Abstract
To investigate how a protein kinase interacts with its protein substrate during extended, multi-site phosphorylation, the kinetic mechanism of a protein kinase involved in mRNA splicing control was investigated using rapid quench flow techniques. The protein kinase SRPK1 phosphorylates approximately 10 serines in the arginine-serine-rich domain (RS domain) of the SR protein SRSF1 in a C-to-N-terminal direction, a modification that directs this essential splicing factor from the cytoplasm to the nucleus. Transient-state kinetic experiments illustrate that the first phosphate is added rapidly onto the RS domain of SRSF1 (t1/2 = 0.1 sec) followed by slower, multi-site phosphorylation at the remaining serines (t1/2 = 15 sec). Mutagenesis experiments suggest that efficient phosphorylation rates are maintained by an extensive hydrogen bonding and electrostatic network between the RS domain of the SR protein and the active site and docking groove of the kinase. Catalytic trapping and viscosometric experiments demonstrate that while the phosphoryl transfer step is fast, ADP release limits multi-site phosphorylation. By studying phosphate incorporation into selectively pre-phosphorylated forms of the enzyme-substrate complex, the kinetic mechanism for site-specific phosphorylation along the reaction coordinate was assessed. The binding affinity of the SR protein, the phosphoryl transfer rate and ADP exchange rate were found to decline significantly as a function of progressive phosphorylation in the RS domain. These findings indicate that the protein substrate actively modulates initiation, extension and termination events associated with prolonged, multi-site phosphorylation.
Splicing is a critical posttranscriptional modification that occurs at a macromolecular complex (spliceosome) composed of several RNAs and numerous polypeptides (1). Among the latter group, the SR proteins have emerged as essential splicing factors that control both early and late events in spliceosome development (2). SR proteins are composed of one or two N-terminal RNA recognition motifs (RRMs)1 that bind exonic splicing enhancer sequences and help establish the 5′ and 3′ splice sites in precursor mRNA. SR proteins derive their name from a C-terminal arginine-serine-rich (RS) domain that varies in length from 50 to over 300 residues. The RS domains contain numerous arginine-serine dipeptide repeats that are extensively phosphorylated in vivo by the SRPK family of serine protein kinases (3). The SRPKs possess a traditional kinase domain that does not require activation loop phosphorylation and whose conserved ATP and substrate binding lobes are bifurcated by a large spacer insert domain (250 aa). This insert domain is intrinsically disordered and interacts with co-chaperones maintaining cytoplasmic localization of the enzyme (4, 5). SR protein phosphorylation is important for recruitment of spliceosomal components (e.g., U1, U2AF) at the exon-intron boundaries and dephosphorylation generates a catalytically active spliceosome (6-8). While several studies suggest that the phosphorylation state of the RS domains may play a role in alternative splicing of some genes (9), phosphorylation is best known to regulate subcellular localization of SR proteins. For example, multi-site phosphorylation of the RS domain of the SR protein SFSR1 (aka ASF/SF2) by cytoplasmic SRPK1 is essential for nuclear import of this splicing factor (10-12).
SRPK1 can rapidly phosphorylate about 10 serines in the N-terminal portion of the RS domain (RS1 segment) of SRSF1 (13). This phosphorylation is essential for initiating interactions with a β1 transportin protein (transportin-SR), nuclear entry and speckle formation (14, 15). Kinetic and proteolytic mapping experiments indicate that the active site initially binds at the C-terminal end of RS1 (initiation region) with very high affinity (Kd=20-50 nM) (4, 16) before sequentially modifying serines in the N-terminal direction (17) (Fig.1A). The X-ray structure of the kinase domain of SRPK1 (lacking the insert domain & N-terminus) and a truncated form of SRSF1 (RRM2-RS1) reveal that an electronegative docking groove in the large lobe of the SRPK1 kinase core binds the N-terminal portion of RS1 (18) (Fig.1B). While a short stretch of Arg-Ser repeats is visible in the docking groove, the remainder of RS1 leading up to the active site is not defined. A small ATP impurity in the nucleotide analog AMPPNP allowed for single-site phosphorylation in the crystal complex and identification of an electropositive pocket (P+2) that binds a C-terminal RS1 phosphoserine just outside the active site (Fig.1C). To catalyze extensive, multi-site phosphorylation, a threading mechanism has been proposed in which the RS domain slides from the docking groove into the active site. Along this trajectory, conformational changes in one of the RRMs (RRM2), detected using cross-linking and circular dichroism experiments, allow a portion of this domain to serve as a recognition element in the docking groove for later phosphorylation steps. Mutagenesis data indicate that the stabilization of the growing phosphoserine chain by the P+2 pocket is important for efficient phosphorylation of later N-terminal serines in RS1 (13).
Figure 1. Structure and Phosphorylation Mechanism of SRSF1.
A) SRSF1 Domain Structure. SRSF1 has two RRMs and a C-terminal RS domain composed of RS1 and RS2 segments. B) Structure of SRPK1 core with AMPPNP and the N-terminal portion of RS1 segment (R204SRSRSR) bound to the docking groove in the large lobe. Arginines are colored blue and serines are colored red. A hydrogen bond is formed between the hydroxyl of Ser207 and the backbone carbonyl of Ser209. RRM2 is not displayed in this depiction. C) Phosphorylation Model. SRPK1 binds SRSF1 with high affinity using an electronegative docking groove that initially recognizes the N-terminal portion of RS1. Upon progressive phosphorylation, RS1 is threaded through the docking groove and into the active site in a C-to-N-terminal direction. In later phosphorylation stages, β4 from RRM2 unfolds and binds in the docking groove and phosphoserines in RS1 are stabilized by an electropositive site (P+2 site)
For many protein kinases the association/dissociation and phosphorylation of protein and peptide targets are fast events and the rate-limiting step in turnover is ADP release (19). In these cases, the kinase often engages in a ‘hit and release’ mechanism in which the protein quickly dissociates after phosphorylation and is free to adopt a new conformation and functionality induced by the phosphorylation event. In contrast, SRPK1 inserts numerous phosphates onto a very narrow polypeptide stretch while engaging the SR protein target as it undergoes phosphorylation-dependent structural changes (Fig.1). These chemical and structural events raise questions regarding the reaction free energy of SR protein activation. Does the close apposition of kinase and substrate during this extensive, multi-site reaction constrain the phosphotransfer steps and introduce unexpected rate-determining events? To address this question, rapid quench flow studies were performed so that the rates of phosphoryl transfer and nucleotide exchange could be monitored in the SRPK1-SRSF1 complex. Transient-state kinetic experiments initiated at various states of RS domain phosphorylation demonstrate that the chemical transfer event is fast during multi-site phosphorylation and ADP release limits overall SRSF1 turnover. However, progressive phosphorylation destabilizes the kinase-SR protein complex and lowers the rates of the phosphoryl transfer and nucleotide exchange steps, thereby providing a means for termination of multi-site phosphorylation and SR protein release. These findings provide a conceptual shift in how we view the static mechanism of protein phosphorylation. Rather than considering the kinase architecture as the principle determinant of phosphorylation efficiency, the kinetic data suggest that the substrate can modify the reaction coordinate in a phosphorylation-dependent manner. Thus, the kinase and substrate engage in active cross-talk and effectively alter the shape of the free energy coordinate as the chemical reaction progresses.
Material and Methods
Materials
Adenosine triphosphate (ATP), 3-(N-morpholino)propanesulphonic acid (Mops), 2-[N-morpholino]ethanesulphonic acid (MES), Tris (hydroxymethyl) aminomethane (Tris), MgCl2, NaCl, EDTA, glycerol, sucrose, acetic acid, Phenix imaging film, BSA, and liquid scintillant were obtained from Fisher Scientific. [γ-32P] ATP was obtained from NEN Products, a division of Perkin-Elmer Life Sciences.
Expression and Purification of Recombinant proteins
SRSF1 was expressed from a pET19b vector containing a 10xHis Tag at the N terminus (15). Mutations in SRPK1 and SRSF1 were described previously (13, 14, 17). The plasmids for wild-type and mutant forms of SRSF1 and SRPK1 were transformed into the BL21 (DE3) E. coli strain and the cells were then grown at 37°C in LB broth supplemented with 100 mg/ml ampicillin or 50 mg/ml kanamycin depending on the type of plasmid vector. Protein expression was induced with 0.4 mM IPTG at room temperature for 5 hours for SRSF1 constructs and 12 hours for SRPK constructs. All SRPK1 constructs were purified by Ni-resin affinity chromatography using a published procedure (16). All SRSF1 constructs were refolded and purified using a previously published protocol (17).
