Abstract
Huntington's disease (HD) is a progressive neurodegenerative disorder caused by a CAG repeat expansion in exon 1 of huntingtin (HTT). Relatively little attention has been directed to the genomic features of the antisense strand at the HD locus, though the presence of a transcript from this strand has been suggested by a survey of the entire transcriptome and the existence of several EST tags. In this study, we identified huntingtin antisense (HTTAS), a natural antisense transcript at the HD repeat locus that contain the repeat tract. HTTAS is 5′ capped, poly (A) tailed and contains three exons, alternatively spliced into HTTAS_v1 (exons 1 and 3) and HTTAS_v2 (exons 2 and 3). Exon 1 includes the repeat. HTTAS_v1 has a weak promoter, and is expressed at low levels in multiple tissue types and throughout the brain. Reporter assays indicate that while efficient promoter activity requires a short repeat, repeat expansion reduces promoter efficiency. Consistent with the reporter assays, levels of HTTAS_v1 are reduced in human HD frontal cortex. In cell systems, overexpression of HTTAS_v1 specifically reduces endogenous HTT transcript levels, while siRNA knockdown of HTTAS_v1 increases HTT transcript levels. Minigene constructs of the HD locus confirm the regulatory effect of HTTAS_v1 on HTT, and demonstrate that the effect is dependent on repeat length and is at least partially Dicer dependent. Together, these findings provide strong evidence for the existence of a gene antisense to HTT, with properties that include regulation of HTT expression.
INTRODUCTION
Huntington's disease (HD) is an autosomal dominant neurodegenerative disorder caused by a CAG trinucleotide repeat expansion in exon 1 of huntingtin (HTT) on chromosome 4p16.3. Onset is usually in mid-life, with relentless progression characterized by abnormalities of movement, emotion and cognition (1–3). Prominent neuropathological findings include cortical and striatal atrophy, with a dorsal-to-ventral gradient loss of medium spiny neurons in the caudate and putamen (4). The expanded CAG repeat tract is translated into polyglutamine resulting in protein aggregation, another hallmark of HD pathology (5,6). While ample evidence supports a major role for polyglutamine-induced neurotoxicity in HD, the complete explanation for HD pathogenesis remains elusive, and no therapy has yet been demonstrated to stop or delay disease onset or progression (7,8).
The function of HTT is complicated and not well understood, though substantial evidence supports interrelated roles in intracellular trafficking, transcription regulation and regulation of trophic factors, especially BDNF (9,10). Even less is known about the regulation of HTT expression. Analysis of the HTT promoter suggests that important regulatory factors are present between nucleotides −324 and +20 (numbered relative to the HTT translation start site), and that polymorphisms in this region may influence HTT expression levels (11). Two potential transcription factors, HDBP1 and HDPB2, bind to a 7 bp (GCCGGCG) triplicated sequence in this region; mutating the CCCGCG sites prevents HTT expression (12). At the level of translation, expression may be impeded by a 5′ UTR upstream ORF (13).
Relatively, little attention has been directed to the genomic features of the antisense strand at the HD locus, though a survey of the entire transcriptome detected a transcript in the antisense orientation to HTT (14) and several EST tags also suggest the expression of a transcript from the antisense strand at the HTT locus (EST#DA153759, BF896464, CD511189) (15). This is consistent with recent evidence demonstrating that antisense transcripts are common throughout the genome (16–18). Further, antisense transcripts with potential pathogenic significance have been detected at the causative loci of other repeat expansion disorders, and may be present at nearly all repeat loci (19). The CGG repeat expansion associated with the Fragile X syndrome silences not only the FMR1 promoter, but also promoter activity for a transcript antisense to FMR1, ASFMR1/FMR4, that appears to be translated and promote cell proliferation (20,21). At the spinocerebellar ataxia type 8 (SCA8) locus on 13q21 (22), a transcript from an untranslated gene in one direction (ATXNOS) contains a CUG repeat expansion, while the gene in the opposite direction expresses a transcript in which a CAG repeat appears to encode polyglutamine (23). Evidence from cell and mouse models, and human brain tissue, suggests that both transcripts contribute to neurotoxicity (24). The causative mutation of myotonic dystrophy 1 (DM1) is a CTG expansion in the 3′ UTR of DMPK (25). An antisense transcript at this locus is converted into 21 nt small interfering RNA (siRNA) that induces heterochromatin formation, favoring transcription of the flanking gene SIX5 (26). In the presence of the repeat expansion, there is a loss of binding to adjacent CTCF sites, leading to abnormal spreading of heterochromatin and consequent repression of SIX5. Other mechanisms of antisense regulation of gene expression include transcriptional collision, genomic rearrangements, changes in mRNA stability and formation of endogenous siRNAs (27).
These studies raised the possibility that functionally significant antisense expression may also occur at the HD locus. Here, using HD and control brain tissue and a series of cell models, we confirm the expression of an antisense transcript at the HD locus, map the antisense gene in relation to HTT and demonstrate repeat length and Dicer-dependent regulation of HTT expression by the antisense transcript.
RESULTS
Detection of an antisense transcript at the HD locus
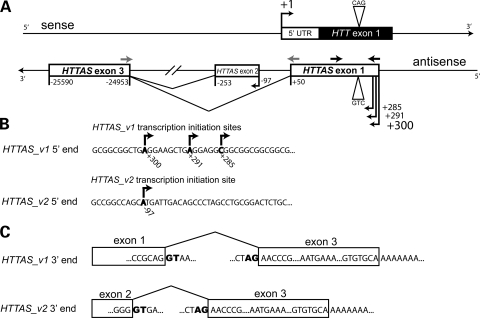
To confirm that an antisense transcript is expressed at the HD repeat locus, we performed strand-specific reverse transcriptase polymerase chain reaction (SS-RTPCR) using total RNA extracted from HD and control frontal cortex. Total RNA was reverse transcribed using an antisense strand-specific primer that incorporated a linker sequence (26). Nested RT–PCR was performed using primers that spanned the repeat (Fig. 1A, black arrows); two rounds of RT–PCR were required to detect an antisense transcript on agarose gel. Sequencing confirmed the specificity of the product and a repeat of the expected length. In keeping with HUGO guidelines, we named the gene expressing this transcript huntingtin antisense, HTTAS.
Figure 1.
HTTAS map. (A) Genomic structure of HTTAS. Numbers are relative to the transcription start site of HTT as defined in NCBI build 36.1. Dark arrows indicate primers used for strand-specific RT–PCR, gray arrows indicate primers used for qPCR. See Figure 2C for further detail of the 5′ flanking region of HTTAS. (B) 5′ region of HTTAS. Nested PCR showed four alternative transcription start sites at +300, +291, +285 and −97 bp (shown in bold) relative to the transcription start site of HTT. The transcription start site at +300 bp was most frequently observed during sequencing. (C) 3′ region of HTTAS. Two rounds of PCR yielded two splicing variants. HTTAS_v1 contains exons 1 and 3 and HTTAS_v2 contains exons 2 and 3 (consensus splicing donor/acceptor sites are shown in bold). Exon 3 contains an alternative poly (A) signal, AATGAA.
