Summary
Objectives
Many patients with influenza have more than one viral agent with co-infection frequencies reported as high as 20%. The impact of respiratory virus copathogens on influenza disease is unclear. We sought to determine if respiratory virus co-infection with pandemic H1N1 altered clinical disease.
Methods
Respiratory samples from 229 and 267 patients identified with and without H1N1 influenza respectively were screened for the presence of 13 seasonal respiratory viruses by multiplex RT-PCR. Disease severity between coinfected and monoinfected H1N1 patients were quantified using a standardized clinical severity scale. Influenza viral load was calculated by quantitative RT-PCR.
Results
Thirty (13.1%) influenza samples screened positive for the presence of 31 viral copathogens. The most prominent copathogens included rhinovirus (61.3%), and coronaviruses (16.1%). Median clinical severity of both monoinfected and coinfected groups were 1. Patients coinfected with rhinovirus tended to have lower clinical severity (median 0), whereas non-rhinovirus co-infections had substantially higher clinical severity (median 2). No difference in H1N1 viral load was observed between coinfected and monoinfected groups.
Conclusions
Respiratory viruses co-infect patients with influenza disease. Patients coinfected with rhinovirus had less severe disease while non-rhinovirus co-infections were associated with substantially higher severity without changes in influenza viral titer.
Keywords: Influenza, Influenza co-infection, Co-infection, Dual infection, Respiratory virus co-infection, Viral co-infection, Pneumonia, Respiratory disease
Introduction
Respiratory tract disease is a major cause of morbidity and mortality throughout the world accounting for approximately 4 million deaths worldwide.1, 2 While viruses such as influenza, parainfluenza, respiratory syncytial virus (RSV), and adenovirus are well described as major respiratory pathogens, advancements in molecular and genomic techniques have increased detection and identification of new respiratory viruses.3, 4, 5, 6, 7, 8, 9, 10, 11 There is also renewed attention on respiratory viruses including the human rhinovirus and coronaviruses as there is mounting evidence that these viruses can also lead to clinically severe disease.5, 12, 13, 14, 15, 16, 17, 18
Currently, the majority of practitioners approach respiratory viral infections assuming a single-agent etiology. However, our group and others have realized that a substantial number of patients with respiratory tract disease have more than one viral pathogen.19, 20, 21 The frequency of respiratory virus co-infections varies widely in the literature but is often reported between 10 and 20%22, 23, 24, 25 and in one report as high as 60%.26 This is understandable as many respiratory viruses circulate at similar times often with a winter time predominance in temperate climates. Recently, Brunstein et al. provided statistical evidence that co-infection with certain pathogens occurs more frequently than expected if co-infection was random.25 Influenza has often been identified with co-infecting viruses.20 Several groups speculate that early spread of influenza may be altered by co-circulation of traditional seasonal viruses.27, 28, 29, 30
There is increasing evidence in the literature for the importance of polymicrobial infections. Virus–virus interactions have been recognized both directly, indirectly and through immunological interactions.31 Yet the clinical relevance of respiratory virus copathogens in association with disease is unclear. Several studies have reported viral co-infections being associated with increased morbidity. These studies show greater hospitalization rates and admission to intensive care associated with co-infection.24, 32, 33 However, this remains controversial as several other reports describe co-infections having no increase in patient morbidity.13, 23, 34, 35, 36, 37, 38
One explanation for the disagreement is the assumption that disease severity associated with dual infection is independent of the copathogen involved. Aberle et al. found that dual infections with rhinovirus and RSV was associated with reduced IFN-γ response and a more severe clinical course than other co-infection combinations.22 In animal models, specific dual respiratory infections resulted in either enhanced clinical manifestations or viral interference.39, 40, 41 These findings suggest certain copathogen pairings may be more clinically relevant than others.
There remain gaps in our knowledge and understanding of respiratory virus co-infections. Previous investigations of influenza co-infection either combine viral copathogens together in analysis20 or do not address clinical outcomes.26, 27, 28, 29, 30, 42 Studies quantifying the effect of viral co-infection are lacking yet such lines of investigation will be paramount to our understanding of viral pathogenesis. In this study we identify the prominent viral copathogens occurring with pandemic H1N1 influenza and compare the resulting clinical disease. This is the first study which critically looks at how specific copathogens alter resulting influenza disease and the effect on influenza viral load.