Phosphorylation Reactions-Manual Mixing
The phosphorylation of wild-type and mutant forms of SRSF1 by SRPK1 was carried out in the presence of 50 mM Mops (pH 7.4), 10 mM free Mg2+, 5 mg/mL BSA, and 100 μM [γ-32P]ATP (4000-8000 cpm pmol−1) at 23 °C according to previously published procedures (17). Reactions were typically initiated with the addition of SRSF1 or SRPK1 in a total reaction volume of 10 μL and then were quenched with 90 μL 30% acetic acid. Phosphorylated SRSF1 was separated from unreacted ATP by a filter binding assay (20). A portion of each quenched reaction (50 mL) was spotted onto a phosphocellulose filter disk and was washed 3 times with 0.5% phosphoric acid. The filter disks were rinsed with acetone, dried, and counted on the 32P channel in liquid scintillant. The total amount of phosphate incorporated into the substrate was then determined by considering the specific activity (cpm/min) of the reaction mixture, and the background retention of 32P ATP in the absence of SRPK1. Full retention of the phosphorylated product on the filters was confirmed by running quenched reaction samples on SDS-PAGE and counting the bands.
Viscosity Experiments
The steady-state phosphorylation of SRSF1 was monitored using the filter binding assay as described above in the presence of 0-30% sucrose. The relative solvent viscosity (ηrel) of the buffer (50 mM Mops, pH 7.4) containing 0-30% sucrose was measured using an Ostwald viscometer and a previously published protocol (21). Values of 1.44, 1.83, 2.32, and 3.43 for ηrel were measured for buffers containing 10, 20, 25, and 30% sucrose at 23 °C, respectively.
Rapid Quench Flow Experiments
Phosphorylation of SRSF1 by SRPK was monitored using a KinTek Corp. Model RGF-3 quench flow apparatus. The apparatus consists of three syringes driven by a stepping motor. Typical experiments were performed by mixing equal volumes of the SRPK1-SRSF1 complex in one reaction loop and 32P-ATP (5000-15000 cpm/pmol) in the second reaction loop in 50 mM Mops (pH 7.4), 10 mM free Mg2+, 5 mg/mL BSA. The reaction was quenched with 30% acetic acid in the third syringe and the reactions were analyzed using the filter binding assay described above. Control experiments lacking SRPK1 were run to define a background correction. Pre-phosphorylated forms of SRSF1 were obtained by adding low concentrations of 32P-ATP (1-4 μM) to an enzyme-substrate complex and allowing to react for 20 minutes before further reaction in the rapid quench flow machine using high concentrations of 32P-ATP (100 μM).
Data Analysis
The CPMs from the rapid quench flow experiments corrected for background were converted to concentrations of phosphoproduct using the specific activity of ATP. In single turnover experiments ([E]>[S]), the time-dependent production of phosphoproduct was fit to either single or double exponential functions. In pre-steady-state kinetic experiments ([S]>[E]), the time-dependent production of phosphoproduct (normalized to the total enzyme concentration) is fit to equation (1)
| (1) |
where α, kb, and kL are the ‘burst’ phase amplitude, ‘burst’ phase rate constant and the linear phase rate constant, respectively. The initial velocity data in manual mixing experiments were fit the Michaelis-Menten equation to obtain Km and Vmax. The Vmax values were converted to kcat using the total enzyme concentration determined from a Bradford assay (kcat = Vmax/Etot).
Results
Phosphorylation of SRSF1 Using Rapid Quench Flow Methods
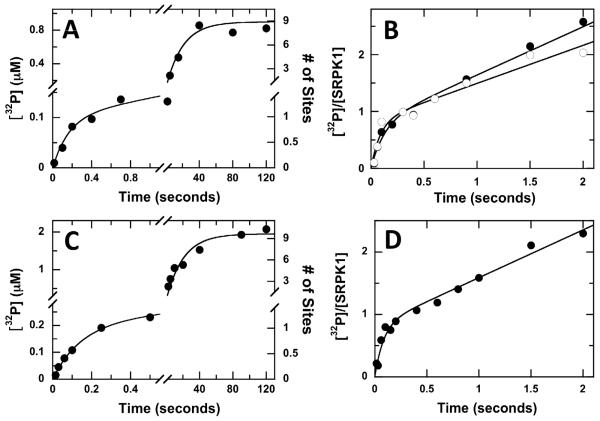
We showed previously that SRPK1 rapidly phosphorylates the first 10 serines in the RS domain of SRSF1 within 2 minutes (14). Since the rate constant for this reaction is about 1-5 min−1, the first phosphorylation event cannot be monitored in single turnover, manual mixing experiments. To circumvent this problem and gain new mechanistic insights, we studied the phosphorylation of SRSF1 using rapid quench flow methods that allow us to measure the phosphorylation events in the millisecond time frame. Using single turnover conditions (0.1 μM SRSF1, 1 μM SRPK1), we found that the incorporation of phosphates into SRSF1 is biphasic over the first 120 sec (Fig.2A). The rate constants for the fast (kp = 6 sec−1) and slow (kmp = 0.042 sec−1) phases are separated by over 2 orders of magnitude and are well-defined in the progress curve. The amplitude of the fast phase (αp = 0.1 μM) is equivalent to the substrate concentration whereas the slower phase (αmp = 0.8 μM) is 8-fold larger. These amplitudes are consistent with the rapid incorporation of a single phosphate in the RS domain within 500 msec followed by the addition of about 8 phosphates in the second phase. These incorporation levels after 2 minutes are consistent with prior manual mixing experiments and MALDI-TOF analyses (13). Also, a substrate-stoichiometric value for αp is consistent with a high affinity complex between SRPK1 and SRSF1 as previously described (13, 16). Overall, the phosphorylation of SRSF1 by SRPK1 can be kinetically partitioned into a fast, single-site phosphorylation (kp) followed by a slower, multi-site phosphorylation (kmp) of the RS1 segment.
Figure 2. Phosphorylation of Full-Length and Truncated SRSF1 Using Rapid Quench Flow Methods.
A&C) Single Turnover Experiments. A complex of SRPK1 (2 μM) and SRSF1 (0.2 μM) (A) or RS domain (0.4 μM) (C) in one syringe is mixed with ATP (200 μM) in the second syringe in the rapid quench flow machine. Final concentrations are 1 μM SRPK1, 100 μM ATP, 0.1 μM SRSF1 in (A) or 0.2 μM RS domain in (C). For SRSF1, the data are fit to values of 6 ± 2 sec−1 for kp, 0.042 ± 0.010 sec−1 for kmp, 0.10 ± 0.03 μM for αp and 0.80 ± 0.10 μM for αmp. For the RS domain, the data are fit to values of 7 ± 3 sec−1 for kp, 0.048 ± 0.010 sec−1 for kmp, 0.20 ± 0.04 μM for αp and 1.8 ± 0.10 μM for αmp. B&D) Pre-Steady-State Kinetic Experiments. A complex of SRPK1 (0.25 or 0.5 μM) and SRSF1 (1 μM) in (B) or SRPK1 (0.5 μM) and RS domain (5μM) in (D) is mixed with ATP (200 μM) in the rapid quench flow machine. Final concentrations in (B) are 0.125 (●) or 0.25 μM (○) SRPK1, 0.5 μM SRSF1 and 100 μM ATP. The data for SRSF1 phosphorylation at 0.125 and 0.25 μM SRPK1 are fit to equation (1) to obtain the respective values of 8 ± 2 and 11 ± 5 sec−1 for kb, 0.79 ± 0.11 and 0.82 ± 0.13 for α, and 0.85 ± 0.10, and 0.82 ± 0.13 sec−1 for kL. Final concentrations in (D) are 2.5 μM RS domain, 0.25 μM SRPK1 and 100 μM ATP. The data for RS domain phosphorylation are fit to equation (1) to obtain the respective values of 12 ± 3 sec−1 for kb, 0.80 ± 0.10 for a, and 0.75 ± 0.08 for kL.
Since SRPK1 rapidly adds the first phosphate onto SRSF1, we wished to determine whether this fast phase is an active-site phenomenon. To address this, we performed rapid quench flow experiments in which, unlike the single turnover experiments, the concentration of SRPK1 is not in excess of SRSF1. We found that the phosphorylation of SRSF1 using limiting SRPK1 observes classic ‘burst’ kinetics with a rapid, exponential phase followed by a slower linear phase that could be fit by equation (1) over a 2 second time frame (Fig.2B). At 0.125 μM SRPK1, the ‘burst’ rate constant (kb) is 8 sec−1, a value close to kp in the single turnover experiment. The linear rate constant (kL) represents the steady-state portion of the reaction and its value (0.8 sec−1) is consistent with kcat measurements from manual mixing assays (vide infra). Owing to poor substrate solubility above 1 μM, these experiments were performed using SR protein concentrations that are 2-4-fold higher than the enzyme. Nonetheless, steady-state conditions are achieved since each substrate molecule contains 10 phosphorylation sites and less than 3% of the available sites are modified in 2 seconds of the assay. The amplitude of the ‘burst’ phase, normalized to the total enzyme concentration is close to the upper limit of 1 expected in pre-steady-state kinetic experiments (22, 23) (α = αobs/[E]= 0.8). The enzyme-normalized burst amplitude does not change at higher SRPK1 concentrations indicating that the fast, initial phase represents an active-site phenomenon (Fig. 1B). Finally, that a large ‘burst’ amplitude is observed in these experiments indicates that the phosphoryl transfer step for this event is not only fast but also highly favorable with very little back reaction (i.e.- ADP phosphorylation). Overall, these observations indicate that the addition of the first phosphate in SRSF1 is much faster than the addition of subsequent phosphates.
