Poly ADP-ribosylation polymerases are necessary for recruitment and/or retention of Ku at double-strand breaks during nonhomologous end-joining DNA repair.
Abstract
Poly adenosine diphosphate (ADP)–ribosylation (PARylation) by poly ADP-ribose (PAR) polymerases (PARPs) is an early response to DNA double-strand breaks (DSBs). In this paper, we exploit Dictyostelium discoideum to uncover a novel role for PARylation in regulating nonhomologous end joining (NHEJ). PARylation occurred at single-strand breaks, and two PARPs, Adprt1b and Adprt2, were required for resistance to this kind of DNA damage. In contrast, although Adprt1b was dispensable for PARylation at DSBs, Adprt1a and, to a lesser extent, Adprt2 were required for this event. Disruption of adprt2 had a subtle impact on the ability of cells to perform NHEJ. However, disruption of adprt1a decreased the ability of cells to perform end joining with a concomitant increase in homologous recombination. PAR-dependent regulation of NHEJ was achieved through promoting recruitment and/or retention of Ku at DSBs. Furthermore, a PAR interaction motif in Ku70 was required for this regulation and efficient NHEJ. These data illustrate that PARylation at DSBs promotes NHEJ through recruitment or retention of repair factors at sites of DNA damage.
Introduction
Poly ADP-ribosylation (PARylation) of proteins by poly ADP-ribose (PAR) polymerases (PARPs) is one of the earliest responses to DNA damage (Amé et al., 2004). The best-characterized role of PARPs in the DNA damage response (DDR) is in repair of DNA single-strand breaks (SSBs; Caldecott, 2008). Although PARP1 and PARP2 PARylate proteins at SSBs, PARP1 is the principle ADP-ribosyltransferase (Adprt) required for their repair (Schreiber et al., 2002; Le Page et al., 2003; Fisher et al., 2007). However, the observation that parp1−/− parp2−/− mice are not viable suggests shared functions between these enzymes in maintaining genome stability or other pathways required for cell viability (Ménissier de Murcia et al., 2003). Although the mechanisms by which PARPs regulate SSB repair remain unclear, they may promote recruitment of repair factors at DNA lesions (El-Khamisy et al., 2003; Okano et al., 2003; Bekker-Jensen et al., 2007; Kanno et al., 2007; Rulten et al., 2008).
PARPs also become activated in response to DNA double-strand breaks (DSBs), which can be repaired by homologous recombination (HR) or nonhomologous end joining (NHEJ; Haber, 2000). Although PARP1 interacts with NHEJ proteins, including Ku and the DNA-dependent protein kinase catalytic subunit (Ariumi et al., 1999; Galande and Kohwi-Shigematsu, 1999), classical NHEJ is normal in murine PARP1−/− cells (Yang et al., 2004). However, PARP1 is required to promote end joining by alternative NHEJ (A-NHEJ; Audebert et al., 2004; Robert et al., 2009) and has been implicated in HR to promote replication restart at damaged replication forks (Yang et al., 2004; Sugimura et al., 2008; Bryant et al., 2009).
Recently, we and others initiated a study of DNA repair in Dictyostelium discoideum and found it contains orthologues of NHEJ and other repair proteins absent in other invertebrates (Block and Lees-Miller, 2005; Hudson et al., 2005; Hsu et al., 2006; Zhang et al., 2009). This suggests that D. discoideum will prove a useful model to study certain repair pathways that show limited conservation in other genetically tractable organisms. In this regard, PARP activity is evident in D. discoideum, and Adprts have been isolated from this organism (Rickwood and Osman, 1979; Kofler et al., 1993). Here, we analyze the role of D. discoideum Adprts in DNA repair and find that, similar to other organisms, multiple Adprts are required for D. discoideum to tolerate SSBs. Furthermore, we exploit D. discoideum to uncover a third PARP that is required for DSB repair and illustrate that PARylation promotes NHEJ through retention of repair factors at damage via a PAR interaction domain present in Ku70.
Results and discussion
D. discoideum Adprts are required for tolerance to SSBs
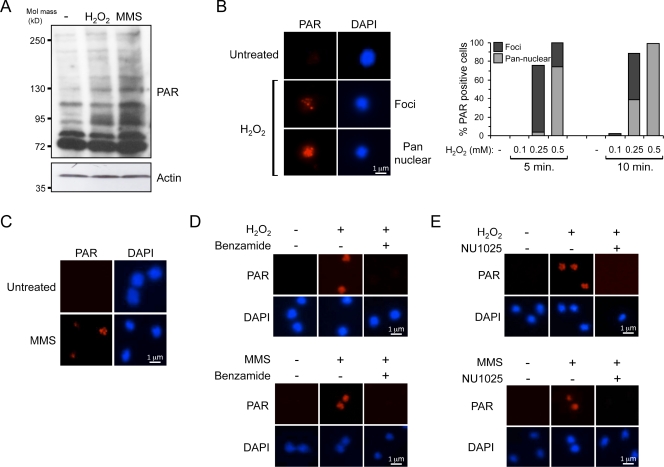
Given that vertebrate PARPs are required for SSB repair, we wished to establish whether Adprt enzymes perform a similar function in D. discoideum. To achieve this, we assessed whether PARylation is induced after SSBs in this organism. Cells were exposed to H2O2 or methanesulfonate (MMS), and PARylation was assessed by Western blotting using a PAR antibody. Consistent with activation of PARPs, PARylated proteins become evident after treatment with both agents (Fig. 1 A).
Figure 1.
PARylation is induced in D. discoideum after SSBs. (A) Ax2 cells were untreated (−) or exposed to 0.5 mM H2O2 for 10 min or 5 mM MMS for 30 min. Whole-cell extracts were analyzed by Western blotting using the indicated antibodies. (B) Ax2 cells were treated with H2O2 as indicated. Coverslips were subjected to immunofluorescence using PAR antibodies. The percentages of PAR-positive cells were scored from a population of >200 cells. Cells were categorized into those that exhibit pannuclear staining or PAR nuclear foci. Data are representative of three independent experiments. (left) Representative images are shown. (C) Ax2 cells were untreated or treated with 5 mM MMS for 30 min. Coverslips were processed for immunofluorescence and stained with PAR antibodies. (D) Ax2 cells were treated with carrier (ethanol) or 5 mM benzamide and exposed to 0.5 mM H2O2 for 10 min (top) or 5 mM MMS for 30 min (bottom). Coverslips were processed for immunofluorescence using PAR antibodies. (E) Ax2 were treated with carrier (DMSO) or 1 mM NU1025 and analyzed as in D.
