Abstract
Heme Nitric oxide and/or OXygen binding (H-NOX) domains have provided a novel scaffold to probe ligand affinity in hemoproteins. Mutation of isoleucine 5, a conserved residue located in the heme-binding pocket of the H-NOX domain from Thermoanaerobacter tengcongensis (Tt H-NOX), was carried out to examine changes in oxygen (O2)-binding properties. A series of I5 mutants (I5F, I5F/I75F, I5F/L144F, I5F/I75F/L144F) was investigated to probe the role of steric bulk within the heme pocket. The mutations significantly increased O2 association rates (1.5–2.5 fold) and dissociation rates (8–190 fold) as compared to wild type Tt H-NOX. Structural changes that accompanied the I5F mutation were characterized using X-ray crystallography and resonance Raman spectroscopy. A 1.67 Å crystal structure of the I5F mutant indicated that introducing a phenylalanine at position 5 resulted in a significant shift of the N-terminal domain of the protein, causing an opening of the heme pocket. This movement also resulted in an increased amount of flexibility at the N-terminus and the loop covering the N-terminal helix as indicated by the two conformations of the first six N-terminal amino acids, high B-factors in this region of the protein, and partially discontinuous electron density. In addition, introduction of a phenylalanine at position 5 resulted in increased flexibility of the heme within the pocket and weakened hydrogen bonding to the bound O2 as measured by resonance Raman spectroscopy. This study provides insight into the critical role of I5 in controlling conformational flexibility and ligand affinity in H-NOX proteins.
Hemoproteins are involved in many crucial cellular processes, including oxidative transformations (P450s), O2 delivery and transport [myoglobin (Mb), hemoglobin (Hb)], and diatomic gas signaling [soluble guanylate cyclase (sGC), CooA]. Attempts to understand the factors that control reactivity versus reversible ligand binding in heme proteins have been the subject of intense study for many decades. While some causes of differences in reactivity are more straightforward, such as thiolate- versus histidyl-ligated hemes, the method by which a protein controls the reactivity of the iron and reversible ligand binding is not fully understood. The majority of the previous work undertaken to elucidate factors responsible for ligand affinity and selectivity in reversible O2-binding heme proteins has been performed on proteins with a globin fold, primarily myoglobin (Mb) (1, 2) and hemoglobin (Hb) (3, 4). These studies on the globins have provided a wealth of information as to how the globin fold modulates ligand affinity and O2 reactivity. Through this work, it has become apparent that many factors affect ligand affinity, including the distal ligand, heme pocket polarity, off-heme binding sites, and iron-histidine bond strength (1, 2, 5–7). Understanding all of the factors involved in controlling ligand binding within heme proteins should allow for the rational engineering of novel proteins for a variety of biotechnological and biomedical applications. However, whether the factors controlling O2 binding within globin domains are specific to this fold or are universal to all heme protein folds has only begun to be investigated.
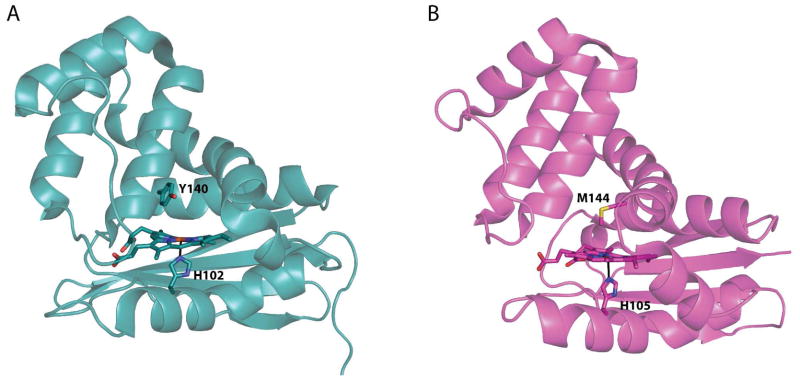
The recent discovery of a new class of heme proteins, termed Heme Nitric oxide/OXygen (H-NOX) binding domains (8, 9), has provided additional insight into how heme proteins control ligand affinity. Soluble guanylate cyclase (sGC), the mammalian receptor for nitric oxide (NO), which regulates functions as diverse as vasodilation, neuropotentiation, and egg fertilization, contains an H-NOX domain that slectively senses NO (10). To date, H-NOX domains from mammalian sGCs and aerobic bacteria do not form stable complexes with O2, either by limiting access of O2 to the heme or by tuning the heme pocket such that O2 binding is extremely unfavorable (11–13). In contrast, H-NOX domains characterized from anaerobic bacteria, such as T. tengcongensis, have been found to tightly bind O2 (Kd ~48 nM), [(11); EEW and MAM, unpublished data]. Structures of both O2- and non-O2-binding H-NOX proteins exhibit the same protein fold (Figure 1, RMSD 1.88 Å, (14, 15)), making this class of proteins ideal for dissecting the factors that allow a single protein fold to exhibit widely varying ligand affinities and specificities. In addition, as Tt H-NOX is amenable to mutagenesis (11, 16–18) and exhibits a fold that is structurally distinct from the globins, it provides a useful model for probing the effects of various mutations on ligand affinity and reactivity, as well as identifying factors that are independent of protein fold.
Figure 1.
Structures of the (A) O2-binding WT Tt H-NOX (1U55, chain A) in teal (15) and (B) a non-O2-binding H-NOX from Nostoc in magenta (14). Hemes and coordinating histidines as well as the distal pocket residue about the heme are shown in sticks. A global alignment of these two structures results in an RMSD of 1.88 Å (using Cα − Cα).