RRMs Do Not Affect the Phosphorylation Mechanism
To determine whether the RRMs can affect the observed phosphoryl transfer rate constant for the first catalytic step in SRSF1, we studied the phosphorylation of the separate RS domain by SRPK1. Previous mass spectrometric studies showed that SRSF1 and the separate RS domain are phosphorylated to the same extent suggesting that the RRMs may not impact accessibility of SRPK1 for serines in the RS domain (13). Similar to the full-length protein substrate, we found that the RS domain is phosphorylated in a biphasic manner at approximately the same rates as SRSF1 in single turnover experiments (Fig.2C). The amplitude terms for the kinetic transient imply that SRPK1 adds about 10 phosphates onto the RS domain in 120 seconds. In rapid quench flow experiments experiments where the substrate exceeds the enzyme concentration, the phosphorylation of the RS domain observes classic ‘burst’ kinetics similar to that for SRSF1 (Fig.2D). These findings indicate that the nature of the mechanism for RS domain phosphorylation is largely unaffected by the presence of the two RRMs.
Docking Groove Is Essential for Fast Phosphoryl Transfer
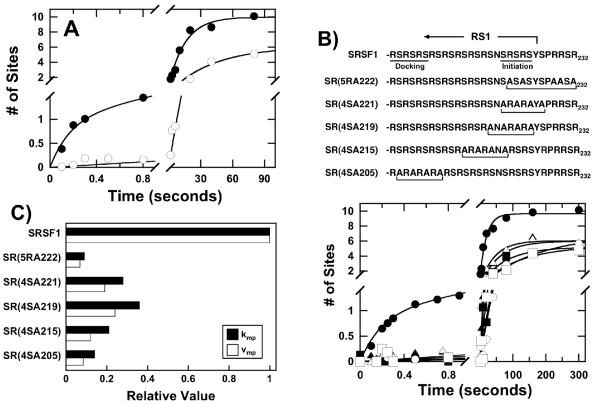
Having shown that the first serine in the RS domain of SRSF1 is rapidly phosphorylated in the SRPK1 active site, factors that control this event were addressed. The X-ray structure of SRPK1 with a truncated form of SRSF1 (RRM2-RS2) reveals a docking groove in the large lobe of the kinase domain that binds the N-terminal portion of RS1 (Fig.1B). Prior studies showed that 6 charge-to-alanine mutations in this groove [SRPK1(6M)] disrupts directional phosphorylation suggesting that the docking groove is essential for the C-to-N-terminal mechanism (13). In single turnover experiments, SRPK1(6M) does not efficiently phosphorylate the first serine in SRSF1 (Fig.3A). This observation is not the result of weak binding as it was shown previously that the Kd for SRPK1(6M) is only 2-fold higher than wild-type SRPK1 and a sufficient amount of enzyme is used in the kinetic experiments to saturate SRSF1 (13). Although only 6 phosphates are modified in the first 2 minutes in rapid quench flow experiments, previous studies showed that SRPK1(6M) can phosphorylate SRSF1 to the same level as the wild-type enzyme at longer time frames (13). The phosphorylation rate constant for these phosphates (kmp) is about 3-fold lower than the rate constant for the second phase in the wild-type enzyme (Fig.3A). These findings indicate that the docking groove in SRPK1 is important for facilitating fast, initial phosphorylation and for maintaining the efficient multi-site mechanism.
Figure 3. Enzyme-Substrate Interactions Support Phosphorylation Rates.
A) Docking Groove. The phosphorylation of SRSF1 by a form of SRPK1 containing 6 mutations in the docking groove [SRPK1(6M)] was studied. The data for wild-type SRPK1 (●) are fit to values of 5.7 sec−1 for kp, 0.063 for kmp, 1 site for αp, and 9 sites for αmp. For SRPK1(6M) (○) the data are fit to single exponential function with values of 0.023 sec−1 for kmp and 6.1 sites for αmp. B) RS Domain. The phosphorylation of multiple Arg-to-Ala and Ser-to-Ala mutants in the RS domain of SRSF1 by SRPK1 were studied. For wild-type SRSF1 (●), values of 5 sec−1 for kp, 0.050 sec−1 for kmp, 1 site for αp and 8.7 sites for αmp are obtained. The data for the mutants are fit to single exponential functions with values of 0.0057 sec−1 (kmp) and 6.9 sites (αmp) for SR(5RA222) (○), 0.018 sec−1 (kmp) and 6.0 sites (αmp) for SR(4SA221) (▲), 0.023 sec−1 (kmp) and 6.0 sites (αmp) for SR(4SA219) (△), 0.013 sec−1 (kmp) and 5.2 sites (αmp) for SR(4SA215) (■), 0.009 sec−1 (kmp) and 5.3 sites (αmp) for SR(4SA205) (□). C) Multi-Site Phosphorylation. The bar graph displays the ratio of kmp and vmp (kmp x αmp) for the mutants compared to SRSF1.
Phosphorylation Rates Are Dependent on RS Domain Content
While the N-terminal portion of RS1 rests in the docking groove at reaction start (Fig.1), it is unclear how the remaining residues in the RS domain contact the enzyme and support phosphorylation since the X-ray structure does not possess sufficient electron density for these residues. To determine whether the observed fast kinetics is strictly dependent on charged residues in and around the initiation box, 5 arginines around the RS1/RS2 border were mutated to alanine [SR(5RA222)] (Fig.3B). We found that SR(5RA222) lacks a rapid phase in rapid quench flow experiments (Fig.3B). Previous studies showed that this mutant has similar affinity to SRPK1 as the wild-type substrate and is phosphorylated to the same level (13) suggesting that these kinetic effects are not the result of an unstable enzyme-substrate complex. In addition to a loss of the rapid phase, kmp and the initial velocity for multi-site phosphorylation (vmp = αmp x kmp) of SR(5RA222) is about 10-fold lower than that for the second phase in the wild-type substrate (Fig. 3C). These findings indicate that charged residues near the initiation box are important for optimal positioning of the RS domain.
Previous studies have shown that SRPKs strongly prefer to phosphorylate serines with flanking arginines in long Arg-Ser dipeptide repeats (14, 24). However, the role of serines in these repeat regions is not well understood. We previously showed that the removal of blocks of 4 serines in RS1 does not alter the affinity of SRSF1 for SRPK1 and only lowers phosphoryl content according to the number of serines removed (17). We wished to determine whether these serines play any role in positioning the RS domain for optimal phosphorylation. In single turnover experiments, the removal of serines at various positions in RS1 leads to a loss of the fast, initial phase (Fig.3B). Furthermore, the introduction of these mutations likewise diminishes kmp and vmp by as much as 10-fold fold compared to wild-type SRSF1 (Fig.3C). The degree of rate reduction appears to follow modestly the position of the serine block with those mutations closest to the N-terminal part of RS1 having the greatest effect. Since only a very limited portion of the RS1 segment is visible in the X-ray structure, it is unclear whether these serines form contacts with SRPK1 or possibly with consecutive serines. In the docking groove, a hydrogen bond between a hydroxyl (Ser-207) and a backbone carbonyl (Ser-209) is observed within RS1 (Fig.1B). To examine the role of a more limited hydrogen-bonding network in the docking groove, we made a double alanine substitution at positions 205 and 207 in SRSF1 [SR(2SA205)]. Like the other serine-to-alanine substitutions, the phosphorylation of SR(2SA205) can be fit by a single exponential and observes no rapid, initial phase (data not shown). Overall, these findings indicate that the RS domain is likely to rely on numerous hydrogen bonding contacts either within the RS domain itself or with the kinase to rightly position the RS1 segment and support fast phosphorylation kinetics.
