Abstract
Background
During previous influenza pandemics, many deaths were associated with secondary bacterial infection. In April 2009, a previously unknown 2009 influenza A virus (2009 H1N1) emerged, causing a global influenza pandemic. We examined the relationship between circulating 2009 H1N1 and the occurrence of secondary bacterial parapneumonic empyema in children.
Methods
Children hospitalized with parapneumonic empyema from August 2004 to July 2009, including a period when the 2009 H1N1 circulated in Utah, were identified using International Classification of Diseases, Ninth Revision codes. We compared the average number of children diagnosed with influenza A and the number of admissions for empyema per month for the previous 4 seasons to rates of empyema during the 2009 H1N1 outbreak. We identified causative bacteria using culture and polymerase chain reaction (PCR).
Results
We observed an increase in hospitalization of children with pneumonia complicated by empyema during a severe outbreak of 2009 H1N1 during the spring and summer of 2009, compared with historical data for the previous 4 seasons. Streptococcus pneumoniae and Streptococcus pyogenes were the predominant bacteria identified.
Conclusions
Similar to previous pandemics, secondary bacterial infection with S. pneumoniae and S. pyogenes were associated with the 2009 H1N1 outbreak. There is an urgent need to better understand bacterial complications of pandemic influenza. In the interim, influenza vaccines, antiviral agents, and pneumococcal vaccines should be used to prevent cases of secondary bacterial pneumonia whenever possible.
Keywords: 2009 pandemic influenza A (H1N1), parapneumonic empyema, complication, children
In April 2009, a previously unknown influenza A (H1N1) virus strain was isolated from individuals presenting with severe respiratory tract infection in Mexico1 and from children in Southern California.2 In less than 6 months, the novel virus 2009 influenza A (2009 H1N1) had been detected in more than 191 countries, and the World Health Organization declared a global pandemic.3 In the United States, 50,768 confirmed cases with 1544 deaths were reported as of October 2009.4
Many deaths during previous influenza pandemics are thought to have been associated with secondary bacterial infection involving the lower respiratory tract.5 There are few data on secondary bacterial infection associated with the current 2009 influenza A (H1N1) pandemic.6,7 A recent review of 36 deaths in the United States attributable to 2009 H1N1, documented 10 of 23 (43%) deaths for whom culture information was available, were associated with secondary bacterial infection.7 However, it is difficult to diagnose and document secondary bacterial infection in nonfatal cases. In Utah, children have high rates of invasive pneumococcal disease (IPD), and we have demonstrated that IPD, particularly pneumonia and empyema, increases in the weeks after seasonal influenza virus circulation in the community.8
The objective of this study was to examine the relationship between the widespread circulation of the 2009 H1N1 virus in Utah and the occurrence of secondary bacterial pneumonia and parapneumonic empyema in children.
METHODS
Setting and Study Population
We identified children younger than 18 years, hospitalized at Primary Children’s Medical Center (PCMC), Salt Lake City, Utah, for treatment of pneumonia complicated by empyema from August 2004 to July 2009, including a period when the 2009 H1N1 influenza virus circulated in Utah. PCMC is a 278-bed children’s hospital that serves both as a community pediatric hospital for Salt Lake County, Utah, and as a tertiary referral center.
Identification of Parapneumonic Empyema and Confirmed Bacterial Infection
For this study, pneumonia and empyema were identified using International Classification of Diseases, Ninth Revision (ICD-9) codes. We queried a computerized data record system, for cases of empyema among Utah children younger than 18 years, hospitalized at PCMC between August 2004 and July 2009 with ICD-9 discharge diagnosis codes specific for pneumonia (481) and empyema (510). Culture-confirmed infections from blood and pleural fluid with Streptococcus pneumoniae (481), Streptococcus pyogenes (482.31), Staphylococcus aureus (482.41 and 482.42), and Haemophilus influenzae (482.2) were also identified using ICD-9 codes. All identified cases were cross-linked to microbiology records. We excluded all children with pleural effusion after chest surgery. Isolates of S. pneumoniae were serotyped using the capsular swelling method at Baylor College of Medicine. Medical records were reviewed (K.K.A. and A.H.) to confirm the diagnoses. Empyema was defined as documentation of pneumonia with pleural fluid demonstrating pus, a positive bacterial culture or gram-stained smear or abnormal pleural fluid chemistry as defined by Light.9 All cases of empyema were drained by thoracocentesis, chest tube, or surgery. To further define bacterial pathogens responsible for empyema, while 2009 H1N1 virus circulated in Utah, we examined all available pleural fluids from children hospitalized with empyema during the study period, using PCR-based bacterial assays.
