Abstract
Genome-wide association studies (GWAS) have successfully identified genetic variants associated with risk for breast cancer. However, the molecular mechanisms through which the identified variants confer risk or influence phenotypic expression remains poorly understood. Here, we present a novel integrative genomics approach that combines GWAS information with gene expression data to assess the combined contribution of multiple genetic variants acting within genes and putative biological pathways, and to identify novel genes and biological pathways that could not be identified using traditional GWAS. The results show that genes containing SNPs associated with risk for breast cancer are functionally related and interact with each other in biological pathways relevant to breast cancer. Additionally, we identified novel genes that are co-expressed and interact with genes containing SNPs associated with breast cancer. Integrative analysis combining GWAS information with gene expression data provides functional bridges between GWAS findings and biological pathways involved in breast cancer.
Keywords: genome-wide association studies gene expression pathway
Introduction
Breast cancer is one of the leading causes of death among women in the United States and around the world.1 In 2009, an estimated 192,370 new cases of invasive breast cancer were diagnosed among women, as well as an estimated 62,280 additional cases of carcinoma in situ.1 At the same time, an estimated 40,170 women died from breast cancer.1 While considerable progress has been made in reducing mortality rates due to increased screening, digital mammography, specialized care, and the widespread use of therapeutic agents such as selective estrogen receptor modulators, aromatase inhibitors, trastuzumab and others, identifying genetic markers remains an important long-term goal for the development of more effective therapeutic strategies and early interventions. Over the last decade, considerable effort and financial resources have been directed at identifying molecular signatures for breast cancer using gene expression profiles.2,3 At least two of these signatures have proven useful for prognostic purposes in the clinic.4,5 However, although these primary analyses have made great strides in deciphering the molecular basis of breast cancer, they have been unsuccessful in determining which genes have causative roles as opposed to being consequences of the breast cancer state.
Recent advances in genotyping and reduction in genotyping costs have made possible the use of genome-wide association studies to identify single nucleotide polymorphisms (SNPs) associated with risk for breast cancer.6–11 Results from these studies are providing valuable information about the genetic susceptibility architecture of breast cancer. However, to date, data generated from GWAS have not been combined with gene expression data to identify biologically relevant associations beyond the ones that meet the stringent genome-wide significance threshold.12 In addition, the single SNP analysis widely used currently could potentially miss important genes and biological pathways. Moreover, the molecular mechanisms through which the identified variants confer risk or influence phenotypic expression remains poorly understood. From current GWAS findings, relatively few SNPs have P-values reaching genome-wide significance (P < 10−5) to give conclusive evidence of association, and even fewer have been replicated in multiple independent studies.13 Conversely, many hundreds of SNPs with moderate (P∼10−2–10−4) have been reported. Although some of these may be false-positives, others are potentially genuine associations with small effects. The presence of a greater than expected number of associated SNPs in genes of similar biological functions interacting within intricate biological pathways could give a degree of confidence that the associations are potentially genuine, even if none of the SNPs individually are highly significant.14
While GWAS can effectively map loci conferring risk for breast cancer, they offer limited insights about the mechanisms by which the SNPs exert their effects or influence phenotypic expression. In addition, GWAS findings do not always lead directly to the gene or genes because some SNPs as evidenced in this study and other studies6–11 map to intergenic regions close to nearby genes. Consequently, identified SNPs do not typically inform the broader context in which the disease-associated genes operate.12 Genes may be regulated in trans or even in cis by genetic variants that are far away from the implicated structural gene. All of these genetic along with environmental factors could severely affect the function of a gene and the putative biological pathways involving its gene products. These factors are difficult to model using the common single-SNP GWAS analysis and provide limited information about the functional basis of GWAS findings. Therefore, novel and complementary approaches are needed to overcome limitations imposed by GWAS. A systematic approach is needed to study how genes containing SNPs associated with risk for breast cancer interact with one another, and with genes not identified by common single-SNP association analysis, to determine clinical endpoints or disease phenotypes.
The objectives of this study were (a) to investigate the power of combining GWAS information with gene expression data to identify functionally related genes and biological pathways enriched by SNPs associated with risk for breast cancer, and (b) to identify genes and biological pathways that could not be identified using traditional single-SNP GWAS analysis. We hypothesized that genes containing SNPs associated with risk for breast cancer are functionally related and interact with each other and other genes not identified by traditional GWAS in intricate biological pathways. We have tested this hypothesis using GWAS information from 43 genome-wide association studies and three different gene expression data sets representing the Caucasian and the Asian populations. Throughout this analysis, we defined the genes containing SNPs associated with risk for breast cancer as candidate genes, and the genes identified from gene expression data but not containing SNPs reported in GWAS as novel genes. Our analysis assumed the gene and the pathway as the units of association. This holistic approach allowed us to account for all the SNPs including rare variants and those with small to moderate effects mapped to genes in our analysis.
Methods
Source of SNP data
SNP data and gene information were obtained by mining data from published reports on GWAS in breast cancer through Pub Med searches and web-sites containing supplementary data. Our search included terms (GWAS, GWA, WGAS, WGA, genome-wide, genomewide, whole genome, all terms + association, or + scan) in combination with breast cancer from the primary published reports through November, 2010. We catalogued all the SNPs that showed significant (P ≤ 0.05) association with risk for breast cancer. We chose this liberal statistical threshold to allow examination of genes containing border-line SNPs with small effect sizes and to accommodate GWAS of various sizes while maintaining a consistent approach. This threshold level also allowed us to address publication bias (“the winner curse”), a tendency on average to publish SNPs with the smallest P-values (P ≤ 10−5). The SNP and gene names along with their aliases were verified using the dbSNP database based on chromosome report build 3.71. Gene names and aliases were further verified using the Human Genome Nomenclature (HGNC) database. SNPs mapping to intergenic regions were removed from the final data set used in the analysis. SNPs were matched with gene names using SNP IDs (rs-IDs) information in the database (dbSNP). SNPs were then sorted and ranked on the basis of P-values derived from GWAS, number of times the SNP in a particular gene has been replicated in multiple independent studies, and number of SNPs within each candidate gene. All together 43 GWAS studies totaling over 250,000 cases and 250,00 controls were evaluated. Only studies with sample size >500 in patients and controls were considered. From the total, 98% of all the SNPs were identified using GWAS data derived from the Caucasian populations.
Sources of gene expression data
Publicly available gene expression data was downloaded from GEO; http://www.ncbi.nlm.nih.gov/geo/http://www.ncbi.nlm.nih.gov/geo/. The data included three data sets derived from Caucasian and Asian populations. The first data set derived from the Caucasian population involved gene expression data derived from RNA extracted from 143 histologically normal breast tissues obtained from patients harboring breast cancer who underwent curative mastectomy and 42 invasive ductal carcinomas (IDC) of various histological grades (1–3). The samples were obtained from breast cancer patients at Moffitt Comprehensive Cancer Center, Florida United States. The data set has been fully described by the originators.15 Briefly, this data set consisted of histological data. Histologically-normal breast has the potential to harbor pre-malignant changes at the molecular level and thus provides an opportunity for identifying risk markers. We postulated that a histologically-normal tissue with tumor-like gene expression patterns might harbor substantial risk for future cancer development. Thus genes associated with these high-risk tissues would be considered to be malignancy-risk genes. “Normal” breast cancer tissue included histologically normal and benign hyperplasia. The data set was generated using the Affymetrix platform on U133 Plus 2.0 Array containing ∼54,000 probes. The data set was downloaded from GEO accession number GSE10780.16
The second data set involved a multi-ethnic Asian population, consisting of Malaysian breast cancer patients (Malays, Chinese and Indian). The data set involved invasive ductal carcinomas and was very similar to the first data set. The data set has been fully described by.17 Briefly, the data set consisted of a total of 43 IDCs with histological grades 1–3 and 43 patient-matched normal tissues collected from Kuala Lumpur, UKM and Putrajaya Hospitals in Malaysia. The data set was generated using the Affymetrix platform’s U133A Chip containing ∼22,000 probes, and was downloaded from GEO accession number GSE15852.16 Population of Asian descent provide special opportunities for this research because of the emerging evidence that genetic variants may confer population-specific risk.18,19 It is conceivable that expression of genes containing SNPs associated with risk for breast cancer could equally be population-specific. Therefore, the rationale for using this data set was to determine whether genes containing SNPs associated with risk for breast cancer would exhibit similar patterns of expression in the Caucasian and Asian populations. Unfortunately no similar data was found on the Africans or African-American population, therefore, no analysis were attempted in those populations.
