Abstract
We have developed a semi-automatic methodology to reconstruct the phylogenetic species tree in Protozoa, integrating different phylogenetic algorithms and programs, and demonstrating the utility of a supermatrix approach to construct phylogenomics-based trees using 31 universal orthologs (UO). The species tree obtained was formed by three major clades that were related to three groups of data: i) Species containing at least 80% of UO (25/31) in the concatenated multiple alignment or supermatrix, this clade was called C1, ii) Species containing between 50%–79% (15–24/31) of UO called C2, and iii) Species containing less than 50% (1–14/31) of UO called C3. C1 was composed by only protozoan species, C2 was composed by species related to Protozoa, and C3 was composed by some species of C1 (Protozoa) and C2 (related to Protozoa). Our phylogenomics-based methodology using a supermatrix approach proved to be reliable with protozoan genome data and using at least 25 UO, suggesting that (a) the more UO used the better, (b) using the entire UO sequence or just a conserved block of it for the supermatrix produced similar phylogenomic trees.
Keywords: protozoan parasites, phylogenomics, universal orthologs
Introduction
Phylogenomics is being intensively used in the “Post Genomic Era”. Many different phylogenetic trees have been published on the basis of different models of sequence evolution,1 applying different parameter settings and algorithms. Although the rDNA,2–4 COI2,5,6 and other single genes have been extremely valuable for phylogenetic studies, single-gene phylogeny has its limitations.7 Therefore, phylogenomics approaches, corroborated by the use of more representative phylogenetic markers, will allow, in principle, a more reliable and representative inference into the tree of life.
Nowadays, one has the option to concatenate multiple gene sequences to construct trees on the genomic level, such as “genome trees” or also called “ supermatrix trees”, possessing more phylogenetic signals making them less susceptible to the stochastic errors than those built from a single gene.
The other option is the construction of the “ supertree” that involves the concatenation of a set of trees.8–10 However, there are fundamental differences between the ways in which phylogenomic approaches integrate the phylogenetic information. Dutilh et al in 200711 systematically compared alternative methodologies such as gene content, superalignment, superdistance (to construct a supermatrix) and supertree approaches using various algorithms and tree-building methods on the Fungi, the eukaryotic clade with the largest number of sequenced genomes. The phylogenomic trees reproduced many of the clades in accordance with the current taxonomic views. Superalignment (supermatrix) and supertrees reproduced better target fungal phylogeny but they were not a guarantee for a successful phylogenomic tree.11
Phylogenomics involving the use of entire genomes to infer a species tree has become the de facto standard for reconstructing reliable species phylogenies.8,12 While some criticism has been made on the recent superalignment tree8 as being a ‘tree of one percent’ of the genome,13 single-gene phylogenetic trees have shown conflict14 due to a variety of causes. Yet, phylogenomic trees have held the promise of minimizing anomalies by the sheer power of genome-scale data as they are based on the maximum quantity of genetic information. A phylogenomic tree should be the best reflection of the evolutionary history of the species.15,16
There are more than 200,000 named species of unicellular eukaryotes that can be classified as Protozoa, of which approximately 10,000 are parasites, but only a small number are sufficiently important to be mentioned on the pages of Trends in Parasitology.18 The systematics of the Protozoa is a subject that has engaged the attention of protozoologists and evolutionists for some time, and advances in molecular methodology have revealed relationships among Protozoa and between Protozoa and other groups (http://www.ncbi.nlm.nih.gov/RefSeq) that can be used to draw up realistic and natural systems of classification.17–19 Protozoa are currently classified as a paraphyletic group, however the term is still used in several publications that relate it to areas such as systematics17 and taxonomy, besides parasitology,20,21 phylogeny,22–25 evolution26,27 and genomics.28
Difficulty is encountered in seeking a consensus taxonomic definition of several kingdoms. In particular, protozoan species are loosely characterized; deciding whether a species belongs to Protozoa or not is based on morphology and biological properties and also partially by the fact of not belonging to another kingdom.18,19,29 They are not a coherent phylogenetic group like other kingdoms or candidate kingdoms are, eg, stramenopila. Thus, to determine if a species belongs to Protozoa we do not just look at one characteristic (such as the multiparted, tubular flagellar hairs of stramenopila) and decide that. Instead, several criteria have to be met.30
Protozoa are phylogenetically connected to other eukaryotic groups and several eukaryotic groups are most likely derived from Protozoa.31 A molecular similarity has been established between alveolate (Protozoa) and stramenopiles.32 In corroboration, residual plastids in the malarial parasite (Plasmodium, Apicomplexa), and in non-malarial apicomplexans (Toxoplasma), suggest a relationship with dinoflagellate plastids and/or plastids of stramenopilous chromistans.33,34 Molecular data relate Choanoflagellate (Protozoa) to Sponges, thus to Animalia. Interestingly, molecular sequencing also supports a relationship of Choanoflagellates and Fungi.31 Sequences of the small subunit of ribosomal RNA gene (SSU rRNA) point to a grouping of Biliphyta (glaucophytes, Rhodophyta) with Cryptomonads.32 Also, several authors provided molecular evidence that primitive flagellates (protozoan species) occurred in the ancestry of Chlorophyta (green algae).24,35–37
Illnesses caused by parasitic Protozoa are a major cause of disease worldwide, but because they are concentrated in low socioeconomic parts of the world, they receive relatively little attention from the pharmaceutical industry. Of the ten diseases targeted as research priorities by the World Health Organization’s Special Program for Research and Training in Tropical Diseases (http://www.who.int/tdr), four are caused by protozoan parasites (malaria, leishmaniasis, Chagas disease and African trypanosomiasis). These diseases and other less dangerous ones (eg, amoebiasis and trichomoniasis) are having an alarming increase in cases, which are refractory to the main treatment. Treatment failure has potentially a multifactorial origin where drug resistance stands out as one of its major concerns.38–42
Our understanding of the phylogenetic position of protozoan within eukaryotes, as well as their relationships, is mainly based on ribosomal DNA analysis.2–4 At first, this process seemed relatively straightforward: trees generated from a single gene, most commonly a SSU rRNA,43 appeared to provide a basic structure for the topology of eukaryotes, although many branches of the tree remained controversial. The 1990s was a period of deconstruction of this theory since several protein coding gene trees revealed serious discrepancies.44,45 Currently, a hypothesis for the tree of eukaryotes resembles the tree presented by Keeling in 2005.36 The tree is a hypothesis composed from the various types of data, including molecular phylogenies and other molecular characteristics, as well as morphological and biochemical evidence. Five ‘supergroups’ were shown, each consisting of a diversity of eukaryotes, most of which were microbial (mostly protists and algae).
Chaudhary in 200546 showed results on available sequencing data. Phylogenetic analysis defined five supergroups of eukaryotes: i) the plant and red/green algal lineage; ii) a clade comprised of animals, fungi, slime molds and amoebozoans named Unikonta eukaryotes which contained the species included in our study (Acanthamoeba, Entamoeba and Dictyostelium);24 and three supergroups that are entirely Protozoa: iii) chromalveolates, iv) excavates and v) rhizaria.36 The first supergroup of Protozoa (chromalveolates) showed three principal groups which included the parasitic phylum apicomplexa (Theileria, Babesia, Plasmodium, Toxoplasma, Neospora, Eimeria and Cryptosporidium); along with ciliates (Tetrahymena, Paramecium and Oxytricha); diatoms and many taxa for which no complete genomes are available (Diatoms and Phytophthora).37,47–50 The second supergroup (excavates) included kinetoplastid parasites (Trypanosoma and Leishmania) and other lineages (many anaerobic and/or parasitic Giardia, Spironucleus, Trichomonas and Naegleria).51
In this study, we explore the use of multiple genes to present a phylogenomics-based study among Protozoa and its relationship with other very close taxonomic species which are considered as mitochondrial or plastid protozoan according to RefSeq (http://www.ncbi.nlm.nih.gov/RefSeq) taxonomic classification.