For the purpose of the study, influenza-like illness (ILI) was defined as fever (temperature of 100°F or 37.8°C or greater) and a cough and/or a sore throat in the absence of a known cause other than influenza.
Respiratory Viral Testing
The PCMC laboratory has tested for respiratory viruses (influenza virus, respiratory syncytial virus, parainfluenza virus, and adenovirus) using direct fluorescent antibody (DFA) and viral culture since December 2000; human metapneumovirus was added to the panel in 2005. DFA negative specimens undergo viral culture.8 For isolation and cohorting purposes, respiratory viral testing is performed on all children hospitalized with respiratory symptoms (eg, upper or lower respiratory tract infection, bronchiolitis, asthma, or bacterial or viral pneumonia).
From May 1 to July 2009, samples from all children hospitalized at PCMC with DFA-confirmed influenza A infection were further evaluated at the Utah Department of Health Virology Laboratory for influenza A typing by PCR, using protocols developed by the Centers for Disease Control and Prevention (CDC).7 During this time period, the 2009 H1N1 virus was by far the most common respiratory virus identified.
Additional Respiratory Viral Testing by PCR
For patients with empyema who had available nasal-pharyngeal aspirate samples, we performed additional PCR-based testing for respiratory viruses at Idaho Technology Inc. (ITI, Salt Lake City, UT) on an integrated diagnostic platform, the “FilmArray.”10 Influenza A was identified by nested PCR reactions specific for conserved gene segments coding for the matrix and nonstructural proteins. Subtyping was performed by amplification of unique regions of the hemagglutinin gene. Primers to distinguish 2009 H1N1 from seasonal H1 were designed based on alignment of 220 unique full-length sequences available in the NCBI database as of August 2009. The 2009 H1N1 primers were validated by testing of available 2009 H1N1 clinical samples obtained from clinical laboratories in several US regions. Nasal-pharyngeal aspirate samples were stored in M4 transport media and frozen at −80°C until testing.
Pleural Fluid Testing by PCR
Testing of pleural fluid for bacterial nucleic acid by PCR was performed at ITI and the Streptococcus Laboratory, CDC (Atlanta, GA). At ITI, nucleic acids were extracted from pleural fluid using the IT 1-2-3 VIBE kit, a spin column purification process. After extraction, a two-step nested PCR reaction was performed. The first-stage PCR was performed on a block thermocycler. Second-stage real time PCR was performed in glass capillaries in the ITI Joint Biologic Agent Identification and Diagnostic System instrument. Second-stage PCR primers target species-specific sequences within the first-stage amplicon. Real time fluorescence was generated by the melting dye LC Green Plus (ITI), and amplicon identity was confirmed by melt analysis. Targets for amplification included: (1) S. pneumoniae—pneumolysin (ply), autolysin (lytA), and ribonucleic acid polymerase B (rpoB); (2) S. aureus—mecA, nuclease (nuc), and the Panton-valentine leukocidin; (3) S. pyogenes and H. influenzae—rpoB.
At the CDC, DNA was extracted from 50 to 200 μL of pleural fluid as described previously (available at: http://www.cdc.gov/ncidod/biotech/strep/pcr.htm).11 S. pneumoniae and S. pyogenes were detected using real time PCR assays for the lytA and spy genes, respectively. For lytA-positive specimens, pneumococcal serotype deduction was achieved using the sequential multiplex PCR strategy as described previously12 with updates for clinical samples described at http://www.cdc.gov/ncidod/biotech/strep/pcr.htm.
Analysis
We calculated the average number of children diagnosed with confirmed influenza A by DFA and culture and the average number of admissions for empyema at PCMC per month for the 4 seasons from August 2004 to July 2008 and calculated 95% confidence intervals around the mean monthly frequency. We compared the frequency of empyema during the 2009 H1N1 outbreak to historical frequencies during 4 previous spring seasons.