HTTAS map
To identify the 5′ end of the HTTAS transcript, we performed 5′ rapid amplification of cDNA ends (RACE). Total RNA from HD or control frontal cortex was treated with calf intestine phosphatase to dephosphorylate non-RNA and truncated RNA to ensure that only full-length RNA transcripts were reverse transcribed. An RNA oligonucleotide with a linker sequence was then attached to the 5′ end of RNA. Ligated RNA was reverse transcribed by random hexamer; two rounds of PCR were performed to minimize artifacts. Sequence analysis of the final PCR product revealed alternative transcription start sites at positions +285, +291 bp, +300 and −97 bp relative to the HTT transcription start site (Fig. 1B). All four transcription sites were found in both HD and control frontal cortex. To identify the 3′ end of HTTAS, total RNA from HD or control frontal cortex was similarly reverse transcribed using an oligo-dT primer with an attached linker sequence. Sequence analysis after two rounds of PCR product revealed two HTTAS splice variants, HTTAS_v1 and HTTAS_v2 (Fig. 1C). HTTAS_v1 contains exons 1 and 3, and HTTAS_v2 contains exons 2 and 3. They both contain consensus splicing donor and acceptor sites. Exon 3 contains an alternative polyadenylation site, AATGAA, at position −25 576 bp (HTTAS_v1) and a poly (A) tail at position −25 590 bp.
The exon–intron structure in relationship to HTT is depicted in Figure 1A. Conservation of these exons is partial. Exon 1 is 77.5% identical to rhesus and 68% identical to mouse, while exon 2 is 35.4% identical to rhesus and 37% identical to mouse. Exon 3 and its flanking regions consist of LTR and SINE elements, and are only found in humans. Short open reading frames (ORFs) of 201 bp (−549 to −749) and 171 bp (−97 to −267) exist in exons 2 and 3, respectively. Blastp searches (28) revealed that the potential peptides are not present in the Genbank non-redundant protein database, while neither potential peptide contains a conserved domain captured in the Conserved Domain Database (29) or the Protein Clusters database (ProtClustDB) (30).
HTTAS_v1 is widely expressed and has a functional promoter
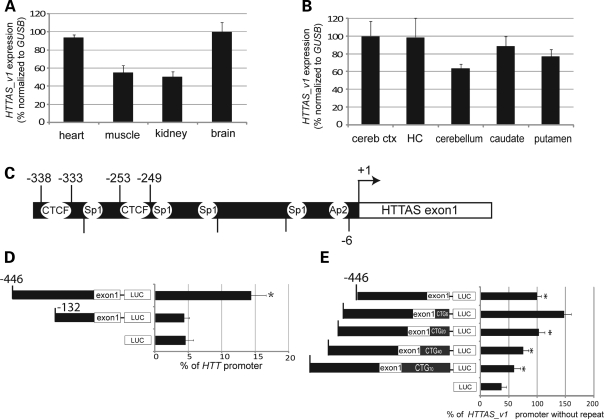
HTTAS_v1 is of particular interest because it includes the CAG/CTG repeat and overlaps with both coding and 5′ UTR of HTT exon 1 (Fig. 1A). We therefore sought to confirm that this transcript variant is expressed. Using forward and reverse qPCR primers in exons 1 and 3, respectively, to generate an amplicon that did not include the repeat (Fig. 1A, gray arrows, see Supplementary Material, Table S3 for qPCR primer sequences), we determined that HTTAS_v1 is expressed in multiple tissues types, with brain and heart expression 2-fold higher than muscle or kidney (Fig. 2A). Within the brain, expression is somewhat lower in the cerebellum than in cerebral cortex, hippocampus, caudate or putamen (Fig. 2B). The putative promoter region of HTTAS contains predicted binding sites for Sp1, Ap2 and CTCF (MapInspector) (31) (Fig. 2C). Both of the CTCF sites and the Sp1 and Ap2 sites are conserved in rhesus, while only one CTCF and neither Sp1 nor Ap2 sites are present in mouse. Consistent with this prediction, CTCF protein binding has been detected in the region between −344 and −152 (32) (see Supplementary Material, Table S2). To test whether the putative HTTAS_v1 promoter is functional, we generated luciferase reporter constructs containing the region between +52 bp and either −446 or −132 bp relative to the most frequently detected transcription start site of HTTAS_v1 (located at +300 on the locus map depicted in Fig. 1B, and +1 on the construct map depicted in Fig. 2C). The nucleotide identity of this 498 bp region is 91.7% to rhesus and 55.7% to mouse. In both HEK293 (Fig. 2D) and SH-SY5Y (Supplementary Material, Fig. S1A) cells, the reporter assay showed that promoter activity requires the region between −132 and −446, and the activity level is ∼10–15% of HTT promoter activity (Fig. 2D).
Figure 2.
HTTAS_v1 expression. (A and B) Relative expression level of HTTAS_v1 in multiple human tissues and brain regions was determined by qPCR using primers that span exons 1 and 3 (gray arrows, Fig. 1A) and normalized to GUSB. cereb ctx, cerebral cortex. HC, hippocampus. Experiment performed in triplicate three times. (C) Genomic region 5′ to the HTTAS_v1 transcription start site, depicting predicted Sp1 and Ap2 transcription factor binding sites, and consensus CTCF sites. (D) HTTAS_v1 promoter activity in HEK293 cells. The putative promoter region was ligated to Pgl-3 vector and promoter activity was measured by luciferase assay. Activity was dependent on the presence of nucleotides −446 to −132 relative to the transcription start site of HTTAS_v1. ANOVA, n = 5, F = 57.463, P < 0.001, Scheffe *P < 0.05 −466 construct versus −132 construct or vector. Performed three times with similar results. Similar results were obtained with transfections performed in SH-SY5Y cells (Supplementary Material, Fig. S1A). (E) Promoter activity is increased by a short repeat, but is reduced by repeat expansion. Promoter activity of constructs containing HTTAS 5′ flanking region and the 5′ region of exon 1, including the repeat region, was assessed using a luciferase assay in HEK 293 cells. ANOVA, n = 4, F = 57.119, P < 0.001. Scheffe *P < 0.05 versus HTTAS_v1 promoter with CAG6. The experiment was performed three times in quadruplicate with similar results. Error bars represent s.d. Assays using SH-SY5Y cells yielded nearly identical results (Supplementary Material, Fig. S1B).
We next investigated the effect of repeat length on HTTAS promoter activity. The insert used to examine promoter activity was expanded to include CTG6, CTG20, CTG40, or CTG70 repeats. Promoter activity was nearly doubled by adding a short repeat (CTG6), but each additional increment in repeat length reduced expression in both HEK293 (Fig. 2E) and SH-SY5Y cells (Supplementary Material, Fig. S1B). Addition of CTG40 reduced expression to the level of the construct with no repeat, and CTG70 reduced expression an additional 30–40%, suggesting that repeat expansion inhibits HTTAS_v1 promoter activity.
HTTAS_v1 is reduced in HD brains
Based on the results of promoter assays, we predicted that expression of HTTAS_v1 in HD brain would be lower than control, primarily due to a loss of expression of the HTTAS_v1 allele containing the expanded repeat. We initially tested this prediction by separately examining HTT and HTTAS_v1 expression in multiple brain regions, using the same sample of extracted cDNA for both HTTAS_v1 and HTT, normalized to GUSB (Fig. 3A and B). To facilitate comparison of HTT and HTTAS_v1 expression, we did not employ nested PCR amplification of HTTAS_v1. As predicted, HTTAS_v1 is expressed at much lower levels than HTT. HTTAS_v1 expression is most prominent in cortex and is almost undetectable in striatum and cerebellum, consistent with the results using a double-amplification protocol shown in Figure 2B. HTT is also more highly expressed in cortex than either cerebellum or striatum. HTTAS_v1 expression in cortex is substantially reduced in HD cases compared with controls; expression in the cerebellum and striatum was too low to detect a difference between HD and control cases.
Figure 3.