Material and Methods
Selection and preparation of marker gene families
We based our methodology on the study by Ciccarelli et al (2006)8 for the selection and construction of the species tree. Thirty-one universal orthologous (UO) genes showing 1:1 orthologous relationships were used (Supplementary Table S1). Those UO were originally identified by Ciccarelli et al (2006)8 showing the following characteristics: i) to be present in all complete genomes available at Genbank until 2006, ii) not to be involved in horizontal transfer, and iii) to be good ones for phylogenomic studies. As those 31 UO have a direct correspondence in the protozoan genome data available at RefSeq, they were mapped to the referred data using (a) the best blast hits (e-value < 1-e50), and (b) manual verification of the annotation (the RefSeq annotation of the best hits needed to match the UO annotation) (Supplementary Table S2). Once mapped, the protozoan protein sequences corresponding to those 31 UO were downloaded in fasta format from RefSeq (ftp://ftp.ncbi.nih.gov/refseq/release/protozoa/) and then aligned using Mafft v5.86152–54 with default parameters.
Selection and preparation of marker gene families for protozoan species trees
Our study used the data (protozoan species) that were obtained based on the taxonomic classification provided in the Release Catalogue of RefSeq (RefSeq-release35-05/04/2009) for Protozoa. This was done as a first tentative to include the different genus and species of Protozoa available in public databases. The full names of species used in our analysis are listed in Supplementary Table S3.
Hidden Markov Models (HMM) profiles55 were constructed for the 31 aligned UO set, then this database (HMM profiles obtained from the alignment of the best hits of UO from protozoan sequences available at RefSeq) served as a seed to search for more UO hits in protozoan sequences available at Genbank and RefSeq. The HMM profiles used as a seed were created (hmmbuild) and calibrated (hmmcalibrate) and the searches (hmmpfam) were done with e-value “1e-5” as cut-off using HMMER version 2.3.2. All protozoan sequences (74 complete and draft genomes) available at RefSeq (ftp://ftp.ncbi.nih.gov/refseq/release/protozoa/) (RefSeq-release35-05/04/2009) and Genbank (http://www.ncbi.nlm.nih.gov/genbank/) (NCBI-Flat File Release 172.0-06/15/2009) were used as a target database.
The best hits (e-value < 1e-5) of the HMMER (hmmpfam) search were added to the multiple alignment that originated the UO HMM profile, then a new multiple alignment was constructed (Mafft v5.861)52–54 containing: a) the original protozoan sequences from RefSeq that originated the UO HMM profile, plus b) the best hmmpfam hits of the UO HMM profiles obtained with protozoan sequences from Genbank and RefSeq. Those new multiple alignments (entire or trimmed) were used: i) to be concatenated and build a supermatrix tree, or ii) to build individual trees, then those trees were concatenated to obtain a supertree (Supplementary Table S3).
Concatenating multiple alignments to build a supermatrix tree
A supermatrix tree was obtained using concatenated multiple alignments, either entire or trimmed:
Entire concatenated alignments (M1): The individual alignments were concatenated using an in-house perl script, resulting in a global supermatrix of 21,260 positions in a total of 74 species (43 Protozoa plus 31 mitochondrial or plastid Protozoa). The 31 plastid or mitochondrial protozoan genomes were included in the study based on the taxonomic classification provided in the Release Catalogue of RefSeq (RefSeq-release35-05/04/2009).
Trimmed concatenated alignments (M2): The individual alignments were trimmed using TrimAl v1.256 (http://trimal.cgenomics.org/) aiming to obtain their most conserved blocks. Those extracted conserved blocks were concatenated using an in-house perl script, resulting in a global supermatrix of 12,807 positions in a total of 74 protozoan species. Positions in the alignment with gaps in more than 10% of the sequences were trimmed with TrimAl.56,57
The resulting supermatrix of M1 or M2 was used to generate separate trees with Phyml 2.4.458,59 using 100 bootstrap replicates and JTT, elected as the best evolutionary model. Each individual alignment was tested for the best evolutionary model using Modelgenerator 0.85. Several models were selected by Modelgenerator 0.85; however, because it is not simple to use multiple models in a single (concatenated) alignment, we decided to adopt JTT that was also the model adopted in the phylogenomics studies of Ciccarelli et al (2006).8 JTT assumed that there were two classes of sites, one class being invariable and the other class being free to change.8
The resulting clades C1, C2, and C3 obtained from the supermatrix tree of M1 or M2 were used to generate the individual trees Ct1, Ct2, and Ct3 with Phyml 2.4.458,59 using 100 bootstrap replicates and the evolutionary models (Supplementary Table S4) obtained with Modelgenerator 0.85.
Building individual trees to obtain a supertree
We also used either entire or trimmed individual alignments to build phylogenetic trees, then we concatenated them to build a supertree: the 31 individual total alignments (M3) and the 31 individual trimmed alignments (M4) were used to construct individual trees with Phyml 2.4.4 using 100 bootstrap replicates and the matrices of the evolutionary models (Supplementary Table S4) obtained with Modelgenerator 0.85.60 The resulting trees of M3 were concatenated to obtain supertrees with Clann 3.1.3.9 The same procedure was adopted for M4 (trimmed alignments).
Test of phylogenetic signal
The content and distribution of the phylogenetic signal of the 64 alignments (2 concatenated and 62 individual) were analyzed. Two statistical approaches, the PTP test and g1 statistics were employed to achieve a measure of the overall signal content. The PTP test (PTP—permutation test probability or permutation tail probability test)61 was implemented in PAUP* (Phylogenetic Analysis Using Parsimony) version 4.0b1062 and it was executed with heuristic search.63,64 G1 statistics was calculated from the characters using the RandTrees function in PAUP.65
Topological test
The Kishino and Hasegawa tests (KH tests)66 were performed in PAUP* 4.0b1062 to assess differences between the most parsimonious trees resulting from the analysis of the full dataset.
Null distribution of the test statistic was simulated using 100 bootstrap replicates likelihoods (full dataset) obtained with Phyml 2.4.4 of the following eight groups: i) Complete total tree (M1), ii) Complete trimmed tree (M2), iii) M1-C1, iv) M2-C1, v) M1-C2, vi) M2-C2, vii) M1-C3 and viii) M2-C3.
Kishino-Hasegawa test66 assumes a null hypothesis where the expected difference in the optimality score between alternative phylogenies is zero.67 This requires that the topologies under comparison must be specified a priori and without reference to the data used for the test. However, nearly all uses of these tests involve comparing alternative topologies to the optimal topology estimated from the data. This application guarantees that the null expectation of difference will always be larger than zero and does not violate any assumption of a normal distribution of differences in optimality scores between topologies.68
Results and Discussion
The vast majority of eukaryotic diversity is Protozoa and most of the sequenced protozoan species are parasites.46 It is important to establish the position of protozoan within the eukaryotic group; despite the taxonomic classification rules are not completely clear. Nowadays, the majority of phylogenies cannot be considered as complete information as they use only single or ribosomal genes.
The evolutionary analysis of the data included several steps. First, we separately evaluated the presence or absence of a significantly structured phylogenetic signal for each data set, and second, we separately compared the trees signalized by the KH test as being “the best” and the trees obtained with Phyml 2.4.4 from the entire concatenated alignments (M1) and the trimmed concatenated alignments (M2).
