Abstract
Background
Invasive ovarian cancer is a significant cause of gynecologic cancer mortality.
Methods
We examined whether this mortality was associated with inherited variation in ~170 candidate genes/regions (993 SNPs) in a multi-stage analysis based initially on 312 Mayo Clinic cases (172 deaths). Additional analyses used The Cancer Genome Atlas (TCGA; 127 cases, 62 deaths). For the most compelling gene, we immunostained Mayo Clinic tissue micro-arrays (TMAs, 326 cases) and conducted consortium-based SNP replication analysis (2,560 cases, 1,046 deaths).
Results
The strongest initial mortality association was in HGF (hepatocyte growth factor) at rs1800793 (HR 1.7, 95% CI 1.3–2.2, p=2.0×10−5) and with overall variation in HGF (gene-level test, p=3.7×10−4). Analysis of TCGA data revealed consistent associations (e.g., rs5745709 [r2=0.96 with rs1800793]: TCGA 2.4, 1.4–4.1, p=2.2×10−3; Mayo Clinic+TCGA 1.6, 1.3–1.9, p=7.0×10−5) and suggested genotype correlation with reduced HGF mRNA levels (p=0.01). In Mayo Clinic TMAs, protein levels of HGF, its receptor MET, and phospho-MET were not associated with genotype and did not serve as an intermediate phenotype; however, phospho-MET was associated with reduced mortality (p=0.01) likely due to higher expression in early-stage disease. In eight additional ovarian cancer case series, HGF rs5745709 was not associated with mortality (1.0, 0.9–1.1, p=0.87).
Conclusions
We conclude that although HGF signaling is critical to migration, invasion, and apoptosis, it is unlikely that genetic variation plays a major role in ovarian cancer mortality; any minor role is not related to genetically-determined expression.
Impact
Our study demonstrates the utility of multiple data types and multiple datasets in observational studies.
Keywords: gynecologic neoplasms, angiogenesis, single nucleotide polymorphism
INTRODUCTION
In the United States, ovarian cancer is the fifth leading cause of cancer death among women (1). Despite clinical responses to combination platinum/taxane-based chemotherapy in most women after surgical debulking, five-year overall survival lingers around 30% and, even with modern chemotherapy, most cases with advanced disease relapse and die of ovarian cancer (2, 3). Inherited variation may influence outcome. For example, cases with rare germline BRCA1 or BRCA2 mutations have improved chemoresponsiveness and survival (4). Common inherited variants may also be prognostic. Notably, we and others have reported evidence for a role of inherited variation in angiogenesis and inflammation genes in ovarian cancer survival (5–7). As initial ovarian cancer genome-wide association studies have not identified common mortality-associated alleles (8), in-depth analysis of additional candidate genes in key biological pathways holds promise for the identification of factors with functional relevance or prognostic utility.
Here, we examined key candidate genes encoding angiogenesis factors (9, 10) mitotic kinases (11), growth stimulatory mediators and stromal factors (12, 13), as well as genes and regions suggested by expression studies (14) and genome-wide association studies (15–17). We first evaluated the association between mortality and inherited single nucleotide polymorphism (SNP) variation among invasive epithelial ovarian cancer patients seen at the Mayo Clinic, and we pursued key findings via analysis of data from The Cancer Genome Atlas (TCGA). We then conducted expression analysis of tissue micro-arrays (TMAs) made from tumors of Mayo Clinic cases, and we examined genetic association in cases from eight additional ovarian cancer case series. In total, a multi-faceted approach integrating tumor and replication studies aimed to provide observational and functional insight into the role of SNPs in ovarian cancer mortality.
METHODS
Candidate Gene Analysis
Initial Study Participants
Recruitment of cases from Mayo Clinic’s gynecologic surgery and medical oncology departments (MAY1) used established protocols approved by the relevant Institutional Review Board (IRB) (5). All participants gave written informed consent. Eligible cases were women aged 20 years or older living in MN, IA, WI, IL, ND, or SD and ascertained within one year of a diagnosis of pathologically-confirmed primary invasive epithelial ovarian cancer. Between December 1999 and March 2006, 328 cases were enrolled; median time from diagnosis to recruitment was five days. Data on vital status through July 31, 2009 was obtained from the National Death Index, computerized medical records, and the Mayo Clinic Cancer Registry which annually follows cases diagnosed or receiving initial treatment at Mayo Clinic. Death certificates were available on 95 of 172 deceased cases, and dates of death were 94.7% concordant with dates obtained via registries (five certificates differed by a median of three days). Of 140 living cases, nine were lost to follow-up more than two years prior. Data on clinical features of disease including histology, surgical outcome, and chemotherapy were abstracted by experienced research nurses with review by gynecologic and medical oncologists. DNA was extracted from 10 to 15 mL fresh peripheral blood using the Gentra AutoPure LS Purgene salting out methodology (Gentra, Minneapolis, MN) and stored at −80°C; samples were bar-coded to ensure accurate processing and plated with duplicates and lab standards. We excluded 12 sequence-confirmed BRCA1 and BRCA2 mutation carriers and four cases with predicted non-European ancestry (Supplemental Figure 1) (18, 19), resulting in 312 analyzed cases (Supplemental Table 1).
Polymorphisms and Genotyping
Key genes within angiogenesis, mitosis, growth and stromal factors, as well as expression-based genes, and genes in key chromosomal regions were identified using published literature (9–17) and the Kyoto Encyclopedia of Genes and Genomes (Supplemental Table 2) (20). For angiogenesis genes, all SNPs with minor allele frequency (MAF) ≥ 0.05 were selected; in 8q24 and 9p24, a combination of region-tagging, gene-tagging, and replication-based SNPs were included (21), and for remaining genes, we selected tagSNPs within 5 kb with MAF ≥ 0.05 based on European linkage disequilibrium (LD) in HapMap v. 22 (22, 23) (r2 ≥ 0.8; expression-based genes r2 ≥ 0.9). Genotyping of 897 samples (including analyzed cases, borderline cases, population controls, and 26 duplicates and laboratory standards including CEPH trios) was performed at the Mayo Clinic using the Illumina GoldenGate BeadArray Assay (24). Of 871 unique MAY1 participants, only one sample failed (call rate < 90%) which was from a control; thus, genotype data were available on all cases. We assessed departures from Hardy Weinberg equilibrium (HWE) using Pearson goodness-of-fit and Fisher exact tests using data from self-reported European-American controls. Manual review of the plots was performed in GenomeStudio (Illumina, San Diego CA) to verify optimal SNP clustering, using both replicate and inheritance information from the CEPH family trio. When data failed to reveal one to three distinct clusters (representing AA, AB and BB genotypes), a SNP was failed. Of 1,152 SNPs attempted (Supplemental Table 3), 25 failed including 15 with call rate < 90%, nine with poor clustering, and one with an unresolved replicate or Mendelian error. We excluded SNPs with MAF < 0.05 (N=123) or HWE p-value < 0.001 (N=11), leaving 993 SNPs in 168 genes. LD plots were created using Haploview v. 4.2 (25) (Supplemental Figure 2).
Statistical Methods
We used Cox proportional hazards regression (26) to estimate hazard ratios (HRs) and 95% confidence intervals (CIs) for association with mortality, adjusted for clinical covariates. We accounted for left truncation using the start-stop counting process style of input within the Cox regression framework; thus, cases did not contribute follow-up time until date of enrollment (27). For each SNP, HRs with 95% CIs were estimated per-allele (i.e., 0, 1, or 2 copies of minor allele), analogous to the Armitage test for trend for binary endpoints (28). As our aim was to evaluate the role of SNPs beyond known prognostic factors, we included as covariates the following clinical variables which were univariately associated with mortality in a stepwise Cox regression model (p < 0.05): age at diagnosis, pre-surgical log10(CA125), tumor stage (I, II, III, IV, unknown), tumor grade (grade 1 or 2, grade 3, grade 4, unknown), volume of residual tumor following debulking surgery (≤ 1 cm, > 1 cm, unknown), and laterality of tumor (right, left, bilateral, unknown). Unadjusted HRs with 95% CIs were also estimated (Supplemental Table 4). The proportional hazards assumption was evaluated using scaled Schoenfeld residuals (29) for covariates and SNPs with p-values < 0.05. We conducted gene-level analyses by testing principal components (30) that explained 90% of SNP variance using multiple degree-of-freedom likelihood ratio tests. Analyses were performed on all MAY1 cases as well as on a subset of 192 cases with serous histology. Due to the exploratory nature of all analyses, no correction for multiple testing was performed.