HTTAS_v1 is reduced in HD brain. (A) Regional HTT expression in HD and control brain. Demonstration of modest reductions of HTT expression in cortex compared with marginal and inconsistent reductions of HTT in striatum and cerebellum. Measurements performed in triplicate, error bars reflect s.d. (B) Regional HTTAS_v1 expression in HD and control brain. Expression levels of HTTAS_v1 are remarkably lower than HTT in all three regions, with little detectable HTTAS_v1 except in control cortex. Error bars reflect s.d. Experiment performed in triplicate. (C) Quantitative comparison of HTTAS_v1 expression in HD and control brain. HTTAS_v1 is 50% lower in HD frontal cortex (N = 8) compared with age-matched control frontal cortex (n = 8). Unpaired t-test, df = 14, t = −2.71, P = 0.017. Three additional experiments with independent RNA extractions all demonstrated ∼50% reduction of HTTAS_v1. (D) Expression of HTTAS_v1 with an expanded repeat is not detected. HTTAS_v1 with an expanded repeat was not detected in HD frontal cortex by SS-RTPCR using primers that span the repeat region (black arrows, Fig. 1A). The same primer pairs, applied to cDNA derived from sense strand transcripts, readily detected both normal and expanded alleles of HTT, though as expected with longer repeats the longer allele was less efficiently amplified (HD141 and HD214). +, reverse transcriptase added. Repeat length is indicated for each HD cortical sample.
To assess the loss of HTTAS_v1 expression in HD brain more quantitatively, we used qPCR of HTTAS_v1 (protocol as described earlier for Fig. 2A and B) in frontal cortex from eight HD brains and eight age-matched controls without known neurodegenerative disease (see Supplementary Material, Tables S4 and S5 for brain details). Consistent with findings from assays of promoter activity in cell lines and from the single amplification protocol, expression of HTTAS_v1 was about 50% lower in HD frontal cortex than in control cortex (Fig. 3C). The levels of HTTAS_v1 in Huntington's disease-like 2 (HDL2) frontal cortex, which neuropathologically closely resembles HD (33–36), was similar to controls, suggesting that the loss of HTTAS_v1 expression is specific to HD (Supplementary Material, Fig. S2).
To determine whether the loss of HTTAS_v1 expression is allele specific, we examined allele expression using SS-RTPCR. We were unable to detect HTTAS_v1 with an expanded repeat in HD brains, even though the same primer pairs on cDNA prepared from the opposite strand readily detected two alleles of HTT in the same brains (Fig. 3D). As expected, amplification of the longer HTT allele is less efficient (37), most notably in the cases with expanded repeat lengths of 48 and 49 triplets. This experiment does not exclude the possibility that this allele is expressed at a level too low for detection even by the nested PCR protocol employed in this experiment. We obtained the same results using CsCl (Supplementary Material, Fig. S3) and guanidinium thiocyanate/glass fiber filter (data not shown) RNA extraction protocols. PCR of HTTAS_v1 in control brains confirmed expression of both normal length alleles (Supplementary Material, Fig. S4). Therefore it appears likely that the overall loss of expression of HTTAS_v1 in HD brain reflects an allele-specific effect of the repeat expansion. In support of this finding, preliminary experiments indicate that expression level of HTTAS_v1 in lymphoblasts is inversely correlated with the number of expanded alleles at the HD locus (data not shown).
HTTAS_v1 negatively regulates HTT expression
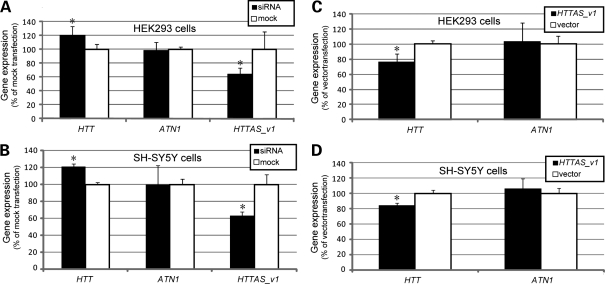
An apparently common function of antisense transcripts is to modulate the expression of complementary sense transcripts. To investigate the hypothesis that HTTAS_v1 modulates HTT expression, we transfected a pool of four siRNA oligonucleotides against exons 1 and 3 to reduce endogenous HTTAS_v1 expression in HEK293 and SH-SY5Y cells (see Supplementary Material, Table S6 for siRNA sequences). The siRNA pool decreased HTTAS_v1 expression by ∼40% in both cell types. There was no effect of the siRNA on expression of ataxin-1, JPH3 or PPP2R2B (Supplementary Material, Fig. S5), associated with, respectively, spinocerebellar ataxia type 1, HDL2 and spinocerebellar ataxia type 12. As predicted, decreasing HTTAS_v1 expression led to a 20% increase in expression of endogenous HTT (Fig. 4A and B). Conversely, overexpression of HTTAS_v1 decreased endogenous HTT expression by ∼25% (Fig. 4C and D). HTTAS_v1 knockdown and overexpression did not affect expression of endogenous ATN1, the CAG-repeat containing gene associated with dentatorubropallidoluysian atrophy, suggesting the effect of HTTAS_v1 is gene specific. These results demonstrate that changes in HTTAS_v1 expression influence levels of HTT.
Figure 4.
HTTAS_v1 regulates endogenous HTT expression. (A) siRNA knockdown of HTTAS_v1 increases levels of endogenous HTT transcript in HEK293 cells without affecting levels of ATN. Assays performed by qPCR. Expression levels were normalized to GUSB. ANOVA, n = 3, F = 95.571, *P < 0.05 versus mock. (B) siRNA knockdown of HTTAS_v1 in SH-SY5Y cells. Experiment as in HEK293 cells. ANOVA, n = 3, F = 94.189, *P < 0.05 versus mock. (C) Overexpression of HTTAS_v1 leads to down regulation of endogenous HTT expression but not ATN1 expression in HEK293 cells. Expression level was normalized to GUSB. ANOVA, n= 3, F = 38.455, *P < 0.05 versus vector. (D) Overexpression of HTTAS_v1 leads to down regulation of endogenous HTT expression in SH-SY5Y cells. Expression level was normalized to GUSB. ANOVA, n = 3, F = 24.808, *P < 0.05 versus vector. All experiments in Figure 5 were performed three times in triplicate with similar results. Error bars represent s.d.
Repeat length and HTTAS_v1 interact to regulate HTT expression
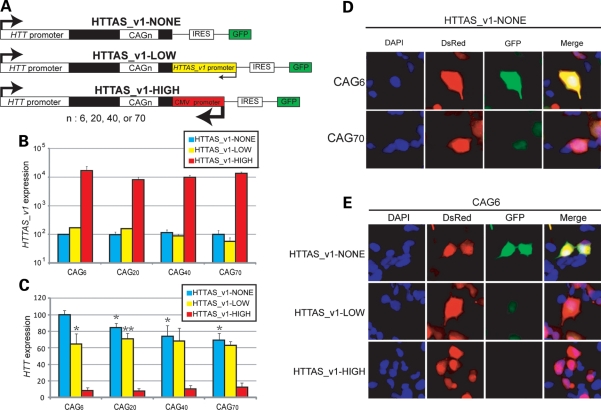
If HTTAS_v1 and HTT expression are inversely correlated, and HTTAS_v1 is downregulated in HD brain, then HTT expression should be elevated in HD brain. However, contrary to this prediction, the available evidence, confirmed by our qPCR data (Fig. 3A), demonstrates that HTT expression in HD and control brain is minimally different. We therefore speculated that the repeat expansion may have a second and counteracting effect on HTT expression. To clarify this issue, we expressed three different sets of minigene constructs in HEK293 cells, enabling us to measure HTT expression in the context of varying levels of HTTAS_v1 expression and varying repeat lengths. Each construct contains the endogenous HTT promoter, HTT exon 1 with CAG6, CAG20, CAG40 or CAG70, an IRES ribosomal entry site, and, in a separate ORF, EGFP, so that the fluorescent intensity of GFP reflects the expression level of the HTT transcript. HTTAS_v1-LOW constructs contain the endogenous HTTAS_v1 promoter. The promoter was deleted to generate HTTAS_v1-NONE constructs, and was replaced by the CMV promoter to generate HTTAS_v1-HIGH constructs (Fig. 5A).