Materials and methods
Sample collection
From September through November 2009, corresponding to the time of the major H1N1 pandemic wave, we archived respiratory specimens which screened positive for influenza. For comparison, we selected influenza negative samples collected during this same period. All samples originated from the emergency department, in patient wards, intensive care units and hospital affiliated primary care outpatient clinics. Samples were submitted to the Core Laboratory at the discretion of the primary medical teams for influenza testing. Influenza screening was performed by either direct immunofluorescence assay (DFA) or real time RT-PCR using standardized techniques.43, 44 All samples were confirmed by RT-PCR using primer sets targeting pandemic H1N1.43
RNA extraction, reverse transcription
Nucleic acid from respiratory samples were extracted using either QIAcube (QIAamp Viral RNA Kit, QIAGEN, Valencia, CA) or MagMAX™-96 Total NA Isolation Kit (Applied Biosystems, Foster City, CA) according to the manufacturer’s protocol. Random hexamer primers (Invitrogen Carlsbad, CA) were used to create a cDNA library for each specimen. Reverse transcription reactions were performed with M-MLV RT (Invitrogen, Carlsbad, CA) according to the manufacturer’s specifications.
Respiratory virus screening
Each cDNA was subsequently screened for the presence of common respiratory viruses including, human parainfluenza 1–3 (HPIV 1–3), human metapneumovirus (MPV), respiratory syncytial virus (RSV), rhinovirus (RV), adenovirus (AdV), and human coronaviruses (HCoV) 229E, OC43, and NL63. Primers and probe sets used for multiplex real.time RT-PCR detection have been described.45 We screened respiratory samples under conditions as described with the exception that chlamydia, mycoplasma, and enterovirus primer sets were omitted. In addition, recently recognized respiratory viruses including coronavirus HKU1, WU Polyomavirus, and human bocavirus were screened using conventional RT-PCR in separate reactions using primer and reaction conditions described.9, 15, 46
Clinical severity score
Medical records of H1N1 positive-individuals were reviewed and recorded on a standard collection form. The clinical severity was scored by criteria as described by Martinello et al.47 This clinical severity score (CSS) was chosen as it is designed specifically around respiratory disease. The CSS ranges from 0 to 6. Two points are assigned if the patient required mechanically ventilator support during the illness, and 1 point is assigned for each of the following: hospital admission, prolonged hospitalization (≥5 days), severe hypoxia ≤87%, and use of supplemental oxygen. H1N1 mono and coinfected individuals were then compared for clinical severity. We defined severe respiratory disease as patients with CSS >3.
Sample collection and chart analysis was approved by the University Hospital-Case Medical Center IRB.
Quantitative real time PCR
Quantitative analysis was performed on a StepOne Plus Taqman Real Time PCR (Applied Biosystems, Branchburg, NJ) using TaqMan Universal PCR Master Mix (Applied Biosystems, Branchburg, NJ), 2ul of cDNA sample, and primers/probes targeting the H1N1 matrix gene.43 A reference standard was prepared using a cDNA fragment of the H1N1 matrix gene and human RNAse P amplified by conventional RT-PCR, gel purified (QIAquick, Qiagen, Valencia, CA), and quantified using a spectrophotometer (Beckman Coulter, Brea, CA).
Statistical analysis
Statistical analysis was performed using R software (version 2.11.1).48 Student t-test was used to analyze the mean log10 viral copy numbers between monoinfected and coinfected groups. Chi-square test of independent proportions was used to analyze clinical severity between mono and coinfected patients. A two-tailed p value <0.05 was considered significant.
Results
A total of 229 H1N1 influenza positive samples and 267 H1N1 negative samples were screened for the presence of common respiratory viruses. Influenza samples originated from 133 (58.0%) adults (age> = 18 yrs) and 96 (42.0%) children (age < 18 yrs) with a median age of 20.3 yrs. Thirty (13.1%) influenza samples screened positive for the presence of 31 viral copathogens including 19 (61.3%) rhinovirus, 5 (16.1%) coronaviruses, 2 (6.4%) adenovirus, 2 (6.4%) human parainfluenza type 2 (HPIV2), and 1 (3.2%) each for WU polyomavirus, HPIV1 and RSV (Table 1 ). One pediatric patient was identified having co-infection with both HPIV2 and coronavirus HKU1 in addition to influenza. Neither hMPV nor HCoV NL63 were detected. With the exception of RSV and HIPV1, The proportion of patients positive for all tested viruses was similar for patients with and without influenza H1N1 infection. Median age for coinfected individuals was 19.2 yrs and included 17 adults (median age 37.7 yrs) and 13 children (median age 7.4 yrs). More adults had rhinovirus co-infection than children (12 vs. 7 respectively).
