Abstract
Expanded polyglutamine (polyQ) tract in the human TATA-box-binding protein (hTBP) causes the neurodegenerative disease spinocerebellar ataxia 17 (SCA17). To investigate the pathological effects of polyQ expansion, we established a SCA17 model in Drosophila. Similar to SCA17 patients, transgenic flies expressing a mutant hTBP protein with an expanded polyQ tract (hTBP80Q) exhibit progressive neurodegeneration, late-onset locomotor impairment and shortened lifespan. Microarray analysis reveals that hTBP80Q causes widespread and time-dependent transcriptional dysregulation in Drosophila. In a candidate screen for genetic modifiers, we identified RBP-J/Su(H), a transcription factor that contains Q/N-rich domains and participates in Notch signaling. Knockdown of Su(H) by RNAi further enhances hTBP80Q-induced eye defects, whereas overexpression of Su(H) suppresses such defects. While the Su(H) transcript level is not significantly altered in hTBP80Q-expressing flies, genes that contain Su(H)-binding sites are among those that are dysregulated. We further show that hTBP80Q interacts more efficiently with Su(H) than wild-type hTBP, suggesting that a reduction in the fraction of Su(H) available for its normal cellular functions contributes to hTBP80Q-induced phenotypes. While the Notch signaling pathway has been implicated in several neurological disorders, our study suggests a possibility that the activity of its nuclear component RBP-J/Su(H) may modulate the pathological progression in SCA17 patients.
INTRODUCTION
Spinocerebellar ataxia 17 (SCA17) is a late-onset, neurodegenerative disease (1,2). It is an autosomal dominant and progressive disease characterized by ataxia, dystonia, parkinsonism, dementia and seizures. Marked cerebellar atrophy and Purkinje cell loss are typical in SCA17 patients (1–3). SCA17 is one of the nine diseases that are associated with expanded tracts of glutamines, referred to as polyglutamine (polyQ) (4). These polyQ diseases include Huntington's disease (HD), spinal bulbar muscular atrophy (SBMA), dentatorubropallidoluysian atrophy (DRPLA) and the other five types of SCA, i.e. 1, 2, 3, 6 and 7. They are characterized by the pathological expansion of the CAG trinucleotide repeat in the coding regions of unrelated genes and comprise one of the most common groups of inherited neurodegenerative conditions (4). For example, the TATA-binding protein (TBP) found in normal human populations has a polyQ tract ranging from 29 to 42 glutamines (2). When this polyQ tract is expanded to 45–63 glutamines, the variant human TBP (hTBP) becomes pathological in inducing SCA17 (4).
Although the nine polyQ diseases are caused by distinct proteins with unrelated biological functions in their non-pathological forms, their phenotypes share some common features. For example, these diseases induce neurodegeneration in a progressive way with symptoms generally developing around midlife (4). The pathological expansion of polyQ in the causative proteins is thought to cause protein aggregates found in patients' brain tissue (4), although it remains controversial whether it is the aggregate, insoluble form or the oligomeric, soluble form that is responsible for neuropathology (5–7). It has been suggested that expanded polyQ sequences can alter how the causative proteins interact with other cellular proteins (referred to as targets) and such aberrant interactions may directly contribute to pathogenesis (8–10). For example, in HD, polyQ expansion induces altered interactions between huntingtin and a range of transcription factors, such as Sp1 and REST/NRSF (11–16). In addition, ataxin1 with an expanded polyQ has been shown to have aberrant interactions with different transcription factors, including LANP, PQBP1, Gfi-1, SMRT, Boat and Sp1 (17–25).
It has been suggested that both the intrinsic toxicity of polyQ and the context of the causative proteins may contribute to the neurodegenerative diseases (26). Among the nine polyQ diseases, only the causative genes for SBMA, SCA6 and SCA17 have, in their non-pathological forms, well-characterized biological functions (27,28). In particular, TBP is a general transcriptional factor that is required for transcription by all three classes of RNA polymerases. TBP is a component of the TFIID complex and provides the DNA-binding specificity for the TATA-box of core promoters of protein-coding genes (29,30). In a previous study, Friedman et al. (31) generated a mouse disease model for SCA17 and found that HSPB1, a small heat shock protein and neuroprotective factor, was significantly down-regulated due to an enhanced interaction between mutant TBP and TFIIB. Shah et al. (32) subsequently found that the mutant TBP interacts with Sp1 more efficiently, an aberrant interaction suggested to influence the normal function of Sp1 as a transcription factor, leading to a reduced expression of its downstream target gene TrkA in the mouse. While these results represent important advances toward understanding the molecular basis of SCA17, the complexities of the protein–protein interaction networks, combined with the fact that TBP plays a role in transcription of virtually all genes, suggest that SCA17, like other polyQ diseases, may reflect altered activities of multiple targets and may involve distinct mechanisms. Since all the polyQ diseases share common pathological features and, furthermore, hTBP immunoreactivity is also detected in the nuclear inclusion in disease brains caused by other polyQ diseases such as HD, SCA1, SCA2, SCA3 and DRPLA (28,33–35), further investigations into the mechanisms of SCA17 should benefit our understanding of these neurodegenerative diseases as a whole.
Suppressor of Hairless, Su(H), the Drosophila homologue of RBP-J (the recombination signal-binding protein for immunoglobulin kappa J region), is a highly conserved transcription factor that participates in Notch signaling (36,37). It also belongs to a group of proteins that contain domains rich in glutamines and asparagines, referred to as Q/N-rich proteins (38). In the absence of Notch, the DNA-bound Su(H) acts to repress transcription (37,39). Upon ligand-induced activation of the Notch receptor, its intercellular fragment, Nicd, is released from the membrane and enters the nucleus, where it directly interacts with Su(H) and induces transcription of its target genes. The Notch signaling pathway is best known for its role in lateral inhibition (40). It is critical to a wide range of developmental processes, such as hematopoiesis, somitogenesis, vasculogenesis and neurogenesis (41). The Notch signaling pathway also plays a role in plasticity-related processes, including patterning of the neurite structure and maintenance of neural stem cells (42,43). Recent studies suggest that deficits in the Notch signaling pathway are involved in several neurodegenerative diseases. For example, mutations in Notch3 have been identified to be causative for cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy, a disease characterized predominantly by neurologic pathology (44). In addition, mutations in Notch4 have been identified as potential causative mutations for Schizophrenia, a complex mental illness (45).
In this study, we establish a Drosophila model (46) to investigate the disease mechanisms for SCA17. We show that mutant hTBP proteins cause defects that are characteristic of SCA17 pathology, including progressive neurodegeneration, late-onset locomotor impairment and early mortality. The severity of these defects is dependent on polyQ lengths. Microarray analysis reveals widespread and time-dependent transcriptional dysregulation in flies expressing a mutant hTBP with a tract of 80 glutamines (hTBP80Q). Our results suggest important contributions of Q/N-rich transcription factors to hTBP80Q-induced transcriptional dysregulation. Using the Drosophila SCA17 model, we identified Su(H) as a genetic modifier for the eye phenotype induced by hTBP80Q. While knockdown of Su(H) expression enhances the hTBP80Q-induced defects in both eye patterning and retinal degeneration, its overexpression suppresses these defects. Genes that contain Su(H)-binding sites at their promoter regions are among those that are dysregulated in flies that express hTBP80Q. We provide evidence suggesting that an altered interaction between hTBP and Su(H) caused by the polyQ expansion reduces the fraction of Su(H) available for its normal biological functions. Together, our results suggest a possibility that a reduced RBP-J/Su(H) activity may contribute to the pathological progression in SCA17 patients.
RESULTS
hTBP is biologically active in Drosophila
As a general transcription factor, TBP is essential for initiation of transcription (29,30). While the C-terminal domains of Drosophila and human TBP proteins (referred to as dTBP and hTBP, respectively) are highly conserved (Supplementary Material, Fig. S1A) and directly involved in DNA binding, the N-terminal divergent domain is suggested to play a role in species-specific interactions (30,47,48). To evaluate the functionality of hTBP in Drosophila and the feasibility of a SCA17 disease model in Drosophila, we obtained from the Bloomington Stock Center a dtbp mutant line (#18301), which has a piggyBac insertion at the 5′ of dTBP. Flies homozygous for this mutant dtbp allele die at the first instar larva stage (Supplementary Material, Fig. S1B), demonstrating that, as expected, dTBP is an essential gene. We constructed rescue vectors expressing from pUAST either the wild-type (WT) dTBP or WT hTBP. Both of these constructs, when driven by Hsp70-GAL4, similarly rescued the homozygous dtbp mutants with flies surviving past the first instar larval stage (Supplementary Material, Fig. S1B). This was only a partial rescue because the flies could not survive past the second instar larval stage, suggesting the importance of proper expression levels and/or patterns of TBP in its full biological functions. The important finding relevant to the current work is that both dTBP and hTBP resulted in a similar rescue, demonstrating that hTBP is biologically active in Drosophila. As shown below, full-length hTBP is stably expressed in Drosophila tissues, further suggesting that Drosophila is a suitable host organism for studying disease mechanisms of SCA17 (46).
hTBP induces eye phenotypes in Drosophila in a manner dependent on polyQ length
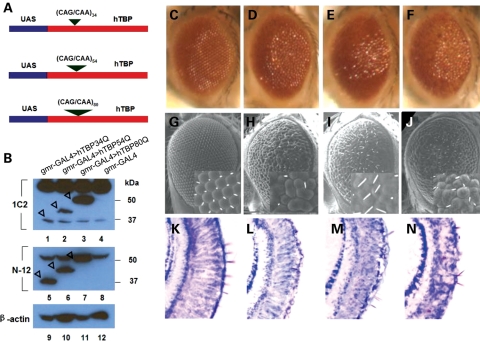
The polyQ tract located in the N-terminal of hTBP is encoded by two homogeneous CAG repeat blocks, one with 8–11 CAG repeats and the other 15–18. The CAG repeats in hTBP are interrupted at four different locations by a CAA triplet, which is suggested to stabilize the repeat sequences (28). Genetic variations, including pathological expansions, generally occur in the second CAG repeat block of the hTBP gene (28). To determine whether polyQ expansion in hTBP can cause neuropathology in flies, we generated hTBP cDNA constructs expressing proteins with varying polyQ lengths. We used hTBP with 34 glutamines as WT hTBP, also referred to as hTBP34Q. We introduced CAG triplets into the second CAG repeat block to generate hTBP mutants encoding proteins with either 54 glutamines (referred to as hTBP54Q) or 80 glutamines (referred to as hTBP80Q) (Fig. 1A). We used the GAL4/UAS system to express these proteins in the eye. The driver gmr-GAL4 directs expression in all differentiated cells of the developing eye, including photoreceptors and accessory pigment cells (49). Western blotting results (Fig. 1B) show that the hTBP proteins with different polyQ lengths were expressed in their full-length forms at comparable levels in fly eyes (see Fig. 1B legend for further details about antibody properties and results).
