Abstract
Insulin-like peptides (ILPs) regulate a multitude of biological processes, including metabolism and immunity to infection, and share similar structural motifs across widely divergent taxa. Insulin/insulin-like growth factor signaling (IIS) pathway elements are similarly conserved. We have shown that IIS regulates reproduction, innate immunity, and lifespan in female Anopheles stephensi, a major mosquito vector of human malaria. To further explore IIS regulation of these processes, we identified genes encoding five ILPs in this species and characterized their expression in tissues. Antisera to ILP homologs in Anopheles gambiae were used to identify cellular sources in An. stephensi females by immunocytochemistry. We analyzed tissue-specific ILP transcript expression in young and older females, in response to different feeding regimens, and in response to infection with Plasmodium falciparum with quantitative reverse transcriptase-PCR assays. While some ILP transcript changes were evident in older females and in response to blood feeding, significant changes were particularly notable in response to hormonal concentrations of ingested human insulin and to P. falciparum infection. These changes suggest that ILP secretion and action may be similarly responsive in Plasmodium-infected females and potentially alter metabolism and innate immunity.
Keywords: insect, mosquito, insulin signaling, malaria, Anopheles, Plasmodium
1. Introduction
Malaria parasites, mosquito vectors, and humans come together in a complex transmission cycle that poses a global challenge in need of novel strategies to lessen disease incidence. Studies of mosquito physiology have demonstrated that conserved metazoan cell signaling pathways regulate processes associated with blood digestion and reproduction [4, 24, 25]. Some of these pathways also regulate mosquito-Plasmodium interactions, indicating that further study of these pathways in this setting may reveal new targets for transmission blocking strategies.
Our work has shown that insulin/insulin-like growth factor signaling (IIS) in Anopheles stephensi is activated by human insulin ingested with a blood meal [11] and that ingested insulin and IIS activation reduce mosquito lifespan and alter the development of the most deadly human malaria parasite, Plasmodium falciparum [6, 8, 11, 28]. Plasmodium falciparum glycosylphosphatidylinositols (GPIs) can activate IIS in the An. stephensi midgut epithelium within 30 min after feeding [14], indicating that GPIs are an important early signal of infection in the mosquito host. Not surprisingly, the effects of IIS on lifespan and immunity are conserved among a variety of invertebrate and vertebrate species [16, 23, 29].
Among mammals, insulin and two insulin-like growth factors bind to dimers of receptor tyrosine kinase isoforms to regulate metabolism and growth [2, 15]. In mammals, insulin exerts a positive effect on its own gene transcription and synthesis via activation of IIS proteins [1, 3, 13], suggesting that the activation of IIS can be amplified by the autocrine effects of endogenous insulin. Seven other insulin-like peptides (ILPs), including relaxin, bind to G-protein coupled receptors that signal through alternate pathways to regulate reproduction and an unidentified range of other processes [10].
Studies to date show that there is a comparable diversity of ILPs and putative receptor types in insects, but only one study has confirmed that an endogenous ILP binds to the single insulin receptor expressed in an insect, the mosquito Aedes aegypti [5, 33]. Eight and seven ILP genes have been identified in the mosquitoes, Ae. aegypti and Anopheles gambiae, respectively [12, 21, 22]. Expression and localization of these ILPs suggested neurohormonal roles related to metabolism and reproduction [15, 21, 22] that were confirmed in subsequent studies [33]. Identification of five ILP genes in the mosquito Culex pipiens led to a study that showed CuILP1 transcription was required for ovarian maturation, suggesting a role in adult diapause [27]. Our previous work implicated IIS in lifespan regulation and anti-parasite immunity in An. stephensi [6, 11]. The goals of this study, therefore, were to extend our significant understanding of IIS in An. stephensi through the identification of genes encoding ILPs in this species and to characterize their tissue-specific gene and peptide expression patterns in response to different feeding regimens and to malaria parasite infection.
2. Materials and Methods
2.1 Mosquito rearing
The Indian wild type strain of An. stephensi, derived from a colony at Walter Reed Army Institute of Research, was maintained at 27°C and 75% relative humidity with a L12:D12 cycle (lights on at 0800). Larvae were provided with a 2% solution of 2:1 Sera Micron®:yeast (Sera North America, Montgomeryville, PA) through day 4 post-hatching and then were reared on high protein, low fat Game Fish Chow® pellets (Purina Mills, St. Louis, MO) until pupation. Mice or hamsters were used as a blood source for colony maintenance in compliance with federal guidelines as implemented by the Animal Care and Use Program at UC Davis.
2.2 ILP sequence identification and phylogenetic analyses
Aedes aegypti and An. gambiae ILP1 and ILP2 cDNA sequences were compared using ClustalW to identify highly conserved regions for primer design. These primers included ILP1-forward (5’-AACTGAGCGACACGTTGG-3’), ILP1-reverse (5’-CGAATGTTGCTACCAAAGCT-3’); ILP2-forward (5’-AAACGCTAGCCTTCCTCTGC-3’); ILP2-reverse (5’-CTGCAAGAAGAGCTGCTCCT-3’). PCR primers based on sequences of AngILP3-5 [12] were used to amplify orthologous target sequences from An. stephensi cDNA derived from heads and midguts. Cycling conditions were modified from [12] to accommodate sequence mismatches: one cycle of 94°C for 5 min; 30 cycles of 94°C for 30 sec, 55°C for 30 sec, 72°C for 30 sec; one cycle of 72°C for 7 min. PCR products were visualized by agarose gel electrophoresis and amplimers were cloned (TOPO-TA cloning kit, Invitrogen, Carlsbad, CA) and submitted for sequencing (Davis Sequencing, Davis, CA) to verify expected homologies with An. gambiae (Ang)ILP sequences. Following sequence confirmation, An. stephensi (Ans)ILP sequence-specific PCR primers were designed to amplify the remaining coding sequences and the 5’ and 3’ untranslated regions (UTRs) using 5’ and 3’ RACE (FirstChoice RLM-RACE Kit, Ambion, Austin, TX).
For phylogenetic analysis, alignments of full-length mosquito and fruit fly (Drosophila melanogaster) ILP propeptide sequences were performed using ClustalW. The sequences were analyzed with PHYLIP Seqboot and Proml to generate 1000 replicates and to estimate the trees by maximum likelihood. PHYLIP Consense generated the majority-rule consensus tree, which was created using Drawgram. Bootstrap consensus values higher than 50% are labeled at the nodes of the trees. The tree was drawn to scale, with branch lengths representing the expected fraction of amino acids changed. A Caenorhabditis elegans ILP was used as the outgroup for all constructed trees.
2.3 RNA extraction and optimization of AnsILP qRT-PCR assays
Heads, thoraces, gut tissues (midgut, hindgut, and Malpighian tubules) or midguts alone, and remaining abdominal body walls without ovaries and other tissues were dissected from female An. stephensi and homogenized in TriZOL reagent (Invitrogen) using pulse sonication. RNA was extracted from homogenates following the manufacturer’s protocol. Contaminating genomic DNA was removed from RNA samples using Turbo DNAse Free (Applied Biosystems, Foster City CA).
Primers and probes were designed based on AnsILP sequences and the ribosomal protein S7 gene using Primer Express software (Applied Biosystems; Supplemental Table 1). RNA samples were analyzed for AnsILP mRNA expression using TaqMan Gold RT-PCR Reagents kit and one step qRT-PCR on an Applied Biosystems 7300 Real-Time PCR System. Amplification conditions were optimized by varying primer concentrations, reagent concentrations and thermal cycling parameters for 10-fold dilution series of An. stephensi genomic DNA. Efficiencies for AnsILP and ribosomal protein S7 amplification were calculated using the equation E=10[−1/slope] [19]. In each reaction plate, every RNA sample was analyzed in triplicate (technical replicates) to confirm amplification consistency (e.g., defined by technical replicates within 0.5 Ct of each other). The Ct for each sample was calculated as the average of technical replicate Ct values. Each plate also included genomic DNA positive controls and no template negative controls. Biological replicates were performed using RNA samples from separate cohorts of An. stephensi.
2.4 Tissue-specific AnsILP transcript expression
A total of 100 female An. stephensi from a single cohort were separated after emergence into a 3.8 L carton and provided with 10% sucrose-soaked cotton pads that were replaced every other day. At days 3–4 and 15–16 after emergence, tissues were dissected from each of 20 females for RNA extraction and analysis. Collected tissues included head, gut tissue (midgut, hindgut, and Malpighian tubules), thorax, and the abdominal body wall after removal of other tissues. Isolated RNA was treated to digest genomic DNA and utilized in the previously described one-step AnsILP qRT-PCR assays. AnsILP expression levels were calculated using the 2−ΔΔCt method [26] relative to ribosomal protein S7 gene expression in the same tissue. Tissue-specific AnsILP expression analyses were performed with tissues from three or four separate cohorts (biological replicates) of An. stephensi.
2.5 AngILP antisera production, specificity, and immunocytochemistry
Peptide synthesis, antigen preparation, and antiserum production were performed by Sigma Genosys (St. Louis, MO). All animal procedures followed federal guidelines as implemented by the Animal Care and Use Program at UC Davis. Based on the predicted processing of their respective prepropeptides [12], A chain fragments of AngILP2 (CCKKSCSYVELRAY) and AngILP5 (CCTRTGCTWEEYAEY) were synthesized and confirmed structurally by mass spectrometry. A composite A chain sequence (CLESCTMDQLLSYCKD) was designed and synthesized as a representative antigen for AngILP1, ILP3, and ILP4 (AngILP1/3/4; Supplemental Fig. 1). The AngILPs were conjugated separately to keyhole limpet hemocyanin with 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide for use as antigens. After collection of preimmune sera, female New Zealand White rabbits were injected subcutaneously with the AngILP antigens (two rabbits per AngILP antigen at 0.5 mg per rabbit in complete Freund's adjuvant) and then boosted at two week intervals (0.5 mg per rabbit in incomplete Freund's adjuvant for a total of 5 boosts). Blood was collected four weeks after the initial immunization and every two weeks thereafter, after which serum was prepared and sent to the laboratory of M.R.B. for processing and storage at −80°C.
