Abstract
During development of the nervous system, axons and growth cones contain mRNAs, such as β-actin, cofilin, and RhoA, that are locally translated in response to guidance cues. Intra-axonal translation of these mRNAs results in local morphological responses, however other functions of intra-axonal mRNA translation remain unknown. Here we show that axons of developing mammalian neurons contain mRNA encoding the cAMP-responsive element (CRE)-binding protein (CREB). CREB is translated within axons in response to NGF and is retrogradely trafficked to the cell body. In neurons that are selectively deficient in axonal CREB transcripts, increases in nuclear pCREB, CRE-mediated transcription, and neuronal survival elicited by axonal application of NGF are abolished, indicating a signalling function for axonally-synthesised CREB. These studies identify a signalling role for axonally-derived CREB, and indicate that signal-dependent synthesis and retrograde trafficking of transcription factors enables specific transcriptional responses to signalling events at distal axons.
An important mechanism by which the protein composition at specific subcellular sites is regulated is by the precise intracellular targeting and translation of certain mRNAs1. This mechanism particularly evident in axons of developing neurons, which contain mRNAs, ribosomes, and other translational machinery2. Most known axonal mRNAs encode proteins that are involved in cytoskeletal regulation, and are involved in mediating the response to guidance cues3–5. Roles for axonal mRNA translation in other processes have not been established.
One critical function of axons during development is to detect signals in the extracellular environment and to transduce these signals into changes in gene expression in the nucleus6–9. In developing sympathetic and sensory axons, growth cones detect nerve growth factor (NGF) synthesised by target cells, resulting in the generation of a retrograde signal that is conveyed to the soma that promotes neuronal survival10.
Here we describe a role for axonal mRNA translation in the regulation of neuronal survival and nuclear transcription elicited by axonal application of NGF. We find NGF triggers axonal protein synthesis, which is required for NGF-mediated retrograde survival. A cDNA library prepared from the axons of developing sensory neurons reveals that CREB mRNA is an axonally-localised transcript. We find that CREB is selectively translated in axons in response to NGF and retrogradely trafficked to the cell body. Furthermore, selective knockdown of axonal CREB mRNA reveals that axonally-synthesised CREB is required for NGF at axons to promote the accumulation of pCREB in the nucleus, transcription of a CRE-containing reporter gene, and neuronal survival. These data identify a role for axonally-synthesised CREB and identify a signalling mechanism involving intra-axonal translation and retrograde trafficking of transcription factors that may have critical roles in signalling from axons to the nucleus.
RESULTS
Retrograde NGF signalling at axon terminals requires protein synthesis
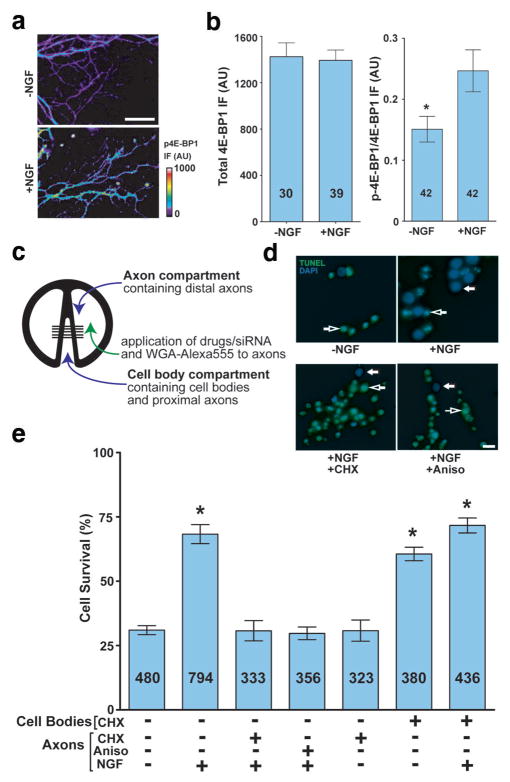
The signalling pathways downstream of NGF in growth cones are unclear. To determine if NGF might regulate local protein synthesis, embryonic day 15 (E15) dorsal root ganglia (DRG) cultures (Fig. S1a,b) were transferred to NGF-free media for 2 h and then stimulated with NGF or vehicle for 1 h. NGF treatment resulted in increased phosphorylation of 4E-BP1 at S64 and T69 in axons (Fig. 1a,b). Multisite phosphorylation of 4E-BP1 is required for mRNA translation11, indicating that NGF may induce local protein synthesis.
Figure 1. Local protein synthesis in axons is required for NGF-dependent survival.
(a) Phospho-4E-BP1 levels increase in growth cones in response to NGF treatment. DRG neurons were incubated with NGF-replete or NGF-free media; phospho-4E-BP1 levels in axons were measured by immunofluorescence. Scale bar, 50 μm. (b) Quantification of total and phospho-4EBP1 in (a) p=0.012. Numbers on bars represent n axons per condition. (c) Schematic diagram of compartmented (Campenot) chambers. E15 dissociated DRG neurons are cultured in the cell body compartment and axons grow under a thinly applied silicone grease layer that seals the chamber with the Permanox® plastic culture slide. (d) Application of protein synthesis inhibitors to axons blocks NGF-mediated retrograde survival. Dissociated DRG neurons were grown in compartmentalised chambers, and vehicle or NGF was added to the axon compartment. 1 μM cycloheximide (CHX) or 40 μM anisomycin (Aniso) were added to the axon compartment concurrently with NGF media. Cell body compartments were kept NGF-free during the course of the experiment. Cells crossing the divider were retrogradely labelled with WGA-Alexa555 and only WGA-positive cell bodies were counted in the data set. Survival was assessed by TUNEL assay. Examples of non-apoptotic and apoptotic are indicated with closed and open arrows, respectively (Blue=DAPI, Green=TUNEL). Scale bar, 20 μm. (e) Quantification of results from (d). *p<0.001. Numbers on bars represent n cells per condition.
To determine if protein synthesis is required for NGF signalling we cultured neurons in compartmented chambers, which permit selective application of NGF to either distal axons or cell bodies12, mimicking the physiologically selective exposure of distal axons to NGF that occurs as axons approach NGF-synthesising target tissues (Fig. 1c, Fig. S1c–f). Axonal application of NGF activates a survival pathway that utilizes CREB, resembling the physiologic requirement for CREB13; bath application appears to utilize different NGF signalling pathways, as it induces survival in the absence of CREB13. Axons crossed the divider by DIV 5, at which point the media in the cell body compartment was replaced with NGF-free media and the media in the axonal compartment is replaced with either NGF-free or NGF-replete media for an additional 48 h. Application of NGF exclusively to the axonal compartment resulted in a significant increase in neuronal survival compared to vehicle-treated axons (Fig. 1d,e). This effect required intra-axonal protein synthesis, as axonal application of cycloheximide or anisomycin, together with NGF, resulted in a significant reduction in survival compared to NGF alone (Fig. 1d,e). The fluidic isolation of treatments was confirmed by our finding that axonal application of translation inhibitors had no effect in the cell body compartment on a membrane-anchored translation reporter (Fig. S1e,f). Furthermore, incubation of translation inhibitors in the cell body compartment supported DRG survival, even in the absence of NGF (Fig. 1e), consistent with previous results that have shown that protein synthesis inhibition in various cells, including DRG neurons, promotes survival due to the requirement for new protein synthesis in apoptotis14–16.
CREB mRNA is localised to axons
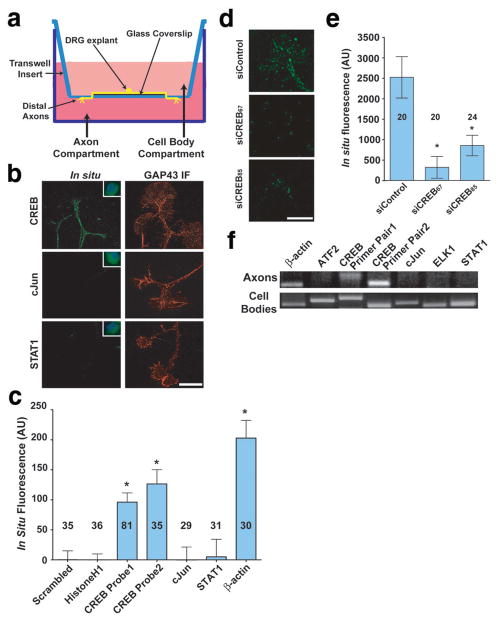
To identify mRNAs that act downstream of NGF in axons, we modified the Boyden chamber technique for isolating axons17 in order to obtain distal axons (Fig. 2a)4. DRG explants were cultured on 12-mm coverslips placed in the centre of Boyden chambers. Axons grow across the coverslip and then grow through the membrane towards the higher NGF concentration in the lower chamber. Axons from six chambers were harvested by scraping the underside of the membrane and used for reverse transcription and cDNA amplification using a protocol designed for unbiased amplification of mRNA from single cells18 (See Supplementary Methods).
Figure 2. CREB mRNA and protein are localised to developing axons of DRG neurons.