To determine the clinical utility of genes containing SNPs associated with risk for breast cancer, we used a third data set involving two disease states. The third data set involved 286 lymphnode-negative primary breast cancer patients from the Caucasian population.20 The data set consisted of 209 ER+ and 77 ER− breast cancer patients. The data set was generated using RNA extracted from fresh-frozen breast cancer tissues. The estrogen receptor alpha (ERα) is a master transcriptional regulator of breast cancer phenotype and the archetype of a molecular marker and therapeutic target. ER+ tumors respond to endocrine therapy.21 Conversely, ER− tumors are generally treated with chemotherapy and do not respond to endocrine therapy. Thus, the objective of this analysis was to determine whether candidate genes could distinguish ER+ from ER− breast cancer patients. The data set was generated using the Affymetrix platform using U133A Gene Chip containing ∼22,000 probes. The data set was downloaded from GEO under accession number GSE2034.16
In each of the data sets described above, entries in the data matrix were expression values generated by Affymetrix’s Microarray Analysis Suite 5.0 (MAS5) statistical algorithm.21 Following normalization and scaling, MAS5 signal values were summarized by Turkey’s biweight estimation of the probe level intensities within each probe set. This was followed by a global normalization (linear scaling) to give all chips the same average intensity. These procedures yield robust weighted means called average-scaled differences that are proportional to the amount of a particular RNA transcript present in the sample after background correction, which we used in this analysis.
Data analysis
Analysis of SNP data: we analyzed SNP and gene expression data using a combination of different analytical techniques described below. For SNP data, the first step was to verify the names of genes to which SNPs map. The gene names were verified as described in the preceding sections. The challenge was how to represent a gene containing SNPs replicated in independent studies, SNPs mapped to different positions within the gene and how to account for correlations among those SNPs. We used a meta-analytical approach using Fisher’s method22 as described below to estimate the overall P-value for SNPs with P-values replicated in multiple independent studies and within a gene. Briefly, assume that the P-values (Pi) of individual SNPs are independent and uniformly distributed under their null hypotheses. It is worth noting here that “independence” here is used conservatively as it could be violated because of linkage disequilibrium among SNPs in the gene and potential correlations among SNPs. Let Pi be the P-value for the corresponding statistic Ti to test the association of the ith SNP with the breast cancer phenotype, where [Pi = P1, P2, …, Pn]T is a vector of P-values obtained by performing independent test statistic [Ti = T1, T2, …, Tn]T on individual SNPs [rsi = rs1, rs2, …, rsn]T. Assuming H as a continuous monotonic function, a transformation of the P-value can be defined as Zi = H−1(1 − Pi).23 Transformation to z allows decorrelating SNPs and treating their P-values as independent. The statistics for combining K independent P-values or for combining information from K SNPs is given by the following equation,22,23
where ZF denotes the sum of zi (Z-scores) of the transformed P-values for the K SNPs. The P-value is obtained by back transforming z.22
Correlations among P-values of SNPs within a gene exist because of linkage disequilibrium among SNPs. Correlations among SNPs will invalidate the existing methods for combining independent P-values.24 Furthermore, the SNPs within a gene may have antagonistic functions which could not be captured by combining P-values. Therefore, a method for combining independent SNPs described above Peng et al24 is an approximation. For multiple SNPs within a gene, Wang et al25 suggested choosing the most significant SNP from each gene as a representative. The limitation of that approach is that genes that contain a number of SNPs jointly having significant risk effects, but individually making only a small contribution, will be missed in such a representation. Therefore, in this study, we combined P-values of SNPs of the same gene reported in multiple independent studies. Instead of relying on correlations among SNPs here we have used gene expression data on genes containing SNPs to guide the correlation structure. This approach allowed us to holistically unravel the genetic susceptibility architecture of breast cancer by jointly considering all common variation including rare variants within the gene and all the genes in the pathway.
To evaluate the pathway as a unit of association we used the hypergeometric test (Fisher’s exact test) to search for an overrepresentation of significantly associated genes in the pathway.22,23 Briefly, let N be the total number of genes presumed to be of interest and S be the number of SNPs significantly associated with risk for breast cancer identified in GWAS. Then let m be the total number of genes in the pathway and let k be the number of significantly associated genes interacting within the pathway. The P-value of observing k significant genes in the pathway was calculated using the following equation;24
Using the above equation, the total effect of the pathway as a unit of association was computed by direct enumeration of the P-values in that pathway.23
Gene expression data analysis: we employed two analysis strategies, supervised and unsupervised analysis, analyzing each data separately. Prior to analysis we normalized the data using the lowess normalization procedure.26 On each normalized data set, we performed supervised analysis comparing gene expression profiles in breast cancer patients and controls using a t-test to identify significantly differentially expressed genes which distinguished breast cancer patients from control subjects; and to identify significantly differentially expressed genes which distinguished ER+ from ER− breast cancer patients. In each data set we employed the permutation test, a re-sampling procedure, to calculate the probability of producing a cross-validated error rate as small as observed given no association between class membership and expression profiles.27,28 We used the Benjamin and Hochberg29 procedure to correct for multiple testing. Probes were ranked on P-values starting with the smallest P-value. Candidate genes and novel genes were identified by matching probes with gene names using the Affymetrix database NetAffx; http://www.affymetrix.com/analysis/index.affx.
To determine whether genes containing SNPs associated with risk for breast cancer are co-expressed and to assess their relationships with each other and with novel genes, we performed unsupervised analysis using hierarchical clustering based on the complete linkage model. We used the correlation coefficient to assess the level of co-expression and to identify genes with similar patterns of expression among SNP-containing genes and novel genes. The correlation coefficient (rXY) between a pair of SNP-containing genes (x and y) or between the SNP-containing gene x and the novel gene (y) were computed using the following equation;
where n is the sample size, Xi and Yi [X̄ and Ȳ] are the expression [mean expression] values for a pair of SNP-containing genes or between a SNP-containing gene and the novel gene, respectively. Prior to clustering we normalized, standardized and centered the data.30 Supervised and unsupervised analyses were performed using Pomello II31 and Gene Pattern software.32
Finally, we performed pathway prediction to determine whether SNP-containing genes interact with each other and with novel genes in biological pathways. For pathway prediction, the input were the genes containing SNPs and novel genes that were found to be associated with cancer or to distinguish disease states in the case of ER+ and ER−. We performed pathway prediction and network modeling using the Osprey System.33 Additional information and validation of predicted pathways and gene regulatory networks was obtained through the literature mining module built in the Osprey System which also provides biological and experimental information about the genes under study and identifies other functionally related genes interacting with input genes. In pathway prediction, genes were represented by nodes and the interactions by vertices. Two genes were considered to share a genetic susceptibility architecture and network properties if they were interconnected as represented by the vertices and were co-expressed as determined by pattern recognition analysis using hierarchical clustering based on the correlation distance as explained in the preceding paragraph. Additional functional assessment was performed using the Gene Ontology (GO) nomenclature.34 Genes with spurious interactions and without interactions were removed from the networks. The rationale being that such genes could be less informative or could distort the reliability of pathway prediction and network modeling.
Results
Characterization of candidate genes and genetic variants
We mined publicly available data on 43 reported GWAS through November 2010 and the accompanying supplementary data posted on websites to identify genetic variants and genes associated with risk for breast cancer. The results of data mining for GWAS information used in this study are summarized in Table A provided as supplementary data. The search yielded 525 SNPs with P-values ranging from 2 × 10−76 to P ≤ 0.05. Out of the genetic variants identified, 113 SNPs mapped to intergenic regions and were not used in further analysis. The remainder 412 SNPs mapped to 150 candidate genes used in this study. A total of 20 genes including ABCC4, CASP8, COL1A1, ECHDC1, ESR1, FGFR2, FBNI, GRIK1, LOC643714, LSP1, PPP2R2B, RAD51L1, SLC4A7, STXBP4, TGFB1, TOX3, BTNL8, H19, MLK and MAP3K1 had SNPs with small P-values (P ≤ 10−5). Forty genes produced SNPs replicated in multiple independent studies. Ten genes including CASP8, ESR1, FGFR2, LSP1, STXBP4, TGFB1, TOX3, LOC643714, SOD2, MAP3K1 contained SNPs with the smallest P-values and have been reproduced in multiple independent studies. Some of the genes including FGFR2 and ESR1 contained multiple SNPs within the gene.