In order to evaluate the statistical significance of any particular branching pattern, statistical tests were performed. Statistical tests in phylogenetics allow the assessment of the degree of confidence in any given tree topology being the true topology or the most consistent that we accept as true. Thus, statistical tests are responsible for the mutual development between the abilities to estimate better trees and to create more realistic models of evolution.69
This was accomplished by conducting exhaustive parsimony searches on each alignment using PAUP* 4.0b262,70 and comparing the resulting g1 statistics of tree-length distribution skewness with critical values published by Hillis and Huelsenbeck in 1992.65 Negatively skewed distributions indicate the presence of trees shorter than what would be expected by chance. As another measure of the phylogenetic robustness of the data, a bootstrap analysis of the combined data set was conducted using 1,000 replicate branch-and-bound parsimony searches. Another popular statistical test that was used in the phylogenetic analysis was the PTP test, which is designed to address whether there is any true phylogenetic signal in any given dataset (alignment).69
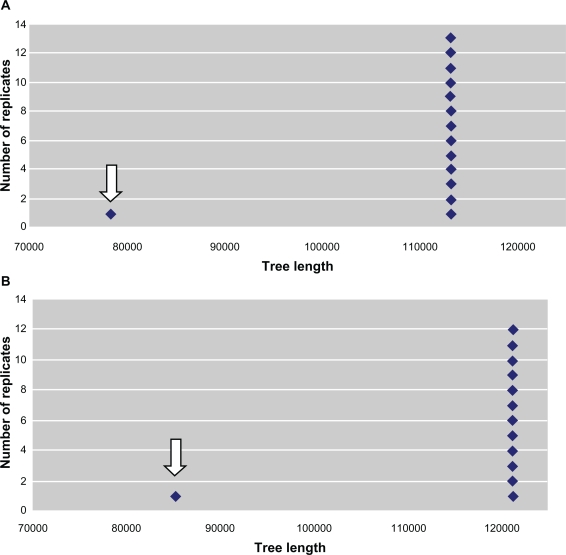
The PTP test indicates that: A) the length value of the most parsimonious tree based on the trimmed concatenated alignments (78,498 steps, P < 0.001) obtained with the original data is distant from the others obtained through the permutation of the data (Figs. 1A and B) the length value of the most parsimonious tree based on the total concatenated alignments (85,268 steps, P < 0.001) obtained with the original data is distant from the others obtained through the permutation of the data (Fig. 1B).
Figure 1.
Permutation Tail Probability Test (PTP) of the concatenated alignments. A) PTP test of the trimmed concatenated alignments of the protozoan UO. (Number of replicates = 1,000, search = heuristic). The gray arrow indicates the most parsimonious tree (TMP = 78,498). p is the probability of getting a more extreme T-value under the null hypothesis of no difference between the two trees (two-tailed test). PTP test indicates significant difference at P < 0.05 between the original (unpermuted) data of AMP and the permuted data. B) PTP test of the total concatenated alignments of the protozoan UO. (Number of replicates = 1,000, search = heuristic). The gray arrow indicates the most parsimonious tree (TMP = 85,268). P is the probability of getting a more extreme T-value under the null hypothesis of no difference between the two trees (two-tailed test). PTP test indicates significant difference at P < 0.05 between the original (unpermuted) data of AMP and the permuted data.
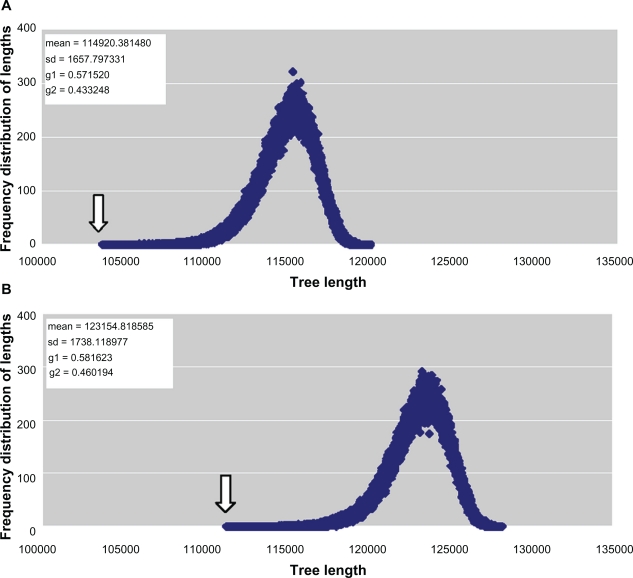
The g1 statistics also indicates that there is phylogenetic signal in the data used in the analysis regarding the tree-length skewness because: A) the tree-length distribution based on the trimmed concatenated alignments showed left skewness (G1 = −0.57, P < 0.001) (Figs. 2A and B) the tree-length distribution based on the total concatenated alignments also showed left skewness (G1 = −0.58, P < 0.001) (Fig. 2B), as the tree-length distribution with significant left skewness contains more phylogenetical signals than more symmetrical or right-skewed distributions.
Figure 2.
The g1 statistics of the concatenated alignments. A) The g1 statistics of the trimmed concatenated alignments of the protozoan UO. (Number of replicates = 1,000,000). The gray arrow indicates the most parsimonious tree (TMP = 103,691). B) The g1 statistics of the total concatenated alignments of the protozoan UO. (Number of replicates = 1,000,000). The gray arrow indicates the most parsimonious tree (TMP = 111,342).
The results of the PTP test for concatenated alignments are (I) Total Characters: i) trimmed: concatenated-12,807 versus single average-412 and ii) total: concatenated-21,260 versus single average-686. (II) Parsimony-Informative Characters: i) trimmed: concatenated-8,586 versus single average-277 and ii) total: concatenated-9,308 versus single average-300 (Tables 1 and 2).
Table 1.
Results of the PTP test for the concatenated alignments of the universal orthologs.
| Concatenated alignments of the universal orthologs | Total characters | Constant characters | Parsimony-uninformative variable characters | Parsimony-informative characters |
|---|---|---|---|---|
| Trimmed | 12807 | 1836 | 2385 | 8586 |
| Total | 21260 | 7479 | 4473 | 9308 |
Table 2.
Results of the PTP test for the 31 single alignments of the universal orthologs.
| Alignments of the universal orthologs | Total characters | Constant characters | Parsimony-uninformative variable characters | Parsimony-informative characters | |
|---|---|---|---|---|---|
| COG0012 | Total | 605 | 153 | 59 | 393 |
| Trimmed | 363 | 11 | 14 | 338 | |
| COG0016 | Total | 964 | 249 | 228 | 487 |
| Trimmed | 578 | 44 | 59 | 475 | |
| COG0048 | Total | 241 | 49 | 58 | 134 |
| Trimmed | 145 | 9 | 6 | 130 | |
| COG0049 | Total | 292 | 103 | 17 | 172 |
| Trimmed | 175 | 6 | 12 | 157 | |
| COG0052 | Total | 339 | 18 | 23 | 298 |
| Trimmed | 203 | 2 | 5 | 196 | |
| COG0080 | Total | 1543 | 1223 | 132 | 188 |
| Trimmed | 926 | 606 | 132 | 188 | |
| COG0081 | Total | 477 | 55 | 112 | 307 |
| Trimmed | 286 | 2 | 14 | 270 | |
| COG0087 | Total | 615 | 115 | 145 | 355 |
| Trimmed | 369 | 16 | 29 | 324 | |
| COG0091 | Total | 522 | 348 | 27 | 147 |
| Trimmed | 313 | 139 | 27 | 147 | |
| COG0092 | Total | 338 | 77 | 38 | 223 |
| Trimmed | 203 | 5 | 7 | 191 | |
| COG0093 | Total | 256 | 84 | 26 | 146 |
| Trimmed | 154 | 2 | 6 | 146 | |
| COG0094 | Total | 242 | 10 | 32 | 200 |
| Trimmed | 148 | 2 | 5 | 141 | |
| COG0096 | Total | 170 | 28 | 16 | 126 |
| Trimmed | 120 | 4 | 2 | 114 | |
| COG0097 | Total | 295 | 75 | 26 | 194 |
| Trimmed | 177 | 1 | 2 | 174 | |
| COG0098 | Total | 298 | 27 | 32 | 239 |
| Trimmed | 179 | 11 | 8 | 160 | |
| COG0099 | Total | 249 | 84 | 21 | 144 |
| Trimmed | 149 | 5 | 10 | 134 | |
| COG0100 | Total | 274 | 114 | 32 | 128 |
| Trimmed | 164 | 9 | 27 | 128 | |
| COG0102 | Total | 805 | 467 | 109 | 229 |
| Trimmed | 438 | 145 | 109 | 229 | |
| COG0103 | Total | 738 | 85 | 421 | 232 |
| Trimmed | 443 | 5 | 206 | 232 | |
| COG0172 | Total | 2065 | 1033 | 522 | 510 |
| Trimmed | 1239 | 207 | 522 | 510 | |
| COG0184 | Total | 540 | 330 | 82 | 128 |
| Trimmed | 324 | 129 | 67 | 128 | |
| COG0186 | Total | 320 | 122 | 45 | 153 |
| Trimmed | 192 | 21 | 18 | 153 | |
| COG0197 | Total | 234 | 31 | 8 | 195 |
| Trimmed | 140 | 5 | 3 | 132 | |
| COG0200 | Total | 666 | 161 | 287 | 218 |
| Trimmed | 400 | 6 | 176 | 218 | |
| COG0201 | Total | 572 | 71 | 69 | 432 |
| Trimmed | 374 | 15 | 12 | 347 | |
| COG0202 | Total | 1033 | 373 | 271 | 389 |
| Trimmed | 620 | 24 | 207 | 389 | |
| COG0256 | Total | 704 | 346 | 78 | 280 |
| Trimmed | 422 | 64 | 78 | 280 | |
| COG0495 | Total | 2307 | 521 | 629 | 1157 |
| Trimmed | 1384 | 27 | 226 | 1131 | |
| COG0522 | Total | 880 | 542 | 79 | 259 |
| Trimmed | 528 | 190 | 79 | 259 | |
| COG0525 | Total | 1611 | 311 | 396 | 904 |
| Trimmed | 967 | 67 | 76 | 824 | |
| COG0533 | Total | 1065 | 274 | 450 | 341 |
| Trimmed | 639 | 57 | 241 | 341 | |
By several approaches, our data (concatenated alignments) showed to be more reliable or informative than single gene phylogenies: A) the two statistical approaches, PTP test and g1 statistics used to achieve a measure of the overall signal content, indicated that the molecular data related to the 64 alignments have phylogenetic signal. B) The use of concatenated alignments offers more Parsimony-Informative Characters in comparison to the use of separated single genes.