(a) Schematic diagram of Boyden chamber. DRGs were cultured in the centre of a glass coverslip placed on top of the microporous membrane. Axons grow across the coverslip and cross through the membrane by DIV4, when they are subjected to experimental conditions and mechanically harvested for analysis. (b) Fluorescent in situ hybridisation (FISH) using riboprobes demonstrated the presence of CREB, but not cJun or STAT1 mRNA transcripts in axons. Insets show labelling in cell bodies of dissociated DRG neuron cultures at 10X magnification to demonstrate efficacy of riboprobes. Counter-stained images show immunofluorescence using anti-GAP-43 antibody (Red). Scale bar, 10 μm. (c) Quantification of FISH data in (b). CREB levels were monitored with two separate probes (Table S1), and CREB levels were comparable to those of β-actin. Background FISH levels were defined as the average signal obtained using a scrambled β-actin riboprobe and subtracted from all other data. *p<0.001. Numbers on bars represent n axons per condition. (d) DRG neurons transfected with non-targeting siRNA demonstrated CREB mRNA FISH signals in axons, while neurons transfected with CREB-specific siRNA demonstrated a near-complete abolishment of CREB mRNA FISH signal. Scale bar, 10 μm. (e) Quantification of data in (d). *p<0.001. Numbers on bars represent n axons per condition. (f) CREB was detected in axonal lysates by RT-PCR using two separate primer pairs (Table S2). RT-PCR fidelity was assayed by concurrent RT-PCR from DRG cell body lysates.
Among the clones in the library, we identified cDNAs encoding specific transcription factors, including CREB. To confirm that CREB transcripts are localised to axons, we performed fluorescent in situ hybridisation (FISH) experiments using E15 DRG explant cultures. CREB-specific riboprobes prominently labelled cell bodies (Fig. 2b), with lower levels detectable in axons (Fig. 2b,c). Probes directed against other transcription factor transcripts, such as cJun and STAT1, resulted in negligible labelling of axons (Fig. 2b,c). CREB FISH signals were markedly reduced in neurons transfected with CREB-specific siRNA (Fig. 2d,e), as well as in DRG neurons cultured from mice homozygous for a hypomorphic CREB allele19 (Fig. S1g,h). RT-PCR using two distinct primer pairs resulted in amplification of CREB transcripts from distal axon preparations from Boyden chambers4, as well as amplification of β-actin, a previously-identified axonal mRNA20, while RT-PCR signals for other transcription factor transcripts were absent (Fig. 2f). The axonal localization of CREB mRNA is reminiscent of a previous report localising CREB mRNA and protein to dendrites21. Together, these data indicate that CREB transcripts are specifically localised to axons of developing DRG neurons.
CREB is synthesised in axon terminals in response to NGF
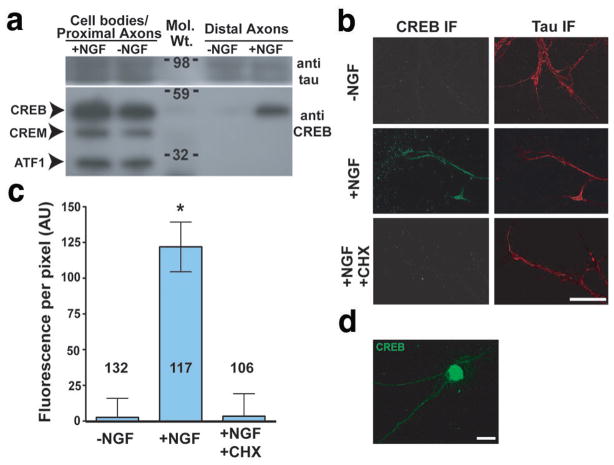
The presence of axonal CREB mRNA raises the possibility of intra-axonal CREB synthesis. To address this, DRG explants were cultured in Boyden chambers; the media in the cell body chamber was replaced with NGF-free media and the media in the axon chamber was replaced with either NGF-replete or NGF-free media for 3 h. Extracts from the upper surface of the Boyden chamber, containing cell bodies and proximal axons, and the lower surface, containing exclusively distal axons4, were harvested, and equal amounts of protein were analysed by Western blot. Western blotting using a CREB family antibody that also recognises CREM and ATF1 indicated that only CREB is present in distal axons, and is dependent on the presence of NGF in the axon compartment (Fig. 3a, Fig. S1i), although all three proteins were detected in the cell body/proximal axon fraction (Fig. 3a).
Figure 3. CREB is specifically translated in axons.
(a) Selective localisation and induction of CREB in distal axons. DRG explants were cultured in Boyden chambers4 and the upper compartment was incubated in 0ng/ml NGF. The lower compartment was incubated in either 0 ng/ml NGF or 100 ng/ml NGF for 3 h. Lysates (25 μg protein) were prepared from the coverslip (cell body/proximal axon) or the underside of the membrane (distal axons) and analysed by Western blot using an antibody that also recognises CREB family members CREM and ATF-1. (b) NGF and protein synthesis are required for CREB localisation in axon terminals. Axons were severed from DIV3 DRG explant cultures, and incubated with 0 or 100 ng/ml NGF, or 100 ng/ml NGF + 1 μM cycloheximide (CHX) for 3 h. CREB was detected by immunofluorescence (IF) using a CREB-specific antibody. Counter-staining shows immunofluorescence using anti-tau antibody (right). Scale bar, 50 μm. (c) Quantification of results in (b). *p<0.0001. Numbers on bars represent n axons per condition. (d) Low-power (20X) image of CREB immunofluorescence (IF) in DIV3 E15 DRGs. The majority of cellular CREB protein is associated with the nucleus, although signals are seen in the cytosol and axon. Scale bar 20 μm.
We also examined the axonal localisation of CREB by immunofluorescence. Axons were severed from cultured DRG explants to rule out potential contributions from anterograde transport of cell body-derived CREB. Immunofluorescence with a CREB-specific antibody revealed axonal CREB protein was present when the media contained NGF, but not when the media was exchanged with NGF-free media for 3 h (Fig. 3b,c), consistent with the Western blot data. Although the majority of CREB protein localises to the nucleus, with substantially lower levels in axons (Fig. 3d), the immunofluorescence signal in axons is specific, as similar immunofluorescence staining is seen using a different CREB-specific antibody that recognises a nonoverlapping epitope, and immunofluorescence staining is substantially reduced in DRG neurons transfected with CREB-specific siRNA, and in DRG neurons prepared from mouse embryos homozygous for a hypomorphic CREB allele19 (Fig. S1j–o). The presence of CREB in severed axons was dependent upon NGF and local translation, as replacement of the media with NGF-free media or NGF-replete media containing cycloheximide resulted in a loss of CREB immunoreactivity (Fig. 3b,c). These data indicate that CREB is found in axons and its levels are dependent on NGF.
CREB mRNA is selectively translated in response to NGF
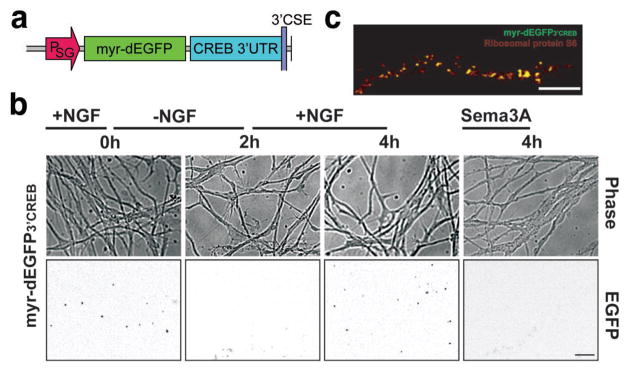
We next examined NGF-dependent axonal CREB translation using a GFP-based reporter assay22. This reporter expresses a transcript encoding a destabilised enhanced GFP with a cellular half-life of 1 h (dEGFP) that enables dynamic changes in translational activity to be reflected by changes in fluorescence intensity22. The dEGFP construct also contains a myristoylation sequence, resulting in reduced diffusion of the reporter in the membrane4,22. As a result of these two features, fluorescence signals reflect newly-synthesised protein near the site of translation as evidenced by their proximity to ribosomes22.
DRG explant cultures were infected with Sindbis virus4,23 expressing an mRNA comprising the myr-dEGFP coding sequence and the 3′UTR of CREB (myr-dEGFP3′CREB) (Fig. 4a). Infection resulted in the appearance of fluorescent puncta throughout infected axons (Fig. 4b), as has previously been found in cultures infected with myr-dEGFP3′RhoA4. No fluorescent signals are detected in axons of DRGs infected with a histone H1f0 myr-dEGFP reporter, an mRNA that is not detectable in axons (Fig. S2a,b). Fluorescent myr-dEGFP3′CREB puncta disappeared following replacement of the media with NGF-free media and reappeared following restoration of NGF, but not Semaphorin 3A (Sema3A), an axonal guidance cue that regulates the translation of axonal RhoA mRNA (Fig. 4b)4. Expressing the RhoA reporter myr-dEGFP3′RhoA led to fluorescent puncta throughout axons that were only slightly affected by removal of NGF (Fig. S2b). These observations indicate that the CREB reporter is responsive to NGF but not to Sema3A, suggesting that the CREB 3′UTR contains a NGF-response element and that axons contain distinct signalling pathways that regulate the translation of different mRNA transcripts. The punctuate myr-dEGFP signals may reflect “hotspots” of protein translation22, as these puncta colocalised with ribosomal protein S6, phospho-eIF4E, and Staufen, but not mitochondria (Fig. 4c, Fig. S2c,d).
Figure 4. NGF dependent translation of a CREB reporter mRNA in axons.
(a) Schematic diagram of the Sindbis viral reporter construct used to monitor CREB translation. The reporter contains a myristoylated, destabilised EGFP (myr-dEGFP) with the 3′UTR of CREB, expressed under control of the Sindbis subgenomic promoter (PSG). (b) E15 DRG explant cultures were infected with Sindbis constructs expressing myr-dEGFP3′CREB on DIV3 and fluorescence (bottom panel) and phase (top panel) images, approximately 1000 μm from the cell body were collected after 24 h. Fluorescence images are shown in inverted contrast in order to more readily visualise puncta. Scale bar, 25 μm. (c) myr-dEGFP3′ CREB puncta colocalise with ribosomal markers. myr-dEGFP3′ CREB-expressing axons were counter-stained by immunofluorescence using a ribosomal protein S6-specific antibody. EGFP fluorescence colocalises with a population of S6-labeled ribosomal clusters. Scale bar, 10μm.