Given that DNA damage-induced nuclear foci are a commonly used marker for posttranslational modifications at or adjacent to sites of DNA damage, we assessed SSB-induced PAR foci formation in D. discoideum nuclei after SSBs. Exposure of cells to H2O2 (Fig. 1 B) or MMS (Fig. 1 C and Fig. S1, A and B) induces PAR-positive nuclei in a dose- and time-dependent manner. PAR staining is pannuclear at high doses of H2O2, whereas a punctate staining pattern is evident at lower doses (Fig. 1 B). Robust induction of γ-H2AX foci is not apparent at the H2O2 and MMS concentrations used, indicating PARylation is not a consequence of secondary DSBs (Fig. S1, C and D). To illustrate staining is a consequence of PAR synthesis, cells were treated with PARP inhibitors. Benzamide has previously been shown to inhibit PARylation in D. discoideum (Rajawat et al., 2007), whereas NU1025 was used as a higher potency alternative. Pretreatment of cells with either agent inhibits PAR nuclear staining in response to H2O2 and MMS (Fig. 1, D and E).
Next, we assessed which Adprt enzymes are required for SSB repair. Vertebrate Adprts can be divided into five subgroups (Otto et al., 2005), with group 1 containing the DNA damage-responsive PARPs, PARP1 and PARP2, in addition to PARP3 and PARP4. D. discoideum contains 13 proteins with putative Adprt catalytic domains, including orthologues of PARPs 1–4 (dictyBase; Otto et al., 2005). Three of these proteins, Adprt1a, Adprt1b, and Adprt2, contain PARP catalytic domains that exhibit a significant homology to PARP1 (Fig. 2 A; Otto et al., 2005).
Figure 2.
D. discoideum Adprt1b and Adprt2 are required for tolerance to SSBs. (A) The domain structure of Adprt1a, Adprt1b, and Adprt2 illustrating the PARP catalytic and regulatory domains, putative zinc finger, pADR, WGR, and BRCA1 C-terminal (BRCT) domains. (B) The indicated strains and their respective parental controls, Ax2 and Ax4, were untreated or exposed to 5 mM MMS for 30 min. Cells were subjected to immunofluorescence using PAR antibodies. The percentages of PAR-positive cells were scored from a population of >200 cells. (C) The indicated strains and their respective parental controls, Ax2 and Ax4, were assessed for their ability to survive exposure to MMS. Error bars represent the SEM from three independent experiments.
To test the role of Adprt enzymes in SSB repair, we generated strains disrupted in the adprt1a or adprt2 genes and obtained strains disrupted in adprt1b (Sawai et al., 2007). None of these strains exhibit a significant defect in PARylation after exposure to 5 mM MMS for 30 min (Fig. 2 B) or shorter times (not depicted). SSB-induced PARylation in the adprt1a− strain is reflected in a lack of sensitivity of cells to MMS (Fig. 2 C). However, adprt1b− and adprt2− strains exhibit increased sensitivity to MMS and H2O2 compared with parental controls (Fig. 2 C and Fig. S1, F–I). The adprt1b− and adprt2− strains are not radiosensitive, arguing against sensitivity to MMS being a consequence of DSBs sustained at the doses of MMS used (Fig. S1 E). Collectively, these data support a role for Adprt1b and Adprt2 in cellular resistance to SSBs and indicate conservation of a PAR-dependent SSB repair pathway in D. discoideum.
Adprt1a is required to promote NHEJ
Having established a role for Adprt enzymes in SSB repair, we assessed their role in DSB repair. Treatment of Ax2 cells with the DSB-inducing agent phleomycin results in PARylation of nuclear proteins, as judged by the appearance of PAR-positive nuclei (Fig. 3 A). Pannuclear PAR staining is observed at high doses of phleomycin, whereas lower doses result in punctate staining (Fig. 3 A). DSB-induced PARylation is inhibited by NU1025, illustrating this event is mediated through PARP activity (Fig. 3 B). PARylation remains intact in adprt1b− cells after DSBs (Fig. 3 C). In contrast to SSBs, a modest reduction of DSB-induced PARylation is observed in adprt2− cells, whereas a significant reduction is evident in the adprt1a− strain (Fig. 3 C and Fig. S2 A). This reduction in PARylation is further decreased by disrupting adprt2 and adprt1a in combination (Fig. 3 C). These observations suggest that although Adprt1b and Adprt2 are required for tolerance to SSBs, Adprt1a and, to a lesser extent, Adprt2 are required for nuclear PARylation after DSBs.
Figure 3.
Adprt1a is required for PARylation after DSBs. (A) Ax2 cells were treated with phleomycin (micrograms per milliliter) and subjected to immunofluorescence using PAR antibodies. Data are representative of three independent experiments. Cells were categorized into those that exhibit pannuclear staining or PAR nuclear foci. (B) Ax2 cells were treated with 1 mM NU1025 or DMSO before exposure to 100 µg/ml phleomycin for 30 min. Cells were subjected to immunofluorescence using PAR antibodies. (C) The indicated strains and their respective parental controls, Ax2 and Ax4, were assessed for PARylation after phleomycin treatment as in B. The percentages of PAR-positive cells were scored from a population of >200 cells. Error bars represent the SEM from three independent experiments. *, P < 0.05 compared with the positive control (Ax2 + phleomycin).