On the strategy to build phylogenetic relationships our study was indeed based on UO originally described by Ciccarelli’s work.8 This choice was made because we believe that this approach is really nice and useful, especially when using genomes with partial or unfinished sequencing. While the strategy is pretty much the same, more data were used in our study. Ciccarelli’s tree8 present only six species of Protozoa (Dictyostelium discoideum, Cryptosporidium hominis, Plasmodium falciparum, Thalassiosira pseudonana, Leishmania major and Giardia lamblia) against 74 (complete and draft genomes) used in our study.
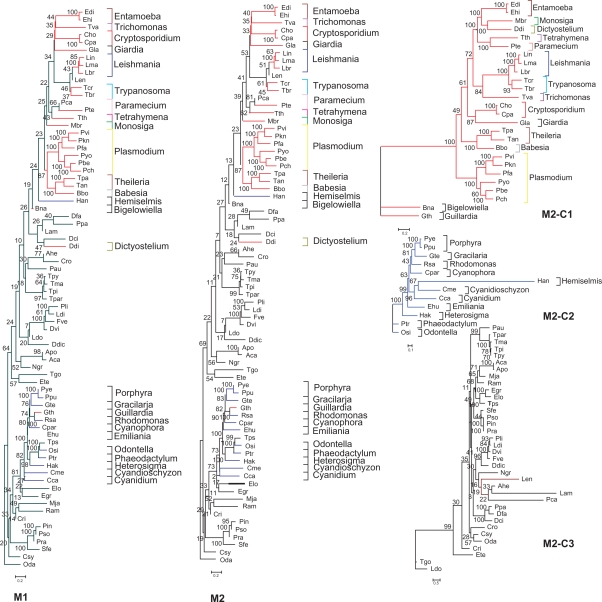
Figure 3 shows the comparison of the trees M1 and M2, using the total (M1) and trimmed (M2) (using TrimAl) concatenated alignments. Both methodologies showed very similar topologies; however, the tree obtained with trimmed alignments showed higher bootstrap values. Also, in Figure 3 (at the right side) are presented the three sub-trees belonging to each of the three clades of the original M2 tree: M2-C1 (Clade 1 of the M2 tree), M2-C2 (Clade 2 of the M2 tree) and M2-C3 (Clade 3 of the M2 tree). Each presented higher bootstrap values when compared to the entire M2 tree. The trees signalized by the KH test as being “the best” (for scores see Supplementary File S3) and the trees M1 and M2 (and their sub-trees M1-C1, M2-C1, M1-C2, M2-C2, M1-C3 and M2-C3) obtained with Phyml 2.4.4 (for loglk see Table 3) presented the same topologies and they are shown in the Supplementary Figure S1 Group A-D, respectively.
Figure 3.
Phylogenomic supermatrix trees of protozoan species using the total and trimmed (using TrimAl) alignments. Supermatrices of 21,260 (M1) and 12,807 (M2) positions were used respectively in 74 protozoan species. Maximum likelihood tree was constructed with Phyml 2.4.4, JTT as evolutionary model and bootstrap 100. The resulting clades of M2: M2-C1 in red, M2-C2 in blue and M2-C3 in black with the evolutionary models obtained with Modelgenerator 0.85 were used to construct three individual trees: M2-Ct1 (Blosum62), M2-Ct2 (RtREV) and M2-Ct3 (WAG).
Table 3.
Kishino-Hasegawa test results.
| Number | Tree | KH test | Phyml likelihood: loglk |
|---|---|---|---|
| 1 | Complete total tree | – | −418,592,428,692 |
| 2 | Complete total tree (KH test) | Best tree of the 100 bootstraps of C1 total: bootstrap 36 | – |
| 3 | Complete trimmed tree | – | −473,396,448,710 |
| 4 | Complete trimmed tree (KH test) | Best tree of the 100 bootstraps of C1 trimmed: bootstrap 91 | – |
| 5 | C1 total | – | −36,241,173,106 |
| 6 | C1 total (KH test) | Best tree of the 100 bootstraps of C1 total: bootstrap 39 | – |
| 7 | C1 trimmed | – | −313,069,182,861 |
| 8 | C1 trimmed (KH test) | Best tree of the 100 bootstraps of C1 trimmed: bootstrap 83 | – |
| 9 | C2 total | – | −5,837,927,846 |
| 10 | C2 total (KH test) | Best tree of the 100 bootstraps of C2 total: bootstrap 20 | – |
| 11 | C2 trimmed | – | −5,457,055,926 |
| 12 | C2 trimmed (KH test) | Best tree of the 100 bootstraps of C2 trimmed: bootstrap 16 | – |
| 13 | C3 total | – | −6,001,226,356 |
| 14 | C3 total (KH test) | Best tree of the 100 bootstraps of C3 total: bootstrap 79 | – |
| 15 | C3 trimmed | – | −5,603,518,000 |
| 16 | C3 trimmed (KH test) | Best tree of the 100 bootstraps of C3 trimmed: bootstrap 61 | – |
The bootstrap values of the M2 tree (containing the three clades: C1, C2 and C3) are weak, and it is probably because not all taxons had the sequences of the 31 UO available at public databases. We tested the possibility to obtain reliable species trees using less than 31 UO, then showed that using at least 80% (25/31) for each taxon provide reliable inferences (most bootstraps equal or higher than 80). The C1, C2, and C3 clades of the M2 tree were re-analysed as separated trees (M2-C1, M2-C2 and M2-C3) (Fig. 3), then better bootstrap values were obtained. The M2-C1 tree showed bootstrap values above 80, except for the values 49, 60, 61, and 72; and the M2-C2 tree also showed bootstrap values above 80, except for the values 43, 63, and 67. Unfortunately bootstrap values could not be enhanced even better either using new/better alignment or more taxa. This weak bootstrap values behaviors has also been noted in Ciccarelli (2006);8 Dutilh (2007)11 and Hartmann (2008)71 using similar data and/or methodology.
The species trees—M1 and M2—presented three major clades. Each clade was related to one of the following groups of data: i) 26 species presenting at least 80% of UO (25/31) in their concatenated alignments called C1, ii) 12 species presenting between 50%–79% (15–24/31) of UO called C2, and iii) 36 species presenting less than 50% (1–14/31) of UO called C3.