Phosphoryl Transfer Rate As A Function of RS Domain Phosphorylation Status
In the preceding rapid quench flow experiments we showed that addition of the first phosphate to the SRSF1 RS domain is fast. To address whether subsequent transfers are slow and potentially rate-limiting, we measured phosphorylation rates at several initial pre-phosphorylation states in SRSF1. We showed previously that SRPK1 catalyzes C-to-N-terminal phosphorylation and that regional phosphoryl content in the RS domain can be controlled through single turnover ATP limitation experiments (17). The addition of limiting ATP to the enzyme-substrate complex allows sequential placement of phosphates starting at the C-terminal end of the RS1 segment of the RS domain. SR proteins have a strong tendency toward aggregation so that studying their kinetic properties in an enzyme-substrate complex with excess SRPK1 is advantageous over purifying partially phosphorylated forms and reconstituting the complex for analysis. Accordingly, we pre-phosphorylated the SRPK1:SRSF1 complex using low levels of 32P-ATP in manual-mixing experiments so that approximately 1-8 serines in the C-terminal end of the RS1 segment of the RS domain are initially phosphorylated (Fig.4A). Starting with these initial phospho-forms (0P, 1P, 4P, 6P & 8P), we then added excess 32P-ATP (100 μM) to re-start the reaction in the rapid quench flow machine. The phosphorylation of the remaining serines is biphasic in all cases although the endpoints are expectedly lower at progressively higher pre-phosphorylation states (Fig.4B). The rate constants for the initial phases (kp) are similar implying that the phosphoryl transfer step is faster than net multi-site phosphorylation in all cases tested.
Figure 4. Positional Analyses of the Phosphoryl Transfer Rate.
A) Pre-phosphorylation of SRSF1. Complexes of SRPK1 (1 μM) and SRSF1 (100 nM) are incubated with limiting 32P-ATP (0-5 μM) for 20 min to attain 0, 1.2, 4, 6.3 and 8.3 phosphates (0P, 1P, 4P, 6P, 8P) onto the RS1 segment. B) Single Turnover Kinetics. Excess 32P-ATP (100 μM) is added to the complexes generated in (A) and the phosphorylation of the remaining sites is monitored. The data are fit to values of 3.6 ± 1.0 sec−1 for kp, 0.050 ± 0.006 sec−1 for kmp, 1.0 ± 0.18 sites for αp and 9.1 ± 0.35 sites for αmp for 0P (●), 3.8 ± 1.2 sec−1 for kp, 0.050 ± 0.004 sec−1 for kmp, 1.1 ± 0.20 sites for αp and 7.5 ± 0.25 sites for αmp for 1P (○), 4.3 ± 1.5 sec for kp, 0.040 ± 0.004 sec−1 for kmp, 1.1 ± 0.14 sites for αp and 4.9 ± 0.35 sites for αmp for 4P (▲), 3.0 ± 1.0 sec−1 for kp, 0.035 ± 0.004 sec−1 for kmp, 0.80 ± 0.05 sites for αp and 2.9 ± 0.08 sites for αmp for 6P (△) and 3.0 ± 1.1 sec−1 for kp, 0.040 ± 0.003 sec−1 for kmp, 0.32 ± 0.05 sites for αp and 1.4 ± 0.25 sites for α mp for 8P (◆). C) Amplitude and Linear Rates. The values of αp and vmp (kmp x αmp) from (B) are plotted as a function of pre-phosphorylation state of SRSF1. D) SRPK1 & ATP Dependence. The 8P complex is generated as described in panel (A) and the reaction is then started with either 400 μM 32P-ATP (●) or 4 μM SRPK1 (○). The data are fit to values of 4.0 ± 0.80 sec−1 (kp), 0.030 ± 0.004 sec−1 (kmp), 0.33 ± 0.06 sites (αp) and 1.7 ± 0.06 sites (αmp) at 400 μM P-ATP and 4.6 ± 0.82 sec−1 (kp), 0.031 ± 0.004 sec−1 (kmp), 0.67 ± 0.037 sites (αp) and 1.3 ± 0.06 sites (αmp) at 4 μM SRPK1. The 0P complex is also reacted with 600 μM 32P-ATP (▲) and fit to values of 25 ± 8 sec−1 (kp), 0.043 ± 0.006 sec−1 (kmp), 0.91 ± 0.16 sites (αp) and 9.0 ± 0.39 sites (αmp)
RS Domain Phosphorylation Regulates SRPK1 Binding Affinity
While the rate constants for the initial phases are fast under all conditions tested, the amplitudes varied as a function of initial phosphorylation state. At low phosphoryl contents (0P, 1P, and 4P), the observed amplitudes for the fast phase (αp) are approximately one site suggesting that all the substrate is initially bound with the enzyme. Above this minimal phosphoryl content, the amplitudes are noticeably lower (Fig.4C). To determine whether this observation for 8P (αp = 0.32 sites) is due to poor binding or an unfavorable phosphoryl transfer event, we measured single turnover kinetics at increased ATP or SRPK1. Although increasing the total ATP concentration (100 to 400 μM) after the pre-phosphorylation step had no effect on the initial phase amplitude (αp = 0.35 sites), increasing the SRPK1 concentration by 4-fold (1 to 4 μM) increased the amplitude by 2-fold (Fig.4D). These findings indicate that the low amplitude at higher, initial phosphoryl content is due to weakened affinity of SRPK1 for the substrate. Although the rate constant for the initial phase does not increase at higher ATP concentrations for 8P, we found that kp increases significantly for 0P at elevated nucleotide concentration without altering the amplitude (Fig4D). These findings imply that there is a change in rate-limiting step in the initial phase from ATP association at 0P to phosphoryl transfer at 8P. We will analyze this phenomenon in greater detail in the following sections. Based on the observed amplitudes at 1 and 4 μM SRPK1 (0.33 & 0.67 sites) and assuming that these amplitudes reflect the relative saturation level of SRSF1 by SRPK1 we can calculate a Kd of about 2 μM for the enzyme-substrate complex at 8P. Also, we can estimate a Kd of about 0.25 μM for the 6P complex using a value of 0.8 for αp. These results imply that the Kd for the enzyme-substrate complex is between 4-10-fold higher at 6P and about 40-100-fold higher at 8P compared to 0P [Kd=20-50nM; (4, 16)]. These calculations suggest that the affinity of SRSF1 for SRPK1 departs sharply from a low, nanomolar value after about 6 phosphorylation events. Finally, we observe a steady decline in vmp as a function of initial phosphorylation status suggesting that multi-site phosphorylation becomes progressively impaired as the phosphoryl content of the RS domain increases (Fig. 4C). This decline occurs even under conditions where the substrate is fully saturated with enzyme (αp = 1 site for 0-4P). Overall, these data indicate that RS domain phosphorylation lowers the affinity of SRPK1 for the SR protein and reduces the observed rate for multi-site phosphorylation.
SRSF1 Phosphorylation By SRPK1 is Viscosity Sensitive
The rapid quench flow studies suggest that the phosphoryl transfer steps are fast implying that other slow events limit complete turnover of SRSF1. To investigate whether the rate-limiting step involves diffusive events we measured the effects of solvent viscosity on the steady-state kinetic parameters. Initial velocities were measured as a function of SRSF1 concentration at a series of sucrose concentrations (0-30%) in steady-state kinetic experiments (Fig.5A). Both kcat and kcat/Km decrease at higher sucrose levels and plots of relative parameters as a function of relative solvent viscosity (ηrel) are linear with slope values [(kcat)η and (kcat/Km)η] close to the theoretical limit of one (Fig.5B). These results imply that diffusion-controlled, bimolecular processes limit both steady-state kinetic parameters. Viscosometric experiments were also performed by varying ATP at several, fixed amounts of sucrose. Both kcat and kcat/Km,ATP decrease as a function of increasing sucrose (Fig.5C) and plots of relative parameters are linear as a function of ηrel with maximal slope values close to one (Fig.5D). To determine whether these observed inhibitory effects are due to structural perturbations, the phosphorylation of a poor substrate was investigated. In a previous study we showed that a mutant form of SRSF1 possessing 10 consecutive Arg-to-Ala mutations starting at Arg-210 in the RS domain [SR(10RA210)] binds with reduced affinity to SRPK1 although it is phosphorylated to the same extent as wild-type SRSF1 (13). We now show that kcat is more than 10-fold lower and the Km is more than 10-fold higher for the Block mutant compared to the wild-type substrate (Fig.5E) and the reduced kinetic parameters are viscosity independent (Fig.5F). Owing to improved solubility, SR(10RA210) concentrations in excess of 1 μM could be easily achieved. The absence of a viscosity dependence on this poor substrate implies that the effects of sucrose on SRSF1 phosphorylation are the result of changes in ligand diffusion in and out of the active site.
Figure 5. Effects of Sucrose on the Steady-State Kinetic Parameters for SRPK1.