Figure 1.
Eye phenotype induced by hTBP in Drosophila. (A) Schematic diagrams of the constructs for UAS:hTBP34Q, UAS:hTBP54Q and UAS:hTBP80Q (not to scale). hTBP34Q is also referred to WT hTBP in the current work. See Supplementary Material, Fig. S1 for an alignment between hTBP and dTBP sequences. (B) Western blots detecting hTBP proteins in the head of control flies (lanes 4 and 8), or flies expressing hTBP34Q (lanes 1 and 5), hTBP54Q (lanes 2 and 6) or hTBP80Q (lanes 3 and 7). hTBP expression was driven by gmr-GAL4 and western blotting was detected by 1C2 (lanes 1–4) or N-12 antibodies (lanes 5–8). Protein bands for hTBP34Q, hTBP54Q and hTBP80Q are pointed by open arrowheads. β-Actin represents loading control (lanes 9–12). While both N-12 and the 1C2 antibodies recognize the N-terminal part of hTBP, 1C2 has the ability to detect more efficiently the pathological proteins with expanded polyQ than WT proteins (74). For this reason, the 1C2 antibody has been used to detect other polyQ expanded proteins involved in several other neurodegenerative diseases like Huntington's disease and SCA2, 3 and 7 (74). As expected, the 1C2 antibody detected more hTBP80Q than hTBP34Q or hTBP54Q proteins in our assays. In contrast, the N12 antibody detected similar amounts of these proteins, indicating that the full-length forms of these proteins are accumulated to similar levels in Drosophila. (C–N) Light microscopic (C–F) and SEM (G–J) images of 1-day-old fly eyes expressing hTBP34Q (D, H, L), hTBP54Q (E, I, M) or hTBP80Q (F, J, N). (C), (G) and (K) represent control flies containing only the gmr-GAL4 driver. Insets for (G)–(J) represent higher magnification of the ommatidia field. (K–N) Staining of sagittal sections of adult eyes with toluidene blue.
The hTBP proteins induced eye-patterning defects in a manner that is dependent on the length of the polyQ tract. Figure 1C and G shows, respectively, the light microscopic and scanning electron microscopic (SEM) images of control eyes of 1-day-old adult flies exhibiting well-organized ommatidia (see also Fig. 1K for toluidine blue-stained section detecting the gross organization of the photoreceptors). Expression of hTBP34Q, driven by gmr-GAL4, caused a relatively mild phenotype in the adult eye with irregular ommatidial morphology (Fig. 1D, H, L). Expression of hTBP54Q led to a more severe (relative to hTBP34Q-expressing flies) eye morphology with noticeable defects in the gross organization of photoreceptors (Fig. 1E, I, M). The most severe disorganization of the ommatidia was observed in fly eyes expressing hTBP80Q (Fig. 1J). Here, we observed a collapse of the eye structure (Fig. 1J), a loss of pigmentation (Fig. 1F) and severely disorganized photoreceptors (Fig. 1N). These abnormalities caused by hTBP80Q are similar to the eye phenotypes observed in the Drosophila model of SCA3 (50). These results suggest that polyQ expansion in hTBP contributes to eye patterning defects in Drosophila.
Progressive retinal degeneration caused by hTBP80Q
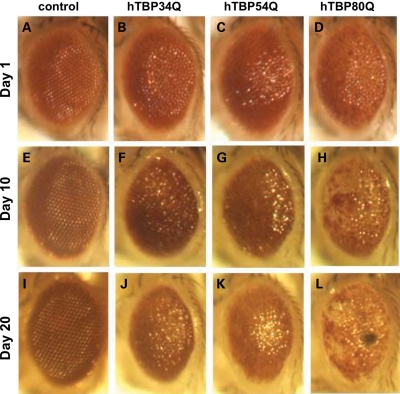
The length of the polyQ tract of the hTBP proteins affected not only the severity of eye morphology as discussed above (Fig. 1) but also phenotypic progression as a function of time. In particular, we compared the eye phenotypes on Day 1, Day 10 and Day 20 in flies that express (from the gmr-GAL4 driver) hTBP proteins with different polyQ lengths. While eye defects worsened over time for each genotype except the WT control (Fig. 2), this phenotypic progression is most dramatic in hTBP80Q-expressing flies. Here, the pigment loss of individual ommatidia was more apparent for 10-day-old hTBP80Q-expressing flies than 1-day-old hTBP80Q-expressing flies (Fig. 2D and H). The loss of pigmentation became even more dramatic on Day 20, with the appearance of spots suggestive of necrosis in the eyes of hTBP80Q-expressing flies (Fig. 2L). These results demonstrate that mutant hTBP causes progressive retinal degeneration in a polyQ-length-dependent manner in flies, further suggesting that these flies represent a good model for SCA17.
Figure 2.
Progressive retinal degeneration induced by hTBP with expanded polyQ. Light microscopic images of 1-day-old (A–D), 10-day-old (E–H) or 20-day-old (I–L) fly eyes expressing hTBP34Q (B, F, J), hTBP54Q (C, G, K) or hTBP80Q (D, H, L). (A)–(D) in this figure are the same as (C)–(F) of Figure 1.
hTBP80Q causes late-onset locomotor impairment and early mortality
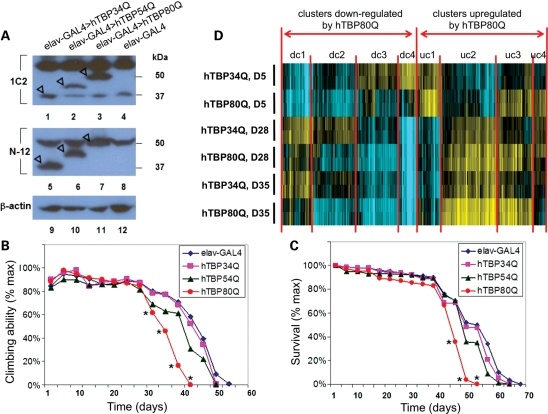
SCA17 is a progressive neurodegenerative disease characterized by ataxia, dystonia, parkinsonism, dementia and seizures in humans (1–3). To determine whether flies expressing mutant hTBP proteins also exhibit this particular aspect of SCA17 pathology, we expressed hTBP proteins with different polyQ lengths in all neurons using the panneuronal driver elav-GAL4 (elav stands for embryonic lethal abnormal visual system). We used these flies to evaluate their climbing performance. Western blotting results further confirmed that these hTBP proteins were expressed in their full-length forms at comparable levels in flies (Fig. 3A, also see Fig. 1B legend for additional details). All flies were backcrossed to w1118 for four generations to minimize background influence on behavior (or lifespan—see below). Figure 3B shows the results of climbing performance tests for adult flies that express hTBP34Q, hTBP54Q and hTBP80Q. While hTBP80Q-expressing flies at young ages performed similarly to other age-matched flies (Fig. 3B), they exhibited a late-onset locomotor impairment. In particular, on Day 31 after eclosion, the percentage of hTBP34Q-expressing flies that could climb to or above the 12 cm mark in 20 s was 78.6%. In contrast, the percentage of hTBP80Q-expressing flies that could perform the same task was only 61.6% (P-value < 0.05, Student's t-test), suggesting that these flies began to show locomotor impairment on Day 31. At later time points, climbing performance of hTBP80Q-expressing flies exhibited a more rapid deterioration and greater differences with flies expressing either hTBP34Q or hTBP54Q (Fig. 3B).
Figure 3.
Late-onset locomotor impairment and early mortality caused by hTBP80Q. (A) Western blots detecting hTBP in the head of control flies (lanes 4 and 8) or flies expressing hTBP34Q (lanes 1 and 5), hTBP54Q (lanes 2 and 6) or hTBP80Q (lanes 3 and 7). hTBP-expression was driven by elav-GAL4. See Figure 1B legend for further details. (B) Cohorts of 200 flies for each genotype were subjected to climbing assays every 3 days. Statistically significant differences between the hTBP80Q-expressing flies and hTBP34Q-expressing flies were indicated by asterisk where P < 0.05 (Student's t-test). (C) Survival curves for hTBP34Q-, hTBP54Q- and hTBP80Q-expressing flies as well as the elav-Gal4 flies. Statistically significant differences between hTBP80Q-expressing flies and hTBP34Q-expressing flies were indicated by asterisk where P < 0.05 (Student's t-test). The genotypes of the flies tested in (B) and (C) are: elav-GAL4/+, elav-GAL4/+; UAS-hTBP34Q/+, elav-GAL4/+; UAS-hTBP54Q/+, and elav-GAL4/+; UAS-hTBP80Q/+. (D) Heatmap representation of transcriptome analyses showing the effects of hTBP alleles on gene expression patterns as a function of animal age. Affymetrix probesets (from left to right) were identified that exhibited differential expression at each of the developmental time points as a function of hTBP allele and subjected to hierarchical clustering. Up- and down-clusters are those transcripts that were activated or repressed, respectively, in their expression by hTBP80Q relative to hTBP34Q at one or more developmental stages (samples are shown from top to bottom). See Supplementary Material, Table S1, for gene identities corresponding to the map and their respective cluster (dc1 = down-regulated by hTBP80Q cluster 1, etc.).