Given the reduced ability of adprt1a− and adprt2− cells to PARylate nuclear proteins after DSBs, we assessed their ability to repair these DNA lesions. To assess NHEJ, we exploited the observation that transfection of linearized plasmid DNA along with the restriction enzyme used for linearization results in stimulation of DNA integration into the genome by restriction enzyme–mediated integration (REMI; Kuspa and Loomis, 1992). The ligation of the vector at endogenous restriction enzyme sites and the fact that vector DNA contains limited sequence homology to the D. discoideum genome suggest that REMI is mediated by NHEJ. Consistent with this, REMI is dependent on Ku70, Ku80, and other components of the NHEJ pathway (Fig. 4 A and Fig. 5 F; Hsu et al., 2011). The adprt1a− strain has a reduced REMI index comparable with ku80− cells, indicating a role for Adprt1a in promoting NHEJ (Fig. 4 A and Fig. S2 B). In contrast, adprt2− cells exhibit a modest reduction in the ability to perform REMI, whereas disruption of adprt2− and adprt1a− in combination does not further exacerbate the REMI defect of adprt1a− cells (Fig. 4 A). These data suggest that although Adprt2 may contribute toward promoting NHEJ, Adprt1a is the principle PARP required to regulate this pathway. In further support for a role of Adprt1a in NHEJ, adprt1a− cells are sensitive to DSBs administered during spore germination (Fig. 4 B and Fig. S2 C), a stage of the D. discoideum life cycle when cells are reliant on NHEJ to tolerate DSBs (Hudson et al., 2005).
Figure 4.
Adprt1a is required to promote NHEJ. (A) REMI efficiency of the indicated strains was assessed as described in Materials and methods. The numbers analyzed were such that experimental strains were compared with >200 colonies from the positive control (Ax2 plus the restriction enzyme). Data are represented as the percentage of REMI induction relative to parental Ax2 controls. *, P < 0.05 compared with the positive control (Ax2). (B) Ax2, adprt1a−, and ku80− spores were germinated before exposure to phleomycin, and cell survival was established as described in Materials and methods. (C) Ax2 and adprt1a− cells were assessed for HR efficiency by measuring targeted integration of the blasticidin resistance cassette at the cdk8 locus. The percentage of HR is the proportion of aggregation-deficient colonies against the total number of blasticidin-resistant colonies. The n number represents to total of blasticidin-resistant colonies analyzed from multiple independent transfections. Error bars represent the SEM from three independent experiments.
Figure 5.
The PBZ domain in D. discoideum Ku70 is required for NHEJ. (A) The indicated strains were exposed to phleomycin as indicated. After 80 min, the indicated cell fractions were analyzed by Western blotting. (B) Alignment of the PBZ domains present in D. discoideum (Dd) Ku70 and vertebrate APLF (APLF1 and APLF2), SNM1, and CHFR. Conserved amino acids are highlighted in black, and similar amino acids are represented in gray. (C) Serial dilutions of GST, GST fused to the C terminus of Ku70 (GST-Ku70C), or GST fused to the same fragment lacking the PBZ domain (GST-Ku70CΔPBZ) were blotted onto nitrocellulose filters. Inclines represent the relative amount of protein blotted onto the filter. After incubation in PAR polymer, Western blotting was performed using the indicated antibodies. (D, left) Whole-cell extracts were prepared from the indicated strains and subjected to Western blotting. The indicated strains were incubated with phleomycin, and Ku was immunoprecipitated (IP) from whole-cell extracts using Ku80 antisera. (right) Myc-Ku70 was confirmed by Western blotting (WB) using Myc antisera. (E) The indicated strains were left untreated or exposed to phleomycin. After 80 min, the indicated cellular fractions were analyzed by Western blotting. (F) REMI efficiency of the indicated strains was assessed as in Fig 4 A. The numbers analyzed were such that experimental strains were compared with >200 colonies from the positive control (ku70− cells expressing Myc-Ku70 plus restriction enzyme). The data are represented as the percentage of REMI induction relative to ku70− cells expressing Myc-Ku70. Error bars represent the SEM from three independent experiments. *, P < 0.05 compared with the positive control (ku70− cells expressing Myc-Ku70). Molecular masses are given in kilodaltons.
Disruption of NHEJ promotes HR in several organisms (Pierce et al., 2001; Zhang et al., 2007; Barlow et al., 2008). Given the requirement for adprt1a in promoting NHEJ, we assessed the impact of disrupting this gene on the ability of cells to perform HR. Accordingly, we assessed targeted HR efficiency at the cdk8 locus in adprt1a− cells and found it to be elevated 4.5-fold compared with Ax2 (Fig. 4 C). Collectively, these data indicate that DSB-induced PARylation by Adprt1a promotes NHEJ at the expense of HR.
PARylation promotes NHEJ through retention of Ku at DSBs
Next, we assessed whether the inability to perform NHEJ in adprt1a− cells reflects a reduced capacity to recruit NHEJ factors to DNA lesions. Subcellular fractionation experiments reveal that NHEJ proteins become enriched in chromatin isolated from vertebrate cells after DSBs (Drouet et al., 2005). Using the same technique in D. discoideum, we observed enrichment of Ku80 in chromatin fractions after DSBs in Ax2 cells. DSB-induced enrichment of Ku80 in chromatin was reduced in adprt1a− cells compared with Ax2, illustrating that Adprt1a is required to recruit and/or retain Ku at DSBs (Fig. 5 A and Fig. S2, D and E).
PAR polymers act as recognition modules to recruit factors to DNA lesions through PAR interaction domains (Kleine and Lüscher, 2009). One such domain, the PAR-binding zinc finger (PBZ) motif, is required for CHFR to fulfill its role in the antephase checkpoint, implicating this motif in regulating the DDR (Ahel et al., 2008). The D. discoideum genome contains several proteins that exhibit a PBZ domain, including Ku70 (Fig. 5 B; Ahel et al., 2008). To determine whether the PBZ domain of D. discoideum Ku70 is capable of interacting with PAR, we expressed and purified the C-terminal 74 amino acids of Ku70 fused to GST (GST-Ku70C) or the same protein lacking the C-terminal 25 amino acids that spans the PBZ domain (GST-Ku70ΔPBZ) and tested their ability to bind PAR. Consistent with the PBZ domain of Ku70 being able to interact with PAR, GST-Ku70C, but not GST-Ku70ΔPBZ, is able to bind PAR in vitro (Fig. 5 C).
