Abstract
Previously, we conducted a genome scan on a population derived from the Dahl salt-sensitive hypertensive (S) and the spontaneously hypertensive rat (SHR) using urinary albumin excretion (UAE) as our primary measure of renal function. We identified 10 quantitative trait loci (QTL) linked to several renal and/or cardiovascular traits. In particular, linkage and subsequent congenic strain analysis demonstrated that the loci on chromosome 2 had a large and significant effect on UAE compared with the S rat. The present work sought to characterize the chromosome 2 congenic strain [S.SHR(2)] by conducting a time-course analysis (week 4–20), including evaluating additional renal parameters, histology, electron microscopy, and gene expression/ pathway analysis. Throughout the time course the congenic strain consistently maintained a threefold reduction in UAE compared with S rats and was supported by the histological findings of significantly reduced glomerular, tubular and interstitial changes. Gene expression/pathway analysis performed at week 4, 12, and 20 revealed that pathways involved in cellular assembly and organization, cellular movement, and immune response were controlled differently between the S and congenic. When all the data are considered, the chromosome 2 congenic appears to attenuate renal damage primarily through an altered fibrotic response. Recombinant progeny testing was employed to reduce the QTL to ~1.5 cM containing several interesting candidate genes. The concordance of this rat QTL with renal disease loci on human chromosome 1q21 demonstrate that elucidating the causative gene and mechanism of the rat QTL may be of particular importance for understanding kidney disease in humans.
Keywords: Dahl salt-sensitive rat, quantitative trait locus, albuminuria, proteinuria, recombinant progeny testing, fibrosis
CHRONIC KIDNEY DISEASE (CKD) is a worldwide healthcare problem with increasing incidence and prevalence (38). It is characterized by a gradual loss of kidney function, usually over years and can culminate in renal failure. Additionally, CKD is an important risk factor for the development of cardiovascular disease and overall mortality. Interestingly, most CKD cases are not associated with primary renal disease, but with systemic conditions like diabetes and hypertension (38, 55). Essential hypertension can cause renal injury, but renal susceptibility genes most likely determine occurrence and severity of renal damage (54).
It has been well established, through familial studies, that CKD has a strong genetic component (2, 18, 19, 49). Investigation of congenital and familial forms of kidney disease has led to the identification of genes required for proper functioning of the glomerular filtration barrier (7). These studies have provided insight into mechanisms of proteinuria and glomerulosclerosis. However, these mutations have not been linked to CKD in the general population, demonstrating that CKD is most certainly a complex trait and as such makes genetic studies in humans more difficult (7). On the other hand, the rat provides a particularly fertile model to study disease because it overcomes some of the limitations associated with using human subjects. There are many well-defined inbred rat strains currently being used to study the genetics of CKD (5, 35, 37, 45–47, 51). In particular, the Dahl salt-sensitive rat (S) was selectively bred as a model to study the genetics of salt-sensitive hypertension (11, 12). However, the S rat also develops progressive proteinuria, severe glomerular, vascular, and tubular lesions, besides the development of hypertension, and consequently can serve as a model of hypertension related renal disease (23, 52).
Recently, a genetic analysis of renal and cardiovascular traits was conducted using the S and the spontaneously hypertensive rat (SHR) by our group (20) and others (42, 48). Linkage analysis identified quantitative trait loci (QTL) on multiple chromosomes (1, 2, 6, 8–11, 13, and 19) for urinary protein excretion (UPE) and/or urinary albumin excretion (UAE) with variable time-course patterns (20). A second study found that several QTL (chromosome 2, 11, and 19) were influenced by salt-loading, presumably a result of changes in blood pressure in response to the high-salt diet (21). Congenic strain analysis established that the QTL on chromosome 2 had a major influence on renal function in the S rat (21).
The first aim of the present work was to characterize the chromosome 2 congenic strain by conducting a time-course analysis and establishing onset and progression of renal disease compared with both parental strains. We employed a comprehensive approach, including evaluation of additional renal parameters, histology, electron microscopy, gene expression analysis, and gene pathway analysis to characterize the strain. A second aim was to employ recombinant progeny testing (RPT) to reduce the QTL to a small genomic region to aid in gene identification.
MATERIALS AND METHODS
Animals
The Dahl salt-sensitive (SS/Jr or S), spontaneously hypertensive rat (SHR/NHsd or SHR), and S.SHR(2) congenic strain were maintained in our animal facility at the University of Toledo, Health Science Campus (UTHSC, formerly the Medical University of Ohio). All experiments had approval of our Institution Animal Care and Use Committee. At 4 wk of age, a group of age-matched males S (n = 50), S.SHR(2) (n = 50), and SHR (n = 50) were weaned to a low-salt diet (0.3% NaCl, TD7034; Harlan Teklad, Madison, WI) and studied for several renal and cardiovascular traits at 4, 5, 6, 8, 12, 16, and 20 wk of age.
At each time point a subset of animals (n = 6 to 12) was euthanized (overdose of pentobarbital sodium) for the collection tissue. Kidney samples were processed for histological, electron microscopy, and microarray analysis. Serum samples were obtained to measure blood parameters. Additionally body, heart, and kidney weights were also measured.
For RPT, a large F1[S × S.SHR(2)] × S backcross population (n = 971) was developed to establish recombinant animals within the introgressed region on chromosome 2. Male S.SHR(2) animals were crossed to S females to produce F1[S × S.SHR(2)] animals. The F1 rats were backcrossed to the S to produce the F1[S × S.SHR(2)] × S population. Additionally, urine parameters were collected on this population to reduce the confidence interval for the QTL region. The 971 rats were handled in six blocks of ~162 animals. Rats in each block were closely age matched, but between blocks the average ages differed by a few days. This allowed urine collections to proceed on all rats at approximately the same ages. Block effects, if any, were removed statistically before further analysis.
Phenotyping
Blood pressure
Blood pressure (BP) was also collected using a telemetry system (Data Sciences International, St. Paul, MN). At 61–63 days of age a transmitter was surgically implanted in S (n = 8) and S.SHR(2) congenic (n = 8) animals. Surgery was performed under 1.5% isoflurane anesthetic in 100% O2. The probe body was placed subcutaneously in the flank, the probe catheter was inserted into the femoral artery, and the tip of the probe was advanced to the lower abdominal aorta as done previously (26). Data on systolic BP, diastolic BP, mean BP, pulse pressure, and heart rate were collected. Readings for each BP parameter was collected for a 24-h period (5-min time intervals for 10 s) at 8, 12, 16, and 20 wk of age.
Urine and blood parameters
To collect urine, animals were kept in metabolism cages (Lab Products, Seaford, DE) for 24 h with free access to water. Sodium azide was added to the collection vials for a final concentration of ~0.01% in the urine as a preservative. UPE was determined colorimetrically using pyrogallol red/molybdate complex (Quantimetrix, Redondo Beach, CA). UAE was determined by rat specific albumin EIA kit (SPI-bio). UPE and UAE are expressed as mg/24 h. Urine creatinine was determined by the Jaffe method (Cayman Chemical, Ann Arbor, MI). Following 24-h urine collection, blood was obtained by cardiac puncture. Blood parameters (Table 1) were determined by standard methods using an Alfa Wassermann ACE automated chemistry analyzer (BioReliance, Rockville, MD).
Table 1.
Comparison of blood and urine parameters for S, S.SHR(2), and SHR at week 20
| S | S.SHR(2) | SHR | |
|---|---|---|---|
| Weight, g | |||
| Body weight | 421±11.1 | 435±10.0 | 339±9.3* |
| Heart weight | 1.3±0.02 | 1.3±0.02 | 1.3±0.01 |
| Total kidney weight | 2.8±0.11† | 2.6±0.07 | 2.6±0.04 |
| Blood | |||
| Total protein | 6.7±0.13 | 6.4±0.17 | 6.4±0.12 |
| Creatinine, mg/dl | 0.54±0.01 | 0.53±0.02 | 0.4±0.02* |
| Blood urea nitrogen, mg/dl | 33.4±1.50† | 29.7±0.49 | 29.3±0.76 |
| Total cholesterol, mg/dl | 114.9±4.3† | 97.3±1.7 | 47.3±7.6* |
| Triglycerides, mg/dl | 106.1±8.5† | 90.9±7.5 | 63.0±7.5* |
| Sodium, meq/l | 149.6±2.36† | 142.8±1.05 | 144.2±0.60 |
| Potassium, meq/l | 4.6±0.13† | 5.4±0.22 | 5.3±0.21 |
| Bicarbonate (CO2), meq/l | 26.6±1.09† | 23.1±0.47 | 23.5±0.16 |
| Urine | |||
| Urine volume | 14.2±1.86 | 22.3±5.1 | 14.2±3.4 |
| Urine pH | 6.6±0.07 | 6.8±0.07 | 7.2±0.10* |
| Creatinine clearance, ml·min−1·g TKW−1 | 0.75±0.018 | 0.78±0.025 | 0.80±0.027 |
| Sodium excretion, meq/day | 0.43±0.03 | 0.45±0.02 | 0.82±0.05* |
Values are means ± SE; n = 6 per group. Heart weight and total kidney weight were adjusted for differences in body weight between the groups. Creatinine clearance was normalized to total kidney weight (TKW) to adjust for differences in animal size.
P < 0.05 vs. Dahl salt-sensitive rat (S) or chromosome 2 congenic strain [S.SHR(2)],
P < 0.05 vs. S.SHR(2) or spontaneously hypertensive rats (SHR).