Locally synthesised CREB is retrogradely trafficked to the nucleus
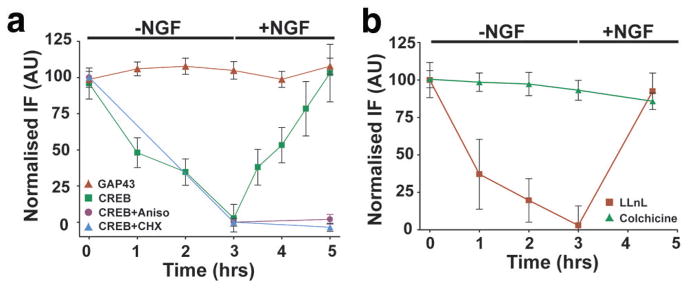
The importin proteins, which bind nuclear-localisation sequences (NLS)24, are present in axons and mediate the retrograde trafficking of axonally-injected fluorescently-labelled NLS peptides25. Since CREB contains a NLS that mediates its nuclear localisation26, axonal CREB may be retrogradely transported to the cell body. To determine if endogenously-expressed CREB is retrogradely trafficked, we examined the time course of CREB reduction in axons upon replacement of media with NGF-free media. CREB levels decreased to baseline within 3 h of NGF removal, and returned to original levels 2 h following restoration of NGF, although significant recovery in CREB levels (~40%) was observed within 30 min of NGF restoration (Fig. 5a). These treatments did not affect GAP-43 (Fig. 5a) or axonal CREB mRNA levels (Fig. S2e). The NGF-dependent restoration of CREB levels was abolished by ribosomal inhibitors (Fig. 5a). The reduction of CREB levels upon removal of NGF was unaffected by the presence of the proteasome inhibitor LLnL (Fig. 5b, Fig. S2f), indicating a proteasome-independent pathway for CREB removal from axons. Application of colchicine, which prevents microtubule-dependent transport, abolished the reduction in CREB levels following removal of NGF (Fig. 5b), indicating a microtubule-dependent process for the loss of CREB protein from axons. The possibility of retrograde transport is supported by the finding that the loss of CREB protein occurs first in distal axons, and then subsequently in medial and proximal axon segments (Fig. S2g).
Figure 5. Axonal translation and retrograde transport of endogenous CREB.
(a) CREB levels in severed axons of E15 DRGs were assayed by immunofluorescence with a CREB-specific antibody. CREB was depleted upon removal of NGF, with near complete loss of fluorescence signals by 3 h. Restoration of NGF resulted in a return of CREB to starting levels within 2 h. Levels of GAP-43 were not significantly affected by NGF removal or by restoration of NGF, indicating that changes in fluorescence intensity were not due to significant changes in axonal volume. Application of 1 μM cycloheximide or 40 μM anisomycin, concurrent with NGF replacement, prevented the restoration of CREB immunofluorescence, indicating that the NGF-dependent increase in CREB levels requires protein synthesis. n≥40 axons per data point. (b) CREB is depleted from axon terminals in a microtubule-dependent manner. CREB levels in severed axons were monitored as in (a) following removal and restoration of NGF, in the presence of LLnL or colchicine. Colchicine, but not LLnL, blocked the reduction in CREB levels following removal of NGF. n≥40 axons per data point.
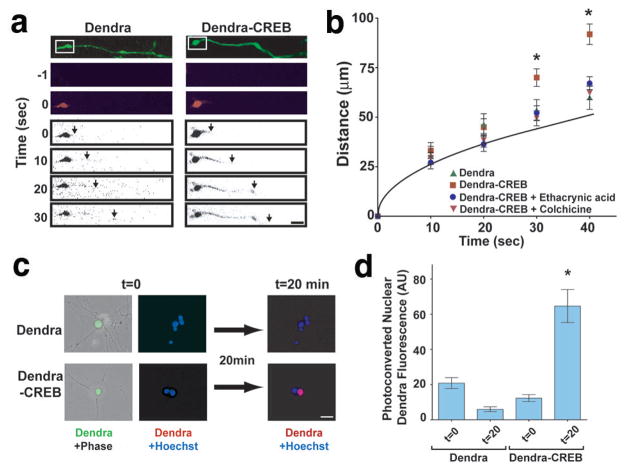
To further examine retrograde trafficking of CREB, we used Dendra, a monomeric GFP relative that converts from green to red fluorescence upon irradiation with blue or ultraviolet light27. DRG explant cultures were infected with Sindbis virus encoding either Dendra or Dendra-CREB transcripts that contained a minimal 3′UTR axon-targeting element28,29. Dendra and Dendra-CREB were photoconverted in selected growth cones (Fig. 6a). The rate of Dendra movement towards the cell body matched the rate predicted by passive diffusion (Fig. 6b). In contrast, photoconverted Dendra-CREB was observed to move at a substantially higher, and constant, rate towards the cell body of 7.8–8.8 mm h−1 (Fig. 6a,b), similar to previously measured rates of retrograde trafficking30,31. Retrograde transport was significantly blocked by colchicine or ethacrynic acid, a dynein inhibitor32, suggesting a microtubule motor-dependent active transport of Dendra-CREB from the axon (Fig. 6b). Photoconversion of a 40-μm section of axon approximately 1000 μm from the cell body was associated with accumulation of photoconverted Dendra-CREB, but not Dendra, in the nucleus within 20 min (Fig. 6c,d).
Figure 6. Retrograde transport of a photoactivatable fluorescent CREB reporter protein.
(a) E15 DRG explant cultures were infected with Sindbis constructs expressing Dendra or Dendra-CREB transcripts. Dendra and Dendra-CREB were visualised as green fluorescence (top panel), and growth cone-localised Dendra[-CREB] was photoactivated to its red form by 50 ms illumination of the boxed regions with a 408 nm laser. Movement of photoactivated Dendra signals was analysed within the axon determined by green Dendra fluorescence mask. The leading edge (arrows) of red fluorescence for photoactivated Dendra-CREB was observed to move along the axon at a significantly faster rate than photoactivated Dendra. Photoactivated Dendra[-CREB] images are shown inverted in order to more readily visualise signals. Scale bar, 10 μm. (b) Quantification of data in (a). Grey line indicates the predicted diffusion rate of photoconverted Dendra-CREB, based on neuronal viscosity measurements49. The expected diffusion rate of Dendra-CREB was calculated at various elapsed time points, using a previously measured diffusion coefficient (D) in neurons49, in the formula x2 = (2Dt), where x is the average displacement. No significant differences in axon diameter or morphology were observed between the neurons assayed. n≥20 axons per data point. *p<0.0001. (c) Dendra or Dendra-CREB was photoactivated by 1 s illumination of a 40 μm axon segment approximately 1 mm from its respective cell body and levels of photoactivated Dendra fluorescence were analysed in the respective cell nucleus. Scale bar, 20 μm. (d) Quantification of data in (c). n=10 cells per data point. *p=0.0012.
TrkA-containing signalling endosomes are trafficked from distal axons to the cell body7 and mediate the activation of Erk5, which is required for CREB phosphorylation in response to axonally-applied NGF33. Punctate regions of phospho-TrkA (pTrkA) immunoreactivity were found along the length of axons of DRG neurons cultured in the presence of NGF (Fig. S2h), in a distribution consistent with previous reports of TrkA-signalling endosomes in axons34. These regions of pTrkA reactivity also contain phosphorylated Erk5 (Fig. S2h). Interestingly, axonal CREB protein exhibits colocalisation with these sites of pTrkA immunoreactivity along axons (Fig. S2h), indicating that axonal CREB may be in proximity to TrkA-signalling complexes in axons.
Axonal CREB is required for the accumulation of pCREB in the nucleus induced by application of NGF to axons
We next asked whether the amounts of CREB synthesised in axons make a substantial contribution to nuclear levels of CREB. Axonal CREB mRNA was knocked down by compartmentalised siRNA transfection35, while BOC-Asp(OMe)-FMK (BAF), a caspase inhibitor, was included in the cell body compartment to prevent neuronal death36. Transfection of CREB-specific siRNA into the axon compartment of dissociated DRG neurons in compartmented chambers resulted in axonal knockdown of CREB protein (72.8 +/− 5.2%) and CREB mRNA (82.5 +/− 4.3%), but did not lead to a reduction in CREB mRNA or protein levels in the cell body compartment (Fig. 7a,b, Fig. S3a,b). Similarly, selective reductions in axonal CREB protein levels are seen by Western blotting (Fig. S3c). These effects are specific, as β-actin mRNA levels in axons or cell bodies were not affected by axonal transfection of CREB-specific siRNA (Fig. S3b).
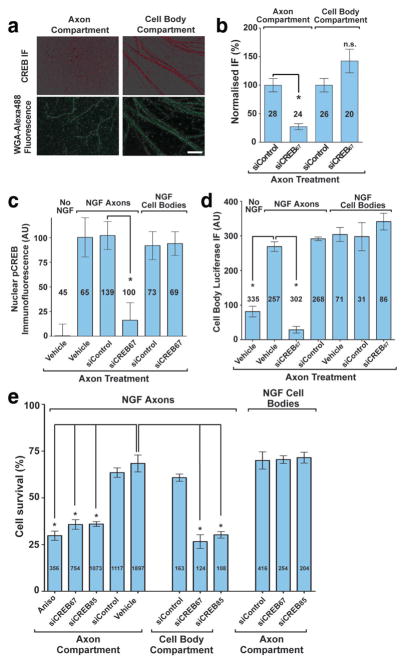
Figure 7. Axonal CREB is required for CRE-dependent transcription and NGF-mediated DRG survival.