RESULTS
2009 H1N1 in Utah and Parapneumonic Empyema
The first confirmed case of the 2009 H1N1 infection was reported in Utah on May 2, 2009, in a patient hospitalized on April 27, 2009 with ILI.13 Between May 1 and July 31, 2009, 607 children were diagnosed with 2009 H1N1 infection at PCMC; 117 (19%) were hospitalized (Fig. 1). The median age of children hospitalized with the 2009 H1N1 infection was 46 months (range 9 days to 196 months).
FIGURE 1.
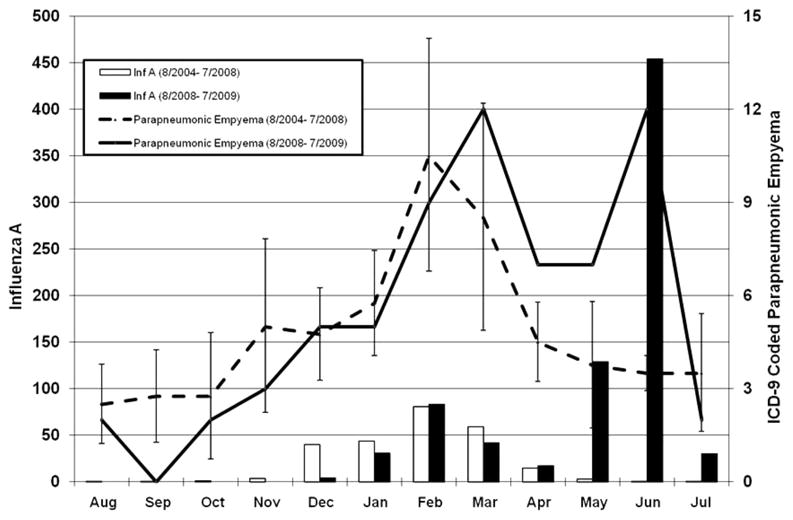
Monthly numbers of confirmed parapneumonic empyema admissions for the 2008–2009 influenza season (solid line) and monthly average with 95% confidence intervals for 4 influenza seasons from August 2004 to July 2008 (dashed line). The solid and open bars indicates influenza A cases by DFA, culture or PCR during the 2008–2009 and August 2004 to July 2008 seasons, respectively. All data are from Primary Children’s Medical Center, Salt Lake City, Utah.
The average historic rate per month of parapneumonic empyema from January through July of 2004 to 2008 was 5.8, 10.5, 8.5, 4.5, 3.8, 3.5, and 3.5 cases per month, compared with 5, 9, 12, 7, 7, 12, and 2 cases per month during the same period in 2009. From May 1 to July 31, 2009, 21 children were admitted to PCMC with confirmed parapneumonic empyema. The historical average (2004 to 2008) cumulative cases during the same period was 10.8 (95% confidence interval 9.5–12; range: 9–12) (P < 0.001; one-sample t test). Twelve cases of empyema occurred during the period of peak 2009 H1N1 in June, compared with a historical rate of 3.5 cases per month in June (95% confidence interval 2.9–4.1; range: 3–4) (P < 0.001; one-sample t test) (Fig. 1). In each of the 4 previous influenza seasons, a peak in hospitalizations for empyema occurred between January and March, correlating with peak seasonal influenza A activity. During the 2008–2009 season, there were 2 periods of peak hospitalization for empyema. The first peak activity was in March coinciding with circulating seasonal influenza A (H1) and influenza B,14 and the second in June, coinciding with 2009 H1N1 activity (Fig. 1).
Clinical and Microbiological Findings Among Children With Parapneumonic Empyema During the 2009 H1N1 Pandemic
The median age of the 21 children with empyema was 37 months (range: 17 to 204 months); 67% were younger than 5 years (Table 1). During the same time period the median age of 114 children hospitalized with 2009 H1N1 infection without empyema was 47 months (range 0.3 to 199 months). Four children (20%) had an underlying medical condition (3 had asthma, and 1 had gastro-esophageal reflux disease).
TABLE 1.