Initial velocities for SRPK1 are plotted as a function of total SRSF1 at fixed ATP (A) or as a function of ATP at fixed SRSF1 (C) using 0% (●), 10% (○), 25% (▲), and 30% (△) sucrose. In the absence of sucrose, the kcat and Km for SRSF1 are 56 ± 5 min−1 and 70 ± 6 nM using fixed ATP (100 μM) and the kcat and Km for ATP are 50 ± 5 min−1 and 12 ± 2 μM using fixed SRSF1 (1 μM). The effects of added sucrose on the relative steady-state kinetic parameters [/kcat and (kcat/Km)°/(kcat/Km)] are plotted as a function of relative solvent viscosity (ηrel) under conditions of varying SRSF1 (B) or ATP (D). For varying SRSF1, and (kcat/Km)η are 1.2 ± 0.16 and 0.90 ± 0.10. For varying ATP, and (kcat/Km)η are 1.2 ± 0.18 and 1.0 ± 0.05. Initial velocities for SRPK1 as a function of Block Mutant are plotted in panel (E) at fixed ATP (100 μM). In the absence of sucrose, the kcat and Km for SRSF1 are 2.4 ± 0.25 min−1 and 2.1 ± 0.60 μM. For varying Block Mutant, and (kcat/Km)η are −.020 ± 0.043 and 0.090 ± 0.010.
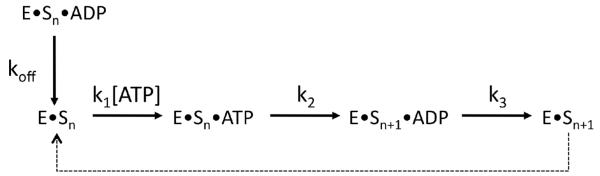
In general, the relative rate constants for bimolecular steps in initial velocity equations are expected to vary in a linear manner as a function of relative viscosity whereas unimolecular processes are expected to be viscosity independent (21, 25). For the simplified general mechanism in Scheme 1, S may represent ATP or SRSF1 when the other is saturating, k2 is the phosphoryl transfer step, and k3 is a diffusion-controlled event and represents the net dissociation rate constant for both products. The slopes of relative steady-state kinetic parameter versus ηrel will lie between the values of 0 and 1 based on equations (2) and (3):
| (2) |
| (3) |
A slope of 1 for (kcat)η implies that the phosphoryl transfer step is fast and the rate-limiting step is a diffusive step, most likely product release (i.e.-k2>>k3). In contrast, a slope of 0 for (kcat)η implies that the phosphoryl transfer step is rate-limiting (i.e.-k3>>k2). Since we observe a slope of 1 for (kcat)η the phosphoryl transfer step is fast and a diffusion-limited step controls maximum turnover (k3=1 sec−1). These results are consistent with the pre-steady-state kinetic experiments and indicate that a rapid phosphoryl transfer step precedes the steady-state linear phase (Fig.2). Using the same analysis for kcat/Km, the slope term allows us to establish the rate of phosphoryl transfer relative to substrate dissociation. Since we observe slopes of 1 for (kcat/Km)η using ATP and SRSF1 as varied substrates, the phosphoryl transfer step is fast relative to the dissociation of either ligand (i.e.- k2>>koff). Also, these maximum slopes indicate that kcat/Km is limited by the association rate constant of ATP (0.09 μM−1sec−1) when SRSF1 is saturating or SRSF1 (14 μM−1sec−1) when ATP is saturating. Overall, the viscosity dependence on the steady-state kinetic parameters allows us to confirm that the phosphoryl transfer rate is fast as predicted by the rapid quench flow data, to show that a product release step limits kcat and that ligand association (SRSF1 or ATP) limits kcat/Km.
Scheme 1.
ATP Dependence on the Observed Phosphoryl Transfer Rate
Since the phosphorylation reactions in the rapid quench flow machine are initiated with ATP whose binding limits kcat/Km, we next wondered whether nucleotide association rather than the phosphoryl transfer step could limit the pre-steady-state kinetic ‘burst’ phase. To address this possibility we performed pre-steady-state kinetic experiments by initiating at low and high ATP concentrations (50 & 500 μM). Although we observed a rapid ‘burst’ in product formation at 500 μM ATP (t1/2 = 35 msec), this phase was considerably slower at 50 M ATP and presented a small ‘lag’ in the first 50 msec (Fig.6A). This substantial kinetic change is not consistent with rapid equilibrium binding but is more in line with rate-limiting ATP association. To analyze the ATP dependence we simulated the transient kinetic data using the program DynaFit (26) and the mechanism in Scheme 2. In this mechanism, the dissociation rate constant for ATP and the reverse phosphoryl transfer rate constant are assumed to be small. The former assumption is in line with the maximum viscosity dependence on kcat/Km for ATP (i.e.- k2>>koff) and the latter assumption is consistent with previous studies on the internal equilibrium constant for protein kinases (23, 27-29) and the large amplitudes in Figs. 2 & 3. Also, in this mechanism, the product of the reaction (Sn+1) becomes the substrate for the second phosphorylation event since SRPK1 catalyzes multi-site phosphorylation of SRSF1. Since SRSF1 binds tightly to SRPK1 for the first several phosphorylation events (Fig.4) and the association rate constant is large (Fig.5) and not expected to limit turnover, we ignored any re-binding steps associated with SRSF1 and only considered ATP cycling. We simulated both curves using values for k1, k2 and k3 of 0.09 μM−1 sec−1, 30 sec−1, and 0.8 sec−1, respectively. The values of k1 and k3 are constrained by kcat/Km for ATP and kcat. In contrast, we found that the ‘burst’ phase is largely defined by k1 and k2. These data indicate that under our reaction conditions (50-500 μM ATP) the association of ATP and the phosphoryl transfer step partially limit the ‘burst’ phase. These findings are also consistent with data in Figure 4A,D which show a clear ATP dependence in the initial phase (kp) for the 0P complex.
Figure 6. Nucleotide Exchange Kinetics in the SRPK1-SRSF1 Complex.
A) ATP-Dependent Kinetic Transients. A complex of SRPK1 (0.5 μM) and SRSF1 (1 μM) in one syringe is mixed with ATP (100 or 1000 μM) in the second syringe in the rapid quench flow machine. Final concentrations are 50 (●) or 500 μM (○) ATP, 0.25 μM SRPK1 and 0.5 μM for SRSF1. The data are simulated using DynaFit (26) and Scheme 2 to obtain values of 0.09 μM−1sec−1, 30 sec−1, and 0.8 sec−1 for k1, k2, and k3, respectively. B) ADP Binding Affinity. Initial reaction velocities are measured as a function of ATP (6-800 μM) at fixed SRSF1 (1 μM) in the absence (●) and presence of 30 (○), 60 (▲), and 120 μM (△) ADP. KI = 10 μM for ADP. C) ADP Catalytic Trapping From 0P. A complex of SRPK1 (0.5 μM) and SRSF1 (1 μM) with (○) or without (●) ADP (120 μM) in one syringe is mixed with ATP (1200 μM) in the second syringe of the rapid quench flow machine. Final concentrations are 600 μM ATP, 0.25 μM SRPK1, 0.5 μM SRSF1 and 60 μM ADP. The data in the absence of ADP are simulated using DynaFit and Scheme 2 to obtain values of 0.09 μM−1sec−1, 30 sec−1, and 1.1 sec−1 for k1, k2, and k3, respectively. These values are then used to simulate the data in the presence of ADP using the mechanism in Scheme 3 and DynaFit to obtain a koff for ADP of 1.1 sec−1. D) ADP Catalytic Trapping from 4P. A complex of SRPK1 (2 μM) and SRSF1 (0.26 μM) is prequibrated for 20 min with 3 μM 32P-ATP to generate a 4P complex. This complex is incubated with (○) or without (●) ADP (120 μM) in one syringe and then mixed with ATP (1200 μM) in the second syringe of the rapid quench flow machine. Final concentrations are 600 μM ATP, 1 μM SRPK1, 0.13 μM SRSF1 and 60 μM ADP. The data in the absence of ADP are simulated using DynaFit and Scheme 2 to obtain values of 0.04 μM−1sec−1, 2.7 sec−1, and 0.10 sec−1 for k1, k2, and k3, respectively. These values are then used to simulate the data in the presence of ADP using the mechanism in Scheme 3 and DynaFit to obtain a koff for ADP of 0.13 sec−1.
Scheme 2.
ADP Release Kinetics Using Catalytic Trapping Experiments
The rapid quench flow experiments indicate that while the first phosphate is rapidly added to SRSF1, subsequent phosphorylation events are slower (Fig.2). Based on the viscosity experiments, this result could be due to slow release of one or both of the products. To address whether ADP dissociation limits turnover, we measured its release from the SRPK1:SRSF1 complex using a catalytic trapping method previously developed in our lab (30). In this experiment, ADP (120 μM) is preequilibrated with a complex of SRPK1 and SRSF1 before a 10-fold excess of ATP is added to start the reaction. Since we showed that ADP is competitive with ATP and has a KI of 10 μM (Fig.6B), all nucleotide-binding sites are saturated with ADP prior to mixing. While the pre-steady-state kinetic transient in the absence of ADP observes a ‘burst’ phase as expected, the transient in the presence of ADP preequilibration has no ‘burst’ and instead displays a ‘lag’ before the linear steady-state phase is achieved (Fig.6C). Since the observed linear production of 32P-SRSF1 after the ‘burst’ phase is the same as that in the presence of ADP (see inset; Fig.6C), sufficient ATP is used in the trapping experiment to displace ADP. The data in the absence of ADP was simulated using the mechanism in Scheme 2 to obtain k1, k2, and k3 of 0.09 μM−1 sec−1, 30 sec−1, and 1.1 sec−1, values similar to those obtained for the kinetic transients in Fig.6A. We then used these rate constants as fixed parameters to simulate the dissociation profile for ADP in Scheme 3. Using this approach, the data in the presence of ADP preequilibration was best simulated using an ADP dissociation rate constant (koff) of 1.1 sec−1. Since koff is identical to k3 (1.1 sec−1), the rate-limiting step for SRSF1 phosphorylation, the release of ADP limits maximum turnover (kcat).