Interestingly, the P-values of replicated SNPs varied with respect to study, presumably due to the genetic and phenotypic heterogeneity, and sample variability. The remainder (majority) of the genes contained SNPs with moderate P-values ranging from P = 0.0001 to P < 0.05. It is conceivable that some of the loci with moderate P-values likely contain several false positives, but may also contain genuine effects of small magnitude. Consequently, in further data analysis, we considered all the 150 candidate genes containing SNPs significantly (P ≤ 0.05) associated with risk for breast cancer. The rationale was that because cancer is a polygenic disease, the presence of SNPs in co-expressed genes with similar biological functions interacting with each other and their downstream targets in biological pathways would give a degree of confidence that the associations are potentially genuine, even if none of the SNPs is individually highly significant.
Association of candidate genes with gene expression
To determine whether genes containing SNPs associated with risk for breast cancer can distinguish breast cancer patients from cancer-free controls, and ER+ from ER−, we analyzed three separate gene expression data sets. We asked the question do genes containing SNPs associated with risk for breast cancer differ in their expression profiles between cancer-free controls and breast cancer patients in the Caucasian and Asian populations; and between ER+ and ER− breast cancer patients? ER+ and ER− data set was not available in the Asian population, therefore this analysis was restricted to the Caucasian population. Secondly, we asked the question does expression in genes containing SNPs differ between Caucasian and Asian populations?
The results showing a list of significantly differentially expressed genes between cases and controls in the Caucasian and Asian populations for genes containing SNPs with the smallest P-values are presented in Table 1. The results of genes containing SNPs replicated in multiple independent GWAS are presented in Table 2. Also presented in the two tables are the results comparing expression for the respective genes between ER+ and ER−; the estimates of P-values for the genes based on gene expression, and the ranges of estimates of P-values for SNPs mapped to respective genes derived from GWAS. A complete list of estimates of P-values for all the genes and FDR for the three data sets used along with information on the biological processes, molecular function and cellular process in which the genes are involved are presented in Table B1 (130 genes) for the Caucasian population, Table B2 (111 genes) for the Asian population and Table B3 (111 genes) for the ER+ and ER− breast cancer patients, provided as supplementary data. The discrepancy between the total number of candidate genes (150) examined in this study and the unique number of candidate genes corresponding with each data set is due to the fact that some genes were not represented on the Chips, presumably due to lack of proper annotation. The differences in the number of genes between data sets is a reflection of differences in Chip density.
Table 1.
List of genes containing SNPs with the largest effect sizes (P ≤ 10−5) estimated from GWAS and the P-values estimated from gene expression values in the Caucasian (EU), Asian and ER+ and ER− (EU) populations.
| Gene name (symbol) | SNP ID (rs-ID) | SNP (P-value) | EU (P-value) | Asian (P-value) | ER+/ER−(P-value) |
|---|---|---|---|---|---|
| ABCC4 | rs1926657 | 1.9 × 10−6 | 0.04 | 0.4 | 0.07 |
| CASP8 | rs1045485 | 1.1 × 10−7 | 0.005 | 0.5 | 0.1 |
| COL1A1 | rs2075555 | 8.3 × 10−8 | 5.00E-06 | 0.0005 | 0.1 |
| ECHDC1 | rs6569480 | 6.1 × 10−8 | 1.00E-05 | 0.001 | 0.01 |
| ECHDC1 | rs7776136 | 6.6 × 10−8 | 1.00E-05 | 0.001 | 0.01 |
| ESR1 | rs3020314 | 8 × 10−5 | 0.7 | 0.0006 | 5.00E-06 |
| FGFR2 | rs2981582 | 2 × 10−76 | 5.00E-06 | 0.1 | 0.49 |
| FGFR2 | rs2981579 | 1.79 × 10−31 | 5.00E-06 | 0.1 | 0.49 |
| FGFR2 | rs2420946 | 3.5 × 10−6 | 5.00E-06 | 0.1 | 0.49 |
| FGFR2 | rs1219648 | 3.2 × 10−6 | 5.00E-06 | 0.1 | 0.49 |
| FGFR2 | rs1078806 | 1.5 × 10−5 | 5.00E-06 | 0.1 | 0.49 |
| FBNI | rs1876206 | 6.0 × 10−6 | 0.6 | 0.2 | 0.03 |
| GRIK1 | rs458685 | 6.0 × 10−6 | 5.00E-06 | 0.7 | 0.02 |
| LOC643714 | rs3803662 | 1 × 10−36 | 1.00E-05 | – | – |
| LSP1 | rs3817198 | 3.0 × 10−9 | 0.6 | 0.1 | 0.01 |
| PPP2R2B | rs9325024 | 1.7 × 10−5 | 0.4 | 0.9 | 0.8 |
| RAD51L1 | rs999737 | 1.74 × 10−7 | 0.5 | 0.1 | 8.00E-05 |
| SLC4A7 | rs4973768 | 4 × 10−23 | 0.8 | 0.1 | 0.6 |
| STXBP4 | rs6504950 | 1.4 × 10−8 | 0.7 | – | – |
| TGFB1 | rs1800470 | 2.8 × 10−5 | 0.0007 | 0.8 | 0.5 |
| TOX3 | rs12443621 | 2 × 10−19 | 0.0001 | 6.50E-05 | 7.00E-05 |
| TOX3 | rs3803662 | 5.9 × 10−19 | 0.0001 | 6.50E-05 | 7.00E-05 |
| TOX3 | rs8051542 | 1.0 × 10−36 | 0.0001 | 6.50E-05 | 7.00E-05 |
| BTNL8 | rs7711990 | 8.4 × 10−5 | 0.5 | 0.1 | 0.2 |
| H19 | rs2107425 | 2 × 10−5 | 0.2 | – | – |
| MAP3K1 | rs889312 | 4.6 × 10−20 | 0.04 | 0.9 | 0.01 |
Notes: EU indicates Caucasians, ER+ and ER− indicate estrogen positive and negative, respectively,—indicates that the gene was not represented on the Chip, thus no estimate of P-values is available.
Table 2.
List of genes containing significantly associated (P ≤ 0.05) SNPs replicated in multiple independent GWAS studies, and the P-values estimated using gene expression data derived from the Caucasian (EU), Asian and ER+ and ER− EU population.
