Abstract
Cancer is caused by multiple genetic alterations leading to uncontrolled cell proliferation through multiple pathways. Malignant cells arise from a variety of genetic factors, such as mutations in tumor suppressor genes (TSGs) that are involved in regulating the cell cycle, apoptosis, or cell differentiation, or maintenance of genomic integrity. Tumor suppressor mouse models are the most frequently used animal models in cancer research. The anti-tumorigenic functions of TSGs, and their role in development and differentiation, and inhibition of oncogenes are discussed. In this review, we summarize some of the important transgenic and knockout mouse models for TSGs, including Rb, p53, Ink4a/Arf, Brca1/2, and their related genes.
Keywords: p53, Rb, Ink4a/Arf, BRCA, tumor suppressor genes, transgenic mice, knockout mice, mouse models, ageing
Introduction
Cancer is induced by genetic and environmental factors. Cancer can be categorized as familial (inherited) or sporadic, the latter comprising the vast majority of cases. Germline mutations predispose an individual to familial cancers. When the function of remaining wild-type allele is lost in a somatic cell, additional mutation(s) on the wild-type locus can accumulate that lead to tumor formation. In sporadic cancers, all tumorigenic mutations are somatic and are present only in the patient’s neoplastic cells. In humans, mutations leading to gain of functions of proto-oncogenes or loss of functions of tumor-suppressor genes (TSGs) predispose to cancer. TSGs can be divided into two groups: “gatekeepers” and “caretakers”.1–3
Gatekeepers regulate the growth of tumors by inhibiting their proliferation or promoting their apoptosis. Caretakers control cellular processes that repair genetic alterations and maintain genomic integrity. Mutations of caretakers can result in an increase in the overall mutation rate in a given cell, an apparent prerequisite for tumorigenesis. The accumulation of mutations in a dysregulated cell favors the subsequent clonal selection of variant progeny with aggressive growth properties leading to malignancy. Familial cancers often result from the initial germline mutation of one allele of a TSG followed later by somatic mutation or loss of the second allele, a process known as loss of heterozygosity (LOH). Knudson’s two-hit cancer model4,5 is based on this repeated observation. Although LOH was originally a prerequisite for the identification of canonical TSGs, the remaining wild-type allele can also be transcriptionally silenced by hypermethylation of its CpG regions. Functional loss then leads to tumorigenesis.
Animal models have been instrumental in the study of genes involved in human cancer initiation and progression. Spontaneous as well as carcinogen-induced malignancies have been studied in dogs, rats, and mice, and TSGs identified in Drosophila have led to the discovery of mammalian orthologues. The availability of null and tissue-specific mouse mutants for tumor-suppressor genes has greatly facilitated our understanding of the mechanisms leading to cancer.
The mouse remains the animal model of choice for several reasons. First, mice and humans have roughly the same number of genes, and intracellular signaling pathways are highly conserved between the two species. Second, the success of genetic engineering in mice has allowed the study of gene functions in vivo. Mice can be genetically manipulated to overexpress (by transgenesis), or not express (by gene targeting), a specific cancer gene. These animals offer unique opportunities to uncover cellular pathways controlled by specific TSGs and to dissect mechanisms underlying malignancies that may closely resemble those in humans. In addition, the interbreeding of an increasing number of mice with specific mutations in both TSGs and oncogenes allows the assessment of how these mutations cooperate to produce cancer.
Tumor models in mice include (1) mice in which cancer is caused by intrauterine or postnatal exposure to chemical mutagens, and mice in which tumors are produced by viral or bacterial infection, (2) xenograft models that were generated by directly implanting cancer cell lines established from human tumors into mice have been widely used for drug discovery, and (3) genetically engineered mouse (GEM: constitutive or conditional transgenic, knockin, and knockout) cancer models. The major limitations of xenograft models are the requirement for an immunocompromized host and the inability of these models to fully recapitulate the complex relationship between the tumor and its microenvironment (eg, angiogenesis). Most importantly, the ability of xenografts to accurately predict drug efficacy in human cancer patients has been disappointing. In contrast, GEM cancer models are becoming more and more sophisticated in their ability to accurately mimic the histology and biological behavior of human cancers.6,7 Numerous tissue-specific GEM models have been developed that exhibit many biologic hallmarks of human cancer, including angiogenesis and stromal interactions, as well as similar histopathologic and genetic abnormalities. The major advantages of GEM models are that (i) the initiating genetic event is known, (ii) the mice are immunocompetent, and (iii) the tumors develop spontaneously in their appropriate tissue compartments. Moreover, GEM cancer models are particularly useful for conducting preclinical studies of rare cancers and for assessing synergy between therapeutic agents since they allow assessment of therapeutic efficacy on a uniform genetic background. They can also potentially provide the tools needed to learn more about the histological and biochemical effects of specific agents prior to human testing.
In this review, we have focused on recent progress in transgenic and knockout mouse models for TSGs involved in apoptosis and the cell cycle, DNA damage repair, cell signaling, and differentiation, and their clinical implications. Special emphasis is placed on conditional models, knock-in models, and transgenic models of TSGs.
p53 models
Mouse knock-in p53 mutation models are beginning to establish clear p53 tumor suppressor genotype-phenotype relationships. Previous studies identified a hot spot variant p53 mutation (R175H) in sporadic human tumors. R175H functions normally in blocking cell cycle progression, but is deficient in promoting apoptosis, similar to the R175L variant. Both p53R270H/+ and p53R172H/+ mice are models for human Li-Fraumeni syndrome, and importantly develop epithelial tumors that are not found in p53−/− mice, including carcinomas in the lung, small intestine, colon, breast, skin, liver, and pancreas, and more frequent endothelial tumors. Thus, point mutant p53 alleles expressed under physiological control have enhanced oncogenic potential beyond the simple loss of p53 function. p53 gain-of-function knock-in models develop carcinomas rather than sarcomas within one year, and are thus more faithfully reproduce human carcinomas than classical p53 knock-out models. These novel p53 knock-in mice will be useful in drug screens for carcinomas. Mutant mice that constitutively express activated p53 showed enhanced resistance to spontaneous tumors compared with wild-type littermates and displayed an early onset of phenotypes associated with ageing.
p53, located on the human chromosome 17p13.1, is one of the most extensively studied TSGs.6–10 Li-Fraumeni syndrome is caused by inherited TP53 mutations and associated with high susceptibility to breast and lung carcinomas, soft tissue sarcomas (STSs) brain tumors, osteosarcoma, and leukemias. The p53 transcript encodes a 393-aa nuclear phosphoprotein of 53 kDa. The p53 protein exists as a tetramer exhibiting its tumor-suppressive effect by binding to DNA at specific sites. p53 activity is modulated by protein stability, regulated largely through interactions with the E3 ligase Mdm2.
Binding of Mdm2 to p53 leads to p53 degradation and loss of activity, a process that can be inhibited by the gatekeeper protein Arf (Fig. 1). Post-translational modifications mediate p53 activity by phosphorylation, acetylation, sumoylation, or glycosylation. p53 activation is triggered by hyperproliferative mitogenic signals mediated through deregulated Myc, Ras, or E2F-1, which in turn mediates the induction of Arf expression which promotes p53 stabilization and activation (Fig. 1).9 p53 activity is also induced in response to DNA damage and involves the regulatory kinases ATM, CHK2, and ATR (Fig. 1).
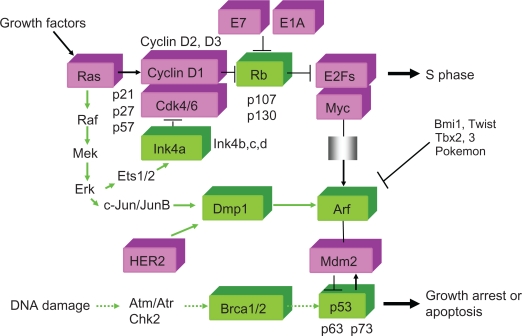
Figure 1.
Molecules that are characterized in this review. Mitogenic signals acting through Ras stimulate the formation of cyclin D/Cdk complexes that phosphorylate RB in mid-late G1 phase. Accentuated by cyclin E/Cdk2 (not shown), RB phosphorylation interrupts its interactions with both histone deacetylase and E2Fs, enabling E2Fs to promote S phase entry. Myc plays a similar role in the sense that it is capable of accelerating S phase entry.181–183 All D-type cyclins are mitogen-responsive, but only cyclin D1 is Ras-responsive. One of the oncogenic effects of adenovirus E1A and papilloma virus E7 is to interfere with RB function. By inhibiting cyclin D-dependent kinase activity, p16INK4a acts as a potent tumor suppressor. p16Ink4a, p19Arf, and tumor suppressor Dmp1 are induced in response to oncogenic Ras-Raf-Mek-Erk signaling. This Arf induction (and also p16Ink4a and Dmp1) quenches inappropriate mitogenic signaling by diverting incipient cancer cells to undergo p53-dependent and independent growth arrest or cell death. Arf is also induced by activated other potentially oncogenic signals stemming from overexpression of oncogenes such as c-Myc, E2F1. Arf expression is repressed by a number of nuclear proteins, such as Bmi1, Twist, Tbx2/3, and Pokemon. Dmp1 (cyclin D binding myb-like protein 1; Dmtf1)116,117,184–186 is unique in that it directly binds and activates the Arf promoter and induces cell cycle arrest in an Arf-dependent fashion. The Dmp1 promoter is also activated by HER2 overexpression.187 Both Dmp1-null and heterozygous mice show hypersensitivity to develop tumors in response to carcinogen DMBA and γ-irradiation,117 suggesting haploid insufficiency. D-type cyclins inhibit Dmp1’s transcriptional activity in a Cdk-independent fashion when E2Fs do not bind to the same promoter; however, D-cyclins cooperate with Dmp1 to activate the Arf promoter. BRCA1/2 proteins are directly or indirectly phosphorylated by ATM/ATR kinases in response to DNA damage, which will interact with p53 to stop cell cycle by activating the p21Cip1/WAF1 promoter. DNA damage (as indicated by the horizontal lines) has been shown to access the Mdm2-p53 machinery independently of Arf. However, Arf-loss enables Mdm2 to work more efficiently in antagonizing p53 function in response to DNA damage.
Activated p53 acts as a regulator of cellular processes such as cell cycle, apoptosis, autophagy, and differentiation. Numerous p53 transcriptional targets have been identified, including p21Cip1/Waf1, Mdm2, Bax, Puma (Bbc), Noxa, Pig8, Gadd45, Pidd, p53R2, DR5, cyclin G, Pten, and CD9.8 Conversely, p53 transcriptionally represses bcl-2, PCNA, and Arf. p53 activation controls the G1 checkpoint of the cell cycle by inducing transcription of the cyclin/Cdk inhibitor p21Cip1/Waf1. Cells with damaged DNA are prevented from entering S phase and replicating the defective chromosomes. p53 controls the G2 checkpoint by regulating expression of p21Cip1/Waf1 and Snk/Plk2.11,12 The arrest of the cell cycle at these checkpoints allows repair of the damaged DNA. Transcriptional activation of pro-apoptotic genes including Bax, Puma, Noxa, PIDD, DR5, and CD95 are dependent on p53.
p53 knockout models
Four strains of mice bearing null mutations of p53 have been created.13–16 Although initial studies of p53−/− mice concluded that p53 had no role in development, subsequent work has revealed that a subset of p53-deficient embryos dies in utero. This embryonic death is associated with exencephaly marked by defects in neural tube closure and overgrowth of neural tissue in the midbrain region.17,18 The finding that Mdm2−/− mutants, which die before E6.5, can be fully rescued in a p53-null background suggests that p53 activity must be tightly regulated by Mdm2 for successful development.19,20
As expected, p53+/− mice are predisposed to tumorigenesis. These animals remain cancer-free for the first 9 months, but half develop osteosarcomas, soft tissue sarcomas, and lymphomas by 18 months of age. Most tumors from p53+/− mice show LOH of the wild-type p53 allele. In p53−/− mice, the onset of tumor development is earlier than in heterozygotes; more than 75% of p53−/− mice develop tumors before 6 months of age. While thymic T cell lymphomas are the major tumor type, p53−/− mice also develop B cell lymphomas, sarcomas, and testicular teratomas.15 The enhanced susceptibility to cancer of p53−/− mice results in their death before 10 months of age. p53-deficient mice are also extremely susceptible to tumorigenesis induced by ionizing radiation (IR) or carcinogens.21,22
Patients with Li-Fraumeni syndrome have germline TP53 mutations, and therefore 50% of them develop malignancies by the age of 30,23 comparable to the tumor incidence in middle-aged (18 months) p53+/− mice. The spectrum of tumors that arise in Li-Fraumeni patients is generally similar to that in p53+/− mice. However, human patients also develop breast cancers and brain tumors, which are rarely observed in p53+/− mice. These differences were puzzling until p53−/− mice were crossed into the BALB/c background. Spontaneous mammary tumors developed in half of the heterozygous females of this strain,24 and the tumors showed the loss of the remaining wild-type p53 allele. Furthermore, when mammary glands from p53−/− BALB/c mice were transplanted into wild-type BALB/c hosts, 75% of the transplanted mice developed mammary tumors. Thus, differences in genetic background can have profound effects on the types of tumors associated with p53 mutation in mice.
In addition to their use as an animal model for p53-associated cancers, p53−/− mice have also been helpful in determining p53 functions in vivo. p53 is required for thymocyte apoptosis induced by double-strand DNA breaks (DSBs) in response to stimuli such as ionizing radiation and adriamycin, but dispensable both for the apoptosis that mediates negative selection of immature thymocytes and thymocyte apoptosis induced in response to ultraviolet irradiation, dexamethasone, or anti-CD95.25,26 With respect to cell cycle control, cells from p53−/− mice show enhanced proliferation in culture compared to controls.27 With respect to maintenance of genomic integrity, p53−/− MEFs cultured in vitro show a high degree of aneuploidy, a hallmark for genomic instability.27 p53 has also been implicated as a component of a spindle checkpoint that ensures the maintenance of diploidy. p53−/− fibroblasts exposed to spindle inhibitors performed multiple rounds of DNA synthesis without completing chromosome segregation, resulting in tetraploid and octaploid cells.28 Increased genomic instability is also seen in mice deficient for Gadd45α, a transcriptional target of p53.29
Tumor suppressors and oncogenes are presumed to cooperate in the induction of tumorigenesis. A p53−/− background dramatically increases tumor frequency and reduces the latency in c-Myc transgenic mice.30 A cooperative effect on transformation between p53 and Rb mutations has also been reported. Mice carrying mutations of both p53 and Rb have reduced viability and exhibit abnormalities not observed with either single mutant, including pinealoblastomas, islet cell tumors, bronchial epithelial hyperplasia, and retinal dysplasia.31
The generation of p53 conditional knockout mice with floxed alleles has shed further light on p53 tumor-suppressor function in vivo.32 Medulloblastomas are among the most common malignancies in childhood, and they are associated with substantial mortality and morbidity. Marino et al generated a mouse model for medulloblastoma by Cre–LoxP-mediated inactivation of Rb and p53 TSGs in the cerebellar external granular layer (EGL) cells. GFAP–Cre-mediated recombination was found both in astrocytes and in immature precursor cells of the EGL in the developing cerebellum. GFAP-Cre; RbLoxP/LoxP; p53−/− or LoxP/LoxP mice developed highly aggressive embryonal tumors of the cerebellum with typical features of medulloblastoma. These tumors were identified as early as 7 weeks of age on the outer surface of the molecular layer, corresponding to the location of the EGL cells during development.32
p53 knock-in mice
Heterozygous mice expressing mutant mouse p53 with an R172H substitution (corresponding to the R175H mutation “hot-spot” in human tumors) develop osteosarcomas, carcinomas, and lymphomas with high metastatic potential.34 LOH studies of p53R172HΔg tumors indicated loss of the wild-type p53 allele in only 1 of 11 tumors. These data indicated clear differences between a p53 missense mutation and a null allele in tumorigenesis in vivo, and suggest that the p53R172HΔg mutant represents a gain-of-function allele.34 Mice with a p53 allele altered at Leu25 and Trp26, residues essential for transcriptional transactivation and Mdm2 binding, synthesized a p53 protein that was stable but did not accumulate after DNA damage.35,36 The mutant p53L25W26 was abundantly expressed, and the level was not affected by DNA damage, but bound to DNA constitutively; however, mutant p53L25W26 showed defects in cell-cycle regulation and apoptosis. Both p53L25W26 mutant and p53-null mouse MEFs were readily transformed by oncogenes, and the corresponding mice were prone to tumors. Thus, the determining pathway for p53 tumor-suppressor function in mice requires the transactivation domain.
