Abstract
Huntington disease (HD) is caused by polyglutamine expansion in the huntingtin (HTT) protein. Huntingtin-interacting protein 14 (HIP14), one of 23 DHHC domain-containing palmitoyl acyl transferases (PATs), binds to HTT and robustly palmitoylates HTT at cysteine 214. Mutant HTT exhibits reduced palmitoylation and interaction with HIP14, contributing to the neuronal dysfunction associated with HD. In this study, we confirmed that, among 23 DHHC PATs, HIP14 and its homolog DHHC-13 (HIP14L) are the two major PATs that palmitoylate HTT. Wild-type HTT, in addition to serving as a palmitoylation substrate, also modulates the palmitoylation of HIP14 itself. In vivo, HIP14 palmitoylation is decreased in the brains of mice lacking one HTT allele (hdh+/−) and is further reduced in mouse cortical neurons treated with HTT antisense oligos (HTT-ASO) that knockdown HTT expression by ∼95%. Previously, it has been shown that palmitoylation of DHHC proteins may affect their enzymatic activity. Indeed, palmitoylation of SNAP25 by HIP14 is potentiated in vitro in the presence of wild-type HTT. This influence of HTT on HIP14 activity is lost in the presence of CAG expansion. Furthermore, in both brains of hdh+/− mice and neurons treated with HTT-ASO, we observe a significant reduction in palmitoylation of endogenous SNAP25 and GluR1, synaptic proteins that are substrates of HIP14, suggesting wild-type HTT also influences HIP14 enzymatic activity in vivo. This study describes an important biochemical function for wild-type HTT modulation of HIP14 palmitoylation and its enzymatic activity.
INTRODUCTION
Palmitoylation is the posttranslational addition of the saturated 16 carbon lipid palmitate via a thioester linkage to specific cysteine residues (1). Many neuronal proteins are palmitoylated, including ion channels, cell adhesion molecules, scaffolding molecules, neurotransmitter release machinery and signaling proteins. Huntingtin (HTT), which, when in its polyglutamine expanded form, is the genetic basis of Huntington disease (HD), is an important palmitoylated protein (2–4). Palmitoylation is mediated by a group of enzymes called palmitoyl acyl transferases (PATs) characterized by a cysteine-rich domain with a defining Asp–His–His–Cys (DHHC) core motif, which is essential for PAT activity both in vitro and in vivo (3,5–7).
One of the first identified mammalian DHHC containing PATs was huntingtin-interacting protein 14 (HIP14 or DHHC-17), which palmitoylates HTT (3,8). Accumulating evidence suggests that aberrant palmitoylation of different proteins may contribute to the pathogenesis of HD (3,9,10). The polyglutamine expansion in mutant HTT results in reduced interaction with HIP14 (9) and, in turn, reduced palmitoylation of HTT, which causes accelerated inclusion formation and enhanced neuronal toxicity and cell death (9). In addition to that of HTT, palmitoylation and function of glial glutamate transporter-1 (GLT-1) are also reduced in the YAC128 mouse model of HD. Impaired GLT-1 palmitoylation is present early in the pathogenesis of HD and may contribute to decreased glutamate uptake, excitotoxicity and, ultimately, neuronal cell death in HD (10).
Multiple studies in cellular and animal model systems indicate that mutant HTT imparts a novel toxic function, and the mechanism of toxicity involves multiple pathogenic pathways (11). However, the normal function of wild-type HTT, an essential cellular protein in higher vertebrates, is not yet well understood (12). HTT is encoded by a single gene and is ubiquitously expressed in mammals. It is primarily a cytoplasmic protein, known to be both vesicle- and microtubule-associated (13,14). Deletion of the HTT gene is lethal early in mouse embryogenesis (15–17). The multi-domain structure of HTT indicates that the protein may have multiple distinct cellular roles, including transcriptional regulation, nucleo-cytoplasmic shuttling, vesicular trafficking and intracellular transport in the cell, synaptic function and anti-apoptotic activity (reviewed in 12,18).
Accumulating data on the roles of wild-type HTT in endocytosis, endosomal motility and axonal transport have led to an emerging model for HTT as an integrator of protein transport along the cellular cytoskeleton (reviewed in 12). HIP14, which is predominantly expressed in the Golgi apparatus, is also found to be associated with endosomal vesicles in the soma, presynaptic terminals and dendritic spines. It is possible that trafficking of these HIP14-containing vesicles to sites where it executes its enzymatic function may require HTT. Given the multi-domain nature of HTT and its strong interaction with HIP14, HTT may also function as an allosteric activator of HIP14. In this study, we tested the hypothesis that wild-type HTT, in addition to serving as a substrate of HIP14, also directly influences HIP14 enzymatic activity.
RESULTS
HIP14 and HIP14L are the two major HTT PATs
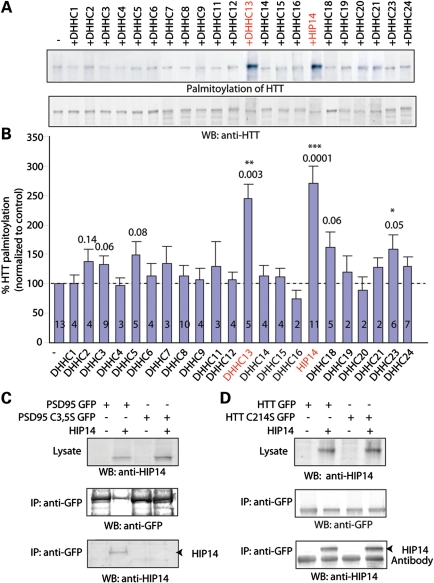
We have previously shown that HTT is palmitoylated by HIP14 and HIP14L (DHHC-13), a HIP14 homolog (3,8,9). In fact, palmitoylation of HTT by HIP14 is essential for its trafficking and function (9). The identification of 23 DHHC proteins in this PAT family raised the question whether other DHHC proteins palmitoylate HTT (5). We therefore applied the acyl biotin exchange (ABE) labeling method to examine the palmitoylation level of HTT upon overexpression of individual DHHC proteins along with HTT in COS cells. We found that HIP14 and HIP14L are the two major PATs that significantly increase palmitoylation of HTT (Fig. 1A and B).
Figure 1.
HIP14 and HIP14L are the two major PATs for HTT. (A) COS cells expressing either HTT alone or HTT with individual DHHC protein. HTT was immunoprecipitated, followed by the biotin palmitoylation labeling assay. Among the indicated DHHC proteins, HIP14L (DHHC-13) and HIP14 (DHHC-17) significantly increased palmitoylation of HTT. (B) Quantification of HTT palmitoylation increase by individual DHHC protein. Palmitoylation of HTT by an individual enzyme was normalized by western blot and compared with control. Number of repeats was indicated in the graph. (C) Interaction between PSD95 and HIP14 requires the presence of palmitoylated cysteines C3, 5. Mutation of cysteines 3 and 5 in PSD95 abolished its interaction with HIP14 detected by co-IP. (D) In contrast, association of wild-type HTT to HIP14 was independent of palmitoylation of HTT.
