Abstract
Cells cope with blockage of replication fork progression in a manner so that DNA synthesis can be completed and genomic instability minimized. Models for resolution of blocked replication involve fork regression to form Holliday junction structures. The human RecQ helicases WRN and BLM (deficient in Werner and Bloom syndromes, respectively) are critical for maintaining genomic stability and postulated to function in accurate resolution of replication blockage. Consistent with this notion, WRN and BLM localize to sites of blocked replication after certain DNA damaging treatments and exhibit enhanced activity on replication and recombination intermediates. Here we examined the actions of WRN and BLM on a special Holliday junction substrate reflective of a regressed replication fork. Our results demonstrate that, in reactions requiring ATP hydrolysis, both WRN and BLM convert this Holliday junction substrate primarily to a four-stranded replication fork structure, suggesting they target the Holliday junction to initiate branch migration. In agreement, the Holliday junction binding protein RuvA inhibits the WRN- and BLM-mediated conversion reactions. Importantly, this conversion product is suitable for replication with its leading daughter strand readily extended by DNA polymerases. Furthermore, binding to and conversion of this Holliday junction is optimal in low MgCl2, suggesting that WRN and BLM preferentially act on the square planar (open) conformation of Holliday junctions. Our findings suggest that, subsequent to fork regression events, WRN and/or BLM could re-establish functional replication forks to help overcome fork blockage. Such a function is highly consistent with phenotypes associated with WRN- and BLM-deficient cells.
RecQ helicases are critical for maintaining genomic stability although their precise molecular functions are still unclear. There are five human RecQ family members (RECQ1, BLM, WRN, RECQ4, and RECQ5) and loss of function of BLM, WRN and RECQ4 proteins is associated with Bloom (BS1), Werner (WS) and Rothmund-Thomson (RTS) syndromes, respectively (1–3). A hallmark feature of all these syndromes is an increased incidence of cancer that is particularly prominent in BS. In addition to an elevated predisposition particularly to soft tissue sarcomas and osteosarcomas, WS is also characterized by early onset and increased frequency of many other age-related phenotypes (4–5). A high incidence of osteosarcoma and abnormalities in skeletal development and skin pigmentation are associated with RTS (2, 6). At the cellular level, loss of BLM, WRN, or RECQ4 function results in increased chromosomal aberrations that likely underlie the elevated cancer incidence of these syndromes. However, BS, WS, and RTS phenotypes differ significantly, suggesting these enzymes have some non-redundant functions.
RecQ members share extensive homology within conserved sequence motifs characteristic of DNA-dependent ATPases and helicases, and most possess 3’ to 5’ nucleic acid unwinding activity. WRN and BLM have very similar DNA substrate specificities, preferring to act on complex DNA structures including forks, bubbles, D-loops, triplexes, Holliday junctions and G-quartets (7). Although not as well-characterized, RECQ4 has weak helicase activity that is stimulated by including complementary DNA strands during unwinding reactions (8). Surprisingly, WRN, BLM, RECQ4 and several other RecQ helicases facilitate the annealing of complementary DNA strands (9–12). Under certain circumstances, RecQ helicases (including WRN and BLM) can coordinate their unwinding and annealing activities to perform strand exchange (13–14). These biochemical studies suggest that at least a subset of human RecQ helicases may be structurally designed and enzymatically suited to act on three- and four-stranded DNA replication and recombination intermediates. In addition to unwinding and annealing activity, WRN is the only human RecQ homolog that also possesses a 3’ to 5’ exonuclease activity with similar substrate specificity for complex DNA structures as described above (15–17).
A significant threat to genome stability is blockage of DNA synthesis that occurs when replication forks encounter DNA damage, abnormal DNA structures or proteins bound to parental DNA. To ensure survival subsequent to such encounters, cells have evolved pathways to deal with replication fork blockage that facilitate replication restart and completion of normal DNA synthesis. RecQ helicases including WRN and BLM are commonly postulated to act in pathways that overcome replication fork blockage (18–20). Consistent with this hypothesis, WRN-deficient cells are hypersensitive to replication blocking agents including hydroxyurea (HU), topoisomerase inhibitors and interstrand crosslinking agents such as mitomycin C, grow slowly in culture with an extended S-phase and exhibit specific replication abnormalities including asymmetry in normal bi-directional replication fork progression (21, 22 Lebel, 1998 #302, 23–24). Furthermore, in normal cells, upon DNA damaging treatments with HU or other agents, WRN is recruited to sites of blocked replication and co-localizes with other replication factors in distinct nuclear foci (25–27). Similarly, BLM-deficient cells are also hypersensitive to mitomycin C, hydroxyurea, and aphidicolin and show replication abnormalities including problems in restart of replication forks after HU and aphidicolin treatment (28–32). Following DNA damaging treatment with HU or the interstrand crosslinking agent psoralen, BLM also relocalizes to sites of ongoing replication that contain PCNA and other replication factors (33–34). Collectively, this evidence suggests that both WRN and BLM may be involved in pathways responding to replication blockage.
According to some models, an initial step in dealing with a blocked replication fork may be fork regression to generate a Holliday junction or “chicken foot” intermediate (20, 35–37). Following fork regression, the block to replication may be overcome by 1) repair of the DNA lesion and reverse branch migration to re-establish a fork structure, 2) use of an extended lagging daughter strand as a template for the synthesis of the leading strand followed by reverse branch migration to bypass the blocking lesion to re-establish the fork, or 3) use of the free double-stranded end of this Holliday junction to initiate RAD51-mediated strand invasion and re-establishment of a functional replication fork. Alternatively, processing or resolution of the Holliday junction could generate a double-strand break that also could be utilized by RAD51-mediated recombinational pathways to restore a functional replication fork. In any case, a coordinated, error-free mechanism for resolution of replication blockage and re-establishment of the replication fork is absolutely critical for maintaining genome stability and promoting cell survival.
As both WRN and BLM bind to and branch migrate Holliday junctions in vitro (25, 38–40), each could potentially convert a regressed fork back to a functional replication fork. To determine if WRN and/or BLM might catalyze this important step in resolution of replication blockage, we constructed a specialized Holliday junction structure and investigated whether WRN or BLM could convert it to a 4-stranded fork structure that could subsequently be subject to DNA replication. Our results demonstrate that both WRN and BLM efficiently catalyze conversion of this Holliday junction to a replication fork structure, with its leading daughter strand capable of being extended in vitro by DNA polymerases. Consistent with the idea that WRN and BLM target the Holliday junction structure to initiate branch migration, this conversion reaction was inhibited by RuvA and more efficient with WRN and BLM compared to the known branch migration enzyme RAD54.Also, WRN (and BLM) show enhanced binding to and action on this Holliday junction substrate at low Mg+2 concentrations, suggesting they preferentially act on the square planar conformation of Holliday junctions. Our results suggest that WRN and/or BLM might act to reset the replication fork subsequent to fork stalling and regression, functions that are highly consistent with the replication abnormalities and specific genomic instability phenotype(s) associated with WS and BS.
EXPERIMENTAL PROCEDURES
Enzymes
All recombinant human WRN proteins (WRN-wt, WRN-E84A, and WRN-K577M) used in these studies were overexpressed in insect cells and purified as previously described (41). WRN-E84A contains a glutamate to alanine mutation that abolishes its 3’ to 5’ exonuclease activity but preserves its normal DNA-dependent ATPase, helicase, and annealing activities; WRN-K577M contains a lysine to methionine mutation that abolishes ATPase and helicase activity but retains exonuclease activity (14, 16, 42). To generate mock preparations, insect cells infected with baculovirus without WRN sequences were lysed and subject to identical purification procedures as WRN proteins. Recombinant human RECQ4 was purified as described (8). Human wild type BLM and BLM-D795A protein containing an aspartate to alanine mutation that abolishes its ATPase and helicase activities were gifts from Joanna Groden (Ohio State University), while human RPA and human (four-subunit) DNA polymerase δ were gifts from Guo-Min Li (University of Kentucky). Purified E. coli UvrD and human RAD54 were obtained from Steve Matson (University of North Carolina) and Alexander Mazin (Drexel University), respectively. Purified E. coli RuvA was purchased from B-Bridge International. Klenow Fragment (3’ to 5’ exo−), from New England Biolabs, is an N-terminal truncation of E. coli DNA Polymerase I that lacks both 5’ to 3’ and 3’ to 5’ exonuclease activities but retains polymerase activity. T4 DNA Polymerase and T4 polynucleotide kinase were purchased from New England Biolabs. Proteinase K was obtained from Invitrogen.
DNA substrate construction
The sequences of oligonucleotides (purchased from Integrated DNA Technologies) used in this study are provided in Supplemental Table 1. LeadD81, LagP70 and 3-way-base62 were radiolabeled (indicated by asterisk hereafter and in figures) at their 5’ ends by standard procedures using 32P-γ-ATP and T4 polynucleotide kinase, and unincorporated nucleotide was removed using a spin column (Roche Applied Science). Unlabeled LagD84 was annealed to *LeadD81 by heating to 95°C for 5 min and slow cooling; in parallel, unlabeled LeadP122 and LagP122 were similarly annealed. The resulting 2-stranded fork structures were annealed to each other for 16 h at 25°C to generate a singly-labeled, static Holliday junction structure (see Fig. 1A, left). To generate the four-stranded replication fork substrate, the heating and slow cooling procedure was used to anneal 1) *LeadD81 with unlabeled LeadP122 and 2) unlabeled LagD84 with unlabeled LagP122, and subsequently the resulting partial duplexes were annealed for 16 h at 25°C. Two-stranded fork (*LeadD81/LagD84 and *3-way-base62/3-way-5’flap), partial duplex (*LeadD81/LeadP122 and *LagP70/LagD30), and 3-way junction (*3-way-base62/3-way-3’flap/3-way-5’flap) DNA substrates were generated by annealing the relevant oligonucleotides using the heating and slow cooling procedure. After formation, all substrates were purified by native polyacrylamide (6%) gel electrophoresis, excised from the gel after autoradiography, eluted for 24 h into buffer containing 10 mM Tris (pH 8.0) and 10 mM NaCl, and stored at 4°C.
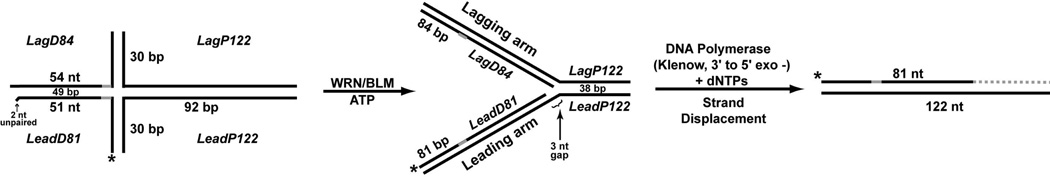
Figure 1. Diagram of Holliday junction substrate structure, conversion and extension products.
Schematic showing putative WRN- or BLM-mediated conversion of our specialized Holliday junction structure (left) to a replication fork (center) with subsequent DNA polymerase-mediated extension from the 3’ end of the labeled (denoted by asterisk) leading daughter strand of the fork; most experiments utilized Klenow fragment, 3’ to 5’ exo− as DNA polymerase, resulting in strand displacement and new DNA synthesis (indicated by dotted gray line) to the end of the parental leading strand template (right). While the vertical arms of this Holliday junction substrate are non-homologous, the left arm is completely homologous to the proximal 51–54 bp of the right arm except for 5 bp of non-homology (denoted by solid gray segment) at the junction. This design ensured a static Holliday junction structure that could be converted to a replication fork structure (by enzymatically branch migrating the junction rightward, overcoming the short non-homologous segment). Note that in the context of the Holliday junction, 2 nt at the 3’ end of the labeled strand were unpaired; upon conversion to replication fork structure these nucleotides are fully base-paired. Names of the oligos used for substrate construction are italicized; nucleotide sequences for these oligos are given in Supplemental Table 1.