To assess a requirement for the PBZ domain of Ku70 in NHEJ, we generated a ku70− strain and expressed full-length wild-type Myc-tagged Ku70 (Myc-Ku70) or Myc-Ku70 lacking the PBZ domain (MYC-Ku70ΔPBZ) in these cells (Fig. 5 D). Importantly, both proteins coimmunoprecipitate with Ku80, indicating they interact with their biologically relevant partner (Fig. 5 D). In Myc-Ku70–expressing cells, Ku80 becomes enriched in chromatin after DSBs (Fig. 5 E). Importantly, Myc-Ku70ΔPBZ–expressing cells exhibit reduced enrichment of Ku80 and Myc-Ku70ΔPBZ in chromatin after DSBs, presumably through an inability of Ku to be recruited and/or retained at DSBs (Fig. 5 E and Fig. S2 F). Although Myc-Ku70 rescues the NHEJ defect of ku70− cells, Myc-Ku70ΔPBZ is unable to do so to the same degree (Fig. 5 F). Collectively, these data suggest the PBZ domain of Ku70 is required to promote NHEJ.
Concluding remarks
Our observations that disruption of adprt1b and adprt2 sensitizes cells to agents that induce SSBs illustrate that the role of PARylation in SSB repair is largely conserved in D. discoideum. The involvement of two PARPs in SSB tolerance is reminiscent of the situation in mammals. PARP2 was initially discovered as a result of residual PARylation in PARP1-deficient mice after DNA damage (Amé et al., 1999). In addition, parp2−/− mice are sensitive to DNA damage and exhibit increased chromosome instability after exposure to alkylating agents and a delay in repair of this variety of DNA damage (Schreiber et al., 2002; Ménissier de Murcia et al., 2003). Our data illustrating that both Adprt1b and Adprt2 respond to SSBs would support the role of multiple PARPs in this response and indicate conservation of a PAR-dependent SSB repair pathway in D. discoideum.
Importantly, we identify Adprt1a as a third PARP that functions in the DDR. Although Adprt1a is dispensable for cells to tolerate SSBs, it is required for NHEJ. We also observe a modest decrease in DSB-induced PARylation in adprt2− cells. This is reflected in a subtle reduction of the REMI index of these cells. Collectively, these data imply that, similar to SSBs, multiple PARPs, namely Adprt1a and Adprt2, are involved in NHEJ. However, NHEJ efficiency in the adprt1a− strain is not reduced further by disrupting adprt2. We therefore believe that although Adprt2 may function in NHEJ, Adprt1a is the principle PARP that promotes this pathway.
Recently, PARP1 has been implicated in promoting end joining by the A-NHEJ pathway (Audebert et al., 2004; Robert et al., 2009). A possibility, therefore, is that Adprt1a may be involved in A-NHEJ. However, we use REMI of plasmid DNA into the genome of D. discoideum as an assay for NHEJ, which occurs accurately at endogenous restriction enzyme sites (Kuspa and Loomis, 1992) and is dependent on Ku70 and Ku80. A-NHEJ is Ku independent and involves extensive processing of DNA termini that will ablate restriction site integrity. We therefore favor that Adprt1a and Adprt2 are involved in classical NHEJ.
Our data suggest that DSB-induced PARylation promotes NHEJ by retention of repair factors at DSBs through the PBZ motif in Ku70. This conclusion is based on our observations that (a) DSB-induced PARylation and NHEJ are compromised in the adprt1a− strain, (b) enrichment of Ku onto chromatin after DSBs is reduced in adprt1a− cells, and (c) recombinant Ku70 lacking the C-terminal PBZ domain exhibits a reduced ability to be enriched in chromatin after DSBs and to promote NHEJ compared with recombinant wild-type Ku70. Given the pivotal role of Ku in initiating NHEJ, we believe the NHEJ defect in adprt1a− or Myc-Ku70ΔPBZ–expressing cells suggests an inability to assemble and/or retain NHEJ factors at DSBs. This is reminiscent of the situation in SSB repair in which PAR polymers promote accumulation of repair proteins at DNA damage (El-Khamisy et al., 2003; Okano et al., 2003; Bekker-Jensen et al., 2007; Kanno et al., 2007; Rulten et al., 2008). Consistent with this hypothesis, human PARP3 has recently been implicated in facilitating NHEJ through promoting the accumulation of APLF and XRCC4–ligase IV at DSBs (Rulten et al., 2011).
Neither vertebrate Ku70 nor Ku80 possess a PBZ domain. However, although the PBZ domain is evident in several D. discoideum DDR proteins, only three human proteins possess this motif (APLF, SNM1, and CHFR; Ahel et al., 2008). We have yet to identify APLF in D. discoideum. Although speculative, an interesting possibility is that the PBZ domain is evident in several D. discoideum proteins to compensate for the absence of APLF. A prediction of this model would be that APLF functions in a variety of DDR pathways, including NHEJ. In this regard, APLF has been implicated in SSB repair, and similar to our observations, its PBZ domain is required for accumulation of NHEJ factors at DSBs (Bekker-Jensen et al., 2007; Iles et al., 2007; Kanno et al., 2007; Rulten et al., 2008, 2011). We have yet to identify the proteins PARylated at DNA lesions in D. discoideum. However, given that PARylation of histones recruits repair factors to strand breaks in vertebrates (Rulten et al., 2008, 2011), it will be interesting to assess whether histones are similarly PARylated in response to DSBs in D. discoideum and act to retain Ku at sites of DNA damage.
Materials and methods
Sequence alignments
D. discoideum nucleic acid and predicted protein sequences were obtained from dictyBase, and other sequences were obtained from the National Center for Biotechnology Information. The MultAlin interface was used for DNA sequence alignment (Corpet, 1988), and ClustalW was used to align protein sequences. Protein domains were identified using InterProScan.
Cell culture and strain generation
D. discoideum strains were grown using standard procedures, either axenically or on SM agar in association with Klebsiella aerogenes. An outline of the strategy to generate the strains used in this study and their verification is illustrated (Fig. S3). To generate the adprt1a disruption strain, DNA fragments upstream (nucleotides 1,070–1,907) and downstream (nucleotides 3,643–4,375) of the adprt1a catalytic domain (nucleotides 2,420–3,404) were generated by PCR from Ax2 genomic DNA. These fragments were cloned on either side of a blasticidin resistance cassette contained within the pLPBLP plasmid (Faix et al., 2004) using KpnI–HindIII and BamHI–NotI, respectively. A similar procedure was followed to disrupt the adprt2 gene by cloning upstream (nucleotides 543–1,500) and downstream (nucleotides 2,482–3,236) DNA fragments flanking the Adprt2 catalytic domain into pLPBLP using KpnI–HindIII and BamHI–NotI, respectively. A ku70− strain was generated by cloning upstream (nucleotides −880 to 27) and downstream (nucleotides 2,357–2,786) DNA fragments into pLPBLP using KpnI–HindIII and PstI–BamHI, respectively. This ku70− disruption construct targeted the entire ku70 gene for deletion. Cells were transfected with the disruption constructs and subjected to selection with 10 µg/ml blasticidin the following day. Blasticidin-resistant clones were isolated using standard procedures, and gene disruption was verified by PCR and Southern blotting. The adprt1b− strain was a gift from T. Cox (Princeton University, Princeton, NJ) and A. Kuspa (Baylor College of Medicine, Houston, TX).