| Gene name (symbol) | SNP ID (rs-ID*, replicated) | Range of SNP (P-value) | EU (P-value) | Asian (P-value) | ER+/ER−(P-value) |
|---|---|---|---|---|---|
| CASP8 | rs1045485** | 0.02–1.1 × 10−7 | 0.005 | 0.1 | 0.1 |
| ESR1 | rs3020314** | 8 × 10−5–8 × 10−5 | 0.7 | 0.0006 | 5.00E-06 |
| ESR1 | rs3020390** | 0.04–0.05 | 0.7 | 0.0006 | 5.00E-06 |
| ESR1 | rs3020394** | 0.003–0.003 | 0.7 | 0.0006 | 5.00E-06 |
| ESR1 | rs1884051** | 0.03–0.03 | 0.7 | 0.0006 | 5.00E-06 |
| ESR1 | rs2228480** | 0.002–0.006 | 0.7 | 0.0006 | 5.00E-06 |
| ESR1 | rs3020396** | 0.003–0.004 | 0.7 | 0.0006 | 5.00E-06 |
| ESR1 | rs3020400** | 0.005–0.005 | 0.7 | 0.0006 | 5.00E-06 |
| ESR1 | rs3020401** | 0.004–0.004 | 0.7 | 0.0006 | 5.00E-06 |
| ESR1 | rs3798577** | 0.04–0.004 | 0.7 | 0.0006 | 5.00E-06 |
| FGFR2 | rs2981582******* | 0.01–2 × 10−76 | 5.00E-06 | 0.1 | 0.4 |
| FGFR2 | rs2981579***** | 0.02–1.79 × 10−31 | 5.00E-06 | 0.1 | 0.4 |
| FGFR2 | rs2420946****** | 0.03–3.5 × 10−6 | 5.00E-06 | 0.1 | 0.4 |
| FGFR2 | rs1219648****** | 0.01–3.2 × 10−6 | 5.00E-06 | 0.1 | 0.4 |
| LSP1 | rs3817198**** | 6.51 × 10−2–3.0 × 10−9 | 0.6 | 0.1 | 0.01 |
| STXBP4 | rs6504950** | 0.04–1.4 × 10−8 | 0.7 | – | – |
| TGFB1 | rs1800470*** | 0.01–2.8 × 10−5 | 0.0007 | 0.8 | 0.5 |
| TOX3 | rs12443621** | 1 × 10−12–2 × 10−19 | 0.0001 | 6.50E-05 | 7.00E-05 |
| ADH1B | rs1042026*** | 0.02–0.03 | 5.00E-06 | 5.0E-06 | 0.04 |
| SORBS1 | rs10450393** | 0.01–0.02 | 0.4 | 0.01 | 0.63 |
| ICAM5 | rs1056538** | 0.05–001 | 0.9 | 0.01 | 0.49 |
| RB1 | rs198580** | 0.02–0.02 | 0.4 | 0.01 | 0.0001 |
| RNF146 | rs2180341** | 0.008–2.9 × 10−8 | 0.7 | – | 0.69 |
| RB1 | rs2854344** | 0.007–0.007 | 0.4 | 0.01 | 0.0001 |
| IGFBP3 | rs2854744**** | 0.06–0.03 | 0.0005 | 0.002 | 0.03 |
| CDKN1A | rs3176336** | 0.003–0.003 | 0.3 | 0.04 | 0.2 |
| CDKN1B | rs34330** | 0.01–0.01 | 0.9 | 0.7 | 5.00E-06 |
| CDKN2A | rs3731239** | 0.01–0.01 | 0.003 | 0.40 | 0.0001 |
| LOC643714 | rs3803662****** | 0.01–1 × 10−36 | 0.00E-05 | – | – |
| EHMT1 | rs4634736** | 0.02–1–0.02 | 7.50E-05 | 0.1 | 0.03 |
| SOD2 | rs4880** | 0.01–0.05 | 0.03 | 0.1 | 0.0008 |
| CCND1 | rs678653** | 0.05–0.002 | 0.81 | 0.3 | 5.00E-06 |
| HCN1 | rs981782** | 10−5–10−2 | 0.008 | – | – |
| CCNE1 | rs997669** | 0.003–0.003 | 5.00E-06 | 0.07 | 5.00E-06 |
| RAD51L1 | rs999737** | 1.74 × 10−7 | 0.5 | 0.1 | 8.00E-05 |
Note:
Indicates the number of times the SNP has been replicated in multiple independent studies.
A comparison between cancer patients and controls in the Caucasian population produced 69 significantly (P ≤ 0.05) differentially expressed candidate genes, which distinguished the two classes. Among the genes identified from this analysis, ten genes ABCC4, CASP8, COL1A1, ECHDC1, FGFR2, GRIK1, LOC643714, TGFB1, TOX3 and MAP3K1 contained SNPs with small P-values (P ≤ 10−5) (Table 1). Twelve genes, CASP8, FGFR2, TGFB1, TOX3, MAP3K1, ADH1B, IGFBP3, CDKN2A, EHMT1, SOD2, HCN1, and CCNE1 contained SNPs replicated in multiple independent GWAS (Table 2). These results confirm our hypothesis that genes containing SNPs associated with risk for breast cancer can distinguish breast cancer patients from cancer-free controls. However, 61 genes containing SNPs associated with risk for breast cancer were not significantly (P > 0.05) differentially expressed between breast cancer patients and cancer-free controls, presumably due to the fact that gene expression can be tissue-specific and breast cancer subtype-specific expressed.3 Under such conditions and given the genetic and phenotypic heterogeneity inherent in breast cancer, such outcome should be expected.
A major concern about GWAS reported thus far, is that majority of these studies have been performed on the Caucasian population. Emerging evidence in the published literature tends to suggest that genetic variants may confer population-specific risk.18,19 To test this hypothesis we compared the expression of candidate genes between cancer patients and cancer-free controls in the Asian population. We asked the question, do genes containing SNPs associated with risk for breast cancer significantly differ in their expression profiles between cancer patients and cancer-free controls in the Asian population? Out of the 111 candidate genes examined, only 35 genes were significantly (P ≤ 0.05) differentially expressed, distinguishing breast cancer patients from cancer-free controls (see Table B2 in the appendix). Four significantly differentially expressed candidate genes including COL1A1, ECHDC1, ESR1 and TOX3 contained SNPs with small P-values (Table 1), whereas eight genes, ESR1, TOX3, ADH1B, SORBSI, ICAM5, RB1, IGFBP3 and CDKN1A contained SNPs replicated in multiple independent GWAS studies (Table 2).
As expected, not all candidate genes associated with gene expression identified in the Caucasian population were found in the Asian population and vice versa. There were 12 genes containing SNPs that were found to be significantly differentially expressed between cases and controls in both the Caucasian and Asian populations. Five of those genes which overlapped between the two populations including TOX3, ECHDC1, COL1A1, ADH1B and IGFBP3 contained SNPs with small P-values and replicated in multiple independent GWAS studies. This tends to suggest that the Caucasians and Asians may have shared genetic susceptibility at some loci. It is not clear why fewer candidate genes were found to be significantly differentially expressed in the Asian population compared to the Caucasian population. There are several plausible reasons for the observed outcome, including the differences in Chip density, within population variation in the Asian sample, diagnostic misclassification, and the environmental conditions to which the two populations were subjected to. All of these factors individually or in combination could affect gene expression.
To determine the clinical utility of genes containing SNPs associated with risk for breast cancer, we compared gene expression between ER+ and ER− breast cancer patients. We asked the question do genes containing SNPs associated with risk for breast cancer differ in their expression between ER+ and ER− breast cancer patients? This analysis produced 70 significantly (P ≤ 0.05) differentially expressed candidate genes, which distinguished ER+ from ER− breast cancer patients (See supplementary Table B3). Eight genes including MAP3K1, TOX3, RAD51L1, LSP1, GRIK1, FBN1, ESR1 and ECHDC1 contained SNPs with the smallest P-values (Table 1), whereas thirteen significantly differentially expressed candidate genes including ESR1, LSP1, TOX3, ADH1B, RB1, IGFBP3, CDKN1B, CDKN2A, EHMT1, SOD2, CCND1, CCNE1 and RAD51L1 contained SNPs replicated in multiple independent GWAS studies (Table 2). The data supports the notion that ER+ and ER− breast cancers have different molecular circuitries and may originate through biologically distinct genetic lesions. Interestingly, five candidate genes (ECHDC1, ESR1, GRIK1, TOX3, MAP3K1) containing SNPs with small P-values and seven candidate genes (ESR1, TOX3, ADH1B, RB1, CDKN2A, EHMT1 and CCNE1) containing SNPs replicated in multiple independent GWAS identified that distinguished ER+ from ER− breast cancer patients also distinguished cases from controls in the Caucasian and Asian populations. This indicates that at least some candidate genes identified through GWAS analysis could serve as potential biomarkers in different populations, though association of these genes with the African and African-American populations remains to be determined.
The variation in gene expression observed in this study is not unique. The results of variability in gene expression among populations observed here are consistent with previous reports. For example, genome-wide association of gene expression variation in humans has been reported,35 though the study used cell lines and did not focus on any particular disease. Variation in gene expression within and among natural populations has also been reported.36 Inconsistent expression of SNP-containing genes has also been reported in prostate cancer.37
Interestingly, majority of the genes distinguishing cases from controls and ER+ from ER− were those which contained SNPs with moderate P-values. This indicates that genes containing SNPs with moderate P-values “often considered not genome-wide significant in traditional single-SNP GWAS analysis” are likely to play a significant role in the pathogenesis of breast cancer.