It has been shown that neurodegenerative diseases can shorten the lifespan of both human patients and disease model organisms, such as flies and mice (31,51). To further evaluate the effect of hTBP mutants, we performed a lifespan assay using adult flies expressing (driven by elav-GAL4) hTBP proteins with different polyQ lengths. Again, hTBP80Q-expressing flies exhibited shortened lifespan than hTBP34Q- or hTBP54Q-expressing flies (Fig. 3C). At the age of 52 days, none of the hTBP80Q-expressing flies survived, while about half of the control flies that expressed hTBP34Q remained alive. These results demonstrate a shortened lifespan of hTBP80Q-expressing flies. Together, our climbing and lifespan assays show that hTBP80Q can cause both late-onset locomotor impairment and early mortality in Drosophila, features characteristic of SCA17 pathology in humans.
Microarray analysis reveals hTBP80Q-induced transcriptional dysregulation and contributions of Q/N-rich transcription factors
Since TBP is a general transcription factor, it is possible that the expansion of its polyQ may cause significant alterations in gene transcription. To investigate this possibility, we performed microarray analysis using RNA samples isolated from fly heads expressing either hTBP80Q or hTBP34Q (driven by elav-GAL4). We analyzed RNA samples at three different time points: Day 5, Day 28 and Day 35. As shown in Figure 3B, Day 5 represents the initial state for both the control flies expressing hTBP34Q and the flies expressing hTBP80Q, each exhibiting a comparable locomotor ability. Day 28 is immediately prior to the onset of locomotor impairment for hTBP80Q-expressing flies, whereas Day 35 represents a state after the onset of the disease. Supplementary Material, Table S1 lists gene transcripts that are either up- or down-regulated by 1.4-fold or greater in hTBP80Q-expressing flies relative to hTBP34Q-expressing flies at each time point. The identification of these genes, referred to as the ‘dysregulated’ genes, demonstrates that polyQ expansion in hTBP causes widespread alterations in transcription, with genes both up- and down-regulated in a time-dependent manner (see Fig. 3D for heatmap for the dysregulated genes). Table 1 shows significantly enriched biological processes, molecular functions and pathways in the dysregulated genes at different time points (P-values from Fisher's exact test; see Supplementary Material, Table S2 for a complete list of enriched features). These results suggest a possible contribution of specific functions of pathways, such as oxidation and mitochondria-related energy metabolism (52–54), to hTBP80Q-induced neuropathology. Our results also revealed that, consistent with data from a mouse SCA17 model (31), Hsp27, the Drosophila homologue of mouse HSPB1 was significantly down-regulated in hTBP80Q-expressing flies relative to hTBP34Q-expressing flies (∼5-fold) on Day 5 and its expression level exhibited a further decrease on both Day 28 and Day 35 relative to Day 5.
Table 1.
Gene set enrichment results for the dysregulated genes using DAVID
| Category | Feature | Name of the feature | No. of genes | P-value (Bonferroni) |
|---|---|---|---|---|
| DAY-5, upregulated | GOTERM_BP_FAT | GO:0006613—cotranslational protein targeting to membrane | 5 | 9.62E−04 |
| GOTERM_CC_FAT | GO:0005784—translocon complex | 4 | 9.96E−04 | |
| GOTERM_CC_FAT | GO:0005783—endoplasmic reticulum | 10 | 5.32E−03 | |
| GOTERM_BP_FAT | GO:0045047—protein targeting to ER | 4 | 4.06E−02 | |
| SP_PIR_KEYWORDS | Oxidoreductase | 11 | 5.43E−02 | |
| DAY-5, downregulated | GOTERM_BP_FAT | GO:0009408—response to heat | 10 | 8.23E−08 |
| SP_PIR_KEYWORDS | Stress response | 7 | 2.14E−07 | |
| SP_PIR_KEYWORDS | Heat shock | 5 | 5.49E−06 | |
| SP_PIR_KEYWORDS | Stress-induced protein | 5 | 5.49E−06 | |
| INTERPRO | IPR001436—alpha crystallin/heat shock protein | 5 | 1.60E−05 | |
| GOTERM_BP_FAT | GO:0009628—response to abiotic stimulus | 11 | 1.97E−04 | |
| SP_PIR_KEYWORDS | Innate immunity | 6 | 9.98E−04 | |
| SP_PIR_KEYWORDS | Immune response | 6 | 1.20E−03 | |
| GOTERM_CC_FAT | GO:0005576—extracellular region | 11 | 4.73E−03 | |
| DAY-35, upregulated | GOTERM_BP_FAT | GO:0055114—oxidation reduction | 21 | 3.41E−05 |
| GOTERM_BP_FAT | GO:0009069—serine family amino acid metabolic process | 6 | 5.51E−05 | |
| GOTERM_BP_FAT | GO:0009161—ribonucleoside monophosphate metabolic process | 6 | 2.48E−04 | |
| KEGG_PATHWAY | dme00670—one carbon pool by folate | 5 | 1.26E−03 | |
| INTERPRO | IPR017973—cytochrome P450, C-terminal region | 8 | 2.84E−03 | |
| GOTERM_BP_FAT | GO:0009167—purine ribonucleoside monophosphate metabolic process | 5 | 3.73E−03 | |
| INTERPRO | IPR017972—cytochrome P450, conserved site | 8 | 4.45E−03 | |
| GOTERM_BP_FAT | GO:0006544—glycine metabolic process | 4 | 1.70E−02 | |
| GOTERM_BP_FAT | GO:0009124—nucleoside monophosphate biosynthetic process | 6 | 2.12E−02 | |
| KEGG_PATHWAY | dme00260—glycine, serine and threonine metabolism | 5 | 3.93E−02 | |
| Day-35, Downregulated | GOTERM_CC_FAT | GO:0005576—extracellular region | 11 | 3.42E−03 |
| SP_PIR_KEYWORDS | Innate immunity | 5 | 1.33E−02 | |
| SP_PIR_KEYWORDS | Immune response | 5 | 1.53E−02 | |
| GOTERM_MF_FAT | GO:0070279—vitamin B6 binding | 4 | 5.27E−02 |
See Supplementary Material, Table S2 for a complete list of enriched features. Dysregulated genes on Day 28 (either up- or down-regulated) did not show any significant enriched features.
Q/N-rich proteins have important biological functions, such as transcription regulation of neurogenesis (55,56). To evaluate whether Q/N-rich transcription factors as a group may differ from their non-Q/N-rich counterparts in hTBP80Q-induced transcriptional dysregulation, we performed a Pearson correlation coefficient analysis. For this analysis, we divided all the experimentally verified transcription factor genes (57) into two classes: those that encode Q/N-rich transcription factors and those that encode the non-Q/N-rich counterparts (38). Our analysis considered all transcription factor genes regardless whether they themselves are dysregulated. Using an absolute correlation coefficient of 0.95 as a cutoff, we obtained 146 highly correlated gene pairs between the 39 Q/N-rich transcription factor genes and the 532 dysregulated genes. In contrast, there are 200 highly correlated pairs identified between the 127 non-Q/N-rich transcription factor genes and the 532 dysregulated genes (Supplementary Material, Table S3). These results show that expression level changes for genes that encode Q/N-rich transcription factors contribute more to hTBP80Q-induced transcriptional dysregulation than their non-Q/N-rich counterparts (P-value <1e−10). There are three Q/N-rich transcription factor genes that are dysregulated, and they account for 72 of the 146 correlations (see Supplementary Material, Table S3, dysregulated transcription factor genes are highlighted). In contrast, the three non-Q/N-rich transcription factor genes that are dysregulated only account for 6 of the 200 correlations (Supplementary Material, Table S3). Together, these results suggest important contributions of Q/N-rich transcription factors to hTBP80Q-induced transcriptional dysregulation (see Discussion for further information related to this issue).
Knockdown of Su(H) enhances the hTBP80Q-induced defects in eye patterning and retinal degeneration
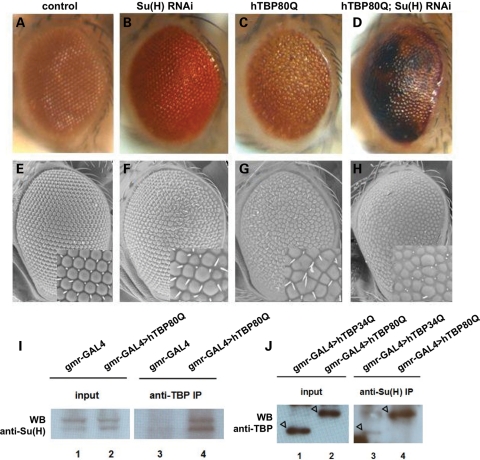
To further study the molecular basis of hTBP80Q-induced defects in Drosophila, we performed a candidate screening for genetic modifier in gmr-GAL4 > hTBP80Q flies. We monitored the eye phenotype in our candidate screen and focused on proteins that contain Q/N-rich domains and have functions related to neurons (see Supplementary Material, Table S4 for a list of genes tested and their effects on phenotypic modification). Our RNAi screening identified Su(H), a nuclear component of the Notch signaling pathway (37). Knockdown of Su(H) by RNAi in the eye enhanced the hTBP80Q-induced phenotype. Figure 4C and G shows, respectively, the light microscopic and SEM images of hTBP80Q-expressing adult eyes on Day 1, which exhibit pigment loss and fused ommatidia. The corresponding images for hTBP80Q-expressing eyes with Su(H) knocked down are shown in Figure 4D and H. These eyes had large areas of dark spots that are suggestive of necrosis and retinal degeneration (Fig. 4D). In addition, the shape of the ommatidia became grossly irregular with bristles largely missing (Fig. 4H). These defects were much more severe than those caused by hTBP80Q expression alone (Fig. 4C and G). These results suggest an involvement of Su(H) in neuropathology induced by hTBP80Q in Drosophila.