Interaction of HIP14 and HTT is unique and distinct from other palmitoylation enzyme–substrate interactions
Co-transfection of a palmitoylation substrate protein with its cognate PAT has been found in a number of cases to result in a long-lived enzyme–substrate interaction seen both by co-localization and co-immunoprecipitation (co-IP) (5,8,19). These interactions typically require that both the substrate protein and PAT be competent for palmitoylation, with interactions not seen either with the mutational removal of substrate protein cysteinyl acceptors or when the co-transfected PAT is mutationally inactivated, suggesting that the interaction may reflect a continued association of enzyme and substrate that may persist following the productive palmitoylation (5,8,19). As an example of this phenomenon, here, we find by co-IP that wild-type PSD95, but not the non-palmitoylatable PSD95 C3,5S mutant interacts with its cognate PAT, HIP14 (Fig. 1C). Interestingly, this is not the case for HTT. The strong HTT interaction with HIP14 persists even when HTT palmitoylation is severely impaired by the C214S mutation of the primary HTT cysteinyl acceptor site (9) (Fig. 1D). Thus, in this regard, the HTT–HIP14 interaction appears to be fundamentally different from the interactions of other palmitoylation substrates with their PATs, in that the stable HTT–HIP14 association does not require the palmitoylated HTT cysteine.
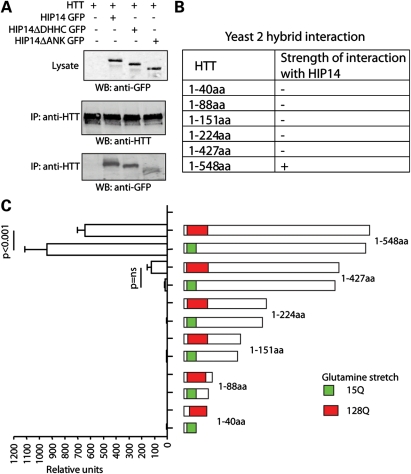
HIP14 and HIP14L are unique among the 23 DHHC proteins in that both contain ankryin repeat domains, mapping within the cytoplasmically disposed N-terminal domains of these two PATs (20). We have constructed two HIP14 deletion mutants that remove either the ankryin repeat domain (HIP14 ΔANK) or the catalytic DHHC domain (HIP14 ΔDHHC). Both mutants were tested for their effects on the HTT–HIP14 interaction (Fig. 2A). While HIP14 ΔANK shows a markedly impaired interaction with HTT, HIP14 ΔDHHC shows a surprisingly robust interaction, indistinguishable from that seen with wild-type HIP14. This lack of reliance on DHHC domain integrity and thus on PAT activity again indicates that the HTT–HIP14 interaction is fundamentally different from those documented for other DHHC PAT–substrate interactions (5,8,19).
Figure 2.
Ankyrin repeats of HIP14 and N-terminus (1–548 amino acids) of HTT are crucial for their interaction. (A) HTT was IPed from COS cells expressing HTT alone or HTT with HIP14 GFP, HIP14 ΔANK GFP or HIP14 ΔDHHC GFP. The amount of HIP14 co-IPed was detected by western blotting with anti-GFP antibody. Deletion of ankyrin repeats significantly reduces the association of HIP14 and HTT. In contrast, deletion of DHHC catalytic domain does not affect the interaction. (B and C) Yeast two-hybrid semiquantitative β-galactosidase assays to map the region of HTT to which HIP14 binds. The strength of interaction is indicated by a semiquantitative scale of colony growth (with +, and − indicating strong and no growth, respectively), indicating that HIP14 interacts specifically with a region of HTT encompassing amino acids 427–548. Interaction between HIP14 and mutant HTT (1–548 amino acids) is significantly decreased compared with that of wild-type HTT.
Next, we used yeast-two-hybrid analysis to delineate the HTT domains that are required for this interaction (Fig. 2B and C). A series of HTT N-terminal fragments were examined. The 548 residue-long N-terminal fragment showed a quite strong interaction. Furthermore, as previously documented (20), reduced interaction is seen for mutant HTT which contains the glutamine expansion. The HTT–HIP14 interaction is quite severely impaired when the HTT fragment is shortened from 548 to 427, indicating that elements within the HTT 427–548 amino acids likely play a key role in this interaction. With further shortening, HIP14–HTT interaction was fully abolished (Fig. 2B and C). Note that the N-terminal HTT fragment 1–88 amino acids that contain exon 1 is unable to interact with HIP14.
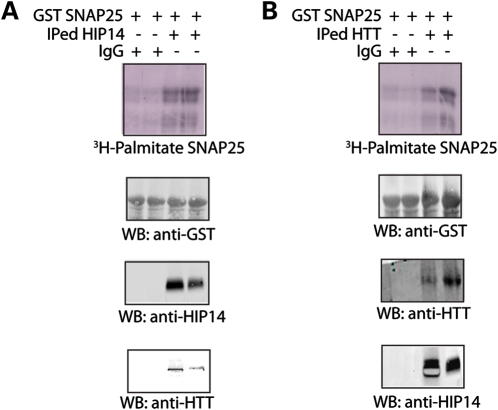
HTT and HIP14 form a tight complex that has enzymatic activity
We have previously showed that HIP14 is a PAT for SNAP25 (3). Consistent with this, immunoprecipitated (IP) HIP14 from rat brain lysate exhibited palmitoyl-transferase activity toward GST-SNAP25 in vitro (Fig. 3A). When HTT was IPed from rat brain lysate, the resulting protein surprisingly also increased palmitoylation of GST-SNAP25 (Fig. 3B). This suggested to us that HTT's binding partner, HIP14, was co-IPed resulting in PAT activity. Western blots (Fig. 3A and B, bottom panels) confirmed that, in all IPs, the respective binding partner was co-IPed. These results suggest that HTT and HIP14 form a tight complex that is enzymatically active.
Figure 3.