To examine the mechanistic nature of p53 missense mutations found in individuals with Li-Fraumeni syndrome, Lang et al37 generated mice harboring a G-to-A substitution at nucleotide 515 of p53 (p53+/515A) corresponding to the p53R175H hot spot mutation in human cancers. Although p53+/515A mice display a similar tumor spectrum and survival curve as p53+/− mice, tumors from p53+/515A mice metastasized with high frequency. Correspondingly, MEFs from the p53515A/515A mutant mice displayed enhanced cell proliferation, DNA synthesis, and transformation. They also reported that disruption of p63 and p73 in p53−/− cells increased transformation capacity and re-initiated DNA synthesis to levels observed in p53515A/515A cells. Additionally, p63 and p73 were functionally inactivated in p53515A cells. Their results provided in vivo validation for the gain-of-function properties of certain p53 missense mutations and suggested a mechanistic basis for these phenotypes.
Similar phenotypes were found in p53 knock-in mice created by Olive et al.38 To ascertain the physiological effects of p53 point mutation, the structural mutant p53R172H and the contact mutant p53R270H (corresponding to codons 175 and 273 in humans) were engineered into the endogenous p53 locus in mice. Both p53R270H/+ and p53R172H/+ mice are models of Li-Fraumeni syndrome; they developed allele-specific tumor spectra distinct from p53+/− mice. In addition, p53R270H/− and p53R172H/− mice developed novel tumors that were not found in p53−/− mice, including carcinomas in the lung, small intestine, colon, breast, skin, liver, and pancreas, and more frequent endothelial tumors.38 Dominant effects that varied by allele and function were observed in primary cells derived from p53R270H/+ and p53R172H/+ mice. These results demonstrate that point mutant p53 alleles expressed under physiological control have enhanced oncogenic potential beyond the simple loss of p53 function.
Transgenic overexpression models for p53
Lavigueur et al39 established p53-transgenic mice driven by the endogenous promoter using murine p53 genomic fragments isolated from a Friend cell line CB7 or BALB/c mouse liver DNA. Mutations in Arg to Pro in 193 or Ala to Val in 135 in p53 gene resulted in multi-organ neoplasias including lymphomas, osteosarcomas, and lung adenocarcinomas. Godley et al33 created transgenic mice expressing wild-type murine p53 under the control of the mouse mammary tumor virus terminal repeat (MMTV-LTR). The p53 transgene was unexpectedly expressed in the kidney, which underwent progressive renal failure due to abnormal kidney development. Similar phenotypes were observed in two transgenic lines that express wild-type p53 within the ureteric bud, but not in transgenic animals expressing a dominant-negative p53 mutant allele. Defective differentiation of the ureteric bud, as evidenced by altered marker expression during development, was accompanied by expression of the p53 transgene. At E17.5–18.5, metanephric mesenchymal cells underwent high rates of apoptosis, and fewer cells than normal were converted to tubular epithelium.33 Proteinuria was observed as early as 2 weeks of age, with azotemia began at 6 weeks. As a result, kidneys in these animals grew to only half of their expected size and contained about half of the normal number of nephrons, with compensatory hypertrophy of the glomeruli. Most affected glomeruli showed global glomerulosclerosis. In this setting, abnormally high levels of wild-type p53 altered cellular differentiation in embryonic ureteric buds, and caused secondary effects (apoptosis and inefficient conversion to epithelium) in the adjacent undifferentiated mesenchyme.33
p53 activation vs. overexpression models and ageing
The role of p53 on ageing has also been studied. Tyner et al40 generated mice with a deletion mutation in the first six exons of the p53 gene that express a truncated RNA capable of encoding a carboxyl-terminal p53 fragment. This mutation confers phenotypes consistent with activated, rather than inactivated, p53. Mutant (p53+/m) mice showed enhanced resistance to spontaneous tumors compared with wild-type (p53+/+) littermates. As p53+/m mice aged, they displayed an early onset of phenotypes associated with ageing, including reduced longevity, osteoporosis, generalized organ atrophy and a diminished stress tolerance.40 By contrast, the phenotypes of p53−/m mice were in distinguishable from those of p53−/− mice, suggesting that a normal p53 allele is required for the mutant p53 gene to have an effect. The tumor resistance of p53+/m mice was considered to be intrinsic to the individual cells of the animals, since fibroblasts from p53+/m were more resistant to transformation by activated Ras plus Myc oncogenes (although this does not rule out the possibility of systemic hormonal, immunological or growth factor changes). The association of early ageing and tumor resistance in the p53+/m mice is consistent with the idea that senescence is a mechanism of tumor suppression. They suggested that ageing-related reduction in stem cell proliferation might have a more important role in longevity than previously recognized.40,41
Garcia-Cao et al42 established and characterized mice carrying supernumerary copies of the wild-type p53 gene as large genomic transgenes. They showed that the p53 transgenic allele (p53-tg), when present in a p53-null genetic background, behaves as a functional replica of the endogenous gene. “Super p53” mice that carry p53-tg alleles in addition to the two endogenous alleles exhibited an enhanced response to DNA damage and were significantly protected from cancer when compared with normal mice. It is conceivable that constitutive or highly frequent activation of p53, such as under chronic exposure to stress, could result in accelerated aging as demonstrated by Tyner et al.40 In contrast, “super p53” mice have a normal aging process despite having clearly increased p53 functionality.42
A critical characteristic of the “super p53” mice is that basal levels of p53 activity are not affected. Attempting to further increase the gene dosage of p53 may eventually reveal a threshold at which deleterious effects will be noticeable, probably as defective tissue regeneration, growth atrophies, and premature aging. This suggests that increases in normally regulated p53, as in the “super p53” mice, could confer cancer protection without affecting ageing, whereas constitutive levels of active p53 provides cancer protection, but promote aging. Thus, cancer resistance could be enhanced by a simple genetic modification of p53 in the absence of undesirable effects.
p73 and p63 models
There is no convincing clinical evidence that p73 or p63 plays a significant role in human tumorigenesis. The combined absence of p63 and p73 severely impaired the induction of p53-dependent apoptosis in response to DNA damage in oncogene-expressing cells and in the developing central nervous system in mice, which was explained by the inability of p53 to bind the promoters of apoptosis-associated target genes and to upregulate their transcription. Thus, there are two classes of p53-family target genes. One class includes genes such as Mdm2 and p21Cip1/Waf1, which p53 regulates in the presence or absence of p63 /p73, and the other group includes genes Perp, Bax, and Noxa that require p63/p73 for p53 to be recruited and function properly. Thus, p63 and p73 are important components of the cellular response to DNA damage, and may portend a greater role for these proteins in tumor suppression and chemosensitivity. The validation of p73 and p63 as novel tumor suppressors may offer novel therapeutic approaches to enhance the chemosensitivity of tumor cells harboring mutated or inactivated p53.
p73 and p63 are members of the p53 family of proteins and thus attractive candidates for TSGs.43,44 Both p73 and p63 share significant amino acid identity with p53 in the transactivation domain, the DNA-binding domain, and the oligomerization domain. Like p53, p73 and p63 can recognize canonical p53 DNA-binding sites and, when overproduced, can activate p53-responsive target genes and induce apoptosis.43 p73 is localized to chromosome 1p36.3, a region of frequent aberrations in human cancers, such as thyroid, colorectal, and breast cancers, pheochromocytomas, and brain tumors.45 However, there is no convincing clinical evidence that p73 and p63 play a significant role in human tumorigenesis. E2F-1 directly activates transcription of p73 leading to activation of p53-responsive target genes and apoptosis.46–49 Moreover, synergistic cooperation between YY1 and E2F-1 through physical interaction was reported in the regulation of the p73 promoter.50
Disruption of p73 function inhibits E2F-1-induced apoptosis in p53-deficient cells, suggesting a role for p73 in p53-independent apoptosis. Similarly, T cell receptor activation-induced cell death, which is p53-independent, is inhibited in the absence of p73.47 Interestingly, a truncated isoform of p73 that is expressed in developing neurons appears to have an anti-apoptotic function.51 However, other evidence weighs against a tumor-suppressive role for p73. Knockout mice null for p73 lacked an increased incidence of tumors up to 15 months of age.52 Several p73 isoforms and two different p73 promoters have been described. Mice deficient for specific isoforms of p73 remain to be reported. Mice deficient for p63 show defects in limb and epidermal morphogenesis and die within hours of birth,53,54 but did not show increased incidence of tumor formation. The combined absence of p63 and p73 severely impaired the induction of p53-dependent apoptosis in response to DNA damage in E1A-expressing cells and in the developing central nervous system in mice.55 This was explained by the inability of p53 to bind the promoters of apoptosis-associated target genes and to upregulate their transcription in p63−/− ; p73−/− ; E1A cells and the developing central nervous system.
These data support the notion that there might be two classes of p53-family target genes. One class includes genes such as Mdm2 and p21Cip1/Waf1, which p53 regulates in the presence or absence of p63 and p73. The other group includes genes such as Perp, Bax, and Noxa that require p63 or p73 for p53 to be recruited and function properly. These data show that p63 and p73 are important components of the response to DNA damage, and may portend a greater role for these proteins in tumor suppression and chemosensitivity. The same group later conducted a detailed study to determine whether p63 or p73 are involved in tumor suppression using aging mice heterozygous for mutations in all p53 family genes.56 They used p73+/− mice for tumor observation, since only 25% of p73−/− mice survived to adulthood in their colony. Spontaneous tumors developed in p63+/− ; p73+/− mice, including mammary and lung adenocarcinomas, thymic and squamous cell carcinoma, and myeloid leukemia. Loss of p63 and p73 can also cooperate with loss of p53 in tumor development. Mice heterozygous for mutations in both p53 and p63 or p53 and p73 displayed higher tumor burden and metastasis compared to p53+/− mice. These findings provide evidence for a broader role for the p53 family in tumor suppression than has been previously reported.56
Retinoblastoma models
The retinoblastoma gene family is composed of three members: the product of the retinoblastoma gene (pRb), and two related proteins, pRb2/p130 and pRb3/p107, which have been shown to be structurally and functionally similar to pRb. The three retinoblastoma family members show growth suppressive properties, although the growth arrest mediated by each of the three pocket regions of the proteins is not identical. Although the three pRB members complement each other, they are not functionally redundant. Among these proteins, pRb and p130 are tumor suppressor proteins, and thus are candidate targets for gene therapy. Mutations commonly found in human small cell lung carcinomas (SCLC) are inactivation of RB and TP53. Recent study established a mouse model for neuroendocrine lung tumors by conditional inactivation of both Rb and p53 in mouse lung epithelial cells. These murine small cell lung carcinoma models may be beneficial for novel drug screening of SCLC.
Retinoblastoma (RB) is an eye tumor that occurs in children. The RB gene is located on chromosome 13q14.2, and mutations of RB have been found in both inherited and sporadic cases.57 RB mutations can also predispose individuals to osteosarcomas, and prostate and breast cancers. In adults, human papilloma virus (HPV) is thought to initiate cervical carcinoma and squamous cell carcinoma of the head and neck, at least in part by inactivating RB through expression of the E7 oncoprotein (Fig. 1). Viral oncoproteins such as adenovirus E1A and SV40 large tumor antigen can bind to the RB protein and inactivate its function (Fig. 1). In liver cancer, hepatitis C virus induces E6 associated protein (AP) degradation of RB.58 Moreover, RB is inactivated in more than 90% of human small-cell lung carcinomas (SCLC),59 and mouse genetic studies have confirmed that Rb plays crucial roles in preventing the initiation of SCLC.60
The RB gene encodes a 928-amino acid (aa) nuclear protein of 105 kDa.61–64 Phosphorylation of the RB protein, which is critical for regulation of its function, is mediated by cyclin/cyclin dependent kinase (CDK) complexes in vivo. Phosphorylation of RB is thus cell cycle-dependent, with the hypophosphorylated, active form being present in G0/G1, and the hyperphosphorylated, inactive form dominant in late G1 of the cell cycle (Fig. 1).62–64 Inactivation of RB by germline mutation of one allele and LOH of the second allele is often found in RB-associated cancers. However, functional inactivation of the RB protein in the absence of RB mutation can also lead to tumorigenesis. For instance, amplification of the 11q13 region results in cyclin D1 overexpression, which, in turn, leads to activation of cyclin D-CDK4/6 activity and hyperphosphorylation and inactivation of RB.65 Functional inactivation of RB can also occur from deficient p16INK4a function caused by gene deletion, promoter methylation, or point mutation.66
The function of RB is to repress the transcription of genes required for DNA replication and cell division. At least two different mechanisms may be involved. Binding sites for the transcription factor E2F are present in the promoters of many genes whose expression is essential and limiting for entry into S phase.67,68 RB binds members of the E2F family (E2F1–4), forming a complex that inhibits transcriptional activation. The RB-E2F complex can also actively repress transcription of genes further downstream.69 In addition to cell cycle regulation, RB plays a role in apoptosis. Increased apoptosis is observed in gene-targeted Rb-deficient (Rb−/−) embryos, and MEFs from Rb−/− mice show activation of E2F-responsive genes and apoptosis.70,71 Published studies have indicated specific roles of E2F1 in apoptosis among the E2F proteins.72 E2F1 activates p73 transcription,46,47 which, in turn, stimulates the transcription of p53-responsive target genes and induces apoptosis.49 Published studies found that activation of E2F2 or E2F3a could also lead to cell death, albeit not to the same degree as E2F1 expression.73,74
Since both E2F2 and E2F3a can activate pro-apoptotic gene transcription, at least when overexpressed, the unique role of E2F1 to induce apoptosis in certain conditions may depend on other specific properties, such as E2F1-specific interacting proteins. Intriguingly, E2F3a did not cause apoptosis in the absence of E2f1.75 Thus, the accumulation of crucial levels of E2F1 activity, and not total E2F activity, appears essential for the induction of apoptosis in response to a deregulated RB pathway.