Two independent clones of the adprt1a− and adprt2− strains were analyzed (adprt1a−.w and adprt1a−.z; adprt2− and adprt2−.2). Although only adprt1a−.w and adprt2− are illustrated in the main figures on the manuscript, data from both clones are illustrated in Fig. S1 and Fig. S2.
The blasticidin resistance cassette was removed from the adprt1a− and ku70− strains as described previously (Faix et al., 2004). In brief, strains were transformed with the pDEX-NLS-cre plasmid, and transformants were transiently selected with 20 µg/ml G418 before being grown in media containing no antibiotics. Surviving colonies were screened for sensitivity to 10 µg/ml blasticidin and 20 µg/ml G418. Cells sensitive to both blasticidin and G418 were cloned on SM agar containing a K. aerogenes lawn, and clones were subjected to PCR to confirm the removal of the blasticidin resistance cassette.
The ku70− strain was complemented with either full-length Ku70 or Ku70 containing a 25–amino acid C-terminal deletion that includes the PBZ domain contained within the pDXA-3C expression vector (Manstein et al., 1995). In both cases, the N-terminal Ku70 primer contained a KpnI restriction site and Myc tag, and the C-terminal primer contained an XbaI restriction site. Strains were transformed with the pDXA-3C complementation vectors and the pREP helper plasmid and subjected to selection with 10 µg/ml G418.
Immunoblotting and antibodies
Whole-cell extracts were prepared by washing cultures in KK2 before boiling cells in Laemmli buffer for 20 min (Hudson et al., 2005). Antibodies were obtained from the following sources: PAR (Trevigen), γ-H2AX (Abcam), Ku80 (Hudson et al., 2005), actin (Santa Cruz Biotechnology, Inc.), and Myc (Cell Signaling Technology).
Immunoprecipitation of Ku80
Exponentially growing cells were seeded at 5 × 106 cells/ml and treated with 100 µg/ml phleomycin for 30 min with rotation. Cells were washed twice in KK2, resuspended to a final density of 107 cells/ml in lysis buffer (50 mM Tris, pH 8.0, 200 mM NaCl, 1% Triton X-100, 1 mM DTT, 1 mM EDTA, 50 U/ml benzonase [EMD], 10 mM sodium butyrate, 1 mM NaF, 20 mM β-glycerophosphate, 1 mM sodium orthovanadate, 1 µM microcystin, and protease inhibitor cocktail [Roche]), and incubated with rotation for 30 min at 4°C. Cell lysates were centrifuged at 14,000 rpm for 10 min, and the supernatant was isolated and precleared with protein G–Sepharose beads at 4°C for 1 h before incubation with Ku80 serum (Hudson et al., 2005) for 30 min at 4°C, followed by an incubation with protein G–Sepharose beads for 45 min at 4°C, with rotation. The beads were pelleted at 500 rpm for 2 min and washed three times with lysis buffer before resuspending the beads in SDS loading buffer containing 100 mM DTT. Samples were analyzed by Western blotting (Hudson et al., 2005).
Protein expression and purification
A fragment of the ku70 gene that encodes the C terminus of Ku70 spanning amino acids 835–909 or the same fragment lacking the last 26 amino acids that encode the PBZ domain was inserted into the pGEX6P-1 vector (GE Healthcare) via BamHI and XhoI sites (termed GST-Ku70C and GST-Ku70CΔPBZ, respectively). GST-tagged proteins were expressed and purified according to the manufacturer’s instructions.
PAR-binding assay
Twofold serial dilutions of proteins ranging in concentration from 2.5 to 0.625 pmol were slot blotted onto a nitrocellulose membrane, which was subsequently blocked with 5% milk in TBST buffer (Tris-HCL, pH 7.5, 150 mM NaCl, and 0.05% Tween 20). After incubation with the PAR polymer (1:5,000 dilution) in TBST for 1 h, membranes were washed once with TBST followed by TBST containing 1 M NaCl. After a final wash in TBST, filters were incubated in anti-PAR or GST antibodies diluted in 5% milk in TBST buffer and incubated at 4°C overnight. Filters were washed three times in TBST before incubation with HRP-conjugated secondary antibody (Dako). After a final three washes in TBST, signals were detected with ECL Western blotting reagent.
Immunofluorescence analysis of DNA damage-induced staining
Cells were allowed to adhere to coverslips for 1 h before being incubated in media containing the indicated concentrations of phleomycin, MMS, or H2O2 for the times stated in the figure. Where indicated, coverslips were mock treated or preincubated for 1 h in media containing 1 mM NU1025 (Enzo Life Sciences) or 5 mM benzamide (Enzo Life Sciences) before addition of the DNA-damaging agent. Coverslips were incubated for 5 min in ice-cold nuclear extraction buffer (10 mM Pipes, pH 6.8, 300 mM sucrose, 3 mM MgCl2, 20 mM NaCl, and 0.5% Triton X-100) before being washed in TBS and fixed with 70% ethanol for a further 5 min. Coverslips were finally rinsed in 100% methanol and washed twice with TBS.
Coverslips were blocked for 1 h at room temperature in 10% swine serum before incubation in the relevant antibody for 1 h at room temperature. Coverslips were washed three times in TBS, incubated with TRITC-conjugated secondary antibody (Dako) for 1 h at room temperature, and washed three more times with TBS. Coverslips were mounted onto glass slides with VECTASHIELD mounting media containing DAPI (Vector Laboratories) and visualized with a microscope (1X71; Olympus) at a magnification of 100× with a lens (Olympus) and immersion oil (Lenzol). All microscopy was performed at room temperature. All images were acquired on a camera (C10600-10B-H; Hamamatsu Photonics) using HCImage Acquisition (Hamamatsu Photonics) image software and processed in Photoshop (Adobe).