(a) DRG axons in compartmented cultures were transfected with CREB siRNA in the axon compartment only, and CREB levels were detected using a CREB antibody. Axons crossing the compartment divider were retrogradely labelled with WGA-Alexa488. All compartments were maintained in NGF-containing media throughout the experiment. Scale bar, 50 μm. (b) Quantification of data in (a). Immunofluorescence levels in each compartment were normalised to fluorescence signals from cultures treated with non-targeting siRNA. *p<0.0001. Numbers on bars represent n axons per condition. (c) DRGs in compartmented cultures were incubated in NGF-free media, supplemented with BAF to suppress apoptosis. Axon compartments were treated with CREB-specific or non-targeting siRNA. After 48 h, 30 ng/ml NGF was added to the axon compartment for 20 min, after which pCREB levels in nuclei were quantified by immunofluorescence. *p=0.0004. Numbers on bars represent n cells per condition. (d) DRGs in compartmented chambers were infected with adenovirus encoding luciferase under the control of a CRE transcriptional element and treated with NGF as in (c). *p<0.001. Numbers on bars represent n cells per condition. (e) E15 dissociated DRG were cultured in compartmented chambers as in Fig. 1c. NGF-induced neuronal survival at DIV7 was assayed following transfection of control or CREB-specific siRNA into the axon compartment at DIV5. *p<0.001. Numbers on bars represent n cells per condition.
Unlike CREB, which is readily detected in the nucleus, Ser133-phosphorylated CREB (pCREB). is present at negligible levels in the nuclei of unstimulated sensory neurons37,38. The low basal level of pCREB makes neurons highly responsive to increases in pCREB levels, which occurs upon application of neurotrophin to axons38 (Fig. 7c, Fig. S3d). Treatment of axons with NGF resulted in increased levels of pCREB in the nucleus within 20 min, which was unaffected by axonal transfection of a control siRNA. Axonal transfection of CREB-specific siRNA significantly reduced the ability of axonally-applied NGF to induce this rapid increase in nuclear pCREB levels (Fig. 7c, Fig. S3d), but did not have a significant effect on cell body accumulation of pTrkA or pERK5 (Fig. S3e,f), suggesting that this effect is not due to inhibition of retrograde transport of signalling endosomes. Knockdown of axonal CREB mRNA also did not affect nuclear pCREB accumulation induced by stimulation of cell bodies with NGF (Fig. 7c), demonstrating that axon-specific CREB knockdown does not have a general inhibitory effect on NGF signalling or CREB phosphorylation. The appearance of pCREB in the nucleus within 20 min of NGF treatment is consistent with the time required for CREB to be transported across the 1-mm divider, based on the trafficking rates measured for Dendra-CREB (Fig 6b). Thus, while axonal synthesis does not contribute a substantial portion to the total amount of nuclear CREB, these data indicate that the axonally-synthesised pool of CREB accounts for the majority of the pCREB that appears in the nucleus upon stimulation of distal axons with NGF.
Axonal CREB mediates the induction of CRE-dependent transcription
We next asked if axon-derived CREB is capable of affecting CRE-dependent transcription. Axons of dissociated DRG neurons in compartmented chambers were subjected to either NGF-replete or NGF-free media and siRNA at 5 DIV. BAF was included in the cell body compartment to prevent neuronal death. At 6 DIV, cell bodies were infected with adenovirus encoding a CRE-luciferase reporter, and cellular luciferase levels were measured 24 h later. Bath application of NGF to DRG neurons lead to a dose-dependent increase in luciferase immunofluorescence, but did not affect a control protein (Fig. S3g). Axonal application of NGF increased luciferase levels, which was prevented by axon-specific transfection of CREB-specific siRNA, but not control siRNA (Fig. 7d). Knockdown of axonal CREB mRNA did not affect luciferase transcription induced by NGF applied to the cell bodies (Fig. 7d), indicating that axonal CREB knockdown does not non-specifically inhibit the reporter. Axon-derived CREB is thus necessary for CRE-dependent transcription induced by application of NGF to distal axons.
Axonal CREB is required for NGF-induced retrograde survival
A role for CREB in DRG neuron survival is reflected in the loss of ~75% of these neurons in CREB null mice13. To determine the role of the axonal CREB in neuronal survival, we cultured DRG neurons in compartmented chambers, and axons were transfected with either control or CREB-specific siRNA. Neuronal survival induced by axonal application of NGF was unaffected by control siRNA, but was markedly impaired by transfection with either of two CREB-specific siRNAs (Fig. 7e). Axon-specific transfection of CREB-specific siRNA did not affect cell survival elicited by application of NGF to cell bodies (Fig. 7e), indicating that NGF signalling at cell bodies does not require axonal CREB. The impairment in survival seen following knockdown of axonal CREB mRNA was comparable to that seen when CREB mRNA was knocked down throughout both cell bodies and axons by transfection of cell bodies with CREB-specific siRNA (Fig. 7e), and similar to the levels of survival seen when both cell bodies and axons are deprived of NGF (Fig. 1e, Fig. S1d)33. These data indicate that axonal CREB translation is required for survival elicited by NGF signalling in distal axons.
DISCUSSION
Our studies reveal a role for intra-axonal mRNA translation in mediating communication between distal axons and the nucleus. CREB mRNA is localised to axons of DRG neurons and translated in response to NGF signalling. Axon-derived CREB is the source of the pCREB that appears in the nucleus following exposure of distal axons to NGF, and is required for the increase in CRE-dependent transcription seen upon stimulation of distal axons with NGF. Furthermore, neuronal survival elicited by NGF signalling at distal axons requires axon-derived CREB. These data indicate that the retrograde signal generated upon axonal application of NGF includes axonally synthesised CREB (Fig. 8). Our findings identify a novel function for local translation involving the translation and retrograde trafficking of transcription factors from the axon to the neuronal nucleus. The regulation of local protein synthesis within axons adds to the previously described signalling pathways downstream of NGF/TrkA. NGF signalling is selective as it leads to the induction of the CREB mRNA translational reporter, but not the RhoA reporter, which is regulated by Sema3A4. This selectivity suggests the presence of sequence elements in the CREB 3′UTR that specifically confer NGF-responsiveness.
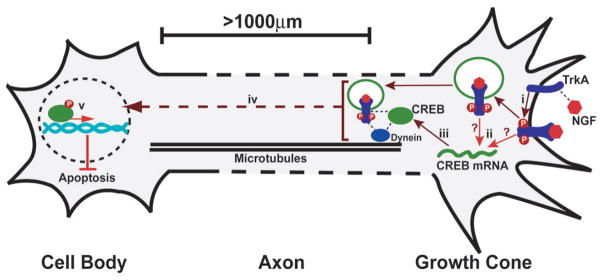
Figure 8. Local translation and retrograde transport of CREB mediates neuronal survival.
(i) NGF binds to TrkA receptors causing dimerisation and autophosphorylation. (ii) TrkA activation leads to translation of axonal CREB mRNA and (iii) the production of CREB protein. NGF-bound, activated TrkA receptors are internalised into endosomes and initiate formation of a signalling complex containing downstream effectors and the motor protein dynein. Axonally translated CREB protein associates with this NGF-pTrkA signalling endosome, which is required for downstream activation of CREB signalling in the cell body. (iv) CREB is retrogradely transported to the nucleus via microtubules and is phosphorylated at S133 downstream of internalised TrkA signalling endosomes, via a kinase cascade including Mek5/Erk533. (v) Axonally-derived pCREB initiates the transcription of anti-apoptotic genes in the nucleus, leading to neuronal cell survival.
A common feature of many types of growth factor signalling pathways, including NGF signalling, is the occurrence of intracellular “signalling platforms” that function as localised signal transduction units39. NGF-mediated TrkA signalling can occur through TrkA at the plasma membrane as well as TrkA localised to endosomes that form upon internalisation of NGF/TrkA complexes40,41. These distinct platforms are characterized by unique cohorts of proximally localized TrkA effectors40,41. Retrograde trafficking of TrkA signalling endosomes, containing both catalytically-active TrkA as well as specific TrkA effectors, is associated with an increase in pCREB levels in the nucleus in a Mek5 and Erk5-dependent pathway33. However, whether CREB is similarly compartmentalised into an effector pool that is preferentially regulated by the signalling endosome has not previously been addressed. We find that CREB is found colocalised with pTrkA in axons: since both CREB and TrkA-signalling endosomes are retrogradely trafficked, the proximity of the pool of axonally-derived CREB may make it preferentially accessible to phosphorylation by TrkA effectors such as Erk533. As TrkA kinase activity in the cell body is required for CREB phosphorylation37, several models could explain how CREB phosphorylation is regulated: (1) CREB is not phosphorylated until it arrives in the cell body; (2) CREB is readily dephosphorylated, and TrkA activity is required to maintain CREB in a phosphorylated state when it arrives in the cell body; or (3) TrkA activity is required to inactivate a cell body phosphatase.
Transcriptional effects elicited by axonal signalling require that an axon-derived signal be conveyed to the cell body. An inherent requirement in this type of signalling is that the axon-derived signal must somehow be distinguished from what would presumably be a much larger amount of similar molecules in the cell body. The low basal level of pCREB in the nucleus of unstimulated neurons33 may allow small increases in the amount of pCREB derived from the axon to result in a substantial fold elevation in pCREB-dependent transcriptional activity. Our results indicate that axonally-synthesised CREB is capable of exerting transcriptional effects in the nucleus by serving as the source of the pCREB that appears in the nucleus following axonal application of NGF. Because the transcriptional effects of CREB are affected by its phosphorylation at sites other than Ser13342,43, as well as by protein-protein interactions43, axon-specific CREB modifications may also impart axonally-synthesised CREB with unique transcriptional effects that differ from cell body-localised CREB.