Enzymatic assays
For studying the conversion of Holliday junction to fork DNA, Holliday junction substrate (~2 fmol) was incubated for the specified times at 37°C with WRN-E84A, WRN-wt, WRN-K577M, BLM, BLM-D795A, RAD54, RECQ4 or UvrD (at concentrations indicated in the figure legends) in WRN reaction buffer (20 µl) containing 40 mM Tris-HCl (pH 8.0), 0.1 mg/ml BSA, 5 mM dithiothreitol, 0.1% Nonidet P-40 and supplemented with MgCl2 (0.5–8 mM, as indicated) and either ATP (0.125–2 mM) or ATPγS (1 mM). Prior to incubation at 37°C in specific experiments, 1) RuvA (4–1000 fmol) was pre-incubated with Holliday junction substrate for 5 min at 4°C before addition of WRN-E84A or BLM, or 2) RPA (2.5–80 fmol) was added subsequent to WRN-E84A while the reactions were kept at 4°C. Unwinding assays using 2-stranded fork and partial duplex substrates were performed similarly; however, assays with RECQ4 and 3-way junction substrate also contained a 25-fold excess of unlabeled 3-way-base62 oligomer. Reactions were terminated by the addition of 5 mM EDTA; then, 10 µg Proteinase K was added and the samples were incubated at 37°C for 10 min. Following addition of one-sixth volume of loading dye (30% glycerol, 0.25 % bromphenol blue, 0.25% xylene cyanol), samples were subjected to native polyacrylamide (5–6%) gel electrophoresis (PAGE) in 1X TBM (90 mM Tris-borate, pH 8.0, 5 mM MgCl2) at 125 V for 3.5 h. The gels were dried and the radioactivity associated with the DNA products was visualized and quantitated by phosphorimaging analysis (Storm 860 phosphorimager and ImageQuant software, GE Healthcare). Conversion of Holliday junction to four-stranded replication fork was measured as a percentage of the radioactivity associated with replication fork to that of the total DNA, after subtraction of low background levels of DNA species present in control reactions without enzyme. Similarly, substrate unwinding was quantified as the percentage of radioactivity associated with the unwound product with respect to the total DNA in the reaction, again correcting for any background level of the product species in reactions without enzyme. To assess the 3’ to 5’ exonuclease activity inherent in WRN-wt and WRN-K577M, DNA products from reactions performed as above were separated by denaturing PAGE (10%); after gel drying, exonucleolytic degradation from the 5’ end-labeled *LeadD81 strand of the substrate/products was visualized by phosphorimaging.
Assays were also performed to study the action of DNA polymerases on the conversion product as well as various control substrates. For these studies, Holliday junction, four-stranded replication fork, and two-stranded fork (*LeadD81/LagD84) substrates (2–4 fmol), were incubated with or without either WRN-E84A, mock preparation of WRN, BLM or BLM-D795A (at concentrations specified in the figure legends) in WRN reaction buffer (20 µl) containing 4 mM MgCl2, either 1 mM ATP or 1 mM ATP γS and 100 µM dNTPs. Following incubation at 37°C, Klenow (3’ to 5’ exo−) (0.01– 0.001units/rxn), T4 DNA polymerase (0.005 U), or human DNA polymerase δ (60 fmol) was added with further incubation at 37°C (times indicated in figure legends). These reactions were stopped by withdrawing a 10 µl aliquot from the reaction and adding an equal volume of formamide loading buffer (95% formamide, 20 mM EDTA, 0.1% bromphenol blue, and 0.1% xylene cyanol). The DNA products were subsequently heated at 90°C for 5 min and separated by denaturing PAGE (6%) in 1× TBE at 40–45 W for 1.5 h. The gels were dried and analyzed by phosphorimaging as described above. The percentage of extension (by Klenow) of the labeled leading daughter strand (of four-stranded forks converted from Holliday junctions) was calculated by dividing the combined signal for all products of higher molecular weight (than the original unextended strand) to the total radioactive signal from the entire lane (amount of unextended leading daughter strand + combined extended products).
Electrophoretic Mobility Shift Assay (EMSA)
Holliday junction substrate (~4 fmol) was incubated with WRN-E84A (0–12 fmol) in WRN reaction buffer (20 µl) supplemented with either 1 or 8 mM MgCl2 for 10 min at 25°C. Subsequently, one-sixth volume of 30% glycerol was added and samples were separated by native PAGE (3.5%, acrylamide:bisacrylamide crosslinking ratio of 37.5:1) in 0.5× TBE at 100 V for 2 h. The gels were dried and subjected to phosphorimaging analysis as described above. WRN-E84A binding was assessed by comparing the amount of bound DNA with the total DNA for each reaction [% DNA bound = (DNA bound/total DNA) × 100].
RESULTS
Specialized Holliday junction substrate design
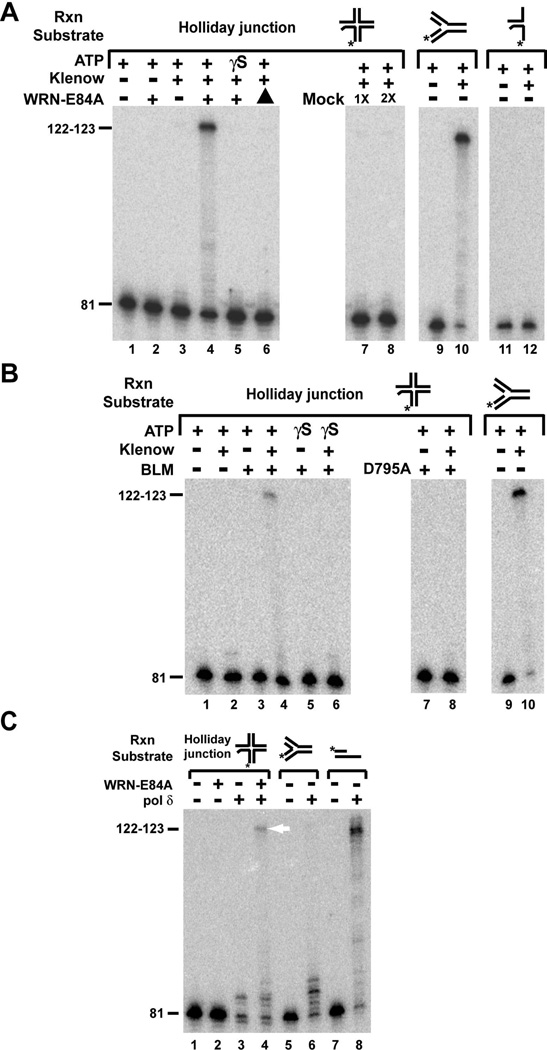
It has been postulated that, as part of a pathway to overcome replication blockage, some RecQ helicases including WRN and BLM may reset the replication fork from Holliday junctions (or chicken foot structures) generated by fork regression (18, 20). To examine a potential role for WRN or BLM in re-establishing a functional replication fork following regression, we constructed a special Holliday junction substrate (Fig. 1, left) using a two-step process: independently annealed *LeadD81/LagD84 and LeadP122/LagP122 species were subsequently annealed to one another. The resulting Holliday junction substrate contained two 30 bp arms (pictured vertically in Fig. 1) that were completely non-homologous; the left arm was completely homologous to the proximal 51–54 bp of the right arm, except for 5 non-homologous bp at the junction. As constructed, this structure was static but could potentially be converted to a four-stranded replication fork (Fig. 1, center) by branch migration of the Holliday junction (rightward in this orientation) through the region of non-homology. In the event this conversion occurred, this replication fork structure would contain a 3 nt single-stranded gap on the leading arm at the fork junction. In the original Holliday junction substrate, the strand that would become the leading daughter strand following conversion to fork was labeled so that DNA polymerase-mediated extension from its 3’ end could be monitored. In the context of the Holliday junction structure, the last 2 nucleotides on the 3’ end of this strand were unpaired; this ensured that this strand could not be extended by a DNA polymerase unless conversion occurred (or without removal of the unpaired nucleotides by a 3’ to 5’ exonuclease). The design of this substrate allowed us to monitor enzyme-mediated conversion to the four-stranded replication fork structure by assaying 1) a structure-dependent change in migration by native PAGE and 2) extension from the 3’ end of the labeled strand by DNA polymerases, analyzed by denaturing PAGE. Notably, strand displacement activity inherent in Klenow fragment (and, to a lesser degree, in human DNA pol δ) allowed polymerization of the labeled leading daughter strand of the four-stranded fork structure (Fig. 1, center) beyond the fork junction to the end of the leading parental strand template to generate labeled extension products of up to 122–123 nt (Fig. 1, right). The following sections detail how purified WRN and BLM proteins act on this specialized Holliday junction substrate.