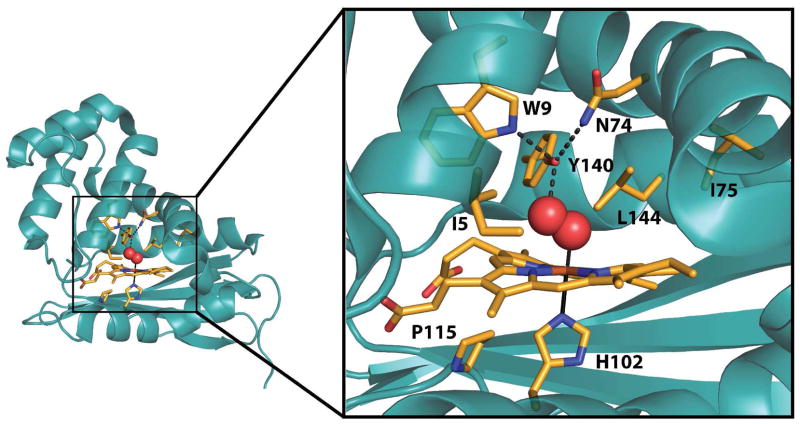
Previous mutational studies on H-NOX domains (11, 16, 18) have found that the presence of hydrogen bonding residues within the distal pocket is a major determinant of O2-binding (Figure 2) (15). In Tt H-NOX, the hydrogen-bonding triad consists of Y140, which directly forms a hydrogen bond to the bound O2, as well as W9 and N74, which orient Y140 within the binding pocket via H-bonds. Removal of Y140 results in diminished O2 affinity (Y140L), while mutation of both Y140 and W9 results in a Tt H-NOX mutant with no measurable O2 binding (Y140L/W9F) (11). Conversely, introduction of hydrogen bond donors into the distal heme pocket of full length sGC does not result in stable O2 binding (19, 20), suggesting that there are factors in addition to hydrogen bonding involved in controlling reversible O2 binding in H-NOX domains.
Figure 2.
Key residue sidechains and the heme within the Tt H-NOX heme pocket are shown in stick representation with carbon atoms in orange, oxygens in red, and nitrogens in blue using chain A of PDB 1U55 (15).
Another feature of H-NOX domains that affects ligand affinity is the highly distorted heme cofactor (15). Flattening the heme, through mutation of P115 (P115A), resulted in increased O2 affinity (16). This increased affinity was attributed to a change in the tilt of the proximal histidine, allowing for increased orbital overlap between the O2 and heme iron, thereby strengthening the Fe-O2 bond (16). In addition, introduction of steric bulk in the form of phenylalanine residues to the heme distal pocket of Tt H-NOX, analogous to the phenylalanine found within the sGC heme pocket (2), was found to rearrange the heme pocket (I75F/L144F) (18). Interestingly, the heme in the I75F/L144F mutant was found to be flatter than in WT Tt H-NOX, similar to the P115A mutant; however, unlike the P115A mutant, the oxygen affinity decreased in the I75F/L144F mutant (18). Thus, many factors control ligand affinity and selectivity within the H-NOX family, and further studies are necessary to fully understand how each of these factors contribute to produce a finely-tuned O2 binding protein.
In this work, we examined the effect of increased steric bulk and structural flexibility in the distal pocket of Tt H-NOX on O2-binding properties and kinetics. The I5 position was chosen for mutagenesis because a medium-sized, hydrophobic amino acid (I, L, V) is highly conserved and within van der Waals contact with the heme (Figure 2), suggesting an important role in controlling ligand affinity and protein structure. Changes in protein structure and ligand binding were investigated in a panel of I5 mutations using time-resolved UV–visible spectroscopy, X-ray crystallography, and steady–state resonance Raman spectroscopy. In addition, the interplay between the I5F mutation and the previously described I75F, L144F, and I75F/L144F mutations was investigated. Together these data point to the critical role of I5 in controlling flexibility and ligand affinity within the H-NOX family.
Materials and Methods
Materials and general methods
Unless otherwise noted, all reagents were purchased in the highest available purity and used as received.
Protein expression and purification
Expressions and purifications of H-NOX proteins were performed as previously described (18). Briefly, cultures were grown at 37 °C to an OD600 of 0.6–1 in media (45 g yeast extract, 1.6 g KH2PO4, 11.5 g K2HPO4 · 3H2O, 1.3% glycerol per 1 L) and cooled to 18 °C prior to induction. Isopropyl β -D-thiogalactopyranoside (Research Products International Corp.) was added to 10 μM and aminolevulinic acid (Cosmo Bio Co. Ltd.) was added to 1 mM. Cultures were grown overnight for 18–24 h and then harvested. Cells were lysed using a homogenizer in 50 mM TEA (pH 7.5), 300 mM NaCl, 10 mM imidazole (Buffer A) with Pefabloc (Centerchem Inc.) and benzamidine (Sigma Aldrich) added to 1 mM. Lysates were heat denatured at 70 °C for 40 min and then pelleted at 130,000 x g. Supernatants were applied to HisPure columns (Pierce) pre-equilibrated with Buffer A. The protein-loaded columns were then washed with 25–30 column volumes of Buffer A. H-NOX proteins were eluted in Buffer A containing 150 mM imidazole and concentrated to < 2 mL. The proteins were then desalted into 50 mM TEA (pH 7.5), 20 mM NaCl, 5% glycerol (Buffer B) using PD10 columns (GE Lifesciences) and stored at −80 °C.
UV–visible spectroscopy
All spectra were recorded on either a Cary 3E spectrophotometer equipped with a Neslab RTE-100 constant temperature bath or a Cary 300Bio spectrophotometer equipped with a Peltier accessory. Preparation of complexes was carried out in Buffer B as previously described (9, 11, 18).
O2 dissociation rate
O2 dissociation rate experiments were performed as previously described with the following modifications (11, 18). Protein and dithionite traps were prepared in Buffer B and saturating CO was not used as part of the trap due to a rate dependence on CO concentration. Dithionite concentration did not affect the O2 dissociation rate. The dissociation of O2 from the heme was monitored and fit globally using SpecFit32 (HiTech Scientific) (11, 18).