Table 1.
Presence of respiratory viruses in H1N1 and non H1N1 positive respiratory samples.
| Virus | H1N1 Positive (N = 229) |
H1N1 Negative (N = 267) |
|---|---|---|
| # (%) | # (%) | |
| RhinoVirus | 19 (8.3%) | 35 (13.1%) |
| Coronavirusesa | 5 (2.2%) | 3 (1.1%) |
| Adenovirus | 2 (0.9%) | 6 (2.2%) |
| HPIV1 | 1 (0.4%) | 16 (5.9%) |
| HPIV2a | 2 (0.9%) | 1 (0.4%) |
| HPIV3 | 0 (0.0%) | 1 (0.4%) |
| WU Polyoma | 1 (0.4%) | 0 (0.0%) |
| RSV | 1 (0.4%) | 13 (4.9%) |
| HBoV1 | 0 (0.0%) | 2 (0.7%) |
| Totala | 30 (13.1%) | 77 (28.8%) |
1 Patient coinfected with HPIV2 and coronavirus.
Charts for 196 (98.0%) monoinfected and 30 (100%) coinfected patients were available for chart review and clinical severity was calculated. Median clinical severity of both monoinfected and coinfected groups were 1 (Table 2 ). However, influenza patients coinfected with rhinovirus tended to have lower clinical severity (median 0), whereas non-rhinovirus co-infections had substantially higher clinical severity (median 2). As a group, coinfected individuals had a higher proportion of severe disease (CSS > 3) compared to influenza monoinection (26.7% vs 11.2%, P < 0.05), the majority of which is derived from non-rhinovirus co-infections (62.5%). Coinfected adults and children had severe disease at similar rates (29.4% vs 23.0%). The majority of influenza monoinfected patients with severe disease were adults (17, 15.2%) compared with only 5 (6.0%) children.
Table 2.
Clinical severity scores (CSS) in H1N1 mono and coinfected patients.
| Median severity score | Severe disease (CSS> = 3) | |
|---|---|---|
| H1N1 Monoinfection (N = 196) | 1 | 22 (11.2%) |
| H1N1 Co-infection (N = 30) | 1 | 8 (26.7%) |
| Rhinovirus (N = 19) | 0 | 3 (15.8%) |
| Non-Rhinovirus (N = 11) | 2 | 5 (45.4%) |
| aCoronavirus (N = 5) | 4 | 3 (60.0%) |
| HPIV1 (N = 1) | 1 | 0 (0.0%) |
| aHPIV2 (N = 2) | 2 | 1 (50.0%) |
| RSV (N = 1) | 6 | 1 (100%) |
| WU Polyoma (N = 1) | 5 | 1 (100%) |
| Adenovirus (N = 2) | 1 | 0 (0.0%) |
1 Patient coinfected with HPIV2 and coronavirus.
Influenza patients with rhinovirus co-infection tended to have less admission to the hospital (52.6% vs. 69.8%) and oxygen use (21.1% vs. 31.0%) (Table 3 ). Non-rhinovirus co-infections had significantly higher ventilator use (45.5% vs. 8.5%) and severe hypoxia (36.4% vs. 9.0%) compared to influenza monoinfected patients (P < 0.05). Coronavirus co-infection had a surprisingly high CSS (Median 4) with 80% being admitted to the hospital and requiring oxygen use as well as 60% requiring mechanical ventilation.
Table 3.
Breakdown of clinical severity score attributes between mono and coinfected H1N1 patients.
| Vent | Admit | Admit ≥ 5d | O2 ≤ 87% | O2 Given | |
|---|---|---|---|---|---|
| H1N1 Monoinfected (N = 199) | 8.5% | 69.8% | 28.1% | 9.0% | 31.0% |
| H1N1 with co-infection (N = 30) | 23.3%∗ | 60.0% | 26.7% | 20.0% | 33.3% |
| Non-rhinovirus co-infections (N = 11) | 45.5%∗ | 72.7% | 27.3% | 36.4%∗ | 54.5% |
| Rhinovirus (N = 19) | 10.5% | 52.6% | 26.3% | 10.5% | 21.1% |
∗P < 0.05 compared to H1N1 monoinfection.