WRN and BLM convert Holliday junction substrate to four-stranded replication fork
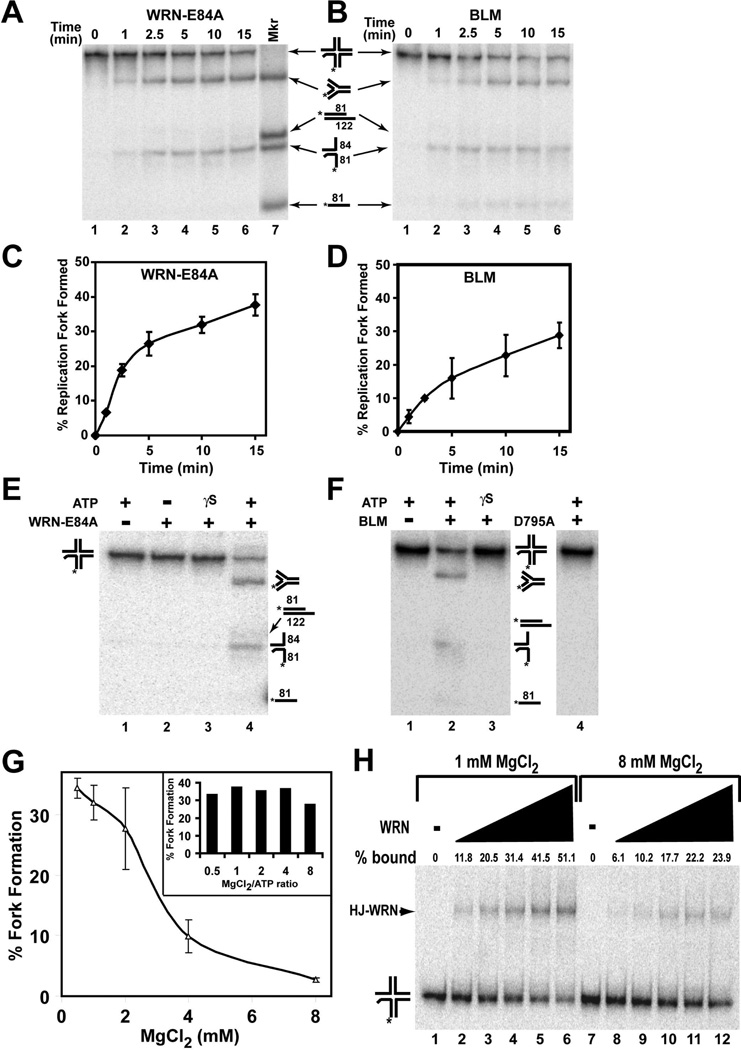
To test the hypothesis that RecQ helicases may be able to act on a Holliday junction structure reflecting a regressed fork and re-establish a functional replication fork, our Holliday junction substrate was incubated with or without purified enzymes. For most WRN-containing reactions, exonuclease-deficient WRN-E84A mutant was used so that the reaction products could be analyzed without complication from possible digestion of the substrate or the products; importantly, use of this mutant also eliminated the possibility that the 3’ to 5’ exonuclease activity of WRN could remove the 2 unpaired nucleotides on the 3’ end of the labeled strand of the original Holliday junction substrate. Under standard conditions of electrophoresis using native PAGE with 1× TBE as the running buffer, the mobilities of the Holliday junction substrate and the four-stranded fork DNA marker representing the conversion product were quite similar. Since it has been shown previously that the mobility of Holliday junctions in polyacrylamide gels is influenced by divalent cation concentration (43–44), we included 5 mM MgCl2 (and omitted EDTA) in both our gels and running buffers. Under these conditions, we obtained adequate separation between the Holliday junction and the four-stranded fork DNA, the former migrating substantially slower than the latter. Theoretically, unwinding of a 4-stranded Holliday junction by a helicase could yield a number of 1-, 2-, or 3-stranded products; in fact, WRN and BLM have been previously shown to unwind partly mobile Holliday junction substrates to generate partial duplex as well as single-stranded products (39–40). However, when we incubated our specialized Holliday junction substrate with either WRN-E84A or BLM in the presence of 4 mM MgCl2 and 1 mM ATP, the primary product generated over time was the four-stranded replication fork structure (Fig. 2A–D); little or no further unwinding of this fork to a partial duplex (*LeadD81/LeadP122) was detected. In addition to the conversion of Holliday junction to 4-stranded fork, some unwinding of the Holliday junction to form the 2-stranded *LeadD81/LagD84 fork species was also observed in these reactions (Fig. 2A and B). Conversion of Holliday junction to four-stranded fork by both WRN-E84A and BLM was direct—i.e., formation of the fork-stranded fork did not result from separate unwinding and annealing steps; note that the *LeadD81/LagD84 species could not be a relevant intermediate in fork formation and no significant amounts of other products were detected at early time points in kinetic assays (Fig. 2A and B). Notably, this conversion reaction requires ATP hydrolysis, as fork formation was not detected in reactions lacking ATP or containing ATPγS, a non-hydrolyzable analog of ATP (Fig. 2E and F). Furthermore, use of ATPase- and helicase-deficient BLM-D795A or WRN-K577M mutant proteins in these reactions also did not cause any detectable conversion of Holliday junction to fork product (Fig. 2F and Supplemental Fig. 1), confirming that these ATPase- and helicase-dependent conversion reactions are specifically catalyzed by BLM and WRN. Notably, the Holliday junction substrate is subject to degradation by the 3’ to 5’ exonuclease activity retained in WRN-K577M (Supplemental Figure 1, lower panel).
Figure 2. Direct conversion of specialized Holliday junction to a replication fork by WRN-E84A and BLM: ATP and Mg+2 Dependence.
Holliday junction substrate (~2 fmol) was incubated with A) WRN-E84A (18 fmol) or B) BLM (12.5 fmol) in MgCl2 (4 mM) and ATP (1 mM) at 37°C for the indicated times. DNA products (lanes 1–6) and marker substrates (lane 7: 4-stranded replication fork, *LeadD81/LeadP122 partial duplex, *LeadD81/LagD84 fork, and *LeadD81 oligomer, migration positions depicted at right) were analyzed by native PAGE as described in Experimental Procedures. Relative positions of migration of these marker substrates are applicable for panels B, E and F and elsewhere. C) The percentage of replication fork formation (calculated as described in Experimental Procedures) mediated by WRN-E84A is plotted versus time for ATP-containing reactions carried out as in A. Data are the mean and S.E. of 3 independent experiments. D) Data from experiments performed as in B were calculated and presented as in C. E) Holliday junction substrate (~2 fmol) was incubated for 30 min at 37°C with WRN-E84A (15 fmol) with or without ATP or ATPγS (1 mM) as indicated and the DNA products analyzed by native PAGE as in A. Positions of migration of specific DNA structures are depicted here at right and also in panel F F) Holliday junction substrate (~2 fmol) was incubated with BLM (12.5 fmol) or BLM-D795A (ATPase- and helicase-deficient mutant) (37.5 fmol) with ATP or ATPγS (1 mM) as indicated and incubated at 37°C for 30 min, then analyzed as in A. G) Holliday junction substrate (~2 fmol) was incubated with WRN-E84A (18 fmol) in the presence of ATP (1 mM) and varying concentrations of MgCl2 (0.5, 1, 2, 4 and 8 mM) for 30 min at 37°C and the reaction products analyzed by native PAGE as for Fig. 1. For 3 independent experiments, the mean percentage (±standard deviation) of conversion of Holliday junction to fork structure is plotted versus the MgCl2 concentration. (bar graph inset) Similarly, reactions containing Holliday junction substrate (~2 fmol), WRN-E84A (18 fmol), 1 mM MgCl2 and varying concentrations of ATP (0.125, 0.25, 0.5, 1 and 2 mM) were incubated for 30 min at 37°C and the percentage of conversion to fork structure is depicted for each MgCl2/ATP ratio. H) Holliday junction substrate (4 fmol) was incubated with WRN-E84A (0, 1.5, 3, 6, 9, and 12 fmol) in the absence of ATP and in the presence of either 1 or 8 mM MgCl2 as indicated for 10 min at 25°C and protein-DNA complexes resolved from free DNA by EMSA as described in Experimental Procedures. The percentage of Holliday junction bound by WRN-E84A is calculated as described in Experimental Procedures and denoted above each lane. Positions of free Holliday junction substrate and Holliday junction-protein (HJ-WRN) complexes are noted at left.
The precise structure of Holliday junctions is known to be variable dependent upon Mg+2 ion concentration, with a stacked X-structure becoming favored over a square planar structure as Mg+2 concentration increases (43–44). Since our results and previous reports show that WRN and BLM readily act on Holliday junction structures, it is highly relevant to also determine the effect of MgCl2 concentration on WRN- and BLM-mediated conversion of our Holliday junction to replication fork product, keeping in mind that Mg+2 is a required cofactor in these reactions. In the presence of a fixed amount of ATP (1 mM), the amount of conversion of a Holliday junction to fork product by WRN-E84A was highly dependent on the MgCl2 concentration (Fig. 2G), being optimal between 0.5 (the minimum concentration tested) and 2 mM MgCl2. WRN-E84A-mediated conversion declined sharply at 4 mM MgCl2 and even more drastically at 8 mM MgCl2.
In order to determine whether it is the absolute concentration of MgCl2, or the MgCl2/ATP ratio in the reaction that influences this activity, reactions were set up in which ATP concentration was varied over a wide range (0.125–2 mM) in the presence of 1 mM MgCl2, an optimal concentration in the experiment above. Thus, the MgCl2/ATP ratio in these reactions varied between 0.5 and 8. The results of this experiment (Fig. 2G, inset) reveal no substantial difference in the conversion of Holliday junction to fork DNA by WRN-E84A over the range of MgCl2/ATP ratios tested, with conversion percentages ranging between 28–38%. Given that the inhibitory effect of increasing MgCl2 concentration from 2 mM to either 4 or 8 mM on fork formation was much greater, our results suggest that a higher absolute amount of Mg+2 was inhibitory to the WRN-mediated conversion process. In contrast, unwinding of a 3’ overhang partial duplex by WRN-E84A was not significantly inhibited by MgCl2 concentrations up to 4 mM (Supplemental Fig. 2) suggesting that the marked inhibition of the Holliday junction conversion reaction at Mg+2 concentrations of 4 mM and above was not due to general loss of WRN catalytic activity. The effect of MgCl2 concentration on BLM-mediated conversion followed the same general pattern as WRN-E84A, although the inhibitory effect at 4 and 8 mM MgCl2 was much less pronounced compared to WRN-E84A (Supplemental Fig. 3A). These findings are consistent with an effect of higher Mg+2 concentration on the structure of the Holliday junction substrate that lowers the ability of WRN-E84A and BLM to catalyze the conversion reaction.
To further explore whether the nature of Holliday junction structure influenced enzyme function, we used EMSA to determine if there was any difference in the binding affinity of WRN-E84A to the Holliday junction substrate under low (1 mM) and high (8 mM) MgCl2 conditions. It has been shown previously that some RecQ helicases including WRN and BLM exhibit high affinity binding and enhanced catalytic activities on 3- and 4-stranded DNA structures including Holliday junctions and forks as compared to single-stranded and duplex DNA (7, 39, 45–46). When binding reactions were performed in the presence of 1 mM MgCl2, WRN-E84A formed stable and specific complexes with Holliday junction substrate as observed after EMSA (Fig. 2H). The amount of enzyme-DNA complex was dependent on WRN-E84A with >50% of the substrate bound at the highest concentration. Although discrete and stable binding was observed between WRN-E84A and Holliday junction DNA when binding reactions contained 8 mM MgCl2, the level of binding was significantly reduced at each concentration of WRN-E84A with less than 25% of the original DNA stably bound at the highest concentration (Fig. 2H). This clearly demonstrates that the initial binding of WRN to Holliday junction substrate or stability of the protein-DNA complexes is higher at 1 mM than at 8 mM MgCl2. Taken together, our results indicate that increasing Mg+2 concentrations (>2 mM) inhibit the binding and conversion activity of WRN on our Holliday junction substrate. Furthermore, these findings suggest that these enzymes preferentially bind to and act on the square planar conformation of Holliday junctions compared to the stacked X form.