Summary of Demographic, Clinical, and Microbiologic Data of 21 Children Hospitalized at PCMC, May 1, 2009 to July 31, 2009
| Case | Age (mo) | Sex | Medical History | Time From Onset of ILI until Diagnosis of Empyema (d) | Chest Tube vs. VATS | Length of Hospital Stay | Influenza A by PCR | Blood and/or Pleural Fluid Bacterial Culture Results (Serotype) | Bacterial PCR of Pleural Fluid at CDC (Serotype) |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 100 | F | Healthy | 10 | VATS | 4 | Detected (seasonal) | No growth | S. pyogenes |
| 2 | 85 | F | Asthma | 3 | CT | 6 | Not detected | No growth | S. pneumoniae (7F/7A) |
| 3 | 181 | M | Healthy | 6 | CT | 5 | Not detected | No growth | No specimen |
| 4 | 204 | F | Autism | 7 | VATS | 21 | Not detected | No growth | S. pneumoniae (7F/7A) |
| 5 | 23 | M | Healthy | 18 | CT | 4 | Not detected | No growth | No specimen |
| 6 | 24 | F | GERD | 12 | CT | 6 | Not detected | No growth | S. pneumoniae (3) |
| 7 | 40 | M | Healthy | 5 | CT | 7 | No specimen | S. pyogenes* | S. pyogenes |
| 8 | 91 | M | Healthy | 11 | CT | 10 | No specimen | No growth | S. pneumoniae (7F/7A) |
| 9 | 37 | F | Asthma | 4 | CT | 12 | Detected (2009 H1N1) | S. pneumoniae (7F)† | S. pneumoniae (7F/7A) |
| 10 | 127 | F | Healthy | 14 | CT | 7 | No specimen | No growth | Negative |
| 11 | 36 | F | Healthy | 7 | CT | 7 | Not detected | No growth | S. pneumoniae (7F/7A) |
| 12 | 35 | F | Healthy | 8 | VATS | 11 | Detected (2009 H1N1) | S. pyogenes* | S. pyogenes |
| 13 | 31 | M | Healthy | 11 | VATS | 12 | No specimen | No growth | No specimen |
| 14 | 17 | F | Healthy | 14 | CT | 10 | Not detected | No growth | S. pneumoniae (7F/7A) |
| 15 | 138 | F | Healthy | 10 | VATS | 9 | Not detected | S. pneumoniae (1)* | S. pneumoniae (1) |
| 16 | 19 | F | Healthy | 5 | CT | 26 | Not detected | S. pneumoniae (3)† | No specimen |
| 17 | 25 | M | Healthy | 14 | T | 9 | Not detected | No growth | Negative |
| 18 | 72 | M | Healthy | 5 | VATS | 7 | Detected (2009 H1N1) | S. pneumoniae (7F)*† | S. pneumoniae (7F/7A) |
| 19 | 27 | M | Healthy | 4 | CT | 7 | Detected (2009 H1N1) | S. pneumoniae (7F)* | S. pneumoniae (7F/7A) |
| 20 | 37 | M | Healthy | 5 | CT/VATS | 9 | No specimen | No growth | No specimen |
| 21 | 32 | M | Asthma | 10 | VATS | 5 | No specimen | No growth | No specimen |
Pleural fluid sample.
Blood sample.
M indicates male; F, female; T, thoracocentesis; CT, chest tube; VATS, video-assisted thoracoscopic surgery; PCR, polymerase chain reaction; GERD, gastro-esophageal reflux disease.
All children hospitalized during the study period with empyema had a history of an ILI, a median of 8 days (range 3 to 18 days), before the diagnosis of empyema. Five children (24%) were treated with oseltamivir at a median of 4 days (range 1 to 9 days) after the onset of their ILI. Only 1 received treatment with oseltamivir within 2 days of onset of ILI symptoms. The median length of hospital stay was 7 days (range 4 to 26 days). In contrast, the median length of stay of children hospitalized with 2009 H1N1 infection without empyema was 3 days (range 1 to 31 days) (P < 0.001). At the time of hospitalization for empyema, respiratory viral testing by DFA and culture was performed for 18 (86%) children. Influenza A virus was detected in 1 of 18 (6%). Residual nasopharyngeal specimens were available for viral testing by PCR in 15 children; 5 (33%) were positive for influenza (4—2009 H1N1, 1—seasonal influenza A).