Scheme 3.
Phosphorylation-Dependent ADP Release
Although ADP release limits kcat based on catalytic trapping experiments, this value is derived from a non-phosphorylated SRPK1-SRSF1 complex (0P). To determine whether reaction progress affects the rate of product release, we measured the dissociation rate constant for ADP at higher, initial phosphorylation of SRSF1. For these experiments, we pre-phosphorylated the SRPK1-SRSF1 complex at 4 sites (4P) using ATP limitation experiments and then incubated the complex with ADP (120 μM) prior to reaction initiation with excess ATP in the rapid quench flow machine. Since only 3 μM 32P-ATP is used to generate the 4P complex, any residual, unreacted 32P-ATP will be displaced by excess ADP in the sample loop prior to reaction start with additional ATP in the rapid quench flow machine. In the absence of ADP pre-incubation, we obtained a biphasic progress curve (Fig.6D). In comparison, ADP preequilibration results in a loss of the initial phase with no apparent change in the linear phase rate, similar to the results of catalytic trapping experiments in the absence of a pre-phosphorylation phase (Fig.6C). The data were simulated in the absence of ADP using DynaFit (26) and Scheme 2 over a 10 second time frame to obtain values of 0.04 μM−1sec−1, 2.7 sec−1 and 0.10 sec−1 for k1, k2 and k3, respectively. These values, which are lower than those obtained at 0P by 2-, 10-, and 10-fold for k1, k2 and k3, respectively, suggest that pre-phosphorylation impacts the phosphorylation of subsequent sites in the RS domain. We then simulated the data in the presence of ADP preequilibration using these constants and Scheme 3 and found that the progress curve was best modeled when koff for ADP is 0.13 sec−1, a value close to k3. These findings indicate that ADP limits the phosphorylation of subsequent sites in the 4P complex and that nucleotide exchange slows as the reaction progresses.
Progress Curve Analysis of Multi-Site Phosphorylation
Using rapid quench flow methods we monitored the attachment of 10 phosphates onto SRSF1 and showed that several individual steps become increasingly more difficult as a function of phosphorylation. To determine whether these changes in step-wise processing can describe SRSF1 activation, we simulated the progress curves for the complete phosphorylation of the 0P and 4P forms. The data were fit to a sequential mechanism (0P→1P→2P→…→10P) in which each species converts to the next phospho-state by irreversible, net rate constants (k0P, k1P, k2, up to k9P). The data for both curves obtained from Fig.4B were fit using a series of rate constants (Fig.7A). The values of the rate constants for the start of the reaction [k0P for 0P (5 sec−1)], and the re-start of the reaction [k4P for 4P (3 sec−1)] are fast relative to the other steps reflecting the rapid phosphorylation of the complexes in the initial phase. While nucleotide association (at 100 μM ATP) mostly limits the initial phase for the 0P complex, the phosphoryl transfer step mostly limits the initial phase for the 4P complex. For both complexes, the simulations model the progress curves with declining rate constants for each progressive phosphorylation event that parallel the experimental data. For example, the release of ADP from 0P measured in trapping experiments is rate-limiting and the same as k1P. Also, the catalytic trapping data indicate that the conversion of 4P→5P should be limited by ADP release at 0.13 sec−1 (Fig.6D). Interestingly, k4P (0.2 sec−1) in the progress curve simulation is close in value to this rate constant (Fig.7A). The value of k4P for the progress curve starting at 4P is much larger than k4P for the 0P complex since the latter reflects rate-limiting ADP release in the slow, multi-site phase and the former reflects rate-limiting phosphoryl transfer step in the initial, fast phase. In the experimental data, the affinity of SRPK1 declines after 4P, a factor that is likely to play an additional role in the progressive decrease in simulated rate constants. Furthermore, the simulated rate constants for the later stages of the reaction for both curves (k5P through k9P) are very similar as expected. These similarities explain why the analytical fitting for the multi-site phases in 0P and 4P (kmp) are not significantly different (Fig.4B). In both cases, the multi-site phases are dominated by the same steps (k5P-k9P) and subtle differences in individual steps earlier in the reaction (k2P-k4P) cannot be easily dissected in these phases with analytical fitting.
Figure 7. Effects of Phosphorylation Progress on Individual Steps.
A) Simulation of Progress Curves. The progress curves for 0P (○) and 4P (△) (data from Fig.4B) were simulated using DynaFit and a sequential mechanism in which 0P converts step-wise to 10P. The irreversible, net rate constants for each individual step are defined by reactant phosphorylation state (k0P, k1P, k2P,…k9P). Simulations for 0P include k0P through k9P whereas those for 4P include k4P through k9P. Values of 5, 1, 0.6, 0.45, 0.2, 0.15, 0.12, 0.1, 0.07, and 0.06 sec−1 for k0P, k1P, k2P, k3P, k4P, k5P, k6P, k7P, k8P, and k9P, respectively, were used to simulate 0P. Values of 3, 0.5, 0.1, 0.06, 0.04, and 0.02 sec−1 for k4P, k5P, k6P, k7P, k8P, and k9P, respectively, were used to simulate 4P. For comparison, the analytical fits to both curves are shown as dotted lines and their parameter fits are included in the legend of Fig. 4B. B) Change in Free Energy Reaction Coordinate Diagram As A Function of Phosphorylation. SRPK1 has high affinity for SRSF1, rapidly phosphorylates the first serine in the RS domain and slowly releases ADP (solid line). After several rounds of phosphorylation (6-8P), the affinity of the substrate declines significantly and the phosphoryl transfer and ADP release rates decrease (dashed line).
Discussion
Most protein kinases catalyze single-site phosphorylation and utilize interactions in the kinase domain to recognize complementary residues in the substrate protein/peptide. In general, these static contacts provide optimal orientation of the phosphate-accepting hydroxyl for rapid phosphoryl transfer (19). Some protein kinases carry out sequential, multi-site phosphorylation using inducible contacts. Glycogen synthase kinase-3 [GSK-3] was shown to phosphorylate four serines in an ordered manner where the phosphoserine in the P+4 position, added by a second protein kinase (e.g., casein kinase II), serves as a binding determinant for upstream phosphorylation at the P position (31). SRPK1 also catalyzes a C-to-N-terminal reaction but, unlike GSK-3, the reaction is much more robust (i.e.- more sites modified), takes place in a more congested environment where serines are separated by 1 rather than 3 residues and prior phosphorylation is not mandatory for upstream modifications (17). While GSK-3 uses an electropositive pocket for upstream phosphorylation, SRPK1 directionality is likely to be driven by the close juxtaposition of the active site and an electronegative docking groove that measure out a specific length of Arg-Ser dipeptide repeats, orients the active site toward the C-terminal end of the RS domain and expels phosphoserines in a threading mechanism (Fig. 1). Using rapid quench flow methods, we found that despite the complex nature of SR protein activation, the phosphoryl transfer step in SRPK1 is fast and exceeds turnover throughout numerous rounds of phosphorylation (Fig.4). Furthermore, the enzyme mechanism appears highly optimized to phosphorylate and re-position the RS domain for subsequent reactions such that the rate-limiting step (up to 4P) is ADP release.
Multiple Contacts Are Necessary for Efficient SR Protein Turnover
Our transient-state kinetic studies indicate that the rate of the first phosphoryl transfer step in SRSF1 is about 30-fold faster than rate-limiting ADP release (30 vs. 1 sec−1). Similar differences between the chemical transfer and product release steps have been observed in other protein kinases (19) raising the question of whether there could be an advantage to a fast phosphoryl transfer rate. Recent studies have demonstrated that rapid phosphoryl transfer can enhance substrate recognition through a chemical clamping mechanism (32). For example, the protein kinase Sky1p binds its physiological target Npl3, an mRNA carrier protein, with poor affinity but owing to a fast phosphoryl transfer step, the Km is lowered by 20-fold relative to Kd (33). A similar clamping mechanism is also operative for PKA and a peptide substrate (Kemptide) whose Km is significantly lower than Kd due to fast phosphoryl transfer (23). However, SRPK1 appears to depart from this kinetic model since its Kd for SRSF1 is lower (20-50 nM), not higher, than its Km (4, 16). What then is the importance of a fast phosphoryl transfer step for this splicing kinase? Through an analysis of sequence determinants in SRSF1 and SRPK1 we showed that removal of contacts along the RS1 segment in the docking groove or active site lowers significantly the phosphoryl transfer rate (Fig.3). These large reductions suggest that even subtle alterations in positioning of the RS domain at any point along the kinase limits both phosphorylation initiation and extension through a change in rate-limiting step from ADP release to phosphoryl transfer. Thus, it appears that rapid phosphoryl transfer provides not only a recognition advantage (ie.- Km<Kd) for poor binding substrates as in the cases of Sky1p and PKA but also a net turnover advantage for kinases such as SRPK1 that rely on precise geometry for multi-site phosphorylation.