O2 association rate
O2 association to the heme was observed using flash photolysis and transient absorption spectroscopy as previously described (18). Briefly, the FeII–CO complex was generated in an anaerobic chamber, as described in an earlier publication (11), and diluted to ~8 μM with anaerobic buffer in a 1 cm pathlength quartz cuvette. The Fe–CO bond was photolyzed by excitation with 8 ns pulses of 532 nm from a doubled Nd:YAG laser, using an apparatus described elsewhere (21). The cuvette was opened to air and stirred with a steady stream of air blown into the mouth of the cuvette to fully saturate the solution for measurement of the O2 association rates. Spectra were taken at ~20 °C. Data points were collected at a rate of 1× 109 s−1 using a LeCroy digital oscilloscope.
Purification of untagged I5F TT H-NOX for crystallization
Non-His6-tagged I5F Tt H-NOX was expressed as described above. Cells were resuspended in 50mM TEA (pH 7.5), 50 mM NaCl and were lysed by passage through a homogenizer at 10,000 psi. The lysate was heat-denatured at 70ºC for 40 min and pelleted at 130,000x g. The supernatant was passed over a SuperQ-650M anion-exchange column (Toyopearl) and the flow-through was collected. The flow-through was buffer exchanged into 50 mM HEPES (pH 6.5), 5 % glycerol using a G25 size exclusion column and then loaded onto a CM-650M cation-exchange column (Toyopearl). Protein was eluted using a linear gradient from 0–500 mM NaCl. Fractions containing the highest purity H-NOX protein were pooled and passed over a S75 16/60 size-exclusion column (Pharmacia) that had been equilibrated with buffer B. Only fractions containing H-NOX protein with a ratio of A417nm/A280nm > 1.6 were pooled, concentrated, and flash-frozen in liquid nitrogen for storage at −80 °C.
Crystal Structure of I5F TT H-NOX
As-purified non-His tagged I5F Tt H-NOX protein was thawed, buffer exchanged into 20 mM TEA (pH 7.5) using a PD-10 column and then concentrated to 26 mg/mL. The protein was crystallized via sitting drop vaper diffusion by mixing 1 μL of protein with 1 μL of precipitant solution (0.2 M Ammonium sulfate, 20% PEG 3350) and equilibrating the drop against 400 μL of precipitant. Crystals formed within two days, were cryoprotected by a brief soak in well solution containing 25 % ethylene glycol, and then frozen in liquid nitrogen for storage.
X-ray diffraction data were collected at the Advanced Light Source (ALS) at Lawrence Berkeley National Laboratory (Berkeley, CA) on beamline 5.0.1 (λ = 0.977408 Å). Crystals diffracted to 1.67 Å and data were processed in HKL2000 (22) with space group P21.
The protein crystallized with two protein monomers in the asymmetric unit. The protein component (heme and water molecules removed) of the WT structure (PDB ID 1U55) was used for molecular replacement in PHASER (23). The resulting structure was refined in Phenix (24) against 1.67 Å data with alternate cycles of positional and ADP refinement with manual refitting in Coot (25). A TLS model was incorporated towards the end of refinement. The final model contains 2 chains with 185 residues in chain A and 183 residues in chain B. 153 water molecules and and two heme molecules were also modeled to produce the final refinement parameters of Rwork of 19.7 % and Rfree of 24.7 %.
Resonance Raman Spectroscopy
FeII–O2 Tt H-NOX samples were prepared and all spectra were acquired as previously described (9, 26).
Results and Discussion
Spectroscopic and Kinetic Effects of the I5F Mutation
To determine how mutation of I5 affects ligand affinity in Tt H-NOX, the O2 binding properties of the leucine and phenylalanine mutants were investigated with steady–state and time-resolved UV–visible spectroscopy. In addition to the I5L and I5F single mutants, the I5F mutation was introduced in combination with the previously described I75F, L144F, and I75F/L144F mutants (26) to probe the role of distal pocket steric bulk. This resulted in the following panel of I5F Tt H-NOX mutants: I5F, I5F/I75F, I5F/L144F, and I5F/I75F/L144F. The steady–state UV–visible spectra of the mutants were very similar to the wild type protein. As the number of phenylalanine residues within the distal pocket increased, a small shift toward longer wavelengths was observed in the Soret absorbance maximum, with a concomitant shift of the α/β bands toward shorter wavelengths, which is potentially due to a structural change within the heme pocket. In addition, a mixture of 5- and 6-coordinate FeII–NO complexes was observed for most of the mutants, as was previously described for the I75F, L144F, and I75F/L144F mutants (Table S1). However, two exceptions to this observed 5/6-coordinate mixture were the I5L (26) and I5F single mutants, which were exclusively 6-coordinate. This suggests that changes to the heme electronics and structure differ when steric bulk is added at the I5 versus the I75 and L144 positions, which are on the opposite side of the Tt H-NOX heme pocket (Figure 2).
The kinetics of O2 binding were investigated to determine the effect of adding steric bulk at the I5 position, as well as the effect of additional bulk at positions 75 and 144. It was previously found that introduction of phenylalanine residues at positions 75 and 144 resulted in weaker O2 affinities (18). The I75F, L144F, and I75F/L144F mutants all exhibited faster O2 dissociation rates, while the L144F and I75F/L144F mutations resulted in slower O2 association rates. This had a net effect of significantly decreasing the O2 affinities (10–230 fold) for all of the I75 and L144 mutants (Table 1).
Table 1.
Kinetic constants for ligand binding to Tt H-NOX domains.