Analysis of Top Hits using Public Data
For SNPs with p < 0.001, we analyzed publicly-available germline genotype and mortality data on white non-Hispanic invasive serous ovarian cancer cases (TCGA1, N=127, 62 deaths) from TCGA (31). These cases were enrolled at seven sites and genotyped with the Illumina 1MDuo panel; Mayo Clinic TCGA cases were excluded. Call rates ranged from 95.7% to 98.3%. As above, Cox proportional hazards regression assuming an ordinal model was used to assess risk of death associated with genotype at each SNP. Analyses were adjusted for study site, age at diagnosis, stage (II, III, IV, unknown), and grade (grade 2, grade 3 or 4, unknown); combined analyses of MAY1 and TCGA1 were adjusted for study site, age, stage (I, II, III, IV), and grade (grade 1 or 2, grade 3 or 4, unknown). Where results were consistent, we obtained TCGA data on additional SNPs within each gene as well as data on tumor mRNA levels acquired via the Affymetrix Human Genome U133 GeneChip Array (using the probe for most the highly-expressed transcript based on prior reports in normal ovarian tissue (32)). Genotype was correlated with tumor mRNA levels using an ANOVA comparison of means test in the R software program (33), and tumor mRNA levels were examined in relation to survival using Cox proportional hazards models adjusted for study site, age at diagnosis, stage (II, III, IV, unknown), and grade (grade 2, grade 3 or 4, unknown).
HGF Tissue Microarray Expression Analysis
Study Participants and Tissue Blocks
TMAs were created from formalin-fixed paraffin-embedded tumors of 326 Mayo Clinic cases enrolled through January 2009. All participants provided written informed consent for an IRB-approved protocol. We used an automated Beecher Instruments ATA-27 arrayer (Reutlingen, Germany) following gynecologic pathologist review indicating tumor location. Three 0.6 mm cores were removed from each case paraffin block and placed in a recipient paraffin block according to a randomized electronic TMA map. Recipient blocks were sliced into 5 µm sections and mounted on charged slides. Characteristics of cases with and without protein expression and HGF genotype data are shown in Supplemental Table 5.
Antibodies, Immunohistochemistry, and Scoring
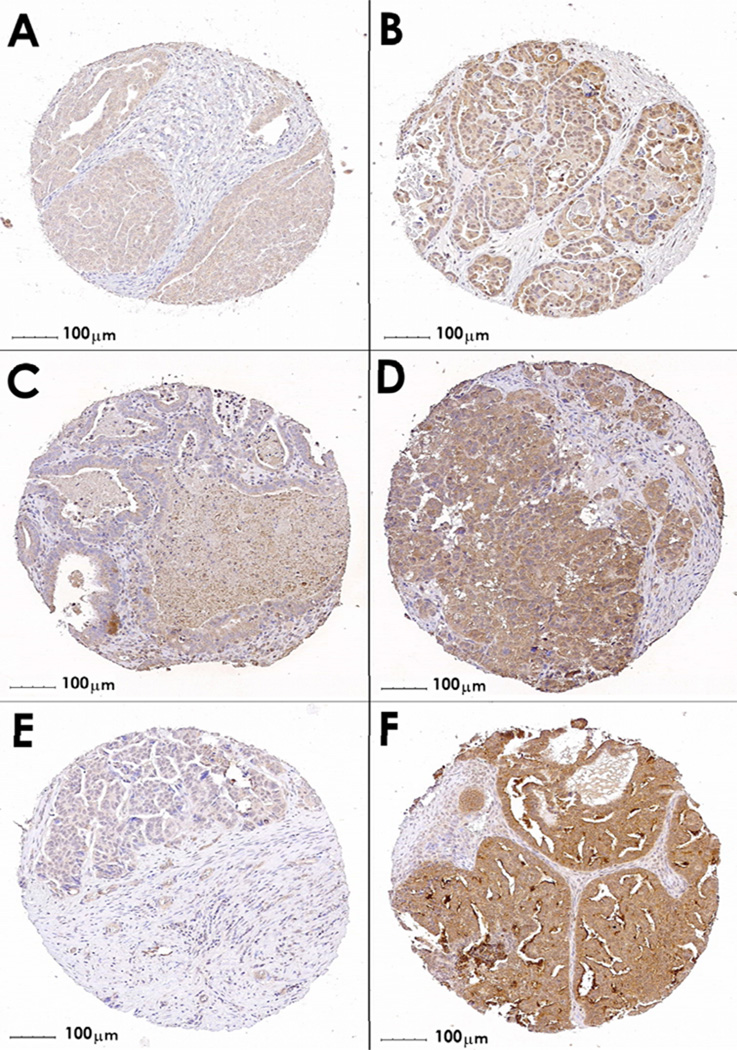
TMA slides were immunostained with primary antibodies recognizing HGF (polyclonal H-145, Santa Cruz; Santa Cruz, CA), MET (monoclonal clone 3D4 Zymed-Invitrogen; Carlsbad, CA), and phospho-MET (polyclonal Y1003) Upstate/Millipore; Charlottesville, VA) after optimizing staining conditions on positive control tissues colorectal cancer and normal liver (34). Negative controls included a non-specific isotype match (for MET) or rabbit IgG (HGF and pMet); conditions were appropriate in that no background staining was observed without the specific primary antibodies. Slides were prepared according to manufacturer’s instructions (Dako; Carpinteria, CA), and digital images were created as described previously (35). Imaging instrument: Bacaus Laboratories Inc. Slide Scanner (BLISS) system, utilizing a Zeiss Axioplan 2 microscope; 20× objective magnification was used during scanning; however, provided as 10× via Figure 1; Tracer Version 0.91 acquisition software was used and no additional techniques done; temperature during scanning was approximately 23C; no oil immersion was used, therefore imaging medium was dry; and no fluorochromes were use. The viewing software used: WebSlide Enterprise, utilizing ActiveX in conjunction with the Slide Tray v5.0 program. The saved format for the scanned pictures is SlideScan.ini, which means that, for these particular core scans, each original image actually is composed of twelve 752×480 pixel jpg scans. Extent was defined as the percentage of epithelial tumor cells staining positive for each antigen (negative, 0%–10% of cells expressing; positive, >10% of cells expressing), and intensity was defined as the strength of antibody staining in epithelial tumor cells (absent, weak, moderate, strong). Slides were scored by two reviewers, and discrepancies were resolved by a gynecologic pathologist. Extent and intensity measures for each core were combined as negative (extent negative), weak (intensity weak; Figure 1), moderate (intensity moderate), and strong (intensity strong; Figure 1), and the strongest protein expression value over the multiple cores for each case was used.
Figure 1. HGF, MET, and phospho-MET staining in epithelial ovarian cancer TMAs.
Immunohistochemical staining of a tissue microarray of epithelial ovarian cancer samples. Representative images are shown of weak (panels A, C, and E) and strong (B, D, and F) staining. Panels A and B show cores stained with antibodies recognizing HGF; Panels C and D represent MET staining, and Panels E and F represent phospho-MET staining.