Figure 5.
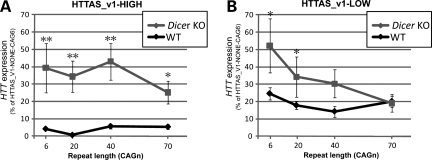
Effect of HTTAS_v1 is repeat length dependent. (A) Minigene constructs. (B) HTTAS_v1 transcribed from HTTAS_v1-NONE, HTTAS_v1-LOW, or HTTAS_v1-HIGH constructs was measured by qPCR and normalized to DsRed expression. HTTAS_v1 is expressed about 100× fold higher when driven by a CMV promoter compared with the background level and about 2× fold higher when driven by an endogenous HTTAS_v1 promoter with short repeats (CAG6, CAG20). Longer repeats (CAG40, CAG70) reduced the HTTAS_v1 expression below the background level. (C and D) Expression of HTT with no HTTAS_v1 promoter (HTTAS_v1-NONE, black bar) reveals an inverse relationship between repeat length and HTT expression. (C and E) Simultaneous expression of HTT and HTTAS_v1 under an endogenous promoter (HTTAS_v1-LOW, yellow bar) eliminates the effect of repeat length on net HTT expression. Overexpression of HTTAS_v1 (HTTAS_v1-HIGH, red bar) markedly reduces HTT expression. 4 × 3 ANOVA n = 10, F(repeat) = 4.47, P(repeat) = 0.008, F(promoter) = 249.77, P(promoter) < 0.001. Scheffe *P < 0.05 versus HTTAS_v1-NONE-CAG6. **P < 0.05 versus HTTAS_v1-NONE-CAG20. Error bars represent s.d.
First, we measured the expression level of HTTAS_v1 transcribed from HTTAS_v1-NONE, HTTAS_v1-LOW and HTTAS_v1-HIGH constructs by qPCR (Fig. 5B). As expected, HTTAS_v1 was expressed about 100× fold higher when driven by a CMV promoter than in the absence of a promoter, regardless of repeat length. HTTAS_v1 under the endogenous promoter is expressed about 2× fold higher than without a promoter in the presence of normal length repeats (CAG6, CAG20). However, longer repeats (CAG40, CAG70) reduced the HTTAS_v1 expression to below the background level.
Next, we measured the level of HTT transcripts from each constructs by quantifying the fluorescent intensity of GFP (Fig. 5C). In the absence of the HTTAS_v1 promoter, HTT expression decreased as repeat length increased, such that expression was 30% less with an expanded repeat (Fig. 5C and D). A similar inverse association between CAG repeat length and transcript levels has been reported for the androgen receptor gene AR (38). Expression of HTTAS_v1 under the endogenous HTTASv1 promoter (HTTAS_v1-LOW construct) relatively reduced HTT expression at shorter but not expanded repeat lengths, with the net effect that HTT expression remained constant across all repeat lengths, consistent with the observations that repeat length does not detectably alter the level of HTT in human brain. In the presence of high HTTAS_v1 expression (HTTAS_v1-HIGH), HTT expression was reduced by about 90% (Fig. 5C and E).
HTTAS_v1 inhibition is partially Dicer dependent
The RNA-induced silencing complex (RISC) pathway involving Dicer is a critical component of several RNA-induced silencing mechanisms. To determine if this pathway plays a role in the silencing of HTT induced by HTTAS_v1, we transfected the HTTAS_v1-NONE, HTTAS_v1-LOW and HTTAS_v1-HIGH constructs into mouse Dicer null or wild-type embryonic stem cells and measured HTT expression. To facilitate comparison, all values were set relative to HTT expression with HTTAS_v1-NONE and a CTG6 repeat. As expected from the experiments with HEK293 cells, high levels of HTTAS_v1 expression reduced expression of HTT to nearly undetectable levels in wild-type embryonic cells (Fig. 6A, black line). Lower level of HTTAS_v1 expression in wild-type cells less markedly reduced HTT expression, and, as in HEK293 cells, the reduction was not repeat length dependent (Fig. 6B, black line). The absence of Dicer markedly increased HTT expression in the presence of high HTTAS_v1 expression in all repeat lengths (Fig. 6A, gray line). However, in the presence of low HTTAS_v1 expression, the absence of Dicer significantly increased HTT expression only in normal repeat lengths (Fig. 6B, gray line), consistent with our data that HTTAS_v1 expression decreases at long expansions. For instance, at low HTTAS_v1 expression and CTG70 repeat, there was no difference between wild-type and Dicer null cells, presumably reflecting the absence of HTTAS_v1 expression with expanded repeat. We therefore conclude that the effect of HTTAS_v1 on HTT expression is at least partly dependent on the RISC pathway.
Figure 6.
Dicer knock down reduces inhibitory effect of HTTAS_v1. (A) With high level of HTTAS_v1 expression, Dicer knock down abolishes inhibitory effect of HTTAS_v1 regardless of repeat lengths. 4 × 2 ANOVA n = 10. F(repeat) = 0.59, P(repeat) = 0.624 F(cell type) = 36.57, P(cell type) < 0.001. Scheffe *P < 0.05 versus wild-type. **P < 0.001 versus wild-type. Error bars represent s.d. (B) With low level of HTTAS_v1 expression, Dicer knock down significantly decreases inhibitory effect of HTTAS_v1 only with short repeats (CAG6 and CAG20). 4 × 2 ANOVA n = 10. F(repeat) = 2.17, P(repeat) = 0.099, F(cell type) = 6.83, P(cell type) = 0.01. Scheffe *P < 0.05 versus wild-type.
DISCUSSION
In this study, we identified HTTAS, a natural antisense transcript at the HD repeat locus. HTTAS is 5′ capped, poly (A) tailed, and contains three exons, alternatively spliced into HTTAS_v1 (exons 1 and 3) and HTTAS_v2 (exons 2 and 3). Exon 1, in HTTAS_v1, includes the repeat. HTTAS_v1 has a weak promoter, and is expressed at low levels in multiple tissue types and throughout the brain. Reporter assays in neuronal and non-neuronal cell lines indicate that while efficient promoter activity requires a short repeat, the repeat expansion reduces promoter efficiency. Consistent with the reporter assays, levels of HTTAS_v1 are reduced in human HD frontal cortex. In cell systems, overexpression of HTTAS_v1 specifically reduces endogenous HTT transcript levels, while siRNA knockdown of HTTAS_v1 increases HTT transcripts. Minigene constructs containing exon 1 of HTT and the HTT promoter, as well as HTTAS_v1 driven by promoters of various strengths, confirm the regulatory effect of HTTAS_v1 on HTT, and demonstrate that the effect is dependent on repeat length and is at least partially Dicer dependent. Together, these findings provide strong evidence for the existence of a gene antisense to HTT that functions to regulate the level of HTT expression.
While we detected three exons forming two splicing variants, we cannot exclude the existence of additional exons and other splice variants. We chose to focus on the function of HTTAS_v1 since exon 1 contains the repeat and preliminary experiments (data not shown) suggest that overexpression of HTTAS_v2 does not alter levels of HTT transcription. Exons 2 and 3 include 55 and 66 amino acid ORFs, respectively. There is no evidence of these putative proteins in the GenBank non-redundant protein database, nor do these putative proteins contain known motifs. Nonetheless, it remains possible that these ORFs do encode short proteins with a function of potential relevance to HD, as in ASFMR1/FMR4 at the Fragile X locus (20,21). Further, the recent discovery of protein expression arising from repeats without an AUG translation initiation codon (39) suggests the possibility of alternative cryptic protein expression from the HD locus.