Sera from different bleeds of the rabbits were tested by immunodot blots to assess recognition of the respective AngILPs, after which selected antibodies were affinity purified with the SulfoLink column kit (Thermo Scientific, Waltham, MA). To confirm specificity of the AngILP antibodies, AngILP A chain peptides were spotted onto nitrocellulose (Protran 0.1 µm; Whatman, Piscataway, NJ) as 0.1, 1, 10 and 100 ng in 1 µl (1:1 acetonitrile and water) and dried overnight. The blots were treated with ECL blocking agent (Amersham, Piscataway, NJ) for 1 h at room temperature (RT) and then incubated with a 1:1000 dilution of the AngILP antibody in blocking solution overnight at 4°C. Blots were washed and then treated with goat anti-rabbit IgG-peroxidase conjugate (1:20,000 dilution; Sigma-Aldrich) in fresh blocking solution for 4 h at RT. Membranes were washed with TBS-T (20 min each) and developed for chemiluminescent detection (ECL Advance kit, Amersham) with the GeneGnome imaging system (Syngene, Frederick, MD).
Brains, alimentary tracts and abdominal body walls with exposed ventral nerve cords of 4-day-old females of the two mosquito species were dissected in 4% paraformaldehyde/ 0.1 M phosphate buffered saline (PBS; pH 7.4) and fixed for 1–3 h at RT. These tissues were permeabilized in 4% paraformaldehyde/PBS with 0.2% Triton for 15 min and then washed in PBS. The tissues were blocked in PBS with goat serum (5%) and 0.1% Tween 20 (PBS-GS-T) at 4°C for 1 h. Affinity purified antibodies in solution were added to the tissues in blocking solution for overnight incubation at 4°C at the following dilutions: 1:8 anti-AngILP2 (R-88) and 1:90 anti-AngILP1/3/4 (R-726). Tissues were washed with PBS-GS-T and incubated with a 1:2000 dilution of goat-anti rabbit Alexa Fluor 488F® (Molecular Probes, Eugene, OR) in PBS-GS-T overnight. Labeled tissues were washed with PBS-T and then mounted in 1:1 PBS/glycerol. Specimens were viewed with the DM RXE/TCS SP2 confocal microscope and images captured with a DFC 420 camera (Leica, Bannockburn, IL). For controls, affinity purified antibodies were preabsorbed with 1 × 10−4M solutions of their respective antigens overnight before use in immunocytochemistry.
2.6 Diet-specific AnsILP transcript expression
A total of 3–6 separate cohorts (biological replicates) of An. stephensi were used for analyses of tissue-specific AnsILP mRNA expression in female mosquitoes held on four dietary regimens. The feeding regimens were designed to assess the effects of 24 h deprivation (no water or sugar) or 24 h water (no sugar) pre-treatment on AnsILP expression patterns at 6 h or 24 h following subsequent access to either sugar or blood. As such, the four dietary regimens included (i) 24 h deprivation followed by provision of 10% sucrose, (ii) 24 h deprivation followed by provision of an artificial blood meal of washed red blood cells in saline (washed RBCs; see below), (iii) 24 h water pre-treatment followed by provision of 10% sucrose, and (iv) 24 h water pre-treatment followed by provision of washed RBCs.
For each dietary regimen study, 100 newly emerged female An. stephensi were transferred to 3.8 L cartons. During emergence and after transfer to the study cartons, mosquitoes were provided with 10% sucrose-soaked cotton pads. Cotton pads were removed from the study cartons 3–4 days after adult emergence and either not replaced (deprivation) or replaced with cotton pads soaked only with water for 24 h. At the end of the 24 h deprivation or water pre-treatment (0 h), head, gut tissue (midgut, hindgut, and Malpighian tubules) and remaining abdominal body wall after other tissues were removed were dissected from a total of 20 deprived or water-treated females for AnsILP mRNA expression analyses. These samples served as baseline controls. The remaining females were provided with 10% sucrose-soaked cotton pads or provided with a meal of washed RBCs in saline (15 mmol l−1 NaCl, 10 mmol l−1 NaHCO3, 1 mmol l−1 ATP, pH 7.0) via a Hemotek circulation system (Discovery Workshops, Accrington, UK). At 6 h and 24 h after provision of 10% sucrose or a meal of washed RBCs, tissues as above were dissected from a total of 20 females in each of the four treatment groups. Isolated RNAs were treated to remove genomic DNA and utilized in the previously described one-step AnsILP qRT-PCR assays. AnsILP expression levels for each tissue were calculated using the 2−ΔΔCt method [26] and analyzed as fold changes relative to AnsILP expression in the respective matched tissue control of deprived or water-treated mosquitoes at 0 h.
2.7 Ingested insulin-dependent AnsILP transcript expression
For each study, 120 newly emerged female An. stephensi were transferred to 3.8 L cartons and maintained according to the 24 h water pre-treatment regimen described above. Females were then provided with either a meal of washed RBCs supplemented with 170 pM human insulin (Sigma-Aldrich, St. Louis, MO) or with an RBC meal with an equivalent volume of diluent buffer (15 mmol l−1 NaCl, 10 mmol l−1 NaHCO3, 1 mmol l−1 ATP, pH 7.0) as a control meal. Normal human blood insulin levels range from 17 pM at fasting to 590 pM without fasting [7], indicating that 170 pM insulin could be ingested by feeding mosquitoes. At 30 min and 120 min after feeding on an insulin-treated or control meal of RBCs, heads, midguts and abdominal body walls after removal of other tissues were dissected from 20 females. Midguts were selected for these assays rather than gut tissue (midgut, hindgut, Malpighian tubules) to facilitate direct comparisons with previous observations of IIS activation in this tissue [11, 28]. Isolated RNAs from the dissected tissues were treated with DNase to remove contaminating genomic DNA and utilized in the previously described AnsILP qRT-PCR assays. AnsILP mRNA expression levels for each tissue were calculated using the comparative Ct method [26] and analyzed as fold changes relative to AnsILP expression in the respective matched tissue control from mosquitoes fed an RBC meal without insulin. These analyses were performed with tissues from three separate cohorts (biological replicates) of An. stephensi.
2.8 Plasmodium falciparum culture, An. stephensi infection, and infection-dependent AnsILP transcript expression
The NF54 strain of P. falciparum was initiated at 1% parasitemia in 10% heat-inactivated human serum and 6% washed RBCs in RPMI 1640 with HEPES (Gibco/Invitrogen, Carlsbad, CA) and hypoxanthine. At days 15–17, stage V gametocytes were evident and exflagellation was evaluated the day before and day of feeding by observation of blood smears before addition of fresh media at 200X magnification with phase-contrast or modified brightfield microscopy. Exflagellation rates of 4–6 parasites per field were observed in all three cohorts, providing confidence in the consistent presence of P. falciparum gametocytes in these infected, artificial blood meals. For each infection study, 100 newly emerged female An. stephensi were transferred to 3.8 L cartons. During emergence and after transfer to the study cartons, mosquitoes were provided with 10% sucrose-soaked cotton pads. At 3–4 days after emergence, cotton pads were replaced with cotton pads soaked only with water for 24 h prior to blood feeding. Experimental females were provided with an infected, artificial blood meal that consisted of P. falciparum culture described above diluted in human RBCs and heat-inactivated serum and immediately fed via a Hemotek circulation system (Discovery Workshops, Accrington, UK). Control females received an identical, artificial blood meal under the same conditions without parasites. At 6 h, 24 h, and 48 h after the infected and control blood meals, heads, midguts, and abdominal body walls after removal of other tissues were dissected from each of 30 females and pooled for RNA isolation. Midguts were selected for these assays rather than gut tissue for the reasons stated above (Section 2.7). Isolated RNAs from control and infected females were treated for genomic DNA removal and utilized in one-step AnsILP qRT-PCR assays. AnsILP mRNA expression levels for each tissue from parasite-infected An. stephensi were calculated using the 2−ΔΔCt method [26] and analyzed as fold changes relative to AnsILP expression in the respective matched, uninfected control tissue at the same timepoint. These analyses were performed with tissues from three separate cohorts (biological replicates) of P. falciparum-infected An. stephensi. At 10 days after P. falciparum infection, 10 An. stephensi midguts from each cohort were dissected to count parasite oocysts. In cohort 1, the prevalence of infected mosquitoes (at least 1 oocyst per midgut) was 70% and the average number of oocysts per dissected midgut was 5.5. In cohort 2, the prevalence of infected mosquitoes was 80% and the average number of oocysts per dissected midgut was 1.8. In cohort 3, the prevalence of infected mosquitoes was 70% and the average number of oocysts per dissected midgut was 2.0.
2.9 Data and Statistics
All qRT-PCR expression data were non-normally distributed as determined by the Kolmogorov-Smirnov test. These data sets, therefore, were log-transformed and analyzed by ANOVA (α = 0.05) for overall significance followed by Neuman-Keuls multiple comparison post-test for all pairwise comparisons of means from significant data sets (α = 0.05; GraphPad Prism version 5.02). Biological replicates of samples analyzed by qRT-PCR (n = 3–6 replicates as defined above in Sections 2.4, 2.6–2.8) are shown as individual non-transformed data points in Figs. 2, 5–7 and Supplemental Figs. 3A–C, 4. Each data point shown is the mean of three technical replicates for each biological sample.