Histology
Kidneys were fixed in 10% neutral buffered formalin, embedded in paraffin, cut into 3-μm sections, and stained with hematoxylin and eosin or Masson’s trichrome. Two central longitudinal sections from each kidney (n = 6, each group) were examined in a blinded fashion. Percent glomerular injury was established by using the number of glomeruli exhibiting glomerulosclerosis and/or mesangial expansion divided by the total number of glomeruli evaluated, on average 1,000 per group. Percent interstitial injury was determined by evaluation of slides stained with Masson’s trichrome. We evaluated 20 random regions of each slide using Image Pro 5.1 (Media Cybernetics) to quantify the percent fibrosis (blue staining) compared with background. Vascular and tubular injury was evaluated separately on a semiquantitative scale from 0 (normal) to 4 (severe). Vascular compartments were assessed for vessel wall thickening, sclerotic involvement, and vasculitis/perivascular inflammation. Tubules were evaluated for the presence of necrosis, hydropic change, and/or tubular casts.
Electron microscopy
For transmission electron microscopy, a 1-cm-square region of kidney cortex was obtained from each animal (n = 3, each group). Samples were fixed in 3% glutaraldehyde for 1.5 h and washed three times for 10 min each with 0.2 M sodium cacodylate buffer, postfixed for 1 h with 1% OsO4 followed by 1 h with saturated uranyl acetate. Dehydration was carried out by a graded series of chilled ethanol solutions (30–100%) and a final dehydration with 100% acetone. Cortical samples were infiltrated overnight in Spurr’s resin (Electron Microscope Sciences) and ultrathin sections were obtained and collected on copper 200-mesh support grids. Sections were poststained with uranyl acetate and lead citrate, and examined using a Philips CM 10 transmission electron microscope.
Microarray and Analysis
DNA microarray analysis was performed using Affymetrix Gene-chip Rat Genome 230 2.0 array at three time points. The chip contains 31,000 probe sets from >30,000 transcripts on one array. Three male S and three male S.SHR(2) congenic were selected at random from each group at weeks 4, 12, and 20. Kidney was cut into ~0.5-cM-size cubes, suspended in RNAlater (Ambion, Austin, TX) and stored overnight at 4°C. RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) and purified using Mini RNeasy kit (Qiagen, Valencia, CA) according to manufacturer’s protocols. RNA quality was assessed by an OD260/280 ratio >2.0 and visually by ethidium bromide staining on an agarose gel.
Biotinylated cRNA was synthesized from 10 μg of total RNA using the One-Cycle Target Labeling Kit (Affymetrix, Santa Clara, CA) as directed by the user manual. cRNA quality for each sample was assessed by hybridization of 5 μg of adjusted kidney cRNA to Affymetrix Genechip Test3 array. Subsequently, 15 μg of adjusted kidney cRNA was hybridized to the Genechip Rat 230 2.0 array. Hybridized chips were automatically washed, stained, and scanned at the UTHSC Genomics core facility using Affymetrix equipment. Data obtained from these gene expression studies are deposited in the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) with the GEO accession number GSE6208.
Microarray analysis was performed using the commercially available GeneSifter.net software platform (http://www.genesifter.net). Expression values were generated by probe level analyses using GC-RMA (robust multiarray averaging with an adjustment for GC content). The GC-RMA method converts the intensities from multiple probes from a probe set into a single expression value and uses both perfect-match and mismatch probe information. The data are normalized by quantile normalization and log2 transformed. Following the normalization process, differentially expressed gene were statistically identified by t-test using two methods at each time point: 1) family-wise error rate procedure, P < 0.05 and fold-change ± 1.5 or greater; and 2) a more stringent procedure, using Benjamini and Hochberg FDR (false discovery rate) which corrects for multiple comparison, using P < 0.05, and fold-change ± 1.5 or greater. Additionally, a two-way ANOVA using a P < 0.01 was performed to identify strain × time interaction effects between the S and S.SHR(2) from week 4 to week 20.
Gene networks and functional analysis were generated through the use of Ingenuity Pathways Analysis (Ingenuity Systems, www.ingenuity.com). Gene identifiers and corresponding expression values from data sets analyzed at week 4, 12, 20 or the strain × time interaction from week 4 to 20 from GeneSifter.net was uploaded into in the application. Each gene identifier was mapped to its corresponding gene object in the Ingenuity Pathways Knowledge Base. These genes, called focus genes, were overlaid onto a global molecular network developed from information contained in the Ingenuity Pathways Knowledge Base. Canonical pathways analysis identified the pathways from the Ingenuity Pathways Analysis library of canonical pathways that were most significant to the data set. The significance of the association between the data set and the canonical pathway was measured in two ways: 1) a ratio of the number of genes from the data set that map to the pathway divided by the total number of genes that map to the canonical pathway; and 2) A Fisher’s exact test was used to calculate a P value determining the probability that the association between the genes in the dataset and the canonical pathway is explained by chance alone.
Expression differences for Cct3 and Sfrp2 were confirmed by quantitative real-time PCR using SYBR-green dye chemistry on a Bio-Rad MyIQ Real-Time PCR machine (n = 6 each strain). Gene expression between the congenic strain and the S control were evaluated using the comparative method (ΔΔCt) of gene expression using β-actin expression as the control gene (22).
Genotyping
Genomic DNA was obtained by tail biopsy from F1[S × S.SHR(2)] × S backcross and RPT animals and prepared using Wizard SV 96 Genomic DNA kit (Promega, San Luis Obispo, CA). Genotyping was done by multiplexing (4 primer sets per PCR reaction) using a fluorescent-based approach on a Beckman Coulter CEQ8000 XL capillary sequencer. In brief, each primer is labeled with a fluorescent dye (dye D4-PA; Beckman Coulter, Fullerton, CA), which produces a PCR product that can be detected using the CEQ system. Given only one fluorescent dye was utilized, primers for multiplex PCR were selected to vary in size and allow multiple PCR products to be identified during any given CEQ run. A total of 24 markers spaced at ~5-cM intervals on chromosome 2 were used to identify recombinant animals in the F1[S × S.SHR(2)] × S backcross population, which were subsequently used for RPT.
PCR reactions were prepared as follows in a 10-μl reaction: 1× buffer containing 1.5 mM MgCl2 (Promega, Madison, WI), 0.2 mM dNTPs (Sigma, St. Louis, MO), 2.5 pmol D4-PA each forward primer (Sigma-Proligo, Boulder, CO), 2.5 pmol each reverse primer, and 0.25 U Taq DNA polymerase (Promega). PCR amplification was performed as follows: an initial step at 95°C for 3 min and continued for 35 cycles of 94°C for 40 s, 55°C for 40 s, 72°C for 1 min. After amplification, the PCR products were diluted 1:10 in water, 1–3 μl of diluted multiplex PCR product was added to 40 μl of deionized formamide containing DNA size standard (Beckman Coulter), loaded on CEQ sequencer, and then analyzed using CEQ fragmentation analysis software.
Statistical Analysis
Linkage analysis and QTL localization were performed using Map Manager QTX (http://www.mapmanager.org) (34). The approximate confidence internal of the chromosome 2 QTL peak was estimated using the bootstrap method of the QTX program. S, S.SHR(2), and SHR time-course data was evaluated by one-way ANOVA followed by post hoc multiple comparisons using Tukey’s test (SPSS, Chicago, IL). RPT data were evaluated by an independent t-test using SPSS. All data are presented as means ± SE.
RESULTS
Characterization of S, S.SHR(2), and SHR
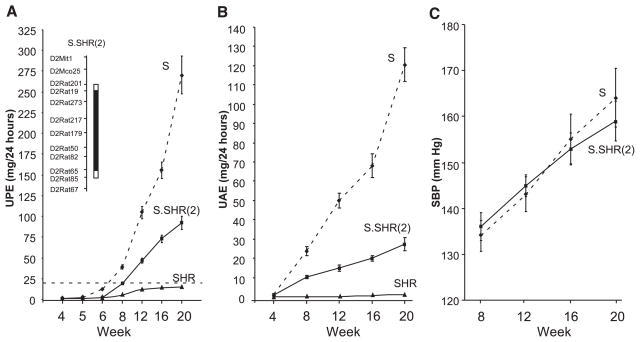
A time course for UPE, UAE, and BP of each group of rats is presented in Fig. 1. No detectable difference in UPE (Fig. 1A) was observed between the groups at week 4. Starting at week 5 and continuing through week 20, the S rat had significantly higher UPE compared with either the S.SHR(2) congenic or SHR (P < 0.0001). The S.SHR(2) congenic consistently maintained at least a twofold reduction in UPE compared with the S rat. The S rat rapidly progressed beyond the “proteinuria” threshold (>20 mg/24 h) between week 6 and 8, while the congenic reached this point between week 8 and 12. The SHR never progressed beyond this threshold. At week 20 S rat UPE was 270 ± 22.9 mg/24 h compared with 92.3 ± 8.3 in the congenic. Differences between groups for UAE (Fig. 1B) were similar to UPE, although the congenic maintained a greater fold reduction (3-fold) compared with the S rat. Despite the large difference between the S and congenic with respect to UPE, no detectable difference in systolic BP (Fig. 1C) or associated parameters, such as diastolic BP, pulse pressure, or heart rate was observed (data not shown).
Fig. 1.