HIP14 and HTT form a tight complex, exhibiting palmitoyl-transferase activity. (A) IPed HIP14 from rat whole brain exhibited palmitoyl-transferase (PAT) activity toward GST-SNAP25. Unimmunized rabbit serum (control) or HIP14 antibody bound to sepharose beads was used to IP HIP14 from rat whole brain lysate. Then the beads containing IgG control or IPed HIP14 were added to reaction mix containing 3H-palmitoyl-CoA to palmitoylate GST-SNAP25 in vitro. As previously shown, IPed HIP14 is capable of increasing palmitoylation of SNAP25 compared with serum control (top panel). HIP14 IP was confirmed by western blot with HIP14 antibody and co-IPed HTT was detected by HTT antibody (bottom two panels). (B) IPed wild-type HTT from rat whole brain also exhibited PAT activity toward GST-SNAP25. Similar to (A), unimmunized mouse serum (control) or HTT antibody bound to sepharose beads was used to IP HTT from rat whole brain lysate. IPed HTT was confirmed by western blot with HTT antibody and co-IPed HIP14 was detected by HIP14 antibody (bottom two panels).
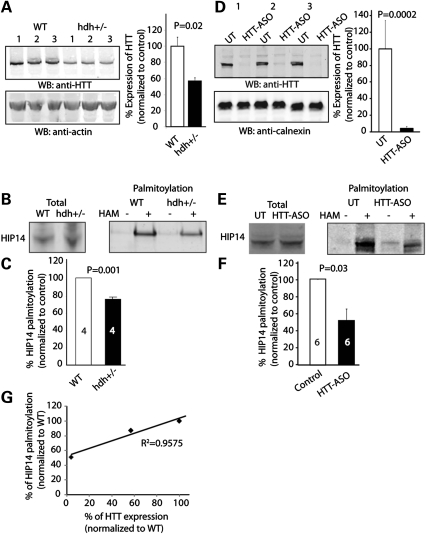
Wild-type HTT influences palmitoylation of HIP14 itself
DHHC PATs themselves are palmitoylated, which is crucial for their enzymatic activity (5,19,21). To determine the effect of wild-type HTT on the catalytic activity of HIP14, we first set out to test whether wild-type HTT may affect palmitoylation of HIP14 itself in vivo using the following two approaches: (i) using hdh+/− mice that lack one allele of the HTT gene and express approximately half of the endogenous HTT protein (Fig. 4A) and (ii) primary mouse cortical neurons treated with HTT antisense oligos (HTT-ASO), in which endogenous HTT expression is reduced by over 95% (Fig. 4D). In both approaches, palmitoylation of HIP14 itself was significantly decreased, with more evident reduction shown in neurons treated with HTT-ASO (Fig. 4B, C, E, F). These results suggest wild-type HTT modulates palmitoylation of HIP14 itself in a dose-dependent manner (Fig. 4G).
Figure 4.
Palmitoylation of HIP14 itself is reduced in hdh+/− mice and HTT-ASO-treated neurons. (A) Western blot and quantification of endogenous HTT expression in brain extract of hdh +/− mice compared with that of WT mice. (B and C) After ABE palmitoyl-protein purifications of wild-type and hdh+/− mice (Materials and Methods), extracted palmitoylated proteins were subject to protein electrophoresis and western blotting with HIP14 antibody. Representative image and quantification showing palmitoylation of endogenous HIP14 was reduced in brain extract of hdh+/− mice compared with that of wild-type mice (87.1 ± 0.7%). Number of repeats was indicated in the graph. (D) Western blot and quantification of endogenous HTT expression in HTT-ASO-treated cortical neurons. (E and F) Same as described in (B) and (C), representative image and quantification showing palmitoylation of endogenous HIP14 is reduced in HTT-ASO-treated cortical neurons compared with untreated controls (UT) (50.9 ± 12.5%). Number of repeats was indicated in the graph. (G) Wild-type HTT modulates palmitoylation of HIP14 itself in a dose-dependent manner. Correlation coefficient of the linear regression R2= 0.96.
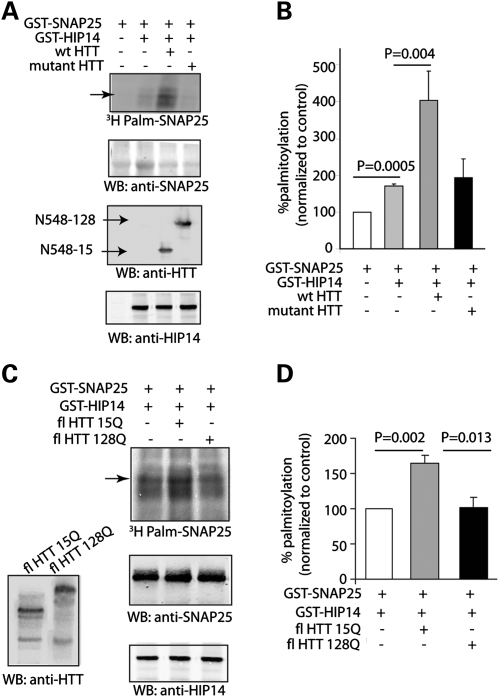
Palmitoylation of GST-SNAP25 by GST-HIP14 is potentiated in vitro in the presence of wild-type but not mutant HTT
Several lines of evidence show that palmitoylation of PATs is required for their enzymatic activity (3,5,21). To examine whether HIP14's activity toward other substrates is affected by wild-type HTT, we first used an in vitro palmitoylation assay, in which both substrate (SNAP25) and enzyme (HIP14) were fused to GST and purified using bacteria. In vitro, HIP14 alone significantly enhanced SNAP25 palmitoylation by ∼70%. In the presence of COS cell lysate expressing wild-type N-terminal fragment HTT (amino acids 1–548), palmitoylation of GST-SNAP25 by HIP14 was further enhanced. In contrast, mutant N-terminal fragment HTT was not capable of potentiating HIP14 activity (Fig. 5A and B). These results were reproduced with full-length HTT (Fig. 5C and D). In addition to SNAP25, palmitoylation of PSD95 (1-PDZ2) by GST-HIP14 in vitro could also be further enhanced by wild-type full-length HTT (Supplementary Material, Fig S1A and B). Taken together, these results point to the ability of wild-type HTT to modulate palmitoylation of HIP14 itself and thereby influencing its enzymatic activity toward other neuronal substrates.
Figure 5.
Wild-type HTT increases HIP14 activity to palmitoylate GST-SNAP25 in vitro. (A) GST-HIP14 enhanced palmitoylation of GST-SNAP25 in the presence of N terminal fragment wild-type HTT (1–548 amino acids), but not mutant HTT (128Q). Both substrate (SNAP25) and enzyme (HIP14) were fused to GST and purified using bacteria. COS cell lysate overexpressing wild-type or mutant HTT was added to the reaction mix containing 3H-palmitoyl-CoA. Western blot shows equal amount of SNAP25, HTT or GST-HIP14 is present (second to bottom panel). (B) In vitro, HIP14 alone significantly enhanced SNAP25 palmitoylation by about 70.4 ± 6.7%. In the presence of COS cell lysate expressing wild-type HTT (amino acids 1–548), palmitoylation of GST-SNAP25 by HIP14 was further enhanced. In contrast, mutant N-terminal fragment HTT was not capable of potentiating HIP14 activity. (C and D) Similar to experiments in (A) and (B), full-length wild-type HTT and not mutant HTT also enhanced palmitoylation of GST-SNAP25 by GST-HIP14 by 34.2 ± 6.0%. All quantifications were done with three to six repeats.