Knockout models for Rb, p107, and p130
Gene-targeted Rb−/− embryos die in utero between E13.5-E15.5.76–78 Increased apoptosis in the nervous system is seen as early as E11.5 and is particularly evident in the hindbrain, spinal cord, and trigeminal and dorsal root ganglia. Ectopic mitoses were also observed, especially in the hindbrain. In addition to defective neurogenesis, Rb−/− embryos exhibit defective hematopoiesis, manifested as an increased number of immature nucleated erythrocytes. Interestingly, apoptosis in lens fiber cells deficient for Rb is dependent on p53, since Rb−/− ; p53−/− embryos show a complete suppression of apoptosis.79 Analyses of viable chimeric mice derived from Rb−/− embryonic stem cells revealed that the Rb-deficient cells contribute widely to adult tissues.80,81 The chimeric erythroid and central nervous system compartments appeared normal, but the developing retina was defective and displayed ectopic mitoses.
The effects of E2f-1 mutation on Rb mutant phenotypes in mice have been examined. Rb−/− ; E2f1−/− embryos die in utero at approximately E17 with anemia and defective skeletal muscle and lung development.82 Significant tissue-specific suppression of apoptosis, S phase entry, and p53 activation was observed in Rb; E2f1 knockout cells. The fact that mutation of E2f1 did not fully rescue the Rb developmental defects indicates that these abnormalities are not entirely E2f1-dependent. Genomic deletion of either E2f1 or E2f3 suppresses both increased proliferation and apoptosis in the lens and nervous system of Rb−/− embryos, suggesting that both E2f1 and E2f3 are required for induction of apoptosis due to loss of Rb.82,83 Intriguingly, a portion of Rb−/−; E2f3+/− embryos exhibited suppression of increased apoptosis, but not ectopic proliferation in the peripheral nervous system.83 This suggests that E2f3 has a primary role in induction of apoptosis due to Rb-loss, rather than simply contributing to abnormal proliferation and resultant apoptosis of Rb-null cells. More recently, de Bruin et al reported that many defects in the Rb−/− embryos are indirectly due to an essential role of Rb in placental development; however, the lens defects and peripheral nervous system abnormalities appear to be cell-autonomous, suggesting that E2f may indeed play a direct role in regulation of proliferation and apoptosis in these tissues.84 Id2 is a dominant-negative antagonist of basic helix-loop-helix transcription factors and proteins of the Rb family. Id2;Rb double knockout (DKO) embryos survive to term with minimal or no defects in neurogenesis and hematopoiesis, but they die at birth from severe reduction of muscle tissue.85
Unlike humans, the Rb chimeras develop pituitary gland tumors rather than retinoblastomas.77 A similar phenotype was observed in Rb+/− mice that, at age 8–10 months, developed tumors in the brain and pituitary gland. These tumors exhibited LOH of the remaining wild-type Rb allele, demonstrating that Rb is a TSG in mice as well as in humans. Although Rb+/− mice are tumor-prone, they do not accurately recapitulate the tumor spectrum observed in humans with RB, since other members of the Rb family can compensate for Rb-loss in murine eyes. Chimeras possessing cells deficient for both Rb and its family member p107 developed retinoblastomas during the early postnatal months.86 Only Rb−/− ; p107−/− chimeras, but not Rb+/− ; p107−/− chimeras, developed retinoblastomas, suggesting that the low number of target cells in the murine retina precludes the acquisition of the required number of mutations to inactivate the remaining Rb allele. Mice homozygous for loss of p107 or p130 are viable, fertile, and healthy.62,78 However, p107−/− ; p130−/− mice experience neonatal lethality,62 and most Rb+/− ; p107−/− mice are growth-retarded. Increased mortality of these mice is observed within the first three weeks of birth.78 Although Rb+/− ; p107−/− pups that survive to adulthood do not show an increased cancer predisposition compared to Rb+/− mice, they develop multiple dysplastic lesions of the retina.78 Thus, unlike Rb, p107 and p130 are not required for embryonic development, and p107+/−, p130+/−, p107−/−, and p130−/− mice do not show increased incidence of tumor development.62 Embryonic stem cells with a simultaneous deficiency of Rb, p107, and p130 (triple knockout, TKO) have normal growth characteristics, but impaired differentiation capacity.87,88 Rb, p107; p130 TKO MEFs have a shorter cell cycle compared to controls and can spontaneously immortalize. TKO mouse embryonic fibroblasts are also resistant to G1 arrest following DNA damage, contact inhibition or serum starvation.
Conditional mutant mice for Rb32 have been generated using the Cre-LoxP system and should prove useful in defining stage- and tissue-specific roles of Rb. Mutations commonly found during the pathogenesis of human lung cancer are inactivation of RB and TP53; and have been identified in up to 90% of human SCLCs. Meuwissen et al53 established a mouse model for neuroendocrine lung tumors by conditional inactivation of Rb and p53 in mouse lung epithelial cells. Mice carrying conditional alleles for both Rb and p53 frequently developed aggressive lung tumors with striking morphologic and immunophenotypic similarities to human SCLC. Most of these tumors, which they designated MSCLC (murine small cell lung carcinoma), diffusely spread through the lung and gave rise to extrapulmonary metastases. In this model, inactivation of both Rb and p53 was a prerequisite for the pathogenesis of SCLC.
Mutation of RB and TP53 tumor suppressors is associated with the development of human osteosarcomas. To establish a mouse model of osteosarcomas in mice, Berman et al89 used conditional mouse strains to inactivate Rb and/or p53 specifically in osteoblast precursors. The resulting Rb; p53 DKO animals were viable, but developed early onset osteosarcomas with complete penetrance. These tumors displayed many features of human osteosarcomas, including being highly metastatic. Cell lines from the DKO osteosarcomas and were highly proliferative and retained their tumorigenic potential, multipotency, and expression of Sca-1, a marker that is typically associated with stem cells/uncommitted progenitors. Tumorigenicity of the osteosarcoma cell lines correlated with the presence of the Sca-1 marker. Finally, loss of Rb and p53 in Sca-1-positive mesenchymal stem/progenitor cells was sufficient to yield transformed cells that can initiate osteosarcoma formation in vivo.
Choi et al90 later inactivated the Rb and p53 genes by Cre-loxP-mediated recombination in mice. More than 90% of mice developed spindle cell/pleomorphic sarcomas after a single subcutaneous injection of adenovirus carrying Cre-recombinase. Similar to human STSs, these sarcomas overexpressed Cxcr4, which contributes to their invasive properties. Sarcomas originated not from bone marrow-derived cells, but from local resident cells. Indeed, dermal mesenchymal stem cells isolated by plastic adherence and low levels of Sca-1 expression (Sca-1low, CD31negCD45neg) have shown enhanced potential for malignant transformation after the conditional inactivation of both p53 and Rb. Sarcomas formed after transplantation of these cells had features typical for undifferentiated high-grade pleomorphic sarcomas. Together, these results indicated that local Sca-1low dermal mesenchymal stem/progenitor cells are preferential targets for malignant transformation associated with deficiencies in both p53 and Rb.
Transgenic model for Rb
Transgenic mice containing 1–7 copies of a human RB cDNA transgene under the transcriptional control of the human RB promoter have been generated.91 Most of these transgenic mice were smaller than non-transgenic littermates, which were found as early as embryonic day 15. The degree of dwarfism correlated roughly with the copy number of the transgene and the corresponding level of RB protein. Transferring the transgene to Rb-deficient mice, which are nonviable, resulted in the development of normal, healthy mice, indicating that the human RB gene can functionally complement the mouse homolog,91 and suggesting that regulation of Rb expression is required for normal development.
Nikitin et al92 generated tetracycline-regulated Rb transgenic mice to explore the potential mechanism of Rb effects on somatic growth, and compared their phenotypes to those of previously established Rb mouse models. By gestational day 12.5, embryos lacking Rb and those expressing twice the normal level of Rb were 15% larger and 10%–30% smaller, respectively, compared with their wild-type littermates. The small mouse (dwarf) phenotype of Rb transgenic mice was associated with increased plasma levels of insulin-like growth factor-I (IGF-I), but not with growth hormone and glucose concentrations.92 Notably, down-regulation of the Rb transgene expression reduced IGF-I plasma concentrations to normal levels and increased somatic growth pre- and postnatally. Consistent with the in vivo results, cells overexpressing Rb required higher thresholds of IGF-I to stimulate proliferation. Thus, the IGF-I pathway is a critical target for Rb to regulate mouse somatic growth and maintenance during ontogenesis.92
Ink4a/Arf models
The frequent mutation or deletion of INK4a/ARF in human tumors as well as the occurrence of tumors in the murine knockout models have identified both p16 and Arf as bona fide tumor suppressors. INK4-ARF deletions frequently occur in clinically aggressive acute lymphoblastic leukemias (Ph(+) ALLs), but are not in more indolent Ph(+) chronic CML or in CML myeloid blast crisis. Thus, although BCR-ABL mutations typify drug resistance in both CML and Ph(+) ALL, loss of INK4-ARF in Ph(+) ALL enhances disease aggressiveness and attenuates the favorable effects of targeted therapy. Super Ink4a/Arf mice that have increased copies of the locus manifest higher resistance to cancer compared to normal, non-transgenic mice. Modest increases in the activity of the Ink4a/Arf tumor suppressor resulted in a beneficial cancer-resistant phenotype without affecting normal viability or aging. Thus, the expression of the Ink4a/Arf tumor suppressor locus is a robust biomarker for tumor indolence and resistance.
The INK4a/ARF locus on chromosome 9p21 is frequently disrupted in human cancers.93 Germline mutations of this locus predispose an individual to familial melanomas, whereas somatic mutations increase the chance of sporadic malignancies of the pancreas and brain. In both mice and humans, the INK4a locus includes two independent, but overlapping genes that encode the gatekeeper proteins p16INK4a and p14ARF (p19Arf in mice).94 Each gene has its own promoter that precedes three coding exons. The first exons for p16INK4a (E1α) and p14ARF (E1β) are specific to each gene. Exons 2 and 3 are shared, although they are read in different frames and produce two different proteins.65,95,96 Most mutations of the INK4a locus were originally thought to inactivate p16INK4a. However, the identification of ARF, and the finding that ARF and p16INK4a share two exons, suggests that some mutations in the INK4a locus may affect only ARF or only p16INK4a, whereas others may affect both proteins. The p16INK4a protein is a cyclin-dependent kinase inhibitor that specifically binds to and inhibits CDK4 and CDK6, proteins that promote the G1/S transition.97 Inhibition of CDKs leads to maintenance of Rb in its active, hypophosphorylated form (Fig. 1). Thus, p16INK4a performs its tumor-suppressor function through the functional inactivation of Rb. Rb represses p16INK4a expression, and upregulation of p16INK4a expression is observed in Rb-deficient cells.98,99 Expression of p16INK4a can also be repressed by the polycomb family member Bmi-1,100 and by the helix-loop-helix protein Id1.90 p16INK4a expression is enhanced by the transcription factors Ets1 and Ets2, two downstream targets of Ras-Raf-MEK signaling.101
p19Arf physically interacts with Mdm2 and promotes its rapid degradation (Fig. 1), leading to p53 stabilization and activation.19,102–107 p53 control Arf expression in return, since cells express high levels of Arf in the absence of functional p53.108 The main activators of Arf expression are oncoproteins such as c-Myc,109 adenovirus E1A,110 Ras, E2F-1, and v-abl, consistent with a role for Arf in sensing hyperproliferative signals. Conversely, Bmi-1,111 Tbx2,112 Twist,113 Pokemon,114 and AML1/ETO115 repress Arf expression, whereas Dmp1 increases it.116,117 Induction of Arf expression by E2F-1116,118 provides a functional link between the p16Ink4a/cyclin D/Cdk4, 6/Rb and the Arf/Mdm2/p53 tumor suppression pathways. In addition to p16Ink4a and p19Arf, the Ink4 family of CKIs includes p15Ink4b and p18Ink4c. These proteins share ∼40% identity, contain four tandem ankyrin motifs, and specifically inhibit Cdk4 and Cdk6.