Two binding sites for CTCF are predicted in the region 5′ to the functional HTTAS_v1 promoter, and CTCF binding had been experimentally demonstrated in this approximate region (32). CTCF is a DNA-binding protein with multiple transcriptional regulatory functions (40). Interestingly, CTCF binding sites have also been detected adjacent to the DM1, SCA2 and SCA7 repeat loci (26,32,41). At the SCA7 locus, CTCF modulates CAG repeat stability by promoting DNA methylation, and experimental elimination of CTCF sites enhances repeat instability (41). Whether similar mechanisms, presumably involving chromatin remodeling, may be influencing sense or antisense transcription at the HD locus remains to be determined.
The HTTAS_v1 transcript contains a CUG repeat; such repeats can be toxic and substantially contribute to disease pathogenesis. In myotonic dystrophy type 1 (DM1), a CUG repeat expansion (>∼50 triplets, and more typically >100 triplets) in the 3′ UTR of DMPK transcripts leads to dysregulation of the splicing factors MBNL1 and CUGBP1 with subsequent missplicing events highly correlated to disease phenotype (42,43). A similar phenomenon has been detected in HDL2, a disorder phenotypically and pathologically very similar to HD (33–35,44). However, we were unable to detect HTTAS_v1 with an expanded repeat in HD by SS-RTPCR, and RNA foci containing long CUG repeats, observed in both DM1 and HDL2 post-mortem brain by in situ hybridization, have not been detected in HD brains (44). Even if expressed, levels of HTTAS_v1 transcript are 100× fold lower than HTT transcript, suggesting that any direct effect of CUG repeats on toxicity would likely be relatively minor.
Promoter activity assays (Fig. 2E) and the minigene experiment (Fig. 5C) both demonstrate that a long repeat reduces HTTAS_v1 expression in cell models. Consistent with this finding, HTTAS_v1 is lower in HD cortex than control cortex (Fig. 3C), with much of the difference arising from loss of expression of the expanded allele. It seems unlikely that loss of expression is an indirect effect of neurodegeneration, as there was no loss of HTTAS_v1 expression in HDL2 frontal cortex (Supplementary Material, Fig. S1). We speculate that the diminished expression reflects impaired transcription efficiency of alleles with increasingly long repeats (45–48), but the mechanisms remain unclear, and possibilities such as loss of transcript stability cannot be excluded.
The inhibitory effect of HTTAS_v1 on HTT expression and the reduced expression of HTTAS_v1 in HD brain suggest that HTT should be upregulated in HD brain. Our minigene experiments demonstrate that in the context of a repeat expansion, HTTAS_v1 expression is indeed decreased, reducing the inhibitory effect of HTTAS_v1 on HTT expression. However, by eliminating expression of HTTAS_v1, we were able to demonstrate that repeat expansion also directly decreases HTT expression. While the mechanism for this effect is not clear, there are many examples of repeat length variations affecting promoter or transcriptional efficiency (49–52). The net effect of repeat expansion on HTT expression in the intact system is therefore almost neutral, as the loss of HTTAS_v1 expression and consequent decrease of HTT inhibition is nearly balanced by the direct effect of the repeat expansion on HTT. This balance is consistent with the similar levels of HTT expression observed in HD and control brain (Fig. 3A). The minigene model also suggests that the effect of HTTASv_1 on HTT levels, at least in cis, depends primarily on the level of HTTASv_1 expression rather than on repeat-mediated changes in other properties of their interaction (e.g. hybridization efficiency).
The mechanism for the relatively selective vulnerability of medium spiny neurons of the striatum (53) and pyramidal neurons of deep cortical layers (54) remains uncertain. Our data demonstrate that HTTAS_v1 in control brain is more prominently expressed in cortex than in either striatum or cerebellum (Fig. 3B), while in HD brain HTTAS_v1 is suppressed in all brain regions. This pattern provides little evidence for a role of HTTAS_v1 in selective neuronal vulnerability, but more definitive negative data await analysis of expression in individual cell types.
HTTAS_v1 overexpression via transient transfection decreased endogenous HTT by about 25% (Fig. 4C), a trans phenomenon, whereas our minigene experiment demonstrated that expression of HTTAS_v1 at low levels from its native promoter in cis with HTT led to ∼35% reduction in HTT expression, and high levels of HTTAS_v1 driven by a CMV promoter in cis suppressed HTT expression by 90% (Fig. 5C). Together, these results suggest that the endogenous regulatory effect of HTTAS_v1 on HTT is potentially of physiological magnitude. Our experiments in Dicer null cells demonstrate that a RISC-mediated process contributes to the cis effect, implying formation of an RNA duplex from sense and antisense transcripts and further supporting a regulatory effect of HTTASv_1 on HTT. The residual cis regulatory effect presumably operates through one of several mechanisms of antisense-mediated transcriptional dysregulation, related to the competitive effects of simultaneous sense and antisense transcription, antisense RNA-mediated transcriptional interference or RNA duplex-mediated epigenetic modifications (55).
Based on our cell experiments and exploration of HTT and HTTAS_v1 expression in HD and control brain, we propose a three-part model of antisense regulation of HTT expression. With a normal length repeat, HTTAS_v1 under control of its endogenous promoter is expressed at low levels, and partially inhibits HTT expression. With an expanded repeat, HTTAS_v1 expression is reduced, removing the inhibitory effect of HTTAS_v1 on HTT. However, this effect is compensated by an HTTAS_v1-independent effect of the repeat expansion on HTT expression. If HTTAS_v1 is overexpressed, in trans and especially in cis, HTT expression is markedly suppressed, regardless of repeat length.
An important approach to HD therapeutics is suppression of mutant HTT expression. It is clearly possible to design siRNA or oligonucelotides that knock down HTT expression (55–58). These approaches face numerous hurdles (59): off target inhibition of other genes, immunostimulation, lack of specificity for the mutant allele, the need to target large regions of the brain and clinically feasible delivery mechanisms. Here we show that overexpression of an endogenous antisense transcript also markedly reduces HTT expression, suggesting that manipulating antisense expression may have clinical implications.
MATERIALS AND METHODS
Primer sequences are listed in Supplementary Material, Table S1. PCR was performed using Platinum Taq DNA Polymerase HF (Invitrogen). RT–PCR was performed using Superscript III RT (Invitrogen).
Total RNA extraction
Brain samples were obtained from the HD samples maintained in the Brain Resource Center of the Johns Hopkins University Department of Pathology. All brain samples were collected under protocols supervised by the Johns Hopkins IRB. See Supplementary Material, Tables S4 and S5 for brain details. Cells or human brain tissues were homogenized by QIAshredder (Qiagen). Genomic DNA was extracted using DNaseI (Invitrogen). Total RNA was extracted using RNeasy kit (Qiagen).
Strand-specific reverse-transcriptase PCR
Total RNA from control (Clontech) and HD brain [expanded allele (CAG)47, normal allele (CAG)21] was reverse transcribed using LK-HD1SHORT(F) (26). First round of PCR was performed using LK-only(F) and HD3(R). Nested PCR was performed using HD1-SHORT(F) and HD3(R). PCR cycle: 95°C (5 min); 95°C (30 s); 67°C (30 s); 68°C (30 s), for 30 cycles, then 68°C (7 min). Sanger sequencing (ABI) was performed after all PCR products were cloned into pCR4-TOPO (Invitrogen).
5′-RACE
RACE-ready mRNA was prepared from total RNA from control (Clontech) and HD frontal cortex using Generacer (Invitrogen) and reverse transcribed using random hexamers (Invitrogen). First round of PCR was performed using Generacer-5′(F) and HTTAS1-1(R) or HTTAS2-1(R). Nested PCR was performed using Generacer-5′nested(F) and HTTAS1-2(R) or HTTAS2-2(R). First round of PCR was performed with a touchdown PCR protocol [95°C (5 min); 95°C (30 s), 72°C (30 s) for five cycles; 95°C (30 s), 70°C (30 s) for five cycles; 95°C (30 s), 67°C (30 s), 68°C (30 s) for 25 cycles; 68°C (7 min)] and nested PCR was performed with the protocol used for SS-RTPCR. PCR products were sequenced as earlier.