Evaluation of functional relationship of candidate genes
To determine whether genes containing SNPs associated with risk for breast cancer are functionally related, we used the Gene Ontology (GO) information and co-expression analysis using gene expression data. The first, GO analysis allows characterization of genes according to the GO nomenclature. The GO Consortium has developed three separate ontologies-molecular function, biological process and cellular component to describe the attributes of gene products. Molecular function defines what a gene product does at the biochemical level without specifying where or when the event actually occurs; biological process describes the contribution of a gene product to a biological objective; and cellular component refers to where in the cell a gene product functions. Here, we have characterized the candidate genes according to all three GO categories. The results are provided in Tables B1 for the Caucasian population, in Table B2 for the Asian population and Table B3 for the ER+/ER− Caucasian population, provided as supplementary data. In all the three cases studied, we found that candidate genes are functionally related and are involved in multiple related functions and biological processes.
The second approach co-expression analysis was conducted to identify candidate genes with similar patterns of expression profiles. We hypothesized that genes containing SNPs associated with risk for breast cancer are co-regulated and have similar patterns of expression profiles. The results showing patterns of expression profiles for candidate genes are shown in Figure A1 and Figure A2 for the Caucasian and Asian population, respectively in the Appendix. The results showing patterns of gene expression profiles for candidate genes in ER+ and ER− breast cancer patients are shown in Figure A3. Only the candidate genes with the most consistent patterns of expression profiles are represented in the figures. In all the three cases, genes containing SNPs associated with risk for breast cancer were co-expressed and produced clusters of genes with similar patterns of expression. Interestingly, genes containing SNPs with moderate P-values were found to be functionally related and co-expressed with candidate genes containing SNPs with small P-values and SNPs replicated in multiple independent studies. This demonstrates the power of combining GWAS information with gene expression data to identify associations beyond the ones that meet the very stringent genome-wide significance threshold, and to determine the functional basis of GWAS discoveries.
Combining GWAS information with gene expression data to identify novel genes
To determine whether genes containing SNPs associated with risk for breast cancer are functionally related and co-expressed with other genes which have not been identified using single-SNP GWAS analysis, we performed supervised followed by unsupervised analysis using hierarchical clustering, as described in the methods section. We asked two fundamentally distinct questions. First, we asked, are there significantly differentially expressed novel genes not identified by GWAS, which distinguish breast cancer from cancer-free controls and ER+ from ER−? Second we asked the question are the novel genes distinguishing breast cancer patients from cancer-free controls functionally related or co-expressed and have similar patterns of expression with candidate genes distinguishing breast cancer from cancer-free controls and ER+ from ER− breast cancer patients?
In all the three cases we identified significantly differentially expressed novel genes not reported in GWAS. For the Caucasians and Asian populations, we identified 62 and 73 highly significant genes which distinguished breast cancer patients from cancer-free controls. We identified 85 highly significantly differentially expressed genes, which distinguished ER+ from ER− breast cancer patients. The novel genes were functionally related. The results showing P-values, FDR and functional relationships for the novel genes are summarized in Table C1 (62 genes) for the Caucasian population, Table C2 (73 genes) for the Asian population and Table C3 (85 genes) for the ER+ versus ER− breast cancer patients, provided as supplementary data.
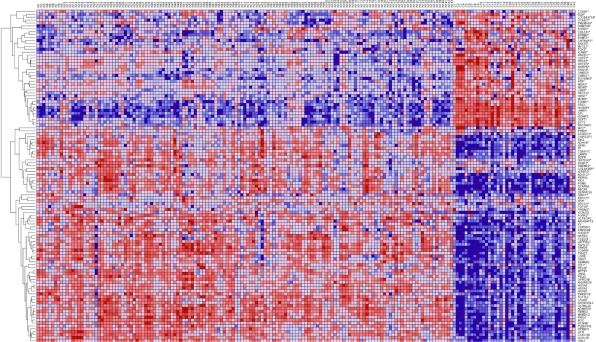
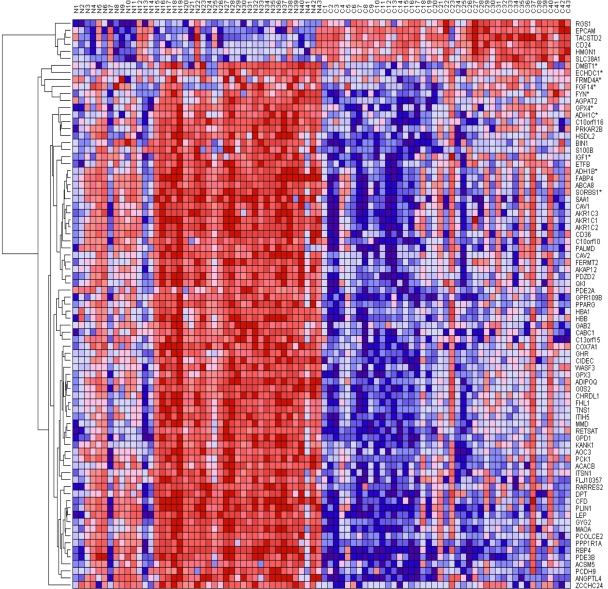
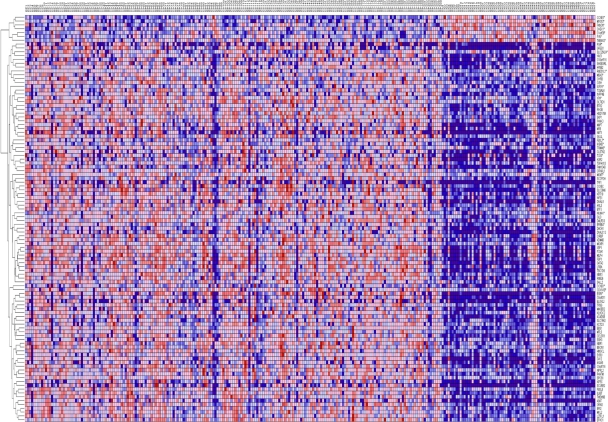
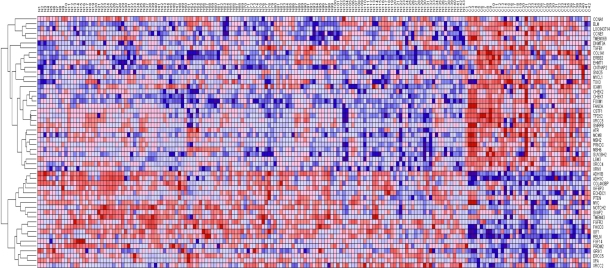
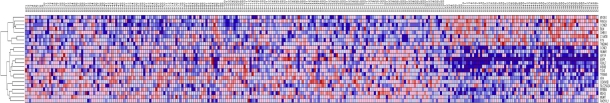
The results showing patterns of gene expression profiles for candidate genes and novel genes are shown in Figure 1 for the Caucasian population, Figure 2 for the Asian population and Figure 3 for the ER+ and ER− breast cancer patients. In all the three cases, genes containing SNPs associated with risk for breast cancer were found to be functionally related, co-expressed and exhibited similar patterns of expression with novel genes not identified in GWAS analysis. These results demonstrate and unequivocally confirm our hypothesis that integrating GWAS information with gene expression data provides a complementary approach to identify novel genes that could not be identified using traditional single-SNP GWAS analysis. Interestingly, genes containing SNPs with moderate P-values and genes containing SNPs not replicated in multiple independent studies were found to be co-expressed and functionally related with novel genes. However, as expected, considerable variation in patterns of expression profiles were observed in all the three cases studied. The identification of novel genes that are functionally related and co-expressed with candidate genes, suggests that the missing variation from GWAS findings could potentially be captured through integration of GWAS information with gene expression profiling.
Figure 1.
Patterns of gene expression profiles for candidate and novel genes in the Caucasian population. The rows represent genes, columns represent 143 cancer-free controls and 42 breast cancer patients. The red and blue colors indicate up and down regulation, respectively.
Figure 2.
Patterns of gene expression profiles for candidate and novel genes in the Asian population. The rows represent genes, columns represent 43 (N) controls and 43 (C) breast cancer patients. The red and blue colors indicate up and down regulation, respectively.
Figure 3.