Knockout models for Ink4a/Arf
A mouse strain (Ink4a/Arfex2–3) in which both Ink4a and Arf are deficient owing to deletion of their common exons 2 and 3 has been created.119 Mice heterozygous for the Ink4a/Arfex2–3 mutation show a moderate increase in fibrosarcomas, lymphomas, squamous cell carcinomas, and angiosarcomas. Forty percent of these tumors exhibit LOH at the Ink4a locus.120 Deletion of exon 1β specific to Arf is also seen in some of these tumors, suggesting that in these cases it is a deficiency of Arf and not p16Ink4a that leads to tumor development in mice. Susceptibility to tumorigenesis induced by the carcinogen DMBA alone or in combination with ultraviolet B irradiation is only slightly increased in Ink4a/Arfex2–3 heterozygous mice compared to controls. Mice homozygous for the Ink4a/Arfex2–3 mutation are viable, suggesting that this locus is not essential for embryonic development or survival. In fact, overexpression of the Ink4a locus can be detrimental to mouse development. Increased expression of p16Ink4a and Arf occurs in Bmi1-deficient mice, which are underdeveloped and have a cerebellum and lymphoid organs of reduced size. Interestingly, these defects are either completely or partially rescued in a homozygous Ink4a/Arfex2–3 background.111
As expected, Ink4a/Arfex2–3−/− mice show an increased susceptibility to cancer, with most developing sarcomas and lymphomas by age 7 months. Earlier onset of these malignancies (at about 13 weeks) is observed in Ink4a/Arfex2–3−/− mice treated with DMBA or ultraviolet B irradiation. Surprisingly, in contrast to humans, mutations of the INK4a locus do not predispose mice to melanoma. However, Ink4a/Arfex2–3−/− mice that overexpress an activated H-rasG12V gene in melanocytes develop cutaneous melanomas with high penetrance by age 5.5 months.121 This finding suggests that a loss of function of the Ink4a locus coupled with a gain of function of ras can result in mice predisposed to melanomas. Although studies of Ink4a/Arfex2–3−/− mice indicate that the Ink4a locus is important for tumor suppression, the specific roles of p16Ink4a and p19Arf cannot be determined in these animals. Mice deficient for p19Arf, but competent for p16Ink4a, have been generated by deleting the Arf-specific exon 1β.122–124 Mice heterozygous for this mutation develop lymphomas, sarcomas, and hemangiomas after a long latency. The tumors show loss of the remaining Arf allele and/or lack of its expression, confirming Arf’s role in tumor suppression. Homozygous Arf−/− mice are viable and fertile, but most develop sarcomas (∼40%) and T cell lymphomas (∼30%) at around 6 months of age and often die by 12 months. Interestingly, Arf−/− mice differ from Ink4a/Arfex2–3−/− mice in several phenotypes. The latency period for tumor formation is shorter in untreated and DMBA-treated Ink4a/Arfex2–3−/− mice (∼32 and ∼12 weeks, respectively) than in untreated and DMBA-treated Arf−/− mice (∼38 and ∼24 weeks, respectively). In addition, Arf−/− mice develop carcinomas and tumors of the nervous system that do not appear in Ink4a/Arfex2–3−/− mice.123
Two different groups created knockout mice specific to p16Ink4a by deleting exon 1α or mutating exon 2, respectively.125,126 Krimpenfort et al126 generated mice carrying a point mutation specifically affecting p16Ink4a. This allele, designated Ink4a*, is silent in the p19Arf reading frame, but introduces a stop codon in p16Ink4a at conserved amino-acid position 101, resulting in deletion of the fourth ankyrin repeat motif. The analogous human allele (CDKN2 A-W110X) is a naturally occurring mutation found in many human tumor types, and results in an unstable protein that poorly inhibits phosphorylation of RB1, and thus, cell-cycle arrest in transfected cells.126 These mice, designated Ink4a*/*, did not show a significant predisposition to spontaneous tumor formation within 17 months. Ink4a*/* mouse embryonic fibroblasts proliferated normally, were mortal, and were not transformed by oncogenic H-RAS. However, Sharpless et al reported that p16Ink4a-null mouse embryonic fibroblasts exhibited an increased rate of immortalization, although less than observed previously for cells null for Ink4a/Arf, Arf, or p53.125 Moreover, T cells deficient in p16Ink4a exon 1α exhibited enhanced mitogenic responsiveness, consistent with the established role of p16Ink4a in constraining cellular proliferation. Furthermore, p16Ink4a deficiency was associated with an increased incidence of spontaneous and carcinogen-induced cancers.125 Ink4a*/Δ2,3 mice that were deficient for Ink4a and heterozygous for Arf spontaneously developed a wide spectrum of tumors, including melanoma. Treatment of these mice with the carcinogen DMBA resulted in an increased incidence of melanoma, with frequent metastasis.126 Thus, the results from these two studies indicated that Ink4a is a tumor-suppressor gene that, when lost, could recapitulate the tumor predisposition seen in humans.
Expression of Arf in tissues of adult mice is difficult to detect, possibly because its induction leads to the arrest or elimination of incipient tumor cells. Zindy et al replaced exon 1β of the mouse cellular Arf gene with a cDNA encoding GFP, thereby producing Arf-null mice in which GFP expression is driven by the endogenous Arf promoter.127 The Arf promoter was induced in several biologic settings previously shown to elicit mouse p19Arf expression. GFP was expressed in cultured mouse embryonic fibroblasts, spontaneously arising tumors, Eμ-Myc-induced lymphomas, and retrolental masses in the vitreous of the eye. This study provided direct evidence that the Arf promoter monitors latent oncogenic signals in vivo.
INK4-ARF genes are epigenetically silenced in hematopoietic stem cells,128 but become ready to respond to oncogenic stress as blood cells differentiate. Thus, inactivation of INK4-ARF provides differentiated cells with an inappropriate self-renewal capacity, a defining feature of cancer cells. In BCR-ABL-induced (Philadelphia chromosome-positive [Ph(+)]) leukemias, INK4-ARF deletions frequently occur in clinically aggressive acute lymphoblastic leukemias (Ph(+) ALLs), but are not seen in more indolent Ph(+) forms of chronic myelogenous leukemia (CML) or in CML myeloid blast crisis. Williams et al129 infected mouse bone marrow cells with retrovirus encoding either of two oncogenic Bcr-Abl isoforms [p210(Bcr-Abl) and p185(Bcr-Abl)] and induced B cell lympholeukemias by transplanting the cells into lethally irradiated mice. When mouse bone marrow cells expressing Bcr-Abl were placed in short-term cultures selectively designed to support the outgrowth of pre-B cells, only those lacking one or two Arf alleles could initiate lympholeukemias when inoculated into immunocompetent recipient mice. Although the ABL kinase inhibitor imatinib mesylate (Gleevec) provides highly effective treatment for BCR-ABL-positive CML, it has proven far less efficacious in the treatment of BCR-ABL-positive acute lymphoblastic leukemias (ALL), many of which sustain deletions of the INK4A-ARF locus. Consistently, mice receiving Arf−/− or Arf+/− p210(Bcr-Abl)-positive pre-B cells did not achieve remission when maintained on high doses of oral imatinib therapy and rapidly developed lympholeukemia.129
Although cells expressing the Bcr-Abl kinase can proliferate in the absence of IL-7, they remain responsive to this cytokine, which can reduce their sensitivity to imatinib.130,131 Treatment of Arf−/−, p210(Bcr-Abl)-positive pre-B cells with imatinib together with an inhibitor of JAK kinases abrogated this resistance, suggesting that this combination may prove beneficial in the treatment of BCR-ABL-positive acute lymphoblastic leukemia. Thus, although BCR-ABL mutations typify drug resistance in both CML and Ph(+) ALL, loss of INK4-ARF in Ph(+) ALL enhances disease aggressiveness and attenuates the favorable effects of targeted therapy. In their following study, Williams et al132 showed that intravenous infusion of 20 Arf−/−, p185(Bcr-Abl)-positive pre-B cells into healthy syngeneic mice induced rapidly fatal, transplantable lymphoblastic leukemias that resist imatinib therapy. However, introduction of BCR-ABL into Arf-null severe combined immunodeficient bone marrow progenitors lacking the cytokine receptor common gamma-chain yielded leukemogenic pre-B cells with greater sensitivity to imatinib in vivo. Hence, cytokines in the hematopoietic microenvironment can facilitate leukemic proliferation and confer resistance to targeted therapy.132
Transgenic models for Ink4a/Arf
Yang et al133 created MMTV-p16Ink4a transgenic mice and studied the effect of p16Ink4a in ErbB2-induced mammary tumorigenesis. They reported that the p16Ink4a tumor suppressor specifically blocks cyclindependent kinase 4 and 6 activity. The p16Ink4a transgene blocked tumorigenesis by ErbB2, demonstrating that deregulation of the cyclin-dependent kinase partner of cyclin D1 is an essential target of ErbB2. ErbB2 overexpression was a determining factor in deregulation of cyclin D1-Cdk4/6 interactions because neither transgenic cyclin D1 nor loss of p16Ink4a accelerated tumorigenesis in MMTV-ErbB2-transgenic mice. ErbB2 was also a deciding factor in deregulation of cyclin D1-Cdk4/6 in human tumors because no loss of Rb or p16Ink4a was found in tumors overexpressing ErbB2, although ErbB2-negative invasive breast adenocarcinomas frequently lacked expression of p16Ink4a or pRb.
Mathew et al134 generated a “super Ink4a/Arf” mouse strain carrying a transgenic copy of the entire Ink4a/Arf locus. Cells derived from super Ink4a/Arf mice had increased resistance to in vitro immortalization and oncogenic transformation. Importantly, super Ink4a/Arf mice manifest higher resistance to cancer compared to normal, non-transgenic mice. Super Ink4a/Arf mice had normal aging and lifespan. Thus, modest increases in the activity of the Ink4a/Arf tumor suppressor resulted in a beneficial cancer-resistant phenotype without affecting normal viability or aging. Matheu et al then crossed “super Ink4a/Arf” mice with “super p53” mice reasoning that Arf/p53 could have anti-ageing activity by alleviating the load of age-associated damage.135 The double transgenic mice showed strong cancer resistance and had decreased levels of ageing-associated damage. These observations extend the protective role of Arf/p53 to ageing, revealing a previously unknown antiageing mechanism and provided a rationale for the co-evolution of cancer resistance and longevity.
The same group later studied the impact of increased dosage of the Ink4a/Arf locus on germ cells.136 Increased gene dosage of Ink4/Arf impaired the production of male germ cells, and in the case of Ink4/Arf-tg/tg mice results in a Sertoli cell-only-like syndrome and a complete absence of sperm. There was a lower incidence of aging-associated cancer proportional to the Ink4/Arf gene dosage. Interestingly, increased Ink4/Arf gene dosage resulted in lower scores in aging markers and in extended median longevity. The increased survival was also observed in cancer-free mice indicating that cancer protection and delayed aging are separable activities of the Ink4/Arf locus. In contrast, mice carrying one or two additional copies of the p53 gene (p53-tg and p53-tg/tg) had a normal longevity despite their increased cancer protection.136 Thus, the Ink4/Arf locus has a global anti-aging effect, possibly by favoring quiescence and preventing unnecessary proliferation.
The p16Ink4a tumor suppressor accumulates in many tissues as a function of advancing age. p16Ink4a is an effector of senescence19,66,124,125 and a potent inhibitor of the proliferative kinase Cdk4, which is essential for pancreatic β-cell proliferation in adult mammals. To study the links between senescence and aging in vivo, Krishnamurthy et al examined Ink4a/Arf expression in mouse models of aging.137 They showed that expression of p16Ink4a and Arf markedly increased in almost all mouse tissues with advancing age, while there was little change in the expression of other Ink4a (p15, p18, and p19) cell cycle inhibitors. Importantly, the increase in expression for Ink4a and Arf occurred in both epithelial and stromal cells. The age-associated increase in expression of p16Ink4a and Arf was attenuated in the kidney, ovary, and heart by caloric restriction, and this decrease correlated with diminished expression of an in vivo marker of senescence, as well as decreased pathology of those organs. Thus, the expression of the Ink4a/Arf tumor suppressor locus is a robust biomarker, and possible effector of mammalian aging.137 To determine the physiological significance of p16Ink4a accumulation on islet function, they assessed the impact of p16Ink4a-deficiency and overexpression with increasing age and in the regenerative response after exposure to a specific beta-cell toxin.138 Transgenic mice that overexpress p16Ink4a to a degree seen with ageing demonstrated decreased islet proliferation. Similarly, islet proliferation was unaffected by p16Ink4a-deficiency in young mice, but was relatively increased in p16Ink4a-deficient old mice. Survival after toxin-mediated ablation of beta-cells, which requires islet proliferation, declined with advancing age; however, mice lacking p16Ink4a demonstrated enhanced islet proliferation and survival after betacell ablation. These genetic data support the view that an age-induced increase of p16Ink4a expression limits the regenerative capacity of beta-cells with ageing.138
p15Ink4b, p18Ink4c, and p19Ink4d models
There is no strong evidence that p15Ink4b, p18Ink4c, and/or p19Ink4d are involved in tumor suppression in humans. However, methylation of INK4C is observed in human gliomas, and the p18INK4C protein is not detectable in 20% of medulloblastoma cases. When combined with Ptc1 mutation, Ink4c shows haplo-insufficiency for tumor suppression. Thus, p18INK4C loss may contribute to medulloblastoma formation in children.
In contrast to p16Ink4a, there is no strong evidence that p15Ink4b, p18Ink4c, and/or p19Ink4d are involved in tumor suppression in humans. Although the p15INK4b locus is often deleted in human tumors, its deletion is concomitant with that of the neighboring INK4a/ARF locus. Gene-targeted mice hemizygous for p18Ink4c or p15Ink4b did not show increased incidence of cancer.139,140 Like other Ink4 family members, p18Ink4c and p15Ink4b are not required for embryonic development. Ablation of these genes, either alone or in combination, did not abrogate cell contact inhibition or senescence of mouse embryo fibroblasts in culture. However, loss of p15Ink4b, but not p18Ink4c, conferred proliferative advantage to these cells and made them more sensitive to transformation by H-Ras oncogenes.140 p18Ink4c−/− mice developed pituitary adenomas (40%, mostly chromophobe adenomas), testicular tumors (12%), and adrenal tumors (10%) that lead to death before 18 months of age. In contrast, the cancer susceptibility of p15Ink4b−/− mice was only slightly increased over controls; angiosarcomas were observed in fewer than 10% of older p15Ink4b−/− mice.140 Ablation of both p15Ink4b and p18Ink4c genes resulted in lymphoproliferative disorders and tumor formation. Moreover, mice lacking p18Ink4c exhibited deregulated epithelial cell growth leading to the formation of cysts, mostly in the cortical region of the kidneys and the mammary epithelium. These results indicate that p15Ink4b and p18Ink4c are tumor suppressor proteins that act in different cellular lineages and/or pathways with limited compensatory roles. Mice with nullizygous or heterozygous mutations of p27Kip1 were predisposed to tumors in multiple tissues when challenged with γ-irradiation or a chemical carcinogen.141 Mice lacking both p18Ink4c and p27Kip1, like chimeric mice for Rb deficiency, developed pituitary tumors and died by 3.5 months of age.139 Hence, p18Ink4c and p27Kip1 mediate two separate pathways to collaboratively suppress pituitary tumorigenesis, likely by controlling the function of Rb.
Overlapping and sustained patterns of expression of two cyclin-dependent kinases, p19Ink4d and p27Kip1, in post-mitotic brain cells suggested that these proteins may be important in actively repressing neuronal proliferation. Animals derived from crosses of Ink4d-null with kip1-null mice exhibited bradykinesia, proprioceptive abnormalities, and seizures, and died as early as 18 days after birth.142 Metabolic labeling of live animals with neuronal markers showed that subpopulations of central nervous system neurons were proliferating in all parts of the brain, including normally dormant cells of the hippocampus, cortex, hypothalamus, pons, and brain stem.142 These cells also expressed G2/M marker phosphorylated histone H3, indicating that neurons were dividing after they had migrated to their final positions in the brain. Increased proliferation was balanced by cell death, resulting in no gross changes in the cytoarchitecture of the brains of these mice. Therefore, the Cdk inhibitors p19Ink4d and p27Kip1 cooperate to maintain differentiated neurons in a quiescent state that is potentially reversible. Thus, inhibiting function of these Cdk inhibitors in the adult brain could provide an avenue for stimulating the growth of neuronal populations lost in degenerative diseases or through traumatic injury.142
Recurrent genetic alterations in human medulloblastoma include mutations in the sonic hedgehog signaling pathway and TP53 inactivation (approximately 25% and 10% of cases, respectively). However, mouse models of medulloblastoma generally depend upon p53 inactivation for rapid onset and high penetrance. The p18Ink4c gene is transiently expressed in mouse cerebellar granule neuronal precursor cells (GNPs) as they exit the cell division cycle and differentiate. Co-inactivation of Ink4c and p53 provided cultured GNPs with an additive proliferative advantage, either in the presence or absence of Shh, and induced medulloblastoma with low penetrance, but with greatly increased the incidence following postnatal irradiation.143 In contrast, mice lacking one or two functional Ink4c alleles and one copy of Patched (Ptc1) encoding the Shh receptor rapidly developed medulloblastomas that retained wild-type p53.143 In tumor cells purified from double heterozygotes, the wild-type Ptc1 allele, but not Ink4c, was inactivated. Therefore, when combined with Ptc1 mutation, Ink4c is haploinsufficient for tumor suppression. Methylation of INK4C was observed in four of 23 human medulloblastomas, and the p18INK4C protein was not detectable in 14 of 73 cases. Hence, p18INK4C loss may contribute to medulloblastoma formation in children.142
BRCA models
A substantial part of hereditary breast cancer cases is caused by BRCA1 germline mutations. Transgenic BRCA1 mice showed delayed development of tumors when challenged with DMBA, relative control mice. Mouse models with conventional and conditional mutations in Brca1/2 have demonstrated critical roles of these proteins in breast cacinogenesis. The most advanced mouse models closely reproduce human BRCA1/2-related breast cancers, and may, therefore be useful for addressing clinically relevant questions including novel drug screens.