| Tt H-NOX | Kd O2 (nM) | koff O2 (s−1) | kon O2 (μM−1s−1) |
|---|---|---|---|
| WT | 48 ± 5 | 1.20 ± 0.02a,b | 25 ± 3 |
| I75F | 497 ± 16b | 11.19 ± 0.12b | 22.5 ± 0.7b |
| L144F | 2360 ± 50b | 16.06 ± 0.21b | 6.8 ± 0.1b |
| I75F/L144F | 11150 ± 330b | 45.7 ± 0.9b | 4.1 ± 0.1b |
| I5L | 413 ± 33 | 9.50 ± 0.64 | 23 ± 1 |
| I5F | 1570 ± 50 | 61.3 ± 1.5 | 39.0 ± 0.5 |
| I5F/I75F | 3730 ± 150 | 233 ± 10 | 62.5 ± 0.1 |
| I5F/L144F | 3750 ± 110 | 39.0 ± 0.8 | 10.4 ± 0.2 |
| I5F/I75F/L144F | 3160 ± 380 | 60.9 ±7.1 | 19.3 ± 0.2 |
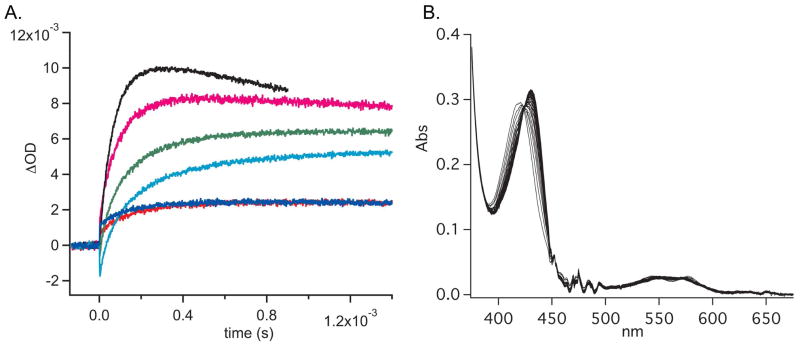
In the present work, the conservative I5L mutation did not affect the O2 association rate, (23 μM−1s−1, compared to 22 μM−1s−1 for WT); however the O2 dissociation rate increased to 9.5 s−1, as compared to 1.20 s−1 for WT (Table 1, Figure 3). Upon introduction of the bulkier I5F mutation, the O2 association and dissociation rates increased to 39 μM−1s−1 and 61.3 s−1, respectively (Table 1, Figure 3). An O2 dissociation rate of 61.3 s−1 is extremely fast for Tt H-NOX, suggesting that these mutations may open the heme pocket to increase both ligand accessibility and dissociation. A more accessible and/or flexibile heme pocket is also consistent with the NO dissociation rate data (Table S2). The I5F mutant was the only mutant to exhibit mono-exponential NO dissociation rates. This may be due to altered dynamics of the I5F mutant that allows more facile entry and exit of ligands from the heme pocket or through cavities within the protein (M. Winter, M. Herzik, MAM, unpublished results).
Figure 3.
Laser photolysis traces of the photolysis of the FeII-CO complex followed by binding of O2 (A) and representative stopped flow kinetic data (B). WT in red; I5L in blue; I5F/L144F in teal; I5F/I75F/L144F in green; I5F in magenta; I5F/I75F in black. Laser photolysis data were analyzed using Igor Pro. Stopped flow data were analyzed using SpecFit32.
In addition to the single I5 mutations, the panel of double and triple mutants was also investigated to determine how increased bulk at both ends of the distal pocket affects O2 binding kinetics. The I5F mutation was found to synergize with the I75F, L144F, and I75F/L144F mutants – in each case resulting in faster O2 association and dissociation rates – when compared to the parent proteins. Of these double and triple mutants, the I5F/I75F mutant was found to have by far the greatest changes in O2 kinetics, exhibiting the fastest O2 association and dissociation rates measured for an H-NOX protein thus far (62.5 μM−1s−1 and 233 s−1, respectively, Table 1 and Figure 3). These rates correspond to a 3-fold increase in association rate and a 195-fold increase in dissociation rate for O2 as compared to WT Tt H-NOX. Therefore it is likely that the changes to the protein flexibility and/or structure differed between the double mutants, resulting in the varied ligand binding kinetics.
X-ray Crystal Structure of the I5F Mutant
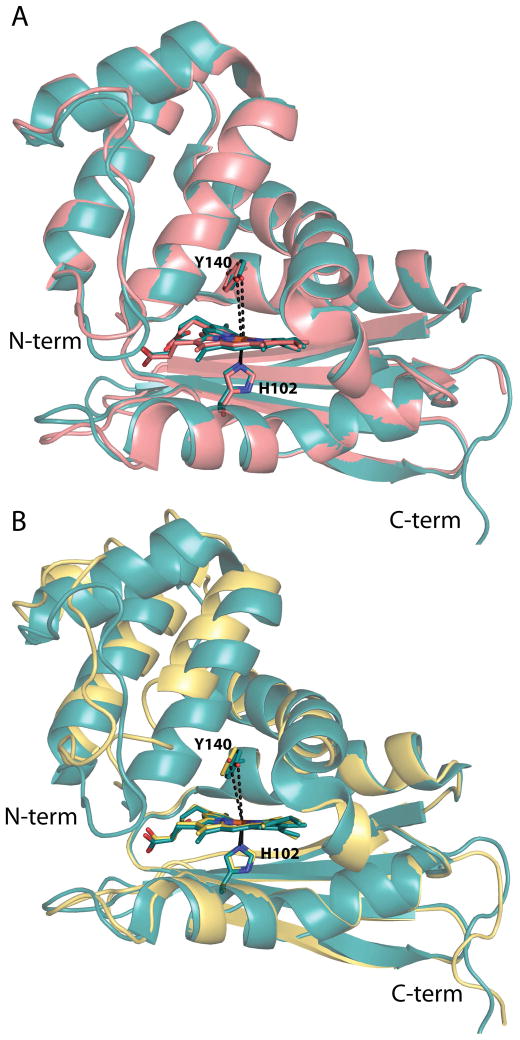
A structure of the conservative I5L mutant has previously been described and only small differences were found when compared with the WT Tt H-NOX structure with a global Cα-Cα RMSD of 0.34 Å (Figure 4A) (17). A slight rotation of the N-terminal subdomain relative to the C-terminal subdomain was observed, as was a small relaxation of the heme distortion. These differences between the WT and I5L structures are likely the cause of the modest changes in the O2 binding kinetics.
Figure 4.
Structural alignments of the WT Tt H-NOX structure with the I5L and I5F mutant structures. (A) Global alignment of WT (teal, PDB ID 1U55) (15) and I5L (salmon, PDB ID 3NVR) (17) Tt H-NOX structures; RMSD 0.34 Å. The Y140–Fe distances are shown in dashed black lines and are 5.0 and 5.1 Å in the WT and I5L structures, respectively. (B) Overlays of the WT (teal, PDB ID 1U55) (15) and I5F (yellow) mutant structures shown following alignment of the last 80 amino acids (C-terminus) of the proteins; RMSD 1.13 Å. The Y140–Fe distances are shown in black dashed lines with the I5F distance being 5.6 Å. H102, Y140, and the heme are shown in sticks for orientation.