Statistical Methods
We assessed associations of HGF genotype with TMA-based protein expression values using linear regression analyses (genotype as exposure and expression value as outcome). We examined associations of protein expression values with mortality using Cox proportional hazards regression analyses, accounting for left truncation as described above (27). HRs and 95% CIs were calculated for each antibody, modeling negative or weak protein expression as the referent group, and, as with genotype analyses, models were fit both unadjusted and adjusted for potential confounding variables. We evaluated whether protein expression values played a role in the association between genotype and mortality by fitting five sets of Cox regression models with genotype as exposure: unadjusted, adjusted for HGF, adjusted for MET, adjusted for phospho-MET, and adjusted for all three protein expression values. Attenuation of the genotype HR estimate due to adjustment for protein expression value(s) was considered evidence that genotype may associate with mortality via expression. All statistical tests were two-sided, and all analyses were carried out using SAS (SAS Institute, Cary, NC).
HGF Replication Analysis
Study Participants and SNP Genotyping
White non-Hispanic invasive epithelial ovarian, fallopian tube, and peritoneal cancer cases from additional independent cases collections (SRO, MAL, BEL, LAX, PVD, BAV, and MAY2) were genotyped at HGF rs5745709 and rs2074725 (a highly-correlated SNP) using germline DNA obtained from peripheral blood lymphocytes. As above, protocols were approved by each IRB, and all participants provided written informed consent. Two studies, BAV and BEL, included prevalent cases thus absolute survival was relatively high and median survival could not be estimated; nonetheless, appropriately accounting for the left-truncated nature of the data enabled analysis (Supplemental Figure 3). Genotyping used a custom Sequenom iPLEX MassArray multiplex assay at the Queensland Institute for Medical Research and included at least eight study duplicates per 384-well plate. Quality control was reviewed for each study requiring ≥ 98% concordance, ≥ 90% call rate for each 384-well plate, ≥ 95% call rate overall, and HWE p-value > 0.05. For each questionable quality control metric, cluster plots were reviewed to ensure appropriate genotype calling; SNPs with appropriate genotype calling showed one to three distinct clusters. In addition, new genotype data on additional invasive serous cases enrolled in the TCGA (TCGA2), as described above, became available and was used. Additional detail on these participants (2,560 cases, 1,046 deaths) is provided in Supplemental Table 1 and Supplemental Table 6.
Statistical Methods
Associations of genotypes with mortality were examined using Cox proportional hazards regression as above, accounting for left-truncation (27) using an ordinal (log-additive) genotypic effect. Analyses were conducted adjusted for study site, age at diagnosis, grade, and stage, as well as for study site and age only. Replication data were analyzed separately as well as combined with initial data (MAY1, TCGA1). We examined heterogeneity in genetic associations across study site by fitting and testing genotype*site interaction terms. As above, all analyses were carried out for all cases, and then subset to high-grade serous cases; all statistical tests were two-sided, and all analyses were carried out using SAS (SAS Institute, Cary, NC).
RESULTS
Characteristics of 312 Mayo Clinic invasive epithelial ovarian cancer cases genotyped in a large-scale candidate gene screen (MAY1) are shown in Supplemental Table 1; 172 cases (55%) died during the study period, including 129 cases with serous subtype. Examination of associations between SNP genotypes and mortality among these cases revealed minor alleles at 29 SNPs in ten genes with p < 0.01 among either case group (all cases or serous subtype only) including 21 SNPs with r2 < 0.9 (Table 1). Gene-level analyses for four genes yielded p < 0.01. There was no violation of the Cox proportional hazards assumption for covariates or SNPs. At HGF (encoding hepatocyte growth factor) rs1800793, the minor allele was associated with increased risk of death (all cases: HR 1.69, 95% CI 1.33–2.16, p=2.0 × 10−5), representing the most statistically-significant single SNP result. Additional correlated SNPs (r2 > 0.9, Supplemental Figure 2) showed this association, as did the less-correlated rs2214825 (r2=0.76; all cases: HR 1.44, 95% CI 1.13–1.84, p=3.3 × 10−3). Gene-level analyses using principal components also detected an association of HGF with mortality (all cases, p=3.7 × 10−4) representing the most statistically-significant gene-level test. Analysis of 12 common HGF haplotypes suggested that the individual SNP results were not a reflection of specific haplotypes (data not shown).
Table 1.
SNPs associated with mortality among Mayo Clinic ovarian cancer cases (MAY1)
| All Cases (N=312) | Serous Subtype (N=192) | ||||||
|---|---|---|---|---|---|---|---|
| Gene/Region | Chr | bp (distance)a | rsidb | HR (95% CI)c | p-trend | HR (95% CI)c | p-trend |
| Angiogenesis | |||||||
| RAF1 | 3 | 12,622,547 | rs17771249 | 0.80 (0.63 – 1.00) | 0.05 | 0.68 (0.52 – 0.89) | 5.0 × 10−3 |
| PLG | 6 | 161,092,932 | rs783173 | 1.24 (0.99 – 1.54) | 0.06 | 1.57 (1.21 – 2.04) | 8.1 × 10−4 |
| HGF | 7 | 81,184,621 | rs1800793 | 1.69 (1.33 – 2.16) | 2.0 × 10−5 | 1.63 (1.20 – 2.21) | 1.8 × 10−3 |
| +24,934 | rs2214825 | 1.44 (1.13 – 1.84) | 3.3 × 10−3 | 1.39 (1.03 – 1.88) | 0.03 | ||
| PLAU | 10 | 75,342,967 | rs2227562 | 1.30 (0.98 – 1.72) | 0.07 | 1.62 (1.17 – 2.26) | 4.0 × 10−3 |
| THBS1 | 15 | 37,665,559 | rs753598 | 1.50 (1.07 – 2.10) | 0.02 | 1.73 (1.16 – 2.56) | 6.5 × 10−3 |
| Mitosis | |||||||
| PRKACB | 1 | 84,307,885 | rs12031680 | 1.26 (1.00 – 1.59) | 0.05 1.47 | 1.47 (1.11 – 1.94) | 7.9 × 10−3 |
| +21,226 | rs12405120 | 0.70 (0.55 – 0.89) | 4.2 × 10−3 0.60 | 0.60 (0.45 – 0.81) | 6.7 × 10−4 | ||
| +43,990 | rs1402694 | 1.51 (1.20 – 1.92) | 5.6 × 10−4 | 1.67 (1.26 – 2.21) | 3.2 × 10−4 | ||
| +28,142 | rs12129768 | 1.68 (1.18 – 2.38) | 3.7 × 10−3 | 2.03 (1.32 – 3.13) | 1.3 × 10−3 | ||
| NEK2 | 1 | 209,909,699 | rs697003 | 0.74 (0.59 – 0.93) | 9.0 × 10−3 | 0.74 (0.57 – 0.96) | 0.02 |
| CUL1 | 7 | 148,098,380 | rs3807446 | 0.42 (0.22 – 0.79) | 6.8 × 10−3 | 0.55 (0.27 – 1.11) | 0.10 |
| WEE1 | 11 | 9,541,835 | rs7929469 | 1.64 (1.15 – 2.34) | 6.4 × 10−3 | 1.91 (1.26 – 2.92) | 2.5 × 10−3 |
| KIAA0999 | 11 | 116,227,036 | rs10047459 | 1.50 (1.13 – 1.99) | 5.2 × 10−3 | 1.27 (0.90 – 1.81) | 0.18 |
| +41,208 | rs499910 | 1.53 (1.18 – 1.99) | 1.5 × 10−3 | 1.37 (1.00 – 1.87) | 0.05 | ||
| DCTN5 | 16 | 23,589,772 | rs12447304 | 1.83 (1.29 – 2.60) | 7.1 × 10−4 | 1.71 (1.13 – 2.58) | 0.01 |
| Growth Factors, Stromal Proteins | |||||||
| PIK3R1 | 5 | 67,605,502 | rs171649 | 0.79 (0.62 – 1.00) | 0.05 | 0.61 (0.45 – 0.83) | 1.8 × 10−3 |
| SHMT2 | 12 | 55,909,088 | rs7301155 | 0.82 (0.63 – 1.05) | 0.11 | 0.67 (0.49 – 0.90) | 9.0 × 10−3 |
| ERBB2 | 17 | 35,119,531 | rs1810132 | 0.70 (0.55 – 0.90) | 4.9 × 10−3 | 0.68 (0.51 – 0.92) | 0.01 |
| Expression-Based | |||||||
| CC2D1A | 19 | 13,905,287 | rs2305778 | 1.50 (0.94 – 2.39) | 0.09 | 1.93 (1.17 – 3.18) | 9.6 × 10−3 |
| Regional | |||||||
| 8q24 | 8 | 128,530,789 | rs10094059 | 0.66 (0.50 – 0.87) | 3.7 × 10−3 | 0.72 (0.52 – 1.00) | 0.05 |
bp on genome build 36.3; within genes, distance is represented in bp from previously-listed SNP (+bp).