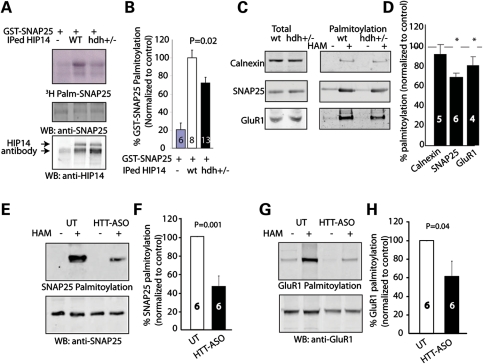
Loss of wild-type HTT leads to palmitoylation defects of SNAP25 and GluR1 in vivo
If HIP14 activity is indeed enhanced by wild-type HTT, then reduced wild-type HTT levels might be expected to impair enzymatic activity of HIP14 toward other substrates in vivo. We first tested the activity of IPed HIP14 from hdh+/− mice toward GST-SNAP25. Palmitoylation of SNAP25 by IPed HIP14 from brains of hdh+/− mice was significantly less than that of wild-type mice (Fig. 6A and B). Next we applied the ABE method to examine whether there is an alteration in palmitoylation of endogenous HIP14 substrates when HTT expression level is reduced. We showed that palmitoylation of endogenous SNAP25 and AMPA receptor subunit, GluR1, was reduced in brains of hdh+/− mice (Fig. 6C and D). Furthermore, in the HTT-ASO-treated cortical neuronal system, further reduction in palmitoylation of SNAP25 and GluR1 was detected (Fig. 6E and F). These results point to the key role of HTT directly modulating palmitoylation of HIP14 substrates in vivo.
Figure 6.
Loss of wild-type HTT leads to palmitoylation defects of SNAP25 and GluR1 in vivo. (A) Representative image shows IPed HIP14 of hdh+/− mice brain exhibited less enzymatic activity to palmitoylate GST-SNAP25, compared with that of wild-type mice. HIP14 antibody bound to sepharose beads was used to IP HIP14 from whole brain lysate of wild-type or hdh+/− mice. Then the beads containing IgG control or IPed HIP14 were added to reaction mix containing 3H-palmitoyl-CoA to palmitoylate GST-SNAP25 in vitro. (B) Quantification revealed that IPed HIP14 of hdh+/− mice brain had ∼30% reduction in its enzymatic activity. (C and D) ABE palmitoylation assay was performed on wild-type and hdh+/− mice brain to determine palmitoylation level of neuronal proteins. After ABE palmitoyl-protein purifications of wild-type and hdh+/− mice, extracted palmitoylated proteins were subject to protein electrophoresis and western blotting with calnexin, SNAP25 or GluR1 antibodies. Quantification and representative blots showed alteration in palmitoylation of SNAP25 (80.6 ± 8.4%) and GluR1 (79.4 ± 3.8%) in hdh+/− brain compared with the wild-type (100%). (E–H) In HTT-ASO-treated mouse cortical neurons, palmitoylation of SNAP25 (46.4 ± 11.8%) and GluR1 (61.6 ± 16.7%) was further reduced as determined by ABE palmitoylation assay described in (C) and (D). Number of repeats was indicated in the graph.
DISCUSSION
It is well established that wild-type HTT is neuroprotective (22–25) and that the loss of the normal function of HTT may contribute to the HD phenotype (26–28). Nonetheless, the normal function of HTT remains poorly defined. Previous work suggests that HTT plays distinct roles in several biological processes, including synaptic transmission, intracellular transport and neuronal transcription (reviewed in 12,29,30).
In this study, we discovered a novel function of wild-type HTT. In addition to serving as a palmitoylation substrate, wild-type HTT also modulates the palmitoylation of HIP14 itself. Using brains of hdh+/− mouse and knockdown of HTT in cultured cortical neurons, we were able to demonstrate that palmitoylation of HIP14 itself is affected by the level of wild-type HTT in a dose-dependent fashion. Reduced HIP14 palmitoylation results in its defective enzymatic activity. In vitro, wild-type HTT and not mutant HTT can potentiate HIP14's enzymatic activity towards SNAP25. In vivo, palmitoylation of SNAP25 and GluR1 [endogenous HIP14 substrates (8)] is significantly reduced in both brains of hdh+/− mice and neurons treated with HTT-ASO. This study describes an important biochemical function for wild-type HTT modulation of HIP14 palmitoylation and enzymatic activity.
There are several mechanisms by which HTT may modulate HIP14 palmitoylation and enzymatic activity.
First, HTT may act as an allosteric activator of HIP14. By binding to HIP14 at the ankyrin repeats site, HTT can regulate the function, structure and/or flexibility of HIP14. It is not uncommon that a substrate of an enzyme can also be an allosteric activator. For instance, full activity of 6-phosphogluconate dehydrogenase is only achieved when carrying its substrate 6-phosphogluconate (31). It is possible that, by influencing the tertiary structure of HIP14, HTT makes the catalytic DHHC domain of HIP14 more readily accessible to substrates. As a result, HTT allows HIP14 to catalyze the palmitoylation of neuronal substrates more effectively.
Secondly, HIP14 may gain access to its neuronal substrates through HTT which binds to a large array of intracellular proteins. Overall, the predicted structure of HTT is consistent with a cellular role as a scaffold protein (32). There are predicted to be up to 36 HEAT repeats which are α helix–loop–α helix motifs that mediate protein–protein interactions, spanning most of the HTT polypeptide (33). By physically bringing substrates to HIP14, HTT may facilitate the palmitoylation of these neuronal proteins by HIP14. Indeed, SNAP25 was identified to interact with HTT in a mass spectrometry analysis of affinity pull-down products of HTT (34). Thus far, no study has shown a direct interaction between GluR1 and HTT. However, GluR1 subunit may interact with HTT indirectly through postsynaptic scaffold proteins Shank and PSD95 (35).
Thirdly, HTT may be crucial for the trafficking of HIP14-containing vesicles to subcelluar sites where HIP14 executes its enzymatic function. Several studies have implicated HTT in the control of intracellular dynamic processes, including secretion and trafficking of vesicles from the Golgi apparatus, vesicular transfer between the actin and microtubule cytoskeleton, transport along microtubules in axons and synaptic endo/exocytic events (36,37). In particular, HTT protein associates with the molecular motor complex that transports organelles along microtubules in axons (38–40). Although we cannot exclude such a mechanism, our in vitro results that HTT can increase the palmitoylation of GST-SNAP25 by GST-HIP14 (Fig. 5) suggest that this mechanism is the less likely.