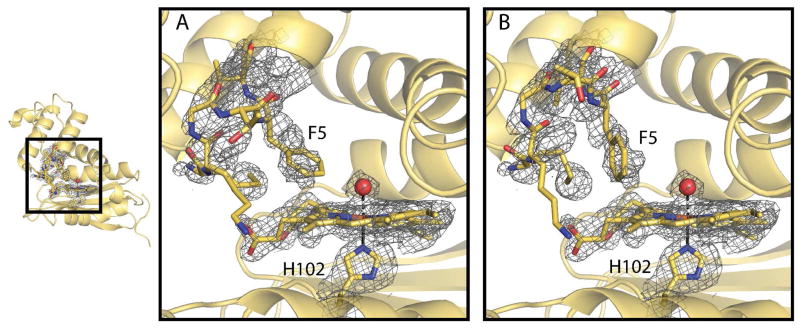
The structure of the sterically bulky I5F mutant was solved to investigate how the mutation caused the markedly increased O2 binding kinetics. A 1.67 Å resolution crystal structure of the mutant indicates that the overall structure of the I5F mutant retains the H-NOX fold (Figure 4B). A well-ordered water molecule is bound above the heme, illustrating that the protein crystallized in the FeIII-H2O state. Alignment of the last 80 C-terminal residues of the I5F and WT structures produced the lowest overall RMSD of 1.13 Å and illustrates that the most notable change introduced by the addition of the I5F mutation is the movement of the N-terminal domain with respect to the C-terminal domain (Figure 4B). The displacement of the N-terminal region of the structure can be traced to increased bulk and two conformations of the phenylalanine at position 5. The two conformations of the phenylalanine result in disorder of the first 6 residues at the N-terminus, which have also been modeled in two conformations (Figure 5). While the two conformations of the F5 residue are clearly represented in a composite omit map, the remaining residues at the N-terminus are less clearly located in the density and suggest an increased amount of disorder in this region of the structure (Figure 5). The amount of disorder in the N-terminal region indicates that this region is flexible and likely dynamic in solution.
Figure 5.
Heme cofactor and disordered N-terminus of the I5F Tt H-NOX structure. Composite omit map at 1.0 σ indicating two conformations (A & B) of the first 6 N-terminal residues. Residues 1 − 6 are shown in sticks with yellow carbon, red oxygen, and blue nitrogen atoms. The heme cofactor, coordinating water molecule, and coordinating histidine 102 are also shown in stick and with corresponding composite omit density for comparison.
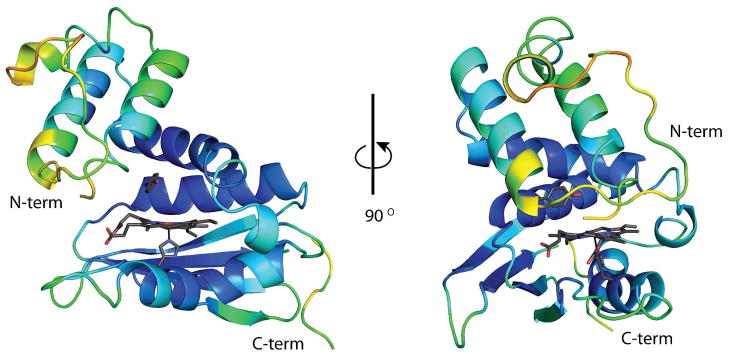
In all previously determined Tt H-NOX structures, the N-terminus is well ordered and clearly present in a single conformation. Thus, the increased flexibility in this region of the structure, which directly covers the heme pocket, allows for greater access to the heme pocket and likely contributes to the rapid O2 association and dissociation rates. In addition to the disorder seen at the N-terminus, the loop covering the N-terminal helix (residues 27–47) is also highly disordered with discontinuous backbone density (Supplemental Figure S3) and increased B-factors with respect to the rest of the structure (49 Å2 vs. 26 Å2; Figure 6). These observations are again consistent with a more dynamic protein with rapid ligand flux in and out of the heme pocket. Phenylalanine 78 lies above the heme on the opposite side of the pocket from F5, and like F5 is in two conformations in the pocket (Supplemental Figure S4). The two conformations of F78 is suggestive of increased flexibility within the heme pocket.
Figure 6.
I5F Tt H-NOX colored in rainbow by B-factor. The areas of the structure with the highest B-factors (70 Å2) are in red and the lowest B-factors (10 Å2) in dark blue. The heme, Y140, and H102 are shown in grey sticks for orientation.
In addition to the disorder at the N-terminus, the Y140–Fe distance was also lengthened, from 5.0 Å for WT (either FeII-O2 or FeIII-H2O structures) to 5.6 Å for the FeIII-H2O I5F mutant (Figure 4B). It was previously found that increasing the distance between the distal pocket hydrogen bond donor and the iron results in a weaker hydrogen bond to the bound O2 and subsequent reduction in O2 affinity (18). The pocket volume has increased slightly due to the N-terminal shift away from the heme and protein core. The increased flexbility and opening of the heme pocket, in concert with the increased Y140–Fe distance, likely results in decreased ligand trapping at the heme, contributing to the decreased O2 affinity of the I5F mutant.