Blood and/or pleural fluid samples were obtained for bacterial culture from 21 children with empyema (Table 1). A pathogen was isolated by culture from 7 (33%) children; 3 from blood and 5 from pleural fluid (1 patient was culture-positive in both blood and pleural fluid). S. pneumoniae was isolated from 5 children and S. pyogenes from 2 children by culture. Serotyping of S. pneumoniae revealed 3 isolates of 7F and 1 isolate each of serotype 1 and 3.
Fifteen pleural samples were available for testing by PCR. S. pneumoniae was detected in 10 (66%) samples and S. pyogenes in 3 (20%) samples at both ITI and CDC. The use of PCR compared with culture alone increased identification of a bacterial pathogen from 40% (6 of 15) to 80% (13 of 15) among those tested by both methods.
PCR identified the same bacterial organism as culture in 6 children with positive blood or pleural fluid cultures. Neither S. aureus nor H. influenzae was detected in pleural fluid samples tested by PCR. Further testing of S. pneumoniae positive samples at CDC using PCR-based capsular serotype deduction12 identified serotype 7F/(7A) in 8 samples, serotype 3 and 1 and in 1 sample each (note that 7A is a rare serotype that is currently not resolvable from 7F in the PCR serotype assay).
DISCUSSION
There are few data on the frequency and spectrum of secondary bacterial infections associated with the recent 2009 H1N1 infection.6,7,15 In this study, we observed an increase in hospitalization of children with pneumonia complicated by empyema during a severe outbreak of 2009 H1N1 during the spring and summer of 2009, compared with historical data over 4 seasons. All children reported an ILI before hospitalization for pneumonia with empyema. Among children hospitalized with empyema, one-third of those tested a median of 8 days after symptom onset ILI had influenza A detected by PCR, 4 of these were 2009 H1N1 influenza. S. pneumoniae and S. pyogenes were the most common cause of empyema during the 2009 H1N1 outbreak. Although a temporal association does not prove causality, the increase in empyema may represent bacterial pneumonia complicating 2009 H1N1 infection. Further supporting evidence comes from an increase in IPD among young adults in the Denver area coincident with an increase in influenza-associated hospitalizations in October of 2009 (CDC, unpublished data, available at: http://www.cdc.gov/h1n1flu/in_the_news/pneumococcal.htm, accessed December 3, 2009).
The association between pandemic and seasonal influenza and pneumococcal infection has been appreciated for decades.5,16,17 Morens et al16 reviewed autopsy specimens from the victims of the 1918–1919 pandemic and the microbiologic literature of the time, and concluded that the majority of influenza deaths were due to secondary bacterial pneumonia. Outbreaks of severe pneumococcal disease after infection with seasonal influenza have been described and several studies have reported correlation between seasonal influenza and IPD.8,18–20 In vitro and animal and human studies suggest that there is synergy between respiratory viral infections and bacterial infections, but the mechanisms remain poorly understood.21,22
In previous pandemics, before the use of pneumococcal conjugate vaccine or the emergence of community-acquired methicillin-resistant S. aureus (MRSA), S. pneumoniae, S. pyogenes, and less commonly S. aureus pneumonia were the dominant causes of secondary bacterial infection.16 Bacteriologic data are extremely limited during the current pandemic. Using newer molecular methods, we could demonstrate a bacterial etiology in 80% of children with empyema complicating pneumonia during the peak of 2009 H1N1 activity. Each of these children had either S. pneumoniae or S. pyogenes infections. Despite the use of several molecular targets for detection of S. aureus, we did not observe any staphylococcal empyema. Our findings are somewhat similar to a recent review of 77 postmortem lung specimens from adults and children with pneumonia during the 2009 H1N1 outbreak.7 Twenty-two (29%) specimens had histopathologic, immunohistochemical, and molecular evidence of coinfection with an identified bacterium. S. pneumoniae or S. pyogenes either alone or in combination with other bacteria were identified in 16 of 22 (72%) of positive specimens, whereas S. aureus alone or in combination with other bacteria were identified in 7 of 22 (32%) lung specimens.6 In contrast, a recent review of 36 children who died with 2009 H1N1 infection, a bacterial infection was documented in 10 of 23 (43%) in whom pathology or microbiology results were available. Half were due to S. aureus (including 3 MRSA), 3 were S. pneumoniae, and one each were S. pyogenes and Streptococcus constellatus.7 Infection with MRSA has increasingly been reported as a cause of severe and often fatal pneumonia after seasonal influenza.23 Because MRSA pneumonia was relatively uncommon in Utah before the arrival of 2009 H1N1,17 the absence of MRSA as an etiology of pneumonia among the cases described here may reflect preexisting regional differences and predominance of S. pneumoniae as the etiology of bacterial pneumonia in Utah.