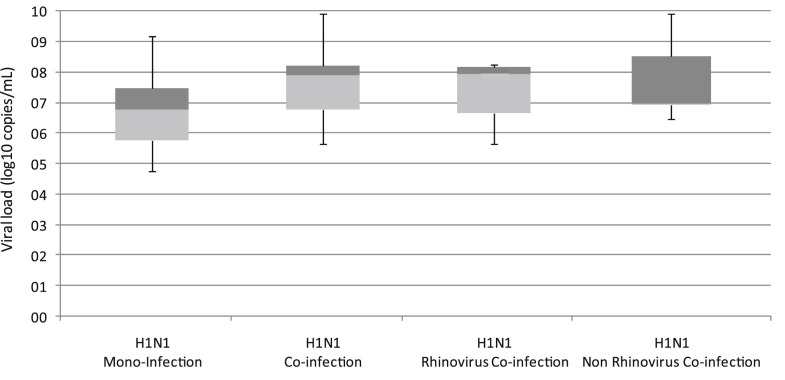
Influenza viral load was calculated by quantitative RT-PCR (Fig. 1 ). No significant difference in mean viral load was observed between coinfected or monoinfected groups (P = 0.36). Influenza titer originating from samples coinfected with rhinovirus was similar to influenza samples coinfected with other viruses.
Fig. 1.
Quantitative RT-PCR of influenza in mono and coinfected patients. Boxplots demonstrate lower and upper quartiles (light and dark shaded respectively) as well as minimum and maximum values (bars) for each group.
Discussion
There are several reports where viral interactions have measurable outcomes on the course of infections. However, these interactions are relatively unexplored given the number of viral illness which occur. Most investigations focus on lifelong viral infections such as HIV, Epstein–Barr, and hepatitis C. Due to the relatively short duration of viral respiratory infections, co-infection is thought to be less of a concern.31 We are only now beginning to recognize the diversity of the nasopharyngeal microbiome in the normal and diseased states.49, 50 With this comes the realization that co-detection of pathogens may become the norm, making our understanding of clinically relevant pathogen pairings essential. This is one of the first investigations to quantify clinical severity of influenza disease based on specific copathogens.
Our co-infection rate of 13.1% is similar to those reported in other studies.24, 38, 42 Difference in patient population, time of sample collection, selection of pathogens to be screened and screening methodologies all contribute to variance in co-infection rates between studies. However, with more sensitive diagnostic methods becoming widely used combined with identification of previously unrecognized viral pathogens, we should expect the number of influenza co-detections to increase.
Similar to other investigations the predominant pathogen co-infecting with influenza was rhinovirus.24, 26, 42 Rhinovirus has historically been considered ubiquitous but clinically mild. Yet a growing number of reports now challenge this notion.16, 17, 18 Rhinovirus co-infections have been associated with worse disease when co-infecting with RSV.13 Our findings demonstrate this is not true regarding influenza. Rhinovirus co-infection had little impact on influenza disease, in fact they had milder findings in several key areas. The opposite was true in patients with coronavirus/influenza co-infection. Comparable to other studies,38, 42 we observe the total number of influenza and coronavirus co-infections is low (1.12%). But we find that the resulting disease was substantially more severe. This highlights the importance of recognizing respiratory virus pairings as distinct clinical entities.
Viral interference of seasonal viruses (especially rhinovirus) with pandemic influenza has been suggested recently. Much of this is based on surveillance and inferred relationships of rhinovirus and influenza detection.27 However our study demonstrates that influenza viral titer was not altered by viral co-infection suggesting that obstruction of influenza growth, either directly or indirectly, is not occurring. Explanation as to why disease severity was altered without changes in viral titer may lie in the resulting immunologic response. Investigation of host cytokine and cellular response associated with co-infections is a logical next step.