Resonance Raman Characterization of I5F Heme Flexibility
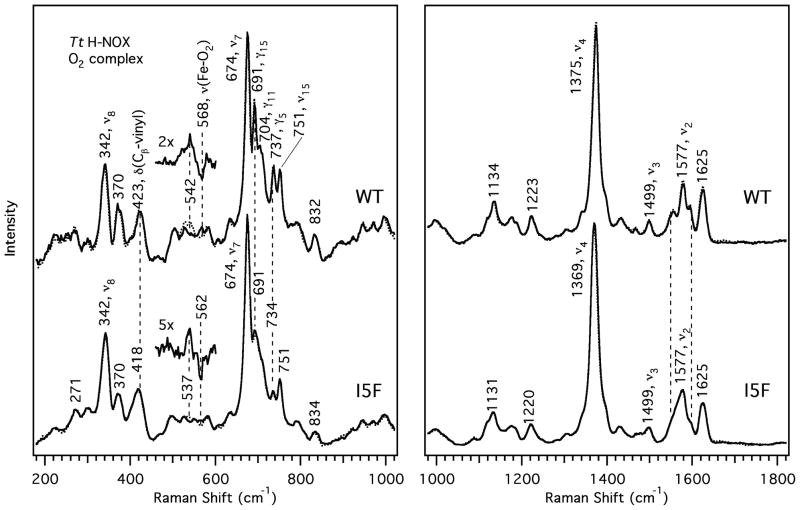
To further investigate how introduction of the I5F mutation changes the local heme environment, steady–state resonance Raman (RR) spectra of the FeII–O2 protein were obtained. Previous work has found that the conformation of the heme and protein are intricately linked (12, 15), suggesting that the multiple confomers of the protein are most likely transmitted into additional flexibility of the heme. Resonance Raman spectroscopy is highly sensitive to changes in the heme environment and structure, providing detailed information about the changes introduced by the mutations in solution (27–30). The main heme skeletal markers in the high frequency region can be used to assess differences in the heme macrocycle core size (ν4) and conformation (ν3, ν2, andν10), in addition to the spin and coordination states. Furthermore, heme deformation modes (e.g. δ(Cβ-vinyl), γ15,ν8) and the FeII–O2 stretching mode in the low frequency region can reveal details about the heme structure and FeII–O2 bond strength.
Previous characterizations of the I5L mutant by UV–visible and RR spectroscopy (26) illustrate that the I5L protein has very few spectral differences with comparison to WT Tt H-NOX (9) (Supporting Information Table S3). However, there were some broadened heme skeletal modes and slight decreases in the RR intensity of heme out-of-plane modes in the RR spectra. These features demonstrate that there is some overall conformational flexibility and a slight relaxation toward a more planar geometry of the heme in the I5L mutant (26).
When compared to WT Tt H-NOX, the heme vibrational modes of the I5F mutant were found at the same frequencies as in the WT spectra, but were considerably broadened, suggesting markedly increased flexibility (Figure 8; Supporting Information Table 3) (9, 26). In addition, the low frequency heme out-of-plane modes were also evident in the RR spectra, albeit at slightly reduced intensities compared to the WT protein. Therefore, the heme deformation observed for the WT protein in the RR spectra and crystal structures is likely conserved in the I5F mutant, but to a lesser extent. This is consistent with the preservation of heme deformation in the I5F crystal structure. However, the broadening of these peaks suggests that the heme is significantly more dynamic in the I5F mutant than in the I5L or WT Tt H-NOX proteins. In addition, the high frequency skeletal markers, ν3, ν2, and ν10, do not display changes in frequency, but exhibit peak broadening, further suggesting dynamic motion of the heme upon introduction of the I5F mutation. This dynamic motion of the heme is likely due to the interconversion of the F5 conformations that alter the orientation of the N-terminus and F78 conformations at the opposite end of the heme pocket.
The FeII–O2 stretch shifted from 568 cm−1 in WT Tt H-NOX (9) to 562 cm−1 in the I5F mutant, which corresponds to a weakening of the FeII–O2 bond. This decrease is consistent with the increased distance between the distal Y140 and the iron observed in the crystal structure. In addition, there was a decrease in the vibrational frequency of ν4 in the I5F mutant spectra (by ~6 cm−1) relative to the WT protein. This reduction corresponds to a decrease in electron density at the porphyrin core and a slightly larger macrocycle core size (27). These results are also consistent with a more dynamic porphyrin as would be expected due to the I5F mutation resulting in the protein N-terminus adopting two conformations and expanding the heme-binding cavity, accounting for the frequency change in ν4 and the broadening of ν3, ν2, ν10, as well as the heme deformation modes. The decreased FeII–O2 stretching frequency is also consistent with the larger heme pocket and the increased distance between the iron and Y140 observed in the crystal structure. An increase in the Fe–Y140 distance is likely the cause of the increase in the O2 dissociation rate for the I5F mutant, as well as all of the double and triple mutants containing the I5F mutation. Without the strong hydrogen bond to trap the O2 at the iron, O2 to dissociation and exit from the heme pocket is much more facile.
Conclusions
In summary, it is clear that the I5 position in Tt H-NOX is very important for both heme and protein conformational stability. This position is conserved as a medium sized (V, I, L), hydrophic amino acid, further emphasizing the role of this residue in controlling protein structure and ligand affinity. Although the conservative I5L mutant shows modest changes, the I5F mutation results in large decreases in O2 affinity due to extremely rapid O2 association and dissociation rates (Table 1). In contrast, mutation of I5 to alanine results in a mutant with a decreased O2 dissociation rate, suggesting an increased O2 affinity (koff = 0.82 s−1). Therefore, the contacts made by the amino acid at position 5 are likely required for tuning the ligand affinity in H-NOX domains. In addition to the single mutants, the I5F mutation synergizes with the previously described I75F and L144F mutations, resulting in proteins with significantly faster O2 association and dissociation rates, as compared to WT Tt H-NOX. Resonance Raman and X-ray crystallographic studies have found that the I5F mutation also results in multiple conformations of the N-terminus and increased flexibility of the heme, suggesting that this residue is a key factor in maintaining structural rigidity. This rigidity may be important for signaling as it has previously been found that N-terminal movement of an H-NOX domain from S. oneidensis affects the activity of its cognate kinase (31). Conformational stability is likely also a method by which the H-NOX family can ensure high affinity ligand binding, even at the elevated temperatures (70–90 °C) at which the T. tengcongensis bacterium lives. These results provide additional evidence for the heme pocket of Tt H-NOX being tightly packed to exquisitely tune ligand–binding properties. In addition, this work further emphasizes the myriad factors that control ligand affinity within heme proteins, as well as the extent to which these factors are interlinked. An improved understanding of the interplay between the factors controlling ligand affinity should allow for the design of novel heme proteins that can be used for a variety of sensing and delivery applications in biotechnology and medicine. Engineered flexibility may be a novel strategy for controlling O2 affinity and binding kinetics to rationally tune the affinity of heme proteins.