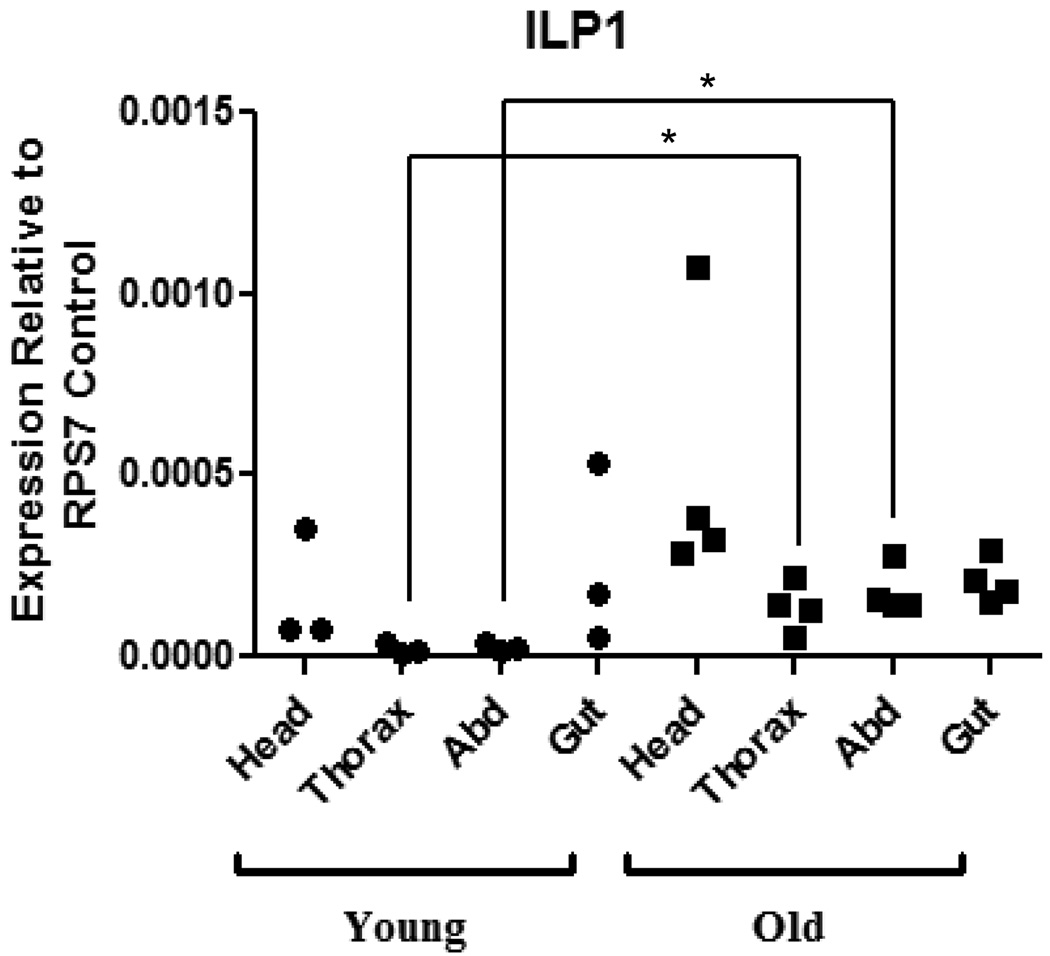
Figure 2. AnsILP1 expression is increased in the thorax and abdomen of older An. stephensi females.
Total RNAs from the head, thorax, abdominal body wall (Abd), and gut tissue (midgut, hindgut, and Malpighian tubules) of females were isolated on days 3–4 (Young) and days 15–16 (Old) post-emergence. AnsILP expression in each tissue was quantified by qRT-PCR. Expression levels are shown relative to expression of ribosomal protein S7 (RPS7) in the same sample. For statistical analysis, data were log-transformed and analyzed by ANOVA followed by Newman-Keuls post-test (α = 0.05).
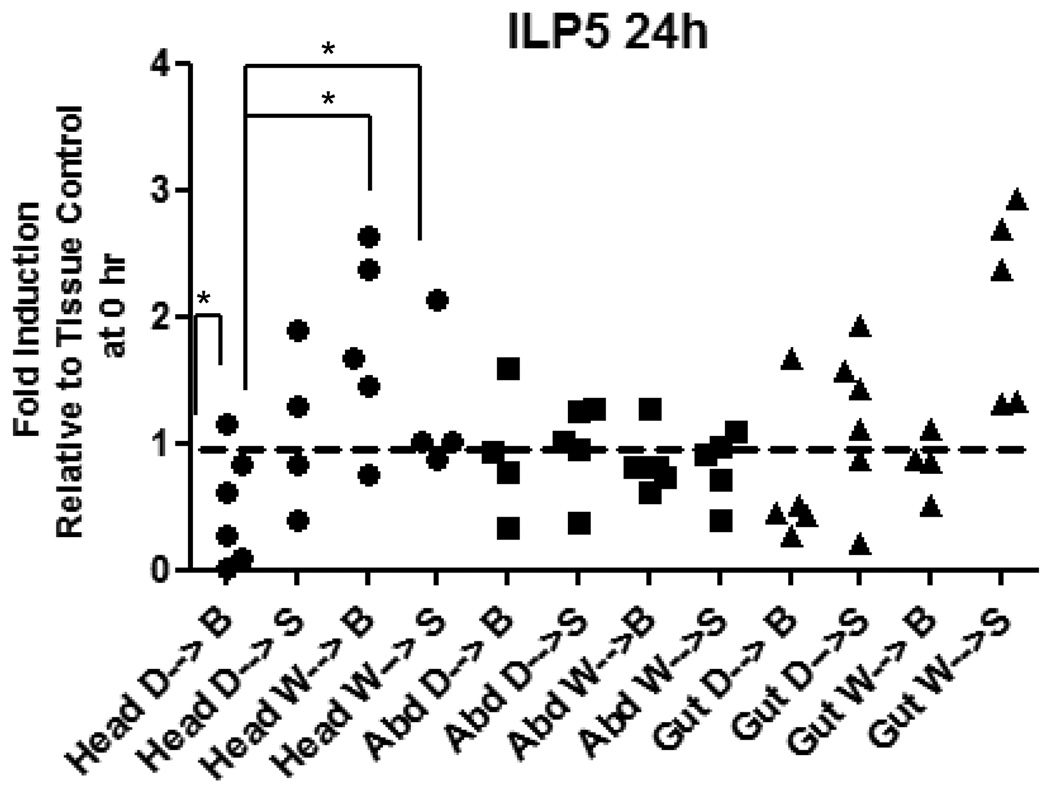
Figure 5. Expression of AnsILP5 is induced in the head of female An. stephensi following blood and sugar ingestion, a response that is blunted by prior deprivation.
Females were given water only (W) or deprived of both sugar and water (D) for 24 h prior to feeding on a sugar solution (S) or washed red blood cells (B). Total RNAs from the head, abdominal body wall (Abd), and gut tissue (midgut, hindgut, Malpighian tubules) were isolated prior to feeding (0 h control) and at 24 h after feeding for qRT-PCR analysis of AnsILP expression. AnsILP expression levels are shown relative to 0 h expression in the matched tissue control, which is set at “1” (dashed line). For statistical analysis, data were log-transformed and analyzed by ANOVA followed by Newman-Keuls post-test (α = 0.05).
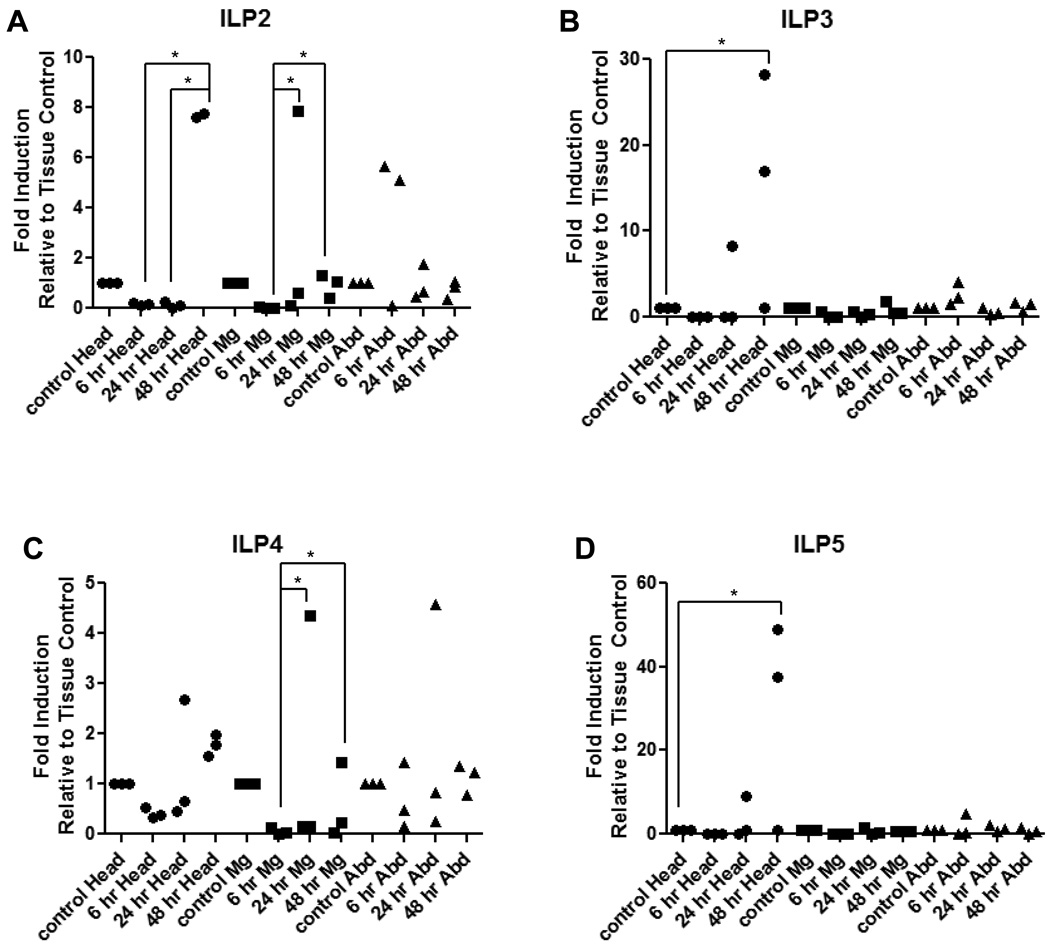
Figure 7. Expression levels of AnsILPs 2–5 are induced in female An. stephensi following infection with P. falciparum.