We should recognize that the absence of specific co-infections may be as important as those which are observed. Both RSV and HPIV1 were significantly absent from influenza samples greater than expected if co-infection occurred by chance alone. This pattern is often noted with RSV, influenza and parainfluenza: when one of these epidemic respiratory viruses reached a peak the others seemed to be relatively inactive.29, 52 One wonders if the reasons behind this pattern involve direct or indirect viral interactions including changes in tissue permissiveness, viral replication, or host immunologic response.
Our study has several limitations. First, not all possible viral pathogens were targeted. The co-infection rate may be even higher were we to expand the number of pathogens to include enterovirus, HPIV4, and other influenza virus types (B and C). Time to presentation also varied within the groups which may affect the sensitivity of viral detection. Advancing molecular modalities including MassTag PCR, ViroChip and Deep sequencing will ultimately provide a more complete picture of co-infection frequency. It has been well described that severe clinical illness in the 2009 H1N1 pandemic was associated with underlying co-morbidities including obesity, diabetes, or pregnancy. Collection and screening of samples as well as the use of the clinical severity score were independent of any patient’s underlying co-morbidities. In addition to patient co-morbidities, the severity of illness may also be associated to other factors, such as delayed medical attention and lack of antiviral therapy. Larger studies that include multivariate analysis of these parameters should be undertaken.
It is important to note, the morbidity and mortality of the 2009 pandemic influenza virus differs from seasonal influenza viruses. Therefore, it is not clear that what happened with the 2009 pandemic influenza virus can be extrapolated to seasonal influenza viruses. Also, relative humidity affects both influenza virus transmission and influenza virus survival.51 Comparisons of findings when influenza is spread mainly by contact (similar to rhinovirus at times of higher absolute humidity) and when it is spread as small particle aerosol (during the peak and lower humidity) may differ. This may explain some differences in our findings compared to similar studies.38 Furthermore, while we point out that disease severity likely depends on the type of the co-infecting pathogen, the numbers of certain co-detected pathogens in the H1N1 infected patients are too small for more detailed analysis. Continued investigation over multiple seasons will be required to answer these questions. Despite these limitations, we demonstrate that clinical relevance of influenza viral co-infection varies greatly and may be associated with the co-infecting pathogen involved.
The recognition of respiratory viral co-infections is increasing with advancement of sensitive multiplex screening of respiratory samples. As the sensitivity of diagnostic methods increase, the number of viral co-infections will continue to rise. Further investigation quantifying the effect of viral co-infection and identification of relevant copathogen pairings will be paramount to our understanding of viral pathogenesis.
Ethical approval
Internal Review Board: Collection of specimens and clinical data were approved by the University Hospitals Human Investigation Committee.
Conflicts of interest
None of the authors report conflicts of interest.
Acknowledgments
This study is supported by the National Institute of Allergy And Infectious Diseases K23 AI065829. We are indebted to the staff of the Department of Pathology Core Laboratory and the Molecular Diagnostics Laboratory at University Hospitals Case medical Center.
Footnotes
Presented in part: IDSA, Vancouver, BC, Canada October, 2010.
References
- 1.Murray C.J.L., Lopez A.D., Mathers C.D., Stein C. Global Programme on Evidence for Health Policy World Health Organization; 2001. The Global burden of disease 2000 project: aims, methods and data sources. [Google Scholar]
- 2.Merrill C.T.E.A. Agency for Healthcare Research and Quality; Rockville, MD: 2002. Hospitalization in the United States. HCUP Fact Book No 6 2005;Publication number 05–0056. [Google Scholar]