C1 showed excavates represented by kinetoplastids, trichomonads and diplomonads. Kinetoplastids, a group of uncertain affinity,45,72,73 was characterized by the presence of a monophyly formed by the paraphyletic groups Leishmania and Trypanosoma. L. major was found to be more closely related to L. infantum than L. brasiliensis. T. brucei and T. cruzi were the representatives of Trypanosoma. Previous phylogenetic analyses36,47 confirmed a closer relationship between diplomonads (Giardia) and trichomonads (Trichomonas), which was confirmed by our results. Diplomonads were closely related to Monosiga, Entamoeba, Apicomplexa alveolates: Cryptosporidium and ciliates: Tetrahymena and Paramecium. Other Apicomplexa as Plasmodium, Theileria and Babesia were also closely related. Toxoplasma was wrongly placed in C3, separated from other Apicomplexa alveolates, which probably occurred because of insufficient phylogenetic information (less than 50% of UO) in the concatenated alignments.
Ciccarelli et al (2006)8 showed that despite a highly resolved and robust tree, they could not exclude a few uncertainties in tree topology due to biased species sampling or Long Branch Attraction (LBA). This LBA was suggested to account for the placement of Diplomonadida (G. lamblia) as the most basal eukaryal taxon and as the most external taxon of Protozoa, followed by the Kinetoplastida (L. major), placing both of them as related to Chromoalveolatas (T. pseudonana, P. falciparum and C. hominis). Our results (Fig. 3, M2-C1) showed different/additional relationships, with the most relevant being that G. lamblia is more closely related to Cryptosporidium than L. major, and that the Giardia-Cryptosporidium clade is closely related to the Trypanosoma-Leishmania-Trichomonas clade. Also, since our M2-C1 tree is more reliable than M2-C2 and M2-C3, our results differ from Ciccarelli’s8 showing that the most basal protozoans could be the genus Bigelowiella and Guillardia forming together a clade. The use of more data from Protozoa in our study (than the original data used by Ciccarelli’s) gave us the opportunity to infer additional and more complete relationships than initially described by those authors. However, a highly resolved protozoan species tree is still a challenge, that will be better addressed by the inclusion of more UO from taxons forming M2-C2 and M2-C3 trees (Fig. 3), and also more genomic data from additional species, especially from basal genus as Bigelowiella and Guillardia.
Animals and their unicellular relatives (together termed ‘Holozoa’) show a strong affinity with Fungi (together termed ‘opisthokonts’) providing strong evidence in a relationship with Protozoa (eg, Monosiga). Our tree showed this relationship: M. brevicollis was found closely related to C1. The opisthokont lineage is supported by insertions in the elongation factor-1a and enolase,74 as well as by many individual2,74 and concatenated gene phylogenies.47,75,76 On the other hand, amoebozoa are supported by a group of several individual and concatenated gene phylogenies,76 partially by the presence of fused genes encoding cytochrome oxidase 1 and 2 in the mitochondrial DNA of slime molds and lobose amoebae.77 ‘Unikont’ is used as the name for the union of two individually well supported groups: amoebozoans and opisthokonts.24 Overall, unikonts include animals and fungi, some amoebae (eg, Entamoeba), slime molds (eg, Dictyostelium), and a few parasitic protists.
The analysis of multiple nuclear genes strongly supports the sharing of a common ancestry of choanoflagellates represented by Monosiga with animals with the exclusion of fungi and the other sampled eukaryotes;47 this is in agreement with single-gene studies2,78,79 and with a mitochondrial multi-gene phylogeny.80
In our phylogenomic trees, C2 was formed by four groups: i) rhodophyta: Porphyra and Gracilaria; ii) cryptophyta: Guillardia and Rhodomonas; glaucocystophyceae: Cyanophora, and haptophyceae: Emiliania (The cryptophyta Hemiselmis was wrongly placed closer to the apicomplexa Theileria and Babesia); iii) stramenopiles: Odontella, Phaeodactylum and Heterosigma; iv) rhodophyta: Cyanidioschyzon and Cyanidium.
C3 was formed by the protozoan euglenozoa/kinetoplastida: L. amazonensis, L. donovani, L. enriettii; the alveolata/apicomplexa: T. gondii and E. tenella; the amoebozoa: A. castellanii, A. polyphaga, A. healyi, D. citrinum, D. fasciculatum and Polysphondylium pallidum; the alveolata/ciliophora: P. aurelia, P. caudatum, T. malaccensis, T. pigmentosa, T. pyriformis and T. paravorax and the euglenozoa/euglenida: E. gracilis, E. longa. Also, other eukaryotes were found in this clade: the stramenopiles: Cafeteria roenbergensis, Chrysodidymus synuroideus, Desmarestia viridis, Dictyota dichotoma, Fucus vesiculosus, Laminaria digitata, Ochromonas danica, Phytophthora infestans, P. sojae, P. ramorum, Pylaiella littoralis, Saprolegnia ferax and T. pseudonana; the rhodophyta: Chondrus crispus, the malawimonadidae: Malawimonas jakobiformis, the heterolobosea: Naegleria gruberi and the jakobidae Reclinomonas americana.
Our phylogenomic analysis showed, as expected, the phylogenetic relationships and the monophyly of Protozoa (Fig. 3) that is in good agreement with previous studies.36,46,47,75,76,80 Unfortunately, because of the use of several incomplete genomes in this study, C3 phylogeny is not reliable, mainly because it is formed by a group of species containing less than 50% (1–14/31) of the 31 UO found. This could be also interpreted as the concatenated alignment having too many gaps (in this case a gap would be a missing UO or whole protein, not only a missing amino acid or nucleotide sequence), thus contributing to the low robustness of the clade or tree. We hypothesize that each missing UO can be treated as a gap in the concatenated alignment, then the more gaps (or less UO) the less reliable trees. It appears logical to expect an inverse relationship between the proportion of gapped sites in an alignment and the accuracy of the inferred phylogeny,71,81,82 particularly if the gaps are not treated as reflective of distinct evolutionary events,83 and thus, containing distinct phylogenetic signal.
The supertree approach did not work in our hands because when Clann 3.1.3 was used for the supertree reconstruction (trees’ concatenation), only a Neighbor-Joining tree (the initial step to produce a supertree) was obtained. The next two steps (heuristic search or the bootstrapping) were not executed because the software showed an error message informing that the individual trees used as inputs could not be concatenated into a one supertree. Nevertheless, Supplementary Figure S2 shows the Neighbor-Joining supertree for the individual total alignments (M3) and for the individual trimmed alignments (M4) and what could be appreciated is the fact that they presented similar topologies when compared to the supermatrix tree (M1 and M2) in Figure 3.
While most of the publicly available eukaryote genome sequence data were obtained with Sanger technology (medium to low coverage), second and third generation sequencing technologies will be probably used to sequence a larger number of species but at low coverage; hence the approach to use partial sequences from several genes (especially UO) appears to be a good option for future phylogenomics-based studies.
Conclusions
We have presented a phylogenomics-based overview for Protozoa. Relationships between protozoan groups are in agreement with previous studies, supporting monophyly. On the other hand, phylogenetic information inferred from C3 is not reliable due to incomplete information (missing UO in those genomes), suggesting that the use of less than 15 UO for phylogenomic reconstruction is not reliable. The inclusion of more data (UO) is necessary to obtain a robust tree in C3. Our phylogenomics-based methodology using a supermatrix approach proved to be reliable with protozoan genome data, suggesting that (a) the more UO used the better, and (b) that the use of the entire UO sequence or just a conserved block of it produce similar reliable results. The highest bootstrap values were obtained when the trees of the clades C1, C2 and C3 were constructed separately. The results of the supertree were obtained only for the Neighbor-Joining tree and were not conclusive. Finally, we need to further investigate if this methodology could be extrapolated or reproduced to other taxonomic groups.
Supplementary Data
Figure S1 Group A. Comparison between the complete trees (total and trimmed) signalized as “the best” by the KH test and the trees M1 and M2 obtained with Phyml. The trees are: i) M1-Complete total tree (M1-Phyml), ii) Complete total tree (KH test), iii) M2-Complete trimmed tree (M2-Phyml), iv) Complete trimmed tree (KH test). All trees were constructed with Phyml 2.4.4.