29 SNPs yielded per-allele p < 0.01 in either case group; only 21 SNPs with r2 < 0.90 are shown.
Hazard ratios (HR) represent the estimated increase (or decrease) in the risk of death for each one-copy increase in the number of minor alleles carried by an individual adjusted for age at diagnosis, pre-surgical log10(CA125), stage (I, II, III, IV, unknown), grade (grade 1 or 2, grade 3, grade 4, unknown), volume of residual tumor following debulking surgery (≤ 1 cm, > 1 cm, unknown), and laterality of tumor (right, left, bilateral, unknown).
Three other genes had SNP p-values < 0.001 including PRKACB (encoding protein kinase, cAMP-dependent, catalytic, beta) with rs1402694 (all cases HR 1.51, 95% CI 1.20–1.92, p=5.6 × 10−4; serous subtype HR 1.67, 95% CI 1.26–2.21, 3.2 × 10−4). Genotypes at DCTN5 (dynactin 5 (p25)) rs12447304 were associated with mortality among all cases (HR 1.83, 95% CI 1.29–2.60, p=7.1 × 10−4), thus we also conducted regional analysis with neighboring gene PLK1 and found that in combination, DCTN5 and PLK1 SNPs were associated with differential survival (all cases p=1.6 × 10−3; serous subtype 4.4 × 10−3). PLG (encoding plasminogen) rs783173 was not associated with survival among all cases, but an association was observed with serous subtype (HR 1.57, 95% CI 1.21–2.04, p=8.1 × 10−4) (Table 1). Additional genes with suggestive SNP associations (p < 0.01) are listed in Table 1, and full gene-level results are provided in Supplemental Table 7.
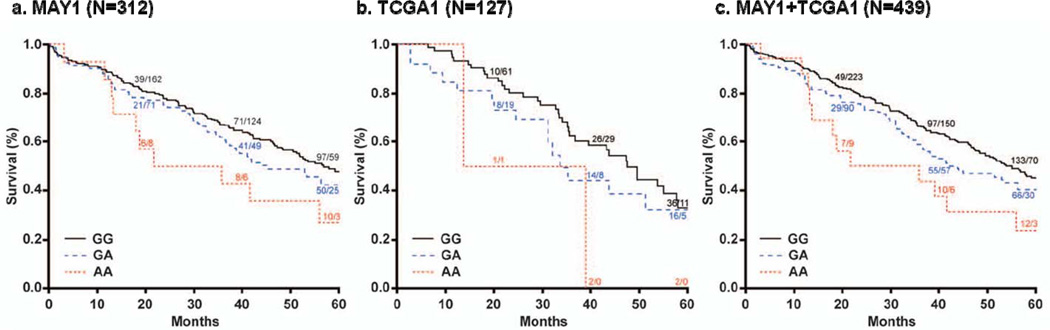
Using publicly-available data on 127 genotyped serous invasive cases of the TCGA (TCGA1, 62 deaths), we analyzed SNPs with p < 0.001 in MAY1 analysis. Results were not consistent at PRKACB rs1402694 (HR=1.13, 95% CI 0.76–1.67, p=0.54) or at PLG rs783173 (HR=0.70, 95% CI 0.48–1.00, p=0.05); no genotypes were available at DCTN5 rs12447304. However, at several HGF SNPs with MAY1 p < 0.001, compelling, consistent associations with mortality were observed including rs5745709 (HR 2.36, 95% CI 1.36–4.09, p=2.2 × 10−3) and the modestly-correlated rs2214825 (r2=0.72; HR 2.27, 95% CI 1.38–3.72, p=1.2 × 10−3; Table 2). Combining these Mayo Clinic and TCGA data (MAY1+TCGA1, N=439), the strongest association with mortality was observed for rs5745709 (HR 1.56, 95% CI 1.25–1.94, p=7.0 × 10−5, Table 2, Figure 2); this SNP is highly correlated with the initially most-significant SNP in MAY1, rs1800793 (r2=0.96; Supplemental Figure 2), which was not genotyped in TCGA. Similar results were observed when analyses were performed only on cases with advanced-stage disease (data not shown). To evaluate a possible mechanism for this association, we then explored HGF ovarian tumor mRNA levels in TCGA1 cases. Carriers of minor alleles at HGF SNPs had reduced HGF mRNA levels (e.g., rs2214825 p=0.03; Supplemental Figure 4). Although sample size was small and HGF mRNA levels themselves were not directly associated with mortality (HR=0.39, 95% CI 0.02–3.79, p=0.52), these results suggested further tumor and germline analysis of HGF in ovarian cancer mortality.
Table 2.
HGF SNPs and mortality in initial analyses
| MAY1 (N=312) | TCGA1 (N=127) | MAY1+TCGA1 (N=439) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| rsid | r2 to next | alleles | MAF | HR (95% CI) | p-trend | MAF | HR (95% CI) | p-trend | HR (95% CI) | p |
| rs2074725 | 0.96 | C>A | 0.20 | 1.59 (1.24 – 2.04) | 2.1 × 10−4 | 0.17 | 1.63 (1.05 – 2.55) | 0.03 | 1.47 (1.19 – 1.81) | 2.9 × 10−4 |
| rs1800793 | 0.97 | G>A | 0.20 | 1.69 (1.33 – 2.16) | 2.0 × 10−5 | --- | --- | --- | --- | --- |
| rs5745720 | 0.99 | A>G | 0.20 | 1.63 (1.28 – 2.08) | 9.0 × 10−5 | 0.17 | 1.87 (1.11 – 3.15) | 0.02 | 1.52 (1.23 – 1.88) | 1.3 × 10−4 |
| rs5745709 | 0.93 | G>A | 0.19 | 1.66 (1.29 – 2.13) | 9.0 × 10−5 | 0.17 | 2.36 (1.36 – 4.09) | 2.2 × 10−3 | 1.56 (1.25 – 1.94) | 7.0 × 10−5 |
| rs2887069 | 0.01 | A>G | 0.19 | 1.59 (1.24 – 2.03) | 2.1 × 10−4 | --- | --- | --- | --- | --- |
| rs5745687 | 0.01 | G>A | 0.06 | 0.78 (0.48 – 1.25) | 0.3 | --- | --- | --- | --- | --- |
| rs2214825 | --- | G>A | 0.22 | 1.44 (1.13 – 1.84) | 3.3 × 10−3 | 0.20 | 2.27 (1.38 – 3.72) | 1.2 × 10−3 | 1.44 (1.17 – 1.78) | 6.3 × 10−4 |
r2 based on LD in MAY1 cases, the first five SNPs listed were in strong LD (r2 > 0.92) and in modest LD with rs2214825 (r2 > 0.76), rs5745687 was independent (r2=0.01); alleles column indicates major>minor allele; MAY1 analyses adjusted for age at diagnosis, pre-surgical log10(CA125), stage (I, II, III, IV, unknown), grade (grade 1 or 2, grade 3, grade 4, unknown), volume of residual tumor following debulking surgery (≤ 1 cm, > 1 cm, unknown), and laterality of tumor (right, left, bilateral, unknown); TCGA1 analyses adjusted for age at diagnosis, grade (grade2, grade 3, unknown), stage (II, III, IV, unknown), and study site; combined analysis adjusted for study site, age at diagnosis, grade (grade 1 or 2, grade 3 or 4, unknown ), and stage (I, II, III, IV, unknown); hazard ratios (HR) represent the estimated increase (or decrease) in the risk of death for each one-copy increase in the number of minor alleles carried by an individual; sorted by position on genome build 36.3.