Substrate-Induced Mechanisms for Phosphorylation Termination
While the rapid quench flow studies describe how extensive protein-protein interactions facilitate multi-site phosphorylation initiation and extension, they also outline three stopgap measures for terminating phosphorylation. First, the affinity of SRPK1 and the SR protein declines abruptly as a function of phosphorylation, thereby, derailing the enzyme from the RS domain. We estimate an affinity decline of up to 100-fold after eight rounds of phosphorylation (Fig.4). These findings explain why GST-SRSF1 robustly pulls down SRPK1 in the absence of ATP but very weakly in its presence when the RS domain is phosphorylated (34, 35). Second, the exchange rate of ADP for ATP slows significantly as a function of phosphorylation progress. We measured a decrease in the rate constant for ADP release in trapping experiments from 1 to 0.1 sec−1 after only 4 rounds of phosphorylation (Fig.6). This decrease is likely to be the dominant factor explaining the decline of multi-site phosphorylation rates (vmp) under conditions where the substrate is still saturated with enzyme (0→4P; Fig. 4C). Third, the rate constant of the phosphoryl transfer step declines from 30 to 3 sec−1 after 4 rounds of phosphorylation, thus, providing an increasing barrier for subsequent phosphorylation steps (Fig.6). Overall, progressive phosphorylation appears to raise the ground and transition state energies of several key steps in the phosphorylation mechanism (Fig.7B). These phosphorylation-coupled changes in individual steps would have the result of reducing net SR protein phosphorylation irrespective of the subcellular availabilities of the enzyme and substrate and whether the system functions under steady-state or pre-steady-state control. These findings contrast starkly with GSK-3 where pre-phosphorylation enhances upstream phosphorylation. For SRPK1, phosphorylation progress introduces barriers that enhance termination.
Biological Implications of Phosphorylation-Coupled Termination
SRPK1 phosphorylates SRSF1 in the cytoplasm providing a signal for transportin-SR binding and passage through the nuclear pore (12, 36). Although largely cytoplasmic, SRPK1 can also enter the nucleus prior to M-phase or under stress signaling (3, 4). While the large cytoplasmic pool results from chaperone interactions (5), the inability to uncover nuclear localization or export signals raises the possibility that SRPK1 could enter the nucleus in complex with SR proteins using a piggy-back mechanism (4). The high affinity observed between SR proteins and SRPK1 strongly supports such a mechanism (4, 16). We now show that the SRSF1 Kd increases by about one order of magnitude upon the addition of only 2 phosphates when converting from the 6P to 8P forms. This net level of phosphorylation (8P) is significant since it was shown previously that 8-10 Arg-Ser dipeptide repeats are sufficient to direct SRSF1 to the nucleus (37, 38). If these serines are fully phosphorylated and provide a minimal consensus for transportin-SR binding, then it is likely that only a small amount of SRPK1 is substrate-bound and can be drawn into the nucleus after each round of SR protein transport. These findings do not prohibit SRPK1 piggy-backing but instead place some constraints on its efficiency. Small amounts of nuclear SRPK1 could be sufficient for splicing regulation or multiple rounds of SR protein shuttling could slowly accumulate a functional level of the kinase particularly if export is sluggish.
Phosphorylation-Dependent Protein-Protein Cross-Talk
The results of many kinetic studies over the years have uncovered a simple mechanism for single-site phosphorylation of peptides and proteins by protein kinases. In general, the substrate is bound with moderate-to-weak affinity and is rapidly phosphorylated and released from the active site (19). While a deep appreciation for the flexibility of protein kinases has emerged over this time (39, 40), the kinetic processing of substrates is largely viewed from the perspective of static contacts in the active site and neighboring docking regions in the kinase. Through the analysis of multi-site processing of an SR protein, we now show that this simple view does not adequately describe the reaction coordinate for the splicing kinase SRPK1 where several discrete steps become modified in a phosphorylation-dependent manner (Fig. 7). The affinity and phosphorylation rate of this SR protein is coordinately down-regulated as the reaction progresses. Of particular interest, increasing phosphoryl content reduces the exchange rate of ADP, a step that has been shown to limit substrate turnover in most protein kinases (19). While the ADP dissociation rate constants have been shown to differ by about one order of magnitude for many kinases (23, 33), these differences are thought to be a function of the architecture of the nucleotide pocket and not due the nature of the substrate. Our new data show that in one example, a protein substrate can function as an effector of catalysis. While the causes of this effector role await further structural characterization, we can speculate upon how substrate contacts may impact kinetic processing. The RS domain is phosphorylated in a directional manner in the active site, a process that is accompanied by changes in RRM2 structure and docking groove and active-site occupancy (Fig.1). This threading mechanism is likely to be associated with multiple changes in contact partners between enzyme and substrate that could impact phosphorylation and nucleotide exchange rates. Overall, the transient kinetic studies presented here shift perspective from the static view of kinase-directed phosphorylation to a new, more dynamic view incorporating substrate-induced phosphorylation guidance.
Footnotes
This work was supported by an NIH grant GM67969 and ARRA supplement to J.A.A.
Abbreviations: RRM, RNA recognition motif; RS domain, domain rich in arginine-serine repeats; SR protein, splicing factor containing arginine-serine repeats; SRPK1, SR-specific protein kinase 1; SRPK1(6M), SRPK1 containing 6 alanine substitutions at D548, E557, E558, D564, E571, & K615; SRSF1, SR protein splicing factor 1 (also ASF/SF2).
References
- 1.Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003;12:5–14. doi: 10.1016/s1097-2765(03)00270-3. [DOI] [PubMed] [Google Scholar]
- 2.Blencowe BJ, Bowman JA, McCracken S, Rosonina E. SR-related proteins and the processing of messenger RNA precursors. Biochem Cell Biol. 1999;77:277–291. [PubMed] [Google Scholar]
- 3.Gui JF, Lane WS, Fu XD. A serine kinase regulates intracellular localization of splicing factors in the cell cycle. Nature. 1994;369:678–682. doi: 10.1038/369678a0. [DOI] [PubMed] [Google Scholar]
- 4.Ding JH, Zhong XY, Hagopian JC, Cruz MM, Ghosh G, Feramisco J, Adams JA, Fu XD. Regulated cellular partitioning of SR protein-specific kinases in mammalian cells. Mol Biol Cell. 2006;17:876–885. doi: 10.1091/mbc.E05-10-0963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhong XY, Ding JH, Adams JA, Ghosh G, Fu XD. Regulation of SR protein phosphorylation and alternative splicing by modulating kinetic interactions of SRPK1 with molecular chaperones. Genes Dev. 2009;23:482–495. doi: 10.1101/gad.1752109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu JY, Maniatis T. Specific interactions between proteins implicated in splice site selection and regulated alternative splicing. Cell. 1993;75:1061–1070. doi: 10.1016/0092-8674(93)90316-i. [DOI] [PubMed] [Google Scholar]
- 7.Kohtz JD, Jamison SF, Will CL, Zuo P, Luhrmann R, Garcia-Blanco MA, Manley JL. Protein-protein interactions and 5′-splice-site recognition in mammalian mRNA precursors. Nature. 1994;368:119–124. doi: 10.1038/368119a0. [DOI] [PubMed] [Google Scholar]