Figure S1 Group B. Comparison between the C1 trees (total and trimmed) signalized as “the best” by the KH test and the trees M1 and M2 obtained with Phyml. The trees are: i) M1-C1 total (M1-Phyml), ii) C1 total (KH test), iii) M2-C1 trimmed (M2-Phyml), iv) C1 trimmed (KH test). All trees were constructed with Phyml 2.4.4.
Figure S1 Group C. Comparison between the C2 trees (total and trimmed) signalized as “the best” by the KH test and the trees M1 and M2 obtained with Phyml. The trees are: i) M1-C2 total (M1-Phyml), ii) C2 total (KH test), iii) M2-C2 trimmed (M2-Phyml), iv) C2 trimmed (KH test). All trees were constructed with Phyml 2.4.4.
Figure S1 Group D. Comparison between the C3 trees (total and trimmed) signalized as “the best” by the KH test and the trees M1 and M2 obtained with Phyml. The trees are: i) M1-C2 total (M1-Phyml), ii) C3 total (KH test), iii) M2-C3 trimmed (M2-Phyml), iv) C2 trimmed (KH test). All trees were constructed with Phyml 2.4.4.
Figure S2. Neighbor-Joining supertree of protozoan species built with Clann 3.1.3 using the total and trimmed (using TrimAl) alignments.
Table S1. Universal orthologs mapped to KOGs found in protozoan.
Table S2. RefSeq accession numbers of the protozoan sequences mapped from COGs and used in the phylogeny (for the sequences see Supplementary File S1).
Table S3. List of the species analyzed in the study. (C1 is red, C2 blue and C3 black).
Table S4. Amino acid evolutionary models obtained with Modelgenerator 0.85 of the individual and concatenated alignments of UO and the clades C1, C2 and C3.
File S1. Fasta sequences used in the phylogeny divided by the UO clusters.
File S2. Fasta sequences and alignments used as input to construct the supermatrices (total and trimmed) and the supertrees (total and trimmed).
File S3. Scores obtained with the Kishino-Hasegawa test over the eight bootstrapped trees: M1, M2, C1-M1, C2-M1, C3-M1, C1-M2, C2-M2 and C3-M2.
Acknowledgments
We thank the following phylogenomic and bioinformatics experts for the constructive comments: Dr. Tobias Doerks (EMBL-Heidelberg), for mapping those universal orthologs into NCBI KOGs; Dr. Kimmen ölander and Dr. Bryan Kolaczkowski, for preliminary insights on phylogenomics of Protozoa using Phylofacts; Dr. João Setubal and Dr. Carlos G. Schrago for critical reading and suggestions about phylogenomics and phylogenetic analysis.
Abbreviations
- UO
Universal Orthologs
- HMM
Hidden Markov Models
- KH test
Kishino and Hasegawa test
Footnotes
Disclosure
This manuscript has been read and approved by all authors. This paper is unique and it is not under consideration by any other publication and has not been published elsewhere. The authors and peer reviewers report no conflict of interest. The authors confirm that they have permission to reproduce any copyrighted material.
References
- 1.Brochier C, Philippe H. Phylogeny: a non-hyperthermophilic ancestor for bacteria. Nature. 2002;417(6886):244. doi: 10.1038/417244a. [DOI] [PubMed] [Google Scholar]
- 2.Wainright PO, Hinkle G, Sogin ML, et al. Monophyletic origins of the metazoa: an evolutionary link with fungi. Science. 1993;260(5106):340–2. doi: 10.1126/science.8469985. [DOI] [PubMed] [Google Scholar]
- 3.Aguinaldo AM, Turbeville JM, Linford LS, et al. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature. 1997;387(6632):489–93. doi: 10.1038/387489a0. [DOI] [PubMed] [Google Scholar]
- 4.Mallatt J, Winchell CJ. Testing the new animal phylogeny: first use of combined large-subunit and small-subunit rRNA gene sequences to classify the protostomes. Mol Biol Evol. 2002;19(3):289–301. doi: 10.1093/oxfordjournals.molbev.a004082. [DOI] [PubMed] [Google Scholar]
- 5.Heraty JM, Woolley JB, Hopper KR, et al. Molecular phylogenetics and reproductive incompatibility in a complex of cryptic species of aphid parasitoids. Mol Phylogenet Evol. 2007;45(2):480–93. doi: 10.1016/j.ympev.2007.06.021. [DOI] [PubMed] [Google Scholar]
- 6.Sainz AC, Mauro LV, Moriyama EN, et al. Phylogeny of triatomine vectors of Trypanosoma cruzi suggested by mitochondrial DNA sequences. Genetica. 2004;121(3):229–40. doi: 10.1023/b:gene.0000039842.82574.02. [DOI] [PubMed] [Google Scholar]
- 7.Wu M, Eisen JA. A simple, fast, and accurate method of phylogenomic inference. Genome Biol. 2008;9(10):R151. doi: 10.1186/gb-2008-9-10-r151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ciccarelli FD, Doerks T, von Mering C, et al. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311(5765):1283–7. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- 9.Creevey CJ, McInerney JO. Clann: investigating phylogenetic information through supertree analyses. Bioinformatics. 2005;21(3):390–2. doi: 10.1093/bioinformatics/bti020. [DOI] [PubMed] [Google Scholar]
- 10.Huerta-Cepas J, Dopazo H, Dopazo J, et al. The human phylome. Genome Biol. 2007;8(6):R109. doi: 10.1186/gb-2007-8-6-r109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dutilh BE, van Noort V, van der Heijden RT, et al. Assessment of phylogenomic and orthology approaches for phylogenetic inference. Bioinformatics. 2007;23(7):815–24. doi: 10.1093/bioinformatics/btm015. [DOI] [PubMed] [Google Scholar]
- 12.Daubin V, Gouy M, Perriere G. A phylogenomic approach to bacterial phylogeny: evidence of a core of genes sharing a common history. Genome Res. 2002;12(7):1080–90. doi: 10.1101/gr.187002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dagan T, Martin W. The tree of one percent. Genome Biol. 2006;7(10):118. doi: 10.1186/gb-2006-7-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teichmann SA, Mitchison G. Is there a phylogenetic signal in prokaryote proteins? J Mol Evol. 1999;49(1):98–107. doi: 10.1007/pl00006538. [DOI] [PubMed] [Google Scholar]
- 15.Doolittle WF. Phylogenetic classification and the universal tree. Science. 1999;284(5423):2124–9. doi: 10.1126/science.284.5423.2124. [DOI] [PubMed] [Google Scholar]
- 16.Ge F, Wang LS, Kim J. The cobweb of life revealed by genome-scale estimates of horizontal gene transfer. PLoS Biol. 2005;3(10):e316. doi: 10.1371/journal.pbio.0030316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cavalier-Smith T. Protist phylogeny and the high-level classification of Protozoa. European Journal of Protistology. 2003;39(4):338–48. [Google Scholar]
- 18.Cox FE. Systematics of the parasitic Protozoa. Trends Parasitol. 2002;18(3):108. doi: 10.1016/s1471-4922(01)02196-1. [DOI] [PubMed] [Google Scholar]
- 19.Cavalier-Smith T. Kingdom protozoa and its 18 phyla. Microbiol Rev. 1993;57(4):953–94. doi: 10.1128/mr.57.4.953-994.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stensvold CR, Lebbad M, Verweij JJ. The impact of genetic diversity in protozoa on molecular diagnostics. Trends Parasitol. doi: 10.1016/j.pt.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 21.Klokouzas A, Shahi S, Hladky SB, et al. ABC transporters and drug resistance in parasitic protozoa. Int J Antimicrob Agents. 2003;22(3):301–17. doi: 10.1016/s0924-8579(03)00210-3. [DOI] [PubMed] [Google Scholar]
- 22.Kutuzov MA, Andreeva AV. Protein Ser/Thr phosphatases of parasitic protozoa. Mol Biochem Parasitol. 2008;161(2):81–90. doi: 10.1016/j.molbiopara.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 23.O’Neil RH, Lilien RH, Donald BR, Stroud RM, et al. Phylogenetic classification of protozoa based on the structure of the linker domain in the bifunctional enzyme, dihydrofolate reductase-thymidylate synthase. J Biol Chem. 2003;278(52):52980–7. doi: 10.1074/jbc.M310328200. [DOI] [PubMed] [Google Scholar]