Figure 2. Kaplan-Meier ovarian cancer survival curves by HGF rs5745709 genotype, initial Mayo Clinic and TCGA analysis.
Genotype-specific Kaplan-Meier survival curves based on (a) MAY1, (b) TCGA1, and (c) MAY1+TCGA1. Numbers superimposed on curves represent genotype-specific cumulative number of deaths/number remaining at risk
We therefore performed immunohistochemical analysis of HGF, MET, and phospho-MET on TMAs created from 326 Mayo Clinic cases. We examined these three antigens because HGF binds to the transmembrane tyrosine kinase receptor MET, which, in turn, results in conversion of MET to its activated form, phospho-MET. This signaling initiates the ERK1/2, PI3K/AKT, and p38 mitogen-activated protein kinase (MAPK) cascades resulting in regulation of cell proliferation, apoptosis, migration, and invasive growth in normal ovarian surface epithelium and in ovarian cancers (36). Representative immunostained cores are provided in Figure 1. The distribution of protein expression values is provided in Table 3; as expected, there were no cases with activated phospho-MET in the absence of MET (data not shown). We assessed whether protein expression values were associated with mortality and found a suggestion that stronger phospho-MET protein expression correlated with decreased mortality (p=0.01; moderate v negative/weak HR 0.77, 95% CI 0.39–1.52; strong v negative/weak HR 0.55, 95% CI 0.27–1.10; Table 3). As known biology predicts that stronger protein expression of phospho-MET would correlate with increased (rather than decreased) mortality, we examined additional clinical characteristics and observed stronger protein expression of phospho-MET among cases with early-stage disease (p=0.003); inclusion of stage in phospho-MET Cox models attenuated the HR. Thus, the association between phospho-MET and decreased mortality is likely driven by an inverse association between phospho-MET and stage. In 255 cases with genotypes (MAY1 or MAY2), we evaluated whether rs5745709 genotype was associated with HGF, MET, and phospho-MET protein expression values, and no significant associations were observed (p ≥ 0.41 Table 3), although trends were consistent with TCGA1 mRNA data. Results were similar for other HGF SNPs (data not shown). We then evaluated relationships among HGF rs5745709 genotype, HGF/MET/phospho-MET protein expression, and mortality; inclusion of protein expression values, alone or in combination, did not at all attenuate the association between genotype and mortality (data not shown); results were similar considering time to recurrence. These results, therefore, suggest that the observed genetic association in the Mayo Clinic dataset is not a result of modified gene expression.
Table 3.
Immunostaining of Mayo Clinic cases
| Antibody | ||||
|---|---|---|---|---|
| HGF | MET | Phospho-MET | ||
| Protein expression values, N (%) | Negative (0) | 1 (0.3%) | 1 (0.3%) | 2 (0.6%) |
| Weak (1) | 52 (16.1%) | 6 (1.9%) | 13 (4.0%) | |
| Moderate (2) | 209 (64.7%) | 124 (38.9%) | 165 (51.4%) | |
| Strong (3) | 61 (18.6%) | 188 (58.9%) | 141 (43.9%) | |
| Association of protein expression with mortality, HR (95%CI) | Negative/Weak | Ref. | Ref. | Ref. |
| Moderate | 1.26 (0.83– 1.91) | 1.44 (0.45 – 4.56) | 0.77 (0.39 – 1.52) | |
| Strong | 1.21 (0.73 – 2.01) | 1.35 (0.43 – 4.25) | 0.55 (0.27 – 1.10) | |
| p-value | 0.48 | 0.90 | 0.01 | |
| Protein expression value by HGF rs5745709 genotype, mean (S.D.) | GG | 2.01 (0.60) | 2.57 (0.53) | 2.35 (0.58) |
| GA | 2.01 (0.60) | 2.56 (0.55) | 2.42 (0.60) | |
| AA | 1.92 (0.49) | 2.55 (0.52) | 2.42 (0.51) | |
| p-value | 0.80 | 0.89 | 0.41 | |
Due to core drop-off, data were unavailable for three cases on HGF, seven on MET, and five on phospho-MET.
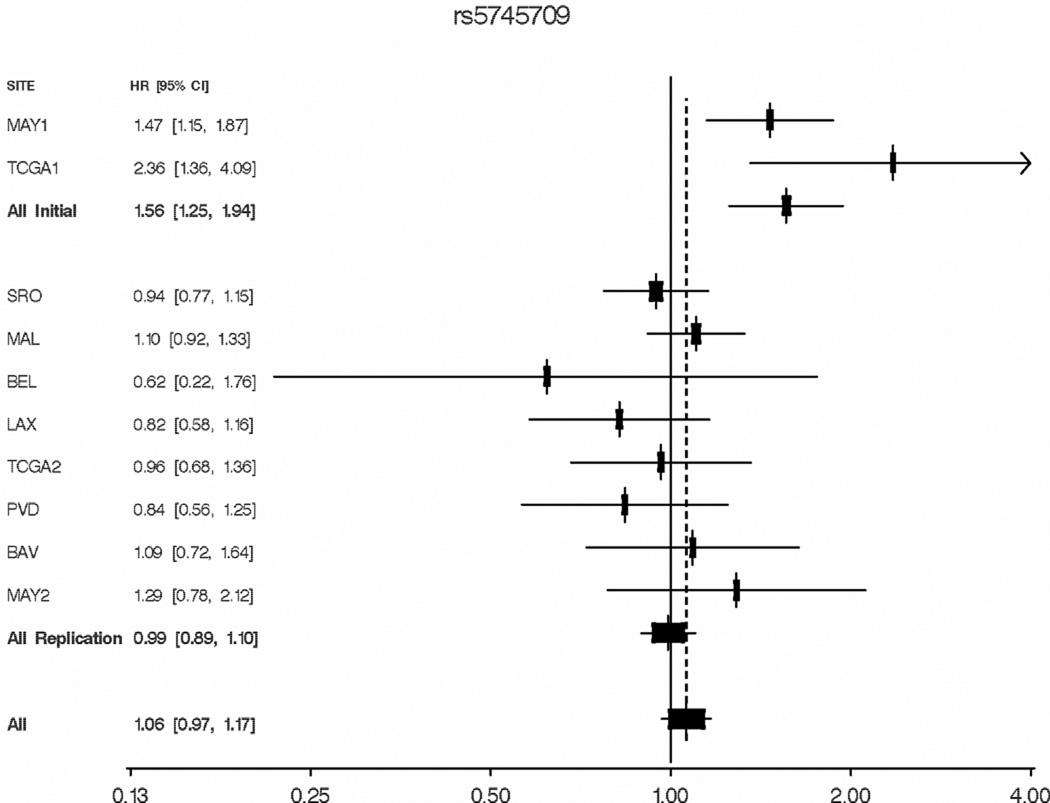
Finally, we characterized HGF SNP associations with mortality in additional ovarian cancer cases populations, including six new studies as well as new cases enrolled at the Mayo Clinic and in the TCGA (Supplemental Table 1, Supplemental Table 6). rs5745709 was successfully genotyped in each population, and MAFs were similar to that expected (range, 0.17–0.25); 284 MAY1 cases were genotyped in both laboratories and were 99.6% concordant (duplicates subsequently excluded). Analyses of the replication set adjusted for site, age, grade, and stage (as done in MAY1+TCGA1 analysis) revealed no association with ovarian cancer survival (HR 0.99, 95% CI 0.89–1.10, p=0.87; Table 4). Figure 3 displays study-specific covariate-adjusted HRs for all cases and reveals the borderline-significant heterogeneity between initial and replication studies (p=0.06). A large number of sensitivity analyses were conducted to examine the root of such heterogeneity. Results were similar when adjusted only for site and age and when restricted to high-grade serous cases; results were also similar at rs2074725 due to high LD (r2= 0.97). Data on time to recurrence were available on some studies (MAY1, SRO, MAL, MAY2), and results were similar. When cases with peritoneal or fallopian tube cancer were excluded and when cases were right-censored at five years, results were also similar. Although we note that other SNP effects appear to exhibit themselves only in the context of optimal debulking (37) and that both Mayo Clinic series included a relatively large proportion of optimally-debulked patients (78% and 87% in MAY1 and MAY2, respectively), analysis of 1,139 optimally-debulked replication cases (BAV, BEL, LAX, MAL, MAY2, SRO) also yielded null results (HR 0.95, 95% CI 0.78–1.15, p=0.60). Finally, no compelling common feature of study populations with risk estimates greater than 1.0 (MAY1, TCGA1, MAL, BAV, and MAY2) could be identified. Thus, we conclude that the initial Mayo Clinic/TCGA HGF association a due to the winner’s curse phenomenon or unexplained heterogeneity.