Our finding that the ability of HTT to modulate enzymatic activity of HIP14 is inversely related to the length of the polyglutamine tract provides direct association between the mutation in HD and HIP14 enzymatic activity. The interaction of mutant HTT with HIP14 in the brain is much less compared with that of wild-type HTT (9). Therefore, it is possible that mutant HTT is less capable of presenting its interacting proteins to HIP14. Alternatively, the mutation may inhibit the allosteric property of HTT. Mutant HTT also forms inclusions and loses the ability to facilitate vesicular transportation along the cytoskeleton. Thus, it is also possible that the trafficking of HIP14-containing vesicles is disturbed. Instead, HIP14 may be aggregated into mutant HTT-containing proteasome degrading machinery. Nonetheless, it will be critical to test whether palmitoylation of HIP14 itself and its neuronal substrates are affected in transgenic mouse models of HD (Singaraja R and Hayden MR, manuscript in preparation).
Another interesting finding is that first 548 amino acids of wild-type HTT are required for the interaction of HTT and HIP14 (Fig. 2). Shorter fragments, such as exon 1 and N171, do not interact with HIP14. Therefore, proteolytic cleavage of mutant HTT in vivo at multiple sites generate fragments that may lose interaction with HIP14 either partially or completely, further compromising the ability of mutant HTT to modulate HIP14 activity. This result suggests that the loss of normal HTT function plays a role in the pathogenesis of HD. However, this proposed mechanism does not contradict current evidence that HD arises predominantly from gain of a toxic function from these mutant HTT fragments. Smaller fragments detected in human post-mortem tissue and in mouse models (41–44) can be highly toxic in vivo, as seen in the R6/2, N171-82Q and YAC128 mouse models. Conversely, transgenic mice expressing mutant HTT resistant to caspase 6 cleavage have strikingly less pathology (45). Our result that mutant HTT lacks the ability to modulate HIP14 activity due to polyglutamine expansion and aberrant proteolysis does not contradict current gain of toxic function hypothesis and suggests that HD pathology may in fact arise from both a gain of toxic function and a loss of wild-type HTT function.
It has been previously shown that some DHHC enzymes must form complexes with other proteins in order to be functional PATs. The yeast DHHC protein Erf2 has to form a complex with Erf4 in order to palmitoylate the yeast Ras (6). In mammals, DHHC9 and GCP16 display the properties of a functional human ortholog of the yeast Ras palmitoyltransferase (46). This raised the question as to whether wild-type HTT is indispensible for HIP14 activity. HIP14 can palmitoylate a number of neuronal proteins including SNAP25 and GluR1 exogenously expressed in COS cells without wild-type HTT, suggesting that HIP14 alone is still functional (8). In our study, even when >95% of endogenous wild-type HTT expression was decreased, a significant portion of HIP14 itself was still palmitoylated. This residual palmitoylation may be sufficient for HIP14 to carry on some basal level of enzymatic activity in vivo. Indeed, ∼50% of SNAP25 and GluR1 palmitoylation was retained. It is also possible that other DHHC PATs, such as HIP14L, may compensate for the defect of HIP14 activity as DHHC PATs exhibit some overlap in their substrate specificity (8). Another significance of our finding is that binding partners of DHHC proteins may determine the substrate specificity, enzymatic kinetics, subcellular localization and stability of these DHHC proteins.
Our study also revealed that, HIP14L (DHHC13), a homolog of HIP14, also increases palmitoylation of HTT strongly in COS cells. The similarity in structures of HIP14 and HIP14L promotes further experiments to determine whether HTT interacts with HIP14L also in poly-glutamine length-dependent manner and whether HTT can modulate palmitoylation and enzymatic activity of HIP14L.
In summary, our study describes an important biochemical function for wild-type HTT and suggests that the loss of the wild-type HTT capacity to modulate palmitoylation of HIP14 neuronal substrates may contribute to the pathogenesis of HD.
MATERIALS AND METHODS
Plasmids
All DHHC enzymes were obtained as previously described (47) (generous gift by Dr Akio Kihara, Hokkaido University). Full-length HIP14 was subcloned into pCI-neo as described earlier (48). HIP14 GFP was generated by fusing enhanced green fluorescent protein (Clontech, CA, USA) at its C-terminus. HIP14 ΔDHHC GFP was generated by deletion of nucleotides encoding amino acids 440–487. HIP14 ΔANK GFP was generated by deletion of the ankyrin repeats, nucleotides encoding amino acids 89–257. Generation of GST-SNAP25 and HIP14-GST was described previously (3). All mutated DNA constructs were sequence confirmed.
Antibodies and chemicals
The antibodies used in this study were: HTT antibody 2166 (Millipore, 1:250 for immunoprecipitation, 1:1000 for western blotting), HIP14 anti-rabbit antibody (Sigma H7414, 1:200 for western blotting), GFP anti-rabbit antibody (developed in-house, 1:100 for immunoprecipitation), GFP anti-rabbit antibody (Synaptic System, 1:2000 for western blotting), SNAP25 (Covance, 1:1000 for western blotting and 1:500 for immunoprecipitation), GluR1 (provided by Dr Yutian Wang, the University of British Columbia) and Calnexin (Sigma C4731 1:5000 for western blotting).
Western blotting analysis
Primary antibodies were applied for an hour at room temperature in Odyssey blocking buffer (Li-COR Bioscience); the secondary antibodies (anti-rabbit IRD800 Rockland, Anti-Mouse Alexa Fluor 680, Invitrogen) were used at a ratio of 1:10000. The fluorescence was scanned and quantified using Odyssey Infrared Imaging System (Li-COR Bioscience). A distribution profile was calculated with Excel software (Microsoft Office) as the average value ± standard error from at least three independent experiments. Data were analyzed by Student's t-test, analysis of variance or nonparametric test.
Primary neuronal culture and htt-ASO treatment
Embryos were dissected from E15.5–E17.5 wild-type FVB/N pregnant. Cortical tissue was isolated and cultured, essentially as described (3). Pan-HTT ASO (Htt-387916; ISIS Pharmaceuticals) was resuspended in sterilized phosphate buffered saline (PBS) to a concentration of 1–5 mm. Neurons were fed with 200 μl neurobasal media with B27 on DIV3. For antisense oligonucleotide treatment, ASO stock was added to supplementary media to create a final concentration of 0.5 μm. Neurons were harvested on DIV18.