Several examples of transcription factors or transcriptional regulators localised to dendrites, and less frequently, developing axons have been described; these include CREB21 and NF-κB44 in dendrites, and nervy in axons45. These transcription factors may have non-nuclear functions; for example, in axons, nervy acts as an adapter protein for signalling from Plexin receptors45. In the case of dendritic NF-κB, a role for transcriptional regulation has been proposed44; however, these studies have not been able to differentiate the role of the somatic and dendritic pool of NF-κB. Similarly, while it is clear that CREB can be synthesised in dendrites21, the inherent difficulties in selectively abolishing the dendritic CREB pool have prevented a thorough elucidation of the exact role of dendritically-synthesised CREB in neuronal signalling. By selectively abolishing axonal CREB mRNA, the data presented here supports a nuclear role for extrasomatic pools of CREB.
During development, axons encounter a variety of signals that affect multiple aspects of neuronal development, such as axonal elongation, branching, and pathfinding, as well as synaptogenesis and neuronal differentiation9,46,47. Increasing evidence suggests that many of these processes involve retrograde signals that affect gene transcription. Translation and retrograde trafficking of axonally localised transcription factor mRNAs in response to target derived signalling molecules could therefore constitute a general mechanism by which signalling at growth cones can selectively and temporally regulate gene transcription during neuronal development.
METHODS
Primary cell culture
E15 rat or E13 mouse embryonic dorsal root ganglion (DRG) explants were plated on glass-bottom culture dishes (MatTek) or glass coverslips pre-coated with 33 μg/ml poly-D-lysine and 1 μg/ml laminin. CREBα/Δ −/+ animals used to generate CREBα/Δ −/− embryos were from Jackson Labs. E15 dissociated DRG neurons were prepared as described4. DRGs were cultured in B27/Neurobasal medium (Invitrogen) supplemented with 100 ng/ml nerve growth factor (NGF) and 20 μM 5′-fluorodeoxyuridine (5-FdU) for 3 days. siRNA-mediated knockdown in DRG neurons has been described previously48 and was performed using siRNAs listed in Table S3. For measurements of CREB levels in isolated axons, axons were severed from cell bodies by removing the explant with a flame-sharpened Pasteur pipette4. Modified Boyden chambers were based on the procedure of Twiss17 and modified to obtain distal axons as described4. mRNA from harvested axons4 was used to prepare a cDNA library using a modified, unbiased single cell protocol18, as described in Supplementary Methods. Compartmented (Campenot) cultures were prepared as described12 (see Supplementary Methods)
In situ hybridisation
Sense oligonucleotides (Table S1) were synthesised with a T7 promoter site at their 3′ end. Antisense riboprobes were in vitro transcribed from the sense oligonucleotides using the MEGAscript T7 transcription kit (Ambion) with digoxigenin-conjugated UTP. DRGs (DIV3) were fixed overnight at 4°C in 4% paraformaldehyde in cytoskeleton buffer (CSB: 10 mM MES pH 6.1, 138 mM KCl, 3 mM MgCl2, 2 mM EGTA, 0.4 M sucrose). Washes were performed in TBST (20 mM Tris pH 8.0, 150 mM NaCl, 0.1% Triton X-100) for 3 × 5 min. DRGs were permeabilised in 0.5% Triton X-100/TBS for 10 min and post-fixed in 4% PFA/TBS for 5 min, followed by fresh acetylation buffer (0.25% acetic anhydride, 0.1 M HEPES) for 10 min, and equilibration with 4X SSC/50% formamide for 30 min. Cultures were incubated with 15 ng riboprobes (Table S1) in 15 μl hybridisation buffer (10% dextran sulfate, 4X SSC, 1X Denhardt’s Solution, 40% formamide, 20 mM ribonucleoside vanadyl complex, 10 mM DTT, 1 mg/ml yeast tRNA, and 1 mg/ml salmon sperm DNA) at 37°C overnight. The coverslips were washed with 40% formamide/1X SSC at 37°C for 20 min, and three times each with 1X SSC and 0.1X SSC at RT for 5min. Neurons were blocked with blocking buffer (100 mM Tris-HCl pH 8.0, 150 mM NaCl, 8% formamide, 5% BSA, 2.5% normal horse serum, and 2.5% normal goat serum) for 30 min. Hybridisation was detected with anti-digoxin antibody (Table S4), which was precleared with rat embryo power in blocking buffer for 2 h at 25°C. Mean fluorescence intensity elicited by the scrambled probe was subsequently deducted from all FISH data to produce specific labelling intensity for each probe.
Quantification of CREB protein levels
Images and measurements of signals in axons were taken from the terminal 50 μm of the axon, except where indicated. Analysed axons were a minimum of 2000 μm for all experiments. DRGs were fixed with 4% PFA in CBS overnight at 4°C, permeabilised with 0.5% Triton X-100/TBS, and blocked in 4% BSA/TBS for 1 h. DRGs were labelled with antibodies (Table S4) in 2% BSA/0.1% Triton X-100/TBS overnight at 4°C. For image acquisition details, see Supplementary Methods.
Generation and infection of recombinant viruses
We used a modified Sindbis vector, pSinRep5, containing a point mutation in nsP2 (P726S) that reduces cytotoxicity in neurons23, and the helper plasmid DH-BB (S. Schlesinger, Washington University, St. Louis) as described previously4. Reporter experiments utilised the myr-dEGFP system22, except that a d1EGFP variant (i.e., an EGFP with a 1 h half-life) was used, as described previously4. pSinRep5-myr-dEGFP3′CREB and pSinRep5-myr-dEGFP3′RhoA, contained the full-length 3′UTR of the human CREB mRNA or the 3′UTR of human RhoA mRNA, fused to the viral 3′CSE. For Sindbis virus encoding Dendra constructs, the virus contained the open reading frame (ORF) of Dendra and, in the case of pSinRep5-Dendra-CREB, the full ORF of human CREB, followed by a 54 nt minimal axonal targeting element28. In the case of IRES-driven Sindbis virus, the pSinRep constructs contained a human encephalomyocarditis viral IRES element from vector pIRES-hyg (BD Biosciences). Sindbis pseudoviruses were prepared according to the manufacturer’s instructions (Invitrogen), purified on a sucrose gradient, concentrated on YM-100 microcon columns, resuspended in Neurobasal medium, and titered using BHK-21 cells. We generated a CRE-luciferase adenovirus reporter by subcloning the complete CRE-Luc reporter gene from vector pCRE-Luc (BD Biosciences) into pAd/PL-DEST (Invitrogen). Virus production and amplification was performed in HEK293A cells, according to manufacturer’s instructions. Adenovirus was purified using the ViraKit AdenoMini-4 system (Virapur), and titered using HEK293T cells. DRGs were infected with equal infectious units of recombinant virus at DIV6 and luciferase levels were measured 24 h later.
Data analysis and presentation
All statistical p values in this study were determined using ANOVA from experiments repeated a minimum of three times, unless stated otherwise. All data are presented as mean +/− s.e.m. n values are represented on all graphs and defined in legends, unless stated otherwise. See Supplementary Methods for more details on data acquisition and handling.
Supplementary Material
(a) Immunofluorescence analysis of 3 DIV DRG cultures +/− 5-FdU. Inclusion of 5-FdU effectively abolishes Schwann cell proliferation in DRG cultures. Scale bar, 50 μm. (b) Immunofluorescence analysis of 3 DIV DRG cultures demonstrates strong labelling of cell bodies and all projections with axon marker GAP43. Dendrite/cell body marker MAP2 is restricted to cell bodies, indicating the absence of MAP2-staining dendrites in DRG cultures. Scale bar, 50 μm. (c) Immunofluorescence analysis of compartmented cultures (montage micrographs). Retrogradely labelled (WGA-Alexa555, red) DRG axons fasciculate and grow under the 1 mm divider into the axon compartment. Axons defasciculate as they enter the axon compartment. Cell bodies (DAPI, blue) are restricted to the cell body compartment. Only WGA-Alexa555 labelled cell bodies are included in data sets, as they comprise the population of neurons extending axons across the divider. (d) Dissociated DRG neurons (5 DIV) in compartmented cultures were treated with NGF in either or both compartments. NGF was capable to supporting DRG survival when applied to either axons or cell bodies individually, corroborating the presence of functional TrkA complexes throughout the developing neuron. Numbers on bars represent n cells per condition. (e,f) DRG axons in compartmented cultures were infected with a Sindbis virus encoding myr-dEGFP under the control of an IRES to both cell body and axonal compartments. Addition of cycloheximide or anisomycin for 48 hrs to the axonal compartment elicited a loss of myr-EGFP fluorescence in the axonal compartment, with no significant effect on myr-EGFP levels in the cell body compartment. All compartments were maintained in identical NGF-containing media throughout the course of the experiment. Scale bar, 50 μm (g) DIV3 E13 DRG neurons from CREBα/Δ +/− mouse embryos demonstrated robust CREB mRNA FISH signal in axons, while neurons from CREBα/Δ −/− embryos demonstrated an 86.3% loss of CREB mRNA FISH signal. Incomplete abolishment of FISH signals, relative to siRNA-treated axons, reflects residual alternatively-spliced CREBβ transcripts in these hypomorphic animals19. Additionally, axonal mRNAs are trafficked as mRNA-ribosome complexes, and impaired CREBβ mRNA-ribosome interactions due to the poor Kozak site19 in this transcript may lead to inefficient trafficking. Scale bar, 10 μm. (h) Quantification of data in (g). *p<0.0001. Numbers on bars represent n axons per condition. (i) Similar to the experiment in Fig. 3a, except equivalent numbers of cell bodies and axon terminals were used for Western blotting. DRG explants were cultured in Boyden chambers, and the upper compartment was incubated in 0 ng/ml NGF. The lower compartment was incubated in either 0 ng/ml NGF or 100 ng/ml NGF for 3 h. Axon lysates (~104 cells) were prepared from the underside of the membrane and analysed by Western blot. As in Fig. 3a, only CREB was detected in axons, and CREB localisation in axons was dependent on NGF in the culture media. (j) DRG cultures were transfected with CREB siRNA and CREB levels were detected using a CREB antibody. Neurons transfected with non-targeting siRNA exhibited CREB immunoreactivity in axons, while neurons transfected with CREB-specific siRNAs exhibited a near-complete abolishment of CREB immunoreactivity. Scale bar, 20 μm. (k) Quantification of data in (j). *p<0.001. Numbers on bars represent n axons per condition. (l) E15 DRG neuronal lysate (10 μg) was subjected to immunoblotting with the CST-9192 CREB antibody. (m) CREB was detected by immunofluorescence using CREB antibody sc-186 (see i, Fig. 3a, and Table S4). Scale bar 10 μm. (n) DIV3 E13 DRG neurons from CREBα/Δ +/− embryos exhibited robust CREB immunoreactivity in axons, while neurons from CREBα/Δ −/− embryos demonstrated an 84.8% loss of CREB IF signal. The greater degree of reduction in CREB immunofluorescence following siRNA treatment (see k) than in the CREB mutant mice likely reflects the more complete abolition of CREB following siRNA transfection than in the hypomorphic animal. Scale bar, 10 μm. (o) Quantification of data in (m). *p<0.0001. Numbers on bars represent n axons per condition.