Table 4.
HGF rs5745709 and ovarian cancer mortality
| Adjusted for site, age, grade, stage | Adjusted for site and age only | |||||||
|---|---|---|---|---|---|---|---|---|
| Histology | Group | N Cases (N Deaths) | HR (95% CI) | p-trend | p-het | HR (95% CI) | p-trend | p-het |
| All Histologies | Initial | 435 (234) | 1.56 (1.25–1.94) | 6.9 × 10−5 | 0.83 | 1.45 (1.16–1.81) | 1.1 × 10−3 | 0.82 |
| Replication | 2,560 (1,046) | 0.99 (0.89–1.10) | 0.87 | 0.63 | 1.01 (0.91–1.12) | 0.92 | 0.97 | |
| Combined | 2,995 (1,280) | 1.06 (0.97–1.17) | 0.21 | 0.06 | 1.07 (0.97–1.17) | 0.17 | 0.38 | |
| High-Grade Serous | Initial | 296 (179) | 1.60 (1.23–2.09) | 5.4 × 10−4 | 0.90 | 1.62 (1.25–2.11) | 3.0 × 10−4 | 0.95 |
| Replication | 1,061 (422) | 0.99 (0.84–1.17) | 0.91 | 0.70 | 1.01 (0.86–1.20) | 0.89 | 0.62 | |
| Combined | 1,357 (601) | 1.13 (0.98–1.30) | 0.10 | 0.25 | 1.14 (0.99–1.31) | 0.07 | 0.22 | |
Initial includes MAY1, TCGA1; Replication includes SRO, MAL, BEL, LAX, TCGA2, PVD, BAV, and MAY2.
Figure 3. rs5745709 and ovarian cancer mortality, all studies.
Hazard ratios and 95% confidence intervals by study site, study phase (initial vs. replication) and overall, from Cox proportional hazards regression analysis. Site-specific analyses adjust for age at diagnosis, tumor stage, and tumor grade. Combined initial, replication and overall analyses adjust additionally for study site.
DISCUSSION
Although less productive than hoped for, the study of candidate genes in cancer epidemiology has yielded a handful of replicated consistent associations (e.g., TERT, CASP8, and NAT1 in ovarian (38), breast (39), and bladder cancers respectively (40)), even in the GWAS era. In ovarian cancer, candidate gene survival analyses have suggested angiogenesis, inflammation, and other pathways as drivers of genetically-determined variation in outcome (5–7). Here, we examined approximately 170 genes and regions and found evidence for association between variants in HGF and mortality among cases enrolled in Mayo Clinic and TCGA studies. With consistency of results and evidence suggesting expression as a mechanism, we then stained ovarian tumors for the primary signaling molecules, and we expanded our association analysis. In these important follow-up studies, we did not observe a clear relationship between SNP genotype, expression of the relevant proteins, and outcome; we also did not replicate the genetic association in a broader collection of samples.
In many ways, this work exemplifies the challenges of modern molecular epidemiologic investigation. A biologically-based hypothesis was comprehensively examined with regard to both the number of genes and the coverage of inherited variants within them. We capitalized initially upon a homogeneous patient population with detailed clinical data with 80% power to detect a HR as small as 1.46, assuming MAF=0.20, α= 0.05, and dominance. Prior to committing additional expenditure, we conducted in silico analysis of the top hits using publicly-available data including exploration of a possible intermediate phenotype (41). In addition to the statistical significance of the combined association, the biology of the most-significant gene was compelling (as is always the case in candidate gene studies). Here, HGF signaling plays a key role in ovarian cancer cell growth, migration, and invasion (36), and tumor expression has been shown to associate with outcome (36, 42). Some studies have reported improved chemoresponsiveness (43–45), consistent with the associations we observed between minor alleles, reduced expression, and increased mortality.
To follow-up in terms of depth, we created and immunostained TMAs from a large group Mayo Clinic cases, most of whom had genotype and outcome data, for protein expression of HGF, MET, and phospho-MET. Cores were randomized with respect to both genotype and survival, removing a source of potential bias in TMA analysis. In contrast to the initial microarray-based TCGA mRNA analyses, no genotype-expression association was seen. We must note that there are complexities in measuring HGF with immunohistochemistry, particularly because it is a soluble ligand; positive staining for the protein may reflect HGF generated at a distant source which bound to the tumor cells only in situ. Random measurement error of HGF protein levels could have attenuated the HRs. In contrast, with mRNA expression studies, mRNA is clearly isolated from the relevant tissue source (tumor or tumor stroma) used in the study. Our inclusion of MET and phospho-MET in some ways addresses these concerns about cellular specificity and also revealed null results. We also note that while genetic variation in HGF does not appear to account for dramatic changes in protein expression, other functional roles for HGF SNPs can not be ruled out as alterations in binding affinity, efficacy of signal transduction, or turnover rate are most likely not detectable using this methodology. We also analyzed protein expression among Mayo Clinic cases restricted to high-grade serous (similar to TCGA eligibility), and results remained null. In construction of TMAs, preferential sampling of cores from the periphery of ovarian tumors has been recommended due to loss of expression centrally (46). Because we did not do this, it is possible that our TMA-based mean protein expression levels (and variance estimates) are lower than they are in truth, perhaps resulting in some loss of statistical power. However, our distribution of protein expression values were similar to those reported in other studies (47, 48).
In terms of breadth, we expanded our initial study of the HGF SNP association with ovarian cancer mortality via an international consortium. Use of consortia has become standard in SNP-oriented molecular epidemiology in order to minimize the false discoveries of smaller sample sizes, to increase precision of true risk allele effect estimates, and to evaluate generalizability (49, 50). Indeed, the large sample size of our replication set provided 80% power to detect relatively small effect sizes of 1.21 in the replication set and 1.19 in the combined set of participants (assuming MAF=0.20, α= 0.05, and dominance), minimizing the possibility of a false negative result. Genotyping was centrally done and included a subset of the cases initially studied; the quality of the data appeared good suggesting laboratory issues were not a concern. Further challenges lie in aligning clinical data across studies. We used a consistent data dictionary with thorough examination of outlying observations and differences in eligibility or follow-up across studies; in addition, sensitivity analyses considering clinical and study design factors yielded consistent results. Factors such as covariate adjustment, restriction to certain clinical subsets, and consideration of time to enrollment or follow-up did not explain the discrepancy of results between initial and replication analyses. Unlike the two Mayo Clinic case collections, results differed between TCGA datasets; however, no systematic differences in clinical or follow-up features appear to exist across TCGA sets. We note that if data on all TCGA cases had been available at the time of our initial investigation, our follow-up strategy would likely have differed. Thus, this work serves as a cautionary reminder of the fluidity of public data. In addition, because the majority of cases were treated with standard chemotherapies, the likelihood is low that a true association exists only in a certain treatment group. Although an unanalyzed factor may account for differences in association across studies, we observed that the majority of data are null and thus variation in risk estimates around unity is likely random.