- 3.van den Hoogen B.G., de Jong J.C., Groen J., Kuiken T., de Groot R., Fouchier R.A. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001 Jun;7(6):719–724. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Hoek L., Pyrc K., Jebbink M.F., Vermeulen-Oost W., Berkhout R.J., Wolthers K.C. Identification of a new human coronavirus. Nat Med. 2004 Apr;10(4):368–373. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esper F., Weibel C., Ferguson D., Landry M.L., Kahn J.S. Evidence of a novel human coronavirus that is associated with respiratory tract disease in infants and young children. J Infect Dis. 2005 Feb 15;191(4):492–498. doi: 10.1086/428138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fouchier R.A., Hartwig N.G., Bestebroer T.M., Niemeyer B., de Jong J.C., Simon J.H. A previously undescribed coronavirus associated with respiratory disease in humans. Proc Natl Acad Sci U S A. 2004 Apr 20;101(16):6212–6216. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Woo P.C., Lau S.K., Chu C.M., Chan K.H., Tsoi H.W., Huang Y. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005 Jan;79(2):884–895. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Allander T., Tammi M.T., Eriksson M., Bjerkner A., Tiveljung-Lindell A., Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci U S A. 2005 Sep 6;102(36):12891–12896. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gaynor A.M., Nissen M.D., Whiley D.M., Mackay I.M., Lambert S.B., Wu G. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 2007 May 4;3(5):e64. doi: 10.1371/journal.ppat.0030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allander T., Andreasson K., Gupta S., Bjerkner A., Bogdanovic G., Persson M.A. Identification of a third human polyomavirus. J Virol. 2007 Apr;81(8):4130–4136. doi: 10.1128/JVI.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chua K.B., Crameri G., Hyatt A., Yu M., Tompang M.R., Rosli J. A previously unknown reovirus of bat origin is associated with an acute respiratory disease in humans. Proc Natl Acad Sci U S A. 2007 Jul 3;104(27):11424–11429. doi: 10.1073/pnas.0701372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rakes G.P., Arruda E., Ingram J.M., Hoover G.E., Zambrano J.C., Hayden F.G. Rhinovirus and respiratory syncytial virus in wheezing children requiring emergency care. IgE and eosinophil analyses. Am J Respir Crit Care Med. 1999 Mar;159(3):785–790. doi: 10.1164/ajrccm.159.3.9801052. [DOI] [PubMed] [Google Scholar]
- 13.Papadopoulos N.G., Moustaki M., Tsolia M., Bossios A., Astra E., Prezerakou A. Association of rhinovirus infection with increased disease severity in acute bronchiolitis. Am J Respir Crit Care Med. 2002 May 1;165(9):1285–1289. doi: 10.1164/rccm.200112-118BC. [DOI] [PubMed] [Google Scholar]
- 14.Arden K.E., Nissen M.D., Sloots T.P., Mackay I.M. New human coronavirus, HCoV-NL63, associated with severe lower respiratory tract disease in Australia. J Med Virol. 2005 Mar;75(3):455–462. doi: 10.1002/jmv.20288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esper F., Weibel C., Ferguson D., Landry M.L., Kahn J.S. Coronavirus HKU1 infection in the United States. Emerg Infect Dis. 2006 May;12(5):775–779. doi: 10.3201/eid1205.051316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chidekel A.S., Rosen C.L., Bazzy A.R. Rhinovirus infection associated with serious lower respiratory illness in patients with bronchopulmonary dysplasia. Pediatr Infect Dis J. 1997 Jan;16(1):43–47. doi: 10.1097/00006454-199701000-00010. [DOI] [PubMed] [Google Scholar]
- 17.Kim J.O., Hodinka R.L. Serious respiratory illness associated with rhinovirus infection in a pediatric population. Clin Diagn Virol. 1998 May 1;10(1):57–65. doi: 10.1016/s0928-0197(98)00004-x. [DOI] [PubMed] [Google Scholar]
- 18.Malcolm E., Arruda E., Hayden F.G., Kaiser L. Clinical features of patients with acute respiratory illness and rhinovirus in their bronchoalveolar lavages. J Clin Virol. 2001 Apr;21(1):9–16. doi: 10.1016/s1386-6532(00)00180-3. [DOI] [PubMed] [Google Scholar]