Patterns of gene expression profiles for candidate and novel genes in the ER+ and ER−Caucasian population. The rows represent genes, columns represent 209 ER+ and 77 ER− breast cancer patients, respectively. The red and blue colors indicate up and down regulation, respectively.
Predicted pathways and gene regulatory networks
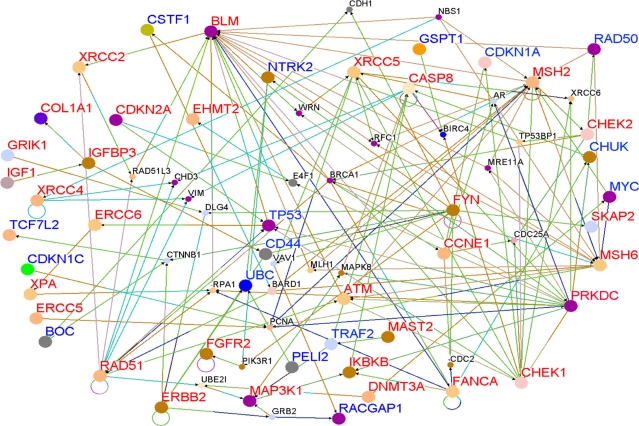
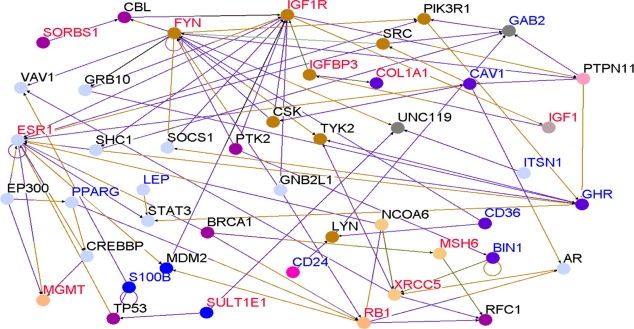
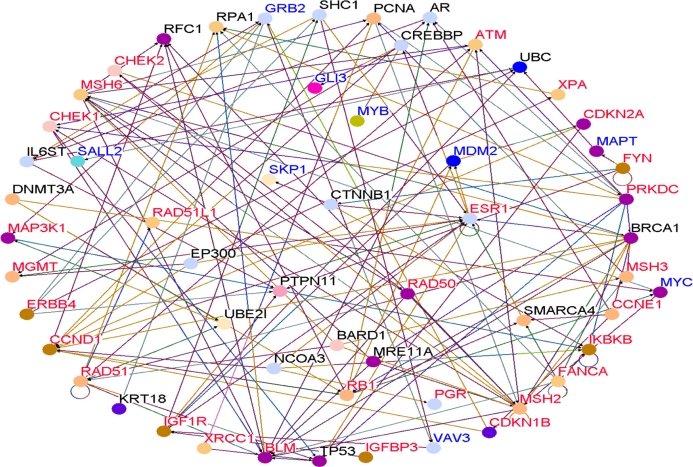
To determine whether genes containing SNPs associated with risk for breast cancer interact with each other and with novel genes in biological pathways and gene regulatory networks, we performed pathways analysis and gene network modeling. We hypothesized that candidate genes and novel genes are functionally related, interact with each other in putative biological pathways and gene regulatory networks associated with breast cancer. Figure 4 presents the color codes showing the biological process in which the genes are involved as depicted in predicted pathways and gene regulatory networks. The results showing predicted pathways and gene regulatory networks using candidate and novel genes are presented in Figure 5 for the Caucasian population, in Figure 6 for the Asian population and in Figure 7 for the ER+/ER− Caucasian population.
Figure 4.

Color code indicating the biological process in which the genes in predicted biological pathways and the regulatory networks are involved.
Figure 5.
Gene interaction networks for genes containing SNPs (Red) and novel genes (Blue) identified using a threshold (P < 10−6) and other functionally related genes (in black) correlated with candidate genes and novel genes in the Caucasian population only. The size of the nodes: Large indicate SNP-containing and Novel genes identified through differential expression analysis, whereas small nodes indicate genes experimentally confirmed in the literature and through co-expression analysis that are functionally related and interact with SNP-containing and novel genes.
Figure 6.
Gene interaction networks for genes containing SNPs (Red) and novel genes (Blue) identified using a threshold (P < 10−6) and other functionally related genes (in black) correlated with candidate genes and novel genes in the Asian population only.
Figure 7.
Gene interaction networks for genes containing SNPs (Red) and novel genes (Blue) identified using a threshold (P < 10−6) and other functionally related genes (in black) correlated with candidate genes and novel genes in the ER+ and ER− breast cancer patients only.
In all the three cases studied, genes containing SNPs associated with risk for breast cancer (shown in red) and novel genes (shown in blue) were found to interact with each other in intricate biological pathways and gene regulatory networks. Additionally, genes experimentally confirmed to interact with genes containing SNPs and novel genes were also identified (genes marked in black font) through co-expression and pathway analysis and network modeling. Our analysis also revealed, as depicted by the color codes, that genes containing SNPs associated with risk for breast cancer are involved in the same biological processes with novel genes identified by gene expression analysis that could not be identified using traditional GWAS alone. Pathway prediction and network modeling confirmed that integrating genetic with gene expression data is a powerful approach to identifying putative biological pathways and gene regulatory networks that could not be identified using traditional GWAS alone.
Interestingly, genes containing SNPs with moderate P-values in GWAS studies were found to interact with novel genes, genes containing SNPs with the smallest P-values and genes containing SNPs replicated in multiple independent studies. Among the biological pathways identified included the P53, MAP kinase, apoptosis, insulin-like growth factor, DNA repair and estrogen receptor pathways, all of which have been implicated in breast cancer. The identification of multiple multi-gene biological pathways tends to suggest that apart from gene-gene interactions, pathway crosstalk may be involved in the development and progression of cancer.
Consistent with our analysis, a wealth of clinical and preclinical information exists on the functional correlations between the genes and pathways identified in this study. ESR1, the gene for ERα containing SNPs replicated in many independent GWAS studies, is the most important therapeutic target in ERα-positive breast cancers. PGR, the progesterone receptor A, another gene containing SNPs replicated in many independent studies, is a direct transcriptional target of ESR1, and provides information on estrogen receptor activity. XRCC2, XRCC3, CHEK1, CHEK2, TP53 and ATM are well-known for their involvement in DNA repair. ERBB2, the gene for Her2/Neu is over expressed in 25% of breast cancers and the target for highly effective agents such as trastuzumab and lapanitib. MAP3K1 also replicated in many independent studies is an upstream component of the MAP kinase cascade that is activated by both ERBB2 and by ESR1. CASP8 is involved in the apoptosis pathway. Both the MAP kinase pathway and PI3K pathway are activated downstream of ERBB2 as well as ESR1, producing survival signals that counteract pro-apoptotic signals mediated by CASP8 and/or by failure of DNA repair and consequent hyper-activation of TP53. The reason for “hyper” is that low-level activation of TP53 actually causes cell growth arrest and survival while DNA repair is underway. It is only high level activation of TP53 that causes apoptosis when DNA repair fails. Thus, the DNA repair genes such as XRCC2, XRCC3, CHEK1, CHEK2 and ATM if experimentally confirmed could serve as potential targets for early therapeutic intervention.
Discussion
We report a novel integrative genomics approach that combines GWAS information with gene expression data to identify functionally related genes and biological pathways enriched by SNPs associated with risk for breast cancer. Our results demonstrate that integrative analysis combining gene expression data with GWAS information has the ability to identify novel genes not identified in single-SNP GWAS analysis. The integrative genomics approach presented here offers several remarkable features.
First, the novel paradigm associates genes containing SNPs associated with risk for breast cancer with expression and demonstrates the predictive value of SNP-containing genes by distinguishing breast cancer patients from controls in the Caucasian and Asian populations; and distinguishing ER+ from ER− breast cancer patients. This is a critical step to translating GWAS findings into clinical practice. Second, the results demonstrate unequivocally that an integrative genomics approach can add structure to data by combining GWAS information with gene expression data, allowing us to gain insights about the broader biological context in which SNP-containing and novel genes operate, and a deeper understanding of the functional basis of GWAS findings. Third, the approach demonstrates that genes containing SNPs associated with risk for breast cancer are functionally related and interact with each in putative biological pathways and gene regulatory networks. This is a significant finding given that traditional single-SNPs GWAS analysis is underpowered to uncover complex biological interactions. The results provide insight into the biological processes underlying breast cancer. Fourth, the integrative approach identified novel genes and established their functional relationship with genes containing SNPs with small and moderate P-values. This is a significant finding given that relatively few SNPs have P-values sufficiently small to give conclusive evidence of association.