3′-RACE
Total RNA from control (Clontech) and HD frontal cortex was reverse transcribed using the GeneRacer oligo-dT primer (Invitrogen). First round of PCR was performed using Generacer-3′(R) and HTTAS1-3(F) or HTTAS2-3(R). Nested PCR was performed using Generacer-3′nested(R) and HTTAS1-4(F) or HTTAS2-4(F). First and nested PCR were performed with the touchdown PCR protocol used for 5′-RACE. The PCR products were sequenced as earlier.
Promoter assay
To construct HTTAS446 and 132-LUC, the region between +52 and either −446 or −132 bp relative to the HTTAS1 transcription start site was amplified using HD3-5-HindIII(R) and either HTTAS446-KpnI(F) or HTTAS132-KpnI(F), respectively, digested with KpnI and HindIII, and cloned into pGL3-basic (Promega). To construct HTTAS446-CAG6, 20, 40 and 70-LUC, HTT-exon 1 (courtesy of CA Ross) with 6, 20, 40 or 70 triplets was amplified with HD1SHORT4(F) and HD3(R). HTTAS446-LUC construct was further amplified with HTTAS446(F) and HD3R(R). The two PCR products were then amplified using HTTAS446-KpnI(F) and HD1SHORT2-HindIII(R). PCR products were digested with KpnI and HindIII and cloned into pGL3-basic. To clone HDpromoter-LUC, human DNA was amplified using Httpromoter1-Kpn1(F) and Httpromoter1-HindIII(R). The product was digested with KpnI and HindIII and cloned into pGL3-basic. 5000 HEK293 and 2500 SH-SY5Y cells were aliquoted into 96-well plates in media without FBS. Twenty-four hours after plating, constructs were transfected into cells using lipofectamine2000 (Invitrogen). Renilla was co-transfected in a 1:20 ratio for internal control. Promoter activity was measured 48 h after transfection using Dual Luciferase Reporter Assay (Promega).
Bidirectional construct cloning
HDpromoter-LUC was amplified using Httpromoter1-BglII(F) and HDSHORT2(R) and the product was digested with TaqI and BglII. HTTAS446-CAG6, 20, 40 and 70-LUC were amplified using HD1SHORT4(F) and 446-SalI(R) and digested with TaqI and SalI. Digested fragments were ligated with T4 ligase (Invitrogen) to generate HTTAS1-LOW inserts. HTTAS1-LOW inserts were amplified using Httpromoter1-BglII(F) and 132-SalI(R) to generate HTTAS1-NONE inserts. The ligated products were cloned into Pires2-EGFP (Clontech) as modified by H. Cui and colleagues (60).
Quantitative PCR
qPCR reaction was performed on a GeneAmp 7900 HT using 30–60 ng of diluted cDNA, 5 μl TaqMan master mix and 0.5 μl TaqMan probes (ABI, see Supplementary Material, Table S3 for primer sequences).
Cell culture
Human embryonic kidney 293 (HEK293; Invitrogen) cells and SH-SY5Y (ATCC) cells were maintained in DMEM with high glucose (GIBCO), supplemented with 10% fetal bovine serum (FBS; GIBCO) and 1% Penicillin–Streptomycin solution (P/S; GIBCO). Wild-type or Dicer null mouse embryonic stem cells provided by G. Hannon (61) were maintained in DMEM KO media containing 90 ml ES cell qualified FBS (GIBCO), 6 ml 100× Beta-merc (GIBCO), 3 ml 100× P/S, 60 μl ESGRO-LIF (Millipore) and 6 ml 100× l-glutamine (GIBCO).
HTTAS1 knockdown and overexpression
A total of 500 000 HEK293 and 250 000 SH-SY5Y cells were plated in six-well plates in media without FBS. Twenty-four hours after plating, pooled siRNA (see Supplementary Material, Table S6) or scrambled siRNA (Dharmacon) was transfected using lipofectamine2000. Total RNA was extracted 72 h after transfection and qPCR was performed using TaqMan probes. pcDNA 3.1 (Invitrogen) containing a full-length HTTASv_1 cDNA was transfected into HEK293 and SH-SY5Y cells using lipofectamine2000. Total RNA was extracted 48 h after transfection and qPCR was performed using TaqMan probes.
GFP quantification assay
A total of 100 000 HEK293 cells, or wild-type or Dicer null mouse embryonic stem cells were plated in 24-well plates in media without FBS. Twenty-four hours after plating, HTTAS1-NONE, HTTAS1-LOW and HTTAS1-HIGH constructs were transfected using lipofectamine2000. DsRed-N1 (Clontech) was co-transfected in a 1:20 ratio. Forty-eight hours after transfection, cells were fixed with 4% paraformaldehyde. Photomicrographs of 10 random fields were taken at 20× using Axiovert 100 (Zeiss). GFP and Ds-Red intensity was measured by Axiovision 4.4 (Zeiss).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health (NS16375 to C.R., NS061099 and NS066111 to R.L.M., NS064138 to D.D.R.).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Greg Hannon for wild-type and Dicer knock out mouse stem cells, Hengmi Cui for modified Pires2-GFP constructs, Olga Pletnikova and Juan Troncoso for HD and HDL2 brain samples, Chengxiu Zhang for technical assistance and Christopher Ross for conceptual input.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Huntington G. On chorea. Med. Surg. Rep. 1872;26:317–321. [Google Scholar]
- 2.Ross C.A., Margolis R.L., Rosenblatt A., Ranen N.G., Becher M.W., Aylward E. Huntington disease and the related disorder, dentatorubral-pallidoluysian atrophy (DRPLA) Medicine (Baltimore) 1997;76:305–338. doi: 10.1097/00005792-199709000-00001. doi:10.1097/00005792-199709000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Bates G., Harper P., Jones L. Huntington's Disease. Oxford: Oxford University Press; 2002. [Google Scholar]
- 4.Vonsattel J.P., Myers R.H., Stevens T.J., Ferrante R.J., Bird E.D., Richardson E.P., Jr Neuropathological classification of huntington's disease. J. Neuropathol. Exp. Neurol. 1985;44:559–577. doi: 10.1097/00005072-198511000-00003. doi:10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- 5.DiFiglia M., Sapp E., Chase K.O., Davies S.W., Bates G.P., Vonsattel J.P., Aronin N. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. doi:10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 6.Ross C.A., Poirier M.A. Protein aggregation and neurodegenerative disease. Nat. Med. 2004;10(suppl.)):S10–S17. doi: 10.1038/nm1066. doi:10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 7.Ferrante R.J. Mouse models of huntington's disease and methodological considerations for therapeutic trials. Biochim. Biophys. Acta. 2009;1792:506–520. doi: 10.1016/j.bbadis.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ross C.A., Shoulson I. Huntington disease: pathogenesis, biomarkers, and approaches to experimental therapeutics. Parkinsonism Relat. Disord. 2009;15(Suppl. 3)):S135–S138. doi: 10.1016/S1353-8020(09)70800-4. doi:10.1016/S1353-8020(09)70800-4. [DOI] [PubMed] [Google Scholar]