Females were held on water only for 24 h prior to infection with P. falciparum via an artificial blood meal. Head, midgut (Mg), and abdominal body wall (Abd) tissues were isolated for qRT-PCR expression analysis of (A) AnsILP2, (B) AnsILP3, (C) AnsILP4, and (D) AnsILP5. AnsILP expression in response to infection is shown relative to expression in matched tissue controls (set at “1”) from mosquitoes fed an uninfected blood meal. For statistical analysis, data were log-transformed and analyzed by ANOVA followed by Newman-Keuls post-test (α = 0.05).
3. Results
3.1 Identification and phylogenetic analysis of AnsILPs
Sequences encoding full-length transcript open reading frames (ORFs) and 5’ and 3’ UTRs of AnsILP 1–5 were identified using primers specific for AngILP sequences followed by species-specific 5’ and 3’ RACE. BLASTP alignments of predicted AnsILP sequences with ILP sequences from An. gambiae, Ae. aegypti, and Cu. pipiens were performed (Supplemental Fig. 1). All AnsILP sequences encoded a start methionine and included a stop codon and putative polyadenylation signal (not shown). Each predicted prepropeptide included signal, B, C, and A peptides characteristic of the insulin superfamily (Supplemental Fig. 1). The signal peptides were identified based on predicted processing sites (SignalP 3.0) and ranged in size from 19–31 amino acids. The B, C, and A peptides were identified by the putative proteolytic cleavage sites (1–3 basic amino acids – arginine and lysine) that flanked the C peptide. The predicted C peptides ranged in length from 39–82 residues. Two conserved cysteine residues in the AnsILP B peptides and four in the A peptides (Supplemental Fig. 1) likely form the two disulfide bonds linking the A and B peptides and a third intra-A peptide bond, which together form the common tertiary structure of the insulin superfamily. AnsILP4 and the highly similar AngILP4 appear not to have orthologs among the ILPs in Ae. aegypti or Cu. pipiens. AnsILP5, like orthologs in other dipterans, possesses an additional amino acid between the second and third cysteine of the A-chain.
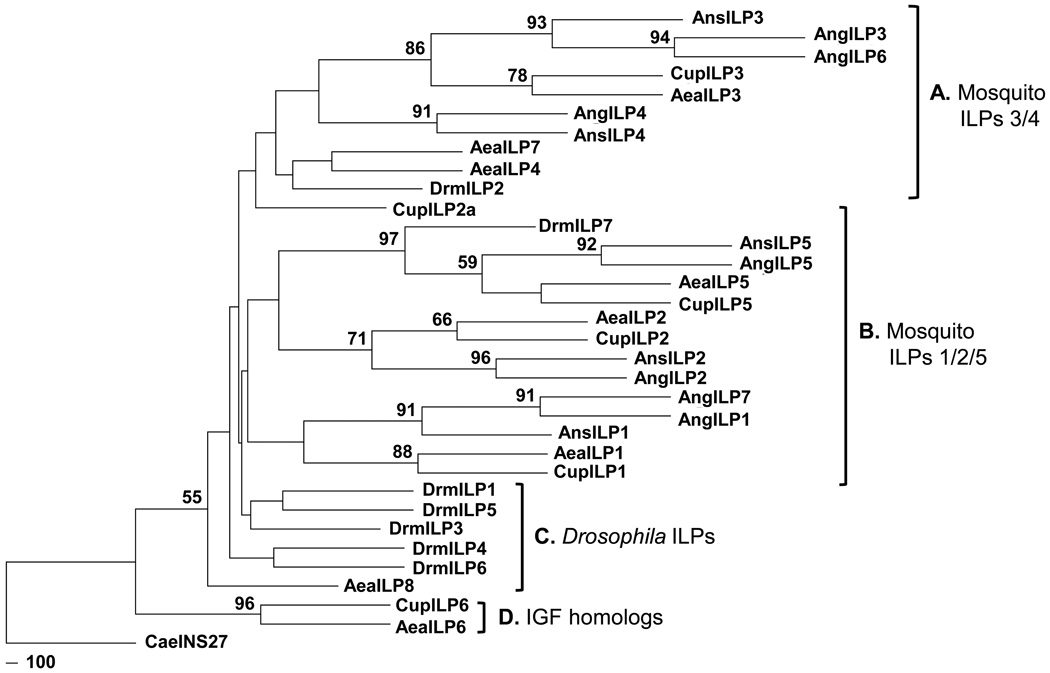
Phylogenetic analysis showed strong support for the groupings of AnsILP1, AnsILP2, and AnsILP5 as a single clade with the corresponding ILPs from other mosquito species (Group B, Fig. 1). Similarly, AnsILP3 and AnsILP4 grouped closely with ILP3 and ILP4 sequences from other mosquito species, with CupILP2A as a notable exception, in a second clade (Group A, Fig. 1). Krieger et al. [12] identified duplicated genes encoding AngILP1/7 and AngILP3/6 based on an early annotation of the An. gambiae genome, but it is not known whether these ILPs are duplicated in An. stephensi. The higher nodal support for Group B clade relative to Group A clade suggest that AnsILP1, AnsILP2, and AnsILP5 share a closer lineage than do AnsILP3 and AnsILP4.
Figure 1. Phylogenetic tree of mosquito and D. melanogaster insulin-like peptides (ILPs) generated with the maximum likelihood method and bootstrap analysis.
Nodes with bootstrap support higher than 50% are shown at the base of the branches and represent the percentage of times that grouping was supported. A–D represents the clade classifications based on the groupings of different ILPs. Abbreviations and NCBI accession numbers of species indicated are as follows: Ae. aegypti (AeaILP1 DQ845750; AeaILP2 DQ845752; AeaILP3 DQ845751; AeaILP4 DQ845753; AeaILP5 DQ845758; AeaILP6 DQ845755; AeaILP7 DQ845757; AeaILP8 DQ845754), An. gambiae (AngILP1 AY324307; AngILP2 AY324308; AngILP3 AY324309; AngILP4 AY324310; AngILP5 AY324312; AngILP6 AY324313; AngILP7 AY324314), An. stephensi (AnsILP1 HM030813; AnsILP2 HM030814; AnsILP3 HM030815; AnsILP4 HM030816; AnsILP5 HM030817), Cu. pipiens (CupILP1 6051901; CupILP2 6051914; CupILP2a 6051902; CupILP3 6051906; CupILP5 6032673; CupILP6 6034782), and D. melanogaster (DrmILP1 NP_648359; DrmILP2 NP_524012; DrmILP3 NP_648360; DrmILP4 NP_648361; DrmILP5 NP_996037; DrmILP6 NP_570000; DrmILP7 NP_570070). INS27 from C. elegans (CaeINS27 Z82082) was used as an outgroup.
Branch lengths indicated low amino acid substitution rates during the early divergence of the dipteran ILPs. The majority of the D. melanogaster ILPs clustered together with the exception of DrmILP2 and DrmILP7 (Group C, Fig.1). This close and basal grouping suggested that the DrmILPs are less divergent and more homologous to each other than to the mosquito ILPs. The grouping of AeaILP6 and CupILP6, putative IGF homologs, was the most basal clade (Group D, Fig.1). Although this would suggest more ancient origins of these ILPs, the position of this clade may be more a reflection of using C. elegans INS27 as an outgroup than an indication of actual evolutionary origins.
3.2 Tissue-specific patterns of AnsILP transcript expression
The five AnsILP transcripts were expressed to varying degrees in all female tissues examined. The ganglia of the nervous system are distributed ventrally along the thorax and abdominal body wall and are the likely source of AnsILP transcripts detected in these body regions. AnsILP1 was expressed at the lowest level in all tissues (Fig. 2) and AnsILP5 had the highest expression (Supplemental Fig. 3A; note scales). AnsILP2-5 expression was generally higher in tissues from younger females relative to patterns observed in older females (Supplemental Fig. 3A), but only AnsILP1 expression was significantly greater in the thorax and abdominal body wall of older females (15–16 days post-emergence; Fig. 2).
3.3 Whole tissue immunocytochemistry
The affinity-purified antibody to AngILP1/3/4 recognized only the corresponding antigen on immunodot blots, but the AngILP2 antibody recognized both its and the AngILP5 antigen (Supplemental Fig. 2). Although the AngILP antibodies were produced before the identification of the AnsILP orthologs, the AngILP2 is identical to the AnsILP2 sequence and the AngILP1/3/4 antigen shares >50% identity with the respective AnsILP sequences (Supplemental Fig. 1). Given these sequence similarities and AngILP antibody specificity, we used the antibodies to immunostain cells expressing the ILPs in whole tissues of female An. gambiae and An. stephensi at 4 days post-emergence, the age at which the majority of AnsILPs (Fig.2, Supplemental Fig. 3A) and AngILPs [12] are highly expressed. Further, by utilizing both Anopheles species we were able to compare ILP distribution patterns and verify the specificity of An. gambiae ILP antibodies in An. stephensi. The AngILP5 affinity purified antibody failed to stain cells in any of the tissues processed as above for both species, although it specifically recognized its antigen on immunodot blots.