Palmitoylation assay
[3H]palmitoyl-CoA was synthesized enzymatically from [9,10-3H(N)]palmitic acid (5 mCi/ml; Perkin Elmer Life Sciences) as described previously (3). GST fusion proteins were produced in E. coli using the pGEX 6p3 expression system (Amersham) and isolated using Glutathione Sepharose 4B (Amersham). To generate a free N terminus of PSD-95, GST-PSD-95 protein was cleaved using thrombin [50 mm Tris–HCl (pH 8.0), 150 mm NaCl, 2.5 mm CaCl2, 0.1% 2-mercapto-ethanol). Palmitoylation reaction (60 μl) contained 5 µCi of [3H]palmitoyl-CoA, 0.33 µg/µl substrate protein, 1 mm adenosine triphosphate, 50 mm 2-(N-morpholino)ethanesulfonic acid, pH 6.4, 0.2 mg/ml bovine liver lipids and 20 µl of PAT enzyme. After 15 min incubation at 37°C, sample buffer was added with final concentration of 5 mm DTT was added and samples were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis.
ABE palmitoyl-protein purifications
Brains or cell pellets were harvested and immediately snap-frozen in liquid nitrogen and then stored in −80°C. In brief, frozen brains or cell pellets were homogenized into 500 μl lysis buffer (150 mm NaCl, 50 mm Tris, 5 mm ethylenediaminetetraacetic acid, pH 7.4) with 10 mm N-ethylmaleimide (NEM). Following the initial Triton X-100 detergent solubilization and NEM blockade steps, each sample was divided into two portions for treatment with, or as a control, without hydroxylamine (i.e. + or − HAM), which then is processed in parallel through the subsequent biotin-N-[6-(biotinamido)hexyl]-3′-(2′-pyridyldithio)-propionamide biotinylation and streptavidin agarose steps (49,50). Total palmitoyl proteins, purified from whole brain or cell pellets either in the presence (+) or absence (−) of HAM, were subjected to western analysis with antibodies specific to the different candidate proteins being tested, as described previously (49,50).
IP-ABE palmitoylation assay
COS cells overexpressing HTT alone or HTT with individual DHHC proteins were lysed and processed for immunoprecipitation as described above. The beads were then washed with wash buffer (PBS, containing 1% Triton X-100) supplemented with 50 mm NEM, followed by treatment with 1m hydroxylamine pH 7.4 for 1 h at room temperature. Samples were then processed as previously described (8,51).
Yeast two-hybrid analysis
HIP14 was identified in the pGAD yeast expression vector during a library screen with HTT (20). The series of truncated HTT constructs were inserted into the yeast expression vector pGBT and sequence verified. Yeast transformations with pGADHIP14 and each pGBT9 HTT constructs were performed using the lithium acetate method (52). To detect the intensity of interaction, liquid β-galactosidase assays were performed by inoculating three individual colonies from each transformation in liquid leu-trp- media. After overnight growth at 30°C, cells were pelleted, washed and resuspended in Z buffer (0.06m Na2HPO4, 0.04 m NaH2PO4, 10 mm KCl) and freeze-thawed three times. Z buffer supplemented with 20% SDS, 1 m DTT, β-mercaptoethanol and O-Nirophenyl β-D-galactopyranoside were added to the tubes and reactions continued until a yellow color appeared. Reactions were stopped by adding 1 m NaCO3 and spun to remove cell debris. The intensity of the reaction was quantified at OD420 and relative units calculated using the formula Relative units = (1000*OD420)/(time*concentration factor*volume of lysate*yeast cell density).
SUPPLEMENTARY MATERIAL
FUNDING
S.S.S. is supported by Canadian Institutes of Health Research and Michael Smith Foundation for Health Research Graduate Scholarship. F.B.Y. is funded by the Canadian Institutes of Health Research -IG Scriver Family MD/PhD Studentship. N.G.D. is supported by NIH GM65525. M.R.H. is a University Killam Professor and the Canada Research Chair in Human Genetics and Molecular Medicine, and is supported by Canadian Institutes of Health Research, Michael Smith Foundation for Health Research, CHDI Foundation Inc, the Huntington Society of Canada and Huntington Disease Society of America.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Dr Gene Hung from ISIS pharmaceuticals, Inc for sharing HTT-ASOs, Dr Akio Kihara from the Hokkaido University for sharing DHHC protein constructs, Stefanie Butland for helpful discussion and Martin Kang and Crystal Doty for technical assistance.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Resh M.D. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim. Biophys. Acta. 1999;1451:1–16. doi: 10.1016/s0167-4889(99)00075-0. [DOI] [PubMed] [Google Scholar]
- 2.Huang K., El-Husseini A. Modulation of neuronal protein trafficking and function by palmitoylation. Curr. Opin. Neurobiol. 2005;15:527–535. doi: 10.1016/j.conb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Huang K., Yanai A., Kang R., Arstikaitis P., Singaraja R.R., Metzler M., Mullard A., Haigh B., Gauthier-Campbell C., Gutekunst C.A., et al. Huntingtin-interacting protein HIP14 is a palmitoyl transferase involved in palmitoylation and trafficking of multiple neuronal proteins. Neuron. 2004;44:977–986. doi: 10.1016/j.neuron.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 4.A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. The Huntington's Disease Collaborative Research Group. Cell. 1993;72:971–983. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 5.Fukata M., Fukata Y., Adesnik H., Nicoll R.A., Bredt D.S. Identification of PSD-95 palmitoylating enzymes. Neuron. 2004;44:987–996. doi: 10.1016/j.neuron.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 6.Lobo S., Greentree W.K., Linder M.E., Deschenes R.J. Identification of a Ras palmitoyltransferase in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:41268–41273. doi: 10.1074/jbc.M206573200. [DOI] [PubMed] [Google Scholar]