- 24.Cavalier-Smith T. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol. 2002;52(Pt 2):297–354. doi: 10.1099/00207713-52-2-297. [DOI] [PubMed] [Google Scholar]
- 25.Stechmann A, Cavalier-Smith T. Rooting the eukaryote tree by using a derived gene fusion. Science. 2002;297(5578):89–91. doi: 10.1126/science.1071196. [DOI] [PubMed] [Google Scholar]
- 26.Cavalier-Smith T. Kingdoms Protozoa and Chromista and the eozoan root of the eukaryotic tree. Biol Lett. 6(3):342–5. doi: 10.1098/rsbl.2009.0948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sullivan WJ, Jr, Naguleswaran A, Angel SO. Histones and histone modifications in protozoan parasites. Cell Microbiol. 2006;8(12):1850–61. doi: 10.1111/j.1462-5822.2006.00818.x. [DOI] [PubMed] [Google Scholar]
- 28.El-Sayed NM, Myler PJ, Blandin G, Berriman M, et al. Comparative genomics of trypanosomatid parasitic protozoa. Science. 2005;309(5733):404–9. doi: 10.1126/science.1112181. [DOI] [PubMed] [Google Scholar]
- 29.Corliss JO. What are the taxonomic and evolutionary relationships of the Protozoa to the Protista? Biosystems. 1981;14(3–4):445–9. doi: 10.1016/0303-2647(81)90049-6. [DOI] [PubMed] [Google Scholar]
- 30.Blackwell WH, Powell MJ. The Protozoa, a kingdom by default? The American Biology Teacher. 2001;63:483–90. [Google Scholar]
- 31.Vickerman K. The diversity of the kinetoplastid flagellates. In: Lumsden WHED, editor. Biology of the Kinetoplastida. Vol. 1. London: Academic Press Inc; 1976. [Google Scholar]
- 32.Van de Peer Y, De Wachter R. Evolutionary relationships among the eukaryotic crown taxa taking into account site-to-site rate variation in 18S rRNA. J Mol Evol. 1997;45(6):619–30. doi: 10.1007/pl00006266. [DOI] [PubMed] [Google Scholar]
- 33.Cavalier-Smith T. Principles of protein and lipid targeting in secondary symbiogenesis: euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J Eukaryot Microbiol. 1999;46(4):347–66. doi: 10.1111/j.1550-7408.1999.tb04614.x. [DOI] [PubMed] [Google Scholar]
- 34.Blanchard JL, Hicks JS. The non-photosynthetic plastid in malarial parasites and other apicomplexans is derived from outside the green plastid lineage. J Eukaryot Microbiol. 1999;46(4):367–75. doi: 10.1111/j.1550-7408.1999.tb04615.x. [DOI] [PubMed] [Google Scholar]
- 35.Nakayama T, Marin B, Kranz HD, et al. The Basal Position of Scaly Green Flagellates among the Green Algae (Chlorophyta) is Revealed by Analyses of Nuclear-Encoded SSU rRNA Sequences. Protist. 1998;149(4):367–80. doi: 10.1016/S1434-4610(98)70043-4. [DOI] [PubMed] [Google Scholar]
- 36.Keeling PJ, Burger G, Durnford DG, et al. The tree of eukaryotes. Trends Ecol Evol. 2005;20(12):670–6. doi: 10.1016/j.tree.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 37.Stechmann A, Cavalier-Smith T. Phylogenetic analysis of eukaryotes using heat-shock protein Hsp90. J Mol Evol. 2003;57(4):408–19. doi: 10.1007/s00239-003-2490-x. [DOI] [PubMed] [Google Scholar]
- 38.Arango E, Carmona-Fonseca J, Blair S. In vitro susceptibility of Colombian Plasmodium falciparum isolates to different antimalarial drugs. Biomedica. 2008;28(2):213–23. [PubMed] [Google Scholar]
- 39.Arevalo J, Ramirez L, Adaui V, et al. Influence of Leishmania (Viannia) species on the response to antimonial treatment in patients with American tegumentary leishmaniasis. J Infect Dis. 2007;195(12):1846–51. doi: 10.1086/518041. [DOI] [PubMed] [Google Scholar]
- 40.Burri C, Keiser J. Pharmacokinetic investigations in patients from northern Angola refractory to melarsoprol treatment. Trop Med Int Health. 2001;6(5):412–20. doi: 10.1046/j.1365-3156.2001.00725.x. [DOI] [PubMed] [Google Scholar]
- 41.Bansal D, Sehgal R, Chawla Y, Mahajan RC, Malla N. In vitro activity of antiamoebic drugs against clinical isolates of Entamoeba histolytica and Entamoeba dispar. Ann Clin Microbiol Antimicrob. 2004;3:27. doi: 10.1186/1476-0711-3-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sobel JD, Nagappan V, Nyirjesy P. Metronidazole-resistant vaginal trichomoniasis—an emerging problem. N Engl J Med. 1999;341(4):292–3. doi: 10.1056/NEJM199907223410417. [DOI] [PubMed] [Google Scholar]
- 43.Sogin ML. Early evolution and the origin of eukaryotes. Curr Opin Genet Dev. 1991;1(4):457–63. doi: 10.1016/s0959-437x(05)80192-3. [DOI] [PubMed] [Google Scholar]
- 44.Keeling PJ, Doolittle WF. A non-canonical genetic code in an early diverging eukaryotic lineage. Embo J. 1996;15(9):2285–90. [PMC free article] [PubMed] [Google Scholar]
- 45.Simpson AG, MacQuarrie EK, Roger AJ. Eukaryotic evolution: early origin of canonical introns. Nature. 2002;419(6904):270. doi: 10.1038/419270a. [DOI] [PubMed] [Google Scholar]
- 46.Chaudhary K, Roos DS. Protozoan genomics for drug discovery. Nat Biotechnol. 2005;23(9):1089–91. doi: 10.1038/nbt0905-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Philippe H, Snell EA, Bapteste E, Lopez P, Holland PW, Casane D. Phylogenomics of eukaryotes: impact of missing data on large alignments. Mol Biol Evol. 2004;21(9):1740–52. doi: 10.1093/molbev/msh182. [DOI] [PubMed] [Google Scholar]
- 48.Stechmann A, Cavalier-Smith T. The root of the eukaryote tree pinpointed. Curr Biol. 2003;13(17):R665–6. doi: 10.1016/s0960-9822(03)00602-x. [DOI] [PubMed] [Google Scholar]
- 49.Fast NM, Xue L, Bingham S, Keeling PJ. Re-examining alveolate evolution using multiple protein molecular phylogenies. J Eukaryot Microbiol. 2002;49(1):30–7. doi: 10.1111/j.1550-7408.2002.tb00336.x. [DOI] [PubMed] [Google Scholar]
- 50.Harper JT, Waanders E, Keeling PJ. On the monophyly of chromalveolates using a six-protein phylogeny of eukaryotes. Int J Syst Evol Microbiol. 2005;55(Pt 1):487–96. doi: 10.1099/ijs.0.63216-0. [DOI] [PubMed] [Google Scholar]
- 51.Simpson AG. Cytoskeletal organization, phylogenetic affinities and systematics in the contentious taxon Excavata (Eukaryota) Int J Syst Evol Microbiol. 2003;53(Pt 6):1759–77. doi: 10.1099/ijs.0.02578-0. [DOI] [PubMed] [Google Scholar]
- 52.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30(14):3059–66. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Katoh K, Kuma K, Miyata T, Toh H. Improvement in the accuracy of multiple sequence alignment program MAFFT. Genome Inform. 2005;16(1):22–33. [PubMed] [Google Scholar]
- 54.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9(4):286–98. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 55.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14(9):755–63. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 56.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009 doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marcet-Houben M, Marceddu G, Gabaldon T. Phylogenomics of the oxidative phosphorylation in fungi reveals extensive gene duplication followed by functional divergence. BMC Evol Biol. 2009;9:295. doi: 10.1186/1471-2148-9-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML Online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res. 2005;33:W557–9. doi: 10.1093/nar/gki352. (Web Server issue): [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52(5):696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 60.Keane TM, Creevey CJ, Pentony MM, Naughton TJ, McLnerney JO. Assessment of methods for amino acid matrix selection and their use on empirical data shows that ad hoc assumptions for choice of matrix are not justified. BMC Evol Biol. 2006;6:29. doi: 10.1186/1471-2148-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Faith DP, Cranston PS. Could a cladograma this short have arisen by chance alone? On permutation tests for cladistic structure. Cladistics. 1991;7:1–28. [Google Scholar]