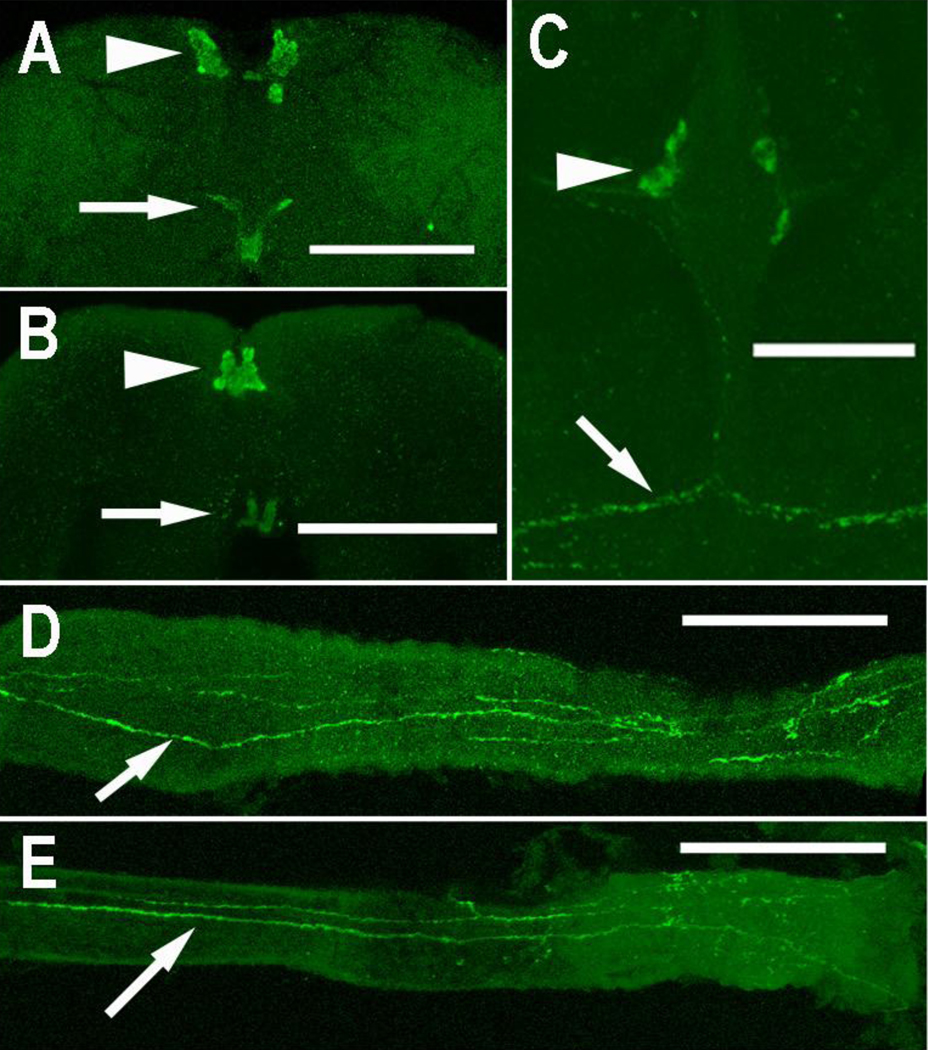
The AngILP1/3/4 antibody stained medial neurosecretory cells (MNCs) in female brains of both species (Figs. 3A, B). Immunostained axons from these cells exited the brain and extended through the corpus cardiacum, a neurohemal organ, and along the anterior midgut (Figs. 3D, E). No other immunostained axons were observed in the brain. Immunostained cells and axons were observed in the abdominal ganglia and along the body wall of An. gambiae (Fig. 3C), but not of An. stephensi. Preabsorption of the AngILP1/3/4 antibody with its antigen peptide blocked all immunostaining in female brains (not shown).
Figure 3. Immunostaining with AngILP1/3/4 antibody.
Medial neurosecretory cells (arrowhead) were immunostained in female An. stephensi (A) and An. gambiae brains (B). Immunostained axons of these cells exited the brain (arrows, A, B) and extended along the anterior midgut (anterior to the right) of female An. stephensi (D) and An. gambiae (E) (D, E scale bars = 100 µm). (C) Paired immunostained cells (arrowhead) are present in an abdominal ganglion of female An. gambiae, and their axons (arrow) extend along the body wall. (A, B, C scale bar = 50 µm).
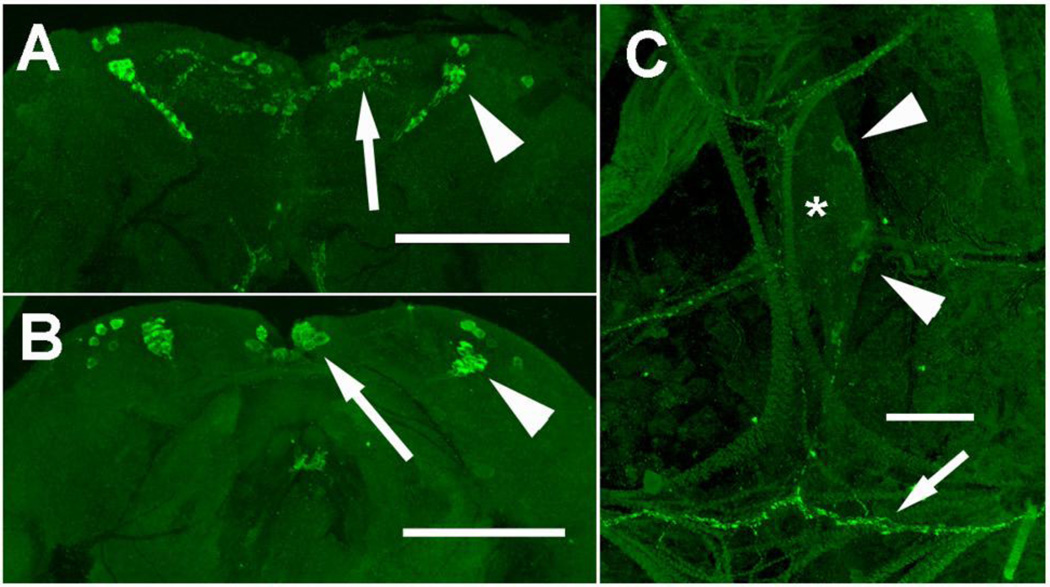
The AngILP2 antibody stained MNCs and bilateral cell clusters in the dorsal lateral regions of brains from both species (Figs. 4A, B). Immunostained axons were observed within the brain, but no staining was observed in the corpus cardiacum or along the anterior midgut. In addition, one or two ILP2 immunostained cells were present in each abdominal ganglion of female An. gambiae (Fig. 4C), and axons extended from these cells along the body wall. No comparable cells were immunostained in An. stephensi. Preabsorption of the AngILP2 antibody with its antigen peptide blocked all immunostaining in female brains (not shown).
Figure 4. Immunostaining with AngILP 2 antibody.
Medial neurosecretory cells (arrow) and lateral neurosecretory cells (arrowhead) were immunostained in female An. stephensi (A) and An. gambiae brains (B) (A,B scale bars = 100 µm). (C) Paired immunostained cells (arrowhead) are present in an abdominal ganglion (*) of female An. gambiae, and their axons (arrow) extend along the body wall. (C; scale bar = 50 µm).
3.4 Diet-dependent patterns of AnsILP transcript expression
AnsILP mRNA expression patterns in heads, abdomens, and guts of female An. stephensi were analyzed in response to four diet regimens reflective of natural conditions: (i) deprivation (no water) for 24 h followed by sugar solution (D→S); (ii) deprivation for 24 h followed by a washed RBC meal (D→B); (iii) water (no sugar) for 24 h followed by sugar solution (W→S); and (iv) water for 24 h followed by a washed RBC meal (W→B; Fig. 5 and Supplemental Figs. 3B, 3C). Tissue samples were prepared for analysis at 6 h or 24 h after provision of a sugar meal or washed RBC meal. At both time points, AnsILP2 exhibited the highest levels of expression (Supplemental Figs. 3B, 3C; note scales). However, analyses of the qRT-PCR data revealed that no significant increases in AnsILP expression were detectable at 6 h after feeding (Supplemental Fig.3B), regardless of deprivation or water pre-treatment. At 24 h, significant changes in expression were noted only for AnsILP5 expression (Fig. 5). In particular, AnsILP5 expression in the heads of deprived females provided a washed RBC meal (D→B; Fig. 5) was significantly less than that in the heads of females taken at the 0 h time point (indicated as the dotted line at 1). Also AnsILP5 expression in the heads of deprived, RBC-fed insects (D→B) was significantly lower than that in the heads of water pre-treated females that were provided either an RBC meal (W→B) or a sugar meal (W→S) (Fig. 5).
3.5 Ingested insulin-dependent AnsILP transcript expression
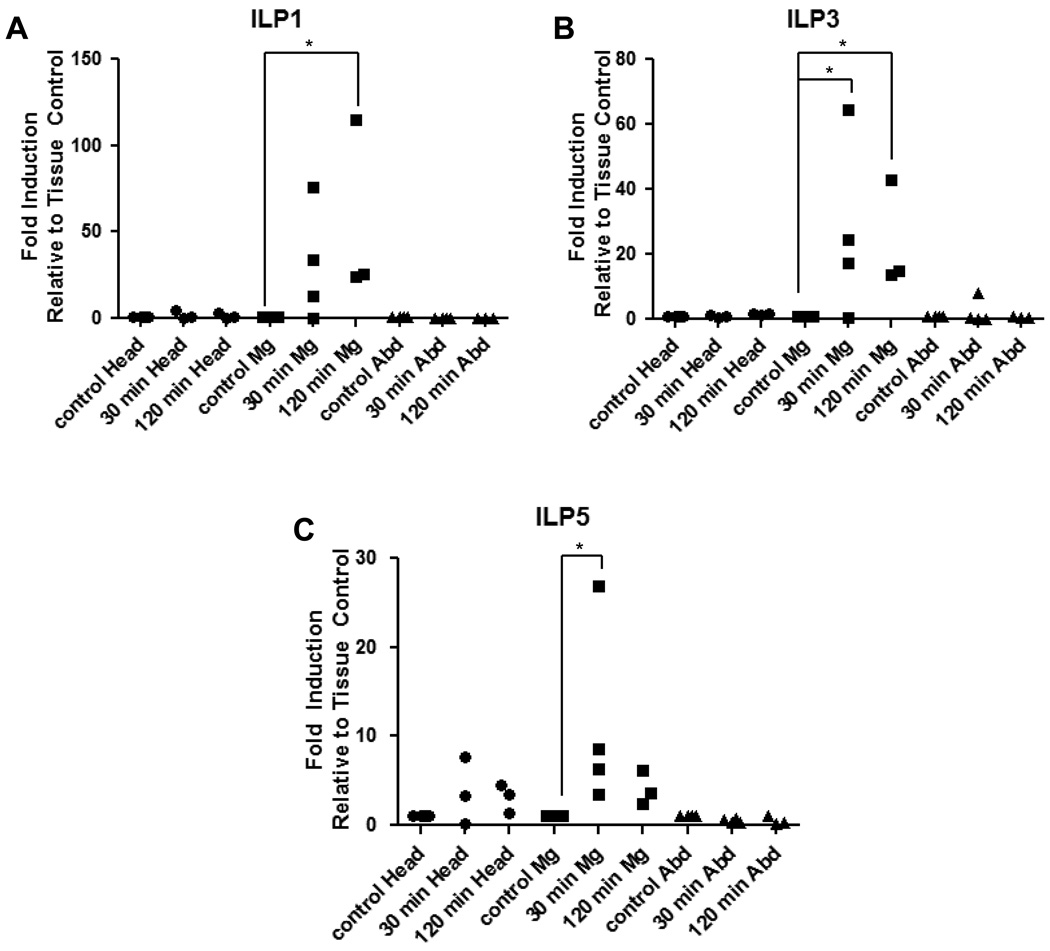
Tissue-specific expression of the AnsILPs in An. stephensi females fed a washed RBC meal supplemented with human insulin was compared to that in females provided only an RBC meal at 30 min and 120 min after feeding. Among the tissues examined, statistically significant increases in insulin-dependent AnsILP1 (up to 115-fold), AnsILP3 (up to 65-fold), and AnsILP5 expression (up to 27-fold) relative to controls were observed only in the midgut (Figs. 6A–C).