- 7.Roth A.F., Feng Y., Chen L., Davis N.G. The yeast DHHC cysteine-rich domain protein Akr1p is a palmitoyl transferase. J. Cell Biol. 2002;159:23–28. doi: 10.1083/jcb.200206120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang K., Sanders S., Singaraja R., Orban P., Cijsouw T., Arstikaitis P., Yanai A., Hayden M.R., El-Husseini A. Neuronal palmitoyl acyl transferases exhibit distinct substrate specificity. FASEB J. 2009;23:2605–2615. doi: 10.1096/fj.08-127399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yanai A., Huang K., Kang R., Singaraja R.R., Arstikaitis P., Gan L., Orban P.C., Mullard A., Cowan C.M., Raymond L.A., et al. Palmitoylation of huntingtin by HIP14 is essential for its trafficking and function. Nat. Neurosci. 2006;9:824–831. doi: 10.1038/nn1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang K., Kang M.H., Askew C., Kang R., Sanders S.S., Wan J., Davis N.G., Hayden M.R. Palmitoylation and function of glial glutamate transporter-1 is reduced in the YAC128 mouse model of Huntington disease. Neurobiol. Dis. 2010;40:207–215. doi: 10.1016/j.nbd.2010.05.027. [DOI] [PubMed] [Google Scholar]
- 11.Orr H.T., Zoghbi H.Y. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 12.Caviston J.P., Holzbaur E.L. Huntingtin as an essential integrator of intracellular vesicular trafficking. Trends Cell Biol. 2009;19:147–155. doi: 10.1016/j.tcb.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutekunst C.A., Levey A.I., Heilman C.J., Whaley W.L., Yi H., Nash N.R., Rees H.D., Madden J.J., Hersch S.M. Identification and localization of huntingtin in brain and human lymphoblastoid cell lines with anti-fusion protein antibodies. Proc. Natl Acad. Sci. USA. 1995;92:8710–8714. doi: 10.1073/pnas.92.19.8710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiFiglia M., Sapp E., Chase K., Schwarz C., Meloni A., Young C., Martin E., Vonsattel J.P., Carraway R., Reeves S.A., et al. Huntingtin is a cytoplasmic protein associated with vesicles in human and rat brain neurons. Neuron. 1995;14:1075–1081. doi: 10.1016/0896-6273(95)90346-1. [DOI] [PubMed] [Google Scholar]
- 15.Nasir J., Floresco S.B., O'Kusky J.R., Diewert V.M., Richman J.M., Zeisler J., Borowski A., Marth J.D., Phillips A.G., Hayden M.R. Targeted disruption of the Huntington's disease gene results in embryonic lethality and behavioral and morphological changes in heterozygotes. Cell. 1995;81:811–823. doi: 10.1016/0092-8674(95)90542-1. [DOI] [PubMed] [Google Scholar]
- 16.Duyao M.P., Auerbach A.B., Ryan A., Persichetti F., Barnes G.T., McNeil S.M., Ge P., Vonsattel J.P., Gusella J.F., Joyner A.L., et al. Inactivation of the mouse Huntington's disease gene homolog Hdh. Science. 1995;269:407–410. doi: 10.1126/science.7618107. [DOI] [PubMed] [Google Scholar]
- 17.Zeitlin S., Liu J.P., Chapman D.L., Papaioannou V.E., Efstratiadis A. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington's disease gene homologue. Nat. Genet. 1995;11:155–163. doi: 10.1038/ng1095-155. [DOI] [PubMed] [Google Scholar]
- 18.Imarisio S., Carmichael J., Korolchuk V., Chen C.W., Saiki S., Rose C., Krishna G., Davies J.E., Ttofi E., Underwood B.R., et al. Huntington's disease: from pathology and genetics to potential therapies. Biochem. J. 2008;412:191–209. doi: 10.1042/BJ20071619. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez-Hernando C., Fukata M., Bernatchez P.N., Fukata Y., Lin M.I., Bredt D.S., Sessa W.C. Identification of Golgi-localized acyl transferases that palmitoylate and regulate endothelial nitric oxide synthase. J. Cell Biol. 2006;174:369–377. doi: 10.1083/jcb.200601051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singaraja R.R., Hadano S., Metzler M., Givan S., Wellington C.L., Warby S., Yanai A., Gutekunst C.A., Leavitt B.R., Yi H., et al. HIP14, a novel ankyrin domain-containing protein, links huntingtin to intracellular trafficking and endocytosis. Hum. Mol. Genet. 2002;11:2815–2828. doi: 10.1093/hmg/11.23.2815. [DOI] [PubMed] [Google Scholar]
- 21.Tsutsumi R., Fukata Y., Noritake J., Iwanaga T., Perez F., Fukata M. Identification of G protein {alpha} subunit-palmitoylating enzyme. Mol. Cell Biol. 2009;29:435–447. doi: 10.1128/MCB.01144-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leavitt B.R., van Raamsdonk J.M., Shehadeh J., Fernandes H., Murphy Z., Graham R.K., Wellington C.L., Raymond L.A., Hayden M.R. Wild-type huntingtin protects neurons from excitotoxicity. J. Neurochem. 2006;96:1121–1129. doi: 10.1111/j.1471-4159.2005.03605.x. [DOI] [PubMed] [Google Scholar]
- 23.Dragatsis I., Levine M.S., Zeitlin S. Inactivation of Hdh in the brain and testis results in progressive neurodegeneration and sterility in mice. Nat. Genet. 2000;26:300–306. doi: 10.1038/81593. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y., Li M., Drozda M., Chen M., Ren S., Mejia Sanchez R.O., Leavitt B.R., Cattaneo E., Ferrante R.J., Hayden M.R., et al. Depletion of wild-type huntingtin in mouse models of neurologic diseases. J. Neurochem. 2003;87:101–106. doi: 10.1046/j.1471-4159.2003.01980.x. [DOI] [PubMed] [Google Scholar]
- 25.Leavitt B.R., Guttman J.A., Hodgson J.G., Kimel G.H., Singaraja R., Vogl A.W., Hayden M.R. Wild-type huntingtin reduces the cellular toxicity of mutant huntingtin in vivo. Am. J. Hum. Genet. 2001;68:313–324. doi: 10.1086/318207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zuccato C., Ciammola A., Rigamonti D., Leavitt B.R., Goffredo D., Conti L., MacDonald M.E., Friedlander R.M., Silani V., Hayden M.R., et al. Loss of huntingtin-mediated BDNF gene transcription in Huntington's disease. Science. 2001;293:493–498. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- 27.Baquet Z.C., Gorski J.A., Jones K.R. Early striatal dendrite deficits followed by neuron loss with advanced age in the absence of anterograde cortical brain-derived neurotrophic factor. J. Neurosci. 2004;24:4250–4258. doi: 10.1523/JNEUROSCI.3920-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strand A.D., Baquet Z.C., Aragaki A.K., Holmans P., Yang L., Cleren C., Beal M.F., Jones L., Kooperberg C., Olson J.M., et al. Expression profiling of Huntington's disease models suggests that brain-derived neurotrophic factor depletion plays a major role in striatal degeneration. J. Neurosci. 2007;27:11758–11768. doi: 10.1523/JNEUROSCI.2461-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reiner A., Dragatsis I., Zeitlin S., Goldowitz D. Wild-type huntingtin plays a role in brain development and neuronal survival. Mol. Neurobiol. 2003;28:259–276. doi: 10.1385/MN:28:3:259. [DOI] [PubMed] [Google Scholar]