(a) Schematic diagram of the Sindbis reporters construct used to monitor RhoA and histone H1f0 translation4. The reporter contains a myristoylated, destabilised EGFP (dEGFP) with the 3′UTR of RhoA or histone H1f0, expressed under control of the Sindbis subgenomic promoter (PSG). (b) E15 DRG explant cultures were infected with Sindbis constructs expressing myr-dEGFP3′RhoA or myr-dEGFP3′H1f0 on DIV3 and fluorescence images were collected after 24h. Explants infected with myr-dEGFP3′RhoA and cultured in the presence of NGF exhibited fluorescent puncta distributed throughout axons (0 h). Following replacement of the media with NGF-free media for 2 h, puncta intensity was not significantly affected (2 h). Explants infected with myr-dEGFP3′H1f0 and cultured in the presence of NGF did not exhibit fluorescent puncta in axons. Scale bar, 25 μm. (c) myr-dEGFP3′CREB-infected axons were counter-stained by immunofluorescence using antibodies specific to translational marker p-eIF4E, RNP-associating protein Staufen, and mitochondrial marker VDAC/Porin1. myr-dEGFP was found in some, but not all ribosomal clusters (Fig. 4c), suggesting that myr-dEGFP-negative clusters are either translationally inactive, that they translate CREB mRNA in response to different stimuli, or that their function involves the translation of other mRNAs. Scale bar, 10 μm. (d) Axons from DIV3 DRGs were analysed for CREB and mitochondrial localisations by immunofluorescence using CREB-specific and VDAC/porin1-specific antibodies. Scale bar, 10 μm. (e) Axons were severed from DIV3 DRG explant cultures, and incubated with 0 or 100 ng/ml NGF for 3 h. CREB mRNA was detected by FISH using a CREB specific riboprobe. CREB mRNA levels did not significantly change during the course of the experiments. Numbers on bars represent n axons per condition.. (f) 40 μM LLnL was added to myr-dEGFP3′CREB-infected axons. Within 5 min, the eGFP fluorescence signal increased twofold, due to inhibition of proteasome-dependent degradation of the destabilised EGFP. Scale bar, 10 μm. (g) 3DIV dissociated DRGs were treated with NGF-free and NGF-replete media as in Fig. 5. CREB levels were assayed by immunofluorescence in contiguous axons: distal segments (450–500 μm), medial segments (250–300 μm) and proximal segments (50–100 μm) from the same axon were determined by their distances from the cell body. n≥10 axons per data point. *p<0.01. (h) Axons of DIV3 DRG explant cultures were analysed by immunofluorescence for NGF effectors CREB, pTrkA and pErk5 at 3 DIV. Scale bar, 5 μm.
(a) Neurons transfected with CREB-specific siRNAs exhibited a specific abolishment of CREB mRNA FISH signal in the axon compartment (“Axon”), but no significant change in CREB mRNA FISH signals in proximal axons in the cell body compartment (“Cell Body”). (b) Quantification of data in (a). FISH levels in each compartment were normalised to fluorescence signals from cultures treated with non-targeting siRNA. β-actin mRNA FISH signals were unaffected by CREB-specific siRNA. *p<0.001. Numbers on bars represent n axons per condition. (c) Transfection was as in Fig. 7a, except CREB knockdown was assessed by Western blot instead of immunofluorescence. Lysates (10 μg protein) were prepared from the axon compartment of CREB-specific siRNA-treated compartmented cultures and analysed by Western blot using a CREB-specific antibody. (d) DRGs in compartmented cultures (5 DIV) were incubated in NGF-free media, supplemented with BAF to prevent apoptosis. Axon compartments were transfected with CREB-specific or non-targeting siRNA. After 48hr, 30 ng/ml NGF was added to the axon compartment for 20min, after which cells were fixed and subjected to immunofluorescence analysis of nuclear pCREB. (e) DRGs in compartmented cultures were treated as in (d, Fig. 7a), after which pTrkA levels in nuclei were quantified by immunofluorescence. (f) DRGs in compartmented chambers were treated as in (d, e, Fig. 7a) and pErk5 levels in nuclei were quantified by immunofluorescence. (g) Dissociated DRG neurons (5 DIV) were infected with equal infectious units of recombinant CRE-luciferase adenovirus and incubated in various concentrations of NGF. After 24hr, cells were fixed and analysed by immunofluorescence for luciferase production. n=36 (0NGF), 29 (1ng/ml NGF), 38 (5ng/ml NGF), 29 (10ng/ml NGF), 35 (20ng/ml NGF), 35 (50ng/ml NGF), 45 (100ng/ml NGF) cells.
Acknowledgments
We thank D. Fischman for suggesting the use of Boyden chambers, K. Wu for advice and assistance, S. Schlesinger (Washington Univ. at St. Louis) and A. Jeromin (U. Texas, Austin) for Sindbis plasmids, R. Ratan for CRE plasmids, and A. North (Rockefeller University) for advice and assistance with Dendra photoactivation. Supported by the Paralyzed Veterans of America (L.J.C), the National Institute of Mental Health, the National Alliance for Autism Research, the Klingenstein Foundation, and a Charles A. Dana foundation grant to Weill-Cornell (S.R.J), and the Russian Academy of Science MCB program and the European Commission (K.A.L).