Figure 6. Expression levels AnsILPs 1, 3, and 5 are induced in the midgut of female An. stephensi midgut in response to ingested human insulin.
Females were held on water for 24 h prior to feeding on washed red blood cells (RBC) supplemented with 170 pM human insulin. Head, midgut (Mg), and abdominal body wall (Abd) tissues were isolated for qRT-PCR analysis of (A) AnsILP1, (B) AnsILP3, and (C) AnsILP5. AnsILP expression in response to ingested insulin/RBC is shown relative to expression in matched tissue controls (set at “1”) from An. stephensi fed on RBC without added insulin. For statistical analysis, data were log-transformed and analyzed by ANOVA followed by Newman-Keuls post-test (α = 0.05).
3.6 Plasmodium infection-dependent AnsILP transcript expression
Induction patterns of AnsILP expression in tissues from P. falciparum-infected An. stephensi were examined relative to tissues from control, uninfected females at 6 h, 24 h, and 48 h after feeding. No significant P. falciparum-dependent changes in expression of the five AnsILPs were detected in any tissue at 6 h relative to controls, and no significant changes were detected for AnsILP1 (Supplemental Fig.4) expression at 24 h or 48 h. However, significant increases in expression of AnsILP2-5 were noted in the head and/or midgut at 24 h and 48 h in infected females (Figs. 7A–D). The highest expression levels were noted for AnsILP3 (Fig. 7B) and AnsILP5 (Fig. 7D), which exhibited significant head-restricted increases above control levels at 48 h after infection. In contrast, significant increases in AnsILP4 expression (Fig. 7C) were restricted to the midgut at 24 h and 48 h relative to 6 h levels in infected insects. The only AnsILP to exhibit significant infection-dependent responses in both the head and midgut – AnsILP2 (Fig. 7A) – was significantly induced in the head at 48 h and in the midgut at 24 h and 48 h relative to earlier time points following infection.
4. Discussion
Our first goal to identify ILP genes in An. stephensi was achieved without the benefit of a sequenced genome. Fortunately, the sequences of the five AnsILPs were nearly identical to their AngILP homologs [12], so the use of primers based on conserved regions of the AngILPs amplified the same AnsILP regions. Up to eight ILPs have been identified in Ae. aegypti and Cu. pipiens, representative species from other Culicinae genera, and the Drosophila spp. complex, a higher dipteran group. The relatedness of mosquito ILPs is clearly supported (Fig. 1). However, no gene encoding an IGF-like peptide with a truncated C peptide, e.g. AeaILP6, has been identified in an Anopheline species [21].
The fact that the predicted AnsILP amino acid sequences were nearly identical to the orthologous AngILP sequences supported the use of antisera generated to the AngILPs for a comparative immunocytochemical study to identify cell sources of ILPs in An. gambiae and stephensi females. These results provided a basis to interpret transcript expression patterns for the AnsILPs and their tissue-specific responses to aging, diet, and malaria parasite infection. We do not know whether significant changes in the AnsILP transcript profiles for given tissues/body regions reflect ILP storage or release, but these results are strongly suggestive that particular ILPs are involved in the regulation of processes associated with blood feeding and Plasmodium infection. In fact, little is known about the cell/tissue specific expression and secretion of ILPs in mosquitoes or any other insect [34], and co-expression of peptide processing enzymes and other specific trafficking and exocytic proteins are required to produce mature, bioactive ILPs. Future efforts to develop quantitative immunoassays for the AnsILPs will allow us to quantify changes in hemolymph ILP levels that reflect storage or release, given the AnsILP expression results discussed below.
In our study, the presence of AnsILP transcripts in tissues of female An. stephensi is best explained by their specific expression in the nervous system, which consists of connected ganglia distributed along the length of the ventral body wall. In particular, the distribution of transcripts for AnsILP1, AnsILP3 and AnsILP4 in the head, thorax, gut/midgut, and abdominal body wall (Figs. 2, 5, 6, and 7) corresponds to AngILP1/3/4 antibody staining of brain neurosecretory cells and axons in both Anopheles species and in cells of the ventral ganglia of An. gambiae (Fig. 3). Identical ganglion cells likely are a source of ILPs in An. stephensi, given that such cells in Ae. aegypti and D. melanogaster are reactive to ILP antibodies [17, 21]. Localization of the immunoreactive axons in the corpus cardiacum, a neuroendocrine organ, and the anterior midgut suggests a neurohormonal function for the endogenous ILPs recognized by this antibody.
Some discordance in peptide and transcript expression patterns was apparent for ILP2. In particular, AnsILP2 transcripts were detected consistently in heads, but higher levels were seen in the thorax and abdomen of An. stephensi females after some experimental manipulations (Fig. 7; Supplemental Figs. 3A–C, 4A). Staining with the AngILP2 antibody, however, revealed many brain neurosecretory cells but no immunoreactivity in the corpus cardiacum (Fig. 4), suggesting that AngILP2 may function in neuronal rather than endocrine signaling. As discussed above, cells in the ventral ganglia of An. stephensi are a likely source of AnsILP2, given the localization in such cells in An. gambiae, and this would be consistent with AnsILP2 expression in the thorax and abdomen.
Detection of AnsILP transcripts in the midgut of An. stephensi (Figs. 6 and 7) suggests that axons from the brain neuroendocrine axis or intrinsic endocrine cells scattered along this tissue may be a likely source. Axons along the anterior midgut were positive for AngILP1/3/4 immunoreactivity (Fig. 3), but AngILP antibodies failed to stain midgut endocrine cells in either Anopheles species. It is possible that transcripts for one or more of the AnsILP genes are present in the axons as observed for neuropeptide mRNAs in other invertebrates [30,31], thus accounting for the qRT-PCR detection. A previous study showed that endocrine cells only in the anterior midgut of female Ae. aegypti were stained with an ILP antibody [21], but a careful study of peptide hormone expression in the midgut of D. melanogaster found that DrmILP3 transcripts were detected only in muscle bands at the anterior end of the midgut [32]. To date, there is little evidence for ILP secretion by the midgut endocrine system in insects, but this intriguing prospect deserves further study given the functional importance of the insect midgut.
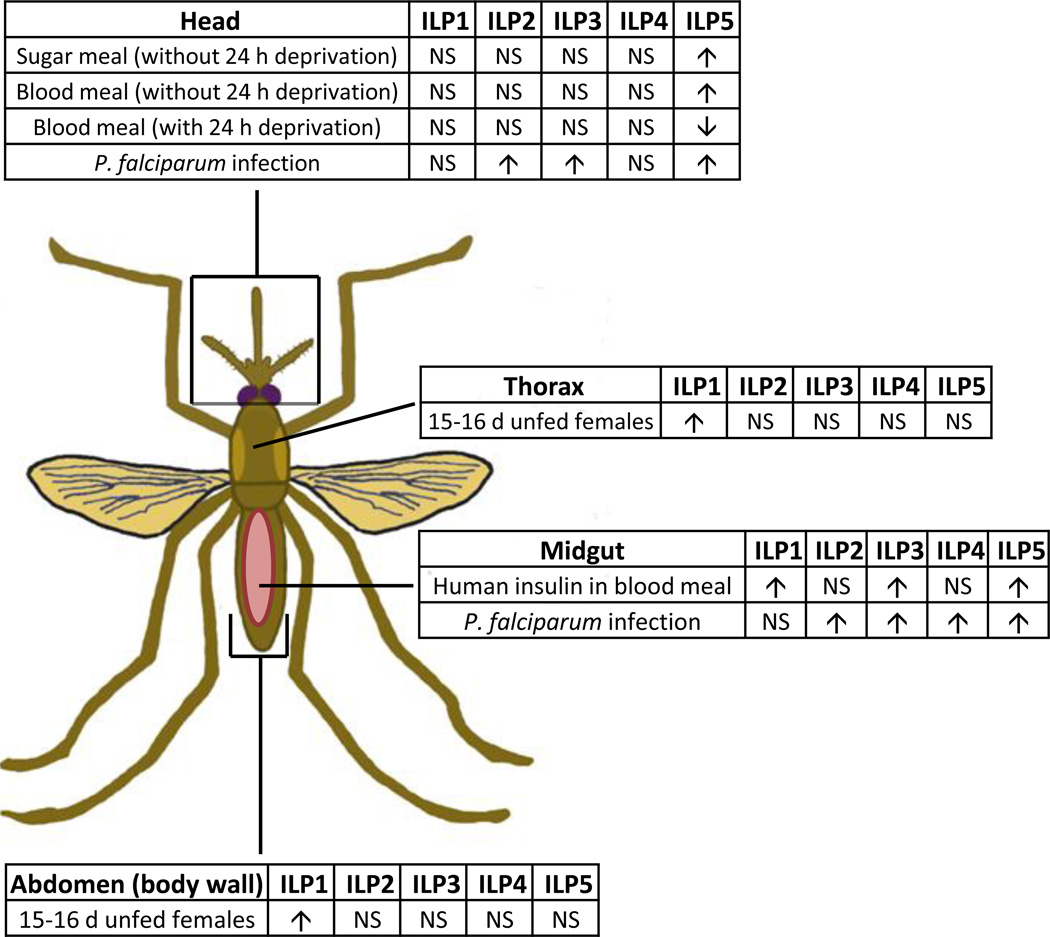
Tissue-specific AnsILP expression patterns in young and old female An. stephensi and in response to various dietary regimens, to ingested human insulin and to infection with P. falciparum are summarized in Fig. 8. Our analysis of AnsILP expression in young and old An. stephensi females spanned a period roughly equivalent to the observed maximum lifespan of this species under field conditions [18, 20]. Although no significant changes in expression were observed, expression levels of AnsILP2-5 were generally decreased in older females (Supplemental Fig.3A, Fig. 8), whereas AnsILP1 expression was significantly higher in older females (Figs. 2, 8). Although further studies are necessary to determine the significance of these changes, these trends suggest a pattern of AnsILP expression that is age-dependent.