Time course for urinary protein excretion (UPE, A); urinary albumin excretion (UAE, B); and systolic blood pressure (SBP, C) in male Dahl salt-sensitive (S), chromosome 2 congenic strain [S.SHR(2)], spontaneously hypertensive (SHR) rats. A schematic of the congenic strain is shown in A, inset. The black bar designates the extent of introgressed SHR alleles on the S background. The open region on each end of the congenic segment represents the recombination interval. In A, the dashed line represents the threshold for “proteinuria” (>20 mg/24 h). Rats were maintained on low-salt diet (0.3% NaCl) for entire course of the experiment. The number of animals at each time varied from n = 50 per group (week 4) to n = 12 per group (week 20). SBP was measured by telemetry (n = 8 per group). Mean values ± SE.
Table 1 shows additional experimental parameters between the groups at week 20. Body weights (BW) between the S and S.SHR(2) congenic were similar but were significantly greater than the SHR. No significant difference in heart weight (adjusted for BW differences) between the groups were observed, but kidneys from the S rat were significantly larger than either the congenic or SHR. Plasma sodium and potassium levels were significantly different in the S compared with either the congenic or SHR. Blood urea nitrogen was also higher in the S rat. Both total cholesterol and triglycerides levels were also significantly decreased in the congenic compared with the S, a common feature of improved renal function. Creatinine clearance, normalized to kidney weight, was not different between the groups, although plasma creatinine was significantly reduced in the SHR.
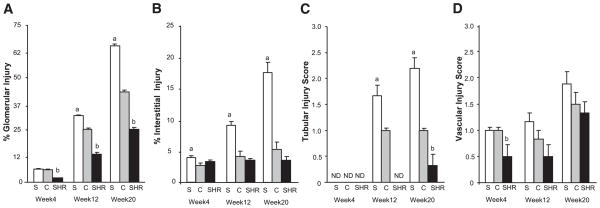
Kidney from S, S.SHR(2), and SHR were examined and evaluated at week 4, 12, and 20 for glomerular, tubular, vascular, and interstitial damage (Fig. 2). Representative histological images for week 20 animals are shown in Fig. 3. As early as week 4, small, but significant interstitial changes were observed between the S and the congenic (Fig. 2B) preceding any significant difference in UPE (Fig. 1). At week 12, glomerular damage of the congenic was significantly lower than the S, although both exhibited focal glomerulosclerosis. Tubular changes were significantly attenuated (~1.5-fold) in the congenic compared with the S (Fig. 2C). No significant vascular changes were seen among any of the groups. At week 20, the S rat had a significantly greater number of damaged glomeruli compared with the congenic (64% vs. 40%). Most notable was that the S rat experienced a significant increase in fibrosis from week 12 (9.2% ± 0.89) to 20 (17.5% ± 1.79), while there was essentially little change in the congenic (4.5% ± 0.24 vs. 6.3% ± 0.44). The SHR showed similar glomerular pathology to the congenic at week 12. Again, no significant vascular changes were seen between groups.
Fig. 2.
Time-course histological examination of male S, S.SHR(2), and SHR rats. Percent glomerular injury (A), percent interstitial injury (B), tubular injury score (C), vascular injury score (D). Bar graphs show the effect of the S.SHR(2) congenic (C) strain compared with both parental strains, S and SHR. Glomerular and interstitial injury were evaluated quantitatively (see MATERIALS AND METHODS). Tubular and vascular injury was graded on a semiquantitative scale from 0 (normal) to 4 (severe). Kidneys from 6 animals were evaluated at each time point. ND, not detectable. a, Significantly different from S.SHR(2) and SHR at P < 0.05; b, significantly different from S and S.SHR(2) at P < 0.05. P values are from a 1-way analysis of variance followed by post hoc multiple comparisons using Tukey’s test. Error bars are SE.
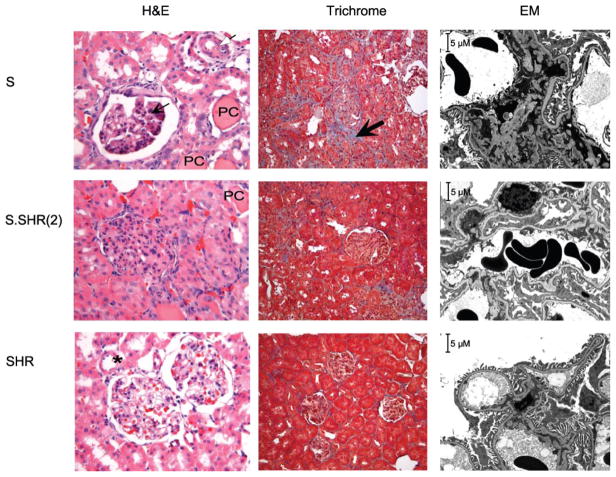
Fig. 3.
Representative light [×40 hematoxylin & eosin (H&E), ×20 Masson’s trichrome] and electron microscopy (EM, ×3,000) images of week 20 low-salt kidney samples from S, S.SHR(2), and SHR. S rats exhibit glomerulosclerosis and mesangial expansion (thin arrow), arterial wall thickening (open arrow), protein casts (PC), and significant fibrosis (thick arrow) compared with S.SHR(2) and SHR. Kidneys from SHR are essentially normal. *Normal arterial vessel. EM of S kidney illustrated significant glomerular disease including evidence of effacement of foot processes, focal mesangial matrix increase, and diffuse mesangial electron-dense deposits. The S.SHR(2) congenic demonstrated significantly less pathology compared with the S. The SHR appeared essentially normal, although there was some evidence of minimal effacement of foot processes (considering all data).
An ultrastructural examination of kidney from S, S.SHR(2), and SHR confirmed the light microscopy data and provided more specific detail on pathological changes between the groups (representative images are shown in Fig. 3 only for week 20). At week 4, little or no pathology was observed between groups. Glomeruli appeared to be within normal limits regarding general appearance of discrete foot processes of the visceral epithelial cells. At week 12, significant differences were seen between the S and congenic. Changes were focal in nature and did not involve all glomeruli. Little or no pathology was seen in the SHR. At week 20, significant glomerular disease including evidence of visceral cell effacement, significant glomerulosclerosis, focal mesangial matrix increase, and diffuse mesangial electron dense deposits were observed in the S (Fig. 3). S.SHR(2) kidneys were also found to have focal visceral cell effacement, focal glomerulosclerosis, focal mesangial matrix increase, and focal mesangial electron dense deposits; however, the pathologic changes were not nearly as severe as the S. SHR had some glomerular pathology including minimal effacement of foot processes, focal mesangial matrix increase, and rare mesangial electron dense deposits.
Expression Profiling and Pathway Analysis
Gene expression profiling was performed at week 4, 12, and 20 using kidney from the S and S.SHR(2) congenic to: 1) identify expression differences of positional candidates within the QTL region; 2) correlate temporal gene expression changes between the S and congenic with degree of renal damage; and 3) identify biochemical pathways potentially involved in the attenuated renal damaged observed in the congenic. Table 2 gives a summary of the number of genes found to be differentially expressed throughout the genome and on chromosome 2, striated by fold-change and statistical significance. On average, depending on fold-change threshold and statistical tests, ~40% of differentially expressed genes map within the congenic interval. A total of 727 protein-coding genes (www.ensembl.org) reside in the congenic interval and are represented on the Affymetrix Genechip, of these, anywhere from 48 or 6.6% (week 4) to 100 or 13.7% (week 20) of genes were found to be differentially expressed.
Table 2.
Number of differentially expressed genes in kidney between S and S.SHR(2) striated by fold-change and statistical significance
| Analyisis Criteria | Time | FWER, P < 0.05
|
FDR, P < 0.05
|
||
|---|---|---|---|---|---|
| All Chromosomes | RN02 | All Chromosomes | RN02 | ||
| Fold-change ±1.5 | week 4 | 168 | 48 | 4 | |
| week 12 | 109 | 50 | 72 | 41 | |
| week 20 | 360 | 100 | 152 | 53 | |
| Fold-change ±1.8 | week 4 | 62 | 26 | 1 | |
| week 12 | 54 | 30 | 42 | 27 | |
| week 20 | 105 | 44 | 54 | 30 | |
| Fold-change ±2.0 | week 4 | 35 | 18 | 1 | |
| week 12 | 44 | 25 | 35 | 22 | |
| week 20 | 58 | 28 | 35 | 21 | |
FWER, family-wiseerror rate; FDR, false discovery rate.
A detailed description of genes found to be differentially expressed and that map within the congenic interval are shown in Table 3. Genome-wide analyses of differentially expressed genes are shown in Supplemental Table S1.1 A total of 37 genes/expressed sequence tags (EST) on chromosome 2 (fold-change ± 2, FDR, P < 0.05) were observed to be differentially expressed between the S and S.SHR(2) throughout the time course. Seven of these genes (Table 3, nos. 31–37) were consistently downregulated at all three time points. In general, genes that were observed to be differentially expressed at one time point were also found to be differentially expressed at an additional time point (serving to confirm the expression data). However, there were many genes observed in entire data set (Supplemental Table S1) where expression differences were time specific.
Table 3.