References
- 1.Czaplinski K, Singer RH. Pathways for mRNA localization in the cytoplasm. Trends Biochem Sci. 2006;31:687–693. doi: 10.1016/j.tibs.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 2.Piper M, Holt C. RNA translation in axons. Annual Review of Cell and Developmental Biology. 2004;20:505–523. doi: 10.1146/annurev.cellbio.20.010403.111746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassell GJ, Zhang H, Byrd AL, Femino AM, Singer RH, Taneja KL, Lifshitz LM, Herman IM, Kosik KS. Sorting of β-actin mRNA and protein to neurites and growth cones in culture. Journal of Neuroscience. 1998;18:251–265. doi: 10.1523/JNEUROSCI.18-01-00251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu K, Hengst U, Cox LJ, Macosko EZ, Jeromin A, Urquhart ER, Jaffrey SR. Local translation of RhoA regulates growth cone collapse. Nature. 2005;436:1020–1024. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piper M, Anderson R, Dwivedy A, Weinl C, van Horck F, Leung KM, Cogill E, Holt C. Signaling mechanisms underlying Slit2-induced collapse of Xenopus retinal growth cones. Neuron. 2006;49:215–228. doi: 10.1016/j.neuron.2005.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haase G, Dessaud E, Garces A, de Bovis B, Birling M, Filippi P, Schmalbruch H, Arber S, deLapeyriere O. GDNF acts through PEA3 to regulate cell body positioning and muscle innervation of specific motor neuron pools. Neuron. 2002;35:893–905. doi: 10.1016/s0896-6273(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 7.Zweifel LS, Kuruvilla R, Ginty DD. Functions and mechanisms of retrograde neurotrophin signalling. Nature Reviews Neuroscience. 2005;6:615–625. doi: 10.1038/nrn1727. [DOI] [PubMed] [Google Scholar]
- 8.Patel TD, Kramer I, Kucera J, Niederkofler V, Jessell TM, Arber S, Snider WD. Peripheral NT3 signaling is required for ETS protein expression and central patterning of proprioceptive sensory afferents. Neuron. 2003;38:403–416. doi: 10.1016/s0896-6273(03)00261-7. [DOI] [PubMed] [Google Scholar]
- 9.Hodge LK, Klassen MP, Han BX, Yiu G, Hurrell J, Howell A, Rousseau G, Lemaigre F, Tessier-Lavigne M, Wang F. Retrograde BMP Signaling Regulates Trigeminal Sensory Neuron Identities and the Formation of Precise Face Maps. Neuron. 2007;55:572–586. doi: 10.1016/j.neuron.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 10.Howe CL, Mobley WC. Long-distance retrograde neurotrophic signaling. Curr Opin Neurobiol. 2005;15:40–48. doi: 10.1016/j.conb.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 11.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 12.Campenot RB. Local control of neurite development by nerve growth factor. Proc Natl Acad Sci U S A. 1977;74:4516–4519. doi: 10.1073/pnas.74.10.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lonze BE, Riccio A, Cohen S, Ginty DD. Apoptosis, axonal growth defects, and degeneration of peripheral neurons in mice lacking CREB. Neuron. 2002;34:371–385. doi: 10.1016/s0896-6273(02)00686-4. [DOI] [PubMed] [Google Scholar]
- 14.Tong JX, Vogelbaum MA, Drzymala RE, Rich KM. Radiation-induced apoptosis in dorsal root ganglion neurons. J Neurocytol. 1997;26:771–777. doi: 10.1023/a:1018566431912. [DOI] [PubMed] [Google Scholar]
- 15.Martin DP, Schmidt RE, DiStefano PS, Lowry OH, Carter JG, Johnson EM., Jr Inhibitors of protein synthesis and RNA synthesis prevent neuronal death caused by nerve growth factor deprivation. J Cell Biol. 1988;106:829–844. doi: 10.1083/jcb.106.3.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wyllie AH, Morris RG, Smith AL, Dunlop D. Chromatin cleavage in apoptosis: association with condensed chromatin morphology and dependence on macromolecular synthesis. Journal of Pathology. 1984;142:67–77. doi: 10.1002/path.1711420112. [DOI] [PubMed] [Google Scholar]
- 17.Zheng JQ, Kelly TK, Chang B, Ryazantsev S, Rajasekaran AK, Martin KC, Twiss JL. A functional role for intra-axonal protein synthesis during axonal regeneration from adult sensory neurons. Journal of Neuroscience. 2001;21:9291–9303. doi: 10.1523/JNEUROSCI.21-23-09291.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tietjen I, Rihel JM, Cao Y, Koentges G, Zakhary L, Dulac C. Single-cell transcriptional analysis of neuronal progenitors. Neuron. 2003;38:161–175. doi: 10.1016/s0896-6273(03)00229-0. [DOI] [PubMed] [Google Scholar]
- 19.Blendy JA, Kaestner KH, Schmid W, Gass P, Schutz G. Targeting of the CREB gene leads to up-regulation of a novel CREB mRNA isoform. EMBO Journal. 1996;15:1098–1106. [PMC free article] [PubMed] [Google Scholar]
- 20.Olink-Coux M, Hollenbeck PJ. Localization and active transport of mRNA in axons of sympathetic neurons in culture. Journal of Neuroscience. 1996;16:1346–1358. doi: 10.1523/JNEUROSCI.16-04-01346.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crino P, Khodakhah K, Becker K, Ginsberg S, Hemby S, Eberwine J. Presence and phosphorylation of transcription factors in developing dendrites. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:2313–2318. doi: 10.1073/pnas.95.5.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. doi: 10.1016/s0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 23.Jeromin A, Yuan LL, Frick A, Pfaffinger P, Johnston D. A modified Sindbis vector for prolonged gene expression in neurons. Journal of Neurophysiology. 2003;90:2741–2745. doi: 10.1152/jn.00464.2003. [DOI] [PubMed] [Google Scholar]
- 24.Goldfarb DS, Corbett AH, Mason DA, Harreman MT, Adam SA. Importin alpha: a multipurpose nuclear-transport receptor. Trends in Cell Biology. 2004;14:505–514. doi: 10.1016/j.tcb.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 25.Hanz S, Perlson E, Willis D, Zheng JQ, Massarwa R, Huerta JJ, Koltzenburg M, Kohler M, van-Minnen J, Twiss JL, Fainzilber M. Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron. 2003;40:1095–1104. doi: 10.1016/s0896-6273(03)00770-0. [DOI] [PubMed] [Google Scholar]
- 26.Waeber G, Habener JF. Nuclear translocation and DNA recognition signals colocalized within the bZIP domain of cyclic adenosine 3′,5′-monophosphate response element-binding protein CREB. Mol Endocrinol. 1991;5:1418–1430. doi: 10.1210/mend-5-10-1431. [DOI] [PubMed] [Google Scholar]
- 27.Gurskaya NG, Verkhusha VV, Shcheglov AS, Staroverov DB, Chepurnykh TV, Fradkov AF, Lukyanov S, Lukyanov KA. Monomeric green-to-red photoconvertible fluorescent protein activated by a visible blue light. Nature Biotechnology. 2006;24:461–465. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- 28.Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. Journal of Cell Biology. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang HL, Eom T, Oleynikov Y, Shenoy SM, Liebelt DA, Dictenberg JB, Singer RH, Bassell GJ. Neurotrophin-induced transport of a beta-actin mRNP complex increases beta-actin levels and stimulates growth cone motility. Neuron. 2001;31:261–275. doi: 10.1016/s0896-6273(01)00357-9. [DOI] [PubMed] [Google Scholar]
- 30.Brimijoin S, Helland L. Rapid retrograde transport of dopamine-beta-hydroxylase as examined by the stop-flow technique. Brain Research. 1976;102:217–228. doi: 10.1016/0006-8993(76)90878-7. [DOI] [PubMed] [Google Scholar]
- 31.Ure DR, Campenot RB. Retrograde transport and steady-state distribution of 125I-nerve growth factor in rat sympathetic neurons in compartmented cultures. Journal of Neuroscience. 1997;17:1282–1290. doi: 10.1523/JNEUROSCI.17-04-01282.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martenson CH, Odom A, Sheetz MP, Graham DG. The effect of acrylamide and other sulfhydryl alkylators on the ability of dynein and kinesin to translocate microtubules in vitro. Toxicol Appl Pharmacol. 1995;133:73–81. doi: 10.1006/taap.1995.1128. [DOI] [PubMed] [Google Scholar]
- 33.Watson FL, Heerssen HM, Bhattacharyya A, Klesse L, Lin MZ, Segal RA. Neurotrophins use the Erk5 pathway to mediate a retrograde survival response. Nature Neuroscience. 2001;4:981–988. doi: 10.1038/nn720. [DOI] [PubMed] [Google Scholar]
- 34.Cui B, Wu C, Chen L, Ramirez A, Bearer EL, Li WP, Mobley WC, Chu S. One at a time, live tracking of NGF axonal transport using quantum dots. Proc Natl Acad Sci U S A. 2007;104:13666–13671. doi: 10.1073/pnas.0706192104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hengst U, Cox LJ, Macosko EZ, Jaffrey SR. Functional and selective RNA interference machinery in axonal growth cones. J Neurosci. 2006;26:5727–5732. doi: 10.1523/JNEUROSCI.5229-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuruvilla R, Zweifel LS, Glebova NO, Lonze BE, Valdez G, Ye H, Ginty DD. A neurotrophin signaling cascade coordinates sympathetic neuron development through differential control of TrkA trafficking and retrograde signaling. Cell. 2004;118:243–255. doi: 10.1016/j.cell.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 37.Riccio A, Pierchala BA, Ciarallo CL, Ginty DD. An NGF-TrkA-mediated retrograde signal to transcription factor CREB in sympathetic neurons. Science. 1997;277:1097–1100. doi: 10.1126/science.277.5329.1097. [DOI] [PubMed] [Google Scholar]
- 38.Watson FL, Heerssen HM, Moheban DB, Lin MZ, Sauvageot CM, Bhattacharyya A, Pomeroy SL, Segal RA. Rapid nuclear responses to target-derived neurotrophins require retrograde transport of ligand-receptor complex. J Neurosci. 1999;19:7889–7900. doi: 10.1523/JNEUROSCI.19-18-07889.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoeller D, Volarevic S, Dikic I. Compartmentalization of growth factor receptor signalling. Curr Opin Cell Biol. 2005;17:107–111. doi: 10.1016/j.ceb.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 40.Howe CL, Valletta JS, Rusnak AS, Mobley WC. NGF signaling from clathrin-coated vesicles: evidence that signaling endosomes serve as a platform for the Ras-MAPK pathway. Neuron. 2001;32:801–814. doi: 10.1016/s0896-6273(01)00526-8. [DOI] [PubMed] [Google Scholar]
- 41.Delcroix JD, Valletta JS, Wu C, Hunt SJ, Kowal AS, Mobley WC. NGF signaling in sensory neurons: evidence that early endosomes carry NGF retrograde signals. Neuron. 2003;39:69–84. doi: 10.1016/s0896-6273(03)00397-0. [DOI] [PubMed] [Google Scholar]