Specificity of the WRN- and BLM-mediated conversion reaction
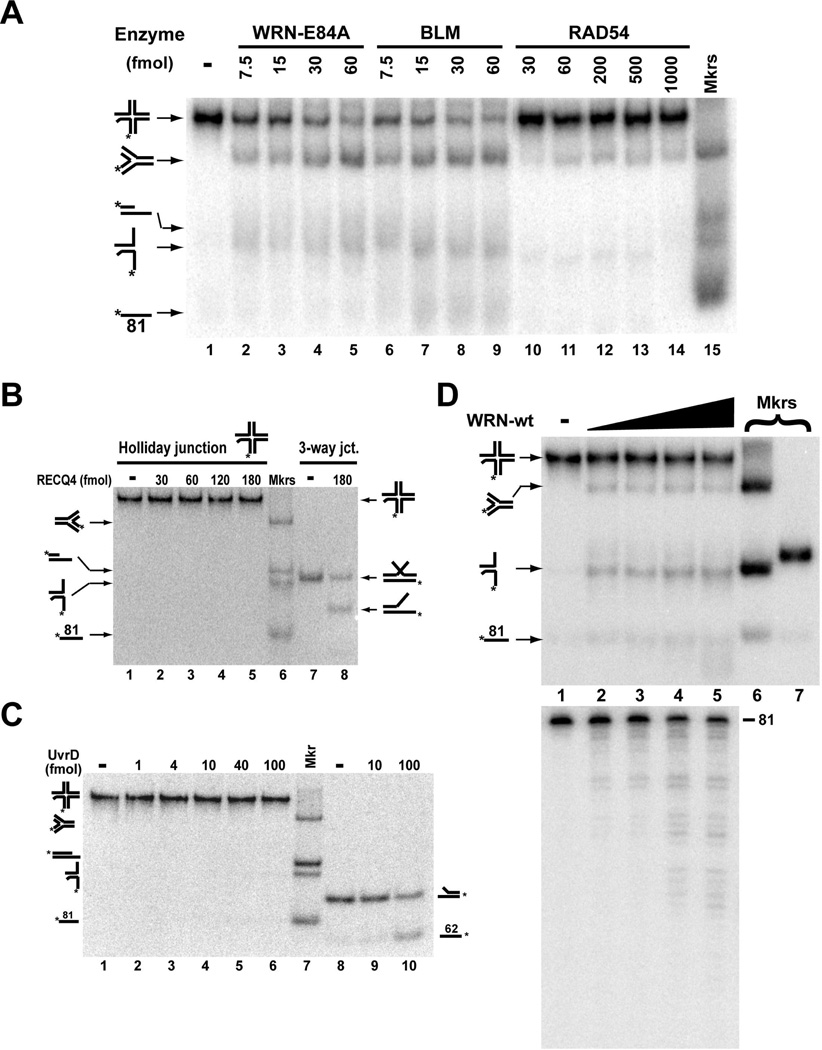
Substantial evidence implicates both WRN and BLM helicases in the response to replication fork blockage and replication restart. Biochemically, each enzyme has previously been demonstrated to have not only helicase activity but also branch migration activity (25, 38). Our conversion reaction, the re-establishment of a replication fork from a (regressed) Holliday junction, would seem to most closely reflect branch migration, and thus it may shed light on the physiological function of these enzymes and perhaps other RecQ helicases. However, we wanted to determine the relative specificity and/or preference of WRN and BLM to catalyze this conversion in comparison with other helicases and branch migration enzymes. Human RAD54 has been demonstrated to branch migrate Holliday junctions and recently has been shown to convert a Holliday junction substrate to replication fork structure, quite similar to our conversion reaction (47–48). Therefore, we directly compared the actions of WRN-E84A, BLM, and RAD54 on our Holliday junction substrate. WRN-E84A and BLM catalyzed similar levels of conversion at equivalent protein concentrations, with the percentage of conversion (compared to the total DNA in the reaction) reaching 42.3 and 36.6%, respectively, at the highest enzyme concentrations (60 fmol) tested (Fig. 3A, lanes 2–9). Although the four-stranded replication fork was detectable when RAD54 was used, the level of conversion was very limited (a maximum of 10.2%) even at much higher protein concentrations (Fig. 3A, lanes 10–14). It is also notable that RAD54 could not detectably catalyze this conversion reaction when a higher MgCl2 concentration (4 mM) was used (Supplemental Fig. 3B). We also tested another RecQ helicase, human RECQ4, and a typical bacterial helicase, E. coli UvrD, in our conversion reaction. Interestingly, RECQ4 unwinding appears to require an “extra” complementary strand (8), suggesting it may catalyze a strand exchange type of reaction instead of typical unwinding. Indeed, we were able to confirm this type of unwinding/strand exchange activity in RECQ4 when using a 3-way junction substrate with a 25-fold excess of complementary strand (Fig. 3B, lane 8). However, RECQ4 had no detectable activity on our Holliday junction substrate (Fig. 3B, lanes 2–5). Similarly, UvrD did not facilitate formation of the fork product or any other DNA species from our Holliday junction substrate over a wide concentration range (Fig. 3C, lanes 2–6), although this helicase could readily unwind a 2-stranded fork substrate (Fig. 3C, lanes 9–10). Taken together, our results indicate that the conversion of our Holliday junction substrate to a four-stranded replication fork involves branch migration activity as opposed to simple unwinding or strand exchange activity. Perhaps more importantly, WRN and BLM preferentially catalyze this specific reaction in comparison to another branch migration enzyme, human RAD54, while this activity is completely lacking in another human RecQ helicase, RECQ4.
Figure 3. WRN and BLM preferentially convert the Holliday junction to replication fork.
A) Holliday junction substrate (~2 fmol) was incubated with WRN-E84A, BLM, or RAD54 at the indicated concentrations for 30 min at 37°C in 1 mM MgCl2 and 1 mM ATP. DNA products of these reactions (lanes 1–14) and markers (lane 15) were analyzed as in Fig. 2A. B) Holliday junction (~2 fmol) or 3-way junction substrate (5 fmol, plus 125 fmol of unlabeled 3-way-base62 trap strand) was incubated with human RECQ4 at the indicated concentrations for 30 min at 37°C in 4 mM MgCl2 and 1 mM ATP. DNA products and markers were analyzed as in A. C) At the indicated concentrations, UvrD was incubated with either 2 fmol of Holliday junction substrate (lanes 1–6) or 5 fmol of two-stranded fork (*3-way-base62/3-way-5’flap) substrate (lanes 8–10) for 30 min at 37°C. Specific pre-formed DNA structures were loaded as markers (lane 7) with their positions of migration depicted on the left and right sides. D) Holliday junction (~2 fmol) was incubated without or with WRN-wt (7.5, 15, 30 and 45 fmol) for 30 min at 37°C in 1 mM MgCl2 and 1 mM ATP. DNA products of these reactions (lanes 1–5) were analyzed both by native PAGE (top) and denaturing PAGE (bottom). Also loaded on native PAGE were markers for replication fork and *LeadD81/LagD84 fork species (lane 6) and for the *LeadD81LeadP122 partial duplex species (lane 7).
WRN is unique among the human RecQ helicases in that it also possesses an inherent 3’ to 5’ exonuclease, the physiological function of which remains unclear. However, previous reports have shown that WRN’s exonuclease activity is highly active on 3’ ends proximal to unusual DNA structures including forks and bubbles (17, 42, 49). Thus, we investigated the processing of our specialized Holliday junction substrate by WRN-wt, using native PAGE to analyze the overall effect on the substrate and denaturing PAGE to assess exonucleolytic degradation from the 3’ end of the labeled strand. When the substrate was incubated with WRN-wt (Fig. 3D, top panel), again the 4-stranded replication fork product was generated, although present in seemingly lower amounts than for WRN-E84A. Also, as compared with WRN-E84A, higher levels of products apparently stemming from *LeadD81/LagD84 fork and single-stranded *LeadD81 were noted, suggestive of significant unwinding. Examination of the same reactions by denaturing PAGE showed substantial digestion from the 3’ end of the radiolabeled strand (Fig. 3D, bottom panel), indicating that WRN’s 3’ to 5’ exonuclease activity could readily target the reaction substrate and/or products. Upon close examination, WRN-mediated exonucleolytic degradation is also noticeable on native PAGE, as evidenced by the slightly faster migration of the 2-stranded fork product than its undigested *LeadD81/LagD84 marker and the faint smear of labeled single-stranded products downward compared to the position of the single-stranded *LeadD81 marker (Fig. 3D, top panel). Importantly, denaturing PAGE cannot reveal degradation of the unlabeled strands of the substrate or products, and it seems clear that the substrate and products are structurally altered by WRN exonuclease activity. The notion that WRN’s exonuclease activity can alter the original Holliday junction structure is confirmed by the 3’ to 5’ digestion of the labeled strand observed in the presence of WRN-K577M, the ATPase- and helicase-deficient mutant that cannot unwind or branch migrate the substrate (Supplemental Fig. 1, bottom panel). Given that the substrate and product structures are in flux during the course of the reaction due to WRN exonuclease activity, it becomes very difficult to accurately assess how the combined helicase and exonuclease activities process this substrate. Nevertheless, it can be concluded that, in the context of the Holliday junction/chicken foot structure, the 3’ end of the leading daughter strand is readily accessible to WRN exonuclease function. We can also conclude that, despite ongoing WRN exonuclease-mediated digestion of the Holliday junction substrate, WRN-wt protein converts a substantial amount of this substrate to 4-stranded replication fork.
Effects of RuvA and RPA on WRN- and BLM-mediated conversion of Holliday junction to replication fork
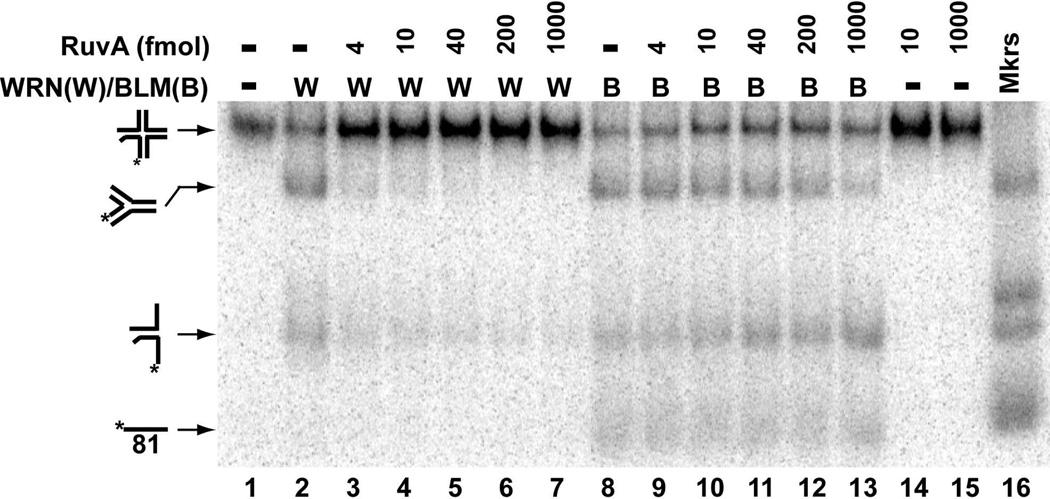
The WRN- and BLM-mediated conversion reaction described above suggests that each enzyme can specifically bind to the Holliday junction structure and initiate branch migration to result in replication fork formation. To determine whether WRN and BLM specifically target the Holliday junction, we tested the effect of E. coli RuvA, a Holliday junction-specific binding protein without catalytic activity, on our conversion reactions. In these experiments, RuvA was pre-incubated with the Holliday junction substrate before either WRN-E84A or BLM was added to initiate the reaction; as in standard unwinding/conversion assays, the DNA products were examined by native PAGE after removal of the proteins. As expected, RuvA alone had no effect on the Holliday junction substrate (Fig. 4, lanes 14–15). As previously, WRN-E84A alone primarily catalyzed conversion of the Holliday junction to 4-stranded replication fork, as did BLM alone (Fig. 4, lanes 2 and 8). Even at the lowest concentration tested, RuvA drastically reduced WRN-mediated production of the replication fork and further inhibited this reaction when higher levels of RuvA were employed (Fig. 4, lanes 3–7). Surprisingly, the effect of RuvA in reactions containing BLM was more modest, with substantial inhibition of BLM-mediated conversion only observed at the highest RuvA concentrations (Fig. 4, lanes 9–13). These results suggest that RuvA binding to the Holliday junction essentially blocks WRN from accessing the substrate to catalyze conversion, but somehow BLM is able to almost completely override RuvA’s effect.
Figure 4. RuvA inhibits WRN- and BLM-mediated conversion of Holliday junction to replication fork.
Holliday junction substrate (2 fmol) was pre-incubated with RuvA (0–1000 fmol, as indicated) in WRN reaction buffer containing 1 mM MgCl2 and 1 mM ATP at 4°C for 5 min; where indicated, WRN-E84A (30 fmol) or BLM (12 fmol) was added and the reactions were incubated for 30 min at 37°C. The DNA products (lanes 1–9), along with markers for four-stranded replication fork, *LeadD81/LeadP122, and *LeadD81/LagD84 (lane 10), were separated by native PAGE and visualized as described in Experimental Methods.