All of the S. pneumoniae identified from children with empyema in our study were of non-PCV7 serotypes, consistent with the increasing role of these serotypes in IPD and parapneumonic empyema in the United States and Europe.24–27 These findings are also consistent with a recent review of the changing epidemiology of IPD among Utah children over a 13-year period. More than 95% of S. pneumoniae isolated from 2005 to 2008 were non-PCV7 serotypes; serotypes 7F, 19A, 1, and 3 were the most common.28 Fortunately, each of these 4 serotypes is included in the 13-valent pneumococcal conjugate vaccine licensed in February 2010 by the Food and Drug Administration. Because receipt of pneumococcal conjugate vaccine has been associated with a 45% reduction in risk of influenza-associated hospitalization,29 the use of 13-valent conjugate vaccine represents an opportunity to reduce influenza-associated morbidity and mortality, either directly for children younger than 5 years or through indirect effects on older children and adults.
Cases similar to those described here are likely to continue until the pandemic is over and thus represent an opportunity for prevention. Children and adults with signs and symptoms of influenza infection should be promptly and empirically treated with antiviral agents according to current recommendations (available at: http://www.cdc.gov/H1N1flu/recommendations.htm). Providers should ensure that children are up to date on pneumococcal conjugate vaccine and, for those with certain high-risk medical conditions who are 2 years of age or older, pneumococcal polysaccharide vaccine (http://www.cdc.gov/H1N1flu/guidance/ppsv_h1n1.htm). Finally, persons with clinical findings consistent with pneumonia should be treated empirically with both antiviral medications and antibacterial agents until additional clinical and laboratory information can be used to guide therapy.
There are a number of limitations to the study. Virologic confirmation of prior 2009 H1N1 infection by PCR or by serology was not performed on all hospitalized children with empyema, and most specimens for molecular viral tests were obtained at least 1 week after the onset of ILI symptoms. However, the hospitalizations for IPD described in this report occurred during a period when other respiratory viruses associated with pneumonia and empyema, ie, RSV and hMPV were not in circulation. Second, although there may be variations in infection with bacterial pneumonia and empyema, from 2004 to 2008 the months of April to July have consistently been low periods for admissions with bacterial pneumonia and empyema. Third, the epidemiology of IPD in Utah is distinctive. A high proportion of circulating serotypes are ones that may have an increased propensity to cause complicated pneumonia, specifically serotype 1, 3, and 7F.24,30 Fourth, not all PCV7 vaccine records were available in children hospitalized with empyema. Finally, although we used strategies to minimize misclassification and performed manual review, ICD-9 coding is subject to error.
CONCLUSIONS
We observed an increase in hospital admissions due to bacterial pneumonia complicated by empyema among children temporally associated with the outbreak of the 2009 H1N1 in Utah. Molecular diagnostic testing improved our ability to identify the etiologic agents with S. pneumoniae and S. pyogenes most frequently isolated. However, when cases of suspected bacterial pneumonia complicating influenza do occur, empiric antibiotic therapy should target S. pneumoniae and S. pyogenes, and MRSA must be considered for severe illness or in regions with substantial circulation of MRSA. There is an urgent need to better understand bacterial complications of pandemic influenza and to enhance all available treatment and prevention strategies.