- 42.Kornhauser JM, Cowan CW, Shaywitz AJ, Dolmetsch RE, Griffith EC, Hu LS, Haddad C, Xia Z, Greenberg ME. CREB transcriptional activity in neurons is regulated by multiple, calcium-specific phosphorylation events. Neuron. 2002;34:221–233. doi: 10.1016/s0896-6273(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 43.Johannessen M, Delghandi MP, Moens U. What turns CREB on? Cell Signal. 2004;16:1211–1227. doi: 10.1016/j.cellsig.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 44.Meffert MK, Chang JM, Wiltgen BJ, Fanselow MS, Baltimore D. NF-kappa B functions in synaptic signaling and behavior. Nature Neuroscience. 2003;6:1072–1078. doi: 10.1038/nn1110. [DOI] [PubMed] [Google Scholar]
- 45.Terman JR, Kolodkin AL. Nervy links protein kinase a to plexin-mediated semaphorin repulsion. Science. 2004;303:1204–1207. doi: 10.1126/science.1092121. [DOI] [PubMed] [Google Scholar]
- 46.Allan DW, St Pierre SE, Miguel-Aliaga I, Thor S. Specification of neuropeptide cell identity by the integration of retrograde BMP signaling and a combinatorial transcription factor code. Cell. 2003;113:73–86. doi: 10.1016/s0092-8674(03)00204-6. [DOI] [PubMed] [Google Scholar]
- 47.Marques G, Haerry TE, Crotty ML, Xue M, Zhang B, O’Connor MB. Retrograde Gbb signaling through the Bmp type 2 receptor wishful thinking regulates systemic FMRFa expression in Drosophila. Development. 2003;130:5457–5470. doi: 10.1242/dev.00772. [DOI] [PubMed] [Google Scholar]
- 48.Higuchi H, Yamashita T, Yoshikawa H, Tohyama M. Functional inhibition of the p75 receptor using a small interfering RNA. Biochemical & Biophysical Research Communications. 2003;301:804–809. doi: 10.1016/s0006-291x(03)00029-9. [DOI] [PubMed] [Google Scholar]
- 49.Bloodgood BL, Sabatini BL. Neuronal activity regulates diffusion across the neck of dendritic spines. Science. 2005;310:866–869. doi: 10.1126/science.1114816. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(a) Immunofluorescence analysis of 3 DIV DRG cultures +/− 5-FdU. Inclusion of 5-FdU effectively abolishes Schwann cell proliferation in DRG cultures. Scale bar, 50 μm. (b) Immunofluorescence analysis of 3 DIV DRG cultures demonstrates strong labelling of cell bodies and all projections with axon marker GAP43. Dendrite/cell body marker MAP2 is restricted to cell bodies, indicating the absence of MAP2-staining dendrites in DRG cultures. Scale bar, 50 μm. (c) Immunofluorescence analysis of compartmented cultures (montage micrographs). Retrogradely labelled (WGA-Alexa555, red) DRG axons fasciculate and grow under the 1 mm divider into the axon compartment. Axons defasciculate as they enter the axon compartment. Cell bodies (DAPI, blue) are restricted to the cell body compartment. Only WGA-Alexa555 labelled cell bodies are included in data sets, as they comprise the population of neurons extending axons across the divider. (d) Dissociated DRG neurons (5 DIV) in compartmented cultures were treated with NGF in either or both compartments. NGF was capable to supporting DRG survival when applied to either axons or cell bodies individually, corroborating the presence of functional TrkA complexes throughout the developing neuron. Numbers on bars represent n cells per condition. (e,f) DRG axons in compartmented cultures were infected with a Sindbis virus encoding myr-dEGFP under the control of an IRES to both cell body and axonal compartments. Addition of cycloheximide or anisomycin for 48 hrs to the axonal compartment elicited a loss of myr-EGFP fluorescence in the axonal compartment, with no significant effect on myr-EGFP levels in the cell body compartment. All compartments were maintained in identical NGF-containing media throughout the course of the experiment. Scale bar, 50 μm (g) DIV3 E13 DRG neurons from CREBα/Δ +/− mouse embryos demonstrated robust CREB mRNA FISH signal in axons, while neurons from CREBα/Δ −/− embryos demonstrated an 86.3% loss of CREB mRNA FISH signal. Incomplete abolishment of FISH signals, relative to siRNA-treated axons, reflects residual alternatively-spliced CREBβ transcripts in these hypomorphic animals19. Additionally, axonal mRNAs are trafficked as mRNA-ribosome complexes, and impaired CREBβ mRNA-ribosome interactions due to the poor Kozak site19 in this transcript may lead to inefficient trafficking. Scale bar, 10 μm. (h) Quantification of data in (g). *p<0.0001. Numbers on bars represent n axons per condition. (i) Similar to the experiment in Fig. 3a, except equivalent numbers of cell bodies and axon terminals were used for Western blotting. DRG explants were cultured in Boyden chambers, and the upper compartment was incubated in 0 ng/ml NGF. The lower compartment was incubated in either 0 ng/ml NGF or 100 ng/ml NGF for 3 h. Axon lysates (~104 cells) were prepared from the underside of the membrane and analysed by Western blot. As in Fig. 3a, only CREB was detected in axons, and CREB localisation in axons was dependent on NGF in the culture media. (j) DRG cultures were transfected with CREB siRNA and CREB levels were detected using a CREB antibody. Neurons transfected with non-targeting siRNA exhibited CREB immunoreactivity in axons, while neurons transfected with CREB-specific siRNAs exhibited a near-complete abolishment of CREB immunoreactivity. Scale bar, 20 μm. (k) Quantification of data in (j). *p<0.001. Numbers on bars represent n axons per condition. (l) E15 DRG neuronal lysate (10 μg) was subjected to immunoblotting with the CST-9192 CREB antibody. (m) CREB was detected by immunofluorescence using CREB antibody sc-186 (see i, Fig. 3a, and Table S4). Scale bar 10 μm. (n) DIV3 E13 DRG neurons from CREBα/Δ +/− embryos exhibited robust CREB immunoreactivity in axons, while neurons from CREBα/Δ −/− embryos demonstrated an 84.8% loss of CREB IF signal. The greater degree of reduction in CREB immunofluorescence following siRNA treatment (see k) than in the CREB mutant mice likely reflects the more complete abolition of CREB following siRNA transfection than in the hypomorphic animal. Scale bar, 10 μm. (o) Quantification of data in (m). *p<0.0001. Numbers on bars represent n axons per condition.
(a) Schematic diagram of the Sindbis reporters construct used to monitor RhoA and histone H1f0 translation4. The reporter contains a myristoylated, destabilised EGFP (dEGFP) with the 3′UTR of RhoA or histone H1f0, expressed under control of the Sindbis subgenomic promoter (PSG). (b) E15 DRG explant cultures were infected with Sindbis constructs expressing myr-dEGFP3′RhoA or myr-dEGFP3′H1f0 on DIV3 and fluorescence images were collected after 24h. Explants infected with myr-dEGFP3′RhoA and cultured in the presence of NGF exhibited fluorescent puncta distributed throughout axons (0 h). Following replacement of the media with NGF-free media for 2 h, puncta intensity was not significantly affected (2 h). Explants infected with myr-dEGFP3′H1f0 and cultured in the presence of NGF did not exhibit fluorescent puncta in axons. Scale bar, 25 μm. (c) myr-dEGFP3′CREB-infected axons were counter-stained by immunofluorescence using antibodies specific to translational marker p-eIF4E, RNP-associating protein Staufen, and mitochondrial marker VDAC/Porin1. myr-dEGFP was found in some, but not all ribosomal clusters (Fig. 4c), suggesting that myr-dEGFP-negative clusters are either translationally inactive, that they translate CREB mRNA in response to different stimuli, or that their function involves the translation of other mRNAs. Scale bar, 10 μm. (d) Axons from DIV3 DRGs were analysed for CREB and mitochondrial localisations by immunofluorescence using CREB-specific and VDAC/porin1-specific antibodies. Scale bar, 10 μm. (e) Axons were severed from DIV3 DRG explant cultures, and incubated with 0 or 100 ng/ml NGF for 3 h. CREB mRNA was detected by FISH using a CREB specific riboprobe. CREB mRNA levels did not significantly change during the course of the experiments. Numbers on bars represent n axons per condition.. (f) 40 μM LLnL was added to myr-dEGFP3′CREB-infected axons. Within 5 min, the eGFP fluorescence signal increased twofold, due to inhibition of proteasome-dependent degradation of the destabilised EGFP. Scale bar, 10 μm. (g) 3DIV dissociated DRGs were treated with NGF-free and NGF-replete media as in Fig. 5. CREB levels were assayed by immunofluorescence in contiguous axons: distal segments (450–500 μm), medial segments (250–300 μm) and proximal segments (50–100 μm) from the same axon were determined by their distances from the cell body. n≥10 axons per data point. *p<0.01. (h) Axons of DIV3 DRG explant cultures were analysed by immunofluorescence for NGF effectors CREB, pTrkA and pErk5 at 3 DIV. Scale bar, 5 μm.
(a) Neurons transfected with CREB-specific siRNAs exhibited a specific abolishment of CREB mRNA FISH signal in the axon compartment (“Axon”), but no significant change in CREB mRNA FISH signals in proximal axons in the cell body compartment (“Cell Body”). (b) Quantification of data in (a). FISH levels in each compartment were normalised to fluorescence signals from cultures treated with non-targeting siRNA. β-actin mRNA FISH signals were unaffected by CREB-specific siRNA. *p<0.001. Numbers on bars represent n axons per condition. (c) Transfection was as in Fig. 7a, except CREB knockdown was assessed by Western blot instead of immunofluorescence. Lysates (10 μg protein) were prepared from the axon compartment of CREB-specific siRNA-treated compartmented cultures and analysed by Western blot using a CREB-specific antibody. (d) DRGs in compartmented cultures (5 DIV) were incubated in NGF-free media, supplemented with BAF to prevent apoptosis. Axon compartments were transfected with CREB-specific or non-targeting siRNA. After 48hr, 30 ng/ml NGF was added to the axon compartment for 20min, after which cells were fixed and subjected to immunofluorescence analysis of nuclear pCREB. (e) DRGs in compartmented cultures were treated as in (d, Fig. 7a), after which pTrkA levels in nuclei were quantified by immunofluorescence. (f) DRGs in compartmented chambers were treated as in (d, e, Fig. 7a) and pErk5 levels in nuclei were quantified by immunofluorescence. (g) Dissociated DRG neurons (5 DIV) were infected with equal infectious units of recombinant CRE-luciferase adenovirus and incubated in various concentrations of NGF. After 24hr, cells were fixed and analysed by immunofluorescence for luciferase production. n=36 (0NGF), 29 (1ng/ml NGF), 38 (5ng/ml NGF), 29 (10ng/ml NGF), 35 (20ng/ml NGF), 35 (50ng/ml NGF), 45 (100ng/ml NGF) cells.