- 62.Swofford DL. In: PAUP* Phylogenetic Analysis Using Parsimony (*and Other Methods) Version 4. S MA, editor. Sinauer Associates; 2002. [Google Scholar]
- 63.Fu J, Murphy RW. Discriminating and locating character covariance: an application of permutation tail probability (PTP) analyses. Syst Biol. 1999;48(2):380–95. doi: 10.1080/106351599260355. [DOI] [PubMed] [Google Scholar]
- 64.Fitzpatrick DA, Logue ME, Stajich JE, Butler G. A fungal phylogeny based on 42 complete genomes derived from supertree and combined gene analysis. BMC Evol Biol. 2006;6:99. doi: 10.1186/1471-2148-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hillis DM, Huelsenbeck JP. Signal, noise, and reliability in molecular phylogenetic analyses. J Hered. 1992;83(3):189–95. doi: 10.1093/oxfordjournals.jhered.a111190. [DOI] [PubMed] [Google Scholar]
- 66.Kishino H, Hasegawa M. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J Mol Evol. 1989;29(2):170–9. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 67.Swofford DLOG, Waddell PJ, Hillis DM. Phylogenetic inference. In: Hillis DMMC, Mable BK, editors. Molecular Systematics. 2nd ed. Sunderland, Massachusetts: Sinauer Associates; 1996. pp. 407–514. [Google Scholar]
- 68.Smith ND. Phylogenetic analysis of pelecaniformes (aves) based on osteological data: implications for waterbird phylogeny and fossil calibration studies. PLoS One. 5(10):e13354. doi: 10.1371/journal.pone.0013354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Felsenstein J. Inferring phylogenies. Sinauer; Sunderland, MA, USA: 2003. [Google Scholar]
- 70.Wilgenbusch JC, Swofford D. Inferring evolutionary trees with PAUP*. Curr Protoc Bioinformatics. 2003 doi: 10.1002/0471250953.bi0604s00. Chapter 6:Unit 6 4. [DOI] [PubMed] [Google Scholar]
- 71.Philippe H, Lopez P, Brinkmann H, et al. Early-branching or fast-evolving eukaryotes? An answer based on slowly evolving positions. Proc Biol Sci. 2000;267(1449):1213–21. doi: 10.1098/rspb.2000.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Philippe H, Germot A, Moreira D. The new phylogeny of eukaryotes. Curr Opin Genet Dev. 2000;10(6):596–601. doi: 10.1016/s0959-437x(00)00137-4. [DOI] [PubMed] [Google Scholar]
- 73.Baldauf SL, Palmer JD. Animals and fungi are each other’s closest relatives: congruent evidence from multiple proteins. Proc Natl Acad Sci U S A. 1993;90(24):11558–62. doi: 10.1073/pnas.90.24.11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Baldauf SL, Roger AJ, Wenk-Siefert I, Doolittle WF. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290(5493):972–7. doi: 10.1126/science.290.5493.972. [DOI] [PubMed] [Google Scholar]
- 75.Bapteste E, Brinkmann H, Lee JA, et al. The analysis of 100 genes supports the grouping of three highly divergent amoebae: Dictyostelium, Entamoeba, and Mastigamoeba. Proc Natl Acad Sci U S A. 2002;99(3):1414–9. doi: 10.1073/pnas.032662799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gray MW, Lang BF, Burger G. Mitochondria of protists. Annu Rev Genet. 2004;38:477–524. doi: 10.1146/annurev.genet.37.110801.142526. [DOI] [PubMed] [Google Scholar]
- 77.King N, Carroll SB. A receptor tyrosine kinase from choanoflagellates: molecular insights into early animal evolution. Proc Natl Acad Sci U S A. 2001;98(26):15032–7. doi: 10.1073/pnas.261477698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Snell EA, Furlong RF, Holland PW. Hsp70 sequences indicate that choanoflagellates are closely related to animals. Curr Biol. 2001;11(12):967–70. doi: 10.1016/s0960-9822(01)00275-5. [DOI] [PubMed] [Google Scholar]
- 79.Lang BF, O’Kelly C, Nerad T, Gray MW, Burger G. The closest unicellular relatives of animals. Curr Biol. 2002;12(20):1773–8. doi: 10.1016/s0960-9822(02)01187-9. [DOI] [PubMed] [Google Scholar]
- 80.Dwivedi B, Gadagkar SR. Phylogenetic inference under varying proportions of indel-induced alignment gaps. BMC Evol Biol. 2009;9:211. doi: 10.1186/1471-2148-9-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hartmann S, Vision TJ. Using ESTs for phylogenomics: can one accurately infer a phylogenetic tree from a gappy alignment? BMC Evol Biol. 2008;8:95. doi: 10.1186/1471-2148-8-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wiens JJ. Missing data and the design of phylogenetic analyses. J Biomed Inform. 2006;39(1):34–42. doi: 10.1016/j.jbi.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 83.Rivas E. Evolutionary models for insertions and deletions in a probabilistic modeling framework. BMC Bioinformatics. 2005;6:63. doi: 10.1186/1471-2105-6-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Group A. Comparison between the complete trees (total and trimmed) signalized as “the best” by the KH test and the trees M1 and M2 obtained with Phyml. The trees are: i) M1-Complete total tree (M1-Phyml), ii) Complete total tree (KH test), iii) M2-Complete trimmed tree (M2-Phyml), iv) Complete trimmed tree (KH test). All trees were constructed with Phyml 2.4.4.
Figure S1 Group B. Comparison between the C1 trees (total and trimmed) signalized as “the best” by the KH test and the trees M1 and M2 obtained with Phyml. The trees are: i) M1-C1 total (M1-Phyml), ii) C1 total (KH test), iii) M2-C1 trimmed (M2-Phyml), iv) C1 trimmed (KH test). All trees were constructed with Phyml 2.4.4.
Figure S1 Group C. Comparison between the C2 trees (total and trimmed) signalized as “the best” by the KH test and the trees M1 and M2 obtained with Phyml. The trees are: i) M1-C2 total (M1-Phyml), ii) C2 total (KH test), iii) M2-C2 trimmed (M2-Phyml), iv) C2 trimmed (KH test). All trees were constructed with Phyml 2.4.4.
Figure S1 Group D. Comparison between the C3 trees (total and trimmed) signalized as “the best” by the KH test and the trees M1 and M2 obtained with Phyml. The trees are: i) M1-C2 total (M1-Phyml), ii) C3 total (KH test), iii) M2-C3 trimmed (M2-Phyml), iv) C2 trimmed (KH test). All trees were constructed with Phyml 2.4.4.
Figure S2. Neighbor-Joining supertree of protozoan species built with Clann 3.1.3 using the total and trimmed (using TrimAl) alignments.
Table S1. Universal orthologs mapped to KOGs found in protozoan.
Table S2. RefSeq accession numbers of the protozoan sequences mapped from COGs and used in the phylogeny (for the sequences see Supplementary File S1).
Table S3. List of the species analyzed in the study. (C1 is red, C2 blue and C3 black).
Table S4. Amino acid evolutionary models obtained with Modelgenerator 0.85 of the individual and concatenated alignments of UO and the clades C1, C2 and C3.
File S1. Fasta sequences used in the phylogeny divided by the UO clusters.
File S2. Fasta sequences and alignments used as input to construct the supermatrices (total and trimmed) and the supertrees (total and trimmed).
File S3. Scores obtained with the Kishino-Hasegawa test over the eight bootstrapped trees: M1, M2, C1-M1, C2-M1, C3-M1, C1-M2, C2-M2 and C3-M2.