- 9.Caviston J.P., Holzbaur E.L. Huntingtin as an essential integrator of intracellular vesicular trafficking. Trends Cell Biol. 2009;19:147–155. doi: 10.1016/j.tcb.2009.01.005. doi:10.1016/j.tcb.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zuccato C., Cattaneo E. Role of brain-derived neurotrophic factor in huntington's disease. Prog. Neurobiol. 2007;81:294–330. doi: 10.1016/j.pneurobio.2007.01.003. doi:10.1016/j.pneurobio.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Coles R., Caswell R., Rubinsztein D.C. Functional analysis of the huntington's disease (HD) gene promoter. Hum. Mol. Genet. 1998;7:791–800. doi: 10.1093/hmg/7.5.791. doi:10.1093/hmg/7.5.791. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka K., Shouguchi-Miyata J., Miyamoto N., Ikeda J.E. Novel nuclear shuttle proteins, HDBP1 and HDBP2, bind to neuronal cell-specific cis-regulatory element in the promoter for the human huntington's disease gene. J. Biol. Chem. 2004;279:7275–7286. doi: 10.1074/jbc.M310726200. doi:10.1074/jbc.M310726200. [DOI] [PubMed] [Google Scholar]
- 13.Lee J., Park E.H., Couture G., Harvey I., Garneau P., Pelletier J. An upstream open reading frame impedes translation of the huntingtin gene. Nucleic Acids Res. 2002;30:5110–5119. doi: 10.1093/nar/gkf664. doi:10.1093/nar/gkf664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He Y., Vogelstein B., Velculescu V.E., Papadopoulos N., Kinzler K.W. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. doi:10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimura K., Wakamatsu A., Suzuki Y., Ota T., Nishikawa T., Yamashita R., Yamamoto J., Sekine M., Tsuritani K., Wakaguri H., et al. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16:55–65. doi: 10.1101/gr.4039406. doi:10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carninci P., Kasukawa T., Katayama S., Gough J., Frith M.C., Maeda N., Oyama R., Ravasi T., Lenhard B., Wells C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. doi:10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 17.Mazo A., Hodgson J.W., Petruk S., Sedkov Y., Brock H.W. Transcriptional interference: an unexpected layer of complexity in gene regulation. J. Cell. Sci. 2007;120:2755–2761. doi: 10.1242/jcs.007633. doi:10.1242/jcs.007633. [DOI] [PubMed] [Google Scholar]
- 18.Werner A., Carlile M., Swan D. What do natural antisense transcripts regulate? RNA Biol. 2009;6:43–48. doi: 10.4161/rna.6.1.7568. doi:10.4161/rna.6.1.7568. [DOI] [PubMed] [Google Scholar]
- 19.Batra R., Charizanis K., Swanson M.S. Partners in crime: bidirectional transcription in unstable microsatellite disease. Hum. Mol. Genet. 2010;19:R77–R82. doi: 10.1093/hmg/ddq132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khalil A.M., Faghihi M.A., Modarresi F., Brothers S.P., Wahlestedt C. A novel RNA transcript with antiapoptotic function is silenced in fragile X syndrome. PLoS One. 2008;3:e1486. doi: 10.1371/journal.pone.0001486. doi:10.1371/journal.pone.0001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ladd P.D., Smith L.E., Rabaia N.A., Moore J.M., Georges S.A., Hansen R.S., Hagerman R.J., Tassone F., Tapscott S.J., Filippova G.N. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum. Mol. Genet. 2007;16:3174–3187. doi: 10.1093/hmg/ddm293. doi:10.1093/hmg/ddm293. [DOI] [PubMed] [Google Scholar]
- 22.Koob M.D., Moseley M.L., Schut L.J., Benzow K.A., Bird T.D., Day J.W., Ranum L.P. An untranslated CTG expansion causes a novel form of spinocerebellar ataxia (SCA8) Nat. Genet. 1999;21:379–384. doi: 10.1038/7710. doi:10.1038/7710. [DOI] [PubMed] [Google Scholar]
- 23.Moseley M.L., Zu T., Ikeda Y., Gao W., Mosemiller A.K., Daughters R.S., Chen G., Weatherspoon M.R., Clark H.B., Ebner T.J., et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat. Genet. 2006;38:758–769. doi: 10.1038/ng1827. doi:10.1038/ng1827. [DOI] [PubMed] [Google Scholar]
- 24.Daughters R.S., Tuttle D.L., Gao W., Ikeda Y., Moseley M.L., Ebner T.J., Swanson M.S., Ranum L.P. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009;5:e1000600. doi: 10.1371/journal.pgen.1000600. doi:10.1371/journal.pgen.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brook J.D., McCurrach M.E., Harley H.G., Buckler A.J., Church D., Aburatani H., Hunter K., Stanton V.P., Thirion J.P., Hudson T. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;69:385. doi: 10.1016/0092-8674(92)90418-c. [DOI] [PubMed] [Google Scholar]
- 26.Cho D.H., Thienes C.P., Mahoney S.E., Analau E., Filippova G.N., Tapscott S.J. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol. Cell. 2005;20:483–489. doi: 10.1016/j.molcel.2005.09.002. doi:10.1016/j.molcel.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 27.Faghihi M.A., Wahlestedt C. Regulatory roles of natural antisense transcripts. Nat. Rev. Mol. Cell Biol. 2009;10:637–643. doi: 10.1038/nrm2738. doi:10.1038/nrm2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. doi:10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchler-Bauer A., Anderson J.B., Chitsaz F., Derbyshire M.K., DeWeese-Scott C., Fong J.H., Geer L.Y., Geer R.C., Gonzales N.R., Gwadz M., et al. CDD: specific functional annotation with the conserved domain database. Nucleic Acids Res. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. doi:10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klimke W., Agarwala R., Badretdin A., Chetvernin S., Ciufo S., Fedorov B., Kiryutin B., O'Neill K., Resch W., Resenchuk S., et al. The national center for biotechnology information's protein clusters database. Nucleic Acids Res. 2009;37:D216–D223. doi: 10.1093/nar/gkn734. doi:10.1093/nar/gkn734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. doi:10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 32.Filippova G.N., Thienes C.P., Penn B.H., Cho D.H., Hu Y.J., Moore J.M., Klesert T.R., Lobanenkov V.V., Tapscott S.J. CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nat. Genet. 2001;28:335–343. doi: 10.1038/ng570. doi:10.1038/ng570. [DOI] [PubMed] [Google Scholar]
- 33.Margolis R.L., O'Hearn E., Rosenblatt A., Willour V., Holmes S.E., Franz M.L., Callahan C., Hwang H.S., Troncoso J.C., Ross C.A. A disorder similar to huntington's disease is associated with a novel CAG repeat expansion. Ann. Neurol. 2001;50:373–380. doi: 10.1002/ana.1312. [DOI] [PubMed] [Google Scholar]
- 34.Holmes S.E., O'Hearn E., Rosenblatt A., Callahan C., Hwang H.S., Ingersoll-Ashworth R.G., Fleisher A., Stevanin G., Brice A., Potter N.T., et al. A repeat expansion in the gene encoding junctophilin-3 is associated with huntington disease-like 2. Nat. Genet. 2001;29:377–378. doi: 10.1038/ng760. doi:10.1038/ng760. [DOI] [PubMed] [Google Scholar]
- 35.Rudnicki D.D., Pletnikova O., Vonsattel J.P., Ross C.A., Margolis R.L. A comparison of huntington disease and huntington disease-like 2 neuropathology. J. Neuropathol. Exp. Neurol. 2008;67:366–374. doi: 10.1097/NEN.0b013e31816b4aee. doi:10.1097/NEN.0b013e31816b4aee. [DOI] [PubMed] [Google Scholar]
- 36.Greenstein P.E., Vonsattel J.P., Margolis R.L., Joseph J.T. Huntington's disease like-2 neuropathology. Mov. Disord. 2007;22:1416–1423. doi: 10.1002/mds.21417. doi:10.1002/mds.21417. [DOI] [PubMed] [Google Scholar]