- 8.Chew SL, Liu HX, Mayeda A, Krainer AR. Evidence for the function of an exonic splicing enhancer after the first catalytic step of pre-mRNA splicing. Proc Natl Acad Sci U S A. 1999;96:10655–10660. doi: 10.1073/pnas.96.19.10655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Massiello A, Chalfant CE. SRp30a (ASF/SF2) regulates the alternative splicing of caspase-9 pre-mRNA and is required for ceramide-responsiveness. J Lipid Res. 2006;47:892–897. doi: 10.1194/jlr.C600003-JLR200. [DOI] [PubMed] [Google Scholar]
- 10.Caceres JF, Misteli T, Screaton GR, Spector DL, Krainer AR. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J Cell Biol. 1997;138:225–238. doi: 10.1083/jcb.138.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kataoka N, Bachorik JL, Dreyfuss G. Transportin-SR, a nuclear import receptor for SR proteins. J Cell Biol. 1999;145:1145–1152. doi: 10.1083/jcb.145.6.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lai MC, Lin RI, Huang SY, Tsai CW, Tarn WY. A human importin-beta family protein, transportin-SR2, interacts with the phosphorylated RS domain of SR proteins. J Biol Chem. 2000;275:7950–7957. doi: 10.1074/jbc.275.11.7950. [DOI] [PubMed] [Google Scholar]
- 13.Hagopian JC, Ma CT, Meade BR, Albuquerque CP, Ngo JC, Ghosh G, Jennings PA, Fu XD, Adams JA. Adaptable Molecular Interactions Guide Phosphorylation of the SR Protein ASF/SF2 by SRPK1. J Mol Biol. 2008;382:894–909. doi: 10.1016/j.jmb.2008.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma CT, Hagopian JC, Ghosh G, Fu XD, Adams JA. Regiospecific phosphorylation control of the SR protein ASF/SF2 by SRPK1. J Mol Biol. 2009;390:618–634. doi: 10.1016/j.jmb.2009.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Velazquez-Dones A, Hagopian JC, Ma CT, Zhong XY, Zhou H, Ghosh G, Fu XD, Adams JA. Mass spectrometric and kinetic analysis of ASF/SF2 phosphorylation by SRPK1 and Clk/Sty. J Biol Chem. 2005;280:41761–41768. doi: 10.1074/jbc.M504156200. [DOI] [PubMed] [Google Scholar]
- 16.Aubol BE, Chakrabarti S, Ngo J, Shaffer J, Nolen B, Fu XD, Ghosh G, Adams JA. Processive phosphorylation of alternative splicing factor/splicing factor 2. Proc Natl Acad Sci U S A. 2003;100:12601–12606. doi: 10.1073/pnas.1635129100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma CT, Velazquez-Dones A, Hagopian JC, Ghosh G, Fu XD, Adams JA. Ordered multi-site phosphorylation of the splicing factor ASF/SF2 by SRPK1. J Mol Biol. 2008;376:55–68. doi: 10.1016/j.jmb.2007.08.029. [DOI] [PubMed] [Google Scholar]
- 18.Ngo JC, Giang K, Chakrabarti S, Ma CT, Huynh N, Hagopian JC, Dorrestein PC, Fu XD, Adams JA, Ghosh G. A sliding docking interaction is essential for sequential and processive phosphorylation of an SR protein by SRPK1. Mol Cell. 2008;29:563–576. doi: 10.1016/j.molcel.2007.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams JA. Kinetic and Catalytic Mechanisms of Protein Kinases. Chemical Reviews. 2001;101:2271–2290. doi: 10.1021/cr000230w. [DOI] [PubMed] [Google Scholar]
- 20.Kemp BE, Graves DJ, Benjamini E, Krebs EG. Role of multiple basic residues in determining the substrate specificity of cyclic AMP-dependent protein kinase. J Biol Chem. 1977;252:4888–4894. [PubMed] [Google Scholar]
- 21.Adams JA, Taylor SS. Energetic limits of phosphotransfer in the catalytic subunit of cAMP-dependent protein kinase as measured by viscosity experiments. Biochemistry. 1992;31:8516–8522. doi: 10.1021/bi00151a019. [DOI] [PubMed] [Google Scholar]
- 22.Fierke CA, Hammes GG. Transient kinetic approaches to enzyme mechanisms. Methods Enzymol. 1995;249:3–37. doi: 10.1016/0076-6879(95)49029-9. [DOI] [PubMed] [Google Scholar]
- 23.Grant BD, Adams JA. Pre-steady-state kinetic analysis of cAMP-dependent protein kinase using rapid quench flow techniques. Biochemistry. 1996;35:2022–2029. doi: 10.1021/bi952144+. [DOI] [PubMed] [Google Scholar]
- 24.Wang HY, Lin W, Dyck JA, Yeakley JM, Songyang Z, Cantley LC, Fu XD. SRPK2: a differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. J Cell Biol. 1998;140:737–750. doi: 10.1083/jcb.140.4.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brouwer AC, Kirsch JF. Investigation of diffusion-limited rates of chymotrypsin reactions by viscosity variation. Biochemistry. 1982;21:1302–1307. doi: 10.1021/bi00535a030. [DOI] [PubMed] [Google Scholar]
- 26.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 27.Cook PF, Neville ME, Jr., Vrana KE, Hartl FT, Roskoski R., Jr. Adenosine cyclic 3′,5′-monophosphate dependent protein kinase: kinetic mechanism for the bovine skeletal muscle catalytic subunit. Biochemistry. 1982;21:5794–5799. doi: 10.1021/bi00266a011. [DOI] [PubMed] [Google Scholar]
- 28.Kong CT, Cook PF. Isotope partitioning in the adenosine 3′,5′-monophosphate dependent protein kinase reaction indicates a steady-state random kinetic mechanism. Biochemistry. 1988;27:4795–4799. doi: 10.1021/bi00413a032. [DOI] [PubMed] [Google Scholar]
- 29.Callaway K, Waas WF, Rainey MA, Ren P, Dalby KN. Phosphorylation of the transcription factor Ets-1 by ERK2: rapid dissociation of ADP and phospho-Ets-1. Biochemistry. 49:3619–3630. doi: 10.1021/bi100199q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou J, Adams JA. Participation of ADP dissociation in the rate-determining step in cAMP-dependent protein kinase. Biochemistry. 1997;36:15733–15738. doi: 10.1021/bi971438n. [DOI] [PubMed] [Google Scholar]
- 31.Fiol CJ, Wang A, Roeske RW, Roach PJ. Ordered multisite protein phosphorylation. Analysis of glycogen synthase kinase 3 action using model peptide substrates. J Biol Chem. 1990;265:6061–6065. [PubMed] [Google Scholar]
- 32.Lieser SA, Aubol BE, Wong L, Jennings PA, Adams JA. Coupling phosphoryl transfer and substrate interactions in protein kinases. Biochim Biophys Acta. 2005;1754:191–199. doi: 10.1016/j.bbapap.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 33.Aubol BE, Ungs L, Lukasiewicz R, Ghosh G, Adams JA. Chemical clamping allows for efficient phosphorylation of the RNA carrier protein Npl3. J Biol Chem. 2004;279:30182–30188. doi: 10.1074/jbc.M402797200. [DOI] [PubMed] [Google Scholar]
- 34.Ma CT, Ghosh G, Fu XD, Adams JA. Mechanism of dephosphorylation of the SR protein ASF/SF2 by protein phosphatase 1. J Mol Biol. 2010;403:386–404. doi: 10.1016/j.jmb.2010.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koizumi J, Okamoto Y, Onogi H, Mayeda A, Krainer AR, Hagiwara M. The subcellular localization of SF2/ASF is regulated by direct interaction with SR protein kinases (SRPKs) J Biol Chem. 1999;274:11125–11131. doi: 10.1074/jbc.274.16.11125. [DOI] [PubMed] [Google Scholar]
- 36.Yun CY, Fu XD. Conserved SR protein kinase functions in nuclear import and its action is counteracted by arginine methylation in Saccharomyces cerevisiae. J Cell Biol. 2000;150:707–718. doi: 10.1083/jcb.150.4.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cazalla D, Zhu J, Manche L, Huber E, Krainer AR, Caceres JF. Nuclear export and retention signals in the RS domain of SR proteins. Mol Cell Biol. 2002;22:6871–6882. doi: 10.1128/MCB.22.19.6871-6882.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ngo JC, Chakrabarti S, Ding JH, Velazquez-Dones A, Nolen B, Aubol BE, Adams JA, Fu XD, Ghosh G. Interplay between SRPK and Clk/Sty Kinases in Phosphorylation of the Splicing Factor ASF/SF2 Is Regulated by a Docking Motif in ASF/SF2. Mol Cell. 2005;20:77–89. doi: 10.1016/j.molcel.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 39.Taylor SS, Yang J, Wu J, Haste NM, Radzio-Andzelm E, Anand G. PKA: a portrait of protein kinase dynamics. Biochim Biophys Acta. 2004;1697:259–269. doi: 10.1016/j.bbapap.2003.11.029. [DOI] [PubMed] [Google Scholar]
- 40.Johnson DA, Akamine P, Radzio-Andzelm E, Madhusudan M, Taylor SS. Dynamics of cAMP-dependent protein kinase. Chem Rev. 2001;101:2243–2270. doi: 10.1021/cr000226k. [DOI] [PubMed] [Google Scholar]
- 41.Huynh N, Ma CT, Giang N, Hagopian J, Ngo J, Adams J, Ghosh G. Allosteric interactions direct binding and phosphorylation of ASF/SF2 by SRPK1. Biochemistry. 2009;48:11432–11440. doi: 10.1021/bi901107q. [DOI] [PMC free article] [PubMed] [Google Scholar]