Time course of genes differentially expressed in kidney between S and S.SHR(2) located on chromosome 2
| Number | Accession# | Gene Name | Fold Change
|
Description | ||
|---|---|---|---|---|---|---|
| Week 4 | Week 12 | Week 20 | ||||
| 1 | AI501417 | Mgst2? | +2.4 | EST | ||
| 2 | BE107853 | +5.4 | +6.4 | EST | ||
| 3 | BF415556 | +5.5 | +4.7 | EST | ||
| 4 | BG374101 | +3.6 | +2.7 | EST | ||
| 5 | AA859982 | +2.6 | +2.2 | EST | ||
| 6 | AW141832 | +2.1 | similar to RIKEN cDNA 2310050P13 (LOC310808) | |||
| 7 | BF551322 | +2.2 | similar to CG5805-PA (LOC365841) | |||
| 8 | BG379771 | +2.8 | EST | |||
| 9 | BF553729 | Wnt2b | +2.7 | EST | ||
| AI555464 | Wnt2b | +2.4 | EST | |||
| 10 | BE097574 | Kcnab1 | +2.4 | EST | ||
| 11 | AA819288 | +2.3 | retinoic acid receptor responder protein 1 | |||
| 12 | BF411765 | Celsr2 | +2.1 | cadherin EGF LAG seven-pass G-type receptor 2 | ||
| 13 | AI715140 | +2.0 | similar to histone protein Hist2 h3c1 (LOC310679) | |||
| 14 | BF284175 | +2.3 | +2.4 | +2.2 | similar to group XII-1 phospholipase A2 (LOC362039) | |
| 15 | BF396545 | Sfrp2* | +2.4 | secreted frizzled-related protein 2 | ||
| 16 | AI511405 | −13.2 | −6.3 | similar to hypothetical protein FLJ22693 (LOC310467) | ||
| 17 | BF522861 | −2.1 | −2.1 | |||
| 18 | BF388224 | −7.4 | −8.7 | |||
| 19 | AI454310 | −2.0 | −2.5 | EST | ||
| 20 | BF402235 | Sars1 | −2.1 | EST | ||
| 21 | AI172217 | −2.9 | similar to protein beta-galactosidase, alpha | |||
| 22 | U86635 | Gstm5 | −2.1 | glutathione S-transferase, mu 5 | ||
| 23 | NM_021584 | −2.0 | −2.8 | activity/neurotransmitter-induced early gene protein 4 | ||
| 24 | BF290410 | −2.2 | EST | |||
| 25 | BE120370 | −2.1 | EST | |||
| 26 | NM_024157 | Cfi | −4.0 | complement factor I | ||
| 27 | NM_012889 | Vcam1 | −2.1 | vascular cell adhesion molecule 1 | ||
| 28 | AF367210 | Il7 | −2.1 | interleukin 7 | ||
| 29 | NM_031120 | Ssr3 | −2.9 | TRAP-complex gamma subunit | ||
| 30 | U05675 | Fgb | −2.3 | fibrinogen, beta polypeptide | ||
| 31 | BI288681 | −7.3 | −6.7 | −3.8 | transcription activator of D-serine dehydratase? | |
| 32 | BE100973 | −5.8 | −8.7 | −8.4 | Rattus norvegicus transcribed sequences | |
| 33 | AI454769 | −4.3 | −4.8 | −3.9 | Rattus norvegicus transcribed sequences | |
| 34 | AI043891 | Cct3* | −2.8 | −2.8 | −2.0 | |
| 35 | AI232217 | −4.2 | −3.7 | −2.2 | Rattus norvegicus transcribed sequences | |
| 36 | NM_031154 | Gstm3 | −5.1 | −4.7 | −3.4 | glutathione S-transferase, mu 3(Yb3) |
| 37 | M28241 | Gstm1 | −3.2 | −3.8 | −3.6 | glutathione S-transferase, mu 1 |
A positive fold change means that the transcript was expressed higher in the S.SHR(2) congenic compared with the S. Conversely, a negative value means that the S transcript was expressed at a higher level than the congenic. Transcripts found to have a fold-change ±2.0 or greater are included here; see Supplemental Table S1 for a complete gene list. Statistical calculations were performed by 2 methods (see MATERIALS AND METHODS FOR MORE DETAIL): P value (FWER) and adjusted P value (FDR using Benjamani and Hochberg correction for multiple corrections). Fold change data shown are all significant at least P < 0.05. For exact P value see Supplemental Table S1.
Both of these genes map to the reduced quantitative trait locus (QTL) interval, differential expression was confirmed using quantitative real-time PCR. Cct3 was found to be differentially expressed at all 3 time points, with a fold-change ranging from −1.20 to −1.35 (at least P < 0.05). Sfrp2 was observed to be differentially expressed at week 4 (confirming the Affymetrix data) and also at week 12 and 20, with a fold-change ranging from +1.60 to +1.79 (at least P < 0.05).
To gain insight into the major biological processes and pathways linked to genes differentially expressed between the S and S.SHR(2) congenic, the gene expression data was evaluated using the Ingenuity Pathway Analysis program (see MATERIALS AND METHODS for details). Multiple networks were discovered in each of the three datasets (week 4, 12, and 20). Cellular movement, cellular growth and proliferation, and connective tissue development and function were recurring top scoring functions from week 4 to 20 (Supplemental Table S2 and Supplemental Fig. S1). Additionally, canonical pathways (which are the most relevant pathways based on the entire dataset) for Wnt/β-catenin signaling, amyloid processing, and antigen presentation were identified in week 4, Wnt/β-catenin signaling pathway and glutathione metabolism in week 12, and nitric oxide signaling in week 20.
A two-way ANOVA was performed on the time-course microarray data to identify genes that were significantly different between strains [S and S.SHR(2)] and over time [week 4, 12, and 20], i.e., strain × time interaction. A total of 186 genes were identified using fold-change ± 1.5 or greater and a P < 0.01 (Supplemental Table S3). Approximately one-third of these genes were not previously observed to be different when examined at each time point individually. Top functions identified involved cellular assembly, cellular movement, and immune related functions (Supplemental Fig. S2).
Fine Mapping the Renal Function Locus
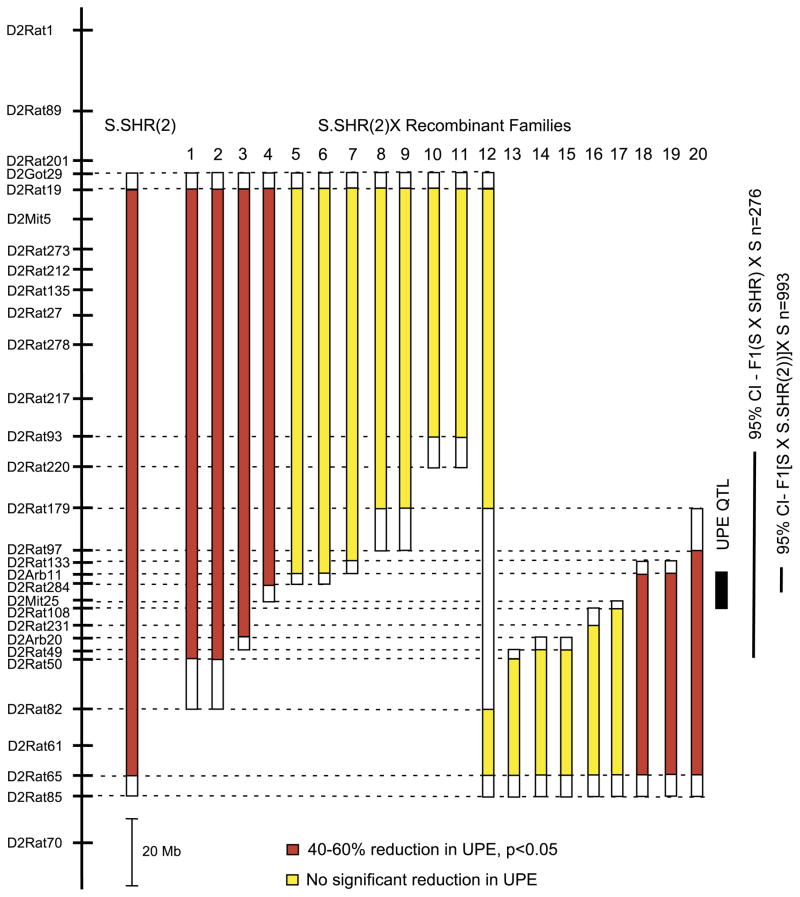
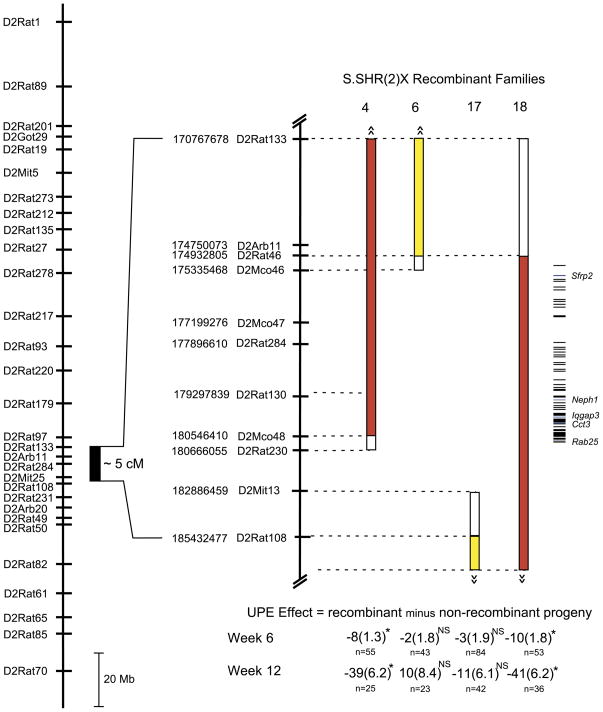
RPT was employed to refine the genomic region containing the QTL responsible for the observed difference in renal function between the S and S.SHR(2) congenic. The basis of RPT is to find recombinants in an interval derived from the S.SHR(2) congenic, to propagate these recombinants, and then to measure the phenotypes of their progeny to determine the genotypic value of each recombinant. A large backcross population (n = 971) was developed from the S.SHR(2) congenic strain and the S rat to identify recombinant animals and to reduce the confidence interval for the QTL region. Bootstrapping analysis of the original linkage analysis (backcross population, n = 276) (20) localized the 95% confidence interval (CI) to a ~14-cM region delimited approximately by markers D2Rat220 and D2Rat50 (Fig. 4). The new backcross population derived from the S.SHR(2) congenic reduced the 95% CI to ~1.5 cM, flanked by D2Arb11 and D2Rat284. A total of 20 recombinant animals were selected from this population for RPT. All recombinant families were evaluated for UPE at 6 wk of age for an initial survey of the families (Fig. 4). Recombinant families 1–4 (shown in red) all demonstrated a significant reduction (40–60%) in UPE, that is, animals that carried the recombinant chromosome (congenic-like) had significantly lower UPE compared with nonrecombinant littermates (S-like). Families 5–17 (shown in yellow) showed no significant effect on UPE, which eliminates the region above D2Arb11, and below D2Rat108, from significantly contributing to the refined QTL. Families 18–20 (also shown in red) demonstrated a significant effect on UPE.