- 37.Stine O.C., Li S.H., Pleasant N., Wagster M.V., Hedreen J.C., Ross C.A. Expression of the mutant allele of IT-15 (the HD gene) in striatum and cortex of huntington's disease patients. Hum. Mol. Genet. 1995;4:15–18. doi: 10.1093/hmg/4.1.15. doi:10.1093/hmg/4.1.15. [DOI] [PubMed] [Google Scholar]
- 38.Choong C.S., Kemppainen J.A., Zhou Z.X., Wilson E.M. Reduced androgen receptor gene expression with first exon CAG repeat expansion. Mol. Endocrinol. 1996;10:1527–1535. doi: 10.1210/mend.10.12.8961263. doi:10.1210/me.10.12.1527. [DOI] [PubMed] [Google Scholar]
- 39.Zu T., Gibbens B., Doty N.S., Gomes-Pereira M., Huguet A., Stone M.D., Margolis J., Peterson M., Markowski T.W., Ingram M.A., et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc. Natl Acad. Sci. USA. 2011;108:260–265. doi: 10.1073/pnas.1013343108. doi:10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phillips J.E., Corces V.G. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. doi:10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Libby R.T., Hagerman K.A., Pineda V.V., Lau R., Cho D.H., Baccam S.L., Axford M.M., Cleary J.D., Moore J.M., Sopher B.L., et al. CTCF cis-regulates trinucleotide repeat instability in an epigenetic manner: a novel basis for mutational hot spot determination. PLoS Genet. 2008;4:e1000257. doi: 10.1371/journal.pgen.1000257. doi:10.1371/journal.pgen.1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee J.E., Cooper T.A. Pathogenic mechanisms of myotonic dystrophy. Biochem. Soc. Trans. 2009;37:1281–1286. doi: 10.1042/BST0371281. doi:10.1042/BST0371281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wheeler T.M., Thornton C.A. Myotonic dystrophy: RNA-mediated muscle disease. Curr. Opin. Neurol. 2007;20:572–576. doi: 10.1097/WCO.0b013e3282ef6064. doi:10.1097/WCO.0b013e3282ef6064. [DOI] [PubMed] [Google Scholar]
- 44.Rudnicki D.D., Holmes S.E., Lin M.W., Thornton C.A., Ross C.A., Margolis R.L. Huntington's disease–like 2 is associated with CUG repeat-containing RNA foci. Ann. Neurol. 2007;61:272–282. doi: 10.1002/ana.21081. doi:10.1002/ana.21081. [DOI] [PubMed] [Google Scholar]
- 45.Kumari D., Biacsi R.E., Usdin K. Repeat expansion affects both transcription initiation and elongation in friedreich ataxia cells. J. Biol. Chem. 2011;286:4209–4215. doi: 10.1074/jbc.M110.194035. doi:10.1074/jbc.M110.194035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Punga T., Buhler M. Long intronic GAA repeats causing friedreich ataxia impede transcription elongation. EMBO Mol. Med. 2010;2:120–129. doi: 10.1002/emmm.201000064. doi:10.1002/emmm.201000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alakurtti K., Virtaneva K., Joensuu T., Palvimo J.J., Lehesjoki A.E. Characterization of the cystatin B gene promoter harboring the dodecamer repeat expanded in progressive myoclonus epilepsy, EPM1. Gene. 2000;242:65–73. doi: 10.1016/s0378-1119(99)00550-8. doi:10.1016/S0378-1119(99)00550-8. [DOI] [PubMed] [Google Scholar]
- 48.Lalioti M.D., Scott H.S., Antonarakis S.E. Altered spacing of promoter elements due to the dodecamer repeat expansion contributes to reduced expression of the cystatin B gene in EPM1. Hum. Mol. Genet. 1999;8:1791–1798. doi: 10.1093/hmg/8.9.1791. doi:10.1093/hmg/8.9.1791. [DOI] [PubMed] [Google Scholar]
- 49.Schmucker S., Puccio H. Understanding the molecular mechanisms of friedreich's ataxia to develop therapeutic approaches. Hum. Mol. Genet. 2010;19:R103–R110. doi: 10.1093/hmg/ddq165. doi:10.1093/hmg/ddq165. [DOI] [PubMed] [Google Scholar]
- 50.Lalioti M.D., Scott H.S., Buresi C., Rossier C., Bottani A., Morris M.A., Malafosse A., Antonarakis S.E. Dodecamer repeat expansion in cystatin B gene in progressive myoclonus epilepsy. Nature. 1997;386:847–851. doi: 10.1038/386847a0. doi:10.1038/386847a0. [DOI] [PubMed] [Google Scholar]
- 51.Sutcliffe J.S., Nelson D.L., Zhang F., Pieretti M., Caskey C.T., Saxe D., Warren S.T. DNA methylation represses FMR-1 transcription in fragile X syndrome. Hum. Mol. Genet. 1992;1:397–400. doi: 10.1093/hmg/1.6.397. doi:10.1093/hmg/1.6.397. [DOI] [PubMed] [Google Scholar]
- 52.Okladnova O., Syagailo Y.V., Tranitz M., Stober G., Riederer P., Mossner R., Lesch K.P. A promoter-associated polymorphic repeat modulates PAX-6 expression in human brain. Biochem. Biophys. Res. Commun. 1998;248:402–405. doi: 10.1006/bbrc.1998.8972. doi:10.1006/bbrc.1998.8972. [DOI] [PubMed] [Google Scholar]
- 53.Cepeda C., Wu N., Andre V.M., Cummings D.M., Levine M.S. The corticostriatal pathway in huntington's disease. Prog. Neurobiol. 2007;81:253–271. doi: 10.1016/j.pneurobio.2006.11.001. doi:10.1016/j.pneurobio.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hedreen J.C., Peyser C.E., Folstein S.E., Ross C.A. Neuronal loss in layers V and VI of cerebral cortex in huntington's disease. Neurosci. Lett. 1991;133:257–261. doi: 10.1016/0304-3940(91)90583-f. doi:10.1016/0304-3940(91)90583-F. [DOI] [PubMed] [Google Scholar]
- 55.Munroe S.H., Zhu J. Overlapping transcripts, double-stranded RNA and antisense regulation: a genomic perspective. Cell Mol. Life Sci. 2006;63:2102–2118. doi: 10.1007/s00018-006-6070-2. doi:10.1007/s00018-006-6070-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boudreau R.L., McBride J.L., Martins I., Shen S., Xing Y., Carter B.J., Davidson B.L. Nonallele-specific silencing of mutant and wild-type huntingtin demonstrates therapeutic efficacy in huntington's disease mice. Mol. Ther. 2009;17:1053–1063. doi: 10.1038/mt.2009.17. doi:10.1038/mt.2009.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu J., Matsui M., Corey D.R. Allele-selective inhibition of mutant huntingtin by peptide nucleic acid-peptide conjugates, locked nucleic acid, and small interfering RNA. Ann. NY Acad. Sci. 2009;1175:24–31. doi: 10.1111/j.1749-6632.2009.04975.x. doi:10.1111/j.1749-6632.2009.04975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pfister E.L., Zamore P.D. Huntington's disease: silencing a brutal killer. Exp. Neurol. 2009;220:226–229. doi: 10.1016/j.expneurol.2009.09.017. doi:10.1016/j.expneurol.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aronin N. Target selectivity in mRNA silencing. Gene Ther. 2006;13:509–516. doi: 10.1038/sj.gt.3302726. doi:10.1038/sj.gt.3302726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu W., Gius D., Onyango P., Muldoon-Jacobs K., Karp J., Feinberg A.P., Cui H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature. 2008;451:202–206. doi: 10.1038/nature06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Murchison E.P., Partridge J.F., Tam O.H., Cheloufi S., Hannon G.J. Characterization of Dicer-deficient murine embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2005;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.