Supplementary Material
Figure 7.
Resonance Raman spectra of the O2 complexes of WT and I5F Tt H-NOX. 18O2 spectra (dotted trace) are overlapped with the 16O2 spectra to indicate the frequency shifts upon isotopic substitution. Difference (18O2-16O2) spectra are magnified and overlaid above each protein for clarity. Spectral intensities were normalized to ν7 and ν4 for the low and high frequencies, respectively.
Table 2.
Statistics for data collection, processing, and structure refinement of the I5F crystal structure.
| Crystal | I5F TT HNOX |
|---|---|
| Space Group | P21 |
| Cell Dimensions | |
| a (Å) | 44.6 |
| b (Å) | 67.1 |
| c (Å) | 66.4 |
| β (°) | 92.0 |
| Wavelength (Å) | 0.977408 |
| Temperature (K) | 100 |
| Unique Reflections | 45085 (2223) |
| Resolution Range (Å) | 50.00 − 1.67 (1.70 − 1.67) |
| Average Redundancy | 3.7 (3.5) |
| Completeness (%)1 | 100.0 (99.9) |
| I/σ(I)1 | 31.1 (2.2) |
| Rsym (%)1,2 | 4.2 (35.5) |
| # Tt HNOX molecules per asu | 2 |
| Rcryst (Rfree) (%)3 | 19.7 (24.7) |
| Average B-factor (Å2) | |
| overall | 30.9 |
| protein | 30.7 |
| heme | 17.9 |
| waters | 37.5 |
| rms deviations | |
| bond lengths (Å) | 0.008 |
| bond angles (°) | 1.024 |
| Ramachandran plot (%)4 | |
| preferred | 97.1 (330) |
| allowed | 2.9 (10) |
| outliers | 0 (0) |
The number in parentheses is for the highest resolution shell.
Rsym = Σihkl |Ii(hkl) − <I(hkl)>|/ Σhkl <I(hkl)>, where Ii(hkl) is the ith measured diffraction intensity and <I(hkl)> is the mean of the intensity for the miller index (hkl).
Rcryst = Σhkl ||Fo(hkl)| − |Fc(hkl)||/Σhkl |Fo(hkl)|. Rfree = Rcryst for a test set of reflections (5%) not included in refinement.
Numbers in parentheses are the number of residues in each category.
Acknowledgments
The authors are grateful to Charlotte Whited, Professor Harry Gray, Dr. Jay Winkler and the Beckman Institute Laser Resource Center at the California Institute of Technology for assistance with O2 association rate measurements, Professor John Kuriyan for use of crystallography equipment, Professor Elizabeth Boon for acquisition of the I5L and I5A O2 dissociation rates, Mark Herzik for assistance with protein crystallography, Michael Winter for helpful discussions, and members of the Marletta laboratory for critical reading of this manuscript. The crystal data was collected at the Advanced Light Source in Berkeley, CA at Beamline 5.0.1, which is supported by the DOE Contract No. DE-AC02-05CH11231.
ABBREVIATIONS
- H-NOX
heme nitric oxide/oxygen binding domain
- Mb
myoglobin
- Hb
hemoglobin
- Tt
Thermoanaerobacter tengcongensis
- RMSD
root mean square deviation
- RR
resonance Ramn spectroscopy
- WT
wild type
- I
isoleucine
- L
leucine
- F
phenylalanine
Footnotes
Financial support provided by NIH grant GM070671, the Rogers Family Fund, and NIH National Heart Lung and Blood Institute Award F32HL090174 (EEW)
ACCESSION CODE
The coordinates have been deposited in the RCSB Protein Data Bank as entry 3SJ5.
SUPPORTING INFORMATION PARAGRAPH
Supporting information can be found on the ACS website at http://pubs.acs.org/. Figure S1 (Comparison of Mb and Tt H-NOX heme pockets), figure S2 (resonance Raman spectra of various steric bulk mutants), figure S3 (composite omit density of the loop 27–47), figure S4 (composite omit density and the two comformations of F78 in the heme pocket), and tables S1 (Electronic absorption spectra), S2 (NO dissociation rates), and S3 (RR skeletal markers) can be found in the Supporting Information.