Fig. 4.
Fine mapping of renal function quantitative trait loci (QTL) using recombinant progeny testing (RPT). The physical map of rat chromosome 2 is shown on the left. The solid bars to the right of the physical map indicate the extent of the SHR-donor regions for each recombinant family. The open region represents the recombination interval. UPE was measured at week 6. The solid black bar denotes the location of the QTL based on the RPT. The 95% confidence interval (CI) obtained from the original linkage analysis (20) and the present study is shown to the right of the figure. Red bars denote that recombinant animals (congenic-like) had significantly (P < 0.05) lower UPE compared with nonrecombinant (S-like) littermates. Yellow bars denote there was not a significant difference in UPE between recombinant and nonrecombinant littermates. The number of animals tested from each recombinant family ranged from 16 to 29.
Figure 5 shows an enlargement of the QTL region and the four recombinant families (4, 6, 17, and 18) important for delimiting the QTL. To confirm the data obtained from the initial survey (Fig. 4), additional animals (n = 43–84) were tested from each family of the four critical families. UPE was evaluated in these families at 6 and 12 wk of age to determine if there was a sustainable effect on UPE in older animals (Fig. 5). Families 4 and 18 demonstrated a significant effect on UPE at week 6, which continued through week 12. For family 4 (week 12), the average UPE for nonrecombinant animals was 95.3 ± 4.94 mg/24 h vs. 56.70 ± 4.45 mg/24 h for recombinant animals. For family 18, the average UPE for nonrecombinant animals was 95.3 ± 4.84 mg/24 h vs. 54.70 ± 4.04 mg/24 h for recombinant animals. No significant effect on UPE was observed for family 5 or 17 at either week 6 or 12, demonstrating that the QTL lies in the interval flanked by D2Rat46 and D2Rat230 (Fig. 5).
Fig. 5.
Enlargement of the QTL region and the 4 recombinant families important in delimiting the QTL region. The physical map of rat chromosome 2 is shown on the left, with an enlargement of the physical map of the QTL region to the right. Map distances are in base pairs (www.ensembl.org, Ensembl v38, Apr 2006). UPE was measured at week 6 and week 12. Data for “UPE Effect” below each bar are UPE of recombinant rats (congenic-like) minus the UPE of nonrecombinant (S-like) littermates. A negative value indicates that recombinant rats had lower UPE than nonrecombinant rats, and when this is significant this indicates that the QTL is in the congenic interval. The numbers of animals tested are shown below the effect. P values are from an independent t-test. *P < 0.0001; NS is not significant. Data are means ± SE.
The refined QTL region spans 1.5 cM or ~5.0 Mb, which contains 64 known and/or predicted genes. The locations of these genes are depicted in Fig. 5 (additional information provided in Supplemental Table S4). Of the 37 differentially expressed genes found to map on RNO2 (Table 3), only two of these map to the refined QTL region. They are secreted frizzled-related protein 2 (Sfrp2) and chaperonin containing TCP1, subunit 3 (Cct3).The region also contains several candidate genes based on known function or renal involvement including, Neph1, Ras-like protein in brain 25 (Rab25), and IQ motif containing GTPase activating protein 3 (Iqgap3).
DISCUSSION
For this study we sought to investigate differences in onset and progression of renal disease in the Dahl S, S.SHR(2) congenic, and SHR using a time course. Our data clearly demonstrate that the locus on chromosome 2 has a major and sustained ability to attenuate renal damage. Not only was the number of damaged glomeruli less in the congenic, but the degree of damage was also significantly less. This is supported by the finding that albumin consistently composed a smaller proportion of the total amount of protein in the congenic compared with the control S. That is to say, a molecule the size (and charge) of albumin will only significantly compose the ultrafiltrate once the glomerular basement membrane/slit diaphragm is sufficiently compromised. This phenomenon also serves to explain why the presence of microalbuminuria is highly correlated with progression of kidney disease in humans. As early as week 4, significant interstitial changes were observed between the S and the congenic that preceded any significant difference in UPE. This suggests that the QTL may attenuate renal damage primarily through controlling fibrosis, as opposed to controlling glomerular permeability. This is further supported by the fact the congenic experienced increased glomerular damage from week 12 to 20, with no significant change in interstitial injury. Additionally, no significant difference in systolic BP was observed between the S and congenic. These data demonstrate that the reduced renal damage in the congenic is not simply a consequence of lower BP compared with the S suggesting the underlying mechanism of the QTL is unlikely to be mediated through BP regulation.
In recent years, several groups have used a combined QTL and gene expression approach to prioritize candidate genes for further study within a QTL region (27, 36, 59), while others have successfully used the approach to expedite the identification of disease-causing genes (1, 17, 28, 44). More recently, two groups have utilized a large-scale integrated approach using either a panel of recombinant inbred strains (25) or chromosome substitution strains (33) with gene expression profiling to gain insight into genes and pathways underlying disease processes. To this end, we sought to utilize gene expression analysis to identify differential expressed genes linked to the QTL and establish biochemical pathways responsible for the renal protective effect of the congenic strain.
Network analysis of differentially expressed genes consistently identified cellular movement and cellular growth and proliferation as top functions associated with gene networks at each week. Additionally, networks linked to genes that were significantly different between strains [S and S.SHR(2)] and over time (week 4, 12, and 20) were cellular assembly and organization, cellular movement, and immune response. The individual analysis provided a “snapshot” of genes differentially expressed at each time point, while the second analysis (strain × time) placed the networks in context of what occurred over time and identified additional genes of interest. The purpose of conducting the gene expression analysis at week 4 before the onset of detectable differences in UPE was to address the issue of “cause and effect.” The assumption being that networks identified at the early time point are more likely to reflect the cause and not the effect of differences in renal function such as networks observed at later time points (week 12 and 20).
At week 4 (and week 12) the Wnt/β-catenin signaling pathway was identified as the most significant pathway based on the entire dataset. Sfrp2 and Wnt2b are genes that participate in this pathway, were differentially expressed, and mapped within the QTL interval (Table 3). Wnts encode secreted proteins that bind members of the Frizzled receptor family, which regulates cell proliferation and differentiation (3, 39). Conventional Wnt signaling is mediated by stabilization of β-catenin, which then translocates into the nucleus and interacts with T-cell factor/ lymphoid enhancing factor family to alter gene expression (39). β-Catenin also mediates the interplay of adherence junctions with the actin cytoskeleton through E-cadherin (39). The Wnt/β-catenin pathway has been implicated in playing in a role in renal fibrosis by driving tubular cells to undergo an epithelial-mesenchymal transition (EMT) into activated fibroblasts (53).
Renal fibrosis represents the pathological event most correlated with loss of renal function considering all forms of renal disease (32, 60). So, does proteinuria in itself cause renal fibrosis? An attractive explanation is that albumin being the principal component of ultrafiltered protein during proteinuria causes tubular cell injury. This hypothesis has been supported by several in vitro studies (15, 61). Another more interesting hypothesis involves the translocation of growth factors accompany proteinuria [including insulin-like growth factor 1; hepatocyte growth factor, and transforming growth factor-β (TGF-β)] into the tubular fluid causing activation of tubular cells and recruitment of circulating fibrocytes (24). The “activated” tubular cells secrete chemokines [namely, monocyte chemoat-tract protein-1 (CCL2) and RANTES (CCL5)] that activate macrophages to secrete TGF-β, promoting tubular EMT and producing myofibroblasts (24). These highly differentiated myofibroblasts cells produce extracellular matrix and in coordination with matrix degrading enzymes result in fibrosis.
The gene expression/network analysis, along with the histology data, seems to support a hypothesis by which the congenic experiences reduced kidney damage primarily through controlling fibrosis. While the congenic does have significantly less UPE compared with the S, it still develops proteinuria and accompanying kidney pathology. This is not surprising given that the congenic still contains other S susceptibility alleles at other QTL (on chromosome 1, 6, 8, 9, 13, and 19, which act to increase UPE). Consistent with the above hypothesis, growth factors make it into the tubular fluid of both the S and S.SHR(2) congenic strain. However, the congenic responds differently to these growth factors and associated signaling pathways, attenuating fibrosis, which results in less renal scarring and associated glomerular damage.