Sensitivity assays
Exponentially growing D. discoideum were seeded at a density of 106 cells/ml. Phleomycin (Sigma-Aldrich), MMS (Sigma-Aldrich), or H2O2 (Sigma-Aldrich) was added at the concentrations indicated in the figure, and cells were incubated at 22°C while being shaken at 100 rpm. After 1 h, cells were diluted to 104 cells/ml in KK2, and 250 cells were plated in duplicate onto two 140-mm Petri dishes containing K. aerogenes in association with SM agar. Petri dishes were incubated in the dark at 22°C, and survival was assessed by counting the number of colonies visible 3, 4, and 5 d after plating. Sensitivity assays using germinated spores were performed as described previously (Hudson et al., 2005). In brief, fruiting bodies were resuspended in KK2 containing 0.1% NP-40, and spores were liberated by passing fruiting bodies through a 19.5-gauge needle. Spores were washed twice in KK2 + 0.1% NP-40 and once in KK2 before being resuspended to 2 × 107 cells/ml in KK2 and divided into 1-ml aliquots. Germination was induced by heat shock at 45°C for 30 min, and the hatched spores were diluted to 106 cells/ml in a 1:5 ratio of HL5/KK2 containing increasing concentrations of phleomycin. Cells were incubated in shaking suspension at100 rpm for 18 h at 22°C before being plated as described in the second sentence of this section, and colonies were counted after 3, 4, and 5 d.
REMI
The vector was prepared by digesting the blasticidin resistance cassette containing a plasmid, pLPBLP, with BamHI and removing the terminal phosphates with calf intestinal phosphatase. The plasmid was purified by phenol/chloroform extraction and ethanol precipitation before being used in transfections.
Exponentially growing D. discoideum were prepared for transfection by washing cells twice in ice-cold H50 buffer (50 mM KCl, 20 mM Hepes, 10 mM NaCl, 5 mM NaHCO3, 1 mM NaH2PO4.H2O, and 1 mM MgSO4.7H2O, pH 7.0) before being resuspended to 5 × 107 cells/ml in H50. Cells were mixed with 2 µg vector with or without 20 U BamHI and electroporated according to the standard procedure (Kuspa and Loomis, 1992). After electroporation, cells were resuspended in HL5 to a density of 5 × 106 cells/ml. After a 15-h incubation without shaking at 22°C, cells were pelleted and mixed with 1,500 µl Escherichia coli B/r (blasticidin-resistant E. coli strain) and divided equally over three 140-mm plates containing LPB agar (2.92 mM lactose, 0.1% bactopeptone, 19 mM Na2HPO4.2H2O, 30 mM KH2PO4, 2% agar, and 10 µg/ml blasticidin S). Plates were incubated at 22°C, and colonies were counted 3, 4, and 5 d after plating. REMI was assessed by measuring the fold induction of the colony number in the presence of BamHI. Before performing REMI in the ku70− strain complemented with Myc-Ku70 or Myc-Ku70ΔPBZ, cells were grown in the absence of G418 for 24 h before being subjected to transfection as described in the previous paragraph.
Targeted HR efficiency at the cdk8 locus
The cdk8 knockout plasmid (Lin et al., 2004) was digested with KpnI and NotI to liberate a fragment of DNA containing regions homologous to the D. discoideum cdk8 gene flanking the BsR resistance cassette and purified using standard procedures. Cells in the exponential phase of growth were transfected with 7 µg DNA using standard procedures, serial diluted in HL5 after a short recovery period, and plated out in 96-well plates. After 24 h, 10 µg/ml blasticidin was added, and plates were incubated in the dark at 22°C for 14 d. After selection, clonal suspensions of blasticidin-resistant transformants were spotted onto SM agar containing a lawn of K. aerogenes. After 5–6 d, plaques were large enough for phenotypic analysis. Integration at the cdk8 locus is indicated by an aggregation-deficient phenotype. Aggregation-proficient or -deficient colonies were randomly selected, and the genomic DNA was analyzed by PCR to confirm targeted and random integration, respectively.
Subcellular fractionation
Subcellular fractionation experiments were performed as described previously (Drouet et al., 2005). In brief, exponentially growing cells were seeded at 5 × 106 cells/ml and exposed to phleomycin or mock treated. At the time points indicated in the figure, cells were washed in KK2 buffer and resuspended in nuclear lysis buffer (50 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 µM microcystin, 1 mM NaF, 2 mM sodium orthovanadate, and protease inhibitor cocktail) supplemented with 0.1% Triton X-100 to a final density of 3 × 107 cells/ml. Cells were incubated for 15 min at 4°C and then centrifuged at 14,000 g for 3 min, giving rise to a pellet and supernatant fraction S1. The pellet was resuspended in nuclear lysis + 0.1% Triton X-100 for a further 15 min at 4°C and centrifuged at 14,000 g for 3 min. The supernatant was pooled with the S1 fraction, and the pellet was resuspended in nuclear lysis buffer + 200 µg/ml RNase A (Sigma-Aldrich), incubated with rotation at room temperature for 30 min, and repelleted as before to give fraction P2. The P2 pellet was resuspended in SDS loading buffer containing 100 mM DTT. Fractions were analyzed by Western blotting using the antibodies indicated in the figure.
Online supplemental material
Fig. S1 further shows that PARylation is stimulated after SSBs and that Adprt1b and Adprt2 are required to tolerate this variety of DNA damage. Fig. S2 further illustrates that Adprt1a is required for NHEJ through PBZ domain–dependent retention of Ku at DSBs. Fig. S3 shows validation of disruption strains. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201012132/DC1.
Acknowledgments
The adprt1b− strain was a gift from T. Cox and A. Kuspa. We are grateful to dictyBase and K. Caldecott for assistance.
This work was supported by Cancer Research UK (grant C1521/A8182), the Biotechnology and Biological Sciences Research Council (grant BB/H009957/1), and the Oxford University Clarendon Scholarship Fund.