Replication protein A (RPA) has been postulated to play an important role in the response to replication blockage and has also been demonstrated to modulate WRN and BLM activities (25, 50–53). It is therefore relevant to determine whether RPA influences the conversion of Holliday junction to replication fork. To this end, our specialized Holliday junction substrate was incubated with WRN-E84A or BLM in the absence or presence of increasing concentrations of human RPA. RPA alone had no effect on the substrate (Supplemental Fig. 4A and B, lanes 9–11). As before, either WRN-E84A or BLM alone primarily generated the four-stranded replication fork (Supplemental Fig. 4A and B, lane 2) Addition of RPA to reactions containing WRN-E84A or BLM had only a very minor effect on these reactions. Specifically, the primary product in both WRN-E84A and BLM-containing reactions remained the four-stranded replication fork with little or no change in its abundance or that of the *LeadD81/LagD84 unwinding product, even at RPA levels that were in 40-fold excess with respect to the substrate (Supplemental Fig. 4A and B, lanes 3–8). In the WRN-E84A reactions, perhaps a very minor amount of the *LeadD81/LeadP122 partial duplex product is barely detectable at the highest RPA concentrations, probably a result of (forward) unwinding of the converted four-stranded replication fork. In the BLM-containing reactions, a modest increase in the single-stranded *LeadD81 product is observed at the highest RPA concentrations, potentially resulting from substrate and/or product unwinding. Taken together, our results suggest that, at least for this reaction, RPA does not significantly shift the reaction in favor of or against replication fork formation, nor does it significantly alter the preference of WRN-E84A or BLM to perform branch migration versus unwinding.
DNA synthesis subsequent to WRN- or BLM-mediated conversion of Holliday junction substrate
To be productive in the context of overcoming replication blockage, enzyme-mediated action on a regressed fork (Holliday junction/chicken foot) should re-establish a functional fork subject to replication by DNA polymerases. To investigate this possibility, we sequentially incubated the Holliday junction substrate with either WRN-E84A or BLM followed directly by various DNA polymerases, under conditions otherwise identical to those of the conversion reactions except with the inclusion of dNTPs. Potential polymerase-mediated extension of the labeled strand was monitored after the reaction products were heated at 90°C and separated by denaturing PAGE. As mentioned above, there are 2 unpaired nucleotides at the 3’ end of the labeled strand in the context of the original Holliday junction substrate; these nucleotides are fully base-paired upon conversion to the 4-stranded fork. Thus, for most reactions, the DNA polymerase used was Klenow fragment, 3’ to 5’ exo−, (hereafter referred to as Klenow) because its lack of 3’ to 5’ exonuclease activity eliminated the possibility of extension from the unpaired 3’ end of the original Holliday junction substrate. This was experimentally confirmed in reactions containing only Holliday junction substrate and Klenow that showed no extension of the labeled strand (Fig. 5A, lane 3 and Fig. 5B, lane 2). As expected, extension was also not detectable when only WRN-E84A or BLM was present (Fig. 5A, lane 2 and 5B, lane 3). Only when Klenow was added to reactions treated with either WRN-E84A or BLM in the presence of ATP was extension of the labeled strand observed (Fig. 5A and B, lane 4). In these reactions, the primary extension product was 122–123 nt, indicating that Klenow had synthesized beyond the fork junction to the end of the template (LeadP122) strand (see Fig. 1, right). The identical product was also observed in control reactions when Klenow alone was incubated with the 4-stranded fork (Fig. 5A and B, lane 10) or the *LeadD81/LeadP122 partial duplex (data not shown). This result indicated that the strand displacement property inherent in Klenow is mostly responsible for extension beyond the fork junction, with possible assistance from WRN-E84A or BLM unwinding activity. The shortest extension products observed may be due to pausing caused by the fork junction while intermediate products are likely pause sites specific for Klenow. Notably, *LeadD81/LagD84, a minor unwinding product in the conversion reaction (Fig. 2A,B,E,F) containing 2 unpaired nucleotides at the 3’ end of the labeled strand, could not be extended by Klenow alone (Fig. 5A, lanes 11 and 12). This result confirmed that all of the extension observed in our Holliday junction-containing reactions takes place on the converted 4-stranded replication fork product. In control reactions containing Klenow, extension products were not observed when 1) ATPγS was substituted for ATP in the presence of WRN-E84A or BLM (Fig. 5A, lane 5 and 3B, lane 6), 2) WRN-E84A was heat-denatured prior to addition (Fig. 5A, lane 6), 3) mock-WRN preparations were substituted for WRN-E84A (Fig. 5A, lanes 7,8), or 4) ATPase- and helicase-deficient BLM-D795A was substituted for wild type BLM (Fig. 5B, lane 8). These findings demonstrate that conversion of our specialized Holliday junction substrate to a replication fork structure is a bona fide activity of WRN and BLM and indicate that ATPase-dependent conversion to four-stranded fork is required for subsequent extension of the labeled (leading daughter) strand by Klenow. It is notable that, although WRN can utilize dATP or dCTP hydrolysis to drive unwinding (50), a 10-fold excess of ATPγS over dATP (1 vs. 0.1 mM, respectively) in these reactions is sufficient to completely inhibit conversion of Holliday junction to four-stranded fork. In WRN-containing reactions, approximately 50% of the labeled strand was extended (to any extent), suggesting that most if not all of the converted substrate (compare to Fig. 2C) was extended partially or fully by Klenow. For BLM and Klenow-containing reactions using the Holliday junction substrate, the percentage of extension of the labeled strand was lower (~25%) than with WRN-E84A, consistent with lower BLM-mediated conversion under these conditions (compare Figs. 2C and D).
Figure 5. Extension of WRN- and BLM-converted Holliday junction substrate by Klenow (3’ to 5’ exo−) and human DNA pol δ.
A) Holliday junction substrate (2 fmol) was incubated with either WRN-E84A (18 fmol) or mock-purified protein preparation (volume normalized, 1X and 2X) (lanes 7,8) in the presence of dNTPs (100 µM) and either ATP or ATPγS (1 mM) as specified for 10 min at 37°C, followed by the addition of Klenow (3’ to 5’ exo−) (0.001U) where indicated and further incubation at 37°C for 20 min. In the reaction depicted in lane 6, WRN-E84A was heat-denatured at 65°C for 5 min before its addition. Similarly, 4-stranded fork substrate (2 fmol, lanes 9 and 10) and *leadD81/lagD84 substrate (2 fmol, lanes 11 and 12) were also incubated without or with Klenow (3’ to 5’ exo−) (0.001 and 0.002 U, respectively) at 37°C for 20 min in the presence of dNTPs (100 µM) and ATP (1 mM). Products were analyzed by denaturing PAGE as described in Experimental Procedures. B) Holliday junction substrate (~2 fmol) was incubated without or with BLM (12.5 fmol) or BLM-D795A (37.5 fmol) in the presence of dNTPs (100 µM) and ATP or ATPγS (1 mM) for 10 min at 37°C, followed by the addition of Klenow (3’ to 5’ exo−) (0.001 U) as indicated and further incubation at 37°C for 20 min. Similar reactions containing ATP (1 mM), dNTPs (100 µM) and 4-stranded fork substrate (2 fmol) without or with Klenow (3’ to 5’ exo−) (0.001 U) (lanes 9,10) were incubated for 20 min at 37°C. Reaction products were analyzed as described in A. C) Holliday junction substrate (2 fmol, lanes 1–4) was incubated with WRN-E84A (18 fmol) in the presence of ATP (1 mM) and dNTPs (100 µM) for 10 min at 37°C, followed by the addition of human DNA pol δ (60 fmol) where indicated and further incubation at 37°C for 20 min. Similarly, 4-stranded fork substrate (2 fmol, lanes 5 and 6) and *LeadD81/LeadP122 substrate (2 fmol, lanes 7 and 8) were also incubated without or with pol δ (15 fmol) at 37°C for 20 min in the presence of dNTPs (100 µM) and ATP (1 mM). Products were analyzed as in A. The white arrow indicates the fully extended 122–123 nt product only in the specific Holliday junction-containing reaction incubated with WRN-E84A followed by pol δ.
We also performed similar reactions on our Holliday junction substrate with WRN-E84A and human polymerase δ (Fig. 5C). As expected, no extension was observed with WRN-E84A alone (lane 2), but some short extension products are visible with pol δ alone (lane 3); these products probably result from removal of the unpaired nucleotides at the 3’ end of the labeled strand by the 3’ to 5’ proofreading function of pol δ followed by fill-in synthesis on the unconverted Holliday junction. Due to this possibility, the origins of similar short extension products in reactions containing both WRN-E84A and pol δ are unclear. Nevertheless, significant amounts of the 122–123 nt full extension product are also observed in these reactions (Fig. 5C, lane 4, denoted by white arrow). As in reactions described above with Klenow, this product reflects synthesis beyond the replication fork junction to the end of the leading parental template strand and its presence confirms that WRN-E84A mediates conversion of the Holliday junction to 4-stranded replication fork and allows access to the 3’ end of the leading daughter strand and DNA synthesis by pol δ. In similar reactions on the Holliday junction substrate, these full extension products were also detected only in reactions containing both BLM and pol δ (Supplemental Fig. 5), but were present at much lower levels than with WRN-E84A and pol δ. Importantly, these experiments confirm that WRN and BLM are capable of re-establishing a functional replication fork from a Holliday junction structure reflective of a regressed fork.
DISCUSSION
RecQ helicases are postulated to participate in pathways that respond to replication fork stalling caused by DNA damage or other circumstances. Accumulating evidence strongly suggests that the human RecQ helicases WRN and BLM (deficient in WS and BS, respectively) function with respect to DNA replication in a manner that maintains genomic stability. In the absence of WRN or BLM function, replication abnormalities are observed including aberrant fork progression dynamics (18, 20, 22, 31) and chromosomal aberrations are dramatically increased. WRN- and BLM-deficient cells have been shown to be hypersensitive to agents that block replication fork progression, including HU, aphidicolin and interstrand crosslinkers (23–24, 28, 30–31). In wild type cells, WRN and BLM are recruited to replication foci in response to certain types of DNA damaging treatments (25–27, 29, 33). Taken together, these studies indicate that WRN and BLM function in response to replication fork blockage. Logically, cells would want to avoid the collapse of replication forks that would result in double-strand breaks and unscheduled recombination events. Indeed, blocked forks seem to be stabilized (in a manner dependent upon S phase checkpoint factor ATR) for a period of time after treatment with the replication inhibitor HU; in the absence of ATR function or after prolonged HU incubation, double-strand breaks eventually form, presumably due to replication fork collapse (54–56). Although the DNA remodeling events that might occur immediately upon replication blockage in vivo remain unclear, some models propose regression of forks involving re-pairing of parental strands and annealing of nascent daughter strands to form a “chicken foot” or Holliday junction intermediate. If so, the seemingly least problematic scenario for replication restart would be for the regressed Holliday junction intermediate to be converted (by “reverse” branch migration) back to a fork structure competent for DNA synthesis. In this study, we have examined whether WRN and/or BLM might function in this process.