BRCA1
In humans, inheritance of one mutated allele of BRCA1 significantly increases the risk of breast or ovarian cancer.144–146 Mutations of the BRCA1 gene, located on chromosome 17q21, are a predisposing factor in approximately 50% of families with hereditary breast cancer and in over 80% of families with hereditary breast-ovarian cancer. Most tumors from predisposed individuals demonstrate LOH for the wild-type BRCA1 allele, and thus it is a classical tumor suppressor. BRCA1 expression is increased in the S phase of the cell cycle, and the BRCA1 protein is phosphorylated in a cell cycle-dependent manner and in response to DNA damage.146
Several lines of evidence support a role for BRCA1 in DNA damage repair. BRCA1 is phosphorylated following activation of ATM, CHK2, and ATR-dependent DNA damage signaling pathways. Furthermore, BRCA1 interacts or forms a complex with multiple proteins involved in DNA damage repair, including RAD51, RAD50-Mre11-p95, MSH2, MSH6, MLH1, ATM, CHK2, BRCA2, and BLM.147 BRCA1 also interacts with proteins that are involved in transcription, such as the RNA polymerase II holoenzyme complex, RNA helicase A, CtIP (CTBP-interacting protein), and CBP/p300. Unlike humans with an inherited BRCA1 mutation, however, mice hemizygous for a Brca1 mutation do not show increased incidence of tumors. Homozygous Brca1 mutations inevitably lead to post-implantation embryonic lethality.148–151 However, phenotypic differences exist among different Brca1−/− strains, such as a range of the onset of lethality from E6.5 to E13.152 Such differences could be due to different types of targeted Brca1 mutations and/or to the frequent alternative splicing of this gene during embryogenesis.
Brca1−/− embryos exhibit defective cellular proliferation and activation of p53-dependent pathways. 153 Mutation of p53 or its transcription target p21Cip1 partially delays the lethality of the Brca1−/− embryos.151,153,154 Hypomorphic Brca1 mutants with a partial loss of Brca1 function show spina bifida and anencephaly accompanied by increased apoptosis in the neuroepithelium and die at E10-E13.148 Hypomorphic Brca1-mutant ES cells and MEFs are hypersensitive to IR155,156 and have a defect in transcription-coupled DNA repair. Human BRCA1 transgenes can rescue the embryonic lethality of Brca1 mutant mice.157,158
Three different conditional Brca1 alleles have been generated, Brca1F5–6, Brca1Co, and Brca1F5–13.159–161 Cre-mediated deletion of Brca1 exons 5–6 or exons 5–13 from the Brca1F5–6 or Brca1F5–13 allele, respectively, induces a frameshift mutation that abrogates the production of all three splicing products (full-length Brca1, Brca1-D11, and Brca1-IRIS). In contrast, deletion of exon 11 of the Brca1Co allele results in a hypomorphic mutation that still allows for expression of Brca1-D11. Mice with a mammary gland-specific hypomorphic Brca1 function showed increased mammary cell apoptosis and abnormal ductal development, including incomplete ductal outgrowth, alveolar differentiation and involution, suggesting that Brca1 function is indispensable for normal mammary gland development.160 These mice developed mammary gland tumors after 10 to 13 months of age that showed genetic instability characterized by aneuploidy and chromosomal rearrangements, or alteration of p53 transcription. This model was later improved by introduction of a single p53-null allele, yielding MMTV-Cre; Brca1Co/Co; p53+/− mice, which developed mammary tumors with reduced latency.162 Cooperation between Brca1 and p53 loss in mammary tumorigenesis was also demonstrated in a K14-Cre; Brca1F5–13/F5–13; p53F2–10/F2–10 mouse model with tissue-specific inactivation of both Brca1 and p53.161 The fact that loss of p53 function accelerates the formation of mammary tumors in female mice with the mammary gland-specific Brca1 mutation paralleled the frequent loss of TP53 in tumors from human BRCA1 patients. Mice with a T cell lineage–specific null mutation of Brca1 exhibit depletion of T lineage cells, abnormal p53 activation, and decreased cell cycle progression, and apoptosis.159
To directly determine the role of BRCA1 in mammary gland development and tumor suppression, a transgenic mouse model of BRCA1 overexpression was developed.163 Using the MMTV promoter/enhancer, transgenic mice expressing human BRCA1 or select mutant controls were generated. Transgenic animals examined during adolescence expressed the human transgene in their mammary glands. The mammary glands of 13-week-old virgin homozygous MMTV-BRCA1 mice showed moderately increased lobuloalveolar development. The mammary ductal trees of both hemizygous and homozygous MMTV-BRCA1t340 were similar to those of control non-transgenic littermates. Interestingly, both hemi- and homozygous transgenic mice expressing a splice variant of BRCA1 lacking the N-terminal RING finger domain (MMTV-BRCA1sv) and exhibited marked mammary lobulo-alveolar development, particularly terminal end bud proliferation. Homozygous MMTV-BRCA1 mice showed delayed development of tumors when challenged with DMBA, relative to non-transgenic and homozygous BRCA1t340 expressing mice. In contrast, homozygous MMTV-BRCA1sv transgenic animals were sensitized to DMBA treatment exhibited a very rapid onset of mammary tumor development and accelerated mortality. MMTV-BRCA1 effects on mortality were restricted to DMBA-induced tumors of the mammary gland. These results demonstrate in vivo roles for BRCA1 in both mammary gland development and in tumor suppression against mutagen-induced mammary gland neoplasia.163
BRCA2
The human BRCA2 gene is located on chromosome 13q12-q13, and encodes a major transcript of 11 kb that is translated into a highly charged 3,418 amino acid protein.146,153 In women, mutations of BRCA2 are responsible for 32% of hereditary breast cancers. BRCA2 mutations are also related to increased breast cancer in men. Like BRCA1, BRCA2 is thought to act as a caretaker involved in DNA damage repair and the maintenance of genomic integrity. BRCA1 and BRCA2 co-immunoprecipitate and co-localize with RAD51 in subnuclear foci and on the axial elements of developing synaptonemal complexes. Furthermore, BRCA2, like BRCA1 and RAD51, relocates to PCNA-positive replication sites following exposure of S phase cells to HU or ultraviolet irradiation.142
Brca2−/− mice die in utero between E7.5 and E9.5.164–166 The onset of abnormalities is seen as early as E5.5 and the mutant embryos remain underdeveloped until death. Defective cell proliferation occurs in vivo and in vitro. Some hypomorphic Brca2 mutants survive to adulthood.167–169 These animals are smaller than their control littermates, show abnormal tissue differentiation, lack germ cells, and are infertile. Brca2 hypomorphs develop lethal thymic lymphomas by 12–14 weeks of age. Mammary gland tumors were not observed, possibly because these animals died early. It is unclear whether Brca2 mutations in mice will accurately reproduce human pathologies. Brca2-deficient cells have defects in their ability to repair DNA damage induced by genotoxic agents such as γ-, X-ray, and ultraviolet irradiation, and MEFs of Brca2 hypomorphic mutants exhibit genomic instability.146,147,169–171
To achieve mammary gland-specific recombination of a Brca2F3–4 conditional allele, Ludwig et al172 created knock-in mice for Brca2F3–4 and crossed with WAP-Cre. Wapcre/+; Brca2ex11/F3–4 female mice developed non-metastatic mammary carcinomas or adenosquamous carcinomas after a relatively long latency of 1.4 years.172 The tumors displayed aneuploidy and chromosomal aberrations, were ErbB2/neu-negative, and ERα and cyclin D1-positive. Cheung et al173 created conditional Brca2F9–10 mice and crossed them with MMTV- or WAP-Cre transgenic lines. Although Brca2 was not required for epithelial expansion in mammary glands of pregnant mice, Brca2-deficient mice developed mammary adenocarcinomas after a long latency (average: 1.6 years). Detailed histopathological analysis of four of these tumors demonstrated that three of them showed abnormal p53 protein expression. Moreover, homozygosity versus heterozygosity for the Brca2 mutation heavily skewed the tumor spectrum toward mammary adenocarcinoma development in p53+/− mice. Thus, although Brca2 is not essential for mammary epithelial development, Brca2-deficiency and hemizygous p53 deletion collaborate to promote mammary tumorigenesis.
Brca2 is also implicated in T cell lymphoma development. A small acceleration of T cell lymphomagenesis was reported for Lck-Cre; Brca2F9–10/F9–10; p53−/− mice, compared to Lck-Cre; p53−/− control animals.174 However, Park and Lee175 found that thymus-specific disruption of Brca2F12/F12 allele in mice that were not crossed to a p53-mutant background also led to development of thymic lymphomas. The tumors were fatal in 25% of these mice from 16 weeks to 66 weeks after birth. The difference between the two studies could be due to different targeting sites between the two types of mice or different genetic backgrounds (129/C57BL/6 vs. FVB). Park and Lee1175 had targeted exon 11 of Brca2, whereas Cheung and colleagues had targeted exon 9 and 10 of Brca2 allele.174 The very large exon 11 of Brca2 contains 6–8 BRC repeats, which are implicated in Rad51-binding and modulating DNA repair. Therefore, it was speculated that targeting exon 11 of Brca2 would yield better survival and less genetic instability. Nevertheless, the results from Park and Lee indicate that Brca2 mutation in T cells predisposed them to mutations that led to cancer.
BRCA2 has also been implicated in the etiology of prostate cancer, but the impact of Brca2 mutations in prostate tumorigenesis is unclear. Francis et al177 showed that deletion of Brca2 specifically in prostate epithelia results in focal hyperplasia and low-grade prostate intraepithelial neoplasia (PIN) in animals over 12 months of age. Simultaneous deletion of Brca2 and p53 in prostate epithelia gave rise to focal hyperplasia and atypical cells at 6 months, leading to high-grade PIN in animals from 12 months.176–177 Epithelial cells in these lesions showed increased DNA damage and had higher levels of proliferation, but also elevated apoptosis. Castration of Brca2; p53 mutant animals led to regression of PIN lesions, but atypical cells continued to proliferate and express nuclear androgen receptor. This study provided evidence that Brca2 can act as a tumor suppressor in the prostate, which could guide the development of new therapeutic approaches to prostate cancer.177
Novel tumor suppressor ARH1
ARHI is an imprinted TSG in breast and ovarian carcinomas; expression of ARHI in cancer cells inhibits cell growth and is associated with down-regulation of the Cyclin D1 promoter activity and induction of p21WAF1/CIP1. Identification of these novel Ras-related GTPase family members has expanded our understanding of the roles of these proteins in cell physiology. ARH1 serves as functionally distinct regulators of as yet to be characterized signaling cascades.
Using differential display PCR, Yu et al identified a gene ARH1 (A Ras homologue member 1; NOEY2) with high homology to ras and rap that is expressed consistently in normal ovarian and breast epithelial cells, but not in ovarian and breast cancers.178 Re-expression of ARH1 by transfection suppresses clonogenic growth of breast and ovarian cancer cells. Growth suppression by ARH1 was associated with down-regulation of the Cyclin D1 promoter activity and induction of p21WAF1/CIP1. LOH of ARH1 was detected in 41% of ovarian and breast cancers. In most cancer samples with LOH, the non-imprinted functional ARH1 allele was deleted, indicating that ARH1 is a classical tumor suppressor.
Xu et al179 developed transgenic mice that overexpress ARH1 under the control of the CMV promoter. Offspring with the transgene weighed significantly less than non-transgenic littermates. In addition, strong expression of the ARH1 transgene was associated with greatly impaired mammary gland development and lactation, failure of ovarian folliculogenesis resulting in decreased fertility, loss of neurons in the cerebellar cortex, and impaired development of the thymus. Decrease in body size and defects in the mammary glands correlated with the level of transgene expression. Immunohistochemical analysis indicated that expression of prolactin (but not growth hormone) was lower in the pituitary glands of mice with defective mammary gland development. The defect in pregnancy-associated mammary tissue proliferation was associated with decreased serum prolactin and progesterone levels. Moreover, lower levels of estrogen receptor and progesterone receptor were observed in postpartum mammary glands and in the ovaries of mice that overexpressed ARH1. Thus, ARH1 can inhibit prolactin secretion and act as a negative regulator in murine growth and development.179
ARH1 is downregulated in more than 60% of human ovarian cancers. Lu et al180 showed that re-expression of ARH1 in multiple ovarian cancer cell lines induced autophagy by blocking PI3 K signaling and inhibiting mammalian target of rapamycin mTOR, upregulating ATG4, and colocalizing with cleaved microtubule-associated protein light chain 3 in autophagosomes. Furthermore, ARH1 was required for spontaneous and rapamycin-induced autophagy in normal and malignant cells. Although ARH1 re-expression led to autophagic cell death when SKOV3 ovarian cancer cells were grown in culture, it enabled the cells to remain dormant when they were grown in mice as xenografts. When ARH1 levels were reduced in dormant cells, xenografts grew rapidly. However, inhibition of ARH1-induced autophagy with chloroquine dramatically reduced re-growth of xenografted tumors upon reduction of ARH1 levels, suggesting that autophagy contributed to the survival of dormant cells.
ARH1 is also frequently lost in pancreatic cancers. ARH1 was re-expressed in pancreatic cancer cells that had lost its expression to demonstrate the molecular mechanisms of cell growth inhibition.180 Flow cytometric analysis indicated that ARH1 blocked cell cycle progression at the G1 phase in pancreatic cancer cells. Re-expression of ARH1 increased the expression of p21WAF1, through the accumulation of p53 protein by the inhibition of PI3K/AKT signaling. In addition, ARH1 enhanced expression of p27KIP1 through the inhibition of PI-3K/AKT signaling. The expression of cyclins A and D1 decreased, followed by decreased activities of CDK2/4. These results suggest that the PI-3K/AKT pathway plays a pivotal role in the pathogenesis of pancreatic cancer and ARH1 exerts its growth-inhibitory effects through modulation of several key G1 regulatory proteins, such as p21WAF1, p27KIP1, CDK2/4, Cyclins A and D1.