- 19.Esper F., Martinello R.A., Boucher D., Weibel C., Ferguson D., Landry M.L. A 1-year experience with human metapneumovirus in children aged <5 years. J Infect Dis. 2004 Apr 15;189(8):1388–1396. doi: 10.1086/382482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng D., Zhao D., Liu J., Wang X., Yang K., Xicheng H. Multipathogen infections in hospitalized children with acute respiratory infections. Virol J. 2009;6:155. doi: 10.1186/1743-422X-6-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waner J.L. Mixed viral infections: detection and management. Clin Microbiol Rev. 1994 Apr;7(2):143–151. doi: 10.1128/cmr.7.2.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aberle J.H., Aberle S.W., Pracher E., Hutter H.P., Kundi M., Popow-Kraupp T. Single versus dual respiratory virus infections in hospitalized infants: impact on clinical course of disease and interferon-gamma response. Pediatr Infect Dis J. 2005 Jul;24(7):605–610. doi: 10.1097/01.inf.0000168741.59747.2d. [DOI] [PubMed] [Google Scholar]
- 23.Subbarao E.K., Griffis J., Waner J.L. Detection of multiple viral agents in nasopharyngeal specimens yielding respiratory syncytial virus (RSV). An assessment of diagnostic strategy and clinical significance. Diagn Microbiol Infect Dis. 1989 Jul–Aug;12(4):327–332. doi: 10.1016/0732-8893(89)90098-9. [DOI] [PubMed] [Google Scholar]
- 24.Drews A.L., Atmar R.L., Glezen W.P., Baxter B.D., Piedra P.A., Greenberg S.B. Dual respiratory virus infections. Clin Infect Dis. 1997 Dec;25(6):1421–1429. doi: 10.1086/516137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brunstein J.D., Cline C.L., McKinney S., Thomas E. Evidence from multiplex molecular assays for complex multipathogen interactions in acute respiratory infections. J Clin Microbiol. 2008 Jan;46(1):97–102. doi: 10.1128/JCM.01117-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Follin P., Lindqvist A., Nystrom K., Lindh M. A variety of respiratory viruses found in symptomatic travellers returning from countries with ongoing spread of the new influenza A(H1N1)v virus strain. Euro Surveill. 2009 Jun 18;14(24) doi: 10.2807/ese.14.24.19242-en. [DOI] [PubMed] [Google Scholar]
- 27.Greer R.M., McErlean P., Arden K.E., Faux C.E., Nitsche A., Lambert S.B. Do rhinoviruses reduce the probability of viral co-detection during acute respiratory tract infections? J Clin Virol. 2009 May;45(1):10–15. doi: 10.1016/j.jcv.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Linde A., Rotzen-Ostlund M., Zweygberg-Wirgart B., Rubinova S., Brytting M. Does viral interference affect spread of influenza? Euro Surveill. 2009;14(40) [PubMed] [Google Scholar]
- 29.Anestad G. Surveillance of respiratory viral infections by rapid immunofluorescence diagnosis, with emphasis on virus interference. Epidemiol Infect. 1987 Oct;99(2):523–531. doi: 10.1017/s0950268800068023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casalegno J.S., Ottmann M., Duchamp M.B., Escuret V., Billaud G., Frobert E. Rhinoviruses delayed the circulation of the pandemic influenza A (H1N1) 2009 virus in France. Clin Microbiol Infect. 2010 Apr;16(4):326–329. doi: 10.1111/j.1469-0691.2010.03167.x. [DOI] [PubMed] [Google Scholar]
- 31.DaPalma T., Doonan B.P., Trager N.M., Kasman L.M. A systematic approach to virus–virus interactions. Virus Res. 2010 Apr;149(1):1–9. doi: 10.1016/j.virusres.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Semple M.G., Cowell A., Dove W., Greensill J., McNamara P.S., Halfhide C. Dual infection of infants by human metapneumovirus and human respiratory syncytial virus is strongly associated with severe bronchiolitis. J Infect Dis. 2005 Feb 1;191(3):382–386. doi: 10.1086/426457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greensill J., McNamara P.S., Dove W., Flanagan B., Smyth R.L., Hart C.A. Human metapneumovirus in severe respiratory syncytial virus bronchiolitis. Emerg Infect Dis. 2003 Mar;9(3):372–375. doi: 10.3201/eid0903.020289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang J.J., Huang T.Y., Huang M.Y., Chen B.H., Lin K.H., Jeng J.E. Simultaneous multiple viral infections in childhood acute lower respiratory tract infections in southern Taiwan. J Trop Pediatr. 1998 Oct;44(5):308–311. doi: 10.1093/tropej/44.5.308. [DOI] [PubMed] [Google Scholar]