RPT was used as an alternative to conventional substitution analysis to reduce the QTL interval. RPT has been suggested as one approach for fine mapping QTL with large and dominant effects (13), both of which are the case for the chromosome 2 QTL (21). Slight variations of this approach (RPT using congenic strains or interval specific congenic strains) have been utilized to substantially narrow QTL regions to small intervals, mostly in mice (8, 9, 17, 31, 40). The main benefit over conventional substitution mapping is rapid development and testing of many recombinant animals and the ability to minimize environmental noise (the phenotypic effect of the recombinant chromosomes is compared with nonrecombinant littermates, instead of between control and experimental groups). A limitation of this approach is that small and/or additive effects may go undetected because the effect of only one allele (heterozygous animals) is evaluated. That is to say, a recombinant family that exhibits no phenotypic difference between genotypes does not necessarily imply it doesn’t contain a QTL. It just means an observable effect may require both alleles to be present.
After one round of RPT, the QTL that contributes the largest and most dominant effect on UPE was significantly reduced to ~1.5 cM, eliminating many of the genes found to be differentially expressed as causative to this QTL. The region still contains many more genes than are feasible to comprehensively evaluate without further work. However, the region does contain several interesting candidate genes, two of which were differentially expressed. These were Sfrp2 and Cct3. SFRPs act as soluble modulators of Wnt signaling, so Sfrp2 could play a role in the proposed QTL mechanism discussed earlier. Cct3 belongs to a protein complex (composed of eight subunits) involved in protein folding of actin and tubulin, suggesting a possible role in maintaining cytoskeleton integrity (6). Additionally, CCT promotes the activation of the anaphase promoting complex, a complex in which inversin (INVS) also interacts (6). Mutations in INVS cause nephronophthisis type 2 (NPHP2), an autosomal recessive form of cystic kidney disease (41). Interestingly, besides kidney cysts, a prominent feature of the disorder is the development of renal interstitial fibrosis (41).
The most interesting gene, Neph1, is structurally related and interacts with Nephrin (NPHS1), a protein that localizes to the slit-diaphragm, and was found to be mutated in congenital nephrotic syndrome type 1 (29). Additionally, mice deficient in Neph1 develop severe proteinuria and die within the first weeks of life (16). Rab25 belongs to a family of small GTPases and are involved in intracellular vesicular transport and trafficking (50). Recently, a null mutation in Rab38 (family member of Rab25) has been shown to be a strong candidate for the development of proteinuria in the fawn-hooded hypertensive rat by potentially altering the tubular reuptake of filtered protein (43). The fact that neither Neph1 nor Rab25 was differentially expressed or contained coding sequence variants between the S and congenic (data not shown) most likely eliminate them being causative to the QTL. Lastly, Iqgap3 belongs to a family of proteins that are integral components of cytoskeletal regulation, which has been linked to E-cadherin-mediated cell-cell adhesion and β-catenin-mediated transcription (4).
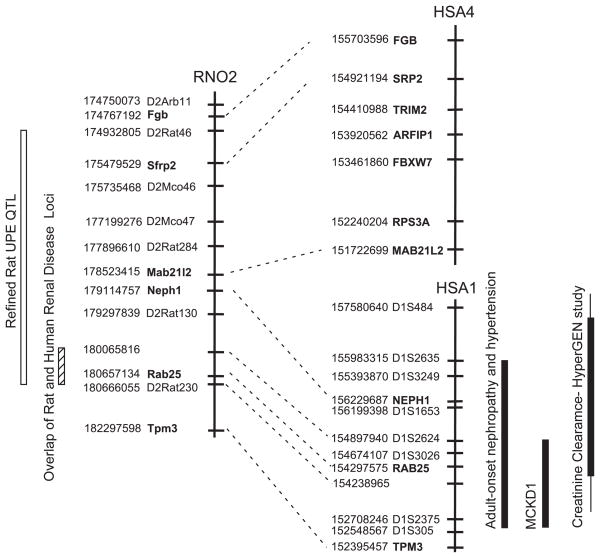
Of importance to the current study is its relevance to human kidney disease. Rat chromosome 2 around the QTL is homologous to both human chromosome 1 and 4 (Fig. 6). The break point (between 178.523 and 179.114 Mb) occurs around the middle of the rat QTL. Interestingly, only human chromosome 1 has been linked to renal disease, whereas human chromosome 4 (homologous region to rat QTL) has not been linked to renal disease. An autosomal dominant form of medullary cystic kidney disease (MCKD1) was mapped to chromosome 1q21 using several kindreds (30, 56, 58). The disease is characterized by tubulointerstitial nephropathy that causes renal salt-wasting and end-stage renal failure (30). Cysts are common but not always present. The presence of proteinuria is variable. Recently, high-resolution haplotype analysis was performed on 16 kindreds with MCKD1 (57). Mutational analysis of 37 genes identified variations in three different genes, including CCT3 (57). Another group mapped an autosomal dominant form of progressive renal failure and hypertension to the same region using a large Israeli family (10). Clinically, all affected members present with hypertension with varying degrees of renal pathology (glomerulosclerosis, interstitial fibrosis, and tubular atrophy). A third study, which conducted a genome scan for renal function among hypertensive individuals (HyperGen study), found linkage of creatinine clearance to the same region on chromosome 1 (14).
Fig. 6.
Comparative map showing overlap of renal susceptibility loci between rat and human. The physical map of the rat QTL is shown on the left. The region in human that is homologous to the rat QTL lies on both human chromosome 1 and 4. Map distances are in base pairs (www.ensembl.org, Ensembl v38, Apr 2006). Adult onset nephropathy and hypertension (10); MCKD1, medullary cystic kidney disease (30, 56, 58); and creatinine clearance (HyperGen study) (14).
The colocalization of multiple renal disease loci clearly demonstrates the importance of this region in renal function. The fact that there are disparate etiologies to these disorders suggests that the gene(s) that underlie each disorder are not the same. However, the possibility does exist that a common gene could underlie all the disorders. Multiple alleles of the “gene” could have a distinct effect on the observed phenotype or the same allele could simply be influenced by the genetic background of the individual, resulting in a distinct disorder. Regardless, colocalization of these renal disease loci provides a means to prioritize the analysis of genes in the QTL for study until subsequent RPT can help delimit the QTL further. So, if one assumes the same gene underlie all the different phenotypes, the QTL can be reduced to 600 Kb containing 18 genes (Fig. 6, Supplemental Table S4).
A limitation of the current study relates to the relatively large size of the congenic interval used to conduct the gene expression analysis. Only two of the 37 differentially expressed genes mapped to the refined QTL. A logical question to ask is to what extent do expression differences in the other 35 differentially expressed genes (or even sequence variants in genes outside the refined QTL) influence the major gene networks observed in this study? That is, are the observed biochemical pathways unrelated to the attenuated renal damage in the S.SHR(2) congenic? Unfortunately, this question can only really be answered once the causative gene is identified and its mechanism understood. So, while the gene expression/ network analysis does provide a clue to a potential mechanism and a starting point for further investigation, the data should be approached with some caution.
We have attempted to use a combined approach using congenic strain analysis (RPT) and gene expression/network analysis to expedite the identification of the gene causative to the QTL on chromosome 2. The concordance of the rat QTL with several human renal disease loci demonstrate that elucidating the causative gene and mechanism of the rat QTL may be of particular importance for understanding human kidney disease.
Supplementary Material
Acknowledgments
The authors thank Dr. Bina Joe for the implantation of the telemetry probes for blood pressure measurements; Dr. David Weaver, director of the genomics core laboratory, for help conducting the Affymetrix Gene chip protocols; and Dr. John Rapp for critical reading of this manuscript.
GRANTS
Support to M. Garrett from National Heart, Lung, and Blood Institute Grant RO1-HL-066998 and from the UTHSC Bioinformatics and Proteomics/ Genomics program is greatly appreciated.
Footnotes
The online version of this article contains supplemental material.