- 30.Cattaneo E., Zuccato C., Tartari M. Normal huntingtin function: an alternative approach to Huntington's disease. Nat. Rev. Neurosci. 2005;6:919–930. doi: 10.1038/nrn1806. [DOI] [PubMed] [Google Scholar]
- 31.Hanau S., Montin K., Cervellati C., Magnani M., Dallocchio F. 6-Phosphogluconate dehydrogenase mechanism: evidence for allosteric modulation by substrate. J. Biol. Chem. 2010;285:21366–21371. doi: 10.1074/jbc.M110.105601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li W., Serpell L.C., Carter W.J., Rubinsztein D.C., Huntington J.A. Expression and characterization of full-length human huntingtin, an elongated HEAT repeat protein. J. Biol. Chem. 2006;281:15916–15922. doi: 10.1074/jbc.M511007200. [DOI] [PubMed] [Google Scholar]
- 33.MacDonald M.E. Huntingtin: alive and well and working in middle management. Sci. STKE. 2003;207:pe48. doi: 10.1126/stke.2003.207.pe48. [DOI] [PubMed] [Google Scholar]
- 34.Kaltenbach L.S., Romero E., Becklin R.R., Chettier R., Bell R., Phansalkar A., Strand A., Torcassi C., Savage J., Hurlburt A., et al. Huntingtin interacting proteins are genetic modifiers of neurodegeneration. PLoS Genet. 2007;3:e82. doi: 10.1371/journal.pgen.0030082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun Y., Savanenin A., Reddy P.H., Liu Y.F. Polyglutamine-expanded huntingtin promotes sensitization of N-methyl-D-aspartate receptors via post-synaptic density 95. J. Biol. Chem. 2001;276:24713–24718. doi: 10.1074/jbc.M103501200. [DOI] [PubMed] [Google Scholar]
- 36.Smith R., Brundin P., Li J.Y. Synaptic dysfunction in Huntington's disease: a new perspective. Cell Mol. Life Sci. 2005;62:1901–1912. doi: 10.1007/s00018-005-5084-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borrell-Pages M., Zala D., Humbert S., Saudou F. Huntington's disease: from huntingtin function and dysfunction to therapeutic strategies. Cell Mol. Life Sci. 2006;63:2642–2660. doi: 10.1007/s00018-006-6242-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gauthier L.R., Charrin B.C., Borrell-Pages M., Dompierre J.P., Rangone H., Cordelieres F.P., De Mey J., MacDonald M.E., Lessmann V., Humbert S., et al. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118:127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 39.Caviston J.P., Ross J.L., Antony S.M., Tokito M., Holzbaur E.L. Huntingtin facilitates dynein/dynactin-mediated vesicle transport. Proc. Natl Acad. Sci. USA. 2007;104:10045–10050. doi: 10.1073/pnas.0610628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gunawardena S., Her L.S., Brusch R.G., Laymon R.A., Niesman I.R., Gordesky-Gold B., Sintasath L., Bonini N.M., Goldstein L.S. Disruption of axonal transport by loss of huntingtin or expression of pathogenic polyQ proteins in Drosophila. Neuron. 2003;40:25–40. doi: 10.1016/s0896-6273(03)00594-4. [DOI] [PubMed] [Google Scholar]
- 41.DiFiglia M., Sapp E., Chase K.O., Davies S.W., Bates G.P., Vonsattel J.P., Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 42.Landles C., Sathasivam K., Weiss A., Woodman B., Moffitt H., Finkbeiner S., Sun B., Gafni J., Ellerby L.M., Trottier Y., et al. Proteolysis of mutant huntingtin produces an exon 1 fragment that accumulates as an aggregated protein in neuronal nuclei in Huntington disease. J. Biol. Chem. 2010;285:8808–8823. doi: 10.1074/jbc.M109.075028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schilling G., Klevytska A., Tebbenkamp A.T., Juenemann K., Cooper J., Gonzales V., Slunt H., Poirer M., Ross C.A., Borchelt D.R. Characterization of huntingtin pathologic fragments in human Huntington disease, transgenic mice, and cell models. J. Neuropathol. Exp. Neurol. 2007;66:313–320. doi: 10.1097/nen.0b013e318040b2c8. [DOI] [PubMed] [Google Scholar]
- 44.Graham R.K., Deng Y., Carroll J., Vaid K., Cowan C., Pouladi M.A., Metzler M., Bissada N., Wang L., Faull R.L., et al. Cleavage at the 586 amino acid caspase-6 site in mutant huntingtin influences caspase-6 activation in vivo. J. Neurosci. 2010;30:15019–15029. doi: 10.1523/JNEUROSCI.2071-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graham R.K., Deng Y., Slow E.J., Haigh B., Bissada N., Lu G., Pearson J., Shehadeh J., Bertram L., Murphy Z., et al. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 2006;125:1179–1191. doi: 10.1016/j.cell.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 46.Swarthout J.T., Lobo S., Farh L., Croke M.R., Greentree W.K., Deschenes R.J., Linder M.E. DHHC9 and GCP16 constitute a human protein fatty acyltransferase with specificity for H- and N-Ras. J. Biol. Chem. 2005;280:31141–31148. doi: 10.1074/jbc.M504113200. [DOI] [PubMed] [Google Scholar]
- 47.Ohno Y., Kihara A., Sano T., Igarashi Y. Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins. Biochim. Biophys. Acta. 2006;1761:474–483. doi: 10.1016/j.bbalip.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 48.Gervais F.G., Singaraja R., Xanthoudakis S., Gutekunst C.A., Leavitt B.R., Metzler M., Hackam A.S., Tam J., Vaillancourt J.P., Houtzager V., et al. Recruitment and activation of caspase-8 by the Huntingtin-interacting protein Hip-1 and a novel partner Hippi. Nat. Cell Biol. 2002;4:95–105. doi: 10.1038/ncb735. [DOI] [PubMed] [Google Scholar]
- 49.Kang R., Wan J., Arstikaitis P., Takahashi H., Huang K., Bailey A.O., Thompson J.X., Roth A.F., Drisdel R.C., Mastro R., et al. Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature. 2008;456:904–909. doi: 10.1038/nature07605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roth A.F., Wan J., Bailey A.O., Sun B., Kuchar J.A., Green W.N., Phinney B.S., Yates J.R., 3rd, Davis N.G. Global analysis of protein palmitoylation in yeast. Cell. 2006;125:1003–1013. doi: 10.1016/j.cell.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Drisdel R.C., Green W.N. Labeling and quantifying sites of protein palmitoylation. Biotechniques. 2004;36:276–285. doi: 10.2144/04362RR02. [DOI] [PubMed] [Google Scholar]
- 52.Gietz R.D., Schiestl R.H., Willems A.R., Woods R.A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.