Figure 8. Summary of AnsILP expression patterns.
All AnsILPs were expressed to some degree in each of the tissues examined (head, thorax, abdomen, midgut/hindgut/Malpighian tubules or midgut alone). Experimental manipulations that produced significant increases (p < 0.05) in the expression of a particular AnsILP in a tissue are denoted by upward facing arrows, while significant decreases are denoted as downward facing arrows. NS= no significant change.
The minimal response to sugar and blood meals suggested that AnsILP transcript expression is not tuned specifically to their ingestion (Supplemental Figs. 3B, 3C, 8), with one exception. Only AnsILP5 expression was induced more than 2-fold in the head at 24 h after sugar or RBC feeding (Fig. 5), and water deprivation blunted the response to both foods. Interestingly, the presence of physiological concentrations of human insulin in the RBC meal was accompanied by earlier and much larger changes in AnsILP5 expression. Specifically, no significant induction in AnsILP5 expression was detected until 24 h after an unsupplemented RBC meal (Fig. 5), but with the addition of insulin to the RBC meal, AnsILP5 expression in the midgut was induced up to 25-fold within 30 min relative to control tissue (Fig. 6C). The insulin-supplemented RBC meal was also accompanied by significant increases in AnsILP1 and AnsILP3 expression in the midgut within 2 h of feeding (Figs. 6A, 6B), suggesting that insulin-dependent changes in AnsILP expression in the context of blood feeding are rapid and specific in the midgut epithelium.
Infection of female An. stephensi with P. falciparum also profoundly affected AnsILP expression (Fig. 8). However, in contrast to the response to insulin that was observed as early as 30 min post-feeding, significant increases in AnsILP expression in response to infection were not detectable until 24 h and 48 h post-infection. The reasons for these differences are not yet clear, but AnsILP expression in response to infection may be dependent on an earlier, primary signaling response that generates a secondary response that activates AnsILP expression. In addition to ingested insulin, parasite GPIs are important signals of infection to the mosquito host. These carbohydrate anchors are released from abundant asexual stage parasites that die in the ingested blood meal and, once released, GPIs can induce activation of IIS in the An. stephensi midgut within 30 min after ingestion [14]. We cannot exclude the possibility that other signaling pathways are activated by parasite GPIs, however, including those that are dependent on the mitogen-activated protein kinases (ERK, JNK, and p38) as shown in mammalian cells [9], and that primary activation of these orthologous, conserved pathways in An. stephensi cells could influence the timing of AnsILP expression.
5. Conclusions
Malaria-induced alterations in the physiology of mammalian hosts are relevant to malaria parasite transmission by mosquitoes. Specifically, the ingested blood meal from the human host contains parasites, parasite-derived factors, and bioactive levels of insulin that can directly alter the development of P. falciparum in the mosquito host. Here, our data suggest that the impact of human insulin on parasite development in An. stephensi is potentiated by the induction of endogenous AnsILPs, which, to our knowledge, is the first example of hormonal crosstalk in a complex vector-borne pathogen cycle. An understanding of the functional effects of changes in AnsILP biosynthesis in An. stephensi in response to malaria parasite infection will fill a significant gap in our understanding of the physiology of parasite infection in the mosquito host.
Research Highlights.
Five insulin-like peptide encoding genes were identified from A. stephensi.
Insulin-like peptide gene expression was evident in all tissues examined.
Anti-ILP antibodies labeled only in cells of the nervous system.
ILP expression was not specifically tuned to feeding on sugar or red blood cells.
ILP expression was increased in response to human insulin and P. falciparum infection.
Supplementary Material
In ILP A and B chain alignments, asterisks denote conserved cysteines. Underlined AngILP A chain sequences were used as peptide antigen for rabbit anti-AngILP2 antibody. In the composite ILP1/3/4 antigen alignment, asterisks denote identical amino acids, whereas double dots and single dots indicate conservative and semi-conservative substitutions, respectively.
An. gambiae ILP A chain peptides were spotted on nitrocellulose at 100, 10, 1 and 0.1 ng and then incubated with AngILP2 or AngILP1/3/4 antibodies as described in the Materials and Methods.
(A) Total RNAs from the head, thorax, abdominal body wall (Abd), and gut tissue (midgut, hindgut, and Malpighian tubules) of females were isolated on days 3–4 (Young) and days 15–16 (Old) post-emergence. AnsILP expression in each tissue was quantified by qRT-PCR. Expression levels are shown relative to expression of ribosomal protein S7 (RPS7) in the same sample. For statistical analysis, data were log-transformed and analyzed by ANOVA followed by Newman-Keuls post-test (α = 0.05). (B, C) Females were given water only (W) or deprived of both sugar and water (D) for 24 h prior to feeding on a sugar solution (S) or washed red blood cells (B). Total RNAs from the head, abdominal body wall (Abd), and gut tissue (midgut, hindgut, Malpighian tubules) were isolated prior to feeding (0 h control) and at 6 h (B) and 24 h (C) after feeding for qRT-PCR analysis of AnsILP expression. AnsILP expression levels are shown relative to 0 h expression in the matched tissue control, which is set at “1” (dashed line). For statistical analysis, data were log-transformed and analyzed by ANOVA followed by Newman-Keuls post-test (α = 0.05). NS = not significant.
(A,B) Females were held on water for 24 h prior to feeding on washed red blood cells (RBC) supplemented with 170 pM human insulin. Head, midgut (Mg), and abdominal body wall (Abd) tissues were isolated for qRT-PCR analysis of (A) AnsILP2 and (B) AnsILP4. AnsILP expression in response to ingested insulin/RBC is shown relative to expression in matched tissue controls (set at “1”) from An. stephensi fed on RBC without added insulin. For statistical analysis, data were log-transformed and analyzed by ANOVA followed by Newman-Keuls post-test (α = 0.05). (C) Females were held on water only for 24 h prior to infection with P. falciparum via an artificial blood meal. Head, midgut (Mg), and abdominal body wall (Abd) tissues were isolated for qRT-PCR expression analysis of AnsILP1. Expression in response to infection is shown relative to expression in matched tissue controls (set at “1”) from mosquitoes fed an uninfected blood meal. For statistical analysis, data were log-transformed and analyzed by ANOVA followed by Newman-Keuls post-test (α = 0.05). NS = not significant.