Figure 4.
Su(H) genetically modifies hTBP80Q-induced defects and physically interacts with hTBP80Q. (A–H) Light microscopic (A–D) and SEM (E–H) images of 1-day-old fly eyes expressing Su(H) RNAi (B and F), hTBP80Q (C and G) or Su(H) RNAi together with hTBP80Q (D and H). (A) and (E) represent control flies containing only the gmr-GAL4 driver. Insets for (E)–(H) represent higher magnification of the ommatidia field. As detailed in main text, knockdown of Su(H) enhanced the hTBP80Q-induced eye phenotype on Day 1. (I) Extracts from fly heads with indicated genotypes were immuno-precipitated by the 1C2 antibody to pull down hTBP and the precipitated products were analyzed by western blots using anti-Su(H) antibody to detect Su(H). (J) Extracts from fly heads with the indicated genotypes were immuno-precipitated by anti-Su(H) antibody to pull-down Su(H) and the precipitated products were analyzed by western blots using N-12 antibody to detect hTBP.
As a general transcription factor, TBP can interact with a variety of transcription factors (29,30). To determine whether Su(H) and hTBPQ80 can interact with each other, we performed a co-immunoprecipitation (co-IP) experiment using extracts of fly heads. We used the 1C2 antibody to pull-down hTBP, and used Su(H) antibody in western blotting to detect the endogenous Su(H) protein in the precipitated products. Our results show that Su(H) was co-precipitated specifically with hTBP80Q (Fig. 4I, lane 4, compared with lane 3 as control), suggesting that hTBP80Q and the endogenous Su(H) protein can interact in fly tissues. To determine whether polyQ expansion may alter how hTBP interacts with Su(H), we performed another co-IP analysis that directly compared hTBP34Q and hTBP80Q in their interactions with Su(H) in fly tissues. In this assay, the Su(H) antibody was used to pull down the endogenous Su(H) protein in head extracts from flies expressing either hTBP34Q or hTBP80Q, followed by western blotting to detect hTBP34Q or hTBP80Q with the N12 antibody. As shown in Figure 4J, the input lanes (1 and 2) show similar amounts of hTBP34Q and hTBP80Q, further demonstrating that these two proteins were expressed in their full-length forms at comparable levels in Drosophila. However, there was a significantly higher level of hTBP80Q in the pull-down products than hTBP34Q (lanes 3 and 4), suggesting that the interaction between hTBP and Su(H) is enhanced by its polyQ expansion.
Su(H) contributes to hTBP80Q-induced transcriptional dysregulation
The effect of Su(H) in modifying hTBP80Q-induced eye phenotype discussed above is consistent with its role in transcriptional dysregulation. Among the 532 dysregulated genes, 215 of them (∼41%; P< 0.05) have putative binding sites for RBP-J/Su(H) in their promoter regions (see Supplementary Material, Table S1). Table 2 lists the top 10 overrepresented transcription factor-binding sites in the promoter regions of the dysregulated genes at each of the time points, and RBP-J/Su(H)-binding site is among them. To evaluate the functional relevance of these binding sites, we used Pearson correlation coefficient to identify pairs of highly correlated genes among the dysregulated genes that have at least one putative RBP-J/Su(H)-binding site in their promoter regions. Out of the 215 dysregulated genes predicted to have RPB-J/Su(H)-binding sites, we identified 473 pairs of genes that are highly correlated, with an absolute correlation coefficient of 0.95 or greater (see Table 3 for a list of top 20 highly correlated gene pairs, and Supplementary Material, Table S5, for a complete list). Since these genes share the common feature of having at least one predicted RPB-J/Su(H)-binding site, the observed correlation represents evidence (58) that Su(H) plays a role in these genes' transcriptional responses to hTBP80Q. Together, our results suggest that Su(H) contributes to hTBP80Q-induced transcriptional dysregulation.
Table 2.
Overrepresented top 10 putative transcription factor-binding sites in the promoter regions of the dysregulated genes
| Dysregulated genes—category | Enriched putative transcription factor-binding sites (top 10) |
|---|---|
| Day 5, upregulated genes | Abd-B, Adf1, Twi, Cf2, Gli, Phdp, Six4, Whn, Vvl, ac-sc |
| Day 5, downregulated genes | Abd-B, Bab1, Twi, Cf2, Onecut, Phdp, Fkhd, mirr, Vvl, Prd |
| Day 28, upregulated genes | Abd-B, Brcz, cf2, Phdp, Su(H), Dsx, Fkhd, Gaf, Vvl, Prd |
| Day 28, downregulated genes | Abd-B, Bab1, Twi, Onecut, Phdp, Odd, Fkhd, mirr, Vvl, Prd |
| Day 35, upregulated genes | Abd-B, Twi, cf2, Elf1, Gli, Phdp, Hsf, Su(H), Dsx, Gaf |
| Day 35, downregulated genes | Abd-B, Bab1, Bcd, cf2, Gcm, Phdp, Kni, Kr, Prd, Brk |
See Supplementary Material, Table S6, for additional details.
Table 3.
Top 20 highly correlated pairs of dysregulated gene that are putative Su(H) target genes
| Gene ID-1 | Gene symbol-1 | Gene ID-2 | Gene symbol-2 | Correlation coefficient |
|---|---|---|---|---|
| FBgn0035510 | Cpr64Aa | FBgn0035553 | CG13722 | 0.997 |
| FBgn0003360 | sesB | FBgn0038820 | CG4000 | 0.996 |
| FBgn0013773 | Cyp6a22 | FBgn0036262 | CG6910 | 0.995 |
| FBgn0036262 | CG6910 | FBgn0038865 | CG10824 | 0.995 |
| FBgn0020521 | pio | FBgn0038820 | CG4000 | 0.994 |
| FBgn0035510 | Cpr64Aa | FBgn0058298 | CG40298 | 0.993 |
| FBgn0001225 | Hsp26 | FBgn0001226 | Hsp27 | 0.993 |
| FBgn0002938 | ninaC | FBgn0038820 | CG4000 | 0.993 |
| FBgn0003360 | sesB | FBgn0030398 | Cpr11B | 0.992 |
| FBgn0032382 | CG14935 | FBgn0040211 | hgo | 0.992 |
| FBgn0001224 | Hsp23 | FBgn0038846 | CG5697 | 0.992 |
| FBgn0013773 | Cyp6a22 | FBgn0038865 | CG10824 | 0.992 |
| FBgn0038172 | Adgf-D | FBgn0040211 | hgo | 0.991 |
| FBgn0035553 | CG13722 | FBgn0058298 | CG40298 | 0.990 |
| FBgn0003360 | sesB | FBgn0020521 | pio | 0.989 |
| FBgn0030398 | Cpr11B | FBgn0038820 | CG4000 | 0.989 |
| FBgn0027348 | bgm | FBgn0034010 | CG8157 | 0.989 |
| FBgn0032940 | Mio | FBgn0040211 | hgo | 0.989 |
| FBgn0001224 | Hsp23 | FBgn0032109 | CG17005 | 0.989 |
| FBgn0037973 | CG18547 | FBgn0040256 | Ugt86Dd | 0.988 |
Rescue of hTBP80Q-induced eye phenotype by Su(H)
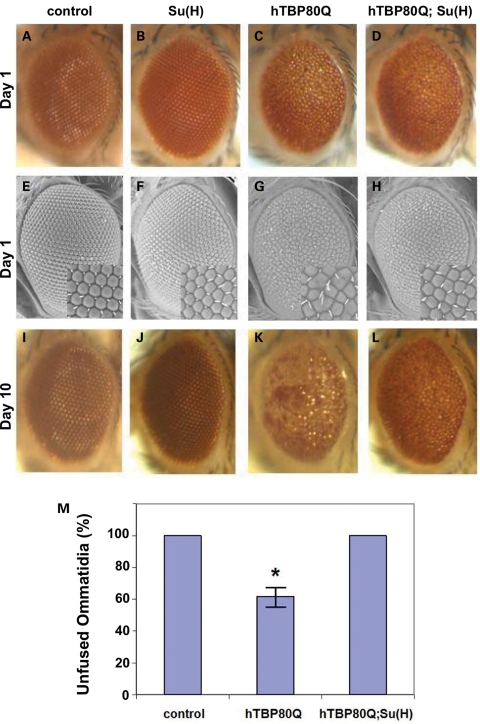
An enhanced interaction between hTBP80Q and Su(H) shown in Figure 4J suggests that, even though the Su(H) expression level itself was unaffected in hTBP80Q-expressing flies relative to hTBP34Q-expressing flies (as observed in our microarray data), the fraction of the Su(H) protein that is available for regulating its downstream target genes may be reduced by the presence of hTBP80Q. This possibility is consistent with our finding that knockdown of Su(H) worsened hTBP80Q-induced eye defects in Drosophila (Fig. 4D and H). To further evaluate the role of Su(H) in hTBP80Q-induced phenotype, we analyzed the dose effect of Su(H). We reasoned that, if hTBP80Q does indeed reduce the biological functions of Su(H), overexpression of Su(H) may rescue hTBP80Q-induced defects. Figure 5C and G shows, respectively, the light microscopic and SEM images of hTBP80Q-expressing adult eyes on Day 1, which exhibit defects of pigment loss and fused ommatidia. Overexpressing Su(H) in hTBP80Q-expressing adult eyes (Day 1) partially rescued the pigment loss, with an improvement in both the shape and organization of ommatidia (Fig. 5D and H). Ommatidial fusion was no longer observed (see Fig. 5M for quantification). The rescue effect became even more pronounced on Day 10 (Fig. 5K and L): while pigment loss was notably more severe in hTBP80Q-expressing fly eyes on Day 10 than on Day 1, Su(H) overexpression almost completely prevented this phenotypic progression. These results demonstrate that hTBP80Q-induced eye defects, in terms of both patterning and retinal degeneration, are suppressible by Su(H) overexpression, further supporting a role of Su(H) in mediating the pathological effects of hTBP80Q in Drosophila.