Acknowledgments
Supported by grants from the National Institute of Allergy and Infectious Diseases (U01 AI074419-01) (to A.J.B., M.P., C.L.B.), (U01A1082482) (to K.A., C.L.B.), (U01AI082184-01) (to A.J.B., A.T.P.), (K23-AI079401-01A1) (to A.J.B.), National Institute of Child Health and Human Development (K-24 HD047249-01A1) (to C.L.B.), and the Centers for Disease Control Prevention (U18-IP000303-01) (to K.A., A.J.B., C.L.B., A.T.P.). This project was further supported by the Children’s Health Research Center at the University of Utah and Primary Children’s Medical Center Foundation. M.P. and C.H. are employees of Idaho Technology Incorporated.
The authors thank Dr Bernard W. Beall, Dr Matt Moore, and Dr Cynthia Whitney of the CDC for their editorial contributions, Garrison Alder at PCMC and staff at Intermountain Health-care Primary Children’s Medical Center (PCMC) microbiology laboratory and Idaho Technology Incorporated laboratory.
Footnotes
Disclaimer: The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
References
- 1.Perez-Padilla R, de la Rosa-Zamboni D, Ponce de Leon S, et al. Pneumonia and respiratory failure from swine-origin influenza A (H1N1) in Mexico. N Engl J Med. 2009;361:680–689. doi: 10.1056/NEJMoa0904252. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention (CDC) Swine influenza A (H1N1) infection in two children–Southern California, March-April 2009. MMWR Morb Mortal Wkly Rep. 2009;58:400–402. [PubMed] [Google Scholar]
- 3. [Accessed October 12, 2009];Pandemic (H1N1) 2009—update 67 (20 September 2009) 2009 Available at: http://www.who.int/csr/don/2009_09_25/en/index.html.
- 4. [Accessed October 12, 2009];FluView (2008–2009 Influenza Season Week 39 ending October 3, 2009) 2009 Available at: http://www.cdc.gov/flu/weekly/
- 5.Morens DM, Fauci AS. The 1918 influenza pandemic: insights for the 21st century. J Infect Dis. 2007;195:1018–1028. doi: 10.1086/511989. [DOI] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention (CDC) Bacterial coinfections in lung tissue specimens from fatal cases of 2009 pandemic influenza A (H1N1)—United States, May–August 2009. MMWR Morb Mortal Wkly Rep. 2009;58:1071–1074. [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention (CDC) Surveillance for pediatric deaths associated with 2009 pandemic influenza A (H1N1) virus infection—United States, April–August 2009. MMWR Morb Mortal Wkly Rep. 2009;58:941–947. [PubMed] [Google Scholar]
- 8.Ampofo K, Bender J, Sheng X, et al. Seasonal invasive pneumococcal disease in children: role of preceding respiratory viral infection. Pediatrics. 2008;122:229–237. doi: 10.1542/peds.2007-3192. [DOI] [PubMed] [Google Scholar]
- 9.Light RW. Parapneumonic effusions and empyema. Proc Am Thorac Soc. 2006;3:75–80. doi: 10.1513/pats.200510-113JH. [DOI] [PubMed] [Google Scholar]
- 10.Poritz MA, Meyers L, Lewis A, et al. Analysis of 250 pediatric NPA samples for 22 respiratory pathogens using an automated nested multiplex PCR platform. Pan American Society for Clinical Virology 24th Clinical Virology Symposium; Daytona Beach. 2008. [Google Scholar]
- 11.Carvalho Mda G, Tondella ML, McCaustland K, et al. Evaluation and improvement of real-time PCR assays targeting lytA, ply, and psaA genes for detection of pneumococcal DNA. J Clin Microbiol. 2007;45:2460–2466. doi: 10.1128/JCM.02498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pai R, Gertz RE, Beall B. Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae isolates. J Clin Microbiol. 2006;44:124–131. doi: 10.1128/JCM.44.1.124-131.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flu Activity and Surveillance. [Accessed September 29, 2009];2009 Available at: http://www.cdc.gov/flu/weekly/fluactivity.htm.
- 14.2008–2009 Weekly Utah Influenza Activity Public Health Professionals and Clinicians Update. [Accessed October 12, 2009];2009 Available at: http://health.utah.gov/epi/diseases/influenza/surveillance/2008-09_Season/PH&C_040809_wk13_flu_posting.pdf.