Acknowledgments
We thank Kong Wai Cheung and Nazzy Pakpour for their assistance with P. falciparum culture and mosquito infection. Funding for these studies was provided by the United States National Institutes of Health National Institute of Allergy and Infectious Diseases (NIH NIAID) grants AI073745 to M.A.R., AI033108 to M.R.B., and AI080799 to S.L. These studies were conducted in a facility constructed with support from Research Facilities Improvement Program Grant Number C06 RR-12088-01 from the United States National Center for Research Resources, NIH. A.G.M. was supported by an Air Force Institute of Technology Scholarship. J.E.P. was supported by fellowship support from an NIH Initiative for Maximizing Student Development grant to UC Davis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Aspinwall CA, Quian WJ, Roper MG, Kulkarni RN, Kahn CR, Kennedy RT. Roles of insulin receptor substrate-1, phosphatidylinositol 3-kinase, and release of intracellular Ca2+ stores in insulin-stimulated insulin secretion in beta -cells. J. Biol. Chem. 2000;275:22331–22338. doi: 10.1074/jbc.M909647199. [DOI] [PubMed] [Google Scholar]
- 2.Belfiore A, Frasca F, Pandini G, Sciacca L, Vigneri R. Insulin receptor isoforms and insulin receptor/insulin-like growth factor receptor hybrids in physiology and disease. Endocr. Rev. 2009;30:586–623. doi: 10.1210/er.2008-0047. [DOI] [PubMed] [Google Scholar]
- 3.Bouche C, Lopez X, Fleischman A, Cypress AM, O'Shea S, Stefanovski D, Bergman RN, Rogatsky E, Stein DT, Kahn CR, Kulkarni RN, Goldfine AB. Insulin enhances glucose-stimulated insulin secretion in healthy humans. Proc. Natl. Acad. Sci. U. S. A. 2009;107:4770–4775. doi: 10.1073/pnas.1000002107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brandon MC, Pennington JE, Isoe J, Zamora J, Schillinger AS, Meisfeld RL. TOR signaling is required for amino acid stimulation of early trypsin protein synthesis in the midgut of Aedes aegypti mosquitoes. Insect. Biochem. Mol. Biol. 2008;38:916–922. doi: 10.1016/j.ibmb.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown MR, Clark KD, Gulia M, Zhao Z, Garczynski SF, Crim JW, Suderman RJ, Strand MR. An insulin-like peptide regulates egg maturation and metabolism in the mosquito Aedes aegypti. Proc. Natl. Acad. Sci. U. S. A. 2008;105:5716–5721. doi: 10.1073/pnas.0800478105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corby-Harris V, Drexler A, Watkins de Jong L, Antonova Y, Pakpour N, Ziegler R, Ramberg F, Lewis EE, Brown JM, Luckhart S, Riehle MA. Activation of Akt signaling reduces the prevalence and intensity of malaria parasite infection and lifespan in Anopheles stephensi mosquitoes. PLoS Pathog. 2010;6:e1001003. doi: 10.1371/journal.ppat.1001003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Darby SM, Miller ML, Allen RO, LeBeau M. A mass spectrometric method for quantitation of intact insulin in blood samples. J. Anal. Toxicol. 2001;25:8–14. doi: 10.1093/jat/25.1.8. [DOI] [PubMed] [Google Scholar]
- 8.Drexler AL, Vodovotz Y, Luckhart S. Plasmodium development in the mosquito: biology bottlenecks and opportunities for mathematical modeling. Trends Parasitol. 2008;24:333–336. doi: 10.1016/j.pt.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gowda DC. TLR-mediated cell signaling by malaria GPIs. Trends Parasitol. 2007;23:596–604. doi: 10.1016/j.pt.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 10.Halls ML, van der Westhuizen ET, Bathgate RA, Summers RJ. Relaxin family peptide receptors--former orphans reunite with their parent ligands to activate multiple signalling pathways. Br. J. Pharmacol. 2007;150:677–691. doi: 10.1038/sj.bjp.0707140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang MA, Mott TM, Tapley EC, Lewis EE, Luckhart S. Insulin regulates aging and oxidative stress in Anopheles stephensi. J. Exp. Biol. 2008;211:741–748. doi: 10.1242/jeb.012955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krieger MJ, Jahan N, Riehle MA, Cao C, Brown MR. Molecular characterization of insulin-like peptide genes and their expression in the African malaria mosquito, Anopheles gambiae. Insect. Mol. Biol. 2004;13:305–315. doi: 10.1111/j.0962-1075.2004.00489.x. [DOI] [PubMed] [Google Scholar]
- 13.Leibiger B, Wahlander K, Berggren PO, Leibiger IB. Glucose-stimulated insulin biosynthesis depends on insulin-stimulated insulin gene transcription. J. Biol. Chem. 2000;275:30153–30156. doi: 10.1074/jbc.M005216200. [DOI] [PubMed] [Google Scholar]
- 14.Lim J, Gowda DC, Krishnegowda G, Luckhart S. Induction of nitric oxide synthase in Anopheles stephensi by Plasmodium falciparum: mechanism of signaling and the role of parasite glycosylphosphatidylinositols. Infect. Immun. 2005;73:2778–2789. doi: 10.1128/IAI.73.5.2778-2789.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu C, Lam HN, Menon RK. New members of the insulin family: regulators of metabolism, growth and now … reproduction. Pediatr. Res. 2005;57:70R–73R. doi: 10.1203/01.PDR.0000159573.55187.CA. [DOI] [PubMed] [Google Scholar]
- 16.Luckhart S, Riehle MA. The insulin signaling cascade from nematodes to mammals: insights into innate immunity of Anopheles mosquitoes to malaria parasite infection. Dev. Comp. Immunol. 2007;31:647–656. doi: 10.1016/j.dci.2006.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miguel-Aliaga I, Thor S, Gould AP. Postmitotic specification of Drosophila insulinergic neurons from pioneer neurons. PLoS Biol. 2008;6:e58. doi: 10.1371/journal.pbio.0060058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quraishi MS, Eshgi N, Faghih MA. Flight range, lengths of gonotrophic cycles, and longevity of P-32-labeled Anopheles stephensi mysorensis. J. Econ. Entomol. 1966;59:50–55. doi: 10.1093/jee/59.1.50. [DOI] [PubMed] [Google Scholar]
- 19.Rassmusen R. Quantification on the LightCycler. In: Meuer S, Wittwer CT, Nakagawara K, editors. Rapid Cycle Real-time PCR, Methods and Applications. Heidelberg: Springer Press; 2001. pp. 21–34. [Google Scholar]
- 20.Reisen WK, Aslamkahn M. A release-recapture experiment with the malaria vector, Anopheles stephensi Liston, with observations on dispersal, survivorship, population size, gonotrophic rhythm and mating behaviour. Ann. Trop. Med. Parasitol. 1979;73:251–269. doi: 10.1080/00034983.1979.11687255. [DOI] [PubMed] [Google Scholar]
- 21.Riehle MA, Fan Y, Cao C, Brown MR. Molecular characterization of insulin-like peptides in the yellow fever mosquito, Aedes aegypti: expression, cellular localization, and phylogeny. Peptides. 2006;27:2547–2560. doi: 10.1016/j.peptides.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 22.Riehle MA, Garczynski SF, Crim JW, Hill CA, Brown MR. Neuropeptides and peptide hormones in Anopheles gambiae. Science. 2002;298:172–175. doi: 10.1126/science.1076827. [DOI] [PubMed] [Google Scholar]
- 23.Rincon M, Rudin E, Barzilai N. The insulin/IGF-1 signaling in mammals and its relevance to human longevity. Exp. Gerontol. 2005;40:873–877. doi: 10.1016/j.exger.2005.06.014. [DOI] [PubMed] [Google Scholar]
- 24.Roy SG, Hansen IA, Raikhel AS. Effect of insulin and 20-hydroxyecdysone in the fat body of the yellow fever mosquito, Aedes aegypti. Insect Biochem. Mol. Biol. 2007;37:1317–1326. doi: 10.1016/j.ibmb.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roy SG, Raikhel AS. The small GTPase Rheb is a key component linking amino acid signaling and TOR in the nutritional pathway that controls mosquito egg development. Insect Biochem. Mol Biol. 2011;41:62–69. doi: 10.1016/j.ibmb.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 27.Sim C, Denlinger DL. A shut-down in expression of an insulin-like peptide, ILP-1, halts ovarian maturation during the overwintering diapause of the mosquito Culex pipiens. Insect Mol. Biol. 2009;18:325–332. doi: 10.1111/j.1365-2583.2009.00872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Surachetpong W, Pakpour N, Cheung KW, Luckhart S. Reactive oxygen species-dependent cell signaling regulates the mosquito immune response to Plasmodium falciparum. Antioxid. Redox Signal. 2010 Dec 2; doi: 10.1089/ars.2010.3401. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tatar M, Bartke A, Antebi A. The endocrine regulation of aging by insulin-like signals. Science. 2003;299:1346–1351. doi: 10.1126/science.1081447. [DOI] [PubMed] [Google Scholar]
- 30.Van Minnen J, Schallig HDFH. Demonstration of insulin-related substances in the central nervous system of pulmonates and Aplysia californica. Cell Tissue Res. 1990;260:381–386. [Google Scholar]
- 31.Van Minnen J. Axon localization of neuropeptide-encoding mRNA in identified neurons of the snail Lymnaea stagnalis. Cell Tissue Res. 1994;276:155–161. doi: 10.1007/BF00354795. [DOI] [PubMed] [Google Scholar]
- 32.Veenstra JA. Peptidergic paracrine and endocrine cells in the midgut of the fruit fly maggot. Cell Tissue Res. 2009;336:309–323. doi: 10.1007/s00441-009-0769-y. [DOI] [PubMed] [Google Scholar]
- 33.Wen Z, Gulia M, Clark KD, Dhara A, Crim JW, Strand MR, Brown MR. Two insulin-like peptide family members from the mosquito Aedes aegypti exhibit differential biological and receptor binding activities. Mol. Cell Endocrinol. 2010;328:47–55. doi: 10.1016/j.mce.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Q, Brown MR. Signaling and function of insulin-like peptides in insects. Annu. Rev. Entomol. 2006;51:1–24. doi: 10.1146/annurev.ento.51.110104.151011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In ILP A and B chain alignments, asterisks denote conserved cysteines. Underlined AngILP A chain sequences were used as peptide antigen for rabbit anti-AngILP2 antibody. In the composite ILP1/3/4 antigen alignment, asterisks denote identical amino acids, whereas double dots and single dots indicate conservative and semi-conservative substitutions, respectively.
An. gambiae ILP A chain peptides were spotted on nitrocellulose at 100, 10, 1 and 0.1 ng and then incubated with AngILP2 or AngILP1/3/4 antibodies as described in the Materials and Methods.
(A) Total RNAs from the head, thorax, abdominal body wall (Abd), and gut tissue (midgut, hindgut, and Malpighian tubules) of females were isolated on days 3–4 (Young) and days 15–16 (Old) post-emergence. AnsILP expression in each tissue was quantified by qRT-PCR. Expression levels are shown relative to expression of ribosomal protein S7 (RPS7) in the same sample. For statistical analysis, data were log-transformed and analyzed by ANOVA followed by Newman-Keuls post-test (α = 0.05). (B, C) Females were given water only (W) or deprived of both sugar and water (D) for 24 h prior to feeding on a sugar solution (S) or washed red blood cells (B). Total RNAs from the head, abdominal body wall (Abd), and gut tissue (midgut, hindgut, Malpighian tubules) were isolated prior to feeding (0 h control) and at 6 h (B) and 24 h (C) after feeding for qRT-PCR analysis of AnsILP expression. AnsILP expression levels are shown relative to 0 h expression in the matched tissue control, which is set at “1” (dashed line). For statistical analysis, data were log-transformed and analyzed by ANOVA followed by Newman-Keuls post-test (α = 0.05). NS = not significant.
(A,B) Females were held on water for 24 h prior to feeding on washed red blood cells (RBC) supplemented with 170 pM human insulin. Head, midgut (Mg), and abdominal body wall (Abd) tissues were isolated for qRT-PCR analysis of (A) AnsILP2 and (B) AnsILP4. AnsILP expression in response to ingested insulin/RBC is shown relative to expression in matched tissue controls (set at “1”) from An. stephensi fed on RBC without added insulin. For statistical analysis, data were log-transformed and analyzed by ANOVA followed by Newman-Keuls post-test (α = 0.05). (C) Females were held on water only for 24 h prior to infection with P. falciparum via an artificial blood meal. Head, midgut (Mg), and abdominal body wall (Abd) tissues were isolated for qRT-PCR expression analysis of AnsILP1. Expression in response to infection is shown relative to expression in matched tissue controls (set at “1”) from mosquitoes fed an uninfected blood meal. For statistical analysis, data were log-transformed and analyzed by ANOVA followed by Newman-Keuls post-test (α = 0.05). NS = not significant.