., Bauer C. E. (2002). Coordination of ubiquinol oxidase and cytochrome cbb3 oxidase expression by multiple regulators in Rhodobacter capsulatus. J. Bacteriol. 184, 2815–2820 10.1128/JB.184.10.2815-2820.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thöny-Meyer L. (1997). Biogenesis of respiratory cytochromes in bacteria. Microbiol. Mol. Biol. Rev. 61, 337–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyofuku M., Nomura N., Fujii T., Takaya N., Maseda H., Sawada I., Nakajima T., Uchiyama H. (2007). Quorum sensing regulates denitrification in Pseudomonas aeruginosa PAO1. J. Bacteriol. 189, 4969–4972 10.1128/JB.00289-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyofuku M., Nomura N., Kuno E., Tashiro Y., Nakajima T., Uchiyama H. (2008). Influence of the Pseudomonas quinolone signal on denitrification in Pseudomonas aeruginosa. J. Bacteriol. 190, 7947–7956 10.1128/JB.00968-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trunk K., Benkert B., Quäck N., Münch R., Scheer M., Garbe J., Jänsch L., Trost M., Wehland J., Buer J., Jahn M., Schobert M., Jahn D. (2010). Anaerobic adaptation in Pseudomonas aeruginosa : definition of the Anr and Dnr regulons. Environ. Microbiol. 12, 1719–1733 [DOI] [PubMed] [Google Scholar]
- Ugidos A., Morales G., Rial E., Williams H. D., Rojo F. (2008). The coordinate regulation of multiple terminal oxidases by the Pseudomonas putida ANR global regulator. Environ. Microbiol. 10, 1690–1702 [DOI] [PubMed] [Google Scholar]
- Unden G., Achebach S., Holighaus G., Tran H. G., Wackwitz B., Zeuner Y. (2002). Control of FNR function of Escherichia coli by O2 and reducing conditions. J. Mol. Microbiol. Biotechnol. 4, 263–268 [PubMed] [Google Scholar]
- Vander Wauven C., Piérard A., Kley-Raymann M., Haas D. (1984). Pseudomonas aeruginosa mutants affected in anaerobic growth on arginine: evidence for a four-gene cluster encoding the arginine deiminase pathway. J. Bacteriol. 160, 928–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasil M. L. (2007). How we learnt about iron acquisition in Pseudomonas aeruginosa : a series of very fortunate events. Biometals 20, 587–601 10.1007/s10534-006-9067-2 [DOI] [PubMed] [Google Scholar]
- Vasil M. L., Ochsner U. A. (1999). The response of Pseudomonas aeruginosa to iron: genetics, biochemistry and virulence. Mol. Microbiol. 34, 399–413 [DOI] [PubMed] [Google Scholar]
- Wang Y., Kern S. E., Newman D. K. (2010). Endogenous phenazine antibiotics promote anaerobic survival of Pseudomonas aeruginosa via extracellular electron transfer. J. Bacteriol. 192, 365–369 10.1128/JB.01188-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams H. D., Zlosnik J. E., Ryall B. (2007). Oxygen, cyanide and energy generation in the cystic fibrosis pathogen Pseudomonas aeruginosa. Adv. Microb. Physiol. 52, 1–71 [DOI] [PubMed] [Google Scholar]
- Winstedt L., von Wachenfeldt C. (2000). Terminal oxidases of Bacillus subtilis strain 168: one quinol oxidase, cytochrome aa3 or cytochrome bd, is required for aerobic growth. J. Bacteriol. 182, 6557–6564 10.1128/JB.182.23.6557-6564.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Bauer C. E. (2010). RegB kinase activity is controlled in part by monitoring the ratio of oxidized to reduced ubiquinones in the ubiquinone pool. MBio 1, e00272–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R. W., Haas D., Ka J.-O., Krishnapillai V., Zimmermann A., Baird C., Tiedje J. M. (1995). Anaerobic activation of the entire denitrification pathway in Pseudomonas aeruginosa requires Anr, an analog of Fnr. J. Bacteriol. 177, 3606–3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S. S., Hennigan R. F., Hilliard G. M., Ochsner U. A., Parvatiyar K., Kamani M. C., Allen H. L., DeKievit T. R., Gardner P. R., Schwab U., Rowe J. J., Iglewski B. H., McDermott T. R., Mason R. P., Wozniak D. J., Hancock R. E. W., Parsek M. R., Noah T. L., Boucher R. C., Hassett D. J. (2002). Pseudomonas aeruginosa anaerobic respiration in biofilms: relationships to cystic fibrosis pathogenesis. Dev. Cell 3, 593–603 [DOI] [PubMed] [Google Scholar]
- Zimmermann A., Reimmann C., Galimand M., Haas D. (1991). Anaerobic growth and cyanide synthesis of Pseudomonas aeruginosa depend on anr, a regulatory gene homologous with fnr of Escherichia coli. Mol. Microbiol. 5, 1483–1490 [DOI] [PubMed] [Google Scholar]
- Zlosnik J. E. A., Tavankar G. R., Bundy J. G., Mossialos D., O'Toole R., Williams H. D. (2006). Investigation of the physiological relationship between the cyanide-insensitive oxidase and cyanide production in Pseudomonas aeruginosa. Microbiology 152, 1407–1415 10.1099/mic.0.28396-0 [DOI] [PubMed] [Google Scholar]
- Zufferey R., Preisig O., Hennecke H., Thöny-Meyer L. (1996). Assembly and function of the cytochrome cbb3 oxidase subunits in Bradyrhizobium japonicum. J. Biol. Chem. 271, 9114–9119 [DOI] [PubMed] [Google Scholar]
- Zumft W. G. (1997). Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61, 533–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumft W. G. (2005). Nitric oxide reductases of prokaryotes with emphasis on the respiratory, heme-copper oxidase type. J. Inorg. Biochem. 99, 194–215 [DOI] [PubMed] [Google Scholar]
- Zumft W. G., Kroneck P. M. H. (2007). Respiratory transformation of nitrous oxide (N2O) to dinitrogen by Bacteria and Archaea. Adv. Microb. Physiol. 52, 107–227 [DOI] [PubMed] [Google Scholar]