- 15.Jain S, Kamimoto L, Bramley AM, et al. Hospitalized patients with 2009 H1N1 influenza in the United States, April–June 2009. N Engl J Med. 2009;361:1935–1944. doi: 10.1056/NEJMoa0906695. [DOI] [PubMed] [Google Scholar]
- 16.Morens DM, Taubenberger JK, Fauci AS. Predominant role of bacterial pneumonia as a cause of death in pandemic influenza: implications for pandemic influenza preparedness. J Infect Dis. 2008;198:962–970. doi: 10.1086/591708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bender JM, Ampofo K, Sheng X, et al. Parapneumonic empyema deaths during past century, Utah. Emerg Infect Dis. 2009;15:44–48. doi: 10.3201/eid1501.080618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Talbot TR, Poehling KA, Hartert TV, et al. Seasonality of invasive pneumococcal disease: temporal relation to documented influenza and respiratory syncytial viral circulation. Am J Med. 2005;118:285–291. doi: 10.1016/j.amjmed.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 19.O’Brien KL, Walters MI, Sellman J, et al. Severe pneumococcal pneumonia in previously healthy children: the role of preceding influenza infection. Clin Infect Dis. 2000;30:784–789. doi: 10.1086/313772. [DOI] [PubMed] [Google Scholar]
- 20.Grabowska K, Hogberg L, Penttinen P, et al. Occurrence of invasive pneumococcal disease and number of excess cases due to influenza. BMC Infect Dis. 2006;6:58. doi: 10.1186/1471-2334-6-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCullers JA. Insights into the interaction between influenza virus and pneumococcus. Clin Microbiol Rev. 2006;19:571–582. doi: 10.1128/CMR.00058-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McAuley JL, Hornung F, Boyd KL, et al. Expression of the 1918 influenza A virus PB1-F2 enhances the pathogenesis of viral and secondary bacterial pneumonia. Cell Host Microbe. 2007;2:240–249. doi: 10.1016/j.chom.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finelli L, Fiore A, Dhara R, et al. Influenza-associated pediatric mortality in the United States: increase of Staphylococcus aureus coinfection. Pediatrics. 2008;122:805–811. doi: 10.1542/peds.2008-1336. [DOI] [PubMed] [Google Scholar]
- 24.Byington CL, Korgenski K, Daly J, et al. Impact of the pneumococcal conjugate vaccine on pneumococcal parapneumonic empyema. Pediatr Infect Dis J. 2006;25:250–254. doi: 10.1097/01.inf.0000202137.37642.ab. [DOI] [PubMed] [Google Scholar]
- 25.Eastham KM, Freeman R, Kearns AM, et al. Clinical features, aetiology and outcome of empyema in children in the north east of England. Thorax. 2004;59:522–525. doi: 10.1136/thx.2003.016105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Obando I, Munoz-Almagro C, Arroyo LA, et al. Pediatric parapneumonic empyema, Spain. Emerg Infect Dis. 2008;14:1390–1397. doi: 10.3201/eid1409.071094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singleton RJ, Hennessy TW, Bulkow LR, et al. Invasive pneumococcal disease caused by nonvaccine serotypes among Alaska native children with high levels of 7-valent pneumococcal conjugate vaccine coverage. JAMA. 2007;297:1784–1792. doi: 10.1001/jama.297.16.1784. [DOI] [PubMed] [Google Scholar]
- 28.Ampofo K, Sheng X, Korgenski EK, et al. The changing age and serotype distribution of invasive pneumococcal disease among Utah children during the PCV7 Era; Implications for the new pneumococcal vaccine (4315) Baltimore: Pediatric Academic Society; 2009. [Google Scholar]
- 29.Madhi S, Klugman K, Group TVT. A role for Streptococcus pneumoniae in virus-associated pneumonia. Nat Med. 2004;10:811–813. doi: 10.1038/nm1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bender JM, Ampofo K, Korgenski K, et al. Pneumococcal necrotizing pneumonia in Utah: does serotype matter? Clin Infect Dis. 2008;46:1346–1352. doi: 10.1086/586747. [DOI] [PMC free article] [PubMed] [Google Scholar]