Although recent findings tend to suggest that common variants could explain most of the variation,38 almost all the known studies reported on breast cancer thus far13 have documented only a small number of loci and provide no putative functional bridges between GWAS findings and genes and biological pathways associated with breast cancer. The presence of SNPs in genes of similar biological functions interacting in biological pathways and gene regulatory networks as demonstrated in this study gives a degree of confidence that the associations could potentially be genuine even if none of SNPs individually is highly significant. Importantly, identification of genes not identified by GWAS could partially explain the missing variation from GWAS findings “also coined as the missing heritability”.13
Firth, genes containing SNPs replicated in multiple independent studies were found to be functionally related and co-expressed with genes containing SNPs not replicated. Because replication is difficult to achieve in current GWAS analysis, this approach may help overcome that limitation. Replication of association findings at the gene or pathway level is potentially much easier than replication at the SNP level. In the published literature, meta-analysis has been carried out as a means of increasing sample size and power.39,40 However, meta-analysis provides no information about the functional basis of GWAS findings or the biological mechanisms underlying the disease. The practical application of the approach and results produced using this approach lies in the fact that it could guide future experimental designs. For example, genes in the pathways identified by this approach could be prioritized for targeted sequencing. Alternatively, SNP-containing genes in the identified pathways could be prioritized for allele-specific expression profiling to understand how genetic variants regulate gene expression and cis regulatory elements. Using in silico analysis of SNP and sequence data, we have recently shown that SNPs tend to disrupt cis regulatory elements (splice sites, silencer and enhancer elements) Chourbanov et al41 potentially affecting transcriptional and post-transcriptional processes that ultimately affects gene expression.
Our results strongly challenge the single-SNP GWAS analysis paradigm, that focuses on SNPs producing the most highly significant P-values. Combining SNP information with gene expression information can identify genes and pathways that may be causally related to breast cancer development and that are still expressed in breast cancers at diagnosis. These genes are most likely to include strong targets for diagnostic and prognostic biomarkers and/or subtype-selective therapeutic targets. For example, identification of the targets of the ESR1 gene. Therefore, this approach has the potential of facilitating translation of GWAS discoveries to the bedside.
Many studies have now attempted pathway-based approaches to dissect the genetic susceptibility architecture of common diseases. This approach has been used in inflammatory diseases,42 bipolar disorder,43 multiple sclerosis,44 breast cancer,45,46 and seven other common diseases.47 To our knowledge, this is the first study to demonstrate the power of combining GWAS information with gene expression data and biological knowledge to identify genes, pathways and gene regulatory networks that could not be identified using traditional GWAS alone.
As mentioned in the preceding sections, the integrative approach has many attractive features. However, limitations must be acknowledged. First, our approach relies on using gene expression data and pathway prediction. Although this holistic approach accounts for all the SNPs in the genes, it provides no information about allele-specific expression. Therefore, it is difficult to discern the effects of individual SNPs on gene expression. The effect sizes (overall P-values) of SNPs were obtained using a meta-analytic approach to combine P-values. This approach can be limited in the presence of positive and negative correlations among SNPs, or where SNPs within the gene have opposite or antagonistic functions, although this limitation is minimized here by considering the gene using expression data and pathway as the units of association. While new methods for aggregating or combining SNPs or rare variants within the gene have been proposed48,49 they equally have limitations and do not take into account the functional information. We used publicly available GWAS and gene expression data, therefore our results could be potentially influenced by factors inherent in such data which are beyond our control. The optimal approach would be to analyze raw data from all the 43 GWAS used in this study. However, for various reasons including data ownership, this was neither practical nor feasible. Our results do not take into account the environmental factors to which the populations under study were subjected. Our study did not include African-Americans. It is conceivable as outlined earlier in this study and other studies18,19 that genetic variants may confer population-specific risk. In this study, we did not examine allele-specific expression and how the risk variants correlate with clinical parameters. However, previous studies have reported allele specific expression and correlations between risk variants and clinical parameters for genes and genetic variants reported in this study. Mayer et al50 reported allele-specific expression in the FGFR2 gene, a gene containing multiple variants extensively replicated in many independent GWAS studies. Huijts et al51 correlated clinical parameters with risk variants mapped to FGFR2, TNRC9, MAP3K1 and LSP1 in a Dutch breast cohort.
Considerable investments have been directed to GWAS over the past five years. The studies contain rich information that could be turned into knowledge to identify targets for early therapeutic interventions. Traditional single-SNP GWAS analysis does not provide the insights about the function basis of GWAS findings and the biological mechanisms underlying the disease. An integrative approach that combines gene expression data with GWAS information provides a complementary approach to single-SNP GWAS analysis and offers the better prospect of facilitating translation of GWAS findings to the bedside. More work is need to determine allele-specific expression and to elucidate the impact of SNPs on gene and protein functions, and to identify cis regulatory elements impacted by genetic variants mapped to genes relevant to breast cancer.
Supplementary Materials
Acknowledgments
This project was support by the University of Mississippi Cancer Institute and the Office of Research, to whom the authors are very thankful.
Appendix.
Gene expression patterns in the three cases studied for the most highly expressed candidate genes only.
Figure A1.
Patterns of gene expression profiles for 52 candidate genes only in the Caucasian population. The roles represent genes, columns represent breast cancer patients and controls. The red and blue colors indicate up and down regulation, respectively.
Figure A2.
Patterns of gene expression profiles for 9 candidate genes only in the Asian population. The roles represent genes, columns represent breast cancer patients and controls. The red and blue colors indicate up and down regulation, respectively.
Figure A3.
Patterns of gene expression profiles for 23 candidate genes only in the ER+ and ER− population. The roles represent genes, columns represent breast cancer patients and controls. The red and blue colors indicate up and down regulation, respectively.
Footnotes
Disclosure
This manuscript has been read and approved by all authors. This paper is unique and is not under consideration by any other publication and has not been published elsewhere. The authors and peer reviewers of this paper report no conflicts of interest. The authors confirm that they have permission to reproduce any copyrighted material.