In summary, we report on a comprehensive analysis of ovarian cancer mortality and SNPs in several genes and regions of interest to cancer biology, and we described a two-pronged approach to follow-up the most promising result with TMA and collaborative studies. Although, in Mayo Clinic cases, HGF SNPs appear associated with increased mortality, and phospho-MET protein expression was associated with early stage disease and reduced mortality, common genetic variation in HGF is unlikely to account for a significant proportion of deaths in ovarian cancer. This body of work is characteristic of the state-of-the-art in molecular epidemiology and demonstrates the importance of incorporating multiple data types and study populations in interpretation of promising genetic associations.
Supplementary Material
ACKNOWLEDGMENTS
We thank Karin Goodman and Melanie Kasner for recruitment and abstraction. The results published here are in whole or part based upon data generated by The Cancer Genome Atlas pilot project established by the NCI and NHGRI. Information about TCGA and the investigators and institutions who constitute the TCGA research network can be found via the following reference (31). The Australian Ovarian Cancer Study (AOCS) Management Group (D. Bowtell, G. Chenevix-Trench, A. deFazio, D. Gertig, A. Green and P.M. Webb) gratefully acknowledges the contribution of all the clinical and scientific collaborators (see http://www.aocstudy.org/). The Australian Cancer Study Management Group (A. Green, P. Parsons, N. Hayward, P.M. Webb, and D. Whiteman) thank all of the project staff, collaborating institutions and study participants.
Financial Support: This work was supported by the National Cancer Institute (R01-CA122443, R01-CA86888, P50 CA136393) and the Fred C. and Katherine B. Andersen Foundation. GC-T is supported by the NHMRC of Australia. We are grateful to the family and friends of Kathryn Sladek Smith for their generous support of Ovarian Cancer Association Consortium through their donations to the Ovarian Cancer Research Fund.
Footnotes
Disclaimers: None
REFERENCES
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Hoskins WJ, Bundy BN, Thigpen JT, Omura GA. The influence of cytoreductive surgery on recurrence-free interval and survival in small-volume stage III epithelial ovarian cancer: a Gynecologic Oncology Group study. Gynecol Oncol. 1992;47:159–166. doi: 10.1016/0090-8258(92)90100-w. [DOI] [PubMed] [Google Scholar]
- 3.McGuire V, Jesser CA, Whittemore AS. Survival among U.S. women with invasive epithelial ovarian cancer. Gynecol Oncol. 2002;84:399–403. doi: 10.1006/gyno.2001.6536. [DOI] [PubMed] [Google Scholar]
- 4.Tan DS, Rothermundt C, Thomas K, Bancroft E, Eeles R, Shanley S, et al. "BRCAness" syndrome in ovarian cancer: a case-control study describing the clinical features and outcome of patients with epithelial ovarian cancer associated with BRCA1 and BRCA2 mutations. J Clin Oncol. 2008;26:5530–5536. doi: 10.1200/JCO.2008.16.1703. [DOI] [PubMed] [Google Scholar]
- 5.Goode EL, Maurer MJ, Sellers TA, Phelan CM, Kalli KR, Fridley BL, et al. Inherited determinants of ovarian cancer survival. Clin Cancer Res. 2010;16:995–1007. doi: 10.1158/1078-0432.CCR-09-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quaye L, Gayther SA, Ramus SJ, Di Cioccio RA, McGuire V, Hogdall E, et al. The effects of common genetic variants in oncogenes on ovarian cancer survival. Clin Cancer Res. 2008;14:5833–5839. doi: 10.1158/1078-0432.CCR-08-0819. [DOI] [PubMed] [Google Scholar]
- 7.Schultheis AM, Lurje G, Rhodes KE, Zhang W, Yang D, Garcia AA, et al. Polymorphisms and clinical outcome in recurrent ovarian cancer treated with cyclophosphamide and bevacizumab. Clin Cancer Res. 2008;14:7554–7563. doi: 10.1158/1078-0432.CCR-08-0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolton KL, Tyrer J, Song H, Ramus SJ, Notaridou M, Jones C, et al. Common variants at 19p13 are associated with susceptibility to ovarian cancer. Nat Genet. 2010;42:880–884. doi: 10.1038/ng.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendiola M, Barriuso J, Redondo A, Marino-Enriquez A, Madero R, Espinosa E, et al. Angiogenesis-related gene expression profile with independent prognostic value in advanced ovarian carcinoma. PLoS One. 2008;3:e4051. doi: 10.1371/journal.pone.0004051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nimeiri HS, Oza AM, Morgan RJ, Friberg G, Kasza K, Faoro L, et al. Efficacy and safety of bevacizumab plus erlotinib for patients with recurrent ovarian, primary peritoneal, and fallopian tube cancer: a trial of the Chicago, PMH, and California Phase II Consortia. Gynecol Oncol. 2008;110:49–55. doi: 10.1016/j.ygyno.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Fredericksen ZS, Vierkant RA, Kosel ML, Pankratz VS, Cerhan JR, et al. Association of genetic variation in mitotic kinases with breast cancer risk. Breast Cancer Res Treat. 2010;119:453–462. doi: 10.1007/s10549-009-0404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Psyrri A, Kassar M, Yu Z, Bamias A, Weinberger PM, Markakis S, et al. Effect of epidermal growth factor receptor expression level on survival in patients with epithelial ovarian cancer. Clin Cancer Res. 2005;11:8637–8643. doi: 10.1158/1078-0432.CCR-05-1436. [DOI] [PubMed] [Google Scholar]
- 13.Skirnisdottir I, Seidal T, Sorbe B. A new prognostic model comprising p53, EGFR, and tumor grade in early stage epithelial ovarian carcinoma and avoiding the problem of inaccurate surgical staging. Int J Gynecol Cancer. 2004;14:259–270. doi: 10.1111/j.1048-891X.2004.014209.x. [DOI] [PubMed] [Google Scholar]
- 14.Hartmann LC, Lu KH, Linette GP, Cliby WA, Kalli KR, Gershenson D, et al. Gene expression profiles predict early relapse in ovarian cancer after platinum-paclitaxel chemotherapy. Clin Cancer Res. 2005;11:2149–2155. doi: 10.1158/1078-0432.CCR-04-1673. [DOI] [PubMed] [Google Scholar]
- 15.Poynter JN, Figueiredo JC, Conti DV, Kennedy K, Gallinger S, Siegmund KD, et al. Variants on 9p24 and 8q24 are associated with risk of colorectal cancer: results from the Colon Cancer Family Registry. Cancer Res. 2007;67:11128–11132. doi: 10.1158/0008-5472.CAN-07-3239. [DOI] [PubMed] [Google Scholar]
- 16.Witte JS. Multiple prostate cancer risk variants on 8q24. Nat Genet. 2007;39:579–580. doi: 10.1038/ng0507-579. [DOI] [PubMed] [Google Scholar]
- 17.Ghoussaini M, Song H, Koessler T, Al Olama AA, Kote-Jarai Z, Driver KE, et al. Multiple loci with different cancer specificities within the 8q24 gene desert. J Natl Cancer Inst. 2008;100:962–966. doi: 10.1093/jnci/djn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 19.Peedicayil A, Vierkant RA, Hartmann LC, Fridley BL, Fredericksen ZS, White KL, et al. Risk of ovarian cancer and inherited variants in relapse-associated genes. PLoS One. 2010;5:e8884. doi: 10.1371/journal.pone.0008884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White K, Sellers TA, Fridley BL, Vierkant RA, Phelan CM, Schildkraut JM, et al. Variation at 8q24 and 9p24 and risk of ovarian cancer. Twin Res and Hum Genet. 2010;13:43–56. doi: 10.1375/twin.13.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The International HapMap Consortium. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelemen LE, Sellers TA, Schildkraut JM, Cunningham JM, Vierkant RA, Pankratz VS, et al. Genetic variation in the one-carbon transfer pathway and ovarian cancer risk. Cancer Res. 2008;68:2498–2450. doi: 10.1158/0008-5472.CAN-07-5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliphant A, Barker DL, Stuelpnagel JR, Chee MS. BeadArray technology: enabling an accurate, cost-effective approach to high-throughput genotyping. Biotechniques. 2002 Suppl:56–58. 60–61. [PubMed] [Google Scholar]
- 25.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 26.Cox DR. Regression models and life tables (with discussion) Journal of Royal Statistical Society, Series B. 1972;34:187–220. [Google Scholar]
- 27.Therneau TM, Grambsch PM, Fleming TR. Martingale-based residuals for survival models. Biometrika. 1990;77:147–160. [Google Scholar]
- 28.Freidlin B, Zheng G, Li Z, Gastwirth JL. Trend tests for case-control studies of genetic markers: power, sample size and robustness. Hum Hered. 2002;53:146–152. doi: 10.1159/000064976. [DOI] [PubMed] [Google Scholar]
- 29.Grambsch P, Louis TA, Bostick RM, Grandits GA, Fosdick L, Darif M, et al. Statistical analysis of proliferative index data in clinical trials. Stat Med. 1994;13:1619–1634. doi: 10.1002/sim.4780131603. [DOI] [PubMed] [Google Scholar]
- 30.Gauderman WJ, Murcray C, Gilliland F, Conti DV. Testing association between disease and multiple SNPs in a candidate gene. Genet Epidemiol. 2007;31:383–395. doi: 10.1002/gepi.20219. [DOI] [PubMed] [Google Scholar]
- 31.The Cancer Genome Atlas (TCGA) 2005 [cited; Available from: http://cancergenome.nih.gov.