References
- 1.Aitman TJ, Glazier AM, Wallace CA, Cooper LD, Norsworthy PJ, Wahid FN, Al-Majali KM, Trembling PM, Mann CJ, Shoulders CC, Graf D, St Lezin E, Kurtz TW, Kren V, Pravenec M, Ibrahimi A, Abumrad NA, Stanton LW, Scott J. Identification of Cd36 (Fat) as an insulin-resistance gene causing defective fatty acid and glucose metabolism in hypertensive rats. Nat Genet. 1999;21:76–83. doi: 10.1038/5013. [DOI] [PubMed] [Google Scholar]
- 2.Bergman S, Key BO, Kirk KA, Warnock DG, Rostant SG. Kidney disease in the first-degree relatives of African-Americans with hypertensive end-stage renal disease. Am J Kidney Dis. 1996;27:341–346. doi: 10.1016/s0272-6386(96)90356-x. [DOI] [PubMed] [Google Scholar]
- 3.Brembeck FH, Rosario M, Birchmeier W. Balancing cell adhesion and Wnt signaling, the key role of beta-catenin. Curr Opin Genet Dev. 2006;16:51–59. doi: 10.1016/j.gde.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Briggs MW, Sacks DB. IQGAP proteins are integral components of cytoskeletal regulation. EMBO Rep. 2003;4:571–574. doi: 10.1038/sj.embor.embor867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown DM, Provoost AP, Daly MJ, Lander ES, Jacob HJ. Renal disease susceptibility and hypertension are under independent genetic control in the fawn-hooded rat. Nat Genet. 1996;12:44–51. doi: 10.1038/ng0196-44. [DOI] [PubMed] [Google Scholar]
- 6.Camasses A, Bogdanova A, Shevchenko A, Zachariae W. The CCT chaperonin promotes activation of the anaphase-promoting complex through the generation of functional Cdc20. Mol Cell. 2003;12:87–100. doi: 10.1016/s1097-2765(03)00244-2. [DOI] [PubMed] [Google Scholar]
- 7.Chow KM, Wong TY, Li PK. Genetics of common progressive renal disease. Kidney Int Suppl. 2005:s41–s45. doi: 10.1111/j.1523-1755.2005.09409.x. [DOI] [PubMed] [Google Scholar]
- 8.Christians JK, Bingham VK, Oliver FK, Heath TT, Keightley PD. Characterization of a QTL affecting skeletal size in mice. Mamm Genome. 2003;14:175–183. doi: 10.1007/s00335-002-3041-2. [DOI] [PubMed] [Google Scholar]
- 9.Christians JK, Keightley PD. Fine mapping of a murine growth locus to a 1.4-cM region and resolution of linked QTL. Mamm Genome. 2004;15:482–491. doi: 10.1007/s00335-004-3046-0. [DOI] [PubMed] [Google Scholar]
- 10.Cohn DH, Shohat T, Yahav M, Ilan T, Rechavi G, King L, Shohat M. A locus for an autosomal dominant form of progressive renal failure and hypertension at chromosome 1q21. Am J Hum Genet. 2000;67:647–651. doi: 10.1086/303051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dahl LK, Heine M, Tassinari L. Effects of chronic excess salt ingestion: Evidence that genetic factors play an important role in the susceptibility to experimental hypertension. J Exp Med. 1962;115:1173–1190. doi: 10.1084/jem.115.6.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dahl LK, Heine M, Tassinari L. Role of genetic factors in susceptibility to experimental hypertension due to chronic excess salt ingestion. Nature. 1962;194:480–482. doi: 10.1038/194480b0. [DOI] [PubMed] [Google Scholar]
- 13.Darvasi A. Experimental strategies for the genetic dissection of complex traits in animal models. Nat Genet. 1998;18:19–24. doi: 10.1038/ng0198-19. [DOI] [PubMed] [Google Scholar]
- 14.DeWan AT, Arnett DK, Atwood LD, Province MA, Lewis CE, Hunt SC, Eckfeldt J. A genome scan for renal function among hypertensives: the HyperGEN study. Am J Hum Genet. 2001;68:136–144. doi: 10.1086/316927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donadelli R, Abbate M, Zanchi C, Corna D, Tomasoni S, Benigni A, Remuzzi G, Zoja C. Protein traffic activates NF-kB gene signaling and promotes MCP-1-dependent interstitial inflammation. Am J Kidney Dis. 2000;36:1226–1241. doi: 10.1053/ajkd.2000.19838. [DOI] [PubMed] [Google Scholar]
- 16.Donoviel DB, Freed DD, Vogel H, Potter DG, Hawkins E, Barrish JP, Mathur BN, Turner CA, Geske R, Montgomery CA, Starbuck M, Brandt M, Gupta A, Ramirez-Solis R, Zambrowicz BP, Powell DR. Proteinuria and perinatal lethality in mice lacking NEPH1, a novel protein with homology to NEPHRIN. Mol Cell Biol. 2001;21:4829–4836. doi: 10.1128/MCB.21.14.4829-4836.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fehr C, Shirley RL, Belknap JK, Crabbe JC, Buck KJ. Congenic mapping of alcohol and pentobarbital withdrawal liability loci to a <1 centimorgan interval or murine chromosome 4: identification of Mpdz as a candidate gene. J Neurosci. 2002;22:3730–3738. doi: 10.1523/JNEUROSCI.22-09-03730.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferguson R, Grim CE, Opgenorth TJ. A familial risk of chronic renal failure among blacks on dialysis? J Clin Epidemiol. 1988;41:1189–1196. doi: 10.1016/0895-4356(88)90023-6. [DOI] [PubMed] [Google Scholar]
- 19.Freedman BI, Spray BJ, Tuttle AB, Buckalew VM., Jr The familial risk of end-stage renal disease in African Americans. Am J Kidney Dis. 1993;21:387–393. doi: 10.1016/s0272-6386(12)80266-6. [DOI] [PubMed] [Google Scholar]
- 20.Garrett MR, Dene H, Rapp JP. Time-course genetic analysis of albuminuria in Dahl salt-sensitive rats on low-salt diet. J Am Soc Nephrol. 2003;14:1175–1187. doi: 10.1097/01.asn.0000060572.13794.58. [DOI] [PubMed] [Google Scholar]
- 21.Garrett MR, Joe B, Yerga-Woolwine S. Genetic linkage of urinary albumin excretion in Dahl salt-sensitive rats: influence of dietary salt and confirmation using congenic strains. Physiol Genomics. 2006;25:39–49. doi: 10.1152/physiolgenomics.00150.2005. [DOI] [PubMed] [Google Scholar]
- 22.Garrett MR, Meng H, Rapp JP, Joe B. Locating a blood pressure quantitative trait locus within 117 kb on the rat genome: substitution mapping and renal expression analysis. Hypertension. 2005;45:451–459. doi: 10.1161/01.HYP.0000154678.64340.7f. [DOI] [PubMed] [Google Scholar]
- 23.Hampton JA, Bernardo DA, Khan NA, Lacher DA, Rapp JP, Gohara AF, Goldblatt PJ. Morphometric evaluation of the renal arterial system of Dahl salt-sensitive and salt-resistant rats on a high salt diet. Lab Invest. 1989;60:839–846. [PubMed] [Google Scholar]
- 24.Hirschberg R, Wang S. Proteinuria and growth factors in the development of tubulointerstitial injury and scarring in kidney disease. Curr Opin Nephrol Hypertens. 2005;14:43–52. doi: 10.1097/00041552-200501000-00008. [DOI] [PubMed] [Google Scholar]
- 25.Hubner N, Wallace CA, Zimdahl H, Petretto E, Schulz H, Maciver F, Mueller M, Hummel O, Monti J, Zidek V, Musilova A, Kren V, Causton H, Game L, Born G, Schmidt S, Muller A, Cook SA, Kurtz TW, Whittaker J, Pravenec M, Aitman TJ. Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat Genet. 2005;37:243–253. doi: 10.1038/ng1522. [DOI] [PubMed] [Google Scholar]
- 26.Joe B, Garrett MR, Dene H, Rapp JP. Substitution mapping of a blood pressure quantitative trait locus to a 2.73 Mb region on rat chromosome 1. J Hypertens. 2003;21:2077–2084. doi: 10.1097/00004872-200311000-00017. [DOI] [PubMed] [Google Scholar]
- 27.Joe B, Letwin NE, Garrett MR, Dhindaw S, Frank B, Sultana R, Verratti K, Rapp JP, Lee NH. Transcriptional profiling with a blood pressure QTL interval-specific oligonucleotide array. Physiol Genomics. 2005;23:318–326. doi: 10.1152/physiolgenomics.00164.2004. [DOI] [PubMed] [Google Scholar]
- 28.Karp CL, Grupe A, Schadt E, Ewart SL, Keane-Moore M, Cuomo PJ, Kohl J, Wahl L, Kuperman D, Germer S, Aud D, Peltz G, Wills-Karp M. Identification of complement factor 5 as a susceptibility locus for experimental allergic asthma. Nat Immun. 2000;1:221–226. doi: 10.1038/79759. [DOI] [PubMed] [Google Scholar]
- 29.Kestila M, Lenkkeri U, Mannikko M, Lamerdin J, McCready P, Putaala H, Ruotsalainen V, Morita T, Nissinen M, Herva R, Kashtan CE, Peltonen L, Holmberg C, Olsen A, Tryggvason K. Positionally cloned gene for a novel glomerular protein–nephrin–is mutated in congenital nephrotic syndrome. Mol Cell. 1998;1:575–582. doi: 10.1016/s1097-2765(00)80057-x. [DOI] [PubMed] [Google Scholar]
- 30.Kiser RL, Wolf MT, Martin JL, Zalewski I, Attanasio M, Hildebrandt F, Klemmer P. Medullary cystic kidney disease type 1 in a large Native-American kindred. Am J Kidney Dis. 2004;44:611–617. [PubMed] [Google Scholar]
