Abstract
Human retrotransposons generate structural variation and genomic diversity through ongoing retrotransposition and non-allelic homologous recombination. Cell culture retrotransposition assays have provided great insight into the genomic impact of retrotransposons, in particular, LINE-1(L1) and Alu elements; however, no such assay exists for the youngest active human retrotransposon, SINE-VNTR-Alu (SVA). Here we report the development of an SVA cell culture retrotransposition assay. We marked several SVAs with either neomycin or EGFP retrotransposition indicator cassettes. Engineered SVAs retrotranspose using L1 proteins supplemented in trans in multiple cell lines, including U2OS osteosarcoma cells where SVA retrotransposition is equal to that of an engineered L1. Engineered SVAs retrotranspose at 1–54 times the frequency of a marked pseudogene in HeLa HA cells. Furthermore, our data suggest a variable requirement for L1 ORF1p for SVA retrotransposition. Recovered engineered SVA insertions display all the hallmarks of LINE-1 retrotransposition and some contain 5′ and 3′ transductions, which are common for genomic SVAs. Of particular interest is the fact that four out of five insertions recovered from one SVA are full-length, with the 5′ end of these insertions beginning within 5 nt of the CMV promoter transcriptional start site. This assay demonstrates that SVA elements are indeed mobilized in trans by L1. Previously intractable questions regarding SVA biology can now be addressed.
INTRODUCTION
Greater than 30% of the human genome has been generated through retrotransposition of LINE elements and other RNA species by the LINE reverse transcriptase (1,2). Retrotransposition is ongoing in human populations as indicated by de novo L1 (3), Alu (4) and SINE-VNTR-Alu (SVA) (5) insertions associated with disease and by the large number of polymorphic insertions (6–12), many of which are at a low allele frequency in human genomes (13). Most of our knowledge regarding human retrotransposons has been accumulated through genomic analyses (1,14–21), cell culture retrotransposition assays (22–36) and mouse models (37–42).
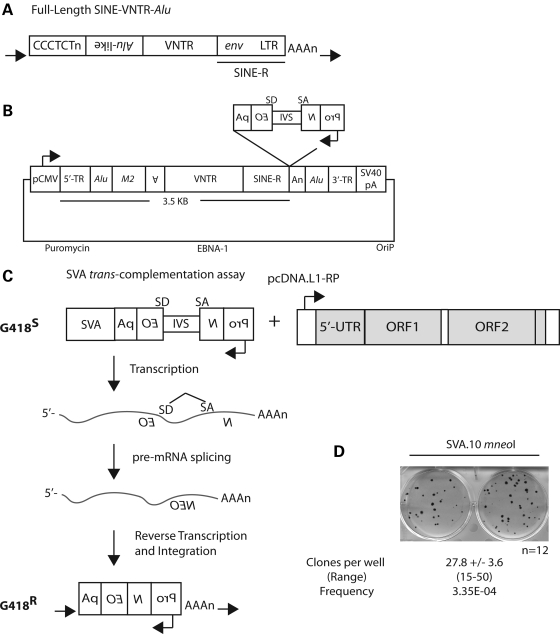
SVAs are hominid specific, generate non-coding RNAs (20) and the youngest active human retrotransposon (5). SVA insertions are associated with eight cases of single-gene disease (43–50). SVAs are composite elements (Fig. 1A) (51–53) consisting of multiple domains, these being in order from the 5′ end: (i) a CCCTCT repeat, (ii) an Alu-like domain, (iii) a GC-rich variable number of tandem repeats (VNTRs) and (iv) an env gene sequence and right LTR from an extinct HERV-K, referred to as SINE-R (5,20,54). The individual SVA domains are present in the genomes of Old World monkeys (55,56), and assembly of these domains presumably occurred primarily by pre-mRNA splicing sometime after the divergence of hominids from Old World monkeys (57).
Figure 1.
A cell-culture SVA retrotransposition assay. (A) A full-length ‘canonical' SVA in the human genome, with the individual domains in order from 5′ to 3′. (i) CCCTCT hexamer; (ii) the Alu-like domain consisting of two antisense-spliced Alu fragments and a sequence of unknown origin; (iii) VNTR; and (iv) SINE-R (env sequence and right LTR from an extinct HERV-K), terminating in a polyA tail (AAAn), with the entire insertion flanked by a TSD (black horizontal arrows). (B) The SVA.10 mneoI construct. A ‘master' SVA locus, SVA.10, from the SVAF1 (MAST2) subfamily containing both 5′ (5′ TR, Alu) and 3′ (Alu, 3′ TR) transductions (TR) marked with the mneoI retrotransposition cassette cloned into the pCEP-Pur plasmid backbone. (C) The rationale of the trans-complementation assay is illustrated. Only if the SVA containing the mneoI reporter undergoes a round of transcription, followed by reverse transcription and integration presumably mediated by a full-length L1 (shown), will the reading frame of the neomycin phosphotransferase reporter be restored, conferring G418 resistance (G418R). (D) G418R foci formation is observed in HeLa HA cells when SVA.10 mneoI is co-transfected with the highly active L1 driver construct, pcDNA.L1-RP. The mean number of clones per well ± SEM and the range of clones across the wells are displayed below. The number of wells (n) assayed for this experiment is shown. The retrotransposition frequency (mean number of clones/number of transfected cells) for SVA.10 mneoI is listed below the range.
SVA genomic insertions contain the hallmarks of L1-mediated retrotransposition, including 5′ truncations, inversions, 3′ transductions, polyA tails and target-site duplications (TSDs) of varying length flanking the insertion site (5,14–16,21,58). However, experimental evidence for mobilization in trans by L1 has not been obtained, despite the efforts by a number of laboratories. Here we experimentally demonstrate mobilization of different SVAs in trans by highly active human L1s in various cell types.
RESULTS
An SVA retrotransposition assay
Recently, a new human-specific SVA subfamily, SVAF1, characterized by the presence of the first exon from the MAST2 gene, and lacking the CCCTCT hexamer along with most of the Alu-like domain, was described (59–61). One element within this family on CH10 (SVA.10) is thought to be the source element for at least 13 additional insertions based upon the presence of shared 5′ and 3′ transductions (59,60), including one SVA that is the progenitor to a disease-causing insertion (46,59). Likewise, a canonical human-specific SVA on CH2 (SVA.2), classified as an SVAD, is thought to be the source element for at least nine insertions in the human genome as indicated by the presence of shared 3′ transductions (21). These two elements potentially represent two of the most active SVAs in humans since the human–chimp divergence (21,59,60). Because both of these loci have produced numerous human-specific SVA insertions, we reasoned that both would be appropriate candidates to test for retrotransposition competency in cultured cells. The entire SVA.10 locus, including the 5′ and 3′ transductions was isolated, marked with the mneoI retrotransposition indicator cassette (24,62,63) and cloned into pCEP4 vector (Fig. 1B) to make SVA.10 mneoI. mneoI consists of a backward neomycin resistance gene, relative to SVA, with a SV40 promoter and thymidine kinase polyA signal. The neo gene is interrupted by an intron (IVS) in the sense orientation (Fig. 1B). Thus, only upon splicing of an SVA transcript followed by reverse transcription and integration into the genome will G418 resistance (G418R) be conferred upon the transfected cell (Fig. 1C). Likewise, the entire SVA.2 locus was isolated, but only the SVA sequence was marked with mneoI, which differs from SVA.10 mneoI, which contains multiple transductions (see Materials and Methods).
To determine whether our constructs were ‘active' in cell culture and L1 was sufficient for trans-mobilization in cultured HeLa HA cells, we transiently co-transfected SVA.10 mneoI, referred to here as the ‘passenger' plasmid, and highly active unmarked L1s, either L1-RP (64) or L1.3 (65,66), which are referred to here as ‘driver' plasmids into HeLa HA cells (Figs 1D and 2A). SVA passengers were cloned into the replication-competent pCEP4 vector, whereas the L1 drivers were cloned into pcDNA6, a non-replicating plasmid in HeLa HA cells, (i) to enable antibiotic selection for both plasmids if need be, and (ii) to preclude the formation of plasmid recombinants, which had been a difficulty in previous attempts to develop an SVA cell culture retrotransposition assay (unpublished data, E.M. Ostertag, J.L.G. and H.H.K., Jr). We used the HeLa HA cell line because Alu displays high levels of L1 trans-mobilization in this HeLa strain (25,67) and thus serves as a robust positive control for our trans-mobilization assay (Supplementary Material, Figs S1 and S2 and data not shown).
Figure 2.
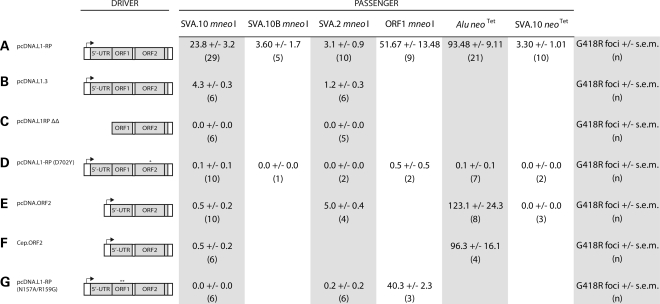
Engineered SVA retrotransposition is mediated by human L1 proteins in HeLa HA cells. Different marked SVAs, ORF1 mneoI and Alu neoTet were co-transfected with various drivers (A–G) to determine the role of L1 proteins in SVA retrotransposition. All transfections were carried out in six-well plates with 1.5 µg of the corresponding ‘driver' plasmid and 0.5 µg of the corresponding ‘passenger' plasmid. Data are presented as the mean number of G418R foci per well ± SEM, with the number of replicates (n) below each mean. Where no data are presented, it means that the experiment was not carried out. ‘Hot' L1s, L1-RP and L1.3 mobilize engineered SVAs (A and B). Removal of the CMV and L1 promoter (5′ UTR) from pcDNA.L1-RP reduces SVA foci formation to background levels (C). Different drivers containing point mutations (D and G) or lacking ORF1 coding sequence (E and F) were co-transfected with SVA. (*) indicates the relative location of the engineered point mutation.
To determine the frequency of SVA.10 mneoI retrotransposition, we transiently co-transfected SVA.10 mneoI with pcDNA.L1-RP (64) (Figs 1D and 2A). The retrotransposition frequency was calculated as the number of foci divided by the number of transfected cells (see Materials and Methods). The mean number [± standard error of the mean (SEM)] of foci/well for SVA.10 mneoI across 12 replicates was 27.8 (± 3.61) (Fig. 1D). The frequency of SVA.10 mneoI retrotransposition driven by pcDNA.L1-RP in this assay is 3.35E − 04 events per transfected cell. SVA.10 mneoI foci formation was significantly less when driven by pcDNA L1.3 (4.33 ± 0.33) (Fig. 2B). This corroborates a previous study which showed that L1-RP is a better driver than L1.3 (23).
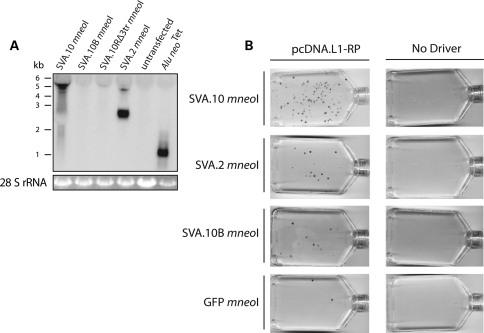
Additional replicates were carried out (Fig. 2A) for SVA.10 mneoI along with SVA.2 mneoI co-transfections with either pcDNA.L1-RP (Fig. 2A) or pcDNA L1.3 (Fig. 2B). SVA.2 mneoI was mobilized by pcDNA.L1-RP (3.1 ± 0.91) and pcDNA L1.3 (1.17 ± 0.31) in trans. Depending on the driver L1, SVA.10 mneoI produces about four to eight times as many foci in HeLa HA cells as SVA.2 mneoI. A few nucleotide substitutions introduced during the cloning process in the SVA.10 3′ transduction (Supplementary Material, Fig. S2) were identified. Therefore, we recloned the 3′ transduction from BAC DNA to make SVA.10B mneoI. Trans-mobilization assays revealed that SVA.10 mneoI produced approximately six times more foci than SVA.10B mneoI (23.79 ± 3.19 versus 3.6 ± 1.69) (Fig. 2A). Although the role of these substitutions is unclear, northern analysis indicates that steady-state levels of SVA.10 mneoI RNA are significantly greater than those of SVA.10B mneoI RNA and a modified SVA.10 construct, SVA.10R mneoI, which lacks the 3′ transduction (Fig. 3A). In contrast, SVA.2 mneoI and Alu neoTet RNA levels appear robust.
Figure 3.
(A) Steady-state levels of spliced RNA from marked SVA constructs differ. HeLa cells were co-transfected with pcDNA.L1-RP and different SVA constructs. Northern analysis used a neo sense probe spanning the intron. A representative northern blot (10 µg of total RNA) is shown. Across the top are the names of the different SVA passenger constructs. Along the left side is a size standard in kilobases (kb). Below is the 28S rRNA loading control. The expected RNA lengths derived from the SVA constructs (kb) including spliced mneoI from the 5′ end of the element to the SV40 polyA signal in pCEP: SVA.10 mneoI = 5.5, SVA.10R mneoIΔ3tr = 4.9, SVA.2 mneoI = 3.5, Alu neoTet = 1.5. (B) Representative T-75 flasks of neo assays carried out are shown. Engineered SVAs or GFP mneoI were co-transfected with pcDNA.L1-RP or without driver plasmid (No Driver). Refer to Table 1 for foci counts and relative activity.
To ensure G418R foci formation represented L1-mediated retrotransposition and not plasmid–plasmid recombination, we co-transfected SVA.10 mneoI (Fig. 2C and Supplementary Material, Fig. S1) and SVA.2 mneoI (Fig. 2C) with pcDNA.L1-RPΔΔ, a modified pcDNA.L1-RP construct lacking both the CMV promoter and the L1 5′ UTR. A driver lacking promoter sequences should not generate G418R foci, unless (i) recombination occurs between the plasmid lacking the promoter and the plasmid containing the promoter (23) or (ii) endogenous RT activity is utilized. The absence of G418R colonies (Fig. 2C) indicates that SVA foci are not a product of plasmid–plasmid recombination (23). Occasional foci are observed when Alu neoTet is transfected with pcDNA.L1-RPΔΔ (data not shown). This result is consistent with low levels of RT activity in this cell line and mobilization of Alu in the absence of driver L1s (27).
Next, to compare SVA.10 retrotransposition activity with that of Alu, we replaced the mneoI reporter in SVA.10 mneoI with the neoTet retrotransposition indicator cassette (68) and refer to this construct as SVA.10 neoTet. neoTet differs from mneoI, in that the neo gene is interrupted by a self-splicing group 1 intron instead of a nuclear mRNA intron. SVA.10 neoTet transfection with pcDNA.L1-RP resulted in 3.3 ± 1.01 foci per well, which corresponds to approximately 1/30 the number of foci produced when Alu neoTet is transfected with the same L1 driver (93.5 ± 9.1, Fig. 2A).
To investigate the retrotransposition activity of SVA relative to processed pseudogene formation, we scaled up our assay from six-well plates to T-75 flasks (see Methods and Materials). We co-transfected cells with pcDNA.L1-RP and the following SVA constructs: SVA.10 mneoI, SVA.10B mneoI, SVA.2 mneoI. Additionally, the progenitor to the SPTA1 (α-spectrin) insertion (5), an SVAE representing another ‘canonical' SVA element, referred to as SRE1, was tested with (SVA.SRE1 mneoI) and without (99 SVA.SRE1 mneoI) an exogenous promoter. We also co-transfected the following control plasmids with pcDNA.L1-RP: (i) ORF1 mneoI-AA (31), a construct containing the ORF1 coding sequence with two point mutations (R261A/R262A) marked with mneoI, and (ii) a marked pseudogene, EGFP mneoI, the coding sequence of EGFP along with mneoI in pCEP4. As a negative control, all passenger constructs were transfected alone (no driver). We normalized the number of G418R foci for each passenger to the number of G418R foci produced by the pseudogene EGFP mneoI. The results are presented in Table 1 (Fig. 3B).
Table 1.
Retrotransposition activity of marked SVAs in HeLa HA cells
| Passenger | pcDNA.L1-RP |
No driver |
|||
|---|---|---|---|---|---|
| n | Number of G418R | Activity | n | Number of G418R | |
| SVA.10 mneoI | 3 | 124.7 ± 17.8 | 54.2 | 2 | 1.0 ± 1.0 |
| SVA.10B mneoI | 3 | 7.3 ± 1.9 | 3.2 | 2 | 0.0 ± 0.0 |
| SVA.2 mneoI | 3 | 14.7 ± 3.5 | 6.4 | 2 | 0.5 ± 0.5 |
| SVA.SRE1 mneoI | 2 | 2.5 ± 1.5 | 1.1 | 2 | 0.0 ± 0.0 |
| 99 SVA.SRE1 mneoI | 2 | 7.0 ± 0.0 | 3.0 | 2 | 0.0 ± 0.0 |
| ORF1 mneoI-AA | 3 | 11.3 ± 2.0 | 4.9 | 2 | 0.0 ± 0.0 |
| EGFP mneoI | 3 | 2.3 ± 0.7 | 1.0 | 2 | 0.0 ± 0.0 |
Transfection of approximately 2 × 106 HeLa HA cells was carried out in T-75 flasks with 8 µg of the driver plasmid DNA and 4 µg of the passenger plasmid DNA. In the column labeled ‘No driver', only 4 µg of the passenger plasmid DNA was transfected. Following G418R selection, flasks were stained and colonies counted. The data are presented as the mean number of G418R colonies across replicates (n ± SEM). Activity is the number of G418R for each passenger divided by the number of EGFP mneoI G418R.
Consistent with the results of performing transfections in six-well plates, SVA.10 mneoI produced the most foci (mean = 124.7 ± 17.8) per T-75 flask and SVA.2 mneoI produced the second greatest number of foci (mean = 14.7 ± 3.5). These engineered SVAs produce foci 1–54 times more often than a marked pseudogene (Table 1). Notably, steady-state RNA levels differ across these constructs (Fig. 2A) and correlate with the retrotransposition activity for SVA.10 mneoI and SVA.10B mneoI.
The role of L1 proteins in SVA trans-mobilization
Both L1 proteins are required in cis to mobilize their own RNA (24). Likewise, processed pseudogene formation requires a functional ORF1p (23,26). However, Alu elements only require L1 ORF2p, although supplementation with L1 ORF1p may enhance Alu retrotransposition (69). To determine the role of L1 ORF2 in SVA trans-mobilization, our SVA constructs were co-transfected with an L1 containing a point mutation (D702Y) in the reverse transcriptase domain, known to abolish RT activity (70) (Fig. 2D). Consistent with a requirement for L1 ORF2p RT activity, few to no colonies were observed for engineered SVAs, ORF1 mneoI and Alu neoTet when co-transfected with L1-RP (D702Y) (Fig. 2D). These data are consistent with SVA trans-mobilization through an RNA intermediate.
Next, we tested whether the L1 RNA-binding protein ORF1p is required for SVA retrotransposition (Fig. 2E and F). Here, we co-transfected either SVA.10 mneoI or SVA.2 mneoI with a construct containing the 5′ UTR and ORF2 coding sequence of L1.3 (65,66,71) cloned into pcDNA6 (pcDNA.ORF2). Consistent with previous reports, Alu mobilization is enhanced when transfected with an ORF2-only construct rather than a full-length L1 (Fig. 2E) (25). Few-to-no colonies were observed when SVA.10 mneoI was co-transfected with pcDNA.ORF2 (0.5 ± 0.22). However, retrotransposition of SVA.2 mneoI co-transfected with pcDNA.ORF2 produces slightly more foci (5 ± 0.4) than transfection with full-length L1 drivers. To further investigate the potential ORF1p requirement of SVA.10 mneoI, co-transfection with ORF2 in a replicating plasmid, pCEP.ORF2, was carried out (Fig. 2F). Similar to what was observed with pcDNA.ORF2, few-to-no colonies (0.5 ± 0.22) were observed in this assay, despite quite robust Alu mobilization (96.25 ± 16.11) (Fig. 2F).
ORF1p is a multi-domain protein that contains coiled-coiled, RRM and C-terminal domains (72,73). To further investigate the role of ORF1p in SVA retrotransposition, we carried out experiments with a driver, pcDNA.L1-RPN157A/R159G, containing the double mutation, N157A/R159G (74), in the RRM domain of ORF1p. This mutation has been shown to abolish engineered L1 retrotransposition in cis, to affect L1 RNP formation and to disrupt both the formation of ORF1p cytoplasmic foci (74) and L1 cytoplasmic foci formation containing ORF2p (75). As a positive control for trans-mobilization, pcDNA.L1-RPN157A/R159G was transfected with ORF1 mneoI (23), as ORF1 mneoI does not require a functional ORF1p in trans (31) for mobilization. ORF1 mneoI transfected with pcDNA.L1-RPN157A/R159G results in slightly fewer foci (40.33 ± 2.33) than ORF1 mneoI driven with pcDNA L1-RP (51.67 ± 13.48) (Fig. 2G). In contrast, transfections of either SVA.10 mneoI or SVA.2 mneoI with pcDNA.L1-RPN157A/R159G resulted in almost no colony formation (0 ± 0 and 0.2 ± 0.2) (Fig. 2G). Therefore, these data suggest that SVA.2 retrotransposition is ORF1p independent, whereas SVA.10 retrotransposition is ORF1p dependent in HeLa HA cells. However, a driver L1 containing the double mutation, N157A/R159G, reduces SVA mobilization to background levels.
Engineered SVAs retrotranspose in multiple cell lines
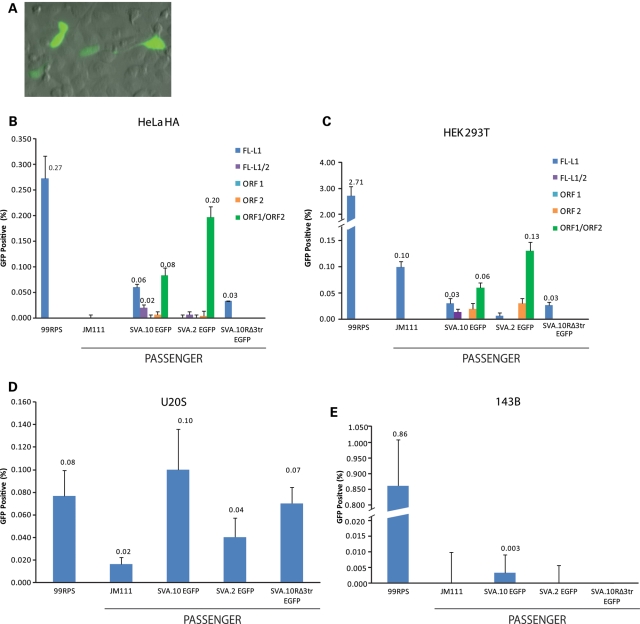
Our understanding of L1 biology has benefited from analysis across various cell types or cell lines of varying origin (reviewed in 34). Likewise, the EGFP retrotransposition indicator cassette (76) has been useful in reducing the amount of time it takes to carry out these assays and in interrogation of phenomena intractable to the neo assay (33). Therefore, we re-engineered SVA.10 and SVA.2 with the EGFP retrotransposition indicator cassette. Likewise, we tested a modified SVA.10 EGFP construct in which we removed the 3′ transduction and restored the 3′ end of the SINE-R to make SVA.10R EGFP. Similar to the mneoI cassette, the EGFP cassette remains non-functional until a round of transcription is followed by integration (76).
To test our new constructs, we transfected our EGFP-marked SVAs into HeLa HA cells, as we have demonstrated these cells are permissive for SVA retrotransposition. As a robust positive control for retrotransposition, we transfected 99 RPS EGFP Pur (76), a construct containing the highly active L1-RP (64) driven by its own promoter. To maintain plasmid DNA to transfection reagent ratios similar to that of the SVA co-transfections, the L1 driver, pcDNA.L1-RP (FL-L1), was co-transfected with 99 RPS EGFP Pur. As a positive control for trans-mobilization, JM111 EGFP Pur (76) and a driver L1 were also co-transfected. JM111 mneoI has been reported to be mobilized in trans at detectable levels in six-well plates by L1 driver constructs (23). SVA retrotransposition, as indicated by EGFP-positive cells, was detectable as early as day 2 under the microscope.
In these assays, we opted to select for the plasmid marked with the EGFP cassette with puromycin. Five days after transfection, cells were counted by flow cytometry to determine the number of EGFP-positive cells. Routinely, at least ∼100 000 transfected cells were counted to obtain reasonable sample size. Retrotransposition frequencies were calculated as the number of EGFP-positive cells per number of transfected cells (see Materials and Methods for more details). All samples were gated on cells co-transfected with SVA.10 EGFP and the pcDNA.L1-RP (D702Y) mutant, as this represents background fluorescence or any effect of endogenous RT activity. Both SVA constructs, SVA.10 EGFP and SVA.2 EGFP, were transfected with an ORF1 driver, pcDNA.ORF1, or an ORF2 driver, pcDNA.ORF2, to confirm our data from the neo assay regarding SVA L1 protein requirements. To examine whether L1 proteins may mobilize SVAs when supplied jointly in trans, and because a previous report suggests that retrotransposition of Alu by ORF2 alone is enhanced when ORF1 is supplemented (69), SVA10 EGFP or SVA.2 EGFP was co-transfected with ORF1 and ORF2 on separate plasmids (pcDNA.ORF1 and pcDNA.ORF2, respectively). To control for the reduction in plasmid containing ORF2 sequence when ORF1 and ORF2 sequences are on different plasmids, additional transfections were carried out in which the amount of L1 driver plasmid was reduced by a factor of 2 (FL-L1/2). All transfections were carried out in triplicate unless indicated otherwise and are presented as the mean %EGFP-positive cells ± 1 standard deviation (Fig. 4).
Figure 4.
Engineered SVAs retrotranspose in multiple cell lines. (A) SVA.2 EGFP-positive foci at day 3 in U2OS cells are shown. SVAs are marked (x-axis) with the EGFP retrotransposition indicator cassette (76) and co-transfected with L1 drivers in HeLa HA (B), HEK 293T(C), U2OS (D) and 143B cells. All transfections were carried out in six-well plates (see Methods and Materials). Five days after transfection, cells were subjected to flow cytometry. Retrotransposition frequency was calculated as the number of events (EGFP-positive cells) relative to the number of cells transfected (y-axis) with the designated passenger plasmids (B–E). Events were gated on cells co-transfected with SVA.10 EGFP and pcDNA.L1-RP (D702Y). 99 RPS, 99 RPS EGFP Pur; JM111, JM111 RPS EGFP Pur; FL-L1, pcDNA.L1-RP (FL = L1/2 refers to a reduction in the amount of pcDNA.L1-RP transfected); ORF1, pcDNA.ORF1; ORF2, pcDNA.ORF2; ORF1/ORF2, co-transfection with pcDNA.ORF1 and pcDNA.ORF2 on separate plasmids. All transfections were performed in triplicate, except SVA.10R EGFP Δ3TR and SVA.10 EGFP/ORF1/ORF2 in HeLa HA cells. Where included, the mean %EGFP-positive cells is given. Error bars represent 1 standard deviation.
In the HeLa HA cell experiment, EGFP-positive cells derived from SVA.10 EGFP were 0.06% of transfected cells, corresponding to 22% of 99 RPS EGFP Pur activity (0.27% ± 0.04). Although some SVA.2 EGFP and JM111 retrotransposition events were seen under an inverted microscope, EGFP-positive events were not detectable above background by flow cytometry (Fig. 4B). As expected, SVA.10 EGFP and SVA.2 EGFP co-transfected with pcDNA.ORF1 produced no events. SVA.10 EGFP and SVA.2 EGFP transfections with pcDNA.ORF2 produced detectable events, although very rarely (Fig. 4B). When SVA.10 EGFP and SVA.2 EGFP were transfected with the ORF1 and ORF2 on different plasmids (pcDNA.ORF1 + pcDNA.ORF2), a noticeable increase in the %EGFP positive cells was observed (Fig. 4B). SVA.10 EGFP is active (0.02% ± 0.01) when co-transfected with an equal amount of ORF2 containing plasmid DNA, but increases to 0.08% ± 0.14 with ORF1 and ORF2 on separate plasmids. SVA.2 EGFP increases from background levels to 0.20% ± 0.02 when co-transfected with ORF1 and ORF2 on separate plasmids. Thus, our SVA EGFP assay was validated and we decided to explore SVA retrotransposition in other cell lines.
Engineered L1s retrotranspose at high levels in the human embryonic kidney (HEK) cell line, 293T (77), and in the osteosarcoma cell line, 143B (78). In addition to these cell lines, we also transfected U2OS cells, another osteosarcoma cell line, with our three SVA EGFP constructs. The same experiment carried out for HeLa Ha cells was replicated in 293T cells, although we only assayed SVA mobilization by full-length L1 in 143B and U2OS cells. Similar to HeLa HA cells, SVA retrotransposition was observed as early as day 2 in 293T cells and day 3 in U2OS cells (Fig. 4A). Flow cytometry data for the 293T and U2OS cells are presented in Figure 4C and D, respectively. In contrast, SVA retrotransposition in 143B cells was low, with only SVA.10 EGFP events detectable by flow cytometry (Fig. 4E) while the other passenger constructs were not above background.
In 293T cells, both JM111 and ORF2-alone trans-mobilization of SVA.10 EGFP and SVA.2 EGFP was increased relative to HeLa HA cells (Fig. 4B). In U2OS cells, 99RPS EGFP Pur activity was reduced relative to HeLa HA or 293T cells. The SVA EGFP constructs exhibit the highest percentage of EGFP cells in U2OS cells, up to 0.1% of transfected cells for SVA.10 EGFP. It is noteworthy that the SVA activity is comparable with the L1 activity in this cell line. These SVA EGFP assays show that multiple cell types are permissive to engineered SVA retrotransposition, support our observations from the neo assay regarding ORF1p independence of SVA.2, and identify at least one cell line where SVA and L1 retrotransposition activity is similar.
SVA insertions display hallmarks of L1-mediated retrotransposition
To confirm that colony formation resulted from authentic retrotransposition events, SVA.10 mneoI, SVA.2 mneoI, SVA.10 mneoIΔ3TR, SVA.SRE1 mneoI and 99 SVA.SRE1 mneoI events were clonally expanded and genomic DNA was extracted from clones. First, we carried out PCR with primers spanning the intron of mneoI (Fig. 5A and data not shown). Consistent with engineered SVAs being mobilized through an RNA intermediate, amplicons representing a spliced mneoI PCR product were observed in genomic DNA isolated from individual clonal lines. Sanger sequencing confirmed that the lower bands represented spliced mneoI PCR products (data not shown).
Figure 5.
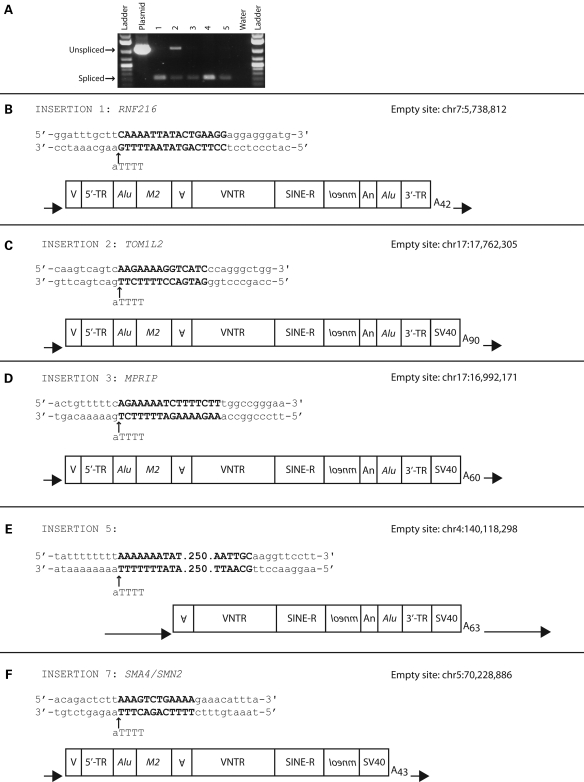
Engineered SVA insertions recovered from genomic DNA resemble SVA genomic insertions and display hallmarks of L1-mediated retrotransposition. Intron-spanning PCR was carried out on genomic DNA isolated from clonal cell lines derived from individual foci produced by different SVAs (A). Unspliced and spliced mneoI bands are indicated by black horizontal arrows. The 5′ and 3′′ ends for SVA.10 mneoI insertions recovered from individual foci (B–F). The genomic coordinates relative to the reference genome assembly (hg19/NCBI37) for each insertion are shown (right corner). The insertion-site nucleotide sequence consisting of the target-site duplication (bold letters), the L1 endonuclease cleavage site of the bottom strand (black vertical arrow) relative to the L1 endonuclease consensus cleavage site (5′-TTTT/a-3′) and 10 nucleotides 5′ and 3′ of the TSD are displayed. Each SVA insertion, including TSDs (black arrows), polyA signal and length of polyA tail, is diagrammed with individual domains annotated as described in Fig. 1B.
To map the location of engineered SVA insertions and characterize the breakpoints, we carried out inverse PCR (iPCR). Genomic DNA from independent foci was digested with restriction enzymes that cut towards the 3′ end of the SINE-R or within the mneoI cassette. This approach should (i) minimize bias in insertion size of SVA inserts and (ii) circumvent PCR amplification problems associated with the SVA GC-rich VNTRs. However, this approach only allows recovery of the 3′ breakpoints of SVA inserts.
We recovered the genomic location of 15 SVA insertions from clonal cell lines (Table 2) (see Material and Methods). Ten out of 15 (66%) engineered SVA insertions are located in genes, including one in the 3′ UTR of a gene (clone 10) and another breakpoint located directly 5′ of the AG splice acceptor of an exon (clone 7). Six out of 10 (60%) of the insertions in annotated genes are on the coding strand. RNA from the SVA.10 mneoI construct has the capability of polyadenylating at a polyA site in the 3′ transduction (59,60) or downstream at the SV40 polyA signal located in the CEP vector, as commonly observed for engineered L1s (24). Four out of five recovered SVA.10 mneoI insertions terminated downstream of the cloned 3′ transduction at the SV40 polyA signal. Therefore, these five 3′ ends represent SVAs that contain 3′ transductions.
Table 2.
SVA insertions recovered from HeLa HA genomic DNA
| Clone | SVA construct | Genomic location | Gene | Strand | Length (kb) (domain)a | TSD (length) | L1 EN site TTTT/a | PolyA lengthb |
|---|---|---|---|---|---|---|---|---|
| 1 | SVA.10 mneoI | CH7p22.1 | RNF216 | Coding | ∼5.4 (full-length) | CAAAATTATACTGAAGG (17) | TTTG/a | ∼42 |
| 2 | SVA.10 mneoI | CH17p11.2 | TOM1L2 | Coding | ∼5.6 (full-length) | AAGAAAAGGTCATC (14) | TCTT/g | ∼90 |
| 3 | SVA.10 mneoI | CH17p11.2 | MPRIP | Coding | ∼5.5 (full-length) | AGAAAAATCTTTTCTT (16) | TTCT/g | ∼60 |
| 4 | SVA.10 mneoI | CH1p36.13 | — | n/a | ∼81 | |||
| 5 | SVA.10 mneoI | CH4q31.1 | — | n/a | ∼4.5 (MAST2) | AAAAAAATAT…(265) | TTTT/a | ∼63 |
| 6 | SVA.10 mneoIΔ3tr | CH5q13.2 | SMA4/SMN2 | Coding | ∼5.0 (full-length) | AAAGTCTGAAAA (12) | CTTT/a | ∼43 |
| 7 | SVA.10 mneoIΔ3tr | CH19q13.2 | MED29 | Coding | ∼67 | |||
| 8 | SVA.2 mneoI | CH2q32.1c | — | n/a | ∼39 | |||
| 9 | SVA.2 mneoI | CH2q32.1c | — | n/a | ∼99 | |||
| 10 | 99 SVA.SRE1 mneoI | CH8p21.3 | AL833246/LOXL2 | Codingd | ∼1.5 (SINE-R) | AAAGAAACGGAGGCTCTTGAA (21) | CTTT/c | ∼31 |
| 11 | 99 SVA.SRE1 mneoI | CH20q13.32 | AK091704/AK054637 | Non-Coding | ∼2.5 (VNTR) | GAAAGACCCATA (12) | TTTC/a | ∼70 |
| 12 | 99 SVA.SRE1 mneoI | CH22q11.21 | PI4KA | Non-coding | ∼2.2 (VNTR) | AAATAAAGTTCCTG (14) | TTTT/a | ∼44 |
| 13 | 99 SVA.SRE1 mneoI | CH1q25.3 | C1orf14 | Non-coding | ∼2.9 (VNTR) | AAAATCGAGTAGTATG (16) | ATTT/a | ∼29 |
| 14 | 99 SVA.SRE1 mneoI | CH14q24.2 | — | n/a | ∼111 | |||
| Meane | — | — | — | — | 3.9 | 43.0 (15.3) | — | 62.1 |
Recovered SVA insertions are listed by clone number (column 1), the SVA construct (column 2) from which the foci was produced, the genomic location of the 3′ junction (column 3), whether it was in a gene (column 4) and orientation relative to the coding strand (column 5). Column 6 lists the length of the total retrotransposed sequence, therefore it includes the length of the mneoI retrotransposition indicator cassette (∼1.1 kb) and the domain in which the insertion 5′ truncates. Column 7 lists the TSD length in nucleotides. Column 8 lists the L1 endoculcease (EN) cleavage site on the bottom strand, with the consensus cleavage EN site listed in parentheses. Column 9 lists the approximate polyA length given in nucleotides.
aThe length of the SVA constructs (kb) including spliced mneoI from the 5′ end of the element to the SV40 polyA signal in pCep: SVA.10 mneoI = 5.5; SVA.10 mneoIΔ3tr = 4.9; SVA.2 mneoI = 3.5; 99 SVA.SRE1mneoI = 4.0 from the CCCTCT hexamer (4.7 including the 5′ flank).
bSVA.10 mneoI contains an additional polyA signals in the 3′ transduction. Despite this, all engineered SVA insertions except clone 1 terminated at the SV40 polyA signal.
cThese SVAs are different insertions. Clone 1 inserted into an L1MA3,chr2:187430675, whereas clone 2 inserted upstream at chr2:185062822.
dThis locus consists of two genes, AL8333246 on the positive strand and LOXL2 on the bottom strand. The insertion breakpoint is located in sequence that is annotated as the 3′ UTR for AL833246.
eThe mean length across engineered SVA insertions where the 5′ end was characterized, mean length of TSDs with and without the 265 nt TSD and polyA tail length are shown. The genomic coordinates and gene annotations are according to hg19 (http://genome.ucsc.edu).
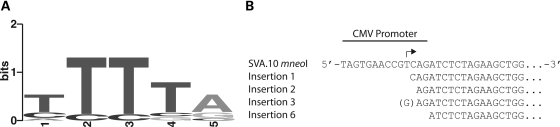
Of the nine insertions for which we obtained both the 5′ and 3′′ breakpoints (Fig. 5 and Table 2), TSDs were observed flanking the insertions (range 12–265 nt, mean excluding the 265 nt TSD = 15.3) (Table 2), insertions occurred at sequences resembling the L1 endonuclease consensus cleavage site (5′-TTTT/A-3′) (Table 2 and Fig. 6A) (22,29,36,79–81), and the insertions contained polyA tails of variable length (range 29–111 nt, mean = 59.2 nt).
Figure 6.
Engineered SVA insertions resemble SVA genomic insertions. (A) A consensus sequence, generated using WebLogo (86), for engineered SVAs insertion sites resembles the L1 endonuclease consensus cleavage site (5′-TTTT/a-3′) (79–81) (Table 2). (B) An alignment of four SVA.10 mneoI insertions, containing 5′ transductions, relative to the SVA.10 mneoI plasmid sequence. The 3′ end of the CMV promoter in CEP labeled along with the known CMV transcriptional start site (black bent arrow). Note that the first base of each insertion is within 5 nt of the CMV transcriptional start site. For insertion 3, a non-templated G at the 5′ breakpoint is displayed as (G).
Four out of five SVA.10 mneoI/SVA.10 mneoIΔ3TR insertions were full-length. The first nucleotide, that is not part of the TSD for these insertions, was located +2 to +5 relative to the transcription start site of the CMV promoter (82) (Fig. 6B), 12–16 nt 5′ of the cloned SVA fragment in pCEP4. One of these insertions, clone 3, contains a non-templated G (Fig. 6B). Thus, these insertions contain short 5′ transductions. The average length of the SVA.10 mneoI/SVA.10 mneoIΔ3TR insertions was ∼5.2 kb with the mneoI cassette and ∼4.0 kb without the cassette. In contrast, all four 99 SVA.SRE mneoI insertions were 5′ truncated, three within the VNTR and one in the SINE-R domain (Table 2). The mean length of the 99 SVA.SRE mneoI insertions was ∼2.3 kb with the mneoI cassette and ∼1.1 kb without the cassette. The combined mean length of the nine insertions, where 5′ and 3′ breakpoints were obtained, was 3.9 kb with the cassette and 2.7 kb without the cassette.
DISCUSSION
SVA is mobilized by human L1s in trans
Here, we provide the first experimental evidence for L1-mediated retrotransposition of SVA elements in cultured cells. SVA elements display all the hallmarks of LINE-1 mobilization; however, the establishment of an SVA retrotransposition assay has been delayed, at least in our hands, due to false-positive insertions and the lack of an appropriate progenitor element. Source elements to L1 disease-causing insertions have proven useful—robust models for L1 retrotransposition; however, this has not been the case for SVA insertions. In this study, we opted to use elements known to be relatively active since the human–chimp divergence, one element from the recently described youngest human-specific SVA subfamily, SVA.10, and one element representing the ‘canonical' SVA, SVA.2.
Consistent with the multiple daughter loci produced from the SVA.10 or SVA.2, both elements are rather active in multiple cell types. In some cell types, our SVA constructs are not as actively trans-mobilized as JM111 EGFP. However, it is interesting that, in U2OS cells, SVA retrotransposition rivals that of one of the most active human L1s, L1 RP. It is possible that SVAs may mobilize sooner in U2OS cells than human L1, and that over a longer time period, engineered L1 retrotransposition would far surpass that of engineered SVAs.
SVA elements are thought to be ‘hot' because numerous recent insertions are associated with Mendelian disease (57,83). SVA activity in our HeLa HA cell culture assays ranges from ∼1 to 54 times that of a pseudogene. Excluding SVA.10 mneoI which contains a few mutations introduced by cloning, SVA activity ranges from approximately one to six times the activity of a pseudogene. It is surprising to us, that SVA.10 and SVA.2 exhibit an ∼8-fold difference in retrotransposition activity (Fig. 2A and Table 1) in HeLa HA cells, whereas in U2OS cells they are not significantly different in activity. Furthermore, it is possible that either the appropriate cell line or precise time in vivo is needed not only for increased SVA activity but also for intrinsic SVA promoter activity to occur. An alternative and equally possible explanation for the modest SVA activity is that SVA elements are not very active overall, and that an unknown ascertainment bias accounts for SVA disease-associated insertions. However, it bears mentioning that engineered L1s are silenced following insertion by deacetylation of the local chromatin (33), and SVA insertions may also be silenced. Along these lines, SVA was originally identified by one laboratory (54) in a screen for densely CpG-methylated loci.
SVA L1 protein requirement
SVA mobilization through an RNA intermediate is supported by our retrotransposition reporter assays along with engineered SVA insertions containing spliced introns and polyA tails. Consistent with these observations, use of an L1 driver containing a point mutation in the RT domain reduces trans-mobilization of SVA and control constructs to background levels. Previous studies of trans-mobilization determined that ORF1p is dispensable for Alu retrotransposition (25) although it is required for pseudogene formation (23,26). Data from our neo assay (Fig. 2E and F) suggest a strict ORF1p requirement for SVA.10 mobilization and little-to-no ORF1p requirement for SVA.2 in HeLa HA cells. Despite an increase in ORF2-alone mobility in EGFP assays, SVA.10 is still mobilized more effectively by the full-length L1. However, it is evident in 293T cells that SVA.2 EGFP is indeed mobilized more efficiently by ORF2 alone relative to full-length L1. Of particular interest is the 10–20-fold increase in SVA EGFP-positive cells when ORF1 and ORF2 are transfected jointly on separate plasmids. It has been reported that Alu retrotransposition is enhanced up to 5-fold when Alu and ORF2-alone transfections are supplemented with ORF1 (69). Why ORF1 and ORF2 delivered on separate plasmids are more active as SVA drivers than full-length L1 is unknown. One possibility is that more ORF2p is synthesized from the monocistronic CMV-driven ORF2-alone construct (75) than ORF2p produced by a full-length L1. Nonetheless, this observation may prove useful for interrogating SVA biology.
The differential ORF1p requirement for SVA.10 and SVA.2 is surprising. This difference may be related to RNA length, as a full-length spliced SVA.10 mneoI RNA is ∼5.5 kb compared with ∼3.5 kb for a full-length spliced SVA.2 mneoI RNA, or perhaps more specifically the length of the 3′ transduction. In addition to size differences, SVA.10 and SVA.2 are quite different at the DNA sequence level, with SVA.10 containing multiple 5′ and 3′ transductions and lacking the CCCTCT hexamer and most of the Alu-like domain.
Engineered SVA insertions
Our recovered SVA insertions display features similar to genomic SVAs: 5′ transductions, 3′ transductions, TSDs, polyA tails and insertion at L1 endonuclease consensus cleavage sites (Table 2, Figs 5 and 6). Four out of five SVA.10 insertions begin at the CMV promoter transcriptional start site, supporting the hypothesis that these elements lack strong promoter sequences and the observation that the majority of SVAs (∼63%) in the human genome reference sequence are full-length (60). In contrast, the 99 SVA.SRE1 mneoI insertions are produced from a vector that contains no CMV promoter. Alternatively, it is possible that a cryptic promoter exists on the plasmid backbone or within the ∼700 bp of 5′ flank cloned with this element. Additionally, it is possible that the SRE1 insertions do not represent 5′ truncations at all, and that SVA transcription initiated within the VNTR as numerous transcription start sites exist throughout the SVA sequence. The locations of the SVA inserts suggest they readily insert into genes (66.6%). If the phenomenon of preferred intragenic insertion that we have seen in our cell culture assays is representative of new SVA insertions in humans, SVAs may indeed contribute to individual variation in gene expression either by mediating alternative splicing or by increasing DNA methylation at a locus. Six out of 10 of the engineered SVA insertions are on the coding strands of genes. This differs from the observation that, in the human reference genome, only 20% of intragenic SVAs are on the coding strand (59).
What is particularly striking is that four out of five SVA.10 mneoI insertions (80%) and four out of nine (44%) of the total recovered insertions are full-length. While this may represent some unknown technical bias, the iPCR approach should limit bias as the restriction enzymes used cut towards the 3′ end of the engineered element. The longer insertions differ greatly from reports for engineered L1 insertions in which the L1 is driven by a heterologous promoter and only ∼5% of the insertions are full-length (22,35,36). Here, it is possible that a size ascertainment bias may have been introduced, as some of these studies relied upon rescue cassette procedures which likely favor easier ligation and smaller insertions. However, a study characterizing transgenic human L1 insertions in mice, where the insertions were recovered by a ligation-independent method, TAIL-PCR, reported that 3/33 (9%) insertions were full-length (40). More recently, another transgenic study that characterized human L1 insertions in the mouse and rat, where the L1 was driven by its own promoter, reported that four out of eight were full-length (38).
The full-length SVA insertions here and in the genome are a testament to the resilience of the L1 RT. Moreover, the ability to reverse-transcribe such a repetitive GC-rich template, as occurs in SVA, argues against a model in which L1 5′ truncations are the consequence of reduced L1 RT processivity and favors the hypothesis that some as-yet unidentified factor is responsible for truncations.
This study provides the first comprehensive analysis of SVA molecular biology to date. Here we extend our current understanding not only of SVA biology but also of L1-mediated retrotransposition. Our assay will not only be useful for comparative studies but may also provide insight into this young, mysterious element.
MATERIALS AND METHODS
Oligonucleotide sequences used in this study are available upon request.
Recombinant DNA plasmids
SVA.10 mneoI: This construct contains a KpnI–AgeI 3.5 kb fragment consisting of the MAST2-SVA (57,60) from the CH10 locus (10q24.2) and its 5′ transductions (including 5′Alu), the mneoI retrotransposition indicator cassette (24,62) and a 0.5 kb SbfI–NotI fragment consisting of the 3′ transduction from the ABCC2 locus, containing the last 23 bp of the SINE-R, SVA polyA signal and polyA tail, the 3′ Alu and the 160 bp transduction, all cloned into pCEP-Pur, a modified pCEP4 (Invitrogen) vector containing the puromycin resistance gene (76).
SVA.10 mneoI Δ3TR: This construct contains a KpnI–AgeI 3.5 kb fragment consisting of the MAST2-SVA (57,60) from the CH10 locus (10q24.2) and its 5′ transductions (including 5′ Alu), and the mneoI retrotransposition indicator cassette (24,62) cloned into pCEP4-Pur.
pcDNA.L1-RP: L1-RPS was liberated from pJCC5(L1RP) (76) as a 6 kb NotI–ApaI fragment and swapped into the NotI–ApaI sites of pcDNA6/myc-hisB (Invitrogen).
pcDNA.L1-RP(D702Y): This construct was generated by site-directed mutagenesis of pcDNA.L1-RP.
pcDNA6.ORF2: The L1.3 5′-UTR-ORF2 was liberated from pCEP 5′-UTR-ORF2 no neo (71) as a 4.8 kb fragment and swapped into pcDNA.L1-RP at NotI–AleI.
pCEP.ORF2: Referred to as pCEP 5′ UTR ORF2 no neo, pCEP.ORF2 has been previously described (71).
Alu neoTet: This construct has been previously described (25).
SVA.2 mneoI: The SVA locus on CH2 was isolated from genomic DNA by PCR and cloned into pBluescript as a 3.9 kb KpnI–HindIII fragment to make pBS SVA.2. The SVA.2 was liberated as a 2 kb KpnI–PpumI fragment and swapped into the KpnI–PpumI sites in pBS SINE-R.10 mneoI, a vector containing the complete SINE-R, derived from SVA.10, to make pBS SVA.2 mneoI. SVA.2 with the mneoI retrotransposition indicator cassette was swapped as a 4.2 kb KpnI–NotI fragment into pCEP-Pur KpnI–NotI sites to make SVA.2 mneoI.
SVA.10.B mneoI: SVA.10 mneoI with the 3′ transduction, consisting of the 3′ transduction from the ABCC2 locus, containing the last 23 bp of the SINE-R, SVA polyA signal and polyA tail, the 3′ Alu and the 160 bp transduction, was recloned from BAC DNA as a 0.5 kb SbfI–NotI fragment.
SVA.10 neoTet: This construct is the same as SVA.10 mneoI except it contains the neoTet retrotransposition indicator cassette (25) cloned in as an AgeI–SbfI fragment.
ORF1 mneoI: This construct has been described previously (23).
ORF1 mneoI-AA: This construct has been described previously (31).
pcDNA.L1.3: L1.3: This construct was liberated from JM101/L1.3 (66) as a 6 kb NotI–AleI fragment and swapped into pcDNA.L1-RP at NotI–AleI sites.
pcDNA.L1-RPN157A/R159G-: This construct was generated by swapping a NotI–BstEII fragment from pEGFP-N3 L1-RPN157A/R159G (74) and swapped into pcDNA.L1-RP.
pcDNA.ORF1-RP: This construct contains the 5′ UTR and ORF1 from L1-RP cloned into NotI–AgeI of pcDNA6 as a 2 kb NotI–XmaI fragment.
pcDNA.L1-RPΔΔ: This construct contains a 5 kb L1-RP sequence lacking the L1 5′ UTR cloned into a modified pcDNA6 vector lacking the CMV promoter.
SVA.SRE1 mneoI: This construct contains the source element (SVA Retrotransposable Element) to the SPTA1 insertion (5). This element is marked with the mneoI retrotransposition indicator cassette (24,62) in pCEP4 and contains ∼700 bp 5′ flanking sequence.
99 SVA.SRE1 mneoI: This construct is the same as SVA.SRE-1 mneoI but is cloned into a modified pCEP vector (24) lacking the CMV promoter.
EGFP mneoI: This construct consists of the EGFP coding sequence from pEGFP-N1 (CLONTECH) cloned into pCEP4 and marked with the mneoI retrotransposition indicator cassette (24,62).
99 RPS EGFP Pur: This construct has been described previously (76).
99 RPS JM111 EGFP Pur: This construct has been described previously (76).
SVA.10 EGFP: This construct is similar to SVA.10 mneoI, except it contains the ∼2.5 kb EGFP retrotransposition indicator cassette (76) cloned in as an AgeI–SbfI fragment.
SVA.2 EGFP: The EGFP retrotransposition indicator cassette was swapped into pBS SVA.2 mneoI as a 2.5 kb SalI–NotI fragment to make pBS SVA.2 EGFP. pBS SVA.2 EGFP was liberated as a 4.5 kb KpnI–NotI and ligated into KpnI–NotI sites in pCEP-Pur.
SVA.10R EGFP Δ3TR: SVA.10 was liberated as a 3.5 kb KpnI–PpumI fragment from pBS SVA.10 mneoI and swapped into pBS SVA.2 EGFP at KpnI–PpumI sites to make pBS SVA.10R EGFP Δ3TR. pBS SVA.10R EGFP Δ3TR was liberated as a 6.2 kb KpnI–NotI and ligated into KpnI–NotI sites in pCEP-Pur.
DNA preparation
Plasmid DNA was prepped using the QIAGEN MaxiPrep Kit (QIAGEN). Genomic DNA was isolated using the DNA Mini Kit (QIAGEN).
Cell culture
HeLa HA, HEK 293T, U2OS and 143B cells were incubated at 37°C with 5% CO2 and 100% humidity in DMEM (Gibco) supplemented with 10% FBS (Hyclone), 1% Penn-Strep (Gibco) and 1% GlutaMax (Gibco).
neo Trans-mobilization assays
Transient retrotransposition assays (84) were carried out similarly to those previously reported for Alu (67,71), with slight modifications. For assays in six-well and T75 flasks, we seeded out ∼2 × 105 or ∼2 × 106 HeLa HA cells per well/flask. The following day, 0.5 µg of the passenger and 1.5 µg of the driver plasmid DNA or 4 µg of the passenger and 8 µg of the driver plasmid DNA for T75 flasks were co-transfected using 6 µl Fugene (Roche) according to the manufacturer's instructions. Seventy-two hours after transfection, G418 was added to the cells. Cells were re-fed routinely with media containing G418. After G418 selection, approximately 14 days, cells were washed, fixed and stained with giemsa.
SVA.10 mneoI retrotransposition frequency was calculated as the number of G418R foci per transfected cell. Transfection efficiency was determined by co-transfecting 0.5 µg of N1-EGFP (CLONTECH) in addition to 0.5 µg of SVA.10 mneoI and 1.5 µg of the driver plasmid using 6 µl Fugene (Roche) into HeLa HA cells in six-well plates. One day after transfection, EGFP-expressing cells were counted using an Accuri C6 flow cytometer. The mean percentage of EGFP-expressing cells from three replicates was calculated.
Northern blot analysis of SVA RNA in HeLa HA cells
HeLa HA cells were seeded in a 10 cm plate at a density to achieve 50% confluency at the time of transfection. Fugene 6 was used to transfect 2 µg of the driver (pcDNA.L1-RP) plasmid DNA and 4 µg of the passenger plasmid DNA into HeLa HA cells. Total RNA was isolated after 48 h using the RNeasy Mini Kit (QIAGEN). RNA was mixed with NorthernMax-Gly Sample Loading Dye (Ambion) at a 1:2 RNA:dye ratio, incubated at 55°C for 1 h, chilled on ice for 5 min before loading. Ten micrograms of total RNA was separated on a 1% denatured agarose gel. The RNA was transferred to a nylon membrane, UV-crosslinked and pre-hybridized at 68°C for 1 h, followed by overnight hybridization at 68°C with a neo sense riboprobe (300 bases), lacking the intron, labeled with digoxygenin (DIG)-11-UTP. The next day, the membrane was washed, immunodetected with anti-DIG-AP Fab fragments (Roche), visualized with the chemiluminescence substrate CDP-star (Roche) and exposed.
EGFP retrotransposition assays
Approximately 2 × 105 HeLa HA, 293T, U2OS or 143B cells were seeded out per well in six-well plates. The following day, 1 µg of the passenger and 1 µg of the driver plasmid (pcDNA.L1-RP, pcDNA.ORF1, pcDNA.ORF2 or pcDNA.L1-RP (D702Y) DNA was co-transfected into cells, using Fugene6 (Roche). 99 RPS EGFP Pur was also co-transfected with the driver plasmid DNA to normalize for co-transfection efficiency. In the experiments labeled FL-L1/2, cells were transfected with 1 µg of the passenger and 0.5 µg of the pcDNA.L1-RP plasmid DNA. For ORF1/ORF2 transfections, 1 µg of the passenger, 0.5 µg of pcDNA.ORF1 and 0.5 µg of pcDNA.ORF2 were co-transfected. Two days after transfection, puromycin was added to the media in order to select for cells transfected with the passenger plasmid. Puromycin was efficient at selecting transfected cells within 2 days after being added to the media. Five days after transfection, cells were harvested, washed and subjected to flow cytometry on a FACSCalibur machine. The gate was set for flow cytometry with cells co-transfected with SVA.10 EGFP and pcDNA.L1-RP (D702Y). We routinely counted approximately 100 000 cells per transfection. All transfections were performed in triplicate, except SVA.10R EGFP Δ3TR and SVA.10 EGFP/ORF1/ORF2 in HeLa HA cells. The %EGFP-positive was calculated as the number of EGFP-positive cells divided by the number of transfected cells.
Inverse PCR
Genomic DNA (0.5 µg) isolated from cell lines generated from individual foci was digested for 4 h to overnight with SacI, PpumI or HindIII (New England Biolabs) in a total reaction volume of 50 µl. The reaction was heat-inactivated by incubating at 65°C for 20 min. The digested DNA was ligated using T4 DNA ligase (New England Biolabs) in a total volume of 500 µl at 16°C for at least 16 h. DNA ligase was heat-inactivated at 65°C for 20 min. The DNA was ethanol-precipitated and resuspended in a total volume of 50 µl. PCR was performed using Ex Taq (Takara) according to manufacturer's instructions with 1 µl from the ligation reaction used as template. Nested PCR was carried out using 1 µl of the first-round PCR diluted 200×. PCR products were resolved on 1% agarose gels. Bands of interest were excised and purified using the QIAquick Gel Extraction Kit (QIAGEN). Purified products were either sequenced directly by the Sanger method or TOPO (Invitrogen) cloned followed by sequencing. DNA sequences were checked for appropriate digestion followed by ligation at the expected restriction site. Genomic locations of SVA inserts were determined relative to the current human genome reference assembly (hg19/NCBI37) using the UCSC genome browser (85).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by a National Institutes of Health grant to H.H.K., Jr. D.C.H was funded in part by a genetics training grant, T32-GM008216-23, from the National Institutes of Health.
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of the University of Pennsylvania School of Medicine and the Core DNA Analysis Facility at Johns Hopkins School of Medicine for DNA sequencing assistance. We would like to thank Dr John Moran for the HA-HeLa cells and CEP L1.3 ORF2 no neo, Dr Thierry Heidmann for the Alu neoTet plasmid and Dr Jose Garcia-Perez for the EGFP mneoI construct. We thank two anonymous reviewers for comments and suggestions that helped improve this manuscript. Likewise, we thank David Sigmon for excellent technical assistance and Adam Ewing for comments on this manuscript and insight throughout the project. Finally, we are grateful to past members of the Kazazian laboratory that worked on SVA over the years, in particular Dr Eric Ostertag, who cloned the SRE.1 mneoI construct.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Lander E., Linton L., Birren B., Nusbaum C., Zody M., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. doi:10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Cordaux R., Batzer M.A. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 2009;10:691–703. doi: 10.1038/nrg2640. doi:10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kazazian H.H., Jr, Wong C., Youssoufian H., Scott A.F., Phillips D.G., Antonarakis S.E. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332:164–166. doi: 10.1038/332164a0. doi:10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- 4.Wallace M.R., Andersen L.B., Saulino A.M., Gregory P.E., Glover T.W., Collins F.S. A de novo Alu insertion results in neurofibromatosis type 1. Nature. 1991;353:864–866. doi: 10.1038/353864a0. doi:10.1038/353864a0. [DOI] [PubMed] [Google Scholar]
- 5.Ostertag E.M., Goodier J.L., Zhang Y., Kazazian H.H., Jr SVA elements are nonautonomous retrotransposons that cause disease in humans. Am. J. Hum. Genet. 2003;73:1444–1451. doi: 10.1086/380207. doi:10.1086/380207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck C.R., Collier P., Macfarlane C., Malig M., Kidd J.M., Eichler E.E., Badge R.M., Moran J.V. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. doi:10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ewing A.D., Kazazian H.H. High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res. 2010;20:1262–1270. doi: 10.1101/gr.106419.110. doi:10.1101/gr.106419.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang C.R.L., Schneider A.M., Lu Y., Niranjan T., Shen P., Robinson M.A., Steranka J.P., Valle D., Civin C.I., Wang T., et al. Mobile interspersed repeats are major structural variants in the human genome. Cell. 2010;141:1171–1182. doi: 10.1016/j.cell.2010.05.026. doi:10.1016/j.cell.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Witherspoon D., Xing J., Zhang Y., Watkins W.S., Batzer M., Jorde L. Mobile element scanning (ME-Scan) by targeted high-throughput sequencing. BMC Genomics. 2010;11:410. doi: 10.1186/1471-2164-11-410. doi:10.1186/1471-2164-11-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kidd J.M., Graves T., Newman T.L., Fulton R., Hayden H.S., Malig M., Kallicki J., Kaul R., Wilson R.K., Eichler E.E. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell. 2010;143:837–847. doi: 10.1016/j.cell.2010.10.027. doi:10.1016/j.cell.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hormozdiari F., Alkan C., Ventura M., Hajirasouliha I., Malig M., Hach F., Yorukoglu D., Dao P., Bakhshi M., Sahinalp S.C., et al. Alu repeat discovery and characterization within human genomes. Genome Res. 2011;21:840–849. doi: 10.1101/gr.115956.110. published in advance 3 December 2010 doi:10.1101/gr.115956.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iskow R.C., McCabe M.T., Mills R.E., Torene S., Pittard W.S., Neuwald A.F., Van Meir E.G., Vertino P.M., Devine S.E. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. doi:10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ewing A.D., Kazazian H.H. Whole-genome resequencing allows detection of many rare LINE-1 insertion alleles in humans. Genome Res. 2011;21:985–990. doi: 10.1101/gr.114777.110. published in advance 27 October 2010 doi:10.1101/gr.114777.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pickeral O.K., Makalowski W., Boguski M.S., Boeke J.D. Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res. 2000;10:411–415. doi: 10.1101/gr.10.4.411. doi:10.1101/gr.10.4.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodier J.L., Ostertag E.M., Kazazian H.H., Jr Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum. Mol. Genet. 2000;9:653–657. doi: 10.1093/hmg/9.4.653. doi:10.1093/hmg/9.4.653. [DOI] [PubMed] [Google Scholar]
- 16.Szak S.T., Pickeral O.K., Makalowski W., Boguski M.S., Landsman D., Boeke J.D. Molecular archeology of L1 insertions in the human genome. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-10-research0052. research0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sen S.K., Huang C.T., Han K., Batzer M.A. Endonuclease-independent insertion provides an alternative pathway for L1 retrotransposition in the human genome. Nucleic Acids Res. 2007;35:3741–3751. doi: 10.1093/nar/gkm317. doi:10.1093/nar/gkm317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srikanta D., Sen S.K., Huang C.T., Conlin E.M., Rhodes R.M., Batzer M.A. An alternative pathway for Alu retrotransposition suggests a role in DNA double-strand break repair. Genomics. 2009;93:205–212. doi: 10.1016/j.ygeno.2008.09.016. doi:10.1016/j.ygeno.2008.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han K., Lee J., Meyer T.J., Remedios P., Goodwin L., Batzer M.A. L1 recombination-associated deletions generate human genomic variation. Proc. Natl Acad. Sci. USA. 2008;105:19366–19371. doi: 10.1073/pnas.0807866105. doi:10.1073/pnas.0807866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang H., Xing J., Grover D., Hedges D.J., Han K., Walker J.A., Batzer M.A. SVA elements: a hominid-specific retroposon family. J. Mol. Biol. 2005;354:994–1007. doi: 10.1016/j.jmb.2005.09.085. doi:10.1016/j.jmb.2005.09.085. [DOI] [PubMed] [Google Scholar]
- 21.Xing J., Wang H., Belancio V.P., Cordaux R., Deininger P.L., Batzer M.A. Emergence of primate genes by retrotransposon-mediated sequence transduction. Proc. Natl Acad. Sci. USA. 2006;103:17608–17613. doi: 10.1073/pnas.0603224103. doi:10.1073/pnas.0603224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Symer D.E., Connelly C., Szak S.T., Caputo E.M., Cost G.J., Parmigiani G., Boeke J.D. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110:327–338. doi: 10.1016/s0092-8674(02)00839-5. doi:10.1016/S0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 23.Wei W., Gilbert N., Ooi S.L., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. doi:10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moran J.V., Holmes S.E., Naas T.P., DeBerardinis R.J., Boeke J.D., Kazazian H.H., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. doi:10.1016/S0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 25.Dewannieux M., Esnault C., Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 2003;35:41–48. doi: 10.1038/ng1223. doi:10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 26.Esnault C., Maestre J., Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 2000;24:363–367. doi: 10.1038/74184. doi:10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 27.Comeaux M.S., Roy-Engel A.M., Hedges D.J., Deininger P.L. Diverse cis factors controlling Alu retrotransposition: what causes Alu elements to die? Genome Res. 2009;19:545–555. doi: 10.1101/gr.089789.108. doi:10.1101/gr.089789.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bennett E.A., Keller H., Mills R.E., Schmidt S., Moran J.V., Weichenrieder O., Devine S.E. Active Alu retrotransposons in the human genome. Genome Res. 2008;18:1875–1883. doi: 10.1101/gr.081737.108. doi:10.1101/gr.081737.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrish T.A., Gilbert N., Myers J.S., Vincent B.J., Stamato T.D., Taccioli G.E., Batzer M.A., Moran J.V. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat. Genet. 2002;31:159–165. doi: 10.1038/ng898. doi:10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 30.Morrish T.A., Garcia-Perez J.L., Stamato T.D., Taccioli G.E., Sekiguchi J., Moran J.V. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature. 2007;446:208–212. doi: 10.1038/nature05560. doi:10.1038/nature05560. [DOI] [PubMed] [Google Scholar]
- 31.Garcia-Perez J.L., Doucet A.J., Bucheton A., Moran J.V., Gilbert N. Distinct mechanisms for trans-mediated mobilization of cellular RNAs by the LINE-1 reverse transcriptase. Genome Res. 2007;17:602–611. doi: 10.1101/gr.5870107. doi:10.1101/gr.5870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia-Perez J.L., Marchetto M.C., Muotri A.R., Coufal N.G., Gage F.H., O'Shea K.S., Moran J.V. LINE-1 retrotransposition in human embryonic stem cells. Hum. Mol. Genet. 2007;16:1569–1577. doi: 10.1093/hmg/ddm105. doi:10.1093/hmg/ddm105. [DOI] [PubMed] [Google Scholar]
- 33.Garcia-Perez J.L., Morell M., Scheys J.O., Kulpa D.A., Morell S., Carter C.C., Hammer G.D., Collins K.L., O'Shea K.S., Menendez P., et al. Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature. 2010;466:769–773. doi: 10.1038/nature09209. doi:10.1038/nature09209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rangwala S.H., Kazazian H.H., Jr The L1 retrotransposition assay: a retrospective and toolkit. Methods. 2009;9:219–226. doi: 10.1016/j.ymeth.2009.04.012. doi:10.1016/j.ymeth.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilbert N., Lutz S., Morrish T.A., Moran J.V. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol. Cell. Biol. 2005;25:7780–7795. doi: 10.1128/MCB.25.17.7780-7795.2005. doi:10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gilbert N., Lutz-Prigge S., Moran J.V. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110:315–325. doi: 10.1016/s0092-8674(02)00828-0. doi:10.1016/S0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 37.Ostertag E.M., DeBerardinis R.J., Goodier J.L., Zhang Y., Yang N., Gerton G.L., Kazazian H.H. A mouse model of human L1 retrotransposition. Nat. Genet. 2002;32:655–660. doi: 10.1038/ng1022. doi:10.1038/ng1022. [DOI] [PubMed] [Google Scholar]
- 38.Kano H., Godoy I., Courtney C., Vetter M.R., Gerton G.L., Ostertag E.M., Kazazian H.H., Jr L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 2009;23:1303–1312. doi: 10.1101/gad.1803909. doi:10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prak E.T., Dodson A.W., Farkash E.A., Kazazian H.H., Jr Tracking an embryonic L1 retrotransposition event. Proc. Natl Acad. Sci. USA. 2003;100:1832–1837. doi: 10.1073/pnas.0337627100. doi:10.1073/pnas.0337627100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Babushok D.V., Ostertag E.M., Courtney C.E., Choi J.M., Kazazian H.H. L1 integration in a transgenic mouse model. Genome Res. 2006;16:240–250. doi: 10.1101/gr.4571606. doi:10.1101/gr.4571606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.An W., Han J.S., Wheelan S.J., Davis E.S., Coombes C.E., Ye P., Triplett C., Boeke J.D. Active retrotransposition by a synthetic L1 element in mice. Proc. Natl Acad. Sci. USA. 2006;103:18662–18667. doi: 10.1073/pnas.0605300103. doi:10.1073/pnas.0605300103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muotri A.R., Chu V.T., Marchetto M.C., Deng W., Moran J.V., Gage F.H. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. doi:10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 43.Kobayashi K., Nakahori Y., Miyake M., Matsumura K., Kondo-Iida E., Nomura Y., Segawa M., Yoshioka M., Saito K., Osawa M., et al. An ancient retrotransposal insertion causes Fukuyama-type congenital muscular dystrophy. Nature. 1998;394:388–392. doi: 10.1038/28653. doi:10.1038/28653. [DOI] [PubMed] [Google Scholar]
- 44.Wilund K.R., Yi M., Campagna F., Arca M., Zuliani G., Fellin R., Ho Y.-K., Garcia J.V., Hobbs H.H., Cohen J.C. Molecular mechanisms of autosomal recessive hypercholesterolemia. Hum. Mol. Genet. 2002;11:3019–3030. doi: 10.1093/hmg/11.24.3019. doi:10.1093/hmg/11.24.3019. [DOI] [PubMed] [Google Scholar]
- 45.Makino S., Kaji R., Ando S., Tomizawa M., Yasuno K., Goto S., Matsumoto S., Tabuena M.D., Maranon E., Dantes M., et al. Reduced neuron-specific expression of the TAF1 gene is associated with X-linked dystonia-parkinsonism. Am. J. Hum. Genet. 2007;80:393–406. doi: 10.1086/512129. doi:10.1086/512129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takasu M., Hayashi R., Maruya E., Ota M., Imura K., Kougo K., Kobayashi C., Saji H., Ishikawa Y., Asai T., et al. Deletion of entire HLA-A gene accompanied by an insertion of a retrotransposon. Tissue Antigens. 2007;70:144–150. doi: 10.1111/j.1399-0039.2007.00870.x. doi:10.1111/j.1399-0039.2007.00870.x. [DOI] [PubMed] [Google Scholar]
- 47.Rohrer J., Minegishi Y., Richter D., Eguiguren J., Conley M.E. Unusual mutations in Btk: an insertion, a duplication, an inversion, and four large deletions. Clin. Immunol. 1999;90:28–37. doi: 10.1006/clim.1998.4629. doi:10.1006/clim.1998.4629. [DOI] [PubMed] [Google Scholar]
- 48.Hassoun H., Coetzer T.L., Vassiliadis J.N., Sahr K.E., Maalouf G.J., Saad S.T., Catanzariti L., Palek J. A novel mobile element inserted in the alpha spectrin gene: spectrin dayton. A truncated alpha spectrin associated with hereditary elliptocytosis. J. Clin. Invest. 1994;94:643–648. doi: 10.1172/JCI117380. doi:10.1172/JCI117380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Legoix P., Sarkissian H.D., Cazes L., Giraud S., Sor F., Rouleau G.A., Lenoir G., Thomas G., Zucman-Rossi J. Molecular characterization of germline NF2 gene rearrangements. Genomics. 2000;65:62–66. doi: 10.1006/geno.2000.6139. doi:10.1006/geno.2000.6139. [DOI] [PubMed] [Google Scholar]
- 50.Akman H.O., Davidzon G., Tanji K., MacDermott E.J., Larsen L., Davidson M.M., Haller R.G., Szczepaniak L.S., Lehman T.J.A., Hirano M., et al. Neutral lipid storage disease with subclinical myopathy due to a retrotransposal insertion in the PNPLA2 gene. Neuromuscul. Disord. 2010;20:397–402. doi: 10.1016/j.nmd.2010.04.004. doi:10.1016/j.nmd.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ono M., Kawakami M., Takezawa T. A novel human nonviral retroposon derived from an endogenous retrovirus. Nucleic Acids Res. 1987;15:8725–8737. doi: 10.1093/nar/15.21.8725. doi:10.1093/nar/15.21.8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu Z.B., Hsieh S.L., Bentley D.R., Campbell R.D., Volanakis J.E. A variable number of tandem repeats locus within the human complement C2 gene is associated with a retroposon derived from a human endogenous retrovirus. J. Exp. Med. 1992;175:1783–1787. doi: 10.1084/jem.175.6.1783. doi:10.1084/jem.175.6.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shen L., Wu L.C., Sanlioglu S., Chen R., Mendoza A.R., Dangel A.W., Carroll M.C., Zipf W.B., Yu C.Y. Structure and genetics of the partially duplicated gene RP located immediately upstream of the complement C4A and the C4B genes in the HLA class III region. Molecular cloning, exon–intron structure, composite retroposon, and breakpoint of gene duplication. J. Biol. Chem. 1994;269:8466–8476. [PubMed] [Google Scholar]
- 54.Strichman-Almashanu L.Z., Lee R.S., Onyango P.O., Perlman E., Flam F., Frieman M.B., Feinberg A.P. A genome-wide screen for normally methylated human CpG islands that can identify novel imprinted genes. Genome Res. 2002;12:543–554. doi: 10.1101/gr.224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han K., Konkel M.K., Xing J., Wang H., Lee J., Meyer T.J., Huang C.T., Sandifer E., Hebert K., Barnes E.W., et al. Mobile DNA in Old World monkeys: a glimpse through the rhesus macaque genome. Science. 2007;316:238–240. doi: 10.1126/science.1139462. doi:10.1126/science.1139462. [DOI] [PubMed] [Google Scholar]
- 56.Kim H.S., Wadekar R.V., Takenaka O., Hyun B.H., Crow T.J. Phylogenetic analysis of a retroposon family in African great apes. J. Mol. Evol. 1999;49:699–702. doi: 10.1007/pl00000083. doi:10.1007/PL00000083. [DOI] [PubMed] [Google Scholar]
- 57.Hancks D.C., Kazazian H.H., Jr SVA retrotransposons: evolution and genetic instability. Semin. Cancer Biol. 2010;20:234–245. doi: 10.1016/j.semcancer.2010.04.001. doi:10.1016/j.semcancer.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ostertag E.M., Kazazian H.H., Jr Twin priming: a proposed mechanism for the creation of inversions in L1 retrotransposition. Genome Res. 2001;11:2059–2065. doi: 10.1101/gr.205701. doi:10.1101/gr.205701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hancks D.C., Ewing A.D., Chen J.E., Tokunaga K., Kazazian H.H., Jr Exon-trapping mediated by the human retrotransposon SVA. Genome Res. 2009;19:1983–1991. doi: 10.1101/gr.093153.109. doi:10.1101/gr.093153.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Damert A., Raiz J., Horn A.V., Lower J., Wang H., Xing J., Batzer M.A., Lower R., Schumann G.G. 5′-transducing SVA retrotransposon groups spread efficiently throughout the human genome. Genome Res. 2009;19:1992–2008. doi: 10.1101/gr.093435.109. doi:10.1101/gr.093435.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bantysh O.B., Buzdin A.A. Novel family of human transposable elements formed due to fusion of the first exon of gene MAST2 with retrotransposon SVA. Biochemistry (Moscow) 2009;74:1393–1399. doi: 10.1134/s0006297909120153. doi:10.1134/S0006297909120153. [DOI] [PubMed] [Google Scholar]
- 62.Freeman J.D., Goodchild N.L., Mager D.L. A modified indicator gene for selection of retrotransposition events in mammalian cells. Biotechniques. 1994;17:46. 48, 49, 52. [PubMed] [Google Scholar]
- 63.Jensen S., Heidmann T. An indicator gene for detection of germline retrotransposition in transgenic Drosophila demonstrates RNA-mediated transposition of the LINE I element. EMBO J. 1991;10:1927–1937. doi: 10.1002/j.1460-2075.1991.tb07719.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kimberland M.L., Divoky V., Prchal J., Schwahn U., Berger W., Kazazian H.H., Jr Full-length human L1 insertions retain the capacity for high frequency retrotransposition in cultured cells. Hum. Mol. Genet. 1999;8:1557–1560. doi: 10.1093/hmg/8.8.1557. doi:10.1093/hmg/8.8.1557. [DOI] [PubMed] [Google Scholar]
- 65.Dombroski B.A., Scott A.F., Kazazian H.H., Jr Two additional potential retrotransposons isolated from a human L1 subfamily that contains an active retrotransposable element. Proc. Natl Acad. Sci. USA. 1993;90:6513–6517. doi: 10.1073/pnas.90.14.6513. doi:10.1073/pnas.90.14.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sassaman D.M., Dombroski B.A., Moran J.V., Kimberland M.L., Naas T.P., DeBerardinis R.J., Gabriel A., Swergold G.D., Kazazian H.H., Jr Many human L1 elements are capable of retrotransposition. Nat. Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. doi:10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- 67.Bogerd H.P., Wiegand H.L., Hulme A.E., Garcia-Perez J.L., O'Shea K.S., Moran J.V., Cullen B.R. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl Acad. Sci. USA. 2006;103:8780–8785. doi: 10.1073/pnas.0603313103. doi:10.1073/pnas.0603313103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Esnault C., Casella J.F., Heidmann T. A Tetrahymena thermophila ribozyme-based indicator gene to detect transposition of marked retroelements in mammalian cells. Nucleic Acids Res. 2002;30:e49. doi: 10.1093/nar/30.11.e49. doi:10.1093/nar/30.11.e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wallace N., Wagstaff B.J., Deininger P.L., Roy-Engel A.M. LINE-1 ORF1 protein enhances Alu SINE retrotransposition. Gene. 2008;419:1–6. doi: 10.1016/j.gene.2008.04.007. doi:10.1016/j.gene.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mathias S.L., Scott A.F., Kazazian H.H., Jr, Boeke J.D., Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. doi:10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 71.Alisch R.S., Garcia-Perez J.L., Muotri A.R., Gage F.H., Moran J.V. Unconventional translation of mammalian LINE-1 retrotransposons. Genes Dev. 2006;20:210–224. doi: 10.1101/gad.1380406. doi:10.1101/gad.1380406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Khazina E., Weichenrieder O. Non-LTR retrotransposons encode noncanonical RRM domains in their first open reading frame. Proc. Natl Acad. Sci. USA. 2009;106:731–736. doi: 10.1073/pnas.0809964106. doi:10.1073/pnas.0809964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Martin S.L. The ORF1 protein encoded by LINE-1: structure and function during L1 retrotransposition. J. Biomed. Biotechnol. 2006;2006:1. doi: 10.1155/JBB/2006/45621. doi:10.1155/JBB/2006/45621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Goodier J.L., Zhang L., Vetter M.R., Kazazian H.H., Jr LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol. Cell. Biol. 2007;27:6469–6483. doi: 10.1128/MCB.00332-07. doi:10.1128/MCB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Doucet A.J., Hulme A.E., Sahinovic E., Kulpa D.A., Moldovan J.B., Kopera H.C., Athanikar J.N., Hasnaoui M., Bucheton A., Moran J.V., et al. Characterization of LINE-1 ribonucleoprotein particles. PLoS Genet. 2010;6:e1001150. doi: 10.1371/journal.pgen.1001150. doi:10.1371/journal.pgen.1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ostertag E.M., Prak E.T., DeBerardinis R.J., Moran J.V., Kazazian H.H., Jr Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 2000;28:1418–1423. doi: 10.1093/nar/28.6.1418. doi:10.1093/nar/28.6.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kubo S., Seleme M.C., Soifer H.S., Perez J.L., Moran J.V., Kazazian H.H., Jr, Kasahara N. L1 retrotransposition in nondividing and primary human somatic cells. Proc. Natl Acad. Sci. USA. 2006;103:8036–8041. doi: 10.1073/pnas.0601954103. doi:10.1073/pnas.0601954103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Brouha B., Schustak J., Badge R.M., Lutz-Prigge S., Farley A.H., Moran J.V., Kazazian H.H., Jr Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl Acad. Sci. USA. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. doi:10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Feng Q., Moran J.V., Kazazian H.H., Jr, Boeke J.D. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. doi:10.1016/S0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 80.Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc. Natl Acad. Sci. USA. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. doi:10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cost G.J., Feng Q., Jacquier A., Boeke J.D. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–5910. doi: 10.1093/emboj/cdf592. doi:10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stenberg R.M., Thomsen D.R., Stinski M.F. Structural analysis of the major immediate early gene of human cytomegalovirus. J. Virol. 1984;49:190–199. doi: 10.1128/jvi.49.1.190-199.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Belancio V.P., Hedges D.J., Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health. Genome Res. 2008;18:343–358. doi: 10.1101/gr.5558208. doi:10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- 84.Wei W., Morrish T.A., Alisch R.S., Moran J.V. A transient assay reveals that cultured human cells can accommodate multiple LINE-1 retrotransposition events. Anal. Biochem. 2000;284:435–438. doi: 10.1006/abio.2000.4675. doi:10.1006/abio.2000.4675. [DOI] [PubMed] [Google Scholar]
- 85.Kent W., Sugnet C., Furey T., Roskin K., Pringle T., Zahler A., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Crooks G.E., Hon G., Chandonia J.-M., Brenner S.E. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. doi:10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.