- 32.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.R Foundation for Statistical Computing. R: A Language and Environment for Statistical Computing. Vienna, Austria: 2009. [Google Scholar]
- 34.Ma PC, Tretiakova MS, MacKinnon AC, Ramnath N, Johnson C, Dietrich S, et al. Expression and mutational analysis of MET in human solid cancers. Genes Chromosomes Cancer. 2008;47:1025–1037. doi: 10.1002/gcc.20604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kelemen LE, Sellers TA, Keeney GL, Lingle WL. Multivitamin and alcohol intake and folate receptor alpha expression in ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:2168–2172. doi: 10.1158/1055-9965.EPI-05-0260. [DOI] [PubMed] [Google Scholar]
- 36.Zhou HY, Pon YL, Wong AS. HGF/MET signaling in ovarian cancer. Curr Mol Med. 2008;8:469–480. doi: 10.2174/156652408785747933. [DOI] [PubMed] [Google Scholar]
- 37.Johnatty SE, Beesley J, Paul J, Fereday S, Spurdle AB, Webb PM, et al. ABCB1 (MDR 1) polymorphisms and progression-free survival among women with ovarian cancer following paclitaxel/carboplatin chemotherapy. Clin Cancer Res. 2008;14:5594–5601. doi: 10.1158/1078-0432.CCR-08-0606. [DOI] [PubMed] [Google Scholar]
- 38.Johnatty SE, Beesley J, Chen X, Macgregor S, Duffy DL, Spurdle AB, et al. Evaluation of candidate stromal epithelial cross-talk genes identifies association between risk of serous ovarian cancer and TERT, a cancer susceptibility "hot-spot". PLoS Genet. 2010;6 doi: 10.1371/journal.pgen.1001016. e1001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cox A, Dunning AM, Garcia-Closas M, Balasubramanian S, Reed MW, Pooley KA, et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat Genet. 2007;39:352–358. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 40.Garcia-Closas M, Malats N, Silverman D, Dosemeci M, Kogevinas M, Hein DW, et al. NAT2 slow acetylation, GSTM1 null genotype, and risk of bladder cancer: results from the Spanish Bladder Cancer Study and meta-analyses. Lancet. 2005;366:649–659. doi: 10.1016/S0140-6736(05)67137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carvajal-Carmona LG. Genetic dissection of intermediate phenotypes as a way to discover novel cancer susceptibility alleles. Curr Opin Genet Dev. 2010;20:308–314. doi: 10.1016/j.gde.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 42.Sawada K, Radjabi AR, Shinomiya N, Kistner E, Kenny H, Becker AR, et al. c-Met overexpression is a prognostic factor in ovarian cancer and an effective target for inhibition of peritoneal dissemination and invasion. Cancer Res. 2007;67:1670–1679. doi: 10.1158/0008-5472.CAN-06-1147. [DOI] [PubMed] [Google Scholar]
- 43.Rasola A, Anguissola S, Ferrero N, Gramaglia D, Maffe A, Maggiora P, et al. Hepatocyte growth factor sensitizes human ovarian carcinoma cell lines to paclitaxel and cisplatin. Cancer Res. 2004;64:1744–1750. doi: 10.1158/0008-5472.can-03-2383. [DOI] [PubMed] [Google Scholar]
- 44.Olivero M, Ruggiero T, Saviozzi S, Rasola A, Coltella N, Crispi S, et al. Genes regulated by hepatocyte growth factor as targets to sensitize ovarian cancer cells to cisplatin. Mol Cancer Ther. 2006;5:1126–1135. doi: 10.1158/1535-7163.MCT-06-0013. [DOI] [PubMed] [Google Scholar]
- 45.Coltella N, Rasola A, Nano E, Bardella C, Fassetta M, Filigheddu N, et al. p38 MAPK turns hepatocyte growth factor to a death signal that commits ovarian cancer cells to chemotherapy-induced apoptosis. Int J Cancer. 2006;118:2981–2990. doi: 10.1002/ijc.21766. [DOI] [PubMed] [Google Scholar]
- 46.Permuth-Wey J, Boulware D, Valkov N, Livingston S, Nicosia S, Lee JH, et al. Sampling strategies for tissue microarrays to evaluate biomarkers in ovarian cancer. Cancer Epidemiol Biomarkers Prev. 2009;18:28–34. doi: 10.1158/1055-9965.EPI-08-0713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tothill RW, Tinker AV, George J, Brown R, Fox SB, Lade S, et al. Novel molecular subtypes of serous and endometrioid ovarian cancer linked to clinical outcome. Clin Cancer Res. 2008;14:5198–5208. doi: 10.1158/1078-0432.CCR-08-0196. [DOI] [PubMed] [Google Scholar]
- 48.Ayhan A, Ertunc D, Tok EC. Expression of the c-Met in advanced epithelial ovarian cancer and its prognostic significance. Int J Gynecol Cancer. 2005;15:618–623. doi: 10.1111/j.1525-1438.2005.00117.x. [DOI] [PubMed] [Google Scholar]
- 49.Fasching PA, Gayther S, Pearce L, Schildkraut JM, Goode E, Thiel F, et al. Role of genetic polymorphisms and ovarian cancer susceptibility. Molecular Oncology. 2009;3:171–181. doi: 10.1016/j.molonc.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boffetta P, Colditz GA, Potter JD, Kolonel L, Robson PJ, Malekzadeh R, et al. Cohorts and consortia conference: a summary report (Banff, Canada, June 17–19, 2009) Cancer Causes Control. 2011 doi: 10.1007/s10552-010-9717-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.