Figure 5.
Rescue of hTBP80Q-induced defects by Su(H). (A–L) Light microscopic (A–D) and SEM (E–H) images of the eyes of 1-day-old flies expressing Su(H) (B and F), hTBP80Q (C and G) or Su(H) and hTBP80Q (D and H). (A) and (E) represent control flies containing only the gmr-GAL4 driver. Insets for (E)–(H) represent higher magnification of the ommatidia field. (I–L) Light microscopic images of eyes of 10-day-old flies that have genotypes corresponding to those shown in (A)–(D), respectively. (M) Quantification of unfused ommatidia in indicated flies. SEM images of adult eyes were used to calculate the percentage of unfused ommatidias in representative areas. The values shown are the mean (n = 3 for each genotype) and standard deviation (shown as error bar). The sign asterisk indicates P < 0.05 (Student's t-test) for comparing hTBP80Q-mediated phenotype either with the gmr-GAL4 control phenotype or with the phenotype obtained when hTBP80Q and Su(H) were co-expressed.
DISCUSSION
Our study presented in this work establishes a first Drosophila disease model for SCA17. We show that hTBP80Q causes progressive retinal degeneration, late-onset locomotor impairment and early mortality, phenotypes characteristic of human SCA17 pathology. Our candidate screen identifies RBP-J/Su(H), a transcription factor with Q/N-rich domains. While the Su(H) transcript level is not affected by hTBP80Q, genes that contain putative Su(H)-binding sites are among those that are dysregulated in hTBP80-expressing flies and, furthermore, the Pearson correlation analysis suggests that these Su(H)-binding sites are functionally relevant. Our biochemical experiments show that polyQ expansion in hTBP enhances its interaction with Su(H), suggesting that a reduction in the fraction of Su(H) available for its normal cellular functions may contribute to hTBP80Q-induced defects. Supportive of this suggestion, we show directly that overexpression of Su(H) alleviates hTBP80Q-induced eye patterning defects and retinal degeneration. It is relevant to note that, in our microarray data, the expression level of Glass, the transcriptional activator for gmr-Gal4, remains at a relatively constant level under all conditions. This finding is consistent with our result that different hTBP proteins were accumulated to similar levels (Fig. 1B). It is also consistent with our result that the hTBP80Q level was insensitive to manipulations of Su(H) expression (Supplementary Material, Fig. S2), suggesting that Su(H)'s role in modifying the eye phenotype is reflective of changes in the levels of its downstream target genes (as opposed to hTBP80Q). Together, our results suggest that an altered interaction between RBP-J/Su(H) and hTBP induced by the polyQ expansion may contribute to neuropathology of SCA17. In mammalian systems (31,32), a mutant TBP with expanded polyQ can alter the biological functions of its targets through abnormal interactions that involve both its aggregate form (with colocalization with its target protein) and its soluble form (without such colocalization). These proposed mechanisms may also explain how hTBP80Q may affect Su(H) function in Drosophila.
The Notch signaling pathway plays an important role in a wide range of developmental processes. Deficits in Notch signaling pathway have been implicated in several neurodegenerative diseases (see Introduction). Costa et al. (59) showed that mice that are heterozygous for either Notch1 or RBP-J/Su(H) have similar spatial learning and memory deficits. Our experiments in Drosophila show that knockdown of Su(H) enhances hTBP80Q-induced defects, while its overexpression rescues such defects. These results demonstrate an important role of Su(H), a nuclear component of the Notch signaling pathway, in SCA17 neuropathology. To determine whether other components in the Notch signaling pathway may have a similar role as Su(H), we analyzed the effects of knockdown or mutant of Notch itself and Kuzbanian, a gene that encodes a protease controlling the proteolytic processing of Notch (60). We found that neither of them had a strong effect on the hTBP80Q-induced eye phenotype (Supplementary Material, Table S4). Although Su(H) is a key downstream component of the Notch signaling pathway, its role is not restricted to mediating Notch-dependent transcriptional activation (37,61,62). In addition to Notch-dependent target genes of Su(H), there are also genes, such as those involved in socket cell differentiation, that require Su(H) but are independent of Nicd (63). Although our study establishes a clear role of Su(H) in mediating hTBP80Q-induced defects, further investigations are needed to elucidate the precise relationship between Su(H) and the Notch signaling pathway during this process.
While the causative disease proteins for the polyQ diseases are ubiquitously expressed, they induce neuropathology selectively. For example, although hTBP is a ubiquitously expressed protein, only cerebellar atrophy and Purkinje cell loss are reported in SCA17 patients (2). How the widely expressed causative proteins, in their pathological forms, lead to selective neuropathology remains an interesting question. Mutant proteins with expanded polyQ tracts may interact abnormally with Q/N-rich proteins to alter their normal cellular functions to contribute to neuropathology (64–66). Both Sp1 and RBP-J/Su(H), which are suggested to play a role in SCA17 neuropathology [(32), and this work], contain Q/N-rich domains, suggesting that, consistent with other polyQ diseases (11–24,31,32), polyQ expansion in hTBP may lead to neurotoxicity through aberrant interactions with multiple target proteins. In addition, our Pearson correlation analysis reveals that changes in the expression levels of Q/N-rich transcription factor genes contribute more to hTBP-induced dysregulation than their non-Q/N-rich counterparts (Supplementary Material, Table S3). Together, these results suggest that hTBP80Q may lead to transcriptional dysregulation through its compounded effects on Q/N-rich transcription factors: (i) it preferentially targets Q/N-rich transcription factors to reduce the fraction these factors available for their normal cellular functions, and (ii) it also preferentially amplifies the effects of the changes in Q/N-rich transcription factor levels on their target genes’ transcription. We suggest that neuropathological selectivity of polyQ diseases may reflect, at least in part, the biological specificity of Q/N-rich transcription factors. Importantly, our results show that RBP-J/Su(H) overexpression can alleviate hTBP80Q-induced phenotypes, suggesting that its function plays a role in modulating the SCA17 pathological outcome.
MATERIALS AND METHODS
Plasmids
WT dTBP cDNA was amplified from a Drosophila cDNA library and inserted into the pUAST vector. WT hTBP cDNA (encoding a protein with 34Q) was amplified from a Flag-hTBP construct (a gift of Dr Jinsong Zhang) and inserted into the pUAST vector. Oligonucleotides of CAG repeats were inserted into its second block of CAG repeat, where the most frequent expansion of CAG triplets in patients occurs, of the WT hTBP cDNA to generate hTBP with expanded polyQ tracts that have either 54 glutamines (hTBP54Q) or 80 glutamines (hTBP80Q) using a previously described method (67). As WT hTBP, the mutant hTBP genes were also based on the pUAST vector.
Drosophila genetics
The UAS:dTBP, UAS:hTBP34Q, UAS:hTBP54Q and UAS:hTBP80Q transgenic flies were generated by P-element-mediated technique using a commercial microinjection service (Rainbow transgenic flies). Independent lines tested for each construct showed comparable functions based on the rescue or eye phenotypic analyses. Rescue experiments were performed using Hsp70-GAL4 to drive UAS:dTBP and UAS:hTBP34Q to ubiquitously express WT Drosophila and human TBP proteins. We used gmr-GAL4 and elav-GAL4 to express UAS:hTBP34Q, UAS:hTBP54Q and UAS:hTBP80Q in the eyes or in all neurons, respectively. RNAi fly lines for the genetic modifier screen were from the Bloomington Stock Center or VDRC and UAS:Su(H) flies were from the Bloomington Stock Center. All phenotypic analyses were performed at 25°C.
Western blot
To determine protein expression levels, adult fly heads from corresponding genotypes were homogenized in 1× sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) loading buffer (50 mm Tris–HCl, pH 6.8, 100 mm dithiothreitol, 2% SDS, 10% glycerol, 0.1% bromophenol blue) and then boiled for 5 min. Proteins were separated by SDS–PAGE and transferred to Immun-Blot™ polyvinylidene fluoride membrane (Bio-Rad) for western blotting using appropriate primary antibodies and an HRP-conjugated second antibody. Western blotting signals were visualized by ECL plus western blotting detection reagents (GE Healthcare) as described previously (68). For western blotting, hTBP proteins with different polyQ lengths were detected by 1C2 (Millipore) or N-12 (Santa Cruz Biotechnology) primary antibodies (see Fig. 1B legend for additional details about the properties of these antibodies); anti-β-actin antibody (Abcam) was used to detect β-actin as loading control.
Histology
For SEM images, whole flies were executed in the steam of chloroform and then analyzed with the scanning electron microscope (TM-1000, HITACHI). For cryosections, adult fly heads were dissected, rinsed in phosphate buffer saline and embedded in the O.C.T. compound (Tissue Tech) and then frozen by emersion in dry ice. Sagittal sections (9 µm) were cut at −20°C and stained with toluidine blue for visualizing the gross organization of photoreceptors.