Biochemical experiments from several groups indicate that WRN and BLM are ATPases and helicases and prefer to bind to and act on complex 3- and 4-stranded DNA structures including Holliday junctions (39–40, 46, 57). Specifically, WRN and BLM bind to and unwind Holliday junctions of limited mobility and are able to perform branch migration over long distances on mobile Holliday junction structures (25, 38–40). Thus, their enzymatic activities, in combination with evidence regarding roles for these proteins in response to replication blockage, implicate WRN and BLM as possible candidate factors that might catalyze this reverse branch migration of “regressed” Holliday junctions to regenerate functional replication forks. Here, the actions of BLM and WRN proteins were examined on a special Holliday junction substrate capable of being converted to a four-stranded replication fork structure. In reactions containing ATP, WRN-wt, exonuclease-deficient WRN-E84A and BLM independently convert this Holliday junction to a replication fork structure. Substitution of ATPγS or use of ATPase- and helicase-deficient BLM-D795A or WRN-K577M mutant protein in these reactions prevented conversion, demonstrating the requirement for ATP hydrolysis for this process specifically by WRN-E84A or BLM. On this particular Holliday junction structure, conversion to the fork is the predominant reaction using either WRN-E84A or BLM; ATP-dependent unwinding/disruption of the Holliday junction to other products occurs at lower levels. Also, the conversion from Holliday junction to four-stranded fork by WRN-E84A and BLM was substantially influenced by the absolute concentration of MgCl2, as was the binding of WRN-E84A to the Holliday junction substrate. Since increasing Mg+2 concentrations favor the stacked X conformation of Holliday junction structures over the square planar conformation (43–44), our results indicate that WRN (and BLM) preferentially bind to and act on the square planar (open) conformation of Holliday junctions. This is also consistent with data showing that spontaneous branch migration of mobile Holliday junctions occurs much faster when these junctions are in the open conformation as compared with the stacked X conformation (58). Notably, the 3’ to 5’ exonuclease activity of WRN-K577M (and WRN-wt) was very evident in reactions performed with the Holliday junction substrate. Since WRN-K577M cannot unwind or branch migrate this substrate, its exonuclease activity is clearly targeted to the 3’ end of the leading daughter strand in the context of this Holliday junction reflective of a “regressed” replication fork. This result suggests that WRN exonuclease activity might be involved in processing the leading daughter strand following fork regression. Such an activity could be useful for removing unpaired nucleotides potentially misincorporated by replicative or translesion polymerases due to obstructions in the template that might be the triggers for fork regression.
This evidence indicates that, when presented with a Holliday junction substrate capable of being either unwound or branch migrated, WRN and BLM preferentially catalyze branch migration. Thus, it was relevant to determine the action of another established branch migration enzyme, human RAD54, on our Holliday junction substrate. Notably, a very recent paper from Mazin and colleagues, using an assay similar to ours, demonstrated that RAD54 catalyzes conversion of a Holliday junction to replication fork, as did BLM (48). Similarly, we show here that RAD54 can convert our Holliday junction substrate to a four-stranded replication fork. However, compared to the low level of conversion we observe with RAD54, WRN and BLM appear to preferentially mediate conversion of our Holliday junction to the four-stranded replication fork species. WRN appears to initially target the Holliday junction structure, as addition of Holliday junction binding protein RuvA markedly inhibits the conversion reaction. We speculate that BLM may have even higher affinity for Holliday junctions than WRN (and even RuvA), because its action is more weakly inhibited by RuvA and higher Mg concentrations. This is also in agreement with studies showing a specific role for BLM, along with TOP3, RMI1, and RMI2, in the processing of Holliday junction structures (59–60). Our results add further support to the concept that, in vivo, ATP hydrolysis by WRN, BLM and perhaps other RecQ family members is primarily utilized to drive branch migration instead of simple duplex unwinding. However, human RECQ4 was not able to catalyze the conversion of Holliday junctions to the replication fork, although it could mediate a complementary strand dependent unwinding or strand exchange reaction. This may suggest the involvement of the RQC and/or HRDC domains (that RECQ4 lacks) in helping mediate this activity, and it certainly further separates the function of RECQ4 with those of WRN and BLM. Although our specific conversion assay seems to show preferential activity by WRN-E84A and BLM, further research is needed to clarify physiological specificity in the putative re-establishment of replication forks from regressed Holliday junctions.
The physiological importance of this conversion process would be to re-establish a viable four-stranded replication fork suitable for resumption of normal bi-directional replication. Therefore, we examined whether the primary product from our WRN-E84A- and BLM-mediated conversion reactions was capable of being acted upon by DNA polymerases. To simplify analysis of the reaction products, we primarily used 3’ to 5’ exonuclease-deficient Klenow fragment in our experiments. While the initial Holliday junction substrate was incapable of being acted upon by Klenow alone, incubation of WRN or BLM followed by Klenow (in the presence of ATP and dNTPs) resulted in extension of the labeled strand. The extent of extension (122–123 nt—i.e., to the end of the leading parental strand template) in these reactions was identical to that observed when Klenow alone was incubated with control fork and partial duplex substrates. We also observed this full length extension product in reactions containing the Holliday junction substrate, WRN-E84A and the replicative T4 DNA polymerase (data not shown). Importantly, we could also readily observe this 122–123 nt extension product in reactions performed on the Holliday junction with WRN-E84A and human DNA pol δ. In similar reactions performed with BLM and DNA pol δ on the Holliday junction substrate, this extension product was detectable but present at very low levels. Taken together, these results 1) confirmed that the product generated from the Holliday junction by WRN and BLM was indeed a replication fork structure and 2) demonstrated that the leading daughter strand of this structure could be fully extended by Klenow and by pol δ. It is not yet clear why better extension was observed with WRN-E84A than with BLM, considering that four-stranded forks would be expected to remain excellent substrates for both BLM and WRN binding (46, 57, 61). However, we speculate that weaker extension of the converted replication fork by both Klenow and pol δ observed in the presence of BLM may be the result of steric hindrance. In agreement, direct extension of the leading daughter strand of four-stranded fork substrate by Klenow was increasingly inhibited by increasing BLM concentrations (data not shown). Also, a physical and functional interaction between WRN and pol δ, as previously reported (62–64), may contribute to the higher level of extension observed in these reactions compared to those with BLM and pol δ. Regardless, our results show that DNA polymerases can act upon four-stranded replication forks re-established by either WRN or BLM from Holliday junctions reflective of fork regression.
Our results suggest that WRN or BLM might act to restore functional replication forks following replication fork blockage and regression to Holliday junction or “chicken foot” structures. It has been suggested that, subsequent to fork stalling in vivo, regression might be a consequence of positive supercoiling built up ahead of the fork (65). However, previous studies in our lab and others suggest that WRN and/or BLM may assist with fork regression (46, 61, 66). Thus, WRN or BLM may be involved in both fork regression and conversion of the regressed fork back to a functional replication fork—i.e., perhaps serving as an ATPase-driven swivel during resolution of replication blockage. In general agreement with this concept, in our experiments with both BLM and WRN, conversion of HJ to four-stranded replication fork appears to plateau at below 50% production of the fork product, suggesting that an equilibrium is reached in vitro between conversion to the fork product and regression back to the Holliday junction structure. This idea is also consistent with the results of Mazin and colleagues on BLM and RAD54 (48) and our experiments showing that WRN binds with high and nearly equal affinity to Holliday junction and four-stranded fork structures, with much lower affinity for partial duplexes and single-stranded DNA (Machwe and Orren, unpublished results).
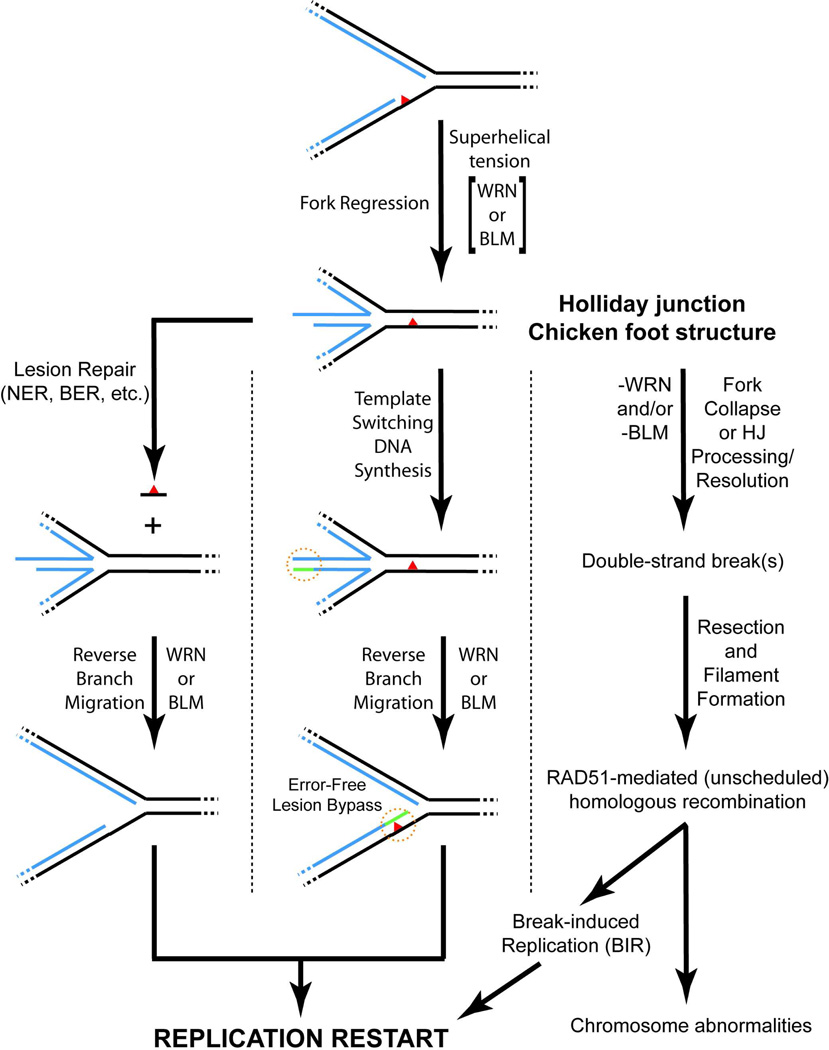
Based upon the existing evidence, we suggest the following model for participation of WRN or BLM in resolution of replication blockage (Fig. 6). Upon stalling of replication forks by DNA lesions or other circumstances, fork regression to generate a Holliday junction occurs (possibly driven by superhelical tension with enzymatic assistance from WRN or BLM). Subsequently, the lesion/obstruction is removed from the reconstituted parental duplex (left pathway) or template switching occurs, in this case using the extended lagging daughter strand as template for synthesis of the leading daughter strand (center pathway). Following these events, WRN or BLM catalyzes “reverse branch migration”—i.e., the conversion of the Holliday junction back to a four-stranded replication fork upon which bi-directional DNA synthesis can be restarted. It should be re-iterated here that all of these events are likely downstream and controlled by the ATR-dependent S phase checkpoint pathway. In the absence of functional WRN or BLM (right pathway), replication blockage results in double-strand break generation, most likely through direct collapse of stalled forks and/or processing/resolution of Holliday junctions formed by fork regression. These double-strand breaks serve as substrates for RAD51-mediated recombination processes that, through the break-induced replication pathway, might restart replication. In agreement, cells deficient in WRN or BLM show increases in (spontaneous or damage-induced) RAD51 foci (23, 67–69) and certain phenotypes of WRN-deficient cells are rescued by introduction of ectopic Holliday junction resolvase enzymes (67, 70). In most instances, the alternate RAD51-mediated pathway would probably be error-free due to the presence of completely homologous partners. However, it might be expected that such recombinational repair processes would be more error-prone if blockage occurred within sequence elements that are highly repeated within the genome. Intriguingly, BLM-deficient cells have a high degree of instability in their repetitive rDNA arrays (71) while WRN- and BLM- deficient cells appear to have substantial telomeric instability (72–74). Even though the phenotypes of WS and BS are distinct, some evidence (including the results presented here) hints at partially redundant roles for WRN and BLM in resolution of replication blockage. However, both WRN and BLM might also participate in other replication- or recombination-related transactions. With regards to BLM, it seems to have both anti- and pro-recombinogenic roles. Its pro-recombinogenic roles may be as a factor enhancing double-strand break resection (75–77) and/or as part of a complex (including TOP3α, RMI1, and RMI2) that acts late in a recombination pathway to suppress sister chromatid exchange (59–60). Additional research is needed to specify and confirm the DNA metabolic steps at which these enzymes act in vivo.