Footnotes
Abbreviations used in this paper:
- Adprt
- ADP-ribosyltransferase
- A-NHEJ
- alternative NHEJ
- DDR
- DNA damage response
- DSB
- double-strand break
- HR
- homologous recombination
- MMS
- methanesulfonate
- NHEJ
- nonhomologous end joining
- PAR
- poly ADP-ribose
- PARP
- PAR polymerase
- PBZ
- PAR-binding zinc finger
- REMI
- restriction enzyme–mediated integration
- SSB
- single-strand breaks
References
- Ahel I., Ahel D., Matsusaka T., Clark A.J., Pines J., Boulton S.J., West S.C. 2008. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 451:81–85 10.1038/nature06420 [DOI] [PubMed] [Google Scholar]
- Amé J.C., Rolli V., Schreiber V., Niedergang C., Apiou F., Decker P., Muller S., Höger T., Ménissier-de Murcia J., de Murcia G. 1999. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J. Biol. Chem. 274:17860–17868 10.1074/jbc.274.25.17860 [DOI] [PubMed] [Google Scholar]
- Amé J.C., Spenlehauer C., de Murcia G. 2004. The PARP superfamily. Bioessays. 26:882–893 10.1002/bies.20085 [DOI] [PubMed] [Google Scholar]
- Ariumi Y., Masutani M., Copeland T.D., Mimori T., Sugimura T., Shimotohno K., Ueda K., Hatanaka M., Noda M. 1999. Suppression of the poly(ADP-ribose) polymerase activity by DNA-dependent protein kinase in vitro. Oncogene. 18:4616–4625 10.1038/sj.onc.1202823 [DOI] [PubMed] [Google Scholar]
- Audebert M., Salles B., Calsou P. 2004. Involvement of poly(ADP-ribose) polymerase-1 and XRCC1/DNA ligase III in an alternative route for DNA double-strand breaks rejoining. J. Biol. Chem. 279:55117–55126 10.1074/jbc.M404524200 [DOI] [PubMed] [Google Scholar]
- Barlow J.H., Lisby M., Rothstein R. 2008. Differential regulation of the cellular response to DNA double-strand breaks in G1. Mol. Cell. 30:73–85 10.1016/j.molcel.2008.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekker-Jensen S., Fugger K., Danielsen J.R., Gromova I., Sehested M., Celis J., Bartek J., Lukas J., Mailand N. 2007. Human Xip1 (C2orf13) is a novel regulator of cellular responses to DNA strand breaks. J. Biol. Chem. 282:19638–19643 10.1074/jbc.C700060200 [DOI] [PubMed] [Google Scholar]
- Block W.D., Lees-Miller S.P. 2005. Putative homologues of the DNA-dependent protein kinase catalytic subunit (DNA-PKcs) and other components of the non-homologous end joining machinery in Dictyostelium discoideum. DNA Repair (Amst.). 4:1061–1065 10.1016/j.dnarep.2005.06.008 [DOI] [PubMed] [Google Scholar]
- Bryant H.E., Petermann E., Schultz N., Jemth A.S., Loseva O., Issaeva N., Johansson F., Fernandez S., McGlynn P., Helleday T. 2009. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 28:2601–2615 10.1038/emboj.2009.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldecott K.W. 2008. Single-strand break repair and genetic disease. Nat. Rev. Genet. 9:619–631 [DOI] [PubMed] [Google Scholar]
- Corpet F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881–10890 10.1093/nar/16.22.10881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drouet J., Delteil C., Lefrançois J., Concannon P., Salles B., Calsou P. 2005. DNA-dependent protein kinase and XRCC4-DNA ligase IV mobilization in the cell in response to DNA double strand breaks. J. Biol. Chem. 280:7060–7069 10.1074/jbc.M410746200 [DOI] [PubMed] [Google Scholar]
- El-Khamisy S.F., Masutani M., Suzuki H., Caldecott K.W. 2003. A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage. Nucleic Acids Res. 31:5526–5533 10.1093/nar/gkg761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix J., Kreppel L., Shaulsky G., Schleicher M., Kimmel A.R. 2004. A rapid and efficient method to generate multiple gene disruptions in Dictyostelium discoideum using a single selectable marker and the Cre-loxP system. Nucleic Acids Res. 32:e143 10.1093/nar/gnh136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher A.E., Hochegger H., Takeda S., Caldecott K.W. 2007. Poly(ADP-ribose) polymerase 1 accelerates single-strand break repair in concert with poly(ADP-ribose) glycohydrolase. Mol. Cell. Biol. 27:5597–5605 10.1128/MCB.02248-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galande S., Kohwi-Shigematsu T. 1999. Poly(ADP-ribose) polymerase and Ku autoantigen form a complex and synergistically bind to matrix attachment sequences. J. Biol. Chem. 274:20521–20528 10.1074/jbc.274.29.20521 [DOI] [PubMed] [Google Scholar]
- Haber J.E. 2000. Partners and pathways repairing a double-strand break. Trends Genet. 16:259–264 10.1016/S0168-9525(00)02022-9 [DOI] [PubMed] [Google Scholar]
- Hsu D.W., Gaudet P., Hudson J.J., Pears C.J., Lakin N.D. 2006. DNA damage signaling and repair in Dictyostelium discoideum. Cell Cycle. 5:702–708 10.4161/cc.5.7.2626 [DOI] [PubMed] [Google Scholar]
- Hsu D.W., Kiely R., Couto C.A., Wang H.Y., Hudson J.J., Borer C., Pears C.J., Lakin N.D. 2011. DNA double-strand break repair pathway choice in Dictyostelium. J. Cell Sci. 124:1655–1663 10.1242/jcs.081471 [DOI] [PubMed] [Google Scholar]
- Hudson J.J., Hsu D.W., Guo K., Zhukovskaya N., Liu P.H., Williams J.G., Pears C.J., Lakin N.D. 2005. DNA-PKcs-dependent signaling of DNA damage in Dictyostelium discoideum. Curr. Biol. 15:1880–1885 10.1016/j.cub.2005.09.039 [DOI] [PubMed] [Google Scholar]
- Iles N., Rulten S., El-Khamisy S.F., Caldecott K.W. 2007. APLF (C2orf13) is a novel human protein involved in the cellular response to chromosomal DNA strand breaks. Mol. Cell. Biol. 27:3793–3803 10.1128/MCB.02269-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno S., Kuzuoka H., Sasao S., Hong Z., Lan L., Nakajima S., Yasui A. 2007. A novel human AP endonuclease with conserved zinc-finger-like motifs involved in DNA strand break responses. EMBO J. 26:2094–2103 10.1038/sj.emboj.7601663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine H., Lüscher B. 2009. Learning how to read ADP-ribosylation. Cell. 139:17–19 10.1016/j.cell.2009.09.018 [DOI] [PubMed] [Google Scholar]