References
- 1.Springer BA, Sligar SG, Olson JS, Phillips GN. Mechanisms of Ligand Recognition in Myoglobin. Chem Rev. 1994;94:699–714. [Google Scholar]
- 2.Dou Y, Maillett DH, Eich RF, Olson JS. Myoglobin as a model system for designing heme protein based blood substitutes. Biophys Chem. 2002;98:127–148. doi: 10.1016/s0301-4622(02)00090-x. [DOI] [PubMed] [Google Scholar]
- 3.Antonini E, Brunori M. Hemoglobin and myoglobin in their reactions with ligands. North-Holland Pub. Co; Amsterdam: 1971. [Google Scholar]
- 4.Lukin JA, Ho C. The structure--function relationship of hemoglobin in solution at atomic resolution. Chem Rev. 2004;104:1219–1230. doi: 10.1021/cr940325w. [DOI] [PubMed] [Google Scholar]
- 5.Olson JS, Phillips GN., Jr Myoglobin discriminates between O2, NO, and CO by electrostatic interactions with the bound ligand. J Biol Inorg Chem. 1997;2:544–552. [Google Scholar]
- 6.Olson JS, Soman J, Phillips GN., Jr Ligand pathways in myoglobin: a review of Trp cavity mutations. IUBMB Life. 2007;59:552–562. doi: 10.1080/15216540701230495. [DOI] [PubMed] [Google Scholar]
- 7.Capece L, Marti MA, Crespo A, Doctorovich F, Estrin DA. Heme protein oxygen affinity regulation exerted by proximal effects. J Am Chem Soc. 2006;128:12455–12461. doi: 10.1021/ja0620033. [DOI] [PubMed] [Google Scholar]
- 8.Iyer LM, Anantharaman V, Aravind L. Ancient conserved domains shared by animal soluble guanylyl cyclases and bacterial signaling proteins. BMC Genomics. 2003;4:5. doi: 10.1186/1471-2164-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karow DS, Pan D, Tran R, Pellicena P, Presley A, Mathies RA, Marletta MA. Spectroscopic characterization of the soluble guanylate cyclase-like heme domains from Vibrio cholerae and Thermoanaerobacter tengcongensis. Biochemistry. 2004;43:10203–10211. doi: 10.1021/bi049374l. [DOI] [PubMed] [Google Scholar]
- 10.Derbyshire ER, Marletta MA. Biochemistry of soluble guanylate cyclase. Handbook Exp Pharmacol. 2009:17–31. doi: 10.1007/978-3-540-68964-5_2. [DOI] [PubMed] [Google Scholar]
- 11.Boon EM, Huang SH, Marletta MA. A molecular basis for NO selectivity in soluble guanylate cyclase. Nature Chem Biol. 2005;1:53–59. doi: 10.1038/nchembio704. [DOI] [PubMed] [Google Scholar]
- 12.Nioche P, Berka V, Vipond J, Minton N, Tsai AL, Raman CS. Femtomolar sensitivity of a NO sensor from Clostridium botulinum. Science. 2004;306:1550–1553. doi: 10.1126/science.1103596. [DOI] [PubMed] [Google Scholar]
- 13.Price MS, Chao LY, Marletta MA. Shewanella oneidensis MR-1 H-NOX regulation of a histidine kinase by nitric oxide. Biochemistry. 2007;46:13677–13683. doi: 10.1021/bi7019035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma X, Sayed N, Beuve A, van den Akker F. NO and CO differentially activate soluble guanylyl cyclase via a heme pivot-bend mechanism. Embo J. 2007;26:578–588. doi: 10.1038/sj.emboj.7601521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pellicena P, Karow DS, Boon EM, Marletta MA, Kuriyan J. Crystal structure of an oxygen-binding heme domain related to soluble guanylate cyclases. Proc Natl Acad Sci USA. 2004;101:12854–12859. doi: 10.1073/pnas.0405188101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Olea C, Boon EM, Pellicena P, Kuriyan J, Marletta MA. Probing the function of heme distortion in the H-NOX family. ACS Chem Biol. 2008;3:703–710. doi: 10.1021/cb800185h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olea C, Jr, Kuriyan J, Marletta MA. Modulating heme redox potential through protein-induced porphyrin distortion. J Am Chem Soc. 2010;132:12794–12795. doi: 10.1021/ja106252b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weinert EE, Plate L, Whited CA, Olea C, Jr, Marletta MA. Determinants of ligand affinity and heme reactivity in H-NOX domains. Angew Chem Int Ed Engl. 2010;49:720–723. doi: 10.1002/anie.200904799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin E, Berka V, Bogatenkova E, Murad F, Tsai A-L. Ligand Selectivity of Soluble Guanylyl Cyclase: Effect of the Hydrogen-Bonding Tyrosine in the Distal Heme Pocket on Binding of Oxygen, Nitric Oxide, and Carbon Monoxide. J Biol Chem. 2006;281:27836–27845. doi: 10.1074/jbc.M601078200. [DOI] [PubMed] [Google Scholar]
- 20.Derbyshire ER, Deng S, Marletta MA. Incorporation of tyrosine and glutamine residues into the soluble guanylate cyclase heme distal pocket alters NO and O2 binding. J Biol Chem. 2010;285:17471–17478. doi: 10.1074/jbc.M109.098269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dmochowski IJ, Winkler JR, Gray HB. Enantiomeric discrimination of Ru-substrates by cytochrome P450cam. J Inorg Biochem. 2000;81:221–228. doi: 10.1016/s0162-0134(00)00111-2. [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromolecular Crystallography, Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.Storoni LC, McCoy AJ, Read RJ. Likelihood-enhanced fast rotation functions. Acta Crystallographica D60. 2004:432–438. doi: 10.1107/S0907444903028956. [DOI] [PubMed] [Google Scholar]
- 24.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D-Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Emsley P, Crowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Tran R, Boon EM, Marletta MA, Mathies RA. Resonance Raman spectra of an O2-binding H-NOX domain reveal heme relaxation upon mutation. Biochemistry. 2009;48:8568–8577. doi: 10.1021/bi900563g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spiro TG, Li XY. Resonance Raman Spectroscopy of Metalloporphyrins. In: Spiro TG, editor. Biological Applications of Raman Spectroscopy: Resonance Raman Spectra of Heme and Metalloproteins. John Wiley & Sons; New York: 1988. pp. 1–37. [Google Scholar]
- 28.Huang Q, Medforth CJ, Schweitzer-Stenner R. Nonplanar heme deformations and excited state displacements in nickel porphyrins detected by Raman spectroscopy at soret excitation. J Phys Chem A. 2005;109:10493–10502. doi: 10.1021/jp052986a. [DOI] [PubMed] [Google Scholar]
- 29.Shelnutt JA, Song XZ, Ma JG, Jia SL, Jentzen W, Medforth CJ. Nonplanar porphyrins and their significance in proteins. Chem Soc Rev. 1998;27:31–41. [Google Scholar]
- 30.Spiro TG, Stong JD, Stein P. Porphyrin Core Expansion and Doming in Heme-Proteins - New Evidence from Resonance Raman-Spectra of 6-Coordinate High-Spin Iron(III) Hemes. J Am Chem Soc. 1979;101:2648–2655. [Google Scholar]
- 31.Erbil WK, Price MS, Wemmer DE, Marletta MA. A structural basis for H-NOX signaling in Shewanella oneidensis by trapping a histidine kinase inhibitory conformation. Proc Natl Acad Sci U S A. 2009;106:19753–19760. doi: 10.1073/pnas.0911645106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.