References
- 1.Breast Cancer Facts and Figures 2009–10. American Cancer Society, Inc; 250 Williams Street, NW, Atlanta, GA: 30303.ic. [Google Scholar]
- 2.Golub TR, Slonin DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–7. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 3.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 4.Van de Vijver MJ, He YD, van’t Veer LJ, et al. A gene expression signature as a predictor of survival in breast cancer. New Eng J Med. 2002;347(25):1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- 5.Weigelt B, Hu Z, He X, et al. Molecular portraits and 70-gene prognosis signature are preserved throughout the metastatic process of breast cancer. Cancer Res. 2005;65(20):9155–8. doi: 10.1158/0008-5472.CAN-05-2553. [DOI] [PubMed] [Google Scholar]
- 6.Hunter DJ, Kraft P, Jacobs KB, Cox DG, Yeager M, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nature Genet. 2007;39(7):870–4. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Easton DF, Pooley KA, Dunning AM, Pharoah PD, et al. Genome-wide association study identifies novel breast cancer susceptibility loci. Nature. 2007;447(7148):1087–93. doi: 10.1038/nature05887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stacey SN, Manolescu A, Sulem P, Rafnar T, Gudmundsson J, et al. Common variants on Chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nature Genet. 2007;39(7):865–9. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 9.Commonly Studied Single-Nucleotide Polymorphisms and Breast Cancer: results from the Breast Cancer Association Consortium. J Natl Cancer Inst. 2006;98(19):1382–96. doi: 10.1093/jnci/djj374. [DOI] [PubMed] [Google Scholar]
- 10.Turnbull C, Ahmed S, Morrison J, Pernet D, et al. Genome-wide association study identifies five new breast cancer susceptibility loci. Nature Genetics. 2010;42(6):504–7. doi: 10.1038/ng.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barnholtz-Sloan JS, Shetty PB, Guan X, et al. FGFR2 and other loci identified in genome-wide association studies are associated with breast cancer in African American and young women. Carcinogenesis. 2010;31(18):1417–23. doi: 10.1093/carcin/bgq128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhong H, Yang X, Kaplan LM, Molony C, Schadt EE. Integrating pathway analysis and genetics in gene expression for genome-wide association studies. Amer J Hum Genet. 2010;86:581–91. doi: 10.1016/j.ajhg.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide assocition loci for human diseases and traits. Proc Nat Acad Sci U S A. 2009 doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holman P, Green EK, Pahwa JS, Ferreira MAR, et al. Gene ontolology analysis of GWA study data sets provides insights into the biology of bipolar disorder. Amer J Hum Genet. 2009;85:13–24. doi: 10.1016/j.ajhg.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen D, Nasir A, Culhane A, et al. Proliferative genes dominate malignancy-risk gene signature in historically-normal breast cancer. Breast Cancer Res Treat. 2009 doi: 10.1007/s10549-009-0344-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucl Acids Res. 2002;30(1):207–10. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ni IBP, Zakaria Z, Muhammad R, et al. Gene expression patterns distinguish breast carcinomas from normal breast tissues: the Malaysia context. Pathology-Res Pract. 2010;206:223–8. doi: 10.1016/j.prp.2009.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Lamason RL, Mohideen MA, Mest JR, Wong AC, et al. SLC24A5, a putative cation exchanger, affects pigmentaion in zebrafish and humans. Science. 2005;310:1782–6. doi: 10.1126/science.1116238. [DOI] [PubMed] [Google Scholar]
- 19.Helgadottir A, Manolescu A, Helgason A, et al. A variant of the gene encoding leukotriene A4 hydrolase confers ethnicity-specific risk of myocardial infarction. Nat Genet. 2006;38:68–74. doi: 10.1038/ng1692. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Klijn JG, Zhang Y, et al. Gene expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365(9460):671–9. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 21.Microarray Analysis Suite 5.0. Affymetrix Inc; Santa Clara, California: [Google Scholar]
- 22.Fisher RA. “Questions and answers #14”. The American Statistician. 1948;2(5):30–1. [Google Scholar]
- 23.Luo L, Peng G, Zhu Y, et al. Genome-wide gene and pathway analysis. Eur J Hum Genet. 2010;18:1045–53. doi: 10.1038/ejhg.2010.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng G, Luo L, Siu H, et al. Gene and pathway-based second-wave analysis of genome-wide association studies. Europ J Hum Genet. 2010;18:111–7. doi: 10.1038/ejhg.2009.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang K, Li M, Bucan M. Pathway-based approaches to analysis of genome-wide association studies. Am J Hum Genet. 2007;81:1278–83. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berger JA, Hautaniemi S, Jarvinen A-K, Edgren H, Mitra SK, Astola J. Optimized lowess normalization parameter selection for DNA microarray data. BMC Bioinformatics. 2004;5:194. doi: 10.1186/1471-2105-5-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radmacher MD, Mcshane LM, Simon R. A paradigm for class prediction using gene expression profiles. J Comput Biol. 2002;9(3):505–11. doi: 10.1089/106652702760138592. [DOI] [PubMed] [Google Scholar]
- 28.Lehmann EL, Stein C. On the theory of some nonparametric hypotheses. Ann Math Stat. 1949;20:28–45. [Google Scholar]
- 29.Benjamini Y, Yosef Hochberg. “Controlling the false discovery rat: a practical and powerful approach to multiple testing”. J Royal Stat Society Series B Methodology. 1995;57(1):289–300. [Google Scholar]
- 30.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Nat Acad Sci U S A. 95:14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morrissey ER, Diaz-Uriarte R. Pomello II: finding differentially expressed genes. Nucl Acids Res. 2009;37:W581–6. doi: 10.1093/nar/gkp366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reich M, Liefeld T, Gould J, Lerner J, Tamayo P. GenePattern 2.0. Nature Genetics. 2006;38(5):500–1. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 33.Breitkreutz B, Stark C, Tyers M. Osprey: a network visualization system. BMC Genome Biology. 2003;4:R22. doi: 10.1186/gb-2003-4-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gene Ontology Consortium Creating the gene ontology resource: design and implementation. Genome Res. 2001;11:1425–33. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stranger BE, Forrest MS, Clark AG, et al. Genome-wide associations of gene expression variation in Humans. PLoS Genetics. 2005;1(6):e78. doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oleksiak MF, Churchill GA, Crawford DL. Variation in gene expression within and among natural populations. Nature Genetics. 2002;32:261–6. doi: 10.1038/ng983. [DOI] [PubMed] [Google Scholar]
- 37.Gorlov IP, Gallick GE, Gorlova OY, et al. GWS meets microarray: are the results of genome-wide association studies and gene expression profiling consistent? Prostate cancer as an example. PLoS One. 2009;4(8):e6511. doi: 10.1371/journal.pone.0006511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park J, Wachoder S, Gail MH, et al. Estimation of effect size distribution from genome-wide association studies and implications for future discoveries. Nature Genetics. 2010;42(7):570–5. doi: 10.1038/ng.610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evangelo E, Maraganore DM. Ioannidis. Meta-analysis in genome-wide association data sets: strategies and application in parkinson disease. PLoS One. 2007;2:e196. doi: 10.1371/journal.pone.0000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ionnidis JP, Patsopoulos NA, Evangelou E. Heterogeneity in Meta-analyses in genome-wide association investigations. PLoS One. 2007;9:e841. doi: 10.1371/journal.pone.0000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Churbanov A, Vorechovsky I, Hicks C. A method for predicting changes in human gene splicing induced by genetic variants in context of cis-acting elements. BMC Bioinformatics. 2010;11:12. doi: 10.1186/1471-2105-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eleftherohorinou H, Wright V, Hoggart C, et al. Pathway analysis of GWAS provides new insights into genetic susceptibility to 3 inflammatory diseases. PLoS One. 2009;4(11):e8068. doi: 10.1371/journal.pone.0008068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Askland K, Read C, Moore J. Pathway based analysis of whole-genome association study data in bipolar disorder reveal genes mediating ion channel activity and synaptic neurotransmission. Hum Genet. 2009;125:63–79. doi: 10.1007/s00439-008-0600-y. [DOI] [PubMed] [Google Scholar]
- 44.Baranzini SE, Galwey NW, Wang J, et al. Pathway and network analysis of genome-wide association studies in multiple sclerosis. Hum Mol Genet. 2009;18(11):2078–90. doi: 10.1093/hmg/ddp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haiman CA, Hsu C, de Bakker PIW, et al. Comprehensive association testing of common genetic variation in DNA repair pathway genes in relationship with breast cancer risk in multiple populations. Hum Mol Genet. 2008;17(6):825–34. doi: 10.1093/hmg/ddm354. [DOI] [PubMed] [Google Scholar]
- 46.Menashe I, Maeder D, Garcia-Closas M, et al. Pathway analysis of breast cancer genome-wide association study highlights three pathways and one canonical signaling cascade. Cancer Res. 2010;7(11):4453–9. doi: 10.1158/0008-5472.CAN-09-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Torkamani A, Topol EJ, Schork NJ. Pathway analysis of seven common diseases assessed by genome-wide association. Genomics. 2008;92:265–78. doi: 10.1016/j.ygeno.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Amer J Hum Genet. 2008;83:311–21. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhatia G, Bansal V, Harismendy O, et al. A covering method for detecting genetic associations between rare variants and common phenotypes. PLoS Comput Biol. 2010;6(10):e1000954. doi: 10.1371/journal.pcbi.1000954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mayer KB, Maia A, O’Reilley M. Allele-specific up-regulation of FGFR2 increases susceptibility to breast cancer. PLoS Biology. 2008;6(5):e108. doi: 10.1371/journal.pbio.0060108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huijts PEA, Vreeswijk MPG, et al. Clinical correlates of low-risk variants in FGFR2, TNRC9, MAP3K1, LSP1, and 8q24 in Dutch cohort of incident breast cancer cases. Breast Cancer Res. 2007;9:R78. doi: 10.1186/bcr1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.