Climbing assays
Climbing assays were performed to determine the locomotor ability as described (69,70) with minor modifications. Twenty flies were placed in a test tube of 15 cm in length and 1.6 cm in diameter. After 30 min recovery from CO2 exposure, flies were gently tapped to the bottom of the test tube. We counted and calculated the percentage of flies that could climb up to or above the 12 cm mark in 20 s. Three trials were performed for each experiment at 1 min intervals, and 10 experiments were carried out for each group of flies with the same genotype.
Survival curve
Two hundred flies from each genotype were monitored for survival. They were maintained in 10 separate vials (each with an initial 20 flies) at 25°C on standard fly food that was changed every 7 days.
Co-immunoprecipitation (co-IP) assays
Fly heads were collected and homogenized in 200 μl of IP buffer (200 mm Tris, pH 7.6, 150 mm NaCl, 10 mm ethylenediaminetetraacetic acid, 1% Triton X-100, 1 mm phenylmethylsulfonyl fluoride, complete protease inhibitor cocktail tablet). Five microliters of 1C2 antibody or Su(H) antibody (Santa Cruz Biotechnology) were added to the lysates after pre-clearing with the protein G Sepharose™ 4 fast flow beads (GE Healthcare) and the mixtures were rocked at 4°C for 1 h. Thirty microliters of protein G Sepharose™ 4 fast flow beads (GE Healthcare) were then added and the mixtures were rocked at 4°C overnight. After being washed six times with the IP buffer, the beads were boiled in 2× SDS–PAGE loading buffer and the Su(H) or hTBP proteins were examined by western blotting with Su(H) antibody or N12 antibody as previously described (68).
Isolation of total RNA and microarray analyses
Two independent samples of total RNA were extracted from 200 fly heads (100 for each sample) of each genotype (elav-GAL4>hTBP80Q and elav-GAL4>hTBP34Q) at each time point (Day 5, Day 28 and Day 35) with the Trizol Reagent (Invitrogen) and then purified with RNeasy columns (Qiagen) following the manufacturers’ instructions. Hybridization for each sample to the GeneChip Drosophila Genome 2.0 Array (Affymetrix) was performed by the Affymetrix Gene Chip Core at the Cincinnati Children's Hospital Medical Center (Cincinnati, OH, USA) using standard protocols. The hybridized arrays were scanned using the Microarray Suite (MAS) Software (Affymetrix). Scanned data were analyzed with GeneSpring 7.1 (Silicon Genetics, Redwood City, CA, USA) using Affymetrix MAS 5.0 cel files subjected to the RMA cel file pre-processor built in to GeneSpring 7.1. The mean expression values from the duplicate samples for each genotype at a time point were used for further analysis. Normalized signal intensities were then used to identify expression changes between hTBP80Q-expressing flies and hTBP34Q-expressing flies at each time point. Transcripts that meet two criteria were identified as being dysregulated: a ≥1.4-fold change in expression level and a paired t-test P< 0.05. Supplementary Material, Table S1 lists 536 dysregulated transcripts from our microarray data, with 524 annotated genes.
Functional enrichment and transcription factor-binding site analyses
The Database for Annotation, Visualization and Integrated Discovery (DAVID) (71) was used for the assessment of biological processes in the dysregulated genes. The promoter sequence for each of dysregulated genes (i.e. the 1000 bp region upstream of the transcription initiation site) was downloaded using the UCSC genome browser (72). MatInspector (73) was then used to identify the putative targets of RBP-J/Su(H). Statistical significance was calculated by comparing with the promoter sequences of all Drosophila genes.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported in part by grants (to J.M.) from the National Institutes of Health (GM072812; GM78381) and the National Science Foundation (IOS-0843424); grants (to B.Z.) from the National Natural Science Foundation of China (30730056) and the 973 program (2007CB914502); a CCTST Methods grant (to L.J.L.); and an exchange student scholarship (to J.R.) from the China Scholarship Council (File No. 2008601047).
Supplementary Material
ACKNOWLEDGEMENTS
We thank members of our groups at CCHMC for discussions and technical assistance, the CCHMC microarray core facility for their service, Jinsong Zhang at the University of Cincinnati for providing the hTBP cDNA plasmid, Baotong Xie in Tiffany Cook's group at CCHMC for assistance in cryosectioning, Wenxia Zhang's lab at Peking University for providing fly facilities, Chuanmao Zhang's lab at Peking University for assistance in SEM imaging and Western blotting, Renjie Jiao at the Institute of Biophysics for comments on the manuscript, and Renjie Jiao's lab for fly stocks and assistance in light microscopic imaging. We also thank the reviewers for their constructive suggestions to improve this manuscript.
Conflitct of Interest statement. None declared.
REFERENCES
- 1.Koide R., Kobayashi S., Shimohata T., Ikeuchi T., Maruyama M., Saito M., Yamada M., Takahashi H., Tsuji S. A neurological disease caused by an expanded CAG trinucleotide repeat in the TATA-binding protein gene: a new polyglutamine disease? Hum. Mol. Genet. 1999;8:2047–2053. doi: 10.1093/hmg/8.11.2047. doi:10.1093/hmg/8.11.2047. [DOI] [PubMed] [Google Scholar]
- 2.Nakamura K., Jeong S.Y., Uchihara T., Anno M., Nagashima K., Nagashima T., Ikeda S., Tsuji S., Kanazawa I. SCA17, a novel autosomal dominant cerebellar ataxia caused by an expanded polyglutamine in TATA-binding protein. Hum. Mol. Genet. 2001;10:1441–1448. doi: 10.1093/hmg/10.14.1441. doi:10.1093/hmg/10.14.1441. [DOI] [PubMed] [Google Scholar]
- 3.Rolfs A., Koeppen A.H., Bauer I., Bauer P., Buhlmann S., Topka H., Schols L., Riess O. Clinical features and neuropathology of autosomal dominant spinocerebellar ataxia (SCA17) Ann. Neurol. 2003;54:367–375. doi: 10.1002/ana.10676. doi:10.1002/ana.10676. [DOI] [PubMed] [Google Scholar]
- 4.Bauer P.O., Nukina N. The pathogenic mechanisms of polyglutamine diseases and current therapeutic strategies. J. Neurochem. 2009;110:1737–1765. doi: 10.1111/j.1471-4159.2009.06302.x. doi:10.1111/j.1471-4159.2009.06302.x. [DOI] [PubMed] [Google Scholar]
- 5.Saudou F., Finkbeiner S., Devys D., Greenberg M.E. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 1998;95:55–66. doi: 10.1016/s0092-8674(00)81782-1. doi:10.1016/S0092-8674(00)81782-1. [DOI] [PubMed] [Google Scholar]
- 6.Koyano S., Iwabuchi K., Yagishita S., Kuroiwa Y., Uchihara T. Paradoxical absence of nuclear inclusion in cerebellar Purkinje cells of hereditary ataxias linked to CAG expansion. J. Neurol. Neurosurg. Psychiatry. 2002;73:450–452. doi: 10.1136/jnnp.73.4.450. doi:10.1136/jnnp.73.4.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arrasate M., Mitra S., Schweitzer E.S., Segal M.R., Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. doi:10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 8.Gatchel J.R., Zoghbi H.Y. Diseases of unstable repeat expansion: mechanisms and common principles. Nat. Rev. Genet. 2005;6:743–755. doi: 10.1038/nrg1691. doi:10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 9.Harjes P., Wanker E.E. The hunt for huntingtin function: interaction partners tell many different stories. Trends Biochem. Sci. 2003;28:425–433. doi: 10.1016/S0968-0004(03)00168-3. doi:10.1016/S0968-0004(03)00168-3. [DOI] [PubMed] [Google Scholar]
- 10.Li S.H., Li X.J. Huntingtin-protein interactions and the pathogenesis of Huntington's disease. Trends Genet. 2004;20:146–154. doi: 10.1016/j.tig.2004.01.008. doi:10.1016/j.tig.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 11.Chen-Plotkin A.S., Sadri-Vakili G., Yohrling G.J., Braveman M.W., Benn C.L., Glajch K.E., DiRocco D.P., Farrell L.A., Krainc D., Gines S., et al. Decreased association of the transcription factor Sp1 with genes downregulated in Huntington's disease. Neurobiol. Dis. 2006;22:233–241. doi: 10.1016/j.nbd.2005.11.001. doi:10.1016/j.nbd.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Dunah A.W., Jeong H., Griffin A., Kim Y.M., Standaert D.G., Hersch S.M., Mouradian M.M., Young A.B., Tanese N., Krainc D. Sp1 and TAFII130 transcriptional activity disrupted in early Huntington's disease. Science. 2002;296:2238–2243. doi: 10.1126/science.1072613. doi:10.1126/science.1072613. [DOI] [PubMed] [Google Scholar]