Conclusions
Mouse models for TSGs, particularly models generated by gene targeting, offer the advantage of studying a genetically modified animal bearing one or only a few mutations. The use of these animals has revealed previously unsuspected developmental roles for TSGs such as Rb, Brca1, and Brca2, and has advanced our overall understanding of tumor-suppressor functions. Mouse models for TSGs have greatly facilitated the identification and characterization of cellular pathways controlled by these genes. This knowledge will be invaluable in choosing pathways or molecules to target for therapy of a specific cancer.
Most mouse models that manipulate TSGs exhibit some degree of predisposition to cancer development. A closer relationship between the murine and human situations may be precluded by several factors. Tumorigenesis associated with homozygous, but not heterozygous, mutation of a mouse TSG strongly suggests that LOH and inactivation of a wild-type allele are rare and limiting events. In addition, the mouse lifespan may be too short to allow the inactivation of the wild-type allele followed by the accumulation of other genetic abnormalities necessary for tumor development. For example, the frequent thymic lymphomas in the mouse model created by Cheung et al174 and Park and Lee175 might preclude these animals from developing other tumors owing to their early death from lymphomas. Finally, different genetic backgrounds give rise to different cancer spectra. Thus, it is not surprising that phenotypic differences exist between human diseases and their mouse models.
Nevertheless, the engineering of mouse models with combinations of mutated TSGs and mice transgenic for different oncogenes will provide powerful systems for identifying synergistic effects of specific mutations/deletions on tumorigenesis. Where the embryonic lethality of a mutation hampers the study of gene function in vivo, conditional mutants can be generated in which deletion or mutation of a TSG can be induced in a tissue of choice. In the future, mouse models in which mutation of both alleles of a TSG is induced in a few somatic cells will replicate sporadic human cancers. The sequencing of the human genome will no doubt lead to the identification of still more TSGs and oncogenes, and thus even more sophisticated mouse models. The study of such models should ultimately be of great benefit to cancer patients.
Acknowledgments
We also thank K. Klein for editing. K. Inoue is supported by NIH/NCI 5R01CA106314, ACS RSG-07-207-01-MGO, and CCCWFU Director’s Challenge pilot award. P. Taneja has been supported by the Susan G. Komen Foundation postdoctoral fellowship KG080179. D. Maglic has been supported by DOD predoctoral fellowship BC100907.
Footnotes
Disclosure
Author(s) have provided signed confirmations to the publisher of their compliance with all applicable legal and ethical obligations in respect to declaration of conflicts of interest, funding, authorship and contributorship, and compliance with ethical requirements in respect to treatment of human and animal test subjects. If this article contains identifiable human subject(s) author(s) were required to supply signed patient consent prior to publication. Author(s) have confirmed that the published article is unique and not under consideration nor published by any other publication and that they have consent to reproduce any copyrighted material. The peer reviewers declared no conflicts of interest.
References
- 1.Kinzler KW, Vogelstein B. Cancer susceptibility genes. Gatekeepers and caretakers. Nature. 1997;386:761–63. doi: 10.1038/386761a0. [DOI] [PubMed] [Google Scholar]
- 2.Sherr CJ. Principles of tumor suppression. Cell. 2004;116:235–46. doi: 10.1016/s0092-8674(03)01075-4. [DOI] [PubMed] [Google Scholar]
- 3.van Heemst D, den Reijer PM, Westendorp RG. Ageing or cancer: a review on the role of caretakers and gatekeepers. Eur J Cancer. 2007;43:2144–52. doi: 10.1016/j.ejca.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 4.Knudson AG. Hereditary cancer: two hits revisited. J Cancer Res Clin Oncol. 1996;122:135–40. doi: 10.1007/BF01366952. [DOI] [PubMed] [Google Scholar]
- 5.Knudson AG. Chasing the cancer demon. Annu Rev Genet. 2000;34:1–19. doi: 10.1146/annurev.genet.34.1.1. [DOI] [PubMed] [Google Scholar]
- 6.Frese KK, Tuveson DA. Maximizing mouse cancer models. Nat Rev Cancer. 2007;7:645–58. doi: 10.1038/nrc2192. Review. [DOI] [PubMed] [Google Scholar]
- 7.Gopinathan A, Tuveson DA. The use of GEM models for experimental cancer therapeutics. Dis Model Mech. 2008;1:83–6. doi: 10.1242/dmm.000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Menendez D, Inga A, Resnick MA. The expanding universe of p53 targets. Nat Rev Cancer. 2009;9:724–37. doi: 10.1038/nrc2730. [DOI] [PubMed] [Google Scholar]
- 9.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro. hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–23. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 10.Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137:413–31. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 11.Chan TA, Hwang PM, Hermeking H. Cooperative effects of genes controlling the G(2)/M checkpoint. Genes Dev. 2000;14:1584–88. [PMC free article] [PubMed] [Google Scholar]
- 12.Burns TF, Fei P, Scata KA. Silencing of the novel p53 target gene Snk/Plk2 leads to mitotic catastrophe in paclitaxel (taxol)-exposed cells. Mol Cell Biol. 2003;23:5556–71. doi: 10.1128/MCB.23.16.5556-5571.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donehower LA, Harvey M, Slagle BL, et al. Mice deficient for p53 are developmentally normal but susceptible to spontaneous tumours. Nature. 1992;356:215–21. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 14.Tsukada T, Tomooka Y, Takai S, Ueda Y, Nishikawa S, et al. Enhanced proliferative potential in culture of cells from p53-deficient mice. Oncogene. 1993;8:3313–22. [PubMed] [Google Scholar]
- 15.Jacks T, Remington L, Williams BO, et al. Tumor spectrum analysis in p53-mutant mice. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 16.Purdie CA, Harrison DJ, Peter A, et al. Tumour incidence, spectrum and ploidy in mice with a large deletion in the p53 gene. Oncogene. 1994;9:603–9. [PubMed] [Google Scholar]
- 17.Armstrong JF, Kaufman MH, Harrison DJ. High-frequency developmental abnormalities in p53-deficient mice. Curr. Biol. 1995;5:931–36. doi: 10.1016/s0960-9822(95)00183-7. [DOI] [PubMed] [Google Scholar]
- 18.Sah VP, Attardi LD, Mulligan GJ, Williams BO, Bronson RT, Jacks T. A subset of p53-deficient embryos exhibit exencephaly. Nat Genet. 1995;10:175–80. doi: 10.1038/ng0695-175. [DOI] [PubMed] [Google Scholar]
- 19.Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–8. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 20.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–6. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 21.Harvey M, McArthur MJ, Montgomery CA, Butel JS, Bradley A, Donehower LA. Spontaneous and carcinogen induced tumorigenesis in p53-deficient mice. Nat Genet. 1993;5:225–9. doi: 10.1038/ng1193-225. [DOI] [PubMed] [Google Scholar]
- 22.Kemp CJ, Wheldon T, Balmain A. p53-deficient mice are extremely susceptible to radiation-induced tumorigenesis. Nat Genet. 1994;8:66–9. doi: 10.1038/ng0994-66. [DOI] [PubMed] [Google Scholar]
- 23.Malkin D. p53 and the Li-Fraumeni syndrome. Cancer Genet Cytogenet. 1993;66:83–92. doi: 10.1016/0165-4608(93)90233-c. [DOI] [PubMed] [Google Scholar]
- 24.Kuperwasser C, Hurlbut GD, Kittrell FS, et al. 2000. Development of spontaneous mammary tumors in BALB/c p53 heterozygous mice. A model for Li-Fraumeni syndrome. Am J Pathol. 2000;157:2151–59. doi: 10.1016/S0002-9440(10)64853-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clarke AR, Purdie CA, Harrison DJ, et al. Thymocyte apoptosis induced by p53-dependent and independent pathways. Nature. 1993;362:849–52. doi: 10.1038/362849a0. [DOI] [PubMed] [Google Scholar]
- 26.Lowe SW, Schmitt EM, Smith SW, Osborne BA, Jacks T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature. 1993;362:847–9. doi: 10.1038/362847a0. [DOI] [PubMed] [Google Scholar]
- 27.Harvey M, Sands AT, Weiss RS, et al. In vitro growth characteristics of embryo fibroblasts isolated from p53-deficient mice. Oncogene. 1993;8:2457–67. [PubMed] [Google Scholar]
- 28.Cross SM, Sanchez CA, Morgan CA, et al. A p53-dependent mouse spindle checkpoint. Science. 1995;267:1353–56. doi: 10.1126/science.7871434. [DOI] [PubMed] [Google Scholar]
- 29.Hollander MC, Sheikh MS, Bulavin DV, et al. Genomic instability in Gadd45 deficient mice. Nat Genet. 1999;23:176–84. doi: 10.1038/13802. [DOI] [PubMed] [Google Scholar]
- 30.Blyth K, Terry A, O’Hara M, et al. Synergy between a human c-myc transgene and p53 null genotype in murine thymic lymphomas: contrasting effects of homozygous and heterozygous p53 loss. Oncogene. 1995;10:1717–23. [PubMed] [Google Scholar]
- 31.Harvey M, Vogel H, Lee EY, Bradley A, Donehower LA. Mice deficient in both p53 and Rb develop tumors primarily of endocrine origin. Cancer Res. 1995;55:1146–51. [PubMed] [Google Scholar]
- 32.Marino S, Vooijs M, van Der Gulden H, Jonkers J, Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 2000;14:994–1004. [PMC free article] [PubMed] [Google Scholar]
- 33.Godley LA, Kopp JB, Eckhaus M, Paglino JJ, Owens J, Varmus HE. Wild-type p53 transgenic mice exhibit altered differentiation of the ureteric bud and possess small kidneys. Genes Dev. 1996;10:836–50. doi: 10.1101/gad.10.7.836. [DOI] [PubMed] [Google Scholar]
- 34.Liu G, McDonnell TJ, Montes de Oca Luna R, et al. High metastatic potential in mice inheriting a targeted p53 missense mutation. Proc Natl Acad Sci U S A. 2000;97:4174–79. doi: 10.1073/pnas.97.8.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chao C, Saito S, Kang J, et al. p53 transcriptional activity is essential for p53-dependent apoptosis following DNA damage. EMBO J. 2000;19:4967–75. doi: 10.1093/emboj/19.18.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jimenez GS, Nister M, Stommel JM, et al. A transactivation-deficient mouse model provides insights into Trp53 regulation and function. Nat Genet. 2000;26:37–43. doi: 10.1038/79152. [DOI] [PubMed] [Google Scholar]
- 37.Lang GA, Iwakuma T, Suh YA, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–72. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 38.Olive KP, Tuveson DA, Ruhe ZC, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–60. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 39.Lavigueur A, Maltby V, Mock D, Rossant J, Pawson T, Bernstein A. High incidence of lung, bone, and lymphoid tumors in transgenic mice overexpressing mutant alleles of the p53 oncogene. Mol Cell Biol. 1989;9:3982–91. doi: 10.1128/mcb.9.9.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tyner SD, Venkatachalam S, Choi J, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 41.Ferbeyre G, Lowe SW. The price of tumour suppression? Nature. 2002;415:26–7. doi: 10.1038/415026a. [DOI] [PubMed] [Google Scholar]
- 42.García-Cao I, García-Cao M, Martín-Caballero J, et al. “Super p53” mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J. 2002;21:6225–35. doi: 10.1093/emboj/cdf595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collavin L, Lunardi A, Del Sal G. p53-family proteins and their regulators: hubs and spokes in tumor suppression. Cell Death Differ. 2010;17:901–11. doi: 10.1038/cdd.2010.35. [DOI] [PubMed] [Google Scholar]
- 44.Zawacka-Pankau J, Kostecka A, Sznarkowska A, Hedström E, Kawiak A. p73 tumor suppressor protein: a close relative of p53 not only in structure but also in anti-cancer approach? Cell Cycle. 2010;9:720–8. doi: 10.4161/cc.9.4.10668. [DOI] [PubMed] [Google Scholar]
- 45.Bagchi A, Mills AA. The quest for the 1p36 tumor suppressor. Cancer Res. 2008;68:2551–6. doi: 10.1158/0008-5472.CAN-07-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Irwin M, Marin MC, Phillips AC, Seelan RS, Smith DI, et al. Role for the p53 homologue p73 in E2F-1-induced apoptosis. Nature. 2000;407:645–48. doi: 10.1038/35036614. [DOI] [PubMed] [Google Scholar]
- 47.Lissy NA, Davis PK, Irwin M, Kaelin WG, Dowdy SF. A common E2F-1 and p73 pathway mediates cell death induced by TCR activation. Nature. 407:642–45. doi: 10.1038/35036608. 200; [DOI] [PubMed] [Google Scholar]
- 48.Waltermann A, Kartasheva NN, Dobbelstein M. Differential regulation of p63 and p73 expression. Oncogene. 2003;22:5686–93. doi: 10.1038/sj.onc.1206859. [DOI] [PubMed] [Google Scholar]
- 49.Stiewe T, Putzer BM. Role of the p53-homologue p73 in E2F1-induced apoptosis. Nat Genet. 2000;26:464–69. doi: 10.1038/82617. [DOI] [PubMed] [Google Scholar]
- 50.Wu S, Murai S, Kataoka K, Miyagishi M. Yin Yang 1 induces transcriptional activity of p73 through cooperation with E2F1. Biochem Biophys Res Commun. 2008;365:75–81. doi: 10.1016/j.bbrc.2007.10.145. [DOI] [PubMed] [Google Scholar]
- 51.Pozniak CD, Radinovic S, Yang A, McKeon F, Kaplan DR, Miller FD. An anti-apoptotic role for the p53 family member, p73, during developmental neuron death. Science. 2000;289:304–6. doi: 10.1126/science.289.5477.304. [DOI] [PubMed] [Google Scholar]
- 52.Yang A, Walker N, Bronson R, et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- 53.Mills AA, Zheng B, Wang XJ, Vogel H, Roop DR, Bradley A. p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature. 1999;398:708–13. doi: 10.1038/19531. [DOI] [PubMed] [Google Scholar]
- 54.Yang A, Schweitzer R, Sun D, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398:714–18. doi: 10.1038/19539. [DOI] [PubMed] [Google Scholar]
- 55.Flores ER, Tsai KY, Crowley D, et al. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002;416:560–4. doi: 10.1038/416560a. [DOI] [PubMed] [Google Scholar]