- Kofler B., Wallraff E., Herzog H., Schneider R., Auer B., Schweiger M. 1993. Purification and characterization of NAD+:ADP-ribosyltransferase (polymerizing) from Dictyostelium discoideum. Biochem. J. 293:275–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuspa A., Loomis W.F. 1992. Tagging developmental genes in Dictyostelium by restriction enzyme-mediated integration of plasmid DNA. Proc. Natl. Acad. Sci. USA. 89:8803–8807 10.1073/pnas.89.18.8803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Page F., Schreiber V., Dherin C., De Murcia G., Boiteux S. 2003. Poly(ADP-ribose) polymerase-1 (PARP-1) is required in murine cell lines for base excision repair of oxidative DNA damage in the absence of DNA polymerase beta. J. Biol. Chem. 278:18471–18477 10.1074/jbc.M212905200 [DOI] [PubMed] [Google Scholar]
- Lin H.H., Khosla M., Huang H.J., Hsu D.W., Michaelis C., Weeks G., Pears C. 2004. A homologue of Cdk8 is required for spore cell differentiation in Dictyostelium. Dev. Biol. 271:49–58 10.1016/j.ydbio.2004.03.020 [DOI] [PubMed] [Google Scholar]
- Manstein D.J., Schuster H.P., Morandini P., Hunt D.M. 1995. Cloning vectors for the production of proteins in Dictyostelium discoideum. Gene. 162:129–134 10.1016/0378-1119(95)00351-6 [DOI] [PubMed] [Google Scholar]
- Ménissier de Murcia J., Ricoul M., Tartier L., Niedergang C., Huber A., Dantzer F., Schreiber V., Amé J.C., Dierich A., LeMeur M., et al. 2003. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 22:2255–2263 10.1093/emboj/cdg206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano S., Lan L., Caldecott K.W., Mori T., Yasui A. 2003. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol. Cell. Biol. 23:3974–3981 10.1128/MCB.23.11.3974-3981.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto H., Reche P.A., Bazan F., Dittmar K., Haag F., Koch-Nolte F. 2005. In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs). BMC Genomics. 6:139 10.1186/1471-2164-6-139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce A.J., Hu P., Han M., Ellis N., Jasin M. 2001. Ku DNA end-binding protein modulates homologous repair of double-strand breaks in mammalian cells. Genes Dev. 15:3237–3242 10.1101/gad.946401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajawat J., Vohra I., Mir H.A., Gohel D., Begum R. 2007. Effect of oxidative stress and involvement of poly(ADP-ribose) polymerase (PARP) in Dictyostelium discoideum development. FEBS J. 274:5611–5618 10.1111/j.1742-4658.2007.06083.x [DOI] [PubMed] [Google Scholar]
- Rickwood D., Osman M.S. 1979. Characterisation of poly(ADP-Rib) polymerase activity in nuclei from the slime mould Dictyostelium discoideum. Mol. Cell. Biochem. 27:79–84 10.1007/BF00218351 [DOI] [PubMed] [Google Scholar]
- Robert I., Dantzer F., Reina-San-Martin B. 2009. Parp1 facilitates alternative NHEJ, whereas Parp2 suppresses IgH/c-myc translocations during immunoglobulin class switch recombination. J. Exp. Med. 206:1047–1056 10.1084/jem.20082468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulten S.L., Cortes-Ledesma F., Guo L., Iles N.J., Caldecott K.W. 2008. APLF (C2orf13) is a novel component of poly(ADP-ribose) signaling in mammalian cells. Mol. Cell. Biol. 28:4620–4628 10.1128/MCB.02243-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulten S.L., Fisher A.E., Robert I., Zuma M.C., Rouleau M., Ju L., Poirier G., Reina-San-Martin B., Caldecott K.W. 2011. PARP-3 and APLF function together to accelerate nonhomologous end-joining. Mol. Cell. 41:33–45 10.1016/j.molcel.2010.12.006 [DOI] [PubMed] [Google Scholar]
- Sawai S., Guan X.J., Kuspa A., Cox E.C. 2007. High-throughput analysis of spatio-temporal dynamics in Dictyostelium. Genome Biol. 8:R144 10.1186/gb-2007-8-7-r144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber V., Amé J.C., Dollé P., Schultz I., Rinaldi B., Fraulob V., Ménissier-de Murcia J., de Murcia G. 2002. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem. 277:23028–23036 10.1074/jbc.M202390200 [DOI] [PubMed] [Google Scholar]
- Sugimura K., Takebayashi S., Taguchi H., Takeda S., Okumura K. 2008. PARP-1 ensures regulation of replication fork progression by homologous recombination on damaged DNA. J. Cell Biol. 183:1203–1212 10.1083/jcb.200806068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.G., Cortes U., Patnaik S., Jasin M., Wang Z.Q. 2004. Ablation of PARP-1 does not interfere with the repair of DNA double-strand breaks, but compromises the reactivation of stalled replication forks. Oncogene. 23:3872–3882 10.1038/sj.onc.1207491 [DOI] [PubMed] [Google Scholar]
- Zhang X.Y., Langenick J., Traynor D., Babu M.M., Kay R.R., Patel K.J. 2009. Xpf and not the Fanconi anaemia proteins or Rev3 accounts for the extreme resistance to cisplatin in Dictyostelium discoideum. PLoS Genet. 5:e1000645 10.1371/journal.pgen.1000645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Hefferin M.L., Chen L., Shim E.Y., Tseng H.M., Kwon Y., Sung P., Lee S.E., Tomkinson A.E. 2007. Role of Dnl4-Lif1 in nonhomologous end-joining repair complex assembly and suppression of homologous recombination. Nat. Struct. Mol. Biol. 14:639–646 10.1038/nsmb1261 [DOI] [PubMed] [Google Scholar]