Figure 6. Model for action(s) of WRN and BLM during resolution of replication blockage.
In the presence of certain lesions/obstructions (denoted by red triangle) on the parental strands, replication fork progression is inhibited by blockage of replicative synthesis of the relevant daughter strand. The blocked fork may then be subject to regression to generate a Holliday junction or chicken foot structure, aided physically by superhelical tension (positive supercoiling) generated ahead of the fork during DNA replication and/or enzymatically by WRN or BLM. Left pathway: the regressed fork regenerates a normal DNA duplex in the vicinity of the lesion, facilitating its recognition and removal by standard repair pathways including nucleotide excision repair (NER) and base excision repair (BER). Reverse branch migration, catalyzed by WRN or BLM, re-establishes the replication fork structure, now without the blocking lesion. Center pathway (regions of particular interest denoted with broken orange circle): Extension of the unblocked daughter strand prior to regression provides for template switching and limited synthesis of the blocked daughter strand (denoted in green) once regression has occurred. In this case, reverse branch migration catalyzed by either WRN or BLM results in error-free lesion bypass (with the lesion presumably repaired at a later time). In these scenarios, WRN- or BLM-mediated re-establishment of the normal replication fork structure facilitates replication restart without replication errors or generation of double-strand breaks. Right pathway: In the absence of WRN or BLM, replication forks collapse either directly or through processing or resolution of regressed Holliday junction structures, resulting in formation of double-strand breaks. These breaks serve as substrates for RAD51-mediated homologous recombination processes that can potentially restart replication through the break-induced replication (BIR) pathway. However, these unscheduled recombination events may be prone to errors, leading to the types of chromosomal aberrations associated with WRN or BLM deficiency. Throughout the diagram, parental and daughter strands are denoted in black and blue, respectively, with continuous regions of DNA indicated by dotted lines.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Drs. Joanna Groden, Guo-Min Li, Alexander Mazin and Steve Matson for contributing reagents essential for these studies and Enerlyn Lozada for critical reading of the manuscript.
This work was supported by grants R01 CA113371 and R01 AG027258 to D.K.O from the National Cancer Institute and the National Institute on Aging, respectively.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Supporting information consists of a table and three figures. Supplemental Table 1 contains the sequences of oligonucleotides used in construction of DNA substrates utilized for experiments in this article. Supplemental Figure 1 shows the action of ATPase- and helicase-deficient WRN-K577M mutant on our specialized Holliday junction substrate. Supplemental Figure 2 depicts the effect of MgCl2 concentration of WRN-E84A-mediated unwinding of a partial duplex. Supplemental Figure 3 shows the effect of increasing MgCl2 concentration on BLM- and RAD54-mediated conversion of Holliday junction to 4-stranded replication fork. Supplemental Figure 4 shows that human RPA has a minimal influence on the conversion of Holliday junction to replication fork by WRN-E84A. Supplemental Figure 5 shows that human DNA polymerase δ can detectably catalyze extension of 4-stranded replication fork converted by BLM. This material is available free of charge via the Internet at http://pubs.acs.org.
Abbreviations used: ATPγS, adenosine 5’-O-(thio)triphosphate; BS, Bloom syndrome; EMSA, electrophoretic mobility shift assay; HU, hydroxyurea; Klenow, Klenow fragment of E. coli. DNA Polymerase I (for these studies, 3’ to 5’ exonuclease-deficient); PAGE, polyacrylamide gel electrophoresis; RTS, Rothmund-Thomson syndrome; WS, Werner syndrome.
REFERENCES
- 1.Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J. The Bloom's syndrome gene product is homologous to RecQ helicases. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 2.Kitao S, Shimamoto A, Goto M, Miller RW, Smithson WA, Lindor NM, Furuichi Y. Mutations in RECQL4 cause a subset of cases of Rothmund-Thomson syndrome. Nat. Genet. 1999;22:82–84. doi: 10.1038/8788. [DOI] [PubMed] [Google Scholar]
- 3.Yu CE, Oshima J, Fu YH, Wijsman EM, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin GM, Mulligan J, Schellenberg GD. Positional cloning of the Werner's syndrome gene. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 4.Martin GM, Oshima J. Lessons from human progeroid syndromes. Nature. 2000;408:263–266. doi: 10.1038/35041705. [DOI] [PubMed] [Google Scholar]
- 5.Goto M. Hierarchical deterioration of body systems in Werner's syndrome: implications for normal ageing. Mech. Ageing Dev. 1997;98:239–254. doi: 10.1016/s0047-6374(97)00111-5. [DOI] [PubMed] [Google Scholar]
- 6.Kitao S, Lindor NM, Shiratori M, Furuichi Y, Shimamoto A. Rothmund-thomson syndrome responsible gene, RECQL4: genomic structure and products. Genomics. 1999;61:268–276. doi: 10.1006/geno.1999.5959. [DOI] [PubMed] [Google Scholar]
- 7.Opresko PL, Cheng WH, Bohr VA. Junction of RecQ helicase biochemistry and human disease. J. Biol. Chem. 2004;279:18099–18102. doi: 10.1074/jbc.R300034200. [DOI] [PubMed] [Google Scholar]
- 8.Xu X, Liu Y. Dual DNA unwinding activities of the Rothmund-Thomson syndrome protein, RECQ4. EMBO J. 2009;28:568–577. doi: 10.1038/emboj.2009.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheok CF, Wu L, Garcia PL, Janscak P, Hickson ID. The Bloom's syndrome helicase promotes the annealing of complementary single-stranded DNA. Nucleic Acids Res. 2005;33:3932–3941. doi: 10.1093/nar/gki712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia PL, Liu Y, Jiricny J, West SC, Janscak P. Human RECQ5beta, a protein with DNA helicase and strand-annealing activities in a single polypeptide. EMBO J. 2004;23:2882–2891. doi: 10.1038/sj.emboj.7600301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macris MA, Krejci L, Bussen W, Shimamoto A, Sung P. Biochemical characterization of the RECQ4 protein, mutated in Rothmund-Thomson syndrome. DNA Repair (Amst) 2006;5:172–180. doi: 10.1016/j.dnarep.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 12.Sharma S, Sommers JA, Choudhary S, Faulkner JK, Cui S, reoli L, Muzzolini L, Vindigni A, Brosh RM., Jr Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J. Biol. Chem. 2005;280:28072–28084. doi: 10.1074/jbc.M500264200. [DOI] [PubMed] [Google Scholar]
- 13.Kanagaraj R, Saydam N, Garcia PL, Zheng L, Janscak P. Human RECQ5{beta} helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res. 2006;34:5217–5231. doi: 10.1093/nar/gkl677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Machwe A, Xiao L, Groden J, Matson SW, Orren DK. RecQ family members combine strand pairing and unwinding activities to catalyze strand exchange. J. Biol. Chem. 2005;280:23397–23407. doi: 10.1074/jbc.M414130200. [DOI] [PubMed] [Google Scholar]
- 15.Huang S, Beresten S, Li B, Oshima J, Ellis NA, Campisi J. Characterization of the human and mouse WRN 3'-->5' exonuclease. Nucleic Acids Res. 2000;28:2396–2405. doi: 10.1093/nar/28.12.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang S, Li B, Gray MD, Oshima J, Mian IS, Campisi J. The premature ageing syndrome protein, WRN, is a 3'-->5' exonuclease. Nat. Genet. 1998;20:114–116. doi: 10.1038/2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Machwe A, Xiao L, Theodore S, Orren DK. DNase I footprinting and enhanced exonuclease function of the bipartite Werner syndrome protein (WRN) bound to partially melted duplex DNA. J. Biol. Chem. 2002;277:4492–4504. doi: 10.1074/jbc.M108880200. [DOI] [PubMed] [Google Scholar]
- 18.Sidorova JM. Roles of the Werner syndrome RecQ helicase in DNA replication. DNA Repair (Amst) 2008;7:1776–1786. doi: 10.1016/j.dnarep.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Atkinson J, McGlynn P. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res. 2009;37:3475–3492. doi: 10.1093/nar/gkp244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petermann E, Helleday T. Pathways of mammalian replication fork restart. Nat. Rev. Mol. Cell. Biol. 2010;11:683–687. doi: 10.1038/nrm2974. [DOI] [PubMed] [Google Scholar]
- 21.Poot M, Hoehn H, Runger TM, Martin GM. Impaired S-phase transit of Werner syndrome cells expressed in lymphoblastoid cell lines. Exp. Cell Res. 1992;202:267–273. doi: 10.1016/0014-4827(92)90074-i. [DOI] [PubMed] [Google Scholar]
- 22.Rodriguez-Lopez AM, Jackson DA, Iborra F, Cox LS. Asymmetry of DNA replication fork progression in Werner's syndrome. Aging Cell. 2002;1:30–39. doi: 10.1046/j.1474-9728.2002.00002.x. [DOI] [PubMed] [Google Scholar]
- 23.Pichierri P, Franchitto A, Mosesso P, Palitti F. Werner's syndrome protein is required for correct recovery after replication arrest and DNA damage induced in S-phase of cell cycle. Mol. Biol. Cell. 2001;12:2412–2421. doi: 10.1091/mbc.12.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poot M, Yom JS, Whang SH, Kato JT, Gollahon KA, Rabinovitch PS. Werner syndrome cells are sensitive to DNA cross-linking drugs. FASEB J. 2001;15:1224–1226. doi: 10.1096/fj.00-0611fje. [DOI] [PubMed] [Google Scholar]
- 25.Constantinou A, Tarsounas M, Karow JK, Brosh RM, Bohr VA, Hickson ID, West SC. Werner's syndrome protein (WRN) migrates Holliday junctions and co-localizes with RPA upon replication arrest. EMBO Rep. 2000;1:80–84. doi: 10.1093/embo-reports/kvd004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karmakar P, Bohr VA. Cellular dynamics and modulation of WRN protein is DNA damage specific. Mech. Ageing Dev. 2005;126:1146–1158. doi: 10.1016/j.mad.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 27.Pichierri P, Rosselli F, Franchitto A. Werner's syndrome protein is phosphorylated in an ATR/ATM-dependent manner following replication arrest and DNA damage induced during the S phase of the cell cycle. Oncogene. 2003;22:1491–1500. doi: 10.1038/sj.onc.1206169. [DOI] [PubMed] [Google Scholar]
- 28.Hook GJ, Kwok E, Heddle JA. Sensitivity of Bloom syndrome fibroblasts to mitomycin C. Mutat. Res. 1984;131:223–230. doi: 10.1016/0167-8817(84)90029-4. [DOI] [PubMed] [Google Scholar]