- 13.Kegel K.B., Meloni A.R., Yi Y., Kim Y.J., Doyle E., Cuiffo B.G., Sapp E., Wang Y., Qin Z.H., Chen J.D., et al. Huntingtin is present in the nucleus, interacts with the transcriptional corepressor C-terminal binding protein, and represses transcription. J. Biol. Chem. 2002;277:7466–7476. doi: 10.1074/jbc.M103946200. doi:10.1074/jbc.M103946200. [DOI] [PubMed] [Google Scholar]
- 14.Zuccato C., Tartari M., Crotti A., Goffredo D., Valenza M., Conti L., Cataudella T., Leavitt B.R., Hayden M.R., Timmusk T., et al. Huntingtin interacts with REST/NRSF to modulate the transcription of NRSE-controlled neuronal genes. Nat. Genet. 2003;35:76–83. doi: 10.1038/ng1219. doi:10.1038/ng1219. [DOI] [PubMed] [Google Scholar]
- 15.Li S.H., Cheng A.L., Zhou H., Lam S., Rao M., Li H., Li X.J. Interaction of Huntington disease protein with transcriptional activator Sp1. Mol. Cell Biol. 2002;22:1277–1287. doi: 10.1128/mcb.22.5.1277-1287.2002. doi:10.1128/MCB.22.5.1277-1287.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhai W., Jeong H., Cui L., Krainc D., Tjian R. In vitro analysis of huntingtin-mediated transcriptional repression reveals multiple transcription factor targets. Cell. 2005;123:1241–1253. doi: 10.1016/j.cell.2005.10.030. doi:10.1016/j.cell.2005.10.030. [DOI] [PubMed] [Google Scholar]
- 17.Matilla A., Koshy B.T., Cummings C.J., Isobe T., Orr H.T., Zoghbi H.Y. The cerebellar leucine-rich acidic nuclear protein interacts with ataxin-1. Nature. 1997;389:974–978. doi: 10.1038/40159. doi:10.1038/40159. [DOI] [PubMed] [Google Scholar]
- 18.Okazawa H., Rich T., Chang A., Lin X., Waragai M., Kajikawa M., Enokido Y., Komuro A., Kato S., Shibata M., et al. Interaction between mutant ataxin-1 and PQBP-1 affects transcription and cell death. Neuron. 2002;34:701–713. doi: 10.1016/s0896-6273(02)00697-9. doi:10.1016/S0896-6273(02)00697-9. [DOI] [PubMed] [Google Scholar]
- 19.Cvetanovic M., Rooney R.J., Garcia J.J., Toporovskaya N., Zoghbi H.Y., Opal P. The role of LANP and ataxin 1 in E4F-mediated transcriptional repression. EMBO Rep. 2007;8:671–677. doi: 10.1038/sj.embor.7400983. doi:10.1038/sj.embor.7400983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsuda H., Jafar-Nejad H., Patel A.J., Sun Y., Chen H.K., Rose M.F., Venken K.J., Botas J., Orr H.T., Bellen H.J., et al. The AXH domain of Ataxin-1 mediates neurodegeneration through its interaction with Gfi-1/Senseless proteins. Cell. 2005;122:633–644. doi: 10.1016/j.cell.2005.06.012. doi:10.1016/j.cell.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 21.Okuda T., Hattori H., Takeuchi S., Shimizu J., Ueda H., Palvimo J.J., Kanazawa I., Kawano H., Nakagawa M., Okazawa H. PQBP-1 transgenic mice show a late-onset motor neuron disease-like phenotype. Hum. Mol. Genet. 2003;12:711–725. doi: 10.1093/hmg/ddg084. doi:10.1093/hmg/ddg084. [DOI] [PubMed] [Google Scholar]
- 22.Tsai C.C., Kao H.Y., Mitzutani A., Banayo E., Rajan H., McKeown M., Evans R.M. Ataxin 1, a SCA1 neurodegenerative disorder protein, is functionally linked to the silencing mediator of retinoid and thyroid hormone receptors. Proc. Natl Acad. Sci. USA. 2004;101:4047–4052. doi: 10.1073/pnas.0400615101. doi:10.1073/pnas.0400615101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mizutani A., Wang L., Rajan H., Vig P.J., Alaynick W.A., Thaler J.P., Tsai C.C. Boat, an AXH domain protein, suppresses the cytotoxicity of mutant ataxin-1. EMBO J. 2005;24:3339–3351. doi: 10.1038/sj.emboj.7600785. doi:10.1038/sj.emboj.7600785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goold R., Hubank M., Hunt A., Holton J., Menon R.P., Revesz T., Pandolfo M., Matilla-Duenas A. Down-regulation of the dopamine receptor D2 in mice lacking ataxin 1. Hum. Mol. Genet. 2007;16:2122–2134. doi: 10.1093/hmg/ddm162. doi:10.1093/hmg/ddm162. [DOI] [PubMed] [Google Scholar]
- 25.Kang S., Hong S. Molecular pathogenesis of spinocerebellar ataxia type 1 disease. Mol. Cells. 2009;27:621–627. doi: 10.1007/s10059-009-0095-y. doi:10.1007/s10059-009-0095-y. [DOI] [PubMed] [Google Scholar]
- 26.Marsh J.L., Walker H., Theisen H., Zhu Y.Z., Fielder T., Purcell J., Thompson L.M. Expanded polyglutamine peptides alone are intrinsically cytotoxic and cause neurodegeneration in Drosophila. Hum. Mol. Genet. 2000;9:13–25. doi: 10.1093/hmg/9.1.13. doi:10.1093/hmg/9.1.13. [DOI] [PubMed] [Google Scholar]
- 27.Cummings C.J., Zoghbi H.Y. Trinucleotide repeats: mechanisms and pathophysiology. Annu. Rev. Genomics Hum. Genet. 2000;1:281–328. doi: 10.1146/annurev.genom.1.1.281. doi:10.1146/annurev.genom.1.1.281. [DOI] [PubMed] [Google Scholar]
- 28.van Roon-Mom W.M., Reid S.J., Faull R.L., Snell R.G. TATA-binding protein in neurodegenerative disease. Neuroscience. 2005;133:863–872. doi: 10.1016/j.neuroscience.2005.03.024. doi:10.1016/j.neuroscience.2005.03.024. [DOI] [PubMed] [Google Scholar]
- 29.Pugh B.F. Control of gene expression through regulation of the TATA-binding protein. Gene. 2000;255:1–14. doi: 10.1016/s0378-1119(00)00288-2. doi:10.1016/S0378-1119(00)00288-2. [DOI] [PubMed] [Google Scholar]
- 30.Davidson I. The genetics of TBP and TBP-related factors. Trends Biochem. Sci. 2003;28:391–398. doi: 10.1016/S0968-0004(03)00117-8. doi:10.1016/S0968-0004(03)00117-8. [DOI] [PubMed] [Google Scholar]
- 31.Friedman M.J., Shah A.G., Fang Z.H., Ward E.G., Warren S.T., Li S., Li X.J. Polyglutamine domain modulates the TBP-TFIIB interaction: implications for its normal function and neurodegeneration. Nat. Neurosci. 2007;10:1519–1528. doi: 10.1038/nn2011. doi:10.1038/nn2011. [DOI] [PubMed] [Google Scholar]
- 32.Shah A.G., Friedman M.J., Huang S., Roberts M., Li X.J., Li S. Transcriptional dysregulation of TrkA associates with neurodegeneration in spinocerebellar ataxia type 17. Hum. Mol. Genet. 2009;18:4141–4152. doi: 10.1093/hmg/ddp363. doi:10.1093/hmg/ddp363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Roon-Mom W.M., Reid S.J., Jones A.L., MacDonald M.E., Faull R.L., Snell R.G. Insoluble TATA-binding protein accumulation in Huntington's disease cortex. Brain Res. Mol. Brain Res. 2002;109:1–10. doi: 10.1016/s0169-328x(02)00450-3. doi:10.1016/S0169-328X(02)00450-3. [DOI] [PubMed] [Google Scholar]
- 34.Uchihara T., Fujigasaki H., Koyano S., Nakamura A., Yagishita S., Iwabuchi K. Non-expanded polyglutamine proteins in intranuclear inclusions of hereditary ataxias–triple-labeling immunofluorescence study. Acta Neuropathol. 2001;102:149–152. doi: 10.1007/s004010100364. [DOI] [PubMed] [Google Scholar]
- 35.Perez M.K., Paulson H.L., Pendse S.J., Saionz S.J., Bonini N.M., Pittman R.N. Recruitment and the role of nuclear localization in polyglutamine-mediated aggregation. J. Cell Biol. 1998;143:1457–1470. doi: 10.1083/jcb.143.6.1457. doi:10.1083/jcb.143.6.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koelzer S., Klein T. Regulation of expression of Vg and establishment of the dorsoventral compartment boundary in the wing imaginal disc by Suppressor of Hairless. Dev. Biol. 2006;289:77–90. doi: 10.1016/j.ydbio.2005.10.008. doi:10.1016/j.ydbio.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 37.Bray S., Furriols M. Notch pathway: making sense of suppressor of hairless. Curr. Biol. 2001;11:R217–R221. doi: 10.1016/s0960-9822(01)00109-9. doi:10.1016/S0960-9822(01)00109-9. [DOI] [PubMed] [Google Scholar]
- 38.Michelitsch M.D., Weissman J.S. A census of glutamine/asparagine-rich regions: implications for their conserved function and the prediction of novel prions. Proc. Natl Acad. Sci. USA. 2000;97:11910–11915. doi: 10.1073/pnas.97.22.11910. doi:10.1073/pnas.97.22.11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma J. Crossing the line between activation and repression. Trends Genet. 2005;21:54–59. doi: 10.1016/j.tig.2004.11.004. doi:10.1016/j.tig.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 40.Greenwald I. LIN-12/Notch signaling: lessons from worms and flies. Genes Dev. 1998;12:1751–1762. doi: 10.1101/gad.12.12.1751. doi:10.1101/gad.12.12.1751. [DOI] [PubMed] [Google Scholar]
- 41.Gridley T. Notch signaling in vertebrate development and disease. Mol. Cell Neurosci. 1997;9:103–108. doi: 10.1006/mcne.1997.0610. doi:10.1006/mcne.1997.0610. [DOI] [PubMed] [Google Scholar]
- 42.Sestan N., Artavanis-Tsakonas S., Rakic P. Contact-dependent inhibition of cortical neurite growth mediated by notch signaling. Science. 1999;286:741–746. doi: 10.1126/science.286.5440.741. doi:10.1126/science.286.5440.741. [DOI] [PubMed] [Google Scholar]