- 31.Liu X, Oliver F, Brown SD, Denny P, Keightley PD. High-resolution quantitative trait locus mapping for body weight in mice by recombinant progeny testing. Genet Res. 2001;77:191–197. doi: 10.1017/s0016672301004943. [DOI] [PubMed] [Google Scholar]
- 32.Liu Y. Renal fibrosis: new insights into the pathogenesis and therapeutics. Kidney Int. 2006;69:213–217. doi: 10.1038/sj.ki.5000054. [DOI] [PubMed] [Google Scholar]
- 33.Malek RL, Wang HY, Kwitek AE, Greene AS, Bhagabati N, Borchardt G, Cahill L, Currier T, Frank B, Fu X, Hasinoff M, Howe E, Letwin N, Luu TV, Saeed A, Sajadi H, Salzberg SL, Sultana R, Thiagarajan M, Tsai J, Veratti K, White J, Quackenbush J, Jacob HJ, Lee NH. Physiogenomic resources for rat models of heart, lung and blood disorders. Nat Genet. 2006;38:234–239. doi: 10.1038/ng1693. [DOI] [PubMed] [Google Scholar]
- 34.Manly KF, Cudmore RH, Jr, Meer JM. Map Manager QTX, cross-platform software for genetic mapping. Mamm Genome. 2001;12:930–932. doi: 10.1007/s00335-001-1016-3. [DOI] [PubMed] [Google Scholar]
- 35.Matsuyama M, Ogiu T, Kontani K, Yamada C, Kawai M, Hiai H, Nakamura T, Shimizu F, Toyokawa T, Kinoshita Y. Genetic regulation of the development of glomerular sclerotic lesions in the BUF/Mna rat. Nephron. 1990;54:334–337. doi: 10.1159/000185890. [DOI] [PubMed] [Google Scholar]
- 36.McBride MW, Carr FJ, Graham D, Anderson NH, Clark JS, Lee WK, Charchar FJ, Brosnan MJ, Dominiczak AF. Microarray analysis of rat chromosome 2 congenic strains. Hypertension. 2003;41:847–853. doi: 10.1161/01.HYP.0000047103.07205.03. [DOI] [PubMed] [Google Scholar]
- 37.Murayama S, Yagyu S, Higo K, Ye C, Mizuno T, Oyabu A, Ito M, Morita H, Maeda K, Serikawa T, Matsuyama M. A genetic locus susceptible to the overt proteinuria in BUF/Mna rat. Mamm Genome. 1998;9:886–888. doi: 10.1007/s003359900888. [DOI] [PubMed] [Google Scholar]
- 38.National Kidney Foundation. K/DOQI clinical practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Am J Kidney Dis. 2002;39:S1–S266. [PubMed] [Google Scholar]
- 39.Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oliver F, Christians JK, Liu X, Rhind S, Verma V, Davison C, Brown SD, Denny P, Keightley PD. Regulatory variation at glypican-3 underlies a major growth QTL in mice. PLoS Biol. 2005;3:e135. doi: 10.1371/journal.pbio.0030135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otto EA, Schermer B, Obara T, O’Toole JF, Hiller KS, Mueller AM, Ruf RG, Hoefele J, Beekmann F, Landau D, Foreman JW, Goodship JA, Strachan T, Kispert A, Wolf MT, Gagnadoux MF, Nivet H, Antignac C, Walz G, Drummond IA, Benzing T, Hildebrandt F. Mutations in INVS encoding inversin cause nephronophthisis type 2, linking renal cystic disease to the function of primary cilia and left-right axis determination. Nat Genet. 2003;34:413–420. doi: 10.1038/ng1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poyan Mehr A, Siegel AK, Kossmehl P, Schulz A, Plehm R, de Bruijn JA, de Heer E, Kreutz R. Early onset albuminuria in Dahl rats is a polygenetic trait that is independent from salt loading. Physiol Genomics. 2003;14:209–216. doi: 10.1152/physiolgenomics.00053.2003. [DOI] [PubMed] [Google Scholar]
- 43.Rangel-Filho A, Sharma M, Datta YH, Moreno C, Roman RJ, Iwamoto Y, Provoost AP, Lazar J, Jacob HJ. RF-2 gene modulates proteinuria and albuminuria independently of changes in glomerular permeability in the fawn-hooded hypertensive rat. J Am Soc Nephrol. 2005;16:852–856. doi: 10.1681/ASN.2005010029. [DOI] [PubMed] [Google Scholar]
- 44.Rozzo SJ, Allard JD, Choubey D, Vyse TJ, Izui S, Peltz G, Kotzin BL. Evidence for an interferon-inducible gene, Ifi202, in the susceptibility to systemic lupus. Immunity. 2001;15:435–443. doi: 10.1016/s1074-7613(01)00196-0. [DOI] [PubMed] [Google Scholar]
- 45.Schulz A, Litfin A, Kossmehl P, Kreutz R. Genetic dissection of increased urinary albumin excretion in the munich wistar fromter rat. J Am Soc Nephrol. 2002;13:2706–2714. doi: 10.1097/01.asn.0000031803.55613.86. [DOI] [PubMed] [Google Scholar]
- 46.Schulz A, Standke D, Kovacevic L, Mostler M, Kossmehl P, Stoll M, Kreutz R. A major gene locus links early onset albuminuria with renal interstitial fibrosis in the MWF rat with polygenetic albuminuria. J Am Soc Nephrol. 2003;14:3081–3089. doi: 10.1097/01.asn.0000100126.62370.25. [DOI] [PubMed] [Google Scholar]
- 47.Shiozawa M, Provoost AP, van Dokkum RP, Majewski RR, Jacob HJ. Evidence of gene-gene interactions in the genetic susceptibility to renal impairment after unilateral nephrectomy. J Am Soc Nephrol. 2000;11:2068–2078. doi: 10.1681/ASN.V11112068. [DOI] [PubMed] [Google Scholar]
- 48.Siegel AK, Kossmehl P, Planert M, Schulz A, Wehland M, Stoll M, Bruijn JA, de Heer E, Kreutz R. Genetic linkage of albuminuria and renal injury in Dahl salt-sensitive rats on a high-salt diet: comparison with spontaneously hypertensive rats. Physiol Genomics. 2004;18:218–225. doi: 10.1152/physiolgenomics.00068.2004. [DOI] [PubMed] [Google Scholar]
- 49.Spray BJ, Atassi NG, Tuttle AB, Freedman BI. Familial risk, age at onset, and cause of end-stage renal disease in white Americans. J Am Soc Nephrol. 1995;5:1806–1810. doi: 10.1681/ASN.V5101806. [DOI] [PubMed] [Google Scholar]
- 50.Stein MP, Dong J, Wandinger-Ness A. Rab proteins and endocytic trafficking: potential targets for therapeutic intervention. Adv Drug Delivery Res. 2003;55:1421–1437. doi: 10.1016/j.addr.2003.07.009. [DOI] [PubMed] [Google Scholar]
- 51.Stella P, Cusi D, Duzzi L, Bianchi G. Relations between hypertension and glomerulosclerosis in first-generation hybrid rats of the Milan strains. Child Nephrol Urol. 1991;11:6–9. [PubMed] [Google Scholar]
- 52.Sterzel RB, Luft FC, Gao Y, Schnermann J, Briggs JP, Ganten D, Waldherr R, Schnabel E, Kriz W. Renal disease and the development of hypertension in salt-sensitive Dahl rats. Kidney Int. 1988;33:1119–1129. doi: 10.1038/ki.1988.120. [DOI] [PubMed] [Google Scholar]
- 53.Surendran K, Schiavi S, Hruska KA. Wnt-dependent beta-catenin signaling is activated after unilateral ureteral obstruction, and recombinant secreted frizzled-related protein 4 alters the progression of renal fibrosis. J Am Soc Nephrol. 2005;16:2373–2384. doi: 10.1681/ASN.2004110949. [DOI] [PubMed] [Google Scholar]
- 54.Tylicki L, Rutkowski B, Horl WH. Multifactorial determination of hypertensive nephroangiosclerosis. Kidney Blood Press Res. 2002;25:341–353. doi: 10.1159/000068698. [DOI] [PubMed] [Google Scholar]
- 55.United States Renal Data System. United States Renal Data System 2003 Annual Data Report. National Institute of Diabetes and Digestive and Kidney Disease; 2003. ( www.usrdr.org) [Google Scholar]
- 56.Wolf MT, Karle SM, Schwarz S, Anlauf M, Glaeser L, Kroiss S, Burton C, Feest T, Otto E, Fuchshuber A, Hildebrandt F. Refinement of the critical region for MCKD1 by detection of transcontinental haplotype sharing. Kidney Int. 2003;64:788–792. doi: 10.1046/j.1523-1755.2003.00161.x. [DOI] [PubMed] [Google Scholar]
- 57.Wolf MT, Mucha BE, Hennies HC, Attanasio M, Panther F, Zalewski I, Karle SM, Otto EA, Deltas CC, Fuchshuber A, Hildebrandt F. Medullary cystic kidney disease type 1: mutational analysis in 37 genes based on haplotype sharing. Hum Genet. 2006;119:649–58. doi: 10.1007/s00439-006-0176-3. [DOI] [PubMed] [Google Scholar]
- 58.Wolf MT, van Vlem B, Hennies HC, Zalewski I, Karle SM, Puetz M, Panther F, Otto E, Fuchshuber A, Lameire N, Loeys B, Hildebrandt F. Telomeric refinement of the MCKD1 locus on chromosome 1q21. Kidney Int. 2004;66:580–585. doi: 10.1111/j.1523-1755.2004.00799.x. [DOI] [PubMed] [Google Scholar]
- 59.Yagil C, Hubner N, Monti J, Schulz H, Sapojnikov M, Luft FC, Ganten D, Yagil Y. Identification of hypertension-related genes through an integrated genomic-transcriptomic approach. Circ Res. 2005;96:617–625. doi: 10.1161/01.RES.0000160556.52369.61. [DOI] [PubMed] [Google Scholar]
- 60.Zeisberg M, Strutz F, Muller GA. Renal fibrosis: an update. Curr Opin Nephrol Hypertens. 2001;10:315–320. doi: 10.1097/00041552-200105000-00004. [DOI] [PubMed] [Google Scholar]
- 61.Zoja C, Morigi M, Remuzzi G. Proteinuria and phenotypic change of proximal tubular cells. J Am Soc Nephrol. 2003;14(Suppl 1):S36–S41. doi: 10.1097/01.asn.0000068626.23485.e0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.