- 29.Sengupta S, Linke SP, Pedeux R, Yang Q, Farnsworth J, Garfield SH, Valerie K, Shay JW, Ellis NA, Wasylyk B, Harris CC. BLM helicase-dependent transport of p53 to sites of stalled DNA replication forks modulates homologous recombination. EMBO J. 2003;22:1210–1222. doi: 10.1093/emboj/cdg114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Phosphorylation of the Bloom's syndrome helicase and its role in recovery from S-phase arrest. Mol. Cell. Biol. 2004;24:1279–1291. doi: 10.1128/MCB.24.3.1279-1291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies SL, North PS, Hickson ID. Role for BLM in replication-fork restart and suppression of origin firing after replicative stress. Nat. Struct. Mol. Biol. 2007;14:677–679. doi: 10.1038/nsmb1267. [DOI] [PubMed] [Google Scholar]
- 32.Lonn U, Lonn S, Nylen U, Winblad G, German J. An abnormal profile of DNA replication intermediates in Bloom's syndrome. Cancer Res. 1990;50:3141–3145. [PubMed] [Google Scholar]
- 33.Bischof O, Kim SH, Irving J, Beresten S, Ellis NA, Campisi J. Regulation and localization of the Bloom syndrome protein in response to DNA damage. J. Cell Biol. 2001;153:367–380. doi: 10.1083/jcb.153.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu D, Muniandy P, Leo E, Yin J, Thangavel S, Shen X, Ii M, Agama K, Guo R, Fox D, 3rd, Meetei AR, Wilson L, Nguyen H, Weng NP, Brill SJ, Li L, Vindigni A, Pommier Y, Seidman M, Wang W. Rif1 provides a new DNA-binding interface for the Bloom syndrome complex to maintain normal replication. EMBO J. 2010;29:3140–3155. doi: 10.1038/emboj.2010.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cox MM. The nonmutagenic repair of broken replication forks via recombination. Mutat. Res. 2002;510:107–120. doi: 10.1016/s0027-5107(02)00256-7. [DOI] [PubMed] [Google Scholar]
- 36.Haber JE. DNA recombination: the replication connection. Trends Biochem. Sci. 1999;24:271–275. doi: 10.1016/s0968-0004(99)01413-9. [DOI] [PubMed] [Google Scholar]
- 37.Kowalczykowski SC. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 38.Karow JK, Constantinou A, Li JL, West SC, Hickson ID. The Bloom's syndrome gene product promotes branch migration of holliday junctions. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6504–6508. doi: 10.1073/pnas.100448097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mohaghegh P, Karow JK, Brosh Jr RM, Jr, Bohr VA, Hickson ID. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sharma S, Otterlei M, Sommers JA, Driscoll HC, Dianov GL, Kao HI, Bambara RA, Brosh RM., Jr WRN helicase and FEN-1 form a complex upon replication arrest and together process branchmigrating DNA structures associated with the replication fork. Mol. Biol. Cell. 2004;15:734–750. doi: 10.1091/mbc.E03-08-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Orren DK, Brosh RM, Jr, Nehlin JO, Machwe A, Gray MD, Bohr VA. Enzymatic and DNA binding properties of purified WRN protein: high affinity binding to single-stranded DNA but not to DNA damage induced by 4NQO. Nucleic Acids Res. 1999;27:3557–3566. doi: 10.1093/nar/27.17.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen JC, Gray MD, Oshima J, Kamath-Loeb AS, Fry M, Loeb LA. Werner syndrome protein. I. DNA helicase and dna exonuclease reside on the same polypeptide. J. Biol. Chem. 1998;273:34139–34144. doi: 10.1074/jbc.273.51.34139. [DOI] [PubMed] [Google Scholar]
- 43.Duckett DR, Murchie AI, Diekmann S, von Kitzing E, Kemper B, Lilley DM. The structure of the Holliday junction, and its resolution. Cell. 1988;55:79–89. doi: 10.1016/0092-8674(88)90011-6. [DOI] [PubMed] [Google Scholar]
- 44.Diekmann S, Lilley DM. The anomalous gel migration of a stable cruciform: temperature and salt dependence, and some comparisons with curved DNA. Nucleic Acids Res. 1987;15:5765–5774. doi: 10.1093/nar/15.14.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brosh RM, Jr, Waheed J, Sommers JA. Biochemical characterization of the DNA substrate specificity of Werner syndrome helicase. J. Biol. Chem. 2002;277:23236–23245. doi: 10.1074/jbc.M111446200. [DOI] [PubMed] [Google Scholar]
- 46.Machwe A, Xiao L, Groden J, Orren DK. The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry. 2006;45:13939–13946. doi: 10.1021/bi0615487. [DOI] [PubMed] [Google Scholar]
- 47.Bugreev DV, Mazina OM, Mazin AV. Rad54 protein promotes branch migration of Holliday junctions. Nature. 2006;442:590–593. doi: 10.1038/nature04889. [DOI] [PubMed] [Google Scholar]
- 48.Bugreev DV, Rossi MJ, Mazin AV. Cooperation of RAD51 and RAD54 in regression of a model replication fork. Nucleic Acids Res. 2011;39:2153–2164. doi: 10.1093/nar/gkq1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen JC, Loeb LA. Werner syndrome exonuclease catalyzes structure-dependent degradation of DNA. Nucleic Acids Res. 2000;28:3260–3268. doi: 10.1093/nar/28.17.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen JC, Gray MD, Oshima J, Loeb LA. Characterization of Werner syndrome protein DNA helicase activity: directionality, substrate dependence and stimulation by replication protein A. Nucleic Acids Res. 1998;26:2879–2885. doi: 10.1093/nar/26.12.2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brosh RM, Jr, Li JL, Kenny MK, Karow JK, Cooper MP, Kureekattil RP, Hickson ID, Bohr VA. Replication protein A physically interacts with the Bloom's syndrome protein and stimulates its helicase activity. J. Biol. Chem. 2000;275:23500–23508. doi: 10.1074/jbc.M001557200. [DOI] [PubMed] [Google Scholar]
- 52.Brosh RM, Jr, Orren DK, Nehlin JO, Ravn PH, Kenny MK, Machwe A, Bohr VA. Functional and physical interaction between WRN helicase and human replication protein A. J. Biol. Chem. 1999;274:18341–18350. doi: 10.1074/jbc.274.26.18341. [DOI] [PubMed] [Google Scholar]
- 53.Machwe A, Lozada E, Wold MS, Li GM, Orren DK. Molecular cooperation between the Werner syndrome protein and replication protein A in relation to replication fork blockage. J. Biol. Chem. 2011;286:3497–3508. doi: 10.1074/jbc.M110.105411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saintigny Y, Delacote F, Vares G, Petitot F, Lambert S, Averbeck D, Lopez BS. Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J. 2001;20:3861–3870. doi: 10.1093/emboj/20.14.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lahkim Bennani-Belhaj K, Buhagiar-Labarchede G, Jmari N, Onclercq-Delic R, Amor-Gueret M. BLM Deficiency Is Not Associated with Sensitivity to Hydroxyurea-Induced Replication Stress. J. Nucleic Acids. 2010;2010 doi: 10.4061/2010/319754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brosh RM, Jr, Driscoll HC, Dianov GL, Sommers JA. Biochemical characterization of the WRN-FEN-1 functional interaction. Biochemistry. 2002;41:12204–12216. doi: 10.1021/bi026031j. [DOI] [PubMed] [Google Scholar]
- 58.Panyutin IG, Hsieh P. The kinetics of spontaneous DNA branch migration. Proc. Natl. Acad. Sci. U.S.A. 1994;91:2021–2025. doi: 10.1073/pnas.91.6.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Y, West SC. More complexity to the Bloom's syndrome complex. Genes Dev. 2008;22:2737–2742. doi: 10.1101/gad.1732808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 61.Machwe A, Xiao L, Lloyd RG, Bolt E, Orren DK. Replication fork regression in vitro by the Werner syndrome protein (WRN): holliday junction formation-the effect of leading arm structure and a potential role for WRN exonuclease activity. Nucleic Acids Res. 2007;35:5729–5747. doi: 10.1093/nar/gkm561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kamath-Loeb AS, Johansson E, Burgers PM, Loeb LA. Functional interaction between the Werner Syndrome protein and DNA polymerase delta. Proc. Natl. Acad. Sci. U.S.A. 2000;97:4603–4608. doi: 10.1073/pnas.97.9.4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shah SN, Opresko PL, Meng X, Lee MY, Eckert KA. DNA structure and the Werner protein modulate human DNA polymerase delta-dependent replication dynamics within the common fragile site FRA16D. Nucleic Acids Res. 2010;38:1149–1162. doi: 10.1093/nar/gkp1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Szekely AM, Chen YH, Zhang C, Oshima J, Weissman SM. Werner protein recruits DNA polymerase delta to the nucleolus. Proc. Natl. Acad. Sci. U.S.A. 2000;97:11365–11370. doi: 10.1073/pnas.97.21.11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Postow L, Ullsperger C, Keller RW, Bustamante C, Vologodskii AV, Cozzarelli NR. Positive torsional strain causes the formation of a four-way junction at replication forks. J. Biol. Chem. 2001;276:2790–2796. doi: 10.1074/jbc.M006736200. [DOI] [PubMed] [Google Scholar]
- 66.Ralf C, Hickson ID, Wu L. The Bloom's syndrome helicase can promote the regression of a model replication fork. J. Biol. Chem. 2006;281:22839–22846. doi: 10.1074/jbc.M604268200. [DOI] [PubMed] [Google Scholar]
- 67.Franchitto A, Pirzio LM, Prosperi E, Sapora O, Bignami M, Pichierri P. Replication fork stalling in WRN-deficient cells is overcome by prompt activation of a MUS81-dependent pathway. J. Cell. Biol. 2008;183:241–252. doi: 10.1083/jcb.200803173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ammazzalorso F, Pirzio LM, Bignami M, Franchitto A, Pichierri P. ATR and ATM differently regulate WRN to prevent DSBs at stalled replication forks and promote replication fork recovery. EMBO J. 2010;29:3156–3169. doi: 10.1038/emboj.2010.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wicky C, Alpi A, Passannante M, Rose A, Gartner A, Muller F. Multiple genetic pathways involving the Caenorhabditis elegans Bloom's syndrome genes him-6, rad-51, and top-3 are needed to maintain genome stability in the germ line. Mol. Cell. Biol. 2004;24:5016–5027. doi: 10.1128/MCB.24.11.5016-5027.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Saintigny Y, Makienko K, Swanson C, Emond MJ, Monnat RJ., Jr Homologous recombination resolution defect in werner syndrome. Mol. Cell. Biol. 2002;22:6971–6978. doi: 10.1128/MCB.22.20.6971-6978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Killen MW, Stults DM, Adachi N, Hanakahi L, Pierce AJ. Loss of Bloom syndrome protein destabilizes human gene cluster architecture. Hum. Mol. Genet. 2009;18:3417–3428. doi: 10.1093/hmg/ddp282. [DOI] [PubMed] [Google Scholar]
- 72.Chang S, Multani AS, Cabrera NG, Naylor ML, Laud P, Lombard D, Pathak S, Guarente L, DePinho RA. Essential role of limiting telomeres in the pathogenesis of Werner syndrome. Nat. Genet. 2004;36:877–882. doi: 10.1038/ng1389. [DOI] [PubMed] [Google Scholar]
- 73.Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 74.Du X, Shen J, Kugan N, Furth EE, Lombard DB, Cheung C, Pak S, Luo G, Pignolo RJ, DePinho RA, Guarente L, Johnson FB. Telomere shortening exposes functions for the mouse Werner and Bloom syndrome genes. Mol. Cell. Biol. 2004;24:8437–8446. doi: 10.1128/MCB.24.19.8437-8446.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gravel S, Chapman JR, Magill C, Jackson SP. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008;22:2767–2772. doi: 10.1101/gad.503108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc. Natl. Acad. Sci. U.S.A. 2008;105:16906–16911. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.