- 35.Lazar I., Weibel C., Dziura J., Ferguson D., Landry M.L., Kahn J.S. Human metapneumovirus and severity of respiratory syncytial virus disease. Emerg Infect Dis. 2004 Jul;10(7):1318–1320. doi: 10.3201/eid1007.030983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Legg J.P., Warner J.A., Johnston S.L., Warner J.O. Frequency of detection of picornaviruses and seven other respiratory pathogens in infants. Pediatr Infect Dis J. 2005 Jul;24(7):611–616. doi: 10.1097/01.inf.0000168747.94999.aa. [DOI] [PubMed] [Google Scholar]
- 37.Richard N., Komurian-Pradel F., Javouhey E., Perret M., Rajoharison A., Bagnaud A. The impact of dual viral infection in infants admitted to a pediatric intensive care unit associated with severe bronchiolitis. Pediatr Infect Dis J. 2008 Mar;27(3):213–217. doi: 10.1097/INF.0b013e31815b4935. [DOI] [PubMed] [Google Scholar]
- 38.Palacios G., Hornig M., Cisterna D., Savji N., Bussetti A.V., Kapoor V. Streptococcus pneumoniae coinfection is correlated with the severity of H1N1 pandemic influenza. PLoS One. 2009;4(12):e8540. doi: 10.1371/journal.pone.0008540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Reeth K., Nauwynck H., Pensaert M. Clinical effects of experimental dual infections with porcine reproductive and respiratory syndrome virus followed by swine influenza virus in conventional and colostrum-deprived pigs. J Vet Med B Infect Dis Vet Public Health. 2001 May;48(4):283–292. doi: 10.1046/j.1439-0450.2001.00438.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Reeth K., Nauwynck H., Pensaert M. Dual infections of feeder pigs with porcine reproductive and respiratory syndrome virus followed by porcine respiratory coronavirus or swine influenza virus: a clinical and virological study. Vet Microbiol. 1996 Feb;48(3–4):325–335. doi: 10.1016/0378-1135(95)00145-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Reeth K., Pensaert M.B. Porcine respiratory coronavirus-mediated interference against influenza virus replication in the respiratory tract of feeder pigs. Am J Vet Res. 1994 Sep;55(9):1275–1281. [PubMed] [Google Scholar]
- 42.Nisii C., Meschi S., Selleri M., Bordi L., Castilletti C., Valli M.B. Frequency of detection of upper respiratory tract viruses in patients tested for pandemic H1N1/09 viral infection. J Clin Microbiol. 2010 Sep;48(9):3383–3385. doi: 10.1128/JCM.01179-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.2009. CDC protocol of realtime RTPCR for influenza A (H1N1)http://www.who.int/csr/resources/publications/swineflu/realtimeptpcr/en/index.html [updated 6 October 2009]; Available from. [Google Scholar]
- 44.Hall C.B., Douglas R.G., Jr. Clinically useful method for the isolation of respiratory syncytial virus. J Infect Dis. 1975 Jan;131(1):1–5. doi: 10.1093/infdis/131.1.1. [DOI] [PubMed] [Google Scholar]
- 45.Brittain-Long R., Nord S., Olofsson S., Westin J., Anderson L.M., Lindh M. Multiplex real-time PCR for detection of respiratory tract infections. J Clin Virol. 2008 Jan;41(1):53–56. doi: 10.1016/j.jcv.2007.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chow B.D., Huang Y.T., Esper F.P. Evidence of human bocavirus circulating in children and adults, Cleveland, Ohio. J Clin Virol. 2008 Nov;43(3):302–306. doi: 10.1016/j.jcv.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martinello R.A., Chen M.D., Weibel C., Kahn J.S. Correlation between respiratory syncytial virus genotype and severity of illness. J Infect Dis. 2002 Sep 15;186(6):839–842. doi: 10.1086/342414. [DOI] [PubMed] [Google Scholar]
- 48.Team R.D.C. 2010. A language and environment for statistical computing.http://www.R-project.org Available from. [Google Scholar]
- 49.Peterson J., Garges S., Giovanni M., McInnes P., Wang L., Schloss J.A. The NIH human microbiome Project. Genome Res. 2009 Dec;19(12):2317–2323. doi: 10.1101/gr.096651.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turnbaugh P.J., Ley R.E., Hamady M., Fraser-Liggett C.M., Knight R., Gordon J.I. The human microbiome project. Nature. 2007 Oct 18;449(7164):804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shaman J., Kohn M. Absolute humidity modulates influenza survival, transmission, and seasonality. Proc Natl Acad Sci U S A. 2009 Mar 3;106(9):3243–3248. doi: 10.1073/pnas.0806852106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hall C.B. Respiratory syncytial virus and parainfluenza virus. N Engl J Med. 2001 Jun 21;344(25):1917–1928. doi: 10.1056/NEJM200106213442507. [DOI] [PubMed] [Google Scholar]