- 56.Flores ER, Sengupta S, Miller JB, et al. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005;7:363–73. doi: 10.1016/j.ccr.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 57.Mastrangelo D, De Francesco S, Di Leonardo A, Lentini L, Hadjistilianou T. The retinoblastoma paradigm revisited. Med Sci Monit. 2008;14:RA231–40. Review. [PubMed] [Google Scholar]
- 58.Munakata T, Liang Y, Kim S, McGivern DR, Huibregtse J, et al. Hepatitis C virus induces E6AP dependent degradation of the retinoblastoma protein. PLoS Pathog. 2007;3:1335–47. doi: 10.1371/journal.ppat.0030139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yokota J, Akiyama T, Fung YK, et al. Altered expression of the retinoblastoma (RB) gene in small-cell carcinoma of the lung. Oncogene. 1998;3:471–5. [PubMed] [Google Scholar]
- 60.Meuwissen R, Linn SC, Linnoila RI, Zevenhoven J, Mooi WJ, Berns A. Induction of small cell lung cancer by somatic inactivation of both Trp53 and Rb1 in a conditional mouse model. Cancer Cell. 2003;4:181–9. doi: 10.1016/s1535-6108(03)00220-4. [DOI] [PubMed] [Google Scholar]
- 61.Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer. 2008;8:671–82. doi: 10.1038/nrc2399. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cobrinik D, Lee MH, Hannon G, et al. Shared role of the pRB-related p130 and p107 proteins in limb development. Genes Dev. 1996;10:1633–44. doi: 10.1101/gad.10.13.1633. [DOI] [PubMed] [Google Scholar]
- 63.Cobrinik D. Pocket proteins and cell cycle control. Oncogene. 2005;24:2796–809. doi: 10.1038/sj.onc.1208619. [DOI] [PubMed] [Google Scholar]
- 64.Giacinti C, Giordano A. RB and cell cycle progression. Oncogene. 2006;25:5220–7. doi: 10.1038/sj.onc.1209615. Review. [DOI] [PubMed] [Google Scholar]
- 65.Sherr CJ. The Pezcoller lecture: cancer cell cycles revisited. Cancer Res. 2000;60:3689–95. [PubMed] [Google Scholar]
- 66.Sharpless NE, DePinho RA. How stem cells age and why this makes us grow old. Nat Rev Mol Cell Biol. 2007;8:703–13. doi: 10.1038/nrm2241. [DOI] [PubMed] [Google Scholar]
- 67.Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat Rev Cancer. 2009;9:785–97. doi: 10.1038/nrc2696. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hallstrom TC, Nevins JR. Balancing the decision of cell proliferation and cell fate. Cell Cycle. 2009;8:532–5. doi: 10.4161/cc.8.4.7609. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol. 2002;3:11–20. doi: 10.1038/nrm714. Review. [DOI] [PubMed] [Google Scholar]
- 70.Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 71.Almasan A, Yin Y, Kelly RE, et al. Deficiency of retinoblastoma protein leads to inappropriate S-phase entry, activation of E2F responsive genes, and apoptosis. Proc Natl Acad Sci U S A. 1995;92:5436–40. doi: 10.1073/pnas.92.12.5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Iaquinta PJ, Lees JA. Life and death decisions by the E2F transcription factors. Curr Opin Cell Biol. 2007;19:649–57. doi: 10.1016/j.ceb.2007.10.006. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vigo E, Muller H, Prosperini E, et al. CDC25A phosphatase is a target of E2F and is required for efficient E2F-induced S phase. Mol Cell Biol. 1999;19:6379–95. doi: 10.1128/mcb.19.9.6379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Moroni MC, Hickman ES, Denchi EL, et al. Apaf-1 is a transcriptional target for E2F and p53. Nat Cell Biol. 2001;3:552–8. doi: 10.1038/35078527. [DOI] [PubMed] [Google Scholar]
- 75.Lazzerini Denchi E, Helin K. E2F1 is crucial for E2F-dependent apoptosis. EMBO Rep. 2005;6:661–8. doi: 10.1038/sj.embor.7400452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Clarke AR, Maandag ER, van Roon M, et al. Requirement for a functional Rb-1 gene in murine development. Nature. 1992;359:328–30. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- 77.Jacks T, Fazeli A, Schmitt EM, Bronson RT, Goodell MA, Weinberg RA. Effects of an Rb mutation in the mouse. Nature. 1992;359:295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- 78.Lee EY, Chang CY, Hu N, et al. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature. 1992;359:288–94. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- 79.Morgenbesser SD, Williams BO, Jacks T, DePinho RA. p53-dependent apoptosis produced by Rb-deficiency in the developing mouse lens. Nature. 1994;371:72–4. doi: 10.1038/371072a0. [DOI] [PubMed] [Google Scholar]
- 80.Maandag EC, van der Valk M, Vlaar M, et al. Developmental rescue of an embryoniclethal mutation in the retinoblastoma gene in chimeric mice. EMBO J. 1994;13:4260–68. doi: 10.1002/j.1460-2075.1994.tb06746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Williams BO, Schmitt EM, Remington L, et al. Extensive contribution of Rb-deficient cells to adult chimeric mice with limited histopathological consequences. EMBO J. 1994;13:4251–59. doi: 10.1002/j.1460-2075.1994.tb06745.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tsai KY, Hu Y, Macleod KF, Crowley D, Yamasaki L, Jacks T. Mutation of E2f-1 suppresses apoptosis and inappropriate S phase entry and extends survival of Rb-deficient mouse embryos. Mol Cell. 1998;2:293–304. doi: 10.1016/s1097-2765(00)80274-9. [DOI] [PubMed] [Google Scholar]
- 83.Ziebold U, Reza T, Caron A, Lees JA. E2F3 contributes both to the inappropriate proliferation and to the apoptosis arising in Rb mutant embryos. Genes Dev. 2001;15:386–91. doi: 10.1101/gad.858801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.de Bruin A, Wu L, Saavedra HI, et al. Rb function in extraembryonic lineages suppresses apoptosis in the CNS of Rb-deficient mice. Proc Natl Acad Sci U S A. 2003;100:6546–51. doi: 10.1073/pnas.1031853100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lasorella A, Noseda M, Beyna M, Yokota Y, Iavarone A. Id2 is a retinoblastoma protein target and mediates signalling by Myc oncoproteins. Nature. 2000;407:592–8. doi: 10.1038/35036504. [DOI] [PubMed] [Google Scholar]
- 86.Robanus-Maandag E, Dekker M, van der Valk M, et al. p107 is a suppressor of retinoblastoma development in pRb-deficient mice. Genes Dev. 1998;12:1599–609. doi: 10.1101/gad.12.11.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dannenberg JH, van Rossum A, Schuijff L, te Riele H. Ablation of the retinoblastoma gene family deregulates G(1) control causing immortalization and increased cell turnover under growth-restricting conditions. Genes Dev. 2000;14:3051–64. doi: 10.1101/gad.847700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sage J, Mulligan GJ, Attardi LD, et al. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 2000;14:3037–50. doi: 10.1101/gad.843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Berman SD, Calo E, Landman AS, et al. Metastatic osteosarcoma induced by inactivation of Rb and p53 in the osteoblast lineage. Proc Natl Acad Sci U S A. 2008;105:11851–6. doi: 10.1073/pnas.0805462105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Choi J, Curtis SJ, Roy DM, Flesken-Nikitin A, Nikitin AY. Local mesenchymal stem/progenitor cells are a preferential target for initiation of adult soft tissue sarcomas associated with p53 and Rb deficiency. Am J Pathol. 2010;177:2645–58. doi: 10.2353/ajpath.2010.100306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bignon YJ, Chen Y, Chang CY, Riley DJ, Windle JJ, et al. Expression of a retinoblastoma transgene results in dwarf mice. Genes Dev. 1993;7:1654–62. doi: 10.1101/gad.7.9.1654. [DOI] [PubMed] [Google Scholar]
- 92.Nikitin AYu, Shan B, Flesken-Nikitin A, Chang KH, Lee WH. The retinoblastoma gene regulates somatic growth during mouse development. Cancer Res. 2001;61:3110–8. [PubMed] [Google Scholar]
- 93.Ruas M, Peters G. The p16INK4a/CDKN2A tumor suppressor and its relatives. Biochim Biophys Acta. 1998;1378:F115–77. doi: 10.1016/s0304-419x(98)00017-1. [DOI] [PubMed] [Google Scholar]
- 94.Dominguez-Brauer C, Brauer PM, Chen YJ, Pimkina J, Raychaudhuri P. Tumor suppression by ARF: gatekeeper and caretaker. Cell Cycle. 2010;9:86–9. doi: 10.4161/cc.9.1.10350. [DOI] [PubMed] [Google Scholar]
- 95.Sherr CJ. The INK4a/ARF network in tumour suppression. Nat Rev Mol Cell Biol. 2001;2:731–7. doi: 10.1038/35096061. Review. [DOI] [PubMed] [Google Scholar]
- 96.Sherr CJ, Bertwistle D, DEN Besten W, et al. p53-Dependent and -independent functions of the Arf tumor suppressor. Cold Spring Harb Symp Quant Biol. 2005;70:129–37. doi: 10.1101/sqb.2005.70.004. Review. [DOI] [PubMed] [Google Scholar]
- 97.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–7. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 98.Liao MJ, Van Dyke T. Critical role for Atm in suppressing V(D)J recombination-driven thymic lymphoma. Genes Dev. 1999;13:1246–50. doi: 10.1101/gad.13.10.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tam SW, Shay JW, Pagano M. Differential expression and cell cycle regulation of the cyclin-dependent kinase 4 inhibitor p16Ink4. Cancer Res. 1994;54:5816–20. [PubMed] [Google Scholar]
- 100.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi- 1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–68. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 101.Ohtani N, Zebedee Z, Huot TJ, et al. Opposing effects of Ets and Id proteins on p16INK4a expression during cellular senescence. Nature. 2001;409:1067–70. doi: 10.1038/35059131. [DOI] [PubMed] [Google Scholar]
- 102.Pomerantz J, Schreiber-Agus N, Liegeois NJ, et al. The Ink4a tumor suppressor gene product, p19Arf, interacts with MDM2 and neutralizes MDM2’s inhibition of p53. Cell. 1998;92:713–23. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 103.Zhang Y, Xiong Y, Yarbrough WG. ARF promotes MDM2 degradation and stabilizes p53: ARF-INK4a locus deletion impairs both the Rb and p53 tumor suppression pathways. Cell. 1998;92:725–34. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 104.Kamijo T, Weber JD, Zambetti G, Zindy F, Roussel MF, Sherr CJ. Functional and physical interactions of the ARF tumor suppressor with p53 and Mdm2. Proc Natl Acad Sci U S A. 1998;95:8292–97. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Stott FJ, Bates S, James MC, et al. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. EMBO J. 1998;17:5001–14. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat Rev Cancer. 2006;6:663–73. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- 107.Dominguez-Brauer C, Brauer PM, Chen YJ, Pimkina J, Raychaudhuri P. Tumor suppression by ARF: gatekeeper and caretaker. Cell Cycle. 2010;9:86–9. doi: 10.4161/cc.9.1.10350. Review. [DOI] [PubMed] [Google Scholar]
- 108.Robertson KD, Jones PA. The human ARF cell cycle regulatory gene promoter is a CpG island which can be silenced by DNA methylation and down-regulated by wild-type p53. Mol Cell Biol. 1998;18:6457–73. doi: 10.1128/mcb.18.11.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–33. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.de Stanchina E, McCurrach ME, Zindy F, et al. E1A signaling to p53 involves the p19(ARF) tumor suppressor. Genes Dev. 1998;12:2434–42. doi: 10.1101/gad.12.15.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jacobs JJ, Scheijen B, Voncken JW, Kieboom K, Berns A, van Lohuizen M. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 1999;13:2678–90. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jacobs JJ, Keblusek P, Robanus-Maandag E, et al. Senescence bypass screen identifies TBX2 which represses Cdkn2a (p19(ARF)) and is amplified in a subset of human breast cancers. Nat Genet. 2000;26:291–99. doi: 10.1038/81583. [DOI] [PubMed] [Google Scholar]
- 113.Maestro R, Dei Tos AP, Hamamori Y, Krasnokutsky S, Sartorelli V, et al. Twist is a potential oncogene that inhibits apoptosis. Genes Dev. 1999;13:2207–17. doi: 10.1101/gad.13.17.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Maeda T, Hobbs RM, Merghoub T, Guernah I, Zelent A, et al. Role of the proto-oncogene Pokemon in cellular transformation and ARF repression. Nature. 2005;433:278–85. doi: 10.1038/nature03203. [DOI] [PubMed] [Google Scholar]
- 115.Linggi B, Müller-Tidow C, van de Locht L, et al. The t(8;21) fusion protein, AML1 ETO, specifically represses the transcription of the p14(ARF) tumor suppressor in acute myeloid leukemia. Nat Med. 2002;8:743–50. doi: 10.1038/nm726. [DOI] [PubMed] [Google Scholar]
- 116.Inoue K, Roussel MF, Sherr CJ. Induction of ARF tumor suppressor gene expression and cell cycle arrest by transcription factor DMP1. Proc Natl Acad Sci U S A. 1999;96:3993–98. doi: 10.1073/pnas.96.7.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Inoue K, Mallakin A, Frazier DP. Dmp1 and tumor suppression. Oncogene. 2007;26:4329–35. doi: 10.1038/sj.onc.1210226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bates S, Phillips AC, Clark PA, Stott F, Peters G, et al. p14 ARF links the tumour suppressors RB and p53. Nature. 1998;395:124–25. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- 119.Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 120.Orlow I, Rabbani F, Chin L, et al. Involvement of the Ink4a gene (p16 and p19arf) in murine tumorigenesis. Int J Oncol. 1999;15:17–24. doi: 10.3892/ijo.15.1.17. [DOI] [PubMed] [Google Scholar]
- 121.Chin L, Pomerantz J, Polsky D, et al. Cooperative effects of INK4a and ras in melanoma susceptibility in vivo. Genes Dev. 1997;11:2822–34. doi: 10.1101/gad.11.21.2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kamijo T, Zindy F, Roussel MF, et al. Tumor suppression at the mouse INK4a locus mediated by the alternative reading frame product p19ARF. Cell. 1997;91:649–59. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 123.Kamijo T, Bodner S, van de Kamp E, Randle DH, Sherr CJ. Tumor spectrum in ARF-deficient mice. Cancer Res. 1999;59:2217–22. [PubMed] [Google Scholar]
- 124.Sharpless NE, Ramsey MR, Balasubramanian P, Castrillon DH, DePinho RA. The differential impact of p16(INK4a) or p19(ARF) deficiency on cell growth and tumorigenesis. Oncogene. 2004;23:379–85. doi: 10.1038/sj.onc.1207074. [DOI] [PubMed] [Google Scholar]
- 125.Sharpless NE, Bardeesy N, Lee KH, et al. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature. 2001;413:86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- 126.Krimpenfort P, Quon KC, Mooi WJ, Loonstra A, Berns A. Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature. 2001;413:83–6. doi: 10.1038/35092584. [DOI] [PubMed] [Google Scholar]
- 127.Zindy F, Williams RT, Baudino TA, Rehg JE, Skapek SX, et al. Arf tumor suppressor promoter monitors latent oncogenic signals in vivo. Proc Natl Acad Sci U S A. 2003;100:15930–5. doi: 10.1073/pnas.2536808100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Randle DH, Zindy F, Sherr CJ, Roussel MF. Differential effects of p19(Arf) and p16(Ink4a) loss on senescence of murine bone marrow-derived preB cells and macrophages. Proc Natl Acad Sci U S A. 2001;98:9654–9. doi: 10.1073/pnas.171217498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Williams RT, Roussel MF, Sherr CJ. Arf gene loss enhances oncogenicity and limits imatinib response in mouse models of Bcr-Abl-induced acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2006;103:6688–93. doi: 10.1073/pnas.0602030103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wong S, Witte ON. The BCR-ABL story: bench to bedside and back. Annu Rev Immunol. 2004;22:247–306. doi: 10.1146/annurev.immunol.22.012703.104753. Review. [DOI] [PubMed] [Google Scholar]
- 131.Drucker BJ, Guilhot F, O’Brien SG, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med. 2006;355:2408–17. doi: 10.1056/NEJMoa062867. [DOI] [PubMed] [Google Scholar]
- 132.Williams RT, den Besten W, Sherr CJ. Cytokine-dependent imatinib resistance in mouse BCR-ABL+, Arf-null lymphoblastic leukemia. Genes Dev. 2007;21:2283–7. doi: 10.1101/gad.1588607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yang C, Ionescu-Tiba V, Burns K, et al. The role of the cyclin D1-dependent kinases in ErbB2-mediated breast cancer. Am J Pathol. 2004;164:1031–8. doi: 10.1016/S0002-9440(10)63190-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Matheu A, Pantoja C, Efeyan A, et al. Increased gene dosage of Ink4a/Arf results in cancer resistance and normal aging. Genes Dev. 2004;18:2736–46. doi: 10.1101/gad.310304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Matheu A, Maraver A, Klatt P, et al. Delayed ageing through damage protection by the Arf/p53 pathway. Nature. 2007;448:375–9. doi: 10.1038/nature05949. [DOI] [PubMed] [Google Scholar]
- 136.Matheu A, Maraver A, Collado M, et al. Anti-aging activity of the Ink4/Arf locus. Aging Cell. 2009;8:152–61. doi: 10.1111/j.1474-9726.2009.00458.x. [DOI] [PubMed] [Google Scholar]
- 137.Krishnamurthy J, Torrice C, Ramsey MR, et al. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114:1299–307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Krishnamurthy J, Ramsey MR, Ligon KL, Torrice C, Koh A, et al. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006;443:453–7. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- 139.Franklin DS, Godfrey VL, Lee H, et al. CDK inhibitors p18(INK4c) and p27(Kip1) mediate two separate pathways to collaboratively suppress pituitary tumorigenesis. Genes Dev. 1998;12:2899–911. doi: 10.1101/gad.12.18.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Latres E, Malumbres M, Sotillo R, et al. Limited overlapping roles of P15(INK4b) and P18(INK4c) cell cycle inhibitors in proliferation and tumorigenesis. EMBO J. 2000;19:3496–506. doi: 10.1093/emboj/19.13.3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fero ML, Randel E, Gurley KE, Roberts JM, Kemp CJ. The murine gene p27 Kip1 is haplo-insufficient for tumour suppression. Nature. 1998;396:177–80. doi: 10.1038/24179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zindy F, Cunningham JJ, Sherr CJ, Jogal S, Smeyne RJ, Roussel MF. Postnatal neuronal proliferation in mice lacking Ink4d and Kip1 inhibitors of cyclin-dependent kinases. Proc Natl Acad Sci U S A. 1999;96:13462–7. doi: 10.1073/pnas.96.23.13462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Uziel T, Zindy F, Xie S, et al. The tumor suppressors Ink4c and p53 collaborate independently with Patched to suppress medulloblastoma formation. Genes Dev. 2005;19:2656–67. doi: 10.1101/gad.1368605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Rahman N, Stratton MR. The genetics of breast cancer susceptibility. Annu Rev Genet. 1998;32:95–121. doi: 10.1146/annurev.genet.32.1.95. [DOI] [PubMed] [Google Scholar]
- 145.Deng CX, Xu X. Generation and analysis of Brca1 conditional knockout mice. Methods Mol Biol. 2004;280:185–200. doi: 10.1385/1-59259-788-2:185. [DOI] [PubMed] [Google Scholar]
- 146.Scully R, Livingston DM. In search of the tumour-suppressor functions of BRCA1 and BRCA2. Nature. 2000;408:429–32. doi: 10.1038/35044000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Deng CX, Brodie SG. Roles of BRCA1 and its interacting proteins. Bio Essays. 2000;22:728–37. doi: 10.1002/1521-1878(200008)22:8<728::AID-BIES6>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 148.Gowen LC, Johnson BL, Latour AM, Sulik KK, Koller BH. Brca1 deficiency results in early embryonic lethality characterized by neuroepithelial abnormalities. Nat Genet. 1996;12:191–94. doi: 10.1038/ng0296-191. [DOI] [PubMed] [Google Scholar]
- 149.Hakem R, de la Pompa JL, Sirard C, et al. The tumor suppressor gene Brca1 is required for embryonic cellular proliferation in the mouse. Cell. 1996;85:1009–23. doi: 10.1016/s0092-8674(00)81302-1. [DOI] [PubMed] [Google Scholar]
- 150.Liu CY, Flesken-Nikitin A, Li S, Zeng Y, Lee WH. Inactivation of the mouse Brca1 gene leads to failure in the morphogenesis of the egg cylinder in early postimplantation development. Genes Dev. 1996;10:1835–43. doi: 10.1101/gad.10.14.1835. [DOI] [PubMed] [Google Scholar]
- 151.Ludwig T, Chapman DL, Papaioannou VE, Efstratiadis A. Targeted mutations of breast cancer susceptibility gene homologs in mice: lethal phenotypes of Brca1, Brca2, Brca1/Brca2, Brca1/p53, and Brca2/p53 nullizygous embryos. Genes Dev. 1997;11:1226–41. doi: 10.1101/gad.11.10.1226. [DOI] [PubMed] [Google Scholar]
- 152.Evers B, Jonkers J. Mouse models of BRCA1 and BRCA2 deficiency: past lessons, current understanding and future prospects. Oncogene. 2006;25:5885–97. doi: 10.1038/sj.onc.1209871. [DOI] [PubMed] [Google Scholar]
- 153.Hakem R, de la Pompa JL, Mak TW. Developmental studies of Brca1 and Brca2 knock-out mice. J Mammary Gland Biol Neoplasia. 1998;3:431–45. doi: 10.1023/a:1018792200700. [DOI] [PubMed] [Google Scholar]
- 154.Deng CX, Brodie SG. Knockout mouse models and mammary tumorigenesis. Semin Cancer Biol. 2001;11:387–94. doi: 10.1006/scbi.2001.0394. Review. [DOI] [PubMed] [Google Scholar]
- 155.Gowen LC, Avrutskaya AV, Latour AM, Koller BH, Leadon SA. BRCA1 required for transcription-coupled repair of oxidative DNA damage. Science. 1998;281:1009–12. doi: 10.1126/science.281.5379.1009. [DOI] [PubMed] [Google Scholar]
- 156.Shen SX, Weaver Z, Xu X, Li C, Weinstein M, et al. A targeted disruption of the murine Brca1 gene causes gamma irradiation hypersensitivity and genetic instability. Oncogene. 1998;17:3115–24. doi: 10.1038/sj.onc.1202243. [DOI] [PubMed] [Google Scholar]
- 157.Chandler J, Hohenstein P, Swing D, et al. Human BRCA1 gene rescues the embryonic lethality of Brca1 mutant mice. Genesis. 2001;29:72–7. doi: 10.1002/1526-968x(200102)29:2<72::aid-gene1007>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 158.Lane TF, Lin C, Brown MA, Solomon E, Leder P. Gene replacement with the human BRCA1 locus: tissue specific expression and rescue of embryonic lethality in mice. Oncogene. 2000;19:4085–90. doi: 10.1038/sj.onc.1203760. [DOI] [PubMed] [Google Scholar]
- 159.Mak TW, Hakem A, McPherson JP, et al. Brca1 is required for T cell lineage development but not TCR loci rearrangement. Nat Immunol. 2000;1:77–82. doi: 10.1038/76950. [DOI] [PubMed] [Google Scholar]
- 160.Xu X, Wagner KU, Larson D, et al. Conditional mutation of Brca1 in mammary epithelial cells results in blunted ductal morphogenesis and tumour formation. Nat Genet. 1999;22:37–43. doi: 10.1038/8743. [DOI] [PubMed] [Google Scholar]
- 161.Liu X, Holstege H, van der Gulden H, et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc Natl Acad Sci U S A. 2007;104:12111–6. doi: 10.1073/pnas.0702969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Brodie SG, Xu X, Qiao W. Multiple genetic changes are associated with mammary tumorigenesis in Brca1 conditional knockout mice. Oncogene. 2001;20:7514–23. doi: 10.1038/sj.onc.1204929. [DOI] [PubMed] [Google Scholar]
- 163.Hoshino A, Yee CJ, Campbell M, et al. Effects of BRCA1 transgene expression on murine mammary gland development and mutagen-induced mammary neoplasia. Int J Biol Sci. 2007;3:281–91. doi: 10.7150/ijbs.3.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Chen J, Silver DP, Walpita D, et al. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol Cell. 1998;2:317–28. doi: 10.1016/s1097-2765(00)80276-2. [DOI] [PubMed] [Google Scholar]
- 165.Sharan SK, Morimatsu M, Albrecht U, et al. Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature. 1997;386:804–10. doi: 10.1038/386804a0. [DOI] [PubMed] [Google Scholar]
- 166.Suzuki A, de la Pompa JL, Hakem R, et al. Brca2 is required for embryonic cellular proliferation in the mouse. Genes Dev. 1997;11:1242–52. doi: 10.1101/gad.11.10.1242. [DOI] [PubMed] [Google Scholar]
- 167.Connor F, Bertwistle D, Mee PJ, et al. Tumorigenesis and a DNA repair defect in mice with a truncating Brca2 mutation. Nat Genet. 1997;17:423–30. [Google Scholar]
- 168.Friedman LS, Thistlethwaite FC, Patel KJ, et al. Thymic lymphomas in mice with a truncating mutation in Brca2. Cancer Res. 1998;58:1338–43. [PubMed] [Google Scholar]
- 169.Patel KJ, Yu VP, Lee H, et al. Involvement of Brca2 in DNA repair. Mol Cell. 1998;1:347. doi: 10.1016/s1097-2765(00)80035-0. [DOI] [PubMed] [Google Scholar]
- 170.Tutt A, Gabriel A, Bertwistle D, et al. Absence of Brca2 causes genome instability by chromosome breakage and loss associated with centrosome amplification. Curr Biol. 1999;9:1107–10. doi: 10.1016/s0960-9822(99)80479-5. [DOI] [PubMed] [Google Scholar]
- 171.Yu VP, Koehler M, Steinlein C, et al. Gross chromosomal rearrangements and genetic exchange between nonhomologous chromosomes following BRCA2 inactivation. Genes Dev. 2000;14:1400–6. [PMC free article] [PubMed] [Google Scholar]
- 172.Ludwig T, Fisher P, Murty V, Efstratiadis A. Development of mammary adenocarcinomas by tissue-specific knockout of Brca2 in mice. Oncogene. 2001;20:3937–48. doi: 10.1038/sj.onc.1204512. [DOI] [PubMed] [Google Scholar]
- 173.Cheung AM, Elia A, Tsao MS, et al. Brca2 deficiency does not impair mammary epithelium development but promotes mammary adenocarcinoma formation in p53(+/−) mutant mice. Cancer Res. 2004;64:1959–65. doi: 10.1158/0008-5472.can-03-2270. [DOI] [PubMed] [Google Scholar]
- 174.Cheung AM, Hande MP, Jalali F, et al. Loss of Brca2 and p53 synergistically promotes genomic instability and deregulation of T-cell apoptosis. Cancer Res. 2002;62:6194–204. [PubMed] [Google Scholar]
- 175.Park PG, Lee H. Development of thymic lymphomas in mice disrupted of Brca2 allele in the thymus. Exp Mol Med. 2008;40:339–44. doi: 10.3858/emm.2008.40.3.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lee H, Trainer AH, Friedman LS, et al. Mitotic checkpoint inactivation fosters transformation in cells lacking the breast cancer susceptibility gene, Brca2. Mol Cell. 1999;4:1–10. doi: 10.1016/s1097-2765(00)80182-3. [DOI] [PubMed] [Google Scholar]
- 177.Francis JC, McCarthy A, Thomsen MK, Ashworth A, Swain A. Brca2 and Trp53 deficiency cooperate in the progression of mouse prostate tumourigenesis. PLoS Genet. 2010;6:e1000995. doi: 10.1371/journal.pgen.1000995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yu Y, Xu F, Peng H, et al. NOEY2 (ARH1), an imprinted putative tumor suppressor gene in ovarian and breast carcinomas. Proc Natl Acad Sci U S A. 1999;96:214–9. doi: 10.1073/pnas.96.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Xu F, Xia W, Luo RZ, et al. The human ARH1 tumor suppressor gene inhibits lactation and growth in transgenic mice. Cancer Res. 2000;60:4913–20. [PubMed] [Google Scholar]
- 180.Lu Z, Luo RZ, Lu Y, et al. The tumor suppressor gene ARH1 regulates autophagy and tumor dormancy in human ovarian cancer cells. J Clin Invest. 2008;118:3917–29. doi: 10.1172/JCI35512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Sexl V, Diehl JA, Sherr CJ, Ashmun R, Beach D, Roussel MF. A rate limiting function of cdc25 A for S phase entry inversely correlates with tyrosine dephosphorylation of Cdk2. Oncogene. 1999;18:573–82. doi: 10.1038/sj.onc.1202362. [DOI] [PubMed] [Google Scholar]
- 182.Cole MD, Nikiforov MA. Transcriptional activation by the Myc oncoprotein. Curr Top Microbiol Immunol. 2006:33–50. doi: 10.1007/3-540-32952-8_2. Review. [DOI] [PubMed] [Google Scholar]
- 183.Lebofsky R, Walter JC. New Myc-anisms for DNA replication and tumorigenesis? Cancer Cell. 2007;12:102–3. doi: 10.1016/j.ccr.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 184.Sugiyama T, Frazier DP, Taneja P, et al. Signal transduction involving the Dmp1 transcription factor and its alteration in human cancer. Clinical Medicine Oncology. 2008;2:209–19. doi: 10.4137/cmo.s548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Inoue K, Sugiyama T, Taneja P, Morgan RL, Frazier DP. Emerging roles of DMP1 in lung cancer. Cancer Res. 2008;68:4487–90. doi: 10.1158/0008-5472.CAN-07-6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Sugiyama T, Frazier DP, Taneja P, Morgan RL, Willingham MC, Inoue K. The role of Dmp1 and its future in lung cancer diagnostics. Expert Rev Mol Diagn. 2008;8:435–48. doi: 10.1586/14737159.8.4.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Taneja P, Maglic D, Kai F, et al. Critical role of Dmp1 in HER2/neu-p53 signaling and breast carcinogenesis. Cancer Res. 2010;70:2084–94. doi: 10.1158/0008-5472.CAN-10-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]