- 43.Hitoshi S., Alexson T., Tropepe V., Donoviel D., Elia A.J., Nye J.S., Conlon R.A., Mak T.W., Bernstein A., van der Kooy D. Notch pathway molecules are essential for the maintenance, but not the generation, of mammalian neural stem cells. Genes Dev. 2002;16:846–858. doi: 10.1101/gad.975202. doi:10.1101/gad.975202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abe K., Murakami T., Matsubara E., Manabe Y., Nagano I., Shoji M. Clinical features of CADASIL. Ann. N Y Acad. Sci. 2002;977:266–272. doi: 10.1111/j.1749-6632.2002.tb04825.x. doi:10.1111/j.1749-6632.2002.tb04825.x. [DOI] [PubMed] [Google Scholar]
- 45.Wei J., Hemmings G.P. The NOTCH4 locus is associated with susceptibility to schizophrenia. Nat. Genet. 2000;25:376–377. doi: 10.1038/78044. doi:10.1038/78044. [DOI] [PubMed] [Google Scholar]
- 46.Marsh J.L., Thompson L.M. Drosophila in the study of neurodegenerative disease. Neuron. 2006;52:169–178. doi: 10.1016/j.neuron.2006.09.025. doi:10.1016/j.neuron.2006.09.025. [DOI] [PubMed] [Google Scholar]
- 47.Gill G., Tjian R. A highly conserved domain of TFIID displays species specificity in vivo. Cell. 1991;65:333–340. doi: 10.1016/0092-8674(91)90166-v. doi:10.1016/0092-8674(91)90166-V. [DOI] [PubMed] [Google Scholar]
- 48.Hampsey M. Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev. 1998;62:465–503. doi: 10.1128/mmbr.62.2.465-503.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ellis M.C., O'Neill E.M., Rubin G.M. Expression of Drosophila glass protein and evidence for negative regulation of its activity in non-neuronal cells by another DNA-binding protein. Development. 1993;119:855–865. doi: 10.1242/dev.119.3.855. [DOI] [PubMed] [Google Scholar]
- 50.Warrick J.M., Paulson H.L., Gray-Board G.L., Bui Q.T., Fischbeck K.H., Pittman R.N., Bonini N.M. Expanded polyglutamine protein forms nuclear inclusions and causes neural degeneration in Drosophila. Cell. 1998;93:939–949. doi: 10.1016/s0092-8674(00)81200-3. doi:10.1016/S0092-8674(00)81200-3. [DOI] [PubMed] [Google Scholar]
- 51.Liu Z., Wang X., Yu Y., Li X., Wang T., Jiang H., Ren Q., Jiao Y., Sawa A., Moran T., et al. A Drosophila model for LRRK2-linked parkinsonism. Proc. Natl Acad. Sci. USA. 2008;105:2693–2698. doi: 10.1073/pnas.0708452105. doi:10.1073/pnas.0708452105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brouillet E., Hantraye P., Ferrante R.J., Dolan R., Leroy-Willig A., Kowall N.W., Beal M.F. Chronic mitochondrial energy impairment produces selective striatal degeneration and abnormal choreiform movements in primates. Proc. Natl Acad. Sci. USA. 1995;92:7105–7109. doi: 10.1073/pnas.92.15.7105. doi:10.1073/pnas.92.15.7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Browne S.E., Beal M.F. Oxidative damage in Huntington's disease pathogenesis. Antioxid. Redox Signal. 2006;8:2061–2073. doi: 10.1089/ars.2006.8.2061. doi:10.1089/ars.2006.8.2061. [DOI] [PubMed] [Google Scholar]
- 54.Chakrabarti L., Zahra R., Jackson S.M., Kazemi-Esfarjani P., Sopher B.L., Mason A.G., Toneff T., Ryu S., Shaffer S., Kansy J.W., et al. Mitochondrial dysfunction in NnaD mutant flies and Purkinje cell degeneration mice reveals a role for Nna proteins in neuronal bioenergetics. Neuron. 2010;66:835–847. doi: 10.1016/j.neuron.2010.05.024. doi:10.1016/j.neuron.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harrison P.M., Gerstein M. A method to assess compositional bias in biological sequences and its application to prion-like glutamine/asparagine-rich domains in eukaryotic proteomes. Genome Biol. 2003;4:R40. doi: 10.1186/gb-2003-4-6-r40. doi:10.1186/gb-2003-4-6-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Butland S.L., Devon R.S., Huang Y., Mead C.L., Meynert A.M., Neal S.J., Lee S.S., Wilkinson A., Yang G.S., Yuen M.M., et al. CAG-encoded polyglutamine length polymorphism in the human genome. BMC Genomics. 2007;8:126. doi: 10.1186/1471-2164-8-126. doi:10.1186/1471-2164-8-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adryan B., Teichmann S.A. FlyTF: a systematic review of site-specific transcription factors in the fruit fly Drosophila melanogaster. Bioinformatics. 2006;22:1532–1533. doi: 10.1093/bioinformatics/btl143. doi:10.1093/bioinformatics/btl143. [DOI] [PubMed] [Google Scholar]
- 58.Ideker T., Thorsson V., Ranish J.A., Christmas R., Buhler J., Eng J.K., Bumgarner R., Goodlett D.R., Aebersold R., Hood L. Integrated genomic and proteomic analyses of a systematically perturbed metabolic network. Science. 2001;292:929–934. doi: 10.1126/science.292.5518.929. doi:10.1126/science.292.5518.929. [DOI] [PubMed] [Google Scholar]
- 59.Costa R.M., Honjo T., Silva A.J. Learning and memory deficits in Notch mutant mice. Curr. Biol. 2003;13:1348–1354. doi: 10.1016/s0960-9822(03)00492-5. doi:10.1016/S0960-9822(03)00492-5. [DOI] [PubMed] [Google Scholar]
- 60.Pan D., Rubin G.M. Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell. 1997;90:271–280. doi: 10.1016/s0092-8674(00)80335-9. doi:10.1016/S0092-8674(00)80335-9. [DOI] [PubMed] [Google Scholar]
- 61.Morel V., Schweisguth F. Repression by suppressor of hairless and activation by Notch are required to define a single row of single-minded expressing cells in the Drosophila embryo. Genes Dev. 2000;14:377–388. [PMC free article] [PubMed] [Google Scholar]
- 62.Klein T., Seugnet L., Haenlin M., Martinez Arias A. Two different activities of Suppressor of Hairless during wing development in Drosophila. Development. 2000;127:3553–3566. doi: 10.1242/dev.127.16.3553. [DOI] [PubMed] [Google Scholar]
- 63.Barolo S., Walker R.G., Polyanovsky A.D., Freschi G., Keil T., Posakony J.W. A notch-independent activity of suppressor of hairless is required for normal mechanoreceptor physiology. Cell. 2000;103:957–969. doi: 10.1016/s0092-8674(00)00198-7. doi:10.1016/S0092-8674(00)00198-7. [DOI] [PubMed] [Google Scholar]
- 64.Furukawa Y., Kaneko K., Matsumoto G., Kurosawa M., Nukina N. Cross-seeding fibrillation of Q/N-rich proteins offers new pathomechanism of polyglutamine diseases. J. Neurosci. 2009;29:5153–5162. doi: 10.1523/JNEUROSCI.0783-09.2009. doi:10.1523/JNEUROSCI.0783-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Doi H., Okamura K., Bauer P.O., Furukawa Y., Shimizu H., Kurosawa M., Machida Y., Miyazaki H., Mitsui K., Kuroiwa Y., et al. RNA-binding protein TLS is a major nuclear aggregate-interacting protein in huntingtin exon 1 with expanded polyglutamine-expressing cells. J. Biol. Chem. 2008;283:6489–6500. doi: 10.1074/jbc.M705306200. doi:10.1074/jbc.M705306200. [DOI] [PubMed] [Google Scholar]
- 66.Yamanaka T., Miyazaki H., Oyama F., Kurosawa M., Washizu C., Doi H., Nukina N. Mutant Huntingtin reduces HSP70 expression through the sequestration of NF-Y transcription factor. EMBO J. 2008;27:827–839. doi: 10.1038/emboj.2008.23. doi:10.1038/emboj.2008.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Michalik A., Kazantsev A., Van Broeckhoven C. Method to introduce stable, expanded, polyglutamine-encoding CAG/CAA trinucleotide repeats into CAG repeat-containing genes. Biotechniques. 2001;31:250–252. doi: 10.2144/01312bm02. 254. [DOI] [PubMed] [Google Scholar]
- 68.Liu J., Ma J. Fates-shifted is an F-box protein that targets Bicoid for degradation and regulates developmental fate determination in Drosophila embryos. Nat. Cell Biol. 2011;13:22–29. doi: 10.1038/ncb2141. doi:10.1038/ncb2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Feany M.B., Bender W.W. A Drosophila model of Parkinson's disease. Nature. 2000;404:394–398. doi: 10.1038/35006074. doi:10.1038/35006074. [DOI] [PubMed] [Google Scholar]
- 70.Coulom H., Birman S. Chronic exposure to rotenone models sporadic Parkinson's disease in Drosophila melanogaster. J. Neurosci. 2004;24:10993–10998. doi: 10.1523/JNEUROSCI.2993-04.2004. doi:10.1523/JNEUROSCI.2993-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dennis G., Jr, Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. doi:10.1186/gb-2003-4-5-p3. [PubMed] [Google Scholar]
- 72.Fujita P.A., Rhead B., Zweig A.S., Hinrichs A.S., Karolchik D., Cline M.S., Goldman M., Barber G.P., Clawson H., Coelho A., et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. doi:10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Werner T. Computer-assisted analysis of transcription control regions. Matinspector and other programs. Methods Mol. Biol. 2000;132:337–349. doi: 10.1385/1-59259-192-2:337. [DOI] [PubMed] [Google Scholar]
- 74.Trottier Y., Lutz Y., Stevanin G., Imbert G., Devys D., Cancel G., Saudou F., Weber C., David G., Tora L., et al. Polyglutamine expansion as a pathological epitope in Huntington's disease and four dominant cerebellar ataxias. Nature. 1995;378:403–406. doi: 10.1038/378403a